Abstract
Trefoil protein 1 (TFF1) is a small secreted protein belonging to the trefoil factor family of proteins, that are present mainly in the gastrointestinal (GI) tract and play pivotal roles as motogenic factors in epithelial restitution, cell motility, and other incompletely characterized biological processes. We previously reported the up-regulation of TFF1 gene in copper deficient rats and the unexpected property of the peptide to selectively bind copper. Following the previous evidence, here we report the characterization of the copper binding site by fluorescence quenching spectroscopy and mass spectrometric analyses. We demonstrate that Cys58 and at least three Glu surrounding residues surrounding it, are essential to efficiently bind copper. Moreover, copper binding promotes the TFF1 homodimerization, thus increasing its motogenic activity in in vitro wound healing assays. Copper levels could then modulate the TFF1 functions in the GI tract, as well as its postulated role in cancer progression and invasion.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0309-7) contains supplementary material, which is available to authorized users.
Keywords: Trefoil factors, TFF1, Copper, Cell motility, Gastrointestinal peptides, Copper binding protein
Introduction
Trefoil protein 1 (TFF1) is a small secreted protein mainly expressed in the epithelial cells of the gastric surface but, as the other members of the TFF protein family, it has been detected in different tissues containing mucus-secreting cells [1]. Moreover, TFF1 is abnormally expressed in inflammatory disorders of the gastrointestinal (GI) tract, thus suggesting its involvement in the protection and repair of mucosal integrity. This biological function is further supported by the observation that TFF1-deficient mice develop gastric adenomas and carcinomas [2, 3] probably as a consequence of a complete lack of mucosal protection. Furthermore, a DNA microarray differential analysis allowed us to identify TFF1 as one of the most up-regulated (≈fivefold) among ≈22,000 rat cDNA probes in copper-deficient rats [4, 5].
The mature protein contains 60 amino acids, and residues spanning from position 7–47 are involved in the formation of the trefoil domain consisting of three packed sequential loops stabilized by three disulphide bonds [6]. Except for two short β-strands, the trefoil stem, formed by juxtaposed N- and C-termini of the protein, remains mainly flexible and unstructured. TFF1 may also form homo- or heterodimers through the formation of an inter-molecular disulphide bridge involving its seventh unbound cysteine (Cys58). It is worth noting that in the homodimer the linker region between the two trefoil domains does not reduce its flexibility, despite the presence of a disulphide bond [7]. As a heterodimer, the only known TFF1 partner is TFIZ1, a secreted protein of 18.3 kDa that was isolated as a peptide bound to TFF1 [8]. On the other hand, it has been shown that TFF1 is a potent chemoattractant for breast cancer cells and that, though the TFF1 dimer and monomer both stimulate the migration of breast cancer cells, the dimer is more potent [9]. The three different isoforms (monomer, homo-, and heterodimer) have different distributions, as well as different and often not fully elucidated functions. The incomplete knowledge about the structure–function relationship of TFF1 oligomers poses several, and still unanswered, questions about the biochemistry of TFF1, as well as the molecular mechanisms driving the formation of its isoforms and the stimuli regulating the equilibria among monomers and dimers.
We recently demonstrated that TFF1 is able to specifically bind copper in vitro, and we suggested the highly conserved and acidic carboxy-terminal tail as the site of metal interaction [4]. Copper is an essential element for all living organisms, because it is involved in several important biochemical processes, although many open questions still do not allow a detailed view of the mechanisms regulating the Cu homeostasis [10]. In these respects, a better understanding of TFF1 biochemistry, as a newly characterized copper-binding protein, could provide further useful elements to elucidate possible unexplored and unknown aspects of the cell biology dealing with the two-faced chemistry of copper as a potentially toxic, but functionally essential element.
Here, we report the fluorimetric and mass spectrometric characterization of the metal binding site carried out on synthetic peptides representative of the putative TFF1 copper-binding site. The results obtained show that the acidic cluster of glutamic acid residues encompassing the last cysteine at the C terminus is essential for copper binding. Further experiments demonstrate that this interaction is able to influence the equilibrium between monomeric and dimeric form, and modulate the well-known motogenic activity of TFF1.
Materials and methods
Peptides
All peptides were synthesized by standard Fmoc [N-(9-Fluorenyl)methoxycarbonyl] chemistry on Tentagel HL resin (Fluka). A solution of 30% piperidine in DMF (dimethyl formamide) was used in the deprotection step (2 × 5 min). Amino acid coupling was performed using a DMF solution of 10 molar excess of Fmoc-amino acid, 9.9 eq HBTU (O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate)/HOBt (Hydroxybenzotriazole) and 20 eq DIPEA (N,N-Diisopropylethylamine) for 60 min followed by a 10-min acetylation step with a solution of acetic anhydride (2 M)/DIPEA (0.55 M)/HOBt (0.06 M). Cleavage from the resin was achieved by treatment with trifluoroacetic acid, triisopropyl silane, water (95; 2.5; 2.5 v/v/v) at room temperature for 3 h. Peptide analysis and purification were performed by RP-HPLC on a C12 Proteo column (Phenomenex). Peptide identity was assessed by ESI mass spectrometry on Thermo Finnigan MSQ LC–MS.
pTFF: Ac-FYPNTIDVPPEEECEF-COOH; MW of 1,969.96 Da
WTFF: Ac-FYPNTIDVPPEEECEW-COOH; MW of 2,008.76 Da
WTFF55: Ac-FYPNTIDVPPAEECEW-COOH; MW of 1,950.86 Da
WTFF56: Ac-FYPNTIDVPPEAECEW-COOH; MW of 1,950.86 Da
WTFF57: Ac-FYPNTIDVPPEEACEW-COOH; MW of 1,950.86 Da
WTFF58: Ac-FYPNTIDVPPEEEAEW-COOH; MW of 1,976.86 Da
WTFF59: Ac-FYPNTIDVPPEEECAW-COOH; MW of 1,950.86 Da
WTFFala: Ac-FYPNTIDVPPAAAAAW-COOH; MW of 1,744.86 Da
WTFFcys/ala: Ac-FYPNTIDVPPAAACAW-COOH; MW of 1,776.81 Da
QK: Ac-KLTWQELYQLKYKGI-COOH; MW of 1,953.31 Da
MALDI-MS analysis of peptides metal-binding
A solution of each peptide (500 nM) in ammonium bicarbonate 5 mM, at variable pHs ranging from 2 to 7.4, was incubated in presence of two- to tenfold molar excess of CuCl2, or tenfold molar excess of ZnCl2, CaCl2, and AgNO3. Experiments with CuCl2 were also carried out in the presence of EDTA (tenfold molar excess with respect to CuCl2 concentration). Mixtures were analyzed by MALDI mass spectrometry in a m/z range of 800–4,500 on a MALDI micro MX™ (Waters, Milford, MA, USA) in reflectron positive ion mode, using α-cyano-4-hydroxycinnamic acid (10 mg ml−1) dissolved in H2O:CH3CN (50/50 v/v) as matrix, without adding TFA.
Nano-LC–MS and RP-HPLC–UV analyses of Cu-induced TFF1 dimerization
A solution of each peptide (2 μM) in ammonium bicarbonate 5 mM, pH 7.4 was incubated in the presence of 0.3- to 10-fold molar excess of CuCl2, or 10-fold molar excess of ZnCl2. Experiments with CuCl2 were also carried out in the presence of EDTA (10-fold molar excess with respect to CuCl2 concentration). Aliquots of the incubation mixtures were collected at different times (from 0 to 36 h) and analyzed by nano-LC–MS and RP-HPLC–UV. Nano-LC–MS runs were carried out on a Q-ToF Premier™ (Waters) equipped with a nanoAcquity Ultraperformance LC system and a nanospray source. Chromatographic elution was carried out on an Atlantis dC18 nanoAcquity Column (100 mm × 75 μm) with a 5 μm Symmetry C18 pre-column (180 μm × 20 mm) by means of a linear gradient from 5 to 65% aqueous ACN containing 0.05% TFA and 1% formic acid, over 35 min at 200 nl min−1. All mass spectra were collected in a m/z range of 600–1,800. RP-HPLC–UV was performed on HP 1100 binary pump (Agilent Technologies, Palo Alto CA, USA) on a Phenomenex C18 narrow-bore column by means of a linear gradient from 5 to 65% aqueous acetonitrile containing 0.05% TFA, over 35 min.
The areas of nano-LC–MS and/or RP-HPLC–UV peaks relative to each peptide in its monomeric and dimeric state were computed by integration and the percentage of dimer formation was calculated by means of the following ratio: Relative dimer abundance (%) = [dimer peak area/(dimer peak area + monomer peak area)] × 100.
Cu(I) detection
Cu(I) formation was detected with the chelator bathocuproine disulfonate (BCS), which complexes cuprous ions as Cu(BCS)3−2 selectively detectable spectrophotometrically at 485 nm (ε = 13,000 M−1 cm−1) [11]. Copper complex formation was followed by adding 30 μM BCS after 2–3 min of incubation of 10 μM WTFF monomer in 20 mM Sodium Phosphate pH 7.4 with 10 μM CuCl2, then absorption at 485 nm was measured in a DU480 Beckman spectrophotometer. The background absorption, not affecting the measures and close to the baseline, was cross-checked by using copper test solutions alternatively lacking BCS or WTFF.
Copper content determination in culture media
Copper was determined in the free serum DMEM medium (Sigma, St. Louis, USA), supplemented with 100 U ml−1 penicillin, 100 μg ml−1 streptomycin (Cambrex), and 600 μg ml−1 neomycin (Sigma), and in the same medium supplemented with 10% (v/v) foetal bovine serum (Cambrex). Analyses were carried out by atomic absorption spectrometry using a Analyst 100 (PerkinElmer, Wellesley, MA, USA), equipped with a AS 800 graphite atomizer apparatus and autosampler (PerkinElmer). Care was taken in all analyses to avoid metal contamination.
Fluorescence spectroscopy
Steady-state fluorescence spectra were recorded on a Perkin-Elmer LS 55 spectrofluorimeter using 5 nm excitation and emission slit widths and a 1-cm cuvette. Emission spectra were collected from 310 and 440 nm (λ ex = 280 nm) and fluorescence intensities at the maximum emission wavelength (λ em = 362 nm) were registered.
The fluorescence quenching on WTFF peptide was analyzed using 20 μM concentration in 20 mM sodium phosphate pH 7.4 with the addition of different concentration of CuCl2 (0–50 μM).
Copper titrations (from 0 to 600 μM) were performed on all synthesized peptides: WTFF, WTFF56, WTFF57, WTFF58, WTFF59, WTFFala, QK.
To assess the metal binding selectivity, WTFF fluorescence intensity was also measured in the presence of 40 μM of different metal quenchers: CoCl2, CsCl2, CdCl2, NiSO4, MnCl2, and AgNO3.
Moreover, copper quenching was evaluated over a range of pH conditions modulated by the following buffers: potassium phosphate (pH = 7.4; 6.5; 6); potassium acetate (pH = 5.6; 5; 4.6; 4; 3.6); HCl (pH = 3; 2.5; 2). Each assay was performed with WTFF 20 μM and CuCl2 40 μM.
Affinity constants
The fluorescence quenching on WTFF peptide was analyzed in 15 μM peptide solutions, dissolved in 20 mM Sodium Phosphate pH 7.4. By adding increasing amounts of CuCl2 to the WTFF (P) solution, the linear decrease of the fluorescence up to a minimum corresponding to a 1:1 copper:peptide stoichiometry was verified, followed by a break in the linearity due to nonspecific interactions of copper with the tryptophan. The experimental conditions corresponding to the lowest fluorescence (ΔF
Max) of the solution was used to challenge the peptide complex with increasing concentrations of glycine or histidine, then following the corresponding increase of fluorescence due to the displacement of copper from the Cu–WTFF complex and the formation of the nonfluorescent Cu–glycine or Cu–histidine.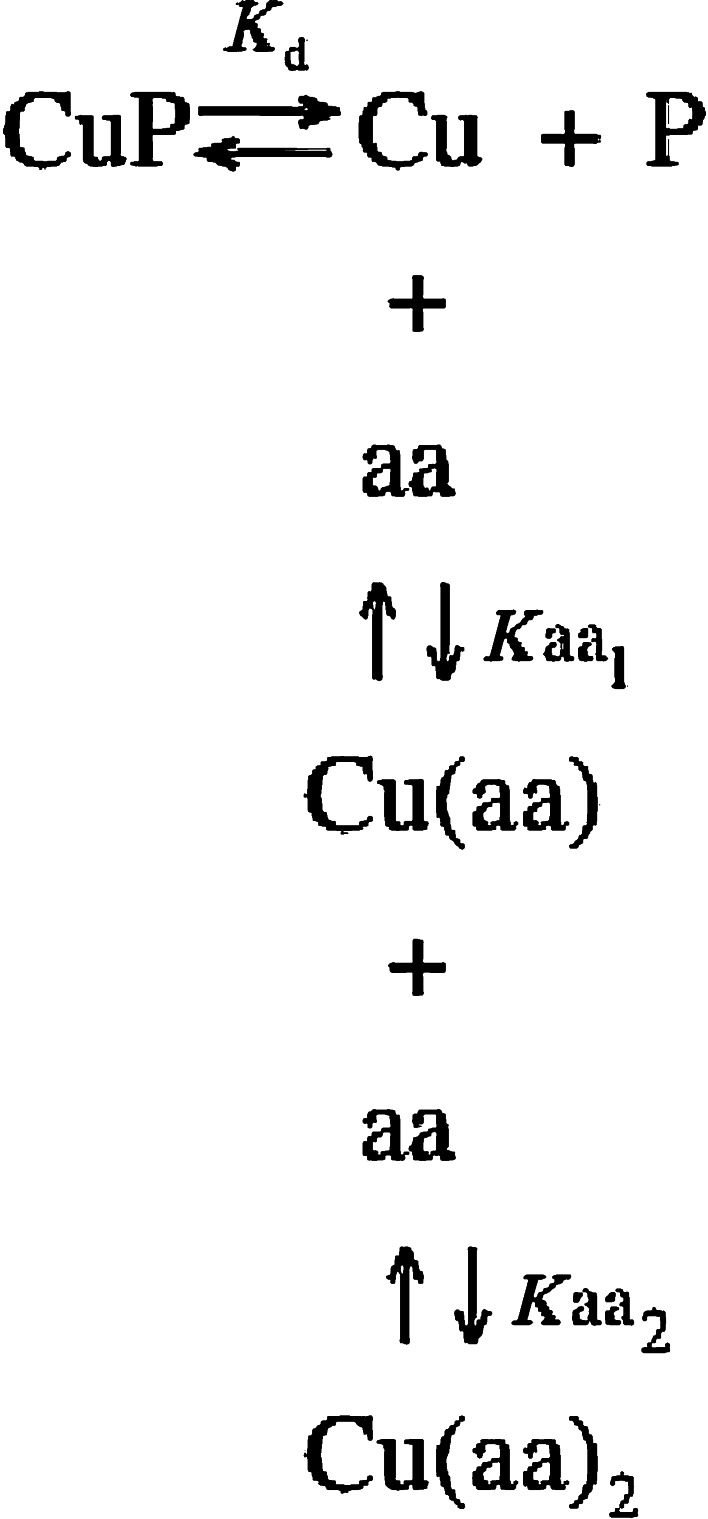
The fraction of copper bound to WTFF was then directly correlated to the tryptophan fluorescence, since the addition of copper reduces linearly the WTFF fluorescence up to the molar equivalence.
Since
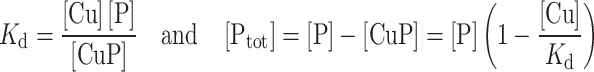 |
when [P] = 1/2 [PTot], [Cu] = K d.
The value of free copper concentration corresponding to the half of the maximum value of fluorescence corresponds to the peptide (P) affinity for copper.
Taking into account all the above described equilibria, it can be demonstrated that the value of free copper concentration can be calculated as follows [12]:
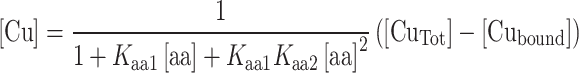 |
where K aa1 = 7.4 × 105 M−1 and K aa2 = 7.4 × 104 M−1 for free glycine, and K aa1 = 2.7 × 108 M−1 and K aa2 = 2.1 × 106 M−1 for free histidine [13]. In fact, either glycine or histidine are able to bind copper with a 2:1 stoichiometry forming Cu(Gly)2 and Cu(His)2 complexes with the above-reported apparent affinity constants.
Cells
The human gastric cancer cell line AGS and the human breast cancer cell line MCF-7 were obtained from the American Type Culture Collection (CRL-1739; Rockville, MD, USA), while the HT29-E12-MTX clone was kindly provided by Dr. Pär Matsson, Department of Pharmacy, Uppsala University, Sweden [14].
Western blot analysis
MCF-7 and HT29-E12-MTX were cultured in DMEM medium (Sigma) supplemented with 10% (v/v) fetal bovine serum (Sigma), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Sigma) at 37°C in a 5% CO2 atmosphere.
MCF-7 cells were cultured up to 80% of confluence, then washed three times with PBS 1X (Sigma) and incubated in serum-free medium supplemented with CuCl2 100 μM to test the effect of copper, in comparison with copper free medium. Conversely, HT29-E12 clone was cultured for 9 days post-confluence and then cells were treated as described above for MCF-7 cultures. After a treatment of 48 h, culture media were collected and clarified by centrifugation at 10,000g and at 4°C for 15 min to remove residual cells.
Next, 100 μl of each supernatant were precipitated with 9 volumes of acetone and then resuspended in Laemmli buffer 1X (2% SDS, 10% glycerol, 0.002% bromo-phenol blue, 62.5 mM Tris–HCl pH 6.7) with or without the addition of reducing agents. Proteins were separated on a 15% SDS–PAGE and then electro-blotted on a Hybond-ECL membrane (GE Healthcare) for 1 h, at 4°C and 100 V. Membranes were blocked with 5% non-fat dry milk (BioRad) for 1 h at room temperature and then incubated with rabbit polyclonal antibody anti-TFF1 (raised against the last 16 aa of the protein) at 4°C overnight. A donkey anti-rabbit (Jackson) was used as secondary antibody and ECL system (GE Healthcare) for detection.
Stable transfection of AGS cells
To produce a stable AGS cell line expressing human TFF1, EcoRI/BamH1 fragment (approximately 280 bp), corresponding to the open reading frame of the human TFF1 cDNA (Accession no. X00474), was inserted into the EcoRI/BamHI site of pUHD 10-3 doxycycline inducible expression vector thus generating the pUHD-hTFF1 plasmid. Since pUHD-hTFF1 was devoid of neomycin resistance, it was co-transfected with the pUHD172.1neo [15]. AGS cells were transfected by calcium-phosphate co-precipitation with the PvuI-linearized pUHD-hTFF1 inducible expression vector as previously described [16]. One clone expressing TFF1 under doxycycline induction (AGS-AC1) was selected.
In vitro wound-healing assay
AGS-AC1 cells were cultured in DMEM medium (Sigma) supplemented with 10% (v/v) foetal bovine serum (Cambrex), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin (Cambrex), and 600 μg ml−1 neomycin (Sigma). Cells were seeded in a 12-well plastic plate at 3 × 105 cells per well, and after 12 h TFF1 expression was induced in 6 wells with 1 μg ml−1 of doxycycline. Then, 12 h later, three induced and three non-induced wells were incubated with 500 μM bathocuproine. The remaining wells were not treated and used as control. After 24 h from the treatments, cells reached 100% confluence and a wound was produced at the centre of the monolayer by gently scraping the adhering cells with a sterile plastic p200 pipette tip. The wounded cell cultures were incubated at 37°C in a humidified and equilibrated [5% (v/v) CO2] incubation chamber of an Integrated Live Cell Workstation Leica AF6000 LX. A 10× phase contrast objective was used to record cell movements. Images were collected with a frequency of 20 min. The migration rate of the front edge of the wounded monolayer was determined by measuring the distances covered by cellular edges from time zero to the following time points (bar of distance tool, Leica ASF software). Three independent replicates for each condition were analyzed. For each wound, three different positions were registered, and for each position, five different points on the cell front were randomly selected to measure the distance covered.
Statistical analyses
Statistical analyses were performed by using the Microsoft Excel™ software. Data were analyzed using unpaired, two-tailed t test comparing two variables. Data are presented as means ± SD. Values <0.05 were considered as significant.
Results
TFF1 carboxy-terminus specifically binds copper
In order to verify the hypothesis of the possible involvement of the acidic C-terminal region of TFF1 in copper binding [4], we synthesized a peptide containing the last 16 amino acids of the protein, except for the last phenylalanine that was changed in a tryptophan (WTFF: Ac-FYPNTIDVPPEEECEW-COOH). This mutation was inserted to obtain a more efficient fluorescent probe close to the residues putatively involved in the metal binding. A similar peptide having the native TFF1 sequence was used as a control, in order to verify possible biases or artifacts affecting the peptide affinity for copper, depending on tryptophan substitution (pTFF: Ac-FYPNTIDVPPEEECEF-COOH). On the other hand, to evaluate the contribution to the binding of the cysteine and the surrounding acidic residues, we synthesized a point mutated peptide array by Ala-scanning, along with two more peptides representing the overall Ala mutations of the entire putative binding site, or only substituting the glutamic residues:
WTFF55: Ac-FYPNTIDVPPAEECEW-COOH
WTFF56: Ac-FYPNTIDVPPEAECEW-COOH
WTFF57: Ac-FYPNTIDVPPEEACEW-COOH
WTFF58: Ac-FYPNTIDVPPEEEAEW-COOH
WTFF59: Ac-FYPNTIDVPPEEECAW-COOH
WTFFala: Ac-FYPNTIDVPPAAAAAW-COOH
WTFFcys/ala: Ac-FYPNTIDVPPAAACAW-COOH
(Numbers refer to the positions in the native sequence)
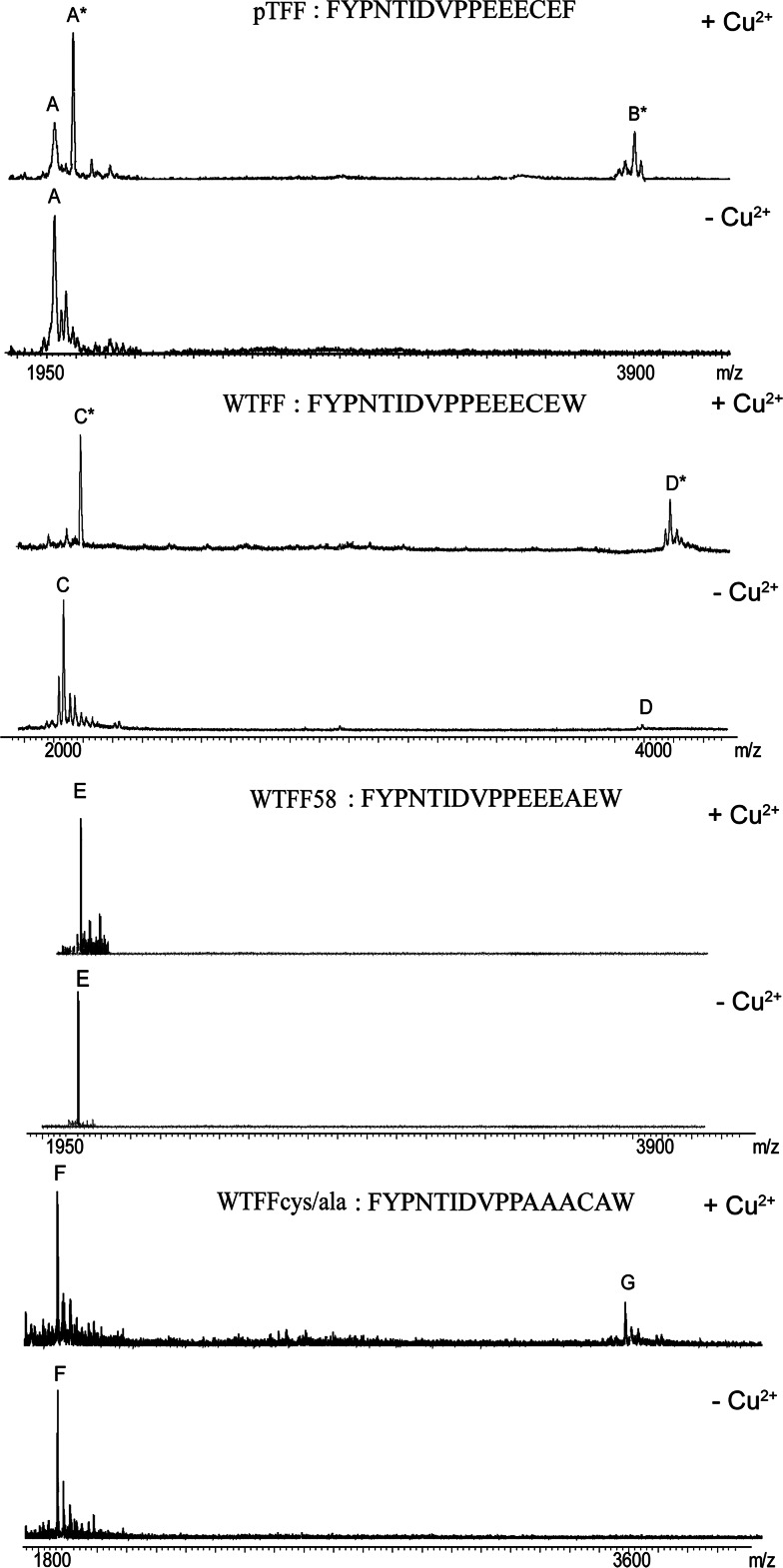
The MALDI mass spectrometry approach allowed a preliminary evaluation of the copper binding ability of each peptide, and the stoichiometry of the complex formation to be determined. Recent reports on complexes of transition metal ions with peptides, or other bio-functional ligands, have demonstrated that the matrix assisted ionization process allows the detection of the intact species raising MALDI mass spectrometry as a suitable technique to monitor the formation of complexes in solution [17, 18]. Briefly, we performed titration experiments collecting mass spectra after the addition of increasing amount of CuCl2 (from 1:2 to 1:10 molar ratio) to each peptide solution (500 nM) in bio-mimetic condition. Figure 1 shows the species observed for pTFF, WTFF, WTFF58, and WTFFcys/ala peptides incubated for 15 min in the presence of a twofold molar excess of CuCl2. The shift of ~64 Da observed for the peak A* (MW of 2,033.65 Da) and C* (MW of 2,072.95 Da) allowed us to identify both species as the Cu2+-bound form of the corresponding monomers (peaks A and C), with a 1:1 stoichiometry.
Fig. 1.
MALDI-MS analysis of pTFF, WTFF, WTFF58, and WCys/ala in the presence and in the absence of twofold CuCl2 molar excess, after 15 min of incubation. Peak A (1,969.52 Da), peak A* (2,033.65 Da), peak B* (4,001.52 Da), peak C (2,008.45 Da), peak C* (2,072.95 Da), peak D (4,015.1 Da), peak D* (4,078.85 Da), peak E (1,976.95 Da), peak F (1,776.83 Da), peak G (3,551.96 Da)
As expected, incubation in the presence of CuCl2, which is an oxidative agent, induced the formation of the covalent dimer of each peptide involving Cys58 [19]. It is worth noting that the dimeric form of WTFF lacking the free SH group of the cysteine involved in the disulphide bridge formation preserves its ability to bind Cu2+ ions. Indeed, as shown in Fig. 1, the MW of the pTFF peak B* (MW of 4,001.52 Da) and the MW of the WTFF peak D* (MW of 4,078.15 Da) correspond to the dimeric forms of the respective peptides bound to one Cu2+ ion. Moreover, titration experiments showed that neither a twofold nor a tenfold molar excess of Cu2+ were able to saturate the binding ability of the peptides. Finally, the incubation in the presence of the metal chelating agent EDTA drastically reduced the intensity of Cu2+ adducts (data not shown). Similar results were obtained for all peptides with point mutations of glutamic residues (electronic supplementary material, ESM, Fig. 1), while WTFF58, WTFFcys/ala, WTFFala and the negative control QK did not show appreciable peaks corresponding to the Cu2+-adduct. Figure 1 shows that no Cu2+ adduct can be detected in WTFF58 and WTFFcys/ala spectra (peak E at m/z of 1,977.95 and peak F at m/z 1,777.83), even in the presence of tenfold molar excess of CuCl2. It is worth noting that if the four Glu residues in the putative binding site are changed to Ala, as in WTFFcys/ala, Cys58 is still able to form disulfide bridges, but the monomeric and dimeric forms are not able to bind Cu2+ (peak G at m/z 3,559.96 Da).
Each peptide was incubated in the presence of tenfold molar excess of ZnCl2, as negative control, and no adducts were detected. On the other hand, besides copper, addition of silver (AgNO3) also results in the formation of a complex, but no dimeric species is evident (ESM, Fig. 2).
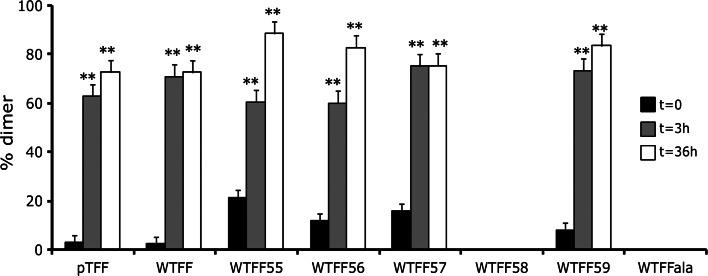
The results obtained by MALDI analysis prompted us to further study the copper induced dimer formation by nano-LC–MS and RP-HPLC–UV techniques. Each peptide was incubated with increasing amounts of CuCl2 and copper adducts formation was followed over time. Figure 2 shows the relative dimer abundance before and after CuCl2 addition, thus pointing out a role of copper as an inducer of dimer formation. Namely, the high resolution mass spectra revealed that the MW of each dimer exactly matches the weight of two monomers linked with a disulfide bridge. The lack of Cys residues in WTFF58 and WTFFala peptides, as expected, prevented the dimer formation.
Fig. 2.
Dimers formation of wild-type and mutant peptides. The increase of dimer formation after CuCl2 incubation for 0, 3 and 36 h is shown for WTFF mutants and pTFF. (3 and 36 h vs 0 h; **p < 0.005)
Finally, in order to verify Cu(II) as the oxidizing species, we monitored the formation of Cu(I) using the chelator bathocuproine disulfonate (BCS), which forms the complex Cu(BCS)3−2 with a maximum of absorption at 485 nm. We first incubated 10 μM WTFF monomeric peptide with 10 μM CuCl2, then we added 30 μM BCS, following spectrophotometrically the complex formation in few minutes after the addition. The solution showed the absorbance value A = 0.105, corresponding to about 8 μM Cu(I). Considering that the assay was performed in the presence of oxygen, the measure is only diagnostic of Cu(I) formation. In fact, Cu(I) is highly unstable and quickly dismutates to metallic Cu and Cu(II) in the presence of oxygen. The absorption increase at 485 nm, diagnostic of the presence of Cu(I), confirms the role of Cu(II) as oxidizing agent in the disulphide bridge formation.
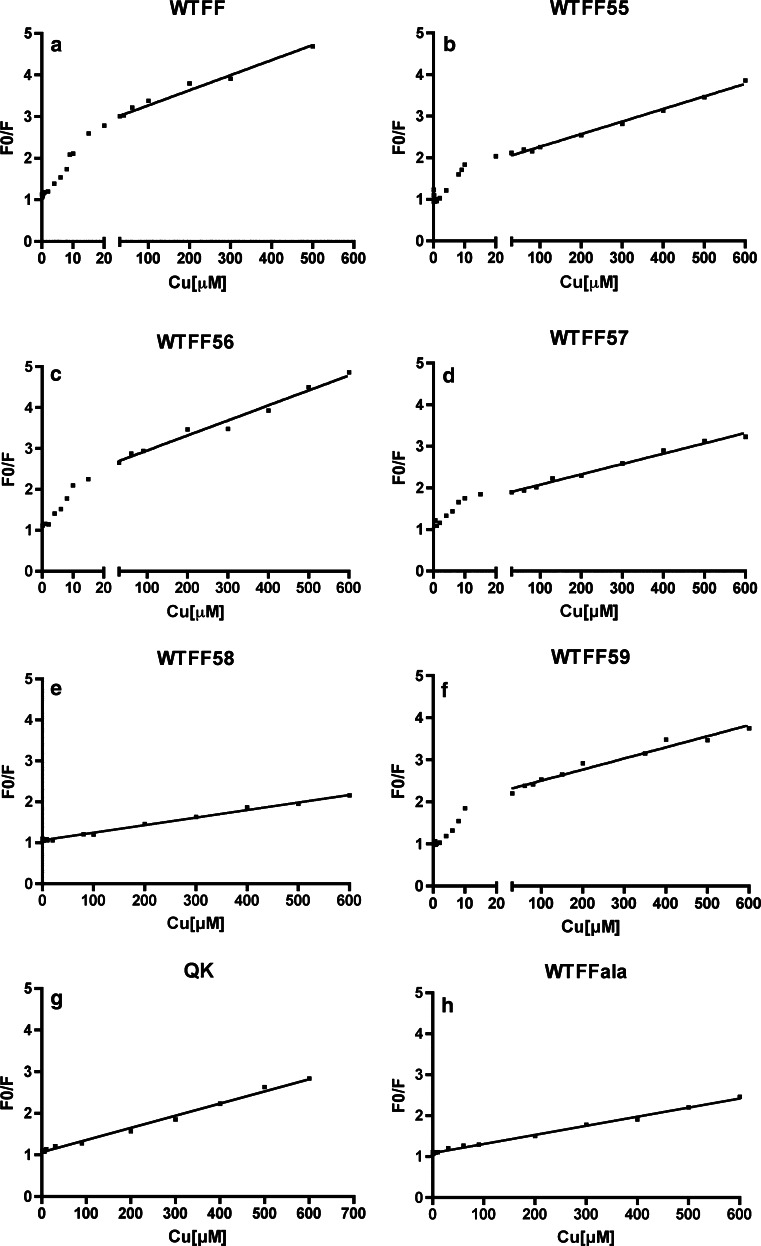
Taking advantage of the well-known fluorescence quenching property of copper, the synthetic peptides were then analyzed for their copper binding ability with a different technique. Fluorescence quenching refers to any process that decreases the fluorescence intensity of a sample. Among the molecular interactions that can result in quenching, static quenching measurements can be a valuable source of information in biochemical applications about binding between the fluorescent sample and the quencher. In static quenching, a complex is formed between the fluorophore and the quencher, and this complex is not fluorescent, while collisional (or dynamic) quenching take into account only the decrease of fluorescence due to the quencher diffused to the fluorophore during the lifetime of the excited state. The titration of WTFF with different copper concentrations (0–600 μM) shows an increasing reduction of tryptophan fluorescence due to the quenching effect of the metal. The mathematical treatment of fluorescence quenching data allows the distinguishing of collisional and/or static quenching through the evaluation of possible slope discontinuity of the Stern–Volmer curves plotted according to the homonymous equation [20]. The plots obtained from the analysis of the peptides are reported in Fig. 3. Tryptophan fluorescence can be quenched either by collisions (dynamic or collisional quenching) or by complex formation with copper (static quenching) [20]. A linear fit of F 0/F versus [Q] of the Stern–Volmer plot (F 0/F = 1 + K SV[Q], where F 0 and F are the fluorescence intensities before and after the addition of the quencher [Q], and K SV is the quenching constant, or Stern–Volmer constant) is diagnostic of pure collisional quenching. Conversely, the peptide WTFF shows a slope change in the Stern–Volmer plot (Fig. 3a), thus indicating the occurrence of a static contribution to the quenching that takes into account of a stable interaction between copper and the peptide [21]. The presence of a static protein–copper interaction can be better evaluated by comparison with the control peptide QK containing a tryptophan in a random sequence of the same length. QK shows a perfect linear trend of the Stern–Volmer plot thus indicating the occurrence of the exclusive collisional component of copper quenching (Fig. 3g) [22]. Quenching analyses demonstrate that the single substitutions of the glutamic residues 55, 56, 57, and 59 do not change the trends of the Stern–Volmer plots (Fig. 3b–d, f) if compared to the native sequence, thus implying the persistence of a static interaction. On the contrary, the mutation of Cys58 (Fig. 3e, h) completely abolishes the interaction with the metal.
Fig. 3.
Stern–Volmer plots of peptides fluorescence quenched by copper. The slope change in a, b, c, d, f are diagnostic of a static component of the fluorescence quenching and a stable copper binding. F 0/F = ratio of fluorescence intensities before (F 0) and after (F) the addition of copper
Stern–Volmer constants for each peptide were calculated using the first region of each plot (range: 0–20 μM copper). Considering that K SV = k qτ, where τ is the excited fluorophore lifetime in the absence of quencher (≈4 ns for tryptophan), we calculated the bimolecular quenching constants k q. As shown in Table 1, k q values for WTFF and all Glu-Ala mutants are considerably higher than the corresponding value of diffusion-limited quenching in water (1010 M−1s−1). On the other hand WTFF58, WTFFala and the negative control QK, show k q values of two orders of magnitude lower, and closer to the diffusion-limited value.
Table 1.
Stern–Volmer constants (K SV) and bimolecular quenching constants (k q) of WTFF and WTFF Ala-scanned peptide array
| Peptide | KSV (M−1) | kq (M−1s−1) |
|---|---|---|
| WTFF | 214,354 | 5.3 × 1013 |
| WTFF55 | 78,996 | 2.0 × 1013 |
| WTFF56 | 81,842 | 2.05 × 1013 |
| WTFF57 | 72,235 | 1.8 × 1013 |
| WTFF58 | 1,778 | 4 × 1011 |
| WTFF59 | 81,815 | 2 × 1013 |
| WTFFAla | 2,900 | 7 × 1011 |
| QK | 2,200 | 7 × 1011 |
QK Tryptophane containing random sequence peptide
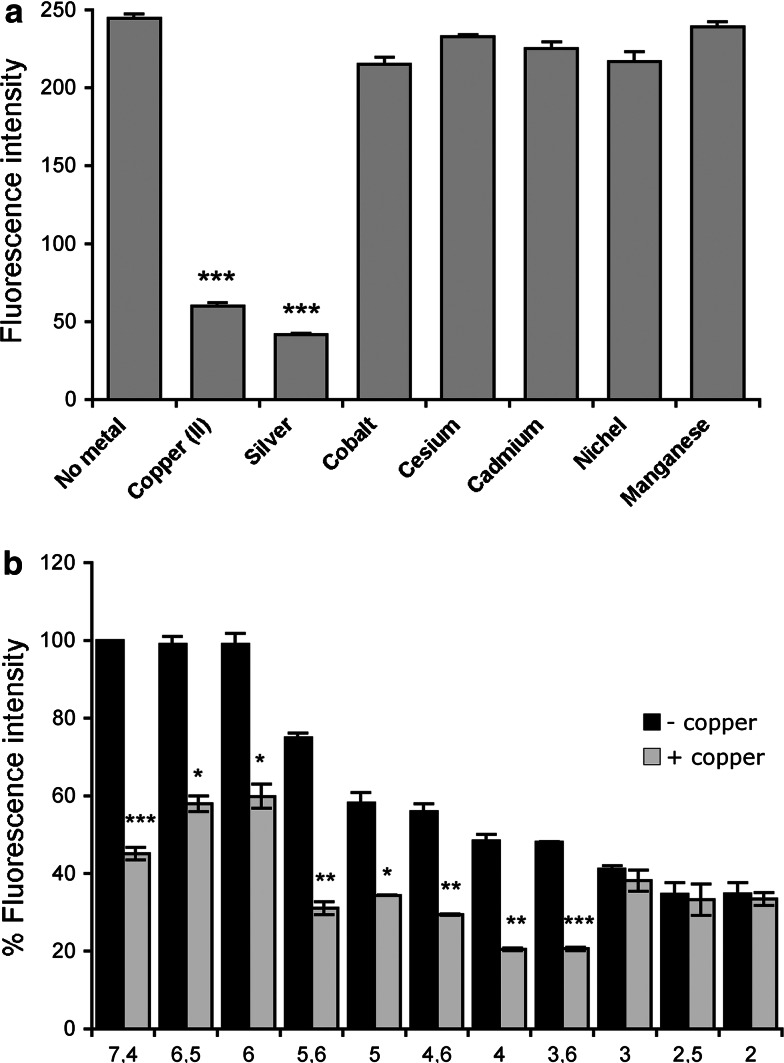
In order to evaluate the binding selectivity for copper and the quenching specificity, we checked the effect of other cationic quenchers on the peptide fluorescence. Figure 4a shows that fluorescence of WTFF peptide is only marginally affected in the presence of cobalt, cesium, nickel, cadmium, and manganese. On the other hand, only silver showed a comparable quenching efficiency on WTFF fluorescence, and a Stern–Volmer plot similar to that obtained for copper (data not shown). As previously reported [23], in some cases the ability to bind silver may be diagnostic of a possible ability of the same protein to bind copper (I). In fact, silver can be more easily manipulated and used as a probe, because cuprous ions are highly unstable. Then, the formation of Ag-TFF1 complexes also suggests a possible Cu(I) binding ability of TFF1 that deserves a targeted investigation and more sophisticated analyses, beyond the scope of the present work.
Fig. 4.
a The effect of metal ions on WTFF fluorescence. The quenching of fluorescence in the presence of Cu(II) and Ag(I) is diagnostic of a selective binding for both metals (***p < 0.0005). b pH dependence of the Cu (II) quenching of WTFF. The absence of quenching below pH 3.6 is indicative of Glu residue titration with a consequent lack of binding [Fluorescence intensities at different pHs were compared in the presence (+) and in the absence (−) of copper; *p < 0.05; **p < 0.005; ***p < 0.0005]
The influence of pH on the quenching effect of copper on WTFF was also evaluated by changing buffers pH values from 7.4 to 2. Figure 4b reports the WTFF fluorescence value at pH 7.4, showing a decrease of fluorescence intensities up to pH 3.6, while lower pH values completely prevent copper to quench fluorescence. Such a drop of the static quenching component at pH values close to the pKa Glu residues led us to infer that a change in the dissociation of glutamic acid abolishes the binding to copper cations.
Copper (II) binding affinity
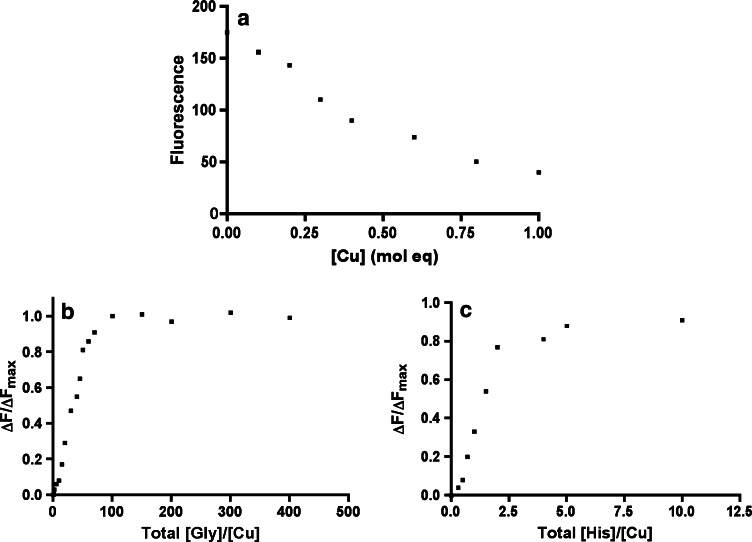
The evaluation of WTFF copper (II) binding affinity is important to speculate on the physiological significance of this interaction. To calculate the affinity constant of WTFF monomers for copper, we used glycine and histidine as competitors in fluorescence quenching experiments. Figure 5a shows a linear decrease of the fluorescence intensity and a minimum value corresponding to the addition of 1 molar equivalent of Cu2+, confirming a 1:1 Cu:WTFF binding stoichiometry, as previously demonstrated by mass spectrometric analysis. The addition of glycine or histidine to the Cu-WTFF displaces copper from the complex, thus allowing the recovery of tryptophan fluorescence up to the values corresponding to the intensity of the unbound (unquenched) levels. Plots in Fig. 5b, c allow to calculate the molar equivalents of both amino acids able to recover half of the maximum tryptophan fluorescence.
Fig. 5.
Cu2+ quenching of WTFF fluorescence: glycine and histidine competition. a WTFF fluorescence in the presence of increasing concentration of Cu2+. Rescue of the tryptophan fluorescence after the addition of glycine (b) or histidine (c). ΔF = F − F 0, ΔF max = F max − F 0, where F 0 is the fluorescence in the presence of 1 molar equivalent of Cu 2+
The half value of ΔF Max was achieved at 28 molar equivalent of glycine and 1.4 molar equivalent of histidine. Considering the concentrations of the solutes ([WTFF] = 15 × 10−6 M, [CuTot] = 15 × 10−6 M; [Cubound] = 7.5 × 10−6 M; [Gly] = 28 × (15 × 10−6) M = 4.3 × 10−4 M; [His] = 1.4 × (15 × 10−6) M = 21 × 10−6 M and substituting the values in the equation reported in “Materials and methods”, we obtained [Cufree] = K d = 0.73 nM using glycine, and [Cufree] = K d = 1.3 nM using histidine. Considering the micromolar average copper concentration in many biological fluids, the nanomolar dissociation constant of WTFF for copper allows us to infer a relevant and strong affinity of TFF1 for copper under physiological conditions.
Copper induces dimer formation in TFF1 native protein
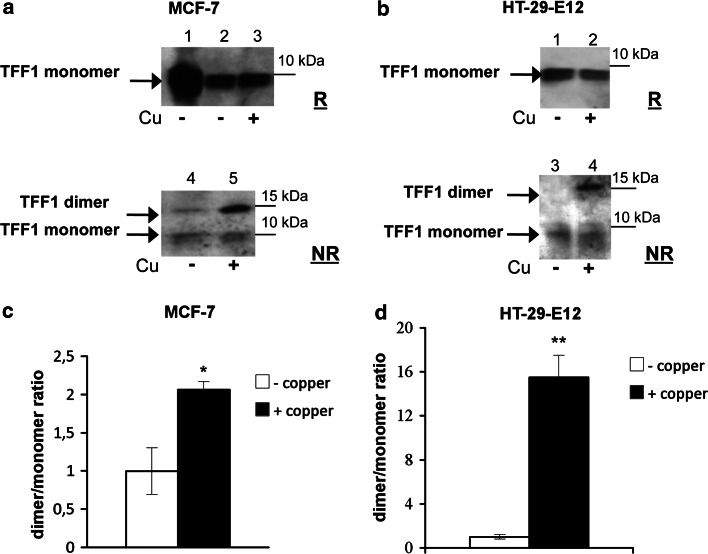
Following MS evidence obtained with synthetic peptides, we analyzed the dimer formation of the native protein after incubation with copper. Analyses on the native protein were carried out by using two different cell lines, selected by considering their high TFF1 production (MCF-7, breast cancer [9]) and/or on the basis of the ability of mimicking the loco regional physiology of the gastrointestinal tract (HT-29 MTX clone E12, goblet cells). HT-29 MTX clone E12 is a mucin-secreting clone, derived from HT-29 colon carcinoma cell line by treatment with methotrexate (MTX) [14], able to express the three trefoil factors and different mucins [24]. Both cell lines were incubated in copper-free medium (without serum) or supplemented with CuCl2 100 μM for 48 h. Treatments of HT29-E12 started the ninth day post-confluence, when the cells produce detectable amounts of TFF1 protein [24]. Secreted TFF1 was analyzed in reducing and non-reducing conditions in order to reveal the presence of the dimeric form in the culture media. It is worth noting that clear results from western blot analyses in non-reducing conditions can be obtained only from serum-free medium. Copper content in culture media, with and without serum, was analyzed by atomic absorption spectroscopy. Media supplemented with serum contained copper 220 ± 20 nM, usually considered an adequate concentration for the correct loading and the physiological function of the overall set of copper-binding proteins present in the cell. On the other hand, serum-free medium can also be considered as copper free, because the copper content was undetectable by atomic absorption analysis. Figure 6a shows the western blot analysis of MCF-7 supernatants, treated (lanes 3–5) and not treated (lanes 2–4) with copper, in reducing (lanes 2 and 3) and non-reducing (lanes 4 and 5) conditions; recombinant TFF1 from Pichia pastoris [25] was loaded in lane 1 as a control. Experimental evidence clearly shows that the presence of copper significantly induces the dimer formation, clearly increasing the dimer/monomer ratio as shown in the panel reporting the protein pattern in non-reducing conditions (compare lane 5 with lane 4). Figure 6b shows that a similar result was obtained with the proteins secreted in the culture medium of HT-29-E12 cells. Finally, Fig. 6c, d show a densitometric analysis of signals of non-reducing western blots of MCF-7 and HT-29-E12 supernatants, carried out at least as triplicate experiments (p < 0.05). In order to verify a possible nonspecific oxidative effect on protein dimerization mediated by other metals, we checked the effects of FeCl3, but no significant changes were observed in western blot analysis of supernatants (data not shown). The apparent molecular weight of the higher band matches the weight of two monomers and is diagnostic of a homodimer formation.
Fig. 6.
Western blot analysis of secreted proteins in reducing (R) and non-reducing (NR) conditions; cells were cultured in copper free culture medium (Cu−) or 100 μM CuCl2 culture medium (Cu+). a MCF-7cells (lane 1 recombinant TFF1 protein). b HT-29-E12 cells. Histograms of the densitometric analyses of MCF-7 (c) and HT-29-E12 (d) cell culture media (−Cu copper free, +Cu 100 μM CuCl2) are representative of the mean signals obtained from western blot analyses of single samples from independent experiments carried out as biological triplicate (*p < 0.05, **p < 0.005)
Copper enhances TFF1 motogenic activity
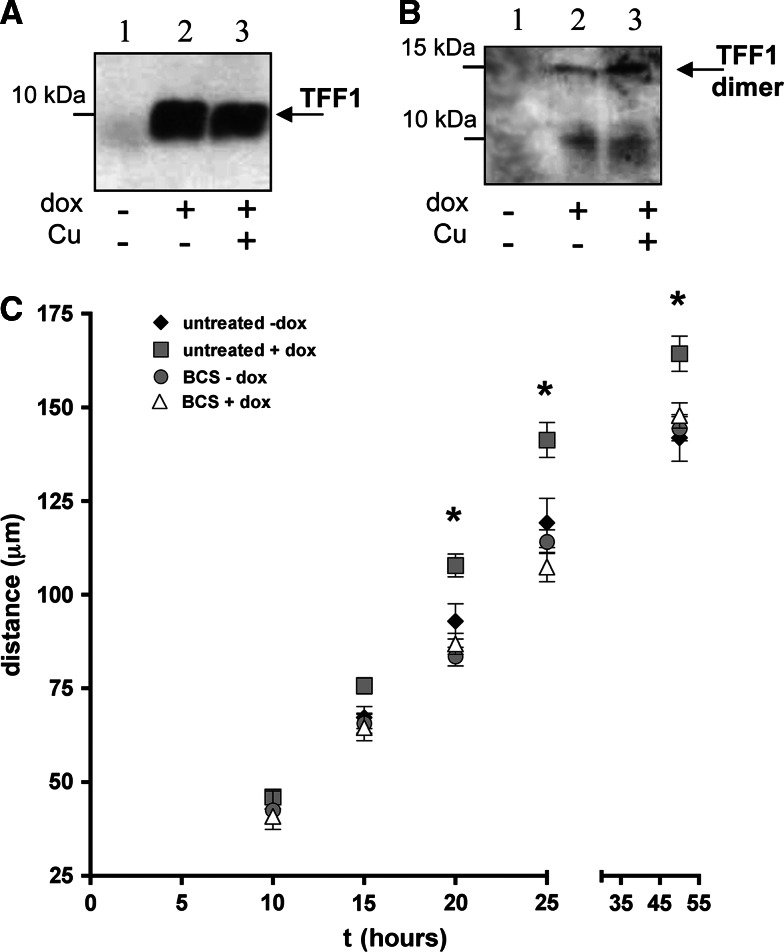
To evaluate the possible biological functions correlated to the metal binding ability of TFF1, we studied the effects of copper binding on the well-described motogenic activity of TFF1 [26, 27]. To this aim, we used an inducible hyper-expressing clone of the gastric carcinoma cell line AGS (AGS-AC1) (Fig. 7a). As well as in MCF-7 and HT-29-E12 cells, the presence of copper induces the increase of the TFF1 dimer/monomer ratio in AGS-AC1 cell cultures (Fig. 7b).
Fig. 7.
A western blot analysis of AGS-AC1 supernatants with anti-TFF1 antibody in reducing (a) and non-reducing (b) conditions: 1 20 μl of supernatant of non-induced cells; 2 20 μl of supernatant after induction with 0.1 mg ml−1 doxycycline; 3 20 μl of supernatant after induction with 0.1 mg ml−1 doxycycline + 100 μM CuCl2. c Time-course of wound-healing assays. Comparison of the migration mean distances of induced or not induced AGS-AC1 cells, treated or not treated with BCS. Diamonds untreated control, squares induction with 0.1 mg ml−1 doxycycline, circles treatment with 500 μM BCS, triangles induction with 0.1 mg ml−1 doxycycline and treatment with 500 μM BCS. p < 0.05 (untreated + dox vs untreated − dox) is marked by asterisks
AGS-AC1 clone, induced or not induced with doxycycline (i.e. expressing or not expressing TFF1), was cultured in the presence of serum. As mentioned above, serum provides an adequate concentration of copper for the physiological functionality of copper proteins, and is also strictly necessary in culture media of migrating cells. Sub-confluent culture plates were then treated with the chelating agent bathocuproine (BCS) and compared to the untreated cells. Then, 24 h later, confluent cells were wounded by scratching the cell layers with a pipette tip, and allowing the cells located on the edge of the wound to migrate into the cleared area. Cell migration was then followed by time-lapse videomicroscopy. A MTT assay was performed to assess cells viability (ESM, Fig. 3).
In Fig. 7c, we report the migration rates of the AGS-AC1 cells expressing and not expressing TFF1, in comparison with cell cultures where the physiological copper concentration was reduced or not reduced by chelation with BCS. The induction of TFF1 hyper-expression with doxycycline (Fig. 7a), increased––as expected––the cell migration rate in untreated cells cultured in the presence of the physiological concentration of copper (gray squares in Fig. 7c), while the incubation with the copper specific chelating agent abolished the gain of migration rate (white triangles in Fig. 7c). Cells not expressing TFF1 did not show any rate change when exposed to BCS (gray circles in Fig. 7c).
Increased TFF1 expression induced by doxycycline produces an appreciable increase in cell migration. This increased migration confirms the cell motogenic activity of TFF1. Copper chelation obtained by the addition of BCS suppresses the benefits of the TFF1 over-expression on cell migration induced by doxycycline in AGS-AC1 cells. These data demonstrate that copper chelation in TFF1-expressing cells is able to reduce the motogenic activity of the protein suggesting a role for TFF1-copper binding in this specific function.
Discussion
In our recent paper, we described a copper-binding property of the gastrointestinal peptide TFF1, suggesting a role for its carboxy-terminal tail in this interaction [4]. To evaluate the involvement of this acidic region in the specific binding with the transition metal, we synthesized a series of peptides containing the last 16 residues of the protein, comprising tryptophan as an endogenous fluorescent probe, that were used for mass spectrometric and fluorescence quenching analyses.
Our results clearly demonstrate a selective and specific binding ability of the native sequence peptide for copper (II); other divalent cations are not able to bind to the peptide.
Point mutations of glutamic acid residues do not substantially change the fluorescence quenching of copper, while the substitution of all four acidic amino acids completely abolishes the static component of quenching, thus suggesting that three glutamic acid residues are sufficient for the interaction, independently from their position. Point mutation of Cys58 dramatically decreases the binding, indicating that this residue is essential for the interaction.
Gastric mucous gel layer enables the stabilization of a pH gradient from acid in the lumen to near neutral at the mucosal surface [28]. The pH dependence of copper binding showed that the native peptide sequence is able to interact with the metal up to pH 3.6. Due to the presence of TFF1 in the mucous layer, our evidence suggests that TFF1–copper complex formation could be regulated by the thickness of the mucus, the local acidic environment, and the luminal availability of copper, thus consequently modulating its functions.
LC–MS or HPLC–MS analyses showed a time-dependent increase of disulphide bond formation for all peptides containing Cys58. It is well known that cysteine-containing peptides can be oxidized to inter-molecular cystine [19], and that oxidation is also influenced by pKa of cysteines, thus suggesting that in our peptides this reaction could be also favored by the presence of the four surrounding glutamates. It is worth noting that both monomers and dimers of wild-type and glutamate point-mutated peptides show a copper binding ability.
The data obtained with the help of synthetic peptides may explain the increase of dimer formation in cultured cells treated with a copper overload. It can be hypothesized that copper binding brings TFF1 monomers closer thereby favoring homodimer formation. As a consequence, copper levels could play a role as a regulator in the equilibrium between the two peptide forms.
Finally, we observed that the increase of cell migration rate in response to the over-expression of TFF1 is lost in copper-free medium. In fact, it has been reported that the TFF1 motogenic activity can be attributable to its homodimeric form, able to stimulate the cell migration [9]. Again, in the absence of copper, TFF1 monomer is the predominant form secreted by our cell system, and the reduced presence of TFF1 homodimers can explain the reduced rate of cell migration.
May et al. [29] recently reported that, in normal human gastric mucosa, TFF1 is present as a heterodimer covalently bound to TFIZ1 while, in pathologic conditions, in which TFIZ1 is absent, heterodimer is not detected. It was postulated that this may favor the production of homodimer that stimulates cancer cell motility and invasion, while copper depletion was also suggested as a therapy able to slow down the cancer invasion. The ancient use of copper in popular medicine as an antiphlogistic and cicatrizing agent, as well as the more recent scientific evidence on its involvement in wound healing processes [30] and angiogenesis [31], can rekindle the interest in the investigation of the still unclear molecular mechanism underlying its functions in physiological and pathological conditions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1: MALDI-MS analysis of the mutant Ala-scanned peptides in the presence and in the absence of five fold CuCl2 molar excess for 15 min. The MW of each species is reported in the figure (TIFF 5871 kb)
Supplementary Figure 2: MALDI-MS analysis of pTFF in the presence and in the absence of two fold AgNO3 molar excess. The pTFF-Ag(I) complex shows a peak at m/z of 2078.12 but no dimer formation is detectable after the incubation with AgNO3 (TIFF 6913 kb)
Supplementary Figure 3: MTT assay of control, BCS- and copper-treated cells. While BCS-treated cells do not show appreciable decrease of viability, copper- treated cells show significantly reduced values of absorbance and for this reason they were not considered in the motility experiment. Mean control absorbance value was set to 100%. (TIFF 6615 kb)
Acknowledgments
This work was supported by the following grants: M.I.U.R.-FISR (“Improvement of lipid and mineral contents of milks to enhance their nutraceutical and safety properties”), Mi.P.A.A.F.-NUME (“MEDITERRANEAN NUTRIGENOMICS: from molecular nutrition to the exploitation of typical products of the mediterranean diet”), and University of Salerno (Intramural ex 60% (FARB) funds).
References
- 1.Kjellev S. The trefoil factor family––small peptides with multiple functionalities. Cell Mol Life Sci. 2009;66:1350–1369. doi: 10.1007/s00018-008-8646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre O, Chenard MP, Masson R, Linares J, Dierich A, LeMeur M, Wendling C, Tomasetto C, Chambon P, Rio MC. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science. 1996;274:259–262. doi: 10.1126/science.274.5285.259. [DOI] [PubMed] [Google Scholar]
- 3.Karam SM, Tomasetto C, Rio MC. Amplification and invasiveness of epithelial progenitors during gastric carcinogenesis in trefoil factor 1 knockout mice. Cell Prolif. 2008;41(6):923–935. doi: 10.1111/j.1365-2184.2008.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosco A, Monti MC, Fontanella B, Rio MC, Gomez-Paloma L, Leone A, Marzullo L. Copper-binding activity of Trefoil factor 1 (TFF1): a new perspective in the study of the multifunctional roles of TFFs. Peptides. 2007;28:1461–1469. doi: 10.1016/j.peptides.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Tosco A, Fontanella B, Danise R, Cicatiello L, Grober OMV, Ravo M, Weisz A, Marzullo L. Molecular bases of copper and iron deficiency-associated dyslipidemia: a microarray analysis of the rat intestinal transcriptome. Genes Nutr. 2010;5:1–8. doi: 10.1007/s12263-009-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polshakov VI, Williams MA, Gargaro AR, Frenkiel TA, Westley BR, Chadwick MP, May FE, Feeney J. High-resolution solution structure of human pNR-2/pS2: a single trefoil motif protein. J Mol Biol. 1997;267:418–432. doi: 10.1006/jmbi.1997.0896. [DOI] [PubMed] [Google Scholar]
- 7.Williams MA, Westley BR, May FE, Feeney J. The solution structure of the disulphide-linked homodimer of the human trefoil protein TFF1. FEBS Lett. 2001;493:70–74. doi: 10.1016/S0014-5793(01)02276-1. [DOI] [PubMed] [Google Scholar]
- 8.Westley BR, Griffin SM, May FE. Interaction between TFF1, a gastric tumor suppressor trefoil protein, and TFIZ1, a brichos domain-containing protein with homology to SP-C. Biochemistry. 2005;44:7967–7975. doi: 10.1021/bi047287n. [DOI] [PubMed] [Google Scholar]
- 9.Prest SJ, May FE, Westley BR. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002;16:592–594. doi: 10.1096/fj.01-0498fje. [DOI] [PubMed] [Google Scholar]
- 10.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 11.Blair D, Diehl H. Bathophenanthroline disulphonic acid and bathocuproine disulphonic acid, water soluble reagents for iron and copper. Talanta. 1961;7:163–174. doi: 10.1016/0039-9140(61)80006-4. [DOI] [Google Scholar]
- 12.Sarell CJ, Syme CD, Rigby SE, Viles JH. Copper(II) binding to amyloid-beta fibrils of Alzheimer’s disease reveals a picomolar affinity: stoichiometry and coordination geometry are independent of Abeta oligomeric form. Biochemistry. 2009;48(20):4388–4402. doi: 10.1021/bi900254n. [DOI] [PubMed] [Google Scholar]
- 13.Dawson RMC, Elliot DC, Elliot WH, Jones KM. Data for biochemical research. Oxford: Clarendon Press; 1986. [Google Scholar]
- 14.Behrens I, Stenberg P, Artursson P, Kissel T. Transport of lipophilic drug molecules in a new mucus-secreting cell culture model based on HT29-MTX cells. Pharm Res. 2001;18:1138–1145. doi: 10.1023/A:1010974909998. [DOI] [PubMed] [Google Scholar]
- 15.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 16.Bossenmeyer-Pourié C, Kannan R, Ribieras S, Wendling C, Stoll I, Thim L, Tomasetto C, Rio MC. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J Cell Biol. 2002;157:761–770. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma QF, Li YM, Du JT, Kanazawa K, Nemoto T, Nakanishi H, Zhao YF. Binding of copper(II) ion to an Alzheimer’s tau peptide as revealed by MALDI-TOF MS, CD, and NMR. Biopolymers. 2005;79:74–85. doi: 10.1002/bip.20335. [DOI] [PubMed] [Google Scholar]
- 18.Nicoll AJ, Miller DJ, Futterer K, Ravelli R, Allemann RK. Designed high affinity Cu2+-binding r-helical foldamer. J Am Chem Soc. 2006;128:9187–9193. doi: 10.1021/ja061513u. [DOI] [PubMed] [Google Scholar]
- 19.Prudent M, Girault HH. The role of copper in cysteine oxidation: study of intra- and inter-molecular reactions in mass spectrometry. Metallomics. 2009;1:157–165. doi: 10.1039/b817061d. [DOI] [PubMed] [Google Scholar]
- 20.Lakowicz JR. Principles of fluorescence spectroscopy. Singapore: Springer; 2006. pp. 277–290. [Google Scholar]
- 21.Kramer ML, Kratzin HD, Schmidt B, Romer A, Windl O, Liemann S, Hornamann S, Kretzschmar H. Prion protein binds copper within the physiological concentration range. J Biol Chem. 2001;276:16711–16719. doi: 10.1074/jbc.M006554200. [DOI] [PubMed] [Google Scholar]
- 22.Syvertsen C, Melo TB, Ljones T. Fluorescence studies on dopamine beta-monooxygenase: effects of salts, pH changes, metal-chelating agents and Cu2+ . Biochem Biophys Acta. 1987;914:6–18. doi: 10.1016/0167-4838(87)90155-5. [DOI] [PubMed] [Google Scholar]
- 23.Palacios O, Polec-Pawlak K, Lobinski R, Capdevila M, González-Duarte P. Is Ag(I) an adequate probe for Cu(I) in structural copper-metallothionein studies? The binding features of Ag(I) to mammalian metallothionein 1. J Biol Inorg Chem. 2003;8:831–842. doi: 10.1007/s00775-003-0481-4. [DOI] [PubMed] [Google Scholar]
- 24.Gouyer V, Wiede A, Buisine MP, Dekeyser S, Moreau O, Lesuffleur T, Hoffmann W, Huet G. Specific secretion of gel-forming mucins and TFF peptides in HT-29 cells of mucin-secreting phenotype. Biochim Biophys Acta. 2001;1539:71–84. doi: 10.1016/S0167-4889(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 25.Kannan R, Tomasetto C, Staub A, Bossenmeyer-Pourié C, Thim L, Nielsen PF, Rio M. Human pS2/trefoil factor 1: production and characterization in Pichia pastoris . Protein Expr Purif. 2001;21(1):92–98. doi: 10.1006/prep.2000.1352. [DOI] [PubMed] [Google Scholar]
- 26.Marchbank T, Westley BR, May FE, Calnan DP, Playford RJ. Dimerization of human pS2 (TFF1) plays a key role in its protective/healing effects. J Pathol. 1998;185:153–158. doi: 10.1002/(SICI)1096-9896(199806)185:2<153::AID-PATH87>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 27.Thuwajit P, Chawengrattanachot W, Thuwajit C, Sripa B, May FE, Westley BR, Tepsiri NN, Paupairoj A, Chau-In S. Increased TFF1 trefoil protein expression in Opisthorchis viverrini-associated cholangiocarcinoma is important for invasive promotion. Hepatol Res. 2007;37(4):295–304. doi: 10.1111/j.1872-034X.2007.00045.x. [DOI] [PubMed] [Google Scholar]
- 28.Allen A, Flemstrom G. Gastroduodenal mucus bicarbonate barrier: protection against acid and pepsin. Am J Physiol Cell Physiol. 2005;288:C1–C19. doi: 10.1152/ajpcell.00102.2004. [DOI] [PubMed] [Google Scholar]
- 29.May FE, Griffin SM, Westley BR. The trefoil factor interacting protein TFIZ1 binds the trefoil protein TFF1 preferentially in normal gastric mucosal cells but the co-expression of these proteins is deregulated in gastric cancer. Int J Biochem Cell Biol. 2009;41:632–640. doi: 10.1016/j.biocel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borkow G, Gabbay J, Zatcoff RC. Could chronic wounds not heal due to too low local copper levels? Med Hypotheses. 2008;70:610–613. doi: 10.1016/j.mehy.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Brewer GJ. Copper lowering therapy with tetrathiomolybdate as an antiangiogenic strategy in cancer. Curr Cancer Drug Targets. 2005;5:195–202. doi: 10.2174/1568009053765807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: MALDI-MS analysis of the mutant Ala-scanned peptides in the presence and in the absence of five fold CuCl2 molar excess for 15 min. The MW of each species is reported in the figure (TIFF 5871 kb)
Supplementary Figure 2: MALDI-MS analysis of pTFF in the presence and in the absence of two fold AgNO3 molar excess. The pTFF-Ag(I) complex shows a peak at m/z of 2078.12 but no dimer formation is detectable after the incubation with AgNO3 (TIFF 6913 kb)
Supplementary Figure 3: MTT assay of control, BCS- and copper-treated cells. While BCS-treated cells do not show appreciable decrease of viability, copper- treated cells show significantly reduced values of absorbance and for this reason they were not considered in the motility experiment. Mean control absorbance value was set to 100%. (TIFF 6615 kb)