Abstract
Triosephosphate isomerase (TIM) is a perfectly evolved enzyme which very fast interconverts dihydroxyacetone phosphate and d-glyceraldehyde-3-phosphate. Its catalytic site is at the dimer interface, but the four catalytic residues, Asn11, Lys13, His95 and Glu167, are from the same subunit. Glu167 is the catalytic base. An important feature of the TIM active site is the concerted closure of loop-6 and loop-7 on ligand binding, shielding the catalytic site from bulk solvent. The buried active site stabilises the enediolate intermediate. The catalytic residue Glu167 is at the beginning of loop-6. On closure of loop-6, the Glu167 carboxylate moiety moves approximately 2 Å to the substrate. The dynamic properties of the Glu167 side chain in the enzyme substrate complex are a key feature of the proton shuttling mechanism. Two proton shuttling mechanisms, the classical and the criss-cross mechanism, are responsible for the interconversion of the substrates of this enolising enzyme.
Keywords: Active site strain, Biocatalysis, Enolising chemistry, Evolution, Protein dynamics, Proton shuttling
Introduction
The previous reviews on triosephosphate isomerase (TIM) (E.C. 5.3.1.1) were written in 1977 and 1991 by Knowles [1, 2]. Since then, there have been specific reviews on the NMR-studies [3] and on the computational studies [4] of the TIM reaction mechanism. Knowles [5] has remarkably advanced our understanding of enzymatic catalysis in general and of TIM in particular, as he used his studies of this enzyme to develop the concept of optimally evolved catalytic power by enzymes through evolutionary pressure. Through this process, non-regulatory enzymes develop optimal catalytic efficiency in vivo and achieve k cat/K m values (for the least stable substrate), which are close to the diffusion encounter limit being 109 M−1 s−1 [6]. Other early classical studies of TIM established the enantioselectivity of the catalytic cycle [7], the importance of preventing the undesirable phosphate elimination side reaction [8], as well as the principle of transition state stabilisation by the active site of TIM [9]. In 1975, the first crystal structure of TIM was reported by Phillips and co-workers [10]. It concerned the structure of chicken muscle TIM (PDB entry 1TIM).
This review will mostly concern the recent advances made in the understanding of the TIM catalytic properties since the 1991 review by Knowles with a focus on the contribution of the structural studies in this respect, as documented in the literature available before 2010. Whenever relevant, the PDB-entry codes of the discussed structural data are mentioned in the text. First, a brief historic perspective of the TIM-research is given in this introduction. Further details will be given in the subsequent sections.
TIM is a dimeric, non-allosteric enzyme of the glycolytic pathway, in which it converts the α-hydroxyketone dihydroxyacetone phosphate (DHAP) stereospecifically into the α-hydroxyaldehyde d-glyceraldehyde-3-phosphate (d-GAP). This is an isomerisation reaction (Fig. 1). The pH optimum is found to be near pH 8 [11]. In the discussions in this review on the mechanism, the conversion of DHAP into d-GAP is considered, which is referred to as the forward reaction. It proceeds without the assistance of any cofactor or metal ion. In this reaction, a C–H bond needs to be broken. In the TIM reaction, as well as in many other reactions of biological relevance, the respective carbon atom is adjacent to a carbonyl group. In the forward TIM reaction, this carbonyl group is the keto group of DHAP, and in the reverse reaction, it is the aldehyde moiety of d-GAP. It intrigued scientists early on how enzymes achieve this activation [7]. This question initiated a search for finding a possible base that was used by the enzyme to abstract the activated proton. Using glycidol phosphate (Fig. 1), as a suicide inhibitor, Rose established that this base was an acidic side chain [12]. This was followed up by Knowles and co-workers [13] using BHAP as a suicide inhibitor (Fig. 1). Using BHAP [13], haloacetol phosphates [14] and glycidol-phosphate [15], the specific acidic residue was identified as Glu167. Schray et al. [15] established that d(S)-glycidol-phosphate is ten times more reactive than l(R)-glycidol-phosphate.
Fig. 1.
The TIM reaction, substrate analogues and suicide inhibitors. TIM catalyses the interconversion of dihydroxyacetone phosphate (DHAP) and d-glyceraldehyde-3-phosphate (d- GAP). Phosphoglycolohydroxamate (PGH), 2-phosphoglycollate (2PG) and 2-(N-formyl-N-hydroxy)-amino-ethylphosphonate (IPP) are reaction intermediate analogues. Bromohydroxyacetone phosphate (BHAP) and d,l-glycidolphosphate (d,l- GOP) are suicide inhibitors. Chiral centers are marked by asterisks
Currently,it has been established from several enzymes what enzymatic rate enhancements have been achieved during evolution when comparing their catalytic rates with the uncatalysed reaction rates [16]. For TIM, this comparison refers to comparing the rate of the chemical catalysis in solution by an organic base with the enzyme catalysed rate which was established to be 109 times faster [17]. Figure 2 visualises the free energy diagram of the catalysed and the uncatalysed reaction. As will be discussed in more detail in the following sections, the active site has developed several geometric tools [2] to stabilise the transition state, such as oxyanion holes [18], precise geometric positioning [2, 19] and specific dynamic properties [20].
Fig. 2.
The free energy profile of the TIM catalysed reaction (dotted line) as well as the buffer catalysed reaction (continuous line) [17]. This free energy profile is constructed from the rates of the relevant reactions; in the case of the second order binding rates for the formation of the enzyme substrate complex, a substrate concentration of 40 μM is assumed [48]. As is also described in the text, each of the steps of the TIM catalysed reaction, visualised in this diagram, namely (1) substrate binding, (2) deprotonation-chemistry (formation of the enediolate), (3) reprotonation-chemistry (formation of product), and (4) release of product, in fact consists of several steps
The comparison of the enzymatic catalysis and the non-enzymatic catalysis also revealed a second, very important functionality of TIM, which concerns the prevention of the undesirable phosphate elimination side reaction [8] (Fig. 3). Later structural studies have shown that this is achieved by tight binding of the phosphate moiety by three loops of the enzyme active site.
Fig. 3.
The formation of the first cis-enediolate intermediate from the substrate DHAP and the undesirable phosphate elimination reaction. The upper part of the figure shows the substrate, the first cis-enediolate intermediate and the product of the TIM catalytic cycle. The lower part visualises the phosphate elimination reaction, which, in solution, occurs easily in case of C3 sugar phosphates [8, 40]. It is favoured when the C1, C2 and C3 π orbitals overlap, as is the case when these orbitals are parallel to each other [29]. In TIM, the phosphate elimination reaction is prevented by the tight hydrogen bonding interactions of the phosphate moiety with the protein loops. Also the C–O(phosphate) bond is in many structures found to be almost in the enediolate plane; for example, the O2–C2–C3–O(phosphate) dihedral is −40° in the atomic resolution structure of the TIM–PGH complex (2VXN). A conformational rearrangement of the intermediates, for example in the case of loose binding of the phosphate moiety [57], will favour the methylglyoxal formation, as visualised in the lower part of the figure
The first crystal structure of TIM [10] revealed for the first time the TIM-barrel topology. This fold consists of a regular eightfold repeating pattern of β-strands and α-helices (Fig. 4). The β-strands form the inner set of eight parallel β-strands, covered on the outside by the subsequent α-helices. It is known that this (βα)8-fold is the most frequent fold of enzymes [21–23], having the active site always located at topologically the same site, being at the C-terminal end of the β-barrel and shaped by the eight βα-loops between these β-strands and the subsequent α-helices, referred to as loop-1, loop-2 to loop-8. Several reviews have summarised the key features of this fold [24, 25].
Fig. 4.
The TIM-barrel fold of triosephosphate isomerase (5TIM). a End-on view; the catalytic residues (Asn11, Lys13, His95 and Glu167) are highlighted. b Side view; the catalytic loops, at the C-terminal end of the β-strands (the catalytic end) are much more extensive than the loops at the N-terminal end of the β-strands (the stability end)
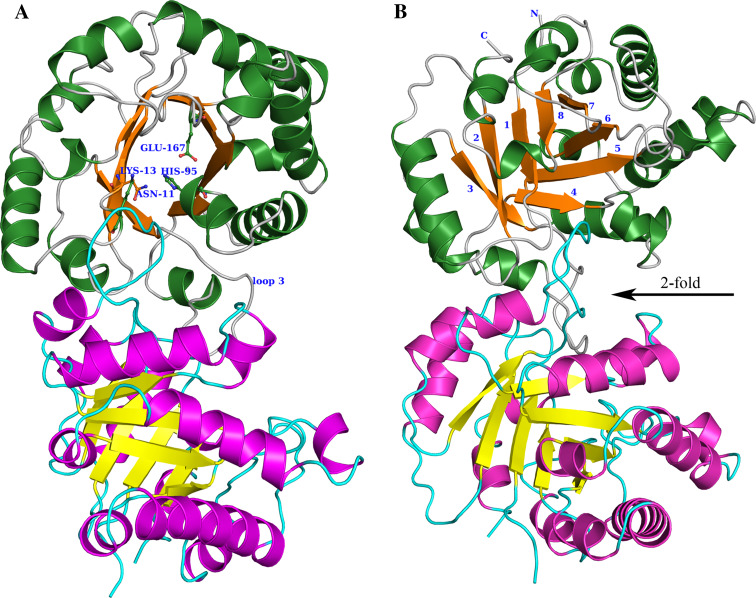
In TIM, three of the eight βα-loops provide the catalytic residues, being loop-1 (Asn11, Lys13), loop-4 (His95) and loop-6 (Glu167). The geometry of the active site is further shaped by a small helix in loop-4, an 11-residue-long flexible loop, which is part of loop-6, and loop-7 and loop-8, which, together with loop-6, provide important main chain hydrogen bond interactions with the phosphate moiety. The flexible loop-6 moves from an unliganded, preferably open, conformation (stabilised by loop-5) to a liganded, preferably closed, conformation (stabilised by loop-7). Loop-2 does not directly contribute to the active site geometry; however, loop-1, loop-2, loop-3 and loop-4 together with loop-8 provide the dimer interface. Only the TIM-dimer (Fig. 5) is fully active, although all catalytic residues of each active site are provided by the same subunit: dimerisation apparently rigidifies each of the two separate active sites to provide full catalytic power.
Fig. 5.
Two views of the wild-type TIM dimer (5TIM). a End-on view; Loop-3 of the lower subunit (cyan) is inserted between loop-1 and loop-4 of the upper subunit. b Side view (rotated by 45°, clockwise around the vertical axis of (a); the twofold dimer-axis is indicated by the arrow. The conformations of loops-8,1,2,3,4 are stabilised by dimer interface interactions. N and C are the N-terminus and C-terminus, respectively
The key components of the active site of TIM are (1) the catalytic base (Glu167), for abstracting the proton, as well as (2) electrostatic stabilisation [26] of the generated oxyanion(s), and (3) exclusion of bulk solvent [27]. The active site must also be able to accommodate the conformational change of the ketone-substrate when being converted into the aldehyde-product. It has been shown that the reactivity of the enzyme bound ketone-carbonyl and aldehyde-carbonyl moiety is different, suggesting different modes of binding of these moieties [28]. Early on, two competitive inhibitors have greatly contributed to the understanding of the TIM reaction mechanism; these molecules are the reaction intermediate analogues 2PG [9, 27, 29] (2YPI) and PGH [27, 30–32] (1TPH) (Fig. 1). These two molecules are the tightest known inhibitors of TIM with inhibition constants of 26 and 8 μM, respectively, for 2PG and PGH, when considering trypanosomal TIM [33]. The TIM inhibitory properties of analogues of these two classical inhibitors have been described more recently [18, 34]. NMR studies by Campbell have shown that 2PG binds in its dianionic form. On binding, the carboxylate of 2PG transfers its proton to Glu167 [35], converting it into a trianion. As shown in Fig. 1, 2PG is one atom shorter than the substrate, but Wolfenden [9] noticed its tight binding and proposed that this tight binding is due to its structural and charge similarity to the putative cis-enediolate-like transition state structure of the TIM catalytic cycle (Fig. 3). Also, PGH appears to bind as a trianion [20].
Physiological role of TIM
TIM is a glycolytic enzyme. In the human genome, there is only one gene encoding for TIM [36]. In most organisms, TIM is located in the cytosol, but in trypanosomes and leishmania, it is found both in the cytosol as well as in the glycosome (which is a peroxisome-like microbody, important for the glycolysis in these organisms) [37]. Also in these organisms only one gene is found [38]. In the glycolytic pathway, TIM comes after fructose-bisphosphate aldolase, which generates DHAP and d-GAP. d-GAP is further metabolised by the subsequent glycolytic enzymes and TIM ensures that the DHAP produced by aldolase can also be further metabolised by the glycolytic enzymes. The TIM activity is of critical importance for the proper functioning of the cell, as severe diseases are associated with point-mutated gene variants which cause a decrease of TIM activity. The disease symptoms are possibly related to the toxicity of DHAP [39].
The glycolytic pathway is interconnected to the pentosephosphate pathway, to lipid metabolism, and to the gluconeogenesis pathway via d-GAP and/or DHAP [36, 40]. The metabolic flow through these pathways will be affected in the case of TIM deficiency, which could also account for some of the disease symptoms.
The E104D-variant of human TIM is the best characterised disease mutation variant; its crystal structure has also been reported [41]. This mutation might have been introduced in one common ancestor, 1,000 years ago [42]. Glu104 is a fully conserved residue [43, 44] (see also Fig. 9, below). In patients homozygous for this mutation, only 2–20% of TIM catalytic activity is left [45]. Heterozygotes carrying this mutation are clinically normal [36, 44]. Homozygous patients with this mutation have severe disorders and usually do not live longer than 5 years [36, 39]. The structure of TIM shows that Glu104 is a dimer interface residue. Interestingly, this E104D-TIM variant when expressed and characterised as a recombinant protein is as equally active as wild-type TIM. Apparently, the E104D mutation prevents efficient folding into the dimer in the cell and/or causes the enzyme to be less stable [41].
Fig. 9.
The alignment of some TIM sequences. Included in this sequence alignment are the sequences of well-studied triosephosphate isomerases: chicken (Gallus gallus), yeast (Saccharomyces cerevisiae), leish (Leishmania mexicana), tryp (Trypanosoma brucei brucei), ecoli (Escherichia coli), and pfal (Plasmodium falciparum). The catalytic residues are highlighted by asterisks, a filled square marks residues which on mutation are known to cause human diseases, the inverted filled triangle identifies the tip of the dimer interface loop-3, and a filled triangle highlights the four NH-groups providing the hydrogen bond donors interacting with the phosphate moiety oxygen atoms
TIM deficiency is the only human glycolytic deficiency disease which is lethal, usually in early childhood [39]. The disease symptoms concern hemolytic anemia but also neurological disorders. Interestingly, it now appears that only certain TIM point mutations are causing the neurological disorders, the classical example being E104D, whereas others, like I170V (human TIM numbering, Ile172 in trypanosomal TIM) are causing the milder symptoms, such as hemolytic anemia, but no neurological disorders. The latter mutation is in loop-6 and is not destabilising the dimer, whereas the E104D mutation is destabilising the dimer. Using various model organisms (yeast, drosophila, mouse), it is now being investigated whether the neurological disorders caused by the E104D mutation should be classified as a conformational disease, related to the formation of amyloid-like precipitates by the misfolded protein in the brain [46].
TIM is a very efficient enzyme
The overall equilibrium constant calculated from the total-[DHAP]/total-[d-GAP] ratio is 22. In solution, DHAP is 60% unhydrated, but d-GAP is only 4% unhydrated [1, 47], which means that the equilibrium constant for the unhydrated species is 340 [1]. The free energy profile of the reaction catalysed by TIM (Fig. 2) has been determined by Knowles and co-workers. These studies have discovered that TIM is not only a very fast enzyme but also a very efficient enzyme [48–50]; for example, the rate of the conversion of d-GAP-to-DHAP is diffusion controlled, meaning that k cat/K m in this direction is close to its maximum possible value of 109 M−1 s−1 [6]. These studies [1] also established that the free energy barriers of the chemical conversion steps and the steps related to binding and conformational changes are not very different. In the forward (physiological, endergonic) direction (DHAP-to-d-GAP), the k cat is near 500 s−1, and in the reverse, exergonic, direction (d-GAP-to-DHAP), it is approximately 5,000 s−1. The K m of DHAP and d-GAP is 1.2 and 0.25 mM, respectively, and for the unhydrated species it is 0.7 and 0.01 mM, respectively [1, 11]. By carrying out the reaction in tritiated solvent and by studying the isotope exchange into the product [1, 48], it appears that in the latter (fastest) direction the rate limiting step is a proton transfer step, related to product formation, and not the DHAP release step. In the forward direction, the rate limiting step could be a conformational change related to the off-dissociation of d-GAP [48], such as the loop opening. In any case the loop opening rates are ligand dependent [51] and the loop opening rate on d-GAP formation appears to be much slower than on DHAP formation.
The free energy profile (Fig. 2) is calculated from measured reaction rates obtained to a great extent from isotope transfer and isotope discrimination experiments [1]. These calculations can be extended to also provide the relative quantities of DHAP, intermediate(s) and d-GAP [47]. The predominant complex is predicted to be the enzyme–DHAP complex (65%), but significant amounts of intermediate(s) and d-GAP are also predicted to be present. The recent NMR experiments by McDermott and co-workers [47] show that in fact only DHAP can be detected in the steady state population, but not the intermediates and d-GAP, suggesting that the reaction intermediates and product complex are short-lived, compared to the turnover time. Apparently, the fine details of the free energy profile are more complicated than is predicted from the kinetic model derived from the isotope studies, for example due to protein dynamics also playing a role in the kinetic landscape [47] (Fig. 2), as also discussed below.
The most difficult step in the forward direction of the catalytic cycle is the initial proton abstraction from DHAP [52, 53]. The catalytic power of the TIM active site for the catalysis of this reaction can be compared with the catalysis of this reaction by a simple base in solution. It turns out that the active site of TIM catalyses this deprotonation reaction approximately 109 times faster than the solution catalysis by a simple base. In terms of free energy barrier, it is calculated from available rate constants that these barriers are, for DHAP as the substrate, 13.5 kcal/mol when catalysed by TIM [48] and 26 kcal/mol for the simple base catalysis [5, 17, 49]. Theoretical studies find that the free energy of the transition state as well as of the intermediate is lowered by the enzyme by approximately 13 and 15 kcal/mol, respectively, as compared to the reaction in solution [54].
Structural studies
There are at least 111 crystal structures of TIMs in the PDB, as listed in Table 1, which is based on a survey completed in February 2010. These structures include the wild-type TIM structures of 22 organisms (Table 2). Three of these structures are at atomic resolution (Table 3). The TIM dimer is the most common quaternary structure, but in thermophilic organisms TIM is known to occur as tetramers. These tetramers are dimers of dimers. Apparently, the assembly of the two dimers into tetramers provides additional thermal stability [43]. Artificial, non-natural TIM monomers have also been extensively characterised by crystallographic (Table 1) and enzymological methods.
Table 1.
A compilation of all TIM PDB-entries
| pdb code | Source | Mutant | Active site ligand | Oligomeric state | Resolution (Å) | Rcryst (%) | Rfree (%) |
|---|---|---|---|---|---|---|---|
| 3KXQ | B. henselae | No | NO3 | Dimer | 1.6 | 15.6 | 18.6 |
| 1BTM | B. stearothermophilus | No | PGA | Dimer | 2.8 | 17.6 | |
| 2BTM | B. stearothermophilus | Yes | PGA | Dimer | 2.4 | 17.6 | 22.0 |
| 1MO0 | C. elegans | No | SO4 | Dimer | 1.7 | 18.3 | 21.3 |
| 1TMH | E. coli | Yes | SO4 | Dimer | 2.8 | 16.6 | |
| 1TRE | E. coli | No | Dimer | 2.6 | 11.9 | ||
| 1M6J | E. histolytica | No | Dimer | 1.5 | 18.4 | 20.6 | |
| 1SPQ | G. gallus | Yes | Dimer | 2.16 | 16.2 | 19.8 | |
| 1SQ7 | G. gallus | Yes | Dimer | 2.85 | 20.5 | 25.5 | |
| 1SSD | G. gallus | Yes | SO4 | Dimer | 2.9 | 15.8 | 19.3 |
| 1SSG | G. gallus | Yes | PGA | Dimer | 2.9 | 20.7 | 24.2 |
| 1SU5 | G. gallus | Yes | PGA | Dimer | 2.7 | 18.7 | 23.6 |
| 1SW0 | G. gallus | Yes | PGA | Dimer | 1.7 | 19.8 | 22.8 |
| 1SW3 | G. gallus | Yes | PGA | Dimer | 2.03 | 14.5 | 17.5 |
| 1SW7 | G. gallus | Yes | PGA | Dimer | 2.22 | 19.7 | 23.6 |
| 1TIM | G. gallus | No | Dimer | 2.5 | |||
| 1TPB | G. gallus | Yes | PGH | Dimer | 1.9 | 18.2 | |
| 1TPC | G. gallus | Yes | PGH | Dimer | 1.9 | 18.2 | |
| 1TPH | G. gallus | No | PGH | Dimer | 1.8 | 18.5 | |
| 1TPU | G. gallus | Yes | PGH | Dimer | 1.9 | 17.6 | |
| 1TPV | G. gallus | Yes | PGH | Dimer | 1.9 | 18.3 | |
| 1TPW | G. gallus | Yes | PGH | Dimer | 1.9 | 20.2 | |
| 8TIM | G. gallus | No | SO4 | Dimer | 2.5 | 17.7 | |
| 2DP3 | G. intestinalis | Yes | SO4 | Dimer | 2.1 | 17.5 | 18.1 |
| 2JGQ | H. pylori | No | PO4 | Dimer | 2.3 | 20.0 | 24.9 |
| 1HTI | H. sapiens | No | PGA | Dimer | 2.8 | 16.7 | |
| 1WYI | H. sapiens | No | Dimer | 2.2 | 26.6 | 29.6 | |
| 2JK2 | H. sapiens | No | Dimer | 1.7 | 22.2 | 25.2 | |
| 2VOM | H. sapiens | Yes | Dimer | 1.85 | 21.9 | 25.2 | |
| 1AMK | L. mexicana | No | PGA | Dimer | 1.83 | 10.7 | |
| 1IF2 | L. mexicana | Yes | 129 | Dimer | 2.0 | 13.6 | 18.1 |
| 1N55 | L. mexicana | Yes | PGA | Dimer | 0.83 | 9.5 | 10.8 |
| 1QDS | L. mexicana | Yes | PGA | Dimer | 2.0 | 15.3 | 19.5 |
| 2VXN | L. mexicana | Yes | PGH | Dimer | 0.82 | 9.2 | 10.3 |
| 2H6R | M. jannaschii | No | Tetramer | 2.3 | 21.3 | 27.8 | |
| 1AW1 | M. marina | No | PGA | Dimer | 2.7 | 19.2 | 21.5 |
| 1AW2 | M. marina | No | SO4 | Dimer | 2.65 | 20.0 | 21.9 |
| 3GVG | M. tuberculosis | No | Dimer | 1.55 | 14.5 | 16.9 | |
| 1R2R | O. cuniculus | No | Dimer | 1.5 | 16.1 | 19.0 | |
| 1R2S | O. cuniculus | No | Dimer | 2.85 | 17.4 | 21.2 | |
| 1R2T | O. cuniculus | No | Dimer | 2.25 | 18.4 | 22.0 | |
| 1LYX | P. falciparum | No | PGA | Dimer | 1.9 | 18.0 | 21.0 |
| 1LZO | P. falciparum | No | PGA | Dimer | 2.8 | 23.2 | 31.1 |
| 1M7O | P. falciparum | No | 3PG | Dimer | 2.4 | 18.3 | 22.5 |
| 1M7P | P. falciparum | No | G3H | Dimer | 2.4 | 17.9 | 21.6 |
| 1O5X | P. falciparum | Yes | 2PG | Dimer | 1.1 | 13.3 | 17.1 |
| 1VGA | P. falciparum | Yes | Dimer | 1.8 | 20.7 | 22.9 | |
| 1WOA | P. falciparum | Yes | G2H | Dimer | 2.8 | 21.9 | 25.2 |
| 1WOB | P. falciparum | Yes | SO4 | Dimer | 2.8 | 18.1 | 22.2 |
| 1YDV | P. falciparum | No | Dimer | 2.2 | 19.8 | 25.5 | |
| 2VFD | P. falciparum | Yes | SO4 | Dimer | 1.4 | 16.6 | 18.8 |
| 2VFE | P. falciparum | Yes | 3PG | Dimer | 2.2 | 22.1 | 27.6 |
| 2VFF | P. falciparum | Yes | Dimer | 1.7 | 19.4 | 22.3 | |
| 2VFG | P. falciparum | Yes | 3PG | Dimer | 1.95 | 21.0 | 24.8 |
| 2VFH | P. falciparum | Yes | 3PG | Dimer | 2.0 | 19.6 | 24.5 |
| 2VFI | P. falciparum | Yes | 3PG | Dimer | 2.25 | 21.2 | 26.0 |
| 1HG3 | P. woesei | No | 3PP | Tetramer | 2.7 | 25.4 | 28.6 |
| 1I45 | S. cerevisiae | Yes | Dimer | 1.8 | 17.5 | 20.8 | |
| 1NEY | S. cerevisiae | Yes | 13P | Dimer | 1.2 | 12.5 | 15.0 |
| 1NF0 | S. cerevisiae | Yes | 13P | Dimer | 1.6 | 20.9 | 26.8 |
| 1YPI | S. cerevisiae | No | Dimer | 1.9 | 21.0 | ||
| 2YPI | S. cerevisiae | No | PGA | Dimer | 2.5 | 17.7 | |
| 3YPI | S. cerevisiae | No | PGH | Dimer | 2.8 | 18.4 | |
| 7TIM | S. cerevisiae | No | PGH | Dimer | 1.9 | 18.3 | |
| 1AG1 | T. brucei brucei | No | PO4 | Dimer | 2.36 | 15.0 | |
| 1DKW | T. brucei brucei | Yes | Dimer | 2.65 | 18.6 | 24.2 | |
| 1IIG | T. brucei brucei | Yes | 3PP | Dimer | 2.6 | 12.5 | |
| 1IIH | T. brucei brucei | Yes | 3PG | Dimer | 2.2 | 14.0 | |
| 1KV5 | T. brucei brucei | Yes | PGA | Dimer | 1.65 | 14.5 | 17.5 |
| 1ML1 | T. brucei brucei | No | PGA | trimer | 2.6 | 23.1 | 24.7 |
| 1MSS | T. brucei brucei | No | Dimer | 2.4 | 19.8 | ||
| 1TPD | T. brucei brucei | No | Dimer | 2.1 | 17.6 | ||
| 1TPE | T. brucei brucei | No | Dimer | 2.1 | 15.5 | ||
| 1TPF | T. brucei brucei | No | DMS | Dimer | 1.8 | 19.9 | |
| 1TRD | T. brucei brucei | No | PGH | Dimer | 2.5 | 14.7 | |
| 1TRI | T. brucei brucei | No | SO4 | Monomer | 2.4 | 16.5 | |
| 1TSI | T. brucei brucei | No | 4 PB | Dimer | 2.8 | 11.5 | |
| 1TTI | T. brucei brucei | Yes | PGA | Monomer | 2.4 | 17.8 | |
| 1TTJ | T. brucei brucei | Yes | PGH | Monomer | 2.4 | 17.8 | |
| 2J24 | T. brucei brucei | Yes | Dimer | 2.1 | 15.3 | 23.0 | |
| 2J27 | T. brucei brucei | Yes | PGA | Dimer | 1.15 | 14.2 | 19.0 |
| 2V0T | T. brucei brucei | Yes | SO4 | Dimer | 2.2 | 18.1 | 23.8 |
| 2V2C | T. brucei brucei | Yes | PGA | Dimer | 1.89 | 14.2 | 18.2 |
| 2V2D | T. brucei brucei | Yes | Trimer | 2.3 | 20.4 | 27.0 | |
| 2V2H | T. brucei brucei | Yes | PGA | Trimer | 1.18 | 13.9 | 18.7 |
| 2V5L | T. brucei brucei | No | SO4 | Dimer | 2.4 | 15.8 | |
| 2VEI | T. brucei brucei | Yes | SO4 | Trimer | 1.89 | 16.8 | 21.5 |
| 2VEK | T. brucei brucei | Yes | CIT | Monomer | 1.6 | 16.7 | 19.3 |
| 2VEL | T. brucei brucei | Yes | PGA | Monomer | 2.3 | 18.4 | 25.7 |
| 2VEM | T. brucei brucei | Yes | BBR | Monomer | 2.2 | 19.6 | 24.7 |
| 2VEN | T. brucei brucei | Yes | CIT | Monomer | 2.0 | 18.8 | 23.7 |
| 2WSQ | T. brucei brucei | Yes | Dimer | 2.1 | 20.8 | 24.4 | |
| 2WSR | T. brucei brucei | Yes | Monomer | 1.65 | 21.5 | 25.5 | |
| 2X16 | T. brucei brucei | Yes | Monomer | 2.13 | 18.4 | 26.1 | |
| 2X1R | T. brucei brucei | Yes | X1R | Monomer | 1.98 | 16.4 | 22.6 |
| 2X1S | T. brucei brucei | Yes | X1S | Monomer | 1.93 | 16.2 | 20.1 |
| 2X1T | T. brucei brucei | Yes | RES | Monomer | 1.83 | 17.4 | 21.9 |
| 2X1U | T. brucei brucei | Yes | SO4 | Monomer | 1.84 | 18.5 | 23.7 |
| 2X2G | T. brucei brucei | Yes | 3PG | Monomer | 1.90 | 17.1 | 22.4 |
| 3TIM | T. brucei brucei | No | Dimer | 2.8 | 13.9 | ||
| 4TIM | T. brucei brucei | No | 2PG | Dimer | 2.4 | 14.9 | |
| 5TIM | T. brucei brucei | No | SO4 | Dimer | 1.83 | 18.3 | |
| 6TIM | T. brucei brucei | No | G3P | Dimer | 2.2 | 13.7 | |
| 1CI1 | T. cruzi | No | Dimer | 2.0 | 18.1 | 23.9 | |
| 1SUX | T. cruzi | No | SO4 | Dimer | 2.0 | 18.3 | 19.6 |
| 1TCD | T. cruzi | No | Dimer | 1.83 | 19.1 | 25.8 | |
| 2OMA | T. cruzi | Yes | SO4 | Dimer | 2.15 | 19.9 | 25.0 |
| 2V5B | T. cruzi | No | Dimer | 2.0 | 21.4 | 25.7 | |
| 1B9B | T. maritima | No | SO4 | Tetramer | 2.85 | 21.1 | 24.9 |
| 2I9E | T. molitor | No | Dimer | 2.0 | 22.1 | 23.9 | |
| 1W0M | T. tenax | No | Tetramer | 2.5 | 19.7 | 22.6 |
123 [2(formyl-hydroxy-amino)-ethyl]-phosphonic acid, 13P 1,3-dihydroxyacetone phosphate, 2PG 2-phosphoglyceric acid, 3PG 3-phosphoglyceric acid, 3PP 3-phosphonopropanoic acid, 3PY 3-hydroxypyruvic acid, 4PBN-hydroxy-4-phosphono-butanamide, ACT acetate ion, BBR (3-bromo-2-oxo-propoxy)phosphonic acid, CIT citric acid, DMS dimethyl sulfoxide, G2H 2-hydroxy-1-(hydroxymethyl)ethyl dihydrogen phosphate, G3H glyceraldehyde-3-phosphate, G3P sn-glycerol-3-phosphate, PGA 2-phosphoglycolic acid, PGH phosphoglycolohydroxamic acid, PO4 phosphate ion, SO4 sulfate ion, X1R 3-(propylsulfonyl)propanoic acid, X1S 3-sulphopropionic acid, RES 4-phospho-d-erythronohydroxamic acid, NO3 nitrate
Table 2.
A compilation of the organisms of which TIM structures are known
| Archaea | Methanocaldococcus jannaschii dsm 2661 |
| Pyrococcus woesei | |
| Thermoproteus tenax | |
| Bacteria | Bartonella henselae |
| Escherichia coli | |
| Bacillus stearothermophilus | |
| Helicobacter pylori | |
| Moritella marina | |
| Mycobacterium tuberculosis | |
| Thermotoga maritima | |
| Eukarya | Caenorhabditis elegans |
| Entamoeba histolytica | |
| Gallus gallus | |
| Giardia intestinalis | |
| Homo sapiens | |
| Leishmania mexicana | |
| Oryctolagus cuniculus | |
| Plasmodium falciparum | |
| Saccharomyces cerevisiae | |
| Tenebrio molitor | |
| Trypanosoma brucei brucei | |
| Trypanosoma cruzi |
Table 3.
The three atomic resolution TIM structures
| PDB code | Source | Resolution (Å) | Ligand |
|---|---|---|---|
| 2VXN | L. mexicana | 0.82 | PGH |
| 1N55 | L. mexicana | 0.83 | 2PG |
| 1NEY | S. cerevisiae | 1.20 | BHAP |
The structural studies have shown that the active site is at the dimer interface, with all catalytic residues for a particular active site coming from the same subunit. The second subunit defines the active site by inserting its loop-3 between loop-1 and loop-4 of the first subunit at the subunit–subunit interface (Fig. 5). In particular, Thr75, at the tip of loop-3, contributes to the hydrogen bonding network of the active site of the other subunit. Several water molecules are an integral part of the dimer interface; six of these water molecules are highly conserved [55].
The structural studies have highlighted the importance of two flexible loops for substrate binding and catalysis. Best known is the closure of loop-6 on substrate binding, as it moves by approximately 7 Å from an open/unliganded, somewhat disordered conformation to a closed/liganded conformation. In the unliganded conformation, loop-6 is interacting with loop-5, whereas in the closed/liganded conformation it interacts with loop-7. Smaller conformational changes are seen for loop-7 when the ligand binds in the active site. The closing of these loops generates a very specific phosphate-dianion binding pocket, in which the phosphate dianion moiety is anchored via four hydrogen bonds to four main chain NH-groups of these loops (1 hydrogen bond to loop-6, 1 hydrogen bond to loop-7 and 2 hydrogen bonds to loop-8) (Fig. 6). Monoanionic sulfonate analogues, like dihydroxyacetone sulfate, are poor TIM binders [56]. Apparently, this geometry of the four main chain NH-groups of loop-6, loop-7 and loop-8 (in the closed conformation) provides a good dianion phosphate binding site, whereas the monoanion analogues bind poorly. Possibly, in the latter case, the repulsive electrostatic interactions between the four spatially close peptide NH-groups of the closed conformation are not sufficiently compensated by the bound monoanionic sulfonate group. A TIM variant in which four residues at the tip of loop-6 have been removed is unable to make four hydrogen bonds with the phosphate moiety, and therefore presumably does not bind the reaction intermediate tight enough to prevent methylglyoxal synthase formation (Fig. 3); this variant has become a methylglyoxal synthase [57].
Fig. 6.
The active site geometry as seen in the structure of the TIM–PGH complex (2VXN) (in stereo). Note that the hydroxamate end of the reaction intermediate analogue is completely dehydrated. The hydroxamate plane mimics the enediolate plane. Four water molecules are hydrogen bonded to the phosphate moiety and two water molecules are hydrogen bonded to the main chain and side chain of the catalytic glutamate. The four hydrogen bonds between the phosphate moiety and the peptide NH-groups of loop-6 (Gly173), loop-7 (Ser214) and loop-8 (Gly234, Gly235) are indicated by dotted lines. Wat-6 is hydrogen bonded to a string of waters which extends to the bulk solvent
Important geometrical features of the active site are visualised in Fig. 6. The catalytic site is completely buried and shielded from bulk solvent. The catalytic site is also very compact, in particular the interactions between the catalytic glutamate side chain (Glu167) and the substrate are very tight. Nevertheless, the side chain of the catalytic glutamate can move in a small hydrophobic pocket above the hydroxamate plane [20]. The key catalytic residue is Glu167, but Asn11, Lys13, and His 95 are also important and therefore fully conserved (see also Fig. 9, below). As shown in Fig. 6, Glu167 is above the hydroxamate plane, whereas Lys13 and Asn11 are below. The imidazole ring of the His95 side chain is coplanar with the cis-enediolate plane. The latter three residues provide an oxyanion hole-like stabilisation for the negatively charged intermediates of the reaction. The importance of Asn11 and Lys13 for electrostatic stabilisation has been described [18, 58, 59]. The His95 side chain is also directly involved in the proton transfer steps, as will be discussed later.
The catalytic end of the substrate is completely dehydrated, but the side chain of the catalytic glutamate is hydrogen bonded to a water molecule (Fig. 6). This water is part of an extensive hydrogen bonding network of waters extending to the bulk solvent [60]. In the apo structure, the active site has more bound water molecules, which are displaced on substrate binding [61].
The three atomic resolution studies of TIM
There are three reported atomic resolution X-ray structures of complexes of TIM. It concerns complexes with the DHAP substrate (1NEY) [19], with the reaction intermediate analogue 2PG (1N55) [62] and with the reaction intermediate analogue PGH (2VXN) [20]. At this high resolution, it becomes possible to obtain information on the anisotropic dynamical properties and the protonation state of the active site components. In the crystal structure of the TIM–DHAP complex (at 1.2 Å), the predominant ligand is DHAP, and the occupancies of other possible conformers, being the intermediate(s) or the product (d-GAP), must be low. Also, NMR studies [47] show that in the presence of DHAP the predominant complex is the enzyme–DHAP complex. Each of these three atomic resolution structures highlights the very tight interactions between the carboxylate group of the catalytic glutamate and the reactive moiety of the substrate. For example, in the TIM–DHAP complex, the carboxylate oxygen is placed at 3 Å from the substrate carbon. As discussed below, the TIM–PGH structure (at 0.82 Å) provides insight into the importance of protein dynamics of the catalytic glutamate for the proton shuttling mechanism. The TIM–2PG structure (at 0.83 Å) revealed that, in the liganded active site of TIM, the proline ring of the Glu167-Pro168 dipeptide is planar [62]. It is speculated that this strained conformation facilitates loop opening and therefore product release, once the reaction has been completed [62, 63].
The TIM-barrel fold
Many catalytic sites, of otherwise totally unrelated enzymes, are now known to be built on the TIM-barrel framework, using the loops emerging out of the C-terminal ends of the eight central β-strands of the TIM-barrel (Fig. 4) [22, 23] for providing the binding and catalytic residues. The minimum symmetry of the TIM-barrel is fourfold [24]. It has been noted that these loops are of variable length. The total number of residues building the TIM-barrel unit varies from 150 to 500 [23], with an average of 300. The TIM barrel is approximately 250 residues. The large variability in size is caused by the variable lengths of the loops at the C-terminal end of the β-barrel (the catalytic end), whereas the loops at the N-terminal end (the stability end) are usually shorter (Fig. 4). Longer TIM-barrel units are sometimes observed in the case where an additional domain has been inserted in the TIM-barrel fold, as observed for example in pyruvate kinase, where an extra domain has been inserted at the C-terminal end of β-strand-3 [64]. A systematic survey [23] lists more examples; also in these cases the inserted domain is after β-strand-3. Apparently, the TIM-barrel fold is a very good framework for many different enzyme active sites. The importance of the electrostatic properties of this fold has also been discussed [65, 66]. Evolutionary studies [22, 25, 67] have shown that the TIM-barrel has possibly evolved from two half barrels [68]. Also, recent enzyme design studies [69, 70], using either directed evolution [71] or computational approaches [72], highlight the importance of the TIM-barrel fold as an important framework for making non-natural enzymes.
The studies of monomeric TIMs
Only the TIM dimer is fully active, and therefore several drug discovery studies have attempted to find inhibitors that interfere with the folding and assembly of monomers into dimers, such that, for example, TIMs from parasitic organisms are selectively inhibited. Two approaches have been reported. Balaram and co-workers [73] have found that a peptide analogue of malarial TIM loop-3 is indeed an inhibitor with an IC50 value of approximately 0.7 μM, when tested on malarial TIM. In drug discovery projects, it is attractive to search for specific inhibitors with binding sites at the protein–protein interfaces of oligomeric enzymes, like TIM, which are only fully active in the oligomeric state, as the sequence conservation of the protein–protein interfaces is much less than at the active site. For example, specific inhibitors binding at the Trypanosoma cruzi TIM dimer interface (1SUX, 2OMA) have been studied [74, 75].
The catalytic residues of TIM are all located on the same subunit. Some of them (Asn11, Lys13, His95) are on loops that are stabilised by the dimer interface interactions. And indeed the enzymatic studies on monomeric TIMs show that the monomers have much reduced catalytic efficiency. Also, the structural properties of several monomeric TIM’s have been studied systematically. The first monomeric TIM, monoTIM (1TRI), was generated after following a careful loop design protocol, using the computational tools implemented in the modelling package ICM [76, 77]. Via this protocol, a 15-residue stretch (loop-3) of the trypanosomal, glycosomal TIM was changed into an 8-residue stretch. The crystal structures of several of these monomeric TIMs (Table 1) have shown that the loops involved in dimerisation in wild-type TIM become disordered or adopt non-wild-type conformations (in particular in the unliganded structures) in the monomeric state [33, 78]. For example, loop-4 is also seen to adopt a new well-ordered outward-oriented conformation in monoTIM (Fig. 7), whereas loop-1 is disordered in most unliganded structures. Loop-8 has an unusual conformation in one of the unliganded structures (1MSS) (Fig. 7). Concerning loop-4, it was found that in monomeric TIMs the wild-type loop-4 conformation is stabilised by the bulky side chain of the A100W-mutation, which was subsequently kept in the later generation of monomeric variants. The mutation A100W does not affect the catalytic properties of these monomeric TIMs [33]. In the TIM-dimer, the dimer interface loops, loop-8, loop-1, loop-2, loop-3 and loop-4 are all very rigid. The structural properties of loop-8, loop-1, loop-3, and loop-4 of the monomeric TIMs suggest that in the wild-type monomeric folding intermediate of TIM these loops are also disordered. Subsequently, in the folding pathway of wild-type TIM towards the mature dimer, these disordered loop regions of the two subunits then interact with each other such that a very rigid dimer interface is generated on dimerisation [79].
Fig. 7.
The comparison of the structures of wild-type and monomeric TIM variants in which only loop-3 is shortened. Three structures are shown, being wild-type TIM (open, unliganded) (5TIM-subunit-A) (green), monoTIM (closed, liganded with sulfate) (1TRI) (purple) and monoTIM-SS (open, unliganded) (1MSS, molecule A) (yellow). In monoTIM-SS, two surface residue point mutations (F45S, V46S) have also been introduced to improve the crystallisation properties; monoTIM and monoTIM-SS have the same catalytic properties [33]. Note the structural variability of loop-8, loop-1, and loop-4; these loops are, in wild-type TIM, very rigid because of the dimer interface interactions, but in the monomeric TIMs they display a large conformational diversity
The monomeric TIMs still have considerable TIM activity having a k cat of approximately 1 s−1 and a K m of approximately 5 mM, which means that the k cat is approximately 1,000-fold lower and the K m is approximately ten times higher than wild-type [33]. Methylglyoxal synthase activity was not detected [33], indicating that the phosphate anchoring mechanism is not disabled by the monomerisation. The monomeric TIMs are somewhat less stable than the wild-type TIM dimer [33, 80]. The pH optimum of these monomeric variants is not known and also the free energy profile of the catalytic cycle has not been determined. However, from the structural studies, it is known that the characteristic open (unliganded) and closed (liganded) conformational states are still present in these variants [78].
The lower catalytic efficiency of the monomeric TIMs can be understood from their structural properties, given the much more flexible catalytic loops, loop-1, loop-4 and loop-8. Also, the active site is much more exposed to bulk solvent. Nevertheless, directed evolution experiments using monoTIM as the starting point [81] have shown that mutations in loop-2 do increase k cat by a factor 11, while reducing K m by a factor of 4.
The first monomeric TIM variants have been used for engineering new substrate specificity by shortening loop-1 (ml1TIM, 1ML1) [82] and subsequently loop-8 (ml8bTIM, 1DKW) [79]. The initial loop-8 deletion variant showed no activity and also poor binding properties, but the V233A variant of this deletion mutant (A-TIM) has much better binding properties [83]. Careful analysis of the high resolution structure of ml8bTIM [83] (2VEI) provided the rational for the importance of the V233A mutation, as it showed that Val233 in the ml8bTIM structure pointed further into the bulk solvent (as compared to wild-type) and it therefore blocked the phosphate binding site, as well as interfering with the wild-type conformation of loop-7. It could be shown that A-TIM has a competent active site, being able to bind the reaction intermediate analogue 2PG (2VEL) (Fig. 8) as well as the suicide inhibitor BHAP (2VEM), although the catalytic activity is below the detection limit of the standard assay [11]. Interestingly, the putative A-TIM active site also binds larger molecules, like citrate, which cannot bind to wild-type TIM [83] (2VEK). In A-TIM, loop-1, loop-3 and loop-8 have been shortened, and there is a point mutation in loop-4 (A100W). These studies show that the loops at the catalytic end of the TIM (βα)8-barrel can be considerably mutated without the framework losing its integrity (Fig. 8). The A-TIM framework is now an attractive starting point for finding new catalytically active variants having a non-natural substrate specificity [83].
Fig. 8.
The comparison of the structures of the closed, 2PG bound complexes of wild-type TIM (1N55) (green), ml1TIM (1ML1, molecule A) (cyan) and A-TIM (2VEL, molecule B) (purple). ml1TIM has a shortened loop-3, and loop-1, as well as the point mutation A100W in loop-4. In A-TIM, loop-8 has also been shortened. There are no structural changes at the stability end of the TIM barrel, while at the catalytic end of the TIM-barrel significant structural changes in the loops are observed, due to the mutations/deletions in these loops
Evolutionary conserved sequence patterns
The best-studied TIMs are chicken TIM, yeast TIM, trypanosomal TIMs, leishmania TIM, and malaria TIM. We refer in this review to the trypanosomal TIM (T. brucei brucei) sequence numbering, in which the active site residues are Asn11, Lys13, His95, and Glu167. Figure 9 compares the sequences of these five TIMs. Important structural and sequence fingerprints are highlighted in this figure.
The four key catalytic residues are fully conserved [43]. Also highly conserved are the residues in loop-6, loop-7 and loop-8. Interestingly, it has been found that in sequences of archea the sequence conservation of loop-6 is different, most likely compensated by sequence differences in loop-7 [84]. Such compensating sequence differences have also been noted for some other unique sequence patterns, such as for Glu65 in leishmania TIM (almost always a glutamine) [85] and for Phe96 in malaria TIM (almost always a serine) [61].
The isomerase reaction
The TIM reaction is initiated by deprotonation of a carbon atom adjacent to a carbonyl moiety. In general, a ketose–aldose isomerisation reaction can be achieved by two possible mechanisms, either by proton transfer or by metal ion-dependent hydride transfer [86]. In solution, at basic pH, both mechanisms occur and are approximately equally efficient [86]. In the catalysis by TIM, the proton transfer mechanism is used. The first step in the reaction mechanism is the abstraction of a proton from a carbon adjacent to a carbonyl function, being either an aldehyde (when the substrate is d-GAP) or a ketone (when DHAP is the substrate). In both cases, the first intermediate formed is a cis-enediolate (Fig. 3). TIM is therefore also referred to as an enolising enzyme [87–89]. TIM is highly specific for the C3-sugar phosphates DHAP and d-GAP. Other sugar phosphate isomerases which use the same proton transfer mechanism [90] are, for example, d-ribose-5-phosphate isomerase (RPI) [91] and phosphoglucose isomerase (PGI) [92]. These two enzymes catalyse the same reaction as TIM, but the substrate is, respectively, a C5-sugar phosphate and a C6-sugar phosphate. There are actually two different RPI enzymes, having completely different folds, but similar reaction mechanisms, referred to as RPI-A [93] and RPI-B [91]. The structures of complexes of the respective hydroxamate reaction intermediate analogues with PGI, RPI-B and TIM have been determined (Fig. 10). Each of these three enzymes uses a glutamate as a catalytic base and in each of these enzymes an oxyanion hole stabilises the negative charge on the cis-enediolate that is formed on proton abstraction. It is interesting to note the different hydrogen bond donors recruited for making the respective oxyanion holes. In TIM, the oxyanion hole for the O2 oxygen is formed by the NZ(Lys13) and NE2(His95), whereas in the complexed RPI-B structure (2BES) the oxyanion hole is formed by the main chain N(Ser71), OG(Ser71) and a water (Wat2064), and in the complexed PGI structure (1KOJ) only water molecules are hydrogen bonded to the carbonyl oxygen atom (Wat241, Wat423, and Wat477). In each of these three enzymes, proton transfer between the two oxygen atoms of the cis-enediolate intermediate is in principle possible, either via a histidine (NE2 (His95)) in TIM (2VXN), or via a water (Wat241) in PGI (1KOJ), or via a serine side chain [OG(Ser71)] in RPI-B (2BES).
Fig. 10.
The comparison of the mode of interactions of the hydroxamate reaction intermediate analogue and the active site residues of three sugar phosphate isomerases: TIM (cyan; 1TPH), RPI-B (green; 2BES) and PGI (magenta; 1KOJ) (in stereo). The hydroxamate moieties of each of the three structures have been superimposed. In each active site, the catalytic base is a glutamate: Glu167 in TIM, Glu75 in RPI-B and Glu357 in PGI. For the shared hydrogen bonding with the O1 and O2 atoms of the substrate and enediolate intermediate in TIM NE2(His95) is important, whereas in RPI-B and PGI the corresponding hydrogen bonding partners are OG(Ser71), and a water molecule (Wat241), respectively. The oxyanion hole for the O2 atom of the enediolate is made by Lys13 and His95 in TIM, by main chain NH-groups of residues Gly70-Ser71, as well as by Wat2064 and OG(Ser71) in RPI-B, and by three water molecules (Wat423, Wat477, Wat241) in PGI
Only in the case of TIM, the phosphate of the substrate is well anchored by interactions with four main chain NH-groups of three different loops, whereas no side chains are involved in this interaction. These phosphate binding loops adopt completely different conformations in the unliganded and liganded conformation of TIM. In PGI, the phosphate has two hydrogen bonds to main chain NH groups, whereas in RPI-B there is no such hydrogen bond to a main chain NH-group. In each of these two structures at least one arginine or lysine side chain contributes to the binding of the phosphate moiety. In RPI-A there also exist hydrogen bonds between the phosphate and side chain lysines and main chain NH-groups. From E. coli RPI-A, the liganded (1O8B) and unliganded (1KS2) [93] structures are available. For this enzyme, a small domain movement is seen, allowing for better hydrogen bonding with the phosphate moiety when comparing the two different states. For RPI-A of Thermus thermophilus [94], only minor structural differences are seen for the liganded (1UJ5) and unliganded structures (1UJ4). For rabbit PGI, the liganded (1KOJ) and unliganded (1HM5) [95] structures are available, and for this enzyme, a small movement of the phosphate binding loop is observed on ligand binding. The phosphate anchoring mechanism in TIM is much tighter and more complicated than in RPI and PGI. This correlates with the notion that in the uncatalysed TIM isomerase reaction the phosphate elimination reaction is favoured (Fig. 3) [8], but not in the RPI and PGI catalysed isomerase reaction, due to the longer carbon skeletal of the substrates, respectively a C5-sugar phosphate and a C6-sugar phosphate.
It has been noted that many sugar isomerases (having as substrates non-phosphorylated sugars) use the metal-dependent hydride transfer mechanism [86], like d-xylose isomerase [96]. Generally, the latter enzymes are less proficient than the enolising isomerases utilising sugar phosphates as substrates [86, 97]. Richard and colleagues have proposed that the phosphate binding energy is used to lower the transition state free energy barrier [98, 99]. A very elegant proof of this is provided by the observation that the conversion of the truncated substrate analogue glycolaldehyde (having no covalently bound phosphate moiety) is activated 700-fold in the presence of the phosphite dianion ((HPO3)2−), a phosphate analogue [99]. Product analysis of the phosphite-activated isomerisation of glycolaldehyde shows that the proton transfer mechanism is the same [100]. Apparently, both reactions occur in the closed form of TIM, and the 700-fold activation by the phosphite dianion is achieved by using its intrinsic binding energy to stabilise this closed conformation. The use of substrate binding interactions to increase catalytic efficiency instead of affinity has also been described for other enzymes [6].
The classical and the criss-cross reaction mechanism
The TIM active site catalyses the interconversion of DHAP and d-GAP in a highly stereospecific manner. The C1 pro-R proton is abstracted when DHAP is the substrate and the C2-R proton is abstracted when d-GAP is the substrate. This chiral specificity can be understood readily from the available crystal structures of the enzyme substrate complexes, but in fact was already proposed by Rose and co-workers [7]. The mode of binding of the substrate with respect to the base is in good agreement with the stereo electronic requirement for efficient enolisation [2, 101], which states that the proton to be abstracted is positioned orthogonal with respect to the enolate plane defined by the O1, C1, C2 and O2 atoms (schematically indicated in Fig. 3).
The first intermediate of the catalytic cycle is generated after the initial proton abstraction by Glu167. For efficient catalysis, it is required that the pK a of the base and substrate are in the same range [27, 88]. In solution, the pK a of the substrate is expected to be in the range of 16–20 [102, 103], whereas for the base, the Glu167 carboxylate moiety, the pK a of the uncomplexed enzyme is approximately 4 [104], which is very similar to the pK a of the glutamate side chain, when dissolved in water [6]. Efficient catalysis requires that these two pK a values will be modified and thereby come closer to each other in the competent complex [88], which is achieved by the buried active site geometry of the closed, liganded structure.
The enzyme achieves a 109 rate enhancement by stabilising the transition state and the intermediate states. Theoretical and experimental [105] studies suggest that tunnelling of the hydrogen atom does not play an important role in the TIM mechanism, as reviewed by Cui and Karplus [4]. The importance of low-barrier hydrogen bonds (LBHBs) for stabilisation of the transition state and intermediates has been addressed [106]. Although NMR [107] and X-ray studies [20] have shown that short hydrogen bonds can be found in the structures of TIM complexed with reaction intermediate analogues, their importance for the catalytic rate enhancement achieved by TIM remains to be investigated further.
Experimental and computational studies suggest that at least two proton transfer pathways can complete the substrate to product conversion: the classical mechanism [2, 108] (Fig. 11) and the criss-cross mechanism [109, 110] (Fig. 12). In the criss-cross pathway, all proton transfer steps are carried out by Glu167, whereas in the classical pathway, the His95 side chain is also involved in the proton transfer. In each of these pathways, the chemistry steps of the simplistic diagram of Fig. 2 are split into several steps. The last step is the reprotonation of the intermediate by Glu167, which then produces the product.
Fig. 11.
The classical reaction mechanism. In this mechanism, the proton transfer steps are carried out by the glutamate as well as by the histidine
Fig. 12.
The criss-cross reaction mechanism. In this mechanism, all proton transfer steps are carried out by the glutamate side chain carboxylate group
Extensive QM/MM calculations on the reaction mechanism of TIM have shown that the two possible reaction mechanisms can contribute to the overall reaction [4, 52]. The NMR studies have found that, in yeast TIM, the classical mechanism contributes at least 3.9% and in chicken TIM at least 12.1% to the overall reaction [110], indicating that there are subtle differences in the reaction mechanisms of these homologues enzymes. Subtle differences in catalytic mechanism between homologous enzymes have also been found for other classes of enzymes, for example in the very elegant transition state analogue studies by Schramm and co-workers [111] of bovine and human purine nucleoside phosphorylase.
In the classical mechanism (Fig. 11), both the Glu167 and the His95 side chains are involved in shuttling protons in the intermediate enediolate complex. This produces a negatively charged histidine side chain [108, 112]. For this proton transfer step, no movement of the histidine side chain seems to be required. The recently published atomic resolution structure of the leishmanial TIM/PGH complex provides a good rationale for how the proton shuttling by the Glu167 side chain in the classical and criss-cross mechanism can be achieved. From the analysis of this structure (Fig. 13), and from an analysis of other ligand-TIM structures (Fig. 14), it appears that the side chain of the catalytic glutamate can move approximately 1 Å in a small cavity above the cis-enediolate intermediate plane. The suggested motion is a sliding motion of the carboxylate moiety in this cavity (Fig. 14), which is achieved by small changes in the main chain and side chain geometry; the side chain dihedral angles of Glu167 remain essentially the same. This movement shuttles the proton between the C1–O1 moiety and the C2–O2 moiety of the cis-enediolate. As described by Knowles [2] and others, the syn-orbital of the carboxylate has the highest affinity for a proton [113] and therefore this orbital is predicted to abstract the proton from C1. In the criss-cross mechanism, when the carboxylate moiety has moved to the other end of its cavity, the proton is delivered to the receiving atom, O2 (Fig. 12). Subsequently, it abstracts the O1-proton and transfers it to the C2-atom, by which the aldehyde product is formed.
Fig. 13.
Visualisation of the anisotropic B-factors of the atoms in the active site of the leishmanial TIM structure complexed with PGH (2VXN) (in stereo). The 70% probability thermal ellipsoids have been drawn. The highest B-factors are observed for OE1(Glu167), OE2(Glu167) and O1(PGH)
Fig. 14.

The comparison of the active site geometry of four liganded high resolution TIM structures, suggesting how a sliding motion of the Glu167 side chain over the catalytic end of the substrate could facilitate the proton shuttling by the catalytic base. The structures concern the complexes with PGH (2VXN) (purple), 2PG (1N55) (green), DHAP (1NEY) (orange), and IPP (1IF2) (cyan). In the PGH complex, the side chain is near Cys126, Ala165, Leu232, and in the IPP complex it is nearest to Ile172
In the physiological DHAP-to-d-GAP conversion, the oxyanion formed on proton abstraction from C1 is the O2-oxygen, which is stabilised by the oxyanion hole formed by the Lys13 and His95 side chains. Subsequently, in the reaction cycle, this negative charge is transferred eventually to the O1-oxygen (Figs. 11, 12). This state actually corresponds to the first intermediate generated on deprotonation of d-GAP when considering the d-GAP-to-DHAP conversion, and the O1 negative charge is proposed to be stabilised by the side chains of His95 and Asn11, as suggested by protein crystallographic studies [18]. Also, NMR studies in D2O suggest the existence of multiple transition states and intermediates, differing in hydrogen bonding at the enediolate O2 and O1 oxyanions formed on deprotonation of DHAP and d-GAP, respectively [114].
Since the first studies by Knowles [1, 115], done with tritiated DHAP substrate, have indicated substantial loss of label from substrate into bulk solvent during catalysis, it has intrigued scientists via what mechanism this is possible [116]. The structure of the active site geometry suggests that this exchange with bulk solvent of the proton abstracted from the substrate by the base is possible via a string of waters forming a hydrogen bonding network between Glu167 and bulk solvent [20] (Fig. 6). More recent NMR studies of unlabeled substrate in D2O show that under these experimental conditions the proton shuttling mechanism transfers the proton much more efficiently directly between the C1 and C2 carbon atom [114], as is the case in the classical mechanism (Fig. 11).
The importance of the flexible loops, loop-6 and loop-7
The free energy diagram of the reaction (Fig. 2) as well as the detailed description describing the chemical steps (Figs. 11, 12) still do not describe all the steps of the complete catalytic cycle, as the substrate binding and product release steps should also be considered. This is also important because such steps can be the rate limiting step, in particular in TIM, in which the chemical steps have become very fast.
The classical view states that loop-6 is open in the unliganded state and closed in the liganded state. Now that there is such a large collection of deposited TIM structures (Table 1), it is apparent that the binding process is a two-step process. For example, a structure (1R2R) is known from rabbit TIM in which the loop-6 is closed, but the structure is unliganded [117], while in a trypanosomal TIM structure [118] (1TSI) as well as in a malaria TIM structure (1LZO) [119], loop-6 is seen in an open conformation, while the ligand is still present. The latter structure can also be referred to as an encounter complex, in which the ligand is bound but not yet having completely reached its competent position. Also, from solution studies [51], it is known that an encounter complex is formed first. In these T-jump experiments, it is found that the formation of the enzyme substrate encounter complex is a very fast bimolecular reaction, followed by a slower uni-molecular loop closure conformational switch. Measuring the loop opening (2,500 s−1) and loop closure (46,700 s−1) rates as a function of temperature provides the activation enthalpies for loop opening and loop closure as 14.1 and 13.8 kcal/mol, respectively, in the case where the ligand is glycerol-3-phosphate [51].
The loop closure conformational changes, in particular of loop-7, allow the substrate to bind deeper into the active site. Jointly with the loop-7 changes, also the loop-6 closes, placing the side chain of the catalytic glutamate near the substrate (the swung-in conformation) (Fig. 15). Therefore, in this closed complex, the substrate is bound competently, such that catalysis of the deprotonation step occurs. The release of product, d-GAP, is expected to also be a multistep process, in which the d-GAP-TIM “departure” complex is formed first (corresponding to the DHAP–encounter complex), before the product is released.
Fig. 15.
The importance of Pro168 for the concerted loop-6/loop-7 closure. The active site geometry of the unliganded (open) conformation (5TIM, subunit A) (green) and the liganded (closed) conformation (1N55) (purple) are compared. The ligand is 2PG. On ligand binding, O(Gly211) (loop-7) rotates 90° upwards, clashing with the Pro168 (loop-6) side chain. Consequently, the Glu167-Pro168 dipeptide also switches to the closed conformation, by which the Glu167 side chain adopts the competent, swung-in conformation. This swung-in conformation is possible, once O(Gly211) has moved upwards
In the conformational change of the loop-6 closure, the tip of this loop moves almost 7 Å on ligand binding. Structural data suggest that the tip of the loop, residues Ile-Gly-Thr-Gly, moves as a rigid body [120, 121]. In the unliganded open form, it contacts loop-5, whereas in the liganded closed form, it moves over to loop-7. Elegant solid state NMR studies show that the loop opening/closing mechanism is not ligand gated, but an intrinsic property of both the unliganded as well as the liganded state [122]. In the unliganded state, loop-6 is more disordered, having high B-factors, whereas in the liganded state, loop-6 is well defined. Specific hydrogen bonding contacts of loop-6 with the YGGS motif of loop-7 exist in this closed form [123], generated by OH(Tyr210) to N(Ala178), and by OG(Ser214) to O(Ala171) and to N(Gly175).
In this closed conformation, a binding site is formed for the phosphate dianion, by four optimally positioned peptide NH-groups (Fig. 6). For this, the conformational change of loop-7 is also important. The conserved sequence of loop-7 (Fig. 9) is a tetrapeptide, YGGS, residues 210–213. It has been found that the Gly211-Gly212 peptide plane rotates by 90°, whereas the Gly212-Ser213 peptide rotates 180° in this conformational switch, allowing N(Ser213) to hydrogen bond to the phosphate moiety. N(Ile172) of loop-6 and N(Gly234), N(Gly235) of loop-8 complete the set of four phosphate binding hydrogen bond donors.
The importance of the tyrosine, Tyr210, of the YGGS-motif has been recognised in early mutagenesis studies [124]. The hydroxyl group of this tyrosine is hydrogen bonded to the main chain of the closed conformation of loop-6. The importance of this tyrosine for the loop dynamics has also been studied [125]. The YGGS-motif is fully conserved in the TIM sequences of eukaryotes and eubacteria (Fig. 9), but not in archeal sequences [84]. Recently, Loria and co-workers [126] replaced this YGGS-motif of chicken TIM by its archeal homologue TGAG, and found, using NMR approaches, that the catalytic properties of this variant were indeed much less efficient.
The proline (Pro168) following immediately after the catalytic glutamate (Fig. 9) is conserved in both the eukaryotic and bacterial sequences, as well as in the archeal sequences [84]. Systematic structural enzymological studies [127] on the properties of the P168A variant have provided the rational for this conservation. The structural data suggest that the ligand-induced conformational change of the YGGS-motif of loop-7 forces the Glu167-Pro168 dipeptide to switch to the swung-in conformation. This switch is apparently caused by the clash of O(Gly211) with the Pro168 side chain, after the 90° rotation of the Gly211-Gly212 peptide plane on ligand binding. Consequently, the Glu167-Pro168 dipeptide has to move, by which the catalytic glutamate adopts its catalytic swung-in position (Fig. 15). In this conformational switch, the χ1 and χ2 side chain dihedrals of the Glu167 side chain do not change very much. The movement of the carboxylate group of Glu167, towards the substrate, is approximately 2 Å.
The loop-6 conformational change appears to be achieved by relatively small changes in the phi/psi values of the N-terminal and C-terminal hinge tripeptides [121]. Interestingly, the much smaller conformational switch of loop-7 is caused by significant changes in the phi/psi values of the two glycines of the YGGS-motif. Because of the peptide flip of the Gly211-Ser212 peptide bond, the phi/psi values of Ser213 change from (−83,118) to (47,51), for which a high energy barrier has to be crossed [120, 128].
The insight on the dynamical properties of (1) loop-6 and loop-7 and of (2) the catalytic glutamate have been summarised above. Further studies [129] will clarify if the overall dynamical properties of the TIM dimer also play an important role in the catalytic cycle, as has been established for other enzymes [130].
Concluding remarks
The recent structural enzymological studies of TIM have been built on the classical reaction mechanism studies by Knowles and others [1, 2], as summarised previously. These recent studies, as reviewed here, have shown the importance of electrostatic stabilisation for the catalytic power of this enzyme. These electrostatic interactions lower the transition state free energy barriers and stabilise the negatively charged cis-enediolate intermediates. A unique property of the TIM active site are the flexible loops, loop-6 and loop-7, which in the liganded state exclude bulk solvent from the catalytic site and sequester the phosphate moiety, thereby preventing the undesirable phosphate elimination reaction. In addition, the binding energy from the phosphate moiety contributes to the transition state stabilisation. In the liganded closed state, the catalytic base, Glu167, at the beginning of loop-6, is switched into its catalytic competent swung-in conformation. The active site geometry of the competent liganded complex provides the side chain of the catalytic glutamate with dynamical properties well suited for shuttling the protons as required for the isomerisation reaction.
Researchers continue to use TIM as a study object for their research, both for computational enzyme mechanism studies as well as for experimental studies. The experimental studies cover a wide range of topics, ranging from its physiological function, including the cause of disease by the deficient TIM variants, to its evolution, as well as on its folding, assembly, dynamics and reaction mechanism. These very different, but complementary, approaches have provided excellent insights into the reaction mechanism of TIM. Further research along these lines will continue to improve the detailed, quantitative biophysical description of all aspects of its catalytic cycle. This reaction mechanism, as developed through evolutionary pressure, highlights the beautiful interplay of the catalytic tools that make the active site of this superb molecular biocatalyst.
Acknowledgments
We thank the Academy of Finland and the Finnish Cultural Foundation for their support. We thank Dr. Annemie Lambeir for critically reading the manuscript.
Abbreviations
- BHAP
Bromohydroxyacetone phosphate
- d-GAP
d-Glyceraldehyde-3-phosphate
- DHAP
Dihydroxyacetone phosphate
- d,l-GOP
d,l-Glycidolphosphate
- IPP
2-(N-formyl-N-hydroxy)-amino-ethylphosphonate
- PGH
Phosphoglycolohydroxamate
- PGI
Phosphoglucose isomerase
- RPI
d-Ribose-5-phosphate isomerase
- TIM
Triosephosphate isomerase
- 2PG
2-Phosphoglycollate
References
- 1.Knowles JR, Albery WJ. Perfection in enzyme catalysis: the energetics of triosephosphate isomerase. Acc Chem Res. 1977;10:105–111. doi: 10.1021/ar50112a001. [DOI] [Google Scholar]
- 2.Knowles JR. Enzyme catalysis: not different, just better. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- 3.Harris TK. The mechanistic ventures of triosephosphate isomerase. IUBMB Life. 2008;60:195–198. doi: 10.1002/iub.43. [DOI] [PubMed] [Google Scholar]
- 4.Cui Q, Karplus M. Catalysis and specificity in enzymes: a study of triosephosphate isomerase and comparison with methyl glyoxal synthase. Adv Protein Chem. 2003;66:315–372. doi: 10.1016/S0065-3233(03)66008-0. [DOI] [PubMed] [Google Scholar]
- 5.Albery WJ, Knowles JR. Efficiency and evolution of enzyme catalysis. Angew Chem Int Ed Engl. 1977;16:285–293. doi: 10.1002/anie.197702851. [DOI] [PubMed] [Google Scholar]
- 6.Fersht AR. Structure and mechanism in protein science. New York: Freeman; 1999. [Google Scholar]
- 7.Rose IA. Mechanism of C–H bond cleavage in aldolase and isomerase reactions. Brookhaven Symp Biol. 1962;15:293–309. [PubMed] [Google Scholar]
- 8.Richard JP. Acid–base catalysis of the elimination and isomerization reactions of triose phosphates. J Am Chem Soc. 1984;106:4926–4936. doi: 10.1021/ja00329a050. [DOI] [Google Scholar]
- 9.Wolfenden R. Transition state analogues for enzyme catalysis. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 10.Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AJ, Milman JD, Offord RE, Priddle JD, Waley SG. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975;255:609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- 11.Lambeir AM, Opperdoes FR, Wierenga RK. Kinetic properties of triose-phosphate isomerase from Trypanosoma brucei brucei. A comparison with the rabbit muscle and yeast enzymes. Eur J Biochem. 1987;168:69–74. doi: 10.1111/j.1432-1033.1987.tb13388.x. [DOI] [PubMed] [Google Scholar]
- 12.Rose IA, O’Connell EL. Inactivation and labeling of triose phosphate isomerase and enolase by glycidol phosphate. J Biol Chem. 1969;244:6548–6550. [PubMed] [Google Scholar]
- 13.De la Mare S, Coulson AF, Knowles JR, Priddle JD, Offord RE. Active-site labelling of triose phosphate isomerase. The reaction of bromohydroxyacetone phosphate with a unique glutamic acid residue and the migration of the label to tyrosine. Biochem J. 1972;129:321–331. doi: 10.1042/bj1290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman FC. Haloacetol phosphates. Characterization of the active site of rabbit muscle triose phosphate isomerase. Biochemistry. 1971;10:146–154. doi: 10.1021/bi00777a021. [DOI] [PubMed] [Google Scholar]
- 15.Schray KJ, O’Connell EL, Rose IA. Inactivation of muscle triose phosphate isomerase by d- and l-glycidol phosphate. J Biol Chem. 1973;248:2214–2218. [PubMed] [Google Scholar]
- 16.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 17.Hall A, Knowles JR. The uncatalyzed rates of enolization of dihydroxyacetone phosphate and of glyceraldehyde 3-phosphate in neutral aqueous solution. The quantitative assessment of the effectiveness of an enzyme catalyst. Biochemistry. 1975;14:4348–4353. doi: 10.1021/bi00690a032. [DOI] [PubMed] [Google Scholar]
- 18.Kursula I, Partanen S, Lambeir AM, Antonov DM, Augustyns K, Wierenga RK. Structural determinants for ligand binding and catalysis of triosephosphate isomerase. Eur J Biochem. 2001;268:5189–5196. doi: 10.1046/j.0014-2956.2001.02452.x. [DOI] [PubMed] [Google Scholar]
- 19.Jogl G, Rozovsky S, McDermott AE, Tong L. Optimal alignment for enzymatic proton transfer: structure of the Michaelis complex of triosephosphate isomerase at 1.2-A resolution. Proc Natl Acad Sci USA. 2003;100:50–55. doi: 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alahuhta M, Wierenga RK. Atomic resolution crystallography of a complex of triosephosphate isomerase with a reaction-intermediate analog: new insight in the proton transfer reaction mechanism. Proteins. 2010;78:1878–1888. doi: 10.1002/prot.22701. [DOI] [PubMed] [Google Scholar]
- 21.Lesk AM, Branden CI, Chothia C. Structural principles of alpha/beta barrel proteins: the packing of the interior of the sheet. Proteins. 1989;5:139–148. doi: 10.1002/prot.340050208. [DOI] [PubMed] [Google Scholar]
- 22.Copley RR, Bork P. Homology among (betaalpha)(8) barrels: implications for the evolution of metabolic pathways. J Mol Biol. 2000;303:627–641. doi: 10.1006/jmbi.2000.4152. [DOI] [PubMed] [Google Scholar]
- 23.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/S0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 24.Wierenga RK. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 2001;492:193–198. doi: 10.1016/S0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 25.Pujadas G, Palau J. Evolution of alpha-amylases: architectural features and key residues in the stabilization of the (beta/alpha)(8) scaffold. Mol Biol Evol. 2001;18:38–54. doi: 10.1093/oxfordjournals.molbev.a003718. [DOI] [PubMed] [Google Scholar]
- 26.Go MK, Koudelka A, Amyes TL, Richard JP. The role of Lys-12 in catalysis by triosephosphate isomerase: a two-part substrate approach. Biochemistry. 2010;49:5377–5389. doi: 10.1021/bi100538b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lolis E, Petsko GA. Transition-state analogues in protein crystallography: probes of the structural source of enzyme catalysis. Annu Rev Biochem. 1990;59:597–630. doi: 10.1146/annurev.bi.59.070190.003121. [DOI] [PubMed] [Google Scholar]
- 28.Webb MR, Knowles JR. The orientation and accessibility of substrates on the active site of triosephosphate isomerase. Biochemistry. 1975;14:4692–4698. doi: 10.1021/bi00692a020. [DOI] [PubMed] [Google Scholar]
- 29.Lolis E, Petsko GA. Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis. Biochemistry. 1990;29:6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- 30.Collins KD. An activated intermediate analogue. The use of phosphoglycolohydroxamate as a stable analogue of a transiently occurring dihydroxyacetone phosphate-derived enolate in enzymatic catalysis. J Biol Chem. 1974;249:136–142. [PubMed] [Google Scholar]
- 31.Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D. Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway. Biochemistry. 1991;30:5821–5826. doi: 10.1021/bi00238a002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Sugio S, Komives EA, Liu KD, Knowles JR, Petsko GA, Ringe D. Crystal structure of recombinant chicken triosephosphate isomerase-phosphoglycolohydroxamate complex at 1.8-A resolution. Biochemistry. 1994;33:2830–2837. doi: 10.1021/bi00176a012. [DOI] [PubMed] [Google Scholar]
- 33.Schliebs W, Thanki N, Eritja R, Wierenga R. Active site properties of monomeric triosephosphate isomerase (monoTIM) as deduced from mutational and structural studies. Protein Sci. 1996;5:229–239. doi: 10.1002/pro.5560050206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonvielle M, Mariano S, Therisod M. New inhibitors of rabbit muscle triose-phosphate isomerase. Bioorg Med Chem Lett. 2005;15:2906–2909. doi: 10.1016/j.bmcl.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 35.Campbell ID, Jones RB, Kiener PA, Waley SG. Enzyme-substrate and enzyme-inhibitor complexes of triose phosphate isomerase studied by 31P nuclear magnetic resonance. Biochem J. 1979;179:607–621. doi: 10.1042/bj1790607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orosz F, Olah J, Ovadi J. Triosephosphate isomerase deficiency: facts and doubts. IUBMB Life. 2006;58:703–715. doi: 10.1080/15216540601115960. [DOI] [PubMed] [Google Scholar]
- 37.Helfert S, Estevez AM, Bakker B, Michels P, Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei . Biochem J. 2001;357:117–125. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michels PA. Evolutionary aspects of trypanosomes: analysis of genes. J Mol Evol. 1986;24:45–52. doi: 10.1007/BF02099950. [DOI] [PubMed] [Google Scholar]
- 39.Orosz F, Olah J, Ovadi J. Triosephosphate isomerase deficiency: new insights into an enigmatic disease. Biochim Biophys Acta. 2009;1792:1168–1174. doi: 10.1016/j.bbadis.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Richard JP. Restoring a metabolic pathway. ACS Chem Biol. 2008;3:605–607. doi: 10.1021/cb800238s. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Almazan C, Arreola R, Rodriguez-Larrea D, Aguirre-Lopez B, de Gomez-Puyou MT, Perez-Montfort R, Costas M, Gomez-Puyou A, Torres-Larios A. Structural basis of human triosephosphate isomerase deficiency: mutation E104D is related to alterations of a conserved water network at the dimer interface. J Biol Chem. 2008;283:23254–23263. doi: 10.1074/jbc.M802145200. [DOI] [PubMed] [Google Scholar]
- 42.Schneider A, Westwood B, Yim C, Cohen-Solal M, Rosa R, Labotka R, Eber S, Wolf R, Lammi A, Beutler E. The 1591C mutation in triosephosphate isomerase (TPI) deficiency. Tightly linked polymorphisms and a common haplotype in all known families. Blood Cells Mol Dis. 1996;22:115–125. doi: 10.1006/bcmd.1996.0019. [DOI] [PubMed] [Google Scholar]
- 43.Maes D, Zeelen JP, Thanki N, Beaucamp N, Alvarez M, Thi MH, Backmann J, Martial JA. The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures. Proteins. 1999;37:441–453. doi: 10.1002/(SICI)1097-0134(19991115)37:3<441::AID-PROT11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Schneider AS. Triosephosphate isomerase deficiency: historical perspectives and molecular aspects. Baillieres Best Pract Res Clin Haematol. 2000;13:119–140. doi: 10.1053/beha.2000.0061. [DOI] [PubMed] [Google Scholar]
- 45.Daar IO, Artymiuk PJ, Phillips DC, Maquat LE. Human triose-phosphate isomerase deficiency: a single amino acid substitution results in a thermolabile enzyme. Proc Natl Acad Sci USA. 1986;83:7903–7907. doi: 10.1073/pnas.83.20.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guix FX, Ill-Raga G, Bravo R, Nakaya T, de Fabritiis G, Coma M, Miscione GP, Villà-Freixa J, Suzuki T, Fernàndez-Busquets X, Valverde MA, de Strooper B, Muñoz FJ. Amyloid-dependent triosephosphate isomerase nitrotyrosination induces glycation and tau fibrillation. Brain. 2009;132:1335–1345. doi: 10.1093/brain/awp023. [DOI] [PubMed] [Google Scholar]
- 47.Rozovsky S, McDermott AE. Substrate product equilibrium on a reversible enzyme, triosephosphate isomerase. Proc Natl Acad Sci USA. 2007;104:2080–2085. doi: 10.1073/pnas.0608876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albery WJ, Knowles JR. Free-energy profile of the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1976;15:5627–5631. doi: 10.1021/bi00670a031. [DOI] [PubMed] [Google Scholar]
- 49.Albery WJ, Knowles JR. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 50.Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988;27:1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 51.Desamero R, Rozovsky S, Zhadin N, McDermott A, Callender R. Active site loop motion in triosephosphate isomerase: T-jump relaxation spectroscopy of thermal activation. Biochemistry. 2003;42:2941–2951. doi: 10.1021/bi026994i. [DOI] [PubMed] [Google Scholar]
- 52.Cui Q, Karplus M. Triosephosphate isomerase: a theoretical comparison of alternative pathways. J Am Chem Soc. 2001;123:2284–2290. doi: 10.1021/ja002886c. [DOI] [PubMed] [Google Scholar]
- 53.Guallar V, Jacobson M, McDermott A, Friesner RA. Computational modeling of the catalytic reaction in triosephosphate isomerase. J Mol Biol. 2004;337:227–239. doi: 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Feierberg I, Åquist J. Computational modeling of enzymatic keto-enol isomerization reactions. Theor Chem Acc. 2002;108:71–84. [Google Scholar]
- 55.Thakur SS, Deepalakshmi PD, Gayathri P, Banerjee M, Murthy MR, Balaram P. Detection of the protein dimers, multiple monomeric states and hydrated forms of Plasmodium falciparum triosephosphate isomerase in the gas phase. Protein Eng Des Sel. 2009;22:289–304. doi: 10.1093/protein/gzp005. [DOI] [PubMed] [Google Scholar]
- 56.Belasco JG, Herlihy JM, Knowles JR. Critical ionization states in the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1978;17:2971–2978. doi: 10.1021/bi00608a005. [DOI] [PubMed] [Google Scholar]
- 57.Pompliano DL, Peyman A, Knowles JR. Stabilization of a reaction intermediate as a catalytic device: definition of the functional role of the flexible loop in triosephosphate isomerase. Biochemistry. 1990;29:3186–3194. doi: 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- 58.Lodi PJ, Chang LC, Knowles JR, Komives EA. Triosephosphate isomerase requires a positively charged active site: the role of lysine-12. Biochemistry. 1994;33:2809–2814. doi: 10.1021/bi00176a009. [DOI] [PubMed] [Google Scholar]
- 59.Belasco JG, Knowles JR. Direct observation of substrate distortion by triosephosphate isomerase using Fourier transform infrared spectroscopy. Biochemistry. 1980;19:472–477. doi: 10.1021/bi00544a012. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Komives EA, Sugio S, Blacklow SC, Narayana N, Xuong NH, Stock AM, Petsko GA, Ringe D. The role of water in the catalytic efficiency of triosephosphate isomerase. Biochemistry. 1999;38:4389–4397. doi: 10.1021/bi9826759. [DOI] [PubMed] [Google Scholar]
- 61.Gayathri P, Banerjee M, Vijayalakshmi A, Balaram H, Balaram P, Murthy MR. Biochemical and structural characterization of residue 96 mutants of Plasmodium falciparum triosephosphate isomerase: active-site loop conformation, hydration and identification of a dimer-interface ligand-binding site. Acta Crystallogr D Biol Crystallogr. 2009;65:847–857. doi: 10.1107/S0907444909018666. [DOI] [PubMed] [Google Scholar]
- 62.Kursula I, Wierenga RK. Crystal structure of triosephosphate isomerase complexed with 2-phosphoglycolate at 0.83-A resolution. J Biol Chem. 2003;278:9544–9551. doi: 10.1074/jbc.M211389200. [DOI] [PubMed] [Google Scholar]
- 63.Donnini S, Groenhof G, Wierenga RK, Juffer AH. The planar conformation of a strained proline ring: a QM/MM study. Proteins. 2006;64:700–710. doi: 10.1002/prot.21006. [DOI] [PubMed] [Google Scholar]
- 64.Allen SC, Muirhead H. Refined three-dimensional structure of cat-muscle (M1) pyruvate kinase at a resolution of 2.6 A. Acta Crystallogr D Biol Crystallogr. 1996;52:499–504. doi: 10.1107/S0907444995016040. [DOI] [PubMed] [Google Scholar]
- 65.Raychaudhuri S, Younas F, Karplus PA, Faerman CH, Ripoll DR. Backbone makes a significant contribution to the electrostatics of alpha/beta-barrel proteins. Protein Sci. 1997;6:1849–1857. doi: 10.1002/pro.5560060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livesay DR, La D. The evolutionary origins and catalytic importance of conserved electrostatic networks within TIM-barrel proteins. Protein Sci. 2005;14:1158–1170. doi: 10.1110/ps.041221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henn-Sax M, Hocker B, Wilmanns M, Sterner R. Divergent evolution of (betaalpha)8-barrel enzymes. Biol Chem. 2001;382:1315–1320. doi: 10.1515/BC.2001.163. [DOI] [PubMed] [Google Scholar]
- 68.Hocker B, Lochner A, Seitz T, Claren J, Sterner R. High-resolution crystal structure of an artificial (betaalpha)(8)-barrel protein designed from identical half-barrels. Biochemistry. 2009;48:1145–1147. doi: 10.1021/bi802125b. [DOI] [PubMed] [Google Scholar]
- 69.Hocker B. Directed evolution of (betaalpha)(8)-barrel enzymes. Biomol Eng. 2005;22:31–38. doi: 10.1016/j.bioeng.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 70.Sterner R, Hocker B. Catalytic versatility, stability, and evolution of the (betaalpha)8-barrel enzyme fold. Chem Rev. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- 71.Claren J, Malisi C, Hocker B, Sterner R. Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds. Proc Natl Acad Sci USA. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 73.Singh SK, Maithal K, Balaram H, Balaram P. Synthetic peptides as inactivators of multimeric enzymes: inhibition of Plasmodium falciparum triosephosphate isomerase by interface peptides. FEBS Lett. 2001;501:19–23. doi: 10.1016/S0014-5793(01)02606-0. [DOI] [PubMed] [Google Scholar]
- 74.Tellez-Valencia A, Olivares-Illana V, Hernandez-Santoyo A, Perez-Montfort R, Costas M, Rodriguez-Romero A, López-Calahorra F, Tuena De Gómez-Puyou M, Gómez-Puyou A. Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface. J Mol Biol. 2004;341:1355–1365. doi: 10.1016/j.jmb.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 75.Olivares-Illana V, Rodriguez-Romero A, Becker I, Berzunza M, Garcia J, Perez-Montfort R, Cabrera N, López-Calahorra F, de Gómez-Puyou MT, Gómez-Puyou A. Perturbation of the dimer interface of triosephosphate isomerase and its effect on Trypanosoma cruzi . PLoS Negl Trop Dis. 2007;1:e1. doi: 10.1371/journal.pntd.0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borchert TV, Abagyan R, Kishan KV, Zeelen JP, Wierenga RK. The crystal structure of an engineered monomeric triosephosphate isomerase, monoTIM: the correct modelling of an eight-residue loop. Structure. 1993;1:205–213. doi: 10.1016/0969-2126(93)90021-8. [DOI] [PubMed] [Google Scholar]
- 77.Borchert TV, Abagyan R, Jaenicke R, Wierenga RK. Design, creation, and characterization of a stable, monomeric triosephosphate isomerase. Proc Natl Acad Sci USA. 1994;91:1515–1518. doi: 10.1073/pnas.91.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borchert TV, Kishan KV, Zeelen JP, Schliebs W, Thanki N, Abagyan R, Jaenicke R, Wierenga RK. Three new crystal structures of point mutation variants of monoTIM: conformational flexibility of loop-1, loop-4 and loop-8. Structure. 1995;3:669–679. doi: 10.1016/S0969-2126(01)00202-7. [DOI] [PubMed] [Google Scholar]
- 79.Norledge BV, Lambeir AM, Abagyan RA, Rottmann A, Fernandez AM, Filimonov VV, Peter MG, Wierenga RK. Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8) of triosephosphate isomerase: toward a new substrate specificity. Proteins. 2001;42:383–389. doi: 10.1002/1097-0134(20010215)42:3<383::AID-PROT80>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 80.Lambeir AM, Backmann J, Ruiz-Sanz J, Filimonov V, Nielsen JE, Kursula I, Norledge BV, Wierenga RK. The ionization of a buried glutamic acid is thermodynamically linked to the stability of Leishmania mexicana triose phosphate isomerase. Eur J Biochem. 2000;267:2516–2524. doi: 10.1046/j.1432-1327.2000.01254.x. [DOI] [PubMed] [Google Scholar]
- 81.Saab-Rincon G, Juarez VR, Osuna J, Sanchez F, Soberon X. Different strategies to recover the activity of monomeric triosephosphate isomerase by directed evolution. Protein Eng. 2001;14:149–155. doi: 10.1093/protein/14.3.149. [DOI] [PubMed] [Google Scholar]
- 82.Thanki N, Zeelen JP, Mathieu M, Jaenicke R, Abagyan RA, Wierenga RK, Schliebs W. Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop. Protein Eng. 1997;10:159–167. doi: 10.1093/protein/10.2.159. [DOI] [PubMed] [Google Scholar]
- 83.Alahuhta M, Salin M, Casteleijn MG, Kemmer C, El-Sayed I, Augustyns K, Neubauer P, Wierenga RK. Structure-based protein engineering efforts with a monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. Protein Eng Des Sel. 2008;21:257–266. doi: 10.1093/protein/gzn002. [DOI] [PubMed] [Google Scholar]
- 84.Kursula I, Salin M, Sun J, Norledge BV, Haapalainen AM, Sampson NS, Wierenga RK. Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase. Protein Eng Des Sel. 2004;17:375–382. doi: 10.1093/protein/gzh048. [DOI] [PubMed] [Google Scholar]
- 85.Williams JC, Zeelen JP, Neubauer G, Vriend G, Backmann J, Michels PA, Lambeir AM, Wierenga RK. Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein Eng. 1999;12:243–250. doi: 10.1093/protein/12.3.243. [DOI] [PubMed] [Google Scholar]
- 86.Nagorski RW, Richard JP. Mechanistic imperatives for aldose-ketose isomerization in water: specific, general base- and metal ion-catalyzed isomerization of glyceraldehyde with proton and hydride transfer. J Am Chem Soc. 2001;123:794–802. doi: 10.1021/ja003433a. [DOI] [PubMed] [Google Scholar]
- 87.Gerlt JA, Gassman PG. Understanding the rates of certain enzyme-catalyzed reactions: proton abstraction from carbon acids, acyl-transfer reactions, and displacement reactions of phosphodiesters. Biochemistry. 1993;32:11943–11952. doi: 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- 88.Frey PA, Hegeman AD. Enzymatic reaction mechanisms. New York: Oxford University Press; 2007. [Google Scholar]
- 89.Pihko P, Rapakko S, Wierenga RK. Oxyanion holes and their mimics. In: Pihko P, editor. Hydrogen bonding in organic synthesis. Weinheim: Wiley; 2009. pp. 43–71. [Google Scholar]
- 90.Rose IA. Mechanism of the aldose-ketose isomerase reactions. Adv Enzymol Relat Areas Mol Biol. 1975;43:491–517. doi: 10.1002/9780470122884.ch6. [DOI] [PubMed] [Google Scholar]
- 91.Roos AK, Burgos E, Ericsson DJ, Salmon L, Mowbray SL. Competitive inhibitors of Mycobacterium tuberculosis ribose-5-phosphate isomerase B reveal new information about the reaction mechanism. J Biol Chem. 2005;280:6416–6422. doi: 10.1074/jbc.M412018200. [DOI] [PubMed] [Google Scholar]
- 92.Arsenieva D, Hardre R, Salmon L, Jeffery CJ. The crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-d-arabinonohydroxamic acid. Proc Natl Acad Sci USA. 2002;99:5872–5877. doi: 10.1073/pnas.052131799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang R, Andersson CE, Savchenko A, Skarina T, Evdokimova E, Beasley S, Arrowsmith CH, Edwards AM, Joachimiak A, Mowbray SL. Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle. Structure. 2003;11:31–42. doi: 10.1016/S0969-2126(02)00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hamada K, Ago H, Sugahara M, Nodake Y, Kuramitsu S, Miyano M. Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi) J Biol Chem. 2003;278:49183–49190. doi: 10.1074/jbc.M309272200. [DOI] [PubMed] [Google Scholar]
- 95.Arsenieva D, Jeffery CJ. Conformational changes in phosphoglucose isomerase induced by ligand binding. J Mol Biol. 2002;323:77–84. doi: 10.1016/S0022-2836(02)00892-6. [DOI] [PubMed] [Google Scholar]
- 96.Fenn TD, Ringe D, Petsko GA. Xylose isomerase in substrate and inhibitor michaelis states: atomic resolution studies of a metal-mediated hydride shift. Biochemistry. 2004;43:6464–6474. doi: 10.1021/bi049812o. [DOI] [PubMed] [Google Scholar]
- 97.Amyes TL, O’Donoghue AC, Richard JP. Contribution of phosphate intrinsic binding energy to the enzymatic rate acceleration for triosephosphate isomerase. J Am Chem Soc. 2001;123:11325–11326. doi: 10.1021/ja016754a. [DOI] [PubMed] [Google Scholar]
- 98.Morrow JR, Amyes TL, Richard JP. Phosphate binding energy and catalysis by small and large molecules. Acc Chem Res. 2008;41:539–548. doi: 10.1021/ar7002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Amyes TL, Richard JP. Enzymatic catalysis of proton transfer at carbon: activation of triosephosphate isomerase by phosphite dianion. Biochemistry. 2007;46:5841–5854. doi: 10.1021/bi700409b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Go MK, Amyes TL, Richard JP. Hydron transfer catalyzed by triosephosphate isomerase. Products of the direct and phosphite-activated isomerization of [1-(13)C]-glycolaldehyde in D(2)O. Biochemistry. 2009;48:5769–5778. doi: 10.1021/bi900636c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Corey EJ, Sneen RA. Stereoelectronic control in enolization-ketonization reactions. J Am Chem Soc. 1956;78:6269–6278. doi: 10.1021/ja01605a013. [DOI] [Google Scholar]
- 102.Richard JP, Amyes TL. Proton transfer at carbon. Curr Opin Chem Biol. 2001;5:626–633. doi: 10.1016/S1367-5931(01)00258-7. [DOI] [PubMed] [Google Scholar]
- 103.Silverman RB. The organic chemistry of enzyme catalyzed reactions (revised edition) London: Academic Press; 2002. [Google Scholar]
- 104.Hartman FC, LaMuraglia GM, Tomozawa Y, Wolfenden R. The influence of pH on the interaction of inhibitors with triosephosphate isomerase and determination of the pKa of the active-site carboxyl group. Biochemistry. 1975;14:5274–5279. doi: 10.1021/bi00695a007. [DOI] [PubMed] [Google Scholar]
- 105.Alston WC, 2nd, Kanska M, Murray CJ. Secondary H/T and D/T isotope effects in enzymatic enolization reactions. Coupled motion and tunneling in the triosephosphate isomerase reaction. Biochemistry. 1996;35:12873–12881. doi: 10.1021/bi960831a. [DOI] [PubMed] [Google Scholar]
- 106.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 107.Harris TK, Abeygunawardana C, Mildvan AS. NMR studies of the role of hydrogen bonding in the mechanism of triosephosphate isomerase. Biochemistry. 1997;36:14661–14675. doi: 10.1021/bi972039v. [DOI] [PubMed] [Google Scholar]
- 108.Bash PA, Field MJ, Davenport RC, Petsko GA, Ringe D, Karplus M. Computer simulation and analysis of the reaction pathway of triosephosphate isomerase. Biochemistry. 1991;30:5826–5832. doi: 10.1021/bi00238a003. [DOI] [PubMed] [Google Scholar]
- 109.Nickbarg EB, Davenport RC, Petsko GA, Knowles JR. Triosephosphate isomerase: removal of a putatively electrophilic histidine residue results in a subtle change in catalytic mechanism. Biochemistry. 1988;27:5948–5960. doi: 10.1021/bi00416a019. [DOI] [PubMed] [Google Scholar]
- 110.Harris TK, Cole RN, Comer FI, Mildvan AS. Proton transfer in the mechanism of triosephosphate isomerase. Biochemistry. 1998;37:16828–16838. doi: 10.1021/bi982089f. [DOI] [PubMed] [Google Scholar]
- 111.Schramm VL. Enzymatic transition state theory and transition state analogue design. J Biol Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 112.Lodi PJ, Knowles JR. Neutral imidazole is the electrophile in the reaction catalyzed by triosephosphate isomerase: structural origins and catalytic implications. Biochemistry. 1991;30:6948–6956. doi: 10.1021/bi00242a020. [DOI] [PubMed] [Google Scholar]
- 113.Gandour RD. On the importance of orientation in general base catalysis by carboxylate. Bioorg Chem. 1981;10:169–176. doi: 10.1016/0045-2068(81)90020-1. [DOI] [Google Scholar]
- 114.O’Donoghue AC, Amyes TL, Richard JP. Hydron transfer catalyzed by triosephosphate isomerase. Products of isomerization of dihydroxyacetone phosphate in D2O. Biochemistry. 2005;44:2622–2631. doi: 10.1021/bi047953k. [DOI] [PubMed] [Google Scholar]
- 115.Herlihy JM, Maister SG, Albery WJ, Knowles JR. Energetics of triosephosphate isomerase: the fate of the 1(R)-3H label of tritiated dihydroxyacetone phosphate in the isomerase reaction. Biochemistry. 1976;15:5601–5607. doi: 10.1021/bi00670a026. [DOI] [PubMed] [Google Scholar]
- 116.Rose IA, Fung WJ, Warms JV. Proton diffusion in the active site of triosephosphate isomerase. Biochemistry. 1990;29:4312–4317. doi: 10.1021/bi00470a008. [DOI] [PubMed] [Google Scholar]
- 117.Aparicio R, Ferreira ST, Polikarpov I. Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity. J Mol Biol. 2003;334:1023–1041. doi: 10.1016/j.jmb.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 118.Verlinde CL, Witmans CJ, Pijning T, Kalk KH, Hol WG, Callens M, Opperdoes FR. Structure of the complex between trypanosomal triosephosphate isomerase and N-hydroxy-4-phosphono-butanamide: binding at the active site despite an “open” flexible loop conformation. Protein Sci. 1992;1:1578–1584. doi: 10.1002/pro.5560011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parthasarathy S, Ravindra G, Balaram H, Balaram P, Murthy MR. Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state. Biochemistry. 2002;41:13178–13188. doi: 10.1021/bi025783a. [DOI] [PubMed] [Google Scholar]
- 120.Noble ME, Zeelen JP, Wierenga RK. Structures of the “open” and “closed” state of trypanosomal triosephosphate isomerase, as observed in a new crystal form: implications for the reaction mechanism. Proteins. 1993;16:311–326. doi: 10.1002/prot.340160402. [DOI] [PubMed] [Google Scholar]
- 121.Joseph D, Petsko GA, Karplus M. Anatomy of a conformational change: hinged “lid” motion of the triosephosphate isomerase loop. Science. 1990;249:1425–1428. doi: 10.1126/science.2402636. [DOI] [PubMed] [Google Scholar]
- 122.Williams JC, McDermott AE. Dynamics of the flexible loop of triosephosphate isomerase: the loop motion is not ligand gated. Biochemistry. 1995;34:8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 123.Berlow RB, Igumenova TI, Loria JP. Value of a hydrogen bond in triosephosphate isomerase loop motion. Biochemistry. 2007;46:6001–6010. doi: 10.1021/bi700344v. [DOI] [PubMed] [Google Scholar]
- 124.Sampson NS, Knowles JR. Segmental motion in catalysis: investigation of a hydrogen bond critical for loop closure in the reaction of triosephosphate isomerase. Biochemistry. 1992;31:8488–8494. doi: 10.1021/bi00151a015. [DOI] [PubMed] [Google Scholar]
- 125.Derreumaux P, Schlick T. The loop opening/closing motion of the enzyme triosephosphate isomerase. Biophys J. 1998;74:72–81. doi: 10.1016/S0006-3495(98)77768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y, Berlow RB, Loria JP. Role of loop-loop interactions in coordinating motions and enzymatic function in triosephosphate isomerase. Biochemistry. 2009;48:4548–4556. doi: 10.1021/bi9002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Casteleijn MG, Alahuhta M, Groebel K, El-Sayed I, Augustyns K, Lambeir AM, Neubauer P, Wierenga RK. Functional role of the conserved active site proline of triosephosphate isomerase. Biochemistry. 2006;45:15483–15494. doi: 10.1021/bi061683j. [DOI] [PubMed] [Google Scholar]
- 128.Yuksel KU, Sun AQ, Gracy RW, Schnackerz KD. The hinged lid of yeast triose-phosphate isomerase. Determination of the energy barrier between the two conformations. J Biol Chem. 1994;269:5005–5008. [PubMed] [Google Scholar]
- 129.Xu Y, Lorieau J, McDermott AE. Triosephosphate isomerase: 15N and 13C chemical shift assignments and conformational change upon ligand binding by magic-angle spinning solid-state NMR spectroscopy. J Mol Biol. 2010;397:233–248. doi: 10.1016/j.jmb.2009.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]