Abstract
Τhe expression of the critical initiator cytokine TNF-α was strongly upregulated in vivo in acute necrotic pancreatitis (AP) in rodents and in vitro in TNF-α activated acinar AR42J cells. Upregulation of tnf-α, inos, icam-1 and il-6 occurred both in TNF-α receptor 1 and 2 knock-out mice, but not in TNF-α knock-out mice, in cerulein-induced acute pancreatitis. Chromatin immunoprecipitation analysis showed that transcriptional factors (ELK-1, SP1, NF-κB and EGR-1) and chromatin modification complexes (HDAC1, HDAC2, GCN5, PCAF and CBP) were recruited and/or released from the promoter in a strictly ordered mechanism. Activation of tnf-α gene was also accompanied by an ordered increased level of histone H3K9, H3K14 and H3K18-acetylation and H3K4 methylation, as well as H4K5 acetylation. A better knowledge of the molecular mechanisms that control tnf-α gene regulation will provide deeper understanding of the initiation and development of the inflammatory processes occurring in acute pancreatitis triggered by TNF-α cytokine.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0272-3) contains supplementary material, which is available to authorized users.
Keywords: Acute necrotic pancreatitis, Chromatin immunoprecipitation, ChIP, Epigenetic, Tumor necrosis factor alpha
Introduction
Acute pancreatitis (AP) is an initially localized inflammation of the pancreatic gland that may lead to local and systemic complications. Mortality due to AP is around 20–30% in the severe forms of the disease because of multiple organ failure [1–5].
The precise mechanisms by which etiological factors induce AP are not yet well known, but when initiated, common inflammatory pathways seem to be involved, with cytokines being their components of major importance [1, 6–8]. TNF-α is a key cytokine initiator of the inflammatory cascade in AP [2, 3, 9, 10], which induces the expression of pro-inflammatory genes such as il-6 [11–13], inos or icam-1 [14–16]. TNF-α interacts with two different surface receptors, TNF-α receptor 1 (TNFR1) and TNF-α receptor 2 (TNFR-2) [2, 17, 18]. In knock-out mice deficient in TNF-α receptors, the rate of mortality due to necrotizing AP decreased significantly, although there was no reduction in the severity of the pancreatic damage [19, 20].
TNF-α promoter contains elements critical for induction of the gene. It has been reported that a unique TNF-α enhancer complex containing Ets, Elk-1, Sp1, ATF-2-Jun, CBP and p300 is assembled on the TNF-α promoter in LPS-stimulated monocytes [21]. Nevertheless, the epigenetic regulation of tnf-α seems to be quite complex because the expression is cell-type specific [21, 22] and also stimulus specific [23, 24]. Thus, different sets of transcriptional factors are recruited in the tnf-α promoter upon ionophore stimulation (ATF-2/c-JUN and NFAT) and virus induction (ATF-2/c-JUN, NFAT and SP1) in fibroblasts or lymphocytes T and B [23].
At present, it is generally accepted that the precise regulation of gene expression by epigenetic mechanisms is required to upregulate cytokine expression in different experimental models. In this study, we provide new insights into the epigenetic mechanisms that control transcriptional activation of tnf-α in the initiation and development of AP.
Materials and methods
Experimental models of AP in animals
Young male Wistar rats and young male mice C57BL/6 were used in the experiments. Animals were cared for and handled in accordance with the European regulations (Council Directive 86/609/EEC), and the studies were approved by the Ethical Research Committee of the University of Valencia. Mice were either wild type, TNF-α receptor 1 knock-out (KO TNF-α R1), TNF-α receptor 2 knock-out (KO TNF-α R2) or TNF-α knock-out (KO TNF-α), and were kindly provided by Horst Bluethmann (Hoffmann-La Roche). Mice were treated with the cholecystokinin analogue cerulein to induce AP [25]. Cerulein was administered in seven injections at hourly intervals, with each injection containing 50 μg/kg body weight, and killed under anesthesia 1 h after the last injection of cerulein. AP was induced in rats by retrograde infusion of sodium taurocholate (3.5%; Sigma) into the biliopancreatic duct, as described by Aho et al. [26]. Rats were killed under anesthesia at 0, 0.5, 1, 3 and 6 h after the infusion of taurocholate or saline. Serum lipase activity was measured, and histological studies were performed to confirm the appropriate induction of pancreatitis.
AR42J pancreatic acinar cell line culture
The pancreatic acinar cell line AR42J (ATCC CRL 1492) was grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL) containing 25 mmol/l glucose, 100 μg/ml penicillin, 100 μg/ml streptomycin and 25 μg/ml fungizone, supplemented with 10% fetal bovine serum (FBS). To activate the tnf-α gene, AR42J cells were treated with 0–100 ng/ml of TNF-α (Sigma) for the indicated times.
Chromatin immunoprecipitation (ChIP) assay and RNApol ChIP
ChIP and RNApol ChIP procedures were performed as previously described [27, 28]. Briefly, rats were killed at the indicated times after taurocholate treatment, and the pancreases were excised and immersed for 10 min, or AR42J cultures were treated for 4 min, in 1% formaldehyde to crosslink the chromatin. After stopping the reaction by adding glycine to a final concentration of 0.125 M, the nuclei were obtained as described [27, 28]. Crosslinked chromatin was disrupted in a Vibra-Cell VCX-500 sonicator (Sonics and Materials) obtaining chromatin fragments of around 500 ± 300 bp. Aliquots of disrupted chromatin (equivalent to 50 μg DNA) were immunoprecipitated using 2 μg of antibodies against: NF-κB (sc-109), AP-1 (sc-1694), C/EBPβ (sc-150), SP1 (sc-59), ELK-1 (sc-355), ETS-2 (sc-351), EGR1 (sc-110), CBP (sc-369), PCAF (sc-8999), GCN5 (sc-6303), HDAC1 (sc-6298), HDAC2 (sc-6296), HDAC3 (sc-11417; from Santa Cruz), H3K4me2 (ab-7766), H3K4me3 (ab-8580), H3K9ac (ab-4441), H3K18ac (ab-1191), H4K5ac (ab-1758; from Abcam), H3R17me2 (07-214), H3K14ac (07-353), H3K27ac (07-360), H4K16ac (07-329; from Upstate) for ChIP assay or RNA pol II (sc-899 from Santa Cruz) for RNApol ChIP. The primer sequences used for target gene PCR analysis of IP samples were: tnf-α (promoter): forward 5′-GGTGAGGACGGAGAGGAGATT-3′ and reverse 5′-TGGGAGTTAGTACCAGGGTGTTC-3′ (277 bp product); tnf-α (coding region, exon 4) forward 5′-CAGCCGATTTGCCATTTCAT-3′ and reverse 5′-TCCTTAGGGCAAGGGCTCTT-3′ (253 bp product); α-actin (promoter): forward 5′-AGGGACTCTAGTGCCCAACACC-3′ and reverse 5′-CCCACCTCCACCCTACCTGC-3′ (186 bp product); α-actin (coding region, exon 2): 5′-AGGATTCCTACGTGGGCGAC-3′ and 5′-TAGAGAGACAGCACCGCCTG-3’ (280 bp product); β-actin (coding region, exon 2): forward 5′-AGAGCAAGAGAGGCA TCCTG-3′ and reverse 5′-GGGTCATCTTTTCACGGTTGG-3′ (245 bp product). PCR fragments were size-fractionated by 2% agarose gel electrophoresis, stained with ethidium bromide and analyzed with the FLA3000 electronic autoradiography system (Fujifilm) and ImageJ software (http://rsbweb.nih.gov/ij/).
RNA extraction and determination of mRNA steady-state levels
A small piece of the pancreas was excised and immediately immersed in 1 ml RNAlater solution (Ambion) to stabilize the RNA. Total RNA was isolated from pancreas and from AR42J cell cultures by the guanidinium thyocianate method [29]. The cDNA used as template for amplification in the PCR assay was constructed by reverse transcription reaction using SuperScript II (Invitrogen) and random hexamers as primers [27]. Real-time PCR was performed using Syber Green PCR Master mix (Applied Biosystems) in an ABI GeneAmp 7000 Sequence Detection System. Each reaction was performed in triplicate, and the melting curves were constructed using Dissociation Curves Software (Applied Biosystems) to ensure that only a single product was amplified. The 18S rRNA was also analyzed as RT-PCR control. The following specific primers were used: tnf-α, forward 5′-CAGCCGATTTGCCATTTCAT-3′, reverse 5′-TCCTTAGGGCAAGGGCTCTT-3′, β-actin: forward 5′-TTCAACACCCCAGCCATGT-3′, reverse 5′-GTGGTACGACCAG AGGCATACA-3; rRNA 18S: forward 5′-AGTCCCTGCCCTTTGTACACA-3′, reverse 5′-GATCCGAGGGCCTCACTAAAC-3′. The threshold cycle (Ct) was determined, and the relative gene expression was expressed as follows: fold change = 2−Δ(ΔCt), where ΔCt = Cttarget−Cthousekeeping and Δ(ΔCt) = ΔCttreated−ΔCtcontrol.
Immunohistochemistry
Pancreas specimens from control rats and rats at 3 h after induction of pancreatitis with taurocholate were fixed in formalin and embedded in paraffin. Immunohistochemistry was performed using the R.T.U. Vectastain Universal Elite ABC kit (Vector Labs, Burlingame, CA) following the manufacturer’s instructions. Briefly, sections were deparaffinized and rehydrated by sequential immersion of slides in xylene followed by graded concentrations of ethanol, and finally with water. Inactivation of endogenous tissue peroxidases was performed by immersing the slides in a solution of 0.5% H2O2 in methanol for 30 min. Sections were incubated in 2.5% horse serum albumin to prevent non-specific binding of antibodies, and rabbit anti-rat-TNF-α antibody (AbD Serotec, Oxford, UK) was probed for 1 h. Afterwards, slides were incubated with biotinylated secondary antibody for 30 min and treated with pre-formed avidin and biotinylated horseradish peroxidase macromolecular complex (Vector Labs, Burlingame, CA) for 30 min. Sections were then washed with water, and peroxidase activity was developed using diaminobenzidine solution (Sigma) with 0.04% H2O2. Finally, tissue was counterstained with Mayer’s hematoxylin solution (Sigma) for 4 min and mounted for light microscopic examination. Slides processed as above but without treatment with primary antibody against TNF-α were used as negative control of staining.
Statistical analysis
Results are expressed as mean ± standard deviation (SD). Statistical analysis was performed in two steps. Firstly, a one-way ANOVA was carried out to find significance in the overall comparison of groups. Then, differences between individual groups were investigated by the Scheffé test. Differences were considered to be significant when P < 0.05.
Results
Expression of pro-inflammatory genes in TNF-α receptor 1 and 2 knock-out mice during AP
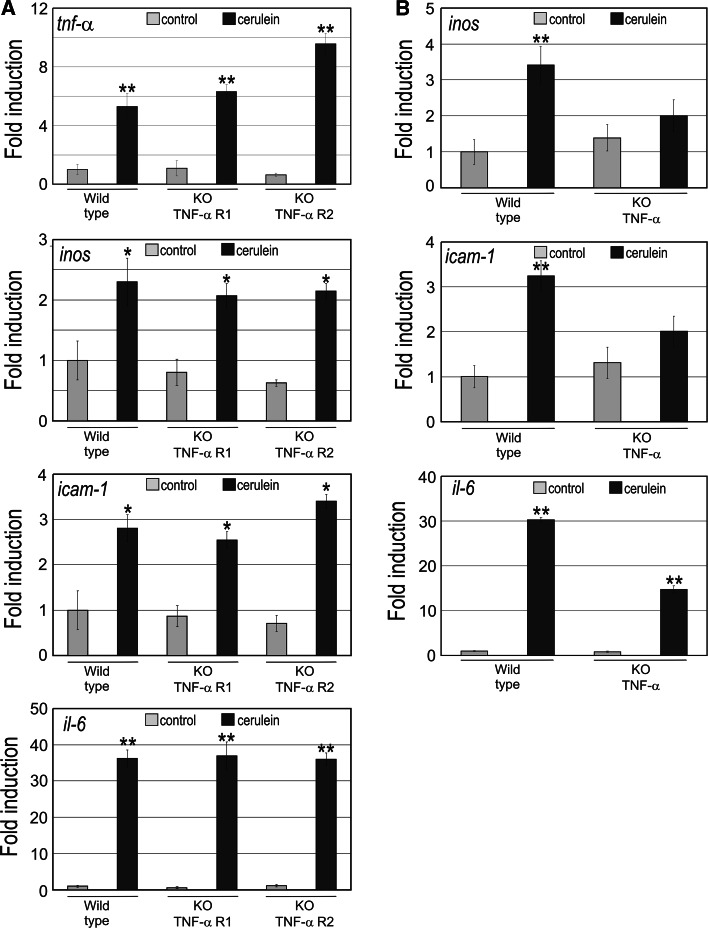
The expression of tnf-α, as well as inducible nitric oxide synthase (inos), intercellular adhesion molecule 1 (icam-1) and interleukin 6 (il-6), was measured in pancreas at 1 h after induction of AP by cerulein in wild-type mice and in TNF-α receptor 1 and 2 knock-out (KO TNF-α R1 and KO TNF-α R2) mice. Figure 1a shows that tnf-α upregulation occurred not only in wild-type mice, but also in mice devoid of either receptor 1 or 2, indicating that the presence of only one of these TNF-α receptors is sufficient to mediate TNF-α positive feed-back and consequently upregulation. Upregulation of inos, icam-1 and il-6 also occurred during AP in all three types of mice (wild type, KO TNF-α R1 and KO TNF-α R2) (Fig. 1a). Hence, either TNF-α receptor 1 alone or receptor 2 alone is sufficient to mediate the inflammatory cascade triggered by TNF-α.
Fig. 1.
Expression of tumor necrosis factor alpha (tnf-α), inducible nitric oxide synthase (inos), intercellular adhesion molecule 1 (icam-1) and interleukin 6 (il-6) in pancreas from a wild-type mice and TNF-α receptor 1 or 2 knock-out mice, and b TNF-α knock-out mice with cerulein-induced AP. Steady-state mRNA levels of tnf-α, inos, icam-1 and il-6 were measured by quantitative RT-PCR in pancreas from mice treated with saline (control) or cerulein as indicated in Methods. The error bars correspond with the standard deviation of 4–5 independent RT-PCR measurements. The statistical significance is indicated as follows: **P < 0.01 or *P < 0.05 vs. control or time 0
Expression of pro-inflammatory genes in TNF-α knock-out mice during AP
The upregulation of inos and icam-1 that occurred in wild-type mice with AP was abrogated in TNF-α KO mice (Fig. 1b). However, the induction of il-6 expression still occurred in TNF-α KO mice with AP, although to a lesser extent, probably because additional cellular signals and pro-inflammatory cytokine Il-1β may be involved in the activation of the gene during AP initiation. Indeed, il-6 expression increased almost 16 times in wild-type mice with AP, whereas it increased up to 8 times in the KO mice with AP (Fig. 1b).
Analysis of tnf-α gene expression in AP
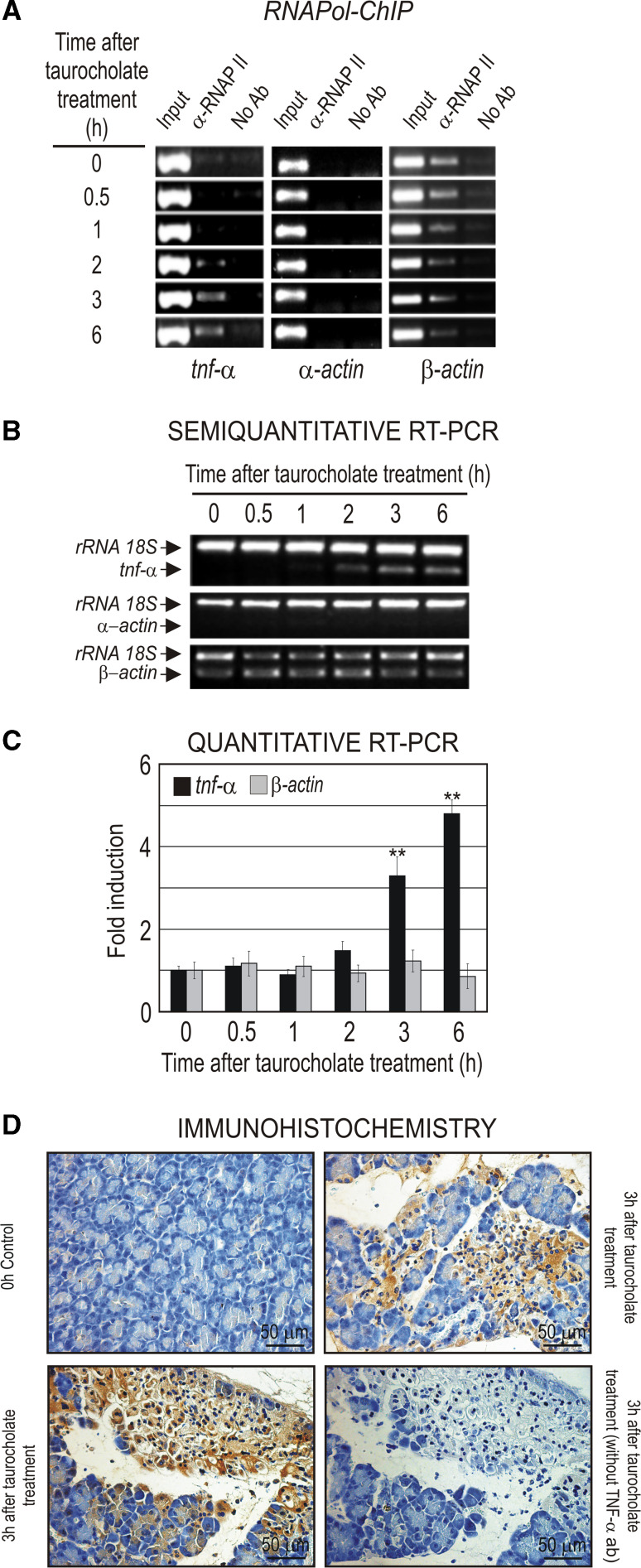
AP induced by taurocholate in rats was used in order to study the regulation of tnf-α expression and particularly the recruitment of transcription factors to tnf-α promoter. This experimental model of AP is well established in rats and provides enough material to carry out the chromatin immunoprecipitation assays and compare the results of the immunoprecipitation assays within the same animal. The expression of the tnf-α gene in AP induced by taurocholate was analyzed by RNApol ChIP assay as well as by semi-quantitative and quantitative RT-PCR (Fig. 2). We have previously described the RNApol ChIP assay as an experimental approach to asses the actual transcription rate [27]. The results indicate that elongating RNApol II was already present in the tnf-α coding region at 2 h of AP induction and thereafter (Fig. 2a), and produces the increase of the steady-state mRNA level at 2–3 h after AP induction, a level that stayed high at least until 6 h post-treatment (Fig. 2b, c). The upregulation of tnf-α expression in pancreas in the early stage of AP seems to be primarily ascribed to acinar cells [28–30]. Accordingly, the immunohistochemical analysis showed that pancreatic acinar cells produce TNF-α at 3 h after induction of AP with taurocholate (Fig. 2d).
Fig. 2.
The tnf-α gene expression in taurocholate-induced AP in rats. a RNApol ChIP analysis. α- and β-Actin genes were used as negative and positive control, respectively, of the RNApol ChIP assay. b Semiquantitative RT-PCR and c quantitative RT-PCR analysis of the tnf-α mRNA level. The rRNA 18S was used as an internal control, and α- and β-actin genes were used as negative and positive control, respectively, of the RT-PCR analysis. In the histogram, the quantitative RT-PCR values obtained for tnf-α or β-actin analysis were normalized against those obtained for rRNA 18S analysis, giving an arbitrary value of 1 to the non-treated sample. The error bars correspond with the standard deviation of at least three independent RT-PCR measurements. The statistical significance is indicated as follows: **P < 0.01 or *P < 0.05 vs. control or time 0. d Immunohistochemical detection of TNF-α production by acinar cells in rat pancreas 3 h after AP induction. Immunostaining negative control with tissue treated in absence of primary TNF-α antibody is also shown
Ordered transcriptional factor recruitment to tnf-α promoter in AP
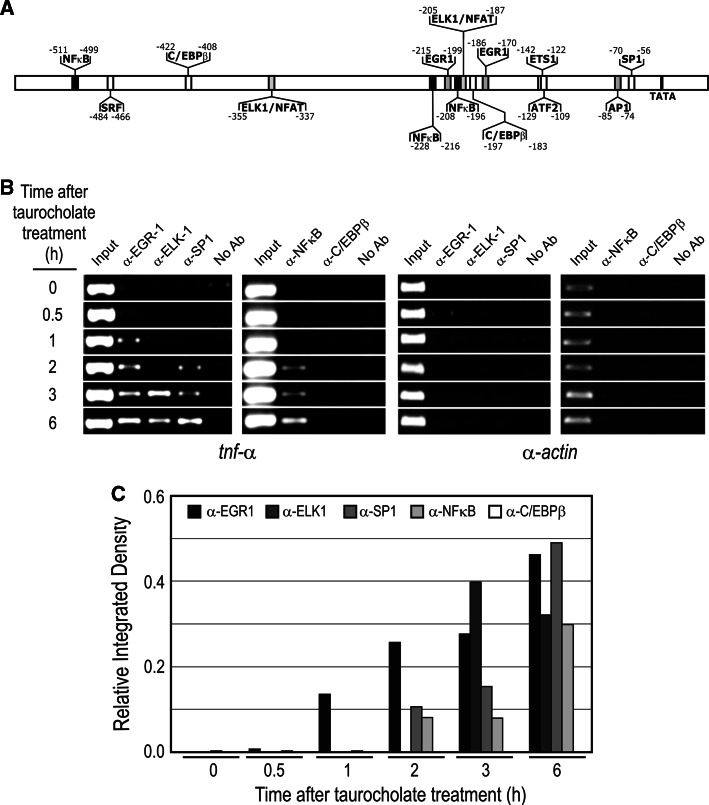
In order to explore the epigenetic regulation of tnf-α in AP, we carried out a kinetic analysis of the recruitment of transcriptional factors to the gene promoter by ChIP assay. First, we analyzed the putative binding sites for transcriptional factors in the promoter by using MatInspector Software (Genomatrix) (Fig. 3a). The analysis showed a complex scenario in which up to nine transcriptional factors might theoretically bind the promoter. To know if they actually bind in vivo, we performed a ChIP assay using the corresponding antibodies (see “Materials and methods”). Figure 3b, c shows that EGR-1, ELK-1, SP1 and NF-κB were bound to the promoter when the gene was activated in AP.
Fig. 3.
ChIP assay of tnf-α promoter occupation in taurocholate-induced AP in rats. a Putative binding sites for transcriptional factors in the tnf-α promoter analyzed by MatInspector Software. b The immunoprecipitated samples with indicated antibodies were analyzed by PCR using primers of the tnf-α promoter region. c ImageJ analysis of PCR signals. α-Actin gene has also been included as a negative control of the ChIP experiment
We further analyzed if an ordered recruitment of these factors could occur during tnf-α activation after AP induction. The factor EGR1 was bound to the promoter at 1 h after AP induction, followed by SP1 and NFκB (2 h) and ELK-1 (3 h) (Fig. 3b, c). The recruitment of the other putative transcriptional factors, C/EBPβ (Fig. 3b, c), ATF-2, AP-1, ETS-1 or SRF, was not detected in our experiments (results not shown).
Analysis of tnf-α gene expression in TNF-α activated AR42J cells
To know the precise order of recruitment of the transcriptional factors to the promoter during tnf-α activation, we used the pancreatic acinar cell line AR42J. The use of cell lines to elucidate the epigenetic mechanism of gene expression provides a more accurate time-dependent profile of the transcriptional factors, chromatin modification complex recruitment and histone post-translational modifications needed for gene activation than AP-induced models. Moreover, the use of cell lines also avoids the response variability to the stimulus of animal models.
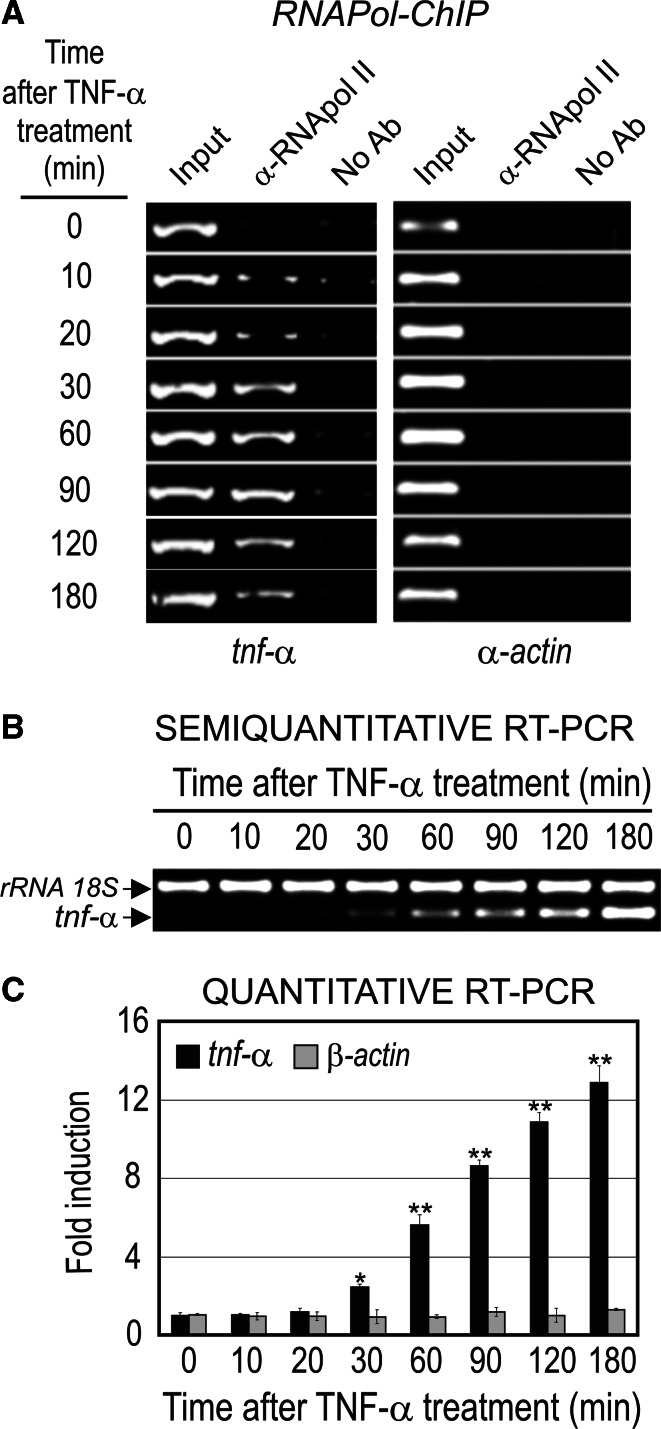
RNApol ChIP showed that RNA polymerase was present in the coding region of tnf-α at 10–20 min, increased significantly at 30 min and stayed bound up to 180 min after treatment of AR42J cells with TNF-α (50 ng/ml) (Fig. 4a). The presence of the elongating RNA polymerase in the coding region of the gene produced tnf-α mRNA accumulation, which is noticeable between 30 or 60 min and 180 min after TNF-α treatment (Fig. 4b, c). The apparent delay in the increase of mRNA level, in comparison with RNApol ChIP data, could be attributed to the accumulation of mRNA since the latter methodology is able to measure steady-state levels of mRNA and not the actual transcriptional rate as RNApol ChIP does.
Fig. 4.
The
tnf-α gene expression in TNF-α-activated AR42J cells. a RNApol ChIP analysis. b Semiquantitative RT-PCR and c quantitative RT-PCR analysis of the tnf-α mRNA level. rRNA 18S was used as an internal control and β-actin as negative control of the RT-PCR analysis and α-actin as negative control of ChIP assay. The statistical significance is indicated as follows: **P < 0.01 or *P < 0.05 vs. control or time 0
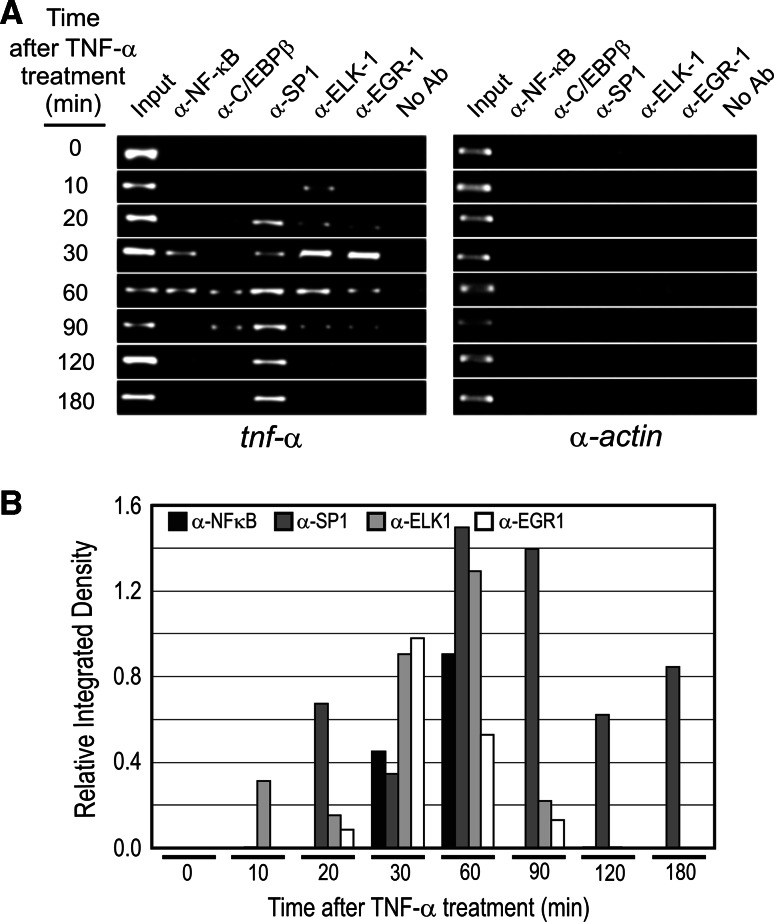
Ordered transcriptional factor recruitment to tnf-α promoter in TNF-α activated AR42J cells
Figure 5 shows that the recruitment of transcriptional factors to tnf-α promoter upon TNF-α activation followed a strict order as occurred with AP models. ELK-1 and SP1 were bound earlier, at 10 to 20 min after TNF-α treatment, NF-κB and EGR-1 were bound later (30 min), and finally C/EBPβ was bound at 60 min. It is noteworthy that when the gene was fully active, from 90 to 180 min (Fig. 5), all the transcriptional factors were released from the promoter with the exception of SP1.
Fig. 5.
a ChIP assay of tnf-α promoter occupation in TNF-α-activated AR42J cells. b ImageJ analysis of PCR signals. The α-actin gene has been included as negative control of the ChIP experiment
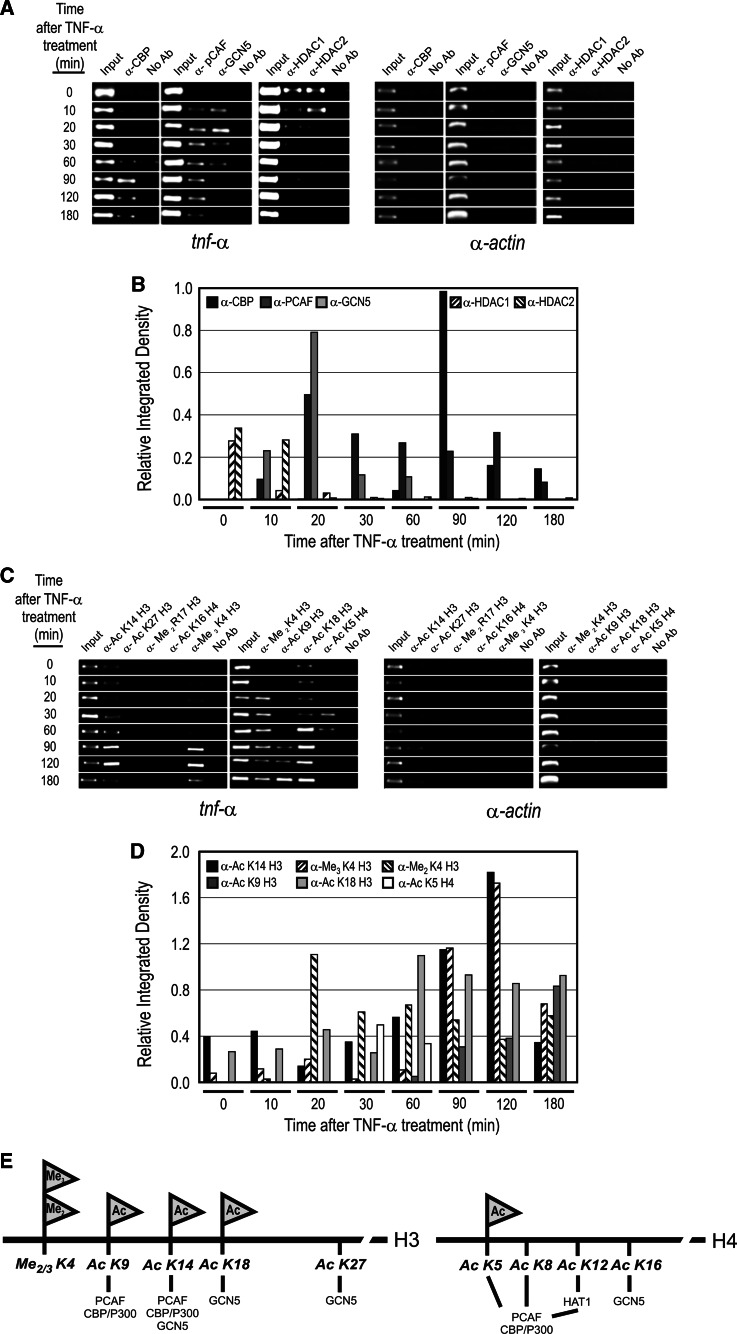
Ordered histone modification pattern in tnf-α upregulation in TNF-α activated AR42J cells
The activation of a specific gene needs a precise pattern of histone modifications, in several cases following an ordered mechanism, to allow the gene to reach a fully active transcriptional state. To establish whether this ordered sequence of epigenetic marks occurs during tnf-α activation, we performed a time-dependent ChIP assay of histone acetyltransferase (HAT) and histone deacetylase (HDAC) complex recruitment, as well as site-specific modification of histones, in AR42J cells treated with TNF-α (Fig. 6). Figure 6a, b shows that, in unstimulated cells, HDAC1 and HDAC2 are bound to tnf-α promoter, maintaining a repressed state of the gene, and that they were released 10 min after TNF-α treatment, when RNA polymerase started to be detectable in the coding region of the gene (Fig. 5). In coincidence with HDAC release, GCN5-containing and PCAF-containing histone acetyltranferase complexes are recruited to the promoter. However, the binding of GCN5 complex displays a transient pattern since 60 min after tnf-α induction the complex is released from the promoter. On the contrary, the PCAF complex remains permanently bound to the promoter during tnf-α activation. The release of GCN5-containing complex was coincident with the recruitment of the HAT CREB-binding protein (CBP) 60 min after TNF-α treatment.
Fig. 6.
ChIP assay of chromatin-modifying complex occupation (a, b) and site-specific histone modifications (c–e) in tnf-α promoter in TNF-α-activated AR42J cells. The α-actin gene has been included as negative control for the ChIP experiment
The recruitment of histone modification complexes is accompanied by an ordered site-specific modification of the histones in tnf-α promoter (Fig. 6c–e). In the repressed state, there is a basal level of histone acetylation in lysine 14 and 18 of the histone H3 (Fig. 6c, d; H3K14ac and H3K18ac), modifications that markedly increase when the gene is fully activated. The significant increase in the acetylation level also occurs in lysine 9 of the histone H3 (Fig. 6c–d; H3K9ac). It is worth nothing that there is a transient increase, from 30 to 60 min of induction, in the acetylation level of lysine 5 in histone H4 (Fig. 6c, d; H4K5ac), which could be produced by GCN5 activity recruited at that time to the promoter of the gene. Finally, the methylation of the lysine K4 seems also to be necessary for tnf-α activation since this residue was di-methylated at 20 min of induction, and tri-methylated at 90 min of induction, when the gene became fully activated.
Discussion
The degree of pancreatic injury in acute pancreatitis correlates directly with TNF-α levels, and this pro-inflammatory cytokine plays a major role in the initiation of AP [33–35]. The use of mice devoid of TNF-α or its receptors allowed us to elucidate the contribution of this primary cytokine to the inflammatory cascade and the receptor(s) involved. So far, the inflammatory effects of TNF-α have been ascribed to its type I receptor [36]. Accordingly, the lack of TFN-α R1 signaling diminished induction of pro-inflammatory cytokines after brain injury [37]. Contrarily, our results show that the upregulation of pro-inflammatory mediators (tnf-α, inos, icam-1 and il-6) in the pancreas during AP also occurs in KO TNF-α R1 mice, and hence it is not exclusively mediated by TNF-α receptor 1. Both TNF-α receptors 1 and 2 seem to be responsible for the pancreatic inflammatory cascade in the initial course of AP, as inos, icam-1, il-6 and tnf-α mRNA levels were similar in TNF-α R1 and TNF-α R2 KO mice. Consequently, the positive feedback responsible for tnf-α induction in the pancreas may be mediated either by receptor 1 or 2. In any case, tnf-α upregulation is critical for the pancreatic inflammatory network since TNF-α KO mice did not exhibit any significant increase in inos and icam-1 mRNAs levels, and only a half increase in il-6 mRNA. The results suggest that upregulation of il-6 seems to be mediated not only by TNF-α, but probably also by the other initiation cytokine IL-1β.
We have also examined the epigenetic mechanisms involved in tnf-α gene activation in vivo in taurocholate-induced AP, as well as in vitro in TNF-α activated acinar AR42J cells. This cell line was selected since acinar cells behave as inflammatory cells and are considered to be a primary source of TNF-α in experimental AP [28–30; Fig. 2d], even before leukocytes are able to increase the cytokine production and propagate the inflammatory cascade [36]. Consequently, the upregulation of pro-inflammatory cytokines in the initiation of AP should be mainly ascribed to acinar cells. In addition, the in vitro experiments on acinar AR42J cells show similar kinetics to AP regarding the gene expression of pro-inflammatory cytokines.
It is well known that TNF-α, IL-1β or IL-6 is critical for the amplification of the inflammatory signal in AP (see reviews in [1–3]). We found that tnf-α was induced when AR42J cells were incubated with TNF-α, but not with IL-1β or IL6 (results not shown). Therefore, we used TNF-α-activated acinar AR42J cells to study the epigenetic mechanism of tnf-α gene expression. When we analyzed the transcriptional factor recruitment during AP or in TNF-α-activated acinar AR42J cells, only ELK-1, SP1, NF-κB, EGR-1 and C/EBPβ of the at least nine putative factors that can be bound to the promoter were implicated in tnf-α induction. Consequently, the transcriptional factors involved in TNF-α upregulation were essentially the same in vivo in AP models and in vitro in acinar AR42J cells. The fact that these transcriptional factors are required for tnf-α gene activation seems to be confirmed when AP is induced in the presence of pentoxifylline, since the anti-inflammatory compound reduced markedly the recruitment of the transcriptional factors NF-κB, SP1 and ELK-1, but not EGR1, to tnf-α promoter and inhibits gene expression (material accessible on line, Fig. S1). Pentoxifylline has been studied as a potential modulator of the immunological response in AP because it inhibits TNF-α production [38, 39] and diminishes the leukocyte infiltrate and edema after taurocholate- or cerulein-induced pancreatitis in rats [39, 40].
Our study goes a step forward by showing that tnf-α expression in vivo and in vitro follows an ordered transcriptional factor recruitment mechanism (Figs. 3 and 5). The factors ELK-1 and SP1 were bound earlier, NF-κB and EGR-1 were bound later, and finally C/EBPβ, at least in the AR42J cell line, was bound to tnf-α promoter. The recruitment of the transcriptional factors to tnf-α promoter in our experiments was, at least in part, distinct from those that have been found in other cell lines under different stimuli. Tsai et al. [21] have described the presence of an enhanceosome, composed by ATF2/c-JUN, ETS1/2, ELK-1, EGR1 and SP1, assembled in tnf-α promoter in the LPS-stimulated monocytic cell line. The differences in recruitment of factors to tnf-α promoter may be due to the gene responding by a cell-specific and/or stimulus-specific mechanism to different pro-inflammatory circumstances as occurs for instance in LPL, viral and chronic or acute pancreatitis inductions.
The strictly ordered mechanism for tnf-α transcriptional activation is also extended to chromatin modification complex recruitment and histone post-translational modifications (Fig. 6). Several authors have also reported that the transcription factors bound to the tnf-α promoter (ELK-1, SP1, NF-κB, EGR-1 and C/EBPβ) physically interact with PCAF, GCN5 and/or CBP/p300 in an inducible manner and, in many cases, its presence in the promoter is a prerequisite for HAT recruitment and activation of target genes [41–46]. In many genes, individual binding sites have only weak affinity for the specific factor, but cooperative binding of adjacent proteins can generate cooperative functional responses allowing the recruitment of coactivators of transcription, for instance CBP/P300, and activating the RNApol II complex (see reviews in [47–49]). The specificity in gene transcription is then achieved by the assembly of these complexes, transcriptional factors and chromatin modifying and remodeling complexes within particular promoters, as seems to occur in tnf-α. The complexes then induce conformational changes in the chromatin structure and epigenetic setting that integrate cellular and environmental signaling and determine the expression of specific pro-inflammatory genes (see reviews in [48, 50–52]).
In our studies we showed that tnf-α promoter is able to integrate the signals occurring during AP through an ordered recruitment of transcriptional factors. The binding and release of these factors may serve as a specific and transitory mark in the promoter for the recruitment of chromatin modifier complexes HAT and HDAC that may produce the district specific histone modification pattern needed to activate the gene (Fig. 6). In this context, Engdahl et al. [53] showed that THP-1 monocytes or PMA-differentiated cells treated with LPS increase histone H3 and H4 acetylation at the TNF-α promoter and that prostaglandin metabolite 15d-PGJ2 is able to block LPS-induced TNF-α expression by decreasing the acetylation level. They also showed that inhibition of HDAC with TSA, or overexpression of CBP, overcomes 15d-PGJ2-mediated repression of the TNF-α promoter. In the same THP-1 cell line, the treatment with a high concentration of glucose activates tnf-α and cox2 genes, not only by recruitment to promoter of NF-κB, CBP/p300 and PCAF, as well as histone acetylation, but also by increasing the transcriptional activity of NF-κB p65 over NF-κB-regulated inflammatory genes [54].
These studies and ours demonstrate the importance of epigenetic regulation in the control of TNF-α expression. These findings may have relevance for inflammatory disorders in which TNF-α is overproduced, as occurs in AP. Further experiments are needed to elucidate if the strict ordered epigenetic mechanism for tnf-α expression is involved in TNF-α-induced remodeling of the chromatin of tnf-α promoter during AP.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suplementary Figure S1. Effect of pentoxyfilline in tnf-α expression and in transcriptional factor recruitment in tnf-α promoter. (A) Quantitative RT-PCR analysis of tnf-α gene expression. β-Actin gene was used as negative control of taurocholate-induced expression. (B) ChIP assay of tnf-α promoter transcriptional factor occupation. α-Actin gene was used as a negative control of the ChIP assay. (C) ImageJ analysis of PCR signals. P: pentoxyfilline treatment; T: taurocholate treatment; P+T: pentoxyfilline plus taurocholate treatment. The statistical significance is indicated as follows: **P < 0.01 vs. control or time 0; ##P < 0.01 vs. taurocholate (TIFF 909 kb)
Acknowledgments
This work was supported by research grants from the Ministerio de Ciencia y Tecnología (BFU2007-63120, CSD2006-49 to G. López-Rodas and SAF2006-06963, SAF2009-09500, CSD2007-00020 to J. Sastre) and from the European Commission FP6 Integrated Project Exogenesis (LSHM-CT-2004-005272 to J.Hidalgo).
References
- 1.Pandol SJ. Acute pancreatitis. Curr Opin Gastroenterol. 2006;22:481–486. doi: 10.1097/01.mog.0000239861.89209.5f. [DOI] [PubMed] [Google Scholar]
- 2.Pereda J, Sabater L, Aparisi L, Escobar J, Sandoval J, Vina J, Lopez-Rodas G, Sastre J. Interaction between cytokines and oxidative stress in acute pancreatitis. Curr Med Chem. 2006;13:2775–2787. doi: 10.2174/092986706778522011. [DOI] [PubMed] [Google Scholar]
- 3.Cosen-Binker LI, Gaisano HY. Recent insights into the cellular mechanisms of acute pancreatitis. Can J Gastroenterol. 2007;21:19–24. doi: 10.1155/2007/930424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan YC, Leung PS. Acute pancreatitis: animal models and recent advances in basic research. Pancreas. 2007;34:1–14. doi: 10.1097/01.mpa.0000246658.38375.04. [DOI] [PubMed] [Google Scholar]
- 5.Pandol SJ, Saluja AK, Imrie CW, Banks PA. Acute pancreatitis: bench to the bedside. Gastroenterology. 2007;132:1127–1151. doi: 10.1053/j.gastro.2007.01.055. [DOI] [PubMed] [Google Scholar]
- 6.Kusske AM, Rongione AJ, Reberl HA. Cytokines and acute pancreatitis. Gastroenterology. 1996;110:639–642. doi: 10.1053/gast.1996.v110.agast960639. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia M, Brady M, Shokuhi S, Christmas S, Neoptolemos JP, Slavin J. Inflammatory mediators in acute pancreatitis. J Pathol. 2000;190:117–125. doi: 10.1002/(SICI)1096-9896(200002)190:2<117::AID-PATH494>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Norton ID, Clain JE. Optimising outcomes in acute pancreatitis. Drugs. 2001;61:1581–1591. doi: 10.2165/00003495-200161110-00005. [DOI] [PubMed] [Google Scholar]
- 9.Norman J. The role of cytokines in the pathogenesis of acute pancreatitis. Am J Surg. 1998;175:76–83. doi: 10.1016/S0002-9610(97)00240-7. [DOI] [PubMed] [Google Scholar]
- 10.Sakorafas GH, Tsiotou AG. Etiology and pathogenesis of acute pancreatitis: current concepts. J Clin Gastroenterol. 2000;30:343–356. doi: 10.1097/00004836-200006000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimada M, Andoh A, Hata K, Tasaki K, Araki Y, Fujiyama Y, Bamba T. IL-6 secretion by human pancreatic periacinar myofibroblasts in response to inflammatory mediators. J Immunol. 2002;168:861–868. doi: 10.4049/jimmunol.168.2.861. [DOI] [PubMed] [Google Scholar]
- 13.Zhou A, Scoggin S, Gaynor RB, Williams NS. Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene. 2003;22:2054–2064. doi: 10.1038/sj.onc.1206262. [DOI] [PubMed] [Google Scholar]
- 14.Hou J, Baichwal V, Cao Z. Regulatory elements and transcription factors controlling basal and cytokine-induced expression of the gene encoding intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1994;91:11641–11645. doi: 10.1073/pnas.91.24.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roebuck KA, Rahman A, Lakshminarayanan V, Janakidevi K, Malik AB. H2O2 and tumor necrosis factor-alpha activate intercellular adhesion molecule 1 (ICAM-1) gene transcription through distinct cis-regulatory elements within the ICAM-1 promoter. J Biol Chem. 1995;270:18966–18974. doi: 10.1074/jbc.270.32.18966. [DOI] [PubMed] [Google Scholar]
- 16.Baker SJ, Reddy EP. Transducers of life and death: TNF receptor superfamily and associated proteins. Oncogene. 1996;12:1–9. [PubMed] [Google Scholar]
- 17.Brockhaus M, Schoenfeld HJ, Schlaeger EJ, Hunziker W, Lesslauer W, Loetscher H. Identification of two types of tumor necrosis factor receptors on human cell lines by monoclonal antibodies. Proc Natl Acad Sci USA. 1990;87:3127–3131. doi: 10.1073/pnas.87.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis M, Tartaglia LA, Lee A, Bennett GL, Rice GC, Wong GH, Chen EY, Goeddel DV. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc Natl Acad Sci USA. 1991;88:2830–2834. doi: 10.1073/pnas.88.7.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastor CM, Frossard JL. Are genetically modified mice useful for the understanding of acute pancreatitis? FASEB J. 2001;15:893–897. doi: 10.1096/fj.00-0672rev. [DOI] [PubMed] [Google Scholar]
- 20.Pastor CM, Matthay MA, Frossard JL. Pancreatitis-associated acute lung injury: new insights. Chest. 2003;124:2341–2351. doi: 10.1378/chest.124.6.2341. [DOI] [PubMed] [Google Scholar]
- 21.Tsai EY, Falvo JV, Tsytsykova AV, Barczak AK, Reimold AM, Glimcher LH, Fenton MJ, Gordon DC, Dunn IF, Goldfeld AE. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol Cell Biol. 2000;20:6084–6094. doi: 10.1128/MCB.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldfeld AE, Maniatis T. Coordinate viral induction of tumor necrosis factor alpha and interferon beta in human B cells and monocytes. Proc Natl Acad Sci USA. 1989;86:1490–1494. doi: 10.1073/pnas.86.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falvo JV, Uglialoro AM, Brinkman BM, Merika M, Parekh BS, Tsai EY, King HC, Morielli AD, Peralta EG, Maniatis T, Thanos D, Goldfeld AE. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol Cell Biol. 2000;20:2239–2247. doi: 10.1128/MCB.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falvo JV, Brinkman BM, Tsytsykova AV, Tsai EY, Yao TP, Kung AL, Goldfeld AE. A stimulus-specific role for CREB-binding protein (CBP) in T cell receptor-activated tumor necrosis factor alpha gene expression. Proc Natl Acad Sci USA. 2000;97:3925–3929. doi: 10.1073/pnas.97.8.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederau C, Ferrell LD, Grendell JH. Caerulein-induced acute necrotizing pancreatitis in mice: protective effects of proglumide, benzotript, and secretin. Gastroenterology. 1985;88:1192–1204. doi: 10.1016/s0016-5085(85)80079-2. [DOI] [PubMed] [Google Scholar]
- 26.Frossard JL, Pastor CM. Experimental acute pancreatitis: new insights into the pathophysiology. Front Biosci. 2002;7:d275–d287. doi: 10.2741/frossard. [DOI] [PubMed] [Google Scholar]
- 27.Sandoval J, Rodriguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, López-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gukovskaya AS, Gukovsky I, Zaninovic V, Song M, Sandoval D, Gukovsky S, Pandol SJ. Pancreatic acinar cells produce, release, and respond to tumor necrosis factor-alpha. Role in regulating cell death and pancreatitis. J Clin Invest. 1997;100:1853–1862. doi: 10.1172/JCI119714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duell EJ, Casella DP, Burk RD, Kelsey KT, Holly EA. Inflammation, genetic polymorphisms in proinflammatory genes TNF-A, RANTES, and CCR5, and risk of pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:726–731. doi: 10.1158/1055-9965.EPI-05-0797. [DOI] [PubMed] [Google Scholar]
- 30.Baumann B, Wagner M, Aleksic T, von Wichert G, Weber CK, Adler G, Wirth T. Constitutive IKK2 activation in acinar cells is sufficient to induce pancreatitis in vivo. J Clin Invest. 2007;117:1502–1513. doi: 10.1172/JCI30876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG, Martinez-Chantar ML, Vina J, Vina JR, Mato JM, Avila MA, Franco L, López-Rodas G, Torres L. Id2 leaves the chromatin of the E2F4–p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398:431–437. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 33.Grewal HP, Mohey el Din A, Gaber L, Kotb M, Gaber AO. Amelioration of the physiologic and biochemical changes of acute pancreatitis using an anti-TNF-alpha polyclonal antibody. Am J Surg. 1994;167:214–218. doi: 10.1016/0002-9610(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 34.Hughes CB, Gaber LW, Mohey el-Din AB, Grewal HP, Kotb M, Mann L, Gaber AO. Inhibition of TNF alpha improves survival in an experimental model of acute pancreatitis. Am Surg. 1996;62:8–13. [PubMed] [Google Scholar]
- 35.Denham W, Yang J, Fink G, Denham D, Carter G, Ward K, Norman J. Gene targeting demonstrates additive detrimental effects of interleukin 1 and tumor necrosis factor during pancreatitis. Gastroenterology. 1997;113:1741–1746. doi: 10.1053/gast.1997.v113.pm9352880. [DOI] [PubMed] [Google Scholar]
- 36.Bazzoni F, Beutler B. The tumor necrosis factor ligand and receptor families. N Engl J Med. 1996;334:1717–1725. doi: 10.1056/NEJM199606273342607. [DOI] [PubMed] [Google Scholar]
- 37.Quintana A, Giralt M, Rojas S, Penkowa M, Campbell IL, Hidalgo J, Molinero A. Differential role of tumor necrosis factor receptors in mouse brain inflammatory responses in cryolesion brain injury. J Neurosci Res. 2005;82:701–716. doi: 10.1002/jnr.20680. [DOI] [PubMed] [Google Scholar]
- 38.Coimbra R, Melbostad H, Hoyt DB. Effects of phosphodiesterase inhibition on the inflammatory response after shock: role of pentoxifylline. J Trauma. 2004;56:442–449. doi: 10.1097/01.TA.0000096642.54111.E8. [DOI] [PubMed] [Google Scholar]
- 39.Pereda J, Sabater L, Cassinello N, Gomez-Cambronero L, Closa D, Folch-Puy E, Aparisi L, Calvete J, Cerda M, Lledo S, Vina J, Sastre J. Effect of simultaneous inhibition of TNF-alpha production and xanthine oxidase in experimental acute pancreatitis: the role of mitogen activated protein kinases. Ann Surg. 2004;240:108–116. doi: 10.1097/01.sla.0000129343.47774.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez-Cambronero L, Camps B, De La Asuncion JG, Cerda M, Pellin A, Pallardo FV, Calvete J, Sweiry JH, Mann GE, Vina J, Sastre J. Pentoxifylline ameliorates cerulein-induced pancreatitis in rats: role of glutathione and nitric oxide. J Pharmacol Exp Ther. 2000;293:670–676. [PubMed] [Google Scholar]
- 41.Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/S1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 43.Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 2003;22:281–291. doi: 10.1093/emboj/cdg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng WG, Zhu Y, Wu KK. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J Biol Chem. 2003;278:4770–4777. doi: 10.1074/jbc.M209286200. [DOI] [PubMed] [Google Scholar]
- 45.Hiroi M, Ohmori Y. The transcriptional coactivator CREB-binding protein cooperates with STAT1 and NF-kappa B for synergistic transcriptional activation of the CXC ligand 9/monokine induced by interferon-gamma gene. J Biol Chem. 2003;278:651–660. doi: 10.1074/jbc.M204544200. [DOI] [PubMed] [Google Scholar]
- 46.Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc Natl Acad Sci USA. 2007;104:2703–2708. doi: 10.1073/pnas.0607378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahman I, Marwick J, Kirkham P. Redox modulation of chromatin remodelling: impact on histone acetylation and deacetylation, NF-kappaB and pro-inflammatory gene expression. Biochem Pharmacol. 2004;68:1255–1267. doi: 10.1016/j.bcp.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 48.Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Revilla Y, Granja AG. Viral mechanisms involved in the transcriptional CBP/p300 regulation of inflammatory and immune responses. Crit Rev Immunol. 2009;29:131–154. doi: 10.1615/critrevimmunol.v29.i2.30. [DOI] [PubMed] [Google Scholar]
- 50.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/S0959-437X(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez M, Rhodes SJ, Bidwell JP. Context-dependent transcription: all politics is local. Gene. 2003;313:43–57. doi: 10.1016/S0378-1119(03)00627-9. [DOI] [PubMed] [Google Scholar]
- 52.Vanden Berghe W, Ndlovu MN, Hoya-Arias R, Dijsselbloem N, Gerlo S, Haegeman G. Keeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigenetics. Biochem Pharmacol. 2006;72:1114–1131. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Engdahl R, Monroy MA, Daly JM. 15-Deoxy-Delta12, 14-prostaglandin J2 (15d-PGJ2) mediates repression of TNF-alpha by decreasing levels of acetylated histone H3 and H4 at its promoter. Biochem Biophys Res Commun. 2007;359:88–93. doi: 10.1016/j.bbrc.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suplementary Figure S1. Effect of pentoxyfilline in tnf-α expression and in transcriptional factor recruitment in tnf-α promoter. (A) Quantitative RT-PCR analysis of tnf-α gene expression. β-Actin gene was used as negative control of taurocholate-induced expression. (B) ChIP assay of tnf-α promoter transcriptional factor occupation. α-Actin gene was used as a negative control of the ChIP assay. (C) ImageJ analysis of PCR signals. P: pentoxyfilline treatment; T: taurocholate treatment; P+T: pentoxyfilline plus taurocholate treatment. The statistical significance is indicated as follows: **P < 0.01 vs. control or time 0; ##P < 0.01 vs. taurocholate (TIFF 909 kb)