Abstract
Huntingtin interacting protein 1 (HIP1) is an accessory protein of the clathrin-mediated endocytosis (CME) pathway, yet its precise role and the step at which it becomes involved are unclear. We employed live-cell imaging techniques to focus on the early steps of CME and characterize HIP1 dynamics. We show that HIP1 is highly colocalized with clathrin at the plasma membrane and shares similar dynamics with a subpopulation of clathrin assemblies. Employing transferrin receptor fused to pHluorin, we distinguished between open pits to which HIP1 localizes and newly internalized vesicles that are devoid of HIP1. Moreover, shRNA knockdown of clathrin compromised HIP1 membranal localization, unlike the reported behavior of Sla2p. HIP1 fragment, lacking its ANTH and Talin-like domains, inhibits internalization of transferrin, but retains colocalization with membranal clathrin assemblies. These data demonstrate HIP1’s role in pits maturation and formation of the coated vesicle, and its strong dependence on clathrin for membranal localization.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0077-4) contains supplementary material, which is available to authorized users.
Keywords: HIP1, Clathrin, Endocytosis, TIRF, Live-cell imaging
Introduction
Clathrin-mediated endocytosis (CME) is a well-studied mechanism involved in many important cell functions, such as the uptake of essential nutrients [1, 2] and modulation of signal transduction [3, 4]. The process of CME involves many accessory proteins, some well characterized, such as the canonical tetrameric endocytic adaptor complex (AP2) and dynamin; others are less so, such as Huntingtin interacting protein 1 (HIP1) (reviewed in [5]).
HIP1 was shown to function at different stages of the CME pathway, but its precise role is still unknown. Based on sequence similarities, it has been shown to contain several binding domains that enable interactions with phosphatidyl inositol 4,5-biphosphate (PIP2), AP2, clathrin’s light and heavy chains (CLC and CHC, respectively) and actin [6, 7]. These findings have been supported by in vitro binding assays, colocalizations and structural analysis of the coiled-coil domain [6, 8–11]. The presumed function of HIP1 in endocytosis is further supported by its influence on the membrane localization levels of various receptors. For example, mice with a targeted mutation in HIP1 have excess surface AMPA receptor following ligand stimulation, suggesting disruption of receptor internalization [12]. The function of NMDA receptor is also affected by HIP1 [13]. In other studies, overexpression of HIP1 in cell culture stabilized EGF receptor levels following EGF stimulation and resulted in increased phosphorylation of downstream effectors [14, 15]. In that case, the prolonged activation of the receptor was interpreted as being due to a block in its movement from early endosomes to late endosomes, rather than to disrupted internalization [14]. Therefore, the exact pathway and step at which HIP1 participates are still under debate.
Members of the Sla2/HIP1 family, which include the yeast Sla2p and the mammalian HIP1R and HIP1, have an overall conserved domain structure. Interestingly, based on this structural similarity, a functional equivalence amongst the proteins is presumed. Thus the different proteins are commonly referred as HIP1/HIP1R or HIP1R/Sla2p. However, accumulating evidence suggests that they segregate in their functions, dynamics and protein–protein interaction partners. The knockout of HIP1 and HIP1R in mice results in different phenotypes [12, 16]. Moreover, the dynamics of Sla2p and HIP1R were shown to substantially differ: while HIP1R was shown to appear and disappear simultaneously with clathrin, Sla2p arrives 25 s later than clathrin and dissociates upon recruitment of actin-related proteins [17, 18]. This difference in dynamics strongly suggests a difference in function. Recently, it was suggested that in yeast, Sla2p serves as a link between the clathrin-coat and actin-remodeling proteins. Indeed, yeast cells devoid of Sla2p display abnormalities in the organization of actin [17]. A significant effect on the actin structure was also described for cells with HIP1R siRNA [19, 20]. Thus, concerning the mediation of actin elongation in endocytosis, these two proteins may represent functional homologues. However, the affinity of HIP1 for actin is 7.5 times lower than HIP1R [21], and no effect on actin organization was so far observed upon alteration in HIP1 levels. In addition, HIP1 also differs from Sla2p and HIP1R in its interaction with clathrin. HIP1 has the ability to directly bind CHC [9], unlike the indirect interaction of Sla2p and HIP1R with CHC through the LC [22, 23]. Thus, it is possible that despite their sequence similarities, HIP1 differs in function from HIP1R and Sla2p. Furthermore, due to the vectorial nature of the initiation, growth, invagination and internalization of the coated pit/vesicle, proteins with different functions are recruited at different stages of the process [24, 25]. In this context, the visualization and description of the dynamics of HIP1 in clathrin-mediated endocytosis are still lacking. In the present study we provide the first quantitative dynamic characterization of HIP1 and suggest that it differs in several aspects from HIP1R and Sla2p.
Here, we addressed the role of mammalian HIP1 in the very early stages of endocytosis. To this end, we employed a two-pronged approach to dissect the role of HIP1 in CME: live-cell imaging aimed at characterizing the dynamic parameters of HIP1 addition to the coated pit, and chemical and genetic perturbations aimed at identifying the regulatory components of HIP1 recruitment to the membrane. We show that the spatiotemporal parameters of the association of HIP1 with the membrane parallel those of clathrin in many respects. The similarities in the dynamics and localization of their recruitment to the membrane are observed in both naïve cells and in response to different manipulations of the coat structure. In addition, reduction in the amount of clathrin in the cells, as a result of shRNA-mediated knockdown, compromises the membranal localization of HIP1, suggesting that clathrin is involved in either recruitment or stabilization of HIP1 at the membrane. We conclude that HIP1 is a dynamic component of the coated pit, but not the internalized vesicle, and functions at the very early phases of endocytosis.
Materials and methods
Cell cultures, plasmids and immunostaining
COS7 and HeLa cells were grown in DMEM supplemented with 10% (v/v) FCS, 2 mM l-glutamine and 100 U/ml penicillin-streptomycin. Transfections were performed using Jet-PEI (PolyPlus Transfection, New York, NY) according to the manufacturer’s instructions using a 1:2 (DNA:reagent) ratio. All plasmids encoding fluorescently labeled clathrin light chain A (CLC) were a kind gift from Prof. Kirchhausen (Harvard Medical School, Boston, MA). Plasmids encoding GFP-fused HIP1 and its 218–604 fragment were a kind gift from Prof. Erich Wanker (Max Delbruck Center, Berlin, Germany). The plasmid encoding tdTomato-HIP1 was created by replacing EGFP in the GFP-HIP1 construct with PCR-amplified tdTomato, using the AgeI and XhoI sites. TfR-pHl plasmid was a kind gift from Prof. Christien Merrifield (MRC Laboratory of Molecular Biology, Cambridge, UK). Clathrin heavy chain silencing and scramble plasmids (CHC-shRNA, SCR-shRNA) were a kind gift from Dr. Eran Bacharach (Tel Aviv University, Tel Aviv, Israel). Immunostaining was performed on 4% (w/v) paraformaldehyde-fixed cells, using mouse anti-clathrin X22 (1:1,000, Novus Biologicals, Littleton, CO) or rabbit anti-HIP1 (1:100, Prof. Erich Wanker), and detection with cy3-conjugated goat anti-rabbit/mouse (Jackson Laboratories, Bar Harbor, ME). Clathrin was silenced as follows: HeLa cells were either transfected with GFP-HIP1 alone or cotransfected with both GFP-HIP1 and one of the silencing constructs: CHC-shRNA or SCR-shRNA. Two days later, the cells were split, plated on glass coverslips, and returned to the incubator for 2 more days. Silencing of the CHC was evaluated as 70% protein reduction using antibody staining (X22 clone, Novus Biologicals; data not shown).
TIRF imaging setup
For imaging purposes, cells were plated on glass coverslips, transfected with plasmids encoding fluorescently tagged proteins and imaged 18–28 h later (for most experiments), or 4 days later (for the shRNA experiments). Imaging medium was similar to growth medium, but consisted of DMEM without phenol red, supplemented with 10% (v/v) HEPES pH 7.4. The imaging setup consisted of an Olympus IX-70 inverted microscope with a 60× (TIRF) objective (Olympus), a T.I.L.L. photonics TIRF condenser (T.I.L.L. photonics, Gräfelfing, Germany), two solid-state lasers (Laser Quantum, Stockport, UK) emitting at 473 and 532 nm, an Andor Ixon 887 EMCCD camera (Andor, Belfast, Northern Ireland), and a dual-view beam-splitting device (Optical Insights, Roper Bioscience, Tuscon, AZ). The equipment was controlled by Metamorph software (Molecular Devices, Downingtown, PA). The decay constant for the evanescent field was calculated according to Lang et al. [26] as 141 nm. The microscope was enclosed in a custom-built temperature-controlled acrylic-glass cage that was set to 32°C, both to provide the cells with adequate temperature and to minimize focus drifts.
Data analysis
Image processing, assemblies identification and tracking
Time-lapse images were pre-processed by subtracting from each image a low-passed version of itself (1/μm spatial frequency) and smoothing the resulting image by low-pass filtering with a spatial frequency of 0.2/μm. Assemblies were identified as diffraction-limited objects that were significantly brighter than the set fluorescence threshold. Tracking was performed using a MATLAB (The MathWorks Inc., Natick, MA) procedure by fitting each assembly with a 2D Gaussian distribution to precisely identify the assembly’s coordinates (for details see [27]).
For the measurements of lifetime and mobility of HIP1 assemblies, time-lapse images were acquired at 0.5 Hz for 4 min. We omitted assemblies that appeared for less than 4 s, assemblies for which we could not establish a start or end point, and those that collided with adjacent assemblies. Tracks were confirmed manually.
Curve fittings
A log-normal distribution was fitted to each data set using MATLAB’s statistical toolbox (MATLAB, The MathWorks Inc., Natick, MA), and the statistical parameters were calculated for each distribution. We estimated the existence of two subpopulations of clathrin assemblies and one population of HIP1 assemblies. One of the populations overlapped with HIP1 (by a fraction q, and corresponding to the degree of colocalization between clathrin and HIP1), and the second subpopulation (CSP; 1 − q) did not colocalize with HIP1. We assumed that the distribution of clathrin’s overall population would thus be equal to the sum of the HIP1 population and the second CSP with a ratio between them of q:(1 − q), respectively. Then we calculated the probability density function of CSP assemblies according to: 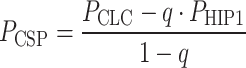 and estimated the statistical parameters of the CSP distribution.
and estimated the statistical parameters of the CSP distribution.
Colocalization calculations
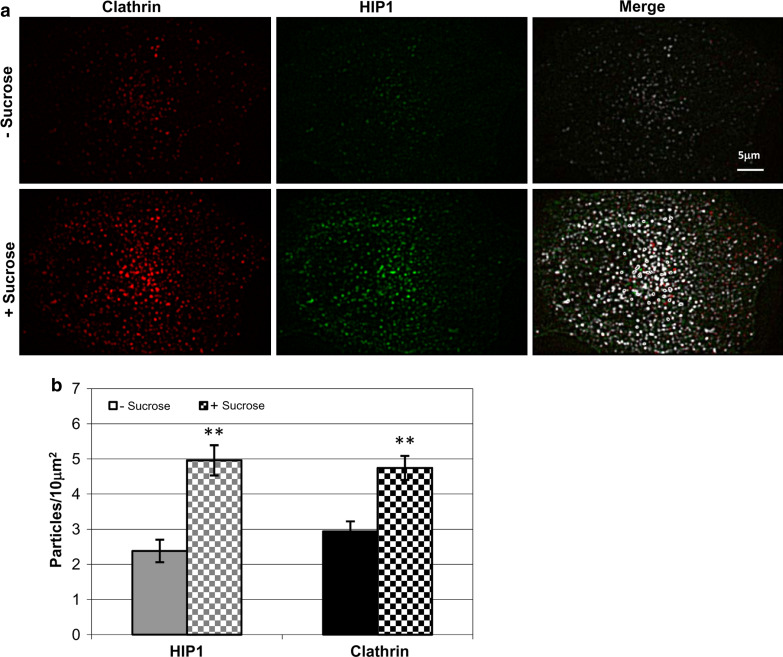
To determine colocalization percentages, we recorded both movies simultaneously, identified the assemblies in each movie separately and then compared the localization of the assemblies in the two movies. We set a maximum distance of 250 nm between the centers of the assemblies to define colocalization. As a control, we flipped HIP1’s images by 180°, and got only ~10% colocalization with the corresponding clathrin image. The merged images, indicating colocalization, in the sucrose experiment (Fig. 4a), were created using the colocalization plugin of WCIF-ImageJ (for details see http://www.uhnresearch.ca/facilities/wcif/imagej/colour_analysis.htm).
Fig. 4.
Hypertonic sucrose solution increases the density of HIP1 and clathrin assemblies. a Filtered TIRF images of a cell coexpressing tdTomato-CLC and EGFP-HIP1, before (upper panel) and after (lower panel) sucrose treatment. Merged images show colocalized pixels in white and others in their relevant color (clathrin in red, HIP1 in green). b Changes in assembly density following sucrose application (mean ± SEM, n = 5, P < 0.005, paired t-test)
Calculations of changes in fluorescence
Cells were imaged by TIRF and recorded at 0.5 Hz for a period of 4 min. The fluorescence-change index was calculated through a series of simple arithmetic manipulations on the images. Briefly, the images were normalized to reduce intensity changes resulting from laser fluctuations. Then, an average intensity value was calculated for the movie, and all pixels that were consistently above that value, i.e., constant elements, were zeroed. The intensity of every pixel was compared to the corresponding one taken from averaging 8–10 frames ahead (HIP1 assemblies’ half life), and if there was a significant change, it was scored 1; otherwise, it was scored 0. These images were subjected to an additional noise-reduction step, where single pixels were erased and only those in the assemblies remained. The changes were summed for each pixel throughout the movie, and a summary image was created. From that image, the fluorescence-change index was calculated as the average value of the highest 10% of the pixels. This index represented a rough estimation of the dynamics of the movie, i.e., the appearance, disappearance, movement and flickering of assemblies.
Clathrin coat manipulations
Disassembly protocol
COS7 cells expressing fluorescently tagged HIP1 and/or CLC were imaged by TIRF. Images were recorded at 0.33 Hz for a period of 6 min. The first 30 s were recorded as a baseline, and then the bath solution was replaced with 2% (v/v) 1-butanol (dissolved in imaging medium) for 1.75 min, after which it was washed away with imaging medium. Images were processed for analysis as described above.
Assembly protocol
COS7 cells coexpressing tdTomato-CLC and EGFP-HIP1 were imaged by TIRF for both fluorophores simultaneously (as described above) and recorded at 0.33 Hz for 6 min. The first 30 s were recorded as a baseline, and then the bath solution was replaced with 0.45 M sucrose (dissolved in imaging medium) for 1.75 min, after which it was washed with imaging medium. Images were processed for analysis as described above. Potassium depletion was achieved as described by Larkin et al. [28]. Briefly, cells were washed with potassium-depleted medium (50 mM Hepes, 100 mM NaCl, pH 7.4), incubated in hypotonic solution (50% DMEM in water) for 5 min and then incubated in potassium-depleted medium for 30 min. Cells expressing TfR-pHl were also washed with potassium-depleted medium in which 50 mM NaCl was replaced with 50 mM NH4Cl before images were taken.
pH-Dependent changes in TfR-pHl
COS7 cells coexpressing tdTomato-HIP1 and TfR-pHl were imaged by TIRF for both fluorophores simultaneously (as described above) and recorded at 0.5 Hz for 4 min. For pH 7.4, bath solution was the imaging medium; for pH 5.5 the solution contained (in mM): NaCl 135, KCl 5, CaCl2 2, MgCl2 0.4, d-glucose 1 and MES 20, with pH adjusted to 5.5. Images were processed for analysis as described above.
Results
To date, a putative role for HIP1 in the early stages of CME has mostly been inferred from binding experiments and in vitro clathrin-assembly assays [6, 29]. However, these experimental approaches do not allow for the characterization of the spatio-temporal parameters of HIP1 recruitment to the coated pit. To investigate the dynamics of HIP1 in the initial stages of CME in vivo, and to correlate them with those of additional components of the CME pathway, we employed total internal reflection fluorescence (TIRF) and live-cell imaging.
HIP1 shows strong colocalization with clathrin at the plasma membrane (PM)
Previous publications have shown colocalization of HIP1 and clathrin [6]; however, detection was performed by confocal microscopy in sections that are usually about 0.7–1 μm thick. In flat fibroblast cells such as COS7, such sections can contain up to 30% of the cell’s depth (data not shown), making it difficult to define whether the colocalization between HIP1 and clathrin is on the PM or in cellular trafficking vesicles [30]. Therefore, we first looked at the localization of fluorescently tagged HIP1 and clathrin (clathrin light chain A, CLC) using dual-view TIRF microscopy. This technique enables visualization of only the lower 200 nm of the cells [31] and thus enables better resolved detection of HIP1 at the PM. Note that even in this thin a section, it is possible to find some inner cell compartments [30, 32]; nevertheless, it provides one of the better representations of PM processes.
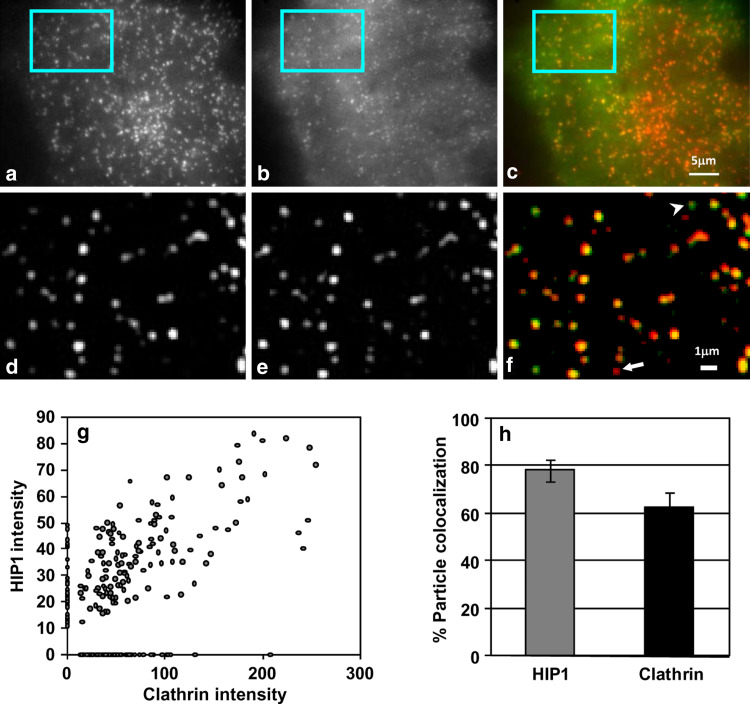
TIRF imaging of COS7 cells expressing EGFP-HIP1 showed that in addition to diffuse staining, EGFP-HIP1 shows a punctuate distribution at the PM. This punctate staining exhibits strong colocalization with clathrin assemblies (Fig. 1a–f). To measure the extent of colocalization, we detected EGFP-HIP1 and clathrin-tdTomato assemblies in cells coexpressing these proteins (n = 171 and 212, respectively) and defined colocalized assemblies as those with adjacent centers (less than 250 nm apart). Based on this calculation, at steady state, we found that 80% (±4.03%) of the HIP1 assemblies colocalized with clathrin and that 63% (±5.33) of the clathrin assemblies colocalized with HIP1 (Fig. 1g, h). In addition, we found the density of both proteins’ assemblies on the PM to be very similar (2.4 ± 0.28 assemblies/10 μm2 for HIP1 and 2.9 ± 0.25 assemblies/10 μm2 for clathrin). This observation demonstrates that HIP1 colocalizes with clathrin assemblies on the PM and suggests HIP1’s involvement in the early stages of CME.
Fig. 1.
HIP1 is colocalized with clathrin at the PM. TIRF images of a cell coexpressing tdTomato-CLC (a) and EGFP-HIP1 (b). c Merged image (clathrin in red, HIP1 in green). d–f Enlarged and filtered images of the regions marked in a–c, respectively. Note that most assemblies are colocalized, though some HIP1-only (arrowhead) or clathrin-only (arrow) spots can be found. g Scatter plot showing the correlation between HIP1 and clathrin assemblies intensity (n = 171 and 212, respectively). h Averaged assemblies colocalization of HIP1 and clathrin shows strong colocalization with clathrin at the PM (mean ± SEM, regions from five cells)
HIP1 dynamics at the PM
The dynamics of clathrin in the formation of coated pits/vesicles have been well studied and are described in several papers [24, 33, 34]. To study the dynamics of HIP1 and determine the spatio-temporal parameters of its recruitment to the coated pit and to correlate these parameters with those of clathrin, we followed EGFP-tagged HIP1 or EGFP- or DsRed-tagged CLC under TIRF illumination for 4 min. Using a tracking software [27], we were able to detect single fluorescent assemblies representing either EGFP-CLC and CLC-DsRed or EGFP-HIP1 clusters, and we followed their location, intensity, appearance and disappearance.
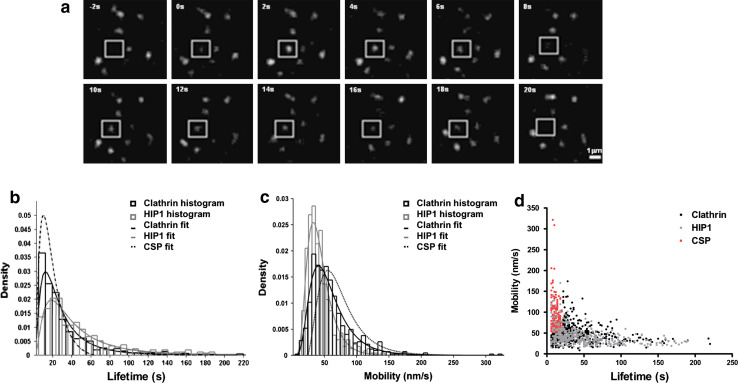
Initially, the lifetime of the assemblies was examined. Only assemblies that appeared and disappeared during the recording period (Fig. 2a) were included in the analysis (about 57% of the clathrin and 44% of the HIP1 assemblies). The lifetimes of the HIP1 and CLC assemblies had similar distributions (Fig. 2b). Median lifetimes were 22 s for CLC assemblies and 34 s for HIP1 assemblies. Taking into consideration the high degree of colocalization among clathrin and HIP1 (Fig. 1h), two possible scenarios can explain the difference in lifetimes; either HIP1 is present for a longer time period in the structures where it colocalizes with clathrin, or the lifetime distribution may not stem from a uniform population of assemblies, in which case only a subpopulation may show colocalization and/or similar dynamics. To discern among these possibilities, we focused our analysis on the predominant category of HIP1 assemblies (with lifetimes of 24–34 s) and compared the time elapsed from initial detection to the achievement of maximum intensity in HIP1 and clathrin pits (clathrin n = 53; HIP1 n = 65). We found that on average, both proteins took the same time to reach peak intensity and then disappear (time to peak—clathrin: 16.18 ± 1.04 s, HIP1: 16.92 ± 0.8 s; time from peak to disappearance—clathrin: 13.62 ± 0.94 s, HIP1: 13.26 ± 0.83 s). The two groups were statistically similar. We therefore hypothesized that clathrin assemblies contain two subpopulations: one that is very similar to that of HIP1 and another that has much shorter lifetimes. To examine this possibility, we fitted our data with log-normal distribution functions (Fig. 2b) and found that clathrin assemblies' distribution could indeed be described as a combination of two subpopulations (see Materials and methods for details). Interestingly, the contribution of each population agreed with our colocalization findings: 60% of the clathrin assemblies shared similar characteristics with HIP1 and 40% belonged to a different clathrin subpopulation (CSP) with shorter lifetimes. Based on that fit, the median lifetime of the shorter lived population was predicted to be 15.2 s, while the median lifetime of the longer lived population was similar to that of HIP1 (34.8 s).
Fig. 2.
HIP1 and clathrin display similar dynamics at the PM. a An example of a short-lived (18 s) HIP1 assembly (box). b Histograms displaying the lifetime distributions for the two proteins. Curves represent log-normal fittings for the data. Hatched curves represent the fit for the predicted shorter lived subpopulation of clathrin assemblies (CSP) (n = 661 and 439; from 10 and 11 cells for clathrin and HIP1, respectively). c Histograms displaying the point-to-point velocity distribution for the two proteins. Curves represent log-normal fittings for the data. Hatched curves represent the fit for the predicted faster-moving subpopulation of clathrin assemblies (CSP). d Correlation plot displaying assemblies velocity vs. lifetime. Note clathrin’s subpopulation of assemblies (CSP) with shorter lifetimes and greater mobility
Clathrin assemblies may differ in their mobility on the plane of the membrane [24, 35, 36]. To further characterize HIP1 and clathrin assemblies identified by our tracking algorithm, we calculated the point-to-point velocity of the assemblies by following their x–y displacements over time. Here too, differences were observed in the median point-to-point velocities of clathrin assemblies (51 nm/s) and HIP1 (38 nm/s) (Fig. 2c).
Based on our identification of two subpopulations of clathrin assemblies that differed in their lifetimes, we hypothesized that the differences observed in the mobility of the assemblies may be an additional indicator of their sub-classification. To test this hypothesis, we performed a similar statistical analysis and found that the distribution of the averaged point-to-point velocity exhibited by clathrin assemblies could be fitted into two subpopulations: one that is similar to HIP1 and another with greater mobility (a median point-to-point velocity of 55.7 nm/s, Fig. 2c).
Having identified subpopulations of assemblies that demonstrate differences in lifetime and mobility, we proceeded to correlate these characteristics in order to identify the nature of the different structures. Displaying all assemblies on a lifetime-velocity correlation graph, we demonstrated that the assemblies with shorter lifetimes are more mobile than longer lived ones. Using the statistical parameters calculated from the curve fittings, we found that 29.8% of the clathrin assemblies belong to both the shorter lifetime and higher mobility subpopulations (Fig. 2d, red dots). Based on its behavior, we suggest that this population of assemblies might consist of the abortive pits described by Ehrlich and colleagues [24]. These results show that HIP1 and clathrin assemblies with a lower degree of mobility, longer lifetimes and nucleation-growth type of behavior present nearly identical dynamics and suggest that HIP1 is localized to active clathrin pits.
HIP1 is part of the clathrin coat structure
Based on the colocalization of HIP1 and clathrin in distinct puncta and on the similarities of the dynamic parameters of HIP1 assemblies with those of a main class of clathrin assemblies, we hypothesized that HIP1 is part of the structure of the clathrin coat. This structure can be manipulated by changing the environmental conditions, resulting in its reversible assembly or disassembly. To determine whether HIP1 is part of the clathrin coat, we challenged the cells with such treatments and followed the dynamics of HIP1 and clathrin.
Application of primary alcohols to live cells causes reversible disruption of the PLD pathway, resulting in depletion of PIP2 from the PM [37]. This, in turn, causes the disassembly of existing clathrin-coated pits and prevents the creation of new ones. Washing away the alcohol relieves the effect and enables reassembly of the coated pits at a higher density, which returns to normal shortly thereafter [35]. We hypothesized that the depletion of PIP2 would affect HIP1 in a similar fashion for two reasons: first, HIP1’s localization at the PM could be dependent on the presence of clathrin, in which case clathrin disassembly would release HIP1 as well; second, HIP1 localization at the PM could be dependent on its ANTH domain, which binds PIP2.
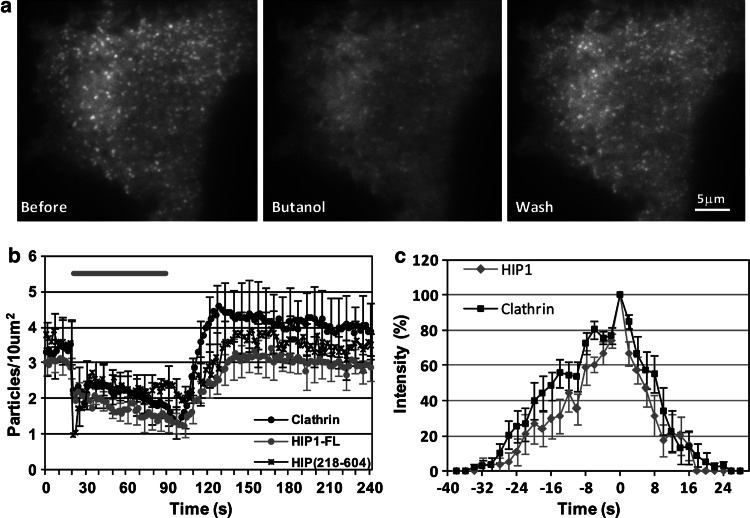
We therefore examined the effect of 1-butanol application and washout on cells expressing tdTomato-CLC or EGFP-HIP1. As predicted, similar to clathrin, HIP1 assemblies faded as 1-butanol was applied and reappeared concomitant with the wash (Fig. 3a). Quantitative analysis of assembly density further supported this observation, though the recovery of HIP1 assemblies seemed to be a bit slower than that of clathrin and lacked the ‘overshoot’ of the latter following the wash (Fig. 3b). This scenario is in line with our previous findings, which indicate that different subpopulations of clathrin assemblies, containing HIP1 or not, coexist at the PM. To further confirm the concomitant presence and similarity of the dynamics of recruitment of clathrin and HIP1 in individual pits, we made use of the ability of 1-butanol to synchronize pit creation as many clusters are formed when 1-butanol is washed away. Accordingly, we applied the 1-butanol incubation/wash procedure to cells coexpressing tdTomato-CLC and EGFP-HIP1. We followed the changes in fluorescence intensity of single assemblies labeled with both proteins using dual TIRF measurement, normalized them to their peaks and overlaid them (Fig. 3c). We found that both proteins arrive and disappear simultaneously. These results clearly demonstrate that HIP1 is a component of the clathrin coat and is recruited at early stages of the coat formation at about the same time as clathrin.
Fig. 3.
Application of 1-butanol dissociates HIP1 from the PM. a Example of changes in TIRF images of HIP1 following 2% butanol application and washout. b Quantification of the changes in assemblies number in response to butanol treatment (clathrin: n = 5; HIP1: n = 7; HIP1218–604: n = 5). Gray bar represents time of butanol application. (Error bars presented for 1/3 of the points.) c Averaged changes in normalized intensity of individual HIP1 and clathrin assemblies that appeared after butanol washout. Minimal intensity for detection was usually about 25%. t = 0 represents maximal intensity (n = 10 pairs)
The observed disassembly of HIP1 clusters upon incubation with 1-butanol and its re-assembly after the 1-butanol washout paralleled those of clathrin both at the population level (Fig. 3b) and at the level of the individual pit (Fig. 3c). This similarity in dynamics did not allow us to address the question of from what the assembly and disassembly of HIP1 is derived in the conditions employed: a direct affinity for PIP2 or a recruitment based on the affinity to other coated pit components such as clathrin itself.
To examine whether the N-terminus ANTH domain of HIP1 is responsible for its interaction with the membrane, we used a truncated form of HIP1 (HIP1218–604), containing only the central domain (amino acids 218–604), which can still bind clathrin without the phospholipid- and actin-binding domains [6]. Despite the loss of its PIP2-binding domain, this fragment can be found at the lower surface of the cell, in small, dynamic clusters, as the full-length HIP1 protein (HIP1-FL) (Supplementary Fig. 1A). However, unlike HIP1-FL, HIP1218−604 is also localized to big, yet unidentified cellular structures [6]. Furthermore, the diffraction limited puncta containing HIP1218−604 showed a high degree of colocalization with clathrin, similar to HIP1-FL (Supplementary Fig. 1B). The PM HIP1218–604 clusters’ response to 1-butanol was similar to that of HIP1-FL clusters (Fig. 3b), implying that HIP1 can localize to clathrin assemblies at the PM independently of its ANTH domain, possibly via its binding to clathrin.
To further characterize the recruitment of HIP1 to endocytic structures, we submitted cells coexpressing tdTomato-CLC and EGFP-HIP1 to hypertonic shock, a condition that induces the formation of clathrin cages. Clathrin cages are structures that retain the overall architecture of a coated pit/vesicle but are devoid of membranes and cargo [38]. Thus, recruitment to these structures is indicative of protein–protein interactions with additional components of the clathrin coat.
Application of 0.45 M sucrose to cells coexpressing tdTomato-CLC and EGFP-HIP1 resulted in a correlated increase in the intensity of both proteins as detected by TIRF microscopy (Fig. 4a). Assembly density of both proteins doubled, from 2.4 and 2.9 assemblies/10 μm2 to 4.9 and 4.7 assemblies/10 μm2 for HIP1 and clathrin, respectively (Fig. 4b, paired t-tests, p < 0.005). Moreover, these assemblies showed a high degree of colocalization (Fig. 4a, merged images). To strengthen our observation that HIP1 is recruited to microcages, we repeated the experiment, this time generating microcages by potassium depletion [28, 38]. Similar to the hypertonic shock experiment, we detected an overall intensity increase of fluorescence signal at the TIRF plane irrespective of the fusion protein (not shown). This could result from changes in cell shape and size that can accompany these manipulations. To specifically address HIP1’s recruitment into microcages, we made use of their nature as defined diffraction limited objects and compared the density of particles prior to or following treatment. As control, we used fluorescently labeled transferrin receptor (TfR). Similar to the hypertonic shock experiment, upon potassium depletion, HIP1 and clathrin were recruited to defined puncta at the plasma membrane (averaged density increase 2.12 ± 0.23, n = 8 and 2.02 ± 0.14, n = 19 for HIP1 and clathrin, respectively). In contrast, no increase in the density of fluorescently labeled puncta was observed for transferrin receptor following similar treatment (averaged density increase 1.22 ± 0.1, n = 15).
Taken together, the experiments described above show that the recruitment and disassembly of HIP1 parallels that of clathrin; a HIP1 fragment lacking membrane and cytoskeleton binding motifs, but containing its clathrin-binding motif showed similar tendencies to the full-length protein and HIP1 is recruited to clathrin cages devoid of membranes and cargo, all of which suggests that protein–protein interaction with clathrin and possibly other coat components are crucial to the recruitment, localization and function of HIP1.
Clathrin silencing alters the association of HIP1 with the membrane
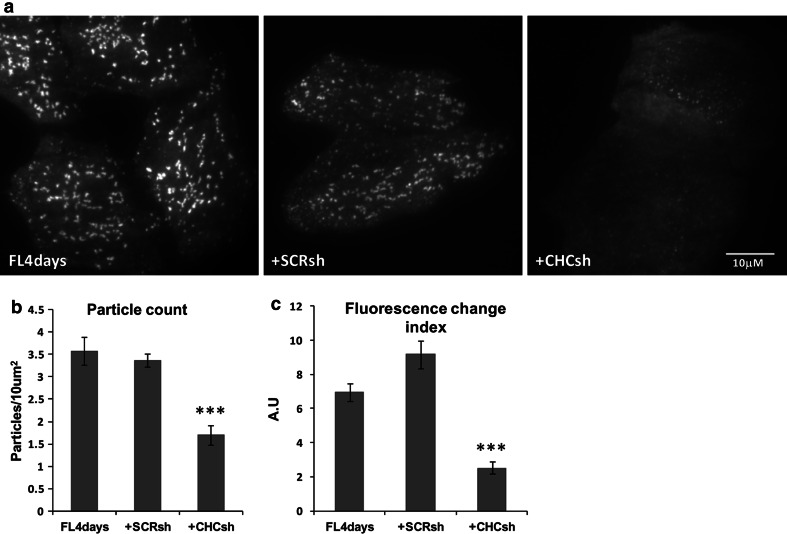
To directly address the influence of clathrin levels on the degree of membrane recruitment of HIP1, we employed shRNA to knockdown clathrin expression. Clathrin content in HeLa cells was reduced by 70% using CHC-shRNA, as determined by immunofluorescence analysis (data not shown). Cells expressing EGFP-HIP1, EGFP-HIP1 with CHC-shRNA or EGFP-HIP1 with scramble-(SCR)-shRNA were imaged by TIRF and analyzed (Fig. 5). We found that down-regulation of CHC causes a twofold reduction in the density of HIP1 assemblies (Fig. 5b), from ~3.5 assemblies/10 μm2 in the control groups to ~1.7 assemblies/10 μm2 in the clathrin-silencing group (for the three groups, one-way ANOVA P < 0.0001; for pairs testing, Tukey’s multiple comparison test: control and SCR-shRNA, P > 0.05, control and CHC-shRNA, P < 0.001, SCR-shRNA and CHC-shRNA, P < 0.001). In addition, we analyzed the recordings using an algorithm that gives scores for changes in fluorescence, indicating movement, flickering, and appearance or disappearance of the assemblies, and ignores the constant elements in the movie. While control (EGFP-HIP1 only) and EGFP-HIP1+SCR-shRNA-expressing cells scored ~7 and ~9 on our index (statistically insignificant), EGFP-HIP1+CHC-shRNA cells got a score of ~2.5 (Fig. 5c; for the three groups, Kruskal-Wallis test, P < 0.0001; for pairs testing, Dunn’s multiple comparison test: control and SCR-shRNA, P > 0.05, control and CHC-shRNA, P < 0.001, SCR-shRNA and CHC-shRNA, P < 0.001). Note that the effects that we measured are probably underestimated, as the cells still have some residual clathrin that might be responsible for some normal activity. These observations suggest that the presence of the clathrin coat is important for either recruitment or stabilization of HIP1 at the membrane, and in its absence, HIP1 is less localized to the PM. Moreover, this rules out a function for HIP1 in the recruitment of clathrin to the membrane.
Fig. 5.
Clathrin silencing reduces the number of HIP1’s assemblies at the PM. a TIRF images of HeLa cells expressing either HIP1-GFP (FL4days) alone, or in combination with scramble shRNA (SCRsh) or clathrin shRNA (CHCsh). b Assembly density is displayed for the three groups (mean ± SEM, regions from 13, 14 and 18 cells, respectively, P < 0.0001, one-way ANOVA). c Fluorescence changes were scored as an indication of vesicle activity (appearance, disappearance and flickering) (mean ± SEM, regions from 37, 36 and 41 cells, respectively, P < 0.0001, Kruskal-Wallis test; difference between FL4days and SCRsh was not significant according to Dunn’s comparison test)
HIP1 localizes with transferrin receptors only at the early stages of CME
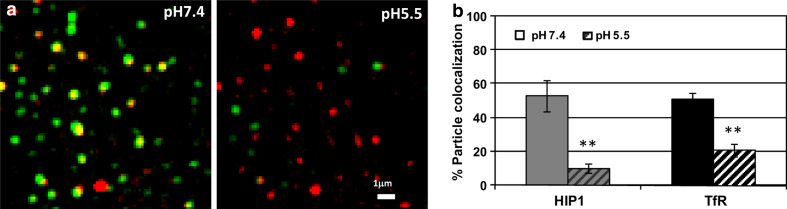
Visualization (by TIRF) together with tracking of fluorescently tagged proteins of the clathrin coat components in dynamic diffraction limited spots is a sensitive and adequate technique to characterize their recruitment to individual coated pits. However, this methodology alone cannot exclude the localization of these coat components to recently internalized vesicles, which retain their proximity to the PM and can thus still be visualized by TIRF [30, 32]. To distinguish between coated pits and internalized vesicles, we used a construct expressing transferrin receptor fused to pHluorin (a pH-sensitive variant of GFP; TfR-pHl) [25]. The pHluorin is located in the extracellular part of the receptor and thus shows fluorescent signal on the PM as long as it is exposed to the bath solution (pH 7.4). Upon internalization, the vesicles acidify and the TfR-pHl fluorescence gradually disappears. Changes in external pH can be utilized to distinguish between open pits and closed vesicles, i.e., changing the bath pH from 7.4 to 5.5 will quench the fluorescence of the TfR-pHl on the membrane, but will not affect the fluorescent signal coming from endocytic vesicles that have just formed (for further details, see [25]).
We coexpressed tdTomato-HIP1 and TfR-pHl in the cells and followed both proteins under TIRF illumination while changing the bath solution pH (Fig. 6a). Under normal pH conditions, about 58% of the EGFP-HIP1 assemblies colocalized with transferrin receptors, and 57% of TfR-pHl colocalized with EGFP-HIP1 (Fig. 6b). Upon lowering the pH, most of the TfR-pHl staining was lost, and there were markedly fewer assemblies representing those that had been recently internalized. While TfR assembly density was ~2.6 assemblies/10 μm2 at physiological pH, it dropped to ~1.1 assemblies/10 μm2 at the lower pH. These leftover vesicles were much more mobile than the membranal assemblies, and their fluorescence rapidly dimmed, supporting the notion that these are freshly formed endocytic vesicles. In a few cases we could detect HIP1 fluorescence on these vesicles (only for the first few seconds after the change in pH), but the vast majority did not show any colocalization with HIP1 assemblies (Fig. 6b). We found a similar trend when this experiment was performed with clathrin (data not shown). These results show that HIP1 rarely localizes with internalized transferrin receptor and that it is shed from the vesicle close to clathrin uncoating. As such, it is probably a component of the coated pits, but not the internalized endocytic vesicles.
Fig. 6.
HIP1 colocalizes with transferrin receptor (TfR). a A region of a cell coexpressing tdTomato-HIP1 (red) and TfR-pHl (green) was visualized in TIRF, under two pH conditions (left, pH 7.4; right, pH 5.5). Both images are filtered under the same conditions, but scaled differently, so that the low-intensity internalized TfR-pHl vesicles would be visible. b Colocalization of HIP1 and TfR assemblies was measured under the two pH conditions (mean ± SEM, regions from seven cells, P < 0.005, paired t-test)
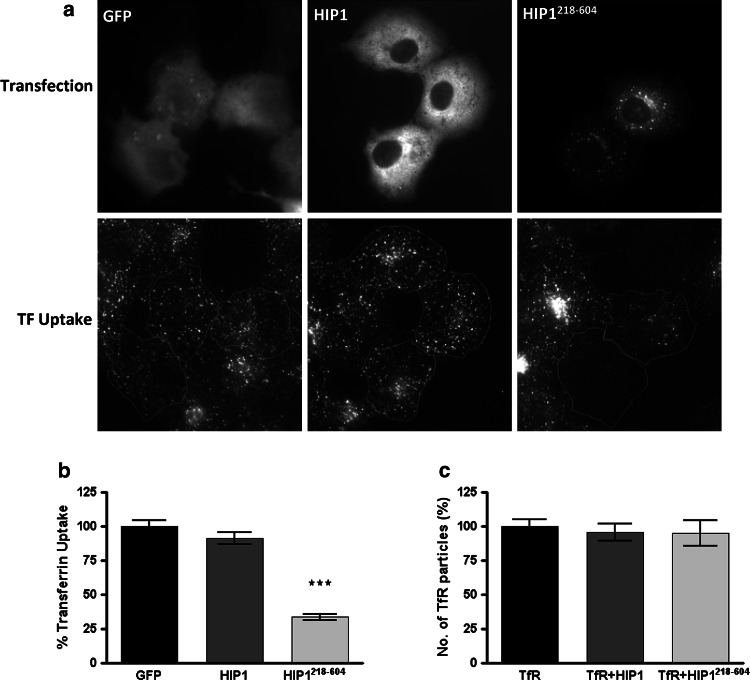
To further pinpoint the stage at which HIP1 functions, we used the HIP1218–604 fragment and investigated at what stage it interferes with endocytosis. We found that HIP1218–604 blocks the uptake of transferrin into COS7 cells (Fig. 7a, b). We then counted the number of transferrin receptor clusters at the PM and found that HIP1218–604 does not change their density (Fig. 7c). Thus, HIP1218–604 does not change the clustering of transferrin receptors, but rather their internalization via coated vesicles. Therefore, HIP1 functions at the late stages of the coated pit maturation step or the formation of the coated vesicles.
Fig. 7.
Expression of HIP1218–604 impairs transferrin uptake, but does not affect the appearance of membranal transferrin receptor assemblies. a COS7 cells expressing either GFP, HIP1-GFP or HIP1218–604-GFP were incubated with 15 μg/ml transferrin-alexa546 for 5 min. Cells expressing the proteins are outlined in the corresponding transferrin uptake image. b Quantification of transferrin accumulation in cells described in a. Bars represent mean ± SEM. Number of cells analyzed: 102, 82 and 74 for GFP, HIP1 and HIP1218–604, respectively, from two independent experiments. P < 0.001. c Transferrin receptor assemblies were counted in TIRF images of cells expressing the TfR-pHlu alone or in combination with HIP1 or HIP1218–604 and found to be similar in these conditions. Bars represent mean ± SEM. Three experiments, total number of cells analyzed 15, 23 and 20 for GFP, HIP1 and HIP1218–604, respectively
Discussion
HIP1, HIP1R and Sla2p form a family of proteins that are involved in clathrin-mediated endocytosis and vesicular trafficking. Based on their sequence and domain similarities, they are frequently related to HIP1/R or HIP1R/Sla2p proteins. However, several lines of evidence point to the fact that in higher eukaryotes these proteins perform non-overlapping functions: (1) The phenotype of the knockout of HIP1 and HIP1R in mice differs, with HIP1 knockouts showing spinal, hematopoietic and testicular defects [39, 40], while HIP1R knockouts grow and develop normally [16]; (2) even though both HIP1 and HIP1R bind clathrin trimers, they do so through different molecular motifs [9, 22, 23], suggesting different mechanisms of regulation. In this work we provide the first quantitative measurements of HIP1 dynamics in live cells and demonstrate that HIP1 plays a role in early steps of endocytosis and differs in several aspects from HIP1R and Sla2p. Thus, HIP1 is recruited and dissociated from pits in parallel to clathrin in untreated cells as well as in sucrose and 1-butanol-treated cells. Moreover, using transferrin receptor conjugated to pHluorin, we located HIP1 to open pits, and not to internalized vesicles. Furthermore, the use of HIP1218–604 as a dominant negative suggests that HIP1 functions at the formation of coated vesicles. In addition, we show that HIP1s’ localization to the PM depends significantly on its interaction with the clathrin coat and to a lesser extent on its ANTH domain as HIP1218–604 that lacks the ANTH domain still associates with the clathrin coat. Nevertheless, the ANTH probably enhances the specificity of HIP1 to a specific population of clathrin structures, as the dynamics of HIP1218–604 parallel those of two clathrin subpopulations (classified according to their lifetime), while full-length HIP1 is probably mainly associated with active clathrin pits (the longer lived clathrin structures). In addition, we found that knocking down clathrin results in less HIP1 localization to the PM, which is in contrast to the recruitment of Sla2p to the PM after clathrin knock down in yeast. These findings place HIP1 as an early component of the CME pathway and suggest that its PM recruitment depends on the presence of a clathrin coat. From the results of the present study, we devised a model of the main steps of HIP1’s involvement in clathrin-mediated endocytosis. This model is illustrated in Fig. 8: HIP1 is recruited to coated pits in parallel to clathrin; this is in contrast to Sla2p, which is recruited later than clathrin [17, 18], but is similar to HIP1R [22]. HIP1 dissociates from the coated pit together with clathrin and is hardly found on coated vesicles. Finally, HIP1218–604 blocks endocytosis following coated pit formation, suggesting that HIP1 functions in the last steps of coated pit maturation and the formation of coated vesicles.
Fig. 8.
A schematic representation of HIP1’s involvement in the initial steps of clathrin-mediated endocytosis. HIP1 dimers are recruited to coated pits, parallel to clathrin and HIP1R [22] arrival. HIP1’s actin-binding capabilities are controversial, therefore marked by a question mark in the scheme. HIP1 dissociates from the coated pit together with clathrin, and it is hardly found on coated vesicles. Additionally, HIP1218–604 blocks endocytosis following coated pit formation, suggesting that HIP1 functions in the last steps of coated pit maturation and the formation of coated vesicles
HIP1 functions in the initial steps of CME
A main objective of this study was to characterize the spatio-temporal parameters of the recruitment of HIP1 to individual plasma membrane-localized clathrin-coated pits. To enable such characterization we opted for TIRF microscopy, which presents the advantage of high resolution in the z-axis and a high signal-to-noise ratio for signals embarking from the PM. Furthermore, we have centered our analysis on diffraction limited objects, which appear and disappear in the course of the time lapse and present for at least three successive frames (6 s). In addition, similarly to a number of live-cell microscopy studies aimed at characterizing the endocytic pathway, we opted for transient expression of fluorescently tagged proteins in COS7 cells as our experimental system [33, 41]. The structures of clathrin and HIP1 identified in this study show dynamics that are very similar to those previously described for clathrin-coated pits by several groups [24, 33, 34]. Interestingly, we could distinguish between two subpopulations of clathrin assemblies: a larger subpopulation (about 60% of the assemblies based on our colocalization results and the statistical calculation) that shares similar dynamics with HIP1 and a smaller subpopulation (less than 40%) that has shorter lifetimes and greater mobility. This latter population might be the abortive pits described by Ehrlich et al. [24]. However, our predicted median lifetime for this population (15.2 s) was shorter than that described in their paper (16–28 s), and the relative percentage was larger in our analysis (40% vs. 25%). These discrepancies could be partially explained by differences in the experimental setups and sampling rates, but mainly by the minimal lifetime considered in each of the experiments. While Ehrlich et al. [24] included assemblies with lifetimes starting at 16 s, we analyzed assemblies with lifetimes as short as 6 s, which inevitably shifts all of the results to smaller values and increases the relative proportion of these assemblies in the general population. Another possible explanation for the shorter lived clathrin population is the presence of inner cell compartments, such as endosomes, at the TIRF level, as has been shown by Keyel et al. [30]. However, this explanation is less likely because of the relatively high percentage of this population in our data. Assuming that the longer lived clathrin population is the active one, it follows that HIP1 is only found in the active pits and might even be their stabilizing factor. This, however, still needs to be experimentally proven. Nonetheless, the finding that HIP1218–604 inhibits internalization of transferrin without interfering with the initial nucleation of pits (Supplementary Fig. 1) or the concentration of cargo (Fig. 7) supports this notion. These observations narrow down HIP1’s function to the maturation of coated pits into coated vesicles.
In addition, utilizing the pH sensitivity of the TfR-pHl fusion protein to distinguish between open coated pits and internalized vesicles, we found HIP1 colocalized with TfR-pHl on open pits, but we could not find it in the internalized vesicles. The same behavioral pattern was observed for clathrin, suggesting that HIP1 disappearance occurs very close to clathrin uncoating from the vesicle. These observations, together with the inhibition of endocytosis by HIP1218–604 fragment, narrow down HIP1’s function to the maturation of coated pits into coated vesicles.
Taken together, we demonstrate that EGFP-HIP1 assemblies colocalize at the PM with clathrin only at the very early steps of CME (Fig. 8). Moreover, these experiments support the concept of HIP1 as a structural coat protein, rather than a cargo-related or later transport protein, as had been previously suggested [12, 42].
What recruits HIP1 to the coated pit?
HIP1 is a multi-domain protein that is endowed with a number of protein–protein and protein–lipid interaction motifs that may mediate its recruitment to the nascent coated pit. To dissect the influence of different interactions we have employed two complementary approaches: (1) treatments that either abrogate or force clathrin assembly through different means; (2) genetic manipulation of HIP1 generating a fragment devoid of its ANTH and talin-like domains.
The abrupt perturbation to the ability of PLD to produce phosphatidic-acid entails a drop in PIP2 and the dispersal of clathrin and AP2 [35]. As expected, HIP1 behaved in a similar manner, indicating that either its ability to directly bind PIP2 is abrogated or that the 1-butanol dispersal of HIP1 interacting proteins leads to its dislodgment from the membrane. Furthermore, upon 1-butanol washout the renewed recruitment of HIP1 to the membrane also paralleled that of clathrin, still preventing the discrimination between these two possibilities. However, three independent lines of evidence point to the role played by protein–protein interactions in general, and clathrin-HIP1 interaction in particular, in the recruitment of HIP1 to the PM: (1) hypertonic shock leads to the recruitment of HIP1 to clathrin microcages, which are devoid of membranes and cargo (Fig. 4); (2) clathrin knockdown by shRNA reduces the amount of HIP1 at the PM (Fig. 5); (3) HIP1218–604, a fragment of HIP1, devoid of its lipid-binding domain (ANTH) and its cytoskeleton association domain (talin-like), is still recruited to coated pits (Supplementary Fig. 1) in untreated cells and upon 1-butanol washout (Fig. 3b). Importantly, the membrane recruitment of this fragment is also abrogated by 1-butanol, indicating that this sensitivity is not dependent on direct lipid binding and probably occurs as a result of protein–protein interaction. A driving factor for the recruitment of HIP1 to the membrane in the absence of its ANTH domain may be the homodimerization of the exogenously expressed fluorescently tagged protein with the endogenous protein. However, although we cannot rule out such a contribution, we propose that the direct interaction with clathrin may be the main driving factor. Support for this notion comes from the findings that a reduction in the amount of clathrin by shRNA reduces the amount of HIP1 recruited to coated pits. Moreover, the recruitment of HIP1218–604 to the coated pit occurs even in cells expressing high amounts of this fragment, the condition in which it would saturate clathrin-binding sites and reduce the probability of the endogenous HIP1 to recruit it through dimerization (data not shown).
It is possible that HIP1 serves in more than one role along the endocytic pathway and that the ANTH domain is needed for some additional function. For example, the binding preferences of HIP1’s ANTH domain, characterized by Hyun and colleagues [14], suggest that HIP1 will bind PIP2 at intracellular vesicles. While Epsin and AP180 bind PI(4,5)P2 and PI(3,4,5)P3 [43, 44], which are enriched at the PM, those authors showed HIP1’s preferential binding to PI(3,4)P2 and PI(3,5)P2 [14], which are enriched in the intracellular vesicles. Their findings support HIP1’s function in later stages of endocytosis, such as EGF receptor degradation post-internalization.
Our results suggest that HIP1 is mainly recruited and/or stabilized at the PM by clathrin or other endocytic proteins. Association of HIP1 with the PM through its ANTH domain might increase the probability for binding clathrin in coated pits or increase the affinity of this interaction, as seen for other endocytic proteins that bind PIP2 [22, 45].
Dynamics of HIP1 are distinct from its yeast homologue Sla2p
Members of the Sla2/HIP1 family share a similar domain structure, and accordingly some of the current views on HIP1’s function are based on results obtained from its yeast homologue, Sla2p. However, in this study we present some discrepancies between HIP1 and Sla2p behaviors.
Newpher and Lemmon [18] described strong recruitment of Sla2p to the membrane and a marked elongation of the Sla2p patches’ lifetimes in CHCΔ cells. In contrast, our results suggest that interfering with clathrin expression in mammalian cells results in reduction of HIP1 at the PM. One possible explanation for this disagreement is HIP1’s ability to directly bind CHC [9], unlike the indirect interaction of Sla2p with CHC through the LC [18]. This indirect interaction was also described for the second mammalian Sla2p homologue, HIP1R [22]. Both Sla2p and HIP1R are missing the clathrin-bindings motifs L(L,I)(D,E,N)(L,F)(D,E), referred to as the “clathrin box,” or the shorter DLL motif found in many proteins that interact with the terminal domain of CHC [46–48]. Moreover, a clear timeline was established for Sla2p, arriving at existing clathrin patches (with a delay of about 25 s) and leaving simultaneously with clathrin during the arrival of the actin-remodeling proteins [17, 18]. Our results show that HIP1 and clathrin appear and disappear simultaneously, as previously described for HIP1R [22]. Thus, we suggest that Sla2p is not a reliable predictor of HIP1 function. Although HIP1R and Sla2p bind CHC through CLC, their PM dynamics are different [18, 22]; thus, each one of these proteins is influenced by different factors that determine its dynamics and steps in endocytosis.
CME involves the assembly of several proteins that interact dynamically with one another. HIP1 is one of the least characterized members of these proteins. In the present study, we provide the first direct and quantitative evidence that HIP1 is part of the clathrin coat and that it is involved in the transition of coated pits to coated vesicles.
Electronic supplementary material
Below is the link to the electronic supplementary material.
HIP1218-604 localizes to small dynamic assemblies at the PM, colocalized with clathrin. A. An example of a short-lived (32 s) HIP1218-604 assembly (box). B. Filtered TIRF images of part of a cell coexpressing HIP1218-604-EGFP and tdTomato-CLC. Merged image demonstrates the high colocalization of the two proteins (HIP1218-604 in green, clathrin in red). (TIFF 651 kb)
Acknowledgments
We thank Prof. Erich Wanker and Dr. Stephanie Waelter for plasmids and discussions during the early stages of this project. We thank Reut Friedrich for her help with the statistical fittings and Ofer Yizhar for providing technical support for the tracking program. This work was supported by a grant from the Israel Science Foundation (grant no. 1211/07 to U.A.). I.G. was supported by the Clore Scholars Program.
References
- 1.Jones AL, Hradek GT, Hornick C, Renaud G, Windler EE, Havel RJ. Uptake and processing of remnants of chylomicrons and very low density lipoproteins by rat liver. J Lipid Res. 1984;25:1151–1158. [PubMed] [Google Scholar]
- 2.Pearse BM. Coated vesicles from human placenta carry ferritin, transferrin, and immunoglobulin G. Proc Natl Acad Sci USA. 1982;79:451–455. doi: 10.1073/pnas.79.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Z, Chen Y, Nissenson RA. The cytoplasmic tail of the G-protein-coupled receptor for parathyroid hormone and parathyroid hormone-related protein contains positive and negative signals for endocytosis. J Biol Chem. 1995;270:151–156. doi: 10.1074/jbc.270.1.151. [DOI] [PubMed] [Google Scholar]
- 4.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 5.Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- 6.Waelter S, Scherzinger E, Hasenbank R, Nordhoff E, Lurz R, Goehler H, Gauss C, Sathasivam K, Bates GP, Lehrach H, Wanker EE. The huntingtin interacting protein HIP1 is a clathrin and alpha-adaptin-binding protein involved in receptor-mediated endocytosis. Hum Mol Genet. 2001;10:1807–1817. doi: 10.1093/hmg/10.17.1807. [DOI] [PubMed] [Google Scholar]
- 7.Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum Mol Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280:6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- 9.Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001;276:39271–39276. doi: 10.1074/jbc.C100401200. [DOI] [PubMed] [Google Scholar]
- 10.Rao DS, Chang JC, Kumar PD, Mizukami I, Smithson GM, Bradley SV, Parlow AF, Ross TS. Huntingtin interacting protein 1 is a clathrin coat binding protein required for differentiation of late spermatogenic progenitors. Mol Cell Biol. 2001;21:7796–7806. doi: 10.1128/MCB.21.22.7796-7806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ybe JA, Mishra S, Helms S, Nix J. Crystal structure at 2.8 Å of the DLLRKN-containing coiled-coil domain of huntingtin-interacting protein 1 (HIP1) reveals a surface suitable for clathrin light chain binding. J Mol Biol. 2007;367:8–15. doi: 10.1016/j.jmb.2006.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson PS, Nagy A, Wang YT, Roder JC, Raymond LA, Hayden MR. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 2003;22:3254–3266. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metzler M, Gan L, Wong TP, Liu L, Helm J, Liu L, Georgiou J, Wang Y, Bissada N, Cheng K, Roder JC, Wang YT, Hayden MR. NMDA receptor function and NMDA receptor-dependent phosphorylation of huntingtin is altered by the endocytic protein HIP1. J Neurosci. 2007;27:2298–2308. doi: 10.1523/JNEUROSCI.5175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyun TS, Rao DS, Saint-Dic D, Michael LE, Kumar PD, Bradley SV, Mizukami IF, Oravecz-Wilson KI, Ross TS. HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J Biol Chem. 2004;279:14294–14306. doi: 10.1074/jbc.M312645200. [DOI] [PubMed] [Google Scholar]
- 15.Rao DS, Bradley SV, Kumar PD, Hyun TS, Saint-Dic D, Oravecz-Wilson K, Kleer CG, Ross TS. Altered receptor trafficking in huntingtin interacting protein 1-transformed cells. Cancer Cell. 2003;3:471–482. doi: 10.1016/S1535-6108(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 16.Hyun TS, Li L, Oravecz-Wilson KI, Bradley SV, Provot MM, Munaco AJ, Mizukami IF, Sun H, Ross TS. Hip1-related mutant mice grow and develop normally but have accelerated spinal abnormalities and dwarfism in the absence of HIP1. Mol Cell Biol. 2004;24:4329–4340. doi: 10.1128/MCB.24.10.4329-4340.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/S0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 18.Newpher TM, Lemmon SK. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Clainche C, Pauly BS, Zhang CX, Engqvist-Goldstein AE, Cunningham K, Drubin DG. A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis. EMBO J. 2007;26:1199–1210. doi: 10.1038/sj.emboj.7601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283:32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154:1209–1223. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17:4343–4352. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Lang T, Wacker I, Wunderlich I, Rohrbach A, Giese G, Soldati T, Almers W. Role of actin cortex in the subplasmalemmal transport of secretory granules in PC-12 cells. Biophys J. 2000;78:2863–2877. doi: 10.1016/S0006-3495(00)76828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yizhar O, Ashery U. Modulating vesicle priming reveals that vesicle immobilization is necessary but not sufficient for fusion-competence. PLoS ONE. 2008;3:e2694. doi: 10.1371/journal.pone.0002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin JM, Brown MS, Goldstein JL, Anderson RG. Depletion of intracellular potassium arrests coated pit formation and receptor-mediated endocytosis in fibroblasts. Cell. 1983;33:273–285. doi: 10.1016/0092-8674(83)90356-2. [DOI] [PubMed] [Google Scholar]
- 29.Mishra SK, Agostinelli NR, Brett TJ, Mizukami I, Ross TS, Traub LM. Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J Biol Chem. 2001;276:46230–46236. doi: 10.1074/jbc.M108177200. [DOI] [PubMed] [Google Scholar]
- 30.Keyel PA, Watkins SC, Traub LM. Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:13190–13204. doi: 10.1074/jbc.M312717200. [DOI] [PubMed] [Google Scholar]
- 31.Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 32.Jaiswal JK, Andrews NW, Simon SM. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol. 2002;159:625–635. doi: 10.1083/jcb.200208154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 34.Rappoport JZ, Simon SM. Real-time analysis of clathrin-mediated endocytosis during cell migration. J Cell Sci. 2003;116:847–855. doi: 10.1242/jcs.00289. [DOI] [PubMed] [Google Scholar]
- 35.Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M. Role of lipids and actin in the formation of clathrin-coated pits. Exp Cell Res. 2006;312:4036–4048. doi: 10.1016/j.yexcr.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris AJ, Frohman MA, Engebrecht J. Measurement of phospholipase D activity. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- 38.Heuser JE, Anderson RG. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khatchadourian K, Smith CE, Metzler M, Gregory M, Hayden MR, Cyr DG, Hermo L. Structural abnormalities in spermatids together with reduced sperm counts and motility underlie the reproductive defect in HIP1-/- mice. Mol Reprod Dev. 2007;74:341–359. doi: 10.1002/mrd.20564. [DOI] [PubMed] [Google Scholar]
- 40.Oravecz-Wilson KI, Kiel MJ, Li L, Rao DS, Saint-Dic D, Kumar PD, Provot MM, Hankenson KD, Reddy VN, Lieberman AP, Morrison SJ, Ross TS. Huntingtin interacting protein 1 mutations lead to abnormal hematopoiesis, spinal defects and cataracts. Hum Mol Genet. 2004;13:851–867. doi: 10.1093/hmg/ddh102. [DOI] [PubMed] [Google Scholar]
- 41.Chetrit D, Ziv N, Ehrlich M. Dab2 regulates clathrin assembly and cell spreading. Biochem J. 2009;418:701–715. doi: 10.1042/BJ20081288. [DOI] [PubMed] [Google Scholar]
- 42.Hyun TS, Ross TS. HIP1: trafficking roles and regulation of tumorigenesis. Trends Mol Med. 2004;10:194–199. doi: 10.1016/j.molmed.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4, 5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 44.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4, 5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 45.Gaidarov I, Keen JH. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dell’Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 47.Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci. 2000;20:8667–8676. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HIP1218-604 localizes to small dynamic assemblies at the PM, colocalized with clathrin. A. An example of a short-lived (32 s) HIP1218-604 assembly (box). B. Filtered TIRF images of part of a cell coexpressing HIP1218-604-EGFP and tdTomato-CLC. Merged image demonstrates the high colocalization of the two proteins (HIP1218-604 in green, clathrin in red). (TIFF 651 kb)