Abstract
Here we show that a small GTPase, Rab32, is a novel protein required for the formation of autophagic vacuoles. We found that the wild-type or GTP-bound form of human Rab32 expressed in HeLa and COS cells is predominantly localized to the endoplasmic reticulum (ER), and overexpression induces the formation of autophagic vacuoles containing an autophagosome marker protein LC3, the ER-resident protein calnexin and endosomal/lysosomal membrane protein LAMP-2, even under nutrient-rich conditions. The recruitment of Rab32 to the ER membrane was necessary for autophagic vacuole formation, suggesting involvement of the ER as a source of autophagosome membranes. In contrast, the expression of the inactive form of, or siRNA-specific for, Rab32 caused the formation of p62/SQSTM1 and ubiquitinated protein-accumulating aggresome-like structures and significantly prevented constitutive autophagy. We postulate that Rab32 facilitates the formation of autophagic vacuoles whose membranes are derived from the ER and regulates the clearance of aggregated proteins by autophagy.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0080-9) contains supplementary material, which is available to authorized users.
Keywords: Rab32, Autophagy, Aggresome, p62/SQSTM1, LC3
Introduction
Autophagy is a degradation pathway that delivers cytoplasmic materials to lysosomes via doubled-membrane organelles termed autophagosomes [1, 2]. Although autophagy can be induced by nutrient deprivation or the absence of growth factors, it also occurs at basal levels in most tissues and contributes to the routine turnover of cytoplasmic components [3–5]. In addition, autophagy is directly or indirectly involved in eliminating aberrant protein aggregates, protecting against tumors and defending against pathogen invasion [6–8]. The autophagic process is initiated when a cup-shaped double membrane structure, termed an isolation membrane or phagophore, engulfs a portion of cytoplasmic materials, including organelles and protein aggregates [9]. The isolation membrane gradually elongates with curvature and finally become enclosed to form an autophagosome [10]. Subsequent fusion of autophagosomes with acid hydrolase-rich lysosomes degrades the sequestered materials. So far, many mammalian homologues of yeast ATG (autophagy-related) genes have been identified and characterized, demonstrating that the molecular mechanisms of autophagy have been conserved from yeasts to mammals [11].
Rab GTPases represent a large family of small GTP-binding proteins that comprises more than 60 known members [12]. In mammalian cells, it is well established that different Rab proteins localize on distinct membrane-bound compartments, where they regulate multiple steps in membrane traffic, including vesicle budding, fusion and movement through cycling between an inactive GDP-bound form and an active GTP-bound form. Akin to the involvement of Rab proteins in the vesicle transport processes, it has been reported that Rab7 and Rab24 function in the autophagic pathway [13–15]. Rab24, which is localized in the ER, cis-Golgi and the ER-Golgi intermediate compartment, is translocated to large autophagic vacuoles under starvation conditions, suggesting the involvement of this protein in autophagy [15]. Although Rab7, a late endosome-specific Rab protein, is implicated in late endocytic transport and lysosome biogenesis [16], two lines of evidence have revealed that Rab7 is required for the progression of autophagy [13] and for the final maturation step in fusion between late autophagic vacuoles and lysosomes [14].
Rab32 contains amino-acid sequences that are shared with only a small number of other Rab proteins. Most conspicuously, threonine in the WDTAGQE sequence, conserved in almost all Rab proteins, is replaced by isoleucine. This WDIAGQE sequence in Rab32 is also found in Rab38 [17], Rab29 [18] and Rab7L1/29 [19], among mammalian Rab proteins identified to date, and is also present in RabE from the slime mold Dictyostelium discoideum [20]. Recently, it has been reported that mouse Rab38 and Rab32 act in a functionally redundant way in regulating skin melanocyte pigmentation and control post-Golgi trafficking of tyrosinase and tyrosinase-related protein 1, thereby suggesting critical roles for melanosome maturation [21]. Most recently, it has been shown that, in Xenopus melanophores, Rab32 is involved in the regulation of melanosome transport by cAMP-dependent protein kinase A [22]. Although Rab32 is expressed in most human tissues [23], little is known about the physiological roles of Rab32 in tissues and cells other than melanocytes. In this paper, using HeLa and COS cells exogenously expressing human Rab32, we show that Rab32 is required for the formation of autophagic vacuoles and the regulation of the clearance of aggregated proteins by autophagy.
Materials and methods
Cell culture and transfection
HeLa cells and COS-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS). Cells were transfected for 48 h using Fugene 6 (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s instructions. For starvation, cells were incubated in Krebs–Henseleit buffer (Sigma-Aldrich, St Louis, MO) at 37°C for 2 h.
DNA constructs
The cDNA for human wild-type Rab32 was obtained from Open Biosystems (Huntsville, AL). Rab32Q85L, Rab32T39N and Rab32N143I in vector pcDNA3.1/Hygro (−) (Invitrogen, Groningen, The Netherlands) were made by PCR-mediated site-directed mutagenesis using Rab32WT in pcDNA3.1 as a template. Briefly, the following primer combinations were used: (Q85L) PCR reaction (1) Rab32 forward primer 5′-AGTGGATCCATGGCGGGCGGAGGAGCC-3′, Q85L reverse primer 5′-AAATCGCTCCAGCCCCGCGATGTC-3′; PCR reaction (2) Q85L site forward primer 5′-GACATCGCGGGGCTGGAGCGATTT-3′ and Rab32 reverse primer 5′-CTTGGTACCTCAGCAACACTGGGATTTGTT-3′. (T39N) PCR reaction (1) Rab32 forward primer: described above, T39N reverse primer 5′-CTTGATGATGCTGTTCTTGCCCACGCC-3′; PCR reaction (2) T39N forward primer 5′-GGCGTGGGCAAGAACAGCATCATCAAG-3′ and Rab32 reverse primer: described above. (N143I) PCR reaction (1) Rab32 forward primer: described above, N143I reverse primer 5′-CTGGTCACATTTGATAGCCAAGAG-3′; PCR reaction (2) N143I forward primer 5′-CTCTTGGCTATCAAATGTGACCAG-3′and Rab32 reverse primer: described above. The reaction products forming (1) and (2) were used as a template in the final PCR reaction, which was amplified using Rab32 forward primer and reverse primer to generate full-length Rab32Q85L, Rab32T39N and Rab32N143I. These were cloned into pcDNA3.1, and digested with BamHI and KpnI. Rab32WT, Rab32Q85L, Rab32T39N and Rab32N143I were cloned in frame with GFP into KpnI/BamHI sites of pEGFP-C1 (Clontech, Palo Alto, CA). For the expression of GST fusion proteins, pGST-Rab32 and pGST-LC3 were obtained by subcloning Rab32 and LC3 cDNAs into the EcoRI/SalI sites of pGEX-KG. FLAG-tagged ubiquitin was kindly provided by Tomoki Chiba [24].
siRNA-mediated silencing of Rab32
HeLa cells were transfected with Oligofectamine (Invitrogen, Carlsbad, CA), in the presence of 10 nM of siRNA specific for human Rab32. The oligonucleotide sequences used for siRNA interference with Rab32 expression corresponded to nucleotides 517–537 (siRNA-1: 5′-CAUUUGAGGCAGUCUUAAAAU-3′) and 483–503 (siRNA-2: 5′-GCUUUUGUAGUCUUUGAUAUA-3′). Control siRNA was constructed with a similar insert where the 21-nucleotide sequence had no homology to any known human gene sequence.
Antibodies
Purified protein Rab32 was generated by cleaving a GST-Rab32 fusion protein with thrombin protease and injected into rabbits to raise anti-Rab32 rabbit polyclonal antibody. An antibody against recombinant Rab32 was purified on an immobilized Rab32-CNBr-activated Sepharose column. Antiserum against GST-LC3 was also raised in rabbits, and anti-GST-LC3 antibody was affinity purified on an immobilized LC3-CNBr-activated Sepharose column. Mouse monoclonal antibodies (mAb) to calnexin, p62 and GM130 were obtained from BD Biosciences (Palo Alto, CA). Mouse monoclonal antibody to Hsp60 was obtained from StressGen Biotechnologies Corp. (Victoria, Canada). FK2 mouse monoclonal antibody was from Nippon Biotest Laboratories (Tokyo, Japan). Anti-Hsc70 mAb was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-FLAG M2 mAb and anti-c-myc mAb were purchased from Sigma-Aldrich and Roche Diagnostics (Mannheim, Germany), respectively. The secondary antibodies used were goat anti-rabbit Alexa488 and goat anti-mouse Alexa488 (Molecular Probes, Eugene, OR), goat anti-mouse Cy3, goat anti-rabbit Cy3 and goat anti-mouse Cy5 (Jackson ImmunoResearch Laboratories, West Grove, PA).
Percoll density gradients
Rab32WT-transfected or untransfected HeLa cells were grown on 60-mm dishes for 48 h. Cells were washed with ice-cold PBS and then suspended in 1 × SHE, which contained 0.25 M sucrose, 10 mM Hepes-KOH, pH 7.2, 1 mM EDTA, plus a protease inhibitor cocktail. Cell suspension was homogenized by 25 passages through a 23-G needle connected to a 1-ml syringe, and then centrifuged at 500×g to obtain a post-nuclear supernatant (PNS). The PNS was overlaid onto a solution of 27% Percoll in 1 × SHE and centrifuged at 20,000×g for 2 h in a HITACHI RP65T rotor at 4°C. The gradients were divided into 16 fractions of 8 ml by downward displacement.
Western blotting
Cells were extracted using RIPA buffer (20 mM Tris–HCl, pH 7.5, 0.15 M NaCl, 0.5% Triton X-100, 0.1% SDS, 1 mM EDTA, 10 mM NaF and 1 mM Na3VO4) and sonicated. The lysates were subjected to SDS-PAGE, and proteins were transferred to a polyvinylidene fluoride membrane (Millipore Corp., Temecula, CA) using a semi-dry blotting system. The membranes were blocked with 5% skimmed milk powder, 0.1% Triton X-100, 20 mM Tris–HCl, pH 7.5, 0.15 M NaCl (blocking buffer) and incubated with the primary antibody (1:2,000) in blocking buffer at 4°C overnight followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000) (Amersham Biosciences, San Francisco, CA). Blots were analyzed using the ECL detection system (Amersham Biosciences), and signals were visualized using Fuji LAS1000.
Immunofluorescence microscopy
Immunofluorescence was carried out as described previously [25]. Cells cultured on coverslips were rinsed with phosphate-buffered saline (PBS) and fixed immediately with 4% paraformaldehyde in PBS, pH 7.4, for 30 min at room temperature, or with cold methanol for 20 min at −20°C (for vimentin staining). Cells were quenched with 50 mM NH4Cl in PBS for 15 min, blocked and permeabilized with 0.05% saponin and 1% bovine serum albumin (BSA) in PBS for 30 min. Cells were then incubated for 1 h with the primary antibody in blocking solution. The primary antibodies were diluted as follows: anti-Rab32 antibody (0.1 μg/ml), anti-GM130 antibody (1:300), anti-FK2 antibody (1:300), anti-Hsc70 antibody (1:100), anti-myc antibody (1:300) and anti-62 antibody (1:300). Cells were washed with PBS and incubated for 30 min with secondary antibodies diluted in blocking solution. For immunostaining of endogenous LC3 in COS-1 cells, cells were stained with the purified anti-GST-LC3 antibody, as previously described [26]. Briefly, cells were washed with PBS and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. After fixation, the cells were permeabilized with 50 μg/ml digitonin in PBS for 5 min, quenched with 50 mM NH4Cl in PBS for 10 min and incubated in blocking buffer (2% goat serum/1% BSA in PBS) for 30 min. Cells were then incubated with the primary antibody in blocking buffer for 1 h, washed with PBS, incubated with the secondary antibody in blocking buffer for 30 min at room temperature and washed with PBS. Finally, cells were mounted in Mowiol (Calbiochem, La Jolla, CA). The cells were observed on a Zeiss confocal microscope (LSM 510 META). The images were processed using Adobe Photoshop CS.
Cotransfection-coimmunoprecipitation
COS-1 cells were transfected by coprecipitates of calcium phosphate with appropriate combinations of pcDNARab32 plasmids and FLAG-ubiquitin plasmid. Forty-eight hours after transfection, cells were lysed in RIPA buffer [20 mM Tris–HCl, pH 7.5, 0.15 M NaCl, 0.5% Triton X-100, 0.1% SDS, 1 mM EDTA, 10 mM NaF, 1 mM Na3VO4 and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan)] and sonicated. Supernatants obtained after centrifugation of cell lysates (13,000×g for 10 min at 4°C) were incubated with protein A-Sepharose preadsorbed with anti-Myc monoclonal antibody (Sigma-Aldrich) or anti-Rab32 antibody (previously described) overnight at 4°C. The precipitates were washed seven times with 0.1% Triton X-100 in PBS, followed by SDS-PAGE and Western blotting.
Analysis of soluble and insoluble Rab32
Triton X-100 soluble and insoluble fractions were prepared as described previously [27, 28]. Briefly, COS-1 cells were transfected using Fugene 6 with pcRab32 plasmids. Forty-eight hours after transfection, cells were washed with ice-cold PBS, and lysed for 30 min on ice with 300 μl 1% Triton X-100 in PBS supplemented with protease inhibitor cocktail and 166 μM MG132. Cell lysates were transferred to 1.5-ml tubes and homogenized on ice by five passages through a 23-G needle connected to a 1-ml syringe. Then, 200 μl of cell lysates was centrifuged at 17,500×g for 15 min at 4°C. The resulting supernatants were saved as Triton X-100-soluble fractions (S), and the pellets were solubilized in 50 μl 10 mM Tris–HCl, pH 7.5 and 1% SDS plus protease inhibitor cocktail for 5 min at room temperature. Subsequently, 150 μl of RIPA buffer II (50 mM Tris–HCl, pH 8.0, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS and 0.05% sodium deoxycholate) was added, and the pellets were then sonicated (I). Equivalent amounts of proteins among different S and I fractions were resolved using SDS-PAGE. Proteins were extracted from cells as previously described [29]. Transfected cells with pcDNARab32 for 48 h on a coverslip were incubated with 6.7 μg/ml of Cy3-conjugated transferrin in serum-free medium for 15 min at 37°C, washed in ice-cold PBS and then treated with 1% Triton X-100-containing buffer (100 mM Hepes-KOH, pH 7.2) for 20 min at 4°C. Cells were rinsed in ice-cold PBS and fixed with 4% paraformaldehyde.
Assay of proteasome activity
Essentially according to a previously described method [30], harvested cells were lysed with proteasome buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 2 mM ATP, 20% glycerol, 4 mM DTT and 0.5% Triton X-100), sonicated and then centrifuged at 13,000×g at 4°C for 10 min. The resulting supernatants (20 μg of protein) were incubated with proteasome activity assay buffer [100 mM Tris–HCl, pH 7.5, 1 mM ATP, 10 mM MgCl2, 1 mM DTT, and 100 μM Suc-Leu-Leu-Val-Tyl-4-methylcoumaryl-7-amide (Suc-LLVY-MCA) (Peptide Institute Inc., Osaka, Japan)] (total volume of reaction mixture: 100 μl) for 1 h at 37°C. The reactions were stopped by adding 1 ml of 5% SDS. Proteasome activity was determined by measuring the fluorescence intensity (excitation, 380 nm; emission, 460 nm) of 7-amino-4-methylcoumarin, one of the reaction products, with fluorescence spectrophotometry (Hitachi F-2500; Hitachi Instruments, Tokyo, Japan).
Inhibitor treatment
Bafilomycin A1 and chloroquine were purchased from Sigma-Aldrich and were used at a concentration of 100 nM for 1 h and 20 μM for 24 h, respectively, at 37°C. Rapamycin was obtained from Calbiochem and was used at a concentration of 0.2 μM for 24 h.
Results
Rab32 localizes to the ER
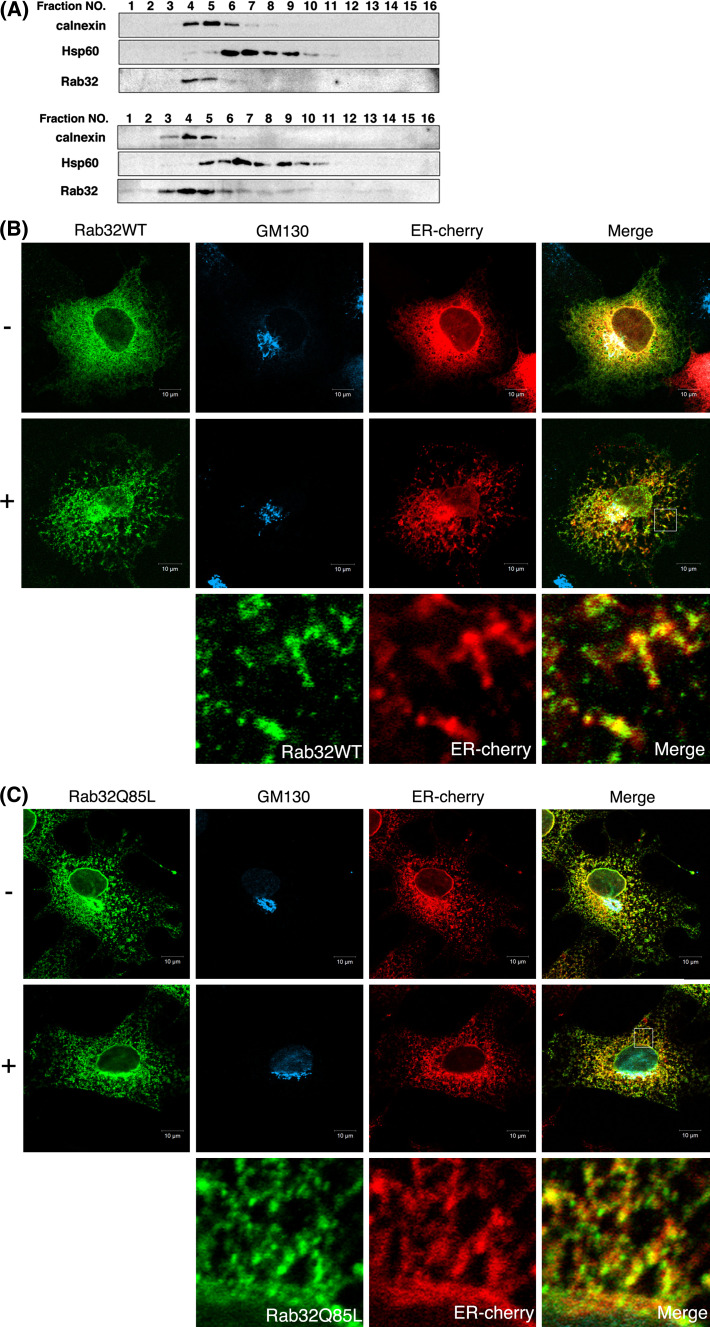
To determine the intracellular localization of Rab32, untransfected HeLa cells or HeLa cells transfected with a construct encoding Rab32 wild-type (WT) were fractionated by Percoll density gradient centrifugation and analyzed by Western blotting. As shown in Fig. 1a, both endogenous Rab32 and exogenously expressed Rab32WT were predominantly recovered in fractions enriched with an ER marker protein, calnexin, but not with a mitochondria marker, Hsp60. Antibodies against human Rab32 produced in this study were able to detect endogenous Rab32 expression in HeLa cells by Western blotting, but not by indirect immunofluorescence; therefore, intracellular localization of Rab32 in immunofluorescence microscopy was examined by the exogenous expression system in COS cells. When Rab32WT was transiently expressed in COS cells, the majority was found exclusively in reticular structures scattered throughout the cytoplasm and juxtanuclear region, where calnexin and a cis-Golgi marker, GM130, respectively, were located (Fig. 1b), whereas Rab32 was not colocalized with a late endosome/lysosome marker, LAMP-2 and an early endosome marker, EEA1 (data not shown). Furthermore, in contrast to expressed mCherry, which localizes to the cytoplasm (Fig. S1), most of the Rab32WT remained in reticular structures and the perinuclear region even after depleting the cytosol by treatment of living cells with digitonin prior to fixation (Fig. 1b). These results apparently indicate that Rab32 primarily localizes to the ER/Golgi, but not to endosomes and lysosomes. Similar intracellular localization was observed in cells in which a GTP-bound form of Rab32, Rab32Q85L, was transiently expressed in COS cells (Fig. 1c).
Fig. 1.
Intracellular location of Rab32. a HeLa cells transfected with (lower panels) or without (upper panels) pcDNA3.1-Rab32WT (pcDNARab32WT) were fractionated by Percoll density gradient centrifugation into 16 fractions. Each fraction was subjected to SDS-PAGE and analyzed by Western blot analysis with affinity-purified rabbit anti-Rab32 antibody, mouse anti-calnexin antibody (ER marker) and mouse anti-Hsp60 antibody (mitochondria marker). b, c COS-1 cells co-transfected pcDNARab32WT (b) or pcDNARab32Q85L (c) and ER-cherry were incubated in 100 mM Hepes-KOH (pH 7.2) (−: upper) or 0.013% digitonin/100 mM Hepes-KOH (pH 7.2) (+: lower) for 2 min at room temperature prior to fixation. Cells were fixed and processed for immunofluorescence using affinity-purified rabbit anti-Rab32 antibody and mouse anti-GM130 antibody. Merged images were generated by combining Rab32 (green), GM130 (cyan) and ER-cherry (red). The panels at the bottom of (b) and (c) show an enlarged view of the boxed regions. Bars are 10 μm
Rab32-associated ER membranes are involved in the generation of autophagosomal membrane structures
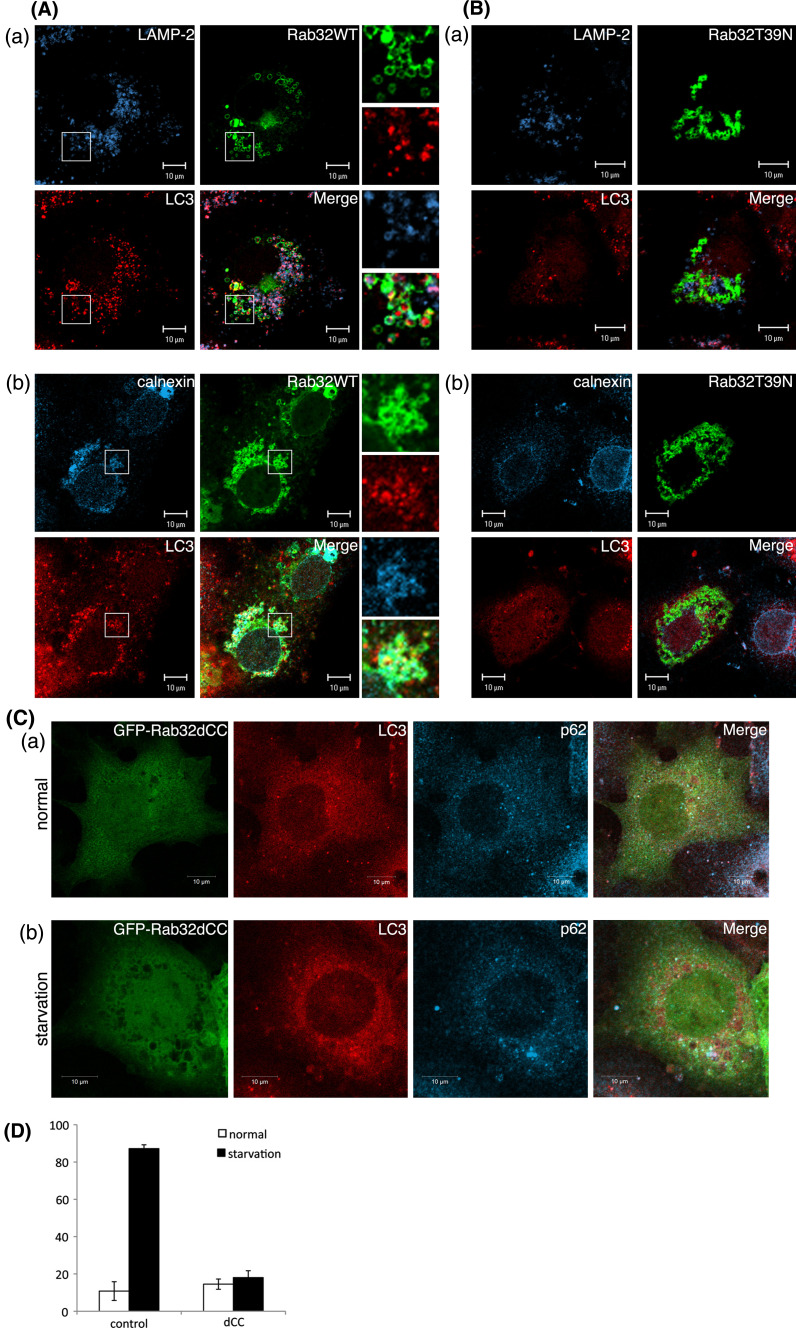
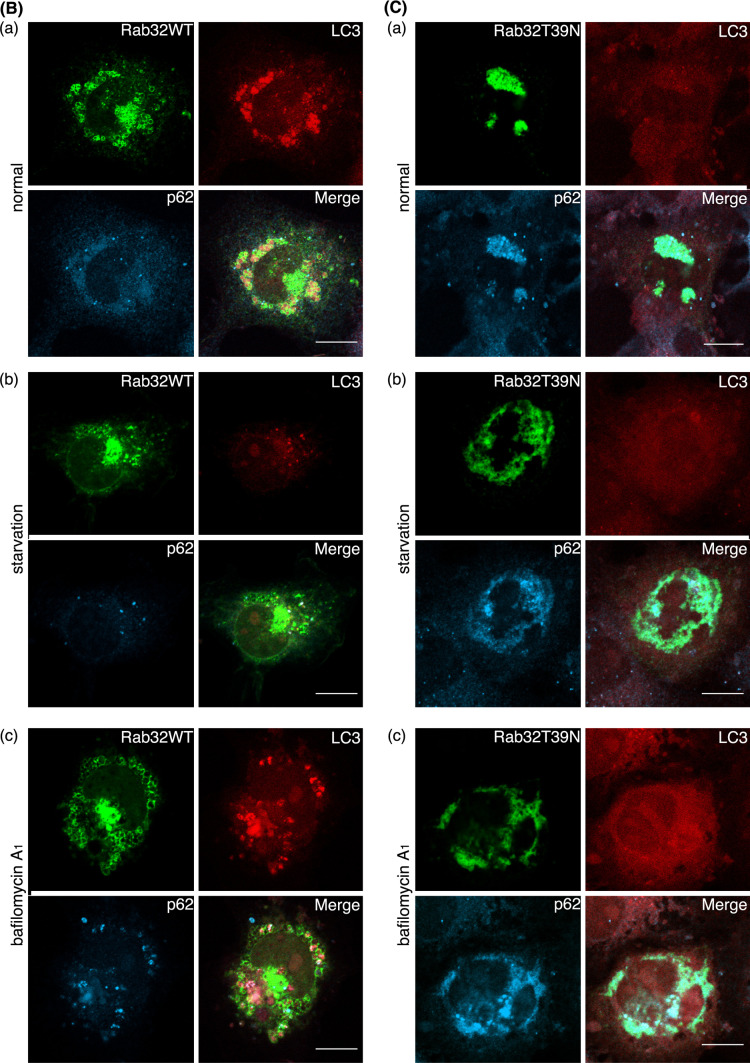
Notably, in cells overexpressing Rab32WT and Rab32Q85L, numerous large spherical structures were occasionally observed in the perinuclear region. These large spherical structures were stained with an antibody against LC3, which is a specific marker of autophagosomes [31], as well as anti-calnexin antibody [Fig. 2a (b) and S2B (b)]. Furthermore, some of these vesicles colocalized with LAMP-2 [Fig. 2A (a) and S2B (a)], thereby suggesting that Rab32WT- and Rab32Q85L-induced spherical structures represent autophagic vacuole-like compartments. These large spherical structures were observed in 54.1% of Rab32WT-expressing cells and 53.8% of Rab32Q85L-expressing cells. By contrast, Rab32T39 N (GDP-bound form)- and Rab32N143I (GTP/GDP-free form)-overexpressing cells showed no obvious change in the distribution patterns of calnexin and LAMP-2 and little punctate LC3 staining (Fig. 2B and S2C). Furthermore, endosomal marker protein EEA1 did not localize to Rab32-positive structures irrespective to GTP- or GDP-bound form (Fig. S3). It is conceivable, therefore, that the GTP-bound form of Rab32 is involved in the formation of autophagic vacuole-like structures containing the ER membrane. In addition to Rab32 activity, the recruitment of Rab32 to the ER membrane was necessary for the formation of autophagic vacuoles, because the expression of a mutant Rab32 deleted two cysteine residues (Rab32dCC) present in the carboxyl-terminal domain, which are essential for the association with membranes [22], did not cause the formation of autophagic vacuole-like structures (Fig. 2c) as observed in Rab32WT overexpression cells (Fig. 2a). The expression of the Rab32dCC mutant also suppressed the formation of LC3 dots induced by starvation (Fig. 2D). Taken together, these results suggest that Rab32 activity and ER localization are required for the formation of autophagic vacuoles. A previous report indicated that human Rab32 expressed in COS cells localizes to mitochondria as an A-kinase anchoring protein (AKAP), and the expression of its GDP-bound form causes the fragmentation of mitochondria [32]. However, we could not confirm these results because in addition to the failure of colocalization between Rab32WT and MitoTracker, there was no effect of Rab32T39 N and Rab32N143I on the morphology of mitochondria (Fig. S4A). Furthermore, since the knockdown of Rab32 by the expression of Rab32-specific siRNA did not influence the morphology of mitochondria (Fig. S4B), it is suggested that Rab32 does not participate in the dynamics of mitochondria.
Fig. 2.
Rab32-associated ER membranes are involved in the generation of autophagosomal membrane structures. (A, B) COS-1 cells were transiently transfected with GFP-Rab32WT (A) or GFP-Rab32T39 N (B) for 48 h. Fixed cells were stained with affinity-purified rabbit anti-LC3, and mouse anti-LAMP-2 (a), or mouse anti-calnexin (b) antibodies. Boxed regions are shown on the right. Bars are 10 μm. (C) GFP-Rab32dCC-transfected COS-1 cells were incubated in normal medium (a) or Krebs–Henseleit buffer (b) for 2 h. After fixation, cells were stained with affinity-purified rabbit anti-LC3 and mouse anti-p62 antibodies. Bars are 10 μm. (D) Untransfected COS-1 cells or GFP-Rab32dCC-transfected COS-1 cells were incubated with normal medium (starvation: −) (white bars) or Krebs–Henseleit buffer (starvation: +) (black bars) for 2 h. Cells with LC3 dots of more than 20 were defined as cells with LC3 dots, because about 10–20 LC3 dots were occasionally visible even in control cells. At least 200 cells expressing each GFP-Rab32 construct were counted at respective experiments and the percentage of cells exhibiting LC3 dots (i.e., autophagic vacuoles) in each GFP-Rab32 construct is quantified as the mean ± SEM (n = 3 separate experiments)
Expression of Rab32T39 N or Rab32N143I forms ubiquitinated protein-containing aggresome-like structures
Interestingly, the expression of Rab32T39 N or Rab32N143I caused the formation of large dot-like or chain-like structures in the perinuclear region (Fig. 2B and S2C). These structures quite closely resembled aggresome-like structures, which are observed when a de-ubiquitinating enzyme (UCH-L1) or presenilin-binding protein-expressing cells were treated with a proteasome inhibitor, MG132 [33, 34]. To investigate whether dot-like and/or chain-like structures appearing in cells expressing Rab32T39 N or Rab32N143I had the characteristics of aggresomes, we examined the colocalization between Rab32T39 N or Rab32N143I and several proteins that are shown to localize to aggresomes, such as ubiquitinated proteins, molecular chaperone Hsc70 and α-synuclein [33, 35, 36]. As shown in Fig. S5A-C, the expression of Rab32T39 N or Rab32N143I resulted in the redistribution of ubiquitinated proteins, Hsc70 and α-synuclein, into dot-like or chain-like structures, from their cytoplasmic distribution, as seen in cells expressing Rab32WT or Rab32Q85L. It has been further shown that aggresomes form at the microtubule-organizing center and are accompanied by the disruption of vimentin intermediate filaments [37]. Cells expressing Rab32T39 N- or Rab32N143I-positive aggresome-like structures, however, did not show a significant rearrangement of vimentin or γ-tubulin (Fig. S5D and E), suggesting that these structures are distinct from classical aggresomes. We conclude that the formation of aggresome-like structures induced by the expression of Rab32T39 N or Rab32N143I is not due to a secondary effect by the overexpression of mutant proteins, because such aggresome-like structures were not observed by the overexpression of dominant negative forms of both Rab5 and Rab7 (data not shown).
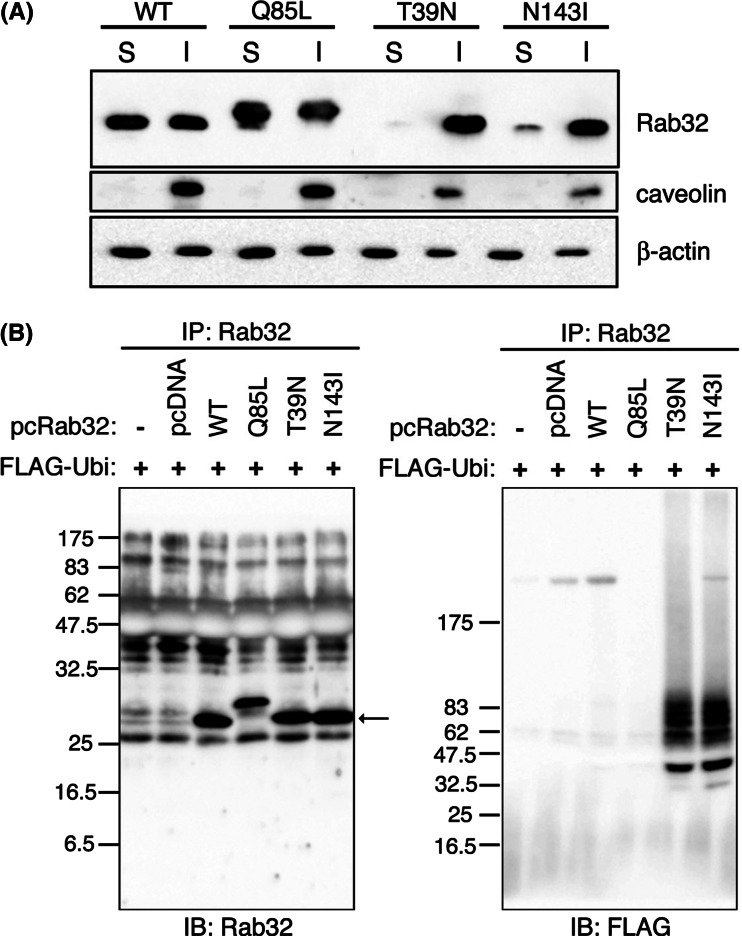
It is well known that proteins deposited within aggresomes are largely detergent insoluble [38]. We found that dot-like and chain-like structures induced by the expression of Rab32T39 N or Rab32N143I were resistant to the treatment of cells with Triton X-100 prior to fixation, in confocal laser microscopy, while under the same conditions internalized Alexa488-labeled transferrin mostly disappeared by Triton X-100 extraction of living cells (Fig. S5F). Western blot analyses revealed that 85-90% of Rab32T39 N or Rab32N143I was recovered in the Triton X-100-insoluble fraction, but 40-50% of each Rab32WT or Rab32Q85L was detected in the Triton X-100-insoluble fraction (Fig. 3a). The treatment of Rab32T39 N- and Rab32N143I-expressing cells with MG132 led to a slight increase in the rate of both mutants in the Triton X-100-insoluble fraction (data not shown), suggesting that part of Rab32T39 N and Rab32N143I may be degraded by proteasomes. These results further led us to speculate that Rab32T39 N and Rab32N143I are ubiquitinated. We then co-expressed wild-type Rab32 or mutant forms of Rab32 and FLAG-tagged ubiquitin. With this procedure, both Rab32T39 N and Rab32N143I, but not Rab32WT and Rab32Q85L, were detected as smear bands with high molecular weight (Fig. 3b), suggesting that both Rab32T39 N and Rab32N143I were polyubiquitinated. These results may suggest, therefore, that both Rab32T39 N and Rab32N143I decreased their solubility by being ubiquitinated, leading to the formation of aggresome-like structures. This is the first example showing that Rab protein is ubiquitinated. It cannot be ruled out, however, that Rab32T39 N and Rab32N143I are not directly ubiquitinated but rather associated with an ubiquitinated protein.
Fig. 3.
Rab32T39 N was ubiquitinated and recovered in the Triton X-100-insoluble fraction. a COS-1 cells were transfected with pcDNARab32WT, pcDNARab32Q85L, pcDNARab32T39 N or pcDNARab32N143I. Forty-eight hours after transfection, cells were lysed for 30 min on ice with 1% Triton X-100 in PBS supplemented with protease inhibitor cocktail and 166 μM MG132, and homogenized. After centrifugation, the resulting supernatants were saved as Triton X-100-soluble fractions (S), and the pellets were solubilized in SDS-containing buffer as Triton X-100-insoluble fractions (I). Equivalent amounts of proteins among different S and I fractions were resolved using SDS-PAGE and analyzed for the solubility of Rab32 by Western blotting. Blots were probed with affinity-purified rabbit anti-Rab32 (upper), rabbit anti-caveolin (middle) and mouse anti-β-actin (lower) antibodies. b COS-1 cells were co-transfected with each pcDNARab32WT or mutant Rab32 (Rab32Q85L, Rab32T39 N or Rab32N143I) plasmids and FLAG-ubiquitin plasmid. Forty-eight hours after transfection, cells were lysed. Supernatants obtained after centrifugation of cell lysates were incubated with protein A-Sepharose preadsorbed with anti-Rab32 antibody. The beads were washed, and bound proteins were subjected to SDS-PAGE and analyzed by Western blotting using affinity-purified rabbit anti-Rab32 and mouse anti-FLAG antibodies. Arrow indicates immunoprecipitated Rab32 proteins. Rab32Q85L migrates slower than other Rab32 proteins on SDS-PAGE. Whereas the biochemical cause of this altered migration is not known, such a situation is similar to that reported in HeLa cells adenovirally infected with HA-rab7Q67L [71]
Autophagy is impaired by the expression of Rab32T39 N and Rab32N143I
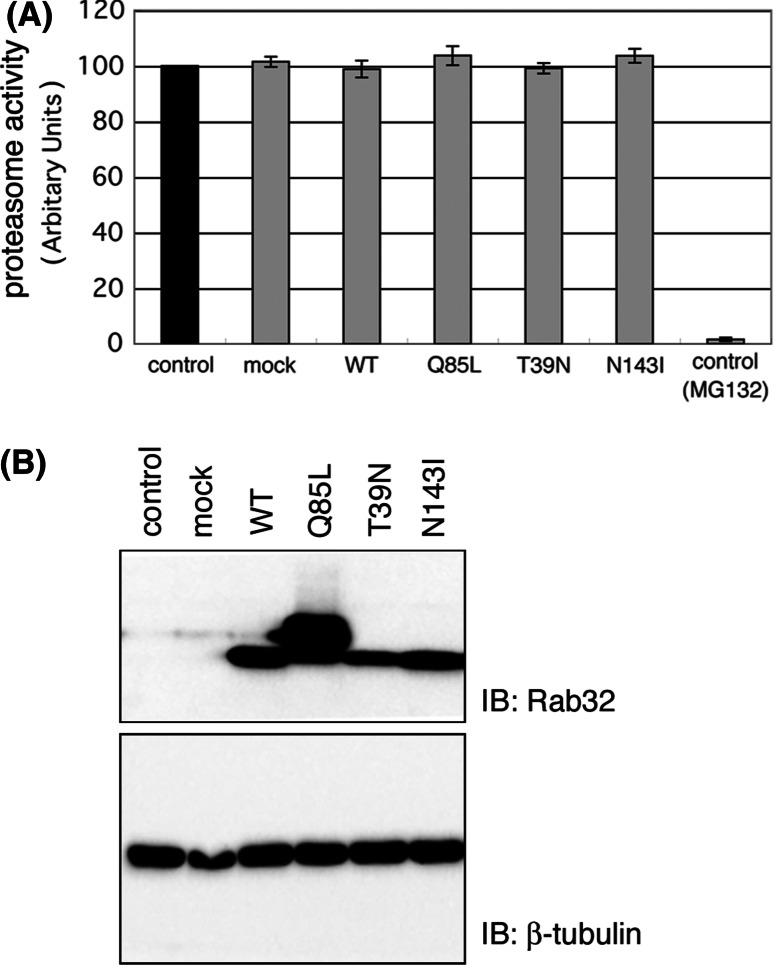
Ubiquitin-proteasome and autophagy-lysosome pathways are the two main routes of protein and organelle clearance in eukaryotic cells [39]. It is conceivable, therefore, that the redistribution of ubiquitinated proteins, Hsc70 and α-synuclein into aggresome-like structure, in both Rab32T39 N- and Rab32N143I-expressing cells, was caused by a defect in the activity of the proteasome degradation pathway and/or autophagic degradation pathway. Therefore, we first investigated whether the formation of aggresome-like structures in Rab32T39 N- and Rab32N143I-expressing cells was caused by the impairment of proteasome activities. When MG132 was used to ensure the specificity of the assays, the activity of proteasome was hardly detected (Fig. 4a). In contrast, there was no significant decrease of proteasome activity in Rab32T39 N- and Rab32N143I-expressing cells as compared to Rab32WT- and Rab32Q85L-expressing cells or untransfected cells, indicating that the accumulation of ubiquitinated proteins in Rab32T39 N- and Rab32N143I-expressing cells is not ascribed to the impairment of proteasome activity. This conclusion is also supported by the result that MG132 inhibited the degradation of Rab32T39 N and Rab32N143I (data not shown).
Fig. 4.
Formation of aggresome-like structures in Rab32T39 N-expressing cells is not caused by proteasome impairment. a The rate of Suc-LLVY-AMC degradation was measured in lysates of COS-1 cells transfected with pcDNA (mock), pcDNARab32WT, pcDNARab32Q85L, pcDNARab32T39 N or pcDNARab32N143I for 48 h. MG132 was used to ensure the specificity of the assays. Relative proteasome activity was expressed in comparison with non-transfected control cells (100% control activity). Fluorescence was determined as described in Materials and Methods using fluorescence spectrophotometry (Ex380/Em460). Data are the means ± SD of six individual experiments. b Western blotting of lysates from pcDNARab32WT-, pcDNARab32Q85L-, pcDNARab32T39 N- or pcDNARab32N143I-transfected COS-1 cells to determine the expression levels of Rab32 in a proteasome activity assay (a); 20 μg of each lysate, equal to the amount used in the proteasome activity assay, was resolved by SDS-PAGE and analyzed by Western blotting with affinity-purified rabbit anti-Rab32 and mouse β-tubulin antibodies
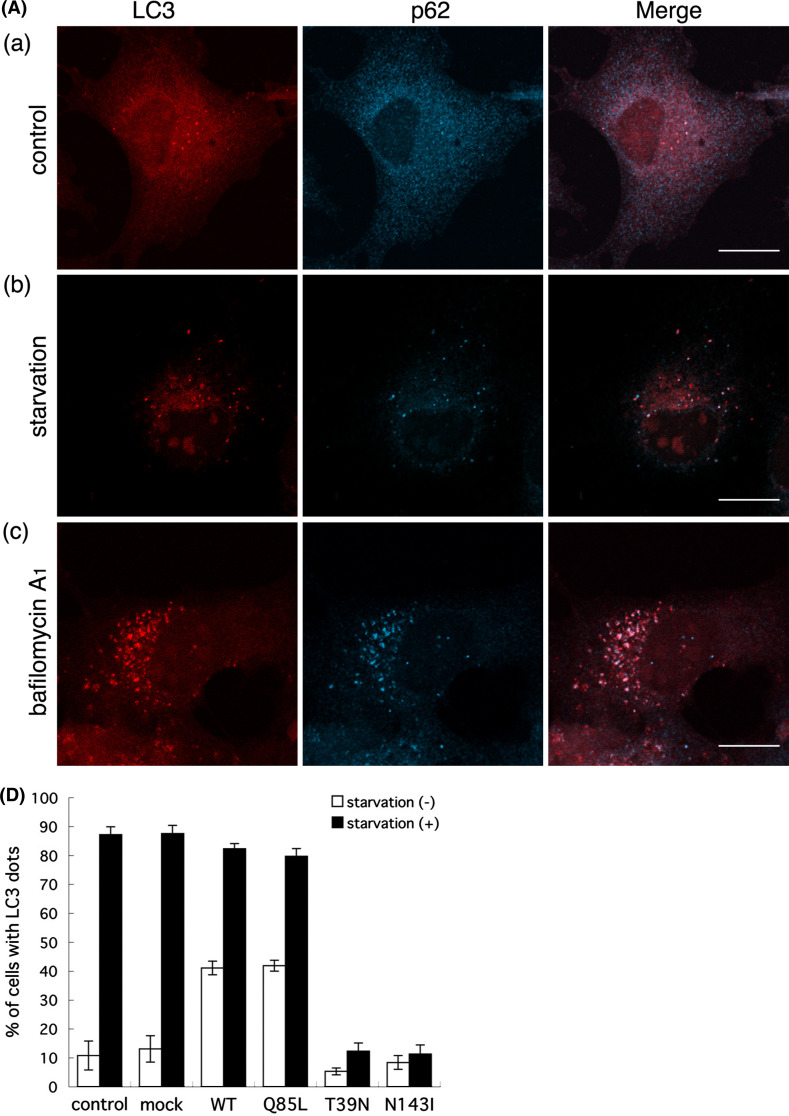
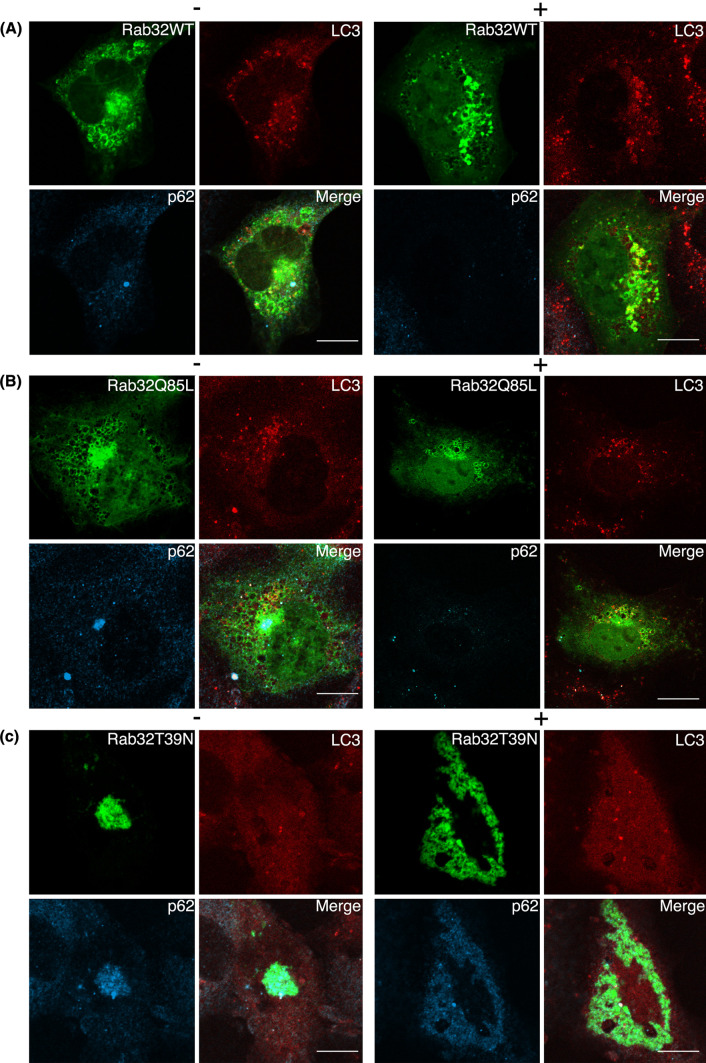
Next, we examined the activity of the autophagy-lysosome pathway in Rab32T39 N- and Rab32N143I-expressing cells by monitoring the disappearance or aggregation of p62/SQSTM1 (sequestosome 1), as well as the formation of LC3-positive structures. p62 is an ubiquitin- and LC3-binding protein [40, 41], and is required for the formation and degradation of ubiquitin-related protein aggregation by autophagy [42]. Because p62 itself is also degraded by autophagy, even at the basal level, deficiency in autophagy results in the accumulation of p62 [43]. As seen in untransfected, Rab32WT- and Rab32Q85L-transfected cells, the induction of autophagy by amino acid starvation led to not only a significant increase in the number of, and fluorescence intensity of LC3-positive dot-like structures (presumably representing autophagosomes), but also the disappearance of diffuse cytoplasmic staining of p62 [Fig. 5A (b), B (b), D, and S6A (b)], indicating the autophagic degradation of p62. Although autophagy is essential for cell survival under starvation conditions, it also has a crucial role in protein degradation under basal constitutive conditions [3–5, 43, 44]. Bafilomycin A1, an H+-ATPase inhibitor [45], blocks autophagosome-lysosome fusion [46], therefore causing the accumulation of not only LC3-positive autophagosomes, but also proteins that are degraded by an autophagic process. LC3 itself is also rapidly degraded by the fusion of autophagosomes with lysosomes [47], which are inhibited by bafilomycin A1 [48]. Indeed, the treatment of untransfected cells, Rab32WT- and Rab32Q85L-transfected cells, with bafilomycin A1 significantly inhibited the fusion of autophagosomes with lysosomes, which proceeds under constitutive conditions, as judged from the accumulation of both LC3 and p62 [Fig. 5A (c), B (c), and S6A (c)]. Interestingly, autophagy was not induced by nutrient deprivation in Rab32T39 N- and Rab32N143I-expressing cells [Fig. 5C (b), D, and S6B (b)]. Rather, the expression of either Rab32T39 N or Rab32N143I induced the massive accumulation of p62 in both Rab32T39 N- and Rab32N143I-positive aggresome-like structures even when cells were cultured with normal medium. Furthermore, cells expressing Rab32T39 N or Rab32N143I did not exhibit LC3-positive dot-like structures when treated with bafilomycin A1, compared with untransfected neighboring cells [Fig. 5C (c), and S6B (c)], thereby suggesting that the expression of Rab32T39 N or Rab32N143I impaired not only starvation-induced, but also basal constitutive autophagy. This assumption was also supported by the accumulation of p62 in Rab32T39 N- and Rab32N143I-expressing cells, although we cannot rule out the possibility that the accumulation of p62 in Rab32T39 N- and Rab32N143I-expressing cells was caused by the sequestration of p62 within aggresome-like structures.
Fig. 5.
Autophagy is impaired by the expression of Rab32T39 N. (A–C) Untransfected COS-1 cells (A), GFP-Rab32WT- (B) or GFP-Rab32T39 N- (C) transfected COS-1 cells were incubated with normal medium (a), Krebs–Henseleit buffer (b) or 100 nM bafilomycin A1-containing normal medium (c) for 2 h. After fixation, cells were stained with affinity-purified rabbit anti-LC3 and mouse anti-p62 antibodies. Merged images were generated by combining GFP-Rab32 (green), LC3 (red) and p62 (cyan). Bars are 10 μm. D Untransfected COS-1 cells or COS-1 cells transfected with the indicated plasmids were incubated with normal medium (starvation: −) (white bars) or Krebs–Henseleit buffer (starvation: +) (black bars) for 2 h. Cells with LC3 dots of more than 20 were defined as cells with LC3 dots, because about 10–20 LC3 dots were occasionally visible even in control cells. At least 200 cells expressing each GFP-Rab32 construct were counted at respective experiments and the percentage of cells exhibiting LC3 dots (i.e., autophagic vacuoles) in each GFP-Rab32-expressing cell is quantified as the mean ± SEM (n = 3 separate experiments)
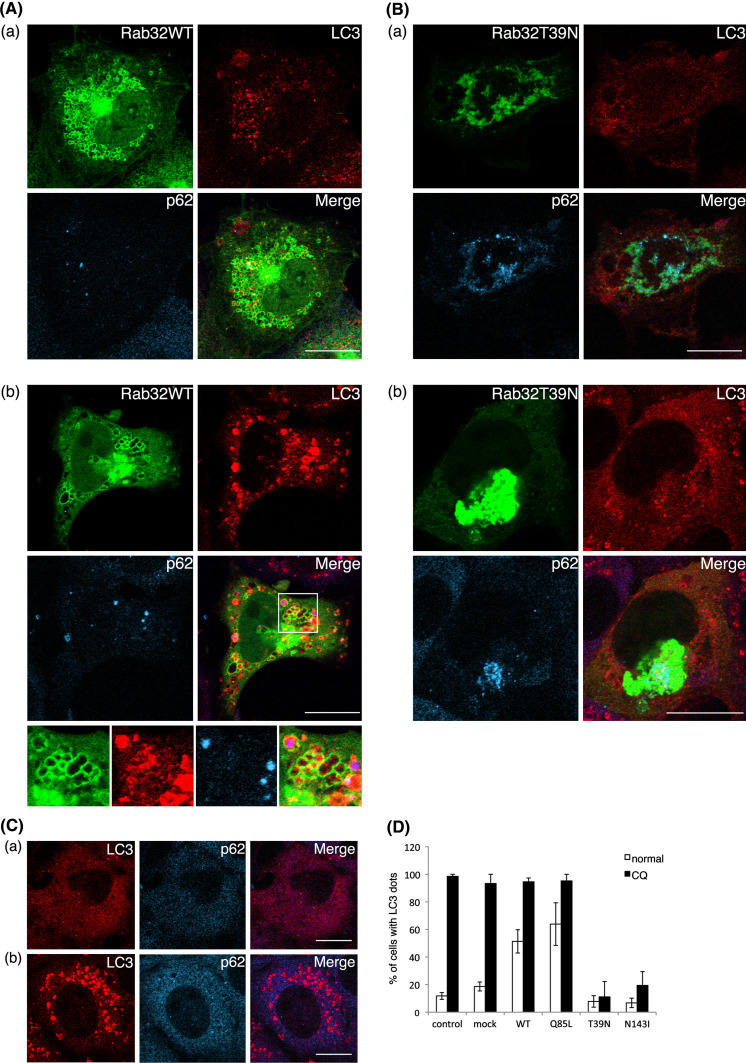
Treatment of rapamycin and chloroquine could not induce the formation of autophagosomes in Rab32T39 N- and Rab32N143I-expressing cells
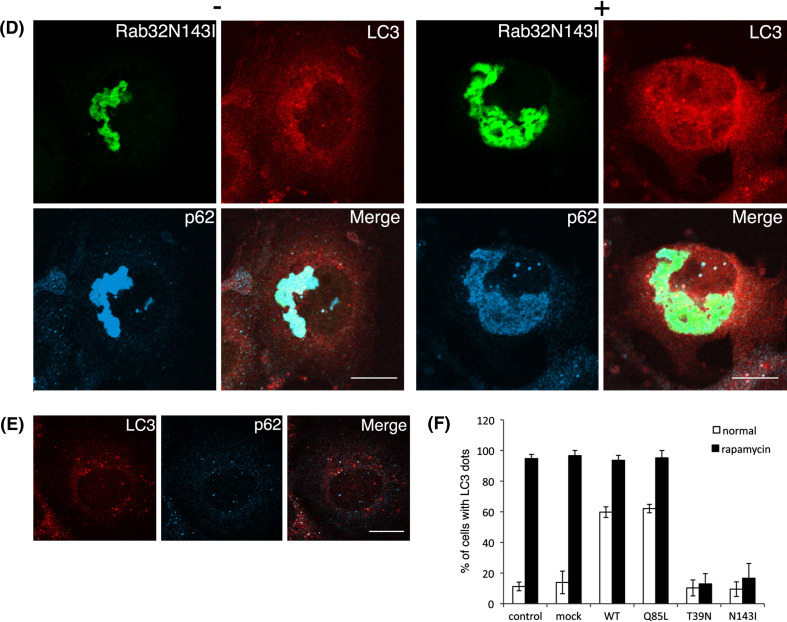
Rapamycin enhances autophagy by inhibiting the mammalian target of rapamycin (mTOR), which negatively regulates this pathway [49, 50]. We then tested whether autophagy could be induced in Rab32T39 N- and Rab32N143I-expressing cells by rapamycin treatment. Rapamycin increased LC3-positive punctate structures in cells expressing Rab32WT or Rab32Q85L, as well as in untransfected cells (Fig. 6a, b and e). In contrast, in Rab32T39 N- and Rab32N143I-expressing cells, most LC3 signals remain to be distributed throughout the cytoplasm (Fig. 6c and d), suggesting that rapamycin could not induce the formation of autophagosomes in Rab32T39 N- and Rab32N143I-expressing cells (Fig. 6f). These results may imply that autophagic vacuole formation in Rab32T39 N- and Rab32N143I-expressing cells was inhibited downstream of the mTOR-dependent autophagic pathway or another pathway. In addition, autophagy can also be induced by treatment with chloroquine [51], which increases both mRNA and protein levels of LC3 [52]. In fact, chloroquine treatment led to a significant increase in the number of LC3-positive punctate structures in non-transfected cells (Fig. 7C and D). By contrast, in Rab32T39 N- and Rab32N143I-expressing cells, most LC3 was distributed throughout the cytoplasm, and few LC3-positive punctate structures were observed (Fig. 7B, D, and S7B). Similar results were obtained with other stimuli of autophagy, such as thapsigargin, ionomycin and ATP [53] (data not shown). Together, these results apparently suggest that the expression of Rab32T39 N or Rab32N143I hampers the formation of autophagic vacuoles by impairing the recruitment of LC3 to, or the generation of, the autophagosomal membrane.
Fig. 6.
Treatment of rapamycin does not induce autophagy in Rab32T39 N-expressing cells. GFP-Rab32WT- (a), GFP-Rab32Q85L- (b), GFP-Rab32T39 N- (c), GFP-Rab32N143I- (d) transfected or untransfected (e) COS-1 cells were incubated with normal medium (−) or 0.2 μM rapamycin-containing normal medium (+) for 24 h. After incubation, cells were fixed and stained with affinity-purified rabbit anti-LC3 and mouse anti-p62 antibodies. Merged images were shown by combining GFP-Rab32 (green), LC3 (red) and p62 (cyan). Bars are 10 μm. e Untransfected COS-1 cells were incubated with 0.2 μM rapamycin-containing normal medium. After fixation, cells were stained with affinity-purified rabbit anti-LC3 and mouse anti-p62 antibodies. Merged images were generated by combining LC3 (red) and p62 (cyan). Bars are 10 μm. f Untransfected COS-1 cells or COS-1 cells transfected with the indicated plasmids were incubated with normal medium (normal) (white bars) or 0.2 μM rapamycin-containing normal medium (rapamycin) (black bars) for 24 h. Cells with LC3 dots of more than 20 were defined as cells with LC3 dots, because about 10–20 LC3 dots were occasionally visible even in control cells. At least 100 cells expressing each GFP-Rab32 construct were counted at respective experiments, and the percentage of cells exhibiting LC3 dots (i.e., autophagic vacuoles) in each GFP-Rab32-expressing cell is quantified as the mean ± SEM (n = 3 separate experiments)
Fig. 7.
Treatment of chloroquine does not induce autophagy in Rab32T39 N-expressing cells. GFP-Rab32WT- (A), GFP-Rab32T39 N- (B) transfected or untransfected (C) COS-1 cells were incubated with normal medium (a) or 20 μM chloroquine-containing normal medium (b) for 1 h. After incubation, cells were fixed and stained with affinity-purified rabbit anti-LC3 (red) and mouse anti-p62 (cyan) antibodies. Merged images were generated by combining Rab32 (green), LC3 (red) and p62 (cyan). The panels at the bottom of A-b show an enlarged view of the boxed regions. Bars are 10 μm. D Untransfected COS-1 cells or COS-1 cells transfected with the indicated plasmids were incubated with normal medium (normal) (white bars) or 20 μM chloroquine-containing normal medium (CQ) (black bars) for 1 h. Cells with LC3 dots of more than 20 were defined as cells with LC3 dots, because about 10–20 LC3 dots were occasionally visible even in control cells. At least 100 cells expressing each GFP-Rab32 construct were counted at respective experiments, and the percentage of cells exhibiting LC3 dots (i.e., autophagic vacuoles) in each GFP-Rab32-expressing cell is quantified as the mean ± SEM (n = 3 separate experiments)
Suppression of Rab32 expression interferes with basal autophagy
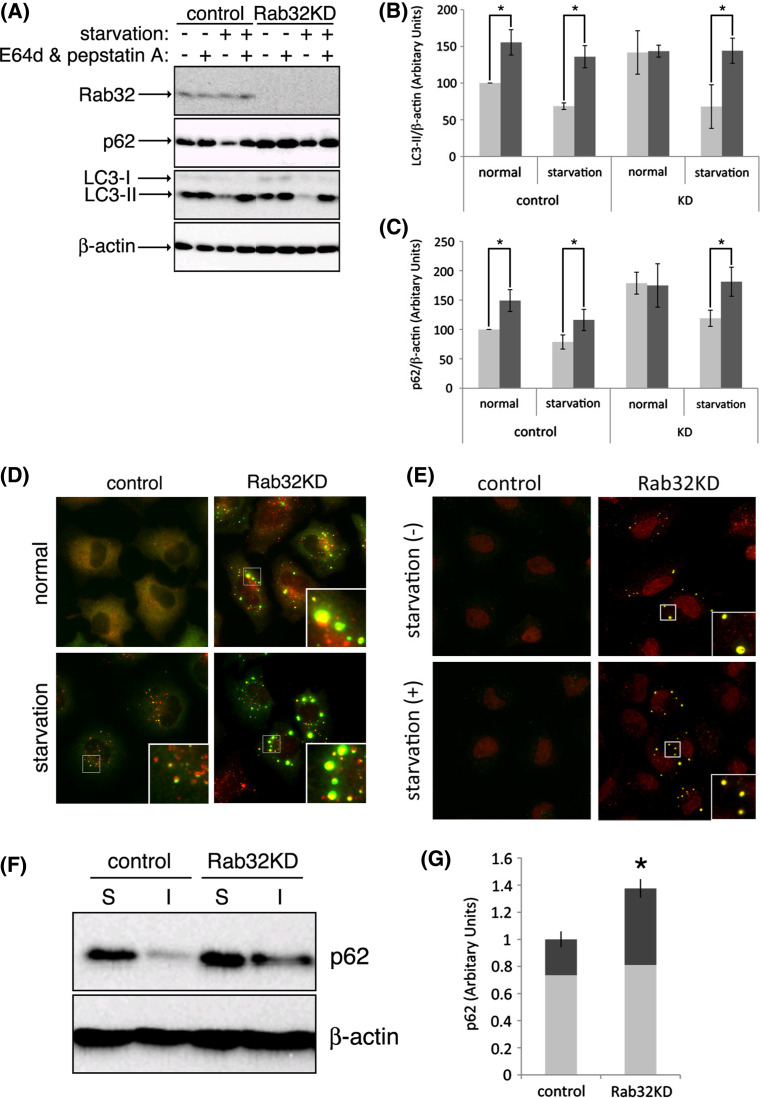
To examine whether Rab32 participates in autophagosome formation, we performed a knockdown experiment using Rab32-specific siRNA. During autophagosome formation, the cytosolic form of LC3 (LC3-I) is conjugated with phosphatidylethanolamine to its C-terminal Gly, producing an autophagosomal membrane-bound form, LC3-II [47]. Although the processing of LC3-I to LC3-II reflects the formation of autophagosomes, LC3-II is rapidly degraded by lysosomal proteases after the fusion of autophagosomes with endosomes/lysosomes. We then measured the effect of the knockdown of Rab32 expression on autophagic activity by monitoring the amount of LC3-II in the presence and absence of lysosomal enzyme inhibitors, E64d and pepstatin A, using Western blot analysis or the formation of LC3-positive vesicular structures using immunofluorescence microscopy. As shown in Fig. 8a, transfection of Rab32-specific siRNA was found to significantly deplete endogenous Rab32 in HeLa cells by Western blot analyses. In cells transfected with control siRNA, amino acid starvation induced autophagy, as judged by the significant decrease of LC3-II and p62 and inhibition of LC3-II and p62 degradation by the supplement of protease inhibitors (Fig. 8a–c).
Fig. 8.
Suppression of Rab32 expression interferes with autophagy. (A–C) HeLa cells were transfected twice with 10 nM of control or Rab32-specific siRNAs at 24-h intervals, and finally kept for 2 h either in normal medium or Krebs–Henseleit buffer (starvation) with (black bars) or without (gray bars) 10 μg/ml E64d and 10 μg/ml pepstatin A. Each cell lysate was subjected to SDS-PAGE and immunoblot analysis to detect endogenous Rab32, p62, LC3 and β-actin (A). Densitometry analysis was performed by normalizing with β-actin levels, using Image J software (NIH) relative to LC3-II (B) or p62 (C) of control siRNA-transfected cells incubated with normal medium, respectively. Data are the means ± SD of five separate experiments, and significance was determined using Student’s t-test. Asterisks indicate significant difference between the values (P < 0.05). D HeLa cells transfected twice with 10 nM of each control or Rab32-specific siRNA at 24-h intervals. Cells were incubated with normal medium (normal) or Krebs–Henseleit buffer (starvation) for 2 h. After fixation, cells were stained with anti-p62 (green) and anti-LC3 (red) antibodies. E HeLa cells transfected twice with 10 nM of each control or Rab32-specific siRNA at 24-h intervals were incubated with normal medium [starvation (−)], Krebs–Henseleit buffer [starvation (+)] and then incubated with 1% Triton X-100 in PBS for 30 min on ice prior to fixation. Fixed cells were stained with mouse anti-p62 (green) and rabbit anti-LC3 (red) antibodies. (F) HeLa cells transfected twice with 10 nM of each control or Rab32-specific siRNA at 24-h intervals were lysed for 30 min on ice with 1% Triton X-100 in PBS and homogenized. After centrifugation, the resulting supernatants were saved as Triton X-100-soluble fractions (S), and the pellets were solubilized in SDS-containing buffer as Triton X-100-insoluble fractions (I). Equivalent amounts of proteins among different S and I fractions were resolved using SDS-PAGE and analyzed for Rab32 solubility by Western blotting. Blots were probed with mouse anti-p62 and mouse anti-β-actin antibodies. (G) Densitometry analysis was performed by normalizing with β-actin levels, using Image J software (NIH), relative to p62 in the soluble fraction (gray) and insoluble fraction (black), respectively. Error bars indicate SD of data from three separate experiments, and significance was determined using Student’s t test. Asterisks indicate significant difference between the values (P < 0.05)
Rab32 knockdown resulted in the starvation-induced degradation of LC3-II and p62, which is sensitive to protease inhibitors treatment, with comparable levels to control siRNA-transfected cells. By contrast, the constitutive degradation of LC3-II and p62 was significantly impaired by the knockdown of Rab32 compared with control siRNA-treated cells (P < 0.05) (“normal” in Fig. 8a–c). These results suggest, therefore, that Rab32 expression may be required for constitutive, but not starvation-induced, autophagy. Notably, Rab32 knockdown caused the appearance of p62-positive large dot-like structures, where LC3 was also colocalized, even under nutrient-rich conditions (Fig. 8d). These dot-like structures morphologically resembled aggresomes rather than autophagosomes, which are frequently observed as ring-like structures, as seen in cells transfected with control siRNA (Fig. 8d, magnifications). Indeed, they were also stained with anti-FK2 antibody, which recognizes ubiquitinated proteins (Fig. S8), and were not significantly increased by starvation (Fig. 8d).
We next examined whether these p62-positive large dot-like structures were aggresome-like compartments. As shown in Fig. 8e, LC3- and p62-positive large dot-like structures in Rab32 siRNA-treated cells were resistant to the treatment of cells with Triton X-100 prior to fixation. Western blot analysis revealed that in Rab32 siRNA-treated cells, about 40% of total amount of p62 was collected into the Triton X-100-insoluble fraction, and that the total amount of p62 in Rab32 siRNA-treated cells was increased to about 1.4-fold of that in control cells (P < 0.05) (Fig. 8f and g). In Rab32 siRNA-treated cells, however, we could not detect Triton X-100-insoluble LC3. Together, these results indicate that the impairment of autophagosome formation by the depletion of Rab32 enhances the formation of aggresomes, in agreement with the results obtained in both Rab32T39 N- and Rab32N143I-expressing cells. We used two kinds of siRNA (siRNA-1 and siRNA-2; see Materials and Methods) for suppression of Rab32 expression and obtained the same results with both siRNAs (data not shown).
Discussion
There is a long-standing debate concerning from where the autophagosomal membrane is derived. So far, two possibilities have been proposed: it arises from pre-existing organelles, such as the ER or Golgi, or from de novo formation [reviewed by Juhasz and Neufeld [54] and references therein]. In this report, we have presented evidence that human Rab32 is predominantly localized to the ER membrane and regulates the formation of autophagic vacuoles, in a nucleotide binding-state-dependent manner. In this case, the formation of autophagic vacuoles apparently depended on the expression levels of Rab32: overexpression of Rab32WT or Rab32Q85L triggered autophagic vacuole formation even in normal culture medium, suggesting that autophagy was activated by Rab32. Notably, Rab32WT- and Rab32Q85L-induced LC3-positive large spherical vesicles contain the ER-resident protein calnexin. Together with the failure of autophagic vacuole formation by the expression of membrane association-deficient mutant (Rab32dCC), we conclude that targeting the Rab32 active form to the ER is prerequisite for the formation of autophagic vacuoles. Alternatively, the active form of Rab32 attached to the ER membrane may be involved in the expansion and completion of autophagosomes by promoting the addition of ER-derived vesicles to a nascent autophagosome membrane.
Rab32 functions as a regulator of melanosomal protein trafficking from the TGN to melanosomes and melanocyte pigmentation, although it is not directly involved in the formation of melanosome [21]. A recent study has further indicated that Xenopus Rab32 is required for melanosome transport as an A-kinase anchoring protein (AKAP) by recruiting cAMP-dependent protein kinase A (PKA) on melanosome membranes [22]. It is noteworthy here that the PKA signaling pathway is also involved in the regulation of autophagy [55–58]. In this context, it is suggested that RIα, which regulates the phosphorylation and activity of mTOR kinase, associates with mTOR on Rab7- and LC3-positive membranes, and promotes the maturation of autophagosomes [59]. Interaction between Rab32 and RIα, however, remains to be elucidated.
Conversely, the expressions of Rab32T39 N and Rab32N143I caused the formation of aggresome-like structures and impaired the formation of LC3-positive autophagic vacuoles even under autophagy-stimulated conditions, such as starvation, rapamycin or chloroquine treatment. This suggests that both Rab32T39 N and Rab32N143I function as dominant negative mutants by sequestering factors involved in the relative early stage of autophagic vacuole formation. Since in addition to the absence of direct interaction between Rab32 and LC3 (data not shown), LC3 in both Rab32T39 N- and Rab32N143I-expressing cells was diffusely distributed throughout the cytoplasm without being sequestrated into aggresome-like structures, it is conceivable that Rab32 participates in autophagosome biogenesis at an early stage; therefore, the molecular mechanism underlying the formation of autophagic vacuoles by Rab32 clearly differs from that by Rab7 and Rab24, which are involved in a relatively late maturation step, i.e., fusion of late autophagic vacuoles and lysosomes [13–15]. It is noteworthy that the expression of mutant Rab24 (Rab24D123I) triggers the accumulation of intracellular inclusion [60, 61], suggesting that Rab24 may participate in autophagic degradation of misfolded cellular proteins.
Aggregation-prone proteins cause many late-onset neurodegenerative diseases [39]. Although many proteins that cause proteinopathies are partly dependent on the ubiquitin-proteasome pathway for their clearance [62, 63], treatment with inhibitors of the autophagy-lysosome pathway or genetic interference that perturbs this pathway also causes the accumulation of aggregated-prone proteins [64–69]. Furthermore, inhibition of autophagy by gene knockout of Atg7 or Atg5 has been shown to cause the accumulation of ubiquitin-positive aggregates in liver hepatocytes or in neural cells, respectively [3, 4]. Therefore, it is now considered that loss of basal autophagy leads to the pathogenesis of neurodegenerative disorders even in disease-associated mutant proteins [3, 5].
Consistent with this notion, our results also show that the knockdown of Rab32 by the expression of Rab32-specific siRNA leads to the formation of aggresome-like structures containing ubiquitinated proteins and p62 under normal growth conditions. Notably, LC3 was also recruited into p62-positive aggresome-like structures by the knockdown of Rab32, which was not inhibited by 3-methyladenine (data not shown). These results can further account for the loss of basal autophagic activity in Rab32 siRNA-treated cells and suggest that LC3 and p62-positive dots induced by Rab32 knockdown are derived from aggresomes, but not autophagic vacuoles. Thus, these results imply the physiological importance of Rab32 in the cellular clearance of aggregated proteins by basal constitutive autophagy. By contrast, in cells suppressed Rab32 expression, amino acid starvation significantly decreased the amount of LC3-II, suggesting that Rab32 does not contribute to starvation-induced autophagy. This was in contrast to the expressions of Rab32T39 N and Rab32N143I, which inhibited not only constitutive autophagy, but also starvation-induced autophagy. One possible explanation for this difference is that the factors required for starvation-induced autophagy were trapped within aggresome-like structures formed by the expression of Rab32T39 N and Rab32N143I. This hypothesis might be supported by more extensive aggregated structures in Rab32T39 N- and Rab32N143I-expressing cells than those in Rab32 siRNA-treated cells. Nevertheless, in contrast to cells transfected with control siRNA upon amino acid starvation, p62 degradation was not significantly induced in Rab32 siRNA-treated cells. Moreover, in this case, part of LC3 appeared in p62-positive aggresome-like dots. Recently, it has been shown that LC3 could be an aggregation-prone protein and is incorporated into aggregates in an autophagy-independent manner [70]. Therefore, although we cannot eliminate the possibility that LC3- and p62-positive dots observed in Rab32 siRNA-treated cells represent autophagy-independent aggresome-like induced structures, as previously described [70], a reasonable explanation is that Rab32 may be specifically required for the autophagic degradation of p62. This hypothesis could account for the elevated levels of p62 in Rab32 siRNA-treated cells. We cannot rule out the possibility, however, that Rab32 participates in autophagy-independent p62 degradation.
The effect of Rab32 on the regulation of autophagy may have important implications for opening up an interesting avenue to explore the origin of autophagosome membranes as well as the molecular mechanism of autophagy. Therefore, the identification of effector molecules and/or interacting proteins should help to shed light on the molecular mechanisms by which Rab32 regulates autophagy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Drs. Ikuo Wada (Fukushima Medical University) and Tomoki Chiba (University of Tsukuba) for providing the ER-cherry and FLAG-tagged ubiquitin constructs, respectively. This study was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Sciences and Technology of Japan. Yuko Hirota is a Research Fellow of the Japan Society for the Promotion of Science.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 3.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 6.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 8.Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn WA. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;15:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 12.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 14.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role of Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 15.Munafó DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 16.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jäger D, Stockert E, Jäger E, Güre AO, Scanlan MJ, Knuth A, Old LJ, Chen Y-T. Serological cloning of a melanocyte rab guanosine 5′-triphosphate-binding protein and a chromosome condensation protein from a melanoma complementary DNA library. Cancer Res. 2000;60:3584–3591. [PubMed] [Google Scholar]
- 18.Fujikawa K, Satoh AK, Kawamura S, Ozaki K. Molecular and functional characterization of a unique Rab protein, RABRP1, containing the WDIAGQE sequence in a GTPase motif. Zool Sci. 2002;19:981–993. doi: 10.2108/zsj.19.981. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu F, Katagiri T, Suzuki M, Watanabe TK, Okuno S, Kuga Y, Nagata M, Fujiwara T, Nakamura Y, Takahashi E. Cloning and chromosome assignment to 1q32 of a human cDNA (RAB7L1) encoding a small GTP-binding protein, a member of the RAS superfamily Cytogenet. Cell Genet. 1997;77:261–263. doi: 10.1159/000134591. [DOI] [PubMed] [Google Scholar]
- 20.Norian L, Dragoi IA, O’Halloran T. Molecular characterization of rabE, developmentally regulated dictyostelium homolog of mammalian rab GTPases. DNA Cell Biol. 1999;8:59–64. doi: 10.1089/104454999315628. [DOI] [PubMed] [Google Scholar]
- 21.Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, Seabra MC. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park M, Serpinskaya AS, Papalopulu N, Gelfand VI. Rab32 regulates melanosome transport in Xenopus melanophores by protein kinase A recruitment. Curr Biol. 2007;17:1–5. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao X, Faris AE, Jang EK, Haslam RJ. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur J Biochem. 2002;269:259–271. doi: 10.1046/j.0014-2956.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, Cho JM, Seol JH, Baek SH, Kim KI, Chiba T, Tanaka K, Bang OS, Chung CH. SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat Cell Biol. 2006;8:1424–1431. doi: 10.1038/ncb1512. [DOI] [PubMed] [Google Scholar]
- 25.Hirota Y, Kuronita T, Fujita H, Tanaka Y. A role for Rab5 activity in the biogenesis of endosomal and lysosomal compartments. Biochem Biophys Res Commun. 2007;364:40–47. doi: 10.1016/j.bbrc.2007.09.089. [DOI] [PubMed] [Google Scholar]
- 26.Asanuma K, Tanida I, Shirato I, Ueno T, Takahara H, Nishitani T, Kominami E, Tomino Y. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. FASEB J. 2003;17:1165–1167. doi: 10.1096/fj.02-0580fje. [DOI] [PubMed] [Google Scholar]
- 27.Saliba RS, Munro PMG, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;15:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Tan JMM, Ho MWL, Zaiden N, Wong SH, Chew CLC, Eng PW, Lim TM, Dawson TM, Lim KL. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson’s disease-linked point mutations. J Neurochem. 2005;93:422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Mosser DD, Bag J. Induction of HSP70 expression and recruitment of HSC70 in the nucleus reduce aggregation of a polyalanine expansion mutant of PABPN1 in HeLa cells. Hum Mol Genet. 2005;14:3673–3684. doi: 10.1093/hmg/ddi395. [DOI] [PubMed] [Google Scholar]
- 30.Sawada MT, Morinaga C, Izumi K, Sawada H. The 26S proteasome assembly is regulated by a maturation-inducing hormone in Starfish Oocytes. Biochem Biophys Res Commun. 1999;254:338–344. doi: 10.1006/bbrc.1998.9948. [DOI] [PubMed] [Google Scholar]
- 31.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alto NM, Soderling J, Scott JD. Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardley HC, Scott GB, Rose SA, Tan NGS, Robinson PA. UCH-L1 aggresome formation in response to proteasome impairment indicates a role in inclusion formation in Parkinson’s disease. J Neurochem. 2004;90:379–391. doi: 10.1111/j.1471-4159.2004.02485.x. [DOI] [PubMed] [Google Scholar]
- 34.Namekata K, Nishimura N, Kimura H. Presenilin-binding protein forms aggresomes in monkey kidney COS-7 cells. J Neurochem. 2002;82:819–827. doi: 10.1046/j.1471-4159.2002.01039.x. [DOI] [PubMed] [Google Scholar]
- 35.Ardley HC, Scott GB, Rose SA, Tan NGS, Markham AF, Robinson PA. Inhibition of proteasomal activity causes inclusion formation in neuronal and non-neuronal cells overexpressing parkin. Mol Biol Cell. 2003;14:4541–4556. doi: 10.1091/mbc.E03-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu L, Gao Y, Tousson A, Shah A, Chen T-LL, Vertel BB, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of parkin. Cell. 2001;105:891–902. doi: 10.1016/S0092-8674(01)00407-X. [DOI] [PubMed] [Google Scholar]
- 39.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 40.Komatsu M, Waguri S, Koike M, Sou Y, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamasaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 41.Pankiv S, Høyvarde T, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p/62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 42.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 44.Ogier-Denis E, Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochim Biophys Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- 45.Gagliardi S, Gatti PA, Belfiore P, Zocchetti A, Clarke GD, Farina C. Synthesis and structure-activity relationships of bafilomycin A1 derivatives as inhibitors of vacuolar H+-ATPase. J Med Chem. 1998;41:1883–1893. doi: 10.1021/jm9707838. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 47.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:e2–e9. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, Yuan J. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blommaart EFC, Luiken JJFP, Blommaart PJE, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 50.Codogono P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki T, Nakagawa M, Yoshikawa A, Sasagawa N, Yoshimori T, Ohsumi Y, Nishino I, Ishiura S, Nonaka I. The first molecular evidence that autophagy relates rimmed vacuole formation in chloroquine myopathy. J Biochem. 2002;131:647–651. doi: 10.1093/oxfordjournals.jbchem.a003147. [DOI] [PubMed] [Google Scholar]
- 52.Kimura N, Kumamoto T, Kawamura Y, Himeno T, Nakamura K, Ueyama H, Arakawa R. Expression of autophagy-associated genes in skeletal muscle: an experimental model of chloroquine-induced myopathy. Pathobiology. 2007;72:169–176. doi: 10.1159/000103376. [DOI] [PubMed] [Google Scholar]
- 53.Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jäättelä M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of autophagy process in Saccharomyces cerevisiae . J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mavrakis M, Lippincott-Schwartz J, Stratakis CA, Bossis I. Depletion of type IA regulatory subunit (RIα) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum Mol Genet. 2006;15:2962–2971. doi: 10.1093/hmg/ddl239. [DOI] [PubMed] [Google Scholar]
- 57.Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae . Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mavrakis M, Lippincott-Schwartz J, Stratakis CA, Bossis I. mTOR kinase and the regulatory subunit of protein kinase A (PRKAR1A) spatially and functionally interact during autophagosome maturation. Autophagy. 2007;3:151–153. doi: 10.4161/auto.3632. [DOI] [PubMed] [Google Scholar]
- 60.Maltese W, Soule G, Gunning W, Calomeni E, Alexander B. Mutant Rab24 GTPase is targeted to nuclear inclusions. BMC Cell Biol. 2002;3:25. doi: 10.1186/1471-2121-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu M, Yin G, Zhao X, Ji C, Gu S, Tang R, Dong H, Xie Y, Mao Y. Human Rab24, interestingly and predominantly distributed in the nuclei of COS-7 cells, is colocalized with cyclophilin A and GABARAP. Int J Mol Med. 2006;17:749–754. [PubMed] [Google Scholar]
- 62.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb JL, Ravikumar B, Atkins J, Skeppers JN, Rubinsztein DC. α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 64.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 65.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qin Z-H, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, Yoder J, Aronin N, DiFiglia M. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12:3231–3244. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- 67.Ravikumar B, Duden R, Rubinsztei DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 68.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntingtin disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 69.Shibata D, Mori Y, Cai K, Zhang L, Yin J, Elahi A, Hamelin R, Wong YF, Lo WK, Chung TKH, Sato F, Karpeh MS, Jr, Meltzer SJ. RAB32 hypermethylation and microsatellite instability in gastric and endometrial adenocarcinomas. Int J Cancer. 2006;119:801–806. doi: 10.1002/ijc.21912. [DOI] [PubMed] [Google Scholar]
- 70.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 71.Ceresa BP, Bahr SJ. Rab7 activity affects epidermal growth factor: epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem. 2006;281:1099–1106. doi: 10.1074/jbc.M504175200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.