Abstract
The inhibitor of growth (ING) family of tumor suppressors has five members and is implicated in the control of apoptosis, senescence, DNA repair, and cancer progression. However, little is known about ING activity in the regulation of cancer progression. ING members and splice variants seem to behave differently with respect to cancer invasion and metastasis. Interaction with histone trimethylated at lysine 4 (H3K4me3), hypoxia inducible factor-1 (HIF-1), p53, and nuclear factor kappa-B (NF-κB) are potential mechanisms by which ING members exert effects on invasion and metastasis. Subcellular mislocalization, rapid protein degradation, and to a lesser extent ING gene mutation are among the mechanisms responsible for inappropriate ING levels in cancer cells. The aim of this review is to summarize the different roles of ING family tumor suppressors in cancer progression and the molecular mechanisms involved.
Keywords: ING, Tumor suppressor, Cancer progression, Invasion, Metastasis
Introduction
Members of the inhibitor of growth (ING) family of proteins are thought to be type II tumor suppressors. There are five members, including ING1–5, as well as a number of splice variants within the member groups, and differences in their biological functions with respect to control of cancer initiation and progression are apparent among groups and splice variants [1]. All ING members have a highly conserved C-terminal PHD finger that interacts with methylated histones [2, 3]. The PHD finger is composed of approximately 60 amino acids that are zinc-binding, and this domain is often implicated in chromatin remodeling [4]. On the other hand, each member has a unique N-terminal domain. For example, the ING1 N-terminus binds to part of the Sin3-HDAC complex [5]. Also, ING members have a domain with an unknown function called the novel conserved region (NCR) [4]. Members also possess a nuclear localization signal (NLS) or multiple nuclear localization signals, which are important with respect to the subcellular localization and subsequent potential for loss of function of ING proteins [2, 4]. The leucine zipper-like (LZL) region is also present in a number of the ING members forming a hydrophobic N-terminal structure [4].
ING proteins have a number of biological functions related to apoptosis, senescence, cell growth, DNA repair and replication, and cancer progression including the regulation of invasion and metastasis [2, 6, 7]. For example, ING1 increases the transcription of p21Waf1 via p53, and p21Waf1 is known to inhibit cyclin-dependent kinase resulting in cell cycle arrest at the G1/S transition [6, 8]. ING1 is also involved in the regulation of the G2/M checkpoint, and other ING members may have similar activity [6, 9]. A number of ING members have also been found to mediate cellular apoptotic responses [10–13]. They have also been linked to DNA repair in response to UV-induced DNA damage. p33ING1b enhances the repair of UV-damaged DNA in a p53-dependent manner by inducing histone H4 acetylation, chromatin relaxation, and by recruiting xeroderma pigmentosum A (XPA) to photolesion sites [14, 15]. ING2 is involved in DNA repair as well and mediates p53–p300 interaction [16, 17], and also regulates DNA replication in the absence of UV-induced DNA damage [7].
ING family members either increase or decrease histone acetylation levels, which regulate chromatin structure. They modify the activity of both histone acetyltransferase (HAT) and histone deacetylase (HDAC) complexes, and are components of these complexes [2, 5, 6]. ING proteins bind to methylated histones, recruiting HAT or HDAC to specific histone regions, modifying chromatin structure and subsequent gene expression. ING1 and ING2 present the mSin3a-HDAC1/2 complex; ING3 is part of the Tip60/NuA4 complex; ING4 is a subunit of the HBO1 complex; and ING5 is found as a component of both HBO1 and MOZ/MORF [18, 19].
The detailed roles of ING proteins in cell senescence and apoptosis have been summarized in recent reviews [20, 21]. Although many studies have investigated the role of ING proteins in cancer cell migration, invasion and angiogenesis in vitro, and cancer progression and metastasis in vivo, there is no review article available on this topic. The purpose of this review is to gain a better understanding of the molecular mechanisms behind the action of ING proteins in cancer progression.
ING1 in cancer progression
ING1, the founding member of the ING family, is mapped to chromosome 13q33–34 [2]. The ING1 gene contains three exons and two introns, and encodes three isoforms based on alternative splicing, p47ING1a, p33ING1b, and p24ING1c [3]. ING1b is the most abundant isoform [3, 22], and most of the tumor suppressive functions are attributed to this isoform. ING1 expression is reduced in a number of cancers, including colorectal, astrocytoma, breast, stomach, hepatocellular carcinoma (HCC), acute lymphoblastic leukaemia (ALL), and gastric cancer [23–30]. Also, ING1 variants have been shown to have unique and sometimes opposing functions. p33ING1b and p24ING1c but not p47ING1a were reduced in HCC [31]. p33ING1b and p24ING1c activated p21Waf1 and Bax, while p47ING1a inactivated the p21Waf promoter [31]. Overexpression of p33ING1b but not p47ING1a results in increased acetylation of H3 and H4, and p47ING1a inhibits this acetylation [32]. p33ING1b has been shown to affect proliferating cell nuclear antigen (PCNA)-p300 interaction by binding to PCNA via its PCNA-interacting protein (PIP) domain [33]. Furthermore, p33ING1b but not p24ING1c associates with Sin3, HDAC1, SAP30, and RbAp48, and p33ING1b expression is associated with transcriptional repression via HDAC1 [34]. Moreover, the mouse ing1 gene codes for two variants of the protein, and these variants have opposite functions with respect to p53 suppressing/activating activity [35]. With respect to the mutational status of the ING1 gene, a number of studies have revealed that ING1 mutation is relatively infrequent [2]. Additionally, mutation of the ING1 gene is not the main cause of ING1 inactivation, and other mechanisms of ING1 inactivation including mislocalization and reduced mRNA stability are probably more important [2].
A number of in vivo investigations have shown that ING1 expression is related to both lymph node and distant metastasis [23, 36], whereas another study provided evidence against the involvement of ING1 in cancer progression [37]. Toyama et al. [36] found that 58% of breast cancers with low ING1 mRNA expression had metastasized to local lymph nodes, whereas only 9% of the high ING1 expressing cancers did so. Ding et al. [23] found that p33ING1 expression was lower in stomach cancer biopsies compared to precarcinomatous tissues, and its expression was related to tumor growth, distant metastasis, and tumor differentiation in 103 stomach cancers.
In contrast, no correlation between p33ING1b nuclear protein expression and metastasis-free survival in colorectal cancer patients was found [37]. Samples from 41 patients with Dukes’ C colorectal cancer were examined to determine whether p53 mutation status and p33INGb expression could be used to predict which colorectal cancer patients might respond to chemotherapy following curative surgical resection of the primary tumor. Although out of the 28 patients with positive nuclear p33ING1b expression, 39% developed metastases, whereas 46.1% of the patients with negative nuclear p33ING1b expression developed metastases; no significant correlation was found between p33ING1b nuclear expression and metastasis-free survival following surgical resection and chemotherapy. A trend was observed, however, as patients with low p33ING1b expression had shorter overall survival times compared to patients with higher p33ING1b expression (P = 0.27). In addition, the time to develop metastasis did not differ between patients with positive or negative p33ING1b nuclear expression. Despite these results, it is possible that ING1 nuclear expression is related to metastasis since the small sample size (n = 41) may have contributed to the insignificant findings [37]. However, nuclear ING1 expression might not be the optimal prognostic marker following surgery and chemotherapy in colon cancer patients.
Studies by Nouman et al. [38] and Zhang et al. [39] examined the relationships between subcellular localization of ING1 protein and cancer progression. In 67 melanocytic lesions ranging from benign to invasive, the expression of nuclear ING1 was determined using immunohistochemistry [38]. Malignant melanoma samples had a loss of nuclear p33ING1b protein compared to normal melanocytes or melanocytes from benign nevi. Among benign nevi, no case had complete loss of nuclear ING1, whereas 47.06% of cases with invasive melanoma showed complete nuclear loss of ING1. Additionally, strong or intermediate ING1 cytoplasmic staining was observed in 35.29% of invasive malignant melanomas, while none of the benign nevi had this much cytoplasmic expression. Since ING1 has a nuclear localization signal, it most likely exerts its tumor-suppressive effects within the nucleus, and a shift into the cytoplasm might reduce its activity [38].
In the other study examining the role of ING1 subcellular localization in cancer invasion and metastasis, immunohistochemistry was used to determine p33ING1b nuclear and cytoplasmic expression in 49 oral squamous cell carcinomas and 20 normal mucosa [39]. Ninety percent of the normal specimens showed nuclear expression, and none of the normal specimens showed cytoplasmic expression. On the other hand, 24% of the carcinoma specimens showed cytoplasmic expression of p33ING1b, and 76% of the carcinoma specimens showed no p33ING1b expression at all. With respect to metastasis, positive cytoplasmic expression was related to increased lymph node metastasis. Interestingly, cancer cases positive for both p33ING1b and particularly interesting new cysteine-histidine rich protein (PINCH) in the cytoplasm had lymph node metastasis the most often, cases that were negative for both PINCH and p33ING1b in the cytoplasm had lymph node metastasis the least often, and cancer cases positive for only one protein or the other in the cytoplasm had an intermediate level of lymph node metastasis. This suggests that p33ING1b may cooperate with PINCH to exert effects on cancer invasion and metastasis. This interaction may occur at the site of cellular adhesion during cancer progression [39]. In general, the literature suggests that transportation of ING1 from the nucleus into the cytoplasm enhances metastasis, at least in melanoma and oral squamous cell carcinoma, although the mechanism behind the shift is currently unknown. However, Gong et al. suggested that 14-3-3, a family of proteins implicated in cell cycle regulation, may mediate the subcellular mislocalization of ING1 [40]. 14-3-3 binds to p33ING1b, and there is a potential 14-3-3 binding site on ING1. Also, p33ING1b phosphorylation is required for the binding of 14-3-3 to p33ING1b and 14-3-3 directs ING1b to the cytoplasm. Finally, 14-3-3η prevents the upregulation of p21Waf1 by p33ING1b, suggesting that the nuclear to cytoplasmic ING1 shift mediated by 14-3-3 affects ING1 transcriptional regulation activity related to cancer development.
Despite the mounting evidence that the ING1 tumor suppressor reduces cancer progression, ING1 does not seem to inhibit MMP expression. We examined matrix metalloproteinase (MMP) levels in melanoma cells transfected with vector, p33ING1b or antisense p33ING1b, and no difference in the protein levels of MMP1, MMP2, and MMP9 was found among the groups [41]. However, it is not known if p33ING1b affects the activity of MMPs.
ING2 in cancer progression
The second member of the ING family of tumor suppressors, ING2, is mapped to chromosome 4q35.1 [2] and also interacts with p53. Overexpression of ING2 has been linked to apoptosis induction and cell cycle arrest in melanoma and colon carcinoma cell lines [42, 43]. On the other hand, simultaneous knockdown of ING2a and b isoforms induces cell cycle arrest and apoptosis in lung adenocarcinoma, osteosarcoma, and glioma cell lines [44]. It seems that ING2 activity is dependent on the specific cellular environment as it has opposite functions in various cancers.
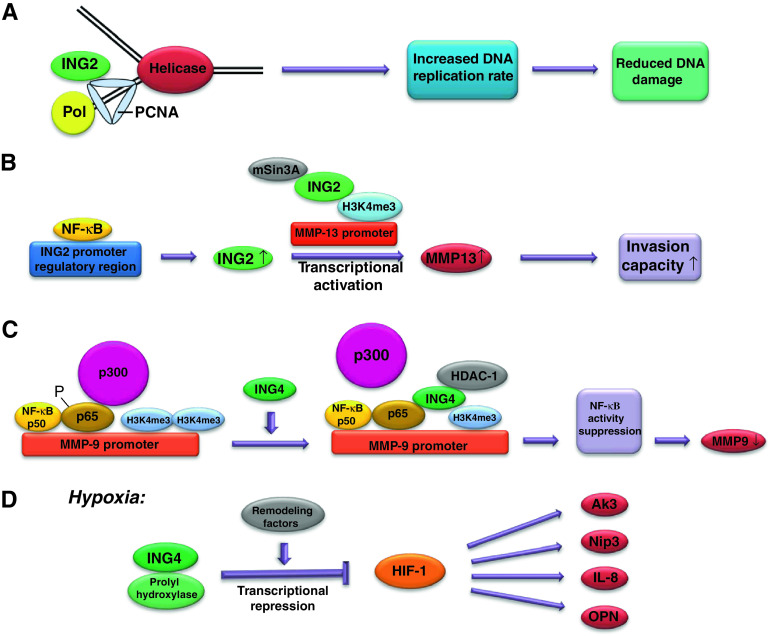
We have shown that p33ING2 enhances nucleotide excision repair of UV-induced DNA damage in melanoma cells and that ING2 is necessary for the recruitment of XPA to photolesions, for chromatin relaxation, and for histone H4 acetylation induced by UV [17]. The leucine zipper-like (LZL) motif is required for ING2 DNA repair activity [16]. ING2 also promotes genomic stability and DNA replication in the absence of UV since bromodeoxyuridine incorporation was reduced in ING2 knockdown U2OS osteosarcoma cells [7]. The decreased DNA replication rate observed in ING2 knockdown cells was likely due to slowed replication fork progression (Fig. 1a). Additionally, there was less chromatin-bound PCNA in ING2 knockdown cells, and ING2 interacted with PCNA, allowing for continued DNA replication. ING knockdown cells also had greater sister chromatid exchange [7]. This promotion of genomic instability and the possible effects on progression-related genes brought about by loss of ING2 may be one explanation for how reduced ING2 levels seem to promote early cancer progression in some cases. Ythier et al. also found that a large proportion of non-small cell carcinoma samples lost ING2 expression at stage I. This suggests that ING2 expression may play an early role in cancer development [45].
Fig. 1.
Potential mechanisms for ING transcriptional regulation of genes involved in cancer progression. a ING2 interacts with PCNA during DNA replication, allowing normal cell proliferation to maintain genomic stability. b In colon cancer, NF-κB binds to the ING2 promoter regulatory region, increasing the expression of ING2. ING2 recruits mSin3a to the MMP-13 promoter and binds to H3K4me3 resulting in MMP-13 transcriptional activation. c ING4 binds to NF-κB at the promoter of NF-κB-regulated genes and suppresses NF-κB activity via reducing H3K4me3 and inhibiting NF-κB p65 phosphorylation. ING4 decreases p65–p300 interaction in favor of HDAC-1 recruitment. d During hypoxia, ING4 interacts with the prolyl hydroxylase and recruits chromatin-remodeling factors to HIF target gene promoters, suppressing HIF-1-regulated gene expression
ING2 expression is upregulated in colorectal cancer [46], but is decreased in melanoma, hepatocellular carcinoma, and non-small cell lung carcinoma [45, 47, 48], and in the majority of lung cancer cell lines with a p53 mutation [49]. Frequent deletion of ING2 locus was found in head and neck squamous cell carcinoma (HNSCC) and basal cell carcinoma (BCC) [47, 50, 51]. In addition, allelic loss of ING2 in HNSCC was associated with advanced tumor stage [47]. Other studies have provided evidence for ING2 as a tumor suppressor, although its expression may be more related to cancer initiation rather than invasion and metastasis [48]. Both primary and metastatic melanomas have reduced nuclear ING2 expression compared to dysplastic nevi. However, no relationships have been observed between ING2 nuclear expression and AJCC stage in melanomas as well as between ING2 nuclear expression and survival in metastatic melanomas [48]. This suggests that ING2 may play a greater role in cell cycle regulation and DNA repair since these events are central to the initiation of cancer rather than to cancer progression.
Although ING2 functions as a tumor suppressor in melanoma, HNSCC and BCC, it seems to act as an oncogene in colon cancer [46]. ING2 has a greater ability (than other ING family members) to bind to trimethylated histone H3 at lysine 4 (H3K4me3), and methylation at lysine 4 is associated with transcriptional activation, whereas methylation at other histone residues is associated with transcriptional suppression. In colon cancer, ING2 mRNA was higher in cancerous tissue compared to nonmalignant mucosa, and ING2 protein was found predominantly in the nucleus of colon cancer cells [46]. An NF-κB binding sequence was also found in the ING2 promoter regulatory region and NF-κB binds to ING2. After treatment with NF-κB inhibitors, RKO and SW837 cells had decreased ING2 expression, whereas when HCT116 and WiDr cells were treated with NF-κB activators, ING2 levels were upregulated. ING2 was also found to upregulate MMP-13 as determined by a microarray analysis and confirmed by RT–PCR, which would enhance cancer invasion and metastasis. To further elucidate the mechanism by which ING2 interacts with MMP-13, relationships between ING2, HDAC1, MMP-13, and mSin3A were considered since it has been shown that ING2 interacts with HDAC1 and mSin3A during chromatin remodeling [52]. HDAC1 and ING2 overexpression increased MMP-13 expression, whereas mSin3A overexpression had no effect. However, cells with both ING2 and mSin3A overexpression had increased levels of MMP-13, greater than the increase observed with ING2 overexpression alone. Furthermore, invasion ability was greater for ING2 overexpressing cells compared to controls and this increased ability was lost with the addition of MMP-13 inhibitor. Overall, these results do not support the role of ING2 as a tumor suppressor in colorectal cancer, and instead suggest that ING2 increases MMP expression via chromatin modification allowing for increased invasion capability.
ING3 in cancer progression
Mapped to chromosome 7q31.3 [2, 39], family member ING3 has also been found to regulate the cell cycle and programmed cell death [1]. Overexpression of p47ING3 reduced the number of RKO cells in S phase and induced apoptosis, and these effects were dependent on p53 [12]. Decreased expression of the Caenorhabditis elegans homolog of ING3 is also linked to inhibition of p53-dependent, radiation-induced germ cell apoptosis in C. elegans [53]. In melanoma cells, ING3 promotes UV-induced apoptosis through activation of the Fas/caspase-8 pathway [54].
Using clinical samples, we and others have found that ING3 is a tumor suppressor in melanoma and HNSCC. We found that nuclear ING3 expression is significantly reduced in primary melanoma compared to dysplastic nevi (P < 0.001) and further reduced in metastatic melanoma (P < 0.001) [55]. This confirms that decreased expression of ING3 in the nucleus is not only implicated in cancer initiation, but also in tumor progression. Furthermore, reduced nuclear ING3 expression is significantly correlated with poorer 5-year disease-specific survival of melanoma patients. Additionally, reduced nuclear ING3 expression was related to increased cytoplasmic expression in many of the primary and metastatic melanomas, implying that nuclear-to-cytoplasmic translocation may be partially responsible for the decreased nuclear expression observed.
However, nuclear-to-cytoplasmic shift of ING3 may not be the only mechanism responsible for its decreased expression in cancer and subsequent increases in invasion and metastasis potential. The Skp-2-dependent rapid degradation of ING3 by the ubiquitin-proteasome pathway may also be responsible for its decreased levels in melanoma [56], and this pathway has been implicated in the control of cancer progression [57–71]. In the degradation pathway, polyubiquitination of the substrate protein results in protein degradation by the 26S proteasome. The E3 ligase provides protein specificity, and the SCF E3 ligase complex (Skp1-Cullin-F-box protein complex) is responsible for ING3 degradation in melanoma [56]. Skp2 knockdown in various melanoma cell lines reduced ING3 ubiquitination, and we found an inverse relationship between Skp2 and ING3 expression in melanoma cell lines [56].
ING3 mRNA expression is decreased in HNSCCs [72]. Although the exact mechanism is unknown, deletion of the ING3 locus may be responsible for reduced ING3 expression in HNSCC. In one study, 49 specimens of HNSCC were screened for loss of heterozygosity (LOH) at the chromosome 7q31 region and two regions, D7S643 and D7S486 had high levels of allelic deletion [73]. Since ING3 is found within 100 kb of D7S643, the group further investigated the role of ING3 in HNSCC. Fifty percent of tumors showed decreased or absent ING3 mRNA expression compared to controls. However, no relationships were observed between ING3 expression level and metastasis-related clinicopathological parameters including distant and lymph node metastasis. Several years later, ING3 was examined as a potential marker of tumor aggressiveness in HNSCC [72]; 52.1% of primary tumors had either decreased or no expression of ING3 compared to controls (n = 71), and reduced ING3 expression was an independent prognostic factor for poor overall survival. There was a trend (although not statistically significant) between ING3 mRNA expression and tumor-node-metastasis (TNM) stage, with lower ING3 expression in more advanced tumors. Thirty-nine percent of early TNM stage tumors had low ING3 expression, whereas 60% of late TNM stage tumors had low ING3 expression (P = 0.19). However, no relationship was observed between lymph node metastasis and ING3 expression. Curiously, ING3 expression was related to overall survival, but not to disease-free survival.
Since investigations into ING3 and cancer progression have been largely in vivo, little is known about the mechanistic detail surrounding the regulation of invasion and metastasis by ING3. For example, even though cytoplasmic and nuclear ING3 expression were inversely related [55], suggesting that ING3 is translocated from the nucleus to the cytoplasm in cancer cells, the in vivo nature of the study did not allow for the determination of the route by which ING3 is lost from the nucleus. Also, the Skp2-dependent degradation pathway may explain improper ING3 expression in cancers, but no in vitro study has been conducted to investigate how this degradation pathway relates to cancer progression specifically. Another problem with the in vivo analyses conducted is small samples sizes resulting in insignificant findings as seen in HNSCC [73]. Further investigations should focus on ING3 overexpression and knockdown in cancer cell lines and subsequent invasion and migration assays.
ING4 and cancer progression
Besides ING1, ING4 is the family member of the ING proteins that is most well-studied [2]. It is mapped to chromosome 12q13.3 [2] and has many variants due to alternative splicing. ING4 expression is reduced in stomach adenocarcinoma [74], glioma [75], melanoma [76, 77], and HNSCC [78]. ING4 expression mediated the growth suppression of U87MG glioma cells [75], M14 melanoma cells [77], and A549 lung carcinoma cells [79, 80], and decreased HepG2 cell growth via induction of G2/M arrest [81]. Additionally, decreased ING4 expression was related to poor prognosis in hepatocellular carcinoma [82]. Allelic loss in the 12p12–13 region containing the ING4 locus was found in 66% of informative HNSCC cases [78], and ING4 was deleted in 10–20% of breast cancer cell lines and primary breast cancers [83].
By far, ING4 is the member of the ING family of tumor suppressors most implicated in the regulation of cancer invasion and metastasis. ING4 was identified in a screen for potential genes that suppress the loss of contact inhibition due to the overexpression of MYCN [83]. Ectopic expression of ING4 also inhibited the growth of T47D breast cancer cells in soft agar, but did not inhibit their growth in standard culture conditions. The mechanism of action is unknown, but it is possible that ING4 directly inhibits the function of the MYC protein, or that ING4 exerts effects on downstream targets of MYC, altering MYC’s transcriptional control. In addition, ING4 suppressed spontaneous loss of contact inhibition in NIH3T3 cells [84].
ING4 has also been shown to directly inhibit anchorage-independent T47D cell growth which is related to tumor invasiveness [85]. This is dependent on the interaction between the ING4 PHD finger and H3K4me3 [85]. The ING4 PHD finger preferentially binds with H3K4me3. Hung et al. [85] conducted experiments to determine if the interaction of ING4 with H3K4me3 is required for the observed effects of ING4 on cell growth in soft agar. As expected, HBO1 is required for acetylation on histone H4, and ING4 is required for HBO1 to acetylate on H4 as well as on H2A, so the complex of HBO1 and ING4 may be responsible for histone acetylation and modification of chromatin.
ING4 and HIF-1
Relationships between ING4 and other factors have elucidated some of the mechanistic detail behind ING4 and cancer progression. HIF-1 is responsible for sustaining cellular proliferation under hypoxic conditions, including in the hypoxic interior of tumors. Inappropriate expression and activity have been linked to the induction of the expression of genes promoting cancer invasion and metastasis [86]. ING4 associates with the HIF prolyl hydroxylase and has been found to affect the activity of HIF-1 [87]. Under normoxic conditions, the von Hippel-Lindau tumor suppressor gene (pVHL) contributes to the degradation of the α-subunit of HIF-1, whereas during hypoxia, this does not occur and HIF-1 levels build up [87]. pVHL causes degradation of HIF-1α during normoxia through the hydroxylation of proline residues, which are modified by HIF prolyl hydroxylases. Ozer et al. [87] found that Nip3 and adenylate kinase 3 (AK3) expression, two genes regulated by HIF-1, increased under hypoxic conditions, but increased to a greater extent when ING4 was knocked down. ING4 affected HIF-1 activity since nuclear HIF-1 levels did not differ between knockdowns and controls. When ING4 and the HIF prolyl hydroxylase were expressed in bacteria and incubated together, they migrated through a size exclusion column in a complex. Also, they compared the effect of ING4 siRNA knockdown in cells either stably or transiently transfected with an HIF-responsive luciferase reporter driven by HIF-responsive elements. ING4 knockdown cells stably transfected with the HIF-responsive luciferase reporter had increased HIF-responsive luciferase reporter activity under hypoxic conditions compared to controls, while no differences were observed between ING4 knockdown cells and control cells transiently transfected with the HIF-responsive luciferase reporter. This suggests that ING4 may suppress HIF-1 activity by recruiting chromatin-remodeling factors since chromatin-remodeling factors would not have any effect on the transcription of transiently transfected DNA, but would influence the transcription of stably transfected (chromosomal) DNA [87]. This was further confirmed by chromatin immunoprecipitation (CHIP) assay and ING4, the prolyl hydroxylase, and a HIF subunit associated with the HIF-responsive luciferase reporter during hypoxia. Additionally, ING family members are known to recruit chromatin-remodeling factors for their transcriptional regulatory activity [5, 6]. Overall, during hypoxia, HIF prolyl hydroxylases may recruit chromatin remodeling factors to HIF target gene promoters through ING4 [87].
Similar results were found in a study examining the effect of ING4 on proteins implicated in angiogenesis [88]. ING4 knockdown increased HIF-1α nuclear activity during hypoxia and resulted in increased Nip3 expression, but did not alter HIF-1α mRNA expression or nuclear protein levels. In HIF-1α/ING4 double knockdowns, the previously observed effects of increases in the expression of HIF-1α target genes interleukin 8 (IL-8) and osteopontin (OPN) upon ING4 knockdown during hypoxia were not observed. This suggests that ING4 acts through HIF-1 to regulate the expression of HIF-1 target genes. Similar to the findings by Ozer and colleagues [86], Colla et al. [88] demonstrated through co-immunoprecipitation that ING4 directly interacts with the HIF prolyl hydroxylase in the nucleus. Thus, ING4 has a suppressive effect on HIF-1 activity during hypoxia by inducing chromatin modification via binding to HIF prolyl hydroxylase. This is thought to be the potential second function of the prolyl hydroxylase, which is also involved in regulating HIF-1 degradation depending on oxygen status. The HIF prolyl hydroxylase recruits ING4 to HIF-1. ING4 then recruits transcriptional repressors to HIF-1, and these repressors exert effects during HIF-1 regulation of HIF-1 target gene expression. Alternatively, ING4-prolyl hydroxylase interaction might stabalize HIF-1-prolyl hydroxylase interaction, physically blocking recruitment of HIF-1 coactivators [86].
ING4, NF-κB, and MMPs
ING4 has been found to interact with proteins other than HIF-1 that are involved in cancer invasion and metastasis and the mechanism by which ING4 influences cancer progression is likely dependent on the specific cellular environment. ING4 directly interacts with the p65 subunit of NF-κB in glioma cells [89]. Like HIF-1, NF-κB regulates a number of the key factors implicated in tumor progression. Garkavtsev et al. found that reducing the expression of ING4 in U87 glioma cells increased the expression of IL-6, IL-8, Cox-2, and colony-stimulating factor 3, which are responsive to NF-κB [89]. Furthermore, NF-κB-responsive genes were overexpressed in ING4 knockdowns in vivo. Increased IL-8 protein was found in tumor xenografts from mice injected with ING4 knockdown U87 cells. ING4 was also found to physically interact with NF-κB by co-immunoprecipitation [89], suggesting that ING4 may act as a repressor for the transcriptional factor NF-κB.
ING4-H3K4me3 binding may be responsible for this transcriptional repression of NF-kB-responsive genes by ING4 [90]. ING4 and NF-κB bind at the promoter sites of NF-κB-regulated genes [90]. Moreover, this binding is associated with decreased acetylated histones and H3K4me3 in glioma cells. These results suggest that ING4 binds to, but does not affect the activation or DNA-binding ability of NF-κB. By binding to NF-κB, ING4 decreases the activity of NF-κB, reducing the expression of NF-κB inducible genes responsible for cancer progression. RT-PCR confirmed that ING4 inhibits Cox-2 and MMP-9 mRNA expression [90]. ING4 expression did not affect the ability of NF-κB p65 to bind to Cox-2 and MMP-9 promoters, but increased ING4 expression reduced the amount of phosphorylated p65 at the Cox-2 promoter and inhibited p65–p300 interaction while promoting HDAC-1 recruitment. ING4 is also present at the Cox-2 and MMP-9 promoters and reduces histone acetylation at both promoters. Interestingly, this effect is opposite to what was observed by Hung et al. [85] who found that ING4 stimulated histone acetylation relating to the fact that ING4 seems to act as a transcriptional activator to inhibit anchorage-independent growth in breast cancer cells while it acts as a transcriptional repressor during NF-κB interaction in glioma cells [90]. In addition, ING4 inhibits RNA Pol II phosphorylation most likely through a reduction of pTef-b since expression of the two subunits of pTef-b, cyclin T1 and CDK9, was inhibited by ING4 expression at NF-κB-regulated promoters. CHIP assay confirmed that ING4 was present at promoters containing H3K4me3 and also, at the Cox-2 promoter, increased ING4 binding was associated with decreased H3K4me3, and the extent of histone methylation can regulate gene transcription. Taken together, these results suggest that ING4 disrupts NF-κB activity by modifying local chromatin through the reduction of histone acetylation via HDAC-1 recruitment and also through the reduction of histone methylation via decreased H3K4me3.
Two other groups provided evidence for the effect of ING4 expression on MMP levels [76, 79]. Xie et al. [79] examined the effect of adenovirus-mediated delivery of ING4 in A549 human lung carcinoma cells as a potential lung cancer therapy. Apart from ING4s effect of downregulating CD34 expression and the expression of proangiogenic cytokines IL-6 and IL-8, ING4 overexpression resulted in a reduction in the number of invasive A549 cells in a Transwell assay compared to control vector expressing A549 cells. Furthermore, ING4-overexpressing A549 cells had decreased MMP-2 and MMP-9 expression compared to controls as determined by RT–PCR, implying that cells with lower ING4 expression have a greater capacity for degradation of the basement membrane and subsequent metastasis. We also found that increased ING4 expression decreased the activity of MMP-2 and MMP-9 in melanoma cells [76]. Zymography revealed that MMP-2 activity was reduced by 25% in ING4-overexpressing cells and MMP-9 activity was reduced by 61%. Using the Boyden chamber assay, we indeed found that cells with high ING4 had a 43% reduction in invasion compared to vector transfected controls.
Cell migration is a key step for cancer invasion and metastasis. We found that ING4 inhibits melanoma cell migration [76]. Using the wound healing assay, we found that control MMRU melanoma cells healed the wound after 24 h, while ING4-overexpressing cells could not migrate to heal the wound. ING4 overexpression decreased MMRU cell migration by 63%. We also performed experiments to elucidate mechanistic detail of the processes involved through the examination of the RhoA-ROCK pathway, implicated in stress fiber reorganization. Rho GTPases are involved in controlling cellular protrusions during cell migration [91]. As determined by the RhoA pull-down assay, ING4 overexpressing cells had decreased RhoA expression, and this was related to decreased stress fiber formation. Also, treatment with a ROCK (Rho-associated coil-containing protein kinase) inhibitor eliminated this effect. Since we previously demonstrated that NF-κB p50 enhances melanoma cell migration by increasing RhoA activity [92], it is reasonable to assume that ING4 may inhibit RhoA/ROCK-dependent cell migration by suppressing NF-κB activity.
ING4 and p53
ING4 may inhibit cancer invasion and metastasis by interacting with p53 as it has been shown that ING4 binds to p53 and enhances its activity [93]. p53 inhibits cancer cell migration by regulating the RhoA-ROCK pathway. Gadea et al. [91] found that p53-deficient fibroblasts displayed greater amoeboid-like motion, indicative of enhanced RhoA activity, and GTP-bound RhoA expression levels were increased in these cells. This was dependent on the activity of ROCK, and p53-deficient cells lost the ability to invade Matrigel upon either RhoA or ROCK inhibition. Similar results were seen in melanoma. Since ING4 increased p53 Lys-382 acetylation and interacted with p53 in vivo [93], it is possible that ING4 may exert effects on p53 to regulate cell migration via RhoA-ROCK signaling. However, Kim et al. [83] found that ING4 inhibited T47D breast cancer cell growth in soft agar as previously mentioned, and this cell line has non-functional p53. Therefore, ING4 also acts independently of p53 with respect to cancer progression.
ING5 and cancer progression
Perhaps the least is known regarding the fifth member of the ING family of tumor suppressors, mapped to chromosome 2q37.3 [2] in cancer invasion and metastasis. One study found that ING5 is located within a preferentially deleted region in oral cancer specimens [94]. Along with ING4, Shiseki et al. [93] found that p28ING5 physically interacts with p53 and p300 and enhances p53 activity. Since p53 regulates a number of proteins responsible for aspects of invasion and metastasis, it is likely that ING5 expression may also influence these factors through p53. ING5 enhanced p53 acetylation at Lys-382 and since post-translational modifications are thought to modify p53 function, ING5 may modify p53 via Lys-382 acetylation. ING5 was found to bind to p300, part of the HAT complexes that are responsible for both histone acetylation and the acetylation of other proteins such as p53. Also, ING5 overexpression activated the p21Waf1 promoter in RKO cells expressing p53, but not in RKO-E6 cells. Since p21Waf1 is a p53-regulated gene, ING5 expression influences p53 activity.
Mechanisms of action
ING1 behaves as a tumor suppressor in cancers of breast [36], stomach [23], leukemia [25], liver [27], as well as melanoma [38], although ING1 expression was not correlated with overall and metastasis-free survival in patients with Dukes’C colorectal cancer [37]. ING2 behaves as an oncogene in colon cancer [46], but as a tumor suppressor in HNSCC [47], melanoma [48] and BCC [51]. Investigations in HNSCC and melanoma have revealed ING3 as a potential invasion and metastasis suppressor [55, 72], and ING4 was found to be a tumor progression suppressor in myeloma, breast cancer, melanoma, glioma, lung carcinoma, cervical cancer, and colon carcinoma [76, 79, 83, 87–89, 95]. Finally, ING5 status in cancer invasion and metastasis is less clear, and no study directly relates ING5 expression to cancer, although it has been linked to p53 in colon carcinoma [93]. The role of each ING protein in cancer progression is summarized in Table 1 . It seems that the status of ING proteins in cancer invasion and metastasis is dependent on the type of malignancy, subcellular localization, extent of degradation, interaction with other proteins including HIF-1, NF-κB and p53, and to a lesser extent, ING gene mutation.
Table 1.
Summary of ING family effects on cancer progression
| Member | ING expression | Progression implication | Study type | Cancer type | Reference |
|---|---|---|---|---|---|
| ING1 | Low | Lymph node metastasis | In vivo | Breast | [36] |
| Low | Tumor progression | In vivo | Melanoma | [38] | |
| Low | Distant metastasis | In vivo | Stomach | [23] | |
| High | Lymph node metastasis | In vivo | OSCC | [39] | |
| ING2 | Low | Metastasis | In vivo | Melanoma | [48] |
| High | Invasion | In vitro | Colorectal | [46] | |
| ING3 | Low | Metastasis | In vivo | Melanoma | [55] |
| Low | TNM stage | In vivo | HNSCC | [72] | |
| ING4 | Low | NF-κB activity | In vitro/in vivo | Glioma | [89] |
| Low | HIF-1 activity | In vitro | Cervical | [87] | |
| Low | HIF-1 activity | In vitro | Myeloma | [88] | |
| Low | Migration | In vitro | Colon | [95] | |
| Low | Metastasis | In vivo | Melanoma | [76] | |
| High | p53 Activity | In vitro | Colon | [93] | |
| High | Metastasis | In vitro | Breast | [83] | |
| High | Migration | In vitro | Colon | [95] | |
| High | Invasion, migration | In vitro | Melanoma | [76] | |
| High | Invasion | In vitro | Glioma | [90] | |
| High | Invasion | In vitro | Lung | [79] | |
| ING5 | High | p53 Activity | In vitro | Colon | [93] |
OSCC oral squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma
ING interaction with H3K4me3, HIF-1, NF-κB, and p53
ING family members exert effects on invasion and metastasis both through interaction with nuclear proteins and subsequent transcriptional regulation as well as through interaction with cytoplasmic proteins. ING proteins regulate transcription through H3K4me3 binding via NF-κB, through HIF-1 during hypoxia, and by influencing p53/RhoA-ROCK activity. NF-κB binds to the ING2 promoter regulatory region, increasing ING2 expression. ING2 then binds to H3K4me3 at the promoter sites of NF-κB-regulated genes, recruiting chromatin remodeling factors to the promoters, thus activating gene transcription (Fig. 1b) [46]. ING2 acts as an oncogene in this case, and MMP-13 expression as well as cellular capacity for invasion and metastasis increases. This may be related to the physical structure of ING2 as ING2 has a greater ability to bind H3K4me3 which is associated with transcriptional activation. On the other hand, the PHD finger of ING4 is also capable of binding to H3K4me3, but this tends to suppress invasion and metastasis. ING4 binds to H3K4me3 and HBO1 resulting in H4 acetylation and subsequent inhibition of anchorage-independent growth [85]. Surprisingly, ING4 seems to have the opposite effect during interaction with NF-κB at NF-κB-regulated promoters. Interaction between NF-κB and ING4 at the Cox-2 and MMP-9 promoters results in decreased H3K4me3 and subsequent reduced Cox-2 and MMP-9 expression (Fig. 1c). ING4 decreases p300–p65 interaction in favor of HDAC-1 recruitment and reduces the amount of phosphorylated p65 at NF-κB-regulated promoters resulting in a less metastatic cellular phenotype [90]. Next, ING4 recruits chromatin-remodeling factors to HIF-1 via the HIF prolyl hydroxylase during hypoxia repressing transcription (Fig. 1d) [87]. Also, the p53/RhoA-ROCK pathway implicated in stress fiber formation and metastasis may be dependent on ING4 and ING5 [93]. p53 inhibits RhoA-ROCK activity, which decreases stress fiber formation, amoeboid motion, and ultimately cellular migration. Since ING4 also inhibits RhoA-ROCK activity [76] and ING4/ING5 enhance p53 activity [93], these two ING members may suppress cell migration via p53.
ING mutation, allelic loss, and degradation
Other potential mechanisms implicated in the control of cancer progression include ING protein degradation and to a lesser extent, ING gene mutation and allelic loss. ING1 mutations do not occur often and do not greatly contribute to the loss of functional cellular ING1 [2]. Loss of an ING2 locus in HNSCC is associated with worse tumor stage [50], relating ING2 gene alteration to cancer progression. Loss of ING2 may also have an effect on genomic stability and DNA repair [7], and there is the potential for disruption of progression-related genes in cells poorly expressing ING2 (Fig. 1a). Also, frequent LOH in the ING3 gene region is observed in HNSCC [73]. Allelic loss of ING4 is observed in HNSCC biopsies, breast cancer cell lines, and primary breast tumors [78, 83], and the ING5 locus may be preferentially deleted in oral cancer biopsies [94].
The degradation of ING proteins may also be responsible for the loss of ING protein observed in many cancers, and this may be related to cancer progression. This is especially true for ING3 as Skp2-dependent ING3 degradation by the ubiquitin-proteasome pathway is responsible for the loss of cellular ING3 in melanoma cells [56].
Subcellular localization and mislocalization
Since the physical structure of the ING family of tumor suppressors suggests that their activity takes place in the nucleus, increased nuclear expression of ING family members would be expected to inhibit cancer invasion and metastasis. Loss of nuclear ING1 and increased cytoplasmic ING1 expression is related to increased metastasis, suggesting that shuttling of ING1 from the nucleus to the cytoplasm may be implicated in cancer progression [38, 39]. ING1 interacts with PINCH in the cytoplasm at sites of cellular adhesion [39]. This has the effect of increasing invasion and metastasis potential. Additionally, loss of nuclear ING2 and ING3 is associated with increased metastatic phenotype in melanoma and this was related to increased cytoplasmic expression of ING3 suggesting the shuttling of ING3 from the nucleus into the cytoplasm in cancer cells [48, 55].
Despite the nature of the physical structures of ING family members, not all of their cancer progression suppressive actions are observed in the nucleus. It has been found that increased cytoplasmic ING expression in certain cancer types reduces cancer metastasis. Specifically, this is true for ING2 and ING4. ING4 splice variants without complete NLSs may exert suppressive effects on cancer progression in the cytoplasm [96]. Moreover, ING4 has been found to interact with cytoplasmic proteins implicated in cell motility and cancer metastasis including liprin α1, and ING4 interacts with liprin α1 in the cytoplasm to decrease progression capacity [95]. In fact, liprin α1 may play a role in shuttling nuclear ING1 into the cytoplasm, although this is only speculation. ING2 was found primarily in the nucleus in colon cancer and acts as an oncogene involved in chromatin remodeling [46]. Overall, it seems that increased nuclear expression of ING1, ING3, and ING4 reduce cancer invasion and metastasis potential. However, cytoplasmic expression of ING4 may also suppress cancer progression.
Conclusion
The role of the ING family of tumor suppressors in cancer invasion and metastasis is important to consider in addition to their role in cell cycle regulation, apoptosis, and DNA repair, and has implications for novel cancer therapy development. Although continued investigation is required, especially for ING2, 3, and 5, significant differences do exist among ING family members and among different isoforms. The effect of ING proteins in cancer progression depends on subcellular localization, extent of degradation, cancer type, general mechanism of action including either interaction with cytoplasmic proteins or proteins involved in chromatin modification, and specific mechanism of action including whether the member acts as a transcriptional activator or suppressor.
Since many of the differences observed among ING family members as well as the differential activity of a number of the ING members in different cancer types are unexplained, investigations examining the expression level of ING proteins in large numbers of tumor biopsies, in vitro studies dissecting the molecular pathways involved in cancer cell migration and invasion, and in vivo animal model investigations of cancer progression should be carried out. These systemic approaches would give some insight as to the exact role and the molecular mechanism of ING proteins in cancer invasion and metastasis. Elucidation of the mechanistic details behind the ING family of proteins and cancer progression could potentially lead to the design of enhanced cancer risk screening protocols and treatment regimens.
Acknowledgments
This work was supported by Canadian Institutes of Health Research (MOP-84559 and MOP-93810) and Canadian Dermatology Foundation to G. Li. B. Piche is a recipient of the trainee award from CIHR Skin Research Training Centre.
References
- 1.Unoki M, Kunamoto K, Takenoshita S, Harris CC. Reviewing the current classification of inhibitor of growth family proteins. Cancer Sci. 2009;100:1173–1179. doi: 10.1111/j.1349-7006.2009.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer. 2008;123:1483–1490. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]
- 3.Gong W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int J Biochem Cell Biol. 2005;37:1054–1065. doi: 10.1016/j.biocel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Coles AH, Jones SN. The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol. 2009;218:45–57. doi: 10.1002/jcp.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Campos E, Chine MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PubMed] [Google Scholar]
- 7.Larrieu D, Ythier D, Binet R, Brambilla C, Brambilla E, Sengupta S, Pedeux R. Ing2 controls the progression of DNA replication forks to maintain genome stability. EMBO Rep. 2009;10:1168–1174. doi: 10.1038/embor.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M, Seki N, Ozaki T, Kato M, Kuno T, Nakagawa T, Watanabe K-i, Miyazaki K, Ohira M, Hayashi S, Hosoda M, Tokita H, Mizuguchi H, Hayakawa T, Todo S, Nakagawara A. Identification of the p33ING1-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line NMuMG. Cancer Res. 2002;62:2203–2209. [PubMed] [Google Scholar]
- 10.Shinoura N, Muramatsu Y, Nishimura M, Yoshida Y, Saito A, Yokoyama T, Furukawa T, Horii A, Hashimoto M, Asai A, Kirino T, Hamada H. Adenovirus-mediated transfer of p33ING1 with p53 drastically augments apoptosis in gliomas. Cancer Res. 1999;59:5521–5528. [PubMed] [Google Scholar]
- 11.Shimada H, Liu TL, Ochiai T, Shimizu T, Haupt Y, Hamada H, Abe T, Oka M, Takiguchi M, Hiwasa T. Facilitation of adenoviral wild-type p53-induced apoptotic cell death by overexpression of p33ING1 in T.Tn human esophageal carcinoma cells. Oncogene. 2002;21:1208–1216. doi: 10.1038/sj.onc.1205176. [DOI] [PubMed] [Google Scholar]
- 12.Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, Yokota J, Harris CC. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene. 2003;22:343–350. doi: 10.1038/sj.onc.1206115. [DOI] [PubMed] [Google Scholar]
- 13.Cheung KJ, Li G. p33ING1 enhances UVB-induced apoptosis in melanoma cells. Exp Cell Res. 2002;279:291–298. doi: 10.1006/excr.2002.5610. [DOI] [PubMed] [Google Scholar]
- 14.Cheung K-J, Jr, Mitchell D, Lin P, Li G. The tumor suppressor candidate p33ING1 mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–4977. [PubMed] [Google Scholar]
- 15.Kuo W-HW, Wang Y, Wong RPC, Campos EI, Li G. The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp Cell Res. 2007;313:1628–1638. doi: 10.1016/j.yexcr.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Wang J, Li G. Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett. 2006;580:3787–3793. doi: 10.1016/j.febslet.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Chin MY, Li G. The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res. 2006;66:1906–1911. doi: 10.1158/0008-5472.CAN-05-3444. [DOI] [PubMed] [Google Scholar]
- 18.Doyon Y, Cayrou C, Ullah M, Landry A-J, Côté V, Selleck W, Lane WS, Tan S, Yang X-J, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/MCB.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menendez C, Abad M, Gomez-Cabello D, Moreno A, Palmero I. ING proteins in cellular senescence. Curr Drug Targets. 2009;10:406–417. doi: 10.2174/138945009788185077. [DOI] [PubMed] [Google Scholar]
- 21.Shah S, Riabowol K. Signaling pathways of the ING proteins in apoptosis. Curr Drug Targets. 2009;10:385–391. doi: 10.2174/138945009788185103. [DOI] [PubMed] [Google Scholar]
- 22.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 23.Ding H, Yang D, Wu FR, Li H, Feng K, Zheng SJ, Ni CR, Yu GZ. P33(ING1) gene expression and mutation in stomach cancer tissues and precarcinomatous tissues. Zhonghua Yi Xue Za Zhi. 2003;83:320–323. [PubMed] [Google Scholar]
- 24.Chen L, Wei JB, Zhou YC, Zhang S, Liang JL, Cao YF, Tang ZJ, Zhang XL, Gao F. Genetic alterations and expression of inhibitor of growth 1 in human sporadic colorectal cancer. World J Gastroenterol. 2005;11:6120–6124. doi: 10.3748/wjg.v11.i39.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nouman G, Anderson JJ, Wood KM, Lunec J, Hall AG, Reid MM, Angus B. Loss of nuclear expression of the p33ING1b inhibitor of growth protein in childhood acute lymphoblastic leukaemia. J Clin Pathol. 2002;55:596–601. doi: 10.1136/jcp.55.8.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouman G, Anderson JJ, Crosier S, Shrimankar J, Lunec J, Angus B. Downregulation of nuclear expression of the p33ING1b inhibitor of growth protein in invasive carcinoma of the breast. J Clin Pathol. 2003;56:507–511. doi: 10.1136/jcp.56.7.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohgi T, Masaki T, Nakai S, Morishita A, Yukimasa S, Nagai M, Miyauchi Y, Funaki T, Kurokohchi K, Watanabe S, Kuriyama S. Expression of p33(ING1) in hepatocellular carcinoma: relationships to tumour differentiation and cyclin E kinase activity. Scand J Gastroenterol. 2002;37:1440–1448. doi: 10.1080/003655202762671332. [DOI] [PubMed] [Google Scholar]
- 28.Oki E, Maehara Y, Tokunaga E, Kakeji Y, Sugimachi K. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett. 1999;147:157–162. doi: 10.1016/S0304-3835(99)00288-8. [DOI] [PubMed] [Google Scholar]
- 29.Tallen G, Kaiser I, Krabbe S, Lass U, Hartmann C, Henze G, Riabowol K, von Deimling A. No ING1 mutations in human brain tumours but reduced expression in high malignancy grades of astrocytoma. Int J Cancer. 2004;109:476–479. doi: 10.1002/ijc.11715. [DOI] [PubMed] [Google Scholar]
- 30.Tokunaga E, Maehara Y, Oki E, Kitamura K, Kakeji Y, Ohno S, Sugimachi K. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett. 2000;152:15–22. doi: 10.1016/S0304-3835(99)00434-6. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z, Luo Z, Li Y, Ni C, Li H, Zhu M. Human inhibitor of growth 1 inhibits hepatoma cell growth and influences p53 stability in a variant-dependent manner. Hepatology. 2009;49:504–512. doi: 10.1002/hep.22675. [DOI] [PubMed] [Google Scholar]
- 32.Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P, Pastyryeva S, Meijer M, Johnston RN, Bazett-Jones DP, McMahon S, Cole MD, Young D, Riabowol K. Human ING1 proteins differentially regulate histone acetylation. J Biol Chem. 2002;277:29832–29839. doi: 10.1074/jbc.M200197200. [DOI] [PubMed] [Google Scholar]
- 33.Scott M, Bonnefin P, Vieyra D, Boisvert F-M, Young D, Bazett-Jones DP, Riabowol K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]
- 34.Skowyra D, Zeremski M, Nezanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- 35.Zeremski M, Hill JE, Kwek SSS, Grigorian IA, Gurova KV, Garkavtsev IV, Diatchenko L, Koonin EV, Gudkov AV. Structure and regulation of the mouse ING1 gene. J Biol Chem. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]
- 36.Toyama T, Hirotaka I, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, Riabowol K. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed I, Kelly S, Anderson J, Angus B, Challen C, Lunec J. The predictive value of p53 and p33ING1b in patients with Dukes’C colorectal cancer. Colorectal Dis. 2007;10:344–351. doi: 10.1111/j.1463-1318.2007.01317.x. [DOI] [PubMed] [Google Scholar]
- 38.Nouman G, Anderson JJ, Mathers ME, Leonard N, Crosier S, Lunec J, Angus B. Nuclear to cytoplasmic compartment shift of the p33ING1b tumour suppressor protein is associated with malignancy in melanocytic lesions. Histopathology. 2002;40:360–366. doi: 10.1046/j.1365-2559.2002.01369.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Wang DW, Li QX, Zhu ZL, Wang MW, Cui DS, Yang YH, Gu YX, Sun XF. Nuclear to cytoplasmic shift of p33ING1b protein from normal oral mucosa to oral squamous cell carcinoma in relation to clinicopathological variables. J Cancer Res Clin Oncol. 2008;134:421–426. doi: 10.1007/s00432-007-0305-y. [DOI] [PubMed] [Google Scholar]
- 40.Gong W, Russell M, Suzuki K, Riabowol K. Subcellular targeting of p33ING1b by phosphorylation-dependent 14–3-3 binding regulates p21WAF1 expression. Mol Cell Biol. 2006;26:2947–2954. doi: 10.1128/MCB.26.8.2947-2954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung K, Li G. The tumour suppressor p33ING1 does not regulate migration and angiogenesis in melanoma cells. Int J Oncol. 2002;21:1361–1365. [PubMed] [Google Scholar]
- 42.Chin MY, Ng KCP, Li G. The novel tumor suppressor p33ING2 enhances UVB-induced apoptosis in human melanoma cells. Exp Cell Res. 2005;304:531–543. doi: 10.1016/j.yexcr.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 43.Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci USA. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unoki M, Kumamoto K, Robles AI, Shen JC, Zheng Z-M, Harris CC. A novel ING2 isoform, ING2b, synergizes with ING2a to prevent cell cycle arrest and apoptosis. FEBS Lett. 2008;582:3868–3874. doi: 10.1016/j.febslet.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ythier D, Brambilla E, Binet R, Nissou D, Vesin A, de Fraipont F, Moro-Sibilot D, Lantuejoul S, Brambilla C, Gazzeri S, Pedeux R (2009) Expression of candidate tumor suppressor gene ING2 is lost in non-small cell lung carcinoma. Lung Cancer (in press) [DOI] [PubMed]
- 46.Kumamoto K, Fujita K, Kurotani R, Saito M, Unoki M, Hagiwara N, Shia H, Bowman ED, Yanaihara N, Okamura S, Nagashima M, Miyamoto K, Takenoshita S, Yokota J, Harris CC. ING2 is upregulated in colon cancer and increases invasion by enhanced mmp13 expression. Int J Cancer. 2009;125:1306–1315. doi: 10.1002/ijc.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borkosky S, Gunduz M, Nagatsuka H, Beder LB, Gunduz E, Ali MAL, Rodriguez AP, Cilek MZ, Tominaga S, Yamanada N, Shimizu K, Nagai N. Frequent deletion of ING2 locus at 4q35.1 associates with advanced tumor stage in head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135:703–713. doi: 10.1007/s00432-008-0507-y. [DOI] [PubMed] [Google Scholar]
- 48.Lu F, Dai DL, Martinka M, Ho V, Li G. Nuclear ING2 expression is reduced in human cutaneous melanomas. Br J Cancer. 2006;95:80–86. doi: 10.1038/sj.bjc.6603205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okano T, Gemma A, Hosoya Y, Hosomi Y, Nara M, Kokubo Y, Yoshimura A, Shibuya M, Nagashima M, Harris CC, Kudoh S. Alterations in novel candidate tumor suppressor genes, ING1, and ING2 in human lung cancer. Oncol Rep. 2006;15:545–549. [PubMed] [Google Scholar]
- 50.Cetin E, Cengiz B, Gunduz E, Gunduz M, Nagatsuka H, Beder LB, Fukushima K, Pehlivan D, Ozaslan M, Yamanada N, Nishizaki K, Shimizu K, Nagai N. Deletion mapping of chromosome 4q22–35 and identification of four frequently deleted regions in head and neck cancers. Neoplasma. 2008;55:299–304. [PubMed] [Google Scholar]
- 51.Sironi E, Cerri A, Tomasini D, Sirchia SM, Porta G, Rossella F, Grati FR, Simoni G. Loss of heterozygosity on chromosome 4q32–35 in sporadic basal cell carcinomas: evidence for the involvement of p33ING2/ING1L and SAP30 genes. J Cutan Pathol. 2004;31:318–322. doi: 10.1111/j.0303-6987.2004.0187.x. [DOI] [PubMed] [Google Scholar]
- 52.Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo J, Shah S, Riabowol K, Mains PE. The caenorhabditis elegans ING-3 gene regulates ionizing radiation-induced germ-cell apoptosis in a p53-associated pathway. Genetics. 2009;181:473–482. doi: 10.1534/genetics.107.080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Li G. ING3 promotes UV-induced apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem. 2006;281:11887–11893. doi: 10.1074/jbc.M511309200. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Dai DL, Martinka M, Li G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin Cancer Res. 2007;13:4111–4116. doi: 10.1158/1078-0432.CCR-07-0408. [DOI] [PubMed] [Google Scholar]
- 56.Chen G, Wang Y, Garate M, Zhou J, Li G (2009) The tumor suppressor ING3 is degraded by SCFSkp2-mediated ubiquitin-proteasome system. Oncogene (in press) [DOI] [PubMed]
- 57.Hung W-C, Tseng W-L, Shiea J, Chang H-C (2009) Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett (in press) [DOI] [PubMed]
- 58.Wang X, Wu YP, Ye B, Lin DC, Feng YB, Zhang ZQ, Xu X, Han YL, Cai Y, Dong JT, Zhan QM, Wu M, Wang MR. Suppression of anoikis by Skp2 amplification and overexpression promotes metastasis of esophageal squamous cell carcinoma. Mol Cancer Res. 2009;7:12–22. doi: 10.1158/1541-7786.MCR-08-0092. [DOI] [PubMed] [Google Scholar]
- 59.Liao Q, Zhong D, Chen Q. Protein expression of Skp2 in osteosarcoma and its relation with prognosis. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2008;33:606–611. doi: 10.3736/jcim20080612. [DOI] [PubMed] [Google Scholar]
- 60.Fang F-M, Chien C-Y, Li C-F, Shiu W-Y, Chen C-H, Huang H-Y. Effect of S-phase kinase-associated protein 2 expression on distant metastasis and survival in nasopharyngeal carcinoma patients. Int J Rad Oncol Biol Phys. 2009;73:202–207. doi: 10.1016/j.ijrobp.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 61.Zhao J, Yang CL, Zhao SY. Expression of Skp2 protein in lung carcinoma and its implication for prognosis. Zhonghua Zhong Liu Za Zhi. 2007;29:289–292. [PubMed] [Google Scholar]
- 62.Einama T, Kagata Y, Tsuda H, Morita D, Ogata S, Ueda S, Takigawa T, Kawarabayashi N, Fukatsu K, Sugiura Y, Matsubara O, Hatsuse K. High-level Skp2 expression in pancreatic ductal adenocarcinoma: correlation with the extent of lymph node metastasis, higher histological grade, and poorer patient outcome. Pancreas. 2006;32:376–381. doi: 10.1097/01.mpa.0000220862.78248.c4. [DOI] [PubMed] [Google Scholar]
- 63.Sui L, Dong Y, Watanabe Y, Yamaguchi F, Sugimoto K, Tokuda M. Clinical significance of Skp2 expression, alone and combined with Jab1 and p27 in epithelial ovarian tumors. Oncol Rep. 2006;15:765–771. [PubMed] [Google Scholar]
- 64.Harada K, Supriatno K, Kawaguchi S, Kawashima Y, Itashiki Y, Yoshida H, Sato M. High expression of S-phase kinase-associated protein 2 (Skp2) is a strong prognostic marker in oral squamous cell carcinoma patients treated by UFT in combination with radiation. Anticancer Res. 2005;25:2471–2475. [PubMed] [Google Scholar]
- 65.Kamata Y, Watanabe J, Nishimura Y, Arai T, Kawaguchi M, Hattori M, Obokata A, Kuramoto H. High expression of Skp2 correlates with poor prognosis in endometrial endometrioid adenocarcinoma. J Cancer Res Clin Oncol. 2005;131:591–596. doi: 10.1007/s00432-005-0671-2. [DOI] [PubMed] [Google Scholar]
- 66.Li H, Jiang X, Zhou X. Expression of Skp2 and p27 proteins in laryngeal squamous cell carcinoma and their significance. Lin Chuang Er Bi Yan Hou Ke Za Zhi. 2005;19:409–411. [PubMed] [Google Scholar]
- 67.Zheng W-Q, Zheng J-M, Ma R, Meng F-F, Ni C-R. Relationship between levels of Skp2 and p27 in breast carcinomas and possible role of Skp2 as targeted therapy. Steroids. 2005;70:770–774. doi: 10.1016/j.steroids.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 68.Takanami I. The prognostic value of overexpression of Skp2 mRNA in non-small cell lung cancer. Oncol Rep. 2005;13:727–731. [PubMed] [Google Scholar]
- 69.Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of Skp2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am J Pathol. 2004;165:175–180. doi: 10.1016/S0002-9440(10)63286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, Masaki T, Kubo A, Fujita J, Dixon DA, Beauchamp RD, Ishida T, Kuriyama S, Imaida K. Correlation of Skp2 with carcinogenesis, invasion, metastasis, and prognosis in colorectal tumors. Int J Oncol. 2004;25:87–95. [PubMed] [Google Scholar]
- 71.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M. S-phase kinase-associated protein 2 expression in laryngeal squamous cell carcinomas and its prognostic implications. Oncol Rep. 2003;10:321–325. [PubMed] [Google Scholar]
- 72.Gunduz M, Beder LB, Gunduz E, Nagatsuka H, Fukushima K, Pahlivan D, Cetin E, Yamanaka N, Nishizaki K, Shimizu K, Nagai N. Downregulation of ING3 mRNA expression predicts poor prognosis in head and neck cancer. Cancer Sci. 2008;99:531–538. doi: 10.1111/j.1349-7006.2007.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, Nagai N, Nishizaki K, Shimizu K. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31 in human head and neck cancers. Oncogene. 2002;21:4462–4470. doi: 10.1038/sj.onc.1205540. [DOI] [PubMed] [Google Scholar]
- 74.Li M, Jin Y, Sun W-j, Yu Y, Bai J, Tong D-d, Qi J-p, Du J-r, Geng J-s, Huang Q, Huang X-y, Huang Y, Han F-f, Meng X-n, Rosales JL, Lee K-Y, Fu S-b. Reduced expression and novel splice variants of ING4 in human gastric adenocarcinoma. J Pathol. 2009;219:87–95. doi: 10.1002/path.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X, Cai L, Chen H, Zhang Q, Zhang S, Wang Y, Dong Y, Cheng H, Qi J. Inhibitor of growth 4 induces growth suppression and apoptosis in glioma U87MG. Pathobiology. 2009;76:181–192. doi: 10.1159/000218334. [DOI] [PubMed] [Google Scholar]
- 76.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–1379. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- 77.Cai L, Li X, Zheng S, Wang Y, Wang Y, Li H, Yang J, Sun J. Inhibitor of growth 4 is involved in melanomagenesis and induces growth suppression and apoptosis in melanoma cell line M14. Melanoma Res. 2009;19:1–7. doi: 10.1097/CMR.0b013e32831bc42f. [DOI] [PubMed] [Google Scholar]
- 78.Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K, Beder L, Hirohata S, Ninomiya Y, Nishizaki K, Shimizu K, Nagai N. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene. 2005;356:109–117. doi: 10.1016/j.gene.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 79.Xie Y, Zhang H, Shen W, Xiang J, Ye Z, Yang J. Adenovirus-mediated ING4 expression suppresses lung carcinoma cell growth via induction of cell cycle alteration and apoptosis and inhibition of tumor invasion and angiogenesis. Cancer Lett. 2008;271:105–116. doi: 10.1016/j.canlet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 80.Li X, Cai L, Liang M, Wang Y, Yang J, Zhao Y. ING4 induces cell growth inhibition in human lung adenocarcinoma A549 cells by means of Wnt-1/beta-Catenin signaling pathway. Anatomical Rec. 2008;291:593–600. doi: 10.1002/ar.20685. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X, Xu L-S, Wang Z-Q, Wang K-S, Li N, Cheng Z-H, Huang S-Z, Wei D-Z, Han Z-G. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 82.Fang F, Luo LB, Tao YM, Wu F, Yang LY. Decreased expression of inhibitor of growth 4 correlated with poor prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:409–416. doi: 10.1158/1055-9965.EPI-08-0575. [DOI] [PubMed] [Google Scholar]
- 83.Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci USA. 2004;101:16251–16256. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S. HuntING4 new tumor suppressors. Cell Cycle. 2005;4:516–517. doi: 10.4161/cc.4.4.1584. [DOI] [PubMed] [Google Scholar]
- 85.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozer A, Bruik RK. Regulation of HIF by prolyl hydroxylases: recruitment of the candidate tumor suppressor protein ING4. Cell Cycle. 2005;4:1153–1156. doi: 10.4161/cc.4.9.2040. [DOI] [PubMed] [Google Scholar]
- 87.Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc Natl Acad Sci USA. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colla S, Tagliaferri S, Morandi F, Lunghi P, Donofrio G, Martorana D, Mancini C, Lazzaretti M, Mazzera L, Ravanetti L, Bonomini S, Ferrari L, Miranda C, Ladetto M, Neri TM, Neri A, Greco A, Mangoni M, Bonati A, Rizzoli V, Guiliani N. The new tumor-suppressor gene inhibitor of growth family member 4 (ING4) regulates the production of proangiogenic molecules by myeloma cells and suppresses hypoxia-inducible factor-1 alpha (HIF-1 alpha) activity: involvement in myeloma-induced angiogenesis. Blood. 2007;110:4464–4475. doi: 10.1182/blood-2007-02-074617. [DOI] [PubMed] [Google Scholar]
- 89.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 90.Nozell S, Laver T, Moseley D, Nowoslawski L, DeVos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN. The ING4 tumor suppressor attenuates NF-κB activity at the promoters of target genes. Mol Cell Biol. 2008;28:6632–6645. doi: 10.1128/MCB.00697-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gadea G, deToledo M, Anguille C, Roux P. Loss of p53 promotes Rhoa-ROCK-dependent cell migration and invasion in 3D matrices. J Cell Biol. 2007;178:23–30. doi: 10.1083/jcb.200701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao K, Dai DL, Martinka M, Li G. Prognostic significance of nuclear factor-kappaB p105/p50 in human melanoma and its role in cell migration. Cancer Res. 2006;66:8382–8388. doi: 10.1158/0008-5472.CAN-05-4402. [DOI] [PubMed] [Google Scholar]
- 93.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC. P29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 94.Cengiz B, Gunduz M, Nagatsuka H, Beder L, Gunduz E, Tamamura R, Mahmut N, Fukushima K, Ali MAS, Naomoto Y, Shimizu K, Nagai N. Fine deletion mapping of chromosome 2q21–37 shows three preferentially deleted regions in oral cancer. Oral Oncol. 2007;43:241–247. doi: 10.1016/j.oraloncology.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 95.Shen J, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, Pedeux R, Harris CC. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with novel binding partner, liprin alpha 1. Cancer Res. 2007;67:2552–2558. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Unoki M, Shen JC, Zheng ZM, Harris CC. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J Biol Chem. 2006;281:34677–34686. doi: 10.1074/jbc.M606296200. [DOI] [PubMed] [Google Scholar]