Abstract
P2X4 and P2X7 receptors are ATP-gated ion channels that are co-expressed in alveolar epithelial type I cells. Both receptors are localized to the plasma membrane and partly associated with lipid rafts. Here we report on our study in an alveolar epithelial cell line of the molecular organization of P2X7R and P2X4R receptors and the effect of their knockdown. Native gel electrophoresis reveals three P2X7R complexes of ~430, ~580 and ~760 kDa. The latter two correspond exactly in size to signals of Cav-1, the structural protein of caveolae. Interestingly knockdown of P2rx7 affects protein levels, the intracellular distribution and the supramolecular organization of Cav-1 as well as of P2X4R, which is mainly detected in a complex of ~430 kDa. Our data suggest upregulation of P2X4R as a compensatory mechanism of P2X7R depletion.
Keywords: P2X protein complexes, Caveolin-1, Caveolae, Epithelial lung cells, Native PAGE
Introduction
P2X receptors (P2XR) are ATP-gated, non-selective cationic channels that are divided into seven subtypes (P2X1–P2X7). They are widely distributed throughout many tissues, including neurons, glia, muscle, bone, hemopoietic tissues, endothelial and epithelial cells [1–3]. P2XR subunits form homo- as well as heterotrimeric assemblies, including P2X1/2, P2X1/4, P2X1/5, P2X2/3, P2X2/6, P2X4/6 and P2X4/7 [4–10]. P2X4R and P2X7R form mainly homotrimers in the plasma membrane of microglia and macrophages [11, 12], but evidence for heteromeric P2X4/7R has been obtained in human embryonic kidney (HEK) 293 cells and in primary cultures of bone marrow-derived macrophages [10, 13]. The formation of P2X4R and P2X7R heteromers provides an important mechanism for the modulation of P2X7R and P2X4R functions under different physiological conditions [13].
The distribution of P2X receptors in lung cells was extensively investigated [14–17]. The predominant P2X receptors in the alveolar epithelial type I (ATI) cells are P2X4R and P2X7R. Co-expression of both receptors is also observed in immune cells such as microglia, monocytes and macrophages and various other tissues [12, 18, 19]. However, little is known about the function of P2X receptors in ATI cells regarding involvement in apoptosis, proliferation, differentiation or migration and injury repair [20].
In previous studies we have shown the association of P2X4R and P2X7R with detergent-resistent membranes (DRMs) in the plasma membrane of ATI like cells [14, 15]. This association may be involved in regulation of alveolar epithelial metabolism and physiological functions [20]. DRMs (lipid rafts) are sphingolipid- and cholesterol-rich membrane microdomains that are involved in receptor signaling, vesicle traffic and protein sorting [21, 22]. The localization in DRMs was also reported for P2X3R in neurons and for P2X1R in smooth muscle cells [23, 24], as well as for P2X7R in mouse T cells, rat submandibular gland and macrophages [25–27]. Caveolae are specialized microdomains in the ATI cells, which form flask-like invaginations of the plasma membrane [22]. Caveolae are involved in trans- and endocytosis, regulation of cholesterol homeostasis and clustering of signaling proteins, e.g., G-protein coupled receptors, acetylcholin receptors and growth factor receptors [22]. In the lung the protein caveolin-1 (Cav-1) is responsible for the typical caveolae formation in ATI cells. Cav-1 co-localizes with caveolin-2 (Cav-2), and both proteins interact with different signaling proteins in the caveolae. Our previous results suggest that part of P2X7R interacts with Cav-1 [14]. So far, no experimental data are available that indicate an association of P2X4R with caveolae. However, the observation that shRNA-mediated downregulation of cav-1 in an alveolar epithelial cell line results in a strong decrease of P2X7R and P2X4R [14, 15] could hint at such an interaction.
Given the multiple and pronounced functions of P2X7R in alveolar epithelial cells and the importance of P2X4Rs and P2X7Rs as therapeutic targets, it is crucial to clarify their interactions and subunit composition [20, 28]. In this study, we used blue native- and high-resolution clear native-PAGE as effective methods to isolate membrane multi-protein complexes without chemical cross-linking [29–32] in order to visualize the molecular organization of native P2X4R and P2X7R. Furthermore, we investigated the effects of shRNA-mediated downregulation of P2X7R and P2X4R proteins in the mouse alveolar epithelial cell line E10.
Materials and methods
Cell line and reagents
The mouse E10 lung cell line was kindly provided by M. Williams (Pulmonary Center, Boston University School of Medicine, Boston, MA). DMEM/Ham’s F12 was acquired from Gibco (Invitrogen, Karlsruhe, Germany). Fetal bovine serum, l-glutamine and trypsin/EDTA were purchased from Biochrom AG Seromed (Berlin, Germany).
Cell culture
Cells were cultured in DMEM/Ham’s F12 medium (1:1) supplemented with 10% fetal bovine serum and 2.5 mM l-glutamine. They were grown at 37°C in a 5% CO2 atmosphere. Cells were seeded at a density of 3 × 104 cells/ml and passaged continuously.
Western blot analysis
Protein concentrations were determined using the Quick Start™ Bradford protein assay (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s guidelines. The amidoschwarz protein assay [33] was used to define the protein concentration for downregulation experiments. SDS-PAGE and Western blot analyses were performed as previously described [14] using the following antibodies: monoclonal mouse anti-Cav-1 (clone 2297, dilution 1:500 v/v and 1:250 v/v; BD Biosciences, Pharmingen, San Jose, CA), polyclonal rabbit anti-P2X4R (dilution 1:500 v/v and 1:1,500 v/v; Sigma-Aldrich, Inc., St Louis, MO), polyclonal rabbit anti-P2X7R (dilution 1:500 v/v and 1:1,500 v/v; Alomone Labs, Ltd., Jerusalem, Israel), monoclonal mouse anti-γ-tubulin (clone GTU-88, dilution 1:2,000 v/v; Sigma-Aldrich) and monoclonal hamster anti-T1α (1:1,500, M. Williams, Pulmonary Center, Boston University School of Medicine, Boston, MA).
shRNA synthesis and transduction
shRNA synthesis and lentiviral transduction were performed as previously described [14]. The detailed sequences of the three nucleotide sense strands of the shRNAs designed from P2rx4 cDNA are CATTACCACCTCCTACCTCAA, TTACCACCTCCTACCTCAAGT and GCACACTCACCAAGGCATATG, respectively. The sequences of P2rx7 nucleotide sense strands are AGCCATGTTCTCTGACTTCCA, TGACGAAGTTAGGACACAGCA and CTACAACTTCAGATATGCCAA. The negative controls (scrambled shRNAs) were designed with siRNA wizard v2.6 (InvivoGen). Scramble shRNAs of P2rx4 have the following sequences: CTCCCCTCAACCTAACACTAT, CTGCCCTTCCCTACCAATAAT and GAACGACCACTCCGGATCATA; scramble P2rx7 shRNAs sequences are GTTCGCTCATCCCGATTTACA, GAGCAACGGGATCGATACAAT and GACCAACTTCATTCGAAATCA. All constructs were verified by sequencing. The concentration of lentiviruses for all experiments was 10 TU/cell, and the cells were reclaimed after 72 h. Protein expression was verified by Western blot analysis as described above.
Biotinylation of P2X4R and P2X7R at the cell surface of E10 cells
Measurement of cell surface expression by biotinylation was carried out as described in [15]; 500 μg total protein was incubated with M-280 streptavidin-linked Dynabeads® (Invitrogen, Ltd., Paisley, UK) overnight at 4°C.
Co-immunoprecipitation experiments
Native membrane preparations were obtained using the ProteoExtract® Native Membrane Protein Extraction Kit (Calbiochem, San Diego, CA) following the protocol of the manufacturer. Membrane proteins were solubilized with lysis buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40 or 1% digitonin and protease inhibitor cocktail (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany; 0.02 mg/ml pancreas extract, 0.0005 mg/ml thermolysin, 0.02 mg/ml chymotrypsin and 0.002 mg/ml trypsin, 0.33 mg/ml papain).
Dynabeads® Protein G (Invitrogen, Ltd., Paisley, UK) were incubated with 5 μg polyclonal rabbit anti-P2X4R (Sigma-Aldrich, Inc., St. Louis, MO) or polyclonal rabbit anti-P2X7R (Alomone Labs, Ltd., Jerusalem, Israel) for 45 min at room temperature. Thereafter, 250 μg of membrane proteins was added to the antibody–Dynabeads® complex and incubated under rotation for 4 h at 4°C. In the negative control no primary antibody was added to the Dynabeads®. Precipitated immune complexes were washed three times with lysis buffer, and proteins were eluted by boiling for 5 min in the sample buffer described in [14]. Finally, samples were separated by SDS-PAGE and analyzed by Western blot using monoclonal mouse anti-Cav-1, polyclonal rabbit anti-P2X4R and anti-P2X7R as described above.
Materials for native PAGEs
Coomassie blue G-250 was purchased from Serva (SERVA Electrophoresis GmbH, Heidelberg, Germany). Digitonin, n-dodecyl-β-d-maltoside (DDM), 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), imidazole and tricine were from Sigma (Sigma-Aldrich, St. Louis, MO), tris-(carboxyethyl)-phosphine hydrochloride (TCEP) from Pierce (Pierce Biotechnology Inc., Rockford, IL) and sodium deoxycholate (DOC) from Calbiochem (EMD Biosciences, Inc., La Jolla, CA). NativeMark unstained protein standard was purchased from Invitrogen (Invitrogen, Karlsruhe, Germany).
Blue native-PAGE (BN-PAGE) and high-resolution clear native-PAGE (hrCN-PAGE-3)
Native membrane extracts were prepared using the ProteoExtract® Native Membrane Protein Extraction Kit (Calbiochem, San Diego, CA). Solubilization of native membranes on ice was identical for hrCN-PAGE-3 and BN-PAGE. The membrane pellets were incubated with lysis buffer containing 50 mM NaCl, 50 mM imidazole/HCl, 5 mM 6-aminohexanoic acid and 4% digitonin for 20 min on ice. Following centrifugation (20 min at 60,000×g, 4°C) the supernatant was supplemented with glycerol (10%) and red Ponceau S (0.1%); 50 μg of total protein was applied to the respective gradient gel. Then 3–12% Bis–Tris Gels NativePAGE™ Novex® (Invitrogen, Ltd., Paisley, UK) were used for separation of digitonin-solubilized complexes. For the second dimension 10% SDS acrylamide gels were used for identification of the constituent proteins of the separated complexes. For this gels were equilibrated in 1% SDS solution containing 5 mM TCEP and 1 mM AEBSF for 1 h at room temperature, afterwards washed twice with ddH2O and placed in transfer buffer (composition described in [14]). The same anode buffer (25 mM imidazole/HCl, pH 7.0) was used for hrCN-PAGE-3 and BN-PAGE, whereas the cathode buffer (50 mM tricine, 7.5 mM imidazole) was differently supplemented. For BN-PAGE the anionic dye Coomassie blue G-250 (0.02% or 0.002%), and for hrCN-PAGE-3 the anionic detergent DOC (0.05%) and non-ionic detergent DDM (0.01%) were added. Gels were put on ice, and the initial voltage was set to 80 V. When the samples entered the gel, voltage was raised to 40 V with the current limited to 10 mA. Electrophoresis was running for 14 h and stopped when the red Ponceau S dye passed the gel front. Electrophoresis conditions for the second dimensions were as previously described [14].
In vitro binding assay of P2XR fusion proteins to membrane lipids
The construction, expression and purification of GST-P2X fusion proteins were performed as described in [15]. The subdomains were amplified with the following primers: 5′-TATATAGAATTCCATGTGTATGAAGAAGAGATACTACTACCGG-3′ and 5′-TATATAGCGGCCGCTCACTGGTCCGTCTCTCC-3′ for P2X4R C-terminal domain (358–388) and 5′-TATATAGAATTCCTACTCCAGTGCTTTCTGCAGGTC-3′ and 5′-TATATAGCGGCCGCTCAGTAGGGATACTTGAAGCCACTATAC-3′ for the P2X7R C-terminal domain (358–595). To minimize formation of inclusion bodies 0.8 M d-sorbitol and 2.5 mM betaine hydrochloride were added to 2× YTA media (1.6% tryptone, 1.0% yeast extract, 0.5% NaCl, 100 μg/ml ampicillin, pH 7.0). The resuspension buffer (1× PBS 1% Triton X-100) was supplemented with 10 μM lysozyme and protease inhibitor cocktail (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany; 0.02 mg/ml pancreas extract, 0.0005 mg/ml thermolysin, 0.02 mg/ml chymotrypsin, 0.002 mg/ml trypsin, 0.33 mg/ml papain). After incubation at 37°C for 15 min the cells were kept on ice and disrupted with a French press. The GST-P2X fusion proteins were eluted from GSH-sepharose beads with elution buffer (50 mM Tris–HCl, 10 mM reduced glutathione, pH 8.0) and purified thoroughly on PD-10 columns Sephadex™ G-25M (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Binding of GST-P2X fusion proteins to membrane lipids was tested with lipid coated-strips (Echelon Biosciences Inc., Salt Lake City, UT). The membranes were blocked with TBS with 3% BSA and afterwards incubated overnight at 4°C in the same buffer containing 1 μg/ml GST-P2X fusion protein or GST-PI(4,5)P2 Grip™ protein (N-terminal GST-tagged recombinant PLC-δ1 PH domain; Echelon Biosciences Inc., Salt Lake City, UT) that served as positive control. GST was used as negative control. Upon washing of the membranes with TBS, they were treated with goat anti-GST antibody (dilution 1:1,000 v/v; Amersham Biosciences UK, Little Chalfont, Buckinghamshire, UK) in TBS with 3% BSA for 1 h at room temperature, washed again with TBS-T and incubated with HRP-conjugated rabbit anti-goat secondary antibody (dilution 1:15,000 v/v; Dianova GmbH, Hamburg, Germany) for 45 min at room temperature. The detection system was as described in [14]. All experiments were carried out three times.
Statistical analysis
Data are presented as mean ± SD. One-way analysis of variance (ANOVA) was used to determine the results of lentiviral shRNA-mediated downregulation experiments. When significance was achieved, it was followed by post hoc Bonferroni test. Statistical analysis was performed using SPSS 16.0 (SPSS, Chicago, IL), and significance was accepted at *p < 0.05 and **p < 0.01. We accomplished a minimal three independent experiments.
Results
Detection of native P2X receptor complexes in the plasma membrane of E10 cells
To investigate the molecular organization of native P2X receptor complexes, digitonin lysates were prepared from cell membranes of mouse alveolar epithelial cells, separated by blue native (BN)-PAGE or high-resolution clear native (hrCN)-PAGE, and analyzed by Western blot with P2X-specific antibodies.
In line with previous data, according to which trimeric P2X receptors have the tendency to form higher order complexes [34–36], we identified three bands of the P2X7R at approximately 430, 580 and 760 kDa (Fig. 1a, b, lane 1). The majority of P2X7R was detected at ~580 kDa. For P2X7R, several interacting proteins including Cav-1 have been identified [15, 37]. Therefore, the presence of Cav-1 in the P2X7R complexes was investigated by probing the Western blots with Cav-1 antibodies. The analysis revealed an almost identical separation profile as observed for P2X7R: the majority of Cav-1 is detected at 580 kDa, with a smaller portion of the protein residing at 760 kDa (Fig. 1a, b, lane 3). These data suggest that both proteins may be constituents of the same 580- and 760-kDa complexes. However, Cav-1 is known to form homotypic high molecular mass oligomers of >400 kDa containing ~14–16 individual molecules [38, 39], which may comigrate with the P2X7R complexes.
Fig. 1.
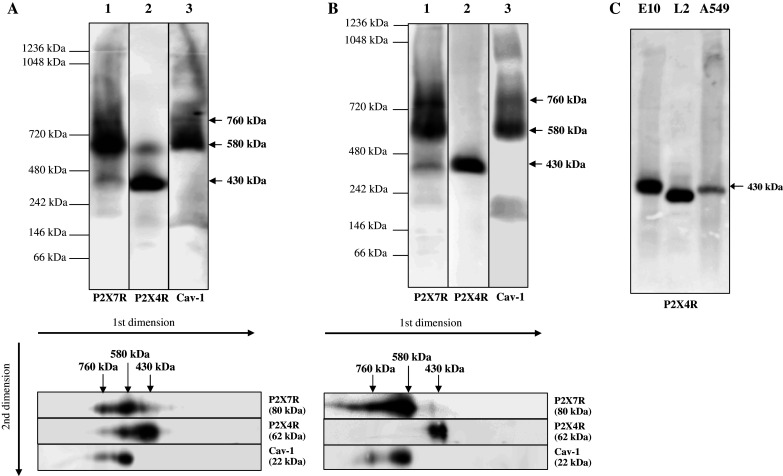
Supramolecular organization of P2X4R, P2X7R and Cav-1. Native membrane extracts from E10 (a–c), L2 (c) and A549 cells (c) were isolated and solubilized with digitonin. Solubilization of native membranes was identical for hrCN-PAGE and BN-PAGE. Proteins were separated by 2D-BN/SDS-PAGE (a) or hrCN-PAGE (b, c), blotted and probed with antibodies against P2X7R, P2X4R and Cav-1. Sizes of the molecular mass standards are indicated
Detection with P2X4R-specific antibodies revealed a major band at 430 kDa and a weaker signal at ~580 kDa, which overlaps with the signals of Cav-1 and P2X7R (Fig. 1a, b, lane 2). This latter complex was only detected by BN-PAGE, but not by hrCN-PAGE. Possibly the presence of the detergent DDM in the cathode buffer interferes with weak protein-protein interactions [32]. This observation is in line with previous findings that trimeric P2X4R channels are able to form higher order complexes and that their sizes depend on the detergent used [12, 36, 40].
Comigration of the two P2XR receptors and Cav-1 in native gels was confirmed by a second dimension BN-/SDS PAGE (Fig. 1a, b, lower panel).
Our results indicate that the ~580 kDa complex harbors concomitantly P2X7R, P2X4R and Cav-1. All three proteins are also constituents of the ~760-kDa complex, although their relative contribution is markedly different. Cav-1 and especially P2X4R are only present in low concentrations. The ~430-kDa complex is dominated by P2X4R, with a minor contribution by P2X7R.
Detection of P2X4R in a native complex of ~430 kDa in the alveolar epithelial cells contrasts data of other groups, which have reported a size of ~230 kDa [34, 36, 40]. To exclude cell-line-specific effects, we analyzed the molecular mass of P2X4R complexes by hrCN-PAGE in two other permanent alveolar epithelial cell lines (L2, A549). Except for some differences in the expression level, we detected a single band at ~430 kDa for the P2X4R in all cell lines (Fig. 1c). The slightly different migration of the complexes may reflect differences in lipid and/or subunit composition. Separation by BN-PAGE revealed an almost identical separation pattern (data not shown), confirming that the complexes are separated in their native state.
Interaction between Cav-1 and P2X7R and the P2X4R and P2X7R in E10 cells
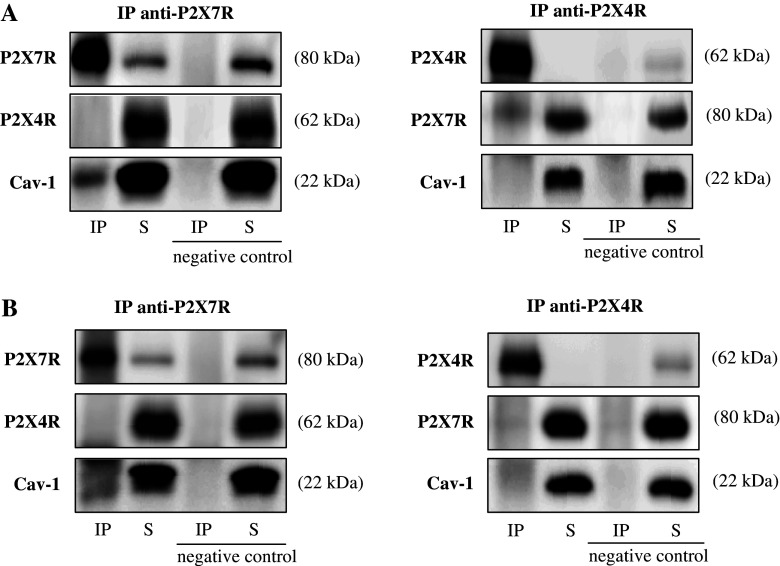
Results obtained from BN-PAGE and hrCN-PAGE indicated a potential association of P2X7R and Cav-1 as well as of P2X7R and P2X4R. We tested the interactions of the proteins by co-immunoprecipitation experiments using cell lysates from E10 cells expressing Cav-1, P2X4R and P2X7R. Immunoprecipitation was carried out with mouse monoclonal antibodies directed against P2X4R and P2X7R, respectively. In the control experiment, the P2XR antibodies were omitted. Membrane proteins were solubilized with 1% NP-40 (Fig. 2a) or 1% digitonin (Fig. 2b). Independent of the detergent used, Cav-1, but not P2X4R was detected upon immunoprecipitation of the receptor complex with the P2X7 antibody (Fig. 2a, b, left). In a previous study [15] we showed that a protein of the expected size of P2X7R (80 kDa) was precipitated with the Cav-1 antibody in the alveolar epithelial cell line E10. Both results are in line with the data obtained by BN- or hrCN-PAGE and strongly suggest that both proteins physically interact. Co-IP with P2X4 antibodies (Fig. 2a, b, right) resulted in a weak detection of P2X7R, especially when NP-40 was used for membrane solubilization. This observation may indicate a weak or transient interaction of both receptors. Cav-1 could not be detected in this experiment.
Fig. 2.
Coimmunoprecipitation of P2X7R and Cav-1 and P2X7R and P2X4R. Native membrane protein extracts were solubilized with 1% NP-40 (a) or 1% digitonin (b) and immunoprecipitated with anti-P2X7R (a, b, left side) or anti-P2X4R antibody (a, b, right side) and analyzed by Western blotting with anti-P2X7R, anti-P2X4R and anti-Cav-1 antibodies. For comparison, immunoprecipitate (IP; lane 1) and supernatant of the immunoprecipitate (S; lane 2) were loaded. Lane 3 shows the immunoprecipitate of the negative control and its supernatant was loaded in lane 4. In addition, the PVDF membrane was probed with anti-P2X7R or anti-P2X4R antibodies to ensure appropriate binding of P2X7R or P2X4R protein to the antibody-Dynabead complex. A representative result from three independent experiments is shown
Downregulation of P2rx7 affects steady-state concentration and supramolecular organization of P2X4R and Cav-1
Previous studies of P2rx7-knockout mice have shown a decreased Cav-1 protein level in lung epithelial type I cells [20]. To analyze the effect of the downregulation of the P2rx7 in the alveolar epithelial cell line E10, we designed three different shRNAs (1–3) targeted to the mouse P2rx7 mRNA. Transfection of all three shRNAs resulted in a significant decrease of the P2X7R protein level to approximately 18% of the wild-type level (Fig. 3a). Interestingly, downregulation of P2rx7 also affects the protein levels of Cav-1 and P2X4R: the level of P2X4R increased to about 220%, whereas that of Cav-1 was reduced to ~60% upon shRNA1 treatment (Fig. 3a). Scrambled P2rx7 shRNAs had no effect on either mRNA or protein levels of Cav-1, P2X4R and P2X7R (Fig. 3b).
Fig. 3.
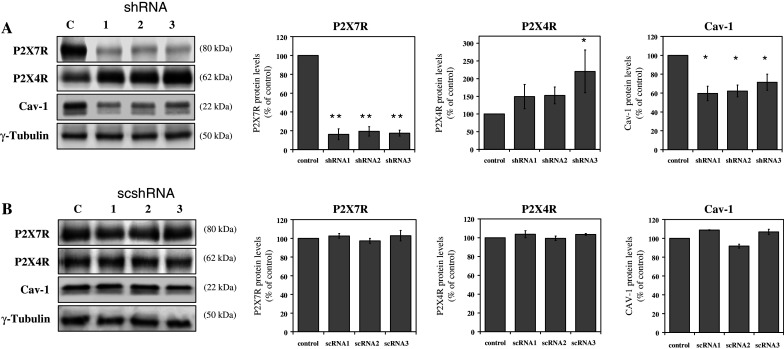
The effect of P2rx7 downregulation in E10 cells. After transfection of E10 cells with three different shRNA constructs targeting P2rx7 (shRNA1-3, a) and three different scrambled shRNAs (scshRNA1-3, b), cells were harvested and P2X7R, P2X4R and Cav-1 protein levels were analyzed by Western blot analysis with rabbit polyclonal anti-P2X7R antibody, rabbit polyclonal anti-P2X4R antibody or mouse monoclonal anti-Cav-1 antibody. γ-Tubulin served as a loading control. Representative data from three separate experiments are shown. Statistical analysis of P2X7R, P2X4R and Cav-1 protein levels are shown on the right side. *p < 0.05; **p < 0.01
Next we addressed the question whether P2rx7 downregulation affects the localization of Cav-1 and P2X4R in the plasma membrane. To this end we performed a cell surface biotinylation analysis. Membrane proteins protruding into the extracellular space of untreated and of P2rx7 knockdown cell were labeled by membrane-impermeable sulfo-N-hydroxysuccinimide ester of biotin. The increase in the biotin-labeled surface fraction of P2X4R is comparable with its overall increase, indicating that the proportion of membrane-associated to cytosolic receptor protein is not severely affected (Fig. 4a). Immunohistochemical data with P2X4R antibodies are in line with this observation, showing a stronger P2X4R fluorescent signal in the plasma membrane and in the cytosol upon P2rx7 downregulation (data not shown). Detection of Cav-1 in the control cells confirmed the known caveolar localization at the cell surface, besides some distinct punctuate cytosolic spots. After P2rx7 knockdown the localization of Cav-1 was no longer restricted to the plasma membrane, but was also distributed intracellularly (Fig. 4b).
Fig. 4.
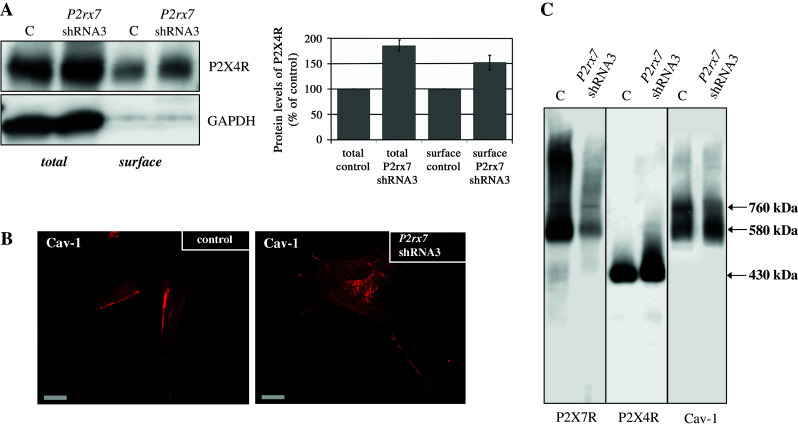
Distribution and localization of P2X4R and Cav-1 after P2rx7 downregulation. a Western blot analysis (using an anti-P2X4R antibody (left side) or an anti-T1α antibody (right side) of biotinylated whole cell lysates (total) and plasma membranes (surface) obtained from untreated cells (control) and after P2rx7 downregulation in E10 cells. A representative Western blot (n = 3) is depicted. The standard protein GAPDH was used to confirm equal protein loading. b Immunofluorescence demonstration of endogenous Cav-1 (Cy3) in untransfected E10 cells (left side) and in E10 cells transfected with P2rx7 shRNA3 (right side). Scale bar corresponds to 20 μm. c Native membrane extracts from untransfected E10 cells and E10 cells transfected with P2rx7 shRNA3 were isolated and solubilized with digitonin. Proteins were separated by hrCN-PAGE, blotted and probed with antibodies against P2X7R, P2X4R and Cav-1
Further, we addressed the question whether the shRNA-mediated knockdown of P2rx7 has an influence on the supramolecular organization of the P2XR/Cav-1 complexes at 430, 580 and 760 kDa. To this end, we compared the hrCN-PAGE profiles in E10 control cells and in E10 cells after shRNA-mediated downregulation of P2rx7.
Figure 4c shows the already described Cav-1 complexes of 580 and 760 kDa in the control cells. Upon P2rx7 knockdown the latter complex is no longer detectable, while the concentration of the 580-kDa complex is increased. This result indicates that P2X7R is a component of the 760-kDa complex and physically associated with Cav-1. For P2X4R we observed an unchanged supramolecular organization; however, the signal at 430 kDa is strongly increased after P2rx7 downregulation (Fig. 4c, middle).
Downregulation of P2rx4 affects the P2X7R protein level and may indicate a crosstalk between P2X7R and P2X4R
To further investigate the relationship among P2X4R, Cav-1 and P2X7R, we performed P2rx4 downregulation experiments in E10 cells (Fig. 5). Three different shRNAs targeted to the mouse P2rx4 mRNA were designed, of which shRNA 3 proved to be most efficient.
Fig. 5.
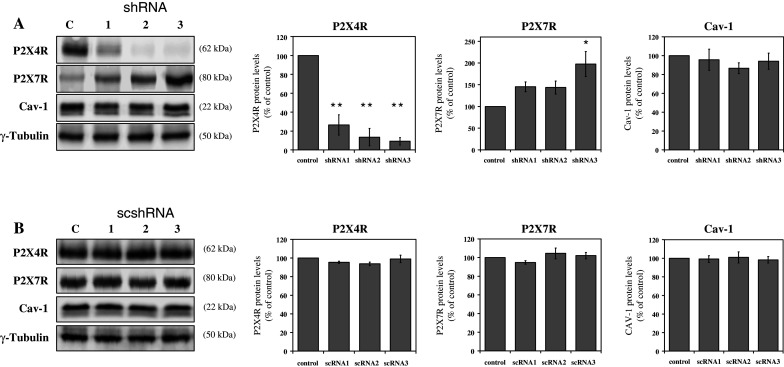
The effect of P2rx4 downregulation in E10 cells. After transfection of E10 cells with three different shRNA constructs targeting P2rx4 (shRNA1-3, a) and three different scrambled shRNAs (scshRNA1-3, b), cells were harvested and P2X4R, P2X7R and Cav-1 levels and were analyzed by Western blot analysis with rabbit polyclonal anti-P2X4R antibody, rabbit polyclonal anti-P2X7R antibody or mouse monoclonal anti-Cav-1 antibody. γ-tubulin served as a loading control. Representative data from three separate experiments are shown. Statistical analysis of P2X4R, P2X7R and Cav-1 protein levels are shown on the right side. *p < 0.05; **p < 0.01
The protein level of P2X4R was significantly decreased to 10% in case of shRNA 3 and to a lesser extent (26 and 14%) by treatment with shRNA 1 and 2. The level of P2X7R increased up to 200% of the wild-type control, whereas the concentration of Cav-1 remained unaffected (Fig. 5a). Scrambled P2rx4 shRNAs had no effect on either mRNA or protein levels of Cav-1, P2X4R and P2X7R (Fig. 5b).
Cell surface biotinylation experiments revealed an increased abundance of P2X7R in the lysate as well as in the cell surface fraction (Fig. 6a). Microscopic examination of the intracellular distribution of P2X7R showed no articulate alteration, except for a slight enrichment in the plasma membrane (data not shown). No apparent changes in the localization of Cav-1 were observed after P2rx4 knockdown (Fig. 6b).
Fig. 6.
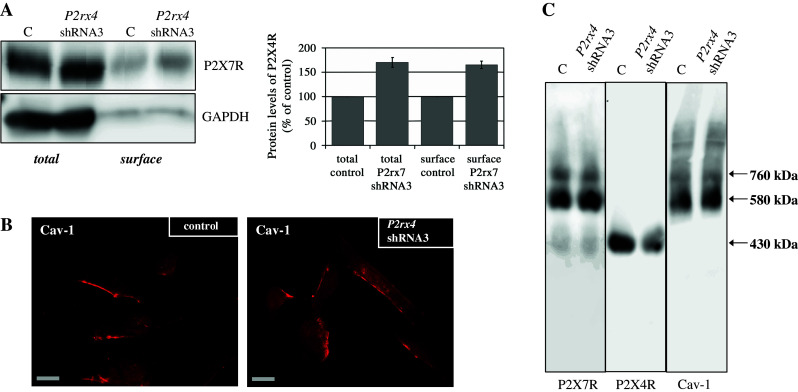
Distribution and localization of P2X7R and Cav-1 after P2rx4 downregulation. a Western blot analysis (using an anti-P2X4R antibody (left side) or an anti-T1α antibody (right side) of biotinylated whole-cell lysates (total) and plasma membranes (surface) obtained from untreated cells (control) and after P2rx4 downregulation in E10 cells. A representative Western blot (n = 3) is depicted. The standard protein GAPDH was used to confirm equal protein loading. b Immunofluorescence demonstration of endogenous Cav-1 (Cy3) in untransfected E10 cells (left side) and in E10 cells transfected with P2rx4 shRNA3 (right side). Scale bar corresponds to 20 μm. c Native membrane extracts from untransfected E10 cells and E10 cells transfected with P2rx4 shRNA3 were isolated and solubilized with digitonin. Proteins were separated by hrCN-PAGE, blotted and probed with antibodies against P2X7R, P2X4R and Cav-1
Figure 6c shows the already described P2X7R complexes of 430, 580 and 760 kDa and Cav-1 complexes at 580 and 760 kDa in the control cells. Upon P2rx4 knockdown we observed an unchanged supramolecular organization for the P2X7R and Cav-1.
In summary, our study demonstrates for the first time that knockdown of P2rx7 in E10 cells affects Cav-1 and P2X4R protein levels and their supramolecular organization in potential Cav-1/P2X7R and P2X4/7R complexes. P2rx4 downregulation affects the P2X7R protein level, but has no apparent effect on Cav-1.
The C-terminal portions of P2X4R and P2X7R interact with negatively charged lipids
In previous studies we reported the localization of P2X4R and P2X7R in DRMs [14, 15]. We used an in vitro lipid-binding assay to test the direct interaction of the P2X4R and P2X7R with several anionic phospholipids. P2XRs possess a characteristic positively charged amino acid sequence in their C-terminal portions [41, 42], which is capable of interacting with negatively charged lipids. The C-terminal domains of the P2X4R (aa 358–388) and of the P2X7R (aa 358–595) were C-terminally fused to GST, heterologously expressed in E. coli and purified by affinity chromatography (data not shown). GST as well as the GST-fusion proteins were tested with an in vitro binding assay for their affinity to various lipids. Interestingly, both fusion proteins showed a similar lipid-binding pattern with higher affinity towards the negatively charged lipids phosphatidic acid (PA), phosphaditylserine (PS), PI(4)P, PI(4,5)P2 and PI(3,4,5)P3. Additionally, we observed binding to 3-sulfogalactosylceramide, phosphatidylglycerol (PG) and cardiolipin (Fig. 7). This result is in agreement with a regulation of P2X7 and P2X4 ion channels by PI(4,5)P2, as described by Zhao et al. [43] and Bernier et al. [41]. Furthermore, the tested negative-charged lipids are most of all involved in signaling pathways, and the association of P2X7R and P2X4R with them proved the important role of P2XRs in signaling cascades.
Fig. 7.
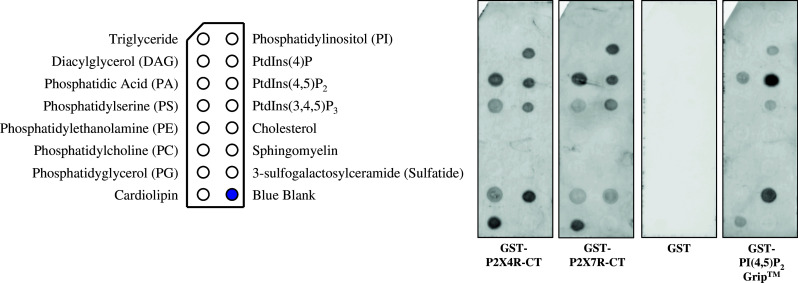
Membrane lipid-binding assay. From left to right: lipid-coated membrane strips were incubated with GST-P2X4R C-terminal fusion protein (GST-P2X4R-CT) and GST-P2X7R C-terminal fusion protein (GST-P2X7R-CT), GST alone and N-terminal GST-tagged recombinant PLC-δ1 PH domain (GST-PI(4,5)P2 Grip™). GST showed no specific binding on immobilized lipids, but the C-terminus of P2X4R and P2X7R bound directly to PA, PS, PG, 3-sulfogalactosylceramide, cardiolipin and the phosphoinositides PI(4)P, PI(4,5)P2 and PI(3,4,5)P3. N-terminal PLC-δ1 PH domain was used as positive control
Discussion
Interrelation between P2X4R and P2X7R
The sequences of human P2X4R and P2X7R are highly homologous: 49% of the amino acids of the transmembrane domain and the extracellular loop are identical, thus exceeding the similarity between P2X7R and other P2XR subtypes [2]. Tandem orientation and the small distance of only 26 kbp on chromosome 5 indicates that the murine genes for P2X7R and P2X4R originated by gene duplication [12, 28].
We noted that the abundance of P2X4R is strongly increased upon P2rx7 downregulation in E10 cells. This increase is accompanied by a more pronounced localization at the plasma membrane compared to the predominant intracellular localization in control cells. P2rx4 downregulation yields complementary results. We observed an increase in the P2X7R protein level and a shift of P2X7R localization to the plasma membrane. At the level of the molecular organization in high molecular mass complexes, we observed an increase of the concentration of P2X4R in complexes of ~430 kDa upon P2rx7 knockdown (Fig. 4c), whereas the small amount of P2X7R in these complexes remains almost unchanged after P2rx4 knockdown (Fig. 6c). Based on these observations we propose that P2X7R depletion results in increased abundance of P2X4R in complexes of ~430 kDa. Eventual functional consequences of the elevated P2X4R concentration remain to be clarified. The situation is less clear in the case of P2X4R depletion. Here we observed an increase of P2X7R in the membrane fraction, again arguing for a compensatory mechanism in the expression of the two receptors. However—unlike in the case of P2rx7 knockdown—this increase was not detected in the ~430-kDa complexes.
Published data on the interaction of P2X7R and P2X4R are conflicting. While Nicke [12] reports that P2X7R and P2X4R form exclusively homomers, which mostly exist as homotrimeric complexes in various tissues, for bone marrow-derived macrophages, P2X4/7R heterotrimers have been described [10, 28]. In rodent immune cells and mouse salivary epithelia cells, an interaction of homotrimeric P2X7R and P2X4R channels was detected [11, 13]. Possibly cell type- or tissue-specific organization can explain these differing data, but the different detergents used for the biochemical analysis should also be taken into account.
Our results provide evidence for a compensatory mechanism of the expression of P2X4R and P2X7R, hinting at a crosstalk and feedback signaling between these receptors in alveolar epithelial cells. Our data provide no experimental evidence for the presence of heterotrimeric P2X4/7R channels, but interactions may be possible between homotrimeric P2X4R and P2X7R channels.
Association of P2X4R and P2X7R with lipids
BN-PAGE reveals that P2X4R is organized in two complexes of 430 and 580 kDa. When analyzed by hrCN-PAGE, we observed only the 430-kDa complex. Trimeric P2X4R channels are able to form higher order complexes, maybe modulated by lipids. Possibly their isolation is dependent on specific detergents [12, 36, 40, 44]. We suppose that DDM, the detergent used in hrCN-PAGE, may influence not only labile protein-protein interactions [32], but also some critical lipid-protein interactions.
P2XRs possess a characteristic positively charged amino acid sequence in their C-terminal portions [41, 42], which is capable of interacting with negatively charged lipids. Our in vitro results demonstrate the direct interaction of fusion proteins between GST and the C-terminal portions of P2X4R or P2X7R with negatively charged lipids. As all these lipids are involved in signaling pathways, their association with P2X7R and P2X4R suggests a role of P2XRs in lipid signaling. Phosphoinositides participate in the association of membrane-bound signaling complexes and in the control of membrane to cytosol interfaces [45]. PI(4,5)P2 is known to act as a membrane anchor that is concentrated in caveolin-enriched detergent-insoluble membrane fractions [46–48]. The activities of a variety of ion channels and transporters, many of which are concentrated in caveolae, are modulated by binding to PI(4,5)P2 [49–54]. Recently, Zhao et al. [43] and Bernier et al. [41] reported the regulation of P2X7R and P2X4R ion channel properties through PI(4,5)P2, confirming our results of the in vitro lipid-binding assay and the partial co-localization of P2X7R and P2X4R in PIP2-rich microdomains.
Unexpectedly the lipid-binding assay provides evidence for an interaction of P2X7R and P2X4R fusion proteins with cardiolipin. This specialized lipid, which is involved in apoptotic processes and discussed as a surface marker of apoptotic cells [55–57], is mainly found in mitochondria [58], but also detected in lysosomes and the endoplasmatic reticulum. A localization of P2X4R predominantly within intracellular compartments, especially in lysosomes, was demonstrated for cultured rat microglia, vascular endothelial cells and freshly isolated peritoneal macrophages [59]. Also in E10 cells P2X4R is mainly localized in intracellular organelles. Our data suggest that P2XR/lipid interactions may be essential determinants for the intracellular localization and function of the purinergic P2X receptors.
Crosstalk between the purinergic receptors P2X7R and P2X4R and Cav-1
Our previous studies of cav-1 knockout mice have shown that the pronounced reduction in the number of caveolae is accompanied by a strong decrease of the concentration of the purinergic receptor P2X7R in the lung AT I cells [20]. Similarly cav-1 downregulation in the alveolar epithelial cell line E10 resulted in a decreased P2X7R protein level and affected its intracellular localization with a more pronounced cytoplasmic distribution [14, 15]. These data suggested that Cav-1 directly or indirectly via its function in caveolae formation may affect the expression, stability and/or organization of P2X7R. A discrete population of P2X7R has been shown to be associated with lipid rafts, as determined by co-fractionation with the raft marker Cav-1 [14]. The finding that part of P2X7R can be co-immunoprecipitated with Cav-1 in alveolar epithelial cells suggests physical interaction of P2X7R with Cav-1.
This study corroborates this view: by analyzing the supramolecular organization of native P2X7R by BN-PAGE and hrCN-PAGE, we demonstrate that it is present in three complexes of ~430, ~580 and ~760 kDa. Cav-1 is detected in two bands, a weaker one at 760 kDa and a stronger one at 580 kDa, both corresponding exactly in size to P2X7R complexes. Upon P2rx7 knockdown the overall concentration of Cav-1 is reduced, which is mainly due to the complete disappearance of the signal at 760 kDa. The intensity of the signal at 580 kDa is even somewhat increased. These results show that the presence of Cav-1 in the 760-kDa complex depends on P2X7R. Moreover, they suggest that under these conditions Cav-1 accumulates in the 580-kDa complex. The stronger Cav-1 signal in the cytosol (Fig. 4c) may result from release of some protein from the membrane. The 580-kDa complex containing both Cav-1 and P2X7R could either represent a discrete independent complex or a precursor form of the 760-kDa Cav-1/P2X7R complex. In this context it may be interesting to note that caveolae are not assembled de novo at the cell surface, but stored in a pre-assembled form as intracellular caveolar vesicles [60]. Lipid raft-associated oligomeric aggregates of caveolin proteins have been proposed as scaffolding complexes that provide a platform for protein-protein interactions between caveolin and raft-associated proteins [22]. Membrane receptors, e.g., TRPC1 and IP3R3, may be targeted to caveolae by their interaction with Cav-1 [61]. Contrary to the significant effects seen after P2rx7 knockdown, Cav-1 is only moderately affected by P2rx4 downregulation. In line with this, Cav-1 is not co-immunoprecipitated with P2X4R. Obviously caveolar localization of P2X4R does not depend on its interaction with Cav-1, but perhaps via its association with P2X7R.
Acknowledgments
We would like to thank Annett Linge, MD, and Sylvia Grossklaus for technical support and Kerstin Pehlke for assistance with molecular biological experiments.
Abbreviations
- P2X
Ionotrophic purinergic receptors
- BN
Blue native
- hrCN
High resolution clear native
- Cav-1
Caveolin-1
- DRM
Detergent resistent membrane
- DDM
n-Dodecyl-β-d-maltoside
- AEBSF
4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride
- TCEP
Tris-(carboxyethyl)-phosphine hydrochloride
- DOC
Sodium deoxycholate
References
- 1.Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442:527–532. doi: 10.1038/nature04886. [DOI] [PubMed] [Google Scholar]
- 2.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 3.Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50:413–492. [PubMed] [Google Scholar]
- 4.Brown SG, Townsend-Nicholson A, Jacobson KA, Burnstock G, King BF. Heteromultimeric P2X(1/2) receptors show a novel sensitivity to extracellular pH. J Pharmacol Exp Ther. 2002;300:673–680. doi: 10.1124/jpet.300.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicke A, Kerschensteiner D, Soto F. Biochemical and functional evidence for heteromeric assembly of P2X1 and P2X4 subunits. J Neurochem. 2005;92:925–933. doi: 10.1111/j.1471-4159.2004.02939.x. [DOI] [PubMed] [Google Scholar]
- 6.Torres GE, Haines WR, Egan TM, Voigt MM. Co-expression of P2X1 and P2X5 receptor subunits reveals a novel ATP-gated ion channel. Mol Pharmacol. 1998;54:989–993. doi: 10.1124/mol.54.6.989. [DOI] [PubMed] [Google Scholar]
- 7.Lewis C, Neidhart S, Holy C, North RA, Buell G, Surprenant A. Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature. 1995;377:432–435. doi: 10.1038/377432a0. [DOI] [PubMed] [Google Scholar]
- 8.King BF, Townsend-Nicholson A, Wildman SS, Thomas T, Spyer KM, Burnstock G. Coexpression of rat P2X2 and P2X6 subunits in Xenopus oocytes. J Neurosci. 2000;20:4871–4877. doi: 10.1523/JNEUROSCI.20-13-04871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le KT, Babinski K, Seguela P. Central P2X4 and P2X6 channel subunits coassemble into a novel heteromeric ATP receptor. J Neurosci. 1998;18:7152–7159. doi: 10.1523/JNEUROSCI.18-18-07152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo C, Masin M, Qureshi OS, Murrell-Lagnado RD. Evidence for functional P2X4/P2X7 heteromeric receptors. Mol Pharmacol. 2007;72:1447–1456. doi: 10.1124/mol.107.035980. [DOI] [PubMed] [Google Scholar]
- 11.Boumechache M, Masin M, Edwardson JM, Gorecki DC, Murrell-Lagnado R. Analysis of assembly and trafficking of native P2X4 and P2X7 receptor complexes in rodent immune cells. J Biol Chem. 2009;284:13446–13454. doi: 10.1074/jbc.M901255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicke A. Homotrimeric complexes are the dominant assembly state of native P2X7 subunits. Biochem Biophys Res Commun. 2008;377:803–808. doi: 10.1016/j.bbrc.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 13.Casas-Pruneda G, Reyes JP, Perez-Flores G, Perez-Cornejo P, Arreola J. Functional interactions between P2X4 and P2X7 receptors from mouse salivary epithelia. J Physiol. 2009;587:2887–2901. doi: 10.1113/jphysiol.2008.167395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth K, Weinhold K, Guenther A, Young MT, Schnittler H, Kasper M. Caveolin-1 influences P2X7 receptor expression and localization in mouse lung alveolar epithelial cells. FEBS J. 2007;274:3021–3033. doi: 10.1111/j.1742-4658.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 15.Barth K, Weinhold K, Guenther A, Linge A, Gereke M, Kasper M. Characterization of the molecular interaction between caveolin-1 and the P2X receptors 4 and 7 in E10 mouse lung alveolar epithelial cells. Int J Biochem Cell Biol. 2008;40:2230–2239. doi: 10.1016/j.biocel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Ma W, Korngreen A, Weil S, Cohen EB, Priel A, Kuzin L, Silberberg SD. Pore properties and pharmacological features of the P2X receptor channel in airway ciliated cells. J Physiol. 2006;571:503–517. doi: 10.1113/jphysiol.2005.103408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tayler AL, Schwiebert LM, Smith JJ, King C, Jones JR, Sorscher EJ, Schwiebert EM. Epithelial P2X purinergic receptor channel expression and function. J Clin Invest. 1999;104:875–884. doi: 10.1172/JCI7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang Z, Burnstock G. Expression of P2X receptors on rat microglial cells during early development. Glia. 2005;52:119–126. doi: 10.1002/glia.20227. [DOI] [PubMed] [Google Scholar]
- 19.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Barth K, Kasper M. Membrane compartments and purinergic signalling: occurrence and function of P2X receptors in lung. FEBS J. 2009;276:341–353. doi: 10.1111/j.1742-4658.2008.06795.x. [DOI] [PubMed] [Google Scholar]
- 21.Pike LJ. Lipid rafts: heterogeneity on the high seas. Biochem J. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev. 2002;54:431–467. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 23.Vacca F, Amadio S, Sancesario G, Bernardi G, Volonte C. P2X3 receptor localizes into lipid rafts in neuronal cells. J Neurosci Res. 2004;76:653–661. doi: 10.1002/jnr.20069. [DOI] [PubMed] [Google Scholar]
- 24.Vial C, Evans RJ. Disruption of lipid rafts inhibits P2X1 receptor-mediated currents and arterial vasoconstriction. J Biol Chem. 2005;280:30705–30711. doi: 10.1074/jbc.M504256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannas P, Adriouch S, Kahl S, Braasch F, Haag F, Koch-Nolte F. Activity and specificity of toxin-related mouse T cell ecto-ADP-ribosyltransferase ART2.2 depends on its association with lipid rafts. Blood. 2005;105:3663–3670. doi: 10.1182/blood-2004-08-3325. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Marcos M, Pochet S, Tandel S, Fontanils U, Astigarraga E, Fernandez-Gonzalez JA, Kumps A, Marino A, Dehaye JP. Characterization and comparison of raft-like membranes isolated by two different methods from rat submandibular gland cells. Biochim Biophys Acta. 2006;1758:796–806. doi: 10.1016/j.bbamem.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Gonnord P, Delarasse C, Auger R, Benihoud K, Prigent M, Cuif MH, Lamaze C, Kanellopoulos JM. Palmitoylation of the P2X7 receptor, an ATP-gated channel, controls its expression and association with lipid rafts. FASEB J. 2009;23:795–805. doi: 10.1096/fj.08-114637. [DOI] [PubMed] [Google Scholar]
- 28.Dubyak GR. Go it alone no more-P2X7 joins the society of heteromeric ATP-gated receptor channels. Mol Pharmacol. 2007;72:1402–1405. doi: 10.1124/mol.107.042077. [DOI] [PubMed] [Google Scholar]
- 29.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 31.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, Di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Dieckmann-Schuppert A, Schnittler HJ. A simple assay for quantification of protein in tissue sections, cell cultures, and cell homogenates, and of protein immobilized on solid surfaces. Cell Tissue Res. 1997;288:119–126. doi: 10.1007/s004410050799. [DOI] [PubMed] [Google Scholar]
- 34.Nicke A, Baumert HG, Rettinger J, Eichele A, Lambrecht G, Mutschler E, Schmalzing G. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. Trimeric architecture of homomeric P2X2 and heteromeric P2X1+2 receptor subtypes. J Mol Biol. 2004;342:333–343. doi: 10.1016/j.jmb.2004.06.092. [DOI] [PubMed] [Google Scholar]
- 36.Torres GE, Egan TM, Voigt MM. Hetero-oligomeric assembly of P2X receptor subunits. Specificities exist with regard to possible partners. J Biol Chem. 1999;274:6653–6659. doi: 10.1074/jbc.274.10.6653. [DOI] [PubMed] [Google Scholar]
- 37.Kim M, Jiang LH, Wilson HL, North RA, Surprenant A. Proteomic and functional evidence for a P2X7 receptor signalling complex. EMBO J. 2001;20:6347–6358. doi: 10.1093/emboj/20.22.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sargiacomo M, Scherer PE, Tang Z, Kubler E, Song KS, Sanders MC, Lisanti MP. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soto F, Garcia-Guzman M, Gomez-Hernandez JM, Hollmann M, Karschin C, Stuhmer W. P2X4: an ATP-activated ionotropic receptor cloned from rat brain. Proc Natl Acad Sci USA. 1996;93:3684–3688. doi: 10.1073/pnas.93.8.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernier LP, Ase AR, Chevallier S, Blais D, Zhao Q, Boue-Grabot E, Logothetis D, Seguela P. Phosphoinositides regulate P2X4 ATP-gated channels through direct interactions. J Neurosci. 2008;28:12938–12945. doi: 10.1523/JNEUROSCI.3038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernier LP, Ase AR, Tong X, Hamel E, Blais D, Zhao Q, Logothetis DE, Seguela P. Direct modulation of P2X1 receptor-channels by the lipid phosphatidylinositol 4, 5-bisphosphate. Mol Pharmacol. 2008;74:785–792. doi: 10.1124/mol.108.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Q, Yang M, Ting AT, Logothetis DE. PIP2 regulates the ionic current of P2X receptors and P2X7 receptor-mediated cell death. Channels. 2007;1:46–55. [PubMed] [Google Scholar]
- 44.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc Natl Acad Sci USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 46.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 47.Pike LJ, Miller JM. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J Biol Chem. 1998;273:22298–22304. doi: 10.1074/jbc.273.35.22298. [DOI] [PubMed] [Google Scholar]
- 48.Waugh MG, Lawson D, Tan SK, Hsuan JJ. Phosphatidylinositol 4-phosphate synthesis in immunoisolated caveolae-like vesicles and low buoyant density non-caveolar membranes. J Biol Chem. 1998;273:17115–17121. doi: 10.1074/jbc.273.27.17115. [DOI] [PubMed] [Google Scholar]
- 49.Mace OJ, Woollhead AM, Baines DL. AICAR activates AMPK and alters PIP2 association with the epithelial sodium channel ENaC to inhibit Na+ transport in H441 lung epithelial cells. J Physiol. 2008;586:4541–4557. doi: 10.1113/jphysiol.2008.158253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edinger RS, Kester L, Weixel KM, Johnson JP. Complex regulation of the epithelial Na+ channel (ENaC) by phosphatidylinsoitol bisphosphate (PIP2) FASEB J. 2007;21:A543–A544. doi: 10.1096/fj.06-6694com. [DOI] [Google Scholar]
- 51.Qin F. Regulation of TRP ion channels by phosphatidylinositol-4, 5-bisphosphate. Handb Exp Pharmacol. 2007;179:509–525. doi: 10.1007/978-3-540-34891-7_30. [DOI] [PubMed] [Google Scholar]
- 52.Voets T, Nilius B. Modulation of TRPs by PIPs. J Physiol. 2007;582:939–944. doi: 10.1113/jphysiol.2007.132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nam JH, Lee HS, Nguyen YH, Kang TM, Lee SW, Kim HY, Kim SJ, Earm YE. Mechanosensitive activation of K+ channel via phospholipase C-induced depletion of phosphatidylinositol 4, 5-bisphosphate in B lymphocytes. J Physiol. 2007;582:977–990. doi: 10.1113/jphysiol.2007.128413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hilgemann DW, Feng S, Nasuhoglu C (2001) The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE 2001:RE19 [DOI] [PubMed]
- 55.Esposti MD. Lipids, cardiolipin and apoptosis: a greasy licence to kill. Cell Death Differ. 2002;9:234–236. doi: 10.1038/sj.cdd.4400997. [DOI] [PubMed] [Google Scholar]
- 56.Sorice M, Circella A, Misasi R, Pittoni V, Garofalo T, Cirelli A, Pavan A, Pontieri GM, Valesini G. Cardiolipin on the surface of apoptotic cells as a possible trigger for antiphospholipids antibodies. Clin Exp Immunol. 2000;122:277–284. doi: 10.1046/j.1365-2249.2000.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalvez F, Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 58.Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39:257–288. doi: 10.1016/S0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 59.Qureshi OS, Paramasivam A, Yu JC, Murrell-Lagnado RD. Regulation of P2X4 receptors by lysosomal targeting, glycan protection and exocytosis. J Cell Sci. 2007;120:3838–3849. doi: 10.1242/jcs.010348. [DOI] [PubMed] [Google Scholar]
- 60.Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- 61.Hardin CD, Vallejo J. Dissecting the functions of protein–protein interactions: caveolin as a promiscuous partner. Focus on “Caveolin-1 scaffold domain interacts with TRPC1 and IP3R3 to regulate Ca2+ store release-induced Ca2+ entry in endothelial cells”. Am J Physiol Cell Physiol. 2009;296:C387–C389. doi: 10.1152/ajpcell.00663.2008. [DOI] [PubMed] [Google Scholar]