Abstract
Poly-ADP-ribose polymerases (PARPs) use NAD+ as substrate to generate polymers of ADP-ribose. We targeted the catalytic domain of human PARP1 as molecular NAD+ detector into cellular organelles. Immunochemical detection of polymers demonstrated distinct subcellular NAD+ pools in mitochondria, peroxisomes and, surprisingly, in the endoplasmic reticulum and the Golgi complex. Polymers did not accumulate within the mitochondrial intermembrane space or the cytosol. We demonstrate the suitability of this compartment-specific NAD+ and poly-ADP-ribose turnover to establish intra-organellar protein localization. For overexpressed proteins, genetically endowed with PARP activity, detection of polymers indicates segregation from the cytosol and consequently intra-organellar residence. In mitochondria, polymer build-up reveals matrix localization of the PARP fusion protein. Compared to presently used fusion tags for subcellular protein localization, these are substantial improvements in resolution. We thus established a novel molecular tool applicable for studies of subcellular NAD metabolism and protein localization.
Keywords: Compartmentation, ADP-ribosylation, NAD metabolism, Mitochondria, Protein import
Introduction
Two major strategies have emerged to determine the subcellular localization of proteins. Cell fractionation has been used to enrich organelles with subsequent identification of the proteins present (“Divide and identify”) [1]. Alternatively, a protein of interest is expressed in fusion with a peptide tag or a fluorescent reporter protein (e.g., green fluorescent protein, GFP). Subcellular localization of the recombinant protein is based on immunocytochemical detection of the peptide tag or the intrinsic fluorescence of the detector protein (“Tag and tell”). Both approaches have been successfully used for large-scale protein localization [2–4]. Unfortunately, these methods do not discriminate between an external association to an organelle and luminal localization, because the detection is independent of suborganellar specifics. In eukaryotic cells, mitochondria pose an additional challenge owing to their two surrounding membranes [5]. To resolve suborganellar localization, elaborate procedures are currently used including protease protection assays following organelle isolation and electron microscopy after immunogold labeling.
We reasoned that different metabolic conditions within subcellular compartments could be exploited to facilitate protein localization. That is, the protein itself would not be detected, but a functional parameter such as enzyme activity. When fused to a protein of interest, the enzyme would be targeted to the native subcellular location of the analyte protein. Application of such an approach would require a readily detectable product which should not be present endogenously at significant concentrations. Moreover, the substrate or any further conversions of the product should have an organelle-specific distribution. As outlined below, these criteria are met using poly-ADP-ribose polymerase-1 (PARP1) as marker enzyme.
Poly-ADP-ribose polymerase-1 generates protein-bound poly-ADP-ribose (PAR) using NAD+ as substrate. It is a nuclear enzyme which preferentially attaches PAR to itself (automodification). The protein-bound PAR chains consist of up to 200 ADP-ribose units and are most often branched [6–8]. These biopolymers, but not NAD or single units of ADP-ribose, are readily detectable by specific antibodies [9]. Under physiological conditions, PAR is not detected in cells, because the catalytic activity of endogenous PARP1 requires binding of the enzyme to DNA strand breaks. Deletion of the DNA binding domain from full-length PARP1 (Fig. 1a) results in a catalytically less, but constitutively active, polymerase in the absence of DNA lesions [6, 10]. PAR can be efficiently degraded to ADP-ribose by PAR glycohydrolase (PARG) whose isoforms are predominantly located within the cytosol and the nucleus [11]. In mammalian cells, transport of NAD+ across intracellular membranes (except the nuclear and outer mitochondrial membranes) has not been detected suggesting the existence of independent, non-exchangeable pools. Therefore, we considered that differential accumulation of PAR provides a reporter system for subcellular protein localization.
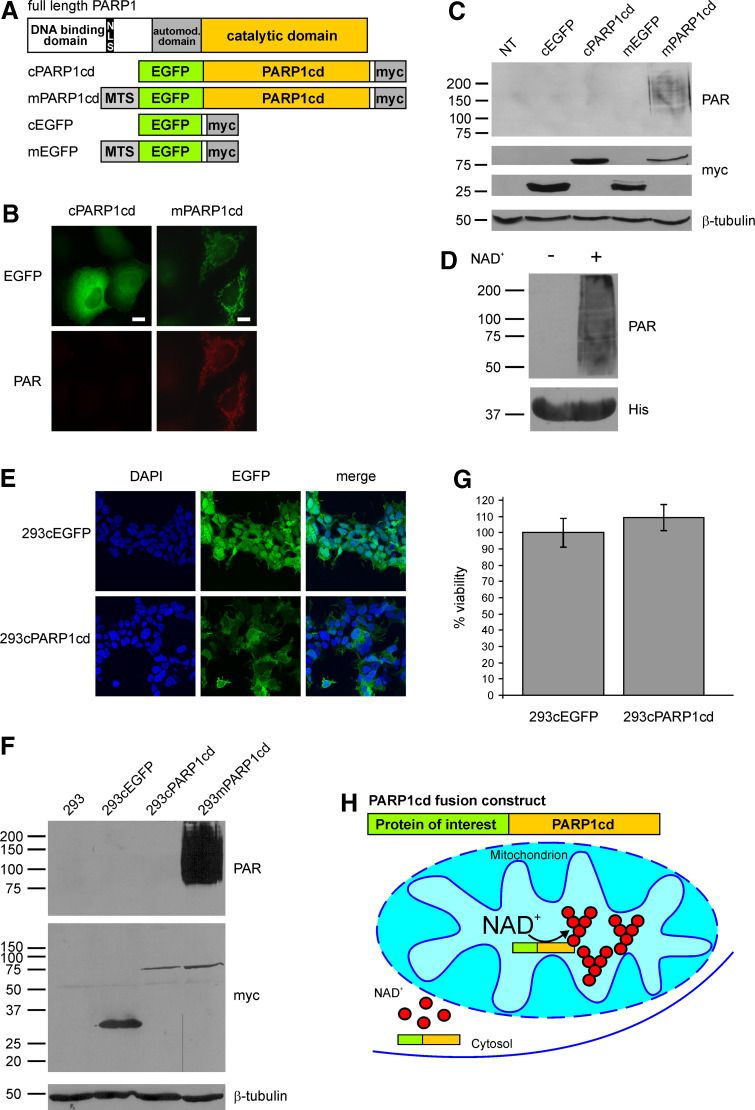
Fig. 1.
Mitochondrial, but not cytosolic overexpression of PARP1cd results in immunodetectable PAR accumulation. a Molecular architecture of poly-ADP-ribose polymerase 1 and the generated PARP1cd and EGFP fusion constructs. MTS Mitochondrial targeting sequence, NLS nuclear localization signal. b Fluorescence micrographs of HeLa S3 cells subjected to PAR immunocytochemistry 24 h after transient transfection with vectors encoding cytosolic or mitochondrial EGFP-PARP1cd (cPARP1cd and mPARP1cd). Protein expression was monitored by the intrinsic fluorescence of the EGFP portion of the constructs. Bar 10 μm. c PAR immunoblot analysis of lysates from HeLa S3 cells expressing cPARP1cd or mPARP1cd or the respective constructs lacking the PARP1cd portion (cEGFP and mEGFP). Overexpression of the proteins was detected by the C-terminal myc-tag. As observed in (b), PAR formation was only detected in cells expressing mPARP1cd. Loading control: β-tubulin. d Addition of NAD+ (1 mM) to bacterially expressed human PARP1cd (amino acids 652–1014) led to automodification of the protein as visualized by PAR immunoblot analysis. The bottom part shows the immunodetection of the protein’s 6xHis-tag. e Stably transfected 293 cells expressing cytosolic EGFP (293cEGFP) or cytosolic EGFP-PARP1cd (293cPARP1cd). Protein expression as monitored by the intrinsic fluorescence of the EGFP portion of the constructs is detectable in all cells. f Immunoblot analyses of lysates from parental and stably transfected 293 cell lines. Expression of cEGFP, cPARP1cd and mPARP1cd was detected by the C-terminal myc epitope. Mitochondrial and cytosolic PARP1cd fusion proteins were expressed at similar levels. PAR immunoblot analysis using 10H antibody revealed detectable PAR formation only in cells expressing mitochondrial PARP1cd. Loading control: β-tubulin. g Constitutive expression of cPARP1cd does not affect cell viability. Cells were seeded into 96-well plates and the viability was determined by MTT assay. Viability of 293cPARP1cd was related to control 293cEGFP cells set to 100%. Data are shown as mean ± SE of four experiments, each performed in triplicate. h Schematic representation of the observed differences in PAR accumulation mediated by targeted overexpression of PARP1cd fusion proteins. Cytosolic PARP1cd does not give rise to immunodetectable PAR. NAD and ADP-ribose are not recognized by the PAR-specific antibodies. The presence of PARP1cd within mitochondria leads to extensive PAR formation
On the other hand, the conversion of NAD+ into immunodetectable polymers would permit the visualizing and study of the subcellular distribution of this nucleotide. Indeed, so far, detection of organellar NAD+ has only been achieved using cell disruption and fractionation followed by nucleotide extraction and analysis. While cytosolic/nuclear and mitochondrial matrix contents have been measured, the presence of NAD in other compartments has not been established, at least in part because of the analytical difficulties. The activities of NAD-dependent dehydrogenases attest the presence of this nucleotide in peroxisomes, while the membranes of these organelles appear impermeable to NAD [12].
Recent research has revealed a tremendous array of regulatory pathways which involve conversions of NAD+ in signaling reactions [13]. For example, NAD+-dependent protein de-acetylation by sirtuins plays a major role in lifespan regulation [14, 15] and energy metabolism [16, 17]. Poly-ADP-ribosylation has important functions in DNA repair [8], telomere maintenance [18], cell division [19], and apoptosis [20]. Several derivatives of pyridine nucleotides including ADP-ribose, cyclic ADP-ribose, and NAADP serve as second messengers in calcium signaling [21, 22]. These signaling processes are compartmentalized. To understand their functions and regulation, it is essential to know the subcellular distribution of NAD+ and the dynamics of its individual pools. NAD(P)H fluorescence has been used to estimate fluctuations of reduced pyridine nucleotides in living cells [23]. However, the sensitivity limits the method to pools of high NAD(P)H concentration, such as mitochondria.
We show here that targeted expression of PARP activity permits the visualizing of individual subcellular NAD+ pools, based on the conversion of the nucleotide to polymers. Moreover, the differential, compartment-specific stability of these polymers provides a novel, easily adaptable technique to establish intra-organellar localization of proteins.
Materials and methods
Chemicals, reagents, and media
All chemicals and reagents were of analytical grade. The following antibodies were used: mouse anti-FLAG and mouse anti-β-Tubulin antibodies (Sigma-Aldrich), rabbit anti-PAR (96-10-04; Alexis Biochemicals), chicken anti-myc and rabbit anti-Pmp70 (Invitrogen), mouse anti-GM130 (BD Biosciences), mouse anti-GFP (JL8; Clontech) and mouse anti-PDI (ab2792; Abcam). Mouse anti-myc (9E10) and mouse anti-PAR (10H) antibodies were from hybridoma cell culture supernatants. Highly cross-adsorbed fluorescent-conjugated secondary antibodies were from Invitrogen/Molecular Probes. ECL reagents and HRP-conjugated goat anti-mouse/goat anti-rabbit antibodies were from Pierce or GE Healthcare/Amersham Biosciences.
Cloning and generation of eukaryotic expression vectors
To generate a vector encoding C-terminally myc-tagged cPARP1cd, the cDNA fragment encoding amino acids (aa) 572–1014 of full-length PARP1 was ligated into pCMV/myc/cyto (Invitrogen) via its PstI/SalI sites. Subsequently, the cDNA encoding enhanced GFP (EGFP; Clontech Laboratories) was inserted into the vector via the PstI site. A vector encoding only the EGFP portion was prepared by replacing the GFP encoding cDNA from pCMV/myc/cyto/GFP (Invitrogen) with the cDNA encoding EGFP using PstI/NotI sites of the vector and PstI/Bsp120I sites for the insert.
For expression of a protein of interest in fusion with PARP1cd, the DNA sequence encoding the C-terminal catalytic domain of PARP1 along with a C-terminal myc-epitope was amplified from an existing vector [24] and ligated into pcDNA3.1(+) (Invitrogen) via EcoRI/XbaI sites. The open reading frames (ORFs) of the proteins of interest were amplified and introduced into this vector via KpnI/EcoRI restriction sites. In order to verify the localization of the proteins of interest by indirect immunocytochemistry, their corresponding cDNA sequences were inserted into pFLAG-CMV-5a (Sigma). Peroxisomal constructs were generated by PCR amplification of the full-length ORFs of EGFP and EGFP-PARP1cd from the above described vector and ligation via KpnI/EcoRI into pcDNA3.1(+); coding for the peroxisomal targeting signal (SKL) was included in the primer sequence. To generate a vector encoding matrix-targeted AIF-PARP1cd, the ORF of AIF-PARP1cd was ligated into pCMV/myc/mito (Invitrogen) using the SalI restriction site. All cloned cDNAs were amplified using Pfu DNA polymerase.
Cell culture
HeLa S3 cells were cultivated in Ham’s F12 medium supplemented with 10% (v/v) FCS and penicillin/streptomycin. Transient transfection of eukaryotic cells was performed for 24–48 h using Effectene reagent (Qiagen) according to the recommendations of the manufacturer. Stably transfected monoclonal 293 lines expressing cEGFP or cPARP1cd were generated as described previously [24] using vectors encoding cEGFP or cPARP1cd. The cell lines, termed 293cEGFP and 293cPARP1cd, were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) FCS, 2 mM glutamine, penicillin/streptomycin, and 100 μg/ml G418.
Cell viability was determined using an MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay as described [24]. Viability was expressed as the ratio of values obtained from cells cultivated for 48 h and cells grown for 24 h. The data obtained for the control 293cEGFP cells were set to 100%.
Immunocytochemistry
Cells grown on cover slips were fixed with ice-cold 4% (v/v) formaldehyde in PBS for 45 min and subsequently permeabilized for 15 min using 0.5% (v/v) Triton X-100 in PBS. In some experiments, cells were treated with 0.2 μM MitoTracker Red CMXRos (Invitrogen) in full medium for 30 min prior to fixation. PAR detection was performed using PAR-specific antibody 96-10-04. Nuclei were stained with DAPI. Images were taken using a Leica DMI6000B epifluorescence microscope (Leica Microsystems) equipped with a ×100 oil immersion objective (numerical aperture, 1.40).
Protein determination, SDS-PAGE and western blot analysis
Protein concentration of cell lysates prepared in 20 mM Tris/HCl pH 7.4, 150 mM NaCl, 2% (w/v) sodium dodecyl sulfate (SDS) and 1 mM EDTA was determined using the bicinchoninic acid method (Pierce). SDS polyacrylamide gel electrophoresis and immunoblotting were carried out according to standard procedures; enhanced chemiluminescence was used for detection.
Results and discussion
PAR accumulation can be observed in the mitochondrial matrix, but not the cytosol
When artificially targeted to the mitochondrial matrix, the catalytic domain of PARP1 (PARP1cd) fused to EGFP gave rise to PAR within the organelles [24]. In contrast, expression of the EGFP-PARP1cd construct in the cytosol did not result in detectable PAR formation (Fig. 1b, c). As a potential reason for the absence of PAR accumulation in the cytosol, the requirement for an endogenous PAR acceptor protein can be ruled out. As shown in Fig. 1d, a bacterially expressed, purified PARP1cd-6xHis protein encompassing amino acid residues 652–1014 of human PARP1 (kindly provided by Dr F. Koch-Nolte, Hamburg, Germany) strongly automodified itself, similar to a previously reported PARP1cd [10].
Poly-ADP-ribose was recently suggested to act as a death signal when accumulating in the cytosol following PARP1 activation during genotoxic stress [20, 25]. However, we can exclude a cytotoxic effect, which could potentially mask PAR build-up by a PARP1cd fusion protein expressed in the cytosol. As shown in Fig. 1b, c, the expression level of cytosolic PARP1cd was at least as high as that of the mitochondrial construct indicating that PARP1cd was equally well tolerated in both compartments. To further validate this point, we generated stably transfected 293 cell lines expressing either just EGFP (293cEGFP cells) or the EGFP-PARP1cd construct (293cPARP1cd cells) in the cytosol. Expression of the proteins was monitored by their intrinsic green fluorescence, which was detected in all cells (Fig. 1e). Immunoblot analyses of 293cPARP1cd cell lysates verified expression of the full-length cPARP1cd protein at a similar level compared to the same construct targeted to mitochondria in stably transfected 293mPARP1cd cells (Fig. 1f). Importantly, no PAR immunoreactivity was observed in lysates from 293cPARP1cd cells, similar to parental 293 or 293cEGFP control cells (Fig. 1f). In addition to the successful generation of these cells per se, the undiminished viability of the 293cPARP1cd cells (Fig. 1g) strongly argues against a possible cytotoxic effect exerted by the cytosolic PARP1cd. Measurements of the total cellular NAD content in 293cPARP1cd cells revealed a significant reduction by about 20% (from 4.8 to 3.9 nmol/mg protein) compared to stably transfected control cells, which constitutively expressed EGFP. This observation further substantiates the notion that the cPARP1cd is catalytically active. Most likely, the polymers synthesized by cPARP1cd are subject to rapid degradation by endogenous PARG, whose isoforms reside predominantly in the cytosol [11]. The high turnover would lead to an overall non-detectable PAR formation, but still cause an appreciable reduction of cellular NAD.
The observations so far indicated that the presence of PARP1cd fusion proteins in either the mitochondria or the cytosol can be distinguished based on the accumulation of PAR, as illustrated in Fig. 1h. Importantly, immunochemical detection of PAR following expression of PARP1cd in the mitochondrial matrix was equally conclusive using western blotting or fluorescence microscopy (Fig. 1b, c).
In mitochondria, PAR accumulation is specific for matrix-localized PARP1cd
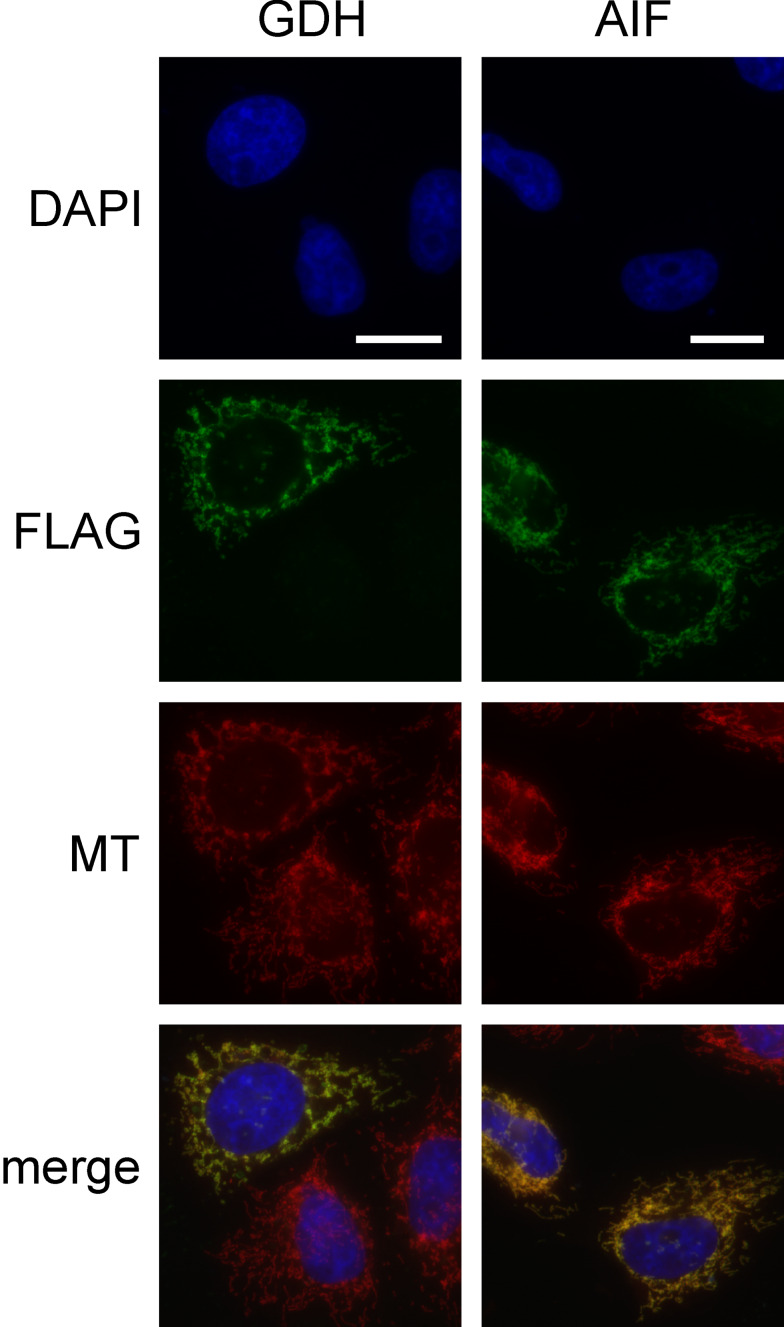
We then compared PAR accumulation for PARP1cd fusion proteins localizing to the matrix or intermembrane space (IMS) of mitochondria. Glutamate dehydrogenase (GDH) is a well-established matrix protein [26] with a typical N-terminal targeting sequence [27]. To direct PARP1cd into the IMS, we used apoptosis inducing factor (AIF) whose targeting is also mediated by its N-terminus [28]. AIF is anchored in the inner mitochondrial membrane facing the IMS [29]. As expected, both GDH and AIF, tagged by a FLAG epitope, similarly co-localized with the mitochondria-specific dye MitoTracker (Fig. 2). However, this common method does not resolve whether a protein is located in the matrix.
Fig. 2.
Association of GDH- and AIF-FLAG constructs with mitochondria. HeLa S3 cells were transiently transfected with vectors encoding GDH and AIF as FLAG tagged proteins. After 24 h cells were stained with MitoTracker (MT) and subjected to FLAG-immunocytochemistry. The fluorescence micrographs show nuclei (DAPI), expressed recombinant proteins (FLAG), and mitochondria (MT). Bar 10 μm
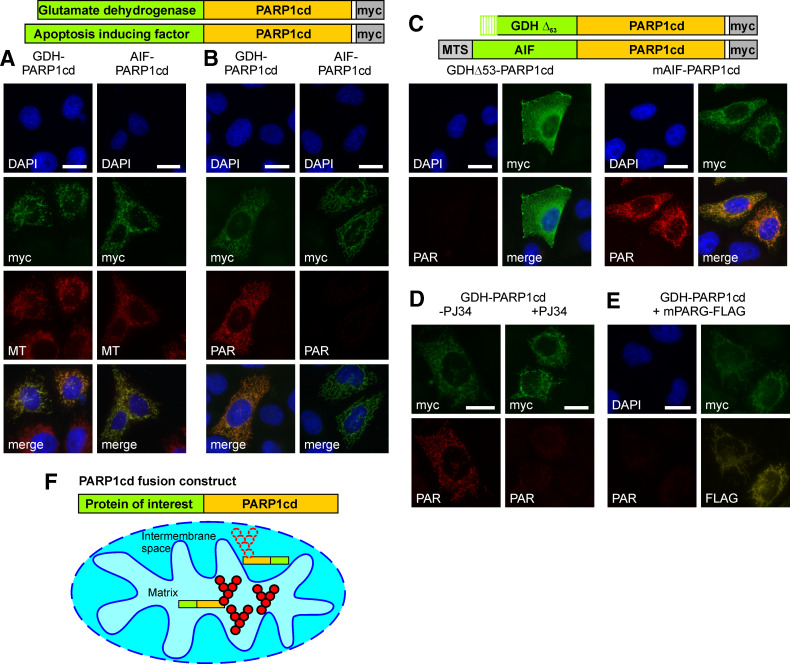
We then transiently transfected HeLa S3 cells with vectors encoding constructs consisting of PARP1cd C-terminally fused to GDH or AIF. Immunodetection of their C-terminal myc-tags demonstrated a cellular distribution just as the FLAG-tagged constructs lacking the PARP1cd (Fig. 3a). Importantly, expression of the GDH-PARP1cd construct resulted in the accumulation of PAR (Fig. 3b, left) confirming that PAR is readily detectable when PARP1cd is directed into the matrix (cf. Fig. 1b, c). In contrast, PAR formation was virtually undetectable in cells expressing IMS-targeted AIF-PARP1cd (Fig. 3b, right). Only a minor portion of these cells showed PAR immunoreactivity, however, and only at the limit of detection and therefore clearly distinguishable from the matrix-localized PAR generated by GDH-PARP1cd.
Fig. 3.
Submitochondrial localization of PARP1cd fusion proteins based on PAR accumulation in the matrix, but not the intermembrane space. a GDH-PARP1cd and AIF-PARP1cd fusion proteins localize to mitochondria. HeLa S3 cells were transiently transfected with vectors encoding the indicated mitochondrial PARP1cd fusion proteins. After 24 h, cells were stained with MitoTracker (MT) and subjected to myc-immunocytochemistry. The fluorescence micrographs show nuclei (DAPI), expressed recombinant proteins (myc), and mitochondria (MT). Bar 10 μm. b Robust PAR accumulation is restricted to the mitochondrial matrix. After transient transfection of vectors encoding GDH-PARP1cd and AIF-PARP1cd, HeLa S3 cells were subjected to immunocytochemistry using myc- and PAR-specific antibodies. The fluorescence micrographs show nuclei (DAPI), expressed recombinant proteins (myc), and generated PAR. Bar 10 μm. c Artificial re-targeting of mitochondrial PARP1cd fusion proteins confirms matrix localization to be essential for PAR accumulation. HeLa S3 cells were transfected with vectors encoding the indicated PARP1cd fusion constructs (matrix-targeted mAIF-PARP1cd and GDHΔ53-PARP1cd lacking the mitochondrial targeting sequence) and subjected to immunocytochemistry as in (b). Bar 10 μm. d PAR accumulation requires PARP1cd catalytic activity. HeLa S3 cells transiently transfected with a GDH-PARP1cd encoding plasmid were cultured in presence (+) or absence (−) of the PARP1 inhibitor PJ34 (5 μM). Cells were subjected to PAR- and myc-immunocytochemistry 24 h post transfection as in (b). Bar 10 μm. e Co-expression of matrix-targeted PARG (mPARG) eliminates GDH-PARP1cd-mediated PAR accumulation. HeLa S3 cells were co-transfected with vectors encoding GDH-PARP1cd and a FLAG-tagged mitochondrial PARG construct (mPARG). The fluorescence micrographs show nuclei (DAPI), GDH-PARP1cd (myc), mPARG (FLAG), and polymers (PAR). Bar 10 μm. f Schematic representation of the principle to distinguish between mitochondrial matrix and intermembrane space protein localization. If PARP1cd fusion proteins are directed into the matrix, PAR accumulation (aggregates of red spheres) is readily observed, whereas PAR is virtually not detectable (dotted spheres) for fusion proteins residing in the intermembrane space
To verify that matrix localization, and not the individual protein fused to PARP1cd, was important for PAR accumulation, the AIF-PARP1cd fusion construct was endowed with a bona fide matrix targeting sequence (mAIF-PARP1cd; Fig. 3c). Conversely, a GDH-PARP1cd fusion protein was expressed which lacked the N-terminal 53 amino acids harboring the matrix targeting sequence of GDH (GDHΔ53-PARP1cd, Fig. 3c). Transiently expressed mAIF-PARP1cd localized to mitochondria and now led to accumulation of PAR (Fig. 3c, right). On the other hand, the GDHΔ53-PARP1cd protein was distributed throughout the cytosol and no PAR was detectable (Fig. 3c, left). Consequently, fusion of mitochondrial proteins to PARP1cd enables to discern their localization in the matrix or IMS (Fig. 3f).
We also verified that the PAR immunoreactivity observed for matrix-targeted PARP1cd indeed relied on the enzymatic activity borne by PARP1cd. As exemplified for GDH-PARP1cd, the presence of the PARP inhibitor PJ34 [30] precluded PAR accumulation (Fig. 3d).
Matrix-targeted expression of PARG eliminates PAR accumulation
Poly-ADP-ribose glycohydrolase degrades PAR to ADP-ribose, thereby preventing accumulation of PAR. Indeed, when GDH-PARP1cd was co-expressed with a matrix-targeted PARG [24], the PAR signal was essentially eliminated (Fig. 3e). Therefore, targeted PARG expression provides an additional tool to verify the suborganellar localization of PARP1cd fusion proteins.
PAR accumulation reveals NAD+ pools and luminal localization of PARP1cd fusion proteins in other cell organelles
We then investigated whether the localization assay could be extended to other membrane-coated compartments. This would require availability of the PARP substrate, NAD+, which could be anticipated for peroxisomes. However, whether NAD+ is present in other organelles has remained unknown, but was suggested for the endoplasmic reticulum (ER) [31].
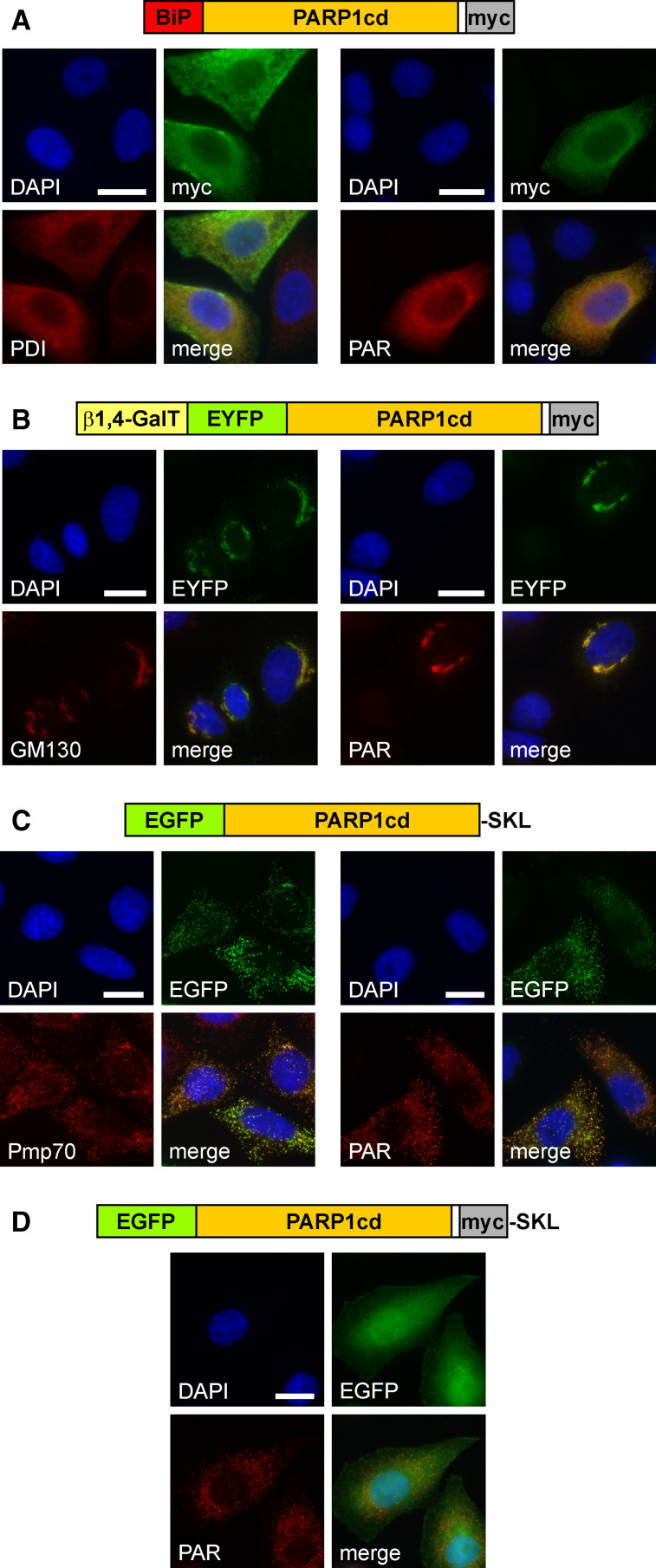
To direct PARP1cd into the ER, it was fused to the N-terminal 100 amino acids of the luminal ER marker binding immunoglobulin protein [32]. As shown in Fig. 4a, this hybrid protein indeed localized to the ER (left) and led to robust accumulation of PAR (right). Interestingly, the C-terminal retention signal (KDEL) was dispensable to retain the fusion protein within the ER, while its presence did not influence localization or PAR formation (not shown).
Fig. 4.
Targeted expression of PARP1cd to the lumen of the endoplasmic reticulum, the Golgi apparatus or peroxisomes results in detectable PAR formation within these organelles. a HeLa S3 cells transfected with a vector encoding the first 100 amino acids of binding immunoglobulin protein (BiP) N-terminally fused to PARP1cd (BiP-PARP1cd) were subjected to immunocytochemistry after 24 h. The overexpressed protein (myc) co-localized with the ER marker protein disulfide isomerase, PDI (left), and mediated PAR accumulation (right). Bar 10 μm. b HeLa S3 cells were transfected with a vector encoding a Golgi-targeted EYFP-PARP1cd fusion construct, gEYFP-PARP1cd. The intrinsic EYFP-fluorescence of the overexpressed recombinant protein localized to the Golgi complex as revealed by immunostaining of the Golgi-specific marker protein GM130 (left). PAR accumulation (right) was observed in cells overexpressing gEYFP-PARP1cd. Bar 10 μm. c HeLa S3 cells were transiently transfected with a vector encoding EGFP-PARP1cd harboring the C-terminal tripeptide SKL for peroxisomal targeting. Cells were subjected to immunocytochemistry after 24 h using antibodies against the peroxisomal marker protein Pmp70 (left). The overexpressed protein led to PAR accumulation within peroxisomes (right). Expression of the recombinant protein was monitored by its intrinsic EGFP fluorescence. Bar 10 μm. d An additional myc epitope, immediately preceding the C-terminal SKL signal, perturbed peroxisomal localization and led to predominant cytoplasmic distribution of the overexpressed protein. However, accumulation of PAR in peroxisomes revealed that the protein was still partially localized within the organelles
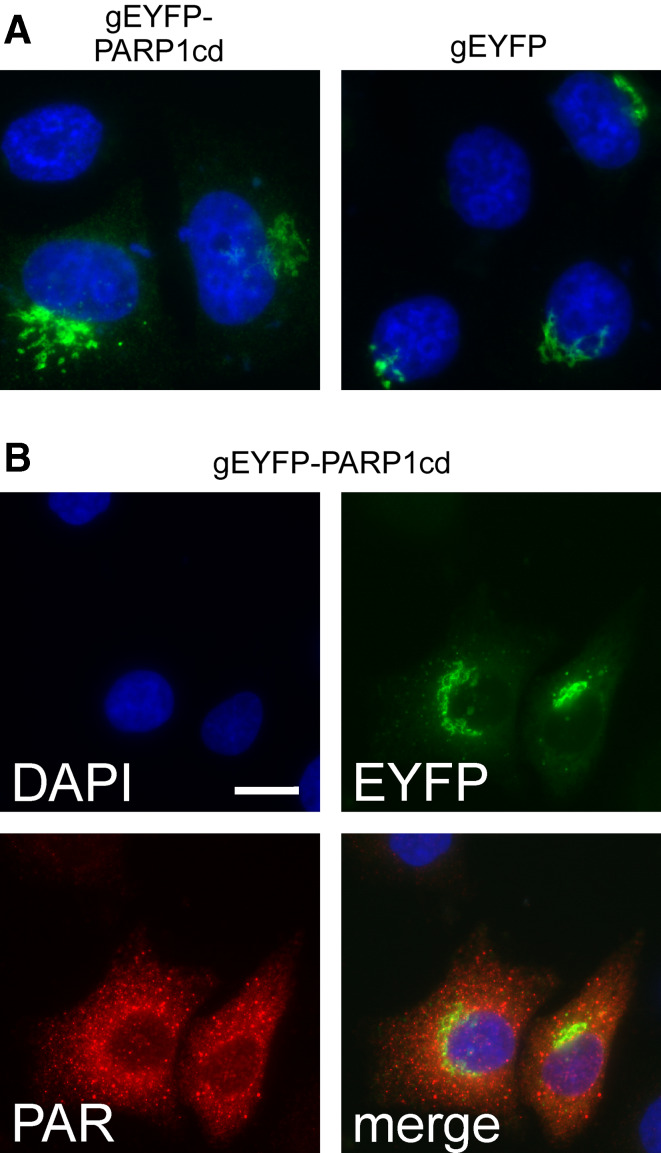
To direct an EYFP-PARP1cd construct to the Golgi complex we used the N-terminal targeting sequence of β1-4 galactosyltransferase [33]. As expected, the fusion protein exhibited a subcellular distribution similar to an established Golgi-targeted EYFP carrying the same targeting sequence (Fig. 5a). Golgi localization of the overexpressed EYFP-PARP1cd was further verified by co-localization with the endogenous marker protein GM130 (Fig. 4b). Strikingly, the overexpressed PARP1cd fusion protein led to PAR accumulation within the Golgi complex (Fig. 4b, right). We noted, however, that only in less than 20% of the transfected cells did the PAR signal coincide with the protein location. More often, the PAR signal was observed in cytoplasmic vesicles of unknown nature (Fig. 5b). Possibly, these vesicles represent post-Golgi compartments (such as lysosomes) in which the protein, but not the polymers, had already been degraded. Importantly, the visualization of PAR, which was generated in living cells, for the first time conclusively demonstrated the presence of NAD+ in the ER and the Golgi complex.
Fig. 5.
Golgi-targeted PARP1cd fusion proteins give rise to PAR-positive cytoplasmic vesicles. a The Golgi-targeted EYFP-PARP1cd fusion protein exhibits the same subcellular distribution as an established Golgi-EYFP construct carrying the same targeting sequence. The proteins were detected by their intrinsic fluorescence, nuclei were stained with DAPI. b HeLa S3 cells expressing a Golgi-targeted PARP1cd fusion construct displayed consistent PAR immunoreactivity as revealed by immunocytochemistry. While the protein was detected in Golgi structures by its intrinsic EYFP fluorescence, the PAR signal most often did not co-localize with the EYFP signal, but was detected in cytoplasmic vesicles. Note that in part of the transfected cells both the protein and the PAR co-localized within the Golgi complex (cf. Fig. 3b). Bar 10 μm
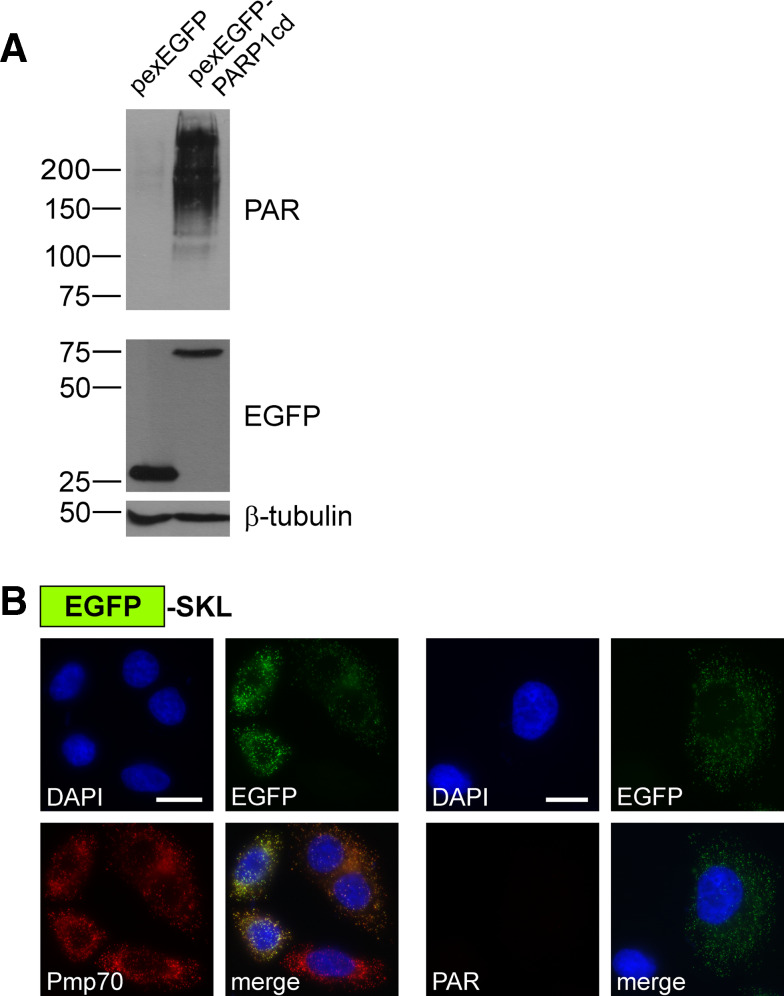
Finally, we targeted the PARP1cd into peroxisomes using peroxisomal targeting signal 1, the tripeptide Ser-Lys-Leu (SKL) [34]. We designed a potential peroxisomal fusion protein consisting of EGFP N-terminally fused to PARP1cd which, in turn, was followed by the SKL sequence (pexEGFP-PARP1cd). As shown in Fig. 4c (left), the recombinant protein indeed co-localized with the peroxisomal marker Pmp70 [35]. Moreover, expression of pexEGFP-PARP1cd led to the formation of readily detectable PAR within peroxisomes (Fig. 4c, right). As exemplified before for mitochondrial matrix-targeted PARP1cd (Fig. 1c), PAR formation within peroxisomes was also readily visualized by western blotting (Fig. 6a). Expression of a peroxisomal EGFP control protein lacking the PARP1cd portion did not confer immunoreactivity for antibodies recognizing PAR, as expected (Fig. 6a, b). Collectively, these results support the conclusion that the ER, the Golgi complex and peroxisomes contain sufficient NAD+ for PARP1cd-mediated PAR synthesis. In addition these organelles contain only low, if any, PAR degrading activity thereby enabling protein localization based on PAR accumulation.
Fig. 6.
Detection of peroxisomal protein localization. a PAR immunoblot analysis of lysates from HeLa S3 cells transiently expressing EGFP or EGFP-PARP1cd targeted to peroxisomes as indicated. PAR accumulation was detected only in lysates from cells expressing the pexEGFP-PARP1cd protein. Each lane was loaded with 70 μg of total protein. The overexpressed proteins were detected using an antibody recognizing the EGFP portion, β-tubulin served as loading control. b HeLa S3 cells transiently expressing EGFP targeted to the peroxisomes by a C-terminal SKL sequence were subjected to immunocytochemistry to detect the endogenous marker Pmp70 (left) or PAR (right). As monitored by its intrinsic EGFP fluorescence the protein co-localized with peroxisomal structures (Pmp70). However, the cells were negative for PAR immunoreactivity. Bar 10 μm
Partial luminal localization of PARP1cd fusion proteins is readily detected, even if most of the overexpressed protein resides in the cytosol
It is not uncommon that addition of tags to proteins may affect subcellular localization. Indeed, an almost identical pexEGFP-PARP1cd fusion protein, which was extended by just a myc epitope immediately upstream of the SKL tripeptide, did not exhibit an apparent peroxisomal localization, but was rather distributed throughout the cytoplasm (Fig. 4d). Strikingly, overexpression of this construct still caused PAR immunoreactivity, but only in structures resembling a peroxisomal pattern (Fig. 4d), which was verified by co-localization with Pmp70 (not shown). Possibly, the negatively charged myc-tag had weakened the recognition of the peroxisomal targeting signal, thereby leading to a preferential cytosolic accumulation of the protein. Nevertheless, the presence of the PARP1cd moiety permitted to reveal an only partial intra-organellar localization which, most likely, would have been overlooked using any of the currently available localization techniques. These observations also strongly reinforce the notion that PAR accumulation is only detectable, if the PARP1cd fusion protein resides within membrane-coated organelles, but not in the cytosol. Here, the protein present in the cytosol was identical to that within the peroxisomes.
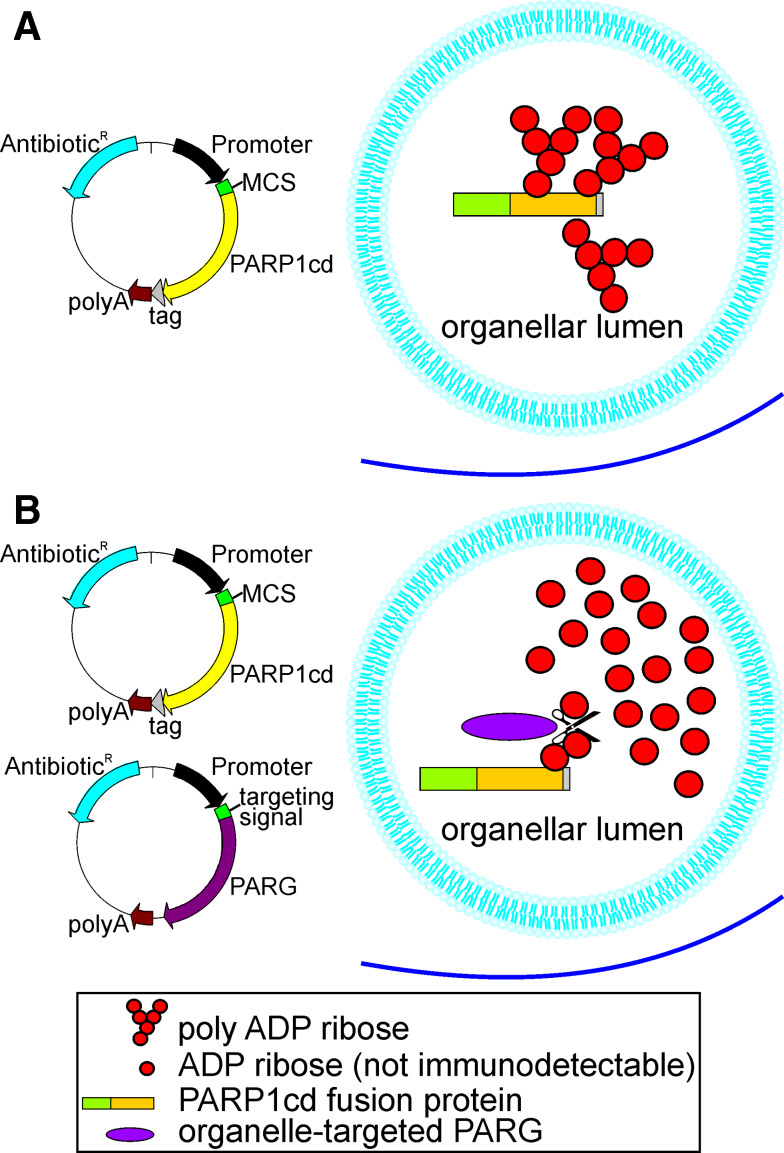
In conclusion, we have developed a PAR-Assisted Protein Localization AssaY, PARAPLAY, which is based on intra-organellar poly-ADP-ribosylation of PARP1cd fused to a protein of interest. The principle of this new method is illustrated in Fig. 7. It is also shown how targeted PARG expression can be applied to verify the localization of the PARP1cd fusion protein. The usefulness of this additional tool has been demonstrated for mitochondrial matrix localization of GDH-PARP1cd (Fig. 3e).
Fig. 7.
Schematic overview of Poly-ADP-Ribose Assisted Protein Localization AssaY (PARAPLAY). a To generate a construct encompassing the protein of interest in fusion with PARP1cd an expression vector (left) can be used harboring a multiple cloning site (MCS) to insert the cDNA encoding the protein of interest upstream of the PARP1cd cDNA. An optional C-terminal tag for protein detection may be included. Transfection of cells with the resultant vector leads to robust PAR accumulation, if the overexpressed protein is targeted to an organellar lumen (right). b Co-transfection of cells with the PARP1cd fusion construct and an organelle-targeted PARG may be conducted to confirm the results. PARG cleaves PAR to ADP ribose monomers, which are not detectable by PAR immunochemistry. Therefore, if the PARP1cd fusion protein resides within the same organelle as the targeted PARG, PAR accumulation is not detectable any more
The method we propose is based on a qualitative rather than quantitative detection of polymers. It was comprehensively tested for submitochondrial protein localization, but is also applicable to other organelles, provided they contain NAD+. Technically, the assay relies only on standard procedures of molecular and cellular biology, similar to the use of peptide tags or GFP-fusion proteins. However, it provides significant improvement in resolution and sensitivity compared to current standard methods. First, PARAPLAY enables to demonstrate intra-organellar residence of proteins (rather than just co-localization with an organelle). Second, once the organellar association of a protein is known, its luminal residence may be established simply by western blotting following overexpression of a PARP1cd fusion construct (Figs. 1c and 6a). Detection of the polymers by western blotting increases sensitivity and accuracy, because it reveals the cumulative signal from many cells, not just individual cells as in fluorescence microscopy. Third, even only partial luminal localization is revealed as experienced with a modified peroxisomal construct, which localized predominantly to the cytosol (Fig. 4d).
Another major achievement of the present study is the unveiling of distinct subcellular NAD+ pools in living cells. The detection of NAD+ in the ER and the Golgi complex suggests that the subcellular distribution of NAD+-dependent processes may be even more complex than so far thought. Recent discoveries of NAD+-dependent signaling pathways have prompted detailed investigations of NAD+ metabolism and its subcellular compartmentation. However, determination of NAD+ and its changes in cells, let alone organelles, is very demanding. Targeted overexpression of PARP1cd, thereby converting the dinucleotide into immunodetectable PAR in vivo, provides a promising approach to study NAD+-dependent processes. Future studies will be aimed at extending this new experimental tool to detect changes in individual NAD+ pools in response to metabolic alterations.
Acknowledgments
Christian Dölle, Marc Niere, and Mathias Ziegler are named as inventors in a pending patent application on this issue. The patent application is owned by Bergen Teknologioverføring AS.
Footnotes
C. Dölle and M. Niere contributed equally to this work.
References
- 1.Simpson JC, Pepperkok R. The subcellular localization of the mammalian proteome comes a fraction closer. Genome Biol. 2006;7:222. doi: 10.1186/gb-2006-7-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 3.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 4.O’Rourke NA, Meyer T, Chandy G. Protein localization studies in the age of ‘Omics’. Curr Opin Chem Biol. 2005;9:82–87. doi: 10.1016/j.cbpa.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 6.Bürkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 7.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 9.Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- 10.Simonin F, Menissier-de Murcia J, Poch O, Muller S, Gradwohl G, Molinete M, Penning C, Keith G, de Murcia G. Expression and site-directed mutagenesis of the catalytic domain of human poly(ADP-ribose)polymerase in Escherichia coli. Lysine 893 is critical for activity. J Biol Chem. 1990;265:19249–19256. [PubMed] [Google Scholar]
- 11.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 12.Antonenkov VD, Sormunen RT, Hiltunen JK. The rat liver peroxisomal membrane forms a permeability barrier for cofactors but not for small metabolites in vitro. J Cell Sci. 2004;117:5633–5642. doi: 10.1242/jcs.01485. [DOI] [PubMed] [Google Scholar]
- 13.Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 15.Taylor DM, Maxwell MM, Luthi-Carther R, Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci. 2008;65:4000–4018. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 19.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 20.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 21.Fliegert R, Gasser A, Guse AH. Regulation of calcium signalling by adenine-based second messengers. Biochem Soc Trans. 2007;35:109–114. doi: 10.1042/BST0350109. [DOI] [PubMed] [Google Scholar]
- 22.Guse AH, Lee HC. NAADP: a universal Ca2+ trigger. Sci Signal. 2008;1:re10. doi: 10.1126/scisignal.144re10. [DOI] [PubMed] [Google Scholar]
- 23.Mayevsky A, Rogatsky GG. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol. 2007;292:C615–C640. doi: 10.1152/ajpcell.00249.2006. [DOI] [PubMed] [Google Scholar]
- 24.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawajiri K, Harano T, Omura T. Biogenesis of the mitochondrial matrix enzyme, glutamate dehydrogenase, in rat liver cells. I. Subcellular localization, biosynthesis, and intracellular translocation of glutamate dehydrogenase. J Biochem. 1977;82:1403–1416. doi: 10.1093/oxfordjournals.jbchem.a131828. [DOI] [PubMed] [Google Scholar]
- 27.Rosso L, Marques AC, Reichert AS, Kaessmann H (2008) Mitochondrial targeting adaptation of the hominoid-specific glutamate dehydrogenase driven by positive Darwinian selection. PLoS Genet. 4:e1000150 [DOI] [PMC free article] [PubMed]
- 28.Loeffler M, Daugas E, Susin SA, Zamzami N, Metivier D, Nieminen AL, Brothers G, Penninger JM, Kroemer G. Dominant cell death induction by extramitochondrially targeted apoptosis-inducing factor. FASEB J. 2001;15:758–767. doi: 10.1096/fj.00-0388com. [DOI] [PubMed] [Google Scholar]
- 29.Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med. 2001;7:255–260. [PubMed] [Google Scholar]
- 31.Bublitz C, Lawler CA. The levels of nicotinamide nucleotides in liver microsomes and their possible significance to the function of hexose phosphate dehydrogenase. Biochem J. 1987;245:263–267. doi: 10.1042/bj2450263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas IG. BiP (GRP78), an essential hsp70 resident protein in the endoplasmic reticulum. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- 33.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramani S, Koller A, Snyder WB. Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- 35.Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. The 70-kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem. 1990;265:4534–4540. [PubMed] [Google Scholar]