Abstract
The recent release of several basidiomycete genome sequences allows an improvement of the classification of fungal glutathione S-transferases (GSTs). GSTs are well-known detoxification enzymes which can catalyze the conjugation of glutathione to non-polar compounds that contain an electrophilic carbon, nitrogen, or sulfur atom. Following this mechanism, they are able to metabolize drugs, pesticides, and many other xenobiotics and peroxides. A genomic and phylogenetic analysis of GST classes in various sequenced fungi—zygomycetes, ascomycetes, and basidiomycetes—revealed some particularities in GST distribution, in comparison with previous analyses with ascomycetes only. By focusing essentially on the wood-degrading basidiomycete Phanerochaete chrysosporium, this analysis highlighted a new fungal GST class named GTE, which is related to bacterial etherases, and two new subclasses of the omega class GSTs. Moreover, our phylogenetic analysis suggests a relationship between the saprophytic behavior of some fungi and the number and distribution of some GST isoforms within specific classes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0104-5) contains supplementary material, which is available to authorized users.
Keywords: Phanerochaete chrysosporium, Glutathione S-transferase, Etherase, Omega class GST, Ure2p, GTT
Introduction
Glutathione S-transferases (GSTs; EC 2.5.1.18) constitute a complex and widespread enzyme superfamily that has been subdivided into an ever-increasing number of classes based on a variety of criteria, including amino acid/nucleotide sequence comparisons, and immunological, kinetic, and structural properties. Four main subfamilies are generally recognized: the cytosolic GSTs, the microsomal GSTs (MAPEG), the mitochondrial (also known as kappa class GST), and the bacterial fosfomycin-resistance GSTs [1, 2]. A common feature of this superfamily is the ability of these proteins to bind a broad range of ligands, and particularly hydrophobic ligands, explaining their potential role as class II biotransformation enzymes that function in the detoxification of xenobiotics and endogenous toxicants [2, 3]. In plants and animals, GSTs are the principal phase II enzymes involved in metabolic detoxification processes. Their main chemistry is to catalyze the conjugation of the tripeptide glutathione (GSH) with compounds containing an electrophilic center (carbon, nitrogen, or sulfur) to form more soluble, non-toxic peptide derivatives, ready to be excreted or compartmentalized by phase III enzymes [4]. A recent remarkable study has highlighted the capacity of the tau class GSTs to bind glutathionylated lipids with high specificity and affinity and also to catalyze the linkage of the glutathione moiety to the acyl group [5].
Some GSTs, however, possess functions which overlap with those of thiol-dependent peroxidases (peroxiredoxins and glutathione peroxidases), reducing peroxides and other products resulting from oxidative stress. In human, a GST has been recognized as one of the predominant enzymes responsible for the metabolism of both 4-hydroxy-2-nonenal (HNE) enantiomers [6]. HNE is a toxic aldehyde generated upon lipid peroxidation, and its GST-dependent conjugation with GSH influences many signal transduction pathways and modulates the activity of transcription factors [2]. Other GSTs can function in prostaglandin and steroid synthesis, or degradation of aromatic amino acids [2], and many of them have been considered as biomarkers of several human diseases, mainly cancer [7–9]. A rat liver membrane-bound microsomal GST contributes to transition pore opening controlling the mitochondrial permeability [10]. In bacteria, GSTs could display various functions such as protection against antibiotics [11] or lignin degradation by cleaving β-aryl ether linkages [12]. They could also be potential useful markers for polycyclic aromatic hydrocarbon (PAH) pollution [13]. In Saccharomyces cerevisiae, the role of ScGTO1 in the peroxisomes could be related to the redox regulation of the Str3 cystathionine β-lyase protein [14].
Cytosolic GSTs are soluble dimeric proteins with a relatively conserved N-terminal thioredoxin-like domain bearing a βαβαββα topology that is responsible for GSH binding and a more variable C-terminal domain. This canonical GST fold is observed extensively in nature, being sometimes associated with biological functions unlinked to GSTs. This is the case for instance of the bacterial stringent starvation protein A (SspA) or the intracellular chloride ion channel (CLIC1) [15].
A common feature in the GST superfamily is the presence of two binding sites for each of the substrates, a G-site for GSH and an H-site for the hydrophobic electrophile. Generally, GSTs are functionally active as homodimers or heterodimers, but in any case GST subunits are able to interact only with subunits of the same class [16]. Usually, within the classes, the N-terminal part of the proteins is the most conserved since it encloses an important part of the active site, namely the glutathione binding area, the G-site [17]. On the other hand, the H-site is found primarily in the C-terminal domain and its structure varies among GSTs. The large diversity among GSTs could thus be explained by the vast spectrum of electrophilic toxic or non-toxic compounds. Usually, the binding of GSH to the enzyme induces a lowering of the pKa value of its thiol group from 9 to about 6.2–6.6 [18], allowing a facilitated deprotonation of the thiol to form the thiolate required for the nucleophilic attack. The catalytic residue of the G-site differs depending on the class considered and it is usually either Ser, Tyr, Phe, or Cys. Furthermore, in the URE2p class, an Asn seems to be essential for the activation of the thiolate of GSH [19]. In addition, a model for GSH activation which involves a water molecule and the GSH glutamyl α-carboxylate group has been proposed [20].
Bacterial and fungal GSTs are so far poorly characterized in terms of functions and diversification in comparison with their plant and animal counterparts [21, 22]. Although nearly 10 isoforms can be found in proteobacteria for instance, the functions of these enzymes have remained obscure until now. The bacterial GSTs that have been investigated are usually involved in degradation pathways of recalcitrant chemicals which can be used for growth by host bacteria [23, 24]. Fungal GSTs have also been poorly studied, the few available data, concerning mainly yeast, indicate that they are potentially involved in protecting cells against damage resulting from oxidative stress, heavy metals, and antifungal compounds, thus highlighting the functional diversity of these enzymes [25–27]. Taking advantage of the recent release of several basidiomycete and ascomycete genome sequences, this review will deal with the description of the main fungal cytosolic GSTs classes focusing mostly on the wood-degrading basidiomycete Phanerochaete chrysosporium. In addition to making an overview of GSTs in fungi, this study aims at establishing a relationship between the way of life of the fungi and the occurrence of specific GSTs, searching for putative environmental biomakers.
Glutathione S-transferases diversity in fungi
We have investigated the diversity of GSTs in fungi focusing to the genomes of P. chrysosporium, Postia placenta, Trichoderma reseei, Neurospora crassa, Aspergillus sp., Magnaporthe grisea, Botrytis cinerea, Sclerotinia sclerotiorum, Laccaria bicolor, Coprinus cinereus, Ustilago maydis, Melampsora larici-populina, Fusarium sp., Mycosphaerella sp., Chaetomium globosum, Stagonospora nodorum, Schizosaccharomyces pombe, Candida albicans, Cryptococcus neoformans, Sporobolomyces roseus, Rhizopus oryzae and Phycomyces blakesleeanus. The methodology used for mining the different genomes as well as the different accession numbers are given in the electronic supplementary material. The choice of these fungi has been made in order to investigate, in a phylogenetic analysis, different ascomycetes, zygomycetes, and basidiomycetes and different ways of life, saprophytic, symbiotic, and pathogenic, in comparison with the well-known yeast S. cerevisiae. Indeed, it appears in this phylogenetic analysis that the total number of GST-related sequences differs strongly from one organism to another (from 7 to 46 sequences), these differences not being related to the evolutionary tree, to the size of the genome or to the gene content, but rather to the physiology of the fungi (Table 1). Organisms containing a high number of GSTs are those corresponding to ascomycetes and basidiomycetes of high complexity that are able to degrade many organic compounds (P. chrysosporium, F. oxysporum, C. cinereus, T. reesei and P. placenta), while yeasts exhibit few GSTs (S. pombe, S. cerevisieae and C. albicans). These differences in GST number could be explained by an overrepresentation of GSTs inside a class and/or the appearance of new classes or subclasses.
Table 1.
Comparative analysis of glutathione S-transferase genes in various sequenced zygomycetes, ascomycetes and basidiomycetes
| Genome size (Mb) | Gene models | GTT1 | GTT2 | URE2p | Omega | EFBγ | MAK16 | GTE | Others | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Schizosaccharomyces pombe | Asc (yeast) | 12.50 | 5,027 | 1 | 0 | 2 | 1 | 2 | 1 | 0 | 0 | 7 |
| Cryptococcus neoformans | Bas (pathogen) | 18.87 | 6,967 | 0 | 1 | 1 | 3 | 1 | 1 | 0 | 2 | 9 |
| Sporobolomyces roseus | Bas | 21.20 | 5,536 | 0 | 1 | 3 | 3 | 1 | 1 | 1 | 0 | 10 |
| Melampsora laricis-populina | Bas (pathogen) | 101.10 | 16,694 | 0 | 1 | 4 | 1 | 1 | 1 | 0 | 2 | 10 |
| Ustilago maydis | Bas (pathogen) | 19.68 | 6,522 | 2 | 0 | 2 | 3 | 1 | 1 | 1 | 0 | 10 |
| Saccharomyces cerevisiae | Asc (yeast) | 11.74 | 5,695 | 1 | 1 | 1 | 3 | 4 | 1 | 0 | 0 | 11 |
| Neurospora crassa | Asc | 39.23 | 9,826 | 1 | 0 | 2 | 3 | 3 | 1 | 1 | 1 | 12 |
| Chaetomium globosum | Asc (saprophyte) | 34.89 | 11,124 | 1 | 0 | 5 | 3 | 2 | 1 | 1 | 0 | 13 |
| Magnaporthe grisea | Asc (pathogen) | 41.70 | 11,074 | 1 | 0 | 2 | 3 | 3 | 1 | 1 | 3 | 14 |
| Rhizopus oryzae | Zyg | 45.26 | 17,459 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | 5 | 14 |
| Mycospherella fijiensis | Asc (pathogen) | 73.40 | 10,327 | 2 | 1 | 4 | 6 | 1 | 0 | 2 | 0 | 16 |
| Candida albicans | Asc (yeast) | 14.30 | 6,177 | 5 | 2 | 2 | 1 | 5 | 1 | 0 | 0 | 16 |
| Phycomyces blakesleeanus | Zyg | 55.90 | 14,792 | 0 | 1 | 0 | 2 | 2 | 1 | 9 | 1 | 16 |
| Mycosphaerella graminicola | Asc (pathogen) | 39.70 | 10,952 | 2 | 0 | 6 | 4 | 1 | 1 | 3 | 0 | 17 |
| Aspergillus clavatus | Asc (pathogen) | 27.86 | 9,121 | 2 | 1 | 4 | 5 | 3 | 1 | 2 | 0 | 18 |
| Aspergillus nidulans | Asc (pathogen) | 30.07 | 10,701 | 1 | 0 | 3 | 5 | 5 | 1 | 3 | 0 | 18 |
| Sclerotinia sclerotiorum | Asc (pathogen) | 38.33 | 14,522 | 3 | 1 | 6 | 5 | 1 | 1 | 1 | 0 | 18 |
| Stagonospora nodorum | Asc (pathogen) | 37.10 | 15,983 | 3 | 1 | 6 | 5 | 1 | 1 | 2 | 0 | 19 |
| Aspergillus terreus | Asc (pathogen) | 29.33 | 10,406 | 1 | 2 | 5 | 4 | 3 | 1 | 4 | 0 | 20 |
| Botrytis cinerea | Asc (pathogen) | 42.66 | 16,448 | 4 | 1 | 5 | 5 | 2 | 1 | 3 | 1 | 22 |
| Fusarium graminearum | Asc (pathogen) | 36.45 | 13,332 | 2 | 2 | 4 | 5 | 2 | 1 | 3 | 3 | 22 |
| Laccaria bicolor | Bas (mycorrhizal) | 64.90 | 20,614 | 1 | 11 | 1 | 3 | 2 | 1 | 3 | 1 | 23 |
| Aspergillus fumigatus | Asc (pathogen) | 29.38 | 9,887 | 4 | 1 | 4 | 5 | 3 | 1 | 2 | 3 | 23 |
| Fusarium verticillioides | Asc (pathogen) | 41.78 | 14,179 | 3 | 4 | 3 | 5 | 2 | 1 | 3 | 3 | 24 |
| Phanerochaete chrysosporium | Bas (saprophyte) | 35.10 | 10,048 | 0 | 3 | 9 | 8 | 1 | 1 | 5 | 0 | 27 |
| Fusarium oxysporum | Asc (pathogen) | 61.36 | 17,735 | 3 | 6 | 4 | 6 | 1 | 1 | 4 | 2 | 27 |
| Coprinus cinereus | Bas (saprophyte) | 36.29 | 13,392 | 4 | 5 | 2 | 4 | 1 | 1 | 14 | 1 | 32 |
| Trichoderma reesei | Asc (saprophyte) | 34.10 | 9,129 | 1 | 2 | 5 | 7 | 3 | 1 | 4 | 10 | 33 |
| Postia placenta | Bas (saprophyte) | 90.90 | 17,173 | 2 | 5 | 17 | 8 | 2 | 1 | 11 | 0 | 46 |
The classification into the different classes is based on a phylogenetic analysis using MEGA4 software
Details and accession numbers are given in the electronic supplementary material
Asc Ascomycete, Bas basidiomycete, Zyg zygomycete
In S. cerevisiae, omega [26], GTT [27], Ure2p [28], MAK16 [29], and EFBγ [30] classes have been independently identified and characterized. Moreover, by screening 67 GST-like sequences from 21 fungal species, essentially ascomycetes, McGoldrick and coauthors have identified three well-known GST classes (EFBγ, URE2p, and MAK16) and two clusters that they named clusters 1 and 2 [22]. Cluster 1 includes GTT1 from S. cerevisieae and cluster 2 includes GSTA from A. nidulans, which could be related to the URE2p class.
Based on a phylogenetic analysis and sequence comparisons with S. cerevisiae GSTs, six known classes have been extrapolated to all the fungi considered in our study: URE2p-like, GTT2, EFBγ, GTT1, omega, and MAK16. Interestingly, we have highlighted a new class that we named GTE (glutathione transferase etherase-related) since the sequences show homology with bacterial etherases [12].
The number of isoforms in each class differs considerably according to the investigated genome. For instance, in C. albicans and A. nidulans, five sequences are related to EFBγ class, whereas only one is detected in P. chrysosporium and C. cinereus. In contrast, the URE2p-like class is overrepresented in P. chrysosporium (nine sequences) whereas only one is present in L. bicolor and C. neoformans for example.
Some fungi, especially saprophytic fungi, exhibit more GST-coding sequences than other fungi and this is mainly due to an extension of the omega, GTT, Ure2p-like, and the newly identified GTE classes. We have thus focused our analysis on these classes.
The GTT classes (GTT1 and GTT2)
Saccharomyces cerevisiae possesses one of the simplest GST-related equipment of the investigated fungi. Among the different identified genes, two GTTs (glutathione transferase) have been characterized [27], one GTT1 and one GTT2. Strains lacking GTT1 and GTT2 are viable and unaffected in growth during normal aerobic conditions [31]. ScGTT1 is associated with the endoplasmic reticulum [27]. Both proteins exhibit activity against classical GST substrates as 1-chloro-2,4-dinitrobenzene (CDNB), but they probably have different physiological functions. Involved in cadmium detoxication, ScGTT2 catalyzes the formation of glutathione-Cd conjugates [32], while ScGTT1 catalyzes the reduction of hydroperoxides, in particular cumene hydroperoxide [33]. ScGTT1 and ScGTT2 have overlapping functions with glutaredoxins (ScGrx1 and ScGrx2), these latter also exhibiting classical glutathione transferase activities [31]. The involvement of glutaredoxins in xenobiotic conjugation could explain why the gtt1 gtt2 mutant does not show any increased sensitivity to CDNB. Moreover, ScGTT1 and ScGTT2 seem to be crucial in the response to H2O2 stress [34], and ScGTT2 acts as a general protective factor involved in quinone detoxification [35].
Among the investigated fungi, the number of ScGTT1-related genes ranges from 0 to 5 isoforms depending on the fungus, this class representing one-third of the total GSTs of C. albicans. In contrast, no GTT1 isoform has been detected in P. chrysosporium.
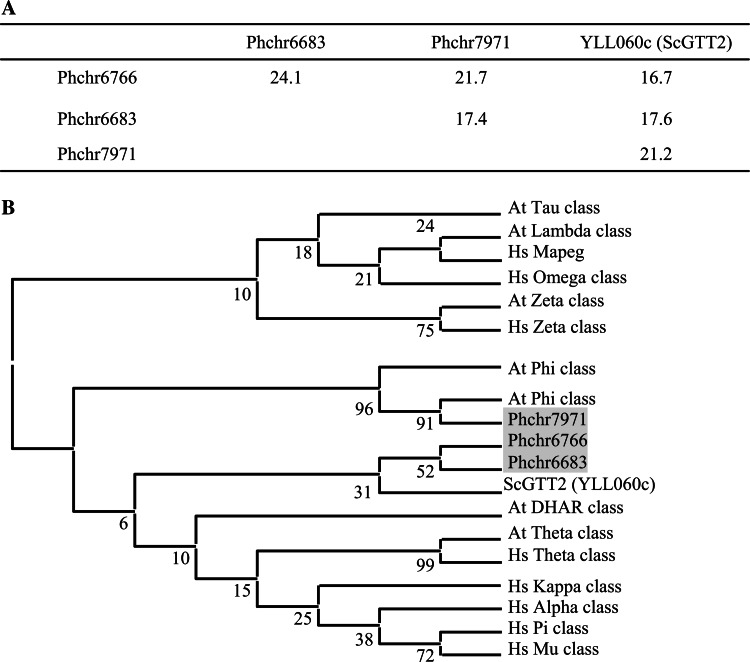
The number of GTT2 isoforms varies from 0 for some ascomycetes to 11 for L. bicolor (Table 1). Although L. bicolor possesses the highest gene content among the fungi studied, the striking difference in the number of GTT2 isoforms in this fungus might not be related to that property, since GTT2 isoforms represent nearly half (48%) of its total GST content. It is not due either to a difference in the thiol-dependent antioxidant systems. In fact, for instance, five and four Grxs could be found in L. bicolor and A. nidulans, respectively [36], while no GTT2 was detected in A. nidulans. P. chrysosporium possesses 3 GTT2-related sequences with few identities between each other and compared to yeast GTT2 (Fig. 1a). In a phylogenetic analysis, Phchr6683 and Phchr6766 sequences cluster with ScGTT2 (Fig. 1b). The recombinant Phchr6766 protein has been produced in Escherichia coli. It exhibits a strong activity with organic peroxides (kcat = 181 s−1 and kcat = 141 s−1 using ter-butyl peroxide and cumene peroxide, respectively) (Morel et al., unpublished). The RasMol representation of Phchr6766 was based on the crystal structure of the glutathione S-transferase-like domain of EF1Bγ from S. cerevisiae [37]. The structure is overall very similar to the glutathione S-transferase proteins and contains a binding pocket highly homologous to those observed in glutathione S-transferase enzymes (data not shown). The third Phanerochaete GTT2-related isoform (Phchr7971) clusters with plant phi GSTs (Fig. 1b). Accordingly, its RasMol structure model could be built from the 3D structure of a phi class GST from Arabidopsis thaliana in complex with an herbicide [38] (data not shown). Phi class is a plant-specific class, which is one of the largest in A. thaliana with 13 members. The proteins possess GST activity against 4-hydroxynonenal (HNE), a naturally occurring lipid peroxidation product [39]. A phi class GST from Oryza sativa (OsGSTF5) appears to have a role in herbicide conjugation and to possess glutathione peroxidase activity [40]. By selectively co-reducing the expression of several of the major phi class GSTs, their role in limiting metabolic changes that arise from oxidative stress has been proposed [41]. The P. chrysosporium isoform (Phchr7971) is newly expressed upon the addition of benzoic acid in a proteomic analysis carried out on P. chrysosporium [42]. Nothing else is known concerning this protein in fungi.
Fig. 1.
GTT2-related sequence comparisons. a Percentage of identity between GTT2-related sequences from P. chrysosporium and S. cerevisiae (ScGTT2), based on global alignment determined by Lalign software. (http://www.ch.embnet.org/software/LALIGN_form.html). b Phylogenetic distribution of GTT2-related sequences of P. chrysosporium among GST classes from Homo sapiens, A. thaliana, and S. cerevisiae. The diagram has been drawn based on a phylogenetic analysis carried out with ClustalW and MEGA4 software. The analyses were conducted using the neighbor-joining (NJ) method implemented in MEGA, with the pairwise deletion option for handling alignment gaps, and with the Poisson correction model for distance computation. Bootstrap tests were conducted using 1,000 replicates. Branch lengths are proportional to phylogenetic distances. Sequences from human, A. thaliana and S. cerevisiae were obtained from NCBI databases (http://www.ncbi.nlm.nih.gov/). Sequences from P. chrysosporium were obtained from JGI (http://genome.jgi-psf.org). Hs Homo sapiens, At A. thaliana, Sc S. cerevisiae and Pc P. chrysosporium. Accession numbers for A. thaliana proteins are: At5g41210; At5g41240; NP_198938; X68304; At4g02520; At2g02930; At1g02950; At1g02940; At1g02930; At1g02920; At2g47730; At2g30860; At2g30870; At3g03190; At5g17220; At3g62760; At1g49860; Q9ZVQ3; Q9ZVQ4; At2g29490; At2g29480; At2g29470; At2g29460; At2g29450; At2g29440; At2g29420; At3g09270; At5g62480; At1g74590; At1g69930; At1g69920; At1g27130; At1g27140; At1g59670; At1g59700; At1g10370; At1g10360; At1g78380; At1g78370; At1g78360; At1g78340; At1g78320; At1g17170; At1g17180; At1g17190; At3g43800; At1g53680; At5g02790; At5g02780; At3g55040; and for human proteins are: AAB96392; AAC13317; AAA60963; AAA70226; NP_899062; NP_665735; CAA33508; Q9Y2Q3
The URE2p class
In the wood-decomposing fungi P. chrysosporium and P. placenta, the URE2p-like class represents about one-third of the total identified GSTs, corresponding to 9 and 17 sequences, respectively (Table 1). P. chrysosporium sequences are quite homologous between each other showing between 25.1 and 83.2% identity (Table 2). In particular, PcURE2p4, 6 and 7 exhibit between 62 and 83% identity suggesting recent duplication events of these sequences. A comparison analysis using blast search suggests that these 3 sequences are related to GSTII of S. pombe (SPCC965.07). A GSTII-lacZ fusion has been constructed in S. pombe and shows that (1) GSTII is basically more expressed than the 2 other GSTs of the fungus, and (2) GSTII gene expression is increased by various stress agents such as sodium nitroprusside, ter-butylhydroquinone, and L-buthionine-[S,R]-sulfoximine [43].
Table 2.
Percentage of identity between URE2p and cluster 2 sequences of Phanerochaete, based on global alignment determined by Lalign software
| PcUre2p3 | PcUre2p4 | PcUre2p5 | PcUre2p6 | PcUre2p7 | PcUre2p8 | PcUre2p9 | Phchr 503 | |
|---|---|---|---|---|---|---|---|---|
| PcUre2p2 | 64.3 | 65.1 | 43.0 | 65.4 | 53.3 | 40.6 | 52.8 | 29.9 |
| PcUre2p3 | 62.6 | 41.3 | 60.9 | 50.2 | 39.3 | 49.3 | 24.9 | |
| PcUre2p4 | 45.8 | 83.2 | 61.9 | 45.8 | 53.3 | 26.2 | ||
| PcUre2p5 | 42.1 | 29.4 | 25.1 | 27.2 | 20.9 | |||
| PcUre2p6 | 68.3 | 47.7 | 52.9 | 29.0 | ||||
| PcUre2p7 | 57.8 | 58.4 | 34.6 | |||||
| PcUre2p8 | 43.9 | 29.5 | ||||||
| PcUre2p9 | 34.8 |
All of the nine URE2p-like sequences of P. chrysosporium could have a specific role in the fungus physiology since they are all transcribed (data not shown). One can wonder how this expanded class in P. chrysosporium and P. placenta is linked to the saprophytic properties of these fungi.
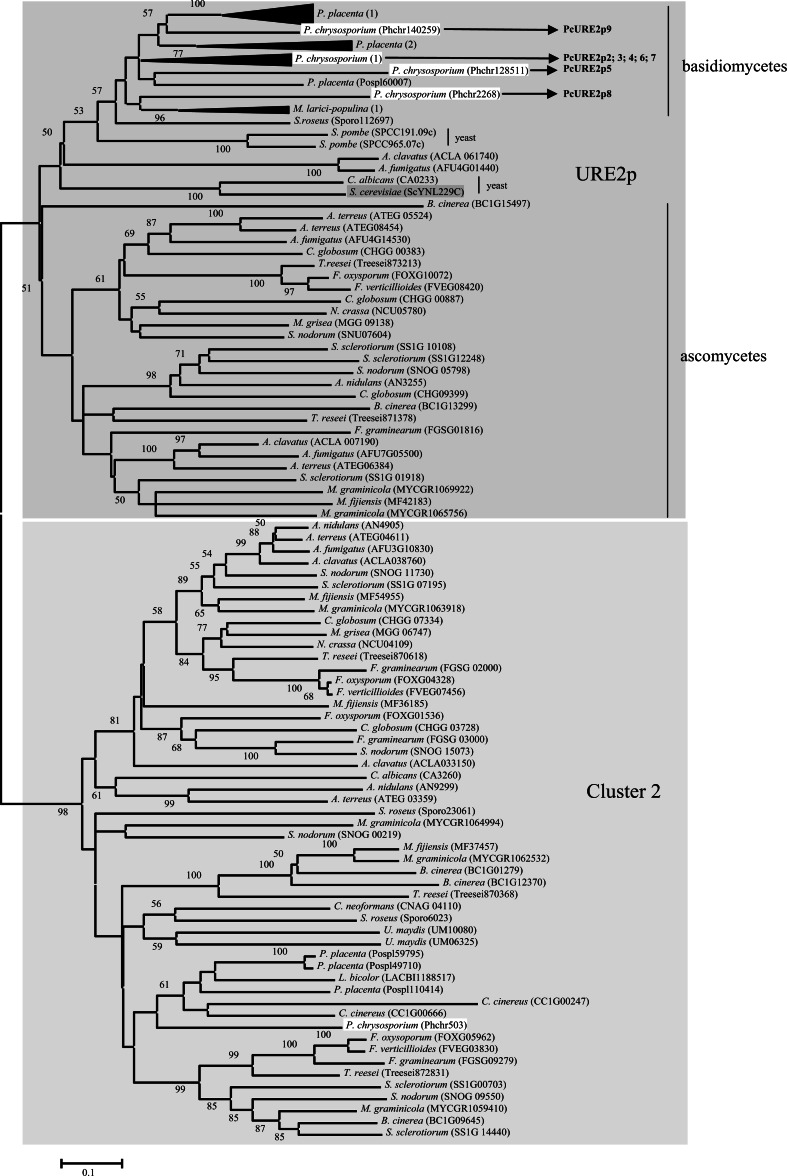
The phylogenetic analysis of the sequences belonging to the URE2p-like class revealed the presence of two subclasses (Fig. 2): one containing the yeast URE2p homologue and URE2p2 to URE2p9 from P. chrysosporium, and a second one containing GSTA (AN4905) from A. nidulans. This protein appeared in a cluster defined by McGoldrick et al. [22] and named cluster2. We have kept this name to define the corresponding subclass.
Fig. 2.
Phylogenetic analysis of URE2p-like proteins. Fungi analysed are those described in Table 1. The alignment was performed with CLUSTALW and phylogenetic tree with MEGA4 software. P. placenta (1) sequences are Pospl121705; Pospl120102; Pospl86808; Pospl9572; Pospl9064; Pospl97355; Pospl91142; Pospl10142; Pospl91146. P. placenta (2) sequences are Pospl93639; Pospl88909; Pospl 27082; Pospl96818. Sequences from P. chrysosporium (1) are: PcURE2p2: Phchr140156; PcURE2p3: Phchr140271; PcURE2p4: Phchr137250; PcURE2p6: Phchr2269; PcURE2p7; Phchr2266. Sequences from M. larici-populina (1) are: Mellp72597; Mellp40377; Mellp90288
For GST classification, it has been admitted for mammalian sequences that, in the same class, GSTs share at least 60% identity in the primary structure and that those with less than 30% identity are assigned to a different class [44]. For bacterial sequences, it has been proposed that proteins belong to the same class with >40% identity, whereas GSTs of different classes share <25% identity [45]. Based on these criteria, we cannot consider cluster2 being part of an independent class from URE2p.
In S. cerevisiae, it has been known for a long time that ScURE2p from the URE2p subclass is involved in the nitrogen catabolite repression (NCR) by preventing gene expression by ScGLN3p in cells grown on the preferred source of nitrogen. Indeed, ScURE2p is able to bind GLN3p and retains it in the cytoplasm. The shift from the preferred source of nitrogen (ammonia or glutamine) to the nonpreferred source of nitrogen (proline) results in the release of ScGLN3p and its entry into the nucleus where it activates the transcription of nitrogen-regulated genes. It is therefore likely that ScURE2p senses the decline in the intracellular glutamine concentration [46]. Yeast URE2p possesses prion-like characteristics. The prion module in the N-terminal domain contributes to the function and stability of the ScURE2p protein and also to the transformation of Ure2p into the Ure3p proteolysed prion form. In addition, it may also influence the interactions between ScURE2p and other nitrogen regulatory proteins [47]. Moreover, ScURE2p exhibits primary sequence and three-dimensional homologies to known glutathione S-transferases. It participates in heavy metal ion and oxidant detoxification when ammonia is used as sole nitrogen source [28]. The protein is composed of two clearly divergent domains: the prion domain is in the N-terminal position and the GST-like domain on the C-terminal side [48]. By comparing URE2p sequences from various species of the genus Saccharomyces, it has been noticed that the N-terminal ~40 residues are largely conserved, but there are a number of differences in the following ~50 residues of the prion domain. Conversely, the C-terminal parts of the molecule are nearly invariant [49]. ScURE2p displays GSH-dependent peroxidase activity and Asn124 has a key role in the catalytic mechanism by functioning in a way similar to that of the catalytic Ser of typical GST enzyme in activation of GSH [19]. The mutation of this Asn into Ala or Val (but not into a Ser, Tyr, or Cys) restores the GST activity of URE2p towards CDNB. The authors thus suggest that mutations have allowed URE2p to diversify and acquire additional functions as a prion and a repressor of nitrogen catabolism. It has been recently shown that ScUREp also shows thiol-disulfide oxidoreductase activity similar to that of glutaredoxins even if it does not possess cysteine residue [50]. Concerning the investigated fungi of our study, only C. albicans and S. cerevisiae exhibit the N-terminal prion domain; however, many other fungal sequences with shorter URE2p and deprived of the prion domain cluster are present in this class (Fig. 2). The distribution of ScURE2p seems to follow the evolution tree since both ascomycete and basidiomycete groups can be distinguished. Surprisingly, yeast sequences are closer to basidiomycetes sequences rather than to ascomycetes.
In our study, cluster 2 is an expanded subclass (Fig. 2) containing A. nidulans GSTA (AN4905) and a sequence from P. chrysosporium (Phchr 503). GSTA lacks the nitrogen metabolite repression activity of URE2p, but contributes to heavy metal and xenobiotic resistance [51]. Additionally to GSTA, another sequence of A. nidulans has been identified (AN9299) showing 50.7% identity with it. Up to now nothing is known about it. The sequence of P. chrysposporium (Phchr503) exhibits 44% identity with GSTA from A. nidulans and between 29.9 and 40.6% identity with the other P. chrysosporium URE2p sequences (Table 2). The characterization of this protein in relation with its putative role in oxidative stress response is an interesting point, which is currently under investigation in our laboratory. The first results show that the recombinant protein exhibits thiol-transferase and reductase activities using β-hydroxyethyl disulphide (HED) and dehydroascorbate (DHA) as substrates, suggesting a putative role in stress response (Anak-Ngadin et al., unpublished).
The omega class
The omega class of GSTs has been identified only recently in fungi, most investigations having been conducted on human proteins [52]. This class is represented in humans by two functional genes, named GTO1 and GTO2. Both proteins exhibit thioltransferase, dehydroascorbate reductase, and monomethylarsonate reductase activities, and their activity is dependent on an active-site cysteine residue [53]. In addition to its ability to act as a glutathione-dependent thioltransferase, it was proposed that HsGTO1-1 can reduce the S-thiol adduct formed between GSH and cysteine residues of proteins under stress, restoring their enzymatic functions [52]. HsGTO1-1 catalyzes the rate-limiting step in the biotransformation of arsenic [54], and it is also involved in various important biological processes. Indeed, human HsGTO1-1 can modulate ryanodine receptors, which are calcium channels in the endoplasmic reticulum of various cells [55], or catalyze the reduction of S-(Phenacyl) glutathione, an intermediate in the degradation of the toxic α-haloketone, to yield non-toxic acetophenones [56]. Moreover, S-(4-nitrophenacyl) glutathione (4NPG) has a high turnover with GSTO1-1 but negligible activity with GSTO2-2 and other members of the glutathione transferase superfamily [57]. Transgenic Caenorhabditis elegans overexpressing GSTO-1 were generated exhibiting an increased resistance to juglone-, paraquat-, and cumene hydroperoxide-induced oxidative stress, while specific silencing of the GSTO-1 by RNAi created worms with an increased sensitivity to several prooxidants, arsenite, and heat shock [58].
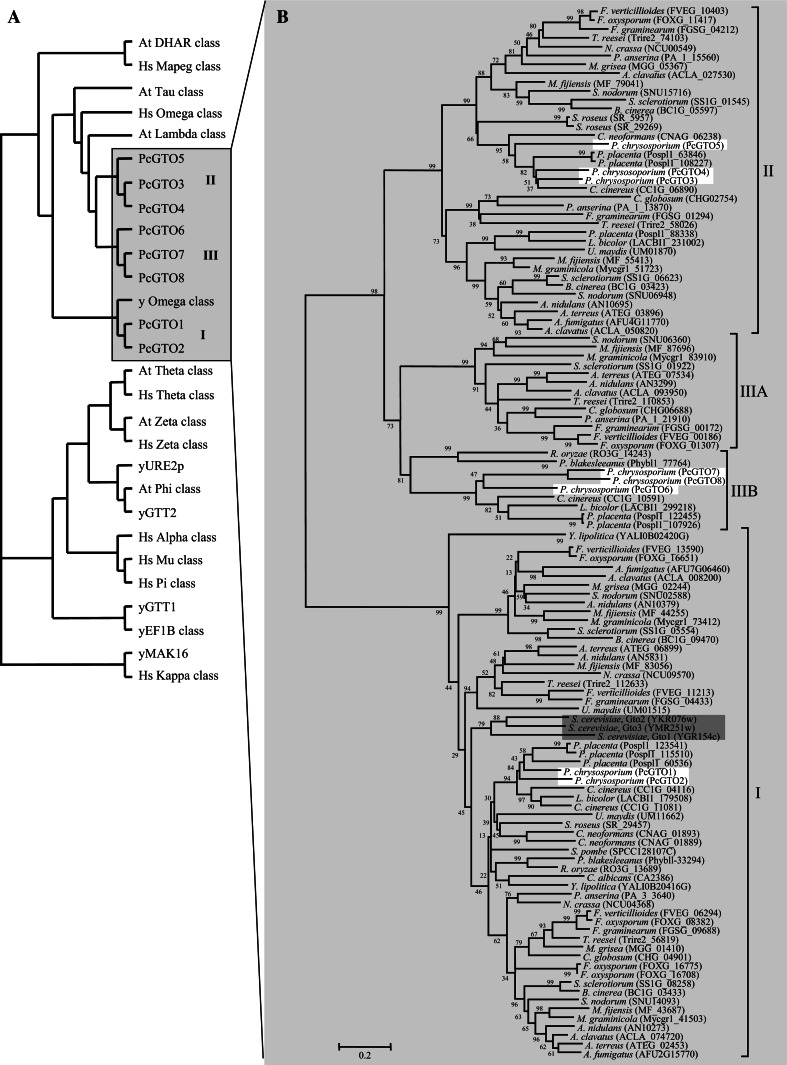
The P. chrysosporium genome possesses eight sequences, named PcGTO1 to PcGTO8, which exhibit homology with GSTs of the omega class. Based on a phylogenetic analysis and the percentage of identity between the omega-related sequences, we show that these eight proteins cluster into three distinct subclasses: The yeast GTO-related subclass (subclass I), and 2 new subclasses, that we named subclass II and III (Figs. 3 and 4a). If we take into account the bacterial classification, which determined that proteins belong to the same class with >40% identity, whereas GSTs of different classes share <25% identity, these 3 subclasses could be considered as independent classes. However, nothing has been established concerning fungal GST classification and since the sequences from subclasses II and III are clearly related to human GTO, we choose to qualify them as subclasses from the omega class.
Fig. 3.
Phylogenetic distribution of omega proteins. a Distribution of omega GSTs of P. chrysosporium among GSTs from H. sapiens, A. thaliana, and S. cerevisiae. The diagram has been drawn based on a phylogenetic analysis carried out with ClustalW and MEGA4 software. Sequences were obtained as described in Fig. 1 and accession numbers are given in the legend of Fig. 1. Accession numbers for S. cerevisiae are given in brackets in Fig. 3b and JGI accession numbers of P. chrysosporium sequences are: PcGTO2: 126388; PcGTO4: 7168; PcGTO5: 7169; PcGTO6: 3911; PcGTO7: 6880; PcGTO8: 6881. PcGTO1 and PcGTO3 have been manually corrected and deposited to NCBI under the accession numbers: EU791894 and EU791893, respectively. b Phylogenetic tree of omega sequences from various ascomycetes (Aspergillus clavatus, Aspergillus fumigatus, Aspergillus terreus, A. nidulans, Mycosphaerella graminicola, Mycosphaerella fijiensis, Fusarium oxysporium, Fusarium verticillioides, Fusarium graminearum, Podospora anserina, Chaetomium globosum, Magnaporthe grisea, Botrytis cinerea, Trichoderma reesei, Sclerotinia sclerotiorum, Septoria nodorum, Schizosaccharomyces pombe, Yarrowia lipolytica, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae), basidiomycetes (Phanerochaete chrysosporium, Laccaria bicolor, Coprinus cinereus, Postia placenta, Cryptococcus neoformans, Ustilago maydis, Sporobolomyces roseus) and zygomycetes (Rhizopus oryzae and Phycomyces blakesleeanus). Sequences were obtained and identified using the transcript or protein accession number, in genomes from the BROAD institute (http://www.broad.mit.edu/annotation/), the JGI (http://genome.jgi-psf.org/euk_home.html), Podospora anserina (http://podospora.igmors.u-psud.fr/index.html), Schizosaccharomyces pombe (http://www.sanger.ac.uk/Data Search/blast.shtml), Yarrowia lipolitica (http://www.ncbi.nlm.nih.gov/projects/genome/seq/) using alternatively the S. cerevisiae or the P. chrysosporium sequences as template. The alignment was performed with CLUSTALW and phylogenetic tree with MEGA4 software
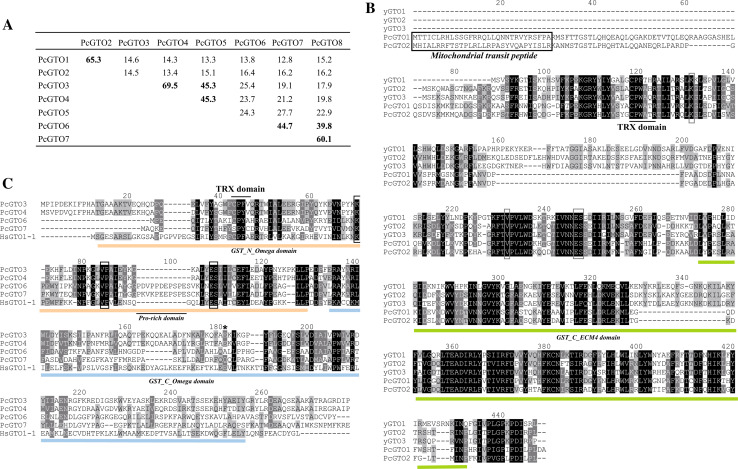
Fig. 4.
Omega GST sequence comparisons. a Percentage of identity between omega GST sequences of P. chrysosporium, based on global alignment determined by Lalign software. b Alignment of P. chrysosporium GST omega proteins from subgroup I in comparison to S. cerevisiae GTOs. Alignments have been performed using ClustalW. Amino acids involved in GSH binding and mitochondrial targeting sequences are framed. C. Alignment of P. chrysosporium GSTs from subclasses II and III previously defined in Fig. 3, in comparison with human GTO1-1. PcGTO5 and PcGTO8 are not represented in the figure because the sequences are not firmly delineated. The conserved amino acids involved in GSH binding are framed. The asterisk localizes Glu155 of HsGTO1-1 sequence
The subclass I encloses PcGTO1 and PcGTO2, subclass II encloses PcGTO3 to PcGTO5, and subclass III encloses PcGTO6 to PcGTO8. Sequences from subclass I are close to yeast GTOs, while subclasses II and III are related to plant tau and lambda proteins, and to human GTOs. Interestingly, while the S. cerevisiae GTO orthologues (subclass I) are identified in all ascomycete and basidiomycete genomes analyzed, sequences from subclasses II and III were not identified in S. cerevisiae, S. pombe, and C. albicans (Fig. 3b). These results suggest acquisition of new functions for the omega-related proteins within the evolution of this phylum.
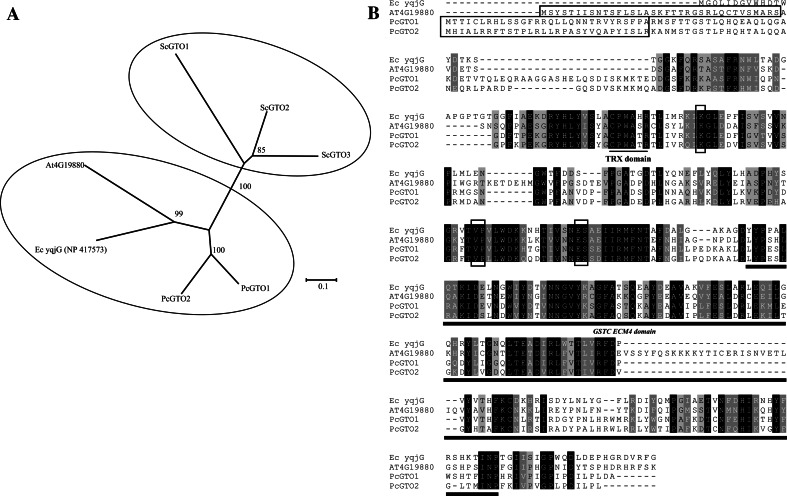
Subclass I is composed of proteins related to the three S. cerevisiae GTOs (ScGTO1, ScGTO2, and ScGTO3). Two P. chrysosporium GSTs (PcGTO1 and PcGTO2) belong to this subclass. Both display a CPWATR active site compatible with the CP(W/F)(A/T)(H/Q)R motive found in ScGTOs (Fig. 4b). In yeast, the GTOs have been partially characterized. The three ScGTOs exhibit glutaredoxin-like activities, being able to reduce HED and DHA, and all three GTO genes are induced by agents causing oxidative stress suggesting a function in the defense against oxidants [33]. ScGTO1 has been shown to be located in peroxisomes and involved in sulfur metabolism. The other fungal orthologs, and in particular PcGTO1 and PcGTO2, clustering in this omega class differ from their yeast counterparts in particular at the H-site and are, in contrast, strongly related to bacterial GSTs. Moreover, according to the prediction softwares, both P. chrysosporium orthologs (PcGTO1 and PcGTO2) possess a signal peptide targeting to mitochondria (respectively 92 and 97% of probability). Surprisingly, we found that PcGTO1 and PcGTO2 exhibit strong homology with some plant lambda and bacterial proteins (Fig. 5a). In E. coli, the homologue of PcGTO1 (yqjG) belongs to an operon containing a gene coding for a quinol oxidase, which is a key energy-transducing respiratory enzyme in microorganisms. This is of particular interest since PcGTO1 is predicted to be localized in the mitochondria. In bacteria, a tetrachlorohydroquinone dehalogenase appeared to be a member of the GST superfamily and, particularly, related to PcGTO1. This enzyme converts tetrachlorohydroquinone to dichlorohydroquinone using GSH as reducing equivalent [59]. In P. chrysosporium, this reductive dehalogenation requires two enzymes, a glutathione S-transferase for glutathionylation and an enzyme called glutathione conjugate reductase for deglutathionylation (GCR) [60]. Subsequent results have shown that some GSTs are able to deglutathionylate aromatic compounds [56], and thus one can wonder whether this GCR might also belong to the GST superfamily. Nothing is known concerning the Arabidopsis homologue (At4G19880). Data from Geneinvestigator (https://www.genevestigator.com/gv/index.jsp) [61] show that the gene is essentially expressed in roots of young plantlets, and during hypoxia and hormonal stress. The multiple alignment of these fungal, bacterial, and plant sequences (Fig. 5b) shows different amino acid identities especially at the active site and the residues important for the interaction with GSH. Similarly to PcGTO1 and PcGTO2, the A. thaliana sequence exhibits a signal peptide for mitochondria (92.5% probability). Moreover, it exhibits a lysine-rich extension at the H-site compared to the other sequences.
Fig. 5.
Subgroup I omega GSTs. a Phylogenetic relationship between omega GST proteins from P. chrysosporium, S. cerevisiae, and A. thaliana (At4G19880) and E. coli (yqjG) homologues. The alignment was performed with CLUSTALW and phylogenetic tree with MEGA4 software. b Alignment of P. chrysosporium GTO1 and GTO2, E. coli (yqjG) and A. thaliana homologues (At4G19880). Amino acids involved in GSH binding and mitochondrial targeting sequences are framed
The analyzed fungi exhibit 1–3 sequences belonging to subclass II, except S. pombe, M. larici-populina, S. cerevisiae, R. oryzae, C. albicans and P. blakesleeanus, which do not possess any sequences of this subclass. Similarly, proteins from subclass III are not represented in all fungi and their distribution follows evolution, i.e. sequences from ascomycetes and basidiomycetes cluster independently (Fig. 3b). In our study, one sequence was identified in the two zygomycetes, some pathogenic ascomycetes and all basidiomycetes, with P. chrysosporium and P. placenta exhibiting 3 and 2 sequences, respectively.
GTOs are known to contain the G-site, which contains a conserved cysteinyl residue essential for activity. Amino-acid alignments of sequences from subclass I (Fig. 4b) and subclasses II and III (Fig. 4c) show the presence of this conserved residue in all described PcGTOs except in PcGTO7. This conserved cysteine aligns with Cys-32 of human GTO1-1, which is located at the beginning of the first α-helix, in a position similar to the thiol group of the catalytic cysteine of the CxxC motif of thioredoxin and glutaredoxin. This suggests that GSTs from subclasses II and III, except PcGTO7, could exhibit the same glutathionylation and deglutathionylation properties exhibited by HsGTO1-1. Moreover, the adjacent proline, which promotes optimal positioning of the Cys32 thiol for stabilisation of the thiolate form in HsGTO1-1, is absolutely conserved [62]. Other residues required for the interaction with GSH in HsGTO1-1 (Lys59, Val72, Glu85, Ser86) [52] are also conserved in the different PcGTOs. The C-terminal part of the proteins, corresponding to the hydrophobic H-site involved in xenobiotic binding in the different GSTs classes [52], exhibits a larger variability. PcGTO6 and PcGTO7 possess a proline rich domain not present in PcGTO3 and PcGTO4. This sequence could play a role either in its own folding or else be required for catalytic activity. It has been recently shown that the deletion of Glu155 of the human GTO1-1 causes a deficiency of the protein possibly due to an induced instability of the variant protein [63]. However, this mutation does not alter sensitivity to arsenic trioxide and other cytotoxic drugs. This residue (marked by an asterisk in Fig. 4c) is conserved in PcGTO3 and PcGTO4 but not in PcGTO6 nor in PcGTO7. PcGTO3 and PcGTO4, but also PcGTO7 and PcGTO8, are present in tandem in the P. chrysosporium genome. They show 69.5% and 60.1% amino acid identity, respectively (Fig. 4a) suggesting quite recent duplication events.
The GTE class
A new class of GSTs that we have called GTE has been highlighted by this phylogenetic analysis. These proteins are related to bacterial Lig proteins. In Sphingomonas paucimobilis, three genes, LigF, LigE, and LigG, are localized in an operon called LigDFEG. They are all members of the GST superfamily. However, a weak activity with the universal substrate for GST (CDNB) was detected in E. coli extracts overexpressing Lig proteins [64]. LigE and LigF are enantioselective GSTs involved in cleavage of β-aryl ether compounds, which is the most important step in lignin degradation. LigG is the glutathione lyase for the glutathione conjugate produced by LigF [12]. In a phylogenetic analysis using representative prokaryotic and eukaryotic members of the GST superfamily, S. paucimobilis LigF and LigG group together, not so far from the omega class, but distant from LigE which is more closely related to zeta class enzymes [45].
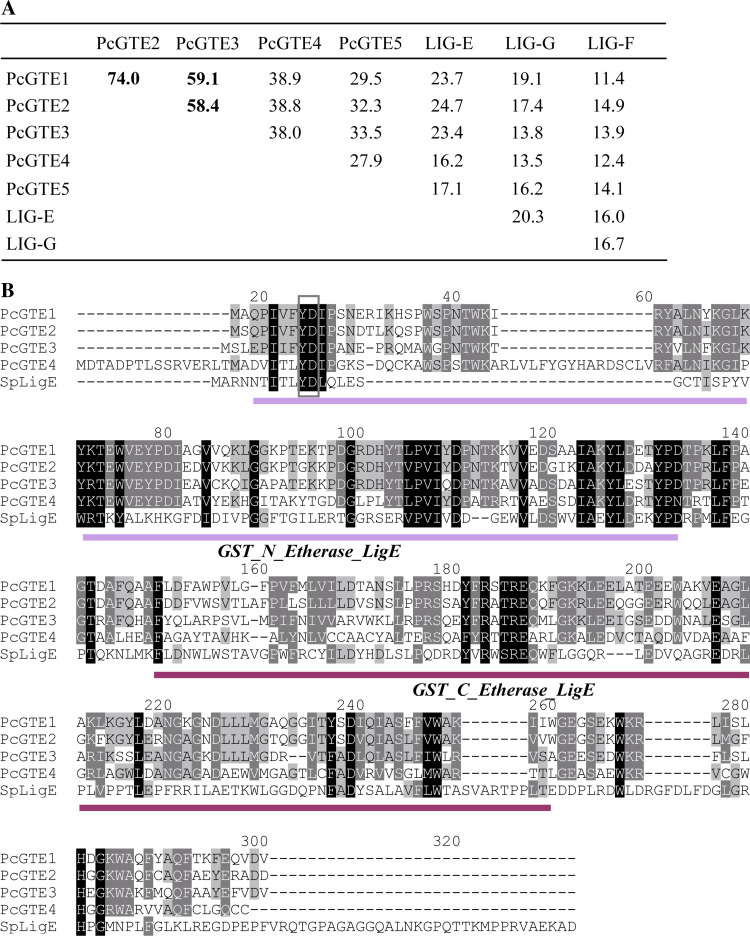
The homologies between the four P. chrysosporium GTE members and bacterial Lig proteins are quite weak, ranging from 11.4 to 23.7% identity (Fig. 6a). PcGTE1, PcGTE2, and PcGTE3 are rather related to LigE, and sequence analysis revealed that PcGTEs possess the specific GST-N and GST-C domains of the LigE sequence (Fig. 6b). These sequences exhibit a conserved tyrosine in the N-terminal region (G-site) putatively involved in catalysis as postulated for their bacterial counterparts [24]. LigE catalyzes the reductive cleavage of the β-aryl ether linkage of guaiacylglycerol β-O-4-methylumbelliferone (GOU) to produce β-hydroxypropiovanillone and 4-methylumbelliferone in vitro [65]. The recombinant GTE3 of P. chrysosporium is not active using this substrate (Morel et al., unpublished results). However, other substrates have to be tested, since a high substrate specificity of the β-aryl ether cleavage activity has been proposed [65]. In S. paucimobilis, LigE is tightly associated with cell membranes [66]. Conversely, the predictions of P. chrysosporium enzymes localization revealed that one could be excreted (PcGTE4), the others being cytosolic (PcGTE1, PcGTE2, and PcGTE3) or mitochondrial (PcGTE5).
Fig. 6.
GTEs and Lig GST sequence comparisons. a Percentage of identity between GTE sequences of P. chrysosporium and Lig sequences from S. paucimobilis (LigE: BAA02032; LigG: BAA77216; LigF: BAA02031), based on global alignment determined by Lalign software. b Amino acid sequence alignments of GTEs and LigE proteins from Sphingomonas paucimobilis. Alignments have been performed using ClustalW. Amino acids putatively involved in catalysis are identified by a frame
Fungal genome analysis revealed that hemiascomycetes such as S. cerevisiae, C. albicans, and S. pombe do not possess GTE proteins. In contrast, depending on their physiology, all filamentous fungi analyzed present a variable number of GTE-like sequences as evidenced by their analogy to the LigE sequence. Most animal and plant pathogenic fungi exhibit 1–3 putative GTEs, while 3–14 genes were found in saprophytic fungi (Table 1). In particular, the basidiomycetes P. placenta and C. cinereus exhibit 11 and 14 GTE sequences, respectively. Moreover, 3 or 4 sequences were identified in necrotroph plant pathogens such as Aspergillus sp., Fusarium sp., or B. cinerea. These fungi are predominately saprophytes and grow on dead or live plant and animal tissues in the soil. For this reason, these enzymes could be very important in nutrient recycling, or for their infectious cycle. These observations highlight an apparent relationship between these GTEs and the capacity of the fungi to degrade organic matter. This is in accordance with their homology with the bacterial Lig proteins. A better characterization of these enzymes is of great interest in understanding wood and xenobiotic degradation processes. Moreover, using a complete expression dataset of L. bicolor genes available at (http://www.ncbi.nlm.nih.gov/geo/) as series GSE9784 [67], it appeared that an etherase-like gene (LbGTE3) is upregulated by about 20-fold in ectomycorrhizas compared to free-living mycelium. This suggests putative functions of GTE proteins, both saprophytic and mycorrhizal.
A surprising result is the occurrence of nine sequences related to GTEs in the zygomycete fungus P. blakesleeanus, corresponding to more than half of its total GST number. This fungus is known for the variety and sensitivity of its responses to light and the regulation of the biosynthesis of the pigment beta-carotene. However, it does not seem to have saprophytic properties, and thus the physiological significance of these GTE-like sequences in this fungus is still an open question.
Conclusion
In summary, this study has highlighted the occurrence of new GST-proteins in fungi, belonging to the omega class (subclasses II and III) and a newly identified GTE class. Globally, saprophytic fungi exhibit more GST-coding sequences than the other fungi, and this is mainly due to an extension of the omega, GTT, Ure2p, and GTE classes.
Proteins from omega, Ure2p, and GTE classes are highly represented in P. chrysosporium. This opens many perspectives to better understand how the fungus is able to oxidize and detoxify a broad range of toxic chemical pollutants. This fungus uses both an extracellular oxidative system (Lignin peroxidases) and GSTs and/or cytochrome P450 mono-oxygenases. Cytochrome P450 mono-oxygenases are involved in phase I detoxification metabolism. Interestingly, P. chrysosporium possesses the highest number of P450 sequences among fungi (1% of the total genome) [68]. Thus, the high diversity of GSTs could be related to this very large number of cytochrome P450 mono-oxygenases. This study also shows for the first time that some basidiomycetes possess GSTs known to have β-etherase activity in bacteria (GTE class). Moreover, we can make a link between the representation of this class among total GSTs and the saprophytic properties of the fungi. These proteins could thus have a role in organic matter degradation and in particular in wood degradation for P. chrysosporium and P. placenta. Through these properties, they can be good actors in xenobiotic oxidation processes and represent a versatile tool with a variety of biotechnological applications such as bioremediation. So far, all GST engineering experiments have been carried out with bacteria [45], mouse, and Drosophila melanogaster (for a review, see [2]). Recently, the effectiveness of gene silencing by RNA interference has been demonstrated in P. chrysosporium [69], opening new perspectives, by using this genetic tool, to study the function of GSTs in detoxification purposes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgment
The research was supported by an ANR project (ANR-06-BLAN-0386).
References
- 1.Frova C. Glutathione transferases in the genomics era: new insights and perspectives. Biomol Eng. 2006;23:149–169. doi: 10.1016/j.bioeng.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- 4.Coleman JOD, Randall R, Blake-Kalff MMA. Detoxification of xenobiotics in plant cells by glutathione conjugation and vacuolar compartmentalization: a fluorescent assay using monochlorobimane. Plant Cell Env. 1997;20:449–460. doi: 10.1046/j.1365-3040.1997.d01-93.x. [DOI] [Google Scholar]
- 5.Dixon DP, Edwards R (2009) Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J Biol Chem (in press). doi:10.1074/jbc.M109.020107 [DOI] [PMC free article] [PubMed]
- 6.Balogh LM, Roberts AG, Shireman LM, Greene RJ, Atkins WM. The stereochemical course of 4-hydroxy-2-nonenal metabolism by glutathione S-transferases. J Biol Chem. 2008;283:16702–16710. doi: 10.1074/jbc.M801725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalhoff K, Buus Jensen K, Enghusen Poulsen H. Cancer and molecular biomarkers of phase 2. Methods Enzymol. 2005;400:618–627. doi: 10.1016/S0076-6879(05)00035-2. [DOI] [PubMed] [Google Scholar]
- 8.Usami H, Kusano Y, Kumagai T, Osada S, Itoh K, Kobayashi A, Yamamoto M, Uchida K. Selective induction of the tumor marker glutathione S-transferase P1 by proteasome inhibitors. J Biol Chem. 2005;280:25267–25276. doi: 10.1074/jbc.M501014200. [DOI] [PubMed] [Google Scholar]
- 9.Haas S, Pierl C, Harth V, Pesch B, Rabstein S, Brüning T, Ko Y, Hamann U, Justenhoven C, Brauch H, Fischer HP. Expression of xenobiotic and steroid hormone metabolizing enzymes in human breast carcinomas. Int J Cancer. 2006;119:1785–1791. doi: 10.1002/ijc.21915. [DOI] [PubMed] [Google Scholar]
- 10.Hossain QS, Ulziikhishig E, Lee KK, Yamamoto H, Aniya Y. Contribution of liver mitochondrial membrane-bound glutathione transferase to mitochondrial permeability transition pores. Toxicol Appl Pharmacol. 2009;235:77–85. doi: 10.1016/j.taap.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Allocati N, Favaloro B, Masulli M, Alexeyev MF, Di Ilio C. Proteus mirabilis glutathione S-transferase B1-1 is involved in protective mechanisms against oxidative and chemical stresses. Biochem J. 2003;373:305–311. doi: 10.1042/BJ20030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masai E, Ichimura A, Sato Y, Miyauchi K, Katayama Y, Fukuda M. Roles of the enantioselective glutathione S-transferases in cleavage of beta-aryl ether. J Bacteriol. 2003;185:1768–1775. doi: 10.1128/JB.185.6.1768-1775.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd-Jones G, Lau PC. Glutathione S-transferase-encoding gene as a potential probe for environmental bacterial isolates capable of degrading polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 1997;63:3286–3290. doi: 10.1128/aem.63.8.3286-3290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreto L, Garcera A, Jansson K, Sunnerhagen P, Herrero E. A peroxisomal glutathione transferase of Saccharomyces cerevisiae is functionally related to sulfur amino acid metabolism. Eukaryot Cell. 2006;5:1748–1759. doi: 10.1128/EC.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley AJ. Glutathione transferases: new functions. Curr Opin Struct Biol. 2005;15:716–723. doi: 10.1016/j.sbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 17.Vararattanavech A, Ketterman AJ. A functionally conserved basic residue in glutathione transferases interacts with the glycine moiety of glutathione and is pivotal for enzyme catalysis. Biochem J. 2007;406:247–256. doi: 10.1042/BJ20070422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caccuri AM, Antonini G, Board PG, Parker MW, Nicotra M, Lo Bello M, Federici G, Ricci G. Proton release on binding of glutathione to alpha, Mu and Delta class glutathione transferases. Biochem J. 1999;344(Pt 2):419–425. doi: 10.1042/0264-6021:3440419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZR, Bai M, Wang XY, Zhou JM, Perrett S. “Restoration” of glutathione transferase activity by single-site mutation of the yeast prion protein Ure2. J Mol Biol. 2008;384:641–651. doi: 10.1016/j.jmb.2008.09.047. [DOI] [PubMed] [Google Scholar]
- 20.Dourado DF, Fernandes PA, Mannervik B, Ramos MJ. Glutathione transferase: new model for glutathione activation. Chemistry. 2008;14:9591–9598. doi: 10.1002/chem.200800946. [DOI] [PubMed] [Google Scholar]
- 21.Vuillemier S, Pagni M. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl Microbiol Biotechnol. 2002;58:138–146. doi: 10.1007/s00253-001-0836-0. [DOI] [PubMed] [Google Scholar]
- 22.McGoldrick S, O’Sullivan SM, Sheehan D. Glutathione transferase-like proteins encoded in genomes of yeasts and fungi: insights into evolution of a multifunctional protein superfamily. FEMS Microbiol Lett. 2005;242:1–12. doi: 10.1016/j.femsle.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Vuillemier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem. 2007;71:1–15. doi: 10.1271/bbb.60437. [DOI] [PubMed] [Google Scholar]
- 25.Veal EA, Toone WM, Jones N, Morgan BA. Distinct roles for glutathione S-transferases in the oxidative stress response in Schizosaccharomyces pombe . J Biol Chem. 2002;277:35523–35531. doi: 10.1074/jbc.M111548200. [DOI] [PubMed] [Google Scholar]
- 26.Garcera A, Barreto L, Piedrafita L, Tamarit J, Herrero E. Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem J. 2006;398:187–196. doi: 10.1042/BJ20060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JH, Lou W, Vancura A. A novel membrane-bound glutathione S-transferase functions in the stationary phase of the yeast Saccharomyces cerevisiae . J Biol Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 28.Rai R, Tate JJ, Cooper TG. Ure2, a prion precursor with homology to glutathione S-transferase, protects Saccharomyces cerevisiae cells from heavy metal ion and oxidant toxicity. J Biol Chem. 2003;278:12826–12833. doi: 10.1074/jbc.M212186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickner RB, Koh TJ, Crowley JC, O’Neil J, Kaback DB. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast. 1987;3:51–57. doi: 10.1002/yea.320030108. [DOI] [PubMed] [Google Scholar]
- 30.Koonin EV, Mushegian AR, Tatusov RL, Altschul SF, Bryant SH, Bork P, Valencia A. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain—study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collinson EJ, Grant CM. Role of yeast glutaredoxins as glutathione S-transferases. J Biol Chem. 2003;278:22492–22497. doi: 10.1074/jbc.M301387200. [DOI] [PubMed] [Google Scholar]
- 32.Adamis PD, Gomes DS, Pinto ML, Panek AD, Eleutherio EC. The role of glutathione transferases in cadmium stress. Toxicol Lett. 2004;154:81–88. doi: 10.1016/j.toxlet.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Herrero E, Ros J, Tamarit J, Belli G. Glutaredoxins in fungi. Photosynth Res. 2006;89:127–140. doi: 10.1007/s11120-006-9079-3. [DOI] [PubMed] [Google Scholar]
- 34.Mariani D, Mathias CJ, da Silva CG, Herdeiro Rda S, Pereira R, Panek AD, Eleutherio EC, Pereira MD. Involvement of glutathione transferases, Gtt1and Gtt2, with oxidative stress response generated by H2O2 during growth of Saccharomyces cerevisiae . Redox Rep. 2008;13:246–254. doi: 10.1179/135100008X309028. [DOI] [PubMed] [Google Scholar]
- 35.Castro FA, Mariani D, Panek AD, Eleutherio EC, Pereira MD. Cytotoxicity mechanism of two naphthoquinones (menadione and plumbagin) in Saccharomyces cerevisiae . PLoS ONE. 2008;3(12):e3999. doi: 10.1371/journal.pone.0003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morel M, Kohler A, Martin F, Gelhaye E, Rouhier N. Comparison of the thiol-dependent antioxidant systems in the ectomycorrhizal Laccaria bicolor and the saprotrophic Phanerochaete chrysosporium . New Phytol. 2008;180:391–407. doi: 10.1111/j.1469-8137.2008.02498.x. [DOI] [PubMed] [Google Scholar]
- 37.Jeppesen MG, Ortiz P, Shepard W, Kinzy TG, Nyborg J, Andersen GR. The crystal structure of the glutathione S-transferase-like domain of elongation factor 1Bgamma from Saccharomyces cerevisiae . J Biol Chem. 2003;278:47190–47198. doi: 10.1074/jbc.M306630200. [DOI] [PubMed] [Google Scholar]
- 38.Prade L, Huber R, Bieseler B. Structures of herbicides in complex with their detoxifying enzyme glutathione S-transferase—explanations for the selectivity of the enzyme in plants. Structure. 1998;6:1445–1452. doi: 10.1016/S0969-2126(98)00143-9. [DOI] [PubMed] [Google Scholar]
- 39.Gronwald JW, Plaisance KL. Isolation and characterization of glutathione S-transferase isozymes from sorghum. Plant Physiol. 1998;117:877–892. doi: 10.1104/pp.117.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho HY, Lee HJ, Kong KH. A phi class glutathione S-transferase from Oryza sativa (OsGSTF5): molecular cloning, expression and biochemical characteristics. J Biochem Mol Biol. 2007;40:511–516. doi: 10.5483/bmbrep.2007.40.4.511. [DOI] [PubMed] [Google Scholar]
- 41.Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, Singh KB. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009;58:53–68. doi: 10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- 42.Matsuzaki F, Shimizu M, Wariishi H. Proteomic and metabolomic analyses of the white-rot fungus Phanerochaete chrysosporium exposed to exogenous benzoic acid. J Proteome Res. 2008;7:2342–2350. doi: 10.1021/pr700617s. [DOI] [PubMed] [Google Scholar]
- 43.Kim HG, Kim BC, Park EH, Ahn K, Lim CJ. Differential regulation of three genes encoding glutathione S-transferases in Schizosaccharomyces pombe . Mol Cells. 2004;18:332–339. [PubMed] [Google Scholar]
- 44.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 46.Magasanik B. The transduction of the nitrogen regulation signal in Saccharomyces cerevisiae . Proc Natl Acad Sci USA. 2005;102:16537–16538. doi: 10.1073/pnas.0507116102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae . Genetics. 2007;176:1557–1565. doi: 10.1534/genetics.107.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. Conservation of the prion properties of Ure2p through evolution. Mol Biol Cell. 2003;14:3449–3458. doi: 10.1091/mbc.E03-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–1167. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang ZR, Perrett S. Novel glutaredoxin activity of the yeast prion protein Ure2 reveals a native-like dimer within fibrils. J Biol Chem. 2009;284:14058–140067. doi: 10.1074/jbc.M901189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fraser JA, Davis MA, Hynes MJ. A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance. Appl Environ Microbiol. 2002;68:2802–2808. doi: 10.1128/AEM.68.6.2802-2808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK, Danley DE, Hoth LR, Griffor MC, Kamath AV, Rosner MH, Chrunyk BA, Perregaux DE, Gabel CA, Geoghegan KF, Pandit J. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J Biol Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 53.Whitbread AK, Masoumi A, Tetlow N, Schmuck E, Coggan M, Board PG. Characterization of the omega class of glutathione transferases. Methods Enzymol. 2005;401:78–99. doi: 10.1016/S0076-6879(05)01005-0. [DOI] [PubMed] [Google Scholar]
- 54.Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, Liebler DC, Aposhian HV. Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily. Chem Res Toxicol. 2001;14:1051–1057. doi: 10.1021/tx010052h. [DOI] [PubMed] [Google Scholar]
- 55.Dulhunty A, Gage P, Curtis S, Chelvanayagam G, Board P. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J Biol Chem. 2001;276:3319–3323. doi: 10.1074/jbc.M007874200. [DOI] [PubMed] [Google Scholar]
- 56.Board PG, Anders MW. Glutathione transferase omega 1 catalyzes the reduction of S-(phenacyl)glutathiones to acetophenones. Chem Res Toxicol. 2007;20:149–154. doi: 10.1021/tx600305y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Board PG, Coggan M, Cappello J, Zhou H, Oakley AJ, Anders MW. S-(4-Nitrophenacyl)glutathione is a specific substrate for glutathione transferase omega 1-1. Anal Biochem. 2008;374:25–30. doi: 10.1016/j.ab.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 58.Burmeister C, Luersen K, Heinick A, Hussein A, Domagalski M, Walter RD, Liebau E. Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1) FASEB J. 2008;22:343–354. doi: 10.1096/fj.06-7426com. [DOI] [PubMed] [Google Scholar]
- 59.McCarthy DL, Navarrete S, Willett WS, Babbitt PC, Copley SD. Exploration of the relationship between tetrachlorohydroquinone dehalogenase and the glutathione S-transferase superfamily. Biochemistry. 1996;35:14634–14642. doi: 10.1021/bi961730f. [DOI] [PubMed] [Google Scholar]
- 60.Reddy GV, Gold MH. Purification and characterization of glutathione conjugate reductase: a component of the tetrachlorohydroquinone reductive dehalogenase system from Phanerochaete chrysosporium . Arch Biochem Biophys. 2001;391:271–277. doi: 10.1006/abbi.2001.2417. [DOI] [PubMed] [Google Scholar]
- 61.Hruz T, Laule O, Szabo G, Wessendrop F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform [DOI] [PMC free article] [PubMed]
- 62.Bousset L, Belrhali H, Melki R, Morera S. Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds. Biochemistry. 2001;40:13564–13573. doi: 10.1021/bi011007b. [DOI] [PubMed] [Google Scholar]
- 63.Schmuck E, Cappello J, Coggan M, Brew J, Cavanaugh JA, Blackburn AC, Baker RT, Eyre HJ, Sutherland GR, Board PG. Deletion of Glu155 causes a deficiency of glutathione transferase Omega 1-1 but does not alter sensitivity to arsenic trioxide and other cytotoxic drugs. Int J Biochem Cell Biol. 2008;40:2553–2559. doi: 10.1016/j.biocel.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 64.Masai E, Kubota S, Katayama Y, Kawai S, Yamasaki M, Morohoshi N. Characterization of the C alpha-dehydrogenase gene involved in the cleavage of beta-aryl ether by Pseudomonas paucimobilis . Biosci Biotechnol Biochem. 1993;57:1655–1659. doi: 10.1271/bbb.57.1655. [DOI] [PubMed] [Google Scholar]
- 65.Otsuka Y, Sonoki T, Ikeda S, Kajita S, Nakamura M, Katayama Y. Detection and characterization of a novel extracellular fungal enzyme that catalyzes the specific and hydrolytic cleavage of lignin guaiacylglycerol beta-aryl ether linkages. Eur J Biochem. 2003;270:2353–2362. doi: 10.1046/j.1432-1033.2003.03545.x. [DOI] [PubMed] [Google Scholar]
- 66.Masai E, Katayama Y, Kawai S, Nishikawa S, Yamasaki M, Morohoshi N. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves beta-aryl ether. J Bacteriol. 1991;173:7950–7955. doi: 10.1128/jb.173.24.7950-7955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kües U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouzé P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 68.Yadav JS, Doddapaneni H, Subramanian V. P450ome of the white rot fungus Phanerochaete chrysosporium: structure, evolution and regulation of expression of genomic P450 clusters. Biochem Soc Trans. 2006;34:1165–1169. doi: 10.1042/BST0341165. [DOI] [PubMed] [Google Scholar]
- 69.Matityahu A, Hadar Y, Dosoretz CG, Belinky PA. Gene silencing by RNA Interference in the white rot fungus Phanerochaete chrysosporium . Appl Environ Microbiol. 2008;74:5359–5365. doi: 10.1128/AEM.02433-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.