Abstract
Isoprenoids form an extensive group of natural products involved in a number of important biological processes. Their biosynthesis proceeds through sequential 1′-4 condensations of isopentenyl diphosphate (C5) with an allylic acceptor, the first of which is dimethylallyl diphosphate (C5). The reactions leading to the production of geranyl diphosphate (C10), farnesyl diphosphate (C15) and geranylgeranyl diphosphate (C20), which are the precursors of mono-, sesqui- and diterpenes, respectively, are catalyzed by a group of highly conserved enzymes known as short-chain isoprenyl diphosphate synthases, or prenyltransferases. In recent years, the sequences of many new prenyltransferases have become available, including those of several plant and animal geranyl diphosphate synthases, revealing novel mechanisms of product chain-length selectivity and an intricate evolutionary path from a putative common ancestor. Finally, there is considerable interest in designing inhibitors specific to short-chain prenyltransferases, for the purpose of developing new drugs or pesticides that target the isoprenoid biosynthetic pathway.
Keywords: Isoprenoid, Prenyltransferase, Short-chain isoprenyl diphosphate synthase, Geranyl diphosphate synthase, Farnesyl diphosphate synthase, Geranylgeranyl diphosphate synthase
Introduction
With more than 55,000 compounds identified in bacteria, archea, and eukaryotes, isoprenoids are the most abundant and structurally diverse natural products. They play essential roles in numerous biochemical pathways: as quinones in electron transport chains, as components of membranes (prenyllipids in archaebacteria, and as sterols in eubacteria and eukaryotes), in subcellular targeting and regulation (prenylation of proteins), as photosynthetic pigments (carotenoids, side chain of chlorophyll), as hormones (gibberellins, brassinosteroids, abscisic acid, retinoic acid, juvenile hormone), and as semiochemical compounds in plants and insects (monoterpenes, sesquiterpenes, diterpenes) [1, 2].
The universal precursors of isoprenoids generated by the mevalonate pathway are synthesized by sequential 1′-4 condensations of isopentenyl diphosphate (IPP, C5) with an allylic diphosphate, in reactions catalyzed by enzymes commonly referred to as prenyltransferases or isoprenyl diphosphate synthases (IPPS). Each member of this enzyme family is classified according to the stereochemistry of the double bonds formed during product elongation and the length of the final product [3]. In general, trans-prenyltransferases synthesize products up to C50 in length, with trans (E) double bonds, while most cis-prenyltransferases characterized to date generate longer products featuring cis (Z) double bonds. Although trans- and cis-prenyltransferases catalyze similar reactions, they are evolutionary and structurally distinct [4]. Trans-prenyltransferases can be further divided into short-chain (scIPPS; C10–C25), medium-chain (mcIPPS; C30–C35), and long-chain (lcIPPS; C40–C50) prenyltransferases [3]. Whereas most short-chain prenyltransferases are active as homodimers, both homo- and heteromers have been observed among the medium- and long-chain enzymes.
Enzymes of the short-chain prenyltransferase subfamily
Geranyl diphosphate synthase (GPP synthase or GPPS)
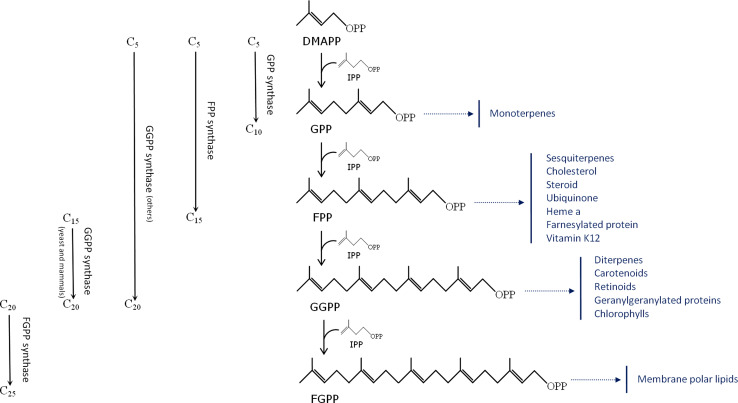
GPPS catalyzes the condensation of two C5 co-substrates, IPP and dimethylallyl diphosphate (DMAPP), to produce GPP, the precursor of all monoterpenes (Fig. 1). Despite the multiple functions of monoterpenes, which are found most commonly in plants and insects where they act as essential oils and pheromones, respectively, our understanding of their biosynthesis remains limited. GPPS genes have been characterized from only a handful of plant species [5–13] and, so far, only one study has reported on the existence of a GPPS in an insect [14]. However, a second putative GPPS has recently been cloned by our group from a longhorn beetle (unpublished data; see “Evolution of short-chain prenyltransferases” for more information), and an enzyme producing both GPP and FPP has been cloned from aphids [15, 16]. Based on sequence comparisons, three different classes of plant GPPSs may be distinguished, one of which contains heterodimeric proteins, while the other two are active as homodimers [13]. In the first heterodimeric GPPS to have been isolated (from Mentha piperita), the two subunits were shown to display very limited similarity to one another, with only one subunit recognized as a typical prenyltransferase [6]. Among the homodimeric plant GPPSs, one class has so far been found only in conifers [13] while the other class has members among diverse plant species, including Arabidopsis thaliana [8]. These two homodimeric GPPS types display only 20% amino acid sequence identity to each other. The first GPPS to be characterized from an animal was isolated from the bark beetle Ips pini, in which it plays a role in aggregation pheromone biosynthesis [14]. Plant and insect GPPSs appear to have no common proximate ancestor (see“Evolution of short-chain prenyltransferases”).
Fig. 1.
Elongation reactions catalyzed by short-chain prenyltransferases, along with examples of final products generated by the geranyl diphosphate (GPP), farnesyl diphosphate (FPP), geranylgeranyl diphosphate (GGPP), and farnesylgeranyl diphosphate (FGPP) precursors
Farnesyl diphosphate synthase (FPP synthase or FPPS)
FPPS is the most extensively studied short-chain prenyltransferase due to its central position in the mevalonate pathway. FPPS catalyzes the condensation of IPP and DMAPP to form the intermediate GPP, which then undergoes a second condensation step to generate FPP (Fig. 1). The FPP product serves as precursor to sesquiterpenes, sterols, dolichols, and some mitochondrial ubiquinones, and is used for protein farnesylation. FPPSs were purified to homogeneity from a number of organisms, including Saccharomyces cerevisiae [17], chicken [18], pig [19], human [20], and green pepper, Capsicum annuum [21]. All these purified enzymes were found to form homodimers, with tightly coupled subunits ranging from 32 to 44 kDa in size. In addition, FPPS cDNAs were cloned from various organisms [22–31]. FPPSs may be divided into two broad categories: type I (eukaryotic) and type II (eubacterial) FPPSs [32, 33]. Recent work on insect FPPSs suggests that the type-I enzymes include various subtypes displaying features that are distinct from those of vertebrate, plant, and yeast FPPSs [27, 34].
In addition to the trans-FPPSs described above, some cis-FPPSs have recently been cloned and characterized from bacteria [35–37] and plants [38]. Whereas the bacterial enzymes use GPP as allylic co-substrate and produce E,Z-FPP, the plant enzyme uses DMAPP and generates Z,Z-FPP. Little is known about the mechanism of product chain length regulation in these enzymes [37, 38].
Geranylgeranyl diphosphate synthase (GGPP synthase or GGPPS)
GGPPS supplies the essential acyclic precursor GGPP for the biosynthesis of a structurally diverse group of compounds that includes diterpenes, carotenoids, chlorophylls, geranylgeranylated proteins, and archaeal ether-linked lipids. In plants and bacteria, GGPP is synthesized by a GGPPS that catalyzes successive additions of IPP to DMAPP, GPP, and FPP (Fig. 1), while yeast and mammalian GGPPSs can only accept FPP as the allylic co-substrate [39]. To date, cDNAs encoding GGPPS have been cloned from various organisms ranging from bacteria to higher eukaryotes [40–46]. GGPPSs have been divided into three categories [32, 33]: type I (archaea and some bacteria), type II (plants and bacteria), and type III (eukaryotes other than plants) (see “Evolution of short-chain prenyltransferases” for further details).
Farnesylgeranyl diphosphate synthase (FGPP synthase or FGPPS)
Tashibana [47] reported on the presence of farnesylgeranyl diphosphate (FGPP) synthase activity, generating C25 isoprenyl diphosphates, in Natronobacterium pharaonis, a haloalkaliphilic archaeon that has C20–C25 diether lipids in its membranes, in addition to the C20–C20 diether lipids commonly found in archaea. The highest activity of this FGPPS was observed when GGPP was used as the allylic substrate. Tashibana et al. [48] observed the same behavior for an FGPPS from the aerobic hyperthermophilic archaeon Aeropyrum pernix, the membrane of which contains only C25–C25 diether lipids. These archeal FGPPSs are very closely related to archeal GGPPSs [48], thus justifying their inclusion here in the group of short-chain prenyltransferases (see also “Evolution of short-chain prenyltransferases”).
Short-chain prenyltransferases displaying “catalytic promiscuity”
Short-chain prenyltransferases displaying dual FPP/GGPP synthase activity have been reported for a protozoan [49], a hyperthermophilic archaea [50], and maize [51]. In the reactions catalyzed by these enzymes, the use of either DMAPP or GPP as allylic co-substrate leads to the formation of both FPP and GGPP, whereas the use of FPP generates GGPP as the sole product. This is in contrast with results obtained for conventional GGPPSs, which tend to generate only GGPP, irrespective of the allylic co-substrate used.
In aphids, Vandermoten et al. [15] identified the first animal short-chain prenyltransferase that can generate both GPP and FPP. Although the recombinant enzyme yielded more GPP than FPP, and the proportion of GPP increased with a rise in the concentration of DMAPP, it generated only FPP if supplied with GPP as the sole allylic co-substrate [15]. In addition, the recombinant aphid enzyme could generate both monoterpenes and sesquiterpenes in linked assays where appropriate terpene synthases were added to the assay buffer [16]. In contrast, the recombinant Drosophila melanogaster FPPS generated almost exclusively FPP in the presence of IPP and DMAPP [15].
In Norway spruce, Picea abies, Shimdt et al. [13] reported on the presence of two GPPSs belonging to two separate groups of homodimeric proteins. While one enzyme produced GPP as its sole in vitro product, the second enzyme produced substantial amounts of FPP and GGPP, in addition to GPP (42% GPP, 33% FPP, and 25% GGPP). In parallel assays performed on a related enzyme from the angiosperm Quercus robur, GPP and FPP were produced in proportions of 55:45, while no GGPP was detected [13].
These dual (or multiple) activities observed for some short-chain prenyltransferases may be examples of “catalytic promiscuity”, an expression that was coined to describe the ability of some enzymes to display an adventitious secondary activity at the active site responsible for the primary activity [52]. Catalytic promiscuity is of interest because an adventitious secondary activity may become useful to the organism at some point, and the enzyme may be recruited to provide the secondary product. For example, the existence of a GPP/FPP synthase in aphids is perhaps not too surprising, given that these insects require both monoterpenes and sesquiterpenes as precursors for sex pheromone, alarm pheromone, and juvenile hormone biosynthesis [15].
Catalytic mechanism
Short-chain prenyltransferases catalyse chain elongation reactions, where the growing chain of an allylic isoprenoid diphosphate (DMAPP, GPP, or FPP) undergoes coupling with IPP through electrophilic alkylation of its carbon–carbon double bond. In all organisms, this reaction occurs following a sequential ionization–condensation–elimination mechanism referred to as head-to-tail condensation. After binding of both co-substrates to the enzyme, a carbocation is formed at the C1’ position of the allylic substrate; this step is activated by divalent cations such as Mg2+ or Mn2+. The carbocation electrophilically attacks the C4 position of IPP resulting in formation of a C–C bond between IPP and the allylic substrate. The product is then released from the active site [53]. The stereochemistry of the 1′-4 condensation was elucidated by Cornforth’s group [54], who established that the allylic carbocation attacks the double bond of IPP from the Si-face. The proR-H at the C2 position of IPP is then removed to form a new allylic trans-double bond.
Structure and active site
Current knowledge about short-chain prenyltransferase 3D-structures is based in large part on the work of Tarshis et al. [55] who reported the crystal structure of unliganded avian FPPS at 2.6-Å resolution, as well as that of the corresponding double F112A/F113S mutant, liganded with DMAPP, GPP, and FPP [56]. More recently, Hosfield et al. [57] reported the crystal structures of two prokaryotic FPPSs at 2.4-Å resolution: the unliganded Staphylococcus aureus FPPS and the Escherichia coli enzyme bound to both IPP and a noncleavable DMAPP analogue. In addition, the structure of the Trypanosoma cruzi FPPS, both unliganded and complexed with substrates and inhibitors, has been elicidated [58]. Furthermore, Kavanaght et al. [59] reported the high-resolution X-ray structures of the human FPPS in complex with two bisphosphonate inhibitors, risedronate and zoledronate, respectively. Finally, Chang et al. [60] reported the first GGPPS structure, which was determined for the yeast Saccharomyces cerevisiae at 1.98-Å resolution.
Altogether, these studies have indicated that short-chain prenyltransferases are typically active as homodimers and exhibit a fold composed of 13 α-helices joined by loops. Ten of these helices form a helical bundle that surrounds a central cavity where the active site is located. Two highly conserved aspartate-rich motifs face each other on opposite walls of this cavity. Using hybrid-type heterodimers of Bacillus stearothermophilus FPPSs, constituting different types of mutated monomers in regions II and IV (see below), Koyama et al. [61] suggested that FPPS subunits interact with each other to form a shared active site in the homodimer structure, rather than an independent active site in each subunit. More recently, the determination of the first mammalian GGPPS crystal structure revealed a novel hexameric quaternary structure [62]. The regions involved in hexamer formation (from three homodimers) are largely conserved for mammalian and insect GGPPSs, but not for plant, bacterial, fungal, or archaeal GGPPSs, suggesting that insect GGPPS may also be hexameric. Unlike the homodimeric FPP or GGPP synthases purified to date, both homo- and heteromeric forms of plant GPPSs have been identified [6, 7, 9].
The deduced amino acid sequences of all short-chain prenyltransferases tend to display a high level of similarity to one another, and multiple sequence alignments have revealed the presence of seven conserved regions [63]. Regions II and VI contain aspartate-rich motifs [DDx(xx)xD; where x represents any amino acid] referred to as the first (FARM) and second (SARM) aspartate-rich motifs, respectively. Site-directed mutagenesis studies targeting the aspartate residues within the FARM have pointed to their involvement in catalysis and binding of the allylic substrate [63, 64]. Binding of IPP, however, involves the C-terminus of the enzyme, which interacts with Lys, Arg, and His residues contacting the IPP pyrophosphate [57]. Among the five residues located upstream from the FARM, some have been shown to be involved in product chain-length determination; this portion of the sequence is therefore referred to as the chain-length determination (CLD) region [33], although additional residues, located on distinct α-helices, can play a role in defining product chain-length [60].
Mechanisms of product chain-length regulation
Although they share the same catalytic mechanism, short-chain prenyltransferases rarely generate a product whose chain length differs from that which is pre-determined for the enzyme. Over the past several years, there has been considerable interest in identifying the mechanisms responsible for product chain-length regulation by scIPPSs. In this context, it has been observed that factors such as salt and metal ion concentrations [65, 66] and the ratio between allylic and IPP co-substrates [15, 67, 68] can, to a limited extent, modulate the product chain-length selectivity of some prenyltransferases. Under certain circumstances, temperature can also have an impact on this variable, as shown by Fujiwara et al. [69] for the dual FPP/GGPP synthase isolated from the hyperthermophilic archea Thermococcus kodakaraensis.
However, the primary molecular mechanism regulating product chain length is dependent upon structural features specific to each subgroup of prenyltransferases. Our current understanding of the mechanisms of product chain-length determination by these enzymes is derived primarily from the elucidation of their 3D structures, combined with several site-directed and random mutagenesis studies. The analysis of the crystal structures of wild-type and mutated avian FPPS suggested that the ultimate product length was dictated by the size of the active site hydrophobic pocket [56]. More specifically, the two aromatic residues Phe112 and Phe113, located at the fourth and fifth positions upstream from the FARM, were shown to form the “floor” of the catalytic pocket, and replacement of these two amino acids with smaller residues directly altered the size of the binding pocket, resulting in the formation of longer products. Additional studies further demonstrated that the product chain-length of wild-type FPP and GGPP synthases was in large part determined by the nature of the amino-acid residues located at both the fourth and fifth positions upstream from the FARM [11, 56, 70, 71]. The side-chains of these amino-acid residues can either prevent or allow additional condensations to take place, with smaller side-chains typically permitting further product elongation. In bacterial FPPSs, which typically have a two-amino-acid insertion within the FARM (i.e., DDxxxxD instead of DDxxD), only the residue at position -5 relative to the FARM seems critical to defining product chain length selectivity [70, 71], possibly because of the structural changes brought about by the two-amino-acid insertion [33].
Residues other than those at positions -4 and -5 relative to the FARM can also play a role in the chain-length selectivity of short-chain prenyltransferases. For example, the Ala116Trp (-1 relative to the FARM) and Asn144Trp (+23 relative to the FARM) avian FPPS mutants displayed a smaller binding pocket for the hydrocarbon chain of the allylic substrate, with altered product selectivity favoring synthesis of shorter products such as GPP [72]. In type-III GGPPSs, in which the fourth and fifth residues upstream from the FARM are either Ser or Ala (Fig. 2), other residues located deeper in the catalytic cavity prevent chain elongation beyond C20. In yeast GGPPS, for instance, Tyr107, Phe108, and His139 seal the bottom of the active site cavity [60].
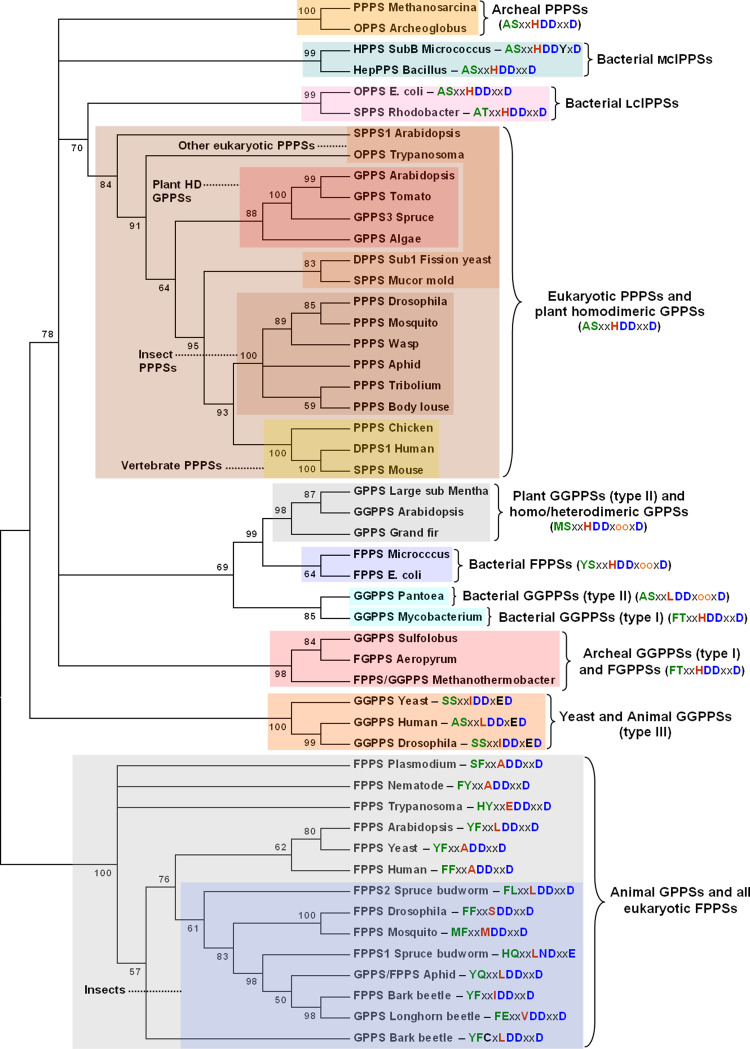
Fig. 2.
Phylogenetic tree (unrooted) of trans-isoprenyl diphosphate synthases from diverse taxa. Nucleotide sequences were retrieved from Genbank and imported into MEGA4 [91], where they were conceptually translated into proteins. Alignments were performed on the amino acid sequences using ClustalW, as implemented by MEGA4, with default settings except for penalties relating to multiple alignment gap openings and extensions, which were set to 3.0 and 1.8, respectively [92]. The back-translated nucleotide alignment was then used to construct a Neighbor-Joining tree using MEGA4’s default settings, except for gap deletions, where the pairwise deletion option was selected. Bootstrapping was performed on 2,000 pseudoreplicates, and all branches with less than 50% boostrap support were collapsed. For each enzyme or enzyme lineage, residues within the chain-length determination (LCD) region [i.e., the first aspartate-rich motif (“FARM” = DDxxD or DDxooxD) and the five residues upstream from it] are shown as a color-coded sequence, where x = L, I, V or M, and o = any amino acid. Residues at position -4 and -5 relative to the FARM are shown in green, while the residue at position -1 is shown in red. Aspartate residues within the FARM are shown in blue. Accession numbers: GPPS: algae, XM_001691017; Arabidopsis, Y17376; bark beetle, AY953508; grand fir, AF513112; longhorn beetle, not yet deposited; Mentha, AF182828; spruce; EU432048; tomato, DQ286930; GPPS/FPPS: aphid, AY968586; FPPS: Arabidopsis, L46350; bark beetle, AY953507; Drosophila, NM_058032; E. coli, D00694; human, NM_002004; Micrococcus, AB003187; mosquito, XM_308653; nematode, NM_060626; Plasmodium-AY219707; spruce budworm-1, AY954920; spruce budworm-2, AY954919; Trypanosoma, AF386102; yeast, NC_001142; FPPS/GGPPS: Methanothermobacter, S75695; Mycobacterium, NC_000962; GGPPS: Arabidopsis, NM_127943; Drosophila, AF049659; human, NM_004837; Pantoea, D90087; Sulfolobus, NC_007181; Yeast, AY692852; FGPPS: Aeropyrum, AB025791; hexaprenyl diphosphate synthase (HPPS; C30): Micrococcus, AB003188; heptaprenyl diphosphate synthase (HepPPS; C35): Bacillus, EF191544; octaprenyl diphosphate synthase (OPPS; C40): Archeoglobus, AE000782; E. coli, U18997; Trypanosoma, XM_799480; solanesyl diphosphate synthase (SPPS): Arabidopsis, NM_001084371; mouse, NM_019501; Mucor mold, AJ496300; Rhodobacter, AB001997; decaprenyl diphosphate synthase (DPPS; C50): fission yeast, NM_001021183; human, NM_014317; PPPS: aphid, XM_001947162; body louse, DS235241; chicken, XM_418592; Drosophila, NM_170546; Methanosarcina, NC_007355; mosquito, XM_565746; Plasmodium, XM_001349505; Tribolium, XM_968226; wasp, XM_001602302
Several insect GPP and FPP synthases do not display an aromatic residue at either or both the fourth and fifth positions upstream from the FARM [27]. For example, the aphid GPP/FPP synthase displays a Gln residue at positions -4 relative to the FARM (YQxxxDDxxD), a feature shared with lepidopteran type-1 FPPSs [27], for which the exact product(s) remain(s) to be identified [73]. Site-directed mutagenesis and molecular dynamics simulations indicated that the dual GPP/FPP synthase activity of the aphid enzyme was largely defined by Gln107 and Leu110, at positions -4 and -1 relative to the FARM, respectively. Although the mechanism responsible for this dual activity involves steric constraints on the ligand, as shown for other short-chain prenyltransferases, two additional factors have been identified, namely ligand stabilization and the disruption of H-bonds formed between Gln107 and residues across the catalytic cavity, which constitutes a novel chain-length determination mechanism for scIPPSs (Vandermoten et al., unpublished data).
Although the mechanism of product chain-length determination of homodimeric plant GPPSs has not been elucidated, that of the heterodimeric GPPS from mint [6] has been shown to be dependent upon the interaction of the small subunit with the large, GGPPS-like subunit [74]. However, this interaction does not appear to result strictly from constraints imposed by the small subunit on the size of the active site cavity in the large subunit [10]. Thus, additional geometric features of the catalytic cavity apparently contribute to product chain-length determination in these GPPSs.
Evolution of short-chain prenyltransferases
Phylogenetic trees of trans-isoprenyl diphosphate synthases were published earlier by other groups [32, 33, 48, 75], but many prenyltransferases have since been cloned and sequenced, including several plant and animal GPPSs, insect FPPSs displaying unusual features and various medium- and long-chain polyprenyl diphosphate synthases, to which short-chain prenyltransferases are related. For this reason, it seemed appropriate to re-examine the phylogeny of this group of enzymes.
All trans-isoprenyl diphosphate synthases are believed to be derived from a common ancestor, and some have argued that archeal GGPPSs are the closest relatives to this common ancestor [33, 75], although this hypothesis has been challenged [48]. Given the high incidence of horizontal gene transfer that has occured among procaryotes [76], it will remain difficult to identify the prenyltransferase that is closest to the root of the tree. For this reason, the present tree (Fig. 2) was left unrooted; as a result, its branching order may not always reflect the evolutionary course that these enzymes have taken. Nevertheless, the analysis clearly indicates that trans-isoprenyl diphosphate synthases form two large and distinct clusters that have presumably originated from a common procaryotic ancestor: animal GPPSs and eukaryotic FPPSs on the one hand, and all other trans-prenyltransferases on the other. Within the first cluster, the residues at positions -4 and -5 relative to the FARM tend to be aromatic amino acids (F or Y), although this is not always the case, particularly among the FPPSs of unicellular organisms such as Plasmodium and Trypanosoma, and the enzymes of insects, where substitutions at these positions can lead to the formation of GPP, instead of FPP, as the principal product [15]. The level of amino acid sequence similarity among these enzymes displays a wide range of variation in pairwise comparisons, even within a species (e.g., spruce budworm FPPS1 and FPPS2). Bark beetle GPPS and FPPS are very divergent and display only a limited degree of similarity to one another, despite a nearly identical CLD region (the GPPS has an uncommon cysteine residue at position -3 relative to the FARM). The nature of the residue at the first position upstream from the FARM, which can have an impact on product chain length [72], also tends to vary considerably within this group (L, M, I, V, A, S, or E). In contrast, all enzymes in the other cluster, with the exception of animal and type-II bacterial GGPPSs, have a histidine residue at the equivalent position. In fact, many of these enzymes, including all eukaryotic polyprenyl diphosphate synthases [PPPSs; i.e., all mcIPPSs and lcIPPSs, including those whose exact product chain-length (C30–C50) is unknown], a sub-group of plant homodimeric GPPSs, archeal PPPSs, and some bacterial medium- and long-chain IPPSs share the same ASxxHDDxxD “FARM signature”. The ubiquity of this histidine residue suggests that it plays an important structural role, although its presence in enzymes displaying vastly different product chain-length selectivities (e.g., plant GPPSs and animal lcIPPSs) points to a limited role in defining product chain length.
Interestingly, animal GGPPSs are closely related to the yeast enzyme (both are type-III GGPPSs), while the latter is clearly distinct from plant GGPPSs (Fig. 2). Type-III GGPPSs are highly divergent from their intra-specific FPPS counterparts. In fact, inter-taxonomic GGPPS-GGPPS comparisons reveal a much higher degree of similarity than that measured for intra-specific FPPS-GGPPS comparisons (e.g., Blastp expect values for human versus Drosophila GGPPS: 1e -103; for human FPPS versus human GGPPS: 1e -007). These observations strongly suggest that the two enzymes have not evolved from one another, but have been acquired independently, possibly through horizontal gene transfer in a common ancestor. A more obvious case of the latter phenomenon is seen with plant GGPPSs and a sub-group of plant GPPSs, which display a CLD region almost identical to that of some bacterial FPPSs and GGPPSs (including the insertion of two extra residues within the FARM), from which they appear to have originated (Fig. 2).
Whereas FPP and GGPP synthases occur nearly ubiquitously in plants, animals, fungi and bacteria, GPPSs appear to have a more limited distribution in nature, having been identified almost exclusively in plants and insects. The present analysis indicates that insect GPPSs have evolved from insect FPPSs while those of plants have evolved once from eukaryotic PPPSs (most homodimeric plant GPPSs) and once from plant GGPPSs (heterodimeric plant GPPSs). The homodimeric grand fir GPPS stands out from the other homodimeric plant GPPSs included in the present analysis, with its distinctive bacterial “FARM signature”. Clearly, trans-prenyltransferases from all organisms share many features that are indicative of common ancestry, but they also seem to have followed a convoluted evolutionary path that blurs some of the branching points in the tree shown here.
Inhibitors of short-chain prenyltransferases and perspectives for applied research
Over the past several years, there has been considerable interest in developing inhibitors that are specific to certain enzymes of the mevalonate pathway. This concept has found widespread clinical use with statins, which are drugs that inhibit hydroxymethylglutaryl-CoA (HMG CoA) reductase and, as a result, reduce cholesterol biosynthesis. Because short-chain prenyltransferases play a pivotal role in the biosynthesis of numerous mevalonate pathway end-products, the inhibition of their activity is also of great interest in applied research. Among this enzyme family, FPPS is a key target, and several groups have investigated the possibility of inhibiting FPPS with nitrogen-containing bisphosphonates (N-BPs) [77–80]. N-BPs are pyrophosphate analogs in which the oxygen bridge between the two phosphorus atoms has been replaced with a carbon atom bearing two side chains, the nature of which affects the compound’s chemical properties, mode of action, and potency.
In mammals, the inhibition of FPPS by N-BPs blocks the formation of FPP, which is required for protein prenylation. A deficit in protein prenylation results in osteoclast apoptosis and inhibition of the bone-destroying action of these cells [81]. Therefore, N-BPs are used to treat disorders characterized by bone resorption such as osteoporosis, Paget disease, hypercalcemia caused by malignancy, tumor metastases in bone, and other ailments [82–85].
A number of N-BPs have recently been shown to have curative effects in in vivo models of visceral [86] and cutaneous [87] leishmaniasis. Their significant activity was also demonstrated against the proliferation of T. brucei and T. cruzi, the causative agents of African trypanosomiasis (sleeping sickness) and Chagas’ disease, respectively. Other N-BPs also show potential as anti-cancer drugs as they can inhibit human GGPPS, which is required for the prenylation of proteins such as Ras involved in cancer development [88].
In the field of agrochemical research, the replacement of the amino group of N-BPs by hydroxyl, ureido, thioureido, or amino moieties leads to compounds inhibiting FPPS and having significant herbicidal properties [89]. Some of the N-BPs that are effective in inhibiting FPP and GGPP synthases also inhibit plant heterodimeric GPPSs [10]; the herbicidal action of N-BPs could therefore result from their inhibitory activity on more than one prenyltransferase. It has also been suggested that the uncommon structural features of some insect GPP and FPP synthases, including those of caterpillars [27] and aphids [15], could be exploited for the design of pest-specific inhibitors displaying insecticidal activity. Since FPPSs are required for the production of juvenile hormone in insects, FPPS inhibition is expected to cause a decrease in the production of this hormone, which would result in a precocious and lethal metamorphosis [90].
Conclusion
Although considerable new information has accumulated on short-chain prenyltransferases during the past decade, with important advances in their cloning and characterization, these enzymes have not yet revealed all their secrets. Indeed, the recent discovery of new types of short-chain prenyltransferases that produce a mixture of products with different chain-lengths points to a new level of complexity in the mechanism regulating product size, which is not always dictated only by steric constraints, as originally suggested. Many of the recently cloned enzymes have yet to be characterized, including several putative insect FPPSs, which could turn out to have properties that differ from those initially inferred from blastp searches, given the sometimes unorthodox nature of their CLD regions. In addition, the mechanism of product chain-length selectivity of homodimeric plant GPPSs, which are in many respects very similar to eukaryotic long-chain prenyltransferases, needs to be examined so as to identify the substitutions that have led the presumed conversion of enzymes that generate long polyprenyl chains into GPPSs; this would help elucidate their exact evolutionary history.
Finally, there are still exciting prospects with respect to the development of inhibitors specific to short-chain prenyltransferases, both in the field of drug discovery and in crop/forest protection. Progress in this area will undoubtedly continue to benefit from sustained efforts in the structural and enzymatic characterization of novel prenyltransferases displaying features that suggest that they might be inhibited in a taxon-selective manner.
Acknowledgments
This work was supported by a PhD scholarship to S.V., awarded by the “Fonds pour la formation à la Recherche dans l’Industrie et l’Agriculture” (Belgium) and an NSERC Discovery Grant to M.C.
References
- 1.Thulasiram HV, Erickson HK, Poulter CD. Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science. 2006;316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 2.Ogura K, Toyama T, Sagami H. Polyprenyl diphopshate synthases. In: Bittman R, editor. Subcellular biochemistry. New York: Plenum; 1997. pp. 57–87. [DOI] [PubMed] [Google Scholar]
- 3.Ogura K, Koyama T. Enzymatic aspects of isoprenoid chain elongation. Chem Rev. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- 4.Ko TP, Chen YK, Robinson H, Tsai PC, Gao YG, Chen APC, Wang AHJ, Liang PH. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J Biol Chem. 2001;276:47474–47482. doi: 10.1074/jbc.M106747200. [DOI] [PubMed] [Google Scholar]
- 5.Croteau R, Purkett PT. Geranyl pyrophosphate synthase: characterization of the enzyme and evidence that this chain-length specific prenyltransferase is associated with monoterpene biosynthesis in sage (Salvia officinalis) Arch Biochem Biophys. 1989;271:524–535. doi: 10.1016/0003-9861(89)90304-4. [DOI] [PubMed] [Google Scholar]
- 6.Burke CC, Wildung MR, Croteau R. Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA. 1999;96:13062–13067. doi: 10.1073/pnas.96.23.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke CC, Croteau R. Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch Biochem Biophys. 2002;405:130–136. doi: 10.1016/S0003-9861(02)00335-1. [DOI] [PubMed] [Google Scholar]
- 8.Bouvier F, Suire C, d’Harlingue A, Backhaus RA, Camara B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 2000;24:241–252. doi: 10.1046/j.1365-313x.2000.00875.x. [DOI] [PubMed] [Google Scholar]
- 9.Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudarevac N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell. 2004;16:977–992. doi: 10.1105/tpc.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke C, Klettke K, Croteau R. Heteromeric geranyl diphosphate synthase from mint: construction of a functional fusion protein and inhibition by bisphosphonate substrate analogs. Arch Biochem Biophys. 2004;422:52–60. doi: 10.1016/j.abb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Narita K, Ohnuma S, Nishino T. Protein design of geranyl diphosphate synthase: structural features that define the product specificities of prenyltransferases. J Biochem (Tokyo) 1999;126:566–571. doi: 10.1093/oxfordjournals.jbchem.a022487. [DOI] [PubMed] [Google Scholar]
- 12.Clastre M, Bantignies B, Feron G, Soler E, Ambid C. Purification and characterization of geranyl diphosphate synthase from Vitis vinifera L. Cv Muscat de Frontignan cell cultures. Plant Physiol. 1993;102:205–211. doi: 10.1104/pp.102.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt, A, Gershenzon J (2008) Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies) Phytochem. 69:49–57 [DOI] [PubMed]
- 14.Gilg AB, Bearfield JC, Titigger C, Welch WH, Blomquist GJ. Isolation and functional expression of an animal geranyl diphosphate synthase, and its role in bark beetle pheromone biosynthesis. Proc Natl Acad Sci USA. 2005;102:9760–9765. doi: 10.1073/pnas.0503277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandermoten S, Charloteaux B, Santini S, Sen SE, Béliveau C, Vandenbol M, Francis F, Brasseur R, Cusson M, Haubruge E. Characterization of a novel aphid prenyltransferase displaying dual geranyl/farnesyl diphosphate synthase activity. FEBS Lett. 2008;582:1928–1934. doi: 10.1016/j.febslet.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 16.Lewis MJ, Prosser IM, Mohib A, Field LM. Cloning and characterisation of a prenyltransferase from the aphid Myzus persicae with potential involvement in alarm pheromone biosynthesis. Insect Mol Biol. 2008;17:437–443. doi: 10.1111/j.1365-2583.2008.00815.x. [DOI] [PubMed] [Google Scholar]
- 17.Eberhardt NL, Rilling HC. Prenyltransferase from Saccharomyces cerevisiae: purification to homogeneity and molecular properties. J Biol Chem. 1975;250:863–866. [PubMed] [Google Scholar]
- 18.Reed BC, Rilling HC. Crystallization and partial characterization of prenyltransferase from avian liver. Biochemistry. 1975;14:50–54. doi: 10.1021/bi00672a009. [DOI] [PubMed] [Google Scholar]
- 19.Yeh LS, Rilling HC. Purification and properties of pig liver prenyltransferase: interconvertible forms of the enzyme. Arch Biochem Biophys. 1977;183:718–725. doi: 10.1016/0003-9861(77)90405-2. [DOI] [PubMed] [Google Scholar]
- 20.Barnard GF, Popjak G. Human liver prenyltransferase and its characterization. Biochim Biophys Acta. 1981;661:87–99. doi: 10.1016/0005-2744(81)90086-3. [DOI] [PubMed] [Google Scholar]
- 21.Hugueney P, Camara B. Purification and characterization of farnesyl pyrophosphate synthase from Capsicum annuum . FEBS Lett. 1990;273:235–238. doi: 10.1016/0014-5793(90)81093-4. [DOI] [PubMed] [Google Scholar]
- 22.Clarke CF, Tanaka RD, Svenson K, Wamsley M, Fogelman AM, Edwards PA. Molecular cloning and sequence of a cholesterol-repressible enzyme related to prenyltransferase in the isoprene biosynthetic pathway. Mol Cell Biol. 1987;7:3138–3146. doi: 10.1128/mcb.7.9.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkin DJ, Kutsunai SY, Edwards PA. Isolation and sequence of the human farnesyl pyrophosphate synthetase cDNA: coordinate regulation of the mRNAs for farnesyl pyrophosphate synthetase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and 3-hydroxy-3-methylglutaryl coenzyme A synthase by phorbol ester. J Biol Chem. 1990;265:4607–4614. [PubMed] [Google Scholar]
- 24.Koyama T, Matsubara M, Ogura K. Isoprenoid enzyme systems of silkworm. II: formation of the juvenile hormone skeletons by farnesyl pyrophosphate synthase II. J Biochem. 1985;98:457–463. doi: 10.1093/oxfordjournals.jbchem.a135300. [DOI] [PubMed] [Google Scholar]
- 25.Castillo-Gracia M, Couillaud F. Molecular cloning and tissue expression of an insect farnesyl diphosphate synthase. Eur J Biochem. 1999;262:365–370. doi: 10.1046/j.1432-1327.1999.00385.x. [DOI] [PubMed] [Google Scholar]
- 26.Kikuchi K, Hirai M, Shiotsuki T. Molecular cloning and tissue distribution of farnesyl pyrophosphate synthase from the silkworm. J Insect Biotech Sericol. 2001;70:167–172. [Google Scholar]
- 27.Cusson M, Béliveau C, Sen SE, Vandermoten S, Rutledge RG, Stewart D, Francis F, Haubruge E, Rehse P, Huggins DJ, Dowling APG, Grant GH. Characterization and tissue-specific expression of two lepidopteran farnesyl diphosphate synthase homologs: implications for the biosynthesis of ethyl-substituted juvenile hormones. Proteins. 2006;65:742–758. doi: 10.1002/prot.21057. [DOI] [PubMed] [Google Scholar]
- 28.Sen SE, Trobaugh C, Beliveau C, Richard T, Cusson M. Cloning, expression and characterization of a dipteran farnesyl diphosphate synthase. Insect Biochem Mol Biol. 2007;37:1198–1206. doi: 10.1016/j.ibmb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Anderson MS, Yarger JG, Burck CL, Poulter CD. Farnesyl diphosphate synthase: molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae . J Biol Chem. 1989;264:19176–19184. [PubMed] [Google Scholar]
- 30.Cunillera N, Arró M, Delourme D, Karst F, Boronat A, Ferrer A. Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem. 1996;271:7774–7780. doi: 10.1074/jbc.271.13.7774. [DOI] [PubMed] [Google Scholar]
- 31.Montalvetti A, Fernandez A, Sanders JM, Ghosh S, Brussel E, Oldfield E, Docampo R. Farnesyl pyrophosphate synthase is an essential enzyme in Trypanosoma brucei: in vitro RNA interference and in vivo inhibition studies. J Biol Chem. 2003;278:17075–17083. doi: 10.1074/jbc.M210467200. [DOI] [PubMed] [Google Scholar]
- 32.Hemmi H, Noike M, Nakayama T, Nishino T. An alternative mechanism of product chain-length determination in type III geranylgeranyl diphosphate synthase. Eur J Biochem. 2003;270:2186–2194. doi: 10.1046/j.1432-1033.2003.03583.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Ohnuma S. Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem Sci. 1999;24:445–451. doi: 10.1016/S0968-0004(99)01464-4. [DOI] [PubMed] [Google Scholar]
- 34.Kinjoh T, Kaneko Y, Itoyama K, Mita K, Hiruma K, Shinoda T. Control of juvenile hormone biosynthesis in Bombyx mori: cloning of the enzymes in the mevalonate pathway and assessment of their developmental expression in the corpora allata. Insect Biochem Mol Biol. 2007;37:808–818. doi: 10.1016/j.ibmb.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Schulbach MC, Brennan PJ, Crick DC. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis . J Biol Chem. 2000;275:22876–22881. doi: 10.1074/jbc.M003194200. [DOI] [PubMed] [Google Scholar]
- 36.Schulbach MC, Mahapatra S, Macchia M, Barontini S, Papi C, Minutolo F, Bertini S, Brennan PJ, Crick DC. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis . J Biol Chem. 2001;13:11624–11630. doi: 10.1074/jbc.M007168200. [DOI] [PubMed] [Google Scholar]
- 37.Ambo T, Noike M, Kurokawa H, Koyama T. Cloning and functional analysis of novel short-chain cis-prenyltransferases. Biochem Biophys Res Commun. 2008;375:536–540. doi: 10.1016/j.bbrc.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 38.Sallaud C, Rontein D, Onillon S, Jabès F, Duffé P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, Causse M, Tissier A. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites . Plant Cell. 2009;21:301–317. doi: 10.1105/tpc.107.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagami H, Korenaga T, Ogura K. Geranylgeranyl diphosphate synthase catalyzing the single condensation between isopentenyl diphosphate and farnesyl diphosphate. J Biochem. 1993;114:118–121. doi: 10.1093/oxfordjournals.jbchem.a124125. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Proteau P, Poulter D, Ferro-Novick S. BTS1 encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae . J Biol Chem. 1995;270:21793–21799. doi: 10.1074/jbc.270.37.21793. [DOI] [PubMed] [Google Scholar]
- 41.Sandmann G, Misawa N, Wiedemann M, Vittorioso P, Carattoli A, Morelli G, Macino G. Functional identification of al-3 from Neurospora crassa as the gene for geranylgeranyl pyrophosphate synthase by complementation with crt genes, in vitro characterization of the gene product and mutant analysis. J Photochem Photobiol. 1993;18:245–251. doi: 10.1016/1011-1344(93)80071-G. [DOI] [PubMed] [Google Scholar]
- 42.Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis . Plant Physiol. 2000;122:1045–1056. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kainou T, Kawamura K, Tanaka K, Matsuda H, Kawamukai M. Identification of the GGPS1 genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim Biophys Acta. 1999;1437:333–340. doi: 10.1016/s1388-1981(99)00028-1. [DOI] [PubMed] [Google Scholar]
- 44.Hojo M, Matsumoto T, Miura T. Cloning and expression of a geranylgeranyl diphosphate synthase gene: insights into the synthesis of termite defence secretion. Insect Mol Biol. 2007;16:121–131. doi: 10.1111/j.1365-2583.2007.00709.x. [DOI] [PubMed] [Google Scholar]
- 45.Ohnuma S, Suzuki M, Nishino T. Archaebacterial ether-linked lipid biosynthetic gene: expression cloning, sequencing, and characterization of geranylgeranyl-diphosphate synthase. J Biol Chem. 1994;269:14792–14797. [PubMed] [Google Scholar]
- 46.Badillo A, Steppuhn J, Deruere J, Camara B, Kuntz M. Structure of a functional geranylgeranyl pyrophosphate synthase gene from Capsicum annum . Plant Mol Biol. 1995;27:425–428. doi: 10.1007/BF00020196. [DOI] [PubMed] [Google Scholar]
- 47.Tachibana A. A novel prenyltransferase, farnesylgeranyl diphosphate synthase, from the haloalkaliphilic archaeon, Natronobacterium pharaonis . FEBS Lett. 1994;341:291–294. doi: 10.1016/0014-5793(94)80475-3. [DOI] [PubMed] [Google Scholar]
- 48.Tachibana A, Yano Y, Otani S, Nomura N, Sako Y, Taniguchi M. Novel prenyltransferase gene encoding farnesylgeranyl diphosphate synthase from a hyperthermophilic archaeon, Aeropyrum pernix: molecular evolution with alteration in product specificity. FEBS J. 2000;267:321–328. doi: 10.1046/j.1432-1327.2000.00967.x. [DOI] [PubMed] [Google Scholar]
- 49.Ling Y, Li Z, Miranda K, Oldfield E, Moreno SNJ. The farnesyl diphosphate/geranylgeranyl diphosphate synthase of Toxoplasma gondii is a bifunctional enzyme and a molecular target of bisphosphonates. J Biol Chem. 2007;282:30804–30816. doi: 10.1074/jbc.M703178200. [DOI] [PubMed] [Google Scholar]
- 50.Chen A, Poulter CD. Purification and characterization of farnesyl diphosphate/geranylgeranyl diphosphate synthase: a thermostable bifunctional enzyme from Methanobacterium thermoautotrophicum . J Biol Chem. 1993;268:11002–11007. [PubMed] [Google Scholar]
- 51.Cervantes-Cervantes M, Gallagher CE, Zhu C, Wurtzel ET. Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol. 2006;140:220–231. doi: 10.1104/pp.106.077008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copley SD. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol. 2003;7:265–272. doi: 10.1016/S1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 53.Koyama T, Ogura K (1999) Isopentenyl diphosphate isomerase and prenyltransferases. In: Barton D, Nakanishi K (eds) Comprehensive natural products chemistry, vol 2. Elsevier, Oxford, pp 69–96
- 54.Cornforth JW, Cornforth RH, Donninger C, Popják G. Studies on the biosynthesis of cholesterol XIX: steric course of hydrogen eliminations and of C–C bond formations in squalene biosynthesis. Proc R Soc Lond B. 1966;163:492–514. doi: 10.1098/rspb.1966.0004. [DOI] [PubMed] [Google Scholar]
- 55.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6 Å resolution. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 56.Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD. Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA. 1996;93:15018–15023. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J Biol Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 58.Gabelli S.B, McLellan J.S, Montalvetti A, Oldfield E, Docampo R, Amzel LM. Structure and mechanism of the farnesyl diphosphate. Synthase from Trypanosoma cruzi: implications for drug design. Proteins. 2005;62:80–88. doi: 10.1002/prot.20754. [DOI] [PubMed] [Google Scholar]
- 59.Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russe RGG, Oppermann U. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc Natl Acad Sci USA. 2006;103:7829–7834. doi: 10.1073/pnas.0601643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang TH, Guo RT, Ko TP, Wang AH, Liang PH. Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J Biol Chem. 2006;281:14991–15000. doi: 10.1074/jbc.M512886200. [DOI] [PubMed] [Google Scholar]
- 61.Koyama T, Gotoh Y, Nishino T. Intersubunit location of the active site of farnesyl diphosphate synthase: reconstruction of active enzymes by hybrid-type heteromeric dimers of site-directed mutants. Biochemistry. 2000;39:463–469. doi: 10.1021/bi991621b. [DOI] [PubMed] [Google Scholar]
- 62.Kavanagh KL, Dunford JE, Bunkoczi G, Russell RG, Oppermann U. The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding. J Biol Chem. 2006;281:22004–22012. doi: 10.1074/jbc.M602603200. [DOI] [PubMed] [Google Scholar]
- 63.Koyama T, Obata S, Osabe M, Takeshita A, Yokoyama K, Uchida M, Nishino T, Ogura K. Thermostable farnesyl diphosphate synthase of Bacillus stearothermophilus: molecular cloning, sequence determination, overproduction, and purification. J Biochem. 1993;113:355–363. doi: 10.1093/oxfordjournals.jbchem.a124051. [DOI] [PubMed] [Google Scholar]
- 64.Song L, Poulter CD. Yeast farnesyl diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domain I and II. Proc Nat Acad Sci USA. 1994;91:3044–3048. doi: 10.1073/pnas.91.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujii H, Sagami H, Koyama T, Ogura K, Seto S, Baba T, Allen CM. Variable product specificity of solanesyl pyrophosphate synthetase. Biochem Biophys Res Commun. 1980;96:1648–1653. doi: 10.1016/0006-291X(80)91363-7. [DOI] [PubMed] [Google Scholar]
- 66.Ohnuma S, Koyama T, Ogura K. Alteration of the product specificities of prenyltransferases by metal ions. Biochem Biophys Res Commun. 1993;192:407–412. doi: 10.1006/bbrc.1993.1430. [DOI] [PubMed] [Google Scholar]
- 67.Ohnuma S, Hirooka K, Ohto C, Nishino T. Conversion from archaeal geranylgeranyl diphosphate synthase to farnesyl diphosphate synthase: two amino acids before the first aspartate-rich motif solely determine eukaryotic farnesyl diphosphate synthase activity. J Biol Chem. 1997;272:5192–5198. doi: 10.1074/jbc.272.1.580. [DOI] [PubMed] [Google Scholar]
- 68.Matsuoka S, Sagami H, Kurisaki A, Ogura K. Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J Biol Chem. 1991;266:3464–3468. [PubMed] [Google Scholar]
- 69.Fujiwara S, Yamanaka A, Hirooka K, Kobayashi A, Imanaka T, Fukusaki E. Temperature-dependent modulation of farnesyl diphosphate/geranylgeranyl diphosphate synthase from hyperthermophilic archaea. Biochem Biophys Res Commun. 2004;325:1066–1074. doi: 10.1016/j.bbrc.2004.10.129. [DOI] [PubMed] [Google Scholar]
- 70.Ohnuma S, Nakazawa T, Hemmi H, Hallberg AM, Koyama T, Ogura K, Nishino T. Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical mutagenesis. J Biol Chem. 1996;271:10087–10095. doi: 10.1074/jbc.271.17.10087. [DOI] [PubMed] [Google Scholar]
- 71.Ohnuma S, Narita K, Nakazawa T, Ishida C, Takeuchi Y, Ohto C, Nishino T. A role of the amino acid residue located on the fifth position before the first aspartate-rich motif of farnesyl diphosphate synthase on determination of the final product. J Biol Chem. 1996;271:30748–30754. doi: 10.1074/jbc.271.48.30748. [DOI] [PubMed] [Google Scholar]
- 72.Fernandez SMS, Kellogg BA, Poulter CD. Farnesyl diphosphate synthase: altering the catalytic site to select for geranyl diphosphate activity. Biochemistry. 2000;39:15316–15321. doi: 10.1021/bi0014305. [DOI] [PubMed] [Google Scholar]
- 73.Sen SE, Cusson M, Trobaugh C, Béliveau C, Richard T, Graham W, Mimms A, Roberts G. Purification, properties and heteromeric association of type-1 and type-2 lepidopteran farnesyl diphosphate synthases. Insect Biochem Mol Biol. 2007;37:819–828. doi: 10.1016/j.ibmb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 74.Burke C, Croteau R. Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem. 2002;277:3141–3149. doi: 10.1074/jbc.M105900200. [DOI] [PubMed] [Google Scholar]
- 75.Chen A, Kroon PA, Poulter CD. Isoprenyl diphosphate synthases: protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure. Protein Sci. 1994;3:600–607. doi: 10.1002/pro.5560030816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng F, Oldfield E. Inhibition of isoprene biosynthesis pathway enzymes by phosphonates, bisphosphonates, and diphosphates. J Med Chem. 2004;47:5149–5158. doi: 10.1021/jm040036s. [DOI] [PubMed] [Google Scholar]
- 78.Dunford JE, Thompson K, Coxon FP, Luckman SP, Hahn FM, Poulter CD, Ebetino FH, Rogers MJ. Structure-activity relationships for inhibition of farnesyl diphosphate synthase in vitro and inhibition of bone resorption in vivo by nitrogen-containing bisphosphonates. J Pharmacol Exp Ther. 2001;296:235–242. [PubMed] [Google Scholar]
- 79.Thompson K, Dunford JE, Ebetino FH, Rogers MJ. Identification of a bisphosphonate that inhibits isopentenyl diphosphate isomerase and farnesyl diphosphate synthase. Biochem Biophys Res Commun. 2002;290:869–873. doi: 10.1006/bbrc.2001.6289. [DOI] [PubMed] [Google Scholar]
- 80.Bergstrom JD, Bostedor RG, Masarachia PJ, Reszka AA, Rodan G. Alendronate is a specific, nanomolar inhibitor of farnesyl diphosphate synthase. Arch Biochem Biophys. 2000;373:231–241. doi: 10.1006/abbi.1999.1502. [DOI] [PubMed] [Google Scholar]
- 81.Hughes DE, Wright KR, Uy HL, Sasaki A, Yoneda T, Roodman GD, Mundy GR, Boyce BF. Bisphosphonates promote apoptosis in murine osteoclasts in vitro and in vivo. J Bone Miner Res. 1995;10:1478–1487. doi: 10.1002/jbmr.5650101008. [DOI] [PubMed] [Google Scholar]
- 82.Rogers MJ, Watts DJ, Russell RG. Overview of bisphosphonates. Cancer. 1997;80:1652–1660. doi: 10.1002/(SICI)1097-0142(19971015)80:8+<1652::AID-CNCR15>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 83.Rodan GA. Mechanisms of action of bisphosphonates. Annu Rev Pharmacol Toxicol. 1998;38:375–388. doi: 10.1146/annurev.pharmtox.38.1.375. [DOI] [PubMed] [Google Scholar]
- 84.Brown DL, Robbins R. Developments in the therapeutic applications of bisphosphonates. J Clin Pharmacol. 1999;39:1–10. doi: 10.1177/00912709922008272. [DOI] [PubMed] [Google Scholar]
- 85.Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- 86.Yardley V, Khan AA, Martin MB, Slifer TR, Araujo FG, Moreno SNJ, Docampo R, Croft SL, Oldfield E. In vivo activities of farnesyl pyrophosphate synthase inhibitors against Leishmania donovani and Toxoplasma gondii . Antimicrob Agents Chemother. 2002;46:929–931. doi: 10.1128/AAC.46.3.929-931.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez N, Bailey BN, Martin MB, Oldfield E, Urbina JA, Docampo R. Radical cure of experimental cutaneous leishmaniasis by the bisphosphonate pamidronate. J Infect Dis. 2002;186:138–140. doi: 10.1086/341074. [DOI] [PubMed] [Google Scholar]
- 88.Chen C, Hudock MP, Zhang Y, Guo RT, Cao R, No JH, Liang PH, Ko TP, Chang TH, Chang SC, Song Y, Axelson J, Kumar A, Wang AH, Oldfield E. Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation. J Med Chem. 2008;51:5594–5607. doi: 10.1021/jm800325y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chuiko AL, Lozinsky MO, Jasicka-Misiak I, Kafarski P. Herbicidal derivatives of aminomethylenebisphosphonic acid. Part IV: hydroxyalkylidenebisphosphonates, iminomethylenebisphosphonates and ureidomethylenebisphosphonates. J Plant Growth Regul. 1999;18:171–174. doi: 10.1007/PL00007066. [DOI] [PubMed] [Google Scholar]
- 90.Palli SR, Cusson M. Future insecticides targeting genes involved in the regulation of molting and metamorphosis. In: Ishaaya I, Nauen R, Horowitz AR, editors. Insecticide design using advanced technologies. Berlin: Springer; 2007. pp. 105–134. [Google Scholar]
- 91.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 92.Hall BG (2008) Phylogenetic trees made easy: a how-to manual, 3rd edn, Sinauer Associates, Sunderland