Abstract
The committed step in the biosynthesis of the phytochrome chromophore phytochromobilin involves the oxidative cleavage of heme by a heme oxygenase (HO) to form biliverdin IXα. Through positional cloning of the photomorphogenic mutant hy1, the Arabidopsis HO (designated AtHO1) responsible for much of phytochromobilin synthesis recently was identified. Using the AtHO1 sequence, we identified families of HO genes in a number of plants that cluster into two subfamilies (HO1- and HO2-like). The tomato (Lycopersicon esculentum) yg-2 and Nicotiana plumbaginifolia pew1 photomorphogenic mutants are defective in specific HO genes. Phenotypic analysis of a T-DNA insertion mutant of Arabidopsis HO2 revealed that the second HO subfamily also contributes to phytochromobilin synthesis. Homozygous ho2-1 plants show decreased chlorophyll accumulation, reduced growth rate, accelerated flowering time, and reduced de-etiolation. A mixture of apo- and holo-phyA was detected in etiolated ho2-1 seedlings, suggesting that phytochromobilin is limiting in this mutant, even in the presence of functional AtHO1. The patterns of Arabidopsis HO1 and HO2 expression suggest that the products of both genes overlap temporally and spatially. Taken together, the family of HOs is important for phytochrome-mediated development in a number of plants and that each family member may uniquely contribute to the phytochromobilin pool needed to assemble holo-phytochromes.
The phytochrome (phy) family of chromoproteins are photoreceptors that play critical roles in mediating photomorphogenesis in higher plants (Neff et al., 2000; Smith, 2000). The functional photoreceptors are homodimers with each subunit containing the linear tetrapyrrole chromophore, (3E)-phytochromobilin (PΦB), attached to an approximately 120-kD polypeptide. PΦB is linked to apo-PHY through a thiol-ether bond to a specific Cys, using a lyase activity intrinsic to the polypeptide. Holo-phys can assume two stable conformations, a red light (R)-absorbing form (Pr) and a far-red light (FR)-absorbing form (Pfr), which are photo-interconvertible by the absorption of R and FR, respectively. By measuring the amount of Pfr and the ratio of Pr to Pfr, plants assess the intensity, duration, and spectral quality of the ambient light environment.
Assembly of holo-phys requires coordination of the pathways that synthesize the PHY polypeptides and the PΦB chromophore. Whereas the synthesis of the apoproteins is directed by a family of nuclear genes (Smith, 2000), the synthesis of PΦB is directed by an enzymatic cascade in the plastid that begins with 5-aminolevulinic acid (Terry et al., 1995; Terry, 1997). The early steps in the PΦB pathway are shared with those required to synthesize chlorophyll and heme. The committed step is the oxidative cleavage of a portion of the heme pool by a heme oxygenase (HO) to form biliverdin IXα (BV). BV is then reduced to (3Z)-PΦB by a ferredoxin-dependent bilin reductase (Frankenberg et al., 2001). Finally, (3Z)-PΦB is isomerized to create PΦB; however the phytochromobilin isomerase activity that is responsible for this 3Z to 3E conversion has not yet been conclusively demonstrated (Terry, 1997). Presumably, PΦB is then exported to the cytoplasm where it binds to the newly synthesized apo-phys.
Photomorphogenic mutants have been isolated in a variety of plant species that individually block either the PHY apoprotein or the PΦB-synthetic pathways. For example, Arabidopsis mutations in four of the five apoprotein-encoding genes have been identified: phyA, phyB, phyD, and phyE (Somers et al., 1991; Parks and Quail, 1993; Aukerman et al., 1997; Devlin et al., 1998, 1999). Analysis of these mutants demonstrated that each phy isoform has distinct and overlapping roles in light-regulated development (Whitelam and Devlin, 1997; Neff et al., 2000). A number of PΦB synthetic mutants also exist, and as predicted, these mutants globally decrease the activity of all phy isoforms. Examples include Arabidopsis long hypocotyl (hy)-1 and 2 (Parks and Quail, 1991), Nicotiana plumbaginifola partly-etiolated-in-white-light (pew)-1 and 2 (Kraepiel et al., 1994), pea phy-chromophore-deficient (pcd)-1 and 2 (Weller et al., 1996, 1997), tomato (Lycopersicon esculentum) aurea (au) and yellow-green (yg)-2 (Terry and Kendrick, 1996), and rice photoperiodic sensitive (se)-5 (Izawa et al., 2000). These mutants have dramatically reduced levels of PΦB and consequently holo-phys, and thus have severely impaired photomorphogenesis.
Analyses of several of the PΦB-synthetic mutants (hy1, se5, pcd1, pew1, and yg-2) suggested that they are specifically deficient in the HO activity(ies) that synthesize BV. hy1 and pew1 plants can be phenotypically rescued by feeding mutant seedlings BV (Parks and Quail, 1991; Kraepiel et al., 1994), whereas etioplast extracts from the pcd1 and yg-2 mutants are unable to convert heme to BV but are fully competent in converting BV into (3Z)-PΦB (Terry and Kendrick, 1996; Weller et al., 1996). By positional cloning of the HY1 locus, Davis et al. (1999) and Muramoto et al. (1999) independently determined that HY1 encodes a HO (designated AtHO1) responsible for much of PΦB synthesis in Arabidopsis. Using the AtHO1 sequence, Izawa et al. (2000) then demonstrated that a specific HO gene (designated here as OsHO1) is also altered in the rice se-5 mutant.
However, it is known that young seedlings of all the PΦB mutants retain residual R/FR sensitivity and, in some cases, they regain much of their phy-regulated responses as they mature, suggesting that other sources of PΦB are available. For example, tomato yg-2 mutants are compromised for most phy responses as young seedlings but respond more similar to wild type as adult plants (Koorneef et al., 1985; Kendrick et al., 1994; van Tuinen et al., 1996; Terry and Kendrick, 1999). These new sources could arise from additional HOs or from alternative pathways for making BV that become more prominent as plants develop.
To help define the importance of PΦB to plant photomorphogenesis, we have continued to characterize the HOs that synthesize the precursor of this bilin. Using the AtHO1 sequence as a query, we show here that most higher plants contain multiple HO genes. In Arabidopsis for example, three more HO genes (AtHO2-4) were identified in addition to AtHO1. Analysis of the tomato yg-2 and Nicotiana plumbaginifolia pew1 mutants demonstrated that each is defective in a specific HO. To help define the roles of the other members of the HO gene family, we isolated a T-DNA insertion mutant of AtHO2, the most divergent member of the Arabidopsis HO family. From phenotypic analysis of ho2-1, we found subtle phenotypes that resemble those seen in the hy1 mutant. Collectively the data indicate that HOs are important for proper photomorphogenesis in a number of plant species and that the family of HOs may function in concert to generate the supply of BV needed for holo-phy assembly.
RESULTS
Identification of HO Genes from Higher Plants
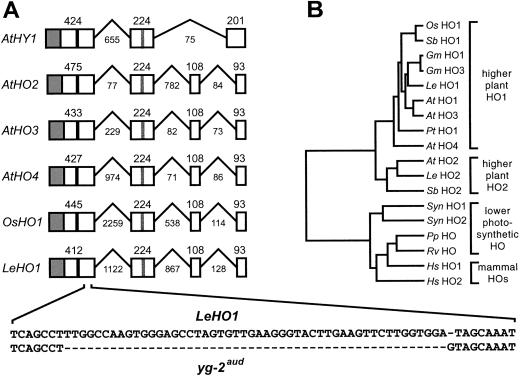
We and others recently identified the HO mainly responsible for PΦB synthesis in Arabidopsis through the positional cloning of the HY1 locus (Davis et al., 1999; Muramoto et al., 1999). (To create a unified HO nomenclature within plants that agrees with that used for HOs from other kingdoms, the various plant genes that encode HOs were given the designation HO with a prefix that identifies the species.) However, preliminary analysis of the Arabidopsis genome indicated that other HO genes are present, including one (designated AtHO2) only approximately 40 kb away from AtHO1 on chromosome II (Davis et al., 1999). To help define the size and organization of the HO family in plants and to identify conserved structural features among the corresponding enzymes, we used the AtHO1 gene as a query to search by blast all available DNA-sequence databases of higher-plant species. Against the essentially complete Arabidopsis genome, we identified numerous related sequences that condensed to four genes, AtHO1 and 2 and two new genes designated AtHO 3 and 4. Putative HO genes were identified from other plant species as expressed sequence tag (EST) cDNAs, including soybean, tomato, wild tomato, potato, ice plant, Medicago truncatula, Lotus joponicus, corn, rice, sorghum, barley, wheat, and pine (Fig. 1; data not shown). The rice HO1 gene, previously called OsHY1 (and SE5) was recently described (Izawa et al., 2000). The cDNAs for soybean GmHO1 and GmHO3, tomato LeHO1 and 2, pine PtHO1, and sorghum SbHO1 and 2 were sequenced in their entirety. With the exception of soybean GmHO1 and 3 and sorghum SbHO1, all these HO cDNAs were predicted to contain the entire coding region. None appeared to be allelic variants, suggesting that each clone represents an independent gene.
Figure 1.
Amino acid sequence comparisons of various plant HOs. Identical and similar residues are in reverse type or gray boxes, respectively. The diamond identifies the predicted cleavage site for the chloroplast-transit peptide. The black triangles denote the beginning of the region in LeHO1 altered in the tomato yg-2aud mutant and the T-DNA insertion site in Arabidopsis ho2-1. The black and white circles mark the histidines considered to be important for heme-iron binding and catalysis and for protein stability, respectively. Sequences include Arabidopsis (ectotype Col) AtHO1 (AF132475), AtHO2 (AF132475), AtHO3 (AF320022), and AtHO4 (AF320023); soybean GmHO1 (AF320024) and GmHO3 (AF320025); rice OsHO1 (C28969); sorghum SbHO1 (AF320026) and SbHO2 (AF320027); pine PtHO1 (AF320030); and tomato (cv Money-Maker) LeHO1 (AF320028) and LeHO2 (AF320029). The XXX in the sequences of GmHO1, GmHO3, and SbHO1 denotes the ends of the inferred polypeptide sequence from their truncated cDNAs.
As shown in Figure 1, amino acid sequence comparisons revealed that all plant HOs have a non-conserved N-terminal extension of various lengths that is followed by a approximately 220-amino acid region of high similarity. The ChloroP algorhythm (Emanuelsson et al., 1999) predicted that many of these N-terminal extensions encode transit peptides (data not shown), consistent with the plastid location of HO activity in plants (Terry et al., 1993; Muramoto et al., 1999). Following the predicted transit-peptide is a region of substantial conservation that is similar to the HO catalytic domain from animals, algae, and cyanobacteria (Ortiz de Montelano and Wilks, 2001). Animal HOs have hydrophobic C-terminal extensions that serve to anchor the enzymes to microsomal membranes (Schuller et al., 1999). No such extensions were evident in the plant counterparts, implying that they behave as soluble proteins.
Close examination of the plant HO sequences revealed that they cluster into two subfamilies related to either AtHO1 or AtHO2 (Figs. 1 and 2B). The HO1 subfamily included Arabidopsis AtHO1, 3, and 4, soybean GmHO1 and GmHO3, tomato LeHO1, sorghum SbHO1, rice OsHO1, and pine PtHO1. Representatives of the AtHO2 subfamily included Arabidopsis AtHO2, tomato LeHO2, and sorghum SbHO2. The main difference between the HO1 and 2 subfamilies is a 34- to 55-amino acid spacer in the HO2s that replaces a small, moderately conserved block of HO1 sequence (Fig. 1). The HO2 spacer is rich in Glu and Asp and/or small amino acids (Gly, Ala, Val, and Ser), suggesting that it forms a flexible, solvent-exposed loop. The HO2 group also contains a potentially important change at the active site. For animal HOs, a positionally conserved His is required for heme-iron binding and subsequent oxidative cleavage (Ortiz de Montelano and Wilks, 2001). This His is also retained in all algal and cyanobacterial HOs and members of the plant HO1 subfamily but is replaced by an Arg in the plant HO2 subfamily (Fig. 1). A second His needed for the structural integrity of animal HOs (Ortiz de Montelano and Wilks, 2001) is present in all higher plant enzymes (Fig. 1).
Figure 2.
Genomic organization and phylogenetic relationships of plant HOs. A, Genomic organization of Arabidopsis AtHO1-4, rice OsHO1, and tomato LeHO1. Boxes and lines denote coding regions and introns, respectively. Intron lengths are not to scale. Numbers refer to the nucleotide-sequence length of each domain. The gray boxes denote the positions of the putative transit peptide sequences. The black and gray vertical lines identify the locations of the histidines required for heme-iron binding and protein structure, respectively. DNA sequence surrounding the position of the yg-2aud mutation is indicated and aligned with the wild-type LeHO1 sequence from the AC cultivar. B, Phylogenetic comparisons of plant HOs with those from animals, algae, and cyanobacteria. The distance along the horizontal axis separating two sequences is proportional to their sequence divergence. Plant HO sequences are from Figure 1. Others include Synechocystis (Syn) HO1 (GenBank accession no. D90091) and HO2 (GenBank accession no. D90912), R. violacea (Rv) HO1 (GenBank accession no. AF000717), P. purpurea (Pp) HO (GenBank accession no. P51271), and human (Hs) HO1 (GenBank accession no. P09601) and HO2 (GenBank accession no. P30519).
Genomic Organization of Plant HO Genes
The genomic sequences of the four Arabidopsis HO genes and the tomato LeHO1 and rice OsHO1 genes were assembled and compared with their respective cDNAs to determine gene organization. The coding regions of four of the five (the exception being AtHO1, which contains only two introns) are interrupted by three introns whose splicing boundaries are positionally conserved (Fig. 2A). Moreover, the coding regions of the last three exons of AtHO2, AtHO3, AtHO4, LeHO1, and OsHO1 are the same length in each gene (224, 108, and 93 bp). Although AtHO1 contains only three exons, its third exon is the exact sum (201 bp) of the third and fourth exons (108 + 93 bp) of the other plant HOs (Fig. 2A). The similarity of this organization from widely divergent plants suggests that plant HO genes arose early in higher plant evolution by gene duplication events. Further, evolutionary pressures have apparently maintained the exact sequence-length conservation seen in the latter exons of plant HOs. Following the duplications and divergence that created the Arabidopsis HO1 subfamily, the AtHO1 gene presumably then lost its third intron.
The Evolutionary Relationships of Plant HOs to Those from Other Organisms
A phylogenetic tree was constructed using all available higher-plant HOs, two HOs from the cyanobacterium Synechocystis sp. PCC 6803 (Cornejo et al., 1998), one each from the alga Porphyra purpurea and Rhodella violacea (Richaud and Zabulon, 1997), and two human HOs (Platt and Nath, 1998). The results from the tree (Fig. 2B) confirmed expectations based on visual inspection of the derived polypeptide sequences (see Fig. 1) that plants have two subfamilies of HO genes, HO1-like and HO2-like. Members of the HO1 subfamily were detected in moncots, dicots, and a gymnosperm with multiple forms present in Arabidopsis, suggesting that this family is widespread in higher plants with each species potentially containing multiple members. In contrast, members of the HO2 subfamily were detected thus far only in angiosperms with a single member present in Arabidopsis, suggesting that this family arose more recently in plant evolution. Plant HOs as a group appear to be distantly related to HOs from animals, alga, and cyanobacteria. Their predicted evolutionary distance from cyanobacterial HOs suggests that, contrary to expectations, plant HOs did not originate from a cyanobacterial precursor during the symbiotic evolution of the chloroplast. Alternative origins include retention of a HO progenitor from the nuclear or mitochondrial genomes.
The Tomato yg-2 and N. plumbaginifolia pew1 Photomorphogenic Mutants Have Lesions in HOs
The tomato yg-2 mutants are phenotypically compromised for a number of phy-regulated photoresponses. In addition to the partially-etiolated phenotype of adult plants (as seen by their yellow-green color; Fig. 3A), young yg-2 seedlings have longer hypocotyls, repressed opening of the apical hook, delayed cotyledon development, and attenuated anthocyanin accumulation in R or white light (Fig. 3A; data not shown; Koorneef et al., 1985; van Tuinen et al., 1996; Terry and Kendrick, 1999). Given the similarity of some yg-2 phenotypes to those of Arabidopsis hy1 and the observations that yg-2 plants lack detectable HO activity (Terry and Kendrick, 1996), we considered it likely that the yg-2 plants contain a lesion in a HO gene. The most likely candidate would be LeHO1, an ortholog of AtHO1 (Fig. 1).
Figure 3.
Phenotypic comparison of the tomato yg-2aud and N. plumbaginifolia pew1 mutants. A, Wild-type (WT) and yg-2aud tomato plants grown for 2 weeks under continuous R (left) or for 4 weeks in a greenhouse under natural lighting (right). B, Complementation of the pew1 mutation by introducing Arabidopsis AtHO1. The pew1 mutant and three T1 lines of pew1 independently transformed with a genomic fragment encompassing AtHO1 (labeled 1–3) were grown for 2 months under continuous white light.
To test whether LeHO1 is defective in the yg-2 mutants, the LeHO1 locus was PCR amplified from genomic DNA isolated from the cv Ailsa Craig (AC) wild type and the AC-derived yg-2aud mutant and the products were directly sequenced. For the coding region, the LeHO1 gene sequence from the AC variety matched exactly with the corresponding LeHO1 cDNA sequence derived from the cv Money-Maker tomato variety presented in Figure 1. Comparing the LeHO1 sequence from the AC wild type with that of the yg-2aud mutant revealed a 51-bp deletion and a 1-bp insertion within the coding region, 303 bp downstream of the initiation codon (Fig. 2A). This alteration in yg-2aud predicted a LeHO1 protein that was missing the C-terminal 178 amino acids (78% of the mature protein) and had a foreign sequence of 23 amino acids (ANWFMILWKELWKRLLFLSVSFS) added. Given the importance of the C-terminal region for the activity of animal HOs (Ortiz de Montelano and Wilks, 2001) and the fact that even less pronounced truncations of AtHO1 abrogate its function in the Arabidopsis hy1 mutants (Davis et al., 1999; Muramoto et al., 1999), the yg-2aud mutant likely represents a null allele of LeHO1.
In many respects, the N. plumbaginifolia pew1 mutant is phenotypically similar to Arabidopsis hy1 and tomato yg-2, including an etiolated stature and a yellow-green color for mature plants grown under photoperiodic or continuous light (Fig. 3B; data not shown; Kraepiel et al., 1994). Because a near wild-type phenotype can be restored when pew1 plants are fed BV, it is likely that the mutation affects one or more HOs (Kraepiel et al., 1994). To test this directly, we tried to PCR-amplify N. plumbaginifolia HO genes from wild type and pew1 using degenerate primers homologous to conserved regions within the plant HO family. Despite repeated attempts using a number of primer combinations with both genomic and reverse transcriptase (RT) DNA preparations, we were unable to amplify any HO sequences.
As an alternative, we examined whether introduction of the Arabidopsis AtHO1 gene would complement the pew1 mutation. A genomic fragment, encompassing the entire AtHO1 coding region and substantial portions of the 5′- and 3′-flanking regions, was introduced into the pew1 mutant, and the resulting T1 plants were phenotypically analyzed. We previously showed that this construction could phenotypically rescue the Arabidopsis hy1 mutant (Davis et al., 1999). As can be seen in Figure 3B, multiple pew1 plants independently transformed with the AtHO1 gene lost their etiolated phenotype and became dark green with compressed internodes, the expected phenotype of wild-type N. plumbaginifolia. This restoration indicated that the pew1 mutant is likely deficient in one or more HOs. Moreover, it showed that the regulatory elements necessary for AtHO1 expression were sufficiently active in N. plumbaginifolia to supply an adequate level of HO protein for normal photomorphogenesis.
Isolation of the ho2-1 Mutation in Arabidopsis
Both the presence of multiple HO genes (Fig. 1) and the retention of some phy responses in the absence of one HO (Kraepiel et al., 1994; Terry and Kendrick, 1996; Davis et al., 1999; Muramoto et al., 1999; Izawa et al., 2000; Figure 3) suggested that one or more additional HOs contribute to PΦB synthesis in plants. To test this possibility, we examined the role of Arabidopsis AtHO2, the most distantly related HO with respect to AtHO1 (Fig. 2B) through characterization of the corresponding mutant. This ho2-1 mutant was identified via a PCR-based search of a population of T-DNA mutagenized Arabidopsis ecotype Wassilewskija (WS) for insertions within the AtHO2 gene (Krysan et al., 1999). The NPTII gene within the T-DNA disrupting AtHO2 conferred kanamycin resistance, thus facilitating future selection of ho2-1 plants. This resistance segregated as a single dominant locus with a 3:1 segregation ratio when heterozygous ho2-1 plants were selfed, indicating that the line likely contained a single insertion. Sequence analysis revealed that the T-DNA interrupted the 98th codon such that translation of the AtHO2 sequence ended 9-residues downstream of the predicted Arg active site (Fig. 1). Given the strong effects caused by similar mutations in the Arabidopsis and tomato HO1 loci hy1 and yg-2aud (see above), we considered it likely that ho2-1 is a null allele.
Effects of the ho2-1 Mutation on Photomorphogenesis
Phenotypic analysis of the ho2-1 mutant revealed that the AtHO2 protein, like AtHO1, also contributes to the proper photomorphogenesis of Arabidopsis, but has a more subtle role. One of the more obvious defects in hy1 plants missing AtHO1, which led to its isolation (Koornneef et al., 1980), is an elongated hypocotyl when grown under continuous R or FR (Parks and Quail, 1991; Davis et al., 1999). As shown in Figure 4, increased fluences of R or FR substantially repressed hypocotyl growth of young wild-type Col seedlings. In contrast, the hy1-100 seedlings (in the Col background) showed near complete insensitivity and continued to elongate regardless of the light fluence. For the ho2-1 mutant, the effect of light on hypocotyl elongation was less dramatic, but reproducible. As compared with the wild-type WS background, ho2-1 seedlings were slightly less sensitive to R and FR, especially at the higher fluences tested (Fig. 4). The etiolated ho2-1 seedlings also retained partially closed cotyledons when grown under low fluences of R or FR (data not shown), a phenotype also observed with hy1 seedlings (Chory et al., 1989).
Figure 4.
Effect of increasing fluence rates of R or FR on hypocotyl elongation of hy1-100 and ho2-1 Arabidopsis. Seedlings were germinated on agar and irradiated with various fluences of continuous R or FR. After 6 d, the hypocotyl lengths were measured. Each bar represents the average hypocotyl length (±sd) of approximately 15 seedlings. The respective wild-type parental ecotype of each mutant is included for comparison (Col for hy1-100 and WS for ho2-1).
As ho2-1 plants developed under white-light growth conditions, it became obvious that they had smaller rosette leaves and were slightly chlorotic as compared with wild-type plants (Fig. 5; Table I). Similar, but more dramatic phenotypes have been well described for hy1 plants (Chory et al., 1989). Chlorosis of hy1 is caused by both a reduction in total chlorophyll (Chl) and an alteration in the ChlA to B ratio, and by a lower number of chloroplasts per cell. To quantitate this chlorotic effect, ho2-1 and hy1-100 plants and their respective WS and Col parents were grown for 3 weeks under a long-day photoperiod, and then the levels of ChlA and B were measured. As expected, hy1 plants had markedly reduced levels of both ChlA and B (15% of wild type) and a skewed ratio of ChlA to B (Table I). Although not affected to the same extent, ho2-1 plants also had reduced levels of total chlorophyll (91% of wild type), but the ratio of Chl A to B was essentially unchanged (Table I). While a lower number of chloroplasts per cell was seen in hy1-100, the number of chloroplasts per cell was unaffected in the ho2-1 plants (Table I), suggesting that the change in Chl was caused primarily by a decrease in Chl content.
Figure 5.
Phenotype of light-grown Arabidopsis hy1-100 and ho2-1 seedlings. The respective wild-type parental ecotype of each mutant is included for comparison (Col for hy1-100 and WS for ho2-1). Plants were grown for 3 weeks under white light with a short-day photoperiod.
Table I.
Phenotypic comparisons among wild-type and HO mutants of Arabidopsisa
| Parameter | WS | ho2-1 | hy1-100 | Col-0 |
|---|---|---|---|---|
| Leaf number at bolting (short day)b | 35 ± 3 | 28 ± 2 | 25 ± 2 | 45 ± 5 |
| Leaf number at bolting (long day)cd | 6.9 ± 0.3 | 5.4 ± 0.3 | 4.4 ± 0.1 | 9.4 ± 1.1 |
| Leaf size (cm)e | 2.6 ± 0.3 | 1.6 ± 0.2 | 1.2 ± 0.4 | 2.9 ± 0.2 |
| Chlorophyll Af | 836 ± 27 | 751 ± 26 | 153 ± 5 | NDg |
| Chlorophyll B | 250 ± 7 | 240 ± 11 | 12 ± 1 | ND |
| Chlorophyll A+B | 1087 ± 33 | 990 ± 36 | 165 ± 5 | ND |
| Chlorophyll A/B | 3.3 ± 0.1 | 3.1 ± 1.0 | 13 ± 1 | ND |
| Chloroplasts per mesophyll cell | 82.6 ± 19.5 | 83.3 ± 19.6 | 46.4 ± 12.4 | 87.4 ± 18.1 |
Except for long-day flowering experiments, all data represents the mean of 12 plants (±sd).
Plants were grown under an 8-h-light/16-h-dark photoperiod.
Plants were grown under a 20-h-light/4-h-dark photoperiod.
Mean ± se of three independent experiments, each with 12 plants; individual experiments gave similar results.
Longest leaf measured on plants grown for 21 d under continuous white light.
Microgram chlorophyll per gram fresh wt tissue.
ND, Not determined.
The transition of vegetative to reproductive development in Arabidopsis is controlled by multiple phys and hence is sensitive to mutations that effect the synthesis of either the PHY apoproteins or PΦB (Murtas and Millar, 2000; Neff et al., 2000; Smith, 2000). For example, it has been well documented that hy1 plants flower earlier than wild-type (Chory et al., 1989) (Table I). Because hy1 and ho2-1 plants grow slower than their respective wild types, floral transition cannot be accurately quantified by time but must be quantified by developmental parameters such as the number of rosette leaves generated before bolting (Amasino, 1996; Michaels and Amasino, 1999). Using this criterion, we found that ho2-1 plants also flowered earlier than wild-type in both long days and short days (Table I). For example, under short days the hy1-100 plants initiated flowering at 25 leaves versus 45 leaves for the Col parent. The ho2-1 plants initiated flowering at 28 leaves versus 35 leaves for the WS parent.
In an attempt to isolate an Arabidopsis mutant deficient in both AtHO1 and AtHO2, we tried to construct a double homozygous line bearing both the hy1-100 and ho2-1 alleles. To overcome the fact that AtHO1 and 2 are within 40 kb of each other on chromosome II (Davis et al., 1999), we generated an F1 hybrid with hy1-100 and ho2-1 in repulsion. Then the F2 progeny were searched for those with long hypocotyls in white light (homozygous for the hy1-100 mutation) and kanamycin resistant (homo- or heterozygous for the ho2-1 T-DNA insertion). Despite repeat attempts and analysis of >500,000 F2 progeny, no lines displaying this phenotype, and thus having the hy1-100 mutation potentially coupled to ho2-1, were identified. Whether this failure indicates that the phenotype of hy1/hy1 ho2-1/+ plants are different from hy1/hy1 plants or that germ cells or plants missing both AtHO1 and 2 are inviable is not yet known.
Assembly of the phyA Holoprotein Is Compromised in ho2-1
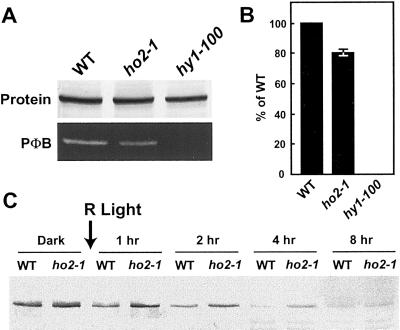
The collective phenotypes of the ho2-1 plants suggested that they, like the hy1 plants, have reduced levels of holo-phys caused by a decrease in PΦB synthesis. For hy1, the reduction is dramatic, as these plants lack detectable holo-phyA, despite containing high levels of the apoprotein (Parks et al., 1989; Parks and Quail, 1991). However, for ho2-1, the effect is likely to be more subtle consistent with the milder phenotypic changes. In an attempt to detect apo-PHYs, crude extracts prepared from etiolated seedlings of the mutant and wild-type lines were subjected to SDS-PAGE followed by an in-gel zinc-induced fluorescence assay to quantitate the amount of holo-phys that contain a bound PΦB chromophore, and by immunoblot analysis to measure the levels of the phyA protein (Bhoo et al., 1997). Because phyA is the predominant phy in etiolated seedlings, its measurement essentially reflects that of the total phy pool (Quail et al., 1995; Whitelam and Devlin, 1997). As shown in Figure 6, A and B, etiolated hy1-100 seedlings had high levels of the PHYA polypeptide but undetectable levels of holo-phy containing bound PΦB. In contrast, ho2-1 seedlings had significant levels of holo-phy containing PΦB, but this level was reduced relative to that of the wild-type WS parent. When equal amounts of phyA protein (as determined by immunoblot analysis) were subjected to SDS-PAGE, a 20% reduction in the level of holo-phy containing PΦB was evident (Fig. 6, A and B). A similar attenuation in the assembly of the other Arabidopsis holo-phys (phyB-E) is expected, but was not examined.
Figure 6.
Detection of apo-phyA in ho2-1 seedlings. A, Detection of the phyA protein and bound PΦB chromophore in wild-type ecotype WS (WT), hy1-100, and ho2-1 seedlings. Crude extracts were prepared from 4-d-old etiolated seedlings and subjected to immunoblot analysis with the anti-phyA monoclonal antibody O73D (top) or assayed for bound PΦB by zinc-induced fluorescence (lower). B, Quantitation of the levels of bound PΦB per phyA protein. Signals obtained from A were quantitated by densitometric scans of the gels. Values (±sd) were expressed relative to those from WT. C, R-light induced degradation of phyA in etiolated seedlings of WT and ho2-1. Etiolated seedlings were irradiated continuously with R. At various times, crude extracts were prepared and subjected to immunoblot analysis with the anti-phyA monoclonal antibody O73D; an equal amount of seedlings (grams fresh weight) was analyzed for each sample.
It has been shown previously that the PHYA apoprotein is stable, but the holoprotein is rapidly degraded when converted to Pfr (Clough and Vierstra, 1997). We consequently used this stability as an indirect test to confirm the presence of apo-PHYs in the ho2-1 background. Dark-grown WS wild-type, ho2-1, and hy1-100 seedlings were irradiated continuously with R, and at various times the amount of phyA polypeptide was assayed by immunoblot analysis. Consistent with previous studies (Parks et al., 1989), phyA was rapidly degraded in wild-type Arabidopsis seedlings with almost all removed within 8 h after the onset of R, whereas apo-PHYA remained stable in the hy1-100 seedlings (Fig. 6C and data not shown). For ho2-1 seedlings, phyA degradation was observed after R, but its overall rate was noticeably slower than that of wild-type (Fig. 6C). This attenuation was especially evident 4 h after the onset of R when the residual levels of the phyA polypeptide were 4 to 5 times higher than that observed for wild type. The slower degradation rate in the ho2-1 mutant background is indicative of a phyA pool containing a mixture of apo- and holo-protein. We also noticed that most of the phyA protein in the ho2-1 seedlings was degraded following prolonged R irradiations (Fig. 6C). This effect may indicate that the pool of apo-PHYA eventually acquired PΦB by recycling previously bound PΦB or from new synthesis.
Expression of Arabidopsis HO1 and 2 and Overlaps
To help understand how both AtHO1 and 2 contribute to PΦB synthesis and thus holo-phy assembly, the expression patterns of the two genes were compared. Fusions of the AtHO1 and 2 promoters with the coding region of β-glucuronidase (GUS) were introduced into Arabidopsis and the expression patterns observed by histochemical staining for GUS (Jefferson et al., 1987; Thoma, et al., 1996). To ensure that all 5′ regulatory sequences were included, the promoter regions used began immediately after the translation stop codon of the upstream annotated gene and ended at the start codon of AtHO1 or 2. The 5′-HO2-GUS reporter was introduced into the wild-type Col and the hy1-100 backgrounds, and the 5′HO1-GUS reporter was introduced into the wild-type WS and the ho2-1 backgrounds to examine the potential effects that each HO (and indirectly the resulting holo-phys) may have on the expression of the other HO gene.
The GUS staining patterns of 5′-HO1-GUS and 5′-HO2-GUS in etiolated and green seedlings are shown in Figure 7. In general, the expression patterns overlapped considerably, but in all cases 5′-HO1-GUS consistently gave higher staining, consistent with the predicted higher expression of the corresponding gene. Etiolated seedlings harboring either transgene stained for GUS in most tissue types including seed coat, cotyledons, apical hook, and hypocotyls (Fig. 7). Highest staining was observed in the vasculature and the hypocotyl/root junction and associated root hairs. Little or no staining was observed in the roots and root tips. This overall pattern was similar to that reported for the overlapping expression patterns of the PHYA and B transcripts with the notable exception that the PHY genes also are actively expressed in roots, especially in the root tips of etiolated plants (Somers and Quail, 1995; Neff et al., 2000). For green seedlings, widespread GUS staining was also observed for AtHO1 and 2 (Fig. 7). Pronounced staining of the leaf and cotyledon vasculature was evident. Highest expression was in the root and tissues surrounding the shoot apex. Whereas, newly initiated roots tips showed high expression, much weaker expression was seen in the tips as the roots elongated (Fig. 7).
Figure 7.
Expression patterns of AtHO1 and 2 using promoter-GUS fusions. The coding region for GUS, under the control of either the AtHO1 or the AtHO2 promoter, was introduced into wild-type WS (AtHO1) or wild-type Col (AtHO2). Plants were stained for GUS activity using the substrate 5-bromo-4-chloro-3-indolyl β-d-GlcUA; staining times for AtHO2 plants were increased to provide staining equivalent to that of AtHO1 plants. Top, Ten-day-old light-grown plants. Insert shows a magnification of a root tip. Bottom, Etiolated 4-d-old seedlings. Arrows point to cotyledons (cot), hypocotyls (hy), leaf (lf), roots (r), root tip (rt), and seed (sd).
When either 5′-HO1-GUS or 5′-HO2-GUS was introduced into the homozygous ho2-1 or hy1-100 backgrounds, respectively, a similar distribution of GUS staining among the various tissues was detected (data not shown). The overall levels of expression for 5′-HO1-GUS were indistinguishable for the WS and ho2-1 backgrounds. However, we noticed from comparing multiple independently transformed lines that expression of 5′-HO2-GUS was markedly reduced in hy1-100 plants as compared with corresponding Col wild type (data not shown), suggesting that AtHO1 (and indirectly phys) is required for optimal AtHO2 expression.
DISCUSSION
HOs are an important family of enzymes required for synthesis of the phy-chromophore PΦB and thus proper photomorphogenesis in plants. We show here that a number of higher plants contain a family of HOs that are highly related to each other, and by a lesser extent, to those in cyanobacteria, algae, and animals. In individual plant species, HOs are encoded by a small gene family; in Arabidopsis, four HO genes are present. Consistent with biochemical data, sequence analysis suggests that all the plant HOs are soluble and likely localized in plastids. The approximately 220-amino acid catalytic domain from each plant enzyme is strongly similar to mammalian HOs, suggesting that plant HOs assume the same three-dimensional structure as that of human HO1 (Schuller et al., 1999).
Because the various higher-plant HO mutants retain some phy responsiveness, especially as the plants mature, it has been proposed that alternative sources of BV exist (Terry and Kendrick, 1996; Davis et al., 1999; Izawa et al., 2000). We can now explain this effect, at least in part, by the presence of multiple HO isoforms, each with potentially different roles or expression patterns. Studies using promoter-GUS fusions indicate that two HO genes from Arabidopsis (AtHO1 and 2) are expressed simultaneously in a pattern that often parallels those of PHYA and B (Somers and Quail, 1995), indicating that they both can be important sources of PΦB for holo-phy assembly. Phylogenetic analyses suggest that higher plant HOs arose from a common progenitor that then generated the various members by gene duplication and divergence. During this evolution, a second type of HO emerged (HO2-like). To date, we have found members of the HO2 subfamily only in moncotyledonous and dicotyledonous species, suggesting perhaps that this HO type is restricted to flowering plants. Although not yet tested, we assume that HO2s will convert heme to BV given their strong similarity to members of the HO1 family and our observations that the Arabidopsis ho2-1 mutant has detectable amounts of apo-phy.
The HO2 subfamily members contain two possibly important changes that could affect their activity, a substitution of an Arg for a His at the putative heme-iron-binding site (Ortiz de Montelano and Wilks, 2001) and an insertion of a hydrophilic and predicted flexible domain just distal to this active site. Using the structure of human HO1 as a template (Schuller et al., 1999), we expect that this insertion in HO2 sequences forms a solvent-exposed extended loop (data not shown). Whether these sequence changes affect the catalytic activity or specificity of the enzyme is not clear. For other HOs, it has been shown that specific types can use different sources of reducing power during cleavage of the heme α-methene bridge. For example, AtHO1 from Arabidopsis and a chloroplast HO from the alga Cyanidium caldarium are ferrodoxin-dependent (Cornejo et al., 1998; Muramoto et al., 1999), whereas the microsomal HOs from animals require NADPH cytochrome-c reductase (Ortiz de Montelano and Wilks, 2001). Thus, it is conceivable that plant HO2s use a different reductant than that used by the HO1-like enzymes. The sequence differences, alternatively, could allow the HO1 and HO2 protein subfamilies to interact with distinct regulatory factors in a way that provides differential control of the enzymes (e.g. activity in the light versus the dark). We also note that the N-terminal extensions for the HO2 sequences are rich in prolines and hydrophillic amino acids, residues not commonly found in transit peptides. Thus the plastid location of HO2s remains in question.
Previous studies by us and others showed that the Arabidopsis hy1 and the rice se5 (Izawa et al., 2000) photomorphogenic mutants are defective in specific HO genes. Here, we extend the importance of HOs with the observations that tomato yg2 and N. plumbaginifolia pew1 mutants are also deficient in HO activity. Whereas, the yg-2aud mutation was mapped to the LeHO1 gene, a defect in one or more HOs in pew1 was inferred by complementation of the pew1 phenotype with wild-type Arabidopsis HO1. Although the pew1 mutation could be within a trans-acting factor that regulates HO expression, a more likely possibility is that the pew1 mutation inactivates a prominent HO gene, based on the similarity of the phenotype to that of HO mutants in other plant species (Chory et al., 1989; Parks and Quail, 1991; Terry and Kendrick, 1996; Izawa et al., 2000). As Arabidopsis HO1 complements pew1, we predict that this prominent N. plumbaginifolia HO is orthologous to AtHO1.
It is interesting that hy1, se5, and yg-2 (and possibly pew1) bear defects in members of the HO1 subfamily, suggesting that this subfamily (and possibly the affected HO1 genes in particular) is responsible for much of the BV synthesis in plants. In support, levels of holo-phy are undetectable in the hy1-100 and se5 backgrounds (Fig. 6; Izawa et al., 2000), indicating that very low levels of PΦB are synthesized in these mutants. A more dominant role also can be inferred from the genomic organization and expression levels of HO1- versus HO2-like genes. Whereas, Arabidopsis contains three members of the HO1 subfamily, it only has a single HO2 gene. The relative abundance of AtHO1 sequences in the EST databases, intensity of mRNA signals by RNA gel-blot analysis, and GUS staining of plants bearing AtHO1- or AtHO2-promoter GUS fusions also indicate that the AtHO1 gene in particular is expressed more strongly than AtHO2 (data not shown; Fig. 7). cDNAs for both AtHO3 and 4 have been detected by RT-PCR of Arabidopsis mRNA, indicating that these members of the Arabidopsis HO1 subfamily are also expressed. Sequence comparisons suggest that the encoded AtHO3 and 4 proteins are also plastid localized, but their relative contributions to total HO activity and their expression patterns are not yet known. The observation that little holo-phyA accumulates in hy1-100 mutant seedlings, lacking just AtHO1, implies that AtHO3 and 4 must have a more specialized role(s) in PΦB synthesis in Arabidopsis. One possibility is that AtHO3 and 4 are expressed in root tips, a tissue known to contain active phys but appears to express AtHO1 and 2 poorly. The identification of mutants in AtHO3 and 4 will be critical toward answering this question.
Although previous data indicated that AtHO1 has a prominent role in PΦB synthesis and proper photomorphogenesis, our data from the Arabidopsis ho2-1 mutant indicates that the AtHO2 subfamily is also important. Subtle, but reproducible attenuation of a variety of photomorphogenic responses controlled by phys was evident in homozygous ho2-1 plants. These defects included reduced hypocotyl photosensitivity, reduced leaf size, accelerated flowering time, and decreased chlorophyll accumulation. These pleiotropic phenotypes were observed in etiolated seedlings, and in young and more mature green plants, indicating that AtHO2 has roles throughout the Arabidopsis life cycle.
The exact role(s) of AtHO2 are not yet clear. That the ho2-1 phenotypes appear similar (although less dramatic) to those of hy1 plants (this study; Chory et al., 1989; Parks and Quail, 1991; Davis et al., 1999; Muramoto et al., 1999), indicates that AtHO1 and 2 may have overlapping functions in the same physiological processes. Certainly the detection of a pool of apo-PHYA not bearing chromophore in ho2-1 plants indicates that AtHO2 also contributes directly or indirectly to the pool of PΦB used to assembly holo-phys. However, given that holo-phys are undetectable in hy1 plants containing a functional AtHO2, it is clear that AtHO2 cannot compensate for a loss of AtHO1 and thus serves a non-redundant function. One possibility is that AtHO1 and 2 have tissue/cell-specific functions. However, we consider this unlikely given the substantial overlap of their expression patterns. Others include distinct subcellular locations, different modes of regulation, and/or biochemical functions other than or in addition to PΦB synthesis. Given that the N-terminal extensions for the HO2 family do not fit the typical transit peptide sequence, it remains possible that HO2s are not plastid localized. Moreover, sequence differences at and surrounding the active site could reflect distinct enzymatic properties. For example, one HO could be activated by or require a reductant produced by the photosynthetic light reactions (e.g. ferrodoxin/thioredoxin) (Schürman and Jacquot, 2000), whereas the other could be constitutively active and/or require a reductant made in both the light and dark. And finally, we cannot rule out the possibility that HO1 and 2 have completely different biochemical functions. For example, one could be responsible for most of PΦB synthesis, whereas the other could play a role in the metabolism of chlorophyll and/or other tetrapyrroles.
The discovery that AtHO1 and 2 have overlapping, yet distinct roles in PΦB biosynthesis and phy-regulated photomorphogenesis illustrates the importance of chromophore abundance in regulating photoreceptor activity. Further characterization of AtHO2 should reveal interesting physiological functions for this enzyme. For instance, it will be worth investigating what role(s) AtHO2 have in other phy-regulated responses, including shade-avoidance, anthocyanin production, setting the circadian clock, and plant senescence (van Tuinen et al., 1996; Neff et al., 2000; Smith, 2000;). One obvious genetic experiment is to phenotypically analyze mutants homozygous for both hy1 and ho2. However, we have been unable to assemble this combination, possibly due to the extremely close linkage of the two genes (Davis et al., 1999). It is clear that the complete contribution of the HO gene family to plant photomorphogenesis will be revealed only through the analysis of loss-of-function alleles at each of the HO loci singly and in combinations.
MATERIALS AND METHODS
Isolation and Analysis of HO Genes from Plants
Plant HO genes were identified by searching the EST databases at http://www.ncbi.nlm.nih.gov/BLAST using the Blast computer algorithm (Altschul et al., 1990) for translated sequences related to AtHO1 (Davis et al., 1999; Muramoto et al., 1999). cDNAs for soybean GmHO1 and 3, tomato LeHO1 and 2, pine PtHO1, and sorghum SbHO1 and 2 were provided by Genome Systems (St. Louis), Clemson University Genomic Institute (Clemson, SC), Dr. Ross Whetten (North Carolina State University, Raleigh, NC), and Dr. Marie-Michéle Cordonnier-Pratt (University of Georgia, Athens, GA), respectively. The complete sequence of each cDNA was determined using Big-Dye Chemistry (Perkin Elmer, Foster City, CA). The genomic sequences for AtHO3 and AtHO4 in Arabidopsis ecotype Columbia-0 (Col) were identified by blast searches at http://www.Arabidopsis.org/blast. Their cDNAs were isolated by RT-PCR from a cDNA pool prepared from rosette-leaf total RNA using the AtHO3 oligonucleotides CTCAATTCGTGTGTGTGTATGTTGCAG and CTATTGAAGATCTGAAGGTTTATGAATC and the AtHO4 oligonucleotides TATTGCAGTTTGCGTGTCTATTAGATTGC and CTTTTTGTTGCCACCGGAAGCTCATCAC. These primers anneal just outside of the predicted coding region. The PCR products were sequenced directly.
Contig assembly and sequence analysis used the fragment assembly system (University of Wisconsin-Genetics Computer Group, Madison). Derived amino acid sequences were compared using the GCG program Pileup, and viewed with Macboxshade (Institute for Animal Health, Pirbright Surrey, UK). Phylogentic analysis of the HO sequences was accomplished by using the Growtree program (UW-GCG).
Characterization of Tomato yg-2
The tomato (Lycopersicon esculentum) lines AC and yg-2aud in the AC background were obtained from Dr. Matthew Terry (University of Southampton, Southampton, UK). Seedlings were grown on filter paper and irradiated continuously with R or grown to adulthood in soil under greenhouse conditions.
To characterize the YG-2 locus (designated here as LeHO1), genomic DNA was prepared (Davis et al., 1999) from the AC wild-type tomato variety and its derived mutant yg-2aud (Terry and Kendrick, 1996; van Tuinen et al., 1996). LeHO1 was PCR amplified from this DNA using the primers, GGTTAATCTGTCCTTTTCACTTCTC and ACCTTTCAACCATCCAATGTACAAG, which annealed immediately before the predicted initiating Met codon and immediately after the predicted translational termination codon of LeHO1, respectively. The resulting approximately 3.0-kb products were gel purified and directly sequenced using gene-specific primers that were designed based on the LeHO1 cDNA sequence.
Transformation of N. plumbaginifolia pew1 with AtHO1
A 3.3-kb genomic fragment of AtHO1 (Davis et al., 1999) encompassing the entire translated region, 1.1 kb of 5′-flanking and 0.6 kb of 3′-flanking region was PCR amplified with Pfu polymerase (Stratagene, La Jolla, CA) from Arabidopsis (Col ectotype) genomic DNA and cloned into the EcoRV site of pPZP211 (Hajdukiewicz et al., 1994). N. plumbaginifolia pew1 was obtained from Dr. Marc Jullien (Institut National de la Recherche Agronomique, Versailles, France) (Kraepiel et al., 1994). AtHO1 was introduced into pew1 by Agrobacterium-mediated transformation of leaf discs with the Agrobacterium tumefaciens strain ABI (Clough et al., 1999). Transformed plants were selected by kanamycin resistance. T1 plants were grown under continuous white light.
Identification of the Arabidopsis T-DNA Mutant ho2-1
DNA pools from The University of Wisconsin Biotechnology-Center T-DNA collection, assembled from approximately 60,000 independent Arabidopsis ecotype WS transformants, were screened by PCR for a T-DNA insertion within AtHO2. PCR reactions included the gene specific primers, AGTGAAGGCAGCGTCTATCTTGGTC-GTCGG (HO2-5′) and CTGGTGCCGGAAACTGTTAACT-TTAAAACC (HO2-3′), in combination with the T-DNA left-border (LB) primer GATGCACTCGAAAATCAGCCAATTTTAGAC and the right border (RB) primer TCCTTCAATCGTTGCGGTTCTGTCAGTTC (Krysan et al., 1996; Krysan et al., 1999). Correct PCR products were confirmed as described (Krysan et al., 1999) by DNA gel-blot analysis using 32P-labeled AtHO2 sequence as the probe. ho2-1 was identified in pool 11B-10 using the RB primer in conjunction with the HO2-3′ primer. An individual ho2-1 was located in plate 2,016. That both the 5′- and 3′-ho2-1 PCR products were amplified with the RB primer indicated that two T-DNA RBs were present, suggestive of a tandem T-DNA insertion in AtHO2 with one insert inverted relative to the other.
The ho2-1 line was backcrossed once to the WS wild type, and in the F2 generation, homozygous ho2-1 plants were identified by PCR. In these ho2-1 plants, an AtHO2 PCR product was produced using the primers HO2-3′ and RB, but not when the HO2-5′ and HO2-3′ primers were used. F3 seeds from several independent F2 homozygous-mutant plants were bulked for phenotypic characterization of ho2-1.
Phenotypic Analysis of ho2-1
Arabidopsis ecotypes Col-0 and WS were obtained from Dr. Richard Amasino (University of Wisconsin, Madison, WI). The hy1-100 line in the Col background (Davis et al., 1999) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). For hypocotyl-growth assays, seeds were plated on solid one-half-strength Murashige and Skoog medium without Suc or vitamins (2.2 g L−1 Murashige and Skoog salts [Life Technologies/Gibco-BRL, Gaithersburg, MD], 2.5 mm 2-[N-morpholino]ethanesulfonic acid, pH 5.7, 0.8% [w/v] agar). These plates were stored in the dark for 3 d at 4°C and then irradiated continuously with R or FR at 22°C as described (Jordan et al., 1996). Hypocotyl lengths after 6 d of growth were measured from computer-scanned images. For analysis of mature plants, seeds were stratified for 4 d at 4°C and then sown directly on soil. For flowering-time measurements, plants were grown at 20°C under approximately 25 μmol m−2 s−1 white light in either a 20-h-light/4-h-dark photoperiod for long days or an 8-h-light/16-h-dark photoperiod for short days. For chlorophyll assays, plants were grown in the long-day photoperiod for 14 d. Chlorophylls were extracted from rosette leaves by methanol (1 mL/100 mg tissue) and quantitated spectrophotometrically by the revised method of Porra et al. (1989).
phyA Quantification
Arabidopsis seeds were placed on one-half-strength Murashige and Skoog agar plates, incubated at 4°C for 3 d, and transferred to approximately 22°C for growth in the dark. After 4 d of growth, the seedlings were frozen at liquid-nitrogen temperatures, ground with a mortar and pestle, and suspended at 2 mL g−1 fresh weight in 37.5% (v/v) ethylene glycol, 75 mm Tris-HCl (pH 8.3, 4°C), 7.5 mm Na4 EDTA, 15 mm NaS2O5, 0.11% (v/v) polyethylenimine, and 1.5 mm phenylmethylsulfonyl fluoride (Jordan et al., 1996). The resulting extract was clarified by centrifugation at 16,000g for 30 min, and the soluble proteins were subjected to SDS-PAGE. Presence of PΦB was detected in the gel by zinc-induced fluoresence (Bhoo et al., 1997). phyA polypeptides were detected by immunoblot analysis, using the monoclonal antibody O73D (Clough et al., 1999) in combination with the goat-anti-mouse immunoglobulin conjugated to alkaline phosphatase (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Levels of PΦB and phyA protein were determined semi-quantitatively by the analysis of scanned images of the gels and blots using the Bio-Rad Gel-Documentation System (Hercules, CA) and their optiquant software. Degradation of phyA was initiated by continuous irradiation of 5-d-old etiolated seedlings with R (Clough et al., 1999).
Expression Patterns of AtHO1 and 2
Fusions of the 5′-flanking sequences of Arabidopsis HO1 and 2 to the GUS coding sequence were created in the vector pCAMBIA1391z (Center for the Application of Molecular Biology to International Agriculture, Canberra, Australia). For AtHO1, approximately 1.1-kb genomic DNA fragment upstream of the AtHO1 open-reading frame was PCR-amplified from the BAC F18A8 with the primers, TCCAATGTCGACATTATGAGTATTATTATTTTTAAATCand CTAAATAGGATCCGGTTTGATCGGAATAG. This PCR product included DNA sequence that began immediately after the proposed stop codon of the upstream gene F18A8.3 and ended at the initiating Met codon of AtHO1. For AtHO2, an approximately 500-bp genomic DNA fragment upstream of the AtHO2 open-reading frame was PCR-amplified from the BAC clone T9J22 with the primers, TCCAATGTCGACCTAAAGAGATCAATTATTG and AA-GAGAGGATCCGGATTCCGACGACCAAG. This PCR product included DNA sequences that began immediately after the proposed stop codon of the upstream gene T9J22.21 and ended at the initiating Met codon of AtHO2. Both products were digested with SalI and BamHI and cloned into pCAMBIA1391z similarly digested, thus creating the vectors 5′-HO1-GUS and 5′-HO2-GUS, bearing the GUS coding region downstream of the 5′ regulatory sequences of AtHO1 and 2.
Transgenic Arabidopsis was generated by the floral-dip method (Clough and Bent, 1998) with the Agrobacterium tumefaciens strain ABI harboring the appropriate vector. 5′-HO1-GUS was introduced into the WS wild-type and ho2-1 backgrounds. 5′-HO2-GUS was introduced into the Col wild-type and hy1-100 backgrounds. T1 seeds were plated on Gamborg B5 medium supplemented with Suc and vitamins, (Life Technologies/Gibco-BRL), 2-[N-morpholino]ethanesulfonic acid (pH 5.7), and 0.7% (w/v) agar. After a 3-d incubation at 4°C, seedlings were either grown for 4 d in darkness or grown under continuous fluorescent lighting (approximately 20 μmol m−2 s−1) for 10 d. Seedlings were stained for GUS activity by incubation in 2 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid as described (Thoma et al., 1996). The resulting blue-stained plants were imaged with a dissecting microscope. Five to seven independent transformants where analyzed for each transgene-genotype combination; in all cases, the staining patterns were similar for each combination.
ACKNOWLEDGMENTS
We thank Marc Jullien and Matthew Terry for supplying the pew1 and yg-2aud mutants, respectively, Ross Wheaton and Marie-Michéle Cordonnier-Pratt for supplying cDNA clones, and Alex Vener for his assistance in generating the GUS constructions.
Footnotes
This work was supported by the Department of Energy Division of Basic Energy Sciences (grant no. DE–FG02–88ER13968), by the Research Division of the College of Agriculture and Life Sciences (grant no. Hatch–N936 to R.D.V.), and by a National Institutes of Health predoctoral fellowship (no. 5 T32 GM07133 to S.J.D.).
LITERATURE CITED
- Altschul S, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amasino RM. Control of flowering time in plants. Curr Opin Genet Dev. 1996;6:480–487. doi: 10.1016/s0959-437x(96)80071-2. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoo SH, Hirano T, Jeong H-Y, Lee J-G, Furuya M, Song P-S. Phytochrome photochromism probed by site-directed mutations and chromophore esterification. J Am Chem Soc. 1997;119:11717–11718. [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell. 1989;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, Gatz C, Vierstra RD. Sequences within both the N- and C-terminal domains of phytochrome A are required for Pfr ubiquitination and degradation. Plant J. 1999;17:155–167. doi: 10.1046/j.1365-313x.1999.00360.x. [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cornejo J, Willows RD, Beale SI. Phytobilin biosynthesis: cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 1998;15:99–107. doi: 10.1046/j.1365-313x.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Von Heijne G. A neural network-based method for predicting chloroplast transit peptides and their cleavage products. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell. 2001;13:425–436. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) Plant J. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RE, Kavanagh TA, Bevan MM. GUS fusions: β-glucuronic acid as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan ET, Cherry JR, Walker JM, Vierstra RD. The amino-terminus of phytochrome A contains two distinct functional domains. Plant J. 1996;9:243–257. doi: 10.1046/j.1365-313x.1996.09020243.x. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Peters JL, Kerckhoffs LH, van Tuinen A, Koornneef M. Photomorphogenic mutants of tomato. Biochem Soc Symp. 1994;60:249–256. [PubMed] [Google Scholar]
- Koorneef M, Cone JW, Kekens RG, O'Herne-Robers EG, Spruit CJP, Kendrick RE. Photomorphogenic responses of long hypocotyl mutants of tomato. J Plant Physiol. 1985;120:153–165. [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-induced hypocotyl elongation in Arabidopsis thaliana L Heynh. Z Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- Kraepiel Y, Jullien M, Cordonnier-Pratt MM, Pratt L. Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant. Mol Gen Genet. 1994;242:559–565. doi: 10.1007/BF00285279. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G, Millar AJ. How plants tell the time. Curr Opin Plant Biol. 2000;3:43–46. doi: 10.1016/s1369-5266(99)00034-5. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Ortiz de Montelano PR, Wilks A. Heme oxygenase structure and mechanism. Adv Inorg Chem. 2001;51:359–407. [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Shanklin J, Koorneef M, Kendrick RE, Quail PH. Immunochemically detectable phytochrome is present at normal levels but is photochemically nonfunctional in the hy1 and hy2 long hypocotyl mutants of Arabidopsis. Plant Mol Biol. 1989;12:425–437. doi: 10.1007/BF00017582. [DOI] [PubMed] [Google Scholar]
- Platt JL, Nath KA. Heme oxygenase: protective gene or Trojan horse. Nat Med. 1998;4:1364–1365. doi: 10.1038/3947. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Richaud C, Zabulon G. The heme oxygenase gene (pbsA) in the red alga Rhodella violacea is discontinuous and transcriptionally activated during iron limitation. Proc Natl Acad Sci USA. 1997;94:11736–11741. doi: 10.1073/pnas.94.21.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat Struct Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- Schürman P, Jacquot J-P. Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants: an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 1995;7:413–427. doi: 10.1046/j.1365-313x.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE. The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J Biol Chem. 1996;271:21681–21686. doi: 10.1074/jbc.271.35.21681. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, McDowell MT, Lagarias JC. (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J Biol Chem. 1995;270:11111–11118. doi: 10.1074/jbc.270.19.11111. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Wahleithner JA, Lagarias JC. Biosynthesis of the plant photoreceptor phytochrome. Arch Biochem Biophys. 1993;306:1–15. doi: 10.1006/abbi.1993.1473. [DOI] [PubMed] [Google Scholar]
- Terry ML. Phytochrome deficient mutants. Plant Cell Environ. 1997;20:740–745. [Google Scholar]
- Thoma S, Sullivan ML, Vierstra RD. Members of two gene families encoding ubiquitin-conjugating enzymes, AtUBC1–3 and AtUBC4–6, from Arabidopsis thaliana are differentially expressed. Plant Mol Biol. 1996;31:493–505. doi: 10.1007/BF00042223. [DOI] [PubMed] [Google Scholar]
- van Tuinen A, Hanhart CJ, Kerckhoffs LHJ, Nagatani A, Boylan MT, Quail PH. Analysis of phytochrome-deficient yellow green-2 and aurea mutants of tomato. Plant J. 1996;9:173–182. [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα. Plant Cell. 1996;8:55–67. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Reid JB, Kendrick RE. The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IX alpha to 3(Z)-phytochromobilin. Plant J. 1997;11:1177–1186. doi: 10.1105/tpc.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsis morphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]