Abstract
Information about the family of protein arginine methyltransferases (PRMTs) has been growing rapidly over the last few years and the emerging role of arginine methylation involved in cellular processes like signaling, RNA processing, gene transcription, and cellular transport function has been investigated. To date, 11 PRMTs gene transcripts have been identified in humans. Almost all PRMTs have been shown to have enzymatic activity and to catalyze arginine methylation. This review will summarize the overall function of human PRMTs and include novel highlights on each family member.
Keywords: Protein arginine methyltransferase, Post-translational modification, Gene family, Transcriptional regulation, Chromatin
Introduction
Posttranslational modification of proteins is a hallmark of signal transduction where cells are able to react quickly to changes or events of the surrounding cells thereby expanding the structural and functional diversity of the proteome. The role of acetylation and phosphorylation of proteins have been extensively studied as highly reversible reactions for fine-tuning gene expression in response to external stimuli or changes in the environmental conditions. Recently, the importance of other types of protein modifications, including ubiquination and methylation, have begun to be recognized [1]. The methylation of proteins and the enzymes that carry out these reactions increased the dimensions of the regulation of gene transcription by marking genes to be or not to be transcribed [2]. Protein methylation can occur on amino acids such as lysine, arginine, histidine, or proline, and on carboxy groups [3]. Arginine methylation of mainly nuclear proteins is an important posttranslational modification process involved in structural remodeling of chromatin, signal transduction, cellular proliferation, nucleocytoplasmatic shuttling, translation, gene transcription, DNA repair, RNA processing, or mRNA splicing [4–7]. Here, I will focus on the family members of the human protein arginine methyltransferases (PRMTs) which modify the characteristics of cellular proteins by transferring methyl residues to the basic amino acid arginine in target proteins and which are important for many benign or malign cellular processes.
The discovery of protein arginine methyltransferases
The proteins included in the PRMT family are evolutionarily conserved between organisms but differ in the number of its members. Arginine methylation activity was discovered in nuclear thymus extracts over 40 years ago and, later, different family members were identified and purified from calf brain, rat liver, and different cell lines [8–12]. Shortly after the discovery of the proteins, the corresponding cDNAs were isolated from mammalian tissues [13–15]. It was only recently that novel proteins of this enzyme family were found using sequence comparison, database searches, or homology studies [16, 17]. From the group around W. K. Paik [18], the discover of the first methylase enzyme, an excellent historical review describing the multifaceted role of protein methylation was published. Currently, the following enzymes have been found: in yeast (Saccharomyces cerevisiae), there are four enzymes (Rmt1/Hmt1, Rmt2, Rmt3, Hsl7/Skb1); in the mold (Aspergillus nidulans), three enzymes (RMTA–C); in the fruit fly (Drosophila melanogster), nine enzymes (DART1–9); in the sea squirt (Ciona intestinalis), six enzymes (Ci1, Ci3–7); and in the zebrafish (Danio rerio), seven PRMT enzymes (Zf1–7). They are described in detail elsewhere [16, 19–22]. As new sequencing projects using many different species have been performed, the combination of relevant database mining with homology searches may reveal novel proteins and extend the PRMT family even further. So far in humans, 11 PRMT proteins have been identified based on differences of the primary protein sequence and of the specificity for distinct substrates. With the exception of PRMT2 and two recently identified putative PRMT genes (PRMT10 and PRMT11), all remaining proteins of the family possess enzymatic and catalytic arginine methylation activity.
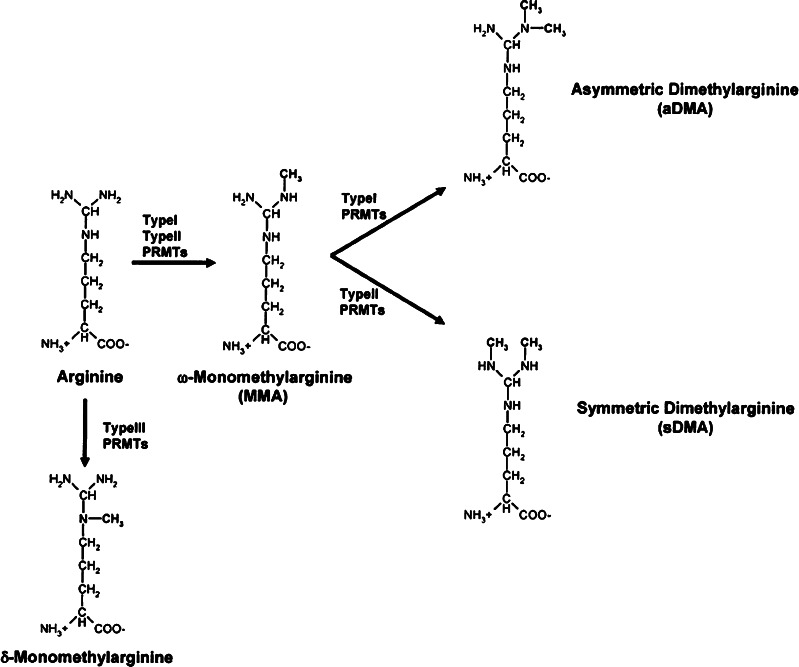
Biochemical reaction of PRMTs
The posttranslational modification of protein arginine methylation is widely appreciated, playing a vital role in cellular function. The PRMT enzymes remove one residue, the methyl group, from the donor molecule S-adenosyl-l-methionine (AdoMet) to generate the product S-adenosyl-l-homocystein (AdoHcy), and hereby transferring the residue to an acceptor molecule which is the terminal nitrogen atom of the guanidinium side chain of an individual arginine residue in the target protein [23, 24]. As there are three nitrogens in the guanidine group, putatively all of them could be methylated; the two ω-guanidino nitrogen atoms and the internal δ-guanidino nitrogen atom [25, 26]. Indeed, mono- and dimethylation reactions of arginine are found to occur in mammalians: ω-N G-monomethylarginine (MMA), symmetric ω-N G,N ′G-dimethylarginine (sDMA), or asymmetric ω-N G,N G-dimethylarginine (aDMA) (Fig. 1). The third methylated arginine is generated by monomethylation of the internal δ-guanidino nitrogen atom of arginine (δ-N-methyl-l-arginine) and has so far been documented only for yeast proteins [27]. According to their methylation status, the PRMT enzymes were classified into different group types. While the type-I PRMT enzymes catalyze the formation of MMA and aDMA, the type-II PRMT enzymes form MMA and sDMA (Fig. 1). The enzymes PRMT1, PRMT3, PRMT4, PRMT6, or PRMT8 belonging to the type-I and PRMT5, PRMT7, or PRMT9 to the type-II enzymes [17, 22, 26]. The type-III PRMT enzymes which catalyzes methylation at the δ-guanidino group in yeast are discussed elsewhere [27, 28]. Type-I and type-II PRMT enzymes were responsible for the arginine methylation of human proteins, and the list of proteins known to be methylated has been growing rapidly over the past decade detecting hundreds of proteins which include methylarginine residues. Interestingly, some of them have implications for disease processes and are involved in certain cancer types, cardiovascular disease, multiple sclerosis, and spinal muscular atrophy [24, 29, 30]. The first arginine methylated proteins identified in humans were histones and heterogenous nuclear ribonucleoproteins (hnRNP) [9, 10]. Recently, many additional proteins such as nucleolin, fibrillarin, and helicases have been found to be methylated by PRMT enzymes [18, 26]. There is also a growing list of biochemical and biological processes, such as signal transduction, proliferation, transcriptional regulation, and RNA splicing, where arginine methylation is involved. Interestingly, for many years it was unknown if the methylation process would be reversible by removing the methyl group from the corresponding amino acid in the target proteins. Only very recently have different enzymes been identified which counteract the methylation process by catalyzing a demethylation step and so remove methyl residues from the target proteins (e.g., LSD1, JMJD6) [31–33]. It should be pointed out that JMJD6, a Jumonji-domain-containing protein, is the only arginine specific demethylase so far identified [32]. These findings clearly indicate that the process of protein methylation is also, like acetylation or phosphorylation, a reversible step in the modification of active and repressed states of chromatin function [34].
Fig. 1.
Methylation of the arginine side chain by PRMTs. Depicted is the amino acid arginine in target proteins. Type-I (TypeI) and type-II (TypeII) enzymes catalyze the formation of MMA by transfer of a methyl group to the ω-guanidino group. In addition, transfer of an additional methyl group results in aDMA (type-I enzymes), or sDMA (type-II enzymes). Type-III (TypeIII) enzymes transfer the methyl group to the internal δ-nitrogen
Structure and functional analysis of the PRMTs
The length of the PRMT proteins vary between 316 and 956 amino acids (Fig. 2, Table 1). All PRMTs have a common catalytic methyltransferase domain which consists of a highly conserved core region of around 310 amino acids and subdomains important for binding to the methyl donor and to the substrate [25]. The individual PRMT family members differ in unique N-terminal regions of variable length and distinct domain motifs (Fig. 2). Some of the enzymes, like PRMT2, PRMT3, or PRMT8, include SH3, zinc finger (ZnF), or myristoylation (Myr) motifs, respectively. In addition, PRMT4 has an unique C-terminal region, PRMT7 or PRMT10, a second catalytic domain, and PRMT9 or PRMT11, an additional module important for protein interaction (F-box). As all the functions of the N- or C-terminal regions of PRMTs are not fully understood, deletion analysis elucidated an impairment in stability as well as a decline in PRMT enzymatic activity [35–37]. Different regions of PRMT5 were able to interact with each other indicating the importance in formation of homomeric protein complexes [38]. Furthermore, an autonomous activation domain in the C-terminus of PRMT4 was important for the coactivator function of the protein [39]. At the time, only the structures of type-I PRMTs were solved using X-ray crystallography studies such as PRMT1, PRMT3, or HMT1p [36, 37, 40].
Fig. 2.
The human PRMT family. The different members are indicated as boxes and the protein length by the numbers of amino acids. Only the longest isoforms are shown. All PRMTs have at least one conserved Catalytic Domain (shaded gray). Different additional domains are highlighted (green): SH3, ZnF zinc finger, Myr myristoylation, F-box, TPR tetratricopeptide, NosD nitrous oxidase accessory protein
Table 1.
The human PRMT family members
| Name | PRMT1 | PRMT2 | PRMT3 | PRMT4 | PRMT5 | PRMT6 | PRMT7 | PRMT8 | PRMT9 | PRMT10 | PRMT11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Other aliases/synonyms |
ANM1 HCP1 IR1B4 HRMT1L2 |
MGC11137 HRMT1L1 |
HRMT1L3 | CARM1 |
JBP1 SKB1 IBP72 SKB1hs HRMT1L5 |
FLJ10559 HRMT1L6 |
FLJ10640 KIAA1933 |
HRMT1L4 |
FBXO11 VIT1 UBR6 FLJ12673 MGC44383 |
LOC90826 FLJ46629 |
FBX10 FBXO10 FLJ41992 MGC149840 |
| Enzyme type | I | Unknown | I | I | II | I | II | I | II | Unknown | Unknown |
| Chromosomal localization | 19q13.3 | 21q22.3 | 11p15.1 | 19p13.2 | 14q11.2 | 1p13.3 | 16q22.1 | 12p13.3 | 2p16.3 | 4q31.23 | 9p13.2 |
| RefSeq transcript(s) | |||||||||||
| Variant 1 | NM_001536 | NM_206962 | NM_005788 | NM_199141 | NM_006109 | NM_018137 | NM_019023 | NM_019854 | NM_025133 | NM_138364 | NM_012166 |
| Variant 2 | NM_198319 | NM_001535 | NM_001039619 | NM_018693 | |||||||
| Variant 3 | NM_198318 | NM_012167 | |||||||||
| Variant 4 | AK022735 | ||||||||||
| RefSeq protein(s) | |||||||||||
| Isoform 1 | NP_001527 | NP_996845 | NP_005779 | NP_954592 | NP_006100 | NP_060607 | NP_061896 | NP_062828 | NP_079409 | NP_612373 | NP_036298 |
| Isoform 2 | NP_938075 | NP_001526 | NP_001034708 | NP_061163 | |||||||
| Isoform 3 | NP_938074 | NP_036299 | |||||||||
| Isoform 4 | BAB14214 | ||||||||||
| Length Isoform 1 (aa) | 371 | 433 | 531 | 608 | 637 | 316 | 692 | 394 | 843 | 845 | 956 |
| Length Isoform 2 (aa) | 347 | 433 | 620 | 686 | |||||||
| Length Isoform 3 (aa) | 353 | 585 | |||||||||
| Length Isoform 4 (aa) | 561 | ||||||||||
| Cellular localization | |||||||||||
| Nucleus | + | + | − | + | − | + | + | − | + | Unknown | Unknown |
| Free cytoplasm | + | + | + | − | + | − | + | − | + | Unknown | Unknown |
| Plasma membrane | − | − | − | − | − | − | − | + | − | Unknown | Unknown |
Listed are the PRMT family members, their aliases and synonyms, the enzyme types, and the chromosomal localization in the human genome. The RefSeq transcripts of the variants and the RefSeq protein isoforms are indicated as described in GenBank. The length of the major isoforms are included with the corresponding amino acids (aa). The cellular localization of the PRMTs are indicated as positive (+) or negative (−) in the nucleus, free cytoplasm, or the plasma membrane
The PRMT family members
Until now, 11 PRMTs have been identified in humans which differ in sequence and length (Fig. 2). In addition, different splice variants coding for the corresponding PRMT isoforms are recognized (Table 1). With the exception of the localization of PRMT1 and PRMT4 on human chromosome 19, all other PRMTs are located on different chromosomes (Table 1). In humans, five PRMTs belong to the type-I and three to the type-II enzymes. For the remaining three proteins (PRMT2, PRMT10, PRMT11) the enzymatic activity as well as the methylation function have not so far been determined (Table 1). PRMT proteins were found in the nucleus and/or the cytoplasm, or even attached to the plasma membrane (Table 1). Phylogenetic studies of the PRMT protein sequences using guide-tree calculation based on the sequence distant method and applying the neighbor-joining algorithm indicated a close relationship between the PRMT isoforms and the different PRMTs (Fig. 3) [41]. So the proteins PRMT1 and PRMT8 were more closely related to PRMT3, whereas PRMT11 and PRMT9 were closer to PRMT5 (Fig. 3). PRMT2, PRMT4 and PRMT6 were more distant to the other PRMTs (Fig. 3). Subsequently, we describe a detailed description of all the individual family members which are known up to now.
Fig. 3.
Phylogenetic analysis of the protein sequences of all PRMTs using guide-tree calculations. The length of the lines indicate the relationship and distance between the PRMT proteins. The sequences for the different PRMTs and the isoforms (indicated by the additional number) are from Table 1
PRMT1
PRMT1 as a type-I enzyme is involved in a variety of processes including gene transcription, DNA repair, and signal transduction [42–44]. PRMT1 was the first protein arginine N-methyltransferase in mammalian cells to be cloned and discovered independently from different groups as a protein interacting with the mammalian intermediate-early TIS21 protein and the leukemia-associated BTG1 protein, or with the intracellular domain of the IFNα receptor [13, 15]. In addition, PRMT1 was found by direct sequence comparison of human expressed sequence tags (ESTs) with the previously cloned homolog protein from yeast (HMT1p) [45]. PRMT1 as the predominant type-I PRMT present in mammalian cells and tissues is expressed in every cell type investigated [46–48]. There are at least three different transcript variants described coding for isoform between 371 and 353 amino acids: isoform 1 (371aa), isoform 2 (347aa), and isoform 3 (353aa) (Table 1). Interestingly, the relative prevalence of alternatively spliced variants were different between normal and cancerous breast tissues making this a valuable tool as a prognostic marker in breast cancer [42]. Close inspection of the genomic organization of the PRMT1 gene revealed up to seven protein isoforms that are expressed in a tissue-specific manner [49]. The variants 4–7 are expressed in distinct tissues such as heart, pancreas, or muscle only, whereas variants 1–3 are expressed ubiquitously. In addition, the different variants are also localized predominantly in the nucleus (variants 1 and 7), the cytosol (variant 2), or both compartments (variants 3–6). Because it is the predominant enzyme in mammalian cells and accounting for most of the cellular activity, PRMT1 was the first arginine methylase discovered. Therefore, it is not surprising that particularly for this enzyme over 40 substrates are already described and discussed elsewhere [26, 48]. The methylation of histone H4 to regulate gene transcription and of the elongation factor SPT5 which regulates its interaction with RNA polymeraseII were two important processes mediated by PRMT1 [11, 50]. Furthermore, PRMT1 methylates proteins involved in RNA processing such as poly(A)-binding proteins and proteins in DNA repair and checkpoint control [51–53]. Besides ribosomal and RNA-binding proteins, a kinase adaptor protein (SAM68) is methylated by PRMT1 indicating a role in cell cycle regulation [54]. Recently, it was shown that PRMT1 played a role in rapid estrogen signaling by methylating arginine residues within the estrogen receptor α (ERα) in breast cells [55]. PRMT1 is also involved in interferon signaling by transferring methyl residues to the protein inhibitor of activated STAT1 (PIAS1) [56]. In addition, PRMT1 was able to cooperate with other PRMTs to regulate gene transcription [57]. The PRMT1 binding proteins such as the antiproliferative protein BTG1 or the deadenylase hCAF1 are able to interact with PRMT1 and thereby regulate the activity of the methylase in a substrate-dependent manner [58, 59]. Interestingly, as most PRMT1 variants are predominantly localized in the nucleus and some in the cytoplasm as well, an independent role of the protein in both compartments is likely [35, 49, 60]. The structure of the mammalian PRMT1 revealed a two-domain structure composed of the AdoMet binding domain and a barrel-like domain letting the active site be situated inbetween [40].
PRMT2
PRMT2 was identified by sequence homology with the human PRMT1 and the yeast methyltransferase [45, 47]. PRMT2 transcripts were detected in most human tissues with an increased expression in heart, prostate, ovary, and the neuronal system [47, 61]. There are two splice variants described in the human genome which differ only in the second exon (Table 1). As the translation start site of PRMT2 is localized in the following exon (exon 3), both variants encoded the identical protein of 433 amino acids. Interestingly, the C-terminus of PRMT2 possessed homology to the SH3 domain of the human SRC oncogene which was essential for the interaction with the heterogeneous nuclear ribonucleoprotein E1B-AP5 (Fig. 2) [62]. Analysis using two-hybrid screening approaches identified PRMT2 as interacting with different nuclear hormone receptors such as the ERα [63] and the androgen receptor (AR) [61]. It is of importance to note that, in contrast to the publication of Qi et al. [63], the binding of PRMT2 to the C-terminal AF2 domain of ERα was dependent on the presence of estrogen [61]. The amplification of ER signaling by PRMT2 strongly depends on the cellular background and differs between neuroblastoma and prostate cells [61]. Furthermore, even if a nuclear localization signal in the PRMT2 protein sequence could not be detected, PRMT2 was found predominantly in the nucleus and to a lower degree also in the cytoplasm of mammalian cells [62]. Furthermore, trafficking of PRMT2 between different cellular compartments was ligand-dependent because an AR agonist was able to translocate PRMT2 together with the nuclear receptor from the cytosol to the nucleus, whereas an AR antagonist could not induce this effect [61]. At the time, the enzymatic activity of PRMT2 could not be demonstrated and therefore no specific substrates were revealed. The high homology between the sequences of PRMT1 and PRMT2 especially in the catalytic core and the substrate binding domain indicated that similar enzymatic activity between both proteins may exist [45]. Also, the interaction and the colocalization of both PRMT2 and the E1B-AP5 protein in the nucleus implied that the protein may be methylated by PRMT2 [62]. In addition, PRMT2 was able to bind its methyl donor indicating that a methylation step would be conceivable [61, 63]. Future studies will have to elucidate this aspect further.
PRMT3
It was suggested that PRMT1 was predominantly present as a component of a polypeptide complex and therefore a two-hybrid screening approach identified PRMT3 as a novel protein binding to PRMT1 [35]. One variant was isolated encoding a protein with 531 amino acids (Table 1). PRMT3 belongs to the type-I enzyme and is expressed widely in human tissues with subcellular localization in the cytoplasm [25, 35]. An important feature of PRMT3 is the ZnF domain in the amino-terminal part of the protein (Fig. 2). Using deletion studies of this motif, it was concluded that this domain confers substrate specificity and appears to be required for the enzyme to bind and methylate target proteins associated with RNA [35]. PRMT3 was able to transfer methyl groups to ribosomal and RNA-binding proteins [65]. Interestingly, the interaction of a tumor suppressor gene important in lung carcinomas (DAL-1/4.1B) with PRMT3 inhibited its ability to methylate cellular substrates and modulated its enzymatic activity negatively. This suggested an important mechanism through which the suppressor gene was able to affect tumor cell growth [66]. The crystal structure of the conserved core revealed a two-domain structure with the substrate binding, and the barrel-like domain provides a mechanism for the methylation reaction and proposing dimer formation of PRMT3 [37].
PRMT4
PRMT4 first described from the laboratory of M. Stallcup as CARM1 (coactivator associated arginine methyltransferase1) was identified using a yeast two-hybrid approach as a novel protein interacting with GRIP1, a member of the p160 family of proteins which enhance transcriptional activation by nuclear receptors [67]. A single variant was isolated encoding a protein with 608 amino acids (Table 1). PRMT4 belongs to the type-I class of PRMT enzymes and its gene is expressed in all tissues investigated with an increased expression in heart, kidney, and testis [67]. The p160 coactivators, a family of three related p160 kDa proteins SRC1, SRC2 (GRIP1, TIF1), and SRC3 (p/CIP, RAC3, ACTR, ABI, TRAM1), serve primarily as coactivators by binding directly to the relevant nuclear receptors, and are able to recruit additional secondary proteins to mediate gene transcription. The PRMT4 protein is able to bind directly to the p160 family of coactivators and in doing so amplifying the nuclear receptor mediated transactivation of target genes [68]. Furthermore, PRMT4 can synergistically enhance the nuclear receptor function and influence gene activation of ER or AR regulated genes [61, 68–70]. In addition, the coactivator function of PRMT4 relies on its ability to transfer the methyl group to the amino-terminal tail of histone H3 after recruitment to the promoter by nuclear receptors and p160 coactivators. This is believed to link the process of methylation directly with transcriptional function [67, 68, 71]. Besides histones, mRNA stabilizing proteins are also methylated by PRMT4 indicating a possible role in post-transcriptional gene regulation [72, 73]. The identification of the methylation of a variety of splicing factors by PRMT4 couples transcription with mRNA processing [74]. A positive regulation of cell cycle gene like cyclin E and an involvement in estrogen-stimulated breast tumors was described for PRMT4 [75, 76]. The methylation of SRC3 by PRMT4 decreased the ER-mediated transactivation suggesting that PRMT4 not only activates transcription but also terminates hormone signaling by disassembly of the coactivator complex [77]. Interestingly, the methyltransferase activity of PRMT4 itself is negatively regulated through phosphorylation at a conserved serine residue in the substrate binding domain of the protein [78]. The nuclear localization of PRMT4 propose an involvement in muscle differentiation where PRMT4 plays a fundamental role during skeletal myogenesis by activating specifically myogenic genes [79]. Embryos with a targeted disruption of PRMT4 were small in size, died perinatally and had a defect during T-cell development [80, 81]. In these animals, estrogen-responsive gene expression was aberrant indicating genetic evidence for an important role of PRMT4 in hormone-mediated transcriptional regulation [80]. Recently, it was shown that PRMT4 is involved in lipid metabolism by promoting adipocyte differentiation, suggesting an important role in adipose tissue biology [82]. PRMT4 has both chromatin and nonchromatin substrates, and methylation can function as a unique transcriptional switch by selectively impairing cAMP-induced transcription while stimulating nuclear receptor target genes [83]. Because these signaling pathways are developmentally and physiologically important, PRMT4 methylation might have unexpectedly broad biological significance [83]. Finally, it was described that PRMT4 was able to cooperate with PRMT1 and be involved in STAT5- and NF-κB-dependent gene expression or in transcriptional activation by the tumor suppressor p53 [57, 84, 85].
PRMT5
PRMT5 was isolated in a two-hybrid search for proteins interacting with the Janus tyrosine kinase (Jak2), implying a role in cytokine-activated transduction pathways [86]. In human tissues, PRMT5 is widely expressed with some higher level in heart, muscle, and testis [86]. Two variants which differ in the first exon are described and code two isoforms with 637 and 620 amino acids in length, respectively (Table 1). PRMT5 was the first type-II enzyme identified that can result in the formation of sDMA residues [87]. PRMT5 plays a significant role in control and modulation of gene transcription, as the proteins methylated by PRMT5 are important in the regulation of genes such as IL-2 and cyclin E1 [88, 89]. PRMT5 was able to methylate histones and transcriptional elongation factors thereby affecting gene transcription [50, 86, 90]. In addition, PRMT5 is involved in the shaping of spliceosomes by forming methylosomes, complexes involved in the assembly of snRNA core particles where proteins were methylated symmetrically to be able to interact properly [91, 92]. In contrast to PRMT1, which is known to be mainly found in the nucleus, human PRMT5 protein is localized in the cytoplasm and able to form homo-oligomers [38]. Recently, it was shown that the histone-binding and selective adaptor protein COPR5 (cooperator of PRMT5), was able to bind PRMT5 thereby modulating the enzyme substrate specificity and influencing the regulation of genes [93]. In addition, PRMT5 was able to transfer methyl residues to the tumor suppressor p53 and help to discriminate between the cell cycle response and the apoptotic response [94]. Furthermore, as arginine methylation has the potential to alter the effects of p53 activation, it may therefore provide a suitable drug target for the manipulation of the p53 pathway [94]. Finally, knock-out studies in animals show that animals heterozygous for a PRMT5 deletion were viable without obvious pathologies whereas homozygous animals lacking PRMT5 led to immediate death of the zygotes [17].
PRMT6
PRMT6 was identified by searching the human genome for PRMT family members using the characteristic structure of the methyltransferase motif [95]. One splice variant expressed predominantly in kidney, brain, and testis was isolated and encodes for a protein with 316 amino acids making PRMT6 the smallest protein in the PRMT family (Table 1). It was shown that PRMT6 displays a strong nuclear localization, as does PRMT4 [95]. PRMT6 belongs to the type-I enzyme of PRMTs catalyzing the formation of aDMA (Table 1). Interestingly, the methylation of selected proteins of the human immunodeficiency virus type-1 (HIV-1) by PRMT6 down-regulated gene expression by acting as a restriction factor for viral replication representing a form of innate cellular immunity [96–98]. In addition, PRMT6 methylated proteins from the high mobility group A (HMGA1a) family of architectural nuclear factors which were important in chromatin dynamics, placing PRMT6 in the context of chromatin structure organization [99, 100]. Recently, PRMT6 methylation of arginine in histone H3 has also been shown to play an important role in post-translational modification [101].
PRMT7
PRMT7 was cloned independently from two groups by sequences homology search that match known PRMT enzymes and by comparison of motifs containing the consensus substrate binding site of PRMT enzymes [102, 103]. One splice variant mainly expressed in dentritic cells, in the thymus and the reproductive system, codes for a protein of 692 amino acids (Table 1) [104]. It was shown that the PRMT7 protein is localized in both cellular compartments: the nucleus and the cytosol of mammalian cells [102]. PRMT7 was initially characterized in hamster cells as a protein that modulates drug sensitivity to DNA-damaging agents [104]. PRMT7 is unusual among the other family members in that two PRMT core domains are present [103]. The gene seems to be derived from a gene duplication event resulting in two putative substrate binding motifs (Fig. 2). For the functionality of the enzyme, both domains are required as each separate domain was unable to function alone [102]. Beside PRMT5, PRMT7 was characterized as a type-II methyltransferase that was able to synthesize sDMA residues in proteins and was able to methylate proteins such as histones, myelin basic protein, fibrillarin, and spliceosomal proteins [17, 102, 103]. PRMT7 is involved in cancer treatment as inhibitor of the enzyme activity sensitizing cancer cells to chemotherapeutics [105].
PRMT8
PRMT8 was revealed by searching of the public human genomic, cDNA, and EST databases. This enzyme shares over 80% sequence identity with the predominant PRMT1 protein making this the highest degree of homology within the enzyme family of PRMTs (Fig. 3) [106, 107]. Expression analysis using Northern analysis revealed a unique tissue-specific expression as PRMT8 transcripts were largely found in human brain [106]. Only a few PRMT8 ESTs were identified in cDNA libraries from lung, testes, pharynx, and kidney origin making it possible that minor expression occurs in these tissues [106]. That feature lacks all the other enzymes of the methyltransferase family as they are more widely expressed in human tissues [106]. The single variant of PRMT8 codes for a protein of 394 amino acids (Table 1). The PRMT8 protein sequence revealed a Myr motif at the N-terminal end of the protein in proximity with some basic amino acids that facilitate electrostatic interactions with membrane lipids (Fig. 2). Indeed, by using PRMT8 fusion or mutation constructs, it could be shown that the cellular localization of PRMT8 is not the nucleus or the cytoplasm but its association with the plasma membrane using the unique Myr motif in the protein [106, 107]. PRMT8 belongs to the type-I enzymes of methytransferases such as PRMT1, PRMT3, PRMT4, and PRMT6. Because of the close sequence homology of PRMT8 with PRMT1, both proteins have some overlapping substrates such as histones, RNA-binding proteins (e.g., hnRNPs) or the EWS protein responsible for causing Ewing sarcoma family tumor [108, 109]. Over 20 proteins able to bind to PRMT8 were identified which contain putative methylation motifs and could be potential substrates for the enzyme, such as the cytoplasmic phosphoprotein caprin which plays a significant role in cell proliferation [109]. In addition, it was shown that the interaction between the methyltransferase PRMT8 and its potential substrate protein is maintained, although the substrate is completely methylated [109]. This suggests that PRMT8 has, besides the methyltransferase activity, another additional functional activity on the plasma membrane [109]. Interestingly, PRMT8 can interact with PRMT1 to form dimers, and its enzymatic activity is regulated by altering the conformation of the protein at the N-terminal end [106, 107].
PRMT9
PRMT9, also known as F-box only protein 11 (FBXO11), was identified by motif search using part of the sequence of the conserved methyl donor binding domain of PRMTs [110]. PRMT9 has little sequence similarities with other PRMTs but the core domains of PRMT9 are more similar to PRMT5 and PRMT7 suggesting that PRMT9 may also belong to the type-II class of the enzyme (Fig. 3). Indeed, biochemical analysis determined that PRMT9 was able to transfer methyl residues to arginine in target proteins [110]. PRMT9 is, like most of the PRMTs, widely expressed and found in a variety of human tissues and associated with pigment loss in a chronic skin disease [111]. There are four major transcripts in the PRMT9 locus in humans encoding four isoforms between 843 and 561 amino acids: isoform 1 (843aa), isoform 2 (686aa), isoform 3 (585aa), and isoform 4 (561aa). All isoforms of PRMT9 contain the putative donor binding domain and, except for the shortest isoform, a F-box in the N-terminal region (Fig. 2) [110]. The F-box is a protein motif of around 50 amino acids that functions as an important side for protein–protein interaction [112, 113]. The longest isoform of PRMT9 includes a ZnF motif at the C-terminus indicating possible interaction with nucleic acids, but no functional information is presently known (Fig. 2) [110]. Only for the shortest PRMT9 isoform 4 is a direct enzymatic activity described, and methylation of histones, the maltose binding protein (MBP) and several peptides, could be demonstrated [110]. Furthermore, PRMT9 has been identified as a type-II methyltransferase as it synthesizes MMA and sDMA [110]. By using transfection experiments to localize PRMT9 in mammalian cells, it was determined that PRMT9 is found throughout the cell including the nucleus and the cytoplasm [110]. Recently, an association between polymorphism in the PRMT9 gene and inflammation of the middle ear was shown, indicating the importance of methylation in disease [114, 115]. Interestingly, besides regulating development, PRMT9 was identified as an adaptor protein to be responsible for posttranslational modification of the tumor suppressor gene p53 by inhibiting the transcriptional activity without affecting its stability [116, 117]. As PRMT9 is a Nedd8 ligase for p53, a direct relationship between neddylation of the suppressor protein and the methyltransferase activity is unknown [116].
PRMT10
PRMT10 was predicted by its homology to PRMT7 [16]. One human transcript encoding a protein with 845 amino acids is described (Table 1). No biochemical activity or substrates have been determined but the resemblance to PRMT7 suggested that PRMT10 may also belong to the type-II arginine methyltransferases [17]. A second catalytic domain at the C-terminus propose similar activity with PRMT7 but no experimental data about enzyme activity, substrate specificity, or function is presently known [16, 17]. In PRMT10, the N-terminal tetratricopeptide repeat (TRP) motif consists of 34 amino acids and, as a protein–protein interaction module, it is involved in different cellular functions such as cell cycle control, transcription, and splicing [118]. The TRP motif is conserved in evolution indicating functional and fundamental importance, but the link with methylation is presently unknown [118].
PRMT11
PRMT11 was found by homology search using the sequence of PRMT9. One variant, encoding for 956 amino acids, is described and makes PRMT11 the longest member of the enzyme family [16, 17]. At the N-terminus of the PRMT11 protein, a F-box motif was identified which enables other proteins to bind, similar as described for PRMT9 (Fig. 2) [114]. GenBank analysis shows a nitrous oxidase accessory protein (NosD) conserved C-terminal motif that may be important for inorganic ion transport and metabolism (Fig. 2). Because of the similarity with PRMT9, it is predicted that PRMT11 also has a methyltransferase activity and may belong to the type-II of PRMT enzymes, but no biochemical data and functional information are known at this time [17].
Concluding remarks
The transfer of methyl groups to the arginine residues in target proteins is a process involved in posttranslational modification and is being recognized as a very important step for the modification of proteins and their subsequent function. At this time, there are 11 family members of the human protein arginine methyltransferase (PRMTs) described which are expressed in many tissues. For most of the proteins, the transfer of methyl residues to the basic amino acid arginine in target proteins can be directly shown and different enzyme types have been classified. The importance of the PRMT protein family is underlined by the presence of additional isoforms in humans and the evolutionary conservation in a wide variety of eukaryotic genomes. Besides the methylation activity, many PRMTs are involved in transcriptional regulation, can interact with other proteins, or can replace each other’s function. Further work will be necessary to elucidate additional novel functions of the enzyme family. Finally, as some new enzymes are already being described, more proteins may be found which reverse the methylation and demethylate arginine residues.
References
- 1.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 3.Lee DJ, Teyssier C, Strahl BD, Stallcup MR. Role of methylation in regulation of transcription. Endocr Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 4.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 5.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lake AN, Bedford MT. Protein methylation and DNA repair. Mutat Res. 2007;618:91–101. doi: 10.1016/j.mrfmmm.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosh SK, Paik WK, Kim S. Purification and molecular identification of two protein methylases I from calf brain. Myelin basic protein- and histone-specific enzyme. J Biol Chem. 1988;263:19024–19033. [PubMed] [Google Scholar]
- 9.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;155:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paik WK, Kim S. Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem Biophys Res Commun. 1967;29:14–20. doi: 10.1016/0006-291x(67)90533-5. [DOI] [PubMed] [Google Scholar]
- 11.Paik WK, Kim S. Protein methylase I. Purification and properties of the enzyme. J Biol Chem. 1968;243:2108–2114. [PubMed] [Google Scholar]
- 12.Rawal N, Rajpurohit R, Paik WK, Kim S. Purification and characterization of S-adenosylmethionine-protein-arginine N-methyltransferase from rat liver. Biochem J. 1994;300:483–489. doi: 10.1042/bj3000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramovich C, Yakobson B, Chebath J, Revel M. A protein–arginine methyltransferase binds to the intracytoplasmatic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A) + -RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin WJ, Gary JD, Yang MC, Clarke S, Herschmann HR. The mammalian intermediate-early TIS21 protein and the leukaemia-associated BTG1 protein interact with a protein–arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 16.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methylatransferases. J Cell Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- 17.Krause CD, Yang Z-H, Kim Y-S, Lee J-H, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutical potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Boulanger MC, Miranda TB, Clarke S, Di Fruscio M, Suter B, Lasko P, Richard S. Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem J. 2004;379:283–289. doi: 10.1042/BJ20031176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung CM, Li C. Identification and phylogenetic analyses of the protein arginine methyltransferase gene family in fish and ascidians. Gene. 2004;340:179–187. doi: 10.1016/j.gene.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 21.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 22.Trojer P, Dangl M, Bauer I, Graessle S, Loidl P, Brosch G. Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemisty. 2004;43:10834–10843. doi: 10.1021/bi049626i. [DOI] [PubMed] [Google Scholar]
- 23.Bedford MT, Richard S. Arginine methylation: an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 25.Bachand F. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot Cell. 2007;6:889–898. doi: 10.1128/EC.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pahlich S, Zakaryan RP, Gehling H. Protein arginine methylation: cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Zobel-Thropp P, Gary JD, Clarke S. δ-N-Methylarginine is a novel posttranslational modification of arginine residues in yeast proteins. J Biol Chem. 1998;273:29283–29286. doi: 10.1074/jbc.273.45.29283. [DOI] [PubMed] [Google Scholar]
- 28.Chern MK, Chang KN, Liu LF, Tam TC, Liu YC, Liang YL, Tam MF. Yeast ribosomal protein L12 is a substrate of protein–arginine methyltransferase 2. J Biol Chem. 2002;277:15345–15353. doi: 10.1074/jbc.M111379200. [DOI] [PubMed] [Google Scholar]
- 29.Aletta JM, Hu JC. Protein arginine methylation in health and disease. Biotechnol Annu Rev. 2008;14:203–224. doi: 10.1016/S1387-2656(08)00008-2. [DOI] [PubMed] [Google Scholar]
- 30.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 31.Anand R, Marmorstein R. Structure and mechanism of lysine specific demethylase enzymes. J Biol Chem. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 32.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 33.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 35.Tang J, Gary JD, Clarke S, Herschman HR. PRMT3. a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 36.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase. Nat Struct Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rho J, Choi S, Seong YM, Cho W-K, Kim SH, Im D-S. PRMT5, which forms distinct homo-oligomers, is a member of the protein—arginine methyltransferase family. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 39.Teyssier C, Chen D, Stallcup MR. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. J Biol Chem. 2002;277:46066–46072. doi: 10.1074/jbc.M207623200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. The neighbour-joining method: a new method for reconstruction phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Iwasaki H, Yada T. Protein arginine methylation regulates insulin signaling in L6 skeletal muscle cells. Biochem Biophys Res Commun. 2007;364:1015–1021. doi: 10.1016/j.bbrc.2007.10.113. [DOI] [PubMed] [Google Scholar]
- 43.Scorila A, Black MH, Talieri M, Diamandis EP. Genomic organization, physical mapping, and expression analysis of the human protein arginine methyltransferase 1 gene. Biochem Biophys Res Commun. 2000;278:349–359. doi: 10.1006/bbrc.2000.3807. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 45.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 46.Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein—arginine methyltransferase from Saccharomyces cerevisiae . J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 47.Katsanis N, Yaspo ML, Fisher EM. Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm Genome. 1997;8:526–529. doi: 10.1007/s003359900491. [DOI] [PubMed] [Google Scholar]
- 48.Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 49.Goulet I, Gauvin G, Boisvenue S, Côté J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- 50.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor R. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 51.Adams MM, Wang B, Xia Z, Morales JC, Lu X, Donehower LA, Bochar DA, Elledge SJ, Carpenter PB. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 2005;4:1854–1861. doi: 10.4161/cc.4.12.2282. [DOI] [PubMed] [Google Scholar]
- 52.Boisvert FM, Déry U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith JJ, Rücknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly (A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 54.Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 55.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 56.Weber S, Maass F, Schuemann M, Krause E, Suske G, Bauer UM. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev. 2009;23:118–132. doi: 10.1101/gad.489409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer U-M. The protein arginine methylatransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berthet C, Guéhenneux F, Revol V, Samarut C, Lukaszewicz A, Dehay C, Dumontet C, Magaud JP, Rouault JP. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells. 2002;7:29–39. doi: 10.1046/j.1356-9597.2001.00497.x. [DOI] [PubMed] [Google Scholar]
- 59.Robin-Lespinasse Y, Sentis S, Kolytcheff C, Rostan MC, Corbo L, Le Romancer M. CAF1, a new regulator of PRMT1-dependent arginine methylation. J Cell Sci. 2007;120:638–647. doi: 10.1242/jcs.03357. [DOI] [PubMed] [Google Scholar]
- 60.Tang J, Kao PN, Herschman HR. Protein—arginine methyltransferase I, the predominant protein—arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–19876. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- 61.Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107:1–14. doi: 10.1016/j.jsbmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Kzhyshkowska J, Schütt H, Liss M, Kremmer E, Stauber R, Wolf H, Dobner T. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem J. 2001;358:305–314. doi: 10.1042/0264-6021:3580305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 64.Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J Biol Chem. 2000;275:32974–32982. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- 65.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004;23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh V, Miranda TB, Jiang W, Frankel A, Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S, Newsham IF. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23:7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- 67.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 68.Stallcup MR, Chen D, Koh SS, Ma H, Lee YH, Li H, Schurter BT, Aswad DW. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem Soc Trans. 2000;28:415–418. [PubMed] [Google Scholar]
- 69.Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 70.Chen D, Huang S-M, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 71.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 72.Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 74.Cheng D, Côté J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 75.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci USA. 2006;103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 77.Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci USA. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat G. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 80.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, Bedford MT. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279:25339–25344. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
- 82.Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. CARM1 promotes adipocyte differentiation by coactivating PPARγ. EMBO Rep. 2008;9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 84.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 85.Hassa PO, Covic M, Bedford MT, Hottiger MO. Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol. 2008;377:668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 86.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 87.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 88.Richard S, Morel M, Cléroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lacroix M, Messaoudi SE, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008;9:452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 95.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 96.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Invernizzi CF, Xie B, Frankel FA, Feldhammer M, Roy BB, Richard S, Wainberg MA. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS. 2007;21:795–805. doi: 10.1097/QAD.0b013e32803277ae. [DOI] [PubMed] [Google Scholar]
- 98.Xie B, Invernizzi CF, Richard S, Wainberg MA. Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J Virol. 2007;81:4226–4234. doi: 10.1128/JVI.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem Biophys Res Commun. 2005;336:831–835. doi: 10.1016/j.bbrc.2005.08.179. [DOI] [PubMed] [Google Scholar]
- 100.Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281:3764–3772. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- 101.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 102.Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280:3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 103.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–22907. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 104.Gros L, Delaporte C, Frey S, Decesse J, de Saint-Vincent BR, Cavarec L, Dubart A, Gudkov AV, Jacquemin-Sablon A. Identification of new drug sensitivity genes using genetic suppressor elements: Protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 2003;63:164–171. [PubMed] [Google Scholar]
- 105.Verbiest V, Montaudon D, Tautu MT, Moukarzel J, Portail JP, Markovits J, Robert J, Ichas F, Pourquier P. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 106.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 107.Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J Biol Chem. 2007;282:36444–36453. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- 108.Kim JD, Kako K, Kakiuchi M, Park GG, Fukamizu A. EWS is a substrate of type I protein arginine methyltransferase, PRMT8. Int J Mol Med. 2008;22:309–315. [PubMed] [Google Scholar]
- 109.Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:25–37. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- 110.Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun. 2006;342:472–481. doi: 10.1016/j.bbrc.2006.01.167. [DOI] [PubMed] [Google Scholar]
- 111.Le Poole IC, Sarangarajan R, Zhao Y, Stennett LS, Brown TL, Sheth P, Miki T, Boissy RE. ‘VIT1’, a novel gene associated with vitiligo. Pigment Cell Res. 2001;14:475–484. doi: 10.1034/j.1600-0749.2001.140608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:3002.1–3002.7. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, Hunter AJ, Cheeseman M, Brown SD. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15:3273–3279. doi: 10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- 115.Segade F, Daly KA, Allred D, Hicks PJ, Cox M, Brown M, Hardisty-Hughes RE, Brown SD, Rich SS, Bowden DW. Association of the FBXO11 gene with chronic otitis media with effusion and recurrent otitis media: the Minnesota COME/ROM family study. Arch Otolaryngol Head Neck Surg. 2006;132:729–733. doi: 10.1001/archotol.132.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, Hutter H, Pagano M, Antebi A. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 118.Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]