Abstract
KLF5 (Kruppel-like factor 5) is a basic transcription factor binding to GC boxes at a number of gene promoters and regulating their transcription. KLF5 is expressed during development and, in adults, with higher levels in proliferating epithelial cells. The expression and activity of KLF5 are regulated by multiple signaling pathways, including Ras/MAPK, PKC, and TGFβ, and various posttranslational modifications, including phosphorylation, acetylation, ubiquitination, and sumoylation. Consistently, KLF5 mediates the signaling functions in cell proliferation, cell cycle, apoptosis, migration, differentiation, and stemness by regulating gene expression in response to environment stimuli. The expression of KLF5 is frequently abnormal in human cancers and in cardiovascular disease-associated vascular smooth muscle cells (VSMCs). Due to its significant functions in cell proliferation, survival, and differentiation, KLF5 could be a potential diagnostic biomarker and therapeutic target for cancer and cardiovascular diseases.
Keywords: KLF5, Proliferation, Survival, Differentiation, Homeostasis, Tumorigenesis, Cardiovascular diseases
Introduction
Transcription factors regulate diverse cellular processes, including proliferation, cell cycle, apoptosis, migration, and differentiation, by controlling gene expression. Accumulating evidence suggests that genetic aberrations of SP/KLF (Kruppel-like factor) transcription factors are involved in the development of various human diseases, including cancer and cardiovascular diseases, as reviewed previously [1–7]. The KLF family consists of ~20 members in humans, and is structurally characterized by three tandem zinc-finger domains at the C-terminus. Several members of the KLF family, such as KLF2 [8], KLF4 [9, 10], KLF5 [11], KLF6 [12, 13], and KLF8 [14], have been demonstrated to play vital roles in the development of various human cancers.
KLF5, also named BTEB2 [15] and IKLF, belongs to the KLF family. KLF5 is widely expressed at varying levels in different tissues. As a basic transcription factor, KLF5 regulates a number of important target genes, such as cyclin D1, cyclin B, PDGFα, and FGF-BP. KLF5 has essential roles in cell cycle regulation, apoptosis, migration, and differentiation. In recent years, the study of KLF5 has been dramatically expanded. This review article comprehensively summarizes the biochemical and molecular aspects of KLF5, including its gene and protein structures, expression patterns, protein posttranslational modifications, interacting proteins, downstream target genes, and upstream regulators. Following that, we review the functions of KLF5 in various physiological and pathological cellular processes, including cell proliferation, survival, migration, differentiation, and stemness. Additionally, we outline all in vivo studies of KLF5 transgenic mouse models. Finally, the relationships of KLF5 and human diseases are summarized and future research directions for KLF5 are proposed.
Biochemistry
The gene and protein structures
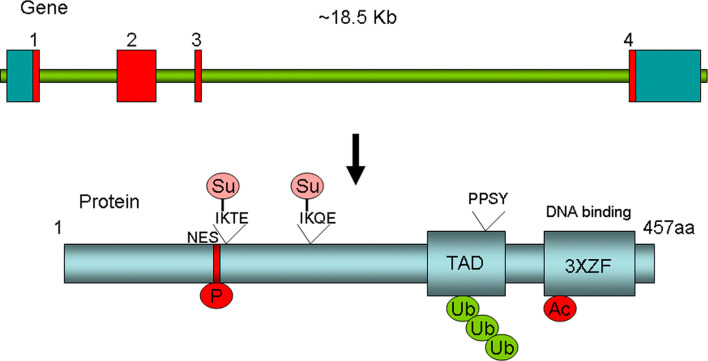
The KLF5 gene is located at 13q21, spanning ~18.5 kb genomic DNA with four exons. The full-length cDNA of human KLF5 consists of 3,350 bp with a 324-bp 5′-untranslated region (UTR), a 1,652-bp 3′-UTR, and a 1,374-bp sequence coding for a 457 amino acid polypeptide (Fig. 1). Similar to other KLFs, the C-terminus of KLF5 protein contains three zinc-finger (ZF) domains, which function in DNA binding. KLF5 has a proline rich transactivation domain (TAD) before the ZF domains [15, 16].
Fig. 1.
The human KLF5 gene and protein structures. The human KLF5 genome contains four exons (exon 1, 585 bp; intron 1, 2,272 bp; exon 2, 874 bp; intron 2, 1,089 bp; exon 3, 60 bp; intron 3, 11,824 bp; and exon 4, 1,831 bp). The KLF5 protein contains three zinc-finger (ZF) domains, one major transactivation domain (TAD) with a PY motif (PPSY328), and a nucleus export signal (NES). The KLF5 protein undergoes different types of posttranslational modifications, including phosphorylation (P at S153), acetylation (Ac at K369), ubiquitination (Ub), and sumoylation (Su at K162 and K209)
Expression
KLF5 is widely expressed at varying levels in different tissues. Based on Northern blot analysis, high levels of KLF5 mRNA are present in the human and mouse digestive tract including intestine, colon, and stomach, and pancreas, placenta, testis, prostate, skeleton muscle, and lung [15, 17, 18]. KLF5 mRNA was also detected in human and rabbit bladder and uterus [19, 20]. Although the expression of KLF5 appears mostly to be epithelial, KLF5 is also expressed in cardiovascular SMCs [19], cornea [21], lymphoid cells [22], and neuronal cells [23]. While most tissues express a 3.3-kb transcript of KLF5, that expressed in the testis is about 1.5 kb [18].
Accumulated evidence suggests that KLF5 is more highly expressed in proliferating cells than in differentiated cells [17]. For example, KLF5 shows temporal changes in expression during embryogenesis [17, 24, 25]. During mouse development, KLF5 mRNA continues to accumulate at a high rate in the basal layer of the epidermis and in the base of the intestinal crypts [24]. Consistently, the KLF5 protein is also exclusively expressed in proliferating epithelial cells at the base of the crypts of the intestine but not in the terminally differentiated epithelial cells in the villi [11].
The KLF5 protein is primarily expressed in the nucleus [18]. However, KLF5 also contains a nuclear export signal (NES) located next to a sumoylation site (Fig. 1) [26]. Du et al. [26] found that sumoylation facilitates KLF5 nuclear localization by inactivating NES.
Posttranslational modifications
KLF5 proteins undergo different posttranslational modifications that modulate the protein level or transactivation activities of KLF5. Such modifications include phosphorylation, acetylation, ubiquitination, and sumoylation (Fig. 1). While KLF5 phosphorylation positively regulates its activity and KLF5 ubiquitination negatively regulates its protein level, the function of acetylation and sumoylation is context-dependent.
KLF5 phosphorylation by PKC at S153 increases the transactivation activities of KLF5 [27]. Zhang et al. [27] reported that a point mutation (S153A) reduces its transactivation function, and that phosphorylation of KLF5 enhances its interaction with CREB-binding protein (CBP). Whether KLF5 is also phosphorylated at other sites by other kinases is still unclear.
KLF5 has been documented to be acetylated by p300 and deacetylated by HDAC1 and SET [28, 29]. Miyamoto et al. [28] reported that the acetyl transferase, p300, acetylates KLF5 at K369, which appears to enhance the transactivation activity of KLF5. The notion was fully confirmed by an independent study: using a KLF5 K369 acetylation specific antibody, Guo et al. [30] demonstrated that TGFβ recruits p300 to acetylate KLF5. The SET histone chaperone was shown to negatively regulate the function of KLF5 in DNA binding and cell proliferation, which is accompanied by an inhibition of KLF5 acetylation [28]. The deacetylase HDAC1 can also interact with KLF5 to inhibit the binding of KLF5 to DNA as well as KLF5-mediated promoter activation through inhibiting the KLF5 acetylation by p300 [29].
KLF5 has been shown to be ubiquitinated. Chen et al. [31] found that KLF5 is degraded through the ubiquitin-proteasome pathway in epithelial cells. Like other crucial transcription factors, such as p53 and c-MYC, KLF5 turns over rapidly. The KLF5 protein half-life is ~1.5 h by pulse chase assays. The destruction domain is within TAD of KLF5. Further investigation revealed that the WWP1 E3 ubiquitin ligase can bind to the PY motif of KLF5 in TAD (Fig. 1), to ubiquitinate and degrade KLF5 [32]. Interestingly, the WWP1 gene is frequently amplified and overexpressed in both breast and prostate cancers [32–34]. The WWP1 protein is highly expressed in ERα positive breast cancer while KLF5 appears to be expressed in ERα negative breast cancer [35, 36]. Our unpublished results suggest that KLF5 may also be ubiquitinated by other E3 ligases. Additionally, degradation of KLF5 by the proteasome pathway can be ubiquitin-independent [37], although the molecular mechanism is not fully understood.
KLF5 has been demonstrated to be sumoylated in two recent studies [26, 38]. Du et al. [39] applied a yeast two-hybrid screen to identify proteins that interact with KLF5 and identified the sumoylation E3 ligase PIAS1 as the potential KLF5-interacting protein. Du et al. [26] further demonstrated that mouse KLF5 is sumoylated at lysine residues 151 and 202, and that sumoylation facilitates nuclear localization and function of KLF5 by inactivating NES located next to K151 (Fig. 1). The sumoylation of human KLF5 was reported in an independent study [38].
KLF5 posttranslational modifications, including sumoylation and acetylation, can switch the function of KLF5. Although KLF5 has no transcription repressor domain, Oishi et al. [38] found that KLF5 directly inhibits transcription of lipid metabolism genes, including Cpt1b, Ucp2, and Ucp3 by recruiting co-repressors. After KLF5 is sumoylated upon agonist stimulation of PPARδ, KLF5 recruits co-activators to induce the Cpt1b, Ucp2, and Ucp3 transcription [38]. Similarly, KLF5 normally suppresses p15 gene transcription. After KLF5 is acetylated upon TGFβ stimulation, KLF5 increases the p15 gene transcription [30]. Thus, KLF5 can regulate the expression of the same set of target genes toward opposite directions in response to environment stimuli.
Transcriptional target genes
KLF5 has been demonstrated to regulate many genes involved in cell proliferation, cell cycle, survival, migration, angiogenesis, stemness, and differentiation (Table 1) in different contexts. Most KLF5 target gene proximal promoters contain one or more GC rich sites [15, 16, 27]. Although KLF5 has been shown to bind to Sp1 sites, GC boxes, and CACCC boxes, there are no strictly conserved consensus core sequences [15, 16, 27].
Table 1.
Functional classification of direct KLF5 target genes
| Functions | Target genes | References |
|---|---|---|
| Cell cycle | Cyclin D1, Cyclin B1, Cdc2, p15, p27 | [30, 40, 49, 75, 79, 128] |
| Angiogenesis | PDGFα, VEGFα, FGF-BP | [40, 41, 55, 59, 129] |
| Inflammation | MCP-1, NF-κB | [56, 124] |
| Apoptosis | Survivin, Pim1 | [45, 80] |
| Migration | ILK, MMP9 | [82, 100] |
| Stemness | Nanog, Tcl1, Oct3/4, Esrrb, Fbxo15 | [90, 92] |
| Differentiation | SMemb/NMHC-B, SM22α, PAI-1, Egr-1, PPARγ, iNOS | [19, 46, 63, 117, 118, 130] |
| Fatty acid metabolism | Cpt1b, Ucp2, Ucp3, FASN | [38, 47] |
| Others | Lactoferrin, TCR Dβ1, γ-globin, MAO-A/B, KLF4, Lama1, DAF, EGFR | [18, 22, 76, 131–137] |
Two genome wide microarray experiments have been performed to systematically identify KLF5-regulated genes. Chen et al. [40] stably expressed KLF5 in the TSU-Pr1 bladder cancer cell line, and performed a microarray analysis. At least 58 genes are differentially expressed between KLF5-positive and KLF5-negative TSU-Pr1 cells [40]. Many of the genes have been validated by different approaches for their differential expression, including HBP17/FGF-BP, B94/TGFAIP2, DUSP1MKP-1, ADRB2, BCAR3, CD24, Dri42, DUSP5, EEF1A2, EMP1, EXT1, ITGA6, Lipocortin III, MN1, p27, PIG12, RAIG1, SAS, Slit, TGFα, TGM2, TIMP2, TMOD, and Wnt7a. Wan et al. [41] generated a lung-specific Klf5 knockout mouse model and performed a microarray analysis. KLF5 regulates the expression of hundreds genes associated with cell cycle, angiogenesis, lipid metabolism, and several paracrine signaling pathways (PDGF-FGF, VEGF, TGFβ, and BMP) [41]. Interestingly, the fibroblast growth factor binding protein 1 (FGF-BP/HBP17) was identified as KLF5 target gene in both studies, suggesting that FGF-BP is regulated by KLF5 in vitro and in vivo. Our unpublished results suggest that KLF5 functions through FGF-BP to promote breast epithelial cell proliferation.
Interacting proteins
To date, about 20 KLF5 interacting proteins have been identified through yeast two-hybrid screening, mass spectrometry, and co-immunoprecipitation (Table 2). Besides the KLF5 posttranslational modifiers described above, KLF5 has been shown to assemble a transcriptional complex at chromosome to regulate gene expression by interacting with multiple proteins of the transcriptional machinery. KLF5 interacts with components of the transcriptional machinery including initiation factors, TFIIB, TFIIEβ, and TFIIFβ as well as the TATA box-binding protein (TBP) [16]. Recently, KLF5 has been demonstrated to bind to PPARδ and co-suppressors NcoR and SMRT at the promoters of Ucp2/3 and Cpt1b [38]. KLF5 can also mediate the modification of histone to regulate chromatin in gene regulation. Munemasa et al. [42] recently found that KLF5 recruits a novel histone chaperone named acidic nuclear phosphoprotein 32B (ANP32B) onto gene promoter, where ANP32B incorporates into promoter-region specific histone to inhibit histone acetylation and repressess the transcription of a KLF5-downstream gene.
Table 2.
KLF5 interacting proteins
| Function | KLF5 interacting proteins | References |
|---|---|---|
| Posttranslational modifiers | WWP1, PKC, p300, HDAC1, SET, PIAS | [27–30, 32, 39] |
| Basic transcription factors and co-activators | TFIIB, TFIIEβ, TFIIFβ, TBP, CBP, p300, ANP32B, NcoR, SMRT | [16, 27, 38, 42] |
| Co-transcription factors | RAR/RXR, NFκB, p53, C/EBPβ/δ, SREBP-1, PPARδ | [38, 44–47, 59, 129] |
| Others | PARP-1 | [48] |
The KLF5 protein has been shown to associate with numerous transcription factors, such as retinoic acid receptor (RAR/RXR), NFκB, p53, C/EBPβ/δ, and sterol-regulatory-element-binding protein-1 (SREBP-1), to regulate gene transcription. Fujiu et al. [43] reported that KLF5 forms transcriptional complexes with RAR/RXR heterodimer on PDGFα promoter. In a yeast one-hybrid screen using a keratinocyte-specific NF-κB binding site as a bait, Sur et al. [44] identified KLF5 as a factor cooperating with NF-κB in epidermal epithelial cells. Zhu et al. [45] found that KLF5 interacts with p53 at the promoter of survivin gene in leukemia. Similarly, KLF5 was shown to interact with C/EBPβ/δ at the PPARγ promoter in adipocyte cells [46]. Most recently, transcriptional regulation of fatty acid synthase (FASN) involves an interaction between KLF5 and SREBP-1 [47]. As an essential effector for the TGFβ pathway, KLF5 has been shown to interact with Smad2/3, Smad4, Myc, Miz-1, and p300 to regulate the expression of p15 [140].
Besides transcription factors, KLF5 also specifically interacts with poly(ADP-ribose) polymerase-1 (PARP-1), a nuclear enzyme important in DNA repair and apoptosis [48]. The protein interaction occurs at the proteolytic fragment of PARP-1. It was believed that the interaction has a pro-survival function through an unknown mechanism [48].
Signaling pathways
KLF5 plays important roles in multiple important growth factor signaling pathways, including Ras-mitogen-activated protein kinase (MAPK), PKC, TGFβ, TNFα, retinoid, and androgen (Fig. 2). As a basic transcription factor, KLF5 could mediate the function of different signaling pathways in different biological processes, including cell proliferation and differentiation, which often involves transcriptional regulation of KLF5.
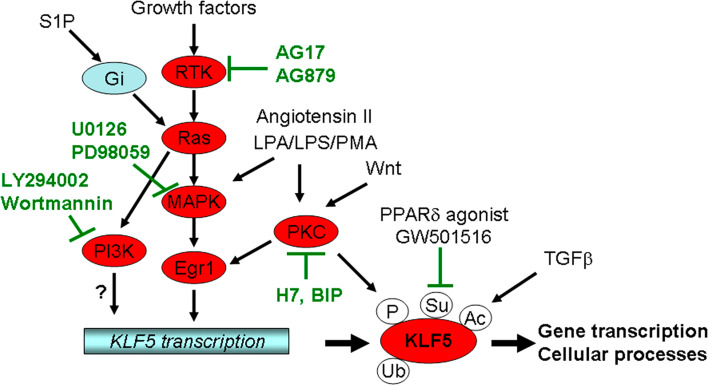
Fig. 2.
The regulatory signaling pathways for KLF5. KLF5 is regulated by multiple signaling pathways at the transcriptional and posttranslational levels. LPA Lysophosphatidic acid, LPS lipopolysaccharide, PMA phorbol 12-myristate 13-acetate, S1P sphingosine 1-phosphate, BIP bisindolyimalimide
First, the notion that KLF5 is induced by the Ras-MAPK signaling pathway is supported by several lines of investigation (Fig. 2). In oncogenic H-Ras-transformed NIH3T3 cells, KLF5 is upregulated at both RNA and protein levels that can be blocked by inhibiting MEK or Egr-1[49]. In SMCs, KLF5 mRNA is rapidly and persistently induced by phorbol 12-myristate 13-acetate (PMA) through Egr-1 [50]. Nandan et al. [51] found that induction of K-Ras (V12G) results in increased expression of KLF5 in IEC-6 intestinal epithelial cells. Consistently, higher levels of KLF5 are expressed in cell lines and primary tumors from human colorectal cancer with mutated K-Ras, and primary colorectal cancers induced by the K-Ras (V12G) mutation. Additionally, KLF5 was found to be upregulated by the NEU/ErbB2 oncogene, which encodes a receptor tyrosine kinase and actions upstream of Ras [52]. Fetal bovine serum, androgen, and fibroblast growth factor have been shown to induce KLF5 expression [53, 54]. Furthermore, sphingosine 1-phosphate (S1P) induces KLF5 in VSMCs and neointimal cells via Gi-protein-Ras-ERK/p38 pathway [55]. Lipopolysaccharide (LPS) is a bacterially-derived endotoxin that also induces KLF5 expression through ERK in the IEC-6 intestinal epithelial cell line [56]. Consistently, MAPK inhibitors PD98059 and U0126 inhibit the KLF5 expression [49]. These results strongly support the idea that KLF5 is an important mediator of the Ras-MAPK pathway.
Second, KLF5 is regulated by Wnt and angiotensin II via the PKC signaling pathway (Fig. 2). In a mouse mammary epithelial cell line C57MG and a clone that overexpresses Wnt-1, cDNA subtractive hybridization screen identified KLF5 as one of the Wnt-responsive genes [57]. Similar results have been detected in mammary tissues from a transgenic mouse overexpressing Wnt-1 [58]. The regulation of KLF5 by the Wnt-1 signaling is β-catenin-Lef/TCF-independent but related to PKC [58]. Angiotensin II, which plays a critical role in cardiovascular remodeling, functions through KLF5, as knockout of KLF5 in mice attenuates angiotensin II-induced cardiac hypertrophy and fibrosis [59]. Consistently, in human venous SMCs treated with angiotensin II, the expression of KLF5 is rapidly upregulated at both the RNA and protein levels [60]. Even in growth-arrested vascular SMCs, angiotensin II still induces KLF5 expression and cell proliferation [61]. A PKC inhibitor can block the KLF5 induction by angiotensin II [61]. Finally, PMA and lysophosphatidic acid (LPA) may also induce KLF5 partially through PKC [50, 62].
Third, KLF5 is also a functional mediator of TGFβ. In the study of SM22α regulation, Adam et al. [63] found that both KLF5 and KLF4 play a role, and that they function by binding to a TGFβ-control element (TCE). The same group further demonstrated an essential role of KLF5 and the TCE in SM22α promoter activity in transgenic mice [64]. More directly, Guo et al. [30] found that KLF5 is essential for TGFβ to inhibit epithelial cell proliferation, although KLF5 alone is necessary for cell proliferation. The underlying mechanism is that TGFβ recruits p300 to acetylate KLF5 (Fig. 2), which in turn mediates the assembly of transcription factors for the regulation of TGFβ target genes.
Additionally, KLF5 could be involved in the retinoid/retinoid receptor signaling. Based on results showing an inhibitory effect of the synthetic RARα-specific agonist Am80 on the activity of KLF5 in SMCs, Fujiu et al. [43] conducted experiments to understand how Am80 inhibits KLF5. They found that Am80 inhibits both the expression and transcriptional function of KLF5. In mouse embryonic stem (ES) cells, KLF5 appears to be repressed by the Hoxa1 protein, as knockout of the Hoxa1 gene increased the expression of KLF5 induced by retinoic acid [65]. Induction of klf5 expression has been detected in additional cell systems receiving different treatments, including cardiac myocytes undergo apoptosis after endothelin-1 treatment [66], bovine endometrium during the oestrous cycle [67], dimethylnitrosamine-induced liver fibrosis in rodents [68], radiation-induced vascular injuries in mouse kidney and rectum [69], tissue factor-triggered blood coagulation [70], and cultured gastric cancer cell line infected with bacteria [71], although the underlying signaling pathways are unknown.
Deletion analyses of the promoter region of rat KLF5 suggest that at least three regions are important for KLF5 transcription, including the GC box, CCAAT box, and NF-1 binding site. Gel mobility shift assays (EMSAs) demonstrated that factors Sp1, CBFa, and NF-1 bind to the KLF5 promoter [72]. A Sp1 site is essential for a basal human KLF5 promoter activity, and the binding of Sp1 to this element has been confirmed by EMSA [54]. An Egr-1 binding site at human KLF5 promoter is essential for the KLF5 induction by PMA [50]. Recently, Bialkowska et al. [73] screened compounds that regulate the 1,959 bp KLF5 promoter and found that phosphoinositide 3-kinase (PI3K) inhibitors (LY294002 and Wortmannin) and that receptor tyrosine kinase (RTK) inhibitors (PDGFR inhibitor AG17 and ErbB2 inhibitor AG879) suppresses the KLF5 transcription in colon cancer cell lines (Fig. 2). Interestingly, AG17 appears to reduce the KLF5 expression through inhibiting the EGR-1 protein expression in colon cancer cell lines [73].
Cellular and physiological function
Cell cycle and proliferation
At present, the best known function of KLF5 is its stimulatory role in the proliferation of different types of cells, including fibroblasts, epithelial cells, and SMCs. Sun et al. found that ectopic expression of KLF5 into NIH3T3 cells significantly increased the rate of cell proliferation and caused a phenotype of transformation in a soft-agar assay [53]. As described above, KLF5 is upregulated in oncogenic H-Ras-transformed NIH3T3 cells [49]. Inhibition of KLF5 expression with KLF5-specific small interfering RNA (siRNA) leads to a decreased rate of proliferation and a significant reduction in colony formation. Similarly, Nandan et al. [51] found that K-Ras(V12G) also induces KLF5 to promote the IEC-6 intestinal epithelial cell proliferation. These findings indicate that elevated KLF5 expression is responsible for the pro-proliferative and transforming activities of oncogenic Ras [49]. Additionally, LPA stimulates cell proliferation through inducing the KLF5 expression in colon cancer cell lines [62].
Retinoids are known inhibitors of epithelial cell proliferation. In the intestinal epithelial cell line IEC-6, treatment with all-trans retinoid acid (ATRA) inhibits cell proliferation due to G1 cell cycle arrest. It was noted that this inhibition is correlated with a decrease in the levels of KLF5 mRNA [74]. On the other hand, stable overexpression of KLF5 in IEC-6 cells abrogates the growth inhibitory effect of ATRA [74]. Furthermore, ATRA appears to inhibit cell proliferation only in human colon cancer cell lines that express a higher level of KLF5 but not in those with low levels of KLF5. These studies suggest that KLF5 is involved in the inhibitory effect of ATRA on intestinal epithelial cell proliferation [74].
KLF5 promotes cell proliferation through accelerating the G1/S and G2/M cell cycle progression. Chen et al. [40] reported that overexpression of KLF5 in the bladder cancer cell line TSU-Pr1 promotes the G1/S cell cycle progression and tumorigenesis. Nandan et al. [51] demonstrated that KLF5 also promotes the G2/M transition in Ras transformed fibroblasts. In addition to inducing cyclin D1 and inhibiting p27 and p15 [40], KLF5 also upregulates cyclin B1 and Cdc2 [75]. In mouse primary cultures of esophageal keratinocytes, KLF5 increases cell proliferation through upregulating EGFR and the MEK/ERK signaling [76].
In contrast, KLF5 has also been found to inhibit cell proliferation in several cancer cell lines in vitro. For example, in the TE2 esophageal cancer cell line, stable expression of KLF5 inhibits cell proliferation [77]. Chen et al. [78] reported that KLF5 inhibits colony formation in the DU145 and 22Rv1 prostate cancer cell lines. KLF5 has shown a growth promoting role in the IEC-18 and IMCE immortalized intestinal cell line but a growth inhibitory role in several colon cancer cell lines [79]. In a recent study by Guo et al. [30], KLF5 was identified as a TGFβ cofactor, and when TGFβ was present, KLF5 plays an inhibitory role in cell proliferation. The inhibitory effect of KLF5 on the proliferation of cancer cells also appears to be TGFβ-dependent [30]. Based on these observations, KLF5 regulates cell proliferation in a context-dependent manner.
Apoptosis
KLF5 has an anti-apoptosis function. In leukemia, Zhu et al. [45] found that KLF5 induces the expression of survivin, a survival factor making cells resistant to apoptosis. Downregulation of KLF5 by siRNA decreases the expression of survivin and thus makes cells prone to doxorubicin-induced apoptosis. Mechanistically, KLF5 binds to the promoter of survivin gene and interacts with p53 to abrogate p53-repressed survivin expression [45]. KLF5 can also modulate apoptosis in a p53-independent manner [80]. When KLF5 is knocked down by RNAi, cells become more sensitive to 5-fluorouracil induced apoptosis, regardless of p53 status. KLF5 siRNA-induced apoptosis is associated with reduced BAD phosphorylation and with downregulation of PIM1, and transfection of wild-type (WT) Pim1 is sufficient to rescue the phenotype [80]. KLF5 can also make SMCs resistant to apoptosis. Suzuki et al. found that KLF5 confers apoptotic resistance in vascular lesions through interacting with PARP-1, a nuclear enzyme important in DNA repair and apoptosis. Acetylation of KLF5 under apoptotic conditions increases the interaction, and the acetylation-deficient mutant of KLF5 loses the capability to inhibit apoptosis [48]. Our recent results suggest that KLF5 can promote breast epithelial cell survival by increasing the MKP-1 protein levels [81].
Migration
KLF5 promotes the migration of epithelial cells, which is an important process in normal epithelial homeostasis in the gut and skin [82]. The mechanism involves the induction of integrin-linked kinase (ILK), which in turn activates Cdc42 and myosin light chain to regulate cell migration and motility [82]. When KLF5 is stably expressed in TSU-Pr1 [40], cell migration is also increased, as shown by a scratch assay (unpublished data).
Differentiation
Based on a large number of published papers, it is becoming evident that KLF5 regulates the differentiation of epithelial cells, SMCs, and adipocytes.
KLF5 plays an essential role in epithelial differentiation and homeostasis. During epithelial homeostasis, stem cells divide to produce progenitor cells, which further proliferate to generate the cell mass for mature epithelia [83]. KLF5 is highly expressed in proliferating epithelial cells such as immortal but untransformed epithelial cell lines and proliferating primary cultures of epithelial cells, which mostly represent progenitor cells [74, 78, 79, 84]. In normal intestine, KLF5 is expressed at a higher level in basal rapidly proliferating cells, but at a lower level in mature and differentiated cells [59], and knockout of one KLF5 allele significantly reduced the size of villi in mouse intestine [59]. In another in vivo study, overexpression of KLF5 in epidermis caused hyperplasia of basal cells but lack of mature skin [85]. In the HaCaT epidermal epithelial cell line treated with TGFβ, which represents a model of TGFβ-induced epithelial differentiation, KLF5 plays an essential role [30].
Although KLF5 is associated with the proliferative phenotype of SMCs, it is induced in activated SMCs to modulate the state of differentiation in response to injury and contribute to vascular regeneration [86–88]. KLF5 upregulates the expression of genes involved in the SMC differentiation, including SMemb/NMHC-B [19] and SM22α [63].
KLF5 is also necessary for adipocyte cell differentiation. In 3T3-L1 pre-adipocyte cells, which differentiate in the presence of micro-molar arsenic, arsenite treatment induces the expression of KLF5 [89]. A role of KLF5 in adipose differentiation was further demonstrated in a mouse model, where neonatal heterozygous KLF5 knockout mice exhibit a marked deficiency in white adipose tissue development [46]. In 3T3-L1 preadipocytes, KLF5 expression is induced at an early stage of differentiation, which is followed by the expression of PPARγ2. Constitutive overexpression of dominant-negative KLF5 inhibits adipocyte differentiation, whereas overexpression of WT KLF5 induces differentiation even without hormonal stimulation. Embryonic fibroblasts from KLF5 (±) mice also have an attenuated adipocyte differentiation [46]. Mechanistically, it appears that KLF5 expression is induced by C/EBPβ and δ, and, in turn, KLF5 acts in concert with C/EBPβ/δ to activate the PPARγ2 expression [46].
Stemness
Accumulated evidence suggests that KLF5 may be essential for self-renewal of embryonic stem cells (ESCs). Jiang et al. [90] reported that Klf5, Klf4, and Klf2 redundantly maintain mouse ESC self-renewal and pluripotency through regulating the Nanog, Tcl1, Esrrb, and Fbxo15 expression, and Klf4 and Klf5 showed the same pattern of expression changes during the differentiation of embryonic stem cells [91]. Indeed, Parisi et al. [92] not only confirmed that Klf5 plays an important role in self-renewal of mouse ESCs but also found that Klf5 knockdown alone causes differentiation of ESCs. Furthermore, constitutive expression of Klf5 attenuates the differentiation of ESCs, likely through direct regulation of genes involved in stem cell renewal including Nanog and Oct3/4 [92]. Consistently, Ema et al. [93] demonstrated that homozygous disruption of Klf5 results in the failure of ESC and early embryonic lethality due to an implantation defect. Klf5 null ESCs show increased expression of several differentiation marker genes and frequent, spontaneous differentiation [93]. Conversely, overexpression of Klf5 in ESCs suppressed the expression of differentiation marker genes and maintained pluripotency in the absence of extrinsic factor LIF [93]. Recently a combination of four transcription factors including KLF4, Oct4, Sox2, and c-Myc was shown to reverse differentiated cells to a pluripotent state [94–96]. Klf5 can replace Klf4 to generate induced pluripotent stem (iPS) cells from differentiated MEF cells in spite of lower efficiency [97].
However, KLF5 appears to be expressed at a lower level in adult stem cells in vivo. A study comparing global gene expression patterns between isolated hair follicle stem cells and non-bulge basal keratinocytes showed that KLF5 is expressed at a lower level in stem cells [98]. Consistently, transgenic expression of Tcf3, a component of the Wnt signaling pathway, represses differentiation of epidermal cells by repressing the expression of KLF5 and other genes and inducing the expression of genes that are associated with an undifferentiated state and shared by embryonic and postnatal stem cells [99].
KLF5 mouse models
Mouse models have been the most definitive way to clarify the physiological role of a gene in development and diseases. Several Klf5 knockout and transgenic mouse models have been established for studying KLF5 function.
Klf5 knockout mice
Based on the studies of Klf5 knockout mouse models, Klf5 has been implicated in pathological and physiological processes including embryonic development, angiogenesis, adipose tissue development, cartilage degradation during skeletal development, energy metabolism, and lung morphogenesis.
Conventional Klf5 homozygous knockout mice died before embryonic day 8.5, indicating that Klf5 is essential for mouse embryonic development. The Klf5 heterozygous knockout mice show diminished levels of arterial-wall thickening, angiogenesis, cardiac hypertrophy, and interstitial fibrosis in response to external stress, suggesting a role of KLF5 in linking external stress and cardiovascular remodeling [59]. The heterozygous KLF5 knockout mice exhibit a marked deficiency in white adipose tissue development [46] and skeletal growth retardation in the perinatal period [100]. While infection of WT mice with bacteria increases the heights of colonic crypt and the expression of KLF5, the KLF5 (±) mice show an attenuated induction of hyperproliferative responses after bacteria infection [101].
Recently, Wan et al. reported a conditional Klf5 knockout mouse model [41]. In this study, Klf5 was found to be essential for lung morphogenesis and function [41]. When Klf5 is specifically knocked out in respiratory epithelial cells in the fetal lung, lung maturation is inhibited during the saccular stage of development, and phenotypic abnormalities occur in different components including the respiratory epithelium, the bronchiolar smooth muscle, and the pulmonary vasculature. Mice with both Klf5 alleles knocked out die of respiratory distress immediately after birth [41]. A set of genes are abnormally regulated by the knockout of KLF5 [41].
Klf5 transgenic (Tg) mice
Besides knockout mouse models, two tissue-specific Klf5 Tg mouse models have been developed [85, 102]. Klf5 has been found to regulate esophageal basal epithelial cell proliferation and stemness of epidermis.
Klf5 has been shown to promote basal epithelial cell proliferation but is not sufficient to produce tumors [102]. Using the ED-L2 promoter of the Epstein–Barr virus, Goldstein et al. expressed KLF5 throughout esophageal epithelia in mice, and examined the role of KLF5 in the proliferation of esophageal cells. While there was no evidence of esophageal dysplasia or cancer, staining for bromodeoxyuridine (BrdU) demonstrated increased proliferation in basal cells but not in suprabasal cells, and did not appear to affect the differentiation of esophageal epithelium [102].
KLF5 has a role in the homeostasis of epidermis and skin morphogenesis. Expression of KLF5 is at relatively higher levels in keratinocytes throughout the adult human epidermis. Expression is stronger in the matrix and the inner root sheath cuticle layer of the hair follicle, sebaceous glands, and sweat glands [44]. Using the keratin 5 promoter, Sur et al. [85] specifically expressed KLF5 in the basal layer of the epidermis, and demonstrated that KLF5 affects epidermal development and disrupts epithelial–mesenchymal interactions necessary for skin adnexae formation as well as craniofacial morphogenesis during embryogenesis. The transgenic mice exhibit exencephaly, craniofacial defects, persistent abdominal herniation, and ectodermal dysplasia. In addition, the epidermis is hypoplastic and undergoes abnormal differentiation with expression of keratin 8, a marker for single-layered epithelia, in the stratified epidermis. Overexpression of KLF5 in adult mice leads to hyperkeratosis, follicle occlusion, and epidermal erosions. Furthermore, a decrease in stem cell population of bulge keratinocytes has been noticed, as characterized by the expression pattern of integrin α6 and CD34 markers.
Taken together, these in vivo studies using the knockout and transgenic techniques support an important role of KLF5 in embryonic and tissue development through regulating cell proliferation, differentiation, and stemness.
KLF5 and diseases
Cancer
Expression of KLF5 was found to be abnormal in many cancer types. Functional studies also support that KLF5 is an important cancer-related gene (Table 3). Based on its positive role in cell proliferation and survival, KLF5 has been suggested to be oncogene. However, some genetic, expression, and functional studies imply that KLF5 could be a tumor suppressor under some scenarios. It is attempting to speculate that KLF5 has context-dependent functions in carcinogenesis.
Table 3.
The role of KLF5 in different types of cancer
| Cancers | Genetic and expression | Function | References |
|---|---|---|---|
| Intestine and colon | mRNA expression is decreased in APCmin mouse adenomas and familial adenomatous polyposis | Promotes cell proliferation | [51, 73, 74, 79, 101] |
| Breast | Gene copy number loss. mRNA is low in ER + cancer cell lines, high in ER-tumors | Promotes MCF7 xenograft growth | [36, 84, 110] |
| Prostate | Gene copy number loss. mRNA is lost in some cancer cell lines | Inhibits DU145 and 22Rv1 colony formation | [78, 111] |
| Overexpression in some prostate cancers | |||
| Bladder | mRNA is upregulated and downregulated in cancer cell lines but not in tumor samples | Promotes TSU-Pr1 xenograft growth | [20, 40] |
| Leukemia | mRNA is upregulated and downregulated in cancer cell lines and acute lymphoblastic leukemia by Northern blot | Promotes EU-4 resistance to doxorubicin | [45] |
| Esophageal | mRNA level is high in stem-like cancer cells by qRT-PCR | Inhibits cell proliferation, survival, and invasion. Promotes basal cell proliferation in KLF5 Tg mice | [77, 102, 112] |
| Salivary gland | Gene copy number gain by CGH | [108] | |
| Gastric cancer | Protein expression is high in early-staged, lymph node metastasis negative, and small sized tumors by IHC | [109] | |
| Nasopharyngeal | mRNA is down-regulated by microarray | [138]. | |
| Melanoma | mRNA is down-regulated in two Ras mutated cancer cell lines by microarray | [139] |
The KLF5 gene undergoes frequent genetic alteration in different cancer types including prostate, breast, and salivary gland tumors. The KLF5 gene is rarely mutated in a large number of prostate and breast cancer cell lines [78, 84]. The KLF5 gene locus (13q21) is the second most frequent deletion in different types of human cancers according to a large number of comparative genomic hybridization (CGH) studies [103]. The deletion of 13q21 associates with metastases and higher tumor grade in prostate cancer [78, 104–107]. Analysis of human tumors demonstrated that KLF5 centers the deletion at 13q21 in prostate cancer. The majority of KLF5 deletions in prostate cancer are hemizygous deletion equivalent to haploinsufficiency [59], which inactivates KLF5 by reducing KLF5 expression. Consistently, KLF5 is excessively degraded in human cancer cell lines by overexpressed WWP1 E3 ligase [31–33]. However, a chromosomal region at 13q spanning the KLF5 locus appears to be amplified in some salivary gland tumors, as detected by CGH [108]. Thus, while deletion at KLF5 is frequent, it can also be amplified in human cancer.
Consistent with the genetic alterations, frequent expression aberrations for KLF5 have been well documented. Chen et al. [78, 84] found that the expression of KLF5 mRNA is frequently reduced or absent in many cell lines from both breast cancer and prostate cancer compared to immortalized cell lines. Similarly, in intestinal tumors and adenomatous polyposis adenomas, reduced expression of KLF5 mRNA occurs [79]. Even in Ras-mediated transformation of IEC-18 and IMCE cells, KLF5 is markedly downregulated [79]. In another study, analysis of 247 gastric carcinomas by immunohistochemical staining showed that KLF5 is expressed in 46% (113/247) of tumor tissues, and expression of KLF5 is more frequently detected in early stage tumors than in late stage tumors (63 vs. 38%), in tumors without lymph node metastasis (54 vs. 40%), and in tumors smaller than 5 cm in size (53 vs. 38%) [109]. In contrast, Tong et al. [110] examined the expression of KLF5 mRNA by qPCR in breast cancer, and found a significant correlation between increased KLF5 expression and reduced disease-free and overall survival in patients with breast cancer. KLF5 expression also appears to positively correlate with HER-2 and MKI67 markers of breast cancer and negatively correlate with patient age at diagnosis [110]. In human prostate cancer tissues, Chaib et al. [111] reported that KLF5 mRNA is upregulated in prostate cancers compared to normal tissues. Stem-like esophageal cancer cells, as defined by the side population, have a higher level of KLF5 expression than other cells [112]. More independent studies in primary tumors with different approaches are required to clarify these inconsistent results. Nevertheless, the expression of KLF5 in tumors appears to be altered during tumor development.
Functional studies in vitro have shown inconsistent results in different cell lines. For example, in the TSU-Pr1 bladder cancer cell line, expression of KLF5 promotes cell proliferation [40]. Consistently, KLF5 promotes survival and drug resistance in the HCT116 colon cancer cell line [80]. Furthermore, knockdown of KLF5 could inhibit the multicellular tumor spheroid formation in vitro [113]. In contrast, KLF5 has shown a growth inhibitory function in vitro in some cancer cell lines tested, including those from the colon [79] and prostate [78]. In the TE2 esophageal cancer cell line, which was derived from a poorly differentiated esophageal squamous carcinoma, stable expression of KLF5 inhibits cell proliferation and invasion, and decreases cell survival [77].
Several in vivo studies suggest that KLF5 promotes tumorigenesis. Chen et al. [40] directly demonstrated that expression of KLF5 promotes TSU-Pr1 bladder cancer cell line tumorigenesis in SCID mice. Similarly, expression of KLF5 in the MCF7 breast cancer cell line also promotes xenograft growth in nude mice in the presence of estrogen (unpublished results). Shindo et al. showed that the angiogenesis of transplanted sarcoma 180 is attenuated in the Klf5 (±) mice [59]. Similarly, the crypt cell proliferation in the colon in response to pathogenic bacterial infection is also reduced in the Klf5 (±) animals [101]. Consistently, Klf5 promotes esophageal basal epithelial cell proliferation although it is not sufficient to produce tumors [102]. Based on these in vivo research results, KLF5 could have context-dependent functions in oncogenesis.
Based on the recent discovery of KLF5 as an essential cofactor for TGFβ in epithelial cells [30], it is likely that the role of KLF5 in tumorigenesis is similar to that of TGFβ signaling, being a tumor suppressor in early stage but a tumor promoter in late stage tumorigenesis [114–116]. We are currently addressing this outstanding question using knockout mouse models.
Cardiovascular diseases
KLF5 has been best studied in the physiology and pathology of VSMCs. Based on a large number of expression and functional analyses, KLF5 contributes to all VSMC-related diseases, such as atherosclerosis, restenosis after angioplasty, cardiac hypertrophy, and hypertension, by regulating the expression of genes including SMemb/NMHC-B, SM22α, Egr-1, PDGF, and others (Table 1).
KLF5 is preferentially expressed in proliferating SMCs but reduced in differentiated cells under physiological and pathological conditions. It has been reported that KLF5 is abundantly expressed in embryonic smooth muscles and is downregulated with vascular development [117, 118]. Consistently, KLF5 is abundantly expressed in fetal but not in adult aortic SMCs of humans and rabbits [119]. The expression of KLF5 is increased in the neointimal smooth muscles in response to vascular injury [118]. In atherectomy specimens from primary and restenotic lesions, predominant expression of KLF5 was detected in SMCs. In addition, restenotic lesions expressing higher levels of KLF5 show higher incidence of restenosis than lesions without [119]. In cultured SMCs from atherectomy specimens obtained from patients with coronary restenosis after angioplasty, Sakamoto et al. [120] also found a correlation between KLF5 expression and SMC activity in outgrowth.
KLF5 also plays a role in the development of cardiac allograft vascular disease. In rats, Ogata et al. performed heterotopic cardiac transplantation, and examined KLF5 expression by IHC in all grafts. From 4 to 8 weeks after transplantation, SMCs showed positive staining for KLF5 in diffusely thickened coronary arteries and the perivascular space, and the level of KLF5 expression was significantly higher in allografts compared to isografts [121]. Using a heterotopic abdominal heart transplant model in monkeys, Wada et al. [122] showed that KLF5 and Egr-1 are induced in VSMCs of rejected cardiac allografts well before morphologic changes, such as intimal thickening, can be detected, suggesting that expression of KLF5 is one of the initial events in allograft angiopathy.
Additional studies provide direct evidence for the role of KLF5 in the modulation of cardiac hypertrophy and hypertension. Angiotensin II plays a critical role in cardiovascular remodeling through KLF5, as knockout of KLF5 in mice attenuates angiotensin II-induced cardiac hypertrophy and fibrosis [59]. Knockout of KLF5 also diminishes levels of smooth muscle and adventitial cell activation [59]. In a spontaneous hypertensive rat model, VSMCs change to the synthetic phenotype, which involves the complement 3 (C3) gene. Yao et al. [123] found that C3 increases the transcription of KLF5 in SMCs.
Other diseases
KLF5 may play a role in inflammatory diseases. The proinflammatory factors TNFα and LPS induce KLF5 expression in human umbilical vein endothelial cells, venous SMCs, and intestinal epithelial cells [60, 124]. The expression induction of monocyte chemoattractant protein-1 (MCP-1) by TNFα depends on KLF5, as knockdown of KLF5 by RNAi inhibits the effect of TNFα on MCP-1 expression [124]. Similarly, knockdown of KLF5 by RNAi reduced the expression of p50 and p65 subunits of NF-κB and its downstream target genes, TNFα and IL-6, in response to LPS [56]. Finally, KLF5 has been shown to directly interact with NF-κB in epidermal epithelial cells [44].
KLF5 may also have a role in obesity. Recently, a role of KLF5 in energy metabolism has been established. Oishi et al. [38] found that mice with one Klf5 allele knocked out are resistant to high fat-induced obesity despite consuming more food than WT mice. This effect appears to be mediated by the function of KLF5 in gene regulation, as expression of genes involved in lipid oxidation and energy uncoupling is upregulated in Klf5 (±) mice.
Involvement of KLF5 in additional pathological processes has also been reported. During intestinal obstruction, which causes dramatic phenotypic changes in intestine smooth muscles, expression of KLF5 initially increases but then decreases [125]. KLF5 is among the genes downregulated in human tissues from ulcerative colitis, an inflammatory bowel disease [126]. In addition, KLF5 could be involved in the pathophysiology of schizophrenia [23].
Conclusions
In summary, KLF5 is an essential transcription factor that regulates the transcription of a large number of genes in different contexts. The transcription of KLF5 itself is also regulated by different signaling pathways, including MAPK, PKC, and PI3 K, generally with an increased expression during cell proliferation and a decreased expression during differentiation, although it is also necessary for signaling-induced differentiation. Different protein modifications including phosphorylation, acetylation, ubiquitination, and sumoylation, which could be mediated by different signaling pathways, occur on KLF5 protein and regulate its transactivation activities and expression levels. Biologically, KLF5 appears to be essential not only for the proliferation of different types of cells including stem and progenitor cells but also for the differentiation of progenitor cells, which likely underlies the function of KLF5 in such physiologic and pathologic processes as epithelial homeostasis, development, tumorigenesis, cardiovascular remodeling, inflammation, and apoptosis. The expression of KLF5 is frequently altered in human diseases, including cancer and cardiovascular diseases.
Perspectives
Although KLF5 is a key transcription factor involved in multiple signaling cascades, whether the KLF5 pathway can be developed as a diagnostic tool and a therapeutic target remains to be elucidated. It is important to further understand KLF5 regulation and mechanisms of action under physiological and pathological conditions, to develop reagents and methods to accurately and conveniently measure KLF5 expression and activity changes in pathological specimens, and to develop treatments targeting the KLF5 pathway.
Although our biochemical and cellular knowledge of KLF5 has rapidly increased in the last decade as reviewed above, the roles of KLF5 in normal tissue development and disease progress are still far from clear. The ubiquitous expression of KLF5 in different types of cells and tissues, the diversified posttranslational modifications and interacting proteins, the numerous context-dependent target genes, and redundancy among the KLF5 family members contribute to the complexity of KLF5 study. It is important to develop inducible and tissue-specific animal models of KLF5 expression for human diseases because KLF5 is temporally and spatially regulated under physiological and pathological conditions.
KLF5 has potential as a biomarker for diagnosis and prognosis in cancers and cardiovascular diseases. The expression of KLF5 mRNA associates with reduced survival in patients with breast cancer [110]. KLF5 expression also appears to correlate positively with a higher incidence of restenosis [119]. Given that IHC is widely used in the clinic for diagnosis and prognosis, it is necessary to establish the standard IHC techniques for KLF5 to evaluate the potential of KLF5 as useful clinical biomarker or as a potential stand-alone prognostic factor. So far, KLF5 protein expression has only been analyzed by IHC in gastric cancer [109].
Although multiple anti-KLF5 antibodies are commercially available, the specificity appears to be unsatisfactory. Future studies focusing on accurately measuring KLF5 expression and activity changes in a large number of pathological specimens will be necessary.
Transcription factors are usually not ideal for targeted therapy using small molecular inhibitors. However, KLF5 directly interacts with RARα to regulate downstream target genes in cardiovascular remodeling [127]. The synthetic RARα agonist Am80 was shown to inhibit the activity of KLF5 [43, 118]. Acyclic retinoid (ACR) also attenuates the functional interaction of KLF5 and RARα [127]. These small molecules have the potential to be developed as drugs for cardiovascular diseases. With the development of siRNA delivery technology, anti-KLF5 siRNAs could also be developed into drugs. Additionally, several RTK and PI3 K small molecular inhibitors (AG17, LY294002, and Wortmannin) were identified as inhibiting KLF5 transcription and colon cancer cell proliferation by cell-based high throughput screening [73]. Once the KLF5 pathway is clearly understood, the positive upstream regulators (e.g., protein kinases) and downstream target proteins (e.g., FGF-BP) could be inhibited by small molecular inhibitors or monoclonal antibodies.
References
- 1.Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/S1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 2.Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/S0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 3.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/S1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 5.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 6.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 9.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, Bland KI, LoBuglio AF, Lobo-Ruppert SM, Ruppert JM. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.CCR-03-0484. [DOI] [PubMed] [Google Scholar]
- 10.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 11.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Hyytinen ER, Sun X, Helin HJ, Koivisto PA, Frierson HF, Vessella RL, Dong JT. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am J Pathol. 2003;162:1349–1354. doi: 10.1016/S0002-9440(10)63930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- 15.Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kojima S, Kobayashi A, Gotoh O, Ohkuma Y, Fujii-Kuriyama Y, Sogawa K. Transcriptional activation domain of human BTEB2, a GC box-binding factor. J Biochem (Tokyo) 1997;121:389–396. doi: 10.1093/oxfordjournals.jbchem.a021600. [DOI] [PubMed] [Google Scholar]
- 17.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27:4807–4815. doi: 10.1093/nar/27.24.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe N, Kurabayashi M, Shimomura Y, Kawai-Kowase K, Hoshino Y, Manabe I, Watanabe M, Aikawa M, Kuro-o M, Suzuki T, Yazaki Y, Nagai R. BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ Res. 1999;85:182–191. doi: 10.1161/01.res.85.2.182. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256. doi: 10.1016/S0006-291X(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 21.Chiambaretta F, De Graeve F, Turet G, Marceau G, Gain P, Dastugue B, Rigal D, Sapin V. Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol Vis. 2004;10:901–909. [PubMed] [Google Scholar]
- 22.Yang XO, Doty RT, Hicks JS, Willerford DM. Regulation of T-cell receptor D beta 1 promoter by KLF5 through reiterated GC-rich motifs. Blood. 2003;101:4492–4499. doi: 10.1182/blood-2002-08-2579. [DOI] [PubMed] [Google Scholar]
- 23.Yanagi M, Hashimoto T, Kitamura N, Fukutake M, Komure O, Nishiguchi N, Kawamata T, Maeda K, Shirakawa O. Expression of Kruppel-like factor 5 gene in human brain and association of the gene with the susceptibility to schizophrenia. Schizophr Res. 2008;100:291–301. doi: 10.1016/j.schres.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi S, Laub F, Matsumoto N, Asaka M, Ramirez F, Yoshida T, Terada M. Developmental expression of the mouse gene coding for the Kruppel-like transcription factor KLF5. Dev Dyn. 2000;217:421–429. doi: 10.1002/(SICI)1097-0177(200004)217:4<421::AID-DVDY9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Moore-Scott BA, Opoka R, Lin SC, Kordich JJ, Wells JM. Identification of molecular markers that are expressed in discrete anterior-posterior domains of the endoderm from the gastrula stage to mid-gestation. Dev Dyn. 2007;236:1997–2003. doi: 10.1002/dvdy.21204. [DOI] [PubMed] [Google Scholar]
- 26.Du JX, Bialkowska AB, McConnell BB, Yang VW. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J Biol Chem. 2008;283:31991–32002. doi: 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196–2208. doi: 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol. 2003;23:8528–8541. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumura T, Suzuki T, Aizawa K, Munemasa Y, Muto S, Horikoshi M, Nagai R. The deacetylase HDAC1 negatively regulates the cardiovascular transcription factor Kruppel-like factor 5 through direct interaction. J Biol Chem. 2005;280:12123–12129. doi: 10.1074/jbc.M410578200. [DOI] [PubMed] [Google Scholar]
- 30.Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J Biol Chem. 2009;284:6071–6078. doi: 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, Zhou Z, Petros J, Frierson HF, Vessella RL, Atfi A, Dong JT. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26:2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121:80–87. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Zhou Z, Sheehan CE, Slodkowska E, Sheehan CB, Boguniewicz A, Ross JS. Overexpression of WWP1 is associated with the estrogen receptor and insulin-like growth factor receptor 1 in breast carcinoma. Int J Cancer. 2009;124:2829–2836. doi: 10.1002/ijc.24266. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Zhou Z, Guo P, Dong JT. Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Lett. 2007;581:1124–1130. doi: 10.1016/j.febslet.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 38.Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat Med. 2008;14:656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 39.Du JX, Yun CC, Bialkowska A, Yang VW. Protein inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor Kruppel-like factor 5. J Biol Chem. 2007;282:4782–4793. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, Zhou Z, Cheng X, Simons JW, Dong JT. KLF5 promotes cell proliferation and tumorigenesis through gene regulation in the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 41.Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munemasa Y, Suzuki T, Aizawa K, Miyamoto S, Imai Y, Matsumura T, Horikoshi M, Nagai R. Promoter region-specific histone incorporation by the novel histone chaperone ANP32B and DNA-binding factor KLF5. Mol Cell Biol. 2008;28:1171–1181. doi: 10.1128/MCB.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujiu K, Manabe I, Ishihara A, Oishi Y, Iwata H, Nishimura G, Shindo T, Maemura K, Kagechika H, Shudo K, Nagai R. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting KLF5. Circ Res. 2005;97:1132–1141. doi: 10.1161/01.RES.0000190613.22565.13. [DOI] [PubMed] [Google Scholar]
- 44.Sur I, Unden AB, Toftgard R. Human Kruppel-like factor5/KLF5: synergy with NF-kappaB/Rel factors and expression in human skin and hair follicles. Eur J Cell Biol. 2002;81:323–334. doi: 10.1078/0171-9335-00257. [DOI] [PubMed] [Google Scholar]
- 45.Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 interacts with P53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711–14718. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- 46.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Lee MY, Moon JS, Park SW, Koh YK, Ahn YH, Kim KS. KLF5 enhances SREBP-1 action in androgen-dependent induction of fatty acid synthase in prostate cancer cells. Biochem J. 2009;417:313–322. doi: 10.1042/BJ20080762. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T, Nishi T, Nagino T, Sasaki K, Aizawa K, Kada N, Sawaki D, Munemasa Y, Matsumura T, Muto S, Sata M, Miyagawa K, Horikoshi M, Nagai R. Functional interaction between the transcription factor Kruppel-like factor 5 and poly(ADP-ribose) polymerase-1 in cardiovascular apoptosis. J Biol Chem. 2007;282:9895–9901. doi: 10.1074/jbc.M608098200. [DOI] [PubMed] [Google Scholar]
- 49.Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 51.Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckers J, Herrmann F, Rieger S, Drobyshev AL, Horsch M, Hrabe de Angelis M, Seliger B. Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114:590–597. doi: 10.1002/ijc.20798. [DOI] [PubMed] [Google Scholar]
- 53.Sun R, Chen X, Yang VW. Intestinal-enriched kruppel-like factor (kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Zhou Y, Zhou Z, Sun X, Otto KB, Uht RM, Dong JT. Regulation of KLF5 involves the Sp1 transcription factor in human epithelial cells. Gene. 2004;330:133–142. doi: 10.1016/j.gene.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 55.Usui S, Sugimoto N, Takuwa N, Sakagami S, Takata S, Kaneko S, Takuwa Y. Blood lipid mediator sphingosine 1-phosphate potently stimulates platelet-derived growth factor-A and -B chain expression through S1P1-Gi-Ras-MAPK-dependent induction of Kruppel-like factor 5. J Biol Chem. 2004;279:12300–12311. doi: 10.1074/jbc.M305025200. [DOI] [PubMed] [Google Scholar]
- 56.Chanchevalap S, Nandan MO, McConnell BB, Charrier L, Merlin D, Katz JP, Yang VW. Kruppel-like factor 5 is an important mediator for lipopolysaccharide-induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 2006;34:1216–1223. doi: 10.1093/nar/gkl014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taneyhill L, Pennica D. Identification of Wnt responsive genes using a murine mammary epithelial cell line model system. BMC Dev Biol. 2004;4:6. doi: 10.1186/1471-213X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziemer LT, Pennica D, Levine AJ. Identification of a mouse homolog of the human BTEB2 transcription factor as a beta-catenin-independent Wnt-1-responsive gene. Mol Cell Biol. 2001;21:562–574. doi: 10.1128/MCB.21.2.562-574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 60.Bafford R, Sui XX, Wang G, Conte M. Angiotensin II and tumor necrosis factor-alpha upregulate survivin and Kruppel-like factor 5 in smooth muscle cells: Potential relevance to vein graft hyperplasia. Surgery. 2006;140:289–296. doi: 10.1016/j.surg.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 61.Gao D, Niu X, Ning N, Hao G. Regulation of angiotensin II-Induced Kruppel-like factor 5 expression in vascular smooth muscle cells. Biol Pharm Bull. 2006;29:2004–2008. doi: 10.1248/bpb.29.2004. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, Bialkowska A, Rusovici R, Chanchevalap S, Shim H, Katz JP, Yang VW, Yun CC. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Kruppel-like factor 5. J Biol Chem. 2007;282:15541–15549. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Sinha S, Owens G. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem. 2003;278:48004–48011. doi: 10.1074/jbc.M301902200. [DOI] [PubMed] [Google Scholar]
- 65.Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–16498. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- 66.Cullingford TE, Butler MJ, Marshall AK, Tham EL, Sugden PH, Clerk A. Differential regulation of Kruppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: Effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta. 2008;1783:1229–1236. doi: 10.1016/j.bbamcr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitko K, Ulbrich SE, Wenigerkind H, Sinowatz F, Blum H, Wolf E, Bauersachs S. Dynamic changes in messenger RNA profiles of bovine endometrium during the oestrous cycle. Reproduction. 2008;135:225–240. doi: 10.1530/REP-07-0415. [DOI] [PubMed] [Google Scholar]
- 68.Ohara F, Nii A, Sakiyama Y, Tsuchiya M, Ogawa S. Pathophysiological characteristics of dimethylnitrosamine-induced liver fibrosis in acute and chronic injury models: a possible contribution of KLF5 to fibrogenic responses. Dig Dis Sci. 2008;53:2222–2232. doi: 10.1007/s10620-007-0112-y. [DOI] [PubMed] [Google Scholar]
- 69.Kruse JJ, te Poele JA, Russell NS, Boersma LJ, Stewart FA. Microarray analysis to identify molecular mechanisms of radiation-induced microvascular damage in normal tissues. Int J Radiat Oncol Biol Phys. 2004;58:420–426. doi: 10.1016/j.ijrobp.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 70.Camerer E, Gjernes E, Wiiger M, Pringle S, Prydz H. Binding of factor VIIa to tissue factor on keratinocytes induces gene expression. J Biol Chem. 2000;275:6580–6585. doi: 10.1074/jbc.275.9.6580. [DOI] [PubMed] [Google Scholar]
- 71.Chiou CC, Chan CC, Sheu DL, Chen KT, Li YS, Chan EC. Helicobacter pylori infection induced alteration of gene expression in human gastric cells. Gut. 2001;48:598–604. doi: 10.1136/gut.48.5.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mori D, Okuro N, Fujii-Kuriyama Y, Sogawa K. Gene structure and promoter analysis of the rat BTEB2 gene. Gene. 2003;304:163–170. doi: 10.1016/S0378-1119(02)01203-9. [DOI] [PubMed] [Google Scholar]
- 73.Bialkowska AB, Du Y, Fu H, Yang VW. Identification of novel small-molecule compounds that inhibit the proproliferative Kruppel-like factor 5 in colorectal cancer cells by high-throughput screening. Mol Cancer Ther. 2009;8:563–570. doi: 10.1158/1535-7163.MCT-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chanchevalap S, Nandan MO, Merlin D, Yang VW. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Kruppel-like factor 5. FEBS Lett. 2004;578:99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 77.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 78.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 79.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Y, Hamza MS, Leong HS, Lim CB, Pan YF, Cheung E, Soo KC, Iyer NG. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1–8. doi: 10.1038/sj.onc.1210625. [DOI] [PubMed] [Google Scholar]
- 81.Liu R, Zheng HQ, Zhou Z, Dong JT, Chen C (2009) KLF5 promotes breast cell survival partially through FGF-BPpERK-mediated MKP-1 protein phosphorylation and stabilization. J Biol Chem In press [DOI] [PMC free article] [PubMed]
- 82.Yang Y, Tetreault MP, Yermolina YA, Goldstein BG, Katz JP. Kruppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem. 2008;283:18812–18820. doi: 10.1074/jbc.M801384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 85.Sur I, Rozell B, Jaks V, Bergstrom A, Toftgard R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. 2006;119:3593–3601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- 86.Walsh K, Takahashi A. Transcriptional regulation of vascular smooth muscle cell phenotype. Z Kardiol. 2001;90(Suppl 3):12–16. doi: 10.1007/s003920170033. [DOI] [PubMed] [Google Scholar]
- 87.Nagai R, Shindo T, Manabe I, Suzuki T, Kurabayashi M. KLF5/BTEB2, a Kruppel-like zinc-finger type transcription factor, mediates both smooth muscle cell activation and cardiac hypertrophy. Adv Exp Med Biol. 2003;538:57–65. doi: 10.1007/978-1-4419-9029-7_5. [DOI] [PubMed] [Google Scholar]
- 88.Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 89.Salazard B, Bellon L, Jean S, Maraninchi M, El-Yazidi C, Orsiere T, Margotat A, Botta A, Berge-Lefranc JL. Low-level arsenite activates the transcription of genes involved in adipose differentiation. Cell Biol Toxicol. 2004;20:375–385. doi: 10.1007/s10565-004-1471-1. [DOI] [PubMed] [Google Scholar]
- 90.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 91.Bruce SJ, Gardiner BB, Burke LJ, Gongora MM, Grimmond SM, Perkins AC. Dynamic transcription programs during ES cell differentiation towards mesoderm in serum versus serum-freeBMP4 culture. BMC Genomics. 2007;8:365. doi: 10.1186/1471-2164-8-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 93.Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, Shimosato D, Uchida K, Suzuki N, Yanagisawa J, Sogawa K, Rossant J, Yamamoto M, Takahashi S, Fujii-Kuriyama Y. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 95.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 96.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 97.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 98.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 99.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 100.Shinoda Y, Ogata N, Higashikawa A, Manabe I, Shindo T, Yamada T, Kugimiya F, Ikeda T, Kawamura N, Kawasaki Y, Tsushima K, Takeda N, Nagai R, Hoshi K, Nakamura K, Chung UI, Kawaguchi H. Kruppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J Biol Chem. 2008;283:24682–24689. doi: 10.1074/jbc.M709857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McConnell BB, Klapproth JM, Sasaki M, Nandan MO, Yang VW. Kruppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology. 2008;134:1007–1016. doi: 10.1053/j.gastro.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1784–G1792. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 103.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong JT, Boyd JC, Frierson HF., Jr Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate. 2001;49:166–171. doi: 10.1002/pros.1131. [DOI] [PubMed] [Google Scholar]
- 105.Hyytinen ER, Frierson HF, Boyd JC, Chung LW, Dong JT. Three distinct regions of allelic loss at 13q14, 13q21–22, and 13q33 in prostate cancer. Genes Chromosomes Cancer. 1999;25:108–114. doi: 10.1002/(SICI)1098-2264(199906)25:2<108::AID-GCC6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 106.Dong JT, Chen C, Stultz BG, Isaacs JT, Frierson HF., Jr Deletion at 13q21 is associated with aggressive prostate cancers. Cancer Res. 2000;60:3880–3883. [PubMed] [Google Scholar]
- 107.Chen C, Brabham WW, Stultz BG, Frierson HF, Jr, Barrett JC, Sawyers CL, Isaacs JT, Dong JT. Defining a common region of deletion at 13q21 in human cancers. Genes Chromosomes Cancer. 2001;31:333–344. doi: 10.1002/gcc.1152. [DOI] [PubMed] [Google Scholar]
- 108.Giefing M, Wierzbicka M, Rydzanicz M, Cegla R, Kujawski M, Szyfter K. Chromosomal gains and losses indicate oncogene and tumor suppressor gene candidates in salivary gland tumors. Neoplasma. 2008;55:55–60. [PubMed] [Google Scholar]
- 109.Kwak MK, Lee HJ, Hur K, Park do J, Lee HS, Kim WH, Lee KU, Choe KJ, Guilford P, Yang HK. Expression of Kruppel-like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134:163–167. doi: 10.1007/s00432-007-0265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- 111.Chaib H, Cockrell EK, Rubin MA, Macoska JA. Profiling and verification of gene expression patterns in normal and malignant human prostate tissues by cDNA microarray analysis. Neoplasia (New York) 2001;3:43–52. doi: 10.1038/sj.neo.7900126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang D, Gao Q, Guo L, Zhang C, Wei J, Li H, Jing WJ, Han X, Shi Y, Shih Hsin L. Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev. 2008;18:465–473. doi: 10.1089/scd.2008.0033. [DOI] [PubMed] [Google Scholar]
- 113.Dardousis K, Voolstra C, Roengvoraphoj M, Sekandarzad A, Mesghenna S, Winkler J, Ko Y, Hescheler J, Sachinidis A. Identification of differentially expressed genes involved in the formation of multicellular tumor spheroids by HT-29 colon carcinoma cells. Mol Ther. 2007;15:94–102. doi: 10.1038/sj.mt.6300003. [DOI] [PubMed] [Google Scholar]
- 114.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 115.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFbeta Signaling Destabilizes Homeostasis and Promotes Squamous Cell Carcinomas in Stratified Epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nagai R, Suzuki T, Aizawa K, Miyamoto S, Amaki T, Kawai-Kowase K, Sekiguchi KI, Kurabayashi M. Phenotypic modulation of vascular smooth muscle cells: dissection of transcriptional regulatory mechanisms. Ann NY Acad Sci. 2001;947:56–66. doi: 10.1111/j.1749-6632.2001.tb03930.x. [DOI] [PubMed] [Google Scholar]
- 118.Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3:1569–1576. doi: 10.1111/j.1538-7836.2005.01366.x. [DOI] [PubMed] [Google Scholar]
- 119.Hoshino Y, Kurabayashi M, Kanda T, Hasegawa A, Sakamoto H, Okamoto E, Kowase K, Watanabe N, Manabe I, Suzuki T, Nakano A, Takase S, Wilcox JN, Nagai R. Regulated expression of the BTEB2 transcription factor in vascular smooth muscle cells: analysis of developmental and pathological expression profiles shows implications as a predictive factor for restenosis. Circulation. 2000;102:2528–2534. doi: 10.1161/01.cir.102.20.2528. [DOI] [PubMed] [Google Scholar]
- 120.Sakamoto H, Sakamaki T, Kanda T, Hoshino Y, Sawada Y, Sato M, Sato H, Oyama Y, Nakano A, Takase S, Hasegawa A, Nagai R, Kurabayashi M. Smooth muscle cell outgrowth from coronary atherectomy specimens in vitro is associated with less time to restenosis and expression of a key transcription factor KLF5/BTEB2. Cardiology. 2003;100:80–85. doi: 10.1159/000073043. [DOI] [PubMed] [Google Scholar]
- 121.Ogata T, Kurabayashi M, Hoshino YI, Sekiguchi KI, Kawai-Kowase K, Ishikawa S, Morishita Y, Nagai R. Inducible expression of basic transcription factor-binding protein 2 (BTEB2), a member of zinc finger family of transcription factors, in cardiac allograft vascular disease. Transplantation. 2000;70:1653–1656. doi: 10.1097/00007890-200012150-00019. [DOI] [PubMed] [Google Scholar]
- 122.Wada Y, Suzuki J, Kawauchi M, Kurabayashi M, Tsukioka K, Zhang T, Endoh M, Takayama K, Nagai R, Takamoto S, Isobe M, Amano J. Early growth-response factor 1 and basic transcriptional element-binding protein 2 expression in cardiac allografts. J Heart Lung Transplant. 2001;20:590–594. doi: 10.1016/S1053-2498(00)00210-2. [DOI] [PubMed] [Google Scholar]
- 123.Yao EH, Fukuda N, Ueno T, Tsunemi A, Endo M, Matsumoto K. Complement 3 activates the KLF5 gene in rat vascular smooth muscle cells. Biochem Biophys Res Commun. 2008;367:468–473. doi: 10.1016/j.bbrc.2007.12.160. [DOI] [PubMed] [Google Scholar]
- 124.Kumekawa M, Fukuda G, Shimizu S, Konno K, Odawara M. Inhibition of monocyte chemoattractant protein-1 by Kruppel-like factor 5 small interfering RNA in the tumor necrosis factor-alpha-activated human umbilical vein endothelial cells. Biol Pharm Bull. 2008;31:1609–1613. doi: 10.1248/bpb.31.1609. [DOI] [PubMed] [Google Scholar]
- 125.Chen J, Chen H, Sanders KM, Perrino BA. Regulation of SRF/CArG-dependent gene transcription during chronic partial obstruction of murine small intestine. Neurogastroenterol Motil. 2008;20:829–842. doi: 10.1111/j.1365-2982.2008.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 127.Kada N, Suzuki T, Aizawa K, Munemasa Y, Matsumura T, Sawaki D, Nagai R. Acyclic retinoid inhibits functional interaction of transcription factors Kruppel-like factor 5 and retinoic acid receptor-alpha. FEBS Lett. 2008;582:1755–1760. doi: 10.1016/j.febslet.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 128.Suzuki T, Sawaki D, Aizawa K, Munemasa Y, Matsumura T, Ishida J, Nagai R. Kruppel-like factor 5 shows proliferation-specific roles in vascular remodeling, direct stimulation of cell growth, and inhibition of apoptosis. J Biol Chem. 2009;284:9549–9557. doi: 10.1074/jbc.M806230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aizawa K, Suzuki T, Kada N, Ishihara A, Kawai-Kowase K, Matsumura T, Sasaki K, Munemasa Y, Manabe I, Kurabayashi M, Collins T, Nagai R. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J Biol Chem. 2004;279:70–76. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 130.Nagai R, Kowase K, Kurabayashi M. Transcriptional regulation of smooth muscle phenotypic modulation. Ann NY Acad Sci. 2000;902:214–222. doi: 10.1111/j.1749-6632.2000.tb06316.x. [DOI] [PubMed] [Google Scholar]
- 131.Teng C, Shi H, Yang N, Shigeta H. Mouse lactoferrin gene. Promoter-specific regulation by EGF and cDNA cloning of the EGF-response-element binding protein. Adv Exp Med Biol. 1998;443:65–78. [PubMed] [Google Scholar]
- 132.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 133.Wong WK, Chen K, Shih JC. Regulation of human monoamine oxidase B gene by Sp1 and Sp3. Mol Pharmacol. 2001;59:852–859. doi: 10.1124/mol.59.4.852. [DOI] [PubMed] [Google Scholar]
- 134.Shih JC, Chen K. Regulation of MAO-A and MAO-B gene expression. Curr Med Chem. 2004;11:1995–2005. doi: 10.2174/0929867043364757. [DOI] [PubMed] [Google Scholar]
- 135.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shao J, Yang VW, Sheng H. Prostaglandin E2 and Kruppel-like transcription factors synergistically induce the expression of decay-accelerating factor in intestinal epithelial cells. Immunology. 2008;125:397–407. doi: 10.1111/j.1365-2567.2008.02847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fang W, Li X, Jiang Q, Liu Z, Yang H, Wang S, Xie S, Liu Q, Liu T, Huang J, Xie W, Li Z, Zhao Y, Wang E, Marincola FM, Yao K. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J Transl Med. 2008;6:32. doi: 10.1186/1479-5876-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, Schadendorf D, Kumar R. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 140.Guo P, Zhao KW, Dong XY, Sun X, Dong J-T (2009) Acetylation of KLF5 alters the assembly of p15 transcription factors in TGFβ-mediated induction in epithelial cells. J Biol Chem (in press) [DOI] [PMC free article] [PubMed]