Abstract
Eukaryotic genomes are replicated with high fidelity to assure the faithful transmission of genetic information from one generation to the next. The accuracy of replication relies heavily on the ability of replicative DNA polymerases to efficiently select correct nucleotides for the polymerization reaction and, using their intrinsic exonuclease activities, to excise mistakenly incorporated nucleotides. Cells also possess a variety of specialized DNA polymerases that, by a process called translesion DNA synthesis (TLS), help overcome replication blocks when unrepaired DNA lesions stall the replication machinery. This review considers the properties of the Y-family (a subset of specialized DNA polymerases) and their roles in modulating spontaneous and genotoxic-induced mutations in mammals. We also review recent insights into the molecular mechanisms that regulate PCNA monoubiquitination and DNA polymerase switching during TLS and discuss the potential of using Y-family DNA polymerases as novel targets for cancer prevention and therapy.
Keywords: Y-family polymerase, PCNA, Replication foci, Translesion DNA synthesis, Mutagenesis, Polymerase switching, Somatic hypermutation
Introduction
The genomes of all living cells are continuously under attack by a variety of endogenous and exogenous genotoxic agents [1]. Multiple DNA repair pathways can remove the majority of DNA lesions [1]. However, some may escape the cellular repair machinery and persist during S-phase, creating the potential of blocked DNA replication and threatening the viability of dividing cells. Eukaryotic cells have evolved multiple strategies for mitigating the lethal effects of arrested DNA replication without prior removal of the offending DNA damage: so-called DNA damage tolerance [2]. Translesion DNA synthesis (TLS) is a mode of DNA damage tolerance that utilizes specialized low-fidelity DNA polymerases to replicate across sites of DNA damage, hence generating mutations. Eukaryotic cells, in particular those from higher eukaryotes, are endowed with multiple such enzymes that can catalyze DNA synthesis past sites of base damage in vitro. These enzymes are devoid of 3′ → 5′ proofreading exonuclease activity and replicate undamaged DNA in vitro with low fidelity and weak processivity [3]. It has been suggested that TLS in eukaryotes may sometimes require the sequential action of two specialized polymerases: an “inserter” and an “extender”. The inserter polymerase is thought to efficiently insert (correct or incorrect) nucleotides directly opposite the arresting lesion, while the extender polymerase is believed to incorporate further nucleotides downstream of the lesion [3].
Some specialized DNA polymerases belong to a novel protein family called the Y-family [4]. These enzymes possess a spacious active site that can physically accommodate a variety of DNA lesions and facilitate their bypass [5]. Members of the Y-family in higher eukaryotic cells include Polκ, Polι, Polη and REV1 [3]. In this article we review the properties of the Y-family of DNA polymerases and their roles in modulating spontaneous and genotoxic-induced mutations in mammals. We also review recent insights into the molecular mechanisms that regulate PCNA monoubiquitination and DNA polymerase switching during TLS. Finally, we discuss the potential for identifying Y-family polymerases as novel targets for cancer prevention and therapy. Additional information on the properties and structures of Y-family DNA polymerases can be found in several recent reviews [3, 5].
Overview of the Y-family of DNA polymerases
Polη
DNA polymerase eta (Polη) encoded by the Polh gene is specifically required for the accurate replicative bypass of cyclobutane pyrimidine dimers (CPDs) in DNA generated by the exposure of mammalian cells to ultraviolet (UV) radiation [1, 6, 7]. Humans (and mice) defective for Polη manifest the symptoms and signs of the skin cancer-prone disease xeroderma pigmentosum (XP). The control of Polη function is presumably highly regulated, since the enzyme replicates undamaged DNA with an error rate of 10−2 to 10−3 and can presumably exhibit highly mutagenic functions if not appropriately controlled [3].
Polh gene expression and regulation
The Polh gene (called Rad30 in yeast and XPV in humans) was originally identified in the yeast Saccharomyces cerevisiae by its sequence homology to the UmuC homologs DinB (PolIV) and REV1 [8, 9]. Polh is exclusively found in eukaryotic organisms, with homologs identified in both vertebrate and invertebrate species. The human Polh gene maps to chromosome 6p21.1 [10]. Human Polh mRNA transcripts are detected in most tissues, but are particularly low or undetectable in peripheral lymphocytes, fetal spleen and adult muscle [11]. The human Polh gene undergoes alternative splicing, most prominently in the testis and fetal liver where exon II is frequently spliced out. Since exon II is the first coding exon of Polh containing the ATG start site, an out of frame protein product is generated from this transcript that encodes a non-functional protein that is unable to complement the UV-sensitivity of XP-V cells [11].
In the yeast S. cerevisiae two sequences indicative of DNA damage-inducibility have been identified in the promotor region of the Rad30 gene. Independent observations agree that the transcript of Rad30 is induced ~3.5-fold in response to DNA damage by UV-irradiation [8, 12]. However, recent reports are contradictory as to whether or not RAD30 protein levels are increased/stabilized in response to UV-irradiation [12, 13]. In contrast to the yeast homolog, induction of mammalian Polh transcripts in response to UV-irradiation has not been observed, although in human cells DNA damage caused by camptothecin (an inhibitor of topoisomerase I that causes double strands breaks) induces the up-regulation of Polh expression in a p53-dependent manner [14]. Indeed, the promotor of Polh contains a p53 response element that can be bound and activated by p53, implicating Polh as a target of p53. However, since induction of Polh is not observed following exposure to UV-irradiation, it is unlikely that p53 has a significant functional role in TLS by Polh. Given that p53-defective cells are proficient for TLS, basal levels of Polη protein are apparently sufficient for its function in the bypass of CPDs [14–16].
Polη enzymatic activity
In yeast, Drosophila, humans, and mice Polη has been shown to replicate past CPDs accurately and efficiently [3]. The incorporation of an A opposite the 3′T and the 5′T of the dimer occurs with nearly the same efficiency and fidelity as opposite the two undamaged Ts [3]. Crystallographic analysis of yeast Polη reveals that Polη lacks the helices “O” and “O1” in the fingers domain and the distinctly open active site of Polη can accommodate both template nucleotides of a CPD [17]. Yeast and human Polη can also replicate DNA containing 7,8-dihydro-8-oxoguanine (8-oxo-G) efficiently and accurately in vitro by inserting C across these lesions and proficiently extending from this base pair [18, 19]. Consistent with in vitro findings, a recent paper from Pfeifer’s lab provides evidence that Polη prevents error-prone bypass of this lesion in human skin fibroblasts [20], although evidence from two other groups indicates that Polη is not essential for the bypass of 8-oxo-G in mammalian cells [21, 22]. Moreover, Polη efficiently replicates past 6O-methyl guanine (m6G) lesions, oxaliplatin and cisplatin GpG adducts in vitro [3, 23]. Data from human cells supports a role of Polη in error-free TLS across cisplatin GpG lesions [24, 25]. Additionally, Polη is remarkably error-prone when bypassing benzo[a]pyrene 7,8-diol 9,10-epoxide (BPDE) deoxyguanosine adducts (BPDE-dG) in vitro, preferring to misincorporate G and A at frequencies 3- to more than 50-fold greater than that for T or the correct base C [26]. In agreement with this, human Polh knockdown cells exhibit decreased mutagenic TLS across BPDE-dG lesions [24].
Polη protein–protein interactions and mechanisms for recruiting Polη to stalled replication foci
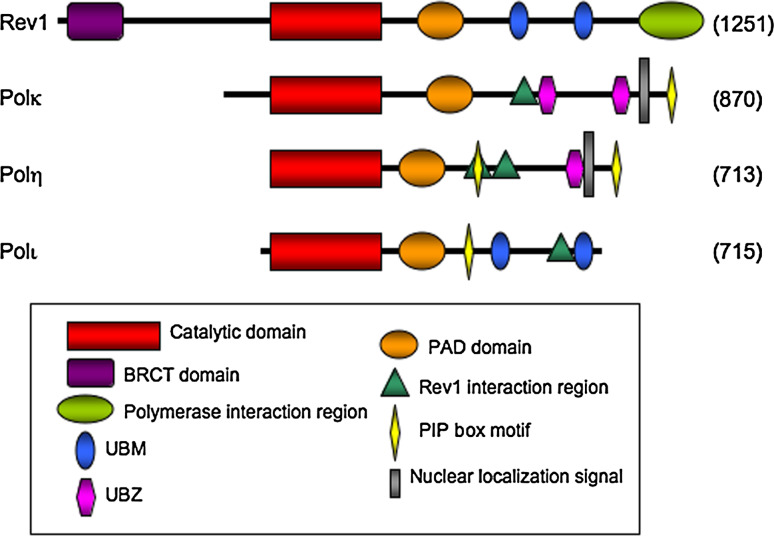
Significant progress has been made toward understanding the molecular mechanisms by which Polη functions during TLS. Mammalian Polη possesses a polymerase catalytic domain located in its N terminus [27, 28] (Fig. 1). A ubiquitin-binding zinc finger domain (UBZ) responsible for mediating an interaction with monoubiquitinated PCNA as well as the monoubiquitination of Polη is located near the C terminus of Polη [29]. Two REV1-binding domains have been identified in human Polη comprising amino acid residues 509–557 and 369–491, respectively [30, 31]. Recently, it was found that two consecutive phenylalanines (483–4FF and 531–2FF) of human Polη are crucial for the Polη–REV1 interaction and for REV1-dependent suppression of spontaneous mutations by Polη [32, 33]. Notably, this function is apparently not conserved in yeast, where an in vivo interaction between REV1 and Polη is not observed [34]. Moreover, a consensus PCNA-interaction peptide (PIP) sequence is located at the extreme C terminus of Polη [35]. A second PIP-like domain was recently identified upstream of the UBZ domain and is required for Polη function in TLS in human cells [36]. Interestingly, the second PIP-like domain is embedded in one of the REV1-binding motifs of Polη (Fig. 1).
Fig. 1.
The structural domains of the Y-family polymerases. Protein size is represented proportionately. BRCT BRCA1 C terminus-like domain, UBM ubiquitin binding motif, UBZ ubiquitin binding zinc finger motif, PAD polymerase associated domain, NLS nuclear localization signal, PIP PCNA interaction peptide
Polη interacts with PCNA and is recruited to replication foci in response to DNA damage caused by UV-irradiation [29, 37–39]. The C terminus of Polη (which includes the PCNA interaction domain) is required for TLS by Polη [28, 35, 40]. Polη manifests increased affinity for monoubiquitinated PCNA [40, 41], a process mediated by the UBZ domain of Polη [29]. The precise mechanistic significance of this interaction, observed both in vivo and in vitro, is not presently known. Zhuang et al. [42] demonstrated that monoubiquitinated PCNA is required for Polη to take over synthesis from stalled Polδ on primed single-stranded M13 circular DNA. Monoubiquitinated PCNA was also reported to stimulate Polη activity in TLS [43], although a conflicting result was obtained by another group using different in vitro primer extension conditions [44].
Studies on the requirements for the localization of Polη in replication foci has further contributed to our understanding of how this DNA polymerase is utilized in cells. Notably, the localization of Polη in such foci appears to be critical for its function, since mutant forms of the protein that do not form foci cannot complement XP-V cells [28]. The UBZs of Polη are essential for the accumulation of Polη into foci [29] and DNA damage-induced focus formation by Polη is dependent upon Rad18 protein [41]. Although these observations implicate monoubiquitinated PCNA as a likely effector in the localization of Polη, a recent report shows that monoubiquitinated PCNA facilitates, but is not essential for the accumulation of Polη in foci [45]. Surprisingly, another study identified some Polη UBZ mutants (H650A, C635A) that are unimpaired for TLS in human cells as determined by foci formation and UV-sensitivity [46]. These data suggest that other UBZ mutants (D652A, H654A, and F655A) may suffer compromised activity due to the loss of an (unknown) function other than the interaction with monoubiquitinated PCNA [46].
Other possible mechanisms may operate for regulating access of Polη to sites of stalled replication. One such mechanism is suggested by the recent demonstration that human Polη is phosphorylated in response to UV-irradiation, and that site-specific mutagenesis of two putative PKC phosphorylation sites prevents the formation of Polη nuclear foci induced by UV-irradiation or other DNA-damaging agents [47]. Another proposed mechanism for the recruitment of Polη to sites of stalled DNA replication may involve interaction with the Y-family polymerase REV1 in mammalian cells [30, 31, 48]. Cellular localization studies demonstrate that REV1 is present with Polη in replication foci and is tightly associated with nuclear structures. These observations suggest that REV1 acts as a scaffold for Polη at sites of stalled replication [30, 48]. Another possible mechanism for the regulation of Polη localization involves p21 protein as a negative regulator of the interaction between Polη and PCNA. p21 is proteolytically degraded when cells are exposed to UV-irradiation; however, when p21 is not degraded it inhibits the interaction of Polη with PCNA and consequently impairs the assembly of Polη in replication foci following exposure to UV-irradiation [49]. Recently, it was found that Polη co-localized with WRN protein (a DNA helicase–exonuclease which is implicated in Werner’s syndrome) in replication foci in response to UV-C radiation. However, foci formation of Polη does not require WRN, and vice versa [50]. Notably, WRN stimulates the polymerase and lesion bypass activity of Polη in vitro [50]. In addition, WRN increases the mutation frequency of Polη without altering its mutation spectra [50]. The functional interaction between WRN and TLS Pols may promote replication fork progression and suppress recombination events at stalled forks, at the expense of increased mutagenesis. Finally, analysis of the subcellular localization of Polη using high-resolution confocal microscopy reveals that the protein is highly mobile within the nucleus, even within individual foci, suggesting that multiple factors may play a role in facilitating Polη localization and function [45].
Polη function(s) in vivo
As noted above, the biological significance of Polη in the bypass of UV radiation-induced lesions is evident from the manifestation of disease in mice and humans lacking normal Polη protein [3, 51–54] (Table 1). Mutations in the Polh gene result in a variant form of the human genetic disorder xeroderma pigmentosum (XP-V), a disease characterized by extreme sunlight sensitivity and an early predisposition to skin cancer [55]. It has been proposed that in the absence of functional Polη another specialized polymerase(s) bypasses CPDs with reduced efficiency and accuracy, resulting in an increased frequency of UV-induced mutagenesis and hence carcinogenesis in XP-V cells [56]. This notion is supported by observations suggesting that the elevated UV-induced mutation frequency in XP-V cells is due to the activity of the highly error-prone Y-family polymerase iota (Polι) [57, 58], which can replicate past CPDs by incorporating G or T opposite the 3′ nucleotide base [59]. The specific requirement for Polη for the replicative bypass of a particular class of DNA damage (CPD in this case) prompts conjecture that each specialized DNA polymerase possesses a “cognate” substrate for which it evolved to bypass accurately.
Table 1.
The functions of Y-family polymerases and their knockout mice phenotypes
| Gene | Protein | Repair pathway | Mutation | Relevant phenotype | References |
|---|---|---|---|---|---|
| Polh | Polη | TLS, HR, SHM | KO | Mice viable and fertile; altered mutational spectrum in Ig genes; mice are prone to skin cancer following exposure to UV radiation | [51–53] |
| Poli | Polι | TLS, BER | Naturally occurring mutation in strain 129 mice | Mice are viable and fertile | [74] |
| REV1 | REV1 | TLS, SHM | KO | Mice display transient growth retardation; strand-biased defect in C/G transversions in hypermutating Ig genes | [130] |
| Polk | Polκ | TLS, NER | KO | Mice are viable and fertile; MEFs sensitive to UV radiation; ES cells sensitive to UV radiation and to B[a]P | [157, 158] |
Modified from reference [54]
If the evolution of Polη was indeed driven by its selection for bypassing UV-induced CPDs during DNA replication, why is it expressed in mammalian tissues that are never exposed to UV-light? The answer to this question may lie in the observed participation of Polη in other aspects of DNA metabolism. In particular, some specialized DNA polymerases have been implicated in the diversification of immunoglobulin (Ig) genes at different stages of the B-cell differentiation pathway, a process known as somatic hypermutation (SHM). Given the inherent low-fidelity and weak processivity of specialized DNA polymerases on undamaged DNA, these proteins are prime candidates for the generation of mutations during SHM. Indeed, Polθ (an A-family TLS polymerase) and Polζ (a B-family TLS polymerase) have been reported to play significant roles in the overall SHM process [60–62]. Several studies also demonstrated that Polη is an A → T mutator during SHM of Ig genes [63, 64]. More recently, Delbos et al. used double MSH2 −/− Polh −/− knockout mice to show that the residual A → T mutagenesis observed in MSH2 −/− mice is contributed solely by Polη [65]. Additionally, decreased levels of Ig gene conversion as well as a reduction in double-strand break-induced homologous recombination (HR) are observed in chicken DT40 cells lacking Polη [66, 67].
Polη is also implicated in the reinitiation of DNA synthesis by HR, presumably at sites of replication fork collapse where double strand breaks may ensue. McIlwraith et al. [68] reported that Polη synthesizes DNA from D-loop recombination intermediates where an invading strand serves as the primer, and showed that cells lacking functional Polη exhibit severely reduced D-loop extension activity [68]. Furthermore, Rad51 recombinase interacts with Polη, and Rad51 stimulates D-loop extension by Polη, observations not made for Polδ or Polι. However, XP-V cells do not manifest defects in double strand break repair, nor does Polη form foci in response to IR suggesting that the role of Polη may be limited to replication intermediates [68].
Polι
Poli, also called Rad30b [1], is a paralog of Polh, with homologs identified in humans, mice and fruit flies [9, 69–71]. The human Poli gene maps to chromosome 18q21.1 [9]. Unlike other Y-family DNA polymerases, structural homologs of Poli have not been identified in bacteria, yeast, or nematodes. Polι exhibits a catalytic function that is likely exercised during TLS in vivo [71, 72]. The in vitro bypass properties of Polι are highly error-prone, with the exception of Drosophila Polι, which exhibits catalytic efficiency and accuracy for CPDs strikingly similar to that of mouse and human Polη [7, 69, 73, 74]. Collectively, these observations lead to the speculation that Poli may have resulted from genetic duplication of the Polh gene shortly before the evolutionary appearance of insects, and that the mammalian homolog of Poli was subjected to evolutionary pressures that altered its biological function(s) [72].
Poli gene expression and regulation
The Poli gene encodes five conserved N-terminal motifs characteristic of all Y-family polymerases. The polymerase active site responsible for the unique catalytic activity of Poli is contained in this region [27] (Fig. 1). A canonical PIP box motif, a peptide that mediates the interaction of Polι with PCNA, is located downstream of this conserved region [75]. Two ubiquitin binding motifs (UBMs) near the C terminus of Polι are required for the interaction of Polι with monoubiquitinated PCNA and with ubiquitin moieties [29].
Tissue-specific expression profiling reveals that human Poli gene is ubiquitously expressed in various adult tissues [76]. Both mouse and human Poli are highly expressed in the testis [9, 74, 76]. Mouse Poli expression in the testis is restricted to post-meiotic round spermatids, indicating a potential role in spermatogenesis [9]. Furthermore, expression of Poli in mammalian cells exhibits strain-specific properties [74]. In contrast to C57BL/6J, BALB/C, and ICR Swiss strains of mice, sequencing or genotypic analysis of genomic DNA from several strains of 129 mice reveals a homozygous nonsense mutation located in codon 27 of exon 2 of Poli, resulting in a truncated protein lacking catalytic function [74]. Furthermore, different alternative splice variants of Poli have been identified in various mouse strains as well as in human cells [57, 77, 78]. Remarkably, extensive differences in sequence conservation between various mouse strains have also been observed. For example, the sequences of Poli in BALB/cJ and A/J mice differ by 25 nucleotide polymorphisms in the coding region accompanied by ten amino acid alterations [77]. Similarly, sequencing Poli cDNAs in the lungs of BALB/cByJ, C57BL/6J, A/J and C3H/HeJ mice revealed 21 BALB/cByJ-specific single-nucleotide polymorphisms (SNPs) in the coding region, as well as seven amino-acid substitutions [78]. Understanding the tissue-specific role of Poli alternative splice variants and the functional contribution of sequence variations of Poli in mice and humans will require further investigation.
Polι enzymatic activity
The function of Polι in mammalian TLS has been primarily characterized in vitro, where the enzyme catalyzes highly error-prone TLS on undamaged or damaged templates, incorporating dGMP opposite thymine 3–10 times more frequently than dAMP in a manner that violates Watson–Crick base pairing [71, 79, 80]. Polι differs most strikingly from replicative DNA polymerases (as well as other Y-family polymerases) in its much higher proficiency and fidelity for nucleotide incorporation opposite template purines than opposite template pyrimidines [71, 79, 80]. Structural analysis of the active site of human Polι reveals that this enzyme uniquely utilizes a Hoogsteen base pairing mechanism for efficient nucleotide incorporation opposite adducted or unadducted purines [81, 82], and the template base is driven to the syn conformation by the incoming dNTP [83]. Furthermore, Polι can support insertion events opposite highly distorting or non-instructional lesions in vitro, such as [6–4] photoproducts and abasic sites, although this DNA polymerase apparently does not have the capacity for extension beyond these insertions [59, 80]. Rather, data suggest that Polι may function in conjunction with another specialized DNA polymerase with extension activity, such as Polζ [80, 84].
It has been reported that the processivity of Polι is enhanced by PCNA in a template-dependent manner [75]. However, these results have been contradicted in a study claiming that the processivity of Polι is not enhanced upon PCNA binding [85]. Rather, the efficiency of nucleotide incorporation (via a reduction in K m) by Polι is improved [85].
Polι binding partners and recruitment to stalled replication foci
Polι co-localizes with the DNA replication machinery in response to UV-irradiation in vivo [86]. Details of the mechanism by which Polι functions in response to UV radiation have surfaced by the identification of several factors affecting the regulation and recruitment of Polι to sites of UV-induced DNA damage. For example, Polι interacts with the C-terminal domain of REV1, and in response to UV-irradiation, Polι co-localizes with REV1 bound to nuclear structures in foci representative of replication factories [30, 31, 48]. A model for DNA polymerase switching at sites of stalled replication implicates a role for REV1 in facilitating a “switch” that likely ensues between Polι and other polymerases such as Polζ [56, 87]. Additionally, Polι interacts with PCNA via a conserved PIP box, an interaction that does not require PCNA to be bound to DNA [85]. A Polι-PIP box mutant fails to accumulate in replication foci, suggesting the importance of the PIP box in recruiting Polι to UV radiation-induced DNA damage [75]. Additionally, Bienko et al. [29] have demonstrated that Polι binds monoubiquitinated PCNA with greater affinity than non-ubiquitinated PCNA through an interaction facilitated by the UBMs of Polι. Mutations in these UBMs abolish the recruitment of Polι into replication foci in response to UV-irradiation, establishing the importance of PCNA and ubiquitin binding for Polι localization in cells [29, 38]. Additional complexity for the recruitment and regulation of Polι is provided by the demonstrated interaction between Polι and Polη. In XP-V cells lacking functional Polη only 10–20% of UV-induced Polι foci are formed, suggesting that the recruitment of Polι is at least somewhat dependent on Polη [86]. The added finding that a PIP box mutant of Polι retains its ability to interact with Polη in vivo suggests that this interaction does not require PCNA and is likely utilized for a subset of presently undefined events [75]. Collectively, these observations suggest that a conglomerate of events are required for Polι localization and function in replication factories generated in response to UV-irradiation.
Polι functions in vivo
Primary mouse fibroblasts derived from 129/J mice harboring a nonsense mutation in exon 2 of Poli exhibit increased sensitivity to UV-irradiation compared to wild-type cells [58] (Table 1). Furthermore, cells deficient for both Polh and Poli are more UV radiation-sensitive than Polh −/− cells alone, and mutagenesis observed in XP-V cells is significantly reduced in Polh −/− Poli −/−cells, suggesting a function for Polι in TLS of UV radiation-induced damage [58]. Wang et al. [57] showed that the high frequency and abnormal spectrum of UV-induced mutations in XP-V cells is not observed when the slower migrating form of human Poli (a product of alternative splicing) is down-regulated. These data support the hypothesis that enhanced UV-induced mutagenesis in the absence of Polη derives from the error-prone activity of Polι. A different study has reported that mouse fibroblasts from 129/J animals do not exhibit sensitivity to UV-irradiation, regardless of Polh status [88]. Yet, Poli deficiency compounded by XP-V in mice leads to mesenchymal tumors not observed in XP-V mice, implicating a role for Polι in UV-induced skin carcinogenesis [88].
In recent years there has been mounting evidence for a role for Polι in carcinogenesis. For example, the overexpression of Poli has been documented in several human breast cancer cell lines and is correlated with the hypermutation observed in these cells [89]. The mouse Poli gene is located within the Par2 (pulmonary adenoma resistance 2) locus on distal chromosome 18, which has been characterized as a major resistance locus with respect to urethane-induced pulmonary adenomas [77, 78]. In addition, among the known mouse Poli alleles, the defective 129X1/SvJ allele is associated with the highest susceptibility to urethane-induced lung tumors [90]. Direct evidence implicating Poli as the gene responsible for the resistance to urethane-induced lung tumors anticipates valuable insight into the biological significance of Polι.
Given the extreme low-fidelity of Polι when copying undamaged DNA, Polι has been proposed as a candidate for SHM [71, 74, 76]. However, the involvement of Poli in the diversification of Ig genes is controversial. In a human Burkitt’s lymphoma cell line in which both alleles of Poli were deleted, stimulated BL2 cells showed a significant reduction in V mutation frequencies [91]. Mutations in BL2 cells were restored by rescuing the activity of Polι with wild-type cDNA, suggesting a role for Poli in SHM. In contrast, the frequency or pattern of mutations observed during SHM in 129/J Poli-deficient mice is not altered, nor does Polι appear to play a role in class switch recombination [74, 92]. In cells doubly deficient for Poli and Polh or Poli −/− and Polk −/−, it does not appear that Polι compensates for the absence of the other polymerase activity, suggesting dispensability of Polι in SHM in mice [92, 93].
Several explanations for these contradictory observations are tenable. If 129/J mice are truly deficient for Polι, it is possible that the mice have compensated for the loss of Polι function over time [92]. However, it is noteworthy that SHM in B-cells isolated from humans and mice is relatively proportionately distributed among G:C and A:T base pairs, yet in the BL2 cell line mutations consist mostly of substitutions of G:C base pairs [74, 91, 92]. These dissimilar results suggest that different proteins are involved in SHM in the BL2 cell line versus animals [92]. As an alternative explanation, human and mouse Poli may be utilized differently for SHM in Ig genes.
Polι is one of several specialized polymerases capable of bypassing 8-oxo-dG [94, 95], a lesion generated during hypoxia/reoxygenation. Recently, Ito et al. [96] reported that hypoxia and hypoxia mimetics enhance the expression of Poli in human tumor cell lines. Furthermore, the hypoxia-inducible factor HIF-1 (which functions as a regulatory transcription factor during hypoxic conditions) binds a consensus sequence in intron 1 of the Poli gene, consequently up-regulating transcription of Poli mRNA in response to hypoxic conditions. Cells exposed to hypoxia show increased point mutations, supporting the notion that Poli may participate in the response to oxidative stress by playing a role in hypoxia-induced mutagenesis [96].
Polι may have functions other than those in TLS and SHM. The enzyme possesses 5′ deoxyribose phosphate (dRP) lyase activity in vitro [97] suggesting the potential involvement of this enzyme in base excision repair (BER) [97]. In an effort to evaluate the BER capacity of Polι, its dRP lyase and DNA polymerase activities were analyzed with BER intermediate substrates and it was observed that Polι can complement the in vitro single-nucleotide BER deficiency of DNA polymerase Polβ-null cell extracts [98]. More recently, it was reported that human MRC5 fibroblasts with stably down-regulated Polι protein exhibit sensitivity to the DNA-damaging agent H2O2. A reduction in BER activity is observed in these cells. Additionally, in wild-type cells Polι accumulates at sites of oxidative damage and interacts with XRCC1 (another protein that participates in BER) [99]. These observations provide support for a role(s) for human Poli in protecting cells against oxidative damage [99].
REV1
The REV1 gene is highly conserved in eukaryotes and plays a central role in promoting mutagenesis from yeast to humans [100]. REV1 protein possesses a unique enzymatic activity in vitro, displaying a marked preference for inserting only dCMP opposite a template G and several DNA lesions. It is thus often referred to as a dCMP transferase [100]. However, since the dCMP transferase activity of yeast REV1 is not required for UV-induced mutagenesis, REV1 is suggested to have non-catalytic roles in TLS [101]. In addition, the expression of REV1 is tightly regulated in cells [102].
REV1 gene expression and regulation
The human REV1 gene is located between the chromosome band 2q11.1 and 2q11.2 [103] and encodes a protein of 1,251 amino acid residues, compared with 1,249 residues in the mouse protein [104]. A human REV1 splicing variant that encodes 1,250 amino acids residues (one amino acid shorter than wild-type REV1 protein) has also been identified [105]. The shorter form of REV1 (REV1S) is expressed similarly to REV1 at the mRNA level and REV1S and REV1 have the same biochemical properties [105]. Comparison of the amino acid sequences of human and mouse REV1 reveals an overall amino acid identity of 84% and similarity of 90%, with all motifs in the human REV1 protein conserved in the mouse counterpart. The REV1 gene is ubiquitously expressed in various human and mouse tissues [103–105] with highest expression of the human REV1 gene in human testis [105, 106]. Relative to other tissues, expression of the mouse REV1 gene is higher in the heart, skeletal muscle, and testis [104].
The human REV1 locus possesses an upstream out-of-frame ATG codon, suggesting that the cellular level of REV1 is probably very low [103]. In S. cerevisiae, REV1 protein levels are 50-fold higher in the G2/M phase of the yeast cell cycle than in S phase [107, 108]. Levels of REV1 mRNA exhibit a pattern of cell cycle regulation similar to that of the protein, peaking slightly before REV1 protein in G2/M [107]. However, the yeast cell cycle-dependent REV1 expression pattern is not conserved in mammalian cells, in which the cellular protein levels of REV1 were unaffected by UV irradiation or cell cycle progression [32].
REV1 enzymatic activity
REV1 is the most extreme of the Y-family DNA polymerases in terms of its nucleotide incorporation specificity [3]. Like yeast REV1, the mammalian protein is a dCMP transferase that specifically inserts a dCMP residue opposite a DNA template G. The REV1 transferase is also able to efficiently and specifically insert dCMP opposite an apurinic/apyrimidinic (AP) site or a uracil residue in vitro, but not opposite a CPD or a [6–4] photoproduct [103–105]. In addition, REV1 protein can incorporate dCMP opposite template A, T, or C and can extend a mismatched terminus by addition of a dCMP residue [104]. This mismatch-extension ability is strongly enhanced by the presence of a guanine residue (but not an AP site) on the template near the mismatched terminus. Kinetic analysis of the dCMP transferase reaction supports the high affinity of dCTP for template G. The crystal structure of the polymerase domain of S. cerevisiae REV1 complexed with a primer-template and incoming dCTP reveals that REV1 uses a novel mechanism of DNA synthesis whereby the incoming dCTP pairs with an arginine rather than the template base, and the template G is evicted from the DNA helix [109].
Single-stranded DNA (ssDNA) inhibits the transferase activity of REV1 due to sequestration of the catalytic site by high affinity binding [110]. The N- and C-terminal domains of REV1 are required for this sequestration. Furthermore, REV1 preferentially utilizes primer-templates that are followed by a long stretch of ssDNA, suggesting that REV1 is targeted specifically to the included primer termini, a property not shared by other DNA polymerases, including human DNA polymerases α, β, and η [110]. This novel activity of REV1 protein may imply a role for ssDNA in the regulation of some modes of TLS.
REV1 protein–protein interactions and mechanisms for recruiting REV1 to stalled replication foci
Mammalian REV1 protein possesses a N-terminal BRCA1 C-terminal (BRCT) domain, a central catalytic domain, a C-terminal region containing two ubiquitin-binding motifs (UBMs) and a special polymerase-binding domain [5] (Fig. 1). The dCMP transferase activity of REV1 is conserved throughout eukaryotic evolution. However, this activity does not account for its role in mutagenesis [3, 100].
The presence of a BRCT domain in REV1 is unique among the Y-family of DNA polymerases [111]. This domain is required for mutagenesis and for resistance to DNA-damaging agents in yeast and mice [102, 112]. BRCT domains have been reported to mediate protein–protein interactions in many cell cycle and DNA repair proteins [113]. A functional BRCT domain is indeed required for a physical interaction between REV1 protein and PCNA [114]. Additionally, the over-expressed REV1 N-terminal fragments REV11–240 and REV11–413 (especially the latter) bind PCNA [114]. However, the possibility that this interaction is indirectly mediated by other protein(s) that bind the REV1 BRCT domain directly cannot be excluded.
In addition to the BRCT domain the UBMs in REV1 are required for its interaction with PCNA after DNA damage [115]. The UBMs mediate an enhanced interaction between REV1 and monoubiquitinated PCNA [115]. At present, conflicting data exist as to whether or not ubiquitination of PCNA increases the REV1 transferase function [43, 44, 116]. The UBMs in REV1 conceivably interact with other ubiquitinated proteins at stalled replication forks. Determining the identities of these proteins will provide further insights into the function of REV1 in vivo.
The C-terminal 100 amino acids of REV1 are required for its interaction with Polκ, Polι, Polη and the noncatalytic REV7 subunit of Polζ in mammalian cells [30, 31, 48, 106]. Additionally, the REV1 C-terminal interaction region is required for resistance to DNA-damaging agents in vertebrates ([117]; C. Guo, E. Sonoda and E. C. Friedberg, unpublished data). The extensive conservation of the REV1 polymerase-binding domain among higher eukaryotes suggests that the REV1-specialized polymerase interaction is conserved in all vertebrates [34]. This C-terminal region in yeast and other lower organisms was previously thought not to be relevant for interaction with REV7 due to poor conservation at the primary sequence level among various eukaryotes. However, a recent study has shown that the interaction between the REV1 C terminus and REV7 is retained in yeast, flies and the nematode C. elegans [34]. Additionally, a comprehensive analysis of S. cerevisiae REV1886–936 which is sufficient for a physical interaction with REV7 in vivo reveals that several novel motifs that, when disrupted, lead to a complete loss of function of the REV1 gene in vivo [118]. Overproduction of a region of the REV1 C terminus containing these motifs confers a dominant-negative effect on survival and mutagenesis after DNA damage [118]. In humans, the REV1–REV7 interaction is stable and results in the formation of a heterodimer [119]. However, neither REV7 nor Polκ affects the transferase activity or stability of REV1 protein in vitro [48, 119]. In addition, affinity purification and mass spectrometry analysis showed that REV1, Polη and Rad18/Rad6 form a complex in HeLa cell nuclear extracts [120]. The interaction of REV1 with Polη on chromatin is enhanced by replication fork arrest caused by nucleotide depletion or DNA lesions [120]. These observations support an important role(s) for REV1 in coordinating the activity of specialized DNA polymerases, possibly by providing a scaffold to facilitate polymerase switching at lesion sites [87].
Ectopically expressed REV1 is distributed homogeneously within the nucleus, although the primary sequence of REV1 does not contain a canonical nuclear localization signal (NLS). However, REV1 appears to be directed to the nucleus via two sequences located in the N-terminal and the C-terminal halves of the protein [30, 121]. In the absence of DNA damage REV1 is localized to replication foci in about 15% of cells [30, 114]. After treatment with UV-irradiation or BPDE, the number of cells containing REV1 foci significantly increases [30, 114, 121, 122]. The distribution of foci containing REV1 is similar to those observed for Polη and Polι [30, 114]. In addition, REV1 co-localizes with PCNA and Polη in replication foci [30]. Deletion or mutational inactivation of the BRCT domain abolishes the targeting of REV1 to replication foci in unirradiated cells [114]. Hence, the interaction between REV1 and PCNA is important for the localization of REV1 to replication foci. Moreover, in cells exposed to UV-irradiation the association of REV1 with replication foci is dependent on functional UBMs [115]. Interestingly, REV1 focus formation is seen not only in S phase but also in the G1 phase [121]. At present, it is not known whether the foci observed in G1 have biological significance. REV1 and activated FANCD2 co-localize in replication foci after replication arrest [123]. The recruitment of REV1 into replication foci depends on an intact FA core complex. Remarkably, FA core complex-dependent REV1 recruitment requires the BRCT domain of REV1 [124]. Furthermore, human Polη and REV7 are not required for ectopically expressed REV1 focus formation [30, 121] but deletion of the C-terminal polymerase-binding domain of mouse REV1 significantly decreases the efficiency of foci formation (C. Guo and E. C. Friedberg, unpublished data). These observations suggest that the C terminus of mouse REV1 is required for the stabilization of other domains that are important for the formation of REV1 foci. Another possibility to be considered is that REV1 focus formation is dependent on its interaction with other specialized DNA polymerases. In support of this, a recent study shows that the localization of endogenous REV1 to UV-irradiated areas in the nucleus is largely dependent on Polη, and that formation of nuclear foci by ectopically expressed REV1 in un-irradiated cells is enhanced by the co-expression of Polη [32].
REV1 functions in vivo
REV1 is important for maintaining genomic integrity by TLS, and together with Polζ is required for most spontaneous and induced mutagenesis in yeast [102]. In DT40 cells REV1 not only facilitates Polζ-dependent bypass, but also modifies the catalytic behavior of Polζ, restraining its synthetic activity to ensure that it incorporates nucleotides in-frame with the damaged template [125]. Human cell lines expressing high levels of human REV1 antisense RNA or ribozyme exhibit a much reduced frequency of 6-thioguanine-resistant mutants induced by UV-irradiation or BPDE [122, 126, 127], indicating that REV1 in higher eukaryotic cells performs functions similar to its yeast homologs. This property has been confirmed in a different experimental system using RNA interference to down-regulate mouse REV1 [128]. However, in contrast with yeast rev1 null mutants or REV1-deficient chicken DT40 cells (which show increased cytotoxicty in response to most of the DNA-damaging agents tested) [129] down-regulation of REV1 in human cells by antisense RNA or ribozyme does not alter sensitivity to UV-irradiation or BPDE [122, 126, 127]. In addition, Polβ-null cells with reduced REV1 expression exhibit slightly enhanced resistance to cisplatin and MMS, but not to UV-irradiation and 4-NQO [128]. Differential survival observed in different species may result from variation in the number of specialized DNA polymerases that can accomplish replication of damaged templates to avoid the collapse of replication forks and cell death. Alternatively, the reduced, but not absent, levels of REV1 in the human cells may be sufficient to rescue cells. Consistent with this suggestion, cells from two different REV1 mutant mice are sensitive to a variety of genotoxic agents [112, 130].
Rev1 B/B (deletion of the REV1 BRCT domain) ES cells display an elevated spontaneous frequency of intragenic deletions at the Hprt locus. Additionally, UV-C light induces delayed progression through late S and G2 phases of the cell cycle and many chromatid aberrations, specifically in a subset of mutant cells. UV-C-induced mutagenesis is reduced and mutations at thymidine–thymidine dimers are absent in Rev1 B/B ES cells, the opposite phenotype of similarly exposed cells from XP-V patients. This suggests that the enhanced UV radiation-induced mutagenesis in XP-V patients may depend on error-prone REV1-dependent TLS [112]. Rev1 −/− mice (with deletion of the REV1 catalytic domain and the polymerase-binding domain) have a strand-biased defect on C/G transversions in the hypermutation of Ig genes: C to G transversions are virtually absent in the non-transcribed strand and reduced in the transcribed strand [130] (Table 1). This defect is associated with an increase of A → T, C → A, and T → C substitutions. These results indicate that REV1 incorporates dCMP during SHM, in agreement with a role of the REV1 catalytic domain in SHM in chicken DT40 cells [129].
Aside from TLS and SHM, REV1 has roles in other DNA damage responses in vertebrates. For example, REV1 participates in Polζ-dependent double strand break repair and Polζ-independent Ig gene conversion in DT40 cells [131]. However, there is no direct evidence to indicate that these additional functions are conserved in mammalian cells.
To date, no REV1-deficient human individuals have been identified. However, SNPs in the human REV1 gene are associated with increased cancer risk. Notably, the REV1–Phe257Ser mutation (downstream of the BRCT domain) is associated with human lung cancer risk [132] and the Phe257Ser heterozygous and Ser257Ser homozygous genotypes are associated with a decreased risk for cervical carcinoma, while Asn373Ser (close to the catalytic domain) and Ser373Ser genotypes are associated with an increased risk [133].
Polκ
Polκ protein is a eukaryotic member of the DinB/Polκ branch of the Y-family of DNA polymerases that is structurally conserved from bacteria to vertebrates [111]. The amino acid sequence of Polκ is different from its homologs Pol IV (Escherichia coli) and DNA polymerase 4 (Dpo4) (Sufolobus solfataricus) by an extension at the N terminus of ~75 amino acids which is indispensable for Polκ activity and is conserved only among eukaryotic Polκ proteins [134]. Polκ shares with Pol IV and Dpo4 a tendency to generate frameshift mutations [3, 135]. In comparison with human Polη and Polι, Polκ is the most resistant to bulky guanine N2-adducts and the most quantitatively efficient in catalyzing dCTP incorporation opposite bulky guanine N2-adducts, particularly the largest (N2-BPDE-dG)[136].
Polk gene expression and regulation
The mouse and human Polk genes were cloned as homologs of the E. coli DinB gene (Pol IV) [111, 137]. The human Polk gene maps to chromosome 5q13 [111]. Mouse Polk is ubiquitously expressed, with highest expression levels in testis [111, 137, 138]. Multiple transcripts are present in this tissue [139] and Polk expression is confined to meiotic spermatocytes and postmeiotic spermatids, suggesting a specific role of Polκ in spermatogenesis [138]. Polk mRNA is also highly expressed in the adrenal cortex in mouse embryos and adult animals [138]. Other cell-specific expression is observed in the epithelium of smaller bronchi and large bronchioles in the adult mouse lung, epithelial cells lining the stomach, the corpus luteum and germinal follicles of the ovary, epithelial cells of the skin, cornea, retina and iris of the eye, and in salivary glands [138]. The physiological explanation of this cell-specific expression is unknown.
Expression of Polk is transcriptionally upregulated in a p53-dependent manner in mouse cells exposed to doxorubicin or UV-irradiation [138], but not in human cells. The mouse Polk gene is developmentally regulated in the testis and utilizes two transcriptional start sites during spermatogenesis, while it utilizes only one site in tissues other than testis [140]. Both the mouse and human Polk genes have two arylhydrocarbon receptor (AhR)-binding sites in their promoter regions and expression of the mouse Polk gene is enhanced upon AhR-activation through its binding with aromatic compounds such as B[a]P and dioxin [140]. Additionally, a stimulating protein-1 (SP1) element and a cyclic AMP-responsive element have been identified in the human Polk promoter and are involved in activation of the Polk promoter [141]. Furthermore, the level of mouse Polκ protein is increased in cells exposed to BPDE or UV-B radiation [142]. Therefore, expression of Polk is regulated by multiple factors, and its inducible expression displays species-specific properties.
Polκ enzymatic activity
In vitro primer extension assays have demonstrated that Polκ can insert nucleotides opposite certain types of base damage in template DNA, including sites of base loss (AP sites) [143], guanine modified with N-acetylaminofluorene or BPDE [144–146], guanine modified with oxidized estrogens [147], 8-oxoG [146], and thymine glycol [148]. A recent study shows that Polκ is particularly efficient in the bypass of bulky N2-guanine minor groove DNA adducts [136]. Additionally, Polκ cooperates with Polζ in error-free TLS across BPDE-dG lesions in human cells [24]. However, Polκ does not support primer extension past CPDs or [6–4] photoproducts generated in DNA exposed to UV radiation, nor past cisplatin intrastrand cross-links and O6-MeG in vitro [3]. Unexpectedly, Polκ and Polζ contribute to a largely error-prone bypass of cisplatin-GG (an intra-strand adduct) in human cells [24]. This role for Polκ in TLS across cisplatin-GG may be enabled by interaction(s) with auxiliary proteins in vivo. Furthermore, the DNA synthetic activity of human Polκ is significantly enhanced by the PCNA/RFC/RPA complex [149], although the processivity of Polκ is not robustly increased in the presence of these protein factors [149]. In addition to its role in TLS, a recent study indicates that Polκ can synthesize DNA during NER [150].
A ternary complex crystal structure of Polκ lacking its C-terminal domain has been determined [151]. The structure reveals almost complete encirclement of the DNA by a unique “N-clasp” at the N terminus of Polκ, which augments the conventional right-handed grip on DNA by the palm, fingers, thumb, and PAD domains and provides additional thermodynamic stability [151]. The constrained active-site cleft in Polκ can accommodate only a single Watson–Crick base pair [151], which explains the high fidelity of Polκ to incorporate dCTP opposite N2-dG adducts [136] as well as the inability of Polκ to insert nucleotides opposite the 3′T of a CPD [3]. Notably, the N-clasp in mammalian Polκ is not present in the error-prone Dpo4.
Polκ protein–protein interactions and mechanisms for recruiting Polκ to stalled replication foci
Like Pol IV in E. coli, mammalian Polκ contains a catalytic domain as well as a conserved motif that forms the unique PAD structure in Y-family polymerases [5, 111]. However, mammalian Polκ differs from its prokaryotic and archaeal counterparts by the presence of unique N-terminal and C-terminal extensions (Fig. 1). The N-terminal region is indispensable for Polκ activity [152]. The C-terminal region shares 60% amino acid identity between mouse and human Polκ [5, 56, 111] and contains a bipartite nuclear localization signal (NLS) [111] as well as a conserved PIP box that contributes to PCNA binding [149]. The Polκ560–615 region interacts with REV1 protein via a highly conserved domain in REV1 [31, 48]. Further analysis of the amino acid sequence of the human Polκ560–615 fragment revealed a novel REV1-interacting region containing two consecutive phenylalanine (FF) residues (FF567–568) that are critical for interaction with REV1 [33]. The FF567–568AA mutant of Polκ that cannot interact with REV1 failed to correct BPDE- and UV-sensitivities of the Polk −/− mouse embryonic fibroblast cells [33]. Additionally, recent studies have shown that the duplicated C2HC Zn-cluster domains in the Polκ C terminus are novel UBZs that mediate the interaction between ubiquitin and Polκ as well as the monoubiquitination of Polκ [29, 142]. The UBZ domains enable Polκ to bind monoubiquitinated PCNA more robustly than non-ubiquitinated PCNA [142, 153].
Polκ is present in microscopically visible foci in cells treated with UV-irradiation or BPDE [142, 154–156]. Similar to the UBZ in Polη and the UBMs in Polι [29, 38], the UBZs in Polκ are critical for the accumulation of Polκ protein in replication foci when cells suffer from DNA damage [142]. In addition, the PIP box and the bipartite NLS are required for Polκ to form nuclear foci after DNA damage [154]. Surprisingly, the fraction of human Polκ foci-positive cells is consistently lower than that observed for mouse Polκ after UV-irradiation [142].
Polκ functions in vivo
Two different Polκ knock-out mouse models have been generated but no significant phenotypes have been reported to date [157, 158] (Table 1). Although Polκ is highly expressed in the testis, Polκ-deficient mice are fertile and viable, demonstrating that the gene is not essential. Polκ-deficient mice also display normal SHM [157]. However, they manifest elevated mutation rates in the male germline [159] and other tissues (J. N. Kosarek, L. D. McDaniel and E. C. Friedberg, unpublished data), suggesting an anti-mutator function in vivo. Some offspring of Polκ mice (any Polk genotype) have been shown to spontaneously manifest various disease states (S. Velasco, L. D. McDaniel and E. C. Friedberg, unpublished observations), suggesting that the absence of Polκ results in a spontaneous mutator phenotype.
Disruption of the Polk gene in mouse cells results in significant sensitivity to killing by BPDE [158], suggesting a specific requirement for Polκ to bypass this planar polycyclic lesion in DNA. Consistent with this finding, BPDE lesion bypass is reduced in Polk-deficient MEFs and is restored by Polk cDNA expression [160]. In addition, the frequency of BPDE-induced mutagenesis is increased in Polk-deficient MEFs. Along these lines, Polκ is downregulated in some human colorectal tumors [141] which may be associated with environmental and dietary exposure to polycyclic aromatic hydrocarbons. Further studies also suggest that Polκ is specifically required for recovery from BPDE-induced S-phase checkpoint arrest [156]. Furthermore, Polκ-deficient mouse embryonic stem and fibroblast cells manifest moderate sensitivity to UV radiation and methyl methanesulfonate [157, 161]. However, Polκ deficiency does not alter cellular sensitivity to ionizing radiation [158].
The over-expression of Polκ can also result in deleterious consequences [162]. Indeed, the over-expression of mouse Polκ in a mouse cell line results in about a tenfold increase in spontaneous mutagenesis [137]. Furthermore, increased levels of Polκ in lung cancers correlate with increased genetic instability, which is detected with not only an enhanced mutation rate but also DNA breaks, increased genetic recombination, loss of heterozygosity, and aneuploidy [162, 163]. Consistent with these dysfunctions, Polκ excess combined with p53 deficiency favors tumorigenesis in nude mice [164]. Hence, spontaneous mutations may be generated by either deficient or excessive Polκ enzyme activity.
Mechanisms of PCNA monoubiquitination and DNA polymerase switching during TLS
Regardless of the specific types of base damage in DNA bypassed by TLS in living cells, a question of considerable interest is how switching between high fidelity DNA polymerases in the replicative machinery and one or more specialized enzymes that support TLS is effected at sites of arrested replication. Recent studies indicate that PCNA provides the central scaffold to which various TLS polymerases can bind in order to access the replicative ensemble stalled at a lesion to execute their roles in lesion bypass [3, 87].
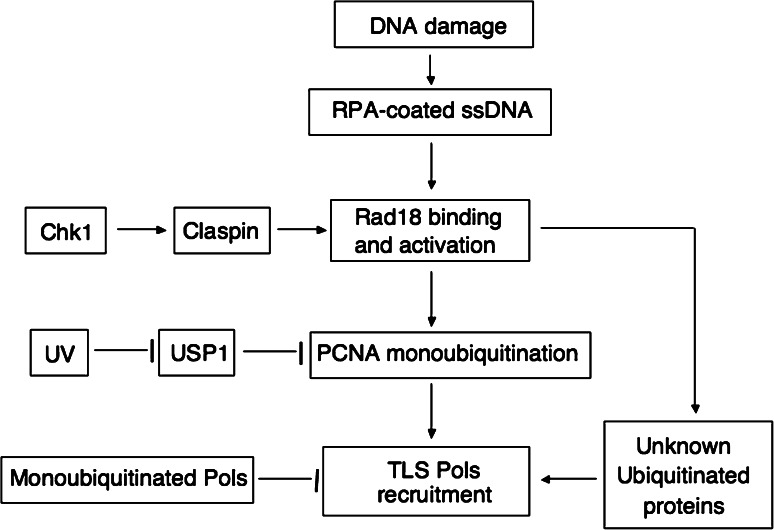
TLS in mammalian cells is apparently promoted (at least in part) by the monoubiquitination of PCNA [56, 87]. PCNA monoubiquitination involves the ubiquitin-conjugating enzyme Rad6 and its cognate ubiquitin ligase Rad18 [165, 166]. In response to UV radiation or BPDE treatment, Rad18 relocalizes to sites of replication stalling and is activated [39]. PCNA is monoubiquitinated at Lys164 in mammalian cells following various DNA damage treatments that cause stalling of the replication fork [37, 56, 167], including UV-irradiation, alkylating and adduct-forming agents, but not those that induce DSBs without associated base damage. The stalled replication intermediates appear to be both necessary and sufficient for activation of PCNA ubiquitination [168]. In addition to Rad18 [37, 39, 167, 168], the ssDNA binding replication protein A (RPA), which can directly interact with Rad18, is also required for DNA damage-induced PCNA ubiquitination in mammalian cells [167, 168]. PCNA ubiquitination also requires the uncoupling of helicase and polymerase activities which can produce stretches of ssDNA [169]. These results suggest that the upstream signal that activates PCNA ubiquitination in vivo is RPA-coated ssDNA at sites of stalled forks, in which RPA targets Rad18 to its sites of action [168] (Fig. 2). Furthermore, Rad18 is a rate-limiting factor for PCNA ubiquitination at stalled replication intermediates. Elevated levels of Rad18 can overcome the requirement for fork stalling and promote ubiquitination of PCNA whenever the clamp is associated with DNA [168]. This observation, together with the finding that the accumulation of RAD18 at DNA damage occurs rapidly and persists for a long period of time in a cell cycle-independent manner, even without DNA replication [170], prompts speculation that the accumulation of Rad18 causes UV radiation-induced PCNA ubiquitination in human cells held in either G0 or G2 [171].
Fig. 2.
A model depicting the regulation of PCNA monoubiquitination and TLS polymerases switching at sites of stalled replication. Following DNA damage treatments, RPA binds to ssDNA at sites of stalled forks, in which RPA targets Rad18 to its sites of action [168] and stimulate PCNA monoubiquitination. Monoubiquitination PCNA enables removal of replication polymerase and recruitment of TLS polymerases to the primer terminus. This process is regulated by Chk1 but not ATR. Claspin, which is stabilized by Chk1, regulates the binding of the ubiquitin ligase Rad18 to chromatin [173]. Meanwhile, UV inactivates USP1 to stimulate accumulation of monoubiquitinated PCNA [174]. Moreover, Rad18/Rad6 complex can also monoubiquitinate some other proteins at sites of stalled replication forks [179, 180], which may recruit TLS polymerases in parallel with monoubiquitinated PCNA. Furthermore, monoubiquitinated Y-family polymerases may contribute to regulation of their compartmentalization in or out of replication factories [29]
It is likely that other cellular factors regulate Rad6/Rad18-dependent PCNA ubiquitination in vivo. In support of this notion, PTIP, Chk1 and the DNA replication proteins Claspin and Timeless are required for efficient PCNA ubiquitination in mammalian cells [172, 173]. Notably, the effect of depleting Chk1 on PCNA ubiquitination is not due to replication fork collapse resulting from comprised ATR signaling, but the reduction of Claspin, which regulates Rad18 chromatin binding [173].
Ubiquitination of PCNA is also regulated by the deubiquitinating enzyme USP1, which is able to remove monoubiquitin from Ub-PCNA [174]. USP1 is inactivated after UV-irradiation, thus enabling monoubiquitinated PCNA to accumulate and to activate TLS [174]. There is a temporal correlation between the disappearance of USP1 and the presence of PCNA ubiquitination after UV-irradiation, but this correlation was not observed after chemical mutagen treatment [167], suggesting that the ability of USP1 to remove monoubiquitin from PCNA might be differentially affected by different DNA-damaging treatments. Interestingly, PCNA ubiquitination can persist for many hours after damage has been removed [167]. The underlying mechanisms for this remain unknown. However, recent work suggests that TLS occurs not only at stalled forks to allow fork progression, but also at the post-replicative gap-filling step [175, 176]. A model of TLS outside of S phase may explain the persistence of Ub-PCNA after DNA damage has been removed [167].
Conjugation of ubiquitin to PCNA may regulate protein–protein interactions at replication foci. Among the known specialized DNA polymerases, Ub-binding domains are confined to the Y-family polymerases [29]. As mentioned above, these domains are required for accumulation of these polymerases in replication foci, as well as their interaction with monoubiquitiated PCNA and TLS in vivo [29, 38, 56, 115, 142]. However, an alternative possibility is that a Ub moiety effects a conformational change on PCNA that destabilizes the PCNA-binding ability of the replicative polymerase and/or other PCNA-associated proteins that otherwise prevent binding of other specialized polymerases to PCNA via their PIP motifs [46]. The underlying mechanism by which ubiquitin on PCNA regulates the access of Y-family polymerases to replication foci remains uncertain.
Recently, Langerak et al. [177] observed that A → T mutations in hypermutated Ig genes were significantly reduced in PCNAK164R B-cells, while G → C mutations were not impaired in the absence of PCNAK164 modification. These observations suggest that not all specialized DNA polymerases depend on PCNA monoubiquitination specifically, but on other ubiquitinated factors [177, 178] such as the heterotrimeric Rad9/Rad1/Hus1 sliding clamp (9-1-1 complex) and replication factor C (which can also be ubiquitinated by the Rad6/Rad18 complex) [179, 180] or unidentified ubiquitinated components in the replication machinery. Recent data showing that ubiquitinated PCNA is not the only ubiquitinated target that drives Polη into foci, support this idea [45]. Notably, all Y-family polymerases can undergo monoubiquitination in vivo [29, 115, 142]. Although the precise role of this post-translational modification remains to be established, it is speculated that this phenomenon may contribute to the regulation of compartmentalization in or out of replication factories [29].
REV1 may also play a role in DNA polymerase switching during TLS. As mentioned above, the essential function of REV1 in TLS does not depend on its dCMP transferase activity, yet the protein is absolutely required for DNA damage-induced mutagenesis. The ability of REV1 to bind all other specialized DNA polymerases as well as to monoubiquitinated PCNA raises the possibility that REV1 might act as a platform for different DNA polymerases during polymerase switching [56]. By employing two complementary assays that measure fork progression and post-replicative gap filling [181] one study observed that the control of TLS differs at stalled replication forks and post-replicative gaps in DT40 cells. TLS at stalled replication forks on UV- or NQO-damaged DNA requires both the polymerase-binding domain and UBMs in the C terminus of REV1. PCNA ubiquitination is not required to maintain normal fork progression on damaged DNA, but is essential for post-replicative gap filling.
As previously suggested, each specialized polymerase may have cognate lesion(s) for TLS. The intrinsic differences in substrate preferences between different polymerases may also regulate polymerase choice and switching during this process [182, 183]. A simple hypothesis is that polymerase switches occur during transitions from preferential to disfavored use of a damaged template primer, and that among multiple possibilities, the polymerase called upon following each successive nucleotide incorporated is the one whose properties ultimately result in the most efficient and least mutagenic bypass [182, 183]. In support of this hypothesis, Polη (which can bypass CPDs with much higher efficiency than that of other eukaryotic DNA polymerases [183, 184]) was found to copy thymine dimers and the flanking bases with higher processivity than undamaged DNA. Polη switched to less processive synthesis when the two damaged base pairs in the duplex primer-template were at positions allowing interaction with the PAD domain [183]. Since the PAD domains of various Y-family polymerases are unique [185] and each TLS enzyme has a different lesion bypass capacity [186], the exact locations of switches before and after TLS may vary depending on the identity of the competing polymerases, lesions, and DNA sequence context [182, 183].
Recently, a study from C. elegans suggested that an active mechanism might be employed in cells to trigger post-TLS polymerase exchange [187]. The study reveals that POLH-1 (the worm ortholog of Polη) undergoes DNA-damage-induced proteolysis and that GEI-17 SUMO E3 regulates the timing of this proteolysis. Upon DNA damage, GEI-17 sumoylates POLH-1 and protects POLH-1 from Cul4–Ddb1–Cdt2-mediated destruction until it has performed its function in TLS [187]. The GEI-17/Cul4–Ddb1–Cdt2-based regulatory system, which controls POLH-1 function, may be important for removing POLH-1 from the replication forks after TLS. At present, it is unknown whether the GEI-17/Cul4–Ddb1–Cdt2-mediated regulation of POLH-1 is conserved in mammalian cells.
The regulation of PCNA monoubiquitination and TLS polymerase switching is complicated (Fig. 2). Several factors involved in multiple pathways apparently participate in this process to keep TLS under strict control. Future work in this area is necessary to more clearly understand how these factors work together to regulate TLS in vivo.
TLS polymerases as therapeutic targets for cancer prevention
Cancer cells exhibit a mutator phenotype [188] and it has long been postulated that mutations in specific genes are central to tumorigenesis. If the frequency of mutagenic events can be reduced despite the continued induction of DNA damage, the risk of tumorigenesis should also be reduced [189]. Since the majority of spontaneous and damage-induced mutations in vivo are caused by error-prone TLS, specialized TLS polymerases may be useful targets for anticancer drugs.
With respect to cancer prevention, strategies to prevent ongoing mutational events in exposed populations are likely to reduce the risk of cancer. Several results indicate that targeting mutagenic specialized polymerases can reduce the frequency of induced mutations [127, 190]. As mentioned above, reducing REV1 levels by expressing REV1-specific ribozymes or anti-sense RNA decreases mutagenic responses without altering cytotoxic responses to several common carcinogens [122, 126, 127]. Since many DNA polymerases have redundant functions, specific inhibition of certain specialized polymerases may be a promising approach to developing anticancer drugs.
In the case of cancer treatment, an obvious strategy is to inhibit DNA polymerases involved in TLS in order to increase the efficacy of chemotherapeutic agents that damage DNA. Proteasome inhibitors can prevent UV- or cisplatin-induced translesion replication in human cancer cells, but not in normal cells [191]. This inhibition is independent of cell origin, histological type or p53 status. In addition, proteasome inhibitors markedly reduce the viability of UV-irradiated or cisplatin-treated cancer cells. In view of the role of the proteasome in protein degradation during cell metabolism, proteasome inhibitors are not suitable drugs for cancer therapy. Nonetheless, since treatment of cells with the general proteasome inhibitor MG132 induces a depletion of the free ubiquitin pool and a concomitant reduction of monoubiquitinated target proteins such as ubiquitinated histones [192], identification of selected monoubiquitinated target(s) may further the development of novel selective chemotherapy drugs.
Developments in recent years have significantly increased our knowledge of the role of Y-family polymerases in TLS. However, many questions remain unsolved. For example: Do the properties of the enzymes as currently studied truly reflect in vivo properties? What are the cognate substrates for Y-family polymerases during TLS in vivo? What other roles do Y-family polymerases have in living cells, especially in spermatogenesis? We anticipate that answers to these important questions will emerge with further study. Finally, the mechanisms that integrate Y-family members into different elements of the DNA damage response should be investigated further in order to better understand this very complex biological response.
Acknowledgment
This work was supported by grant ES11344 (NIEHS) (ECF).
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. Washington DC: American Society for Microbiology; 2005. [Google Scholar]
- 2.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/S1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 8.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- 10.Yuasa M, Masutani C, Eki T, Hanaoka F. Genomic structure, chromosomal localization and identification of mutations in the xeroderma pigmentosum variant (XPV) gene. Oncogene. 2000;19:4721–4728. doi: 10.1038/sj.onc.1203842. [DOI] [PubMed] [Google Scholar]
- 11.Thakur M, Wernick M, Collins C, Limoli CL, Crowley E, Cleaver JE. DNA polymerase eta undergoes alternative splicing, protects against UV sensitivity and apoptosis, and suppresses Mre11-dependent recombination. Genes Chromosomes Cancer. 2001;32:222–235. doi: 10.1002/gcc.1186. [DOI] [PubMed] [Google Scholar]
- 12.Pabla R, Rozario D, Siede W. Regulation of Saccharomyces cerevisiae DNA polymerase eta transcript and protein. Radiat Environ Biophys. 2008;47:157–168. doi: 10.1007/s00411-007-0132-1. [DOI] [PubMed] [Google Scholar]
- 13.Skoneczna A, McIntyre J, Skoneczny M, Policinska Z, Sledziewska-Gojska E. Polymerase eta is a short-lived, proteasomally degraded protein that is temporarily stabilized following UV irradiation in Saccharomyces cerevisiae . J Mol Biol. 2007;366:1074–1086. doi: 10.1016/j.jmb.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleaver JE, Bartholomew J, Char D, Crowley E, Feeney L, Limoli CL. Polymerase eta and p53 jointly regulate cell survival, apoptosis and Mre11 recombination during S phase checkpoint arrest after UV irradiation. DNA Repair (Amst) 2002;1:41–57. doi: 10.1016/S1568-7864(01)00004-0. [DOI] [PubMed] [Google Scholar]
- 16.Cleaver JE, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe JP, Busch DB, Coleman DM, Ziffer DW, Yu Y, Nagasawa H, Little JB. Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- 17.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/S1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7, 8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 19.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 20.Lee DH, Pfeifer GP. Translesion synthesis of 7, 8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busuttil RA, Lin Q, Stambrook PJ, Kucherlapati R, Vijg J. Mutation frequencies and spectra in DNA polymerase eta-deficient mice. Cancer Res. 2008;68:2081–2084. doi: 10.1158/0008-5472.CAN-07-6274. [DOI] [PubMed] [Google Scholar]
- 22.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat Res. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 23.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 24.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bassett E, King NM, Bryant MF, Hector S, Pendyala L, Chaney SG, Cordeiro-Stone M. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64:6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 26.Chiapperino D, Kroth H, Kramarczuk IH, Sayer JM, Masutani C, Hanaoka F, Jerina DM, Cheh AM. Preferential misincorporation of purine nucleotides by human DNA polymerase eta opposite benzo[a]pyrene 7, 8-diol 9, 10-epoxide deoxyguanosine adducts. J Biol Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- 27.Yang W. Damage repair DNA polymerases Y. Curr Opin Struct Biol. 2003;13:23–30. doi: 10.1016/S0959-440X(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 28.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 30.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 32.Akagi JI, Masutani C, Kataoka Y, Kan T, Ohashi E, Mori T, Ohmori H, Hanaoka F (2009) Interaction with DNA polymerase eta is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair (Amst) (in press) [DOI] [PubMed]
- 33.Ohashi E, Hanafusa T, Kamei K, Song I, Tomida J, Hashimoto H, Vaziri C, Ohmori H. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosarek JN, Woodruff RV, Rivera-Begeman A, Guo C, D’Souza S, Koonin EV, Walker GC, Friedberg EC. Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair (Amst) 2008;7:439–451. doi: 10.1016/j.dnarep.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/S1097-2765(04)00259-X. [DOI] [PubMed] [Google Scholar]
- 38.Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae . Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- 42.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabbioneda S, Gourdin AM, Green CM, Zotter A, Giglia-Mari G, Houtsmuller A, Vermeulen W, Lehmann AR. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol Biol Cell. 2008;19:5193–5202. doi: 10.1091/mbc.E08-07-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase {eta} in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YW, Cleaver JE, Hatahet Z, Honkanen RE, Chang JY, Yen Y, Chou KM. Human DNA polymerase eta activity and translocation is regulated by phosphorylation. Proc Natl Acad Sci USA. 2008;105:16578–16583. doi: 10.1073/pnas.0808589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J Cell Sci. 2008;121:3271–3282. doi: 10.1242/jcs.027730. [DOI] [PubMed] [Google Scholar]
- 50.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, Kucherlapati R. Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- 53.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Cleaver JE. Xeroderma pigmentosum: variants with normal DNA repair and normal sensitivity to ultraviolet light. J Invest Dermatol. 1972;58:124–128. doi: 10.1111/1523-1747.ep12538913. [DOI] [PubMed] [Google Scholar]
- 56.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 58.Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci USA. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine–thymine dimers by human DNA polymerase iota. EMBO J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/S1074-7613(01)00142-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A–T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 64.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA, Weill JC. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steele EJ. DNA polymerase-eta as a reverse transcriptase: implications for mechanisms of hypermutation in innate anti-retroviral defences and antibody SHM systems. DNA Repair (Amst) 2004;3:687–692. doi: 10.1016/j.dnarep.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 67.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 68.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Ishikawa T, Uematsu N, Mizukoshi T, Iwai S, Iwasaki H, Masutani C, Hanaoka F, Ueda R, Ohmori H, Todo T. Mutagenic and nonmutagenic bypass of DNA lesions by Drosophila DNA polymerases dpoleta and dpoliota. J Biol Chem. 2001;276:15155–15163. doi: 10.1074/jbc.M009822200. [DOI] [PubMed] [Google Scholar]
- 70.McDonald JP, Tissier A, Frank EG, Iwai S, Hanaoka F, Woodgate R. DNA polymerase iota and related rad30-like enzymes. Philos Trans R Soc Lond B Biol Sci. 2001;356:53–60. doi: 10.1098/rstb.2000.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 72.Vaisman A, Frank EG, McDonald JP, Tissier A, Woodgate R. poliota-dependent lesion bypass in vitro. Mutat Res. 2002;510:9–22. doi: 10.1016/s0027-5107(02)00248-8. [DOI] [PubMed] [Google Scholar]
- 73.Tissier A, Frank EG, McDonald JP, Vaisman A, Fernandez de Henestrosa AR, Boudsocq F, McLenigan MP, Woodgate R. Biochemical characterization of human DNA polymerase iota provides clues to its biological function. Biochem Soc Trans. 2001;29:183–187. doi: 10.1042/BST0290183. [DOI] [PubMed] [Google Scholar]
- 74.McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J Exp Med. 2003;198:635–643. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- 76.Frank EG, Tissier A, McDonald JP, Rapic-Otrin V, Zeng X, Gearhart PJ, Woodgate R. Altered nucleotide misinsertion fidelity associated with poliota-dependent replication at the end of a DNA template. EMBO J. 2001;20:2914–2922. doi: 10.1093/emboj/20.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang M, Devereux TR, Vikis HG, McCulloch SD, Holliday W, Anna C, Wang Y, Bebenek K, Kunkel TA, Guan K, You M. Pol iota is a candidate for the mouse pulmonary adenoma resistance 2 locus, a major modifier of chemically induced lung neoplasia. Cancer Res. 2004;64:1924–1931. doi: 10.1158/0008-5472.CAN-03-3080. [DOI] [PubMed] [Google Scholar]
- 78.Lee GH, Nishimori H, Sasaki Y, Matsushita H, Kitagawa T, Tokino T. Analysis of lung tumorigenesis in chimeric mice indicates the Pulmonary adenoma resistance 2 (Par2) locus to operate in the tumor-initiation stage in a cell-autonomous manner: detection of polymorphisms in the Poli gene as a candidate for Par2. Oncogene. 2003;22:2374–2382. doi: 10.1038/sj.onc.1206387. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/MCB.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 81.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 82.Pence MG, Blans P, Zink CN, Hollis T, Fishbein JC, Perrino FW. Lesion bypass of N2-ethylguanine by human DNA polymerase iota. J Biol Chem. 2009;284:1732–1740. doi: 10.1074/jbc.M807296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 84.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 85.Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2002;21:6246–6256. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 88.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, Masutani C, Hanaoka F. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol Cell Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH. Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004;64:5597–5607. doi: 10.1158/0008-5472.CAN-04-0603. [DOI] [PubMed] [Google Scholar]
- 90.Lee GH, Matsushita H. Genetic linkage between Pol iota deficiency and increased susceptibility to lung tumors in mice. Cancer Sci. 2005;96:256–259. doi: 10.1111/j.1349-7006.2005.00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud CA, Weill JC. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 2002;419:944–947. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- 92.Martomo SA, Yang WW, Vaisman A, Maas A, Yokoi M, Hoeijmakers JH, Hanaoka F, Woodgate R, Gearhart PJ. Normal hypermutation in antibody genes from congenic mice defective for DNA polymerase iota. DNA Repair (Amst) 2006;5:392–398. doi: 10.1016/j.dnarep.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Shimizu T, Azuma T, Ishiguro M, Kanjo N, Yamada S, Ohmori H. Normal immunoglobulin gene somatic hypermutation in Pol kappa–Pol iota double-deficient mice. Immunol Lett. 2005;98:259–264. doi: 10.1016/j.imlet.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 94.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ito A, Koshikawa N, Mochizuki S, Omura K, Takenaga K. Hypoxia-inducible factor-1 mediates the expression of DNA polymerase iota in human tumor cells. Biochem Biophys Res Commun. 2006;351:306–311. doi: 10.1016/j.bbrc.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 97.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 98.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J Biol Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 99.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P. Human DNA polymerase iota protects cells against oxidative stress. EMBO J. 2008;27:2883–2895. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 101.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 102.Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/S1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 103.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Masuda Y, Takahashi M, Fukuda S, Sumii M, Kamiya K. Mechanisms of dCMP transferase reactions catalyzed by mouse Rev1 protein. J Biol Chem. 2002;277:3040–3046. doi: 10.1074/jbc.M110149200. [DOI] [PubMed] [Google Scholar]
- 105.Masuda Y, Takahashi M, Tsunekuni N, Minami T, Sumii M, Miyagawa K, Kamiya K. Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J Biol Chem. 2001;276:15051–15058. doi: 10.1074/jbc.M008082200. [DOI] [PubMed] [Google Scholar]
- 106.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 107.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sabbioneda S, Bortolomai I, Giannattasio M, Plevani P, Muzi-Falconi M. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst) 2007;6:121–127. doi: 10.1016/j.dnarep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 109.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 110.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J Biol Chem. 2006;281:24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 111.Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jansen JG, Tsaalbi-Shtylik A, Langerak P, Calleja F, Meijers CM, Jacobs H, de Wind N. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 2005;33:356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huyton T, Bates PA, Zhang X, Sternberg MJ, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res. 2000;460:319–332. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]
- 114.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 115.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J Biol Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 117.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.D’Souza S, Waters LS, Walker GC. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair (Amst) 2008;7:1455–1470. doi: 10.1016/j.dnarep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 120.Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. [DOI] [PubMed] [Google Scholar]
- 121.Murakumo Y, Mizutani S, Yamaguchi M, Ichihara M, Takahashi M. Analyses of ultraviolet-induced focus formation of hREV1 protein. Genes Cells. 2006;11:193–205. doi: 10.1111/j.1365-2443.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- 122.Mukhopadhyay S, Clark DR, Watson NB, Zacharias W, McGregor WG. REV1 accumulates in DNA damage-induced nuclear foci in human cells and is implicated in mutagenesis by benzo[a]pyrenediolepoxide. Nucleic Acids Res. 2004;32:5820–5826. doi: 10.1093/nar/gkh903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 124.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Szuts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase {zeta} to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clark DR, Zacharias W, Panaitescu L, McGregor WG. Ribozyme-mediated REV1 inhibition reduces the frequency of UV-induced mutations in the human HPRT gene. Nucleic Acids Res. 2003;31:4981–4988. doi: 10.1093/nar/gkg725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Poltoratsky V, Horton JK, Prasad R, Wilson SH. REV1 mediated mutagenesis in base excision repair deficient mouse fibroblast. DNA Repair (Amst) 2005;4:1182–1188. doi: 10.1016/j.dnarep.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 129.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, Sobue T, Kunitoh H, Saito R, Shimizu K, Hirama C, Kimura J, Maeno G, Hirose H, Eguchi T, Saito D, Ohki M, Yokota J. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer. 2005;114:730–737. doi: 10.1002/ijc.20790. [DOI] [PubMed] [Google Scholar]
- 133.He X, Ye F, Zhang J, Cheng Q, Shen J, Chen H. REV1 genetic variants associated with the risk of cervical carcinoma. Eur J Epidemiol. 2008;23:403–409. doi: 10.1007/s10654-008-9251-5. [DOI] [PubMed] [Google Scholar]
- 134.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure (Camb) 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 135.Jarosz DF, Godoy VG, Walker GC. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle. 2007;6:817–822. doi: 10.4161/cc.6.7.4065. [DOI] [PubMed] [Google Scholar]
- 136.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 137.Ogi T, Kato T, Jr, Kato T, Ohmori H. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells. 1999;4:607–618. doi: 10.1046/j.1365-2443.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- 138.Velasco-Miguel S, Richardson JA, Gerlach VL, Lai WC, Gao T, Russell LD, Hladik CL, White CL, Friedberg EC. Constitutive and regulated expression of the mouse Dinb (Polkappa) gene encoding DNA polymerase kappa. DNA Repair (Amst) 2003;2:91–106. doi: 10.1016/S1568-7864(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 139.Guo C, Gao T, Confer N, Velasco-Miguel S, Friedberg EC. Multiple PolK (POLK) transcripts in mammalian testis. DNA Repair (Amst) 2005;4:397–402. doi: 10.1016/j.dnarep.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 140.Ogi T, Mimura J, Hikida M, Fujimoto H, Fujii-Kuriyama Y, Ohmori H. Expression of human and mouse genes encoding polkappa: testis-specific developmental regulation and AhR-dependent inducible transcription. Genes Cells. 2001;6:943–953. doi: 10.1046/j.1365-2443.2001.00478.x. [DOI] [PubMed] [Google Scholar]
- 141.Lemee F, Bavoux C, Pillaire MJ, Bieth A, Machado CR, Pena SD, Guimbaud R, Selves J, Hoffmann JS, Cazaux C. Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase kappa in colorectal cancer. Oncogene. 2007;26:3387–3394. doi: 10.1038/sj.onc.1210116. [DOI] [PubMed] [Google Scholar]
- 142.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem. 2008;283:4658–4664. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 143.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 144.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 145.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suzuki N, Itoh S, Poon K, Masutani C, Hanaoka F, Ohmori H, Yoshizawa I, Shibutani S. Translesion synthesis past estrogen-derived DNA adducts by human DNA polymerases eta and kappa. Biochemistry. 2004;43:6304–6311. doi: 10.1021/bi0360298. [DOI] [PubMed] [Google Scholar]
- 148.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 149.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 151.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 152.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 153.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ogi T, Kannouche P, Lehmann AR. Localisation of human Y-family DNA polymerase kappa: relationship to PCNA foci. J Cell Sci. 2005;118:129–136. doi: 10.1242/jcs.01603. [DOI] [PubMed] [Google Scholar]
- 155.Bergoglio V, Bavoux C, Verbiest V, Hoffmann JS, Cazaux C. Localisation of human DNA polymerase kappa to replication foci. J Cell Sci. 2002;115:4413–4418. doi: 10.1242/jcs.00162. [DOI] [PubMed] [Google Scholar]
- 156.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 157.Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, White CL, Friedberg EC, Rajewsky K, Esposito G. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 158.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Burr KL, Velasco-Miguel S, Duvvuri VS, McDaniel LD, Friedberg EC, Dubrova YE. Elevated mutation rates in the germline of Polkappa mutant male mice. DNA Repair (Amst) 2006;5:860–862. doi: 10.1016/j.dnarep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 160.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 161.Takenaka K, Ogi T, Okada T, Sonoda E, Guo C, Friedberg EC, Takeda S. Involvement of vertebrate Polkappa in translesion DNA synthesis across DNA monoalkylation damage. J Biol Chem. 2006;281:2000–2004. doi: 10.1074/jbc.M506153200. [DOI] [PubMed] [Google Scholar]
- 162.Bavoux C, Leopoldino AM, Bergoglio V, O-Wang J, Ogi T, Bieth A, Judde JG, Pena SD, Poupon MF, Helleday T, Tagawa M, Machado C, Hoffmann JS, Cazaux C. Up-regulation of the error-prone DNA polymerase {kappa} promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res. 2005;65:325–330. [PubMed] [Google Scholar]
- 163.O-Wang J, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, Tagawa M. DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 2001;61:5366–5369. [PubMed] [Google Scholar]
- 164.Bavoux C, Hoffmann JS, Cazaux C. Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase kappa. Biochimie. 2005;87:637–646. doi: 10.1016/j.biochi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 165.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 166.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 167.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 170.Nakajima S, Lan L, Kanno S, Usami N, Kobayashi K, Mori M, Shiomi T, Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J Biol Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- 171.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 172.Gohler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair (Amst) 2008;7:775–787. doi: 10.1016/j.dnarep.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 173.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 175.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 176.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 177.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, Edelmann W, Scharff MD. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 180.Tomida J, Masuda Y, Hiroaki H, Ishikawa T, Song I, Tsurimoto T, Tateishi S, Shiomi T, Kamei Y, Kim J, Kamiya K, Vaziri C, Ohmori H, Todo T. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J Biol Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 182.McCulloch SD, Kokoska RJ, Kunkel TA. Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle. 2004;3:580–583. [PubMed] [Google Scholar]
- 183.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis–syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 184.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/S0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 186.Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 187.Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans . Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Watson NB, Mukhopadhyay S, McGregor WG. Translesion DNA replication proteins as molecular targets for cancer prevention. Cancer Lett. 2006;241:13–22. doi: 10.1016/j.canlet.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 190.Li Z, Zhang H, McManus TP, McCormick JJ, Lawrence CW, Maher VM. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res. 2002;510:71–80. doi: 10.1016/s0027-5107(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 191.Takezawa J, Ishimi Y, Yamada K. Proteasome inhibitors remarkably prevent translesion replication in cancer cells but not normal cells. Cancer Sci. 2008;99:863–871. doi: 10.1111/j.1349-7006.2008.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]