Abstract
The subfamily of WNK protein kinases is composed of four human genes and is characterised by a typical sequence variation within the conserved catalytic domain. Although most research has focussed on the role of WNK1, WNK3 and WNK4 in regulating different ion transporters in both the kidney and extrarenal tissues, there is growing evidence for additional roles of WNK kinases in various signalling cascades related to cancer. Here, we review the connection between WNK kinases and tumorigenesis and describe existing experimental evidence as well as potential new links to major aspects of tumour biology. In particular, we discuss their role in G1/S cell cycle progression, metabolic tumour cell adaptation, evasion of apoptosis and metastasis.
Keywords: Apoptosis, Cell proliferation, Signal transduction, Tumorigenesis, WNK protein kinases
Introduction
Protein phosphorylation is a major mechanism to regulate the activity or function of cellular proteins. Protein kinases mediate this reaction and a total of 518 different protein kinase genes have been identified in the human genome [1–3]. These kinases share highly conserved sequence elements in their catalytic domains and form a gene superfamily. The most typical 428 protein kinases fall into seven major families (TK, CAMK, AGC, CMGC, STE, TKL and CK1), except for 63 kinases that present slight sequence variations in their catalytic domains and have been classified separately as ‘Other’ [3, 4]. This group includes the WNK kinases, a phylogenetically separate protein kinase branch, most closely related to the STE (mammalian homologs of the Saccharomyces cerevisiae STE families of serine/threonine kinases) and TKL (tyrosine kinase-like) family branches [1, 2].
The WNK subfamily of protein kinases
The sequence variation in the catalytic domain that characterises WNK protein kinases is the lack of a highly conserved catalytic lysine in subdomain II, originating their name with no [K] = lysine [5, 6]. Although this lysine is important for the correct positioning of adenosine triphosphate (ATP) within the typical catalytic domain [4], the determination of the recombinant WNK1 kinase domain structure has revealed an alternative lysine from subdomain I that reaches into the position normally occupied by the conserved lysine residue from subdomain II and this confers catalytic activity [5, 7] (Fig. 1a). This sequence variation induces a conformational change in the catalytic domain, likely responsible for unique substrate binding properties of WNK kinases, but also offering a unique target for the development of specific small molecule kinase inhibitors.
Fig. 1.
Sequence features of the WNK subfamily of protein kinases. a Comparison of the sequence in subdomains I and II of the catalytic domain between a typical protein kinase and the WNK domain. A conserved lysine in subdomain II, which binds ATP, is absent in WNK kinases and functionally substituted by another lysine located in subdomain I [5, 7], as indicated. b The invariant WNK signature sequence identified from the sequence alignment shown in (c). c Alignment of subdomains I and II of the catalytic domains of WNK kinases from various species. Species abbreviations are: hs Homo sapiens, mm Mus musculus, at Arabidopsis thaliana, rn Rattus norvegicus, pt Pan troglodytes, clf Canis lupus familiaris, gg Gallus gallus, dm Drosophila melanogaster, ce Caenorhabditis elegans (updated from Veríssimo and Jordan 6)
The characteristic catalytic lysine in subdomain I together with seven other adjacent amino acid residues form an invariant WNK signature sequence [6] (Fig. 1b). Searching the Genbank database using this WNK signature sequence revealed the existence of a variable number of WNK genes in animals and plants but none in unicellular organisms (Fig. 1c). The number of WNK genes increases from one in Caenorhabditis elegans or Drosophila melanogaster to four different WNK genes in mouse and man. The higher plant Arabidopsis thaliana has nine WNK genes [6, 8]. The properties of the four WNK genes in the human genome are compared in Table 1.
Table 1.
Comparison of molecular characteristics of the four human WNK genes and their expression
| Gene | Chromosome | Size (Mb) | Exon nb. | Codons | MW (kDa) | Expression in tissues | Other features | References |
|---|---|---|---|---|---|---|---|---|
| WNK1 | 12p13.3 | 150 | 28 | 2,382 | 251 | Widespread | Alternative splicing in exons 9, 11 and 12. | [5, 6, 73, 103, 113] |
| Multiple Promoters | [102] | |||||||
| WNK2 | 9q22.31 | 136 | 31 | 2,297 | 243 | Brain, heart, colon | Alternative C-terminal exon (either 30 or 31) | [17, 103] |
| WNK3 | Xp11.23–21 | 165 | 24 | 1,743 | 192 | Brain, liver, small intestine | Alternative splicing in exons 18 and 22 | [86, 103, 107, 114] |
| WNK4 | 17q21–q22 | 16 | 19 | 1,243 | 135 | Kidney, skin, colon, liver, lung | Not described | [73, 79, 103] |
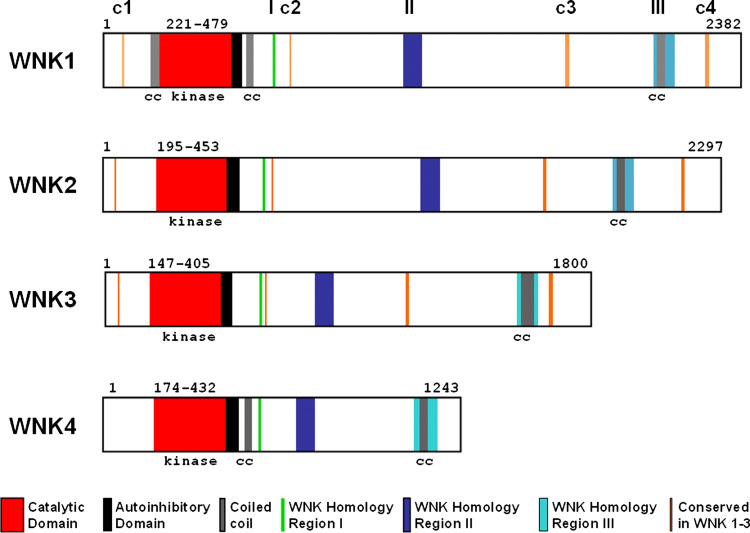
WNK proteins are serine/threonine kinases that undergo autophosphorylation and require phosphorylation of at least one serine residue within the WNK activation loop to become active [9, 10]. This serine has been identified as S382 in WNK1 and is part of an activation site motif (S378FAKS382) that is conserved in MEKK family kinases. Homologous serine residues are found in positions 356, 308 and 332 of WNK2, WNK3 and WNK4, respectively. Another primary level in the regulation of WNK kinase activity is provided by an autoinhibitory domain encompassing about 70 residues just C-terminal to the catalytic domain [9, 11]. The four WNK proteins share high homology within their kinase domains and the adjacent auto-inhibitory domain [9]; however, homology outside their catalytic domains is low except for the presence of three short WNK homology regions (Fig. 2). The most C-terminal homology region III contains a coiled-coil domain, and multiple PXXP motifs, the typical binding sites for Src-homology 3 (SH3) domains [12], are also present in WNK protein kinases. These features together with their large size suggest that these proteins also have a scaffolding function and are involved in multiple cellular signalling processes [6, 13].
Fig. 2.
Schematic representation of the four human WNK proteins. Catalytic, autoinhibitory and coiled-coil domains are shown as well as conserved regions of homology between WNK kinases
Role of WNK kinases in cell cycle progression and proliferation
Sequence alignments using the catalytic domain of WNK1 showed that the closest human homologous kinases are MEKK, Raf and PAK, which all display around 50% sequence homology [6]. They belong to the mitogen-activated protein kinases (MAPKs), the activation of which is intimately linked to the transition from a quiescent to a proliferative state in epithelial cells. The best characterised MAPK pathways in mammals involve the extracellular signal-regulated kinases 1 and 2 (ERK1/2), the p38 isoforms, the c-jun N-terminal kinases (JNK1-3) and ERK5. In general, MAPK cascades consist of three protein kinase layers: MAPKs are phosphorylated and activated by MAPK kinases (MAP2Ks) which are in turn activated by phosphorylation by MAPK kinase kinases (MAP3Ks) in response to growth factor stimulation. Other kinases, including p21-activated protein kinases (PAKs), can act as upstream regulators (MAP4Ks) for these MAPK cascades and connect them with additional signalling pathways. MAPK signalling is deregulated in approximately one-third of all human cancers [14].
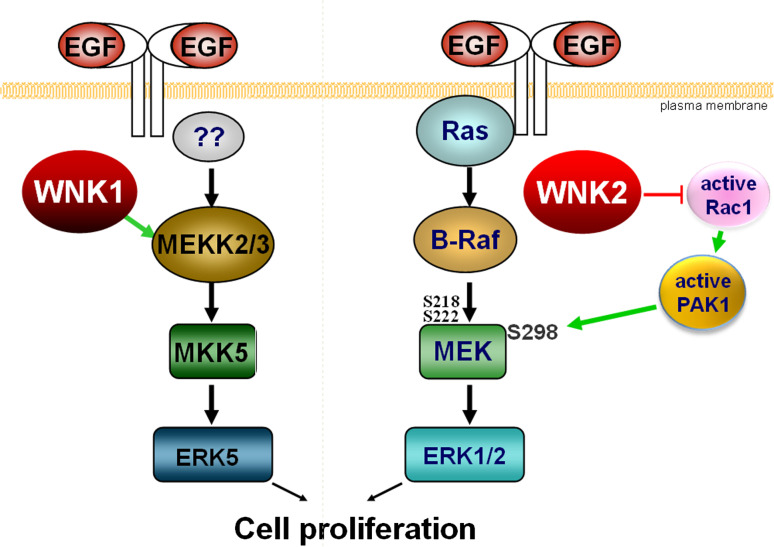
WNK kinases affect various MAPK cascades. WNK1 was reported to be required for EGF-dependent stimulation of ERK5 without affecting the activation of ERK1/2, JNK or p38 MAP kinases [15]. Transfection of cells with either WNK1 or its kinase-dead mutant WNK1K233M stimulates MEKK3 autophosphorylation and activity towards its substrate MEK6. In vitro, WNK1 interacts with and phosphorylates MEKK2 and MEKK3; however, this phosphorylation does not seem to stimulate MEKK2/3 activity. Apparently, WNK1 acts by protein–protein interaction to assemble an ERK5 activation complex and thus acts as an upstream regulator of the ERK5 pathway (Fig. 2). WNK1 was required for activation of ERK5 by EGF in HeLa cells, but high concentrations of EGF can override this effect [15]. In agreement with these findings, the downregulation of WNK1 in C17.2 mouse neural progenitor cells also suppressed activation of ERK5 and greatly reduced cell growth [16].
In contrast to WNK1, human WNK2 has no effect on ERK5 but modulates the activation level of ERK1/2. Experimental depletion of WNK2 or overexpression of a kinase-dead WNK2K207M mutant led to increased phospho-ERK1/2 levels when a basal ERK stimulation was present but not, for example, in serum-free culture conditions [17]. This increase in ERK1/2 activation promoted cell cycle progression through G1/S and sensitised cells to respond to lower concentrations of EGF. From these data, one might predict that loss of WNK2 expression will promote cell cycle progression in tumour cells. Interestingly, WNK2 expression is silenced in a significant percentage of human gliomas ([18, 19]; see also below) suggesting that this pathway may be used in some tumour types to promote cell proliferation. The molecular mechanism through which a reduction in WNK2 expression can increase ERK1/2 activation involves phosphorylation of MEK1 at serine 298, a modification that increases MEK1 affinity towards ERK1/2. Apparently, WNK2 affects PAK1 activation via Rac1, and PAK1 is the kinase responsible for MEK1 S298 phosphorylation [20].
Overexpression of WNK4 was also claimed to increase the phosphorylation of ERK1/2 and p38 MAPKs following EGF stimulation or hyperosmotic stress in HEK293 cells [21]. However, the mechanism of WNK4 action remains unclear and the corresponding effect of endogenous WNK4 was not addressed.
Together, these data demonstrate that WNK kinases modulate cell proliferation and act upon different MAPK cascades (Fig. 3). Intriguingly, their actions are likely to be concerted. This can be deduced from the observation that WNK kinases regulate each other’s activities. For example, the recombinant kinase domain of WNK1 was shown to phosphorylate recombinant WNK2 and WNK4 catalytic domains in vitro, in particular WNK4 on serine 332 in its activation loop [22]. WNK4 can also phosphorylate the catalytic domain of WNK1 and expression of the autoinhibitory domains of WNK1 and WNK4 are able to inhibit each others catalytic activities [11, 22, 23]. Gel filtration profiles further indicate that endogenous WNK1 exists as a tetramer [22], and hetero-complexes of different WNKs have been observed by co-immunoprecipitation ([23], authors’ unpublished data). Most likely, the conserved coiled-coil protein interaction domain present in the four human WNK proteins mediates their interaction. In this way, the activity of individual WNK proteins on different MAPK cascades could become integrated into a coordinated response [13].
Fig. 3.
Schematic representation of the role of human WNK1 and WNK2 in the cellular response to EGF. Left side physiological activation of the ERK5 pathway is enhanced in the presence of WNK1. Right side expression of WNK2 controls a Rho GTPase/PAK-mediated signalling cross-talk that can increase ERK1/2 activation
WNKs furthermore regulate the STE20-related protein kinases that function upstream of the described MAPK pathways (i.e. as MAP4Ks). These include PAK and the germinal centre kinases (GCKs) proline–alanine-rich kinase (SPAK) or oxidative-stress responsive 1 (OSR1, also referred to as OXSR1). Members of the GCK subfamily are primarily known for their role in cell volume sensing and osmotic homeostasis, a process often linked to cell cycle progression [24, 25]. For example, SPAK is a known activator of the p38 and JNK cascades [26–28] and OSR1 acts upstream of CaMKII and p38 in C. elegans [29].
OSR1 and SPAK interact with WNK1, WNK3 and WNK4 [30–36], and WNK1 and WNK4 were shown to phosphorylate these proteins on two residues: T233 and S373 in SPAK and T185 and S325 in OSR1. Residues T233 or T185 are located in the activation loops of these kinases’ catalytic domains and mediate their activation [30]. The molecular details of how the phosphorylation of SPAK and OSR1 by WNK kinases relates to cell proliferation have not yet been investigated.
As mentioned above, WNK2 also acts upon another STE20 member, PAK1, but direct phosphorylation does not seem to be involved. Rather, WNK2 affects activation of Rac1, a known upstream stimulator of PAK1 [20], and active PAK1 phosphorylates MEK1 on S298, leading to increased activity of ERK1/2 [37–41].
There is indirect evidence suggesting that WNKs can also affect growth factor receptors which act upstream of MAP kinases. After ligand binding, most receptors become internalised into signalling-competent endosomes and then either recycle back to the cell surface or become degraded in lysosomes as a mechanism to downregulate receptor signalling in cells [42, 43]. Vesicular traffic can therefore modulate cellular signalling intensity, and a role for WNKs in this process has emerged. WNK1 and WNK4 have been shown to stimulate clathrin-dependent endocytosis of the renal outer medullar potassium 1 channel (ROMK1), thus inhibiting potassium secretion [46–48]. This process involves the interaction of WNK1 and WNK4 with the scaffold protein intersectin 1 (ITSN1), which binds to specific proline-rich WNK motifs via its SH3 domains and does not require WNK kinase activity [48]. Moreover, expression of WNK4 inhibited the activity of the sodium chloride cotransporter NCC by diverting the channel to lysosomal degradation [44, 45]. This effect involved a general adaptor protein (AP) 3-dependent endosomal sorting mechanism so that WNK4 may equally be able to modulate the ratio of degradation of endocytosed growth factor receptors. Additional support for a role of WNKs in endocytosis comes from a genome-wide RNAi screen to determine the effect of human protein kinases on vesicle traffic. In this screen, virus entry was used as a measure of clathrin-mediated (vesicular stomatitis virus, VSV) or caveolin-dependent (simian virus, SV40) endocytosis, and it was found that WNK4 interfered with VSV entrance whereas WNK2 inhibited caveolin-mediated SV40 uptake [49]. These data are highly suggestive for a role of WNKs in growth factor receptor turnover and should be further explored.
Many tissues and derived cancer cells also express receptors for insulin and insulin-like growth factor 1 (IGF1) [50]. Following IGF1 treatment of cells, Akt is activated downstream of phosphatidylinositol 3-kinase (PI3K) and PDK1 and phosphorylates WNK1 on threonine residue 60, although this phosphorylation does not appear to affect WNK1 kinase activity [51–55]. When 3T3-L1 adipocytes were treated with insulin, an increased WNK1 phosphorylation was observed, and RNAi-induced depletion of WNK1 enhanced insulin-stimulated thymidine incorporation by about twofold without significantly affecting insulin-stimulated glucose transport in these cells [52]. This observation suggests a negative regulatory role for WNK1 in insulin-mediated proliferation control.
IGF1 also activates the Akt-homologous serum- and glucocorticoid-induced protein kinase SGK1. Xu and collaborators [53] proposed that WNK1 phosphorylation by Akt contributes to SGK1 activation by IGF1. While SGK1 is known to regulate sodium transport in the kidney, it also promotes degradation of the cyclin-dependent kinase inhibitor protein p27kip in other cell types [56].
Growth inhibitory signals are transmitted by transforming growth factor (TGF) β in many cell types [57]. (TGF) β stimulates the formation of heteromeric complexes of type I and type II serine/threonine kinase receptors which then phosphorylate and activate cytoplasmic transcription activators of the Smad family. WNK1 and WNK4 kinase domains both interacted with and phosphorylated Smad2 in vitro. The siRNA-mediated knockdown of WNK1 increased Smad2/3-dependent transcriptional responses [58]. This suggests that WNK1 imposes an inhibitory constraint on Smad2 and TGFβ signalling. Interestingly, a systematic mapping of the TGFβ receptor interactome revealed a link to another WNK1 substrate, the OSR1 protein kinase [59], but the functional relevance of this finding for cell proliferation has not been tested.
Cancer cell proliferation can further be promoted through an increase in cytoplasmic calcium, which serves as a second messenger for calmodulin- and calcineurin-activated signalling pathways [60, 61]. One mechanism of increasing cytoplasmic calcium is uptake from the extracellular medium by calcium-channels, such as the transient receptor potential vanilloid (TRPV) channels, and TRPV6 overexpression has been reported in carcinomas of the colon, breast, thyroid and ovary [62]. Expression of WNK4 specifically enhanced TRPV5-mediated calcium uptake, which correlates with increased membrane expression of the channel [63], whereas WNK3 stimulated TRPV5 and TRPV6-mediated transport [64]. In contrast, WNK4 and WNK1 decreased cell surface expression of TRPV4, which plays a role in osmoregulation following hypotonic cell swelling [65].
Based on these data, we suggest that WNK kinases can affect multiple signalling pathways involved in the proliferation of cells from various tissue origins. At present, the most solid experimental evidence concerns their role as upstream regulators of MAPK cascades.
WNK kinases and metabolic adaptation of tumour cells
Cell adaptation to the extracellular environment is required during the initiation of tumour formation when cells start to proliferate and face suboptimal supply of oxygen and nutrients due to diffusion limits. In response, cancer cells upregulate aerobic glycolysis leading to the generation of lactate and acidosis of the extracellular medium. Tumour cells have to compensate for this acid-induced toxicity in order to remain viable and continue to grow [66, 67].
One adaptation is to increase glucose transport into malignant cells by overexpression of glucose transporters (GLUTs) [68], and elevated expression of some GLUTs has been described in many cancers.
In 3T3L1 adipocytes, the syntaxin 4-SNARE complex functions in insulin-stimulated GLUT4 vesicle translocation and fusion [69]. WNK1 recruits the syntaxin 4-inhibitory protein Munc18c via its kinase domain without phosphorylating, and thus promotes Glut4 translocation [70]. Interestingly, increased glucose supply decreased the expression of WNK4 in mouse kidneys, indicating the existence of a regulatory mechanism that links WNK expression to extracellular glucose concentrations [71]. In insulin secreting pancreatic β cells, WNK1 also phosphorylates another protein implicated in vesicle fusion events, synaptotagmin [72]. The interaction with Munc18c and synaptotagmins provides a putative mechanism for how WNKs may regulate retention or insertion of plasma membrane glucose transporters.
WNK kinases are also important for maintaining fundamental cell functions related to electrolyte homeostasis. Mutations in the WNK1 and WNK4 genes were initially discovered to cause pseudo-hypoaldosteronism type II (or Gordon’s syndrome), a rare familial form of hypertension [73] characterised by increased renal salt reabsorption accompanied by hyperkalemia and metabolic acidosis due to impaired K+ and H+ excretion. Subsequent studies have shown that WNK1, WNK3 and WNK4 are involved in the regulation of a variety of renal but also extra-renal ion channels. Summarising these studies, evidence links WNKs to the regulation of cell surface expression or channel activity of the cotransporters NCC, NKCC and KCC, the ROMK and Kir1.1 potassium channels, the CFTR chloride channel, the Cl−/HCO3 − exchangers SLC26A6 and A9, the epithelial sodium channel (ENaC), and the TRPV4 and TRPV5 calcium channels (reviewed in [74–77]). Electrolyte homeostasis is an important issue for tumour cells, especially regarding pH regulation. The extracellular environment in the tumour is more acidic than the intracellular pH (6.2–6.9 compared with 7.3–7.4). The increased production and export of glycolytic creates a reversed pH gradient across the tumour cell membrane [78]. Through overexpression of the Na+/H+ exchanger NHE1, the inwardly directed sodium gradient can drive the uphill extrusion of protons and alkalinise intracellular pH but further acidifies the extracellular pH. A supporting activity in pH regulation is provided by the chloride/bicarbonate (Cl−/HCO3−) anion exchangers of the SLC26A family. The combined transport activities of NHE and SLC26A channels lead to electroneutral NaCl absorption with net H+ secretion. Recent findings suggest that the expression of the SLC26A family is regulated by WNKs. In Xenopus oocytes, WNK4 affects the plasma membrane expression of SLC26A6 [79] and WNK1, WNK3 and WNK4 regulate the expression of SLC26A9 [80].
Electrolyte homeostasis is also important for the electrochemical gradient across the plasma membrane which most cells require to drive Ca2+ entry, sustain nutrient uptake or release metabolic products. For example, the electrochemical gradient for Na+ drives the uptake of nutrients and is in part maintained by the negative membrane potential. K+ ions need to be expelled through membrane potassium channels in order to keep the Na+/K+ ATPase going. A solid body of evidence documents that various types of tumours overexpress voltage-gated K+ channels, in particular the ether-a-go-go (EAG) and BK (Ca2+-activated K+ channels) subfamilies. These channels open in response to a depolarisation of the cell membrane, thus allowing an efflux of K+ ions. Experimental overexpression of these channels promotes tumour cell proliferation [81] and potassium ionophores such as salinomycin or nigericin are toxic to some cancer cells [82], indicating an important connection between the membrane potential and cell survival. Although WNK kinases have not yet been linked directly to the regulation of voltage-gated K+ channels, high extracellular concentrations of NaCl or KCl were shown to provoke a marked and reproducible increase in WNK1 activity [5, 9, 10, 22]. Together, these data lead us to predict that voltage-gated K+ channels represent a further link between WNKs and tumour biology.
Role of WNK kinases in the evasion of apoptosis
The apoptotic program is a major barrier to tumour development that must be inactivated to achieve net tumour cell proliferation. A complex interplay between different pro- or anti-apoptotic factors determines whether cells survive or activate the cell death programme [83]. Experimental data and mathematical models have suggested that apoptosis activation depends on molecular threshold values that trigger either the cell death pathway or maintain cell survival [84, 85]. Thus, either over-production of anti-apoptotic factors, or loss of expression of pro-apoptotic factors, or changes in their activation status can shift the balance towards cell survival. WNK3 has been proposed to act on this balance by promoting cell survival in a caspase-3-dependent pathway [86]. Suppression of endogenous WNK3 by RNA interference accelerated the apoptotic response of HeLa cells and promoted the activation of caspase-3. The mechanism of WNK3 action involves interaction with procaspase-3 and heat-shock protein 70. The prosurvival role was only in part dependent on the catalytic activity of WNK3 because a kinase-dead WNK3K159M mutant also interacted with procaspase-3 and increased cell survival to some extent. This indicates an adaptor or scaffold function for WNK3 within a protein complex that controls procaspase-3 activation. Accordingly, the level of WNK3 expression or activity will determine sensitivity or tolerance of cells towards apoptotic stimuli.
At the genome-wide level, the role of protein phosphorylation in apoptosis induction has been recently studied by systematic depletion of each human protein kinase or phosphatase in HeLa cells using RNA interference [87]. This approach identified 73 kinases whose suppression increased the level of apoptosis by at least twofold over control, defining them as survival kinases. In this screen, WNK1 and WNK3 kinases also scored positive (WNK1––1.98-fold; WNK3––1.58-fold) whereas WNK2 and WNK4 had no effect on cell survival (J. Blenis, Boston, personal communication). These results are further supported by a genome-wide screen for cell survival factors in Drosophila melanogaster. The single Drosophila WNK gene (designated CG7177; NM_141072) is most homologous to WNK3 and WNK1, and its depletion by RNAi affected cell survival in fly S2R+ cells [88].
Although WNK1 and WNK3 were detected in these screens, they did not stand out as essential genes for sustaining cell survival. However, these screens scored for spontaneous induction of apoptosis. A different physiological response can be expected when cells are exposed to conditions that challenge cell survival. For example, the single C. elegans WNK kinase (designated C46C2) was found essential for worm survival but only following hyperosmotic stress conditions [89]. The survival of C. elegans under osmotic stress depends on the WNK substrate OSR1 [29], which in mammalian cells regulates ion homoeostasis and volume control [90, 91]. We thus predict that the roles of WNK1 and WNK3 in cell survival will become more evident when cells face metabolic stress situations, including the above-mentioned glycolytic acidosis and cell volume regulation.
WNK kinases in invasion and metastasis
Cancer cells can escape from the physiological stress constraints of the primary tumour and enter into the circulation to reach distant organs and form metastases.
In many solid tumour types, the epithelial tumour cells switch to a highly motile fibroblastoid or mesenchymal phenotype, a process called epithelial–mesenchymal transition (EMT). A well-characterised inducer of EMT is TGFβ signalling [92, 93] that uses Smad-mediated gene expression to induce the transcription factors Snail and Slug. These repress expression of the E-cadherin gene required for epithelial cell adhesion, a hallmark phenotype of EMT [94]. Since WNK1 was shown to phosphorylate Smad2 in vitro and apparently negatively controls Smad2/3-dependent transcriptional responses and TGFβ signalling [58], this suggests that loss of expression or inactivating WNK1 mutations could promote EMT of epithelial tumour cells.
Rho GTPases control the dynamics of the actin cytoskeleton and are important for cell migration and invasiveness [97]. WNK2 controls, through an as yet unknown mechanism, the activation of the small GTPase RhoA, which in a reciprocal way regulates activation of Rac1 [20]. The suppression of WNK2 in cell lines leads to reduced RhoA, but increased Rac1 activation. It remains to be determined whether changes in WNK2 expression or activity affect RhoA/Rac1 activation in tumours, but in this sense, the reported epigenetic silencing of WNK2 in infiltrative gliomas [18] is highly suggestive.
A recent report also links WNK1 to Rho GTPases. In neuronal cells, WNK1 can be isolated in a complex with Rho-GDI and has been proposed to mediate the regulation of Nogo66-induced RhoA activation and neurite outgrowth [98]. Interestingly, in F11 neural tumour cells, WNK1 expression was found to correlate with invasiveness. In particular, experimental suppression of the GD3-synthase gene led to a reduced rate of cell migration and invasiveness [95], conditions under which a dramatic decrease in WNK1 expression was observed [96]. Whether this effect involves Rho-GTPases remains to be determined.
Mutational mechanisms affecting WNK genes in tumours
Considering the emerging evidence describing a role for WNK signalling in tumour development, it is not surprising that genetic alterations affecting either WNK gene expression or their coding sequence have been described. Deregulation of gene expression was reported for the WNK2 gene, which is silenced in a large percentage of human gliomas due to extensive methylation in the CpG island encompassing the 5′-end of this gene [18]. Likewise, the WNK2 promoter was hyper-methylated in 83 and 71% of grade II and III meningiomas, respectively, and this was associated with decreased WNK2 expression in primary tumours. In contrast, promoter methylation was rare in a total of 209 tumours from 13 other tumour types [19]. This finding makes WNK2 a candidate tumour suppressor gene in brain tumours. WNK2 indirectly inhibits MEK1 and restrains growth-promoting signals through the EGF receptor. Thus, it is possible that the epigenetic silencing of WNK2 interacts on a functional level with genetic alteration of EGFR signalling, a common abnormality in glioblastomas especially due to EGFR gene amplifications [99]. It is also interesting to note that solid tumours show frequent hyper-stimulation of the EGFR signalling pathway and that partial clones of WNK2 have previously been isolated as the colon cancer antigen SDCC43 [100] and T cell recognised pancreatic cancer cell antigen P/OKcl13 [101].
The other three human WNK genes share with WNK2 the presence of large CpG islands surrounding their promoter regions and extending into the first exons [6, 18, 102, 103]. This observation suggests that similar hyper-methylation of the WNK1, WNK3 or WNK4 promoters may exist in other tumour types.
Epigenetic changes can also affect promoter usage. In WNK1 intron 4, an intragenic promoter was identified that generates a kidney-specific isoform (KS-WNK1) lacking kinase activity [102] and thus may generate a dominant-negative WNK isoform in tumours, a possibility that remains to be tested as a potential mutagenic mechanism.
An additional unexplored avenue is the role of WNK alternative splicing variants. For example, one WNK1 variant lacks exons 11 and 12 [6] and two WNK2 transcripts exist that differ in their C-terminal exon with different C-terminal sequences [17]. Two WNK3 variants are known that differ in usage of mostly brain-specific exons [86, 103]. Alternative splicing variants can affect transcript turnover or encode protein isoforms. At present, no simple structure/function relationship exists to predict functional differences between two splicing variants from the same gene; however, many examples have been documented showing that resulting variant proteins can significantly differ regarding regulation or signalling activities [104–106]. Indeed, a recent report revealed that the two WNK3 variants have opposite effects on expression of the ion channel NCC [107].
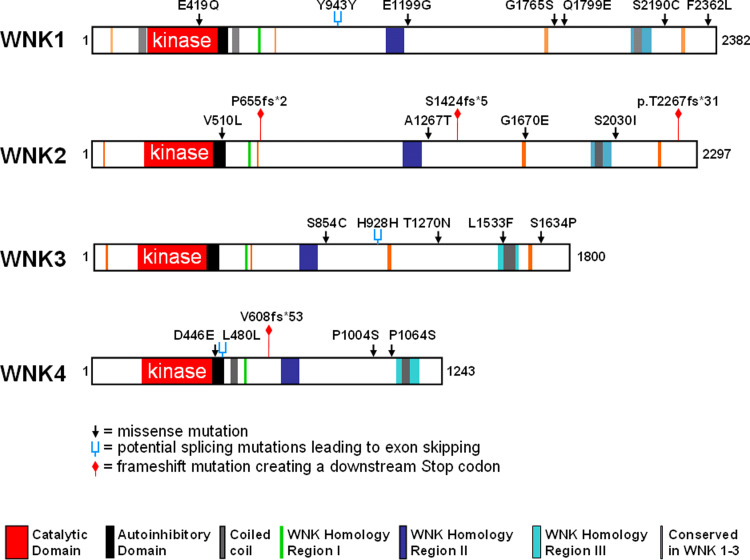
In the last few years, large-scale cancer genome sequencing efforts have been conducted to determine the full spectrum of mutations in selected tumour samples. These efforts revealed a variety of point mutations in several WNK genes that were identified in breast, colon, lung or brain tumours (Table 2; Fig. 4) [108–112]. At present, there is insufficient experimental evidence to decide whether the resulting missense or frameshift mutations have a functional impact on the corresponding WNK proteins. Only one of the mutants, WNK2-G1619E, was tested by transfection into WNK2-negative glioma cells and found to exhibit a reduced growth inhibitory effect compared to wild-type WNK2 [18]. Although the somatic alterations in WNK genes were not considered as tumour-initiating ‘driver’ mutations [108, 111], they may affect functional properties of the corresponding WNK proteins and, in consequence, have been selected for during the process of oncogenic transformation.
Table 2.
List of somatic WNK mutations identified in different unbiased cancer genome sequencing efforts (corresponding references are indicated)
| Tissue | Histology/type | Gene | Zygosity | cDNA | Protein | Mutation | References |
|---|---|---|---|---|---|---|---|
| Breast | Pleomorphic lobular carc. | WNK1 | Hetero | c.1255G>C | p.E419Q | Missense | [109, 111] |
| Breast | IDC | WNK1 | Hetero | c.5395C>G | p.Q1799E | Missense | [109–111] |
| Breast | Pleomorphic lobular carc. | WNK1 | Hetero | c.6569C>G | p.S2190C | Missense | [109, 111] |
| Ovary | Serous carcinoma | WNK1 | Hetero | c.2829C>T | p.Y943Y | Silent | [111] |
| Colon | Colorectal | WNK1 | Hetero | c.3596A>G | p.E1199G | Missense | [110] |
| Brain | Glioblastoma | WNK1 | Hetero | c.5293G>A | p.G1765S | Missense | [112] |
| Lung | Adenocarcinoma | WNK1 | Homo | c.7086C>A | p.F2362L | Missense | [108, 111] |
| Colorectal | Adenocarcinoma | WNK2 | Hetero | c.1964delC | p.P655 fs*2 | Frameshift deletion | [111] |
| Brain | Glioblastoma | WNK2 | Hetero | c.3799G>A | p.A1267T | Missense | [112] |
| Stomach | Adenocarcinoma | WNK2 | Hetero | c.4269delC | p.S1424 fs*5 | Frameshift deletion | [111] |
| Lung | Neuroendocrine carcinoma | WNK2 | Hetero | c.5009G>A | p.G1670E | Missense | [108, 111] |
| Lung | Adenocarcinoma | WNK2 | Hetero | c.6089G>T | p.S2030I | Missense | [108, 111] |
| Ovary | Serous carcinoma | WNK2 | Hetero | c.1528G>T | p.V510L | Missense | [111] |
| Ovary | Mucinous carcinoma | WNK2 | Hetero | c.6798delC | p.T2267 fs*31 | Frameshift deletion | [111] |
| Glioma | Glioblastoma | WNK3 | Hetero | c.2784C>T | p.H928H | Silent | [111] |
| Lung | Squamous cell carcinoma | WNK3 | Hetero | c.2561C>G | p.S854C | Missense | [111] |
| Lung | Large cell carcinoma | WNK3 | Hetero | c.4599G>T | p.L1533F | Missense | [111] |
| Kidney | Clear cell carcinoma | WNK3 | Hetero | c.3809C>A | p.T1270 N | Missense | [111] |
| Kidney | Clear cell carcinoma | WNK3 | Hetero | c.4900T>C | p.S1634P | Missense | [111] |
| Ovary | Mucinous carcinoma | WNK4 | Hetero | c.1338C>G | p.D446E | Missense | [111] |
| Melanoma | Metastatic | WNK4 | Hetero | c.1438C>T | p.L480L | Silent | [111] |
| Melanoma | Not described | WNK4 | Hetero | c.3010C>T | p.P1004S | Missense | [111] |
| Melanoma | Not described | WNK4 | Hetero | c.3190C>T | p.P1064S | Missense | [111] |
| Stomach | Adenocarcinoma | WNK4 | Hetero | c.1822delG | p.V608 fs*53 | Frameshift deletion | [111] |
Note that the nucleotide and codon designations of WNK2 and WNK4 mutations described in [108, 111] required correction because they were based on incomplete protein sequences. Because the sequence context for each of the mutation described in these references is available online at http://www.sanger.ac.uk/perl/genetics/CGP/cgp_viewer?action=bygene&letter=W&mutant=1, the mutations could be unambiguously localised in the full length cDNA and protein sequences, allowing correction of their designations (as full length reference sequences, we used Ensembl gene IDs ENST00000315939 (WNK1), ENST00000297954 (WNK2), ENST00000354646 (WNK3) and ENST00000246914 (WNK4), all available at http://www.ensembl.org/index.html)
Fig. 4.
Graphic representation of the somatic WNK mutations described in Table 2. The positions of the mutations are illustrated with respect to the catalytic, autoinhibitory and coiled-coil domains. Missense mutations are given as black arrow heads and frameshift mutations leading to premature stop codons are marked by red signposts. Silent nucleotide changes are marked as blue forks because they can act at the nucleotide level by interfering with exon recognition during the pre-mRNA splicing process and result in exon skipping. A sequence analysis of the three silent changes using the ASD database (http://www.ebi.ac.uk/asd-srv/wb.cgi?method=8) indeed indicates a potential loss of splicing factor binding sites, namely for SC35 in WNK1 Y943Y, for ASF/SF2 and SC35 in WNK3 H928H, and for SRp40 in WNK4 L480L
Concluding remarks
Considering the available data, a role of WNK kinases in cancer cell signalling is beginning to emerge. At present, experimental evidence is strongest concerning their role as upstream regulators of MAPK cascades, as modulators of Rho-GTPases and inhibitors of apoptosis. During the next few years, the putative role of WNKs in other aspects of cancer biology that we reviewed will probably be clarified. It will be of particular interest to elucidate how their abundantly documented role in the regulation of activity or cell surface expression of ion channels can be related to tumour development. In order to understand the underlying signalling network, it will be important to systematically identify WNK interacting proteins, their physiological substrate proteins, cellular phenotypes following WNK suppression, and the genetic changes affecting WNK genes during tumorigenesis.
Acknowledgments
The authors wish to thank Drs. Fátima Veríssimo (Heidelberg), Karl Kunzelmann (Regensburg) and Jonathan Morris (London) for their helpful suggestions on the manuscript. Work in the authors’ laboratory was supported by the Fundação para a Ciência e a Tecnologia, Portugal (Programa de Financiamento Plurianual do CIGMH, grants POCTI/33221/99, POCTI/56294/04 and fellowship BD 11180/02 to S.M.).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Kostich M, English J, Madison V, Gheyas F, Wang L, Qiu P, Greene J, Laz TM (2002) Human members of the eukaryotic protein kinase family. Genome Biol 3, RESEARCH0043 [DOI] [PMC free article] [PubMed]
- 3.Hanks SK. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 2003;4:111.1–111.7. doi: 10.1186/gb-2003-4-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanks SK, Hunter T. The eucaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 5.Xu BE, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 6.Veríssimo F, Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 7.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T. Compilation and characterization of a novel WNK family of protein kinases in Arabiodpsis thaliana with reference to circadian rhythms. Biosci Biotechnol Biochem. 2002;66:2429–2436. doi: 10.1271/bbb.66.2429. [DOI] [PubMed] [Google Scholar]
- 9.Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH. Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem. 2002;277:48456–48462. doi: 10.1074/jbc.M207917200. [DOI] [PubMed] [Google Scholar]
- 10.Zagórska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, Thastrup J, Deak M, Campbell DG, Morrice NA, Prescott AR, Alessi DR. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol. 2007;176:89–100. doi: 10.1083/jcb.200605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Yang CL, Ellison DH. Comparison of WNK4 and WNK1 kinase and inhibiting activities. Biochem Biophys Res Commun. 2004;317:939–944. doi: 10.1016/j.bbrc.2004.03.132. [DOI] [PubMed] [Google Scholar]
- 12.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 13.Xu BE, Lee BH, Min X, Lenertz L, Heise CJ, Stippec S, Goldsmith EJ, Cobb MH. WNK1: analysis of protein kinase structure, downstream targets, and potential roles in hypertension. Cell Res. 2005;15:6–10. doi: 10.1038/sj.cr.7290256. [DOI] [PubMed] [Google Scholar]
- 14.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 15.Xu BE, Stippec S, Lenertz L, Lee B-H, Zhang W, Lee YK, Cobb MH. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem. 2004;279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Gao L, Yu RK, Zeng G. Down-regulation of WNK1 protein kinase in neural progenitor cells suppresses cell proliferation and migration. J Neurochem. 2006;99:1114–1121. doi: 10.1111/j.1471-4159.2006.04159.x. [DOI] [PubMed] [Google Scholar]
- 17.Moniz S, Veríssimo F, Matos P, Brazão R, Silva E, Kotelevets L, Chastre E, Gespach C, Jordan P. Protein kinase WNK2 inhibits cell proliferation by negatively modulating the activation of MEK1/ERK1/2. Oncogene. 2007;26:6071–6081. doi: 10.1038/sj.onc.1210706. [DOI] [PubMed] [Google Scholar]
- 18.Hong C, Moorefield KS, Jun P, Aldape KD, Kharbanda S, Phillips HS, Costello JF. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci USA. 2007;104:10974–10979. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun P, Hong C, Lal A, Wong JM, McDermott MW, Bollen AW, Plass C, Held WA, Smiraglia DJ, Costello JF. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro Oncol. 2009;11:414–422. doi: 10.1215/15228517-2008-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moniz S, Matos P, Jordan P. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cell Signal. 2008;20:1762–1768. doi: 10.1016/j.cellsig.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Shaharabany M, Holtzman EJ, Mayan H, Hirschberg K, Seger R, Farfel Z. Distinct pathways for the involvement of WNK4 in the signaling of hypertonicity and EGF. FEBS J. 2008;275:1631–1642. doi: 10.1111/j.1742-4658.2008.06318.x. [DOI] [PubMed] [Google Scholar]
- 22.Lenertz LY, Lee BH, Min X, Xu BE, Wedin K, Earnest S, Goldsmith EJ, Cobb MH. Properties of WNK1 and implications for other family members. J Biol Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 23.Yang CL, Zhu X, Wang Z, Subramanya AR, Ellison DH. Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. J Clin Invest. 2005;115:1379–1387. doi: 10.1172/JCI22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 25.Strange K, Denton J, Nehrke K. Ste20-type kinases: evolutionarily conserved regulators of ion transport and cell volume. Physiology (Bathesda) 2006;21:61–68. doi: 10.1152/physiol.00139.2005. [DOI] [PubMed] [Google Scholar]
- 26.Johnston AM, Naselli G, Gonez LJ, Martin RM, Harrison LC, Deaizpurua HJ. SPAK, a Ste20/SPS1-related kinase that activates the p38 pathway. Oncogene. 2000;19:4290–4297. doi: 10.1038/sj.onc.1203784. [DOI] [PubMed] [Google Scholar]
- 27.Polek TC, Talpaz M, Spivak-Kroizman T. The TNF receptor, RELT, binds SPAK and uses it to mediate p38 and JNK activation. Biochem Biophys Res Commun. 2006;343:125–134. doi: 10.1016/j.bbrc.2006.02.125. [DOI] [PubMed] [Google Scholar]
- 28.Yan Y, Nguyen H, Dalmasso G, Sitaraman SV, Merlin D. Cloning and characterization of a new intestinal inflammation-associated colonic epithelial Ste20-related protein kinase isoform. Biochim Biophys Acta. 2007;1769:106–116. doi: 10.1016/j.bbaexp.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon A, Bandhakavi S, Jabbar S, Shah R, Beitel GJ, Morimoto RI. Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics. 2004;167:161–170. doi: 10.1534/genetics.167.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 32.Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, Zhao Y, Cobb MH. WNK1 and OSR1 regulate the Na+, K+, 2Cl− cotransporter in HeLa cells. Proc Natl Acad Sci USA. 2006;103:10883–10888. doi: 10.1073/pnas.0604607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-chloride cotransporters is modulated by the interaction of two kinases: SPAK and WNK4. Am J Physiol Cell Physiol. 2006;290:C134–C142. doi: 10.1152/ajpcell.00037.2005. [DOI] [PubMed] [Google Scholar]
- 34.Vitari AC, Thastrup J, Rafiqi FH, Deak M, Morrice NA, Karlsson HK, Alessi DR. Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1. Biochem J. 2006;397:223–231. doi: 10.1042/BJ20060220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson C, Rafiqi FH, Karlsson HK, Moleleki N, Vandewalle A, Campbell DG, Morrice NA, Alessi DR. Activation of the thiazide-sensitive Na+–Cl− cotransporter by the WNK-regulated kinases SPAK and OSR1. J Cell Sci. 2008;121:675–684. doi: 10.1242/jcs.025312. [DOI] [PubMed] [Google Scholar]
- 36.Ponce-Coria J, San-Cristobal P, Kahle KT, Vazquez N, Pacheco-Alvarez D, de Los Heros P, Juárez P, Muñoz E, Michel G, Bobadilla NA, Gimenez I, Lifton RP, Hebert SC, Gamba G. Regulation of NKCC2 by a chloride-sensing mechanism involving the WNK3 and SPAK kinases. Proc Natl Acad Sci USA. 2008;105:8458–8463. doi: 10.1073/pnas.0802966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost JA, Steen H, Shapiro P, Lewis T, Ahn N, Shaw PE, Cobb MH. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 1997;16:6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coles LC, Shaw PE. PAK1 primes MEK1 for phosphorylation by Raf-1 kinase during cross-cascade activation of the ERK pathway. Oncogene. 2002;21:2236–2244. doi: 10.1038/sj.onc.1205302. [DOI] [PubMed] [Google Scholar]
- 39.Eblen ST, Slack JK, Weber MJ, Catling AD. Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol Cell Biol. 2002;22:6023–6033. doi: 10.1128/MCB.22.17.6023-6033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slack-Davis JK, Eblen ST, Zecevic M, Boerner SA, Tarcsafalvi A, Diaz HB, Marshall MS, Weber MJ, Parsons JT, Catling AD. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162:281–291. doi: 10.1083/jcb.200212141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park ER, Eblen ST, Catling AD. MEK1 activation by PAK: a novel mechanism. Cell Signal. 2007;19:1488–1496. doi: 10.1016/j.cellsig.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorkin A, von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 43.González-Gaitán M. Signal dispersal and transduction through the endocytic pathway. Nat Rev Mol Cell Biol. 2003;4:213–224. doi: 10.1038/nrm1053. [DOI] [PubMed] [Google Scholar]
- 44.Cai H, Cebotaru V, Wang YH, Zhang XM, Cebotaru L, Guggino SE, Guggino WB. WNK4 kinase regulates surface expression of the human sodium chloride cotransporter in mammalian cells. Kidney Int. 2006;69:2162–2170. doi: 10.1038/sj.ki.5000333. [DOI] [PubMed] [Google Scholar]
- 45.Subramanya AR, Liu J, Ellison DH, Wade JB, Welling PA. WNK4 diverts the thiazide-sensitive NaCl cotransporter to the lysosome and stimulates AP-3 interaction. J Biol Chem. 2009;284:18471–18480. doi: 10.1074/jbc.M109.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahle KT, Wilson FH, Leng Q, Lalioti MD, O’Connell AD, Dong K, Rapson AK, MacGregor GG, Giebisch G, Hebert SC, Lifton RP. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet. 2003;35:372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 47.Cope G, Murthy M, Golbang AP, Hamad A, Liu CH, Cuthbert AW, O’Shaughnessy KM. WNK1 affects surface expression of the ROMK potassium channel independent of WNK4. J Am Soc Nephrol. 2006;17:1867–1874. doi: 10.1681/ASN.2005111224. [DOI] [PubMed] [Google Scholar]
- 48.He G, Wang HR, Huang SK, Huang CL. Intersectin links WNK kinases to endocytosis of ROMK1. J Clin Invest. 2007;117:1078–1087. doi: 10.1172/JCI30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 50.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 51.Vitari AC, Deak M, Collins BJ, Morrice N, Prescott AR, Phelan A, Humphreys S, Alessi DR. WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem J. 2004;378:257–268. doi: 10.1042/BJ20031692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang ZY, Zhou QL, Holik J, Patel S, Leszyk J, Coleman K, Chouinard M, Czech MP. Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J Biol Chem. 2005;280:21622–21628. doi: 10.1074/jbc.M414464200. [DOI] [PubMed] [Google Scholar]
- 53.Xu BE, Stippec S, Chu PY, Lazrak A, Li XJ, Lee BH, English JM, Ortega B, Huang CL, Cobb MH. WNK1 activates SGK1 to regulate the epithelial sodium channel. Proc Natl Acad Sci USA. 2005;102:10315–10320. doi: 10.1073/pnas.0504422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu BE, Stippec S, Lazrak A, Huang CL, Cobb MH. WNK1 activates SGK1 by a phosphatidylinositol 3-kinase-dependent and non-catalytic mechanism. J Biol Chem. 2005;280:34218–34223. doi: 10.1074/jbc.M505735200. [DOI] [PubMed] [Google Scholar]
- 55.Sale EM, Hodgkinson CP, Jones NP, Sale GJ. A new strategy for studying protein kinase B and its three isoforms. Role of protein kinase B in phosphorylating glycogen synthase kinase-3, tuberin, WNK1, and ATP citrate lyase. Biochemistry. 2006;45:213–223. doi: 10.1021/bi050287i. [DOI] [PubMed] [Google Scholar]
- 56.Hong F, Larrea MD, Doughty C, Kwiatkowski DJ, Squillace R, Slingerland JM. mTOR-raptor binds and activates SGK1 to regulate p27 phosphorylation. Mol Cell. 2008;30:701–711. doi: 10.1016/j.molcel.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 58.Lee BH, Chen W, Stippec S, Cobb MH. Biological cross-talk between WNK1 and the transforming growth factor beta-Smad signaling pathway. J Biol Chem. 2007;282:17985–17996. doi: 10.1074/jbc.M702664200. [DOI] [PubMed] [Google Scholar]
- 59.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 60.Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ. Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer. 2007;7:519–530. doi: 10.1038/nrc2171. [DOI] [PubMed] [Google Scholar]
- 61.Roderick HL, Cook SJ. Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat Rev Cancer. 2008;8:361–375. doi: 10.1038/nrc2374. [DOI] [PubMed] [Google Scholar]
- 62.Prevarskaya N, Zhang L, Barritt G. TRP channels in cancer. Biochem Biophys Acta. 2007;1772:937–946. doi: 10.1016/j.bbadis.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 63.Jiang Y, Ferguson WB, Peng JB. WNK4 enhances TRPV5-mediated calcium transport: potential role in hypercalciuria of familial hyperkalemic hypertension caused by gene mutation of WNK4. Am J Physiol Renal Physiol. 2007;292:F545–F554. doi: 10.1152/ajprenal.00187.2006. [DOI] [PubMed] [Google Scholar]
- 64.Zhang W, Na T, Peng JB. WNK3 positively regulates epithelial calcium channels TRPV5 and TRPV6 via a kinase-dependent pathway. Am J Physiol Renal Physiol. 2008;295:F1472–F1484. doi: 10.1152/ajprenal.90229.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y, Subramanya A, Rozansky D, Cohen DM. WNK kinases influence TRPV4 channel function and localization. Am J Physiol Renal Physiol. 2006;290:F1305–F1314. doi: 10.1152/ajprenal.00391.2005. [DOI] [PubMed] [Google Scholar]
- 66.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 67.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 68.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 69.Thurmond DC, Pessin JE. Molecular machinery involved in the insulin-regulated fusion of GLUT4-containing vesicles with the plasma membrane. Mol Membr Biol. 2001;18:237–245. doi: 10.1080/09687680110082400. [DOI] [PubMed] [Google Scholar]
- 70.Oh E, Heise CJ, English JM, Cobb MH, Thurmond DC. WNK1 is a novel regulator of Munc18c-syntaxin 4 complex formation in SNARE-mediated vesicle exocytosis. J Biol Chem. 2007;282:32613–32622. doi: 10.1074/jbc.M706591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song J, Hu X, Riazi S, Tiwari S, Wade JB, Ecelbarger CA. Regulation of blood pressure, the epithelial sodium channel (ENaC), and other key renal sodium transporters by chronic insulin infusion in rats. Am J Physiol Renal Physiol. 2006;290:F1055–F1064. doi: 10.1152/ajprenal.00108.2005. [DOI] [PubMed] [Google Scholar]
- 72.Lee BH, Min X, Heise CJ, Xu BE, Chen S, Shu H, Luby-Phelps K, Goldsmith EJ, Cobb MH. WNK1 phosphorylates synaptotagmin 2 and modulates its membrane binding. Mol Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 73.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 74.Kahle KT, Wilson FH, Lalioti M, Toka H, Qin H, Lifton RP. WNK kinases: molecular regulators of integrated epithelial ion transport. Curr Opin Nephrol Hypertens. 2004;13:557–562. doi: 10.1097/00041552-200409000-00012. [DOI] [PubMed] [Google Scholar]
- 75.Gamba G. Role of WNK kinases in regulating tubular salt and potassium transport and in the development of hypertension. Am J Physiol Renal Physiol. 2005;288:F245–F252. doi: 10.1152/ajprenal.00311.2004. [DOI] [PubMed] [Google Scholar]
- 76.Kahle KT, Rinehart J, Ring A, Gimenez I, Gamba G, Hebert SC, Lifton RP. WNK protein kinases modulate cellular Cl-flux by altering the phosphorylation state of the Na–K–Cl and K–Cl cotransporters. Physiology (Bethesda) 2006;21:326–335. doi: 10.1152/physiol.00015.2006. [DOI] [PubMed] [Google Scholar]
- 77.Kahle KT, Ring AM, Lifton RP. Molecular physiology of the WNK kinases. Annu Rev Physiol. 2008;70:329–355. doi: 10.1146/annurev.physiol.70.113006.100651. [DOI] [PubMed] [Google Scholar]
- 78.Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer. 2005;5:786–795. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]
- 79.Kahle KT, Gimenez I, Hassan H, Wilson FH, Wong RD, Forbush B, Aronson PS, Lifton RP. WNK4 regulates apical and basolateral Cl− flux in extrarenal epithelia. Proc Natl Acad Sci USA. 2004;101:2064–2069. doi: 10.1073/pnas.0308434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dorwart MR, Sahcheynikov N, Wang Y, Stippec S, Muallem S. SLC26A9 is a Cl(−) channel regulated by the WNK kinases. J Physiol. 2007;584:333–345. doi: 10.1113/jphysiol.2007.135855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 82.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Igney FH, Krammer P. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 84.Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I. Bistability in apoptosis: roles of bax, bcl-2, and mitochondrial permeability transition pores. Biophys J. 2006;90:1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dogu Y, Díaz J. Mathematical model of a network of interaction between p53 and Bcl-2 during genotoxic-induced apoptosis. Biophys Chem. 2009;143:44–54. doi: 10.1016/j.bpc.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 86.Veríssimo F, Silva E, Morris JD, Pepperkok R, Jordan P. Protein kinase WNK3 increases cell survival in a caspase 3-dependent pathway. Oncogene. 2006;25:4172–4182. doi: 10.1038/sj.onc.1209449. [DOI] [PubMed] [Google Scholar]
- 87.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 88.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 89.Choe K, Strange K. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival following hypertonic shrinkage in Caenorhabditis elegans . Am J Physiol Cell Physiol. 2007;293:C915–C927. doi: 10.1152/ajpcell.00126.2007. [DOI] [PubMed] [Google Scholar]
- 90.Delpire E, Gagnon KB. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem J. 2008;409:321–331. doi: 10.1042/BJ20071324. [DOI] [PubMed] [Google Scholar]
- 91.Richardson C, Alessi DR. The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci. 2008;121:3293–3304. doi: 10.1242/jcs.029223. [DOI] [PubMed] [Google Scholar]
- 92.Thiery JP. Epithelial–mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 93.Padua D, Massagué J. Roles of TGFβ in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 94.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng G, Gao L, Yu RK. Reduced cell migration, tumor growth and experimental metastasis of rat F-11 cells whose expression of ganglioside GD3 was suppressed. Int J Cancer. 2000;88:53–57. doi: 10.1002/1097-0215(20001001)88:1<53::aid-ijc8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 96.Zeng G, Gao L, Xia T, Gu Y, Yu RK. Expression of the mouse WNK1 gene in correlation with ganglioside GD3 and functional analysis of the mouse WNK1 promoter. Gene. 2005;344:233–239. doi: 10.1016/j.gene.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 97.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Z, Xu X, Zhang Y, Zhou J, Yu Z, He C. LINGO-1 interacts with WNK1 to regulate Nogo-induced inhibition of neurite extension. J Biol Chem. 2009;284:15717–15728. doi: 10.1074/jbc.M808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor a, and epidermal growth f actor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 100.Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Türeci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 101.Ito M, Shichijo S, Tsuda N, Ochi M, Harashima N, Saito N, Itoh K. Molecular basis of T cell-mediated recognition of pancreatic cancer cells. Cancer Res. 2001;61:2038–2046. [PubMed] [Google Scholar]
- 102.Delaloy C, Lu J, Houot AM, Disse-Nicodeme S, Gasc JM, Corvol P, Jeunemaitre X. Multiple promoters in the WNK1 gene: one controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol. 2003;23:9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Holden S, Cox J, Raymond FL. Cloning, genomic organization, alternative splicing and expression analysis of the human gene WNK3 (PRKWNK3) Gene. 2004;335:109–119. doi: 10.1016/j.gene.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 104.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 105.Venables JP. Unbalanced alternative splicing and its significance in cancer. BioEssays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 106.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 107.Glover M, Zuber AM, O’Shaughnessy KM. Renal and brain isoforms of WNK3 have opposite effects on NCCT expression. J Am Soc Nephrol. 2009;20:1314–1322. doi: 10.1681/ASN.2008050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, O’Meara S, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh BT, Yuen ST, Lakhani SR, Easton DF, Weber BL, Goldstraw P, Nicholson AG, Wooster R, Stratton MR, Futreal PA. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 109.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O’Meara S, Parker A, Tarpey P, Avis T, Barthorpe A, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh B, Yuen ST, Nicholson AG, Lakhani S, Easton DF, Weber BL, Stratton MR, Futreal PA, Wooster R. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 110.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 111.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Choate KA, Kahle KT, Wilson FH, Nelson-Williams C, Lifton RP. WNK1, a kinase mutated in inherited hypertension with hyperkalemia, localizes to diverse Cl−-transporting epithelia. Proc Natl Acad Sci USA. 2003;100:663–668. doi: 10.1073/pnas.242728499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rinehart J, Kahle KT, de Los Heros P, Vazquez N, Meade P, Wilson FH, Hebert SC, Gimenez I, Gamba G, Lifton RP. WNK3 kinase is a positive regulator of NKCC2 and NCC, renal cation-Cl− cotransporters required for normal blood pressure homeostasis. Proc Natl Acad Sci USA. 2005;102:16777–16782. doi: 10.1073/pnas.0508303102. [DOI] [PMC free article] [PubMed] [Google Scholar]