Abstract
Histone acetyl transferases (HATs) play a crucial role in eukaryotes by regulating chromatin architecture and locus-specific transcription. The GCN5 HAT was identified as a subunit of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) multiprotein complex. Vertebrate cells express a second HAT, PCAF, that is 73% identical to GCN5. Here, we report the characterization of the mammalian ATAC (Ada-Two-A-Containing) complexes containing either GCN5 or PCAF in a mutually exclusive manner. In vitro ATAC complexes acetylate lysine 14 of histone H3. Moreover, ATAC- or SAGA-specific knock-down experiments suggest that both ATAC and SAGA are involved in the acetylation of histone H3K9 and K14 residues. Despite their catalytic similarities, SAGA and ATAC execute their coactivator functions on distinct sets of inducible target genes. Interestingly, ATAC strongly influences the global phosphorylation level of histone H3S10, suggesting that in mammalian cells a cross-talk exists linking ATAC function to H3S10 phosphorylation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0199-8) contains supplementary material, which is available to authorized users.
Keywords: ADA2a, GCN5, ATAC2, Histone acetyltransferase, Immediate early gene, PCAF, H3S10P
Introduction
Post-translational modifications of histones are known to play fundamental roles on the biology of the cell [1]. One of these modifications, the acetylation of lysine residues, has immediate effects on gene regulation, affects the folding of chromatin fibers, and also plays a role in the interaction between histones and other proteins [2–5]. Histone acetylation in general is a mark of active transcription, although the different lysine residues seem to be important for distinct biological processes. For example, it has been reported that histone H3 K9/K14 acetylation defines distinct chromatin regions permissive for gene expression [3, 6]. Furthermore, histone H4K5Ac and H4K12Ac have been suggested to function as deposition marks for newly synthesized histones [7], while acetylation of H4K16 leads to global opening of the chromatin due to changes in the physico-chemical properties of the chromatin fiber [8]. The histone acetyl transferases that add the acetyl group to lysines are classified into several HAT enzyme families and have distinct substrate specificities [9, 10]. They function as co-activators/adapters of transcription, but can also acetylate non-histone substrates, thereby regulating their activity or stability [4, 11]. GCN5 is the founding member of the GNAT family of HATs. While most of the metazoan genomes code for one GCN5 type factor, vertebrates have a second gene encoding PCAF (p300/CBP associated factor), which is highly homologous to GCN5 [4, 11, 12]. GCN5 (or PCAF) is a subunit of the SAGA (Sgf-Ada-Gcn5 containing acetyltransferase) complex that is conserved from yeast to human cells [4, 13, 14]. Eukaryotic SAGA complexes harbor 19 subunits, which include TRRAP (or yTra1), ENY2 (or ySus1), USP22 (or yUbp8), and subunits belonging to the ADA, SPT, TAF, and SGF group of proteins [13]. Recently, a second GCN5-containing complex has been described from metazoans, called ATAC (ADA-Two-A containing) [15–18]. The ATAC complex shares a common core with SAGA, composed of GCN5-ADA3-SGF29 and either ADA2a (in ATAC) or ADA2b (in SAGA). Besides these subunits, the human ATAC complexes contain a second putative HAT, called hATAC2 (or hCSRP2BP), and five other subunits; hYEATS2, hZZZ3, hMBIP, hWDR5, and hNC2β in human cells or their Drosophila orthologues in dATAC. At present, the biological function of the ATAC complex is not well understood.
In vitro, yeast and human GCN5 acetylates mainly histone H3K14. However, when incorporated into the hSAGA complex, although GCN5 still shows a preference for histone H3K14, it also acetylates H3K9 and H3K18 [19, 20]. The Drosophila ATAC possesses different substrate specificity than dSAGA, since it mainly acetylates histone H4 [16, 17, 21]. The H4 specific activity was suggested to result from the presence of the second HAT, ATAC2, in the complex [17]. When testing the HAT activity of different hATAC preparations on free histones, the Flag-MBIP immunoprecipitated (IPed) hATAC acetylated histone H3 and H4, while the Flag-YEATS2 IPed hATAC acetylated only histone H3 [15, 18]. Thus, at present, the function of the distinct metazoan ATAC complexes is not clear, and the physiological targets of the two known subunits with enzymatic activities (ATAC2 and GCN5) await further analysis. Moreover, it is not yet well understood why metazoan cells have two different GCN5-containing HAT complexes and whether these complexes carry out redundant or specific functions.
Here, we report the identification of the endogenous human ATAC complexes. We determined the composition and the histone acetylation specificity of the hATAC complex in vitro. Importantly, we propose that the biological relevance of having two GCN5-containing HAT complexes, hSAGA and hATAC, manifests mainly in vivo. We show in both Drosophila salivary glands and human cells that the SAGA and ATAC HAT complexes respond to different stimuli and thus play a transcriptional coactivator function at distinct sets of target genes. We also provide evidence for ATAC playing a role in the regulation of global histone H3 Ser10 phosphorylation suggesting a cross-talk between histone acetylation and phosphorylation on histone H3 tails.
Materials and methods
Antibodies
The anti-hADA2a monoclonal antibody (2AD2A1) was raised against the peptide MDRLGSFSNDPSDKPP(C) and its specificity tested in immunofluorescent staining (electronic supplementary material, ESM, Fig. 1). To raise polyclonal antibodies for the ATAC subunits, the following pepides were synthetized and coupled to ovalbumine: for the anti-ADA2a antibody (#2619), the MDRLGSFSNDPSDKPP(C) peptide; for the anti-hZZZ3 antibody (#2616), the (C)GNNNGRTTDLKQQSTRESW peptide; for the anti-mADA3 antibody (#2678), the LEGKTGHGPGPGPGRPKSKN(C) peptide; for the anti-ATAC2 antibody (#2734), the IRSHLHRSDPHWTPEPD(C) peptide, for the anti-hYEATS2 antibody (#2783), the LSQHNDFLSDKDNNSNM(C) peptide; for the anti-hMBIP antibody (#2786), the TRPEGIPGSGHKPNSMLR(C) peptide; for the anti-mGCN5 antibody (#2676), the MAEPSQAPTPAPAAQPRPL(C) peptide; and for the anti-mPCAF (#2760) antibody, the MAEAGGAGJPALPPAPPHG(C) peptide. The sera were affinity purified by using the Sulfolink Coupling Gel (Pierce) following the manufacturer's recommendations.
Antibodies against the following proteins have been described earlier: GNC5: 2GC2C11 [22], TAF10: 23TA1H8 [23], TRRAP: 2TRR2D5 [24], TBP: 3G3 [25], USP22: #2391 [26], SPT20 (p38IP): #4112 [27] and #2487 [13], WDR5: [28], HCF-1: N18 [29], NC2α and NC2β [30].
Antibodies used for detection of histone modifications on western blot were: H3 core: ab1791 Abcam; H3K9Ac: #06-942 Millipore; H3S10P: #05-598 Millipore; H3K14Ac: #07-353 Millipore; H4K5Ac: ab51997 Abcam; H4K12Ac: ab1761 Abcam; H4K16Ac: #07-329 Millipore.
Antibodies used for ChIP were: H3 core: ab1791 Abcam; H3K9/K14Ac: #06-599 Millipore.
Drosophila antibodies used for polytene chromosome staining
Primary antibodies were rabbit anti-FLAG (F-7425; Sigma–Aldrich) at 1:50, monoclonal antibody against RNA Polymerase II phosphorylated on Ser5 (H14; COVANCE) at 1:50 and rabbit anti-Ada2b [31] at 1:25 dilution. Secondary antibodies were Texas Red Goat Anti-Mouse IgM, μ chain specific (Jackson ImmunoResearch), and Alexa Fluor® 488 Donkey anti-rabbit IgG (Molecular Probes) at 1:1,000 dilution.
Baculovirus infections and protein overexpression
Overexpression and protein preparation was done as described in [32]. For the HA-ATAC2 construct, the cDNA clone IRAUp969G0838D was purchased from ImaGENES, the coding sequence amplified with the following primers: 5′ AAGAATTCGATGGATAGTAGCATCCACCTGAG 3′; 5′ TCCTCGAGCTGTATTCGCATCAGCGCC 3′, and then cloned to the EcoRI XhoI sites of pCDNA3 vector modified to contain an N-terminal HA tag. The tagged cDNA was further cloned to pVL1393 baculovirus transfer vector to generate recombinant virus. The hGCN5 expressing construct was described in [33].
Immunoprecipitations were carried out as described earlier [33].
Cell growth condition, stress treatments
HeLa cells were grown in Dulbecco’s modified Medium supplemented with 1 g/l glucose, 5% FCS, phenol red, and gentamycine. Before TPA treatment, cells were serum straved O/N (0.5% FCS) and then treated with 50 ng/ml TPA during 1 h. Control treatment was carried out with DMSO.
Chromatin immunoprecipitation (ChIP) was carried out as described in [13].
RNA purification, reverse transcription and qPCR
Total RNA was purified using Trizol reagent (Invitrogen), reverse transcribed by MMLV reverse transcriptase using random hexamers, and analyzed by the quantitative PCR (Q-PCR) machine Roche LightCycler480 with Syber Green (Quiagen) Master mix. All the detected values represented in the manuscript have been normalized to CyclophilinB and represent biologically independent replicates.
siRNA
Negative control (ref. number D-001810-10), anti hZZZ3 siRNA (L-013939-01), anti hATAC2 (L-008481-00), anti hADA2a siRNA (ref number L-017516-00), and anti hSPT20 siRNA (ref number L-013820-00) was purchased from Dharmacon and transfected using Lipofectamine 2000 and OptiMEM serum-free medium following the manufacturer’s recommendations.
Nuclear extract preparation was described in [33].
Purification of histone H3/H4 dimers, octamers, mono- and polynucleosomes was performed as described in [34].
Fly stock and generation of transgenic flies
Fly maintenance and crosses were performed as described previously [26]. The expression constructs of FLAG-tagged D12 and CG10238 in pUAST vector were sent to bestGene (CA, USA) Drosophila embryo injection services for generation of transgenic flies. Dpp-Gal4 (to obtain UAS-FLAG-CG10238/dpp-Gal4 flies) and dpp.blk1-Gal4 (to obtain UAS-FLAG-D12/+;dpp.blk1-Gal4/+ flies) stocks were obtained from the Bloomington Drosophila Stock centre.
Preparation and treatment of polytene chromosomes
Drosophila salivary glands were dissected from third instar larvae in 40% acetic acid and fixed for 5 min in 3.7% formaldehyde, 1% Triton-X 100, 1/2× PBS, and followed by 2 min in 3.7% formaldehyde, 50% acetic acid on poly-L lysine-treated slide glass. For TPA treatment, salivary glands were dissected in 0.7% NaCl, 0.1% NP-40 solution, incubated with 4 nM TPA in Schneider’s Drosophila Medium (1×) (Invitrogene), 10% FBS for 30 min at 25°C, and then fixed. After fixation, polytene chromosomes were squashed by squeezing the slide and siliconized cover slip and were frozen in liquid nitrogen. The cover slip was removed and dehydrated for 10 min in ice-cold 50% acetone, 50% methanol solution. The slide was washed in PBS for 10 min and, for blocking, incubated in 5% skim milk, PBST (PBS with 0.1% Tween 20) for 1 h at RT. The squashed polytene chromosomes were covered overnight at 4°C with the diluted primary antibody in a humid chamber. The slides were washed three times for 5 min in PBST and incubated with the secondary antibody for 2 h at RT. Slides were washed three times for 5 min with PBST and mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories, Burlingame). The specimens were photographed in confocal microscopy LSM510 (Carl Zeiss MicroImaging).
Acetylation assays
Peptide acetylations were performed as described in [13, 35]. Briefly, 1.2 μg of peptide (corresponding to the N-terminal tail of H3 at positions 6–20 or the N-terminal tail of H4 from positions 1–19) was added to the immunopurified protein sample together with 1 μl of H3 Acetyl CoenzymeA (Amersham; 50 μCi/ml) in the reaction buffer (50 mM TrisHCl pH 8, 20 mM KCl, 5 mM DTT, 4 mM EDTA) and incubated at 30°C for 1 h. Samples were spotted on Whatmann P81 nitrocellulose filters, washed 3 times for 10 min in ice-cold 50 mM NaHCO3 pH 9 buffer, and dried. Filters were then dropped into 5 ml of ReadySafe liquid scintillation cocktail (Beckman Coulter) and radioactivity was quantified by an LS6000SC Beckman counter.
Histone acetylation assay: histones were incubated with 14C-Acetyl-CoA and ATAC complex (purified from HeLa nuclear extracts by anti-ADA2a IP followed by elution with peptide) or SAGA complex (purified from HeLa nuclear extracts by anti-USP22 IP followed by peptide) in 1×HAT buffer (50 mM Tris pH 7.9, 10% glycerol, 0,1 mM EDTA, 50 mM KCl, 20 mM Sodium Butyrate, 1 mM DTT and protease inhibitors). The reactions were incubated during 1 h at 30°C, stopped by adding Laemmli buffer with 100 mM DTT and boiled for 10 min. Proteins were then loaded on a 13% SDS–PAGE and analyzed by coomassie brillant blue staining. The gel was then incubated for 20 min in “Amplify” fluorographic reagent (GE Healthcare) and dried. Blank phosphor screen (Fuji) was placed overnight on the gel and the radioactive signal was analysed with phosphorimager scanner Typhoon 8600.
Primers used for the RT qPCR analysis
EGR-1 RT L: ACCTGACCGCAGAGTCTTTTCC;
EGR-1 RT R: CAGGGAAAAGCGGCCAGTATAG;
FRA-1 RT L: CAGGAACCGGAGGAAGGAACT;
FRA-1 RT R: TGCTTCTGCAGCTCCTCAATCT;
c-FOS RT L: GGGGCAAGGTGGAACAGTTATC;
c-FOS RT R: TAGTTGGTCTGTCTCCGCTTGG;
CyclophilinB RT L: CTTCCCCGATGAGAACTTCAAACT;
CyclophilinB RT R: CACCTCCATGCCCTCTAGAACTTT;
GAPDH RT L: ACAGTCCATGCCATCACTGCC;
GAPDH RT R: GCCTGCTTCACCACCTTCTTG.
Primers used in the ChIP quantification reactions
EGR-1 P L: CTAGGGTGCAGGATGGAGGTG;
EGR-1 P R: TATGGGAAGCAGGAAGCCCTAA;
FRA-1 P L: GTTCCCCGAAGTCTCGGAACAT;
FRA-1 P R: GTGGTTCAGCCCGAGAACTTTT;
c-FOS P L: TTGAGCCCGTGACGTTTACACT;
c-FOS P R: TTCTCAGATGCTCGCTGCAGAT;
non-coding L:TGGAACTTCTGGAAGACACTGGAA;
non-coding R: TACACCACTCAAGGGAAACTGGAA.
Results
Composition of the endogenous hATAC complex
To isolate endogenous human ATAC complexes we carried out immunopurification (IP) using a monoclonal antibody raised against ADA2a (ESM, Fig. 1) that is the defining subunit of the complex (ADA-Two-A-Containing) [15–18]. The IP was carried out on a HeLa cell nuclear extract without overexpressing any of the putative ATAC subunits and the proteins that coprecipitated with hADA2a were resolved by SDS–PAGE and visualized by silver nitrate staining (ESM, Fig. 2a). The identification of the components of this endogenous complex by mass spectrometry indicated mostly the presence of the same polypeptides as the ones reported by Guelman and colleagues [15] (ESM, Fig. 2a, and see Table 1). To further verify the presence of all the reported subunits in our endogenous complex, we raised specific antibodies against most of the known subunits of hATAC. In addition, to carry out a comparative examination of the two human GCN5-containing complexes, we have also purified endogenous hSAGA complex by using an antibody raised against the recently identified hSPT20 subunit [13]. Consequent comparison of the two complexes by western blotting analysis revealed the similarities and differences between hSAGA and hATAC. Common subunits present in both complexes are: hGCN5, hADA3 and hSGF29 (Fig. 1a). Human ATAC specific components are: hYEATS2, a large protein containing a so-called YEATS domain with no known function; hZZZ3, a potential transcription factor with zinc finger and SANT domains; hATAC2 containing a putative acetyl transferase domain; hADA2a, a known adaptor protein affecting the activity of hGCN5 [36]; hMBIP that was shown to interact with the MAPK upstream kinase [37]; and hWDR5 also known as a subunit of the MLL complex [38] (Fig. 1a lane 2). Importantly, SAGA specific subunits such as hTRRAP, hUSP22 and hTAF10 did not copurify with hADA2a (Fig. 1a lane 2). As TBP and HCF1 were reported to interact with the anti-Flag-YEATS2 purified hATAC complex [18] (Table 1), we tested the presence of these factors in our endogenous ATAC complex preparations. In good agreement with our mass spectrometry data, neither TBP nor HCF1 could be detected in the complex by western blot analysis (data not shown).
Table 1.
Subunit composition of the known Drosophila and human ATAC complexes
| Drosophila ATAC subunits [16, 17] | Human ATAC purified by anti-flagYEATS2 IP [18] | Human ATAC purified by anti-flagMBIP IP [15] | Human ATAC purified by anti-hADA2a IP (present work) |
|---|---|---|---|
| dGCN5 | hGCN5 | hGCN5 | hGCN5 |
| hPCAF | hPCAF | ||
| dADA2a | hADA2a | hADA2a | hADA2a |
| dADA3 | hADA3 | hADA3 | hADA3 |
| dATAC1 (CG9200) | ZZZ3 | ZZZ3 | ZZZ3 |
| dATAC2 (CG10414) | hATAC2 (hCSRP2BP) | hATAC2 (hCSRP2BP) | hATAC2 (hCSRP2BP) |
| CG30390 | hSGF29 | hSGF29 | hSGF29 |
| dATAC3 (CG32343) | |||
| HCF | HCF1 | ||
| WDS | WDR5 | WDR5 | WDR5 |
| D12 | YEATS2 | YEATS2 | YEATS2 |
| NC2β | NC2β | NC2β | NC2β |
| CG10238 | MBIP | MBIP | MBIP |
| UBAP2L | |||
| CHRAC14 | POLE3 | ||
| POLE4 | |||
| MAP3K7 | |||
| TBP |
Fig. 1.
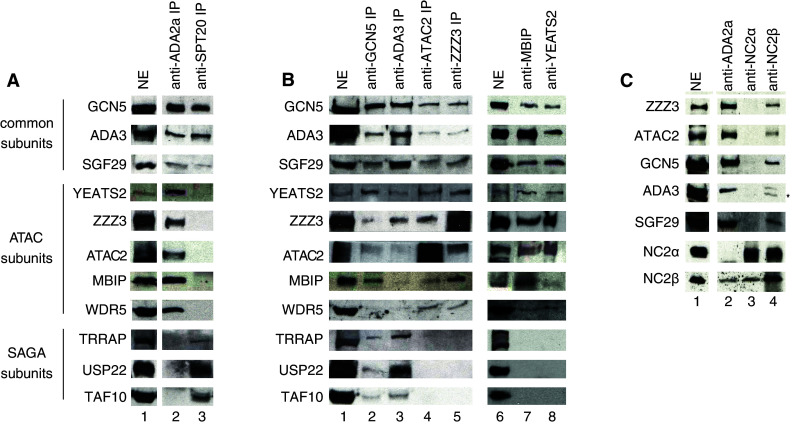
Subunit composition of the endogenous human ATAC complex. a Comparison of an anti-ADA2a and an anti-hSPT20 IP reveals the differences in composition of hATAC and hSAGA, respectively. The two complexes were purified from HeLa nuclear extract (NE) by means of antibodies developed against hADA2a (lane 2) or hSPT20 (lane 3) and the coprecipitated proteins were detected by western blotting. b Different IPs demonstrate the existing interactions between hSAGA and hATAC subunits. The common, the ATAC-specific and the SAGA-specific, subunits are marked on the left side of the figure. c NC2β, but not NC2α is a component of the hATAC complex, as it copurifies with other subunits of the complex both in anti-ADA2a (lane 2) and in anti-NC2β (lane 4) IPs. Asterisk the heavy chain of the antibody
To further confirm our results on the composition of the endogenous hATAC complex, we carried out reciprocal IPs with antibodies raised against the putative ATAC subunits, such as hGCN5, hADA3, hATAC2, hZZZ3, hMBIP, and hYEATS2. Western blot analyses of samples eluted by excess of the antigene peptide show that besides the common subunits (hGCN5, hADA3, and hSGF29), the ATAC specific proteins (hZZZ3, hYEATS2, hATAC2, hMBIP, and hWDR5) copurify with the precipitated proteins in all the cases (Fig. 1b). On the other hand, antibodies against common subunits of SAGA and ATAC (anti-hGCN5 or anti-hADA3) also pull down hSAGA specific subunits, such as hTRRAP, hUSP22, and hTAF10 (Fig. 1b lanes 2, 3). Based on our observations, we conclude that hADA2a, hGCN5, hADA3, hSGF29, hZZZ3, hATAC2, hYEATS2, hMBIP, and hWDR5 are all bona fide subunits of the endogenous ATAC complex in human cells.
As we have also obtained one peptide by mass spectrometry corresponding to the NC2β (also called Dr1; [39–41]) in one of our ATAC purifications, and as NC2β has been reported to be a component of the Flag-purified ATAC complexes [15, 18], we have investigated the interaction between hADA2a and the two subunits of the NC2 complex, NC2α and NC2β [30, 42]. Our results clearly show that only NC2β is associated with the endogenous hATAC complex since an antibody against NC2β coprecipitated the tested hATAC subunits and, vice versa, the anti-ADA2a IP precipitated NC2β (Fig. 1c lanes 2, 4). In contrast, the anti-NC2α antibody coprecipitated only NC2β, but no ATAC subunits (Fig. 1c lane 3). Thus, NC2β is also a bona fide subunit of the endogenous hATAC complex.
In summary, our results provide evidence for the existence of an endogenous ATAC complex containing at least ten subunits (see Table 1).
In vitro hATAC and SAGA acetylate histone H3 K14
After the purification of the endogenous ATAC complex from human cells, we aimed to test its substrate specificity and compare it to that of hSAGA. To this end, we performed in vitro acetylation assays on wild-type and mutated N-terminal histone tail peptides. In each mutant peptide, we changed one acetylable lysine (K) residue to arginine (R) that mimics the non-acetylated form of the amino acid. First, we analyzed the substrate specificity of the two GCN5-containing complexes, ATAC and SAGA, in peptide acetylation assay. Both complexes were purified from HeLa cells by specific IPs and normalized for the content of the common subunits (ESM, Fig. 3a). Surprisingly, in this in vitro peptide acetylation test, we observed no difference between hSAGA and hATAC, as both complexes showed a preference for acetylating histone H3K14 (Fig. 2a, b).
Fig. 2.

Histone acetyltransferase activity of the human GCN5 containing complexes and their HAT subunits. a–d Acetylation activity of endogenous purified hATAC (a), hSAGA (b), recombinant (rec) GCN5 (c), and recombinant ATAC2 (d) on histone tail peptides was measured by liquid scintillography. Histone H3 peptides are shown in gray, histone H4 peptides and the reaction without peptide are in black. Note that, due to the described weak HAT activity of recombinant hATAC2 [15], in (d), ten times more recombinant protein was used than in the reactions shown in (c). e Acetylation activity of endogenous complexes on H3–H4 dimers and histone octamers. f Acetylation activity of hATAC and hSAGA complexes on mono- and polynucleosomes. In e and f, upper panels show the autoradiography and lower panels the corresponding coomassie stained histones
Next, we analyzed the activity of the two ATAC-associated HAT enzymes alone. Both recombinant enzymes (flag-GCN5 and HA-ATAC2) were purified from baculovirus infected SF9 insect cells by immunoprecipitation and consecutive peptide elution (ESM, Fig. 3b). While GCN5 showed specificity toward H3K14 in these reactions (Fig. 2c), ATAC2 had no detectable activity even when ten times more enzyme was added to the reactions (Fig. 2d). Thus, our data suggest that the second putative acetyltransferase subunit of the ATAC complex is inactive in vitro on the tested histone tail peptides.
To exclude the possibility that the use of non-physiological substrates (i.e., short peptides) changes the specificity of the enzymes in the complexes, we tested the HAT activity of ATAC and SAGA on purified full-length histone H3–H4 dimers and histone octamers containing all the four core histones. In these acetyltransferase assays, both SAGA and ATAC complexes acetylated mainly histone H3 (Fig. 2e). Similar results were obtained when we tested the acetyltransferase activity of the two complexes on mono- and polynucleosomes (Fig. 2f). Note, however, that polynucleosomes seemed to be better substrates than mononucleosomes. Our results are in good agreement with those of the Martinez group [18], and suggest that, in spite of the evolutionary conservation of the protein sequences from Drosophila to human, the human complex has no or very weak H4 specificity in vitro. Thus, the functional difference between the human SAGA and ATAC complexes seem not to be related to their histone substrate specificity.
ATAC complexes containing either GCN5 or PCAF exist in mouse fibroblasts
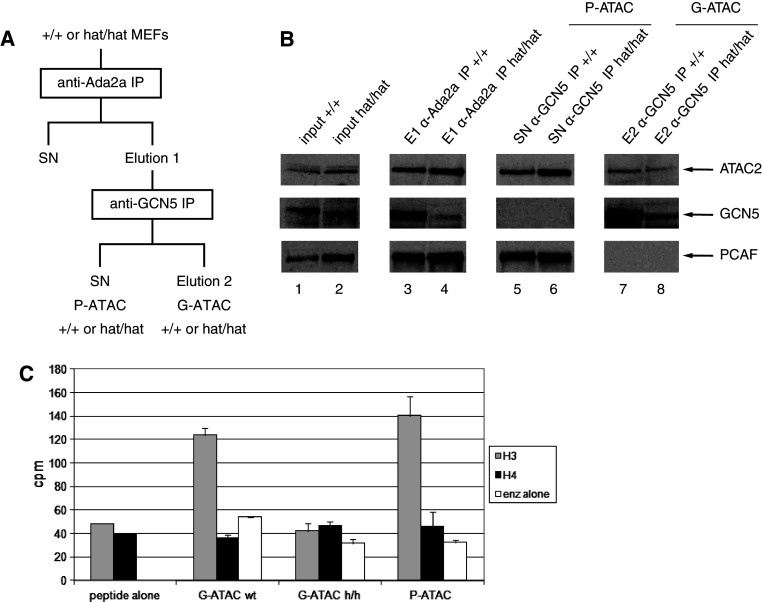
The fact that the ATAC complex contains two HATs (GCN5 and ATAC2) raised the possibility that the two enzymes may have overlapping or redundant activities within the complex. To be able to measure the activity of ATAC2 alone in the context of the intact ATAC complex, we decided to purify ATAC complexes from Gcn5 hat/hat mouse embryo fibroblasts (MEFs), in which the GCN5 catalytic activity was inactivated by double E568A and D608A mutations in the HAT domain [43]. We prepared nuclear extracts from wild-type (+/+) and Gcn5 hat/hat MEFs and carried out IPs using the above described anti-ADA2a antibody (Fig. 3a, elution 1). Surprisingly, in nuclear extracts prepared from Gcn5 hat/hat mutant MEFs, PCAF expression was significantly up-regulated (2- to 3-fold), indicating that, when GCN5 is inactivated, cells compensate the loss of its activity with that of its paralogue (see Introduction and Fig. 3b lane 2). Note that similar results were described in a different cellular system [44]. Moreover, our western blot analysis indicated that ATAC, purified by a simple anti-ADA2a IP, contained both PCAF and GCN5 (Fig. 3b lanes 3, 4). The question thus rose whether these complexes can be separated into fractions containing exclusively GCN5 or PCAF. Alternatively, a given ATAC complex might contain both GCN5 and PCAF. To decide, the ADA2a-containing complexes were re-IPed with an anti-GCN5 antibody (see Fig. 3a). When the GCN5-free supernatant of this second anti-GCN5 IP was compared to the GCN5-containing elution fraction by western blot (elution 2 on Fig. 3a), it became clear that we have separated PCAF-containing ATAC complexes (hereafter called P-ATAC) from GCN5-containing complexes (hereafter called G-ATAC) (Fig. 3b, compare lanes 5–6 with 7–8). These results clearly demonstrate that vertebrate cells contain both G-ATAC and P-ATAC complexes and that the presence of GCN5 or PCAF in these complexes is mutually exclusive.
Fig. 3.
Composition of different ATAC complexes from MEFs and their HAT activity on histone tail peptides. a Purification scheme of the separation of GCN5- (G-ATAC) and PCAF-containing (P-ATAC) ATAC complexes from wild-type (+/+) and mutant (hat/hat) MEFs. b Western blot analysis of the different fractions obtained during the purification shown in (a) using the indicated antibodies. c Acetylation of histone H3 and H4 tail peptides using the indicated purified complexes was measured. The amount of the purified complexes was normalized to their ATAC2 content
The in vitro HAT activities of the different ATAC complexes
To measure the acetyltransferase activity of ATAC2 in ATAC, we have compared the HAT activities of the above purified GCN5- or GCN5hat-containing ATAC complexes on histone tail peptides as previously (see Fig. 2). Surprisingly, in this in vitro peptide acetylation test using either the H3 (aa 6–20) or the H4 peptide (aa 1–19), we observed no activity of the G-ATAChat complex, while wild-type G-ATAC acetylated the H3 peptide as before (Fig. 3c). This result, together with the lack of activity obtained with recATAC2 on histone tails (Fig. 2d), suggests that the mammalian ATAC2 is inactive in vitro in conditions that are appropriate for the GCN5 HAT activity. Moreover, in this in vitro test, the HAT specificity of P-ATAC was similar to that of G-ATAC (Fig. 3c). Thus, our observations suggest that (1) in vertebrates at least two different ATAC complexes exist with very similar composition and in vitro substrate specificity, (2) the mouse or human ATAC2 may have non-histone substrates, and (3) that in vitro no significant differences could be determined between the HAT activities of SAGA and ATAC.
ATAC- or SAGA-specific knock-downs lead to a drop in global histone H3 acetylation on K9 and K14, but do not affect histone H4 acetylation levels
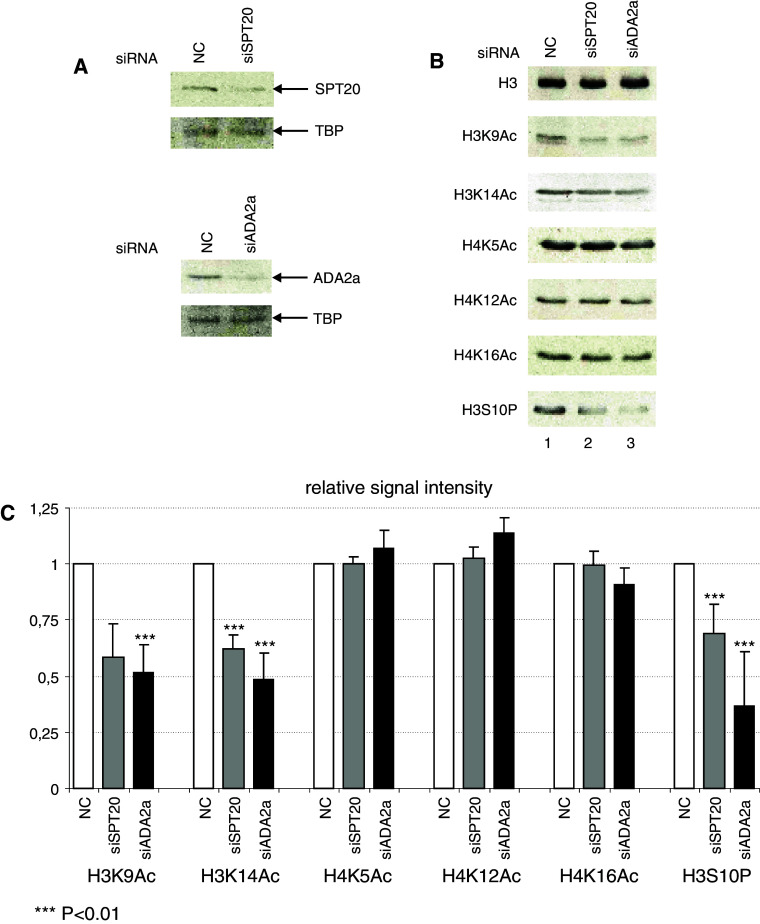
To further analyze the differences between the two human GCN5-containing complexes in vivo, we tested how the global level of post translational modifications of histones are affected in cells where either an ATAC-specific or a SAGA-specific subunit was knocked down by using siRNAs. HeLa cells were transfected with control non-targeting siRNA (NC), anti-SPT20 siRNA (SAGA-specific) or anti-ADA2a (ATAC-specific) siRNA (Fig. 4a). At 48 h post-transfection, cell extracts were prepared and analysed by western blotting. The knock-down of SPT20 or ADA2a was efficient since we obtained a clear reduction in the respective protein levels (Fig. 4a). Next, the amount of histone content in each extract was normalized using an antibody recognizing the core domain of histone H3 (Fig. 4b upper panel). Surprisingly, compared to the control, only the H3K9Ac and the H3K14Ac marks were reduced and this reduction was of similar extent in both siADA2a and siSPT20 treated cells (Fig. 4b, c). On the other hand, the acetylation marks located on histone H4, such as K5Ac, K12Ac, and K16Ac, were unaffected in our siRNA-transfected cells. These experiments show that, similarly to the in vitro experiments, on global histone acetylation levels no significant differences could be determined between the HAT activities of SAGA and ATAC. Also, in HeLa cells, the knock-down of ATAC- and SAGA-specific subunits do not influence globally the histone H4 acetylation.
Fig. 4.
Global changes of post translational histone modifications in cells deficient for ATAC or SAGA. a Transfection of non-targeting control siRNA (NC) or siRNAs directed against SPT20 or ADA2a leads to the specific knock-down of the targeted subunit as compared in western blot. TBP served as a loading conrol. b Changes in the histone acetylation and phosphorylation marks in siRNA-transfected cells. The global levels of the given modifications were analyzed by western blot. Blotting with an antibody recognizing the core domain of histone H3 served as a loading control. c Histone H3 post-translational modifications are perturbed in siRNA-transfected HeLa cells. Quantification of signal intensities of four independent knock-down experiments shows global changes in H3 modifications, while H4 acetylation remains unaffected. The H3S10P signal is strikingly low in siADA2a-transfected cells
ATAC-specific knock-down leads to a drop in global histone H3 phosphoryalation on serine 10
As the Drosophila ATAC complex was shown to influence histone H3S10 phosphorylation [45], we have also tested whether ATAC- or SAGA specific knock-downs affect H3S10 phosphorylation in human cells. Interestingly, we found that in HeLa cells with decreased ADA2a levels the H3S10 phosphorylation mark was reduced to 36% of the control, while the effect of SAGA knock-down was much milder (70%) (Fig. 4b, c). Importantly, this result defines an evolutionarily conserved cross-talk linking ATAC HAT function to H3S10 phophorylation.
dATAC and dSAGA regulate different inducible genes in Drosophila salivary glands in vivo
To explore the in vivo functional differences between ATAC and SAGA complexes, first we examined the localization of the complexes in salivary glands on polytene chromosomes of Drosophila. To be able to visualize the recruitment of dATAC specific subunits and compare the function of dATAC to dSAGA, transgenic flies were generated in which two dATAC-specific subunits, D12 (homologue of hYEATS2) and CG10238 (homologue of hMBIP, see Table 1), were Flag tagged. The localization of ADA2b, a SAGA specific subunit, was followed by an anti-dADA2b antibody labeling. For detecting the polymerase, we used an antibody raised against the serine 5 phosphorylation of the CTD of the large subunit of Pol II (Pol II Ser5P), which is a marker for Pol II incorporating in a functional preinitiation complex (PIC). Interestingly, under non-stimulated conditions, all the bands stained by antibodies against Flag-D12, Flag-CG10238, or ADA2b localized to euchromatic segments giving weak DAPI signal (Fig. 5a, c, e). Although ATAC and SAGA were suggested to function in histone modification and transcriptional regulation, our in vivo results in these non-stimulated conditions showed only rare colocalization of dATAC or dSAGA with RNA Pol II. To establish the role of ATAC in transcription activation, we induced genes by a TPA (12-O-tetradecanoylphorbol-13-acetate)-treatment and tested the recruitment of the dATAC complex to the transcriptionally active loci. Following the TPA-treatment, we observed an increased recruitment of dATAC (visualized by D12 and CG10238) to Pol II positive chromosome regions (see white arrows in Fig. 5b, d). In striking contrast to dATAC subunits, the dSAGA-specific subunit ADA2b showed no recruitment to the active bands following TPA induction (see Fig. 5f). These in vivo results suggest for the first time that SAGA and ATAC regulate different set of genes depending on the cellular stress received.
Fig. 5.
dATAC, but not dSAGA, is recruited to TPA-induced transcription sites on Drosophila polytene chromosomes. Polytene chromosome co-staining of non-treated (a, c, e) and TPA treated (b, d, f) samples is shown. The DAPI staining (white), the anti-flag (a–d) or the anti-ADA2b staining (e, f) (green), and the anti-RNA Pol II Ser5P staining (red) is shown in each panel together with the merged picture. A dramatic increase in RNA Pol II and flag-D12 (homologue of hYEATS2) or flag-CG10238 (homologue of hMBIP) colocalization occurs after TPA treatment (compare a to b and c to d) marked with arrowheads. In contrast, no increase in the colocalization was detected for ADA2b and RNA Pol II (compare e to f)
hATAC is recruited to the promoter of immediate early genes in human cells
The above-described in vivo results obtained on Drosophila salivary gland polytene chromosomes hinted at regulatory mechanisms in which the function of SAGA and ATAC do not overlap. In addition, our recent results showed that human SAGA is not involved in the transcription activation of immediate early (IE) genes [13]. Thus, we tested whether hATAC would participate in the regulation of IE genes following stimulation. To this end, we analyzed the recruitment of the hATAC complex to IE gene promoters after TPA treatment by chromatin immunoprecipitation (ChIP) in HeLa cells. First, we examined the mRNA level of three IE genes (c-FOS, FRA-1, and EGR-1) after 1 h of TPA treatment. As a result, we obtained a 3- to 20-fold stimulation of these mRNA species compared to the control, while the expression of GAPDH mRNA remained unchanged (Fig. 6a). We also carried out a control DMSO treatment, where we obtained no significant effect on the expression of IE genes (not shown). Then, we prepared chromatin from both non-treated and TPA-induced cells and carried out ChIPs by using antibodies raised against RNA Pol II, hATAC subunits (hADA3, hZZZ3), and a hSAGA-specific subunit (hSPT20). The results showed that, together with RNA Pol II, subunits of the hATAC complex (hZZZ3 and hADA3) got recruited to the three IE promoters after TPA treatment in human cells (Fig. 6b–d). At the same time, no increase in the occupation of these loci was observed for hSPT20, which is a hSAGA specific subunit (Fig. 6e). The amount of the IPed control region (a genomic region not harboring any Pol II transcription unit) remained unchanged following the TPA treatment (Fig. 6b–e) and was close to levels obtained with negative control anti-GST ChIP (not shown). Note that TPA did not induce the expression of the tested ATAC or SAGA subunits in the cells (ESM Fig. 4). These results together with those obtained with Drosophila salivary gland stainings show that ATAC, but not SAGA, is recruited to TPA-induced gene promoters. Thus, our observations indicate for the first time a differential recruitment of the two HAT complexes, ATAC and SAGA, to stress-regulated genes in mammalian cells.
Fig. 6.

hATAC subunits get recruited, together with RNA Pol II, to the promoter of immediate early genes after TPA treatment. a Expression level of IE genes c-FOS, EGR-1 and FRA-1. Fold change of mRNA levels normalized by cyclophylinB are shown on the graph after 1 h of TPA treatment. White bars represent the nontreated sample, gray bars show the results obtained after TPA treatment. b–e ChIP results obtained with antibodies against RNA Pol II (b), ZZZ3 (c), ADA3 (d), and SPT20 (e) are shown on three IE gene promoters and the control non-coding region. Non-treated values (% input) are represented as white bars, the treated samples are shown by gray bars. Similar results were obtained in two biological replicates. Results obtained in a representative experiment are shown with SD values calculated from qPCR triplicates for each time point
Knock-down of ATAC subunits leads to defects in the TPA-induced gene expression
Next, we analyzed whether the knock-down of ATAC subunits influences the regulation of the above tested TPA-regulated genes. After transfection of HeLa cells with siRNA against hZZZ3 or hATAC2, we observed a ~50% decrease in the corresponding mRNA levels (Fig. 7a) and a ~ 75% decrease in the protein levels of these two ATAC subunits, respectively (Fig. 7b), when compared to the control siRNA-transfected cells. To analyze the effect of the knock-down of a SAGA specific subunit, we carried out anti-SPT20 RNAi experiments in parallel. Following TPA-treatment in ZZZ3- or ATAC2-siRNA-transfected cells, the transcriptional activation of the tested IE genes was significantly reduced to ~50% of the negative control situation (Fig. 7c–e). At the same time, knock-down of SPT20 had no significant effect on the up-regulation of IE genes. This is in good agreement with our previous observations showing that SAGA is not required for IE gene induction [13]. The siRNA transfections had no effect on the GAPDH mRNA level (Fig. 7f). Altogether, these results confirm that ATAC, but not SAGA, is recruited to the promoters and is required for the induction of the studied TPA-induced genes.
Fig. 7.
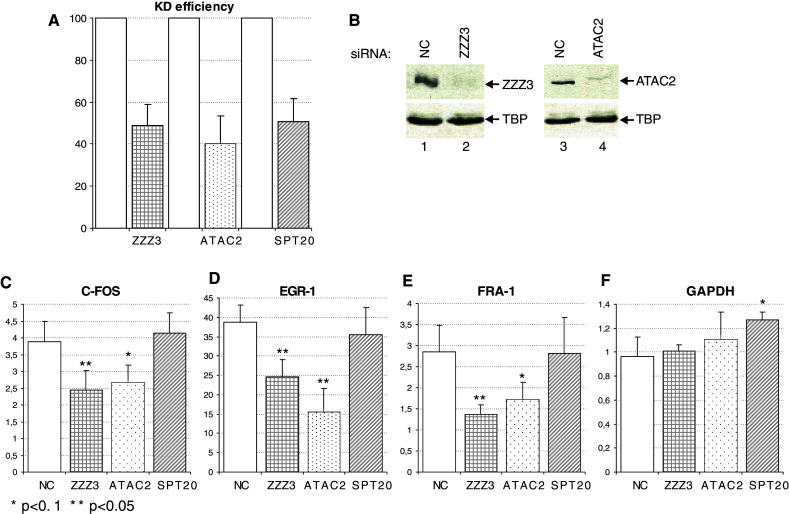
hATAC but not hSAGA is indispensable for correct up regulation of IE genes after TPA treatment. a Knock-down efficiency of the three siRNAs (against ZZZ3, ATAC2 and SPT20) was analyzed by RT qPCR. The amount of residual mRNA is shown (bars with different patterns) compared to the negative control (NC) siRNA-transfected cells (white bars). b Knock-down efficiency of the siZZZ3 (lane 2) and siATAC2 (lane 4) was analyzed by western blotting and compared to negative control (NC) siRNA-transfected cells (lanes 1, 3). As a loading control, the same blot was developed with an anti-TBP antibody. c–f. Induction of IE gene expression after TPA treatment in cells transfected with different siRNAs (indicated on the bottom of the graphs). Results of IE genes or GAPDH mRNA quantification are shown as fold change over the non-treated samples and represent three independent experiments. The different patterns of the bars refer to a
ATAC is indispensable for the correct histone H3 acetylation status of IE gene promoters both under non-induced and activated conditions
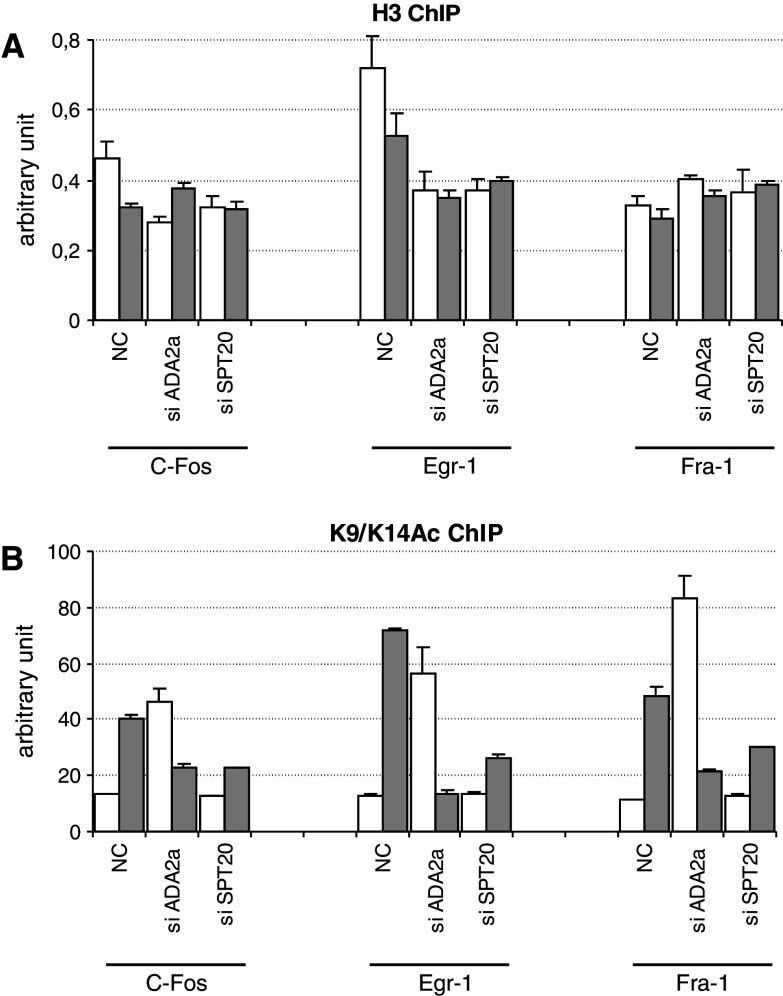
As the above results indicated that the knock-down of ATAC subunits leads to defects in the induction of IE gene expression and that the global levels of both H3K9Ac and H3K14Ac were decreased in cells deficient for either their SAGA or ATAC function (Figs. 4 and 7), we tested whether the knock-down of ATAC- and SAGA-specific subunit(s) influences the acetylation at specific gene loci. Thus, we carried ChIP experiments to test the H3 acetylation at positions K9 and K14 on histone H3 at the promoters of IE genes that we found to be regulated only by ATAC (see Figs. 6 and 7). HeLa cells were transfected either with non-targeting siRNA (NC), or siRNA against ADA2a or SPT20. At 48 h after transfection, the cells were serum starved overnight and then half of them treated with TPA for an hour as above. Chromatin was prepared and subjected to two different IPs. The anti-H3 core IP served as a control, while the IP using the anti-H3K9/K14Ac antibody highlighted the changes of this active chromatin mark at the tested genomic regions. All values obtained were normalized with those obtained at a non-coding region, where no transcriptional regulation takes place, thus the level of the histone marks remains unchanged. No significant changes were obtained in H3 levels at the tested IE gene promoters following TPA treatment (Fig. 8a). As expected, in the control siRNA-treated cells, the histone H3 acetylation level increased considerably (3- to 6-fold) at all the three IE promoters tested (see NC in Fig. 8b). Similarly, an increase in H3K9 and K14 acetylation was also observed in siSPT20-transfected cells; however, the increase was less pronounced (2-fold; Fig. 8b). Surprisingly, in siADA2a-transfected cells, the H3K9 and K14 acetylation pattern was completely deregulated before activation. In the non-treated cells, the H3K9 and H3K14 positions became highly acetylated at the promoters of these three, normally silent, genes. The fact that the presence of the positive H3K9/K14Ac histone mark at the promoter of IE genes does not correlate with the transcriptional status of the genes (Fig. 8, see also Figs. 6a and 7c–e) suggests that the H3K9 and H3K14 acetylation marks alone are not sufficient for recruiting the Pol II transcription machinery (see also Discussion). Additionally, in the siADA2a-transfected cells, following TPA induction, the H3K9/K14 acetylation dropped at the three tested promoters, when compared to the non-treated cells (Fig. 8b). This deregulated acetylation balance at the IE promoters may thus be responsible for the impaired activation of the IE genes in ATAC knock-down cells (see Fig. 7c–e). In all, our observations suggest that ATAC is indispensable for the transcriptional regulation of IE genes both in non-induced and in activated situation, while SAGA is not required.
Fig. 8.
Histone H3 acetylation is strongly affected in ATAC deficient cells at the promoter of IE genes. a ChIP results of siRNA-transfected cells obtained at the promoter of three IE genes (see Fig. 6a for indications) are represented before and after induction with TPA treatment (white and gray bars, respectively). The chromatin was immunoprecipitated with an antibody recognizing the core domain of H3. Results were normalized with those obtained at a non-transcribed region of the genome. The siRNA used for knocking down specific subunits of ATAC or SAGA is marked on the axis. b ChIP results obtained with an antibody specifically recognizing the H3K9/K14Ac signal shows perturbation at the promoter of TPA induced genes in siADA2a-transfected cells compared to the control (NC). In siSPT20-transfected cells, the response to stimulation at the acetylation level seems to be only slightly affected. Results are shown with SD values calculated from qPCR triplicates for each time point
Discussion
Metazoan ATAC complexes are conserved through evolution
Although GCN5 was the first enzyme identified to link histone modification and regulation of gene expression [46], it was only during recent years that data shed light on the existence of several GCN5-containing complexes in vivo in metazoans. The first indication came from the discovery that in metazoans and in plants the yeast Ada2 protein, an adaptor having effect on the activity of GCN5 activity [36, 47], has two orthologues: ADA2a and ADA2b [31, 48, 49]. These two orthologues in plants and Drosophila have distinct biological functions [50, 51]. Both ADA2a and ADA2b are essential in Drosophila [51], and the two proteins associate with dGCN5 in the context of two different complexes. While SAGA is a well-studied transcriptional co-activator complex [4, 52], the second complex, ATAC, was long over-looked in the different studies. The first signs of its presence in metazoans emerged during the determination of size of ADA2a- and ADA2b-containing complexes [16, 31]. The genetic analysis of mutant flies also suggested that two different assemblies are at play on the genome and that the substrate specificities of SAGA and ATAC differ in vivo [21, 51]. Further analysis of the dATAC complex established that a second HAT enzyme, dATAC2, is also part of the complex that seems to acetylate histone H4 at position H4K16 during Drosophila embryogenesis [17]. This observation, however, contradicts the results obtained on the polytene chromosomes prepared from ADA2a mutant flies, which show a clear decrease in acetylated H4K5 and H4K12 levels, while the H4K16 hyperacetylation on the male X chromosome is not altered [21, 53]. The different results of the two systems still await a precise analysis and explanation.
Considering the high conservation of the known subunits of both dSAGA and dATAC through evolution (reviewed in [4, 54]), the identification of an ATAC-like complex in vertebrates was expected. Early publications provided evidence for the interaction between hADA2a and hGCN5 or its mammalian-specific homologue PCAF [14, 55], but results clearly showing that both ATAC and SAGA exist in human cells were lacking until the last year [15, 18]. Our present study validates the results obtained by overexpressing one of the ATAC subunits and using it for consequent immunopurification [15, 18]. At the same time, some of the identified components of the hATAC purified by an anti-flag IP seem to be missing in our endogenous system (see Table 1). Wang and colleagues reported the presence of additional proteins in the ATAC purification associating with the Flag-tagged YEATS2 (i.e., UBAP2L, MAP3K7, POLE4, and TBP) [18]. These subunits or their Drosophila orthologues have not been identified in our endogenous complex or in the other reported ATAC complexes ([15, 17] and our study) (Table 1). Interestingly, dHCF1 and its human homologue were described to be components of the dATAC- and the Flag-tagged hYEATS2-containing hATAC complexes (Table 1); however, in our endogenous ATAC preparations and in that purified by Guelman and colleagues [15], no hHCF1 was identified either by mass spectrometry or by western blot analysis.
A strikingly high level of conservation exist between the human and Drosophila ATAC complexes in composition (see Table 1), though some differences remain. So far no human homologue of dATAC4 (GABPβ2) has been found in human ATAC preparations. Moreover, the Drosophila and the human ATAC complexes seem to vary both in their substrate specificity and their overall size. While the dATAC complex is clearly smaller than dSAGA [17, 31, 48], analysis of the endogenous or the Flag-purified human ATAC highlights a surprisingly large and variable size of the human complexes (from 2 MDa to about 600 kDa) ([15], and our unpublished results). These observations predict the future identification of new subunits that might be human specific, as the sum of the masses of the already identified subunits is only about 800 kDa. Alternatively, hATAC complexes may have heterogeneous stoichiometry of certain subunits or simply heterogeneous shapes possibly due to binding of ATAC to nucleic acids or to other substrates. In contrast, the human SAGA complex seems to have a well-conserved size (of about 2 MDa) and structure.
Our results together with those published recently [15, 18] point to a difference between the human and the Drosophila ATAC complexes. While in the case of the fly ATAC complex a strong histone H4 specific activity was documented, in the case of hATAC this activity is hardly detectable in vitro and in vivo. The in vitro observed differences between the substrate specificity of the Drosophila and the human ATAC complexes could be explained by eventual non-optimal conditions used in the reactions; however, the in vivo differences are more difficult to reconcile. In this respect, it is worth noting that, while the Drosophila ATAC2 enzyme was shown to acetylate H4K16 [17], its human homologue shows no measurable activity on histone tail peptides as substrates in vitro, neither when isolated nor in the context of the ATAC complex (see Figs. 2d and 3c). This discrepancy may be explained by the fact that the Drosophila ATAC2 protein contains a canonical PHD domain on its N-terminal end, while the vertebrate ATAC2 proteins lack key residues in this domain and contain only a putative Zn finger (ESM, Fig. 5). Thus, it is possible that, while dATAC2 is able to bind to histone tails via its PHD domain, as described for the PHD domain of TAF3 [56, 57], the vertebrate proteins are unable to do so. Consequently, the human ATAC complex may bind to other histone marks than the Drosophila complex, for example, via Tudor, WD40, or SANT domains present in its different subunits, and thus could acetylate different histone tail residues than the Drosophila complex. At the same time mice lacking ATAC2 die early in development and possess decreased global histone acetylation levels. However, since mATAC2 seems also to be essential for the integrity of the mammalian ATAC complex [15], future experiments should decide whether the drop in histone acetylation levels in Atac2 knock-out mice is due to the lack of ATAC2 as a HAT enzyme per se, or rather to the lower level of the entire ATAC complex. The mouse ATAC2 ablation in Atac2 knock-down or knock-out systems leads to a decrease in global H3K9, H4K5, H4K12 and H4K16 acetylation levels [15]. The decrease of H3K9Ac is in agreement with our results obtained following ADA2a knock-down in human cells; however, we did not detect any change in the different histone H4 acetylations. Thus, understanding the exact biological function, the precise acetyltrasferase specificity of the mammalian ATAC complexes (G-ATAC and P-ATAC), and the role of the second potential acetyltransferase, ATAC2, in these complexes still awaits further analysis.
ATAC and SAGA regulate different set of stress inducible target genes
Yeast SAGA was suggested to play a role in stress-regulated genes [58] and act as a locus-specific coactivator complex that binds close to the nuclosome-free region formed upstream of the +1 nucleosomes on expressed genes in the yeast genome [59]. Similarly, Drosophila SAGA subunits (TRRAP and GCN5) were detected at the inducible hsp70 gene promoters following heat shock [60]. Human SAGA was shown to play a direct role in the up-regulation of p53-dependent genes following UV-C irradiation [36] and also in the regulation of endoplasmatic reticulum stress-induced genes [13]. In contrast, hSAGA did not seem to be involved in the regulation of IE genes induced by Na-arsenite stress [13], suggesting that hSAGA is not required as a promoter-specific coactivator at every stress-regulated gene promoter.
A stimulation-specific coactivator role of ATAC can be drawn from our new results, which seems to be conserved between Drosophila and human. ATAC gets recruited to TPA-induced transcription puffs on the polytene chromosomes of Drosophila, while these sites are deprived from dSAGA as no ADA2b, a SAGA specific subunit, was detected at the TPA-induced puffs. The same scenario stands for human cells, where hATAC subunits accumulate at the promoter of activated IE genes together with RNA Pol II after TPA induction. In agreement, the induction of the tested human IE genes was seriously compromised when cells harbored decreased level of different ATAC subunits (Fig. 7). Also, when a core ATAC subunit was knocked down by siRNA, the H3K9 and K14 acetylation marks decreased at the promoters following TPA stimulation (Fig. 8). Surprisingly, under “non-activated” conditions, the same acetylation marks were about 3- to 7-fold higher at the tested promoters than in the control cells. This increased basal histone H3 acetlyation level might be the indirect result of the perturbation we observed in the global H3S10P mark (Fig. 4). It is possible that the decreased H3S10 phosphorylation makes the chromatin at the IE promoters more permissive for the recruitment of another HAT complex than ATAC, which normally would not act at these sites in the cell. Thus, our results shed light on a dual function of ATAC at the IE gene promoters. On one hand, ATAC is indispensable for the induced transcription of these genes after stress. On the other hand, the complex is also required, probably indirectly, for the maintenance of the low level of H3K9/K14Ac marks at the same promoters in basal conditions.
Our results show that, in contrast to ATAC, TPA-inducible promoters lack hSAGA, providing evidence for the different recruitment pattern of the two complexes on the genome. In good agreement with this differential coactivator recruitment model, the group of R. Roeder has shown that to UV-stress-regulated gene promoters are occupied only by SAGA, but not ATAC [36]. These observations together demonstrate that two distinct types of GCN5- (or PCAF)-containing HAT complexes with potentially different coactivator activities exist in the cells to regulate different subset of induced genes.
One distinction between IE genes and other inducible genes is that the histones at IE genes get both acetylated and phosphorylated upon induction [61]. In this respect, it is worth noting that dATAC was shown to play a role in histone H3S10 phosphorylation [45]. In ADA2a mutant flies, decreased histone acetylation led to consequent decrease in H3S10 phosphorylation by the JIL kinase [45]. Our novel observations show that the mammalian ATAC complex is also required for global H3S10 phosphorylation establishing a conserved link between ATAC function and H3S10 phophorylation during evolution. Human cells with knock-down levels of ADA2a (hATAC subunit) also possess a strongly decreased global H3S10P level. Thus, ATAC is acting both at global and locus-specific levels on the genome. Our results suggest that in ADA2a knock-down condition the loss of the ATAC HAT complex results in unbalanced H3 tail acetylation at the IE gene promoters, that in combination with the global decrease of the H3S10 phophorylation makes the activation of immediate early genes deficient. Thus, our data demonstrate that ATAC plays a crucial role in the transcriptional regulation of IE genes.
During evolution, the complexity increased not only at the level of gene number and genome size but also at regulatory circuits. Our present understanding on the composition and functioning of GCN5-containing HAT complexes is a nice example for this increase. In the unicellular organism Saccharomyces cerevisiae, Gcn5 is the component of two complexes (ySAGA and yADA) of which the catalytic core remains exactly the same, formed by the Ada2-Ada3-Gcn5 triad [62]. In Drosophila, we find two genes encoding ADA2 paralogues, ADA2a and ADA2b, and, as a consequence, two different complexes, dATAC and dSAGA, have evolved [17, 21, 31]. Furthermore, in dATAC, a second potential acetyltransferase enzyme was identified that brings another activity to the complex [17]. The complexity reaches its maximum in mammalian cells, where on the top of the two ADA2 proteins, two GCN5 homologues are also present (GCN5 and PCAF). Our data suggest, together with that of Gamper and colleagues [36], that both GCN5 and PCAF form SAGA- and ATAC-type complexes and that even all the four possibilities may coexist in one cell (this study, and not shown). At the same time, our results also provide evidence that the functional differences of these complexes materialize mainly in vivo. The numerous subunits surrounding the enzymes (GCN5 or PCAF) in such complexes can on the one hand affect their activity, as shown for hADA2b in the context of hSAGA [36], while on the other hand, these subunits possibly function as interaction surfaces for different transcription activators playing roles in distinct signaling pathways during development or stress response. In vitro dissection of the interactions within the complexes might provide data that will help understanding the biological role of each subunit. However, further in vivo genetic studies are indispensable for the comprehension of function in the cellular context. In the present study, we identified the components of the endogenous hATAC complex, and we also provide evidence that SAGA and ATAC complexes do not regulate the same subset of inducible genes that could be the starting point for future more detailed genome-wide studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to W. Herr, B. Malecova and T. Oelgeschläger for reagents, to R. and J. Conaway for sharing unpublished results, to R. Schneider, K. Kamieniarz, J. Bonnet and G. Lang for suggestions and help with the HAT assays, to the IGBMC core facilities, and to D. Devys, B. Malecova and T. Pankotai for critically reading the manuscript. Z.N. was supported by a fellowship from the European Community grant (HPRN-CT-00504228) and by a fellowship from the Fondation pour la Recherche Médicale (FRM). A.R. and A.K. were supported by fellowships of the Alsace Region. This work was supported by funds from CNRS, INSERM, Université de Strasbourg, the FRM, and European Community (HPRN-CT 00504228, STREP LSHG-CT-2004-502950 and EUTRACC LSHG-CT-2007-037445) and INCA (2008-UBICAN) grants.
Footnotes
Z. Nagy and A. Riss contributed equally to this work.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Hayes JJ, Hansen JC. Nucleosomes and the chromatin fiber. Curr Opin Genet Dev. 2001;11:124–129. doi: 10.1016/S0959-437X(00)00168-4. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 5.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 6.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 9.Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 10.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 11.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/MMBR.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/S0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 15.Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol. 2009;29:1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, Workman JL. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila . Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 18.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- 20.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 21.Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol. 2006;26:9413–9423. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 24.Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, Mandel JL, Tora L, Devys D. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 25.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly JM, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 28.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3–K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malecova B, Gross P, Boyer-Guittaut M, Yavuz S, Oelgeschlager T. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem. 2007;282:24767–24776. doi: 10.1074/jbc.M702776200. [DOI] [PubMed] [Google Scholar]
- 31.Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, Cserpan I, Lebedeva L, Nabirochkina E, Udvardy A, Tora L, Boros I. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol. 2003;23:306–321. doi: 10.1128/MCB.23.1.306-321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leurent C, Sanders SL, Demeny MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Mapping key functional sites within yeast TFIID. EMBO J. 2004;23:719–727. doi: 10.1038/sj.emboj.7600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demeny MA, Soutoglou E, Nagy Z, Scheer E, Janoshazi A, Richardot M, Argentini M, Kessler P, Tora L. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE. 2007;2:e316. doi: 10.1371/journal.pone.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnitzler GR (2001) Isolation of histones and nucleosome cores from mammalian cells. Curr Protoc Mol Biol Ch 21, Unit 21.5 [DOI] [PubMed]
- 35.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/S1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 36.Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol. 2008;28:2517–2527. doi: 10.1128/MCB.01461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuyama K, Yoshida M, Yamashita A, Deyama T, Baba M, Suzuki A, Mohri H, Ikezawa Z, Nakajima H, Hirai S, Ohno S. MAPK upstream kinase (MUK)-binding inhibitory protein, a negative regulator of MUK/dual leucine zipper-bearing kinase/leucine zipper protein kinase. J Biol Chem. 2000;275:21247–21254. doi: 10.1074/jbc.M001488200. [DOI] [PubMed] [Google Scholar]
- 38.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 39.Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 40.Meisterernst M, Roeder RG. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-C. [DOI] [PubMed] [Google Scholar]
- 41.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 42.Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, Meisterernst M, Burley SK. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001;106:71–81. doi: 10.1016/S0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 43.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kikuchi H, Takami Y, Nakayama T. GCN5: a supervisor in all-inclusive control of vertebrate cell cycle progression through transcription regulation of various cell cycle-related genes. Gene. 2005;347:83–97. doi: 10.1016/j.gene.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Ciurciu A, Komonyi O, Boros IM. Loss of ATAC-specific acetylation of histone H4 at Lys12 reduces binding of JIL-1 to chromatin and phosphorylation of histone H3 at Ser10. J Cell Sci. 2008;121:3366–3372. doi: 10.1242/jcs.028555. [DOI] [PubMed] [Google Scholar]
- 46.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 47.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 48.Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hark AT, Vlachonasios KE, Pavangadkar KA, Rao S, Gordon H, Adamakis ID, Kaldis A, Thomashow MF, Triezenberg SJ. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim Biophys Acta. 2009;1789:117–124. doi: 10.1016/j.bbagrm.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, Boros I. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol. 2005;25:8215–8227. doi: 10.1128/MCB.25.18.8215-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carre C, Ciurciu A, Komonyi O, Jacquier C, Fagegaltier D, Pidoux J, Tricoire H, Tora L, Boros IM, Antoniewski C. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep. 2008;9:187–192. doi: 10.1038/sj.embor.7401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 55.Forsberg EC, Lam LT, Yang XJ, Nakatani Y, Bresnick EH. Human histone acetyltransferase GCN5 exists in a stable macromolecular complex lacking the adapter ADA2. Biochemistry. 1997;36:15918–15924. doi: 10.1021/bi971664x. [DOI] [PubMed] [Google Scholar]
- 56.van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure. 2008;16:1245–1256. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 57.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/S1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 59.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lebedeva LA, Nabirochkina EN, Kurshakova MM, Robert F, Krasnov AN, Evgen’ev MB, Kadonaga JT, Georgieva SG, Tora L. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci USA. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/S0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 62.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.