Abstract
A mitogen-activated protein kinase (MAPK), Pfmap2, has been identified in Plasmodium falciparum. However, its bona fide activator remains elusive as no MAPK kinase (MAPKK) homologues have been found so far. Instead, Pfnek3, a NIMA (never in mitosis, Aspergillus)-related kinase, was earlier reported to display a MAPKK-like activity due to its activating effect on Pfmap2. In this study, the regulatory mechanism of Pfnek3 was investigated. Pfnek3 was found to possess a SSEQSS motif within its activation loop that fulfills the consensus SXXXS/T phospho-activating sequence of MAPKKs. Functional analyses of the SSEQSS motif by site-directed mutagenesis revealed that phosphorylation of residues S221 and S226 is essential for mediating Pfnek3 activity. Moreover, via tandem mass-spectrometry, residue T82 was uncovered as an additional phosphorylation site involved in Pfnek3 activation. Collectively, these results provide valuable insights into the potential in vivo regulation of Pfnek3, with residues T82, S221 and S226 functioning as phospho-activating sites.
Keywords: Activation loop, MAPK, Phosphorylation, Plasmodium falciparum, Site-directed mutagenesis
Introduction
Malaria remains a leading cause of morbidity and mortality in many tropical countries despite continuous efforts to eradicate it or control its spread. Each year, this disease affects 350–500 million people worldwide and accounts for over 1 million deaths [1]. The most lethal form of human malaria is caused by infection with Plasmodium falciparum. These parasitic protozoa undergo complex physiological and morphological changes as they develop through their life cycles within the mosquito and human hosts [2]. Although various stages of the P. falciparum life cycle have been defined, little is known about the mechanisms regulating the growth and differentiation of these parasites. With the emergence of P. falciparum resistance against many of the existing anti-malarial drugs, elucidating these mechanisms would enhance our knowledge of the parasite pathogenesis and facilitate the design of novel treatments against malaria.
The mitogen-activated protein kinase (MAPK) signaling pathways are among the most widespread mechanisms involved in cell regulation. They are highly conserved throughout eukaryotic evolution and mediate crucial biological processes including cell proliferation, differentiation, migration, and apoptosis [3]. A distinctive feature of MAPK pathways is the presence of a three-kinase phosphorelay module which includes a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK). In response to various stimuli, such as growth factors and inflammatory cytokines, MAPKKKs activate MAPKKs via phosphorylation of the serine and serine/threonine residues within the SXXXS/T activation site [4]. The dual-specificity MAPKKs in turn phosphorylate the MAPKs on both threonine and tyrosine residues within the TXY motif to activate them. Upon activation, MAPKs exert their influences on different physiological processes through phosphorylation of appropriate downstream substrates [5].
As MAPK signaling pathways play pivotal roles in the cellular processes of many eukaryotes, it is plausible that similar mechanisms might be adopted by P. falciparum to regulate its complex life cycle. To date, only two MAPK homologues (Pfmap1 and Pfmap2) have been identified in these parasites [6–8]. In a previous gene knock-out study, Pfmap2, but not Pfmap1, was found to be essential for P. falciparum to complete its intraerythrocytic development [9]. This study thus shed light on the importance of a MAPK in mediating the growth and proliferation of these parasites. Prior biochemical characterizations of Pfmap2 revealed that it possesses a TSH motif in place of the consensus TXY activation site found in most MAPKs [8]. As such, activation of Pfmap2 in vivo may require phosphorylation within the TSH motif by upstream kinases. However, analyses of the P. falciparum kinome did not uncover sequences that are obvious homologues of typical MAPKKs. In view of this, the identity of a MAPK activator and the existence of a complete MAPK signaling pathway in P. falciparum remain to be established [10, 11].
Nevertheless, attempts to isolate plasmodial MAPKK-related genes using a degenerate oligonucleotide-based PCR method and BLAST analyses of the Plasmodium genome have led to the identification of Pfnek1 (P. falciparum NIMA-like kinase 1) and PfPK7 (P. falciparum protein kinase 7) [12, 13]. Although Pfnek1 shares low-protein sequence identity (10%) with the mammalian MAPKKs, it possesses an activation motif reminiscent of those found in the MAPKKs [12]. On the other hand, the similarity between PfPK7 and MAPKKs is restricted to the C-terminal lobe of the kinase domain [13]. Of these two kinases, only Pfnek1 was capable of phosphorylating Pfmap2 in vitro [12]. Moreover, Pfnek1 and Pfmap2 act synergistically in exogenous substrate phosphorylation [12]. Typically, NIMA-like kinases function as important cell cycle regulators in eukaryotic cells [14]. The ability of Pfnek1 to phosphorylate Pfmap2 suggests the possibility of Pfnek1 as a MAPK regulator, and that NIMA-like kinases may adopt unique functions in the malarial parasite.
Interestingly, a second plasmodial NIMA-like kinase, Pfnek3, was subsequently found to be competent in phosphorylating and activating Pfmap2 [15]. The mechanism by which Pfnek3 activates Pfmap2 was further validated to involve phosphorylation of the conserved threonine residue, T290, within the latter’s TSH activation motif [16]. Despite sharing only 17–19% amino acid sequence identity with the mammalian and yeast MAPKKs, these observations support Pfnek3 as a plasmodial kinase displaying a MAPKK-like activity. Hence, in the absence of a typical MAPKK, Pfnek3 could potentially function as a MAPK regulator in P. falciparum, similar to that of Pfnek1. Currently, the regulatory mechanism of Pfnek3 remains enigmatic. As such, this study aimed to examine the regulation of Pfnek3 activity and assess if it possesses additional biochemical properties that are similar to the MAPKKs.
Materials and methods
Homology modeling of Pfnek3
Tertiary structure of Pfnek3 was predicted using the SWISS-MODEL program (http://www.swissmodel.expasy.org//SWISS-MODEL.html) [17], based on templates which share at least 25% primary sequence similarity. The Pfnek3 amino acid sequence submitted for modeling process was retrieved from the Plasmodium genome database (PlasmoDB; http://www.PlasmoDB.org). The Pfnek3 tertiary model, together with the crystal structure of human MAPKK1 (1S9J) [18], were analyzed and compared using the Swiss-PdbViewer (http://expasy.org/spdbv/) [19].
Construction of Pfnek3 mutants via site-directed mutagenesis
The truncated Pfnek3 gene, encoding a catalytically active form of Pfnek3, was expressed by the GST-encoding pGEX-6P-1 vector [15]. The resultant recombinant expression plasmid was used as the template for site-directed mutagenesis reactions. All GST-Pfnek3 mutants were generated according to the QuikChange™ site-directed mutagenesis protocol (Stratagene), using mutagenic primers summarized in Table 1. Following which, DNA sequencing was performed to verify the authenticity of the respective GST-Pfnek3 mutants and to ensure that no spontaneous nucleotide substitution was introduced during the PCR amplification process. The catalytically inactive recombinant Pfnek3 (GST-ΔPfnek3) harboring a K102M mutation at the ATP binding site was previously constructed and used as a negative control in kinase assays [15].
Table 1.
Mutagenic primers used for the construction of GST-Pfnek3 mutants
| Mutation | Sequence of mutagenic primers (5′-3′) |
|---|---|
| T82A | F: TGTTAGATTTCATGGCATCAGATAGTGAAATTCATTTG |
| R: CAAATGAATTTCACTATCTGATGCCATGAAATCTAACAC | |
| S221A | F: GATTTCGGTATAGCATCAGAACAAAGTTC |
| R: GAACTTTGTTCTGATGCTATACCGAAATC | |
| S221D | F: GATTTCGGTATAGATTCAGAACAAAGTTC |
| R: GAACTTTGTTCTGAATCTATACCGAAATC | |
| S221E | F: GATTTCGGTATAGAATCAGAACAAAGTTC |
| R: GAACTTTGTTCTGATTCTATACCGAAATC | |
| S222A | F: GATTTCGGTATATCAGCAGAACAAAGTTC |
| R: GAACTTTGTTCTGCTGATATACCGAAATC | |
| S225A | F: TCATCAGAACAAGCGTCAAATAATAATTTAGG |
| R: CCTAAATTATTATTTGACGCTTGTTCTGATGA | |
| S226A | F: GAACAAAGTGCAAATAATAATTTAGGAAC |
| R: GTTCCTAAATTATTATTTGCACTTTGTTC | |
| S226D | F: GAACAAAGTGATAATAATAATTTAGGAAC |
| R: GTTCCTAAATTATTATTATCACTTTGTTC | |
| S226E | F: GAACAAAGTGAAAATAATAATTTAGGAAC |
| R: GTTCCTAAATTATTATTTTCACTTTGTTC |
The designations F and R denote the forward and reverse primers, respectively, with the sites of mutagenesis underlined
Heterologous expression and purification of recombinant proteins
Recombinant pGEX plasmids expressing GST-Pfnek3, GST-Pfnek3 mutants, and GST-Pfmap2 (a gift from C. Doerig) were independently electro-transformed into Escherichia coli BL21-CodonPlus™ cells (Stratagene). Transformants obtained were inoculated into 100 ml Luria Bertani broth supplemented with ampicillin and chloramphenicol (100 and 40 μg/ml, respectively) for vector propagation. The cultures were grown at 37°C with continuous agitation at 220 rpm until an OD600 of 0.6–0.8 was reached, after which they were induced with 1 mM isopropyl-beta-d-thiogalactopyranoside (Sigma) and incubated at room temperature overnight with shaking at 220 rpm. Following this, the cells were harvested and lysed by sonication as previously described [16]. Fusion proteins were purified from soluble cell free extracts by affinity chromatography using the Microspin GST Purification Module (GE Healthcare) according to manufacturer’s instructions. The eluted proteins were subsequently analyzed by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Gel images of separated proteins were captured using the Bio-Rad Molecular Imager Gel Doc XR system equipped with the Quantity One® software. Protein concentration of the purified samples was determined by the Bio-Rad protein assay, using bovine serum albumin (BSA; Sigma) as the reference protein standard. In experiments that required cleavage of the GST fusion partner, purified proteins were incubated overnight at 4°C with the PreScission Protease (GE Healthcare).
Protein phosphorylation analysis of GST-Pfnek3 and its mutants
Following bacterial expression and purification, wild-type and mutant GST-Pfnek3 proteins were separated by SDS-PAGE using 10% polyacrylamide gels. After electrophoresis, the gels were stained with Pro-Q Diamond phosphoprotein gel stain (Invitrogen) according to manufacturer’s protocol and visualized on a UV transilluminator. The gels were subsequently stained with Coomassie Blue to ensure uniform loading of protein samples for each lane.
Dephosphorylation of GST-Pfnek3 by lambda phosphatase
For dephosphorylation reactions, 10 μg of wild-type GST-Pfnek3 was treated with 500 U of lambda phosphatase (Millipore) in 40 μl of reaction buffer supplied by the manufacturer. The phosphatase was omitted from mock-treated samples which served as negative controls. Incubations were carried out for 30 min at 30°C before the reaction mixture was supplemented with a 10× phosphatase inhibitor cocktail (Thermo Fisher Scientific) to terminate the phosphatase activity. One-half of the reaction mixture was thereafter subjected to phosphorylation analysis to ensure dephosphorylation of the protein had occurred. The remaining half was used in kinase assays to examine the activity of dephosphorylated GST-Pfnek3.
ELISA-based protein kinase assay
Kinase assays were performed using 96-well microtiter plates (Costar) coated with dephosphorylated myelin basic protein (MBP; Millipore). Each kinase assay reaction mix of 60 μl comprised of 10 μg of wild-type or mutant GST-Pfnek3 enzymes, 500 μM ATP, 40 mM MgCl2 and 50 mM Tris–HCl (pH 7.2). The assay was initiated upon addition of the kinase reaction mix to each well. The microplates were then incubated at 30°C for 30 min before the reactions were quenched by repeated washing with phosphate-buffered saline (PBS). Mixed monoclonal anti-phosphoserine/threonine antibodies (1:500 dilution; Millipore) were subsequently added to each well and incubated at room temperature for 1 h. During this period, any phosphorylation of MBP that arose from the activities of the kinases would be detected by these antibodies. After another round of extensive washing with PBS, the wells were incubated with a peroxidase-conjugated secondary antibody (1:2,000 dilution; Thermo Fisher Scientific) for 45 min. The wells were again rinsed, followed by addition of the SuperSignal West Femto peroxidase substrate (Thermo Fisher Scientific). The resulting chemiluminescent signal arising from the presence of phosphorylated serine and/or threonine residues was measured using the Genios™ microplate reader (Tecan).
For co-incubation kinase assays, the same protocol was adopted except that each MBP-coated well was incubated for 15 min with a kinase reaction mix that contained 5 ng of wild-type or mutant GST-Pfnek3 and 3 μg of GST-Pfmap2.
Liquid chromatography-tandem mass spectrometric (LC-MS/MS) analysis
To determine the phosphorylation sites present in Pfnek3, 10 μg of wild-type GST-Pfnek3 was gel-purified by 10% SDS-PAGE and the coomassie-stained band containing the protein of interest was excised. In-gel digestion with trypsin and the subsequent LC-MS/MS analysis were performed by A*STAR Experimental Therapeutics Centre using API QSTAR Pulsar (Applied Biosystems). The experiment was repeated with the catalytically inactive GST-ΔPfnek3 to ascertain that any phospho-sites detected in the wild-type kinase were indeed due to autophosphorylation and not attributed to the kinase activities of any contaminating bacterial kinases.
All MS/MS samples were analyzed using Mascot (version 2.2.0; Matrix Science, London, UK). Mascot was set up to search the NCBInr_20070223 database, specifying trypsin as the digestion enzyme. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 500 ppm. Scaffold (version Scaffold-01_07_00; Proteome Software, Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [20]. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [21].
Results
The activity of Pfnek3 was abrogated in the absence of phosphorylation
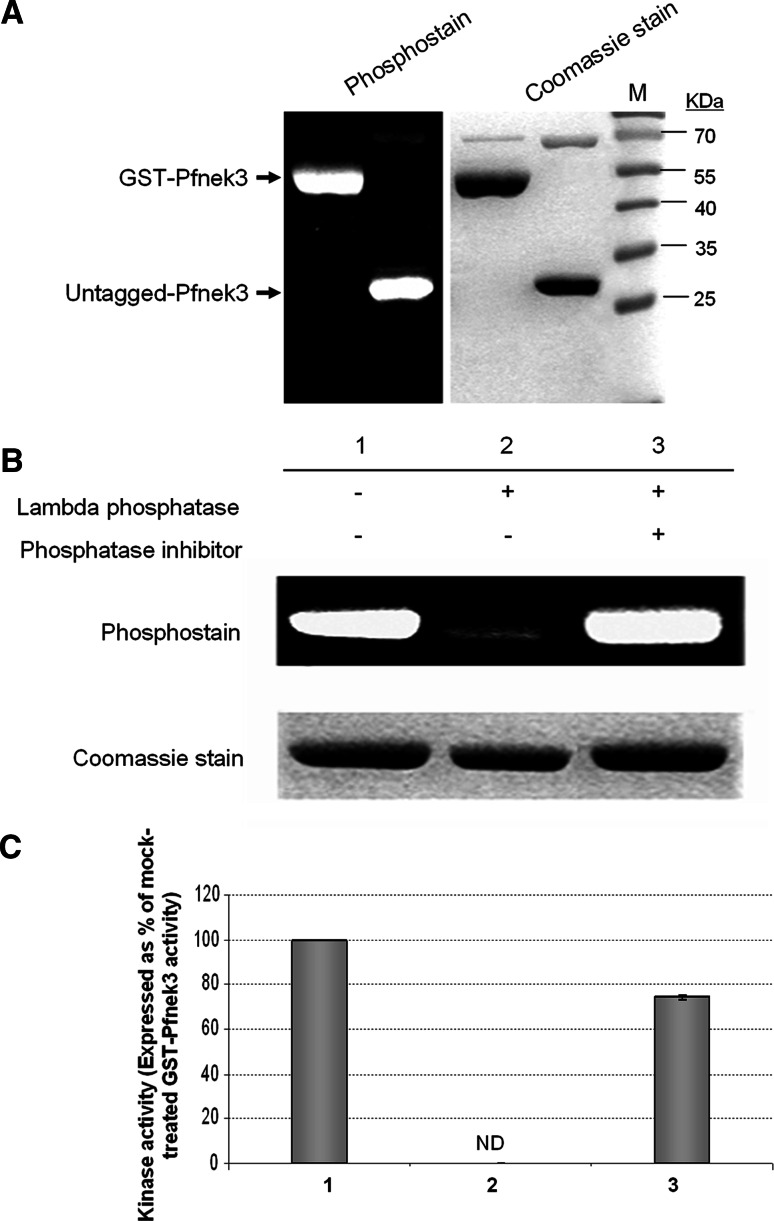
From an earlier study, recombinant wild-type GST-Pfnek3 was observed to display a significant level of phosphorylation [15]. As its kinase inactive counterpart (GST-ΔPfnek3) lacks detectable phosphorylation, Pfnek3 was proposed to exhibit an autophosphorylation property [15]. To exclude the possibility that phosphorylation had occurred solely on the GST moiety, GST-Pfnek3 was subjected to PreScission protease cleavage and the resultant untagged Pfnek3 was examined for its phosphorylation status. As shown in Fig. 1a, the untagged kinase retained a level of phosphorylation that was comparable to GST-Pfnek3, hence confirming the autophosphorylation ability of Pfnek3.
Fig. 1.
Activation of Pfnek3 is correlated with its phosphorylation status. a To authenticate the autophosphorylation ability of Pfnek3, GST-Pfnek3 was subjected to PreScission protease cleavage to remove the GST moiety. The phosphorylation level of the untagged recombinant kinase was subsequently analyzed using the Pro-Q Diamond phosphoprotein gel stain. The untagged Pfnek3 displayed a similar level of phosphorylation as its GST-fused counterpart. Lane M denotes the protein standard marker (Fermentas) used for estimating the protein sizes. b Purified GST-Pfnek3 was treated with lambda phosphatase in the presence (lane 3) or absence (lane 2) of the phosphatase inhibitor cocktail. In the mock-treated sample (lane 1), lambda phosphatase was omitted. c Following lambda phosphatase treatment in (b), an aliquot of the respective reaction mixture was transferred to MBP-coated microtiter plates to assay for the activity of dephosphorylated GST-Pfnek3. ND Kinase activity not detected
To determine whether phosphorylation of Pfnek3 plays a crucial role in regulating its kinase activity, wild-type GST-Pfnek3 was treated with lambda phosphatase to eliminate phosphoryl groups on serine, threonine, and tyrosine residues. Upon incubation with lambda phosphatase, a significant reduction in the phosphorylation level of GST-Pfnek3 was observed (Fig. 1b, lane 2), validating the dephosphorylated status of the kinase. In addition, phosphatase-treated GST-Pfnek3 exhibited a complete loss of kinase activity using MBP as the exogenous substrate (Fig. 1c, lane 2). On the contrary, preincubation of lambda phosphatase with the phosphatase inhibitor cocktail blocked both dephosphorylation and inactivation of GST-Pfnek3 (Fig. 1b, c, lane 3). These results indicate that the loss of kinase activity observed earlier was indeed associated with the absence of phosphorylation and therefore highlight the importance of phosphorylation in the activation of Pfnek3.
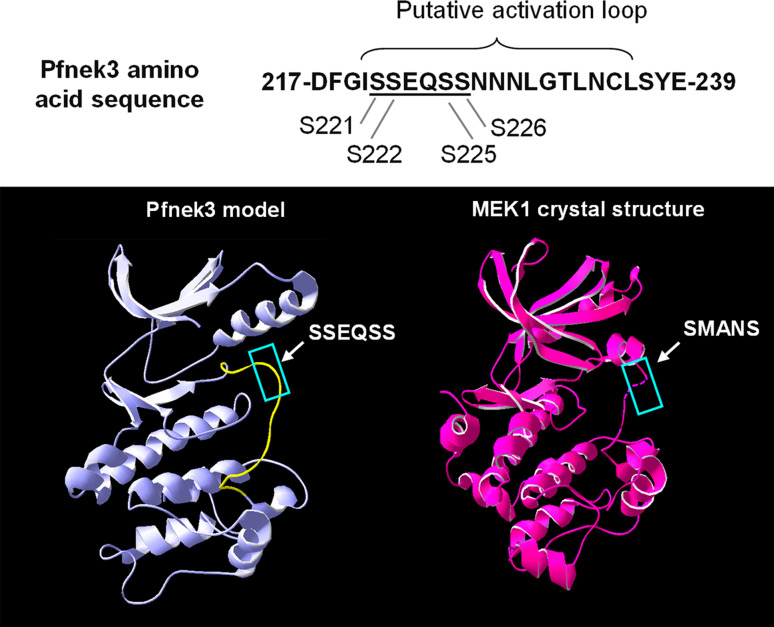
Pfnek3 possesses a SSEQSS motif that is analogous to the activation sites of MAPKKs
Several classes of eukaryotic protein kinases, including the MAPKKs, are regulated via phosphorylation at the activation loop [22]. This segment is commonly defined as the region that spans the DFG and APE motifs within kinase subdomains VII and VIII, respectively [23]. In Pfnek3, the APE motif has been altered to a SYE, and the region that represents its putative activation loop is highlighted in Fig. 2. Interestingly, a SSEQSS motif within this loop fulfills the conserved SXXXS/T phospho-activating sequence found in the MAPKKs. To further assess whether this motif is analogous to the activation sites of MAPKKs in terms of its tertiary structure, the three-dimensional model of Pfnek3 was generated using the SWISS-MODEL program. When the Pfnek3 model was compared with the crystal structure of human MEK1 (PDB identifier: 1S9J), a well-characterized MAPKK, it was observed that the SSEQSS motif of Pfnek3 coincides with the SMANS activation site of MEK1 (Fig. 2). It has been established that phosphorylation of the two serine residues within the SMANS motif is critical for the activation of MEK1 [22]. Hence, these observations provide a strong indication that the SSEQSS motif could adopt a similar function as the SMANS motif in mediating the activation of Pfnek3.
Fig. 2.
Pfnek3 possesses a MAPKK-like activation site within its activation loop. Top The Pfnek3 amino acid sequence corresponding to the putative activation loop is illustrated. The underlined SSEQSS motif bears a resemblance to the SXXXS/T activation sites of MAPKKs. Bottom The Pfnek3 model was generated using SWISS-MODEL and subsequently compared with the crystal structure of human MEK1 (1S9J). The putative activation loop of Pfnek3 is highlighted in yellow. The SSEQSS motif was observed to locate at the surface of this loop and reside in a position analogous to the SMANS activation site of human MEK1 (both boxed up in blue)
Mutations of residues S221 and S226 in the SSEQSS motif affect autophosphorylation and kinase activity of Pfnek3
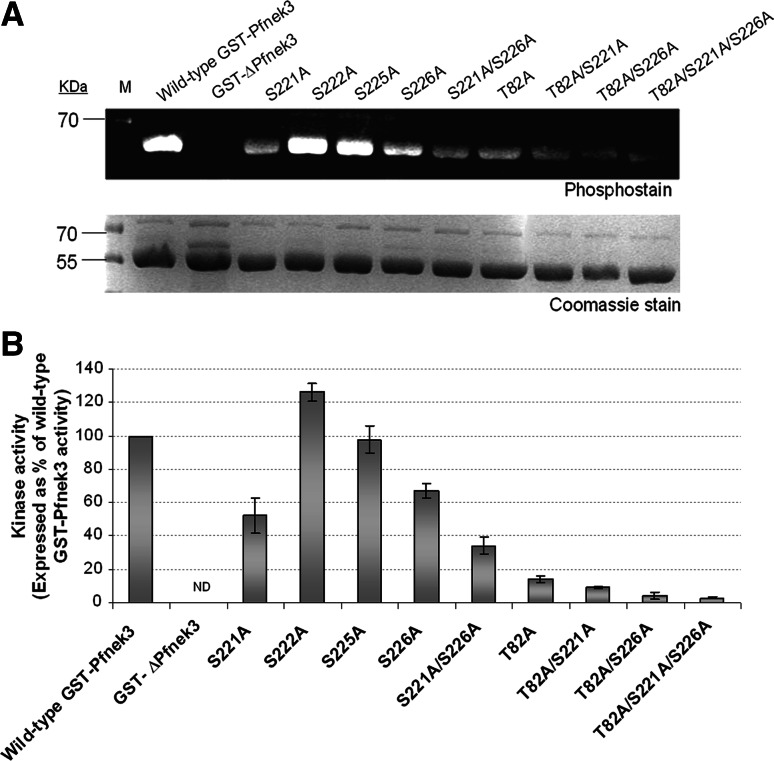
To investigate whether any of the serine residues in the SSEQSS motif could serve as phospho-regulatory sites, they were independently substituted with a non-phosphorylatable alanine via site-directed mutagenesis. Each of these GST-tagged mutants (hereafter known as S221A, S222A, S225A, and S226A) was similarly expressed in E. coli cells and assayed to determine their relative autophosphorylation levels. While both S222A and S225A mutants exhibited a level of autophosphorylation that was similar to the wild-type enzyme, significant reductions in autophosphorylation levels were observed for the S221A and S226A mutants (Fig. 3a). A double mutant (S221A/S226A) was thereafter constructed, and it displayed a much lower autophosphorylation signal as compared to either of the individual single mutants.
Fig. 3.
Autophosphorylation levels and kinase activities of GST-Pfnek3 alanine-substituted mutants. a Purified wild-type GST-Pfnek3 and the indicated mutants were resolved by SDS-PAGE and stained with the phosphoprotein gel stain to assess their autophosphorylation levels. In the protein standard marker (M), the 70-kDa protein band corresponds to a phosphoprotein that could be detected using the phosphostain. Following phosphostaining, the gel was stained with Coomassie Blue to ensure that relatively equal amounts of proteins were loaded. b The in vitro kinase activities of the respective GST-Pfnek3 mutants were subsequently determined by their ability to phosphorylate the MBP substrates. ND Kinase activity not detected
The effect of these mutations on Pfnek3 activity was then analyzed using the microtiter plate kinase assay. Consistent with the phenomenon observed in Fig. 3a, the kinase activities of both S221A and S226A mutants were lower than that of the wild-type kinase (Fig. 3b). Likewise, the activity of the S221A/S226A double mutant was significantly lower as compared to its respective single mutants. The concomitant reductions in autophosphorylation levels and kinase activities of the S221A and S226A mutants indicate the potential of these serine residues to function as crucial phospho-activating sites in Pfnek3.
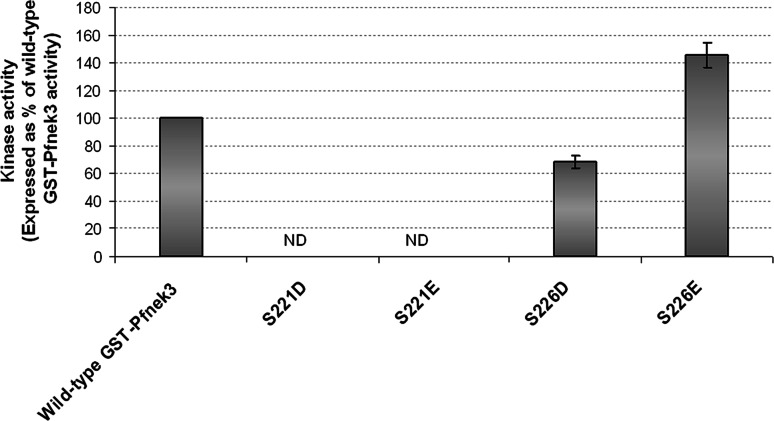
Phospho-mimic mutants (S221D/E and S226D/E) were subsequently constructed in an effort to further examine the above postulation. Aspartates and glutamates are utilized, with varying competency, to simulate the effects of phosphorylation and maintain kinases in the activated states [24]. Replacing S221 with either aspartate or glutamate residues resulted in a loss of Pfnek3 activity (Fig. 4). Conversely, the activity of the S226D mutant was approximately half of the wild-type kinase activity. When S226 was altered to a glutamate residue, the activity of the mutant was increased by approximately 50% as compared to wild-type Pfnek3. This finding supported our speculation that S226 is an activating phospho-acceptor site, whose functionality could be substituted by the negatively charged glutamate. At this point, however, it is difficult to unequivocally rule out the possibility of S221 as a phosphorylation site. Since the catalytic activity of Pfnek3 was completely abrogated upon mutation of S221 to either an aspartate or a glutamate, these mutations may have led to unfavorable conformational changes resulting in the loss of kinase activity.
Fig. 4.
Kinase activities of phospho-mimic GST-Pfnek3 mutants. Residues S221 and S226 were each replaced with either aspartate or glutamate to mimic phosphorylation at these residues. Activities of these mutants were analyzed using the ELISA-based kinase assay as described in “Materials and methods”. ND Kinase activity not detected
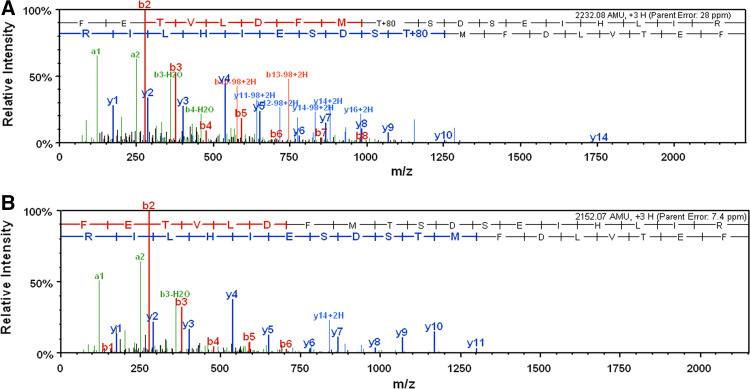
LC-MS/MS analyses revealed amino acid T82 as a crucial phosphorylation site in Pfnek3
The low level of autophosphorylation and kinase activity retained by the S221A/S226A double mutant implies that additional phosphorylation sites are present in Pfnek3, which could potentially be involved in regulating its kinase activity. As such, LC-MS/MS was employed to verify this speculation. In total, seven tryptic peptides were identified which made up 30% of the Pfnek3 amino acid sequence. A single phosphorylation site was detected in the 74FETVLDFMTSDSEIHLIR91 peptide, and amino acid residue T82 was validated as the site of phosphorylation (Fig. 5a). The loss of 80 Da detected on this residue corresponds to the neutral loss of a phosphate group. When LC-MS/MS was repeated using the catalytically inactive GST-ΔPfnek3, no phosphorylation was detected on the same site (Fig. 5b). These observations thus validate that residue T82 is an autophosphorylation site. To elucidate whether phosphorylation of T82 serves as a mechanism to regulate the kinase activity of Pfnek3, the amino acid residue was similarly replaced with an alanine to prevent phosphorylation from occurring at this site. Absence of the T82 phosphorylation site provoked a loss of wild-type activity by over 80% (Fig. 3b), suggesting its importance in modulating the catalytic activity of Pfnek3. A significant drop in autophosphorylation level was also observed for the T82A mutant (Fig. 3a). This finding was somewhat expected since removal of this phospho-site is likely to reduce the overall phosphorylation content of the kinase.
Fig. 5.
Pfnek3 autophosphorylates at residue T82. a After bacterial expression and purification, GST-Pfnek3 was subjected to SDS-PAGE. The desired protein band was in-gel digested with trypsin before analysis by LC-MS/MS. Amino acid T82, which resides within the tryptic peptide 74FETVLDFMTSDSEIHLIR91, was identified as a phosphorylation site. The neutral loss of 80 Da corresponded to the mass of a phosphate group. b LC-MS/MS analysis was repeated with the catalytically inactive GST-ΔPfnek3. The same peptide was devoid of phosphorylation at residue T82, confirming the phosphoryl-group detected in (a) was a result of autophosphorylation reaction
As both the S221A and S226A mutations were earlier found to reduce autophosphorylation level and kinase activity of Pfnek3, T82A/S221A, T82A/S226A, and T82A/S221A/S226A mutants were also constructed to investigate their combined effects on the autophosphorylation status and catalytic activity of Pfnek3. The T82A/S221A mutant displayed a slightly lower catalytic activity as compared to the single T82A mutant (Fig. 3b). On the other hand, activities of both the T82A/S226A and T82/S221A/S226A mutants were nearly eliminated completely (Fig. 3b). It was also found that all these mutants exhibited drastic reductions in autophosphorylation levels as compared to the wild-type kinase (Fig. 3a). Collectively, these data strongly suggest the importance of T82, S221 and S226 in regulating the activity of Pfnek3.
Amino acid residues T82 and S226 are crucial for Pfnek3 to activate Pfmap2 in vitro
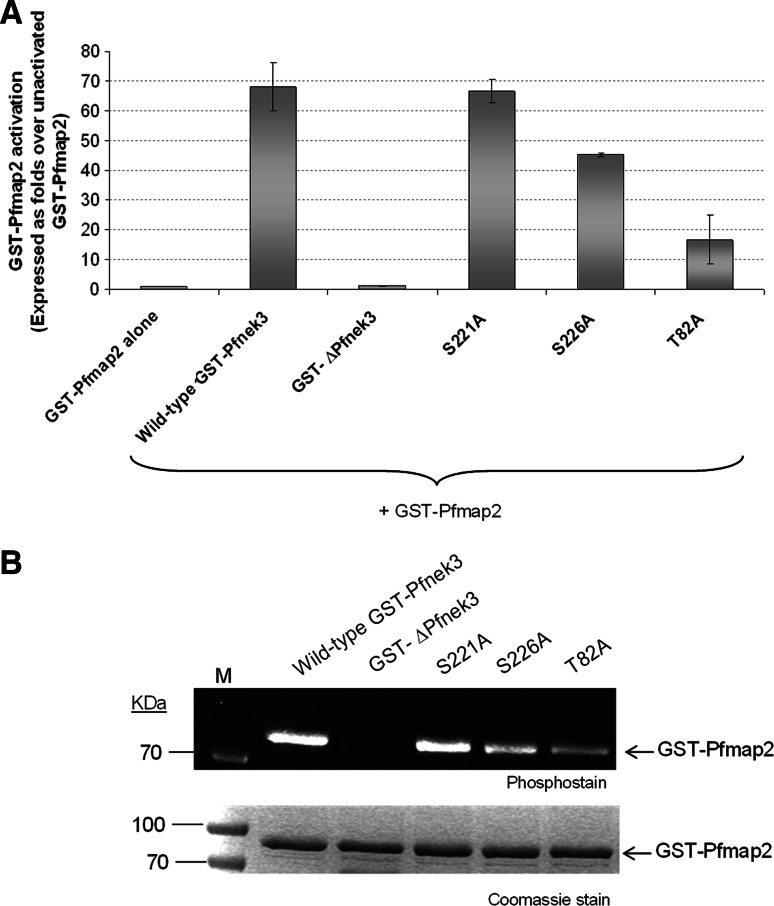
Since Pfnek3 was reported to function as a Pfmap2 activator in vitro [15], we postulate that any alteration in Pfnek3 activity could affect its ability to phosphorylate and trigger Pfmap2 activation. As the S221A, S226A, and T82A mutants displayed reduced activities towards the exogenous MBP substrate, their influence on GST-Pfmap2 activation was also assessed via a co-incubation kinase assay. In this assay format, GST-Pfmap2 was mixed with GST-Pfnek3 or its mutants in the MBP-coated microtiter wells. It was previously noted that, when GST-Pfnek3 activates GST-Pfmap2, a synergistic increase in the phosphorylation of MBP would be observed [15].
GST-Pfmap2 alone, or when mixed with the kinase-dead GST-ΔPfnek3, displayed a modest level of kinase activity (Fig. 6a). The co-incubation of GST-Pfnek3 and GST-Pfmap2 led to the activation of GST-Pfmap2 by approximately 70-fold (Fig. 6a). When residue T82 or S226 was replaced with an alanine, the ability of GST-Pfnek3 to activate GST-Pfmap2 was reduced by at least 50% (Fig. 6a). Surprisingly, the S221A mutant was able to stimulate GST-Pfmap2 to a similar extent as the wild-type GST-Pfnek3. One possible explanation for this outcome could be that S221 is not fundamental for Pfnek3 to phosphorylate and activate Pfmap2. To examine this notion, the phosphorylation status of GST-Pfmap2 was analyzed following incubation with the various GST-Pfnek3 proteins. Indeed, the S221A mutant retained the ability to phosphorylate the GST-Pfmap2 substrate (Fig. 6b). On the other hand, both the S226A and T82A mutants phosphorylated GST-Pfmap2 to a lower extent. These data thus indicate that residues T82 and S226 are essential for Pfnek3 to stimulate the activity of Pfmap2, and we propose phosphorylation at these residues might be responsible for this phenomenon.
Fig. 6.
Effect of T82A, S221A, and S226A mutations on the ability of GST-Pfnek3 to activate GST-Pfmap2. a Adopting the co-incubation kinase assay, 5 ng of wild-type GST-Pfnek3 or the respective mutants were mixed with 3 μg of GST-Pfmap2 in MBP-coated wells. When existing alone, or mixed with the kinase-dead GST-ΔPfnek3 mutant, GST-Pfmap2 displayed a low level of kinase activity (lanes 1 and 3). In the presence of wild-type GST-Pfnek3, the activity of GST-Pfmap2 was stimulated by approximately 70-fold. Only the S221A, but not the S226A or T82A mutants, retained the ability to activate GST-Pfmap2. b After GST-Pfmap2 was incubated with the indicated GST-Pfnek3 proteins in a kinase assay mix, the kinases were resolved by SDS-PAGE and subjected to phosphorylation analysis to determine the phosphorylation status of GST-Pfmap2. The Coomassie staining of the same gel is shown in lower panel. M Protein standard marker
Discussion
Pfnek3 was previously validated to exhibit a MAPKK-like activity due to its activating effect on Pfmap2 via phosphorylation at the signature TSH activation motif. This triggered our interest in investigating the regulation of Pfnek3 activity to elucidate additional properties that might be common between Pfnek3 and the MAPKKs. Kinase activities are commonly modulated by several strategies including changes in cellular localization, binding of regulatory subunits and phosphorylation by upstream kinases or via autophosphorylation [25]. The authentication of Pfnek3 autophosphorylation (Fig. 1a) led us to hypothesize that phosphorylation of the kinase plays a crucial role in regulating its activity. This notion was further supported when GST-Pfnek3 activity was abrogated upon dephosphorylation with lambda phosphatase (Fig. 1c).
For a number of kinases, phosphorylation within specific motifs found in the kinase activation loop is essential for enzyme activation [26]. In NIMA-like kinases, for instance, stimulation of kinase activity requires phosphorylation of the threonine residue within the consensus FXXT motif [27]. However, this motif is absent in Pfnek3. Instead, Pfnek3 was found to possess a SSEQSS motif which resembles the characteristic SXXXS/T activation site of MAPKKs. Similar MAPKK-like activation motif has also been reported in Pfnek1 [12].
A comparison of the Pfnek3 tertiary model with the human MEK1 crystal structure revealed that the SSEQSS motif resides in a position that is analogous to the SMANS activation motif of MEK1 (Fig. 2). Phosphorylation of both serines within the SMANS motif has been validated to be a prerequisite for MEK1 activation [22]. Thus, the co-location of SSEQSS motif with the known activation site of MEK1 highlights its potential to serve as a phospho-regulatory site in Pfnek3. In support of this, our results showed that the S221A and S226A mutants displayed significant reductions in kinase activities that were accompanied by decreases in autophosphorylation levels (Fig. 3). Therefore, out of the four serines within the SSEQSS motif, phosphorylation of S221 and S226 is likely to induce the activation of Pfnek3.
We also attempted to replace S221 and S226 with either an aspartate or a glutamate to mimic phosphorylation at these residues. Among these mutants, only S226E exhibited a considerably higher activity than the wild-type Pfnek3 (Fig. 4), suggesting that phosphorylation of S226 indeed occurs. Additionally, this observation may signify that the phosphorylation state of S226 could only be efficiently mimicked by glutamate, and not aspartate, to maintain Pfnek3 in an active conformation. On the other hand, the inability of the S221D and S221E mutants to activate Pfnek3 cannot completely revoke the potential of S221 as a phospho-activating site. This is because, in certain kinases, negatively charged amino acids may be incompetent in emulating the function of experimentally verified phospho-sites. For instance, although the second serine within the SMANS activation site of human MEK1 has been validated as an important phosphorylation site for kinase activation, its mutation to either glutamate or aspartate did not constitutively activate the kinase [24]. It was suggested that the phospho-mimic mutations may have led to conformational changes in the kinase structure that prevented them from simulating the effect of serine phosphorylation [24]. Likewise, the S221D and S221E mutations may also have led to undesirable conformational changes in the Pfnek3 structure, rendering their inability to simulate the effect of the negative charges arising from phosphorylation at this position. Given that phosphorylation at the activation loop is a widespread mechanism in mediating kinase activation, we are in favor of the proposal that both S221 and S226 serve as phospho-activating residues in Pfnek3.
In this study, amino acid residue T82 is a novel phosphorylation site authenticated via LC-MS/MS analysis (Fig. 5). To evaluate the implication of T82 phosphorylation in regulating the kinase activity of Pfnek3, the threonine residue was mutated to a non-phosphorylatable alanine. The T82A mutant displayed a more drastic reduction in autophosphorylation and kinase activity as compared to the S221A and S226A mutants, suggesting that phosphorylation at this residue plays a more crucial role in stimulating Pfnek3 activity (Fig. 3). Interestingly, residue T82 is located within a unique FMTSDS motif which was earlier reported to replace the highly conserved glycine triad (GXGXXG) in kinase subdomain I [15]. In general, this glycine triad is essential for modulating the activity of eukaryotic protein kinases due to its involvement in anchoring ATP during the phospho-transfer reaction [23]. In the absence of this motif, ATP binding in Pfnek3 might involve an unusual mechanism that potentially requires the phosphorylation of residue T82. Hence, the huge decrease in Pfnek3 activity upon mutation of T82 could be attributed to its importance in triggering efficient ATP binding during phosphorylation reactions. However, the amino acid residues flanking T82 were not visible in the modeled Pfnek3 structure, conceivably due to the low sequence homology between Pfnek3 and the modeling templates at this region. As such, until the structure of Pfnek3 in both the unphosphorylated and phosphorylated state is made available, the bona fide role of T82 phosphorylation in regulating Pfnek3 activity remains debatable.
Since Pfnek3 was earlier reported to modulate the activity of Pfmap2, it was also of interest to examine whether phosphorylation of S221, S226, and T82 was crucial for Pfnek3 to activate Pfmap2. From the results illustrated (Fig. 6), only residues T82 and S226 were found to be essential for Pfnek3 to phosphorylate and trigger GST-Pfmap2 activation. The outcome was unexpected since the absence of these three residues has rendered Pfnek3 less capable of phosphorylating the exogenous MBP substrate. Consequently, this may imply that phosphorylation of T82 and S226 is more crucial for Pfnek3 to recognize the Pfmap2 substrate.
The results of this study have strengthened the notion that Pfnek3 is a MAPKK-like kinase in P. falciparum with an activation mechanism similar to the well-known MAPKKs. On top of this, Pfnek3 activation further requires phosphorylation at residue T82, suggesting the regulation of Pfnek3 activity might involve a more complex mechanism as compared to the MAPKKs. Moreover, since the T82A/S221A/S226A triple mutant retained a low, but observable, level of phosphorylation, it is plausible that additional phosphorylation sites are present that may contribute to the regulation of Pfnek3 activation. It is also noteworthy to mention that, although phosphorylation of T82, S221 and S226 in vitro is a result of Pfnek3 autophosphorylation, the possibility remains that phosphorylation of these residues in vivo may be performed by upstream kinases and/or require binding with regulatory subunits. In view of this, it is imperative to identify interacting partners of Pfnek3 so as to fully understand its regulation in the malarial parasite.
Acknowledgments
We are grateful to Professor Christian Doerig (University of Glasgow) for providing the plasmid carrying the Pfmap2 gene. We would also like to express our sincere gratitude to Dr. Manfred Raida, Ms Ee Kim Huey and Mr Li Rong (A*STAR Experimental Therapeutics Centre) for providing technical expertise in the LC-MS/MS analyses.
References
- 1.World Health Organization (2008) World Malaria Report 2008
- 2.Khan SM, Waters AP. Malaria parasite transmission stages: an update. Trends Parasitol. 2004;20:575–580. doi: 10.1016/j.pt.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Schramek H. MAP kinases: from intracellular signals to physiology and disease. News Physiol Sci. 2002;17:62–67. doi: 10.1152/nips.01365.2001. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Dong C. Regulatory mechanisms of mitogen-activated kinase signaling. Cell Mol Life Sci. 2007;64:2771–2789. doi: 10.1007/s00018-007-7012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/er.22.2.153. [DOI] [PubMed] [Google Scholar]
- 6.Doerig CM, Parzy D, Langsley G, Horrocks P, Carter R, Doerig CD. A MAP kinase homologue from the human malaria parasite, Plasmodium falciparum . Gene. 1996;177:1–6. doi: 10.1016/0378-1119(96)00281-8. [DOI] [PubMed] [Google Scholar]
- 7.Graeser R, Kury P, Franklin RM, Kappes B. Characterization of a mitogen-activated protein (MAP) kinase from Plasmodium falciparum . Mol Microbiol. 1997;23:151–159. doi: 10.1046/j.1365-2958.1997.2071571.x. [DOI] [PubMed] [Google Scholar]
- 8.Dorin D, Alano P, Boccaccio I, Ciceron L, Doerig C, Sulpice R, Parzy D. An atypical mitogen-activated protein kinase (MAPK) homologue expressed in gametocytes of the human malaria parasite Plasmodium falciparum. Identification of a MAPK signature. J Biol Chem. 1999;274:29912–29920. doi: 10.1074/jbc.274.42.29912. [DOI] [PubMed] [Google Scholar]
- 9.Dorin-Semblat D, Quashie N, Halbert J, Sicard A, Doerig C, Peat E, Ranford-Cartwright L. Functional characterization of both MAP kinases of the human malaria parasite Plasmodium falciparum by reverse genetics. Mol Microbiol. 2007;65:1170–1180. doi: 10.1111/j.1365-2958.2007.05859.x. [DOI] [PubMed] [Google Scholar]
- 10.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anamika K, Srinivasan N. Comparative kinomics of Plasmodium organisms: unity in diversity. Protein Pept Lett. 2007;14:509–517. doi: 10.2174/092986607780989949. [DOI] [PubMed] [Google Scholar]
- 12.Dorin D, Le Roch K, Sallicandro P, Alano P, Parzy D, Poullet P, Meijer L, Doerig C. Pfnek-1, a NIMA-related kinase from the human malaria parasite Plasmodium falciparum Biochemical properties and possible involvement in MAPK regulation. Eur J Biochem. 2001;268:2600–2608. doi: 10.1046/j.1432-1327.2001.02151.x. [DOI] [PubMed] [Google Scholar]
- 13.Dorin D, Semblat JP, Poullet P, Alano P, Goldring JP, Whittle C, Patterson S, Chakrabarti D, Doerig C. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum . Mol Microbiol. 2005;55:184–196. doi: 10.1111/j.1365-2958.2004.04393.x. [DOI] [PubMed] [Google Scholar]
- 14.O’Connell MJ, Krien MJ, Hunter T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003;13:221–228. doi: 10.1016/S0962-8924(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 15.Lye YM, Chan M, Sim TS. Pfnek3: an atypical activator of a MAP kinase in Plasmodium falciparum . FEBS Lett. 2006;580:6083–6092. doi: 10.1016/j.febslet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Low H, Lye YM, Sim TS. Pfnek3 functions as an atypical MAPKK in Plasmodium falciparum . Biochem Biophys Res Commun. 2007;361:439–444. doi: 10.1016/j.bbrc.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 17.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, Mueller WT, Delaney A, Omer C, Sebolt-Leopold J, Dudley DT, Leung IK, Flamme C, Warmus J, Kaufman M, Barrett S, Tecle H, Hasemann CA. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 19.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 22.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 24.Huang W, Erikson RL. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc Natl Acad Sci USA. 1994;91:8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engh RA, Bossemeyer D. The protein kinase activity modulation sites: mechanisms for cellular regulation—targets for therapeutic intervention. Adv Enzyme Regul. 2001;41:121–149. doi: 10.1016/S0065-2571(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 26.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/S0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 27.Pu RT, Xu G, Wu L, Vierula J, O’Donnell K, Ye XS, Osmani SA. Isolation of a functional homolog of the cell cycle-specific NIMA protein kinase of Aspergillus nidulans and functional analysis of conserved residues. J Biol Chem. 1995;270:18110–18116. doi: 10.1074/jbc.270.44.26033. [DOI] [PubMed] [Google Scholar]