Abstract
We have developed a method for reconstructing gene association networks and have applied this method to gene profiles from 3T3-L1 cells. Priorization of the candidate genes pinpointed a transcript annotated as APMAP (adipocyte plasma membrane-associated protein). Functional studies showed that APMAP is upregulated in murine and human adipogenic cell models as well as in a genetic mouse model of obesity. Silencing APMAP in 3T3-L1 cells strongly impaired the differentiation into adipocytes. Moreover, APMAP expression was strongly induced by the PPARγ ligand rosiglitazone in adipocytes in vitro and in vivo in adipose tissue. Using ChIP-qPCR and luciferase reporter assays, we show a functional PPARγ binding site. In addition, we provide evidence that the extracellular C-terminal domain of APMAP is required for the function of APMAP in adipocyte differentiation. Finally, we demonstrate that APMAP translocates from the endoplasmatic reticulum to the plasma membrane during adipocyte differentiation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0424-5) contains supplementary material, which is available to authorized users.
Keywords: Adipogenesis, APMAP, PPARγ, Transcriptional regulation, Gene expression, Gene networks
Introduction
Obesity, the excess deposition of adipose tissue, is one of the most pressing health problems on a global scale. The mechanisms controlling the process of preadipocyte differentiation have been extensively studied in cell culture models. Upon appropriate hormonal stimulation, confluent 3T3-L1 and 3T3-F442A cells accumulate lipid and develop the morphology of mature adipocytes with many characteristics of adipose cells found in vivo [1]. Adipogenesis is known to be controlled by a transcriptional cascade, including activators, coactivators and repressors [2]. At the core of this cascade, the early expression of CCAAT/enhancer-binding protein β (C/EBPβ) and delta (C/EBPδ) initializes the expression of the nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ), the master regulator of adipogenesis, which is necessary and sufficient for adipocyte differentiation [3]. Many of the genes which are direct targets of PPARγ and recognized as important in adipogenesis are transcription factors. Among them, C/EBPα is a target gene of PPARγ that is induced late in adipogenesis and most abundantly in mature adipocytes. In terminal differentiation, PPARγ and C/EBPα enhance each other’s expression and together they upregulate many adipogenic genes (for review, see [4]). In addition to PPARγ and C/EBPs, the involvement of several other transcription factors including Kruppel-like factors (KLFs) [5], Krox20 [6], and GATA [7] as well as regulatory proteins (e.g., Wingless and Int-1 proteins (Wnts) [8]) has been demonstrated.
During the last decade, novel high-throughput technologies assisted in the disclosure of new molecular components building the genetic network controlling adipogenesis. Microarrays have been successfully applied to identify candidate genes, such as KLF4 [5] and Krox20 [6]. Additionally, large-scale chromatin immunoprecipitation studies were applied to identify target genes of (adipogenic) key factors and gene–gene interactions [9, 10]. These large-scale studies provided abundant data, but the selection of candidate genes from these studies for further functional characterization is challenging and often results in suboptimal choice. We have therefore developed a novel method based on the calculation of correlation and mutual information to reconstruct gene–gene association networks and prioritize candidate genes for further functional studies. In this paper, we show that the use of this method and data from a previous gene expression study [11] led to the identification of a novel PPARγ-targeted transcript annotated as a RIKEN clone, and subsequently mapped as adipocyte plasma membrane-associated protein (APMAP). We show here that APMAP is upregulated in mouse and human mature adipocytes and in white adipose tissue of genetically obese mice. In addition, we demonstrate that APMAP is required for adipogenesis, is upregulated in vivo and in vitro by the PPARγ ligand rosiglitazone, and is functionally targeted by PPARγ. Furthermore, we provide computational evidence that APMAP is a transmembrane protein with a six-bladed β-propeller and hydrolase activity. Finally, experimental studies demonstrate that APMAP localizes at the endoplasmatic reticulum (ER) in preadipocytes and translocates to the plasma membrane in mature adipocytes, and that the six-bladed β-propeller is necessary for the role of APMAP in adipogenesis.
These results are the first to demonstrate the importance of APMAP in the process of fat cell development and to validate the utility of applying our computational/experimental approach for the elucidation of gene regulatory networks.
Methods and materials
Network reconstruction algorithm
Expression profiles were first discretized as described by Di Camillo et al. [12] assuming an error model with Gaussian distribution of mean 0 and standard deviation estimated from the biological replicates of the original data. Genes were clustered according to their discrete expression profile, and the continuous centroid of each cluster was also calculated. Genes exhibiting no change in expression across the time course were discarded. For network reconstruction, we first ran REVEAL [13] on the discrete expression profiles of the clusters restricted to one regulator. In order to reduce the number of false positives, we only considered those interactions found by REVEAL for which the correlation between the centroids of the clusters was significantly different from 0 with a delay of one time point. Because the time course considered in this study was short (8 time points), significance for the correlation coefficient was calculated based on a permutation test implemented in the R program permax. Cytoscape [14] was used for network visualization. Detailed description of the algorithm with all its functionality and code can be found in the Electronic supplementary material (ESM, file 4).
Promoter analyses
Gene clusters with degree ≥5 from the network analysis were further analyzed. Transcripts involved in transcriptional regulation were identified based on gene ontology (GO:0003700, GO:0045944, GO:0006355) and transcription factors (TF) with available vertebrate position weight matrix (PWM) from TRANSFAC [15] or JASPAR [16] databases (as summarized at http://genome.tugraz.at/Logo) were considered. Promoters (−10 to +10 kb relative to transcription start site) of genes within the cluster including the TF and neighboring clusters were searched for potential binding sites of the respective TF. For this purpose, genomic sequences (mm9) and mouse–human genome alignments (mm9–hg18) were derived from UCSC genome browser [17] using genomic coordinates of the Refseq database [18]. Potential binding sites were identified if the similarity score for each PWM was equal or above a threshold (allowing 1 binding site with 10 kb of mouse coding sequences) based on an implementation of the MatInspector algorithm [19]. Binding sites were counted as conserved if the similarity score was equal or above the threshold in both the mouse and the human sequence from the alignment.
Animal studies
Male C57/Bl6 mice were used for this study. Animals were kept on a 12/12-h light/dark cycle. Mice were put on either chow diet or chow diet with rosiglitazone (0.01% w/w) immediately after weaning and kept on this diet for 7 weeks. Before harvesting the epididymal fat pads, mice were fasted for 4 h. All animal procedures used were approved by the Austrian Bundesministerium für Wissenschaft und Forschung.
Silencing of APMAP using short hairpin RNA lentivirus particles
One control non-targeting shRNA lentivirus and four shRNA lentiviruses directed against APMAP were purchased from Sigma (MISSION™ shRNA lentiviral particles NM_027977). 3T3-L1 cells were seeded into 6-well plates 12 h before transduction using 3 × 104 cells/well (around 30% confluence). Cells were infected o/n with three MOIs (multiplicity of infection) in complete medium containing 8 μg/ml hexadimethrine (Sigma). After 16 h, the infection medium was replaced with fresh medium containing 3 μg/ml puromycin (Sigma). 3T3-L1 cells were selected for stable expression for at least 8 days.
Generation of recombinant retrovirus
The coding sequence of mouse APMAP was amplified by PCR from mouse adipose tissue cDNA using Pfu polymerase (Fermentas). The primers were designed to create XhoI (5′) and EcoRI (3′) restriction sites and the product, containing the whole open reading frame, was ligated into XhoI-EcoRI digested Murine Stem Cell Virus vector (pMSCV puro; BD Biosciences Clontech). To produce infectious but replication-incompetent recombinant retroviruses expressing APMAP, PhoenixEco packaging cells (cultured in DMEM with 10% FBS in 5% CO2) were transfected with pMSCV-APMAP using Metafectene (Biontex Laboratories, Germany). The supernatant containing the viral particles was collected 48 h after transfection. Viral supernatants were supplemented with 8 μg/ml hexadimethrine (Sigma) and added to 3T3-L1 cells (30% confluence) for infections for 18–24 h. Cells were selected with 3 μg/ml puromycin (Sigma), expanded, and seeded for differentiation experiments. For the stable expression of the truncated versions of APMAP, the DNA coding for the N-terminal 60AA or 63AA was inserted into pMSCV puro and the stable cell lines were produced as described above. The empty pMSCVpuro was used as control for all the above-described stable cell lines.
Cell culture, adipocyte differentiation, lipid staining and quantification
3T3-L1 cells (ATCC, American Type cell Culture Collection) and MEFs were grown and maintained in high glucose DMEM containing 10% FBS, 50 μg/ml streptomycin, and 50 U/ml penicillin (Invitrogen). Cells were induced to differentiate 2 days after confluence with 0.5 mM 3-isobutyl-1-methylxanthine (Calbiochem), 1 μM dexamethasone (Sigma) and 2 μg/ml insulin (Sigma). After 3 days, medium was changed to medium containing 1 μg/ml insulin for 48 h and thereafter maintained in DMEM with 10% FBS (Invitrogen) in 5% CO2 until harvested (at the time points indicated in the experiments). Plates were oil-red O stained as described earlier [20]. In experiments where each component is added separately or in groups, the concentrations are as in the standard hormone cocktail.
OP-9 cells were grown and maintained in α-MEM (Invitrogen) containing 20% FBS and Penicillin/Streptomycin (P/S). OP-9 cells were induced to differentiate as described above. After 3 days, medium was changed to maintenance.
SGBS cells were cultured in DMEM/F12 supplemented with 10% FCS, P/S, 8 μg/ml biotin, and 4 μg/ml pantothenic acid. To induce differentiation, cells were grown to confluence and subsequently cultured in serum-free DMEM/F12 medium, containing P/S, 8 μg/ml biotin, and 4 μg/ml pantothenic acid, 0.01 mg/mL transferrin, 1 μM cortisol, 200 pM triiodotyronine (T3), 20 nM human insulin, 0.25 μM DEX, 500 μM IBMX, and rosiglitazone (2 μM until day 4, 1 μM until day 8). Afterwards, the same medium was used without rosiglitazone.
3T3-L1 adipocytes were treated at day 10 with rosiglitazone and GW9662 for the doses and times indicated in the text, figures and figure legends.
For triglyceride (TG) content, fully differentiated 3T3-L1 cells were washed with PBS, collected in 0.5 ml PBS/well and sonicated twice for 20 s. TG content was measured using Infinity Triglyceride Reagent (Thermo). Values were corrected by protein content measurement using the BCA reagent (Pierce Biotechnology).
Western blot analysis
Control (ntc) and APMAP-silenced (sil) 3T3-L1 cells (at several differentiation time points) were harvested for protein analysis by scraping with lysis buffer (50 mM TrisHCl pH 6.8, 10% glycerol, 2.5% SDS, 1× protease inhibitor cocktail, 1 mM PMSF) after two washes with ice-cold PBS. The suspension was incubated at 90°C for 10 min. Benzonase (Merck) was added followed by incubation at room temperature for 1 h. Protein concentration was determined with the BCA protein assay kit from Pierce according to the manufacturer’s instructions. Each lane of a 10% Bis–Tris Gel (NuPAGE, Invitrogen) was loaded with 30 μg of sample. After electophoresis, gels were blotted to Nitrocellulose-membranes (Invitrogen). Blots were blocked and incubated with the following antibodies: anti-APMAP (Eurogentec) (diluted 1:300), and anti-β-actin (1:25,000; Sigma). For chemiluminescent detection, a horseradish peroxidase-conjugated secondary antibody was used (anti-rabbit 1:3,000; Pierce) and ECL component (Pierce) served as a substrate. Developed films were scanned with an HP Scanjet at 600 dpi.
RNA isolation, reverse transcription, and gene expression analysis
Cells were washed with PBS and harvested using the RNA isolation kit from Marcherey-Nagl. Tissue RNA was isolated with the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Expression of genes was assessed by real-time reverse transcriptase-polymerase chain reaction (RT-PCR) using an ABI Prism 7700 Sequence Detector system utilizing SYBR Green PCR master mix (Applied Biosystems). Gene expression was normalized using TFIIβ for murine tissues and cells and β-Actin for human cells as reference genes. Relative mRNA expression levels were calculated using averaged ∆∆C t values for each biological replicate as implemented in [21]. Sequences of primer pairs can be found in ESM, file 5.
Chromatin immunoprecipitation analyzed by qPCR (ChIP-qPCR)
Chromatin immunoprecipitation was performed with anti-PPARγ1+2 antibody (Santa Cruz; N20) on day 10 3T3-L1 adipocytes as described previously [9]. The resulting DNA fragments were analyzed using SYBR green qPCR. Data were normalized to a site near the S18 rRNA gene. Primer pairs (a list is given in ESM, file 6) were designed to cover the proximal promoter and binding regions suggested by a recent genome-wide location analysis [9].
Luciferase reporter assays
The intron1–2/exon2 region of APMAP (~7,400–8,800 bps downstream of the TTS, primer pairs are given in ESM, file 6) was cloned into the pGl4.26 (Promega) luciferase reporter vector containing a minimal promoter (the whole construct was named pGL4.26i12e2). The Renilla reporter vector pGl4.75 (Promega) was cotransfected in all experiments in a ratio of 1:50 to luciferase reporter vectors as a control for varying transfection efficiencies.
Cotransfected PPARγ2 and RXRα containing pCMX expression vectors were kindly provided by M. Schupp. Transfection into COS7 cells was performed in 96-well plates using MetafectenePro (Biontex) according to the provider’s protocol in ratio of 3:1 (μl MFP:μg DNA). 100 ng of luciferase reporter vector and either 50 ng of each, PPARγ2 and RXRα, or 100 ng of the empty pCMX as a control were used. After 48 h cells were lysed and assayed according to the protocol provided with the Dual-luciferase assay system (Promega). Luminescence read-outs were generated with a Berthold Orion II luminometer. Relative luciferase activity was calculated by referring Renilla-normalized values to empty luciferase vector measurements.
Fluorescence microscopy
APMAP-YFP
3T3-L1 cells were seeded and grown on coverslips (Corning #1.5) as described above. A construct coding for APMAP fused to the n-terminus of YFP was cloned between the HindIII and the XbaI restriction sites of the pcDNA3 vector. At 80% confluence, they were transfected with 1 μg of the APMAP-YFP construct using 4.5 μl Metafectene (Biontex Laboratories). For negative controls, an empty vector was transfected. The transfection was performed according to the manufacturer’s manual. Then, 48 h after transfection, the coverslips were removed from the medium and prepared for imaging. For live cell experiments, the cells were mounted on microscopy slides with a drop of growth medium enclosed by a parafilm frame and imaged immediately. For high resolution 3D imaging, the cells were fixed with freshly prepared 3.5% paraformaldehyde in PBS for 15 min and washed 3 times with PBS, nulcei were counterstained for 5 min with 0.05 μg/ml DAPI in PBS, and the cover slips were mounted on microscopy slides with the SlowFade Gold mounting medium (Invitrogen, CA).
Immunofluorescence of APMAP
3T3-L1 and OP9 cells were grown and differentiated on coverslips as described above. Preconfluent (~80% confluence) or differentiated cells (day 10) were fixed for 15 min with freshly prepared 2% paraformaldehyde/0.2% glutaraldehyde/PBS. After fixation, the cells were incubated for 5 min in PBS containing a few drops of 1 M glycine pH 8.5, washed 3 times in PBS, permeabilized for 5 min 0.2% Triton-X 100 in PBS and again washed 3 times with PBS. For immuno detection of APMAP, we used polyclonal antibodies against synthetic peptides consisting of residues 366–381 (Eurogentec) raised in rabbit. The endoplasmatic reticulum was detected by a monoclonal mouse anti-Calreticulin antibody (BD Biosciences). Both antibodies were diluted 1:200 in 4% BSA/0.1% Tween-20/PBS, applied to the cells, and incubated over night at 4°C. After incubation, the cells were washed three times with PBS and incubated for 1 h at room temperature with an anti-mouse antibody and an anti-rabbit antibody coupled to AlexaFluor-488 and AlexaFluor-594, respectively (Invitrogen). The secondary antibodies were diltuted in the same way as described above. After washing three times with PBS, the nulcei were counterstained for 5 min with 0.05 μg/ml DAPI in PBS and the coverslips were mounted on microscopy slides with the SlowFade Gold mounting medium (Invitrogen).
All images were collected on a Zeiss AxioImager Z1 epifluorescence microscope equipped with a Zeiss AxioCam MRm CCD-camera using a ×63 1.4 NA objective lens. To obtain high resolution images, 3D stacks with an axial spacing of 240 nm were recorded and processed by the maximum likelihood estimation 3D-deconvolution algorithm available with the Huygens Software (Scientific Volume Imaging, SVI).
Statistical analysis
If not otherwise stated, results are mean values (±standard deviation) of at least three independent experiments. Statistical significance was determined using the two-tailed Student’s t test.
Results
Reconstruction of a gene association network from microarray data predicts a novel adipogenesis key player
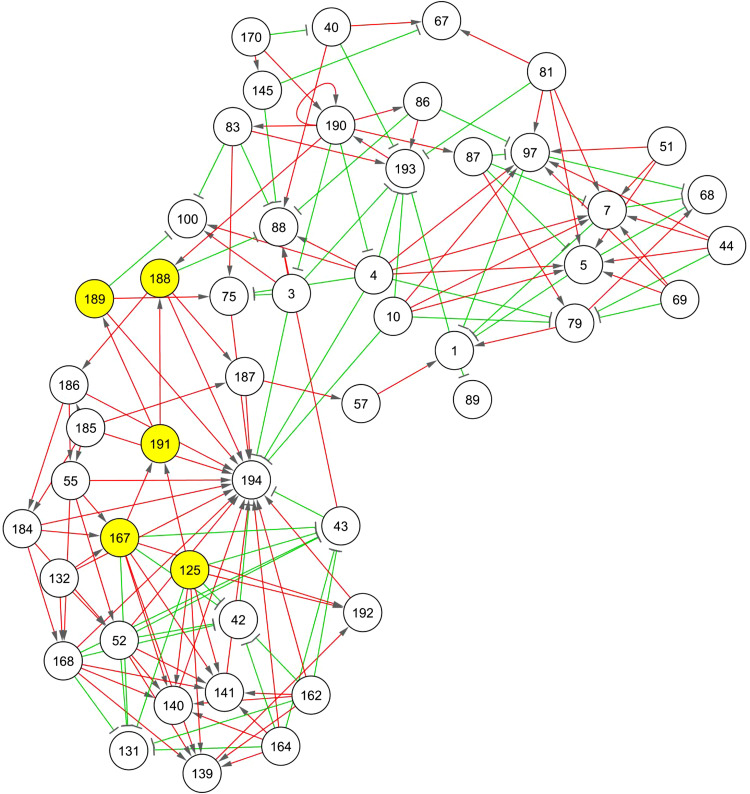
In order to identify previously unknown regulators of adipogenesis, a gene–gene association network was reconstructed from 14,368 gene expression profiles obtained from microarray data of differentiating 3T3-L1 cells [11]. After discretization of their expression profiles [12], 194 gene clusters with different discrete expression profiles were used for network reconstruction, summarizing the dynamic profile of a total of 6,858 expressed sequence tags (ESTs). The complete list of genes within each cluster and the reconstructed network can be found in ESM, file 2. We then focused on the network hubs, i.e. clusters with at least five neighbor nodes (Fig. 1; ESM, file 2). Gene ontology (GO) [22] analysis of these genes revealed over-representation of genes related to metabolic processes (33.99%, adjusted p < 0.001) and to protein binding activity (55.07%, adjusted p < 0.001). In particular, some of these 49 network hubs comprised known adipogenic transcription factors and adipocyte markers: PPARγ in cluster 191, Pnpla2 (adipose triglyceride lipase, ATGL), pyruvate carboxylase (Pcx) in cluster 188 and C/EBPα in cluster 168.
Fig. 1.
Association network built from gene expression data. Only nodes with 5 or more neighbors are displayed. Each node represents a cluster of gene expression profiles with identical discrete profile. The algorithm for network reconstruction distinguishes between repression (green) and activation (red) interactions and also shows the direction of the regulation. Cluster 191 contains PPARγ, a known key player for adipocyte differentiation. The nodes in yellow are direct neighbors of the PPARγ cluster. A clone mapping APMAP is in cluster 167. The network was visualized using Cytoscape [14]
We further focused on the clusters containing known adipogenic transcription factors. Particularly, we investigated the neighbors of the PPARγ cluster (cluster 191, see Fig. 1). Cluster 188 contains a probe for the gene ATGL, already proved to be regulated by PPARγ [23]. Additional promoter analyses of the remaining genes in the neighbor clusters identified an uncharacterized probe in cluster 167 [RIKEN cDNA 2310001A20 gene (2310001A20Rik)] with putative PPARγ and C/EBPα binding sites (ESM, file 3). Thus, based on the results of the network reconstruction and promoter analyses, a novel high confidence candidate gene was prioritized for further functional studies. Sequence analyses of the RIKEN clone showed that this probe represents a protein annotated as APMAP. It was previously shown to be expressed during adipogenesis in 3T3-L1 cells [24, 20, 11]. However, the physiological relevance and precise role during adipogenesis remained elusive.
APMAP expression is upregulated during murine and human adipogenesis
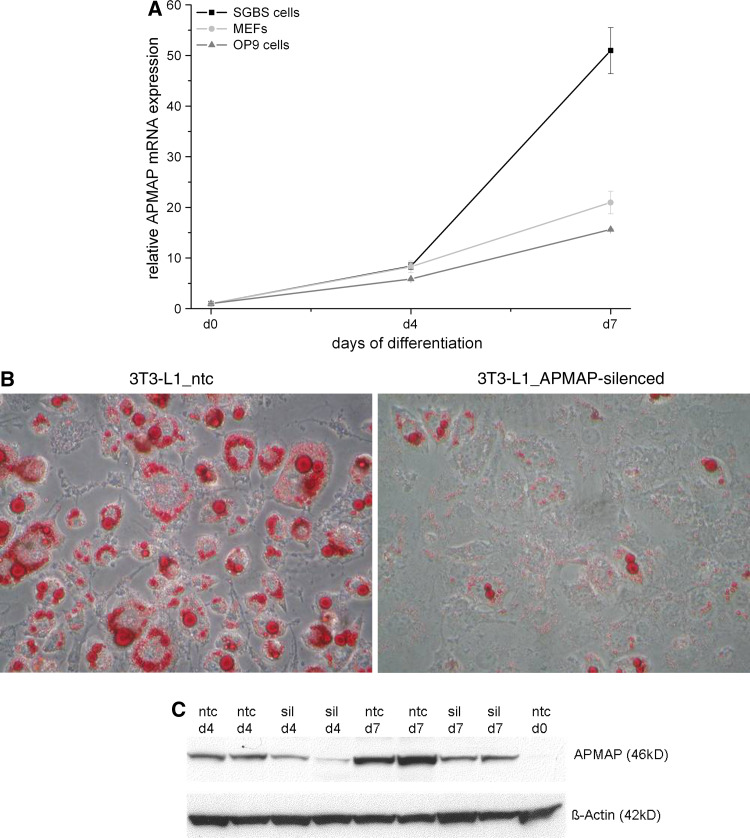
We then assessed the gene expression of APMAP in cell lines other than 3T3-L1 that undergo adipogenesis and in murine white adipose tissue (WAT). APMAP was strongly upregulated during adipogenic differentiation of OP9 cells and mouse embryonic fibroblasts (MEFs) (Fig. 2a). A similar expression profile of the human ortholog gene of APMAP (C20orf3) was observed in the SGBS cells (Simpson-Golabi-Behmel syndrome) (Fig. 2a). In accordance with the APMAP profiles seen in the cell culture models, we found APMAP expression to be twofold increased in WAT of genetically obese mice (ob/ob) at the age of 3–4 months compared to their wild-type littermates (ESM, file 1). However, mice on a high fat diet (40% calories in fat) did not show any change of APMAP expression in WAT, neither on a short-term (4 weeks) nor on a long-term (6 months) challenge on this diet. Overall, these data indicate that the level of APMAP expression is upregulated during adipogenesis in mice and men suggesting a potential role in fat cell biology.
Fig. 2.
Expression and requirement of APMAP for adipogenesis. a APMAP expression is induced during adipocyte differentiation of OP9 cells, MEFs, and human SGBS cells. mRNA expression was analyzed using quantitative real-time PCR. The relative APMAP mRNA expression is shown in comparison to day 0. b APMAP knockdown impairs expression of genes normally upregulated during adipogensis. 3T3-L1 cells were infected with the MISSION™ TRC lentiviral particles coding for APMAP shRNA (3T3-L1_APMAP-silenced) or a non-targeting shRNA as control (3T3-L1_ntc), selected for puromycin resistance, expanded as a mixed population, and induced to differentiate using DMI cocktail. Silencing efficiency of one out of four knockdown lentiviruses against APMAP was determined by qRT-PCR assay and compared to 3T3-L1_ntc cells. For one lentiviral construct APMAP gene expression was 90% reduced in APMAP-silenced cells compared to ntc cells in three independently silenced biological replicates. Expression levels (in %) of PPARγ, C/EBPα, ATGL and aP2 in APMAP-silenced 3T3-L1 cell lines compared to control cells (ntc). Inset shows oil red O staining of control and APMAP-silenced 3T3-L1 cells at day 10 of differentiation. c Protein was harvested at indicated timepoints during 3T3-L1 differentiation of control (ntc) and APMAP-silenced cells (sil) and subjected to western blotting using the anti-APMAP antibody. β-actin served as loading control. APMAP protein expression is strongly decreased in APMAP-silenced 3T3-L1 cells at day 4 and 7 in comparison to control cells
APMAP is a required enhancing factor of adipogenesis
To gain insight into APMAP’s function in adipocytes, we first evaluated whether silencing of APMAP in 3T3-L1 cells has an influence on the differentiation process. For that purpose, we used APMAP-targeting shRNA encoded by lentiviral vectors to generate stably transduced cell lines with constitutive silencing of APMAP expression. The cells were grown to confluence and induced to differentiate using the standard hormonal cocktail (MDI). One of four APMAP-targeting shRNA lentiviral construct showed up to 90% APMAP-silencing capacity in comparison to a non-targeting control (ntc) (see Fig. 2b for mRNA expression). In addition, we controlled whether protein expression is also reduced in these APMAP-silenced cells. As depicted in Fig. 2c, APMAP protein expression is increasing from day 0, over day 4 to day 7 in control cells and the expression is strongly reduced in APMAP-silenced cells at day 4 and day 7 when compared to control cells, respectively. Next, we investigated the differentiation capacity of APMAP-silenced cells in comparison to control cells. As shown as oil red O staining (inset of Fig. 2b) and in Table 1, 10 days after induction of differentiation the APMAP-silenced cells accumulated significantly less lipid than the control cells (3T3-L1_ntc). The triglyceride (TG) content of the APMAP-silenced 3T3-L1 cells was up to 80% reduced. Concomitantly, RNA levels of genes known to be upregulated during adipogenesis, such as PPARγ, C/EBPα, ATGL and aP2 were significantly decreased in APMAP-silenced cells (Fig. 2b). However, stable overexpression of APMAP in 3T3-L1 cells did not promote adipogenesis (data not shown). These data implicate that expression of APMAP is required for fat cell differentiation.
Table 1.
Triglyceride content (given in %)
| 3T3-L1_ntc | 3T3-L1_APMAP-silenced | Addition of |
|---|---|---|
| 100 ± 11 | 20 ± 9***,a | MDI |
| 177 ± 15**,b | 62 ± 15*,c | MDI + rosiglitazone (10 μM) |
The percentage of TG content of control (ntc) and APMAP-silenced 3T3-L1 cells at day 10 after induction of differentiation upon addition of the standard hormonal cocktail (MDI) with or w/o rosiglitazone. Different letters indicate two-tailed Student’s t tests in comparison to 3T3-L1_ntc
*p < 0.05, **p < 0.01, ***p < 0.001
APMAP is a direct and functional PPARγ target gene
APMAP is induced by the PPARγ ligand rosiglitazone in adipocytes and in vivo in adipose tissue
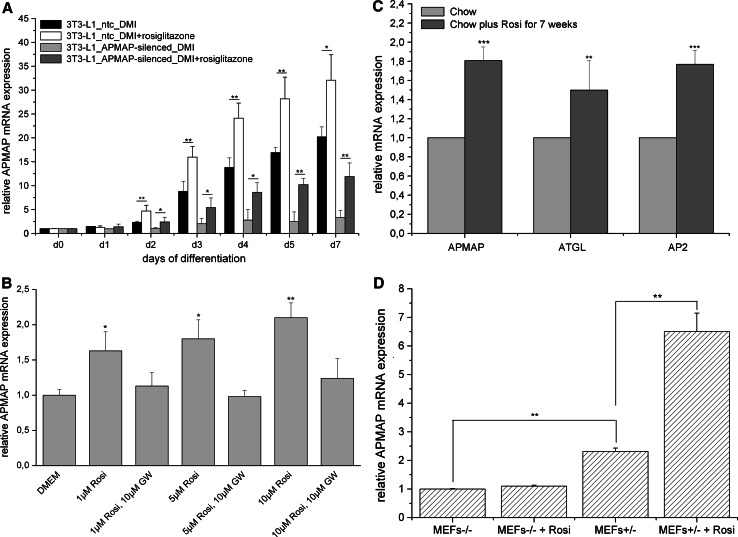
Since the reconstructed network (shown in Fig. 1) and the promoter analysis (ESM, file 3) revealed a putative interaction between PPARγ and APMAP, we next evaluated whether PPARγ regulates APMAP expression. 3T3-L1 cells were differentiated with the standard cocktail (MDI) in the presence of the PPARγ agonist rosiglitazone. APMAP mRNA expression was at least 1.8-fold increased from day 2 on when rosiglitazone was added during differentiation (Fig. 3a, black and white bars). Further, we evaluated whether short-term treatment (24 h) with rosiglitazone also has an influence on the expression of APMAP in fully differentiated 3T3-L1 adipocytes. The results show a dose-dependent (1, 5 and 10 μM) rosiglitazone-mediated induction of APMAP mRNA expression in fully differentiated 3T3-L1 adipocytes that was totally abolished in the presence of the PPARγ antagonist GW9662 (10 μM) (Fig. 3b). Additionally, we examined the effect of rosiglitazone treatment on APMAP mRNA expression in mouse adipose tissue in vivo. For this purpose, male C57/Bl6 received a chow diet supplemented with rosiglitazone (0.01% w/w) for 7 weeks immediately after weaning. Rosiglitazone-treated mice did not gain more weight than their wild-type littermates, only brown adipose tissue weight was significantly elevated compared to control mice (up to threefold, data not shown). Epididymal fat pads were collected after 4 h of daytime fasting, RNA was isolated and subjected to quantitative RT-PCR analysis. As depicted in Fig. 3c, APMAP mRNA expression was significantly induced in rosiglitazone-treated mice in comparison to control mice. As positive control, genes ATGL (adipose triglyceride lipase) and aP2 were used. These results show that APMAP expression is subject to PPARγ-dependent regulation, thus confirming the interaction between PPARγ and APMAP suggested by the reconstructed network in Fig. 1.
Fig. 3.
APMAP expression is induced by the PPARγ ligand rosiglitazone in vitro and in vivo. a APMAP expression during adipocyte differentiation of 3T3-L1 control cells and 3T3-L1 APMAP-silenced cells is increased by adding rosiglitazone (10 μM) to the standard induction cocktail (n = 3). Rosiglitazone was added during the whole differentiation process. b Rosiglitazone-mediated increase of APMAP mRNA expression in fully differentiated 3T3-L1 cells (d10) is abolished by addition of the PPARγ antagonist GW9612 (n = 3). Rosiglitazone and the agonist were both added for 24 h to fully differentiated 3T3-L1 cells and compared to cells in standard growth medium. c APMAP mRNA expression in epididymal fat pads of control or rosiglitazone-treated male C57/Bl6 mice (n = 6). Tissues were collected after 4 h of fasting. As positive controls ATGL (adipose triglyceride lipase) and AP2 are shown. d APMAP mRNA expression in PPARγ−/− and PPARγ+/− MEFs. APMAP is not expressed in PPARγ−/− MEFs (n = 3). APMAP mRNA expression in PPARγ+/− MEFs is increased by addition of the PPARγ agonist rosiglitazone (10 μM). Rosiglitazone was added during the whole differentiation process. Statistical significance was determined using the two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001
PPARγ agonists partly overcome the differentiation defect in APMAP-silenced 3T3-L1 cells
Since the PPARγ agonist rosiglitazone increases APMAP expression in differentiating and fully differentiated adipocytes (Fig. 3a, black and white bars, and b), we evaluated whether addition of rosiglitazone to APMAP-silenced 3T3-L1 cells can overcome the differentiation defect of these cells. Control and APMAP-silenced 3T3-L1 cells were treated with the hormonal standard cocktail (MDI) with and w/o rosiglitazone for 10 days. In the presence of rosiglitazone (10 μM), control and APMAP-silenced cells showed enhanced triglyceride accumulation compared to the according cells w/o rosiglitazone (Table 1). However, the differentiation defect that occurs in the absence of APMAP (20% of the triglyceride content of control cells) could only be partly overcome when the PPARγ agonist was added to APMAP-silenced cells (60% of the TG content of control cells) (Table 1). Rosiglitazone-treated APMAP-silenced cells not only showed enhanced lipid accumulation compared to APMAP-silenced cells differentiated w/o rosiglitazone, but also significantly induced APMAP expression (Fig. 3a, light and dark grey bars). Thus, addition of the PPARγ agonist can partly rescue the differentiation defect of APMAP-silenced cells likely due to an induction of APMAP expression.
PPARγ is required for APMAP expression in MEFs
To further investigate whether PPARγ is required for the expression of APMAP, we used MEFs from PPARγ−/− and PPARγ+/− mice and subjected them to hormone-induced adipocyte differentiation. While PPARγ+/− MEFs accumulated lipid droplets 10 days after induction, PPARγ−/− MEFs did not. RNA from these samples collected for qPCR analysis showed that only PPARγ+/− MEFs express APMAP while PPARγ-ko MEFs do not (Fig. 3d). Addition of rosiglitazone during the whole differentiation process increased APMAP expression in PPARγ+/− MEFs, whereas no increase could be observed in PPARγ−/− MEFs (Fig. 3d). These results show that PPARγ is required for APMAP expression and that, as expected, rosiglitazone can only increase APMAP expression in the presence of PPARγ. To evaluate whether forced expression of APMAP in PPARγ−/− MEFs induces adipocyte differentiation, PPARγ−/− MEFs stably overexpressing APMAP were generated. However, inducing these cells with the appropriate hormonal cocktail did not lead to lipid droplet formation (data not shown). Thus, APMAP cannot induce adipogenesis in the absence of PPARγ.
APMAP is a functional PPARγ target
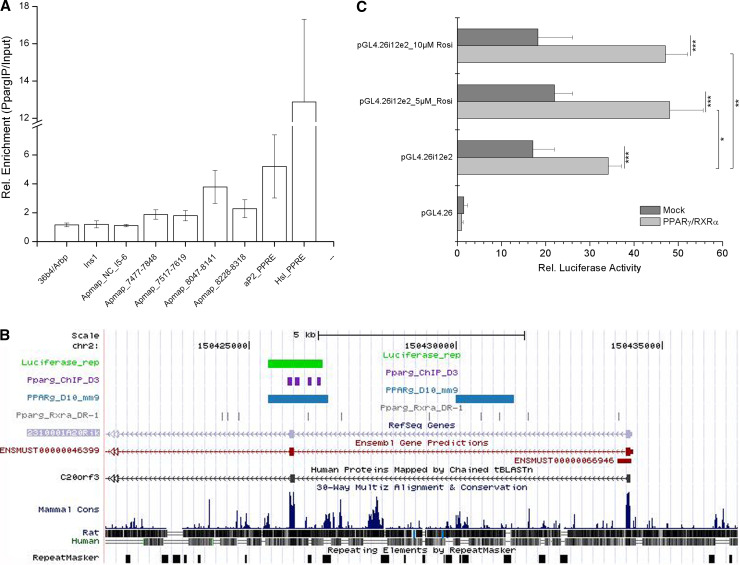
Given that APMAP expression responded to PPARγ activation, we investigated whether PPARγ is physically binding to a cis-regulatory sequence of the APMAP gene. We analyzed the genomic region around the APMAP transcription start site (TSS) using chromatin immunoprecipitation (ChIP) followed by qPCR. Immunoprecipitation using a PPARγ-specific antibody was performed on day 10 using samples from differentiating 3T3-L1 adipocytes. While primer pairs covering the proximal promoter region (up to ~5 kb upstream of the TSS) showed only modest enrichment for PPARγ binding (data not shown), a region in intron1–2 and exon 2 (~8 kb downstream) showed a ~fourfold enrichment (Fig. 4a). These results suggest a genomic region in the vicinity of the APMAP TSS to have functional binding site(s) and this intron1–2/exon2 region also shows high similarity to a PPARγ-RXRα DR-1 [9] consensus motif (similarity score ≥0.90). The regions investigated by ChIP-qPCR are shown in Fig. 4b. To prove the functionality of this PPARγ binding region, we performed luciferase assays in Cos7 cells. For that purpose, the intron1–2/exon2 region was cloned into a luciferase reporter vector (named pGL4.26i12e2) and co-transfected with either the control plasmid pCMX or PPARγ/RXRα in the absence or presence of rosiglitazone. As depicted in Fig 4c, upon cotransfection with PPARγ/RXRα, the luciferase activity of pGL4.26i12e2 was significantly increased when compared to pCMX(mock)-transfected cells (twofold, p = 0.0009). Addition of rosiglitazone (5 or 10 μM) to cells cotransfected with pGL4.26i12e2 and PPARγ/RXRα again significantly increased luciferase activity, if compared to mock-transfected cells (2.2-fold with p = 0.0004 for 5 μM rosiglitazone and 2.6-fold with p = 0.0002 for 10 μM rosiglitazone, respectively) or to cotransfected cells without rosiglitazone (1.4-fold, for both 5 and 10 μM rosiglitazone, respectively) (Fig. 4c). Collectively, these data suggest that the intron1–2/exon2 region of APMAP is a functional enhancer which can be induced by the adipogenic master regulator PPARγ and is responsive to its agonist rosiglitazone.
Fig. 4.
APMAP is a direct and functional PPARγ target gene. a Relative fold-enrichments for PPARγ binding in 3T3-L1 cells 10 days after differentiation start. Fold-enrichments are normalized to 18S rRNA site and related to genomic input DNA (-ddCt). Primer pair coordinates are relative to the APMAP transcription start site (in intron1-2). A region in intron5–6, in the 36b4 promoter, and in the insulin promoter serve as negative controls, while known Pparg-response elements (PPRE) in the HSL and aP2 promoters represent bona fide PPARγ binding sites. Values are mean values (±SD) from three independent immunoprecipitations. b The gene organization around the first intron of Apmap using the UCSC genome browser (NCBI37/mm9). Sites with high similarity to the DR-1 motif (similarity score ≥0.90) (gray), PPARγ binding regions as identified by a previous study [9] (blue), amplicons from ChIP-qPCR (violet), and genomic sequence used for luciferase reporter assay (green) are mapped. c The intron1–2/exon2 region of the APMAP gene was cloned into the luciferase reporter vector pGL4.26 containing a minimal promoter (the whole construct was named pGL4.26i12e) and cotransfected with either PPARγ/RXRα expressing vectors or an empty vector (pCMX) into Cos7 cells. After 48 h, cells were lysed and assayed for luciferase activity using a luminometer. Data was normalized to Renilla luciferase readings to account for differences in transfection efficiencies. Finally, data were related to empty reporter vector measurements. Statistical significance was determined using the two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001
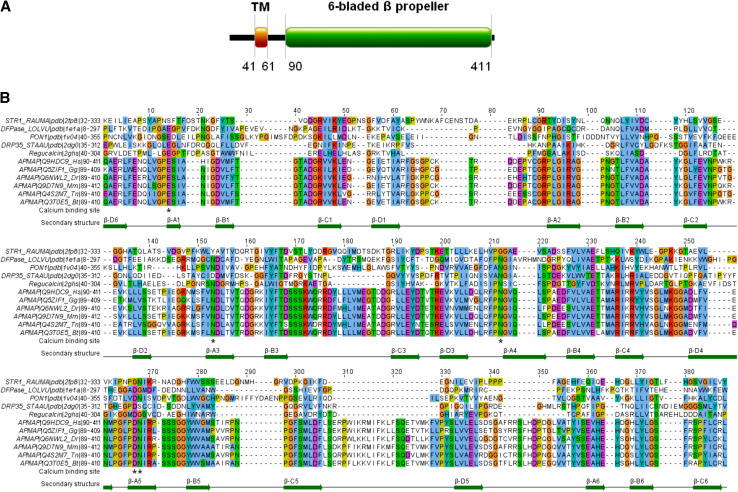
Sequence-analysis-based molecular function prediction of APMAP suggests a six-bladed β-propeller with calcium-dependent hydrolase activity
The analysis of the sequence architecture following a published sequence-analytic recipe [25] with the ANNOTATOR software [26] revealed 4 segments (Fig. 5a). There are two apparently non-globular segments (the region 1–40 with 35% charged residues and the proline-rich linker 62–89). The stretch 41–61 between them is a transmembrane helical region. The segment from residue 90 to the C-terminus appears to be a globular domain. It is hit by two PFAM models of enzymatic domains with conflicting properties: the calcium-binding SGL (superficial gray layer) full domain and the strictosidine synthetase domain fragment, a domain without metal ions.
Fig. 5.
APMAP is a calcium-binding protein with a six-bladed β-propeller domain. a Sequence architecture of human APMAP. See text for details. b Alignment of the C-terminal segments from APMAP orthologues in animal genomes with the sequences of the known structures STR1, PON1, DFPase, Drp35 and regucalcin. Starting with a manually generated sequence alignment of the five known structures, the template was extended for the orthologous APMAP sequences with the alignment program MAFFT [41]. Towards the C-terminus portion, APMAP contains a six-bladed β-propeller domain (positions 90–411). In particular, APMAP has four conserved sites at E103, N201, N260 and D306/N307 that are predicted to be necessary for calcium binding in the active site
To clarify the evolutionary relationships of the APMAP C-terminal domain, a search with fan-like heuristics [25] of successive PSI-BLAST [27] runs was performed to collect the family of APMAPs and its related homologues. Besides orthologous APMAPs for a number of eukaryotes and other proteins, it was interesting to also find the structures of strictosidine synthase (STR1) from Rauvolfia serpentine [28] serum paraoxonase (PON1) from Oryctolagus cuniculus [29], diisopropylfluorophosphatase (DFpase) from Loligo vulgaris [30], Drp35 from Staphylococcus aureus [31], and regucalcin from Agrobacterium tumefaciens with significant E-values. Notably, PON1, DFpase, Drp35 and regucalcin are classified as calcium-dependent phosphotriesterases under the six-bladed β-propeller protein family in the SCOP database [32], whereas the group of strictosidine synthetase-like enzymes does not bind metal ions.
A sequence-to-structure alignment of the orthologous APMAPs of representative eukaryote animals to the six-bladed β-propellers (STR1, PON1, DFPase, Drp35 and regucalcin) is shown in Fig. 5b. We conclude that the two groups have essentially the same hydrophobic pattern. The predicted secondary structure of APMAP (with JPRED; [33]) coincides with the observed secondary structural elements in the structure group of representative eukaryote animals (Fig 5b). Thus, the APMAP C-terminal domain is predicted to be a six-bladed β-propeller. At the same time, APMAP is not a strictosidine synthetase-type enzyme and does not belong to this family. The conservation pattern among the APMAP orthologues for functional residues clearly resembles that of the calcium binding structures and not that of the Rauvolfia strictosidine synthetase known not to bind metal ions. In APMAP, all four sites necessary for coordinating the metal ions are present. In summary, APMAP is predicted to represent a hydrolase with a calcium binding active site.
The C-terminal domain of APMAP is required for the function of APMAP during adipogenesis
As we and others have showed, APMAP is a transmembrane protein (41–60 AA) [24] with the C-terminal domain predicted to be a six-bladed β-propeller [34]. To assess whether the six-bladed β-propeller is necessary for the function of APMAP in adipogenesis, APMAP-silenced cells were stably transduced with two truncated APMAP constructs. The first construct was terminated immediately after the transmembrane region at amino acid 60 (named AA60) and the second construct after 63 amino acids (named AA63), thus both proteins are translated without the six-bladed propeller region. Neither AA 60 nor AA 63 could rescue the loss of differentiation capacity of APMAP-silenced cells whereas expression of the full-length APMAP protein (AA415) in silenced cells rescued the differentiation failure as shown in Table 2 by means of TG content measurements of the cell lysates. These results show that the C-terminal six-bladed β-propeller is required for the role of APMAP in adipogenesis.
Table 2.
Triglyceride content (given in %)
| 3T3-L1_ntc | 3T3-L1_APMAP-silenced | 3T3-L1_APMAP-silenced_AA60 | 3T3-L1_APMAP-silenced_AA63 | 3T3-L1_APMAP-silenced_AA415 | Addition of |
|---|---|---|---|---|---|
| 100 ± 11 | 20 ± 9***,a | 26 ± 12***,b | 19 ± 15***,c | 80 ± 17**,d | MDI |
The percentage of TG content of control (ntc), APMAP-silenced and APMAP-silenced 3T3-L1 cells that are additionally stably expressing either the truncated versions of the APMAP protein (AA60 and AA63) or the whole APMAP protein (AA415) at day 10 after induction of differentiation upon addition of the standard hormonal cocktail (MDI). Letters a, b, c indicate two-tailed Student’s t tests in comparison to 3T3-L1_ntc, letter d indicates two-tailed student’s t tests in comparison to 3T3-L1_APMAP-silenced 3T3-L1 cells
*p < 0.05, **p < 0.01, ***p < 0.001
APMAP translocates from the ER to the plasma membrane during adipogenesis
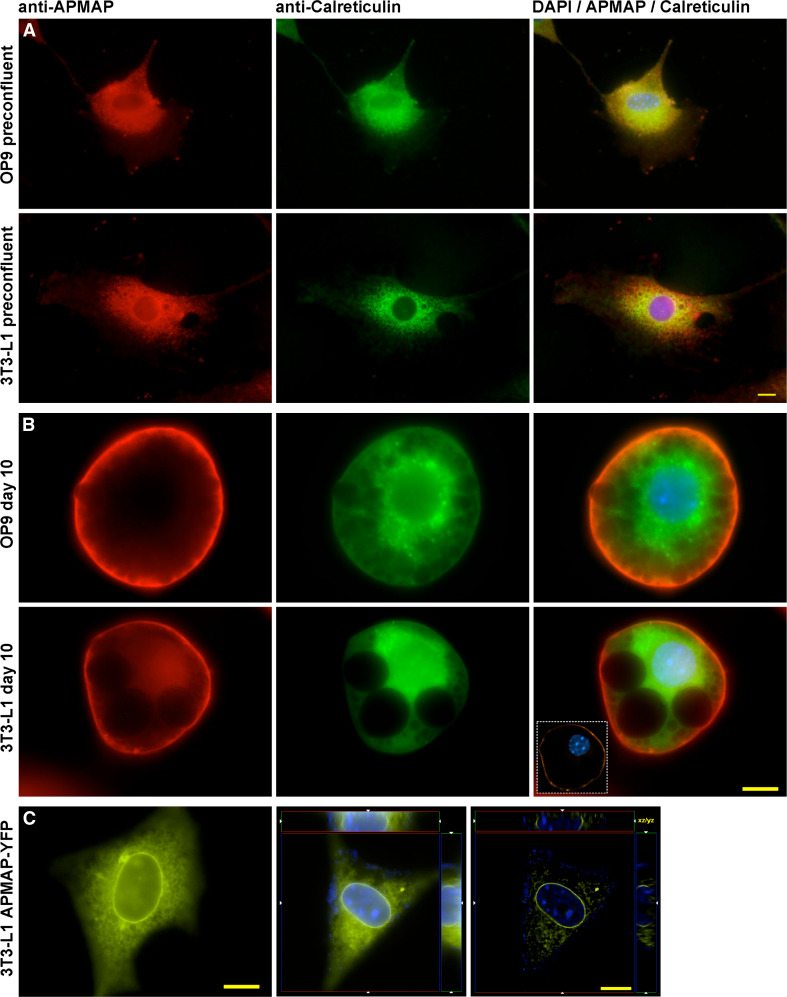
To determine the subcellular localization of APMAP in preadipocytes and mature adipocytes, we performed immuofluorescence experiments in preconfluent and fully differentiated 3T3-L1 and OP9 cells using an antibody against a synthetic peptide consisting of APMAP residues 366–381 (Eurogentec). Our results show that in preconfluent and undifferentiated 3T3-L1 and OP9 cells, APMAP was mainly located at the ER as demonstrated by double immunostaining of APMAP and the ER marker Calreticulin (Fig. 6a). The experiment was repeated in differentiated 3T3-L1 and OP9 cells 10 days after hormonal induction. Mature adipocytes were imaged and APMAP was found to be translocated to the plasma membrane (Fig. 6b).
Fig. 6.
Subcellular localization of APMAP. a Double immunolabeling of APMAP (red) and calreticulin (green), an ER marker, reveals that APMAP localizes at the endoplasmatic reticulum in uninduced (preconfluent) 3T3-L1 and OP9 cells. The merged images show a significant overlap of the green and red signal. b Immunoflurorescence of APMAP and calreticulin (ER) in differentiated (day 10) 3T3-L1 and OP9 adipocytes. APMAP (red) is located in the plasma membrane and does not localize at the ER (green) in mature adipocytes. The inset in the merged image shows a high resolution deconvolved optical section of the same cell. Images were obtained by using an antibody against a synthetic peptide consisting of APMAP residues 366–381. c APMAP-YFP overexpression in uninduced 3T3-L1 cells. The panel presents a live cell image (left), a raw (middle), and the deconvolved 3D image stack (right) of APMAP-YFP overexpressing 3T3-L1 preadipocytes. When overexpressed, APMAP is located in the nuclear membrane and ER. Scale bars 10 μm
Next, we asked if overexpression of APMAP in preconfluent 3T3-L1 cells leads to a translocation of APMAP from the ER to the plasma membrane. Therefore, we used a construct in which APMAP was fused to the N-terminus of YFP and cloned it into the pcDNA3 expression vector. This construct was transiently transfected into preconfluent 3T3-L1 cells growing on coverslips and, 48 h later, we collected images from living cells and 3D image stacks of fixed cells. Raw and deconvolved images of the fluorescent signal showed a pattern resembling the ER rather than the plasma membrane. Moreover, we observed a strong signal delineating the nuclear membrane (Fig. 6c). These results demonstrate that APMAP localizes at the ER in preadipocytes and translocates to the plasma membrane in mature adipocytes, whereas forced high expression alone is not driving APMAP to be located in the plasma membrane.
Discussion
In this study, we present a combined computational-experimental strategy using gene expression data as the starting point to identify new candidate genes for adipogenesis. Priorization of the candidate genes with annotational features pinpointed a transcript annotated as APMAP and potentially targeted by PPARγ. Our functional studies showed that APMAP is upregulated in various cell lines that undergo adipogenesis and in WAT of genetically obese mice. Moreover, we show that APMAP is required for adipogenesis, upregulated upon addition of rosiglitazone in adipocytes in vivo and in vitro and is a functional target of PPARγ, thus validating the hypothesized interactions. We provide sequence-analytic evidence that APMAP is a transmembrane protein with a six-bladed β-propeller and potential hydrolase activity. Further, we experimentally demonstrate that APMAP translocates from the ER to the plasma membrane during adipocyte differentiation and reveal the necessity of the C-terminus harboring the six-bladed β-propeller for its role in adipogenesis. This study has both biological and methodological implications.
Our strategy has provided new biological insights that could only be derived from such a combined computational–experimental approach. We identified another component of the network regulating adipogenesis and show its role on adipocyte biology in both murine and human models: APMAP is expressed in murine and human cell lines, primary cells, and a genetic model of obesity. Recently, we have also reported the expression of the human APMAP ortholog C20orf3 in hMADS (human multipotent adipose-derived stem cells) [35]. In addition, a public dataset of gene expression analysis across human tissues (Gene Expression Atlas) [36] shows elevated expression levels of C20orf3 in adipocytes. These results highlight the biological relevance of the protein in mice and humans.
Furthermore, our data show that APMAP is necessary for adipocyte differentiation. Silencing of APMAP in 3T3-L1 cells leads to strongly reduced differentiation capacity and downregulation of adipocyte markers, such as PPARγ, C/EBPα, and aP2. We also provide several lines of evidence that PPARγ is regulating APMAP and that APMAP is a direct PPARγ target gene: (1) APMAP expression is induced in vitro and in vivo by the PPARγ agonist rosiglitazone and this induction is abolished by an antagonist in vitro; (2) the differentiation defect in APMAP-silenced 3T3-L1 cells can at least partly be overcome by rosiglitazone which upregulates APMAP gene expression; (3) PPARγ-deficient MEFs do not express APMAP; and (4) ChIP-qPCR and luciferase activity assay data demonstrate direct and functional binding of PPARγ to a region in intron1–2/exon2 (8 kb downstream of the APMAP TSS). Recent genome-wide PPARγ location studies in 3T3-L1 cells showed that more than 30% of the identified binding sites are to be found in intronic regions [9, 10], and a subsequent study verified the functionality of distant PPARγ binding (8 kb) in the resistin promoter [37]. Hence, it is likely that the binding of PPARγ to the region in intron1–2/exon2 of APMAP is functionally driving APMAP expression. An intriguing question is whether APMAP plays a role in the insulin-sensitizing and weight-promoting effects of rosiglitazone. Further studies with knock-out animals might provide the answer.
Stable expression of APMAP did not accelerate lipid droplet formation in 3T3-L1 cells when induced with the standard hormonal cocktail and did not promote differentiation of PPARγ-ko MEFs. Overexpression of APMAP in NIH-3T3 cells was also not sufficient for the induction of the differentiation program in these cells. Interestingly, upregulation of APMAP during differentiation correlates with its translocation to the plasma membrane, whereas forced expression of APMAP leads to an accumulation of the protein in the ER without the translocation to the plasma membrane. Thus, the function/role of APMAP in adipogenesis might depend on the protein translocation to the plasma membrane and this could explain why overexpression of APMAP alone does not induce/accelerate adipogenesis.
One interesting finding of this study is that APMAP—a required factor for adipogenesis—is a predicted hydrolase. The majority of the molecules regulating adipogenesis are transcription factors. Only recently, several other classes of genes encoding factors not previously recognized in developmental regulation have been shown to be implicated in adipocyte differentiation. This includes classic enzymes such as retinol saturase, the enzymatic activity of which has been shown to be required for adipogenesis (24), and xanthine oxidoreductase which is necessary for full adipocyte development and PPARγ2 expression in mice (25). Further examples include steroyl-CoA desaturase 2 (SCD2), as a regulator of adipogenesis (26). Also, a phospholipase, namely PLA2 (27), has been found to be required for hormone-induced differentiation of 3T3-L1 preadipocytes. In this study, we added APMAP, another putative enzyme, to the list of components regulating adipogenesis. So far, the substrate for the predicted hydrolase activity of APMAP is still unidentified. For the human APMAP ortholog gene (C20orf3), arylesterase activity with artificial subtrates has been shown in pancreatic and liver cell lines [34]. However, using hydrolase assays with different substrates including p-nitrophenylacetate, p-nitrophenylpalmitate, triarachidin, triolein and phosphatidylcholine, we were not able to show any increased enzymatic activities in APMAP-overexpressing COS7 and 3T3-L1 cells compared to controls under various testing conditions like pH and calcium concentration. Thus, we could neither confirm the arylesterase activity described for the human protein [34] nor identify a different substrate for the mouse homolog. Additional studies will be necessary to identify the natural substrate of APMAP and the role of the enzyme activity of APMAP in adipocyte differentiation. One intriguing speculation is that the hydrolytic activity of APMAP might play a role in the production of a small molecule required for adipogenesis, e.g., an as yet unidentified ligand for PPARγ, as has been shown for xanthine oxidoreductase (25). Regardless of the search for the ligand, it is obvious that the predicted C-terminal six-bladed β-propeller of APMAP (including a globular domain with metal binding sites) is required for the function of APMAP in the context of fat cell differentiation, since overexpression of the C-terminally truncated APMAP cannot rescue the differentiation defect of APMAP-silenced 3T3-L1 cells.
Over and above the biological implications shown here, our study also demonstrated methodological advances. Using a method that combines mutual information and time-lagged correlation, we have been able to confirm many known relationships during the adipocyte differentiation process and to identify a new key regulator. The importance of APMAP during adipocyte differentiation was first suggested in this study by its role as a highly connected hub also linked to the PPARγ cluster in the gene–gene network reconstructed from a microarray experiment monitoring adipogenesis. A variety of statistical and computational methods have already been developed for reverse engineering microarray data including mutual information algorithms [13, 38–40]. Our method has three advantages over these published tools: (1) it can distinguish between regulator and regulated nodes; (2) the algorithm is well-suited for time series experiments; and (3) the algorithm can detect relationships where more than one regulator are acting simultaneously.
Despite the methodological advantages of the mutual information and correlation algorithm, reconstructed networks from gene expression data from experiments like cell development comprise hundreds of clusters of genes. Cell development is a dramatic process in which the cell undergoes biochemical and morphological changes and therefore thousands of transcripts have to be regulated. In our study, signals at every time point could be detected from more than 14,000 ESTs [11]. Thus, additional analysis is necessary to prioritize the candidate genes for further functional studies. Using promoter analyses, we were able to identify and experimentally verify high-confidence gene.
In summary, this work suggests an important role for APMAP in adipocyte biology and demonstrates the utility of the integrated computational and experimental approach in identifying genes not previously suspected to regulate adipogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the Austrian Ministry for Science and Research (GEN-AU projects GOLD and BIN) and the Austrian Science Fund SFB (Project Lipotoxicity). PPARγ-MEFs were a gift from Dr. E. Rosen. OP9 cells were kindly provided by B. Pickel and SGBS cells by Novo Department of Pediatrics and Adolescent Medicine, University of Ulm. We thank David J. Steger and Mitch A. Lazar for providing ChIP material. The authors acknowledge the technical assistance provided by Stephan Seifriedsberger, Florian Stoeger, Martina Schweiger and Marie Loh.
Footnotes
J. G. Bogner-Strauss and A. Prokesch contributed equally to this work.
Contributor Information
Juliane G. Bogner-Strauss, Email: juliane.bogner-strauss@tugraz.at
Zlatko Trajanoski, Phone: +43-512-900371401, FAX: +43-512-900374400, Email: zlatko.trajanoski@i-med.ac.at.
References
- 1.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 2.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-X. [DOI] [PubMed] [Google Scholar]
- 4.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005;1:93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte–adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 8.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 9.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackl H, Burkard TR, Sturn A, Rubio R, Schleiffer A, Tian S, Quackenbush J, Eisenhaber F, Trajanoski Z. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 2005;6:R108. doi: 10.1186/gb-2005-6-13-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Camillo B, Sanchez-Cabo F, Toffolo G, Nair SK, Trajanoski Z, Cobelli C. A quantization method based on threshold optimization for microarray short time series. BMC Bioinformatics. 2005;6(Suppl 4):S11. doi: 10.1186/1471-2105-6-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang S, Fuhrman S, Somogyi R (1998) Reveal, a general reverse engineering algorithm for inference of genetic network architectures. Pac Symp Biocomput 18–29 [PubMed]
- 14.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- 21.Pabinger S, Thallinger GG, Snajder R, Eichhorn H, Rader R, Trajanoski Z. QPCR: application for real-time PCR data management and analysis. BMC Bioinformatics. 2009;10:268. doi: 10.1186/1471-2105-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kershaw EE, Schupp M, Guan HP, Gardner NP, Lazar MA, Flier JS. PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab. 2007;293:E1736–E1745. doi: 10.1152/ajpendo.00122.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albrektsen T, Richter HE, Clausen JT, Fleckner J. Identification of a novel integral plasma membrane protein induced during adipocyte differentiation. Biochem J. 2001;359:393–402. doi: 10.1042/0264-6021:3590393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider G, Neuberger G, Wildpaner M, Tian S, Berezovsky I, Eisenhaber F. Application of a sensitive collection heuristic for very large protein families: evolutionary relationship between adipose triglyceride lipase (ATGL) and classic mammalian lipases. BMC Bioinformatics. 2006;7:164. doi: 10.1186/1471-2105-7-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ooi HS, Kwo CY, Wildpaner M, Sirota FL, Eisenhaber B, Maurer-Stroh S, Wong WC, Schleiffer A, Eisenhaber F, Schneider G. ANNIE: integrated de novo protein sequence annotation. Nucleic Acids Res. 2009;37:W435–W440. doi: 10.1093/nar/gkp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockigt J, Barleben L, Panjikar S, Loris EA. 3D-Structure and function of strictosidine synthase–the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol Biochem. 2008;46:340–355. doi: 10.1016/j.plaphy.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 30.Scharff EI, Koepke J, Fritzsch G, Lucke C, Ruterjans H. Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure. 2001;9:493–502. doi: 10.1016/S0969-2126(01)00610-4. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka Y, Morikawa K, Ohki Y, Yao M, Tsumoto K, Watanabe N, Ohta T, Tanaka I. Structural and mutational analyses of Drp35 from Staphylococcus aureus: a possible mechanism for its lactonase activity. J Biol Chem. 2007;282:5770–5780. doi: 10.1074/jbc.M607340200. [DOI] [PubMed] [Google Scholar]
- 32.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilhan A, Gartner W, Nabokikh A, Daneva T, Majdic O, Cohen G, Bohmig GA, Base W, Horl WH, Wagner L. Localization and characterization of the novel protein encoded by C20orf3. Biochem J. 2008;414:485–495. doi: 10.1042/BJ20080503. [DOI] [PubMed] [Google Scholar]
- 35.Scheideler M, Elabd C, Zaragosi LE, Chiellini C, Hackl H, Sanchez-Cabo F, Yadav S, Duszka K, Friedl G, Papak C, Prokesch A, Windhager R, Ailhaud G, Dani C, Amri EZ, Trajanoski Z. Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genomics. 2008;9:340. doi: 10.1186/1471-2164-9-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomaru T, Steger DJ, Lefterova MI, Schupp M, Lazar MA. Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding proteins. J Biol Chem. 2009;284:6116–6125. doi: 10.1074/jbc.M808407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 39.Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- 40.Castelo R, Roverato A. Reverse engineering molecular regulatory networks from microarray data with qp-graphs. J Comput Biol. 2009;16:213–227. doi: 10.1089/cmb.2008.08TT. [DOI] [PubMed] [Google Scholar]
- 41.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.