Abstract
Cone dystrophies are genetic diseases characterized by loss of cone photoreceptor function and severe impairment of daylight vision. Loss of function is accompanied by a progressive degeneration of cones limiting potential therapeutic interventions. In this study we combined microarray-based gene-expression analysis with electroretinography and immunohistochemistry to characterize the pathological processes in the cone photoreceptor function loss 1 (cpfl1) mouse model. The cpfl1-mouse is a naturally arising mouse mutant with a loss-of-function mutation in the cone-specific Pde6c gene. Cpfl1-mice displayed normal rod-specific light responses while cone-specific responses were strongly diminished. Despite the lack of a general retinal degeneration, the cone-specific functional defect resulted in a marked activation of GFAP, a hallmark of Müller-cell gliosis. Microarray-based network-analysis confirmed activation of Müller-glia-specific transcripts. Unexpectedly, we found up-regulation of the cytokine LIF and the anti-apoptotic transcription factor STAT3 in cpfl1 cone photoreceptors. We postulate that STAT3-related pathways are induced in cpfl1 cone photoreceptors to counteract degeneration.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0376-9) contains supplementary material, which is available to authorized users.
Keywords: cpfl1, Cone photoreceptor function loss 1, STAT3, Cone photoreceptor degeneration, Transcriptomics
Introduction
Cone dystrophies are a group of highly heterogeneous genetic disorders characterized by a specific loss of cone photoreceptor-mediated light responses and slow to fast progressing degeneration of cone photoreceptors, the subclass of photoreceptors that mediate daylight vision. Early onset cone dystrophy is also termed achromatopsia or rod monochromasy. Affected patients are severely visually handicapped and suffer from a combination of very poor visual acuity, nystagmus, photophobia, and lack of color discrimination. Four genes have been linked to achromatopsia, the two genes encoding the cone photoreceptor cyclic nucleotide-gated channel (Cnga3 and Cngb3) [1], the gene encoding the cone-specific transducin (Gnat2) [2], and more recently the gene for the cone-specific α′-subunit of the cGMP-phosphodiesterase (Pde6c) [3, 4]. Most interestingly, the cpfl1 (cone photoreceptor function loss 1) mouse, a naturally arising mouse mutant initially identified because of its complete lack of cone-mediated light responses [5, 6], has been associated with a loss-of-function mutation in the Pde6c gene [3]. In the cpfl1 mouse, a 116-bp insertion (c.864_865ins116) in the Pde6c gene results in a frame-shift and a premature termination codon. This mutation is predicted to lead to the loss of cGMP-phosphodiesterase activity in cone photoreceptors. The functional impairment in the cpfl1 mouse is accompanied by a fast progressing degeneration of cone photoreceptors. In contrast, rod function and morphology is not affected [3]. Injury of photoreceptors or genetic defects that lead to degeneration are thought to activate repair and cell death signaling pathways that involve Müller cells, the main glia of the retina [7]. These pathways were studied for general retinal injury (e.g., light damage or retinal detachment) or genetic diseases that affect the majority of photoreceptors (e.g., retinitis pigmentosa or rod-cone dystrophies) [7, 8]. In contrast, our knowledge on the molecular mechanisms leading to degeneration of cone photoreceptors in cone photoreceptor-specific dystrophies is very limited. In this study we combined microarray-based gene-expression analysis with electroretinography (ERG) and morphological data to characterize the pathological processes in the cpfl1 mouse. We identified, for the first time, activation of signal transducer and activator of transcription 3 (STAT3)-related signaling pathways in degenerating cone photoreceptors.
Materials and methods
Electroretinography
ERG is an established diagnostic technique in clinical ophthalmology as well as in basic research, which allows an objective evaluation of retinal function in vivo. In normal retinas, under dark-adapted conditions, the rod-mediated components dominate the response, whereas under light-adapted conditions, responses are usually cone-mediated [9]. Protocols that take advantage of these sensitivity differences to distinguish rod from cone functions are commonly used for diagnostic purposes in humans [10] as well as in animal models [11–13].
ERGs were recorded as described [12]. In anesthetized mice using a Ganzfeld bowl, a direct current amplifier, and a PC-based control and recording unit (Multiliner Vision; VIASYS Healthcare GmbH, Hoechberg, Germany). ERG recordings were obtained in both scotopic (dark-adapted overnight) and photopic (light-adapted 10 min at 30 cd/m2) conditions. Single white-flash stimulation ranged from −4 to 1.5 log cd s/m2 under scotopic, and from −2.0 to 1.5 log cd s/m2 under photopic conditions. Ten responses were averaged with an inter-stimulus interval of either 5 or 17 s (for 0–1.5 log cd s/m2). For additional photopic bright flash experiments, we used a Mecablitz 60CT4 flash gun (Metz, Germany) added to the Ganzfeld bowl. The intensity used in this photopic bright flash protocol was 4.1 log cd s/m2.
Spectral domain optical coherence tomography (OCT)
For OCT imaging, we used a commercially available Spectralis™ HRA + OCT device from Heidelberg Engineering as previously described [14]. Each two-dimensional B-Scan recorded at 30° field of view consists of 1,536 × 496 pixels, which are acquired at a speed of 40,000 scans/s. Optical depth resolution is ca. 7 μm with digital resolution reaching 3.5 μm [15]. Resulting data were exported as 8-bit color bitmap files and processed in Adobe Photoshop CS2 (Adobe Systems, San Jose, CA, USA).
RNA extraction
Cpfl1 and wild-type mice were euthanized, eyes enucleated and immediately transferred to PBS-buffer (Gibco®, Invitrogen, Karlsruhe, Germany). After removal of the cornea and lens, retinas were gently dissected from the eye cup and placed in 350-μl RLT buffer (Qiagen, Hilden, Germany) + 1% β-mercaptoethanol (Sigma-Aldrich Chemie, Steinheim, Germany). Extraction of total RNA was performed using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions for tissues. QIAshredder mini-spin columns (Qiagen) as well as needle and syringe homogenization were applied. RNA quality was observed with an Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip Kit (Agilent Technologies, Boeblingen, Germany) following the manufacturer’s instructions.
cDNA synthesis and qRT-PCR
One microgram of RNA of three wild-type and three cpfl1 animals was applied to the cDNA synthesis using the QuantiTect® Reverse Transcription Kit (Qiagen), which includes digestion of genomic DNA.
qRT-PCR was performed with the LightCycler480 System (Roche, Mannheim, Germany) using the QuantiTect® SYBR® Green PCR Kit (Qiagen) or LightCycler®480 Probes Master Kit (Roche) according to the manufacturer’s instructions.
Standard curves for each amplified transcript were generated to obtain the PCR efficiency. CP-values were determined by the LightCycler®Software 480 (Roche). Expression levels of each sample were detected in triplicate reactions. Pyruvate dehydrogenase ß-subunit (Pdh) was used as reference gene to calculate the relative expression of each target gene applying the efficiency-corrected equation by Pfaffl [16].
Oligonucleotides for the qRT-PCR were designed using the Primer3 Software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) or the Roche Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). The sequences of the oligonucleotides are listed in Supplementary Table 1.
Table 1.
Stat3-associated transcripts showing a complex deregulation in the cpfl1 expression profile
| Transcript | Regulation | Reference |
|---|---|---|
| Adrb1 | ↓ | [30] |
| Ahr | ↑ | Binding site for Stat3 in the Ahr promotor (MatInspector) |
| Bcl6 | ↑ | [31] |
| C1qa | ↑ | [32] |
| Cd44 | ↑ | [32] |
| Cd9 | ↑ | [33] |
| Cebpd | ↑ | [34] |
| Clu | ↑ | Binding site for Stat3 in the Clu promotor (MatInspector) |
| Cp | ↑ | Binding site for Stat3 in the Cp promotor (MatInspector) |
| Ctss | ↑ | [35] |
| Fgf2 | ↑ | [36] |
| Gadd45b | ↑ | Binding site for Stat3 in the Gadd45b promotor (MatInspector) |
| Gbp2 | ↑ | [37] |
| Gfap | ↑ | [38] |
| Hes5 | ↓ | [39] |
| Hipk2 | ↓ | [40] |
| Il6st | ↑ | [41] |
| Mt2 | ↑ | [42] |
| Opn1mw | ↓ | [43] |
| Osmr | ↑ | [44] |
| Pten | ↓ | [45] |
| Socs3 | ↑ | [46] |
| Tnnt2 | ↑ | Binding site for Stat3 in the Tnnt2 promotor (MatInspector) |
| Vim | ↑ | [47] |
Microarray analysis
Microarray experiments of retina tissue were performed at two different ages (4 and 8 weeks) comparing gene expression of cpfl1 and wild-type animals using the Affymetrix™ platform according to the instructions of the manufacturer. Fragmented and labeled cRNA of three wild-type and three cpfl1 retinas (one retina each) were hybridized on Affymetrix™ Mouse Genome 430 2.0 Arrays, respectively. A probe-level summary was determined using the Affymetrix™ GeneChip Operating Software using the MAS5 algorithm. Normalization of raw data was performed by the Array Assist™ Software 4.0 (Stratagene, La Jolla, Canada), applying a GC-robust multichip average (RMA) algorithm. Significance was calculated using a t test without multiple testing correction (Array Assist™ software), selecting all transcripts with a minimum change in expression level of 1.5-fold together with a p value less than 0.05. Selected changes of transcripts of the Stat3 pathway were validated by qRT-PCR.
Gene regulation networks
Gene regulation networks were generated by Ingenuity Pathways Analysis 3.1 (http://www.ingenuity.com). For that purpose, data sets containing gene identifiers and the corresponding expression and significance values were uploaded into the application. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity.
Functional analyses
Functional analysis identified biological functions and/or diseases that were most significant to the data set. Genes from the data set that met the negative logarithmic significance cut-off of five or higher, and were associated with biological functions and/or diseases in the Ingenuity Pathways knowledge base, were considered for further analyses. Fischer’s exact test was used to calculate a p value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Genomatix-based promoter analysis
We extracted the mouse promoter sequences from the ElDorado database (Genomatix Suite-ElDorado, release 4.8, Mouse Genome build 37, Genomatix, Munich). The GEMS launcher task “FrameWorker” using the available weight matrix library (GEMS launcher version 4.4; Genomatix, Munich; http://www.genomatix.de) was used to generate the model of the promoter framework. The FastM task of GEMS Launcher was used to optimize models. ModelInspector (a GEMS launcher task) was used to search the mouse promoter database (Genomatix promoter database, GPD; Genomatix, Munich) with the optimized model. Additional information about connections between the genes from the initial list and candidate genes found by the model search was taken from BiblioSphere analyses (Genomatix, Munich). Default parameters were used for the initial analyses in all programs, if not indicated otherwise.
Immunohistochemistry
Immunohistochemical experiments were performed on 10-μm vertical cryostat sections as previously described [17]. The sources and working dilutions of primary antibodies are supplied in Supplementary Table 2. FITC-labeled peanut agglutinin (PNA 1:100; Sigma-Aldrich, Deisenhofen, Germany) was used to label the extracellular matrix of cone photoreceptors and glycogen phosphorylase to label cone photoreceptor cell bodies. Secondary detection of the antibodies was performed with Cy2 or Cy3-labeled donkey anti-rabbit, anti-mouse, anti-goat, or FITC-labeled anti-guinea pig IgG (Dianova, Hamburg, Germany). To visualize cell nuclei, we applied 5 μg/ml Hoechst33342 (Molecular Probes). The stained retinal slices were analyzed by confocal microscopy (LSM510 Meta, Carl Zeiss, Germany).
Results
Cone photoreceptor-specific loss of function and degeneration in cpfl1 mice
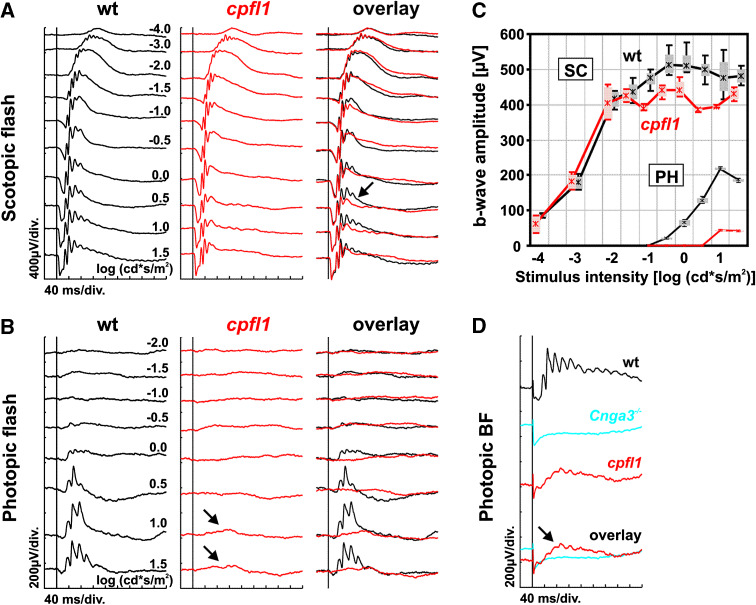
To ascertain baseline functional properties in cpfl1 mice, flash ERGs were recorded from postnatal week (PW) four wild-type and cpfl1 mice under dark-adapted (Fig. 1a) and light-adapted (Fig. 1b) conditions. The entire responses to dim stimuli and the initial part of the responses to bright stimuli were identical to that of the control mice (see overlay in Fig. 1a), indicating that the rod system, which is responsible for these signals [13], was not affected primarily by the disease. In contrast, the responses of the cone system were already strongly reduced to unrecordable at this point. The cone system influences the later part of the dark-adapted responses to bright stimuli (arrow in Fig. 1a) and generates the light-adapted ERGs (Biel et al. [18]), which are practically absent in cpfl1 mutants (arrows in Fig. 1b).
Fig. 1.
Electroretinographic data from wild-type and cpfl1 mice at 4 weeks of age. a Scotopic (dark-adapted) single flash ERG intensity series of a wild-type (left) and a cpfl1 mouse (center), together with an overlay of both signals (right). Vertical line crossing each trace shows the timing of the light flash. The alteration of the trailing edge of the scotopic b-wave at high stimulus intensities is illustrated (arrow in a). b Photopic (light-adapted) single flash ERG intensity series of a wild-type (left) and a cpfl1 mouse (right). The photopic responses are strongly reduced but not completely absent (arrows in b). c Scotopic (SC) and photopic (PH) b-wave amplitudes from wild-type and cpfl1 mice as a function of the logarithm of the flash intensity. Boxes represent the 25–75% quantile range, whiskers indicate the 5 and 95% quantiles, and the asterisk is the median of the data. d Photopic bright flash ERG responses obtained from a wild-type and a cpfl1 mouse, and also from a functionally all-rod mouse (Cnga3 −/−, cone cyclic nucleotide-gated channel deficient). Small response was detected in cpfl1 mice (arrow in d), indicating some remaining cone system function at 4 weeks of age
In summary, the ERG baseline results were comparable to those in a model lacking any cone function like the Cnga3 −/− mutants deficient of a subunit of the cone cyclic nucleotide-gated channel [11, 12]. However, unlike in Cnga3 −/− mice, photopic responses were not completely absent in cpfl1 mice (arrows in Fig. 1b). To confirm that these changes were not due to baseline fluctuation but actually reflected residual cone system function, we performed a photopic bright flash ERG using a light stimulus with 4.1 log cd s/m2 intensity (Fig. 1d). Although the initial part of the response using this protocol is masked by a flash artefact, a small but distinct light-evoked response was typically found (arrow in Fig. 1d), indicating either a very limited ability of the cpfl1 cone system to respond to stimulation by light, or a small number of desensitized rods in the vicinity of affected cones (bystander effect) that might respond at unphysiological light conditions. The same results were obtained for PW8 wild-type and cpfl1 mice (data not shown).
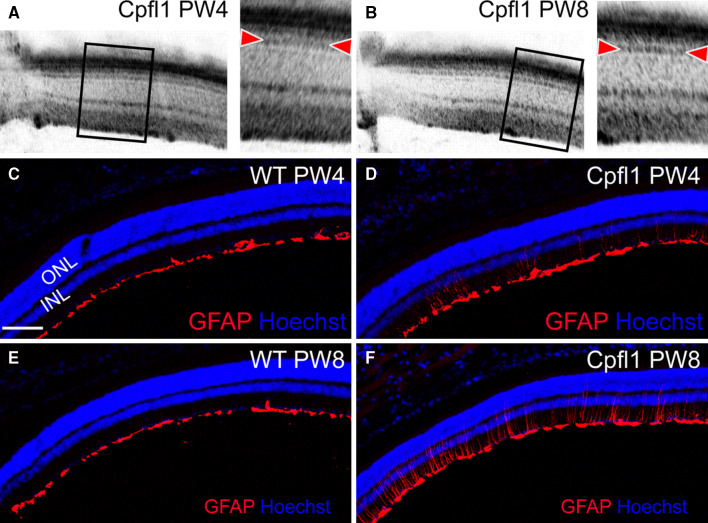
To analyze the morphology of the retina, we first used optical coherence tomography (OCT). OCT sections of the retina of cpfl1 mutant mice at PW4 and PW8 did not reveal any structural changes in retinal layering (Fig. 2a, b). In particular, the band associated with the inner/outer segment border of photoreceptors (indicated by the arrowheads) was not different between PW4 and PW8. However, despite the relatively low percentage of cones among the photoreceptors, the functional loss apparently triggered substantial reactive Müller gliosis characterized by induction of intermediate fiber glial fibrilary acid protein (GFAP) at PW4 and particularly at PW8 (Fig. 2c–f).
Fig. 2.
Invasive and non-invasive cross-sectional retinal imaging in WT and cpfl1 mice at PW4 and PW8. a, b Virtual cross sections using OCT demonstrate intact retinal architecture of cpfl1 retinas. The bands associated with photoreceptor structures, in particular the inner/outer segment border marked by arrowheads, appeared unaltered between PW4 (a) and PW8 (b). c–f Retinal cryosections stained for GFAP (red) and with DAPI (blue) for nuclei. Increasing GFAP staining was evident in the cpfl1 retinas (d, f) indicating progressive gliosis, but not in WT (c, e). Scale bar in c–f 100 μm
Loss of phototransduction-related transcripts in the cpfl1 mouse
The molecular events associated with the loss of cone photoreceptor function and degeneration in the retina of cpfl1 animals are not yet well understood. To gain more insight into the nature of the degeneration, we performed microarray analyses of 4- and 8-week-old cpfl1 and wild-type animals using Affymetrix MOE 430 2.0 microarrays. Expression levels of cpfl1 and wild-type retinas were compared at the different ages. At 4 weeks of age, 337 transcripts (listed in Supplementary Table 3) showed a minimum difference in expression level of 1.5-fold in combination with a p value less than 0.05, with 168 of them being up-regulated and 169 being down-regulated. Using the same criterion for the comparison of the 8-week-old animals, 222 transcripts were found to be differently regulated (specified in Supplementary Table 4), with 77 of those being up-regulated and 145 down-regulated. There was an overlap of 30% between the two age groups.
All data sets of our expression analyses were uploaded into the Ingenuity Pathways knowledge base and were grouped into categories representing selected biological functions and diseases (Fig. 3a). The following pathways were altered in cpfl1 animals compared to wild-type: cell cycle (PW4: 35 differently regulated genes; PW8: 13 differently regulated genes), cell signaling (PW4: 92; PW8: 48), disturbances in development and function of the visual system (PW4: 8; PW8: 12), cancer (PW4: 88; PW8: 38), cellular growth and proliferation (PW4: 75; PW8: 41), cellular assembly and organization (PW4: 28; PW8: 20), cell morphology (PW4: 41; PW8: 18), development and function of the cardiovascular system (PW4: 23; PW8: 17), gene expression (PW4: 40; PW8: 8), development and function of the nervous system (PW4: 75; PW8: 41).
Fig. 3.
a Selection of biological functions and diseases affected in the cpfl1 mutant. This diagram illustrates a selection of biological functions and diseases that appear to be disturbed in the cpfl1 mutant. The particular function is plotted versus the negative logarithm of the significance. Alterations in the 4-week-old animals are indicated by the black bar, changes in the 8-week-old animals by the light bar. b, c Relative gene expression evaluated by qRT-PCR and microarray analysis. Expression levels are displayed as signal log ratio (SLR). All genes evaluated by qRT-PCR show a significance of p ≤ 0.05. b Real-time validation of 4-week-old animals. c Real-time validation of 8-week-old animals
An important part of the data analysis of microarray experiments is the analysis of canonical pathways, which invokes a comparison between the uploaded data set and characterized published signaling pathways. The analysis of canonical pathways affected in the cpfl1 mouse highlighted a distinct dysregulation of phototransduction. We selected a subset of seven phototransduction-related genes that were found to be down-regulated in our microarray analysis for evaluation by qRT-PCR. All identified down-regulations were verified using qRT-PCR (p < 0.05, Fig. 3b, c) confirming the accuracy of our microarray data.
Loss of cone-specific RNA reflects loss of cone photoreceptor cells in the cpfl1 mouse
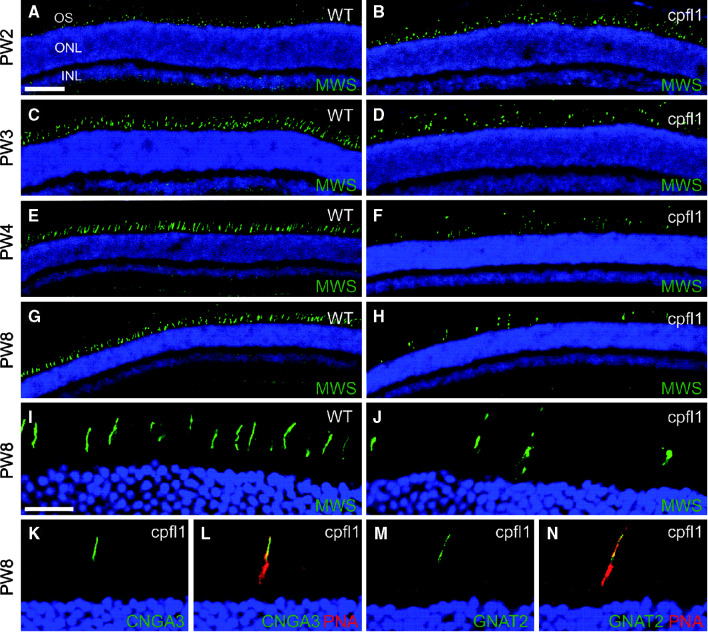
It remained uncertain whether the down-regulation was caused by a specific repression of these genes or rather reflected a general loss of cone photoreceptors. To clarify this issue, we examined the cellular expression of selected proteins in retinal sections. We double-labeled cone photoreceptors with the cone-specific marker peanut agglutinin (PNA) that specifically labels the extracellular matrix of cone photoreceptors and antibodies directed against one of the two mouse cone opsins (medium wavelength-sensitive opsin, MWS). At 2 weeks of age the number and appearance of cone photoreceptors in the cpfl1 retina (Fig. 4a, b) was indistinguishable from that of wild-type controls (not shown). Starting from PW3 we found a progressive loss of cone photoreceptors (Fig. 4c–f). In 8-week-old cpfl1 animals, the number of cone photoreceptors appeared to be drastically reduced (Fig. 4g, h). All cone photoreceptors persisting until late in the course of degeneration still contained significant amounts of opsin (Fig. 4g–j). The same results were obtained for the short wavelength-sensitive opsin (SWS; not shown), the cone cyclic nucleotide-gated channel CNGA3 subunit (Fig. 4k, l) and the cone transducin alpha subunit (GNAT2; Fig. 4m, n). Importantly, all these cone outer segment proteins were normally expressed and localized, suggesting widely preserved outer segment structure. Compared to CNGA3-deficient mice [11, 17], the time course of cone degeneration in cpfl1 mice was much faster. Based on these results, we conclude that the observed down-regulation of phototransduction transcripts in the cpfl1 retina originated from the loss of degenerating cone photoreceptors rather than from real gene repression.
Fig. 4.
Localization of phototransduction proteins in cpfl1 retinal slices. Confocal laser scanning micrographs of retinal sections showing immunolocalization of various markers. a–h Wild-type (a, c, e, g) and cpfl1 (b, d, f, h) retinal slices at different ages (postnatal week, PW 2–8) labeled for the medium wavelength-sensitive opsin (MWS, green). i–n High magnification images showing cone outer segment localization of MWS (i, j, green), CNGA3 (k, green), and GNAT2 (m, green) in retinal slices of PW8 wild-type (i) or cpfl1 (j–n) mice. In the images shown in (l and n) cone photoreceptors were co-labeled with peanut agglutinin (PNA, red). Scale bar for (a–h) is 100 μm (shown in a) and for (i–n) 20 μm (shown in i). Cell nuclei were labeled with Hoechst nuclear dye (blue). INL, inner nuclear layer, ONL, outer nuclear layer, OS, photoreceptor outer segments
Induction of STAT3-related signaling pathways in the degenerating cpfl1 retina
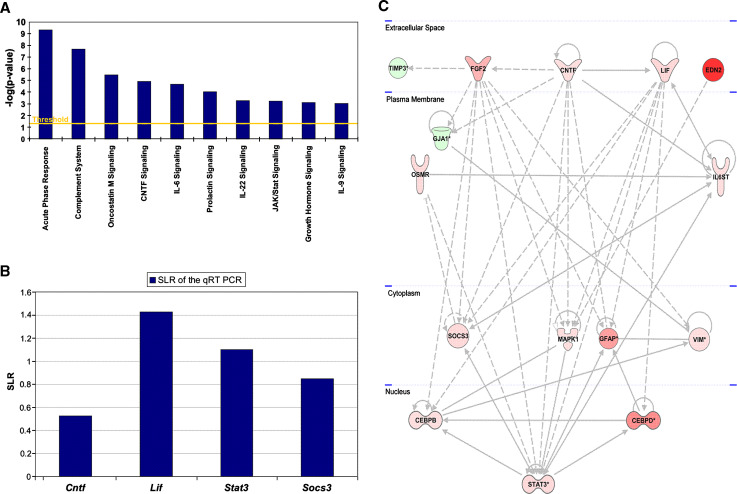
We next hypothesized that genes that were up-regulated in our microarray analysis at an early stage of degeneration in the cpfl1 retina may be involved in cell death mechanisms and/or survival mechanisms that are activated to counteract degeneration. Based on this hypothesis, we focused on the 168 up-regulated genes in 4-week-old cpfl1 animals. At this time point, degeneration of photoreceptors has just started and potential cell death and rescue signaling cascades are presumably activated. Figure 5a displays a selection of the ten primary canonical pathways, which appear to be induced in 4-week-old cpfl1 animals. A closer observation of these canonical pathways showed a remarkable impact of the signal transducer and activator of transcription 3 (Stat3) signaling since Stat3 and several components of its signaling cascade are part of all but one (complement system) of the ten identified canonical pathways. To verify this induction of Stat3-related genes, we selected three of the identified transcripts (Stat3, Socs3, and Cntf) and in addition on leukemia inhibitory factor (Lif), a member of the interleukin-6 family of cytokines, that has been shown to be frequently involved in Stat3 activation in the retina [18, 19], for further analysis by qRT-PCR. Significant up-regulation of all three genes was confirmed by qRT-PCR analysis (p < 0.05) (Fig. 5b). For Lif, we also found significant 3.5-fold up-regulation (Fig. 5b). The inability to detect this up-regulation in the previously performed microarray expression analysis may be explained by suboptimal oligonucleotide probes on the microarray. We next analyzed the full dataset (up- and down-regulated transcripts) from 4-week-old animals including Lif with the Ingenuity Pathways knowledge base matrix to generate gene regulation networks that facilitate the integration of the data into a biological context. Figure 5c shows the regulation network with the highest interaction score. Importantly, Stat3 plays a central role in this signaling network. Fifteen differently regulated genes (13 up-regulated and two down-regulated) were included, which showed 46 interactions. The network is divided into different cellular compartments and includes the down-regulated gene tissue inhibitor of metalloproteinase 3 [Timp3; fold change (FC) −2; p 0.011] as well as the up-regulated genes fibroblast growth factor 2 (Fgf2; FC 5.2; p 0.001), ciliary neurotrophic factor (Cntf; FC 1.4; p 0.01), leukemia inhibitory factor (Lif; FC 2.7; p 0.03), and endothelin 2 (Edn2; FC 27.6; p 0.001) in the extracellular space. The down-regulated gene gap junction protein, alpha-1 (Gja1; FC −3; p 0.001), as well as the up-regulated genes oncostatin-M-specific receptor subunit beta precursor (Osmr; FC 2.5; p 0.001) and interleukin-6 receptor subunit beta precursor (Il6st; FC 1.5; p 0.02) are localized in the plasma membrane. For proteins known to be localized in cytoplasm the up-regulated genes vimentin (Vim; FC 1.7; p <0.001), suppressors of cytokine signaling (Socs3; FC 2.5; p 0.001), mitogen-activated protein kinase 1 (Mapk1; FC 1.6; p 0.003) and glial fibrillary acidic protein (Gfap; FC 6.7; p <0.001) were found. The up-regulated transcription factors CCAAT/enhancer-binding protein, beta (Cebpb; FC 1.6; p 0.014) CCAAT/enhancer-binding protein, delta (Cebpd; FC 7.8; p 0.001) and signal transducer and activator of transcription 3 (Stat3; FC 2.5; p 0.047) are shown in the nuclear compartment.
Fig. 5.
a Selection of canonical pathways altered in the cpfl1 mutant and derived from a subset of all up-regulated genes included in the differential regulated gene list of the 4-week-old animals. The particular canonical pathway is plotted versus the negative logarithm of the significance. b Relative expression of genes associated with Stat3 signaling in cpfl1 mutant relative to wild-type. The diagram indicates the relative gene expression evaluated by qRT-PCR. Expression levels are displayed as signal log ratio (SLR). All genes show a significance of p ≤ 0.05. c Gene regulation network of Stat3 signaling. This network, derived from the data set of the 4-week-old cpfl1 mice, contains 15 genes connected through 46 interactions. The 13 up-regulated genes appear as red symbols, the two down-regulated genes as green icons. The color intensity correlates with the degree of regulation. Direct interactions are indicated as drawn through lines and indirect interactions as dashed connections
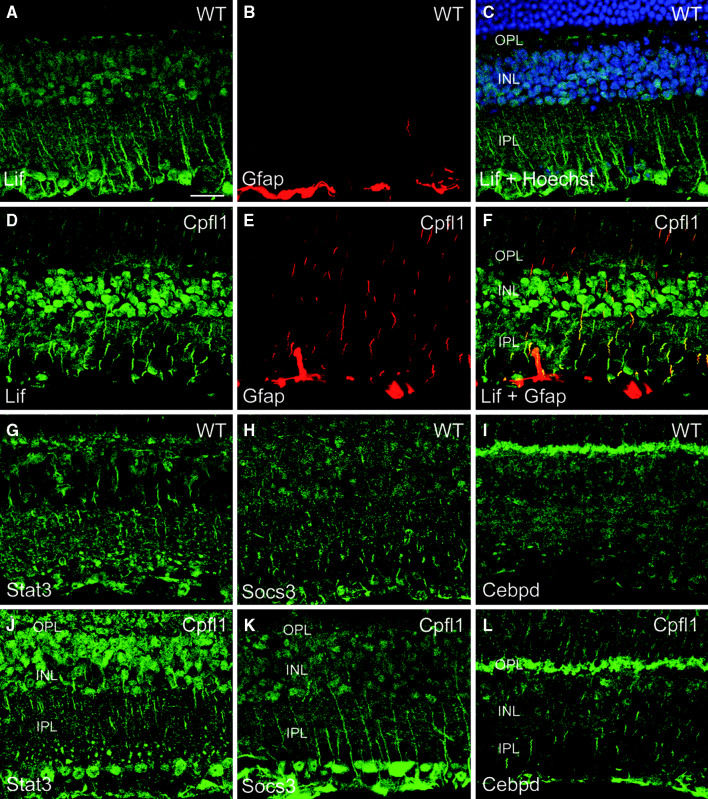
In order to localize the cells that induced Stat3 signaling in the cpfl1 mouse, we performed immunohistochemical stainings for selected proteins of this signaling cascade. LIF protein was found in Müller glia and other inner retinal cells of wild-type control mice (Fig. 6a). LIF was profoundly up-regulated (Fig. 6d) and colocalized with GFAP in the cpfl1 retina (Fig. 6e, f). We also found Müller cell expression of STAT3, SOCS3, and CEBPD (Fig. 6g–l). In accordance with the more moderate fold change in gene expression of these genes, no obvious changes were detected between cpfl1 and wild-type mice for SOCS3 and CEBPD. All four proteins tested were also expressed in cone photoreceptors (Fig. 7) and localized to the cell body, the synapse, and the inner segment (see co-labeling with glycogen phosphorylase, a marker of cone photoreceptors that labels all these structures [20]). Intriguingly, we also observed up-regulation of LIF and STAT3 in cpfl1 cone photoreceptors (Fig. 7a–b’ and c–d’). SOCS3 and CEBPD protein were only slightly if at all elevated in cpfl1 cones (Fig. 7e–f’ and g–h’). To quantify STAT3 expression in cone photoreceptors, we evaluated 14,000 μm2 in dorsal and ventral retinal areas of three wild-type and three cpfl1 animals and found in both conditions more than 90% of cone photoreceptors expressing STAT3.
Fig. 6.
Müller glia cells in the cpfl1 retina induce Lif. Confocal laser scanning micrographs of retinal sections showing immunolocalization of various markers in wild-type (a–c, g–i) and cpfl1 (d–f, j–l) mice. (a–f) GFAP-positive (red) Müller glia induce expression of Lif (green). Stat3 (g, j, green), SOCS3 (h, k, green) and Cepbd (i, l, green) are also expressed in inner retinal neurons and localize to Müller cell fibers in cpfl1 mice. Images (in b and e) show GFAP (red) labeling. In (c), cell nuclei were labeled with Hoechst nuclear dye (blue) to reveal retinal cell layers. The image in (f) represents an overlay (yellow) of the Lif (green) and GFAP (red) signals shown in d and e, respectively. Scale bar is 20 μm. INL, inner nuclear layer, IPL, inner plexiform layer, OPL, outer plexiform layer
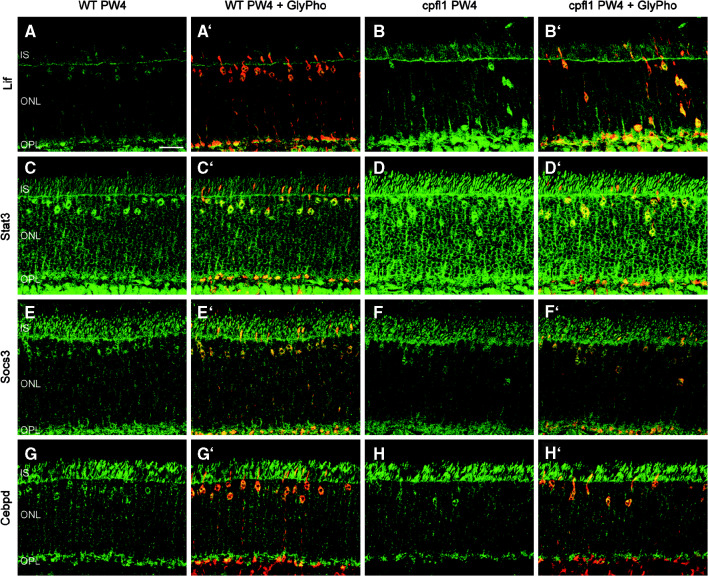
Fig. 7.
Localization of Stat3-related proteins in cpfl1 cone photoreceptors. Confocal laser scanning micrographs of retinal sections showing immunolocalization of various markers (green) in wild-type (a, c, e, and g) and cpfl1 (b, d, f, and h) at PW 4. Merged images of this signal and the labeling for the cone photoreceptor marker glycogen phosphorylase (red) are shown in a’–h’. A clear induction of Lif (b) and Stat3 (d) was observed in cpfl1 mice. Socs3 (e–f) and Cebpd (g–h) also localize to cone photoreceptors, but are only slightly up-regulated in the cpfl1 mouse. Scale bar is 20 μm. IS, photoreceptor inner segments, ONL, outer nuclear layer, OPL, outer plexiform layer
In order to elucidate the impact of increased STAT3 signaling on the expression profile of the cpfl1 mouse model, we used the GenomatixSuite software BiblioShpere and the MatInspector tool to define genes directly or indirectly associated with STAT3 transcriptional activation. The analysis revealed 24 up or down-regulated genes in the cpfl1 expression profile affecting STAT3 activation or being influenced by STAT3 signaling, which are listed in Table 1. All 19 genes showing an elevated expression level include a STAT3 binding site in their promoter region and can thus be activated by STAT3. Importantly, seven of these genes were members of the signaling network shown in Fig. 5c. The majority of the down-regulated genes on the other hand represent negative regulators of STAT3 activation.
Discussion
Neurodegenerative disorders comprise complex processes within primarily affected cells as well as interactions between affected and unaffected cells. The involved pathways are in part specific to cells and in part general, canonical pathways (like apoptosis). Thus, to achieve a more comprehensive understanding, a bridging of physiological, structural, and molecular approaches is necessary. Here, we analyzed the specific retinal neurodegeneration in the cpfl1 mouse. Recent studies have shown that mutations in human Pde6c, analogous to the molecular defect in the cpfl1 mouse, cause autosomal recessive achromatopsia or early onset progressive cone dystrophy [3, 4] making cpfl1 mice a unique murine model for this disorder. In the cpfl1 mouse, cone photoreceptors are primarily affected, leading to a fast and progressive loss of these cells. Because of this specificity and a minimum of secondary degenerative events (in contrast to those associated with rod photoreceptor degeneration), a microarray-based gene expression analysis in combination with functional and morphological data appeared particularly promising to identify cone photoreceptor-related pathologic signaling pathways.
In our ERG analysis of cpfl1 mice we observed that light responses originating from cone photoreceptors were practically non-recordable already at PW4. In contrast, rod-mediated light responses were normal in both PW4 and PW8 animals. OCT-aided morphological analysis showed no apparent differences between wild-type and cpfl1 retina, indicating that, in contrast to rod photoreceptor diseases, no general degeneration of the outer retina occurred. However, despite the low number of cone photoreceptors in the mouse retina (only 3% of all photoreceptors), we found a marked activation of Müller glia. Our microarray analysis identified activation of STAT3 signaling pathways in the cpfl1 retina. Until today, activation of STAT3 signaling has been reported for animal models with general photoreceptor degeneration or light-induced retinal damage [7, 8]. Our results provide new evidence for a combined activation of the STAT3 signaling cascade in both cone photoreceptors and Müller cells in a cone-specific degeneration model.
The activation of these signaling processes in the cpfl1 retina could possibly be initiated by the influence of Edn2, which was found to be significantly up-regulated (28-fold) in the cpfl1 retina. Edn2 is secreted in response to photoreceptor stress and is proposed to signal to Müller cells by binding to its receptor Ednrb [7]. Up-regulation of Edn2 was also observed in light-damaged retinas and in the retinas of prCAD knockout mice, a model of general photoreceptor degeneration [7]. It was also proposed that activation of Edn2 results in up-regulation of Cebpd, Gfap, Osmr, Socs3, and Stat3 in these two conditions of global photoreceptor degeneration [7]. Interestingly, we found up-regulation of all these proteins in the cpfl1 retina as well (see identified gene regulation network in Fig. 3b). The transcriptional analyses of light-damaged retinas additionally revealed an up-regulation of Cd44, Vim, Fgf2, Il6st, and Ahr [7], which were up-regulated in the four 4-week-old animals as shown by our microarray expression analysis.
Two growth factors secreted by damaged retinal neurons and Müller cells [21, 22], fibroblast growth factor 2 (Fgf2; 5.2-fold), and ciliary neurotrophic factor (Cntf; 1.4-fold) were also up-regulated in the cpfl1 retina. These growth factors are known to induce expression of the also up-regulated genes Gfap (FC 6.7) [21–24] and vimentin (Vim; FC 1.7) [25] in Müller cells. We postulate that induction of Gfap most likely derives from a combined action of Fgf2 and Cntf.
Cntf and Lif (FC 2.7), another up-regulated gene within the identified regulation network, are both known to induce Stat3 signaling to counteract induction of apoptosis [26]. In agreement with results from different mouse models of general retinal degeneration [8, 27], Stat3 (FC 2.5) is also up-regulated in the cpfl1 retina. Alternatively, activation of Stat3 signaling can be also promoted by oncostatin-M receptors [28]. This may also be the case in the cpfl1 retina, since oncostatin-M specific receptor subunit beta precursor (Osmr; FC 2.5) and interleukin-6 receptor subunit beta precursor (Il6st; FC 1.5) are also up-regulated in our model of cone-specific degeneration. In agreement with this we found up-regulation of suppressors of cytokine signaling 3 (Socs3; FC 2.5). SOCS3 associates with activated cytokine receptors (e.g., OSMR and IL6ST) probably through inhibiting janus kinases (JAK)-mediated activation of the receptors and thus causing a negative regulation of cytokine signaling. It is assumed that SOCS proteins further recruit ubiquitin ligases, causing degradation of these cytokine receptor signaling complexes [29].
In conclusion, the up-regulation of the genes Lif, Cntf, Osmr, Il6st, Stat3, and Socs3 in the cpfl1-mutant mice underlines the activation of cell rescue signaling pathways in these animals. However, at the same time, induction of this STAT3 signaling does not seem to be sufficient to rescue cone photoreceptors in the cpfl1 mouse. This is in line with observations from mouse models of genetic photoreceptor degeneration [8]. At the age of 8 weeks, the degenerative process in the cpfl1-retina has proceeded strikingly and expression levels of the Fgf2, Cntf, Osmr, Il6st, Stat3, and Socs3 have returned to normal. However, the up-regulation of Gfap and Edn2 remains. This reversal of up-regulation of these genes may be explained by two mechanisms: (1) the progress of degeneration could have proceeded to such extent that attempts of photoreceptor rescue have been abandoned and (2) the cells secreting potential stress signals have diminished to such a degree that the concentration of rescue-stimuli is not sufficient to activate the STAT signaling cascade. However, the up-regulation of Edn2, which is considered to function as stress signal [7], favors the first possibility. To our knowledge, this is the first study linking activation of this STAT3 signaling cascade to cone photoreceptor-specific degeneration and death. This suggests that the endogenous STAT3 system may be commonly activated in degenerating retinas, probably independently of the disease-causing stimulus. Targeting molecules of this signaling pathway by neuroprotective treatments may prove beneficial for the management of a large number of degenerative diseases. Our results also reveal that degenerating cone photoreceptors induce similar signaling pathways as degenerating rod photoreceptors. Since both pathologies share common rescue and cell-death pathways, common treatment strategies may be developed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Bo Chang (Jackson Laboratory) for providing us with cpfl1 animals and Brigitte Pfeiffer-Guglielmi (Univ Tübingen) for the gift of anti-glycogen phosphorylase antibody. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, grants Bo2089/1, Bo2089/2, Se837/5-2, Se837/6-1, Se837/7-1), the German Ministry of Education and Research (BMBF grant 0314106), and the European Union grants LSHG-CT-512036 and EU HEALTH-F2-2008-200234.
References
- 1.Kohl S, Varsanyi B, Antunes GA, Baumann B, Hoyng CB, Jagle H, Rosenberg T, Kellner U, Lorenz B, Salati R, Jurklies B, Farkas A, Andreasson S, Weleber RG, Jacobson SG, Rudolph G, Castellan C, Dollfus H, Legius E, Anastasi M, Bitoun P, Lev D, Sieving PA, Munier FL, Zrenner E, Sharpe LT, Cremers FP, Wissinger B. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet. 2005;13:302–308. doi: 10.1038/sj.ejhg.5201269. [DOI] [PubMed] [Google Scholar]
- 2.Kohl S, Baumann B, Rosenberg T, Kellner U, Lorenz B, Vadala M, Jacobson SG, Wissinger B. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002;71:422–425. doi: 10.1086/341835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang B, Grau T, Dangel S, Hurd R, Jurklies B, Sener EC, Andreasson S, Dollfus H, Baumann B, Bolz S, Artemyev N, Kohl S, Heckenlively J, Wissinger B. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci USA. 2009;106:19581–19586. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiadens AA, den Hollander AI, Roosing S, Nabuurs SB, Zekveld-Vroon RC, Collin RW, De Baere E, Koenekoop RK, van Schooneveld MJ, Strom TM, van Lith-Verhoeven JJ, Lotery AJ, van Moll-Ramirez N, Leroy BP, van den Born LI, Hoyng CB, Cremers FP, Klaver CC. Homozygosity mapping reveals PDE6C mutations in patients with early onset cone photoreceptor disorders. Am J Hum Genet. 2009;85:240–247. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. A mouse model of cone photoreceptor function loss (cpfl1) Invest Ophthalmol Vis Sci. 2001;42:S527. doi: 10.1167/iovs.05-1468. [DOI] [PubMed] [Google Scholar]
- 6.Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–525. doi: 10.1016/S0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 7.Rattner A, Nathans J. The genomic response to retinal disease and injury: evidence for endothelin signaling from photoreceptors to glia. J Neurosci. 2005;25:4540–4549. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samardzija M, Wenzel A, Aufenberg S, Thiersch M, Reme C, Grimm C. Differential role of Jak-STAT signaling in retinal degenerations. Faseb J. 2006;20:2411–2413. doi: 10.1096/fj.06-5895fje. [DOI] [PubMed] [Google Scholar]
- 9.Tanimoto N, Muehlfriedel RL, Fischer MD, Fahl E, Humphries P, Biel M, Seeliger MW. Vision tests in the mouse: functional phenotyping with electroretinography. Front Biosci. 2009;14:2730–2737. doi: 10.2741/3409. [DOI] [PubMed] [Google Scholar]
- 10.Marmor MF, Holder GE, Seeliger MW, Yamamoto S. Standard for clinical electroretinography (2004 update) Doc Ophthalmol. 2004;108:107–114. doi: 10.1023/B:DOOP.0000036793.44912.45. [DOI] [PubMed] [Google Scholar]
- 11.Biel M, Seeliger M, Pfeifer A, Kohler K, Gerstner A, Ludwig A, Jaissle G, Fauser S, Zrenner E, Hofmann F. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci USA. 1999;96:7553–7557. doi: 10.1073/pnas.96.13.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeliger MW, Grimm C, Stahlberg F, Friedburg C, Jaissle G, Zrenner E, Guo H, Reme CE, Humphries P, Hofmann F, Biel M, Fariss RN, Redmond TM, Wenzel A. New views on RPE65 deficiency: the rod system is the source of vision in a mouse model of Leber congenital amaurosis. Nat Genet. 2001;29:70–74. doi: 10.1038/ng712. [DOI] [PubMed] [Google Scholar]
- 13.Jaissle GB, May CA, Reinhard J, Kohler K, Fauser S, Lutjen-Drecoll E, Zrenner E, Seeliger MW. Evaluation of the rhodopsin knockout mouse as a model of pure cone function. Invest Ophthalmol Vis Sci. 2001;42:506–513. [PubMed] [Google Scholar]
- 14.Fischer MD, Huber G, Beck SC, Tanimoto N, Muehlfriedel R, Fahl E, Grimm C, Wenzel A, Remé CE, van de Pavert SA, Wijnholds J, Pacal M, Bremner R, Seeliger MW. Noninvasive, in vivo assessment of mouse retinal structure using optical coherence tomography. PLoS One. 2009;19(4):e7507. doi: 10.1371/journal.pone.0007507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf-Schnurrbusch UE, Enzmann V, Brinkmann CK, Wolf S. Morphologic changes in patients with geographic atrophy assessed with a novel spectral OCT-SLO combination. Invest Ophthalmol Vis Sci. 2008;49:3095–3099. doi: 10.1167/iovs.07-1460. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalakis S, Geiger H, Haverkamp S, Hofmann F, Gerstner A, Biel M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci. 2005;46:1516–1524. doi: 10.1167/iovs.04-1503. [DOI] [PubMed] [Google Scholar]
- 18.Ueki Y, Wang J, Chollangi S, Ash JD. STAT3 activation in photoreceptors by leukemia inhibitory factor is associated with protection from light damage. J Neurochem. 2008;105:784–796. doi: 10.1111/j.1471-4159.2007.05180.x. [DOI] [PubMed] [Google Scholar]
- 19.Joly S, Lange C, Thiersch M, Samardzija M, Grimm C. Leukemia inhibitory factor extends the lifespan of injured photoreceptors in vivo. J Neurosci. 2008;28:13765–13774. doi: 10.1523/JNEUROSCI.5114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeiffer-Guglielmi B, Fleckenstein B, Jung G, Hamprecht B. Immunocytochemical localization of glycogen phosphorylase isozymes in rat nervous tissues by using isozyme-specific antibodies. J Neurochem. 2003;85:73–81. doi: 10.1046/j.1471-4159.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 21.Fischer AJ, Omar G, Eubanks J, McGuire CR, Dierks BD, Reh TA. Different aspects of gliosis in retinal Müller glia can be induced by CNTF, insulin, and FGF2 in the absence of damage. Mol Vis. 2004;10:973–986. [PubMed] [Google Scholar]
- 22.Wahlin KJ, Campochiaro PA, Zack DJ, Adler R. Neurotrophic factors cause activation of intracellular signaling pathways in Müller cells and other cells of the inner retina, but not photoreceptors. Invest Ophthalmol Vis Sci. 2000;41:927–936. [PubMed] [Google Scholar]
- 23.Kahn MA, Huang CJ, Caruso A, Barresi V, Nazarian R, Condorelli DF, de Vellis J. Ciliary neurotrophic factor activates JAK/Stat signal transduction cascade and induces transcriptional expression of glial fibrillary acidic protein in glial cells. J Neurochem. 1997;68:1413–1423. doi: 10.1046/j.1471-4159.1997.68041413.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Smith SB, Ogilvie JM, McCool DJ, Sarthy V. Ciliary neurotrophic factor induces glial fibrillary acidic protein in retinal Müller cells through the JAK/STAT signal transduction pathway. Curr Eye Res. 2002;24:305–312. doi: 10.1076/ceyr.24.4.305.8408. [DOI] [PubMed] [Google Scholar]
- 25.Lewis GP, Erickson PA, Guerin CJ, Anderson DH, Fisher SK. Basic fibroblast growth factor: a potential regulator of proliferation and intermediate filament expression in the retina. J Neurosci. 1992;12:3968–3978. doi: 10.1523/JNEUROSCI.12-10-03968.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 27.Mechoulam H, Pierce EA. Expression and activation of STAT3 in ischemia-induced retinopathy. Invest Ophthalmol Vis Sci. 2005;46:4409–4416. doi: 10.1167/iovs.05-0632. [DOI] [PubMed] [Google Scholar]
- 28.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 29.Krebs DL, Hilton DJ. SOCS physiological suppressors of cytokine signaling. J Cell Sci. 2000;113(Pt 16):2813–2819. doi: 10.1242/jcs.113.16.2813. [DOI] [PubMed] [Google Scholar]
- 30.Yin F, Li P, Zheng M, Chen L, Xu Q, Chen K, Wang YY, Zhang YY, Han C. Interleukin-6 family of cytokines mediates isoproterenol-induced delayed STAT3 activation in mouse heart. J Biol Chem. 2003;278:21070–21075. doi: 10.1074/jbc.M211028200. [DOI] [PubMed] [Google Scholar]
- 31.Arguni E, Arima M, Tsuruoka N, Sakamoto A, Hatano M, Tokuhisa T. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int Immunol. 2006;18:1079–1089. doi: 10.1093/intimm/dxl041. [DOI] [PubMed] [Google Scholar]
- 32.von Gertten C, Flores Morales A, Holmin S, Mathiesen T, Nordqvist AC. Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci. 2005;6:69. doi: 10.1186/1471-2202-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka M, Tagoku K, Russell TL, Nakano Y, Hamazaki T, Meyer EM, Yokota T, Terada N. CD9 is associated with leukemia inhibitory factor-mediated maintenance of embryonic stem cells. Mol Biol Cell. 2002;13:1274–1281. doi: 10.1091/mbc.02-01-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol Cell Biol. 1998;18:2108–2117. doi: 10.1128/mcb.18.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura H, Kamon H, Sawa S, Park SJ, Katunuma N, Ishihara K, Murakami M, Hirano T. IL-6-STAT3 controls intracellular MHC class II alphabeta dimer level through cathepsin S activity in dendritic cells. Immunity. 2005;23:491–502. doi: 10.1016/j.immuni.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Cao H, Dronadula N, Rizvi F, Li Q, Srivastava K, Gerthoffer WT, Rao GN. Novel role for STAT-5B in the regulation of Hsp27-FGF-2 axis facilitating thrombin-induced vascular smooth muscle cell growth and motility. Circ Res. 2006;98:913–922. doi: 10.1161/01.RES.0000216954.55724.a2. [DOI] [PubMed] [Google Scholar]
- 37.Puxeddu E, Knauf JA, Sartor MA, Mitsutake N, Smith EP, Medvedovic M, Tomlinson CR, Moretti S, Fagin JA. RET/PTC-induced gene expression in thyroid PCCL3 cells reveals early activation of genes involved in regulation of the immune response. Endocr Relat Cancer. 2005;12:319–334. doi: 10.1677/erc.1.00947. [DOI] [PubMed] [Google Scholar]
- 38.Sriram K, Benkovic SA, Hebert MA, Miller DB, O’Callaghan JP. Induction of gp130-related cytokines and activation of JAK2/STAT3 pathway in astrocytes precedes up-regulation of glial fibrillary acidic protein in the 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine model of neurodegeneration: key signaling pathway for astrogliosis in vivo? J Biol Chem. 2004;279:19936–19947. doi: 10.1074/jbc.M309304200. [DOI] [PubMed] [Google Scholar]
- 39.Gu F, Hata R, Ma YJ, Tanaka J, Mitsuda N, Kumon Y, Hanakawa Y, Hashimoto K, Nakajima K, Sakanaka M. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res. 2005;81:163–171. doi: 10.1002/jnr.20561. [DOI] [PubMed] [Google Scholar]
- 40.Okuda H, Manabe T, Yanagita T, Matsuzaki S, Bando Y, Katayama T, Wanaka A, Tohyama M. Novel interaction between HMGA1a and StIP1 in murine terminally differentiated retina. Mol Cell Neurosci. 2006;33:81–87. doi: 10.1016/j.mcn.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Selander KS, Li L, Watson L, Merrell M, Dahmen H, Heinrich PC, Müller-Newen G, Harris KW. Inhibition of gp130 signaling in breast cancer blocks constitutive activation of Stat3 and inhibits in vivo malignancy. Cancer Res. 2004;64:6924–6933. doi: 10.1158/0008-5472.CAN-03-2516. [DOI] [PubMed] [Google Scholar]
- 42.Oshima Y, Fujio Y, Nakanishi T, Itoh N, Yamamoto Y, Negoro S, Tanaka K, Kishimoto T, Kawase I, Azuma J. STAT3 mediates cardioprotection against ischemia/reperfusion injury through metallothionein induction in the heart. Cardiovasc Res. 2005;65:428–435. doi: 10.1016/j.cardiores.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Ozawa Y, Nakao K, Kurihara T, Shimazaki T, Shimmura S, Ishida S, Yoshimura A, Tsubota K, Okano H. Roles of STAT3/SOCS3 pathway in regulating the visual function and ubiquitin-proteasome-dependent degradation of rhodopsin during retinal inflammation. J Biol Chem. 2008;283:24561–24570. doi: 10.1074/jbc.M802238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiffen PG, Omidvar N, Marquez-Almuina N, Croston D, Watson CJ, Clarkson RW. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol Endocrinol. 2008;22:2677–2688. doi: 10.1210/me.2008-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E, 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Sylvester R, Tighe AP, Chen S, Gudas LJ. Transcriptional activation of the suppressor of cytokine signaling-3 (SOCS-3) gene via STAT3 is increased in F9 REX1 (ZFP-42) knockout teratocarcinoma stem cells relative to wild-type cells. J Mol Biol. 2008;377:28–46. doi: 10.1016/j.jmb.2007.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Diab I, Zhang X, Izmailova ES, Zehner ZE. Stat3 enhances vimentin gene expression by binding to the antisilencer element and interacting with the repressor protein, ZBP-89. Oncogene. 2004;23:168–178. doi: 10.1038/sj.onc.1207003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.