Abstract
Nuclear distribution gene C homolog (NudC) is a highly conserved gene. It has been identified in different species from fungi to mammals. The high degree of conservation, in special in the nudC domain, suggests that they are genes with essential functions. Most of the identified genes in the family have been implicated in cell division through the regulation of cytoplasmic dynein. As for mammalian genes, human NUDC has been implicated in the migration and proliferation of tumor cells and has therefore been considered a possible therapeutic target. There is evidence suggesting that mammalian NudC is also implicated in the regulation of the inflammatory response and in thrombopoiesis. The presence of these other functions not related to the interaction with molecular motors agrees with that these genes and their products are larger in size than their microbial orthologous, indicating that they have evolved to convey additional features.
Keywords: Dynein, Cytokinesis regulation, Nuclear migration, Inflammatory response, PAF
NudC
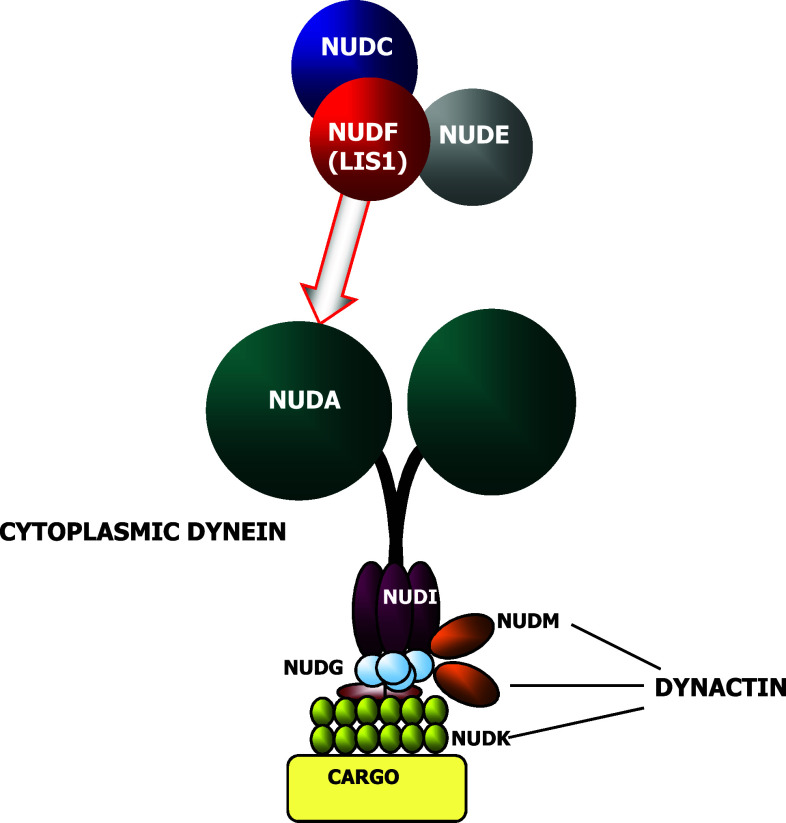
NudC was first identified as a regulator of nuclear movement in the asexual reproductive cycle of the filamentous fungus Emericella nidulans [1]. It is one of the heat-sensitive nud mutants discovered in E. nidulans which prevent nuclear migration into the mycelium; nud colonies are severely restricted for growth and differentiation. In E. nidulans more than 14 nud loci have been identified and nine genes have been cloned. The identification of these genes have revealed that they code for light, intermediate and heavy chains of dynein (an actin-associated molecular motor) [1] and ARP1 and p150 components of dynactin complex [2], which regulates dynein (Fig. 1) [3]. Deletions in nudC gene result in a more severe phenotype than other nuclear distribution mutants, profoundly affecting the morphology and composition of the cell wall and resulting in lethality [4]. Functional studies have revealed its essential role in the synthesis and organization of the cell wall [5]. NudC works through the molecular motor dynein and, together with other Nud genes, regulates dynein-mediated actions such as vesicle axonemal transport in neurons [6, 7], perinuclear localization of Golgi apparatus and lysosomes or endocytic vesicles transport [8], kinetochores localization and bipolar spindle organization [9], phagosome movement [10], nuclear transport [6] and membrane organelles organization [6].
Fig. 1.
Nud gene products and their relationship. NUD proteins that form part of the dynein and dynactin complexes are shown. Other NUD proteins (NUDC, NUDF and NUDE) which regulate cytoplasmic dynein functions but that do not form part of these complexes are also depicted
NudC studies have demonstrated that it is required to maintain the protein levels of NUDF [11]. Thus, some NudC genomic mutations affect negatively NUDF levels and its function while overexpression of NUDF reverts the phenotypic effects caused by NudC mutations [4]. Therefore, NUDC appears as a post-transcriptional regulator of NUDF. It has been suggested that NUDC/NUDF complex, together with another Nud gene product, NUDE [12], plays a role in the regulation of the minus-end molecular motor cytoplasmic dynein [13, 14].
NudC genes
This gene is highly conserved, both structurally and functionally, and several orthologous cDNAs have been cloned. The gene has been found in Caenorhabditis elegans [15], Drosophila melanogaster [16], Pleurodeles waltl [17], Rattus norvegicus [11], Mus musculus [18] or Homo sapiens [19]. All identified orthologous genes are included in Table 1.
Table 1.
Orthologous genes of NudC
| Organism | Similarity to hNUDC (% identity) | NCBI accessions |
|---|---|---|
| Chimpanzee (Pan troglodytes) | 98.59 | 737457 XM_001145962.1 |
| Dog (Canis familiaris) | 92.04 | 487349 XM_544475.2 |
| Rat (Rattus norvegicus) | 90.03 | 29648 NM_017271.1 |
| Mouse (Mus musculus) | 89.53 | 18221 NM_010948.1 |
| Alicante grape (Vitis vinifera) | 81.75 | CF415422.1 |
| Zebrafish (Danio rerio) | 76.25 | 393893 BC045909.1 |
| Chicken (Gallus gallus) | 75.77 | 419578 NM_001006311.1 |
| Tropical clawed frog (Silurana tropicalis) | 75.75 | CF592851.1 |
| Wheat (Triticum aestivum) | 73.72 | BG604268.1 |
| African clawed frog (Xenopus laevis) | 75.70 | CF549165.1 |
| Soybean (Glycine max) | 75.51 | 46752062 |
| Sea squirt (Ciona intestinalis) | 73.34 | BW260335.1 |
| Rice (Oryza sativa) | 73.00 | AK073934.1 |
| African malaria mosquito (Anopheles gambiae) | 63.14 | 1276787 XM_316177.2 |
| Rice blast fungus (Magnaporthe grisea) | 62.80 | 2676687 XM_361061.1 |
| Bread mold (Neurospora crassa) | 61.11 | 2713784 XM_331786.1 |
| Thale cress (Arabidopsis thaliana) | 54.09 | 835421 NM_124719.3 |
| Fission yeast (Schizosaccharomyces pombe) | 50.54 | 2540736 NP_596344.1 |
| Worm (Caenorhabditis elegans) | 49.00 | 1767541 NM_067348.4 |
| Fruit fly (Drosophila melanogaster) | 48.00 | 398791 NM_140666.2 |
All identified genes are included. In each case it is shown the percentage of identity in nucleotide sequences as compared to human NUDC and its NCBI accession number(s)
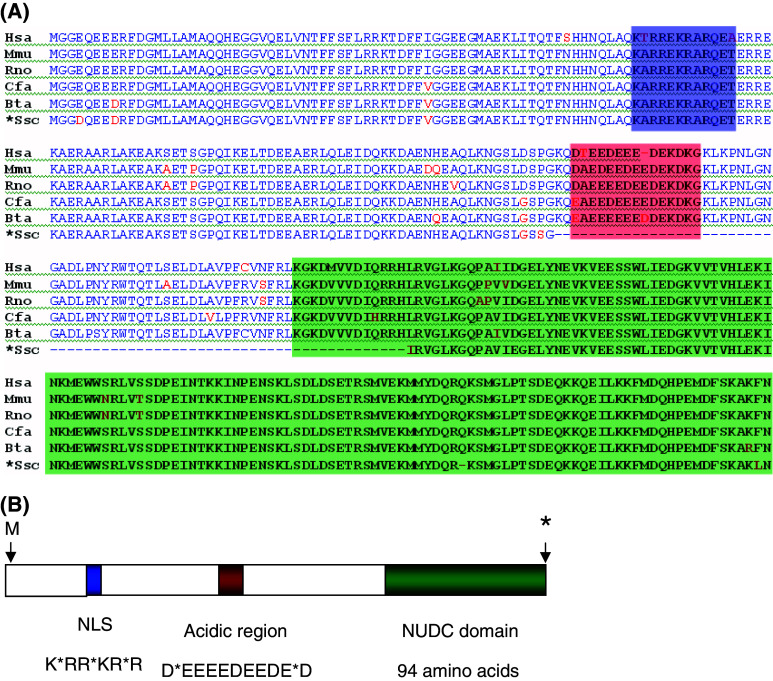
Mammalian NudC cDNAs have been isolated in different experimental conditions. Thus, rat NudC was identified during the determination of genes involved in T lymphocytes proliferation by prolactin [11, 20], mouse NudC (mNudC) was identified in a study about overexpressed genes in activated macrophages [21] and human NUDC (hNUDC) was isolated from lymphoma cells [22]. The isolated cDNAs show a high degree of similarity (around 90% identity). The similarity is maintained throughout all their sequences, which has facilitated the identification of three conserved motifs in their proteins. These motifs, as shown in Fig. 2 are the following: (1) The amino terminal region of mammalian NUDC proteins contains a basic stretch similar to the nuclear localization signal (NLS) found in proteins that translocate to the nucleus. The potential NLS sequence is very similar to that of the steroid hormone receptor NLS consensus sequence, RKWKR/K**R/K [23]. The functionality of this domain is suggested by the fact that NUDC colocalizes with the Golgi apparatus and close to the nucleus [24]. (2) The central region of NUDC contains an acidic region (D*EEEEDEEDE*D) where 11 out of 13 residues are negatively charged. This sequence has been found to be repeated several times in the nuclear protein Nopp 140. This protein is able to move through the nuclear membrane conveying other proteins into the nucleus [25]. This domain is included in the region responsible of the interaction of NUDC with the thrombopoietin receptor Mpl [26]. (3) The carboxy terminal 94 amino acids, comprising from amino acid 239 to amino acid 332 in human NUDC, shows high sequence similarity (about 60% identity) to E. nidulans NUDC domain. Interestingly, although the amino acid sequences in this carboxy terminal region are very similar in the mammalian and E. nidulans proteins, the DNA sequences show a considerable number of differences. This is due to the high number of silent mutations that the mammalian genes have accumulated. Thus, E. nidulans and murine proteins show 64 out of 94 identical amino acids in this carboxy terminal region [11]. However, about 50% of the codons corresponding to these amino acids contain silent mutations. These silent mutations are not only found in the third base position, but they are also found in the more unusual first and second base positions. Since the high similarity between the proteins is confined to the carboxy terminus, it is conceivable that this region of microbial nudC and their mammalian orthologous have been conserved through evolution for an important structural and/or functional role [22, 24].
Fig. 2.
Mammalian NUDC proteins. a Alignment of several NUDC amino acid sequences including human (Hsa), mouse (Mmu), rat (Rno), dog (Cfa), cow (Bta), and pig partial cDNA sequence (Ssc). The three conserved regions are highlighted in blue (NLS), red (acidic region) and green (NudC domain). b Schematic drawing of a mammalian NUDC protein showing the three regions highlighted in a. The consensus amino acid sequences of the NLS and acidic region are shown
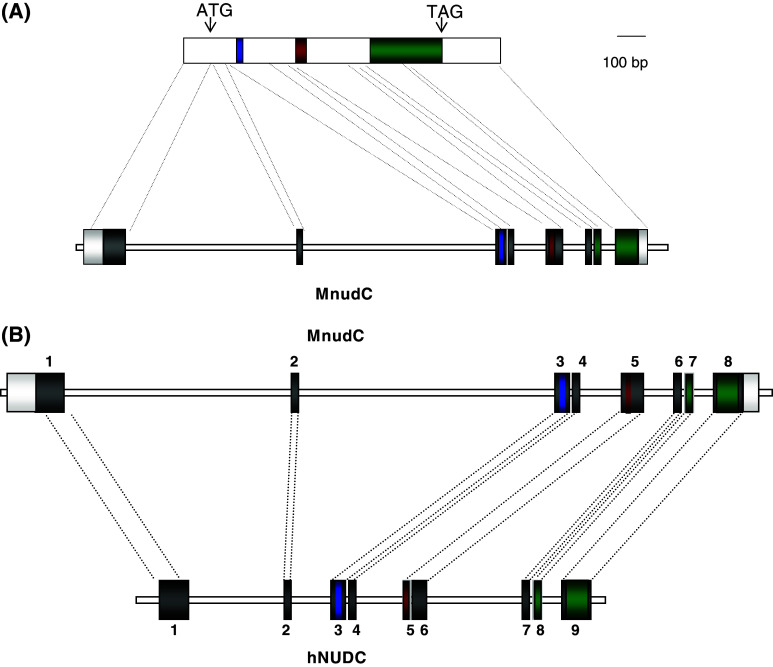
The structures of some mammalian NudC genes have been reported. Matsumoto and Ledbetter have determined that the human NUDC gene contains nine exons and spans about 8 kb in chromosome 1 of human genome [19]. The exons range from 66 to 266 bp in size, while the eight introns range from 92 bp to 2 kb. Together with NUDC, two processed pseudogenes (1.7 and 2.0 kb) were also identified [19]: (1) NUDCD1 (also known as CML66), coding for a broadly immunogenic tumor antigen which is highly expressed in a variety of solid tumors and in leukemias [27] which shares significant similarity with p23 [28, 29]. P23 interacts with the chaperone HSP90 and participates in the folding of different regulatory proteins [30, 31]. Recently, it has been reported that NUDCD1 exhibits chaperone activity in vitro [32]. (2) NUDCL, which codes for a protein essential for dynein stability and cell viability [33]. NUDCL might have a passive chaperone activity by binding to partially folded dynein intermediate chains, preventing their aggregation until they are completely folded [33].
The genomic structure of murine NudC revealed eight exons and seven introns spanning about 15 kb located in chromosome 4 (Fig. 3a). Although both human NUDC and murine NudC encode 332 amino acids, it is interesting that hNUDC gene consists of nine exons, in contrast with the eight exons of mNudC. When comparing the genomic structure of murine NudC and human NUDC, it became apparent that exon 5 of the murine gene corresponds to exons 5 and 6 of the human gene, though the acidic region is coded by exon 5 in both cases. Murine introns 1 and 2 are three- to fourfold larger than the same introns in hNUDC (Fig. 3b). The comparison of the 5′ UTR regions from mammalian genes (human, rat and mouse) shows a high conservation along their sequences, suggesting that their promoter regions share highly conserved transcriptional regulatory mechanisms. In mouse cells and tissues; however, we have observed a single transcript of 1.7 kb which could be detected in all fetal and adult tissues examined [18] while in human tissues a major 2 kb and a minor 1.7 kb mRNAs have been observed. Very likely these differences are the result of alternative splicing models occurring in the two species, given the exon–intron distribution mentioned above. Whether this is related to possible functional differences is unknown yet. The high degree of conservation in gene promoters suggests, however, that all mammalian genes share transcriptional regulatory mechanisms and that they have similar responses to similar stimuli [34].
Fig. 3.
Structure of the mouse and human NudC genes. a Schematic drawing of the mouse NudC cDNA (upper) and the mNudC gene (lower). In the cDNA the three regions highlighted in Fig. 2 are indicated. The relative positions of the initiation (ATG) and termination (TAG) codons are also shown. The NudC gene depicts the exon–intron distribution. The 100-bp scale refers only to cDNA. b Comparison of the murine and human genes showing their exon–intron distribution. The relative sizes of introns in both genes is depicted. The human gene spans for 8 kb while the mouse gene spans for 15 kb
Functions of mammalian nudC
Mammalian NudC gene products appear to be implicated in several functions which are not completely understood. In lower organisms; however, their role in nuclear migration during development is better defined. Thus, the functional complementarity of nudC and nudF was soon established in E. nidulans, in which overexpression of NUDF regenerated the naive genotype of nudC mutants [35]. It was then established their role in the regulation of nuclear migration [36]. Thus, the change in hNUDC levels by either small interfering RNA-mediated gene silencing or adenovirus-mediated overexpression in HeLa and C. elegans cells resulted in multinucleated cells [36]. Both the gain of function (overexpression) and the loss of function (depletion) of hNUDC may interfere with the stabilization of microtubule/dynein/dynactin complex that is required for cytokinesis [36, 37].
As for the mammalian orthologous genes, they have been related to the regulation of nuclear migration during mitosis [38, 39], thus sharing a biological role with their orthologous in E. nidulans, D. melanogaster or Neurospora crassa [1, 16]. The high similarity in the components of dynein and its regulatory complex dynactin between mammals and lower organisms favors the idea of an essential function conserved during evolution.
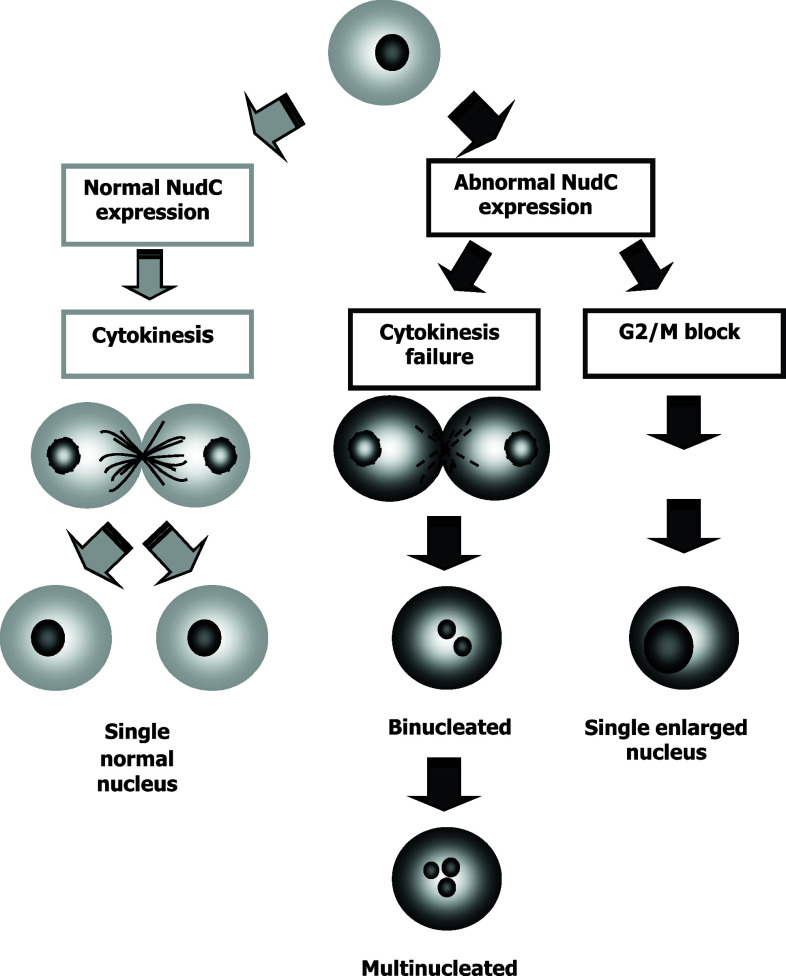
Human NUDC could lead to the completion of cytokinesis and the partition of chromosomes to two daughter cells. Aberrant hNUDC expression produces a failure in cytokinesis, which results in an increase in enlarged nucleus cells, binucleated cells or multinucleated cells. This aberrant hNUDC expression may also lead to other defects, including a G2/M block where cells duplicate their chromosomes but fail to enter mitosis, or a failure in karyokinesis where cells enter mitosis but fail to complete nuclear division, resulting in the accumulation of large cells with a single enlarged nucleus (Fig. 4) [40].
Fig. 4.
Effect of NudC on cell division. Gray arrows indicate the complete cytokinesis route when NudC has a normal expression. Black arrows indicate two consequences of NudC abnormal expression: (1) a cytokinesis failure that renders binucleated or multinucleated cells and (2) the arrest in G2/M phase of cell cycle render single cells with enlarged, undivided nuclei
Human NUDC has also been implicated in neurogenesis and neuronal migration through its relationship with NUDF. Immunohistochemistry studies of human tissues and proteome analysis of stem midbrain cells have shown that hNUDC is upregulated during neuronal differentiation to terminally differentiated neurons [41–43]. There is a high similarity between NudF [44] and LIS1, a gene playing a role in human lissencephaly [45–47]. LIS1 was first identified in mutations or deletions in patients with Miller–Dieker disease. This illness is caused by a deficiency in neuronal migration during brain development which leads to under-development in brain cortex and cerebellum regions. The interaction of Lis1 with NudC has been demonstrated by immunoprecipitation studies in brain tissues. This, together with the characteristics of MDS and ILS phenotypes, raises the possibility that nuclear movement in the ventricular zone is linked to the specification of neuronal fates and thus to cortical architecture [41, 43]. These findings are compatible with the idea that LIS1 and the proteins interacting with it (NudE, NudEL and NudC) play a role in glioma migration and proliferation analogous to their role during brain development [48]. These same authors suggest that these proteins may play a role in tumor growth. Thus, by using immunohistochemistry techniques they have shown the differential expression of LIS1, dynein, dynactin, NudE/NudEL and hNUDC in infiltrated cells and various human neuroectodermal tumors [48]. It is conceivable that these molecules play important roles in the migration and proliferation of tumor cells, analogous to their role in progenitor cell migration and proliferation.
These molecules could be potential therapeutic targets [48]. Thus, Lin et al. have demonstrated that hNUDC overexpression leads to a block in cell division of prostate cancer cells (LNCaP, DU145 or PC-3) which stop at the G2/M phase of the cell cycle, being therefore conceivable new anticancer drugs which target the function of these microtubule motor-associated proteins [40]. Also, hNUDC protein was expressed in nasopharyngeal tissues, its expression being higher in cancerous cells than in nasopharyngeal non-cancerous tissues. Specific antibodies against NUDC could be useful in inhibiting the growth of these carcinogenic cells [49]. hNUDC was also identified in cutaneous T-cell lymphoma by serological identification of antigens which have triggered the immunological response in tumor-bearing patients [50]. hNUDC has also been identified in a proteomic background study of esophageal cancer comparing tumor cells and the matched neighboring normal epithelial cells from surgical specimens. The protein was identified by 2D-DIGE and its identification validated with specific antibodies [51]. An inverse correlation between hNUDC expression levels and nodal metastasis was established [51].
Tang et al. have shown the interaction of hNUDC with thrombopoietin receptor (Mpl) which promotes transcriptional activation as well as proliferation and differentiation in a leukemic cell line [52]. Also, hNUDC has been identified as a gene implicated in human malign hematopoiesis. In normal human bone marrow, hNUDC protein and mRNA are highly expressed in early myeloid and erythroid precursors. The expression declines as these cells are terminally differentiated [53], again suggesting its conserved role in cell division.
Another evidence for the role of hNUDC in nuclear migration derives from its regulation by phosphorylation catalyzed by the polo-like kinase 1 (PLK1). In a cDNA phage display screen, hNUDC was identified as a PLK1 binding protein [54]. In another study it was established the interaction between PLK1 and hNUDC, showing that PLK1 phosphorylates hNUDC at conserved S274 and S326 residues in vitro. It was also shown that hNUDC is a substrate for PLK1 in vivo [54] and that downregulation of NudC by RNA interference results in multiple mitotic defects, including multinucleation and cells arrested at the midbody stage, which can be rescued by ectopic expression of wild-type NudC, but not by NudC with mutations in the Plk1 phosphorylation sites. These findings suggest that hNUDC functions in mitosis and cytokinesis, in part by regulating microtubule organization.
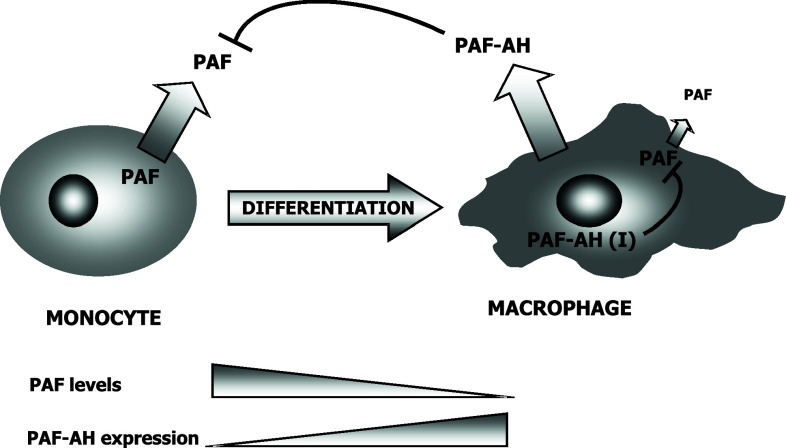
Several studies reveal a novel role for phospho-hNUDC as a spatial regulator of PLK1 and the Kinesin-7 motor (CENP-E) from prometaphase kinetochores [55]. In support of this idea, the localization of hNUDC to the midzone and midbody [55], as well as the hNUDC-dependent targeting of PLK1 to these structures, raises the possibility that the PLK1-hNUDC complex functions during cytokinesis independently of its role at the kinetochore [55]. Another point about the essential role of mammalian NUDC in dynein regulation is provided by a study in which it is described the presence and activity of LIS1, and probably hNUDC, in cilia and flagella. LIS1 and hNUDC could, therefore, be related to the movement of cilia and flagella regulating the axonemal dynein [56]. Finally, there is a newer and somehow unexpected function in which NUDC has been implicated. This is the regulation of platelet-activating factor acetylhydrolase (PAF-AH) through LIS1, which is actually the isoform I of PAF-AH regulatory subunit β (PAFAH1B) [57]. PAH-AH is highly specific and transforms the acetylated PAF in its inactive form, Lyso-PAF. As PAF is an important pro-inflammatory secondary lipidic messenger, PAF-AH(I) acts by regulating these pro-inflammatory functions [58, 59]. The interaction of NUDC with PAF-AH(I) regulatory subunit therefore implies that NUDC functions as a regulator of PAF-AH(I) and, consequently, of PAF-mediated actions. It is interesting that we isolated NudC cDNA from a monocyte-macrophage cell line challenged with inflammatory stimuli [21], which leads to activation as well as differentiation. It has also been described that PAF-AH activity increases during monocytes differentiation [60], indicating that the regulation of PAF is important during monocyte-macrophage activation and differentiation. In macrophages, high levels of extracellular PAF-AH are synthesized. At the same time, NUDC acts by upregulating the activity of intracellular PAF-AH(I). Both mechanisms lead to the inactivation of PAF, thus contributing to a negative regulation of monocyte-macrophage differentiation and/or activation (Fig. 5).
Fig. 5.
Expression of PAH and PAF-AH during macrophage differentiation. Monocytes express low levels of intra- and extracellular PAF-AH, thus permitting high levels of PAF production. As monocytes are activated with inflammatory stimuli and their differentiation to macrophage triggered, both intracellular and extracellular PAF-AH activities increase, the former being dependent of NUDC. High levels of PAF-AH results in a low production of PAF by the differentiated cell and in the inactivation on extracellular PAF
In relationship to the regulation of PAF production, hNUDC has been characterized as a novel accumulator that specifically acts on in vitro megakaryocytopoiesis and in vivo platelet production [61]. Thus, it has been demonstrated that hNUDC significantly increases megakaryocyte maturation and stimulates colony formation [61]. hNUDC is able to bind to the extracellular domain of the thrombopoietin receptor (Mpl). hNUDC may have two functional domains that could induce multinucleation by two independent mechanisms. The analysis in a yeast–two hybrid system indicated that the domain of hNUDC that binds Mpl spanned a portion of internal amino acids from residues 100 to 238 [26], whereas the C-terminal region was shown to associate intracellularly with Lis1 and the dynein motor complex [41].
There is evidence of direct binding of hNUDC to cell surface Mpl, thus promoting megakaryocyte proliferation and differentiation [62]. As previously indicated, native hNUDC and Mpl were localized around the nucleus and in cytoplasm extensions at all stages of megakaryocyte development, including platelet-shedding megakaryocytes [26]. The localization of hNUDC from Golgi to the ER and its ability to associate with tubulin in accordance to morphological data has raised the possibility of colocalization of hNUDC and Mpl with microtubules during the process of secretion. Indeed, the coexpression of Mp1 and hNUDC leads to the release of hNUDC to the culture medium [62]. Thus, NUDC appears to act as a cytokine, triggering many responses similar to that of thrombopoietin through Mpl, such as cell morphological changes and polyploidy. Also, hNUDC induced a sustained activation of the extracellular signal-regulated protein kinases-1 and -2 (ERK1/2) as well as p38 MAPK [52]. Finally, it has been proposed that hNUDC may form a complex with LIS1/dynein/Mpl and functions in part by mediating the organization of the microtubule cytoskeleton during megakaryocyte development process [62].
Concluding remarks
NudC is a highly conserved gene during evolution. All the mammalian genes isolated and characterized up to now share a very similar gene structure. Moreover, the promoter regions in rat, mouse and human genes have highly conserved sequences, indicating that they have binding sites for the same transcription factors and therefore that they are regulated similarly in response to specific stimuli. NudC is a gene implicated in nuclear migration during cell division, the human gene being able to correct by complementarity aberrant nudC mutations in E nidulans. NUDC works by regulating the minus-end molecular motor dynein, a function that may have relevance in cell proliferation. Thus, NUDC has been implicated in a variety of processes such as neuronal development, hematopoiesis or tumor development, all of them related to cellular division. In this regard, NudC could be envisaged as a carcinogenic marker and a possible therapeutic target.
And yet, a novel function of NUDC not related to cellular division has been suggested. It has been shown that NUDC may act on the inflammatory lipid PAF through its regulatory enzyme PAF-AH. The direct interaction of NUDC with NUDF (LIS1) has been shown. NUDF (LIS1) is the regulatory subunit of PAF-AH, PAFAH1B and its interaction with NUDC renders an enzyme with increased catalytic activity. This in turn would have an effect in regulating PAF levels at the cytoplasm. These new functions not related to cellular division might be the consequence of the presence of new motifs in mammalian NudC genes. Thus, E nidulans nudC gene comprises about one-third of the mammalian NudC sequences. These have gained two extra sequences coding for an NLS and an acidic domain whose functions are not yet understood.
References
- 1.Osmani AH, Osmani SA, Morris NR. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans . J Cell Biol. 1990;111:543–551. doi: 10.1083/jcb.111.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang X, Zuo W, Efimov VP, Morris NR. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet. 1999;35:626–630. doi: 10.1007/s002940050461. [DOI] [PubMed] [Google Scholar]
- 3.Xiang X, Osmani AH, Osmani SA, Roghi CH, Willins DA, Beckwith S, Goldman G, Chiu Y, Xin M, Liu B. Analysis of nuclear migration in Aspergillus nidulans . Cold Spring Harb Symp Quant Biol. 1995;60:813–819. doi: 10.1101/sqb.1995.060.01.087. [DOI] [PubMed] [Google Scholar]
- 4.Chiu YH, Morris NR. Extragenic suppressors of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans . Genetics. 1995;141:453–464. doi: 10.1093/genetics/141.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu YH, Xiang X, Dawe AL, Morris NR. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol Biol Cell. 1997;8:1735–1749. doi: 10.1091/mbc.8.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fath KR, Trimbur GM, Burgess DR. Molecular motors and a spectrin matrix associate with Golgi membranes in vitro. J Cell Biol. 1997;139:1169–1181. doi: 10.1083/jcb.139.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/S0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 8.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- 10.Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans . Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris SM, Anaya P, Xiang X, Morris NR, May GS, Yu-Lee L. A prolactin-inducible T cell gene product is structurally similar to the Aspergillus nidulans nuclear movement protein NUDC. Mol Endocrinol. 1997;11:229–236. doi: 10.1210/me.11.2.229. [DOI] [PubMed] [Google Scholar]
- 12.Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann B, Zuo W, Liu A, Morris NR. The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to alpha- and gamma-tubulin. J Biol Chem. 2001;276:38877–38884. doi: 10.1074/jbc.M106610200. [DOI] [PubMed] [Google Scholar]
- 15.Dawe AL, Caldwell KA, Harris PM, Morris NR, Caldwell GA. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans . Dev Genes Evol. 2001;211:434–441. doi: 10.1007/s004270100176. [DOI] [PubMed] [Google Scholar]
- 16.Cunniff J, Chiu YH, Morris NR, Warrior R. Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech Dev. 1997;66:55–68. doi: 10.1016/S0925-4773(97)00085-3. [DOI] [PubMed] [Google Scholar]
- 17.Moreau N, Aumais JP, Prudhomme C, Morris SM, Yu-Lee LY. NUDC expression during amphibian development. Int J Dev Biol. 2001;45:839–843. [PubMed] [Google Scholar]
- 18.Riera J (2004) Aislamiento y caracterización del gen mNudC. Regulación de la acetilhidrolasa del PAF por NUDC. In: Biochemistry and molecular biology. Universidad de Oviedo, Oviedo, pp 112
- 19.Matsumoto N, Ledbetter DH. Molecular cloning and characterization of the human NUDC gene. Hum Genet. 1999;104:498–504. doi: 10.1007/s004390050994. [DOI] [PubMed] [Google Scholar]
- 20.Axtell SM, Truong TM, O’Neal KD, Yu-Lee LY. Characterization of a prolactin-inducible gene, clone 15, in T cells. Mol Endocrinol. 1995;9:312–318. doi: 10.1210/me.9.3.312. [DOI] [PubMed] [Google Scholar]
- 21.Riera J, Rodriguez R, Carcedo MT, Campa VM, Ramos S, Lazo PS. Isolation and characterization of nudC from mouse macrophages, a gene implicated in the inflammatory response through the regulation of PAF-AH(I) activity. FEBS Lett. 2007;581:3057–3062. doi: 10.1016/j.febslet.2007.05.065. [DOI] [PubMed] [Google Scholar]
- 22.Gocke CD, Reaman GH, Stine C, Zhang MY, Osmani SA, Miller BA. The nuclear migration gene NudC and human hematopoiesis. Leuk Lymphoma. 2000;39:447–454. doi: 10.3109/10428190009113375. [DOI] [PubMed] [Google Scholar]
- 23.Thompson PD, Jurutka PW, Haussler CA, Whitfield GK, Haussler MR. Heterodimeric DNA binding by the vitamin D receptor and retinoid X receptors is enhanced by 1, 25-dihydroxyvitamin D3 and inhibited by 9-cis-retinoic acid. Evidence for allosteric receptor interactions. J Biol Chem. 1998;273:8483–8491. doi: 10.1074/jbc.273.14.8483. [DOI] [PubMed] [Google Scholar]
- 24.Morris SM, Yu-Lee LY. Expression of RNUDC, a potential nuclear movement protein, in mammalian cells: localization to the Golgi apparatus. Exp Cell Res. 1998;238:23–32. doi: 10.1006/excr.1997.3822. [DOI] [PubMed] [Google Scholar]
- 25.Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-O. [DOI] [PubMed] [Google Scholar]
- 26.Pan RM, Yang Y, Wei MX, Yu XB, Ge YC, Xu P. A microtubule associated protein (hNUDC) binds to the extracellular domain of thrombopoietin receptor (Mpl) J Cell Biochem. 2005;96:741–750. doi: 10.1002/jcb.20573. [DOI] [PubMed] [Google Scholar]
- 27.Yang XF, Wu CJ, McLaughlin S, Chillemi A, Wang KS, Canning C, Alyea EP, Kantoff P, Soiffer RJ, Dranoff G, Ritz J, et al. CML66, a broadly immunogenic tumor antigen, elicits a humoral immune response associated with remission of chronic myelogenous leukemia. Proc Natl Acad Sci USA. 2001;98:7492–7497. doi: 10.1073/pnas.131590998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felts SJ, Toft DO. p23, a simple protein with complex activities. Cell Stress Chaperones. 2003;8:108–113. doi: 10.1379/1466-1268(2003)008<0108:PASPWC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Ranea JA, Mirey G, Camonis J, Valencia A. p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 2002;529:162–167. doi: 10.1016/S0014-5793(02)03321-5. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri ES, Litwack G, Toft D. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 31.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faircloth LM, Churchill PF, Caldwell GA, Caldwell KA. The microtubule-associated protein, NUD-1, exhibits chaperone activity in vitro. Cell Stress Chaperones. 2009;14:95–103. doi: 10.1007/s12192-008-0061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou T, Zimmerman W, Liu X, Erikson RL. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc Natl Acad Sci USA. 2006;103:9039–9044. doi: 10.1073/pnas.0602916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris NR, Efimov VP, Xiang X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998;8:467–470. doi: 10.1016/S0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang MY, Huang NN, Clawson GA, Osmani SA, Pan W, Xin P, Razzaque MS, Miller BA. Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp Cell Res. 2002;273:73–84. doi: 10.1006/excr.2001.5414. [DOI] [PubMed] [Google Scholar]
- 36.Aumais JP, Williams SN, Luo W, Nishino M, Caldwell KA, Caldwell GA, Lin SH, Yu-Lee LY. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- 37.Willins DA, Xiang X, Morris NR. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans . Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gocke CD, Osmani SA, Miller BA. The human homologue of the Aspergillus nuclear migration gene nudC is preferentially expressed in dividing cells and ciliated epithelia. Histochem Cell Biol. 2000;114:293–301. doi: 10.1007/s004180000197. [DOI] [PubMed] [Google Scholar]
- 39.Helmstaedt K, Laubinger K, Vosskuhl K, Bayram O, Busch S, Hoppert M, Valerius O, Seiler S, Braus GH. The nuclear migration protein NUDF/LIS1 forms a complex with NUDC and BNFA at spindle pole bodies. Eukaryot Cell. 2008;7:1041–1052. doi: 10.1128/EC.00071-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin SH, Nishino M, Luo W, Aumais JP, Galfione M, Kuang J, Yu-Lee LY. Inhibition of prostate tumor growth by overexpression of NudC, a microtubule motor-associated protein. Oncogene. 2004;23:2499–2506. doi: 10.1038/sj.onc.1207343. [DOI] [PubMed] [Google Scholar]
- 41.Aumais JP, Tunstead JR, McNeil RS, Schaar BT, McConnell SK, Lin SH, Clark GD, Yu-Lee LY. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:RC187. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffrogge R, Mikkat S, Scharf C, Beyer S, Christoph H, Pahnke J, Mix E, Berth M, Uhrmacher A, Zubrzycki IZ, Miljan E, Volker U, Rolfs A. 2-DE proteome analysis of a proliferating and differentiating human neuronal stem cell line (ReNcell VM) Proteomics. 2006;6:1833–1847. doi: 10.1002/pmic.200500556. [DOI] [PubMed] [Google Scholar]
- 43.Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee LY. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr Biol. 1998;8:603–606. doi: 10.1016/S0960-9822(98)70232-5. [DOI] [PubMed] [Google Scholar]
- 44.Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 46.Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller–Dieker syndrome. Hum Mol Genet. 1997;6:157–164. doi: 10.1093/hmg/6.2.157. [DOI] [PubMed] [Google Scholar]
- 47.Reiner O, Albrecht U, Gordon M, Chianese KA, Wong C, Gal-Gerber O, Sapir T, Siracusa LD, Buchberg AM, Caskey CT. Lissencephaly gene (LIS1) expression in the CNS suggests a role in neuronal migration. J Neurosci. 1995;15:3730–3738. doi: 10.1523/JNEUROSCI.15-05-03730.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki SO, McKenney RJ, Mawatari SY, Mizuguchi M, Mikami A, Iwaki T, Goldman JE, Canoll P, Vallee RB. Expression patterns of LIS1, dynein and their interaction partners dynactin, NudE, NudEL and NudC in human gliomas suggest roles in invasion and proliferation. Acta Neuropathol. 2007;113:591–599. doi: 10.1007/s00401-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 49.Chen YQ, Li TY, Qu SH, Tang XJ. Expression and effects of nuclear distribution C (NUDC) protein in nasopharyngeal carcinoma cell lines. Ai Zheng. 2006;25:708–712. [PubMed] [Google Scholar]
- 50.Hartmann TB, Mattern E, Wiedemann N, van Doorn R, Willemze R, Niikura T, Hildenbrand R, Schadendorf D, Eichmuller SB. Identification of selectively expressed genes and antigens in CTCL. Exp Dermatol. 2008;17:324–334. doi: 10.1111/j.1600-0625.2007.00637.x. [DOI] [PubMed] [Google Scholar]
- 51.Hatakeyama H, Kondo T, Fujii K, Nakanishi Y, Kato H, Fukuda S, Hirohashi S. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics. 2006;6:6300–6316. doi: 10.1002/pmic.200600488. [DOI] [PubMed] [Google Scholar]
- 52.Tang YS, Zhang YP, Xu P. hNUDC promotes the cell proliferation and differentiation in a leukemic cell line via activation of the thrombopoietin receptor (Mpl) Leukemia. 2008;22:1018–1025. doi: 10.1038/leu.2008.20. [DOI] [PubMed] [Google Scholar]
- 53.Miller BA, Zhang MY, Gocke CD, De Souza C, Osmani AH, Lynch C, Davies J, Bell L, Osmani SA. A homolog of the fungal nuclear migration gene nudC is involved in normal and malignant human hematopoiesis. Exp Hematol. 1999;27:742–750. doi: 10.1016/S0301-472X(98)00074-5. [DOI] [PubMed] [Google Scholar]
- 54.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–138. doi: 10.1016/S1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 55.Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee LY. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 56.Pedersen LB, Rompolas P, Christensen ST, Rosenbaum JL, King SM. The lissencephaly protein Lis1 is present in motile mammalian cilia and requires outer arm dynein for targeting to Chlamydomonas flagella . J Cell Sci. 2007;120:858–867. doi: 10.1242/jcs.03374. [DOI] [PubMed] [Google Scholar]
- 57.Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Miller–Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase (corrected) Nature. 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 58.Peplow PV. Regulation of platelet-activating factor (PAF) activity in human diseases by phospholipase A2 inhibitors, PAF acetylhydrolases, PAF receptor antagonists and free radical scavengers. Prostaglandins Leukot Essent Fatty Acids. 1999;61:65–82. doi: 10.1054/plef.1999.0038. [DOI] [PubMed] [Google Scholar]
- 59.Shmueli O, Cahana A, Reiner O. Platelet-activating factor (PAF) acetylhydrolase activity, LIS1 expression, and seizures. J Neurosci Res. 1999;57:176–184. doi: 10.1002/(SICI)1097-4547(19990715)57:2<176::AID-JNR3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 60.Elstad MR, Stafforini DM, McIntyre TM, Prescott SM, Zimmerman GA. Platelet-activating factor acetylhydrolase increases during macrophage differentiation. A novel mechanism that regulates accumulation of platelet-activating factor. J Biol Chem. 1989;264:8467–8470. [PubMed] [Google Scholar]
- 61.Wei MX, Yang Y, Ge YC, Xu P. Functional characterization of hNUDC as a novel accumulator that specifically acts on in vitro megakaryocytopoiesis and in vivo platelet production. J Cell Biochem. 2006;98:429–439. doi: 10.1002/jcb.20803. [DOI] [PubMed] [Google Scholar]
- 62.Zhang YP, Tang YS, Chen XS, Xu P. Regulation of cell differentiation by hNUDC via a Mpl-dependent mechanism in NIH 3T3 cells. Exp Cell Res. 2007;313:3210–3221. doi: 10.1016/j.yexcr.2007.06.021. [DOI] [PubMed] [Google Scholar]