Abstract
Reverse transcription is a critical step in the life cycle of all retroviruses and related retrotransposons. This complex process is performed exclusively by the retroviral reverse transcriptase (RT) enzyme that converts the viral single-stranded RNA into integration-competent double-stranded DNA. Although all RTs have similar catalytic activities, they significantly differ in several aspects of their catalytic properties, their structures and subunit composition. The RT of human immunodeficiency virus type-1 (HIV-1), the virus causing acquired immunodeficiency syndrome (AIDS), is a prime target for the development of antiretroviral drug therapy of HIV-1/AIDS carriers. Therefore, despite the fundamental contributions of other RTs to the understanding of RTs and retrovirology, most recent RT studies are related to HIV-1 RT. In this review we summarize the basic properties of different RTs. These include, among other topics, their structures, enzymatic activities, interactions with both viral and host proteins, RT inhibition and resistance to antiretroviral drugs.
Keywords: Reverse transcription, Retroviruses, DNA and RNA-dependent DNA synthesis, Ribonuclease H, RT inhibitors, HIV, Drug resistance
Historical perspective and overview
The breakthrough discovery of reverse transcriptase (RT) by Temin and Baltimore in 1970 had an immense impact on life sciences. The identification of a previously unknown new enzyme, an RNA-dependent DNA polymerase in RNA tumor viruses, has markedly challenged the prevailing central dogma, according to which, the genetic information flows from DNA to RNA and subsequently to proteins (or in the case of RNA viruses from RNA to RNA and proteins). Since the new enzyme transcribes nucleic acids from RNA to DNA, which was considered at that time “in reverse” to the existing dogma, it was dubbed “reverse transcriptase.” Following studies showed that RT had in fact three activities: an RNA-dependent DNA polymerase (RDDP), a DNA-dependent DNA polymerase (DDDP) as well as a ribonuclease H (RNase H) that cleaved the RNA strand of an RNA-DNA heteroduplex. As all RT activities could not have been described by a simple designation, the original name of the enzyme was kept. Accordingly, the RT-harboring viruses that were initially called “RNA tumor viruses” were renamed as “retroviruses.” Later on, the retrovirus-related movable elements were also discovered and termed “retroelements” and “retrotransposons” [1–3].
The discovery of RT enabled also a substantially better understanding of malignant transformation that is induced by RNA tumor viruses (retroviruses). A major mechanism suggested for this process was the ability of some retroviruses to acquire a cellular proto-oncogene during reverse transcription (RTN) [1]. RT is also required for the mobility of retroelements into new positions that may be upstream to cellular proto-oncogenes. Both processes can induce cellular genetic changes and may lead to malignancies. Historically, the first oncogenes were discovered as part of the genetic material carried by retroviruses.
During the last decades, RT has become a key tool in molecular biology, for the synthesis of complementary DNA (cDNA) from messenger RNA (mRNA). This application has largely expanded our knowledge of numerous cellular regulatory mechanisms. Moreover, combining RT activity with PCR amplification has long become a gold standard as the first step in cloning the coding region of any gene of interest. Evidently, RTs have been critical in advancing molecular biology, genetics and medicine to their current stage. Even nowadays, the development of biotechnology heavily relies on the availability of efficient RTs, in applications, such as the microarrays of reverse transcribing mRNA that are then analyzed by high throughput techniques.
A revival of RT research was triggered by the discovery, in the early 1980s, of a new human retrovirus that was later named human immunodeficiency virus (HIV), the pathogenic agent of acquired immunodeficiency syndrome (AIDS) in humans [1, 3, 4]. Evidently, the already-known information on retroviruses and RTs has helped enormously in identifying and rapidly advancing the studies of this novel human pathogen. High rates of morbidity and mortality in AIDS patients have directly led to a massive search towards antiretroviral interventions that identified HIV RT as a prime target for virus inhibition. To achieve this aim, an intensive research of HIV RT has been conducted. Indeed, azidothymidine (AZT, or zidovudine by its commercial name), which effectively inhibits the DNA polymerase activity of HIV-1 RT, was the first anti-retroviral drug used (in 1987) to treat HIV-1 carriers. Since then, RT inhibitors have been a major component in the drug combinations used to combat HIV-1. In addition, numerous HIV-1 RT drug-resistant mutations were characterized and extensively studied. This translated into a huge number of publications on the different functions and structures of RTs that turn RT altogether to one of the most studied enzymes.
The enormous potential of reverse transcription processes to affect cell and developmental biology, human genetics and medicine has been recently revealed after sequencing the entire human genome. It is now estimated that nearly 50% of all sequences found in this genome were initially introduced through RTN. Moreover, about 8% of the human genome is composed of endogenous retroviruses.
Due to its outstanding importance, several review articles (for example, [5, 6]) as well as special issues and books (e.g., [1–3]) have been dedicated to RT. In this review, we summarize the functions and structures of different RTs, with a concise and up-to-date description of the major topics surrounding them. As most recent studies on RTs were performed with HIV-1 RT, it will be considered in this review as the prototype RT, and, whenever it is not otherwise stated, we will refer to this RT. Obviously, cardinal contributions were also made prior to the identification of HIV in studies performed with other RTs, mainly those of murine leukemia virus (MLV) and avian sarcoma/leukemia virus (ASLV). A detailed review, written by us, that summarizes the unique properties of RTs other than those of HIV-1 and MLV can be found in the recent special issue dedicated to reverse transcriptase [7].
Introduction
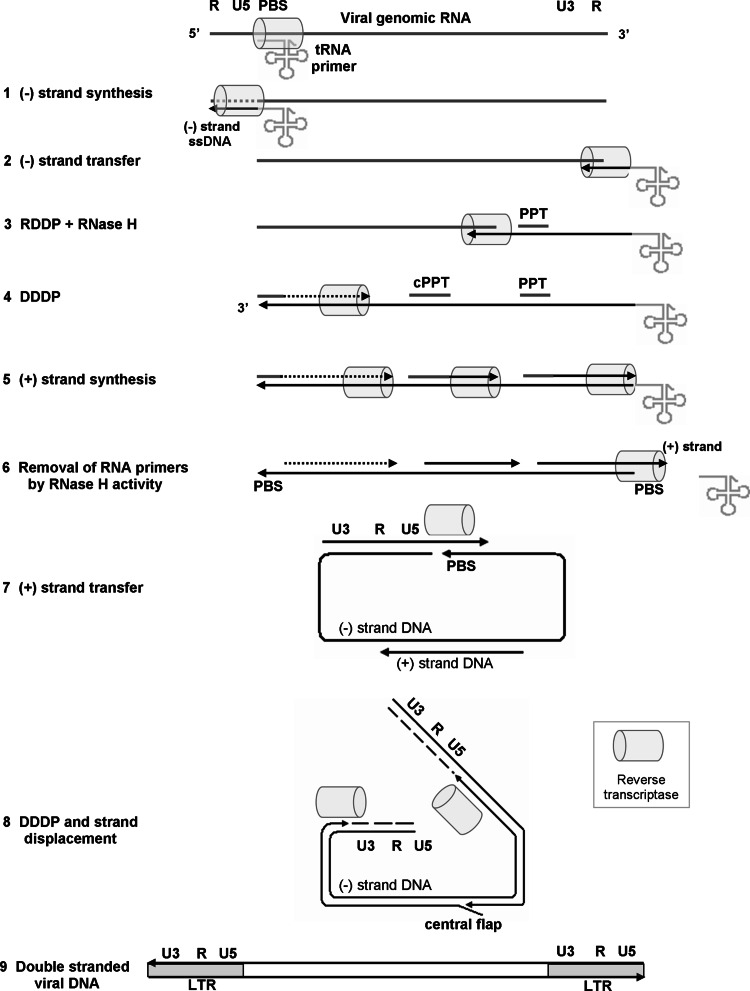
Retroviruses have evolved by selecting a complex and unique strategy to replicate inside target cells. Upon entry into the host cell, the viral single-stranded RNA is copied in the cell cytoplasm by the viral RT into a double-stranded linear viral DNA molecule. Subsequently, the generated DNA is translocated into the nucleus, where it is inserted into the host genome by the retroviral integrase (IN). The integrated viral DNA, designated provirus, can then direct the synthesis of both viral genomic RNA (to be packed in the viral progeny) and viral mRNA species, required for synthesizing all viral proteins. The different RNAs are exclusively synthesized by the host DNA-dependent RNA polymerase II. However, in order to use the synthesized genomic RNA in the next round of infection, RT has to overcome a substantial obstacle. The synthesis of genomic RNA does not transcribe the viral promoter, located at the 5′-end of the long terminal repeat (LTR) of the provirus and the synthesized sequences beyond the poly(A) signal (at the 3′-end LTR) are removed during processing of the mRNA, similar to all other cellular mRNAs (see below and Fig. 2). As a result, the viral RNA genome contains only partial LTR sequences at both ends. These sequences must be extended during the generation of the proviral DNA in the next round of infection to contain the regulatory elements required for transcription and processing of the viral mRNA (Fig. 2). For that reason, retroviral RTs have evolved to assemble these regulatory elements during RTN by duplicating viral sequences found within their RNA genome, and, therefore, their activity is much more complex than simply copying the viral RNA. Generation of the double-stranded DNA is done in several steps. Initially, the RDDP activity copies the viral RNA template into the complementary (−) DNA strand. Concomitantly, the RNA strand of the nascent RNA-DNA heteroduplex is hydrolyzed by the inherent RNase H activity of RT. This is followed by DDDP activity that synthesizes the second (+) DNA strand, using the already synthesized (−) DNA strand as a template. These three interconnected activities enable the transfer of the genetic data stored in the viral genome into a double-stranded DNA and the assembly of the LTR regulatory elements in both sides of the double-stranded viral DNA.
Fig. 2.
The reverse transcription process. A schematic description of the different steps of the RTN process, drawn not to scale. See the text for a detailed description. RNA is depicted in grey and DNA in black. The direction of DNA synthesis is marked by the arrowheads. In various retroviruses (+) DNA synthesis can potentially be initiated from multiple RNA primers. The initiation of (+) DNA synthesis from the cPPT and 3′-PPT is shown in this scheme as straight black arrows (step 7), as these are the main initiation sites of the RTN of HIV-1. An additional potential site is shown upstream as a dotted arrow
Biogenesis of RTs
Retroviruses can be subdivided into several groups (genus) according to their evolutionary relatedness (Table 1). All groups carry the gag, pol and env coding domains, whereas the complex retroviruses (such as the lentiviruses and deltaretroviruses) carry additional genes that partially overlap the former genes and encode (after alternative splicing of the mRNAs) small regulatory proteins. The RT sequences reside within the pol coding domain. RTs from the different groups of retroviruses share similar activities but can differ in various parameters, such as: structure and subunit composition, molecular weights, catalytic properties, biochemical and biophysical characteristics and sensitivity to different inhibitors [1, 2, 7].
Table 1.
Subunit organization of RTs from various groups of retroviruses
| Genus | Representative virus | RT | Additional viruses | Genomea | |
|---|---|---|---|---|---|
| Subunit organization | Putative size (kDa) | ||||
| Alpharetroviruses | ASLV | Heterodimer (the larger β-subunit contains also IN) | 94/62 | RSV, AMV | Simple |
| Betaretroviruses | MMTV | Monomer | 66 | MPMV | |
| Gammaretroviruses | MLV | Monomer | 75 | PERV | |
| Deltaretroviruses | BLV | Monomer | 64 | HTLV-1 | Complex |
| Lentiviruses | |||||
| Primate | HIV-1 | Heterodimer | 66/51 | HIV-2, SIV | |
| Non-primate | BIV | Heterodimer | 64/51 | EIAV, FIV | |
| Spumaviruses | HFV | Monomer | 80–81 | BSV | |
Epsilonretroviruses were omitted due to limited available data
aA simple genome, as in all orthoretroviruses, contains the gag, pro,pol and env genes, encoding for viral proteins. The complex genome of the other groups encodes in addition several viral regulatory proteins
RSV Rous sarcoma virus, AMV avian myeloblastosis virus, MPMV Mason-Pfizer monkey virus, HFV human foamy virus, BSV bovine syncytial virus [1]
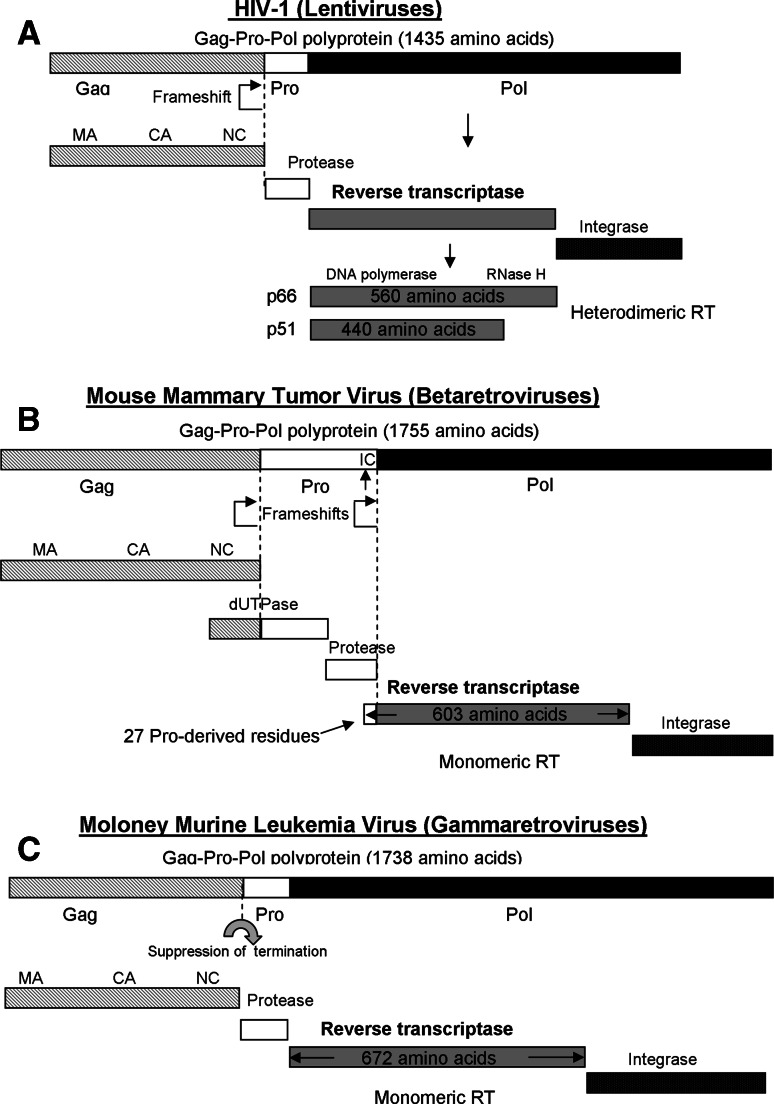
In all retroviruses, the viral pol gene encodes the Pol precursor protein that consists both RT and IN proteins. The fused protein is synthesized as part of a larger Gag-Pro-Pol polyprotein, from which the mature RT and IN proteins are cleaved out during virus assembly by the viral protease (PR) (encoded by the pro gene) (Fig. 1). Although the order of the gag, pro, pol and env genes is invariant in retroviruses, their reading frames are quite different. The Gag and Gag-Pro-Pol polyproteins are translated from identical mRNA, but in the latter there is either a termination suppression or a frameshift by one nucleotide (−1 frameshifting) that tightly regulates the ratio of Gag to Gag-Pro-Pol synthesis. In the most common organization, epitomized by lentiviruses, the pro and pol are in the same reading frame, and synthesis of Gag-Pro-Pol requires only a single frameshift event relative to the gag reading frame. As a result, the PR, RT and IN proteins are in equimolar ratios, and the amount of each is about 20-fold less than the amount of the Gag molecules. In ASLV (an alpharetrovirus) there is also a single −1 frameshift event before the pol gene; therefore, the pro gene is in the same reading frame as gag. Therefore, Pro is synthesized as part of the Gag polyprotein, and the ratio of PR to Pol is much higher compared to lentiviruses (where they are in equimolar ratios). A different arrangement of two consecutive frameshifting-associated events exists in mouse mammary tumor virus (MMTV) (a betaretrovirus) and in human T-cell leukemia virus-1 (HTLV-1) or bovine leukemia virus (BLV) (both are deltaretroviruses). In this case, the pro gene, which stands in its own reading frame, is in a −1 reading frame relative to gag, and the pol gene is in a −1 reading frame relative to the pro gene (Fig. 1). Frameshifting at each site is relatively efficient, with up to 30% of the translations, to ensure that a sufficient level of the final Gag-Pro-Pol polyprotein is made after the two serial shifts. Interestingly, an entirely different frameshift-independent arrangement is found in MLV (epitomizing gammaretroviruses), where the pro and pol genes are in the same reading frame as gag, but are separated by a stop codon (Fig. 1). The synthesis of Pro and Pol proteins is a product of an in-frame read-through suppression of the termination codon at the end of the gag gene. Frameshifting and read-through have probably evolved as strategies to provide the proper ratios of Gag, Gag-Pro and Gag-Pro-Pol polypeptides in the infected cell. Fusion of the viral enzymes to the Gag polyprotein also provides a means for incorporating these enzymes into the new progeny virions during assembly. Finally, spumaviruses (foamy viruses) markedly differ from all other previously described retroviruses, which are termed collectively orthoretroviruses. The RT of spumaviruses is generated from a separate spliced mRNA instead of the Gag-Pro-Pol polyprotein precursor, and infectious particles contain double-stranded DNA, similar to full length cDNA of the orthoretroviruses (see below). This suggests that RTN of spumaviruses takes place during the packaging of viral particles and before the virions enter the new target cells in a process reminiscent of the life cycle of hepadnaviruses [7–9].
Fig. 1.
The biogenesis of RTs in selected retroviral groups. Retroviral RTs are cleaved from the Gag-Pro-Pol polyprotein precursor. In lentiviruses the Gag-Pro-Pol polyprotein is synthesized after a single translational frameshift event. a In HIV-1, the resulting RT, which is cleaved from the polyprotein, is a heterodimer with the molecular weight of p66/p51. b In MMTV, the Gag-Pro-Pol is generated by two subsequent events of translational frameshift. The resulting RT is a monomer with 603 amino acids, of which 576 derived from the pol gene and 27 from the C-terminal portion of the Pro protein. Therefore, there is a putative internal cleavage site (IC) at this position within the Pro. The Gag-Pro polyprotein is generated by a single frameshift event and may be also the source for all the Gag and Pro derived proteins. c MLV uses a completely different strategy in which the Gag-Pro-Pol polyprotein is generated by an in-frame suppression of the stop codon. In the case of MLV, the scheme shows the protein lengths of the Moloney strain. This schematic description was not drawn to scale
Despite the clear separation between the pro and pol genes, there are some variations in the processing of RT by PR in different retroviral groups. These differences affect the final assembly of the proteins, the sequences incorporated into RTs and their molecular arrangements and structures. All known RTs are either monomeric or heterodimeric in structure, as summarized in Table 1. The smaller subunit in the heterodimeric RTs is produced by a proteolytic cleavage of the larger subunit. This activity removes the C-terminus of the large subunit that usually corresponds to the RNase H domain of RT. However, in alpharetroviruses, such as ASLV, the larger subunit (designated β) is a fused RT-IN polypeptide, while the shorter one (designated α) lacks the IN sequences but still retains the RNase H domain. In ASLV virions a free enzymatically active IN is also found [1, 7]. Generation of heterodimeric RT requires cleavage of the Gag-Pro-Pol polyprotein to liberate the large subunits of RT that are then associated into a homodimeric protein (e.g., p66/p66 in the case of HIV-1 RT and β/β in ASLV). A further PR-directed processing yields the final heterodimeric RT versions of p66/p51 and β/α in these two retroviruses, respectively [7, 10, 11]. Interestingly, several isoforms of RT can be found in ASLV virions including the major “mature” β/α, the β/β and the monomeric α isoforms, all of which are fully active, with some notable differences in their enzymatic activity [12, 13]. In HIV-1 RT, only minor differences were reported between the catalytic activities of p66/p51 versus p66/p66 isoforms [14, 15], whereas the p51 subunit by itself is nearly devoid of any enzymatic activity [16].
All monomeric RTs (e.g., those of gammaretroviruses or betaretroviruses) have both DNA polymerase and RNase H domains, and their N-terminal may contain, in some cases, residues from the upstream PR protein. In MMTV the approximately 27 residues that appear at the C-terminus of PR are also present in the RT enzyme (Fig. 1) [17, 18]. This means that cleavages at alternative sites in Gag-Pro-Pol liberate either the full length PR or RT, and the 27-residue segment has a dual function as both the C-terminus of the PR and the N-terminus of RT. In BLV RT, the N-terminal 26-residue segment of RT is also derived from the pro gene (in the second reading frame). However, in this case, the mature PR ends upstream to the 3′-end of the pro gene, and there are not any overlapping sequences between RT and PR [7]. The rest of the BLV RT sequence is derived from the pol gene after −1 frameshifting into the third reading frame [7, 19].
The process of reverse transcription
RTN is initiated after the association of RT with genomic viral RNA and a primer. The primer is required for copying the template RNA by all RTs, and, in most cases, DNA synthesis is initiated from a transfer RNA (tRNA) primer that is supplied by the host cell. A tRNAlys3 is used by lentiviruses and betaretroviruses, tRNApro by gammaretroviruses and tRNAtrp by alpharetroviruses. The tRNA contains 18 nucleotides (nts) at its 3′-end that are complementary to the primer binding site (PBS), located close to the 5′-end of the viral RNA genome (in HIV-1, the PBS is located 180 nts from this end). This enables the hybridization of the tRNA to the PBS [1, 20, 21] (see Fig. 2). In rare cases, as in the LTR retrotransposon Tf1, priming is tRNA-independent and results from an 11-nt self-complementarity between the sequence, located at the 5′-terminus of the genomic RNA and the PBS located further downstream. In this instance, the RNA is nicked at the 3′-end of the primer by the RNase H activity of Tf1 RT, to generate a functional 11-nt self-primer for DNA synthesis [22, 23].
RTN catalyzed by all LTR retroelements and retroviruses is initiated by the synthesis of the first (−) DNA strand that is complementary to the viral (+) strand RNA genome. This synthesis starts by extending the 3′-OH terminus of the RNA primer (either tRNA or self-primer) while copying the 5′-end of the viral RNA genome that contains the unique 5′-end sequence (U5) and the 5′-end repeat (R) region (Fig. 2, step 1). The synthesized DNA strand and the viral RNA produce an RNA–DNA hybrid in which the RNA strand is then hydrolyzed by the RT-associated RNase H activity. This activity, which utilizes the DNA 3′-end-directed cleavage mode to hydrolyze the RNA (see below), leaves the nascent (−) DNA strand as single stranded that is designated (−) strand strong stop DNA (sssDNA). Nonetheless, the RTs of HIV-1 and MLV cleave on average only once for every polymerized 100–120 nts that is insufficient to completely hydrolyze the RNA. Therefore, multiple internal cleavages are probably required to generate gaps that allow a further degradation by the RNA 5′-end-directed mode of RNase H cleavage [1, 2, 24, 25] (see below). The generated (−) sssDNA segment contains a sequence that is complementary to the identical repeat (R) sequences, located at both the 5′ and 3′ -ends of the viral RNA genome (Fig. 2). The length of the R region varies in different retroviruses and ranges from 12 nts (in MMTV) to 235 nts (in BLV), with 98 nts in HIV-1 and 68 nts in MLV [1, 2]. As a result of the duplicated R regions, the (−) sssDNA can hybridize to the R sequences, located at the 3′-end of the viral RNA molecule. Since there are two such RNA molecules in each virion, this template switching step, or first (−) strand transfer (Fig. 2, step 2), can be either intermolecular, by switching to the second RNA molecule, or intramolecular, by switching to the other end of the same RNA molecule. Evidently, these two alternative switches occur at similar frequencies during (−) strand transfer [26, 27]. Interestingly, (−) strand transfer can occur before the synthesis of the (−) sssDNA has been completed. After the nascent DNA hybridizes to 3′-end R region (−) strand DNA synthesis proceeds along the viral RNA, while the RNase H concomitantly degrades the already copied RNA (Fig. 2, step 3).
Most cleavages by RNase H are not sequence-specific; however, specific purine-rich sequences (designated poly-purine tracks, PPTs) show high resistance to RNase H hydrolysis. The major PPT, which is located close to the 3′ end of the viral RNA (hence designated 3′ PPT) and is quite diverse in different retroviruses, serves as a major primer for the second (+) strand DNA synthesis after removing other RNA sequences [28]. Nevertheless, other RNA segments that are left after partial RNA degradation can also serve as minor primers for (+) strand DNA synthesis [2, 6] (Fig. 2, step 4 and 5). Thus, although all retroviruses initiate (+) strand synthesis using the 3′ PPT as a primer [28], some retroviruses, such as HIV-1, equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV) and ASLV, as well as the retrotransposon Ty1, begin (+) strand DNA synthesis from additional RNA priming sites. The most notable site for a second PPT is located in the center of the viral genome in the IN-encoding sequence and termed central PPT (cPPT) [2, 29, 30]. Potentially, all (+) DNA versions, which are primed by different RNA segments, may play some roles in the following steps of RTN (Fig. 2, steps 4 and 5).
Synthesis of (+) DNA strand continues until RT starts copying the tRNA primer (Fig. 2, step 5). The first 18 nts at the 3′-end, which are complementary to the viral PBS, are copied by RT, but the next nucleotide in the tRNA is a modified “rA” and cannot serve as a template for DNA synthesis. The synthesized DNA strand and the 3′-end of the tRNA generates a new RNA-DNA hybrid that can serve as substrate for the RNase H activity. In most retroviruses, the RT-associated RNase H removes the complete tRNA segment, while in HIV-1 the RNase H activity of RT cleaves exactly one nucleotide from the junction between tRNA and DNA, thus leaving an extra “rA” at the 5′ end of the (−) DNA strand. The newly formed second DNA strand is designated (+) strand strong stop DNA (sssDNA).
At this stage, the R region, the U5 sequence and the unique viral RNA sequence, located at the 3′-end (U3), have been brought all together to form the 3′ LTR of the (+) strand DNA (Fig. 2, step 6). The (+) strand is discontinuous and is annealed to nearly a full length (−) DNA strand. Subsequently, a second template switch, or second (+) strand DNA transfer, is catalyzed by RT. This strand transfer is mainly intramolecular [26] and is mediated by annealing of the single-stranded DNA form of the PBS sequence, located at the (+) sssDNA, to the complementary region at the 3′ of the (−) DNA (Fig. 2, step 7). This results in the transfer of the 3′-end of the (+) DNA strand to the 3′-terminus of the (−) DNA strand, or the generation of a circular DNA molecule. Once the second transfer occurs, both DNA strands can be further extended by the DDDP activity of RT (Fig. 2, step 8). DNA synthesis may require the strand displacement activity of RT to remove annealed DNA strands (Fig. 2, step 8, and see below) or possibly may involve the DNA repair activity of host cell enzymes. The complete process eventually leads to a full length, integration-competent, double-stranded DNA with two identical LTRs, one at each end.
Further analysis of unintegrated retroviral DNA in cells infected by different retroviruses has suggested that the generation of the full length (+) DNA strand during RTN may have additional complexities. Both MLV and MMTV appear to have large gaps in their (+) DNA strand. In contrast, (+) DNA strand of HIV-1 contains a central DNA tail or flap in which there are two overlapping (+) strand segments. Such an element is created after the second strand transfer, when (+) strand DNA synthesis that was initiated at the 3′-end PPT proceeds downstream and partially displace an already synthesized (+) DNA strand extending from the cPPT (Fig. 2, step 8). During this process, the cPPT-extended DNA strand is dislocated by the strand displacement activity of RT to allow synthesis of DNA and generation of the flap. The DNA flaps are more abundant in ASLV (+) strands and may also facilitate recombination between two genomic viral RNAs (see below). The size and position of the DNA flaps are determined by the site of DNA initiation and a specific termination sequence located further downstream. The position of this termination sequence in HIV-1 creates a DNA flap (88 or 98 nts-long) that is located downstream to the cPPT, while in Ty1 the flap is up to 130 nts. The viral nucleocapsid (NC) protein (see below) increases the rate of DNA synthesis, as it was suggested that this protein stabilizes structural fluctuations within the central flap [30]. One theory suggests that the central flap is required to introduce flexibility into the center of the DNA duplex, thus helping with the formation of a pre-integration complex (PIC). The PIC, which is composed of the fully complete linear double-stranded viral DNA, the viral proteins, RT, IN, NC, viral protein R (Vpr) and cellular proteins, undergoes nuclear localization in non-dividing cells [31, 32]. Nevertheless, the importance of this single-stranded central flap is yet not fully settled, as an HIV-1 mutant that carries a disrupted central DNA flap can be still pathogenic in vivo [33, 34]. In addition, altering the central flap had little effect on the efficiency of nuclear localization of the PIC [35].
There are still several additional complexities in the RTN process, and we will further highlight some of them. Since the viral DNA, synthesized during RTN, is a substrate for integration by IN, the ends of the linear viral DNA must be reasonably accurate. The RNase H cleavage of the tRNA primer defines the U5 end, and the removal of the PPT primer defines the U3 end of this DNA. Although the RNase H activity does not usually recognize specific sequences, it performs these particular cleavages with a high accuracy; therefore, the ends of the linear viral DNA are well defined to the precise nucleotide. The presence of two identical (or almost identical in case of mutations) copies of the viral RNA genome in each virion may also be important for productive infections. Such a virion can survive even if one of the RNA molecules contains single breaks, as RTN that was initiated on this strand can proceed by transferring the nascent DNA to the second RNA template (by a mechanism designated forced strand transfer, see below [36]). Theoretically, when two copies of the RNA genome are present, a complete DNA copy can be made even if both RNA molecules are extensively nicked, provided that there are no identical nicked sites at both RNA molecules. The effects on genetic recombinations might be another possible reason for the presence of two RNA copies. In host cells that contain two different integrated proviral DNA genomes, virions with two related RNA genomes but with different sequences can be produced. Switches between these two RNA templates can then generate a recombinant genome that is composed of different segments of the two parental sequences. Evidently, in cells that contain only one integrated DNA genome, the two RNA copies will be identical (unless errors occur during transcription), and, therefore, template switches will not bear any noteworthy consequences. This may have fundamental consequences. For instance, as frequently happens in HIV-1, recombination can lead to viruses that are resistant to multiple drugs, in contrast to the parental viruses that are resistant to just a single drug.
The three-dimensional structure of RTs
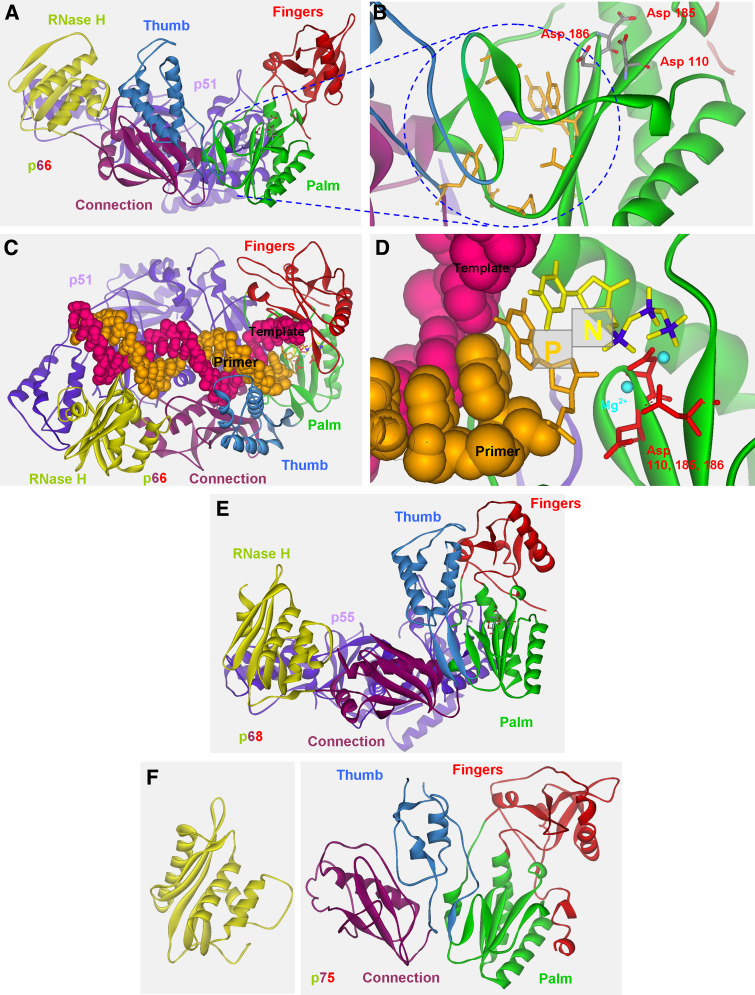
As described above, three enzymatic activities of RTs are required to generate the double-stranded DNA from the viral single-stranded RNA genome. DNA synthesis generates either the (−) or (+) viral DNA, whereas RNA degradation removes both the viral RNA and the tRNA initially used for priming the (−) DNA strand [1, 2]. To enable parallel, multi-functional activity during the RTN process, RT must adopt a well-defined structure in solution. Similar to the structures of all studied DNA polymerases, the DNA polymerase domain of all studied RTs forms a structure resembling a right hand conformation that is linked to the C-terminal RNase H domain (Fig. 3). Both catalytic domains are spatially separated. The polymerase domain of RTs can be further divided into fingers, palm, thumb and connection subdomains (the connection is linked to the RNase H domain), each having a unique role in nucleotide incorporation during DNA synthesis. Most of our knowledge on structural features of RTs is based on the many crystal structures of HIV-1 RT, both the wild-type protein and drug-resistant mutants. Many of these studies determined the crystal structures of HIV-1 RT in complex with the DNA–RNA substrate and/or with RT inhibitors. Additional information is available from a few crystal structures of HIV-2 RT and MLV RT, thus allowing the comparison of structures from different RTs in order to highlight structural elements common to all RTs.
Fig. 3.
The 3-dimensional structures of HIV-1, HIV-2 and MLV RTs. a Spatial structure of wild-type HIV-1 RT (PDB entry 1FK9) with its defined subdomains. HIV-1 RT was co-crystallized with the inhibitor efavirenz and is displayed from the side view after removing the inhibitor. b Magnification of the NNRTI hydrophobic pocket from a along with the three aspartic acids that form the catalytic site of the DNA polymerase domain. a, b Adapted with permission from Herschhorn et al. [201]. c The catalytic complex of HIV-1 RT together with a primer-template and dTTP (PDB entry 1RTD) from a top view. RT is displayed as solid ribbon, the template-primer as CPK (Corey, Pauling, and Kultin), and the 3′ nucleotide of the primer as well as the dTTP are displayed as a stick model. d Magnification of the N and P sites (marked as N and P) adjacent to the DNA polymerase sites from c and their position in relation to the three Asp catalytic residues. In c and d a hydroxyl was added to the 3′ position of the ribose in the P site to illustrate the presence of dNTP during DNA synthesis (the original complex was co-crystallized with ddNTP nucleotide). e The structure of HIV-2 RT (PDB entry 1MU2). f The structure of MLV RT was resolved in detail as two fragments, one for DNA polymerase domain (PDB entry 1RW3, right side) and the second for the RNase H domain (PDB entry 2HB5, left side). In all panels, the fingers subdomain is depicted in red, palm subdomain in green, thumb subdomain in light blue, connection subdomain in purple and RNase H domain in yellow (all of the large subunit or the only subunit in the case of MLV). All structures were displayed with Accelrys Discovery Studio Visualizer v1.6 (Accelrys Software Inc.)
HIV-1 RT
HIV-1 RT is an asymmetric heterodimer that is composed of two subunits. The larger p66 subunit is 560 residues long and exhibits all the enzymatic activities of RT. The DNA polymerase domain of HIV-1 RT adopts in solution the right hand conformation with four defined subdomains [37]. The fingers are located at residues 1–85 and 118–155, the palm at residues 86–117 and 156–236, the thumb at residues 237–318, and the connection to the RNase H is located at residues 319–426 [37, 38]. The RNase H domain extends approximately beyond residue 440. The smaller p51 subunit is about 440 residues long and is identical to the N-terminal portion of the p66. The p51 is cleaved from a p66 subunit by the viral PR after association of two p66 subunits to generate p66/p66 homodimers [11] (see above). For the most part, the p51 subunit stabilizes the heterodimer but may also participate in binding the tRNA primer [1, 2]. This subunit can also affect the RNase H activity [39, 40]. The fingers, palm, thumb and connection subdomains in the p51 subunit (except for the C-terminus of the connection, [1]) fold similarly to the corresponding subdomains in the p66. However, the spatial organization of each subdomain relative to the others differs substantially in the two RT subunits [37]. As a result, p51 is more tightly packed, with the connection rotated to cover the palm and displacing the thumb [1]. Interestingly, though the sequence of p51 is identical to the N-terminal portion of p66, the tertiary structures of the two subunits are considerably different. This unique case shows that similar primary and secondary structures do not necessarily lead to similar tertiary protein structures.
All p66 subdomains, along with the thumb and connection subdomains of p51, participate in binding the nucleic acids, but the RNase H domain plays only a minor role in this binding. The connection subdomains of both subunits, together with the thumb of the p51 portion, form the “floor” of the binding cleft [38]. HIV-1 RT binds the nucleic acid substrate in a configuration that places the substrate in direct contact simultaneously with both the DNA polymerase and RNase H active sites. The distance between the two active sites is about 60 Å [41]. This allows positioning the substrate so that 17 nts (for DNA–DNA) or 18 nts (for RNA–DNA) separate the two sites on the nucleic acid substrate. DNA synthesis is an ordered reaction in which RT first binds its primer-template substrate and places the 3′-OH group of the primer terminus at the priming site (P site) that is located next to the polymerase active site (Fig. 3) [38, 42]. Comparison of crystal structures of the unliganded RT with structures of RT bound to its nucleic acid substrate shows that the binding results in a change of the position of the p66 thumb from “close” to “open” conformation [43, 44]. The flexibility of the thumb allows movements from the “close” position, in which the thumb touches the tip of the fingers to the “open” position, in which the thumb moves about 30° away from the fingers. A contradictory report observed a conformation similar to the “open” state for the unliganded RT, but this was probably due to growing the RT crystals with a weakly binding non-nucleoside RT inhibitor (NNRTI) that was later soaked out from the crystal [45]. The conformational change associated with binding to the primer template is followed by a second binding of a dNTP to the nucleotide binding site (N site) to form a ternary complex (Fig. 5) [42]. The active site environment facilitates the trans-esterification reaction that leads to the incorporation of the incoming dNTP into the primer (see below). The nucleic acid substrate must then translocate relative to RT to free the N site for the next dNTP. Accordingly, RT is actually moving along the nucleic acid substrate and adding the next nucleotide in each cycle of processive synthesis.
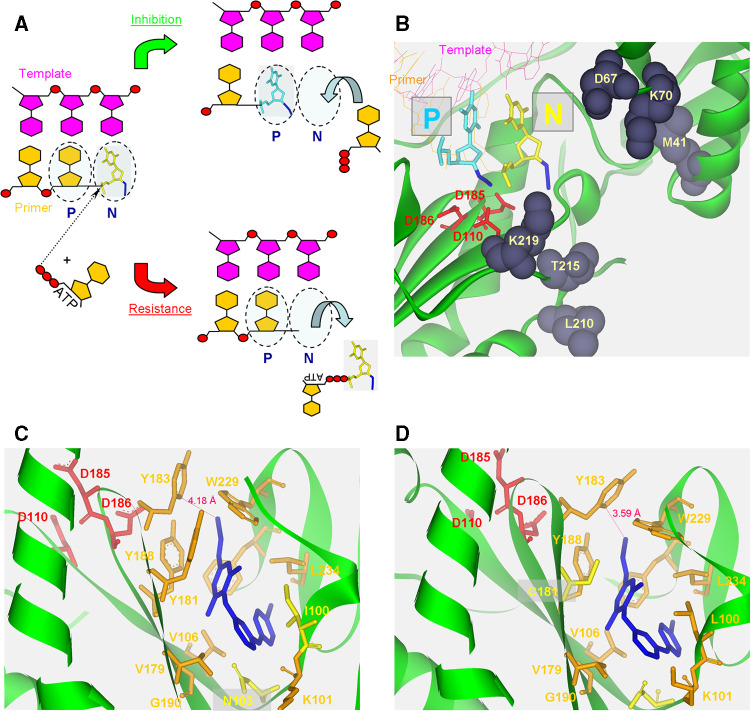
Fig. 5.
HIV-1 RT inhibitors and drug-resistant RT mutants. a Incorporation of AZT-TP into the nascent DNA strand by HIV-1 RT can lead to two alternatives. Translocation of the incorporated NRTI into the P site (P) may be followed by binding of the next dNTP to the N site, resulting in the formation of a dead-end complex and inhibition of any further DNA synthesis. Conversely, excision of the chain terminator, AZT-MP, which is mediated by ATP as a pyrophosphate donor, releases the incorporated NRTI and consequently reverses inhibition. b The three-dimensional structure of the incorporated AZT-MP at the end of the primer at either the N site (N, yellow) or the P site (P, cyan). The pre- and post-translocation structures of HIV-1 RT are from PDB entries 1N6Q and 1N5Y, respectively. The coordinates of AZT-MP at the P site were taken from the post-translocation structure and were used to insert this molecule into the pre-translocation structure. This was done to indicate the general orientation of AZT-MP, and it does not necessary represent the exact position of AZT-MP in the P site. Residues that are associated with increased resistance to AZT are displayed in CPK style and are labeled. The primer and template are displayed in line style and the AZT-MP in stick style (yellow for the N site and cyan for the P site). HIV-1 RT is displayed as a solid ribbon in green. c, d The hydrophobic pocket of HIV-1 RT that binds NNRTIs in two structures of RT mutants. HIV-1 RTs with different mutations, which are associated with resistance to classical NNRTIs, were co-crystallized with the TMC278 NNRTI, a broad-range inhibitor of many mutant RT variants. The double mutant L100I + K103N is shown in (c) and the double mutant K103N + Y181C in (d). The highly potent inhibitor is flexible and can accommodate and interact with different residue in different mutants. Inhibiting the K103N + Y181C double mutant is mediated at least partially by interacting with Y183 that leads to its movement and the repositioning by 1.5 Å in the conserved Y183 MDD [194]. Residues of the hydrophobic pocket are displayed in orange and the mutant ones in yellow. All residues are presented as stick model and labeled. The distance between Y183 and the cyano group of TMC278 in both structures is also shown. All structures were displayed with Accelrys Discovery Studio Visualizer v1.6 (Accelrys Software Inc.)
The DNA polymerase active site is composed of three highly conserved Asp residues at positions 110, 185 and 186, located within the palm subdomain of p66. The carboxylate moieties bind two metal ions, probably Mg+2 in vivo, facilitating the incorporation of new dNTPs into the primer. The precise alignment of the catalytic residues of RT, the 3′-OH group of the primer and the α phosphate of the incoming dNTP allows a nucleophilic attack of the 3′-OH on the α phosphate group, consequently liberating a pyrophosphate molecule that leaves the active site as the fingers open. Structural differences between the pre- and post-translocation complexes have shown that the conserved YMDD loop may act as a “springboard” during translocation to move the primer terminus from the N to the P site after dNMP incorporation [46]. There is also evidence that the pyrophosphate of the added nucleotide is released before translocation takes place [47, 48]. Specific roles in the polymerization reaction are associated with several defined regions and motifs within the DNA polymerase subdomains of HIV-1 RT. The catalytic Asp at positions 185 and 186 are also part of the YMDD motif of HIV-1 RT. This motif is highly conserved among RTs; however, the Met is replaced by Val, Leu or Ala in other RTs. The “primer grip” at the p66 β12–β13 (β-sheet12–β-sheet13, residues 227–235) hairpin in HIV-1 RT facilitates the exact position of the 3′-OH of the primer within the primer site [38]. The “template grip” at β4 (residues 73–77), αB (α-helix B, residues 78–83), β5a (residues 86–90) and the β8–αE (residues 141–174) connecting the loop portions of the p66 palm and fingers is closely associated with the template strand [1]. In addition, the β3–β4 loop is involved in many RT activities including processivity [49], fidelity of DNA synthesis [50] and strand displacement synthesis [51]. The helixes αH (residues 255–268) and αI (residues 278–286) of the p66 thumb extensively contact the primer template [38]. There are also several conserved residues in the N site of HIV-1 RT, including Tyr115, which facilitate deoxyribose ring binding of the incoming dNTP and functions as a steric gate discriminating between rNTPs and dNTPs (see below) [52–54], Arg72 and Lys64, which coordinate the triphosphate group of the incoming dNTP phosphates, and Gln151 that binds the 3′-OH of the incoming dNTP [42]. Many crystal structures of HIV-1 RT complexed with different NNRTIs demonstrate that the inhibitors bind a specific hydrophobic pocket, located about 10 Å underneath the catalytic active site of the DNA polymerase domain (Fig. 3). This pocket is rich in aromatic residues, such as Tyr188 and Tyr181, and will be discussed in detail below (in the section on inhibitors of RT and drug resistant RT variants).
The RNase H activity of HIV-1 RT degrades the RNA strand in the RNA-DNA heteroduplex during DNA synthesis. In addition to the removal of the template RNA and the primer tRNA, it also produces the PPTs for the (+) DNA strand synthesis and eventually hydrolyses them (see above) [55]. The structure of HIV-1 RT RNase H shows a high structural similarity to E. coli RNase H, most remarkably in the catalytic cores [56]. This is quite surprising, since the primary sequence identity between the two proteins is merely about 20% [1]. It should be emphasized that the specific activity of the bacterial enzyme is at least 100-fold higher than that of the complete retroviral RTs. Moreover, the isolated HIV-1 RT RNase H domain is by far less active than the intact RT molecule [57, 58]. This is probably due to the inherent low capacity of the RNase H to bind the primer-template substrate that is mostly bound during synthesis by the DNA polymerase domain. This suggestion is further supported by evidence that the activity of the isolated HIV-1 RNase H domain can be restored. Restoration of this activity can be accomplished by inserting a basic loop, present only in the bacterial RNase H and not in HIV-1 RNase H, into the isolated RNase H domain [59, 60]. Alternatively, the RNase H activity can be restored by mixing in trans the isolated HIV-1 RT RNase H domain with the polymerase domain of the same RT [61, 62] (see also below).
The HIV-1 RT RNase H domain folds into a five-stranded mixed beta sheet, flanked by four alpha helices. The active site is surrounded by a cluster of four conserved acidic residues: Asp443, Glu478, Asp498 and Asp549 [56]. In the original crystal, the acidic residues interacted with two Mn2+ ions located 4 Å from each other, but it is likely that in vivo both metal ions are Mg+2 [6]. The two metal ions are asymmetrically coordinated in the active site and have distinct roles in activating the nucleophile and stabilizing the transition state [55].
The co-crystals of HIV-1 RT, prepared with duplex DNA, showed that the DNA in the vicinity of the polymerase active site is more close in structure to an A-form helix, whereas the DNA near the RNase H site is more structurally close to a B-form, with an angle of about 40°–45° (forming a “kink”) between the helical axes of the two forms [38]. All three RNA-RNA, RNA-DNA and DNA-DNA duplexes are used during RTN and have to be properly positioned in the binding cleft. However, RNA-RNA duplexes cannot assume a B-form structure and, therefore, the DNA substrate in the DNA polymerase site may be constrained to assume the A form to give a relatively uniform structure to all template primers used for synthesis [1]. The nucleic acid structure near the RNase H site has a minor groove about 9 Å wide. This may contribute to the cleavage specificity of RT as substrates with narrower width are poorly cleaved. Cleavage specificity is also controlled by the RNase H primer grip. This structural element consists of several amino acids that interact with the DNA primer strand near the RNase H active site. The specific residues are: Lys395 and Glu396 of p51, Gly359, Ala360 and His361 from the p66 connection subdomain and Thr473, Asn474, Gln475, Lys476, Tyr501 and Ile505 from the p66 RNase H domain [41]. The amino acid distribution in several domains highlights the existence of tight cross interactions between residues located in different domains of HIV-1 RT (in this case, residues in the p51 subunit and residues in the DNA polymerase and the connection of the p66 subunit, affect the RNase H activity).
HIV-2 RT
Like HIV-1 RT and probably all other lentiviral RTs (for example, the RTs of FIV [63, 64], bovine immunodeficiency virus (BIV) [65] and EIAV [15, 66, 67]), HIV-2 RT is an asymmetric heterodimer composed of two subunits. The large p68 subunit, which is 559 residues long, is similar to the p66 of HIV-1 RT, whereas the smaller subunit (that was found in different studies to be either p55 or p54) is analogous to the HIV-1 p51 subunit. The amino acid sequence of HIV-2 RT shows a significant homology to the sequence of HIV-1 RT [68]. The smaller subunit of HIV-2 RT lacks the RNase H domain, similar to HIV-1 RT, and the remaining fingers, palm, thumb and connection subdomains are more tightly packed than there are in the p68 subunit. Interestingly, the HIV-2 RT smaller subunit, produced by HIV-2 protease, is 484 residues long, while the comparable p51 subunit of HIV-1 RT is only 440 residues long, resulting in a significantly longer connection subdomain in HIV-2 RT [69]. In contrast, the large subunits of HIV-2 and HIV-1 RTs differ in length by only a single residue (559 amino acids in HIV-2 RT and 560 in HIV-1 RT). The disparity in the processing of the large subunits of the two HIV RTs to form the smaller subunits is attributed to the distinct specificities of the two HIV proteases. Accordingly, bacterial proteases, with yet a different specificity, have produced a substantially shorter version of the small HIV-2 RT subunit that is only 427 residues long [70]. The structure of HIV-2 RT has the greatest similarity to the unliganded HIV-1 RT structure when comparing the different structures of HIV-1 RT. The thumb subdomain of p68 in unliganded HIV-2 RT is rotated by 8° relative to the unliganded HIV-1 RT [71]. HIV-2 RT also forms a more stable heterodimer than HIV-1 RT does [72].
The most significant structural differences between HIV-2 RT and HIV-1 RT are located in the NNRTI binding pocket, near the DNA polymerase active site (Fig. 3) [71]. The conformation of Ile181 in HIV-2 RT, instead of Tyr181 in HIV-1 RT, can contribute to the resistance of HIV-2 RT to many commonly used NNRTIs. In addition, residue 188, which is a Leu in HIV-2 and Tyr in HIV-1 RT, as well as the lack of the aromatic side chains at positions 181 and 188 of HIV-2 RT, abolish the effect of ring stacking interactions with many NNRTIs. Also, the positions of several conserved residues are modified in HIV-2 RT relative to HIV-1 RT and thus can further affect binding to inhibitors.
MLV RT
MLV RT represents a group of monomeric retroviral RTs that includes also the RTs of MMTV, BLV, porcine endogenous retrovirus (PERV) and of the LTR retrotransposon, Tf1 [7]. The crystal structure of the full length enzyme, which is composed of 671 residues (75 kDa), was determined at a 3 Å resolution [73] and provided the first resolved structure of a monomeric RT. To obtain suitable crystals for structure determination, Das et al. [73] had to improve the solubility of MLV RT by a deletion of the first 23 amino acids and by substituting Leu435 by a Lys residue. This MLV variant still retained full enzymatic activity and required for crystallization the presence of a 14 nts DNA duplex that is rich in A and T nucleotides. The single polypeptide contains the known polymerase subdomains: fingers, palm, thumb and connection, followed by the RNase H domain. The four subdomains were determined from a well-ordered electron density, whereas the RNase H could be only generally positioned, and thus its exact structure was independently determined [74] and its coordinates were used for the whole RT structure. The fingers and palm subdomains of MLV RT fold into a rigid and stable structure that is independent of the rest of the enzyme. The structures of these subdomains, when expressed as the N-terminal portion of the enzyme, are nearly identical for both the unliganded fingers-palm segment and the one complexed with DNA. They are also remarkably similar to the same subdomains in the full-length MLV RT structure [73].
The structures of HIV-1 RT and MLV RT have been recently compared in detail [75], and, therefore, we will only briefly describe the major differences and similarities between the two enzymes. There are several conserved elements that are common to the two enzymes. The polymerase active site in MLV RT is located in the palm subdomain, composed of three aspartic acids at positions 150, 224 and 225 that are analogs to the corresponding residues at positions 110, 185 and 186 of HIV-1 RT. The highly conserved YXDD motif, which includes two of the three catalytic aspartic acids, is similar in both enzymes, where X is a Val in MLV RT and a Met in HIV-1 RT. These two RTs share a conserved positively charged patch on their surface that may contribute to the recognition and binding of the nucleic acid substrate. In addition, the conserved sequence LPQG appears at residues 188–191 in MLV RT and 151–154 in HIV-1 RT. Lys65 and Arg72 of HIV-1 RT, which contribute to dNTPs binding, are located in MLV RT at positions 103 and 110, respectively. The RNase H domain of both enzymes carries identical catalytic residues that are Asp524, Glu562, Asp583 and Asp653 in MLV RT and Asp443, Glu478, Asp498 and Asp549 in HIV-1 RT [75]. In addition, sequence alignment of the RNase H domains of HIV-1, HIV-2 and MLV RTs suggests that residues that form the RNase H primer grip are significantly conserved [41].
There are also considerable differences between the structures of the two RTs. The MLV RT polypeptide is longer by 111 residues than the comparable p66 subunit of HIV-1 RT. Structure-based sequence alignments suggest that these additions are distributed mainly in three different regions of MLV RT [75]. At the N-terminal portion there are an additional 40 amino acids. Another 32 residues that are positioned between the connection and RNase H may represent a highly flexible region with an unstructured pattern. In addition, there is a short segment that folds into a putative C-helix (amino acids 594–601) and a loop region. Both are also missing in HIV-1 RT and the C-helix was deliberately deleted from the isolated MLV RT RNase H domain that was used for crystallization. Interestingly, a similar motif is present in E. coli RNase H, and it is part of the basic helix loop. When this segment was inserted into the RNase H domain of HIV-1 RT, it could restore the RNase H activity of the isolated RNase H domain (as discussed above). This motif may strengthen the binding of the RNase H domain to DNA and contribute to its independent catalytic activity. Both E. coli RNase H and the isolated RNase H domain of MLV are fully enzymatically active. This is in contrast to the HIV-1 RNase H domain that is highly dependent for its catalytic activity on the fused DNA polymerase domain. The relative positions of the thumb, connection subdomains as well as the RNase H domains of MLV RT give rise to a more compact structure with a clamp shape in comparison to the structure of HIV-1 RT [73]. Notably, the polypeptide linking the connection and RNase H is the most significant difference between MLV RT and HIV-1 RT. In both proteins, this linking peptide exits the connection subdomain in a different and opposite direction. Some differences were also observed in the “minor grove binding track” motif that is located within the thumb subdomain and supports the translocation of the substrate during polymerization. Important residues in the “minor grove binding track” of HIV-1 RT such as Gln258, Gly262 and Trp266 are not conserved in MLV and correspond in this enzyme to Arg298, Glu302 and Thr306 according to structure-based alignments [73, 76]. These residues are part of an α helix that has a different path and length in the two RTs [73].
The biochemistry of RTs
All retroviral RTs produce an integration-competent, double-stranded DNA from the single-stranded viral RNA genome, combining both polymerizing and hydrolytic functions to synthesize about 18,000 phosphodiester bonds in a single RTN cycle. As shown above, the RTN cycle requires the precise combination in time and space of both RT-associated DNA polymerase and RNase H activities. Genetic, structural and biochemical studies as well as protein sequence alignments have all indicated that a single DNA polymerase active site is responsible for the RDDP and DDDP functions, while the RNase H activity is provided by a separate active site; still both sites are located on the same polypeptide. The DNA polymerase domain is located in the N terminus of the large subunit of RT, occupying about two-third of the subunit, whereas the RNase H domain is positioned on the C-terminal portion of this polypeptide (Fig. 3). Despite the spatial separation, there are both functional and structural mutual interactions between the two catalytic activities [1, 2].
The first methodical studies of RTs were performed with enzymes purified from relatively large quantities of purified virions, obtained from either infected animals or from virus grown in vitro in cell cultures. Lysis of the virions in buffers containing mild non-ionic detergents, such as Triton X-100 or NP-40, has led to the release of a soluble, enzymatically active and relatively stable enzyme that could be purified to homogeneity by a combination of ion exchange and affinity chromatography columns. In fact, early efforts to show an RDDP activity in virions owe their success, at least in part, to the use of such detergents in the enzyme preparations.
Almost none of the RTs undergo post-translational modifications. The only exception is ASLV RT that undergoes phosphorylation in the IN-related sequence close to the C-terminus of the β subunit (probable on Ser854) and may have some effect on the enzyme’s activity [7, 77, 78] (see also above). Comparative analyses of various RTs did not reveal significant differences between recombinantly expressed RTs and those released from viruses. As the technology to produce recombinant protein became widely available, a variety of highly active RTs could be expressed in substantial amounts as recombinant proteins, mostly in bacteria. Consequently, almost all structural and enzymatic studies of RTs in the last 2 decades were performed with the recombinant proteins, taking advantage of the ability to get large amounts of proteins and the simplicity to generate mutant variants. Consequently, almost all studies mentioned in this review were carried out, unless otherwise stated, with recombinantly expressed RTs.
The DNA polymerase activity of RT
General properties
RTs can be considered for the most part as a DNA polymerase that, based on structure and many of its catalytic features, is similar in many respects to other cellular and viral DNA polymerases. However, unlike other DNA polymerases (that are all DNA-dependent), RTs can copy RNA as efficiently as DNA. As a result of numerous biochemical, genetic and structural studies, which were performed mostly with HIV-1 and MLV RTs, the mechanisms of DNA synthesis of RTs are reasonably well understood. Similar to other DNA polymerases, RTs require both a primer and a template. Thus, they perform a template-directed extension of DNA or RNA primers by dNTPs incorporation according to Watson and Crick base pairing. Polymerization is done by extending the 3′-end hydroxyl of the primer by one nucleotide at a time, while forming a 3′–5′ phosphodiester bond that is accompanied by a pyrophosphate release. Accordingly, primers that lack a free 3′-OH, or nascent DNA molecules with an incorporated dNTP analog lacking the 3′-OH, cannot be extended [1, 3]. Based on this mode of action, many of the nucleoside/nucleotide analogs, routinely used to inhibit HIV-1 RT in AIDS patients, terminate DNA chain elongation as they lack the 3′-OH (see below). The polymerization reaction is reversible, and under special conditions that lead to synthesis arrest, excision of the 3′-terminal nucleotide can also occur in the presence of either pyrophosphate or ATP as phosphate donors (by a process of pyrophosphorolysis that is discussed in detail below).
As described above, the polymerization reaction is ordered in sequence, starting with the binding of RT to the template-primer substrate and followed by binding of the incoming dNTP to the N site. The binding rates of the correct versus incorrect incoming dNTPs are quite similar and are diffusion limited. Yet, the correct dNTP is bound better than the incorrect dNTP, due to the preferential hydrogen bonding with its complementary template nucleotide. Other interactions also participate in selecting the correct nucleotide, including: base stacking and residues that form the N site (see above). In addition, the geometry of the base pair formed was found to be even more important than the correct hydrogen bonding between the incoming dNTP and the template nucleotide [79]. Still, misincorporation of a wrong nucleotide is quite recurrent in RTs, especially in lentiviral RTs (see below). Since all RTs are deficient in the 3′ → 5′ exonucleolytic activity, which is common to many other DNA polymerases, the resulting lack of proofreading largely contributes to the high error rates observed for RTs. The rate-limiting step in a single nucleotide incorporation is the conformational change that accompanies the movement of the finger subdomain of p66 to close down on the incoming dNTP in order to properly align its α-phosphate with the 3′-OH of the primer (or the nascent DNA strand) and the polymerase active site. The incorporated new nucleotide is then translocated from the N to the P site of the RT (for details, see above). Yet, the overall polymerization by RTs is limited by the relatively low dissociation rate from the nucleic acid substrate [80, 81].
All RTs require divalent cations for DNA synthesis; Mg+2 appears to be the preferred one in vitro and probably also in vivo. Nonetheless, the RTs of MLV and other gammaretroviruses show an in vitro preference to Mn++ with several substrates. Two divalent cations molecules are required to coordinate the oxygen atoms of all three phosphates of the incoming dNTP and the side chains of the three catalytic aspartic acid residues (at positions 110, 185 and 186 in HIV-1 RT, Fig. 3). This interaction activates the 3′-OH of the primer and facilitates the nucleophilic attack on the α phosphate of the incoming dNTP. Additionally, the charges of the reaction intermediates are also stabilized by the two divalent cations.
The rates of polymerization in vitro by RTs are fairly slow relative to other DNA polymerases. The in vitro-measured rates depend substantially on the nature of substrate and the assay conditions. Reported rates varied from 1 to 15 nts per second, depending on the conditions of the performed study. Since it takes about 4 h from infection by HIV-1 to the first appearance of a full viral double-stranded DNA (of about 9 kb), even after considering the complexity of the RTN process, RT-directed DNA synthesis is definitely slow. Although the viral heteropolymeric RNA is the natural substrate for RTs, synthetic homopolymeric heteroduplexes of RNA–DNAs serve in vitro as better substrates. Accordingly, poly(rA)-oligo(dT) and poly(rC)-oligo(dG) are highly sensitive template-primers for testing the RDDP activity of almost all RTs, and the poly(2′-O-methyl-C)-oligo(dG) is considered to be highly specific for this activity. The different substrates for assaying DDDP activity are usually less efficiently used, with poly(dA)-oligo(dT) and poly(dC)-oligo(dG) the most efficient ones for in vitro synthesis.
Unlike other DNA polymerases, all RTs show a relatively low specificity to the templates, as they are capable of copying both RNA and DNA. However, similar to other DNA polymerases, RTs are highly specific in synthesizing only DNA and not RNA. Yet, this property can be modified by mutating an aromatic residue of RT, which is located in the nucleotide-binding pocket of the DNA polymerase domain, and this leads to an RT variant with a significant RNA polymerase activity. Consequently, it was proposed that the aromatic rings of F155, Y115 and F119, in MLV, HIV-1 RT and MMTV RT, respectively, act directly as a “steric gate” that allows the incorporation of dNTPs (that have a hydrogen atom at the 2′-position on the ribose) and discriminates against the incorporation of rNTPs with the larger 2′-OH group [53, 82, 83]. Replacing the bulky aromatic side chains of these residues by smaller amino acids causes the “gate” to open up and allows the larger rNTPs to be incorporated into the nascent polynucleotide chain (see above). Nonetheless, even these mutated RTs are at least as effective in DNA synthesis as in RNA synthesis.
As described above for RTN, during DNA synthesis from the viral RNA, RT must pause at four linear template ends (Fig. 2). In addition, the retroviral genomes are frequently nicked. As a result, RTs may encounter multiple template ends during each replication cycle. Several studies have indicated that RTs can further modify these template ends in vitro by adding non-templated nucleotides to the 3′-ends of the nascent DNA strand, similar to other DNA polymerases that lack 3′ → 5′ exonuclease activity [84–86]. However, in the case of retroviral RTs, such polymerization at template ends requires usually high ratios of RT over template primer, and it is achieved at very high dNTP concentrations that are beyond physiological conditions. Moreover, non-templated nucleotides are added poorly by HIV-1 RT under intracellular dNTP concentrations (5–10 μM), in a rate that is at least 10,000-times slower than templated synthesis. It was also suggested that such additions to blunt-ended DNA by HIV-1 RT can lead to a non-specific strand transfer if the formed non-templated 3′-end overhangs are complementary to 3′-end sequences of the acceptor DNA strand. Since this non-specific strand transfer is achieved at a very high excess (70-fold) of the acceptor strand, the biological relevancy of this finding is still questionable [85]. A much higher capacity to perform non-templated addition of nucleotides in vitro has been recently shown for the RT of the LTR retrotransposon Tf1 [86]. A surprisingly large portion of cDNA molecules of Tf1, produced in vivo in cells, have random, presumably non-templated sequences on both U3 and U5 ends [87]. Therefore, it is likely that the ability of Tf1 RT to incorporate non-templated nucleotides into the nascent DNA strands represents a unique and biologically relevant RT feature.
Processivity of DNA synthesis
The processivity in vitro of various RTs is quite poor relative to other DNA polymerases, as RTs do not stay attached to the growing DNA strand for many successive cycles of dNTP additions [88]. Despite their ability to polymerize up to a few hundred nucleotides in a single synthesis round, RTs tend to fall off in vitro at specific sequences or structures, defined as “strong stops.” Although not all of these sequences are well defined, it was found that several RNA structures (e.g., pseudo-knots) are mostly difficult to copy, thus raising their potential use as RT inhibitors [1]. In addition, some mutations in HIV-1 RT were shown to affect the processivity of the enzyme. More interestingly, a correlation was found in several HIV-1 RT mutants between their increased resistance to nucleoside/nucleotide RT inhibitors (NRTIs) and both the decreased processivity and increased fidelity of DNA synthesis in vitro [89, 90]. Specifically, a decrease in the ability to incorporate nucleoside analogs (which is manifested by an increase in drug resistance) is accompanied by a reduction in both the processivity of DNA synthesis and the capacity to misincorporate dNTPs and extend the formed mismatches (see below) [79, 91–94]. Still, it is not clear whether these findings reflect also a low in vivo processivity in infected cells. It is likely that other factors may be more relevant to the in vivo activity, as it is one of the driving forces behind the rapid mutagenic processes that accompany retroviral replication.
Fidelity of DNA synthesis
As mentioned above, the lack of the 3′ → 5′ exonucleolytic proofreading function can contribute by itself to the low fidelity in synthesizing DNA by RTs. However, other features of RT can also increase the tendency to introduce errors while synthesizing DNA. The accuracy of DNA synthesis has been measured in many in vitro studies, employing both synthetic polynucleotide substrates as well as “natural” DNA or RNA substrates [79, 95, 96]. Base substitutions were detected at high frequencies, due to nucleotide misinsersions as well as the subsequent mispair extensions, all revealing that RTs are much more error prone (by factors ranging from 10 to 103) than cellular DNA polymerases. Interestingly, RTs of lentiviruses (including those of HIV-1 and HIV-2) were found to be less accurate than most of other RTs studied [95–100], highlighting the effects of specific RT sequences on the level of fidelity. Certain dNTP-binding residues, various residues that interact with the template or the primer strands, as well as RT residues in the minor groove-binding track, were all shown to play major roles in the fidelity of retroviral RTs [101]. The sensitivity of RT to NRTIs can be seen as a manifestation of the inherent low fidelity of RT, as the enzyme misincorporates NRTIs instead of the correct dNTPs. Hence, drug-resistant variants of HIV-1 RT may evolve to distinguish better than the wild-type RT between these competitive NRTI and the correct dNTPs (see below). Consequently, in several of these mutants (e.g., E89G, M184V or M184L and Q181I), the reduced sensitivity to NRTIs is accompanied by an enhanced discrimination between the mispaired (but unmodified) nucleotides and the correct nucleotides [79, 93]. The type of nucleic acid copied (RNA vs. DNA) does not seem to affect misinsertion and mispair extension frequencies, while the specific sequences by themselves have substantial effects on the fidelity of each RT tested. This suggests that the fidelity of a given RT is sequence-dependent and enzyme related [99].
The frequencies of base substitutions that are formed by a given RT may vary and depend on the created mispairs. However, misinsertions and the resulting mispair extensions are not the only ways to produce errors during RTN. An error can be introduced by a slippage of the primer strand over the template strand that may result in either nucleotide deletions or insertions. More complex alternatives, such as dislocation mutagenesis, were also proposed [95] and can operate over large distances, thus leading to massive consequences. The in vivo rates, by which retroviruses undergo mutations, are estimated to be between 10−4 and 10−5 mutations per nucleotide. Several studies have tested the correlation between in vivo mutation rates and the in vitro fidelity of RT, using the forward mutagenesis assay with either phage or plasmid DNA with a reporter gene [79, 95, 96]. Correlation to the RTN process is only partial, since it is apparent that this process is much more complex, due to the consecutive strand transfer steps (vide supra) as well as the involvements of other viral and cellular proteins (vide infra). Therefore, it is possible that some of the systems utilized to measure in vitro fidelity of DNA synthesis are valid for mainly comparative purposes, i.e., to evaluate how different enzymes and various RT mutants behave under identical conditions.
The viral RT is not the only factor contributing to the high mutation rates observed in retroviruses. Cellular RNA polymerase II, which has a relatively low fidelity and transcribes the viral DNA into RNA of the progeny virions, also affects retroviral mutation rates. Furthermore, other viral and cellular proteins can also indirectly influence retroviral mutations rates. These include the retroviral dUTPase, the regulatory protein Vpr of HIV-1 [102] and the cellular APOBEC3 [103], all of which are described in detail below.
Initiation of RT-directed DNA synthesis
In almost all cases, the initiation of (−) strand DNA synthesis in RTN depends on a tRNA primer (see above). Elaborate inter- and intramolecular tRNA-genomic RNA interactions appear to be the hallmark of initiating (−) strand DNA synthesis [20]. In addition, the involvement of chaperone activity of HIV-1 NC protein in forming the initiation complexes was well documented as part of multimolecular functional interactions between NC, RT and the tRNAlys3 primer [104–106]. Once initiation complex is generated, the RT-directed (−) DNA strand synthesis takes place in two steps. First, there is an initiation step where up to 6 nts are added to the 3′-end of the primer in a non-processive manner. Afterwards, there is an increase in the affinity of RT towards the substrate that is accompanied by a huge increase of the polymerization rate (by approximately 3,000-fold). This represents a transition from the initiation step to elongation that probably depends on structural features of the tRNAlys3, such as modified bases at the anti-codon domain [20]. Concurrent with the synthesis of (−) sssDNA, the RNase H activity of RT degrades the RNA template with the help of the viral NC that can unwind 8–12-nts-long double-stranded sequences [104–106] (see also below). Non-specific initiation events, which are likely also to take place (due to nicks in the genomic RNA), are repressed by NC that coats the RNA and unwinds unstable RNA duplexes.
Synthesis of (+) strand DNA is initiated from the 3′ PPT and some additional RNA primers, which are relatively resistant to the degradation by RNase H during partial removal of the (+) RNA strand (see above). In HIV-1 RT (+) strand synthesis is initiated also from both the cPPT and the 3′ PPT (vide supra). Although the 3′ PPT is the major (+) strand initiation site, in several lentiviruses the presence of the cPPT was shown to be critical for an efficient replication. The (+) strand DNA priming is much more efficient with PPT primers than extending non-PPT RNA primers annealed to DNA. Since PPTs are generated by precise cleavages of the viral RNA that is annealed to the (−) strand DNA by the RNase H activity, the initiation and elongation of (+) strand DNA requires a high coordination between both RT-associated activities, as described below (for review, see [28]).
Strand transfer activity
As outlined above, during RTN there are two strand transfer events, also referred to as template switches (Fig. 2). In both cases, there is a switch of either the 3′-end of the elongated primer, or the 3′-end of the nascent (+) DNA strand to an acceptor template. These switches depend on sequence complementarity usually found at the 3′-end of both the donor and acceptor strands (for a recent comprehensive review, see [36]). Retroviral virions carry two similar (+) RNA copies of the viral genome. Therefore, (−) strand transfer can be potentially either intermolecular or intramolecular, and both types were observed with similar frequencies. As the acceptor template is the (+) RNA genome, which is present in two copies in each virion, equal ratios of inter- or intra-molecular strand transfer may actually represent the probability to encounter a single RNA strand. On the other hand, (+) strand transfer is predominantly intramolecular [26, 27]. This could be expected, as intermolecular strand transfer would require the less likely scenario of two (−) strand DNA copies. Additional strand transfer events were suggested in the forced copy choice model. According to this model, RT initiates (−) DNA strand synthesis on a single genomic RNA molecule, but upon reaching a break in this template it switches to the second RNA molecule, which was co-packaged in the same virion, to continue the synthesis. This mechanism was supported by in vivo studies conducted with viruses containing a relatively large amount of cleaved genomic RNA. Since these viruses were fairly infectious, it is likely that strand transfer rescued the viruses with damaged genomes. A different model is the copy choice model that expands the alternative of (−) strand template switching to include intact templates rather than nicked ones. In both models, there is a requirement for high sequence homology between the donor and acceptor template molecules. Evidently, during DNA synthesis the nascent DNA strand reaches a region, in which the synthesis on the second strand is more favorable than synthesis on the current strand, leading to a strand transfer. This can be enhanced by specific sequences that are mostly pausing sites, but additional factors may also affect strand transfer. Theses include the delicate balance between the DNA polymerase and RNase H activities of RT [36] (see below) and the chaperone activity of the viral NC protein, which facilitates the annealing of the acceptor RNA to the (−) strand DNA via their complementary sequences (see below). An additional model, designated the dynamic copy-choice model for retroviral recombination, was proposed. Here, the steady states between the rates of polymerization and RNA degradation determine the frequency of template switching by RT [107].
Strand displacement activity
This activity is common to all RTs and absent in most other DNA polymerases. Strand displacement is required during (+) DNA strand synthesis, when the synthesis has been initiated from multiple primers (Fig. 2). This activity is mediated by the RT-associated helicase activity that enables the enzyme to unwind the double-stranded DNA during strand displacement synthesis. Displacement of a downstream (+) DNA strand may assist the removal of discontinuous (+) DNA segments in order to generate the complete double-stranded DNA that is vital for a successful integration into the host genome. This activity can also generate the central DNA flap, as described above. Although the rates of both displacement and non-displacement DNA synthesis vary for different sites over the template, strand displacement synthesis is, on the average, slower by a factor of 3–4 than regular synthesis [108, 109]. Displacement activity during (+) strand DNA synthesis was shown in permeabilized virions, supporting its biological significance [110]. RTs have also been shown to displace in vitro RNA segments, which are too long to be removed by the RNase H activity and are annealed ahead of the primer terminus during (+) strand DNA synthesis [108, 111]. Such RNA displacing DNA synthesis may be required for removing stably annealed genomic RNA fragments after (−) strand DNA synthesis. The fingers subdomain of RTs is involved in DNA-DNA displacement synthesis through interactions with either the template or non-templated strands. Mutations in this domain were found to affect the strand-displacement activity of RT but not the processivity of DNA synthesis [51, 112].
The RNase H activity of RT
The RT-associated RNase H activity functions as a processive endonuclease that hydrolyzes the RNA in RNA-DNA heteroduplexes (for recent detailed reviews, see [24, 25]). Unlike conventional ribonuclease, the RNase H activity hydrolyses phosphodiester bonds to produce a 3′-OH and a 5′-phosphate ends. This property is critical for RTN, as the oligo-ribonuceotides formed by RNase H must prime (+) strand DNA synthesis (see above). The RNase H domain of RT is separated from the DNA polymerase domain by the connection domain (Fig. 3); however, there are mutual critical contacts between the two domains. The hydrolytic cleavage by the retroviral RNases H activity involves a two-Mg+2-ion catalytic mechanism, described above [24, 25, 113]. In HIV-1 RT, the RNase H active site is rich in acidic residues, including Asp443, Glu478, Asp498 and Asp549. The catalysis continues in a two-metal ion-dependent trans-esterification reaction, similar to the one that has been proposed for DNA polymerases. However, the RNase H activity is mediated by a nucleophilic attack of a water molecule on the scissile phosphate, breaking the bond between two adjacent ribonucleosides in the RNA strand (instead of the nucleophilic attack of 3′-OH of the primer on the α phosphate of the incoming dNTP during DNA synthesis). This involves activation of the nucleophilic water by one of the divalent cations, with transition-state stabilization apparently mediated by both Mg+2 ions.
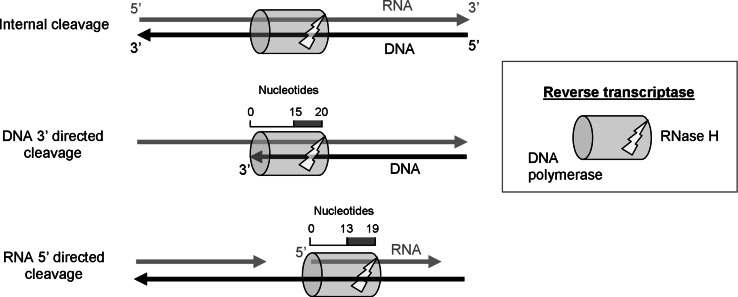
Three distinct modes of RNase H cleavages were characterized based on the interactions between RT and the RNA–DNA substrate (for further details, see Fig. 4 and [24, 25]). The first mode is a DNA 3′-end directed cleavage, where the DNA polymerase domain of RT is bound to the 3′-end of nascent DNA strand, which is annealed to the longer RNA template. This mode positions the RNase H active site towards the 3′-end of the RNA template (behind the 3′-end of the growing strand), so that the two RT domains are separated by 15–20 nucleotides on the RNA strand. Further cleavages of the RNA, at distances corresponding to about 8 nts from the 5′-end of the nascent DNA strand, may take place due to repositioning of the RT on the DNA–RNA substrate. These cleavages can occur either during DNA polymerization or independent of any polymerization. This mode of cleavage takes place during (−) strand DNA synthesis to facilitate strand transfer. When it is tightly linked to DNA polymerization, the DNA 3′-end directed cleavage correlates also with the pausing sites, caused by secondary structures encountered during the RDDP process. However, the polymerization rate of RT is greater than the hydrolysis rate of the enzyme. Therefore, the polymerization-dependent RNase H activity is insufficient to fully remove the genomic RNA template, and other modes of RNA degradation are required as well.
Fig. 4.
A schematic description of the three modes of RT-associated RNase H cleavage. In all heteroduplex substrates for RNase H activity, the RNA strand is depicted in gray and DNA strand in black, and in both strands the 3′ termini are indicated by arrowheads. The viral RNA or host tRNA can be cleaved in either internal sites (top), or relative to the recessed end of either the DNA or RNA (middle and bottom schemes, respectively). When the DNA polymerase domain is associated with the 3′-end of the DNA strand (middle), the cleavage window on the RNA is between 15 and 20 nts opposite to the 3′-end of the DNA strand. Alternatively, in the recessed RNA 5′-end cleavage mode the polymerase active site is positioned on the DNA strand opposite to or near the RNA 5′-end. In this case the cleavage window is between 13 and 19 nts
The second mode of RNase H activity is the RNA 5′-end-directed cleavage, and it is polymerization-independent. In this case, the polymerase domain binds internally the DNA strand in a site that is opposite to or near the 5′-end of the recessed RNA strand in the DNA–RNA heteroduplex. Consequently, the RNase H domain is positioned 13–19 nts from the 5′-end of the RNA. In some cases, the cleavage has been observed as close as 7 nts and as far as 21 nts from the 5′-end of the RNA. This possibly reflects a tendency of the RT to slide back on the RNA strand towards its 5′-end after the initial binding. The accessibility of the 5′-end affects the RNA 5′-end-directed cleavage, since the 5′-end at a nick is not recognized by RT; yet, a gap of two or more nucleotides upstream to the 5′-end of the RNA is adequate for such a cleavage.
The third mode of cleavage by the RT-associate RNase H activity is an internal one that is also polymerization-independent. In this mode, the RT-associated RNase H behaves as a typical endonuclease, cleaving any RNA-DNA hybrids without involving the positioning of the DNA polymerase domain on the ends of either RNA or DNA strands. The most important factor controlling the internal cleavage point is the sequence in its vicinity [24, 25].
Generation of the PPT segment requires a precise cleavage by the RT-associated RNase H activity although this activity does not have any defined sequence specificity. The PPT is extended by (+) strand DNA synthesis and at a later stage is hydrolysed by the RNase H activity. Removal of the PPT at the RNA-DNA junction requires an extension by at least 2–3 deoxynucleotides 3′ to the PPT. In general, the RNase H activity of different RTs strongly prefers to cleave one nucleotide away from the DNA-tRNA junction, and this activity probably depends on the flanking sequences. Accordingly, a single ribonucleotide (rA) is left on the 5′-end of the (−) strand DNA by HIV-1 RT, but in HIV-2 the complete tRNA is removed. Both alternatives were observed for MLV RT [25]. Statistical analyses of nucleotide frequencies in sequences surrounding internal cleavages have revealed that preferred sequences can be found on both sides of the cleavable phosphodiester bonds. The HIV-1 RT-associated RNase H activity prefers certain nucleotides located at positions +1, −2, −4, −7, −12 and −14, while for MLV RT RNase H, the preferred positions are at +1, −2, −6 and −11. For these RTs, the preferred nucleotides at position +1 are either A or U, and at position −2 are C or G. The importance of many of these sequences was confirmed by nuclease footprinting assay, mutagenesis [114, 115] as well as three-dimensional studies of HIV-1 RT co-crystallized with a PPT-hybrid [41]. The structural basis for the sequence preferences remains for the most part obscure; yet the contact between Gln475 in HIV-1 RT and the G at position −2 in the RNA strand probably contributes to the preference at this position [24, 25].
Although the isolated RNase H domain of MLV RT is enzymatically active, it cannot perform the removal of the PPT and tRNA that is so vital for RTN (see above). However, this specific activity of RNase H can be restored by adding the isolated DNA polymerase domain in trans [116]. In HIV-1 RT the DNA polymerase domain also affects the RNase H activity, and many mutations in the polymerase domain substantially influence the RNase H activity. Thus, modifying Trp266 and Phe61 in HIV-1 RT renders the RNase H incapable of either generating the PPT primer or removing it after extending the (+) strand [117, 118]. Residues in the connection subdomain of HIV-1 RT also affect the RNase H activity (i.e., His361 of the p66 subunit, located close to Tyr501 in the RNase H active site, see above the description of the structure of RNase H primer grip). As can be expected, the opposite scenario is also possible and mutations in the RNase H domain also alter the polymerase activity (for example, see [16, 119]).
The RNase H activity of HIV-2 RT is about ten times lower than the same activity of HIV-1 RT (despite similar polymerase activities). This drop in activity was mapped to residue 294 of the small RT subunit, which is a Pro in HIV-1 RT and a Gln in HIV-2 RT (located at the tip of alpha helix I in the thumb subdomain). When the Gln was replaced by a Pro, either in both HIV-2 RT subunits or only in the smaller subunit, the resulting HIV-2 RT showed an about tenfold increase in its RNase H activity in comparison to the wild-type RT [39, 40]. Interestingly, changing Gln294 of HIV-2 RT p55 to residues that represent all amino acid groups (except for a Gln) has also resulted in an increase in the RNase H activity [120]. The interactions between the polymerase and the RNase H domains may be particularly important in the case of HIV-1 and HIV-2 RTs, since the expression of the smaller subunit of the RTs (p51 and p55, respectively), which lacks the entire RNase H domain, results in an enzyme with a very low DNA polymerase activity [16, 121]. In contrast, the comparable DNA polymerase domain of MLV RT is enzymatically active, although a substantial reduction in processivity of DNA synthesis has been observed [122]. In addition, in several cases, mutations of HIV-1 variants that are resistant to NRTIs are located in the connection subdomain and in the RNase H domain (e.g., N348I and G509L, for these two regions, respectively) [123, 124].
The coordination between the DNA polymerase and RNase H activities of RT
To perform an efficient RTN process, RTs require a delicate and regulated balance between the DNA polymerase and the RNase H activities. Biochemical studies showed that RT adopts different orientations to coordinate its DNA polymerase and RNase H activities. This balance is reflected by the fact that the rate by which the template RNA is hydrolysed substantially affects the rate of (−) strand transfer and also the speed by which RT synthesizes (−) strand DNA during RTN. As could be expected from the pivotal role of the RNase H activity in RTN, mutant RTs devoid of RNase H activity fail to carry out (−) strand transfer in vitro and the viruses are replication defective [36]. However, HIV-1 and MLV virions that contain RTs with a partially reduced RNase H activity are still infectious, though they exhibit a decrease in strand transfer frequencies [125, 126]. The rate-limiting effect of the level of RNase H activity on the (−) strand transfer was further confirmed in vitro for HIV-2 RT that has an intrinsic low RNase H activity. This property directly reduces the (−) DNA strand transfer activity of HIV-2 RT in comparison to that of HIV-1 RT. Conversely, a slower DNA synthesis allows more time for an efficient RNase H cleavage of the donor RNA template and thus enhances DNA transfer [39, 40]. It should be also noted that the dynamic copy-choice model mentioned above depends also on the ratio between the rates of DNA synthesis and RNA degradation, thus affecting the frequency of template switching by RT [107].
A reduction in the RNase H activity allows the 3′-end of the nascent DNA strand to spend more time at N site of the DNA polymerase domain [127]. This partial halt in DNA elongation can change the balance between polymerization and the reverse process (pyrophosphorolysis). Pyrophosphorolysis is negligible unless the DNA synthesis is blocked following incorporation of a chain-terminating nucleotide. In this case, the 3′-end nucleotide may be excised in the presence of a pyrophosphate donor, such as ATP or pyrophosphate. In this context, several recent reports have shown that, as predicted, mutations in the RNase H domain confer high-level resistance to thymidine analog NRTIs, due to extended time available for excising of the incorporated NRTIs from 3′-terminated primers. The resistance is even significantly increased when the RNase H mutations are combined with thymidine analog mutants (TAM) of HIV-1 RT [128–130]. An obvious implication of these studies is that inhibitors of HIV-1 RT-associated RNase H, which are currently being identified as antiretroviral agents, may be in fact counterproductive (in case they do not completely block RNase H or if resistance quickly develops) by enhancing resistance to some NRTIs (see below).
The RT molecule must switch between its two-activity modes and functions as either an RNase H or a DNA polymerase. This ability has been recently documented in a single molecule spectroscopy study that provided different snapshots of the conformational dynamics of HIV-1 RT (in contrast to the static picture that is derived from X-ray crystallography) [131, 132]. This novel technique follows individual steps in the RTN process with a significantly greater precision (for a recent comprehensive review, see [48]). The findings explain, at least in part, the flexible adaptations that RT undergoes during RTN, which are largely determined by the RNA content of the primer. Thus, it has been suggested that pre-existing features of the RNA-DNA hybrid dictate whether RT adopts either a hydrolytic or a polymerizing orientation. These recent findings support previously published biochemical data showing that in RNA-DNA hybrids the 5′-end of the RNA plays a critical role in determining both the position and orientation of RT [133, 134]. The initiation of (+) strand DNA synthesis represents an unusual case, in which the PPT RNA primer must be precisely recognized by both DNA polymerase and RNase H catalytic domains of RT. As expected, the new technique suggests that the presence of the incoming dNTP strongly favors a polymerization-competent binding mode. However, in the presence of the NNRTI, nevirapine, there is an increase in the frequency by which RT binds the same substrate in the opposite, RNase H-competent orientation, consequently blocking DNA synthesis [48]. This may explain why NNRTIs were shown to selectively inhibit (+) strand initiation from the RNA PPT primer, under conditions where little or no inhibition of (−) strand synthesis was observed [135].
The involvement of other proteins in the RT-associated activities
Retroviral virions contain, besides the viral proteins, two copies of a single-stranded (+) RNA genome that can also serve as mRNAs for the synthesis of viral proteins. RTN of the genomic RNA requires RT and is further facilitated by other viral proteins, in particular the nucleic acid chaperone, NC, and probably some cellular factors (see below). At least in one case, that of Ty3 LTR retrotransposon, the retroviral IN is also required for initiating RTN [136]. Evidently, the contribution of these factors may be critical to complete this process in vivo, although RT has all the necessary enzymatic activities for RTN.
Retroviral proteins
The viral nucleocapsid protein (NC)
The NC proteins of retroviruses are Gag-derived highly basic nucleic acid-binding proteins that contain, in most retroviruses, two zinc finger domains (the gammaretroviruses NC has only one). These small proteins are less than 100 residues (a p7 in the case of HIV-1 RT, hence designated NCp7, and a p10 in MLV) and have nucleic acids chaperone activities that affect a variety of RT-related functions. These include tRNA annealing, RTN initiation, (+) strand transfer and strand displacement synthesis. In addition to the pivotal involvement in the early stages of viral infection, NC also has critical roles in late stages of virus assembly. These include RNA packaging, the dimerization of genomic viral RNA, tRNA placement onto the genomic RNA and RNA scaffolding (binding of NC to viral RNA drives Gag mulitmerization). Several recent comprehensive reviews summarize the different functions of NC [104–106] as well as the control of NC on the timing of RTN [137]. Therefore, we will briefly mention only the RT-associated NC activities. Since all RT’s functions can be performed to varying extents in the absence of NC, this protein can be considered as a regulator of these activities.
As already mentioned above, the chaperone activity of NC, which supports the formation of the initiation complex for (−) strand DNA synthesis, involves multimolecular interactions between HIV-1 RT and the tRNAlys3 primer and between RT and NC. The NC protein can unwind double-stranded nucleic acids, which are 8–12 nts long, and probably speeds up the RNase H-directed degradation that follows the synthesis of (−) sssDNA [104, 106]. NC also substantially suppresses initiations of non-specific priming of DNA synthesis that can occur from nicked genomic RNA due to self priming. In addition, NC significantly enhances the (−) strand transfer that follows synthesis of (−) sssDNA. This results from both the stimulation of the RNase H activity by NC and accelerating the annealing of the (−) sssDNA to the R region at the 3′-end of the genomic RNA [106, 138–140]. The increase in strand transfer can also explain the reduction in self priming. This potentially destructing event can be caused by the self-folding of the generated cDNA R region. On the other hand, NC was also shown to reduce the efficiency of non-specific strand transfer that results from a non-templated synthesis of DNA [which forms a 3′-end overhang and can be annealed to a complementary sequence in an acceptor DNA strand (vide supra)].
The ability of NC to destabilize the secondary structures of nucleic acids explains its enhancing effect on the processivity of DNA synthesis by RT, as reflected by the reduction in generating partial DNA products. NC also stimulates the (+) strand DNA transfer, although in this case the regions of secondary structures are less complex than those encountered during the (−) strand transfer (the stem loops in the Tat-responsive RNA region, termed TAR, and in the DNA that is complementary to TAR, located in the R region) [139, 141]. The fidelity of (+) strand DNA transfer is also guaranteed by the NC chaperone activity, as NC can block mispriming by non-PPT RNAs [142]. In addition, NC facilitates internal strand transfer, thereby supporting recombination events between the two RNA molecules in the virion [143–145] (see above). NC was also found to significantly enhance DNA strand-displacement synthesis, which is required for completing the RTN process, as mentioned above. Finally, even after RTN is completed, NC as part of the PIC has been suggested to participate in the integration of the viral DNA into the host genome by binding both the viral IN and DNA.
The viral integrase
There are several mechanistic and physical linkages between RT and IN. Not only the RT-generated DNA product is the substrate for IN, but the mature RT and IN proteins are also the proteolytic products of the same Pol polyprotein precursor encoded by the pol gene (see above) [1, 5]. Moreover, in ASLV virions the entire IN protein is found in two forms: either as part of the large RT subunit (the β subunit) or as a free enzymatically active IN protein (pp32) [1, 7]. In addition, the PICs, which are capable of performing in vitro integration, contain the viral DNA, cellular proteins and the viral IN, RT, NC and Vpr proteins [146–148]. Furthermore, the RTs and INs of both HIV-1 and MLV exhibit physical interactions, and direct contacts between HIV-1 RT and IN were also recently confirmed by various methods [149–151]. In the retrotransposon Tf1, a two-hybrid analysis detected interactions between IN and the RNase H domain of the RT [152]. Lastly, recent studies have shown that RT, as well as RT-derived peptides, can inhibit in vitro the enzymatic activities of IN, suggesting that IN activities can be further regulated by functional interactions between the two enzymes [149, 153, 154]. As could be expected from the close RT-IN contacts, the opposite effect of IN on RT was also observed. In an in vivo study with Ty3 LTR retrotransposon, IN was required for initiating RTN [136]. A more recent study on HIV-1 enzymes has identified a peptide, derived from IN, that binds RT and inhibits its DNA-polymerase activities (without affecting the RNase H activity). This inhibition was non-competitive and probably resulted from obstructing the formation of RT-DNA complexes by this peptide [154]. Another study has suggested that HIV-1 IN can promote RTN through specific interactions with the nucleoprotein reverse transcription complex [155].
Viral protein R (Vpr)
During HIV-1 infection, both the RTN complexes and the PICs contain Vpr (in addition to NC, IN and RT), suggesting the existence of mutual interactions between theses different proteins [146–148]. Indeed, recent data showed that peptides derived from Vpr affected the activities of both RT and IN. The experimental data and the consequent molecular modeling suggested that RT and IN are inhibited as a result of steric hindrance or conformational changes of their active sites [156]. Further studies are required to provide additional support for the relevance of these results in the biology of retroviruses.
The dUTP pyrophosphatase (dUTPase)
In contrast to the viral proteins, suggested above to affect RT either directly or indirectly, dUTPases affect the RT-associated DNA polymerase only indirectly. Mutagenesis caused by uracilation of DNA represents a constant threat to the survival of many organisms including viruses (for review, see [157]). Deoxy-uracil may appear in DNA either by misincorporation of dUTP instead of dTTP (since RTs can easily utilize dUTP) or by cytosine deamination. Some retroviruses, such as the betaretroviruses and non-primate lentiviruses, encode a fourth enzyme, dUTPase (additional to RT, IN and PR), that hydrolyzes dUTP and prevents its incorporation by RT into the nascent DNA [158, 159].
Cellular proteins
Uracil DNA glycosylase (UNG2)
In retroviruses that lack dUTPase, such as HIV-1, there is an alternative mechanism to correct dUTP damage after its incorporation into DNA. This is performed by packaging the cellular UNG2 enzyme into the viral particles. However, the exact mechanism for this process is still elusive, as UNG2 may be recruited into the virions either by the HIV-1 Vpr [102] or independently [160]. In addition, UNG2 was suggested to be incorporated into viral particles by specific association with the IN domain of the Gag-Pol polyprotein precursor, with a critical role of IN Leu172 in UNG2 association and hence encapsidation [161]. In HIV-1 lysates, UNG2 can repair G:U mismatched pairs to the correct G:C matched pairs. Therefore, it is likely that HIV-1 counteracts the incorporation of dUTP into viral cDNA by UNG2-mediated uracil excision [162]. Moreover, there are some indications that this is followed an RT-mediated endonucleolytic cleavage, which is dNTP-dependent, and subsequently by strand-displacement polymerization [163]. Apparently, additional studies are required to further explore the interplay between RT and UNG2, especially in light of the selective packaging of UNG2 by HIV-1 but not by either HIV-2 or simian immunodeficiency virus (SIV) [164].
ABOBEC3
The ability of members of the APOBEC3 (A3) family of cellular proteins to confer intrinsic immunity to cells infected by retroviruses has led to a flood of many new studies (for recent reviews, see [103, 165, 166]). A3 proteins are cytidine deaminases that cause hypermutations in the nascent retroviral DNA by deaminating deoxycytidines to deoxyuridines. Although A3 proteins can restrict HIV replication, this inhibition is blocked by the viral infectivity factor (Vif) protein. The inhibitory effects of APOBEC proteins are not limited to HIV-1 but extend also to other retroviruses and LTR retroelements. Interestingly, new data show that A3 proteins can inhibit retroviral replication even in the absence of a cytidine deaminase activity, and therefore their inhibition is not only due to their DNA editing capacity but rather also to another anti-viral mechanism. In the absence of DNA editing, A3 proteins can impair the accumulation of RTN products [167]. In addition, the initiation of HIV RTN and/or processivity of DNA synthesis by RT can be also inhibited by human A3G [168]. Additional studies confirmed that the deaminase-deficient A3 proteins, especially the human A3F member, reduce the accumulation of RTN products in cells [169, 170]. Moreover, A3G inhibited all RT-catalyzed DNA elongation reactions, but not the RNase H activity or the ability of NC to promote annealing [168]. Specifically, human A3G inhibits both (−) sssDNA and (+) sssDNA synthesis (−) and (+) DNA strand transfers and the elongation of either (−) and (+) DNA strands. The presence of A3G resulted in the formation of aberrant viral 3′-LTR DNA ends, suggesting that it may interfere with cleavage and removal of the tRNA primer. Sequence analyses of 2-LTR circle junctions from unintegrated DNA, synthesized in the presence of A3G, indicated that six RNA nucleotides, derived from the 3′-terminus of tRNALys3, were occasionally added to the U5 DNA [171]. This suggested that A3G can cause defects in the tRNA primer removal and thus limits (+) strand DNA transfer, resulting in integration-incompetent viral DNA ends [171]. In all, the inhibition of RTN at each step has a cumulative effect and could explain the reduction in the levels of integrated viral DNA.
Inhibitors of RT and drug-resistant RT variants
Many different RT inhibitors have been identified and further developed until now. Most of them were specifically developed against HIV-1 RT as part of an intensive worldwide effort to discover novel compounds that block the life cycle of HIV-1. Successful candidates were later used as part of the highly active antiretroviral therapy (HAART) offered to HIV-1 carriers. RT inhibitors may block either the DNA polymerase activity, the RNase H activity or both activities. Inhibitors against the DNA polymerase activity can be further divided into two subclasses of compounds: NRTIs and NNRTIs [172–174].
Nucleoside/nucleotide RT inhibitors
NRTIs are competitive inhibitor analogs that can be phosphorylated by cellular kinases and mimic normal dNTPs, but they all lack the free 3′-OH moiety. Therefore, once incorporated into the nascent DNA by RT, NRTIs prevent additional incorporation of nucleotides and hence terminate chain elongation [172–174]. Due to a rapid replication rate, the RT-associated error rate and the flexibility of HIV-1 RT, mutant variants rapidly develop in patients treated with NRTIs. Such mutations appear more slowly in patients treated with a combination therapy of different types of inhibitors. Two mechanisms are currently known for the development of resistance to NRTIs: (1) exclusion or preventing the incorporation of the nucleotide analog into the 3′-end of the nascent DNA strand and (2) excision of the already incorporated analog to release the block of the DNA synthesis at the 3′-end of the DNA.
The first mechanism involves mutations in RT that allow the enzyme to discriminate better between the NRTIs and normal nucleotides. All such mutations are concentrated in the finger and palm subdomains of the RT, where they can affect nucleotide binding. An important mutation with such mechanism was observed at Met184 of HIV-1 RT, which is within the highly conserved YXDD motif (here, X is Met). Mutating this Met to either Leu or Ile leads to a high resistance to both 3TC and FTC. This mutation causes a steric hindrance that impedes the incorporation of the dNTP analog [175, 176]. Interestingly, MLV RT that carries a Val at this position (within the same conserved YXDD motif) is highly resistant to 3TC. Moreover, mutating this Val to either Met (present in the wild-type HIV-1 RT) or to Ile, Ala or Ser, does not change the high resistance of these MLV RT mutants to 3TC [177]. This suggests that determinants outside the YXDD motif in MLV RT substantially contribute to resistance to 3TC.
Resistance of HIV-1 RT variants to NRTIs can be also induced by a second mechanism that increases the capacity of RT to excise the already incorporated analog from the 3′-end of the nascent DNA strand. This is mediated by enhancing the pyrophosphorolytic activity of the RT (Fig. 5). This excision activity, which is the reverse of DNA polymerization, requires a pyrophosphate donor that is most likely to be in vivo an ATP molecule. Similar to the addition of dNTPs (described above), incorporation of NRTI into the growing DNA strand by the wild-type RT leads to a translocation of the primer. This is usually followed by a movement of the primer to position the incorporated 3′-end NRTI at the P site and to liberate the N site, allowing binding of the next nucleotide. A subsequent dNTP binding leads to the formation of an inhibitory state, in which RT binds the template-primer-NRTI and establishes a closed complex with the incoming dNTP (also designated dead-end complex). In contrast, when the translocation of the incorporated NRTI to the P site is reversed or does not occur at all, excision can occur by removing the incorporated NRTI from the N site (Fig. 5a) [3, 48, 178].
Several HIV-1 RT mutations that improve the excision of specific NRTIs from the nascent DNA strand were intensively studied. Isolated HIV-1 RT mutants that are resistant to AZT inhibition carry different combinations of the modification: M41L, D67N, K70R, L210W, T215F/Y or K219E/Q (Fig. 5b). All of these mutations are collectively defined as thymidine-analog mutations (TAMs). Still, additional mutations can enhance the specificity of the pyrophosphorolytic activity of RT to other incorporated NRTIs [6]. TAMs facilitate the excision reaction by increasing the affinity of RT for ATP and, thus, enable the excision at physiological ATP concentrations [178]. The specificity of TAMs-carrying RT variants to excise the 3′-end terminal AZT-MP may involve steric constraints of the long azido group, causing the end of an AZT-MP-terminated primer to preferentially reside at the N site, and this limitation may destabilize the closed complex with the incoming dNTP. However, not all 3′-azido analogs of different dNMPs are excised efficiently by AZT-resistant mutants of HIV-1 RT [179], and even in the absence of dNTPs not all NRTIs can be excised. Thus, other factors such as the identity of nucleotide sequences may also contribute to the efficiency of the reaction.
HIV-1 and HIV-2 RTs differ significantly in the way they evolve to evade AZT inhibition. Although these two enzymes show a high homology of about 60–70% in their amino acids sequences [68], they have still selected different mechanisms to overcome inhibition by AZT [180]. Resistant variants of HIV-1 RT excise AZT from the 3′-end of the primer, whereas resistant HIV-2 RT mutants exclude the inhibitor from the nucleotide binding site and, consequently, do not incorporate the NRTI (as in the case of the Q151M mutant of HIV-2 RT). Biochemical analyses show that both RTs can use PPi to excise in vitro AZT-MP, but HIV-1 RT uses more efficiently ATP as the pyrophosphate donor in comparison to HIV-2 RT. This finding indicates that wild-type HIV-1 RT has some intrinsic ability to bind ATP, and this ability can be further improved by acquiring drug-resistant mutations. Further structural analysis supports this assumption, as prominent differences were shown to exist between the two HIV RTs in regions that form the putative ATP binding pocket. Several residues, including 44, 46, 113, 116 and 117, are positioned differently in the two enzymes. In addition, in HIV-1 RT there is also a hydrogen bond between residues 3 and 117 and additional interaction between residues 2 and 213 [180]. Collectively, these data suggest that it is easier for each virus to acquire mutations that allow it to extend its intrinsic, wild-type properties. HIV-2 RT does not seem to have any intrinsic ability to bind ATP. However, the Q151M mutation makes this enzyme relatively resistant to AZT, and therefore HIV-2 RT mutates to discriminate between AZT-TP and dNTPs. On the other hand, the HIV-1 RT variant that carries the Q151M mutation is still susceptible (although to a lower extent than the wild-type RT) to AZT, but has an intrinsic ability to bind ATP. Therefore, this property is further strengthened by other mutations that increase the affinity of HIV-1 RT towards ATP. The excision reaction of mutant RTs can be further modified by other NRTI or NNRTI-resistant mutations. Mutations that affect the ability of RT to bind its template-primer can also influence the excision reaction. Importantly, both the exclusion and excision mechanisms are involved in several cases in multiple-drug resistance [127].
The development of resistance to NRTIs is still a continuous obstacle to an effective and durable therapy for HIV-1 carriers. One means to effectively overcome resistance is to replace the oxygen of the α-phosphate (the non-bridging oxygen) in the NRTIs by a borano group, thus generating alpha-borano nucleotide analogs [181–183]. These compounds are quite promising, since they can still undergo a second and third phosphorylation reaction by cellular nucleotide kinases and efficiently inhibit drug-resistant HIV-1 RT mutants (as well as the wild-type RT). Notably, modification of the NRTI, 3TC-TP, to an α-borano-3TC-TP suppresses by 30-fold the resistance of M184V HIV-1 RT and by 180-fold the resistance of the double mutant K65R/M184V HIV-1 RT relative to the original 3TC-TP. Moreover, α-borano-ddNTPs and α-borano-AZT-TP repress the resistance of both K65R and Q151M HIV-1 RT mutants. The mechanism by which these drugs affect resistance involves the enhancement of Mg2+ ions binding to the active site of the RT–DNA–dNTP ternary complex, consequently raising the catalytic rate constant of incorporating the α-borano analogs. Thus, while resistance could develop in RT mutants by decreasing the rate of NRTIs incorporation, this property may be suppressed by using the α-borano-NRTIs with an increased incorporation rate. Interestingly, the α-borano-dNTPs compounds were also reported to reduce the excision reaction with either ATP or PPi, when tested with AZT-resistant HIV-1 RT mutants [184, 185].
Another approach for designing novel NRTIs is to lock the ribose conformation in the nucleoside analogs. During DNA synthesis, the ribose ring of all nucleotides must interchange from the N to the S conformation when moving on the elongating chain. This requirement provides an additional step to block RT and, indeed, new compounds with a locked conformation in their ribose ring that still retain a 3′-OH moiety efficiently inhibit HIV-1 RT after a few rounds of successive DNA polymerization [186].
A totally different approach to circumvent resistance is to use mutagenic nucleosides. These compounds are not chain terminators but rather exert their antiviral effects by inducing mutagenesis of the viral genome beyond tolerable levels. KP-1212 is a deoxycytosine analog in which the pyrimidine base was modified to include an additional nitrogen atom in the ring. Once incorporated into the DNA strand, KP-1212 can base-pair with either the correct dG on the complementary strand or mismatch with dA, hence, substantially raising the error rate in the viral genome during second strand DNA synthesis [187]. Thirteen passages of HIV-1 in the presence of 10 μM of KP-1212 resulted in viral suppression, with no viable virus detected. KP-1212-resistant HIV-1 strains have not yet been isolated; the compound has a low genotoxicity profile and is not toxic to mitochondria [188]. Still, due to the mutagenesis potential, safety issues should be further studied before further development of this compound.
Non-nucleotide RT inhibitors
NNRTIs are a collection of structurally diverse hydrophobic inhibitors of HIV-1 RT. Most of them bind to a hydrophobic pocket in RT, located about 10 Å underneath the active site of the DNA polymerase domain [189]. Several residues in the p66 subunit and one residue in the p51 subunit compose the NNRTI hydrophobic pocket (Fig. 5). Mutations of these residues and those positioned around the pocket were identified in variants of HIV-1 RT that are resistant to NNRTIs. Also, this pocket does not exist in HIV-1 RT structures that were crystallized without NNRTI. Although binding of these inhibitors essentially raises the affinity of RT to the DNA substrate, it concurrently dislocates the catalytic residues and consequently halts any additional DNA synthesis by RT [189, 190]. In addition, binding of NNRTI may also distort the position of the RT’s primer grip. NNRTIs do not typically compete with binding of RT to the free nucleotides or to the nucleic acid substrate, but rather lower the maximal velocity of DNA synthesis, catalyzed by HIV-1 RT [1]. In addition, NNRTIs do not usually inhibit the RT-associated RNase H activity in most standard assays, although a few reports suggest that these inhibitors either inhibit or stimulate this activity, depending on the substrate used in the in vitro assay [173].
For several years, the first three clinically approved NNRTIs (nevirapine, delavirdine and efavirenz) have been successfully combined with other anti-HIV-1 drugs (e.g., PR inhibitors and NRTIs) as part of antiretroviral therapy of HIV-1 infection [173]. However, the high specificity of these NNRTIs against only wild-type HIV-1 RT poses a significant obstacle for their continuous use, as their efficacy is drastically reduced against RT mutants that rapidly develop in the population of infectious viruses [190, 191]. In many cases, the rigid structure of these NNRTIs, which is designed to lower the antropy loss upon binding to the hydrophobic pocket, decreases their ability to accommodate themselves inside the hydrophobic pocket of the mutated RT. As a result, usually one or two mutations in HIV-1 RT residues that form the hydrophobic pocket (or even a relatively distal residue, see below) lead to a drastic reduction in the sensitivity to these drugs. Resistance is developed due to changes or loss of interactions, due to steric hindrance or to the formation of new bonds between the mutated residue and other RT residues. Resistance can also be developed as a result of mutating a distal residue (such as V106A in HIV-1 RT), thus changing the position of residues that interact with the NNRTI [191].
High resistance of RT mutants to the approved NNRTIs has constantly led to the search for novel broad-range inhibitors. Etravirine is a recently approved NNRTI that was developed as a flexible inhibitor with a broad-spectrum inhibition profile [192]. This compound efficiently inhibits many drug-resistant HIV-1 RT variants, including those resistant to other NNRTIs [193]. Nevertheless, several mutants have demonstrated some cross resistance between etravirine and efavirenz (e.g., K101P, K101Q, E138Q or M230L mutations), as well as between etravirine and nevirapine (e.g., Y181C and Y181L mutations). TMC278, a derivative of etravirine, is perhaps the most promising NNRTI candidate at the present time for further therapeutic development. This compound is very effective at relatively low doses in clinical trials treating wild-type and drug-resistant HIV-1 infections. Further studies into its mode of action revealed that its structural flexibility enables TMC278 to adapt different modes of inhibition for diverse RT mutants, including the two double mutants, K103N/Y181C and L100I/K103N [194, 195].
The two drug-resistant HIV-1 RT mutants, K103N and Y181C, are the most frequently observed in patients treated with NNRTIs. Diverse mechanisms of resistance have been suggested to explain their ability to efficiently resist the presently used NNRTIs. In the K103N mutant, there is a new hydrogen bond between residues Y188 and N103 that may make it difficult for the NNRTI to enter the hydrophobic pocket [196]. Mutations of Tyr181 can lead to a loss of aromatic interactions with the NNRTI and lower the affinity of the inhibitor towards the mutant RT. Apparently, TMC278 still binds efficiently the HIV-1 RT double mutant, K103N + Y181C, by interacting with Tyr183 to compensate for the loss of the aromatic interactions with the original Y181. This causes a shift of about 1.5 Å of Tyr183 relative to its position in the wild-type RT [194] and of about 0.6 Å relative to the L100I + K103N double mutant (Fig. 5). Such an interaction is particularly important as Tyr183 is part of the YXDD motif that is absolutely conserved in all retroviruses. Additional subtle conformational changes allow TMC278 to adapt to a hydrophobic tunnel in the proximity of Trp229. TMC278 also adapts to the hydrophobic pocket of the double mutant L100I/K103N by shifting away from residue Ile100 toward Asn103 and displacing its position by about 1.5 Å within the hydrophobic pocket. Some residues in the hydrophobic pocket are also rearranged to optimize the inhibitor–protein interaction [194].
Despite the notion that NNRTIs exert their inhibition activity primarily by binding to the NNRTI binding pocket of HIV-1 RT, some of them have acquired additional modes of action. For example, efavirenz inhibits also the late stages of HIV-1 replication by interfering with HIV-1 Gag-Pol polyprotein processing. Others, such as the pyrimidinediones, have a dual function by inhibiting RTN as well as by blocking the cell entry of both HIV-1 and HIV-2 (for a recent review see [197]). Inhibition by NNRTIs is a prominent feature that distinguishes between the RTs of HIV-1 and HIV-2. As described above (in the 3-dimensional structure section), a comparison of the crystal structures of these two highly related RTs reveals that the most significant difference between them lies in the region that can form the hydrophobic pocket that binds the different NNRTIs [71]. Most NNRTIs that inhibit HIV-1 RT do not significantly affect the catalytic activity of HIV-2 RT. An exception is PETT-2, which effectively inhibits both RTs of HIV-1 and HIV-2, with IC50 values of about 5 nM and 2.2 μM for these two enzymes, respectively [198]. Nonetheless, it is apparent that HIV-1 RT is much more sensitive to this compound. Interestingly, PETT-1 that carries a nitrile substituent instead of the chlorine in PETT-2 (on the pyridine ring) inhibits HIV-1 RT as efficiently as PETT-2, but shows only a weak inhibition of HIV-2 RT (IC50 > 50 μM) [198].
The RNase H activity of HIV-1 RT provides an additional target for drug development, but there is not yet any approved inhibitor against this activity for treating HIV infection. In contrast to the DNA polymerase activity, against which numerous inhibitors have been identified, only a few anti-RNase H inhibitors with reasonable in vitro activity have been explored so far. At the moment, the most promising HIV-1 RT-associated RNase H inhibitor is dihydroxy benzoyl naphthyl hydrazone (DHBNH) [199]. DHBNH inhibits in vitro the RNase H activity with an apparent IC50 value of about 0.5 μM without any significant effect on the DNA polymerase activity, except for its ability to inhibit the initiation of RNA-primed DNA synthesis at high concentrations. This compound is effective against a variety of drug-resistant HIV-1 RT mutants and, most importantly, it inhibits effectively HIV-1 infection in a cell-based assay. DHBNH inhibits wild-type HIV-1 replication, with an EC50 of about 5.5 μM, as well as several drug-resistant mutants (including nevirapine, efavirenz and AZT-resistant HIV-1 variants) with similar IC50 values (between 5.6 and 8.2 μM). In addition, DHBNH exhibits no cytotoxic effect at concentrations up to 100 μM. The co-crystallization of HIV-1 RT with DHBNH showed, unexpectedly, that it binds more than 50 Å away from the RNase H active site, near both the polymerase active site and the NNRTI hydrophobic pocket. DHBNH interacts with Asp186 and Trp229 and the backbones of several less conserved residues. Structural insights into the mode of DHBNH binding allowed also the design of bi-functional compounds that may inhibit both the RNase H and the DNA polymerase activities of HIV-1 RT.
As mentioned above, inhibiting the RNase H activity of HIV-1 RT might, nonetheless, exert potential undesirable effects, as mutations in the RNase H and the connection subdomains of RT can potentially enhance the RT resistance to both NRTIs and NNRTIs [129, 200]. Specifically, a lower RNase H activity has been shown to reduce the degradation of the RNA template and, consequently, allowing more time for the excision mechanism to take place, a process that might lead indirectly to an increase in the resistance to AZT [129]. Therefore, the benefit of inhibiting RNase H activity has to be carefully considered and its consequences must be completely analyzed.
Concluding remarks
Forty years of intensive investigations on different RTs have yielded enormous information on many aspects of this critical retroviral enzyme. Nevertheless, RT research is still positioned in the forefront of biomedical research, mainly due to the intensive search for novel and potent anti-HIV/AIDS drugs and the desire to better understand the pivotal role of RT in retroviral infections. Future studies on RTs that use state of the art techniques are likely to uncover additional facets of the exact mechanism of their enzymatic activities, structures, resistance to new drugs and their impact on anti-retroviral therapy. In addition, these new insights will probably advance substantially our knowledge of the unique processes that drive the evolution of retroviruses and retrotransposons.
Acknowledgments
We are grateful to Drs. Iris Oz-Gleenberg and Shoshana Loya for critically reading the manuscript and for helpful suggestions.
Footnotes
A. Hizi: Incumbent of the Gregorio and Dora Shapira Chair for the Research of Malignancies.
References
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Skalka AM, Goff SP. Reverse transcriptase. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. [PubMed] [Google Scholar]
- 3.Menendez-Arias L, Berkhout B (2008) Special issue on: retroviral reverse transcription. Virus Res 134 [DOI] [PubMed]
- 4.Parniak MA (2004) Special issue: molecular biology of HIV. Int J Biochem Cell Biol 36
- 5.Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 6.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hizi A, Herschhorn A. Retroviral reverse transcriptases (other than those of HIV-1 and murine leukemia virus): a comparison of their molecular and biochemical properties. Virus Res. 2008;134:203–220. doi: 10.1016/j.virusres.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Linial M. Foamy viruses. In: Knife DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott Williams & Williams; 2007. pp. 2245–2262. [Google Scholar]
- 9.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 10.Hizi A, Joklik WK. RNA-dependent DNA polymerase of avian sarcoma virus B77. I. Isolation and partial characterization of the alpha, beta2, and alphabeta forms of the enzyme. J Biol Chem. 1977;252:2281–2289. [PubMed] [Google Scholar]
- 11.Sluis-Cremer N, Arion D, Abram ME, Parniak MA. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int J Biochem Cell Biol. 2004;36:1836–1847. doi: 10.1016/j.biocel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Hizi A, Leis JP, Joklik WK. RNA-dependent DNA polymerase of avian sarcoma virus B77. II. Comparison of the catalytic properties of the alpha, beta2, and alphabeta enzyme forms. J Biol Chem. 1977;252:2290–2295. [PubMed] [Google Scholar]
- 13.Hizi A, Leis JP, Joklik WK. The RNA-dependent DNA polymerase of avian sarcoma virus B77. Binding of viral and nonviral ribonucleic acids to the alpha, beta2, and alphabeta forms of the enzyme. J Biol Chem. 1977;252:6878–6884. [PubMed] [Google Scholar]
- 14.Restle T, Muller B, Goody RS. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 15.Wohrl BM, Howard KJ, Jacques PS, Le Grice SF. Alternative modes of polymerization distinguish the subunits of equine infectious anemia virus reverse transcriptase. J Biol Chem. 1994;269:8541–8548. [PubMed] [Google Scholar]
- 16.Hizi A, McGill C, Hughes SH. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci USA. 1988;85:1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taube R, Loya S, Avidan O, Perach M, Hizi A. Reverse transcriptase of mouse mammary tumour virus: expression in bacteria, purification and biochemical characterization. Biochem J. 1998;329(Pt 3):579–587. doi: 10.1042/bj3290579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Entin-Meer M, Avidan O, Hizi A. The mature reverse transcriptase molecules in virions of mouse mammary tumor virus possess protease-derived sequences. Virology. 2003;310:157–162. doi: 10.1016/S0042-6822(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 19.Perach M, Hizi A. Catalytic features of the recombinant reverse transcriptase of bovine leukemia virus expressed in bacteria. Virology. 1999;259:176–189. doi: 10.1006/viro.1999.9761. [DOI] [PubMed] [Google Scholar]
- 20.Le Grice SF. “In the beginning”: initiation of minus strand DNA synthesis in retroviruses and LTR-containing retrotransposons. Biochemistry. 2003;42:14349–14355. doi: 10.1021/bi030201q. [DOI] [PubMed] [Google Scholar]
- 21.Abbink TE, Berkhout B. HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus Res. 2008;134:4–18. doi: 10.1016/j.virusres.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Levin HL. It’s prime time for reverse transcriptase. Cell. 1997;88:5–8. doi: 10.1016/S0092-8674(00)81851-6. [DOI] [PubMed] [Google Scholar]
- 23.Hizi A. The reverse transcriptase of the Tf1 retrotransposon has a specific novel activity for generating the RNA self-primer that is functional in cDNA synthesis. J Virol. 2008;82:10906–10910. doi: 10.1128/JVI.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champoux JJ, Schultz SJ. Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. 2009;276:1506–1516. doi: 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Jetzt AE, Ron Y, Preston BD, Dougherty JP. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]
- 27.van Wamel JL, Berkhout B. The first strand transfer during HIV-1 reverse transcription can occur either intramolecularly or intermolecularly. Virology. 1998;244:245–251. doi: 10.1006/viro.1998.9096. [DOI] [PubMed] [Google Scholar]
- 28.Rausch JW, Le Grice SF. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int J Biochem Cell Biol. 2004;36:1752–1766. doi: 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Charneau P, Alizon M, Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol. 1992;66:2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hameau L, Jeusset J, Lafosse S, Coulaud D, Delain E, Unge T, Restle T, Le Cam E, Mirambeau G. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement dna synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J Virol. 2001;75:3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 32.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limon A, Nakajima N, Lu R, Ghory HZ, Engelman A. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J Virol. 2002;76:12078–12086. doi: 10.1128/JVI.76.23.12078-12086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsden MD, Zack JA. Human immunodeficiency virus bearing a disrupted central DNA flap is pathogenic in vivo. J Virol. 2007;81:6146–6150. doi: 10.1128/JVI.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dvorin JD, Bell P, Maul GG, Yamashita M, Emerman M, Malim MH. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J Virol. 2002;76:12087–12096. doi: 10.1128/JVI.76.23.12087-12096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 38.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sevilya Z, Loya S, Hughes SH, Hizi A. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits. J Mol Biol. 2001;311:957–971. doi: 10.1006/jmbi.2001.4904. [DOI] [PubMed] [Google Scholar]
- 40.Sevilya Z, Loya S, Adir N, Hizi A. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is modulated by residue 294 of the small subunit. Nucleic Acids Res. 2003;31:1481–1487. doi: 10.1093/nar/gkg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 43.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/S0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 44.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 46.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchand B, Tchesnokov EP, Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 48.Gotte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SF (2009) Reverse transcriptase in motion: Conformational dynamics of enzyme-substrate interactions. Biochim Biophys Acta. doi:10.1016/j.bbapap.2009.07.020 [DOI] [PMC free article] [PubMed]
- 49.Kew Y, Olsen LR, Japour AJ, Prasad VR. Insertions into the beta3–beta4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J Biol Chem. 1998;273:7529–7537. doi: 10.1074/jbc.273.13.7529. [DOI] [PubMed] [Google Scholar]
- 50.Kim B, Ayran JC, Sagar SG, Adman ET, Fuller SM, Tran NH, Horrigan J. New human immunodeficiency virus, type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. Accuracy, template binding, and processivity. J Biol Chem. 1999;274:27666–27673. doi: 10.1074/jbc.274.39.27666. [DOI] [PubMed] [Google Scholar]
- 51.Fisher TS, Darden T, Prasad VR. Substitutions at Phe61 in the beta3-beta4 hairpin of HIV-1 reverse transcriptase reveal a role for the Fingers subdomain in strand displacement DNA synthesis. J Mol Biol. 2003;325:443–459. doi: 10.1016/S0022-2836(02)01225-1. [DOI] [PubMed] [Google Scholar]
- 52.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 2000;97:3056–3061. doi: 10.1073/pnas.97.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin-Hernandez AM, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 55.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 56.Davies JF, 2nd, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 57.Evans DB, Brawn K, Deibel MR, Jr, Tarpley WG, Sharma SK. A recombinant ribonuclease H domain of HIV-1 reverse transcriptase that is enzymatically active. J Biol Chem. 1991;266:20583–20585. [PubMed] [Google Scholar]
- 58.Smith JS, Roth MJ. Purification and characterization of an active human immunodeficiency virus type 1 RNase H domain. J Virol. 1993;67:4037–4049. doi: 10.1128/jvi.67.7.4037-4049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keck JL, Marqusee S. Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc Natl Acad Sci USA. 1995;92:2740–2744. doi: 10.1073/pnas.92.7.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stahl SJ, Kaufman JD, Vikic-Topic S, Crouch RJ, Wingfield PT. Construction of an enzymatically active ribonuclease H domain of human immunodeficiency virus type 1 reverse transcriptase. Protein Eng. 1994;7:1103–1108. doi: 10.1093/protein/7.9.1103. [DOI] [PubMed] [Google Scholar]
- 61.Hostomsky Z, Hostomska Z, Hudson GO, Moomaw EW, Nodes BR. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1991;88:1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith JS, Gritsman K, Roth MJ. Contributions of DNA polymerase subdomains to the RNase H activity of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1994;68:5721–5729. doi: 10.1128/jvi.68.9.5721-5729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.North TW, Cronn RC, Remington KM, Tandberg RT, Judd RC. Characterization of reverse transcriptase from feline immunodeficiency virus. J Biol Chem. 1990;265:5121–5128. [PubMed] [Google Scholar]
- 64.Amacker M, Hottiger M, Hubscher U. Feline immunodeficiency virus reverse transcriptase: expression, functional characterization, and reconstitution of the 66- and 51-kilodalton subunits. J Virol. 1995;69:6273–6279. doi: 10.1128/jvi.69.10.6273-6279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avidan O, Bochner R, Hizi A. The catalytic properties of the recombinant reverse transcriptase of bovine immunodeficiency virus. Virology. 2006;351:42–57. doi: 10.1016/j.virol.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 66.Souquet M, Restle T, Krebs R, Le Grice SF, Goody RS, Wohrl BM. Analysis of the polymerization kinetics of homodimeric EIAV p51/51 reverse transcriptase implies the formation of a polymerase active site identical to heterodimeric EIAV p66/51 reverse transcriptase. Biochemistry. 1998;37:12144–12152. doi: 10.1021/bi9731596. [DOI] [PubMed] [Google Scholar]
- 67.Shaharabany M, Rice NR, Hizi A. Expression and mutational analysis of the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochem Biophys Res Commun. 1993;196:914–920. doi: 10.1006/bbrc.1993.2336. [DOI] [PubMed] [Google Scholar]
- 68.Shaharabany M, Hizi A. The catalytic functions of chimeric reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J Biol Chem. 1992;267:3674–3678. [PubMed] [Google Scholar]
- 69.Fan N, Rank KB, Leone JW, Heinrikson RL, Bannow CA, Smith CW, Evans DB, Poppe SM, Tarpley WG, Rothrock DJ, Tomassseli AG, Sharma SK. The differential processing of homodimers of reverse transcriptases from human immunodeficiency viruses type 1 and 2 is a consequence of the distinct specificities of the viral proteases. J Biol Chem. 1995;270:13573–13579. [PubMed] [Google Scholar]
- 70.Bird LE, Chamberlain PP, Stewart-Jones GB, Ren J, Stuart DI, Stammers DK. Cloning, expression, purification, and crystallisation of HIV-2 reverse transcriptase. Protein Expr Purif. 2003;27:12–18. doi: 10.1016/S1046-5928(02)00567-3. [DOI] [PubMed] [Google Scholar]
- 71.Ren J, Bird LE, Chamberlain PP, Stewart-Jones GB, Stuart DI, Stammers DK. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc Natl Acad Sci USA. 2002;99:14410–14415. doi: 10.1073/pnas.222366699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Divita G, Rittinger K, Restle T, Immendorfer U, Goody RS. Conformational stability of dimeric HIV-1 and HIV-2 reverse transcriptases. Biochemistry. 1995;34:16337–16346. doi: 10.1021/bi00050a014. [DOI] [PubMed] [Google Scholar]
- 73.Das D, Georgiadis MM. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure. 2004;12:819–829. doi: 10.1016/j.str.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 74.Lim D, Gregorio GG, Bingman C, Martinez-Hackert E, Hendrickson WA, Goff SP. Crystal structure of the moloney murine leukemia virus RNase H domain. J Virol. 2006;80:8379–8389. doi: 10.1128/JVI.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cote ML, Roth MJ. Murine leukemia virus reverse transcriptase: structural comparison with HIV-1 reverse transcriptase. Virus Res. 2008;134:186–202. doi: 10.1016/j.virusres.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 77.Hizi A, Joklik WK. The beta subunit of the DNA polymerase of avian sarcoma virus strain B77 is a phosphoprotein. Virology. 1977;78:571–575. doi: 10.1016/0042-6822(77)90132-5. [DOI] [PubMed] [Google Scholar]
- 78.Hizi A. Regulation of Rous sarcoma virus RNA-dependent DNA polymerase isoenzymes by in vitro phosphorylation-dephosphorylation. Arch Biochem Biophys. 1982;219:394–400. doi: 10.1016/0003-9861(82)90171-0. [DOI] [PubMed] [Google Scholar]
- 79.Rezende LF, Prasad VR. Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates. Int J Biochem Cell Biol. 2004;36:1716–1734. doi: 10.1016/j.biocel.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 80.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 81.Rittinger K, Divita G, Goody RS. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci USA. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:19759–19767. doi: 10.1074/jbc.M910361199. [DOI] [PubMed] [Google Scholar]
- 83.Entin-Meer M, Sevilya Z, Hizi A. The role of phenylalanine-119 of the reverse transcriptase of mouse mammary tumour virus in DNA synthesis, ribose selection and drug resistance. Biochem J. 2002;367:381–391. doi: 10.1042/BJ20020712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel PH, Preston BD. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Golinelli MP, Hughes SH. Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology. 2002;294:122–134. doi: 10.1006/viro.2001.1322. [DOI] [PubMed] [Google Scholar]
- 86.Kirshenboim N, Hayouka Z, Friedler A, Hizi A. Expression and characterization of a novel reverse transcriptase of the LTR retrotransposon Tf1. Virology. 2007;366:263–276. doi: 10.1016/j.virol.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 87.Atwood-Moore A, Ejebe K, Levin HL. Specific recognition and cleavage of the plus-strand primer by reverse transcriptase. J Virol. 2005;79:14863–14875. doi: 10.1128/JVI.79.23.14863-14875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avidan O, Meer ME, Oz I, Hizi A. The processivity and fidelity of DNA synthesis exhibited by the reverse transcriptase of bovine leukemia virus. Eur J Biochem. 2002;269:859–867. doi: 10.1046/j.0014-2956.2001.02719.x. [DOI] [PubMed] [Google Scholar]
- 89.Avidan O, Hizi A. The processivity of DNA synthesis exhibited by drug-resistant variants of human immunodeficiency virus type-1 reverse transcriptase. Nucleic Acids Res. 1998;26:1713–1717. doi: 10.1093/nar/26.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oude Essink BB, Back NK, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212–3217. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bakhanashvili M, Avidan O, Hizi A. Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett. 1996;391:257–262. doi: 10.1016/0014-5793(96)00747-8. [DOI] [PubMed] [Google Scholar]
- 92.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad VR. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 93.Rubinek T, Bakhanashvili M, Taube R, Avidan O, Hizi A. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74 → Val and Glu89 → Gly. Eur J Biochem. 1997;247:238–247. doi: 10.1111/j.1432-1033.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 94.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 95.Bebenek K, Kunkel TA. The fidelity of retroviral reverse transcriptases. In: Sakaka AM, Goff SP, editors. Reverse transcriptase. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 96.Menendez-Arias L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses. 2009;1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu H, Goodman MF. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J Biol Chem. 1992;267:10888–10896. [PubMed] [Google Scholar]
- 98.Bakhanashvili M, Hizi A. Fidelity of the RNA-dependent DNA synthesis exhibited by the reverse transcriptases of human immunodeficiency virus types 1 and 2 and of murine leukemia virus: mispair extension frequencies. Biochemistry. 1992;31:9393–9398. doi: 10.1021/bi00154a010. [DOI] [PubMed] [Google Scholar]
- 99.Bakhanashvili M, Hizi A. The fidelity of the reverse transcriptases of human immunodeficiency viruses and murine leukemia virus, exhibited by the mispair extension frequencies, is sequence dependent and enzyme related. FEBS Lett. 1993;319:201–205. doi: 10.1016/0014-5793(93)80067-5. [DOI] [PubMed] [Google Scholar]
- 100.Bakhanashvili M, Hizi A. Fidelity of DNA synthesis exhibited in vitro by the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochemistry. 1993;32:7559–7567. doi: 10.1021/bi00080a030. [DOI] [PubMed] [Google Scholar]
- 101.Menendez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog Nucleic Acid Res Mol Biol. 2002;71:91–147. doi: 10.1016/S0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 102.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J Virol. 2000;74:7039–7047. doi: 10.1128/JVI.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aguiar RS, Peterlin BM. APOBEC3 proteins and reverse transcription. Virus Res. 2008;134:74–85. doi: 10.1016/j.virusres.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 104.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 105.Bampi C, Bibillo A, Wendeler M, Divita G, Gorelick RJ, Le Grice SF, Darlix JL. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J Biol Chem. 2006;281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- 106.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hwang CK, Svarovskaia ES, Pathak VK. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc Natl Acad Sci USA. 2001;98:12209–12214. doi: 10.1073/pnas.221289898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kelleher CD, Champoux JJ. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- 109.Whiting SH, Champoux JJ. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J Mol Biol. 1998;278:559–577. doi: 10.1006/jmbi.1998.1720. [DOI] [PubMed] [Google Scholar]
- 110.Boone LR, Skalka AM. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. I. Kinetics of synthesis and size of minus- and plus-strand transcripts. J Virol. 1981;37:109–116. doi: 10.1128/jvi.37.1.109-116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fuentes GM, Rodriguez-Rodriguez L, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J Biol Chem. 1996;271:1966–1971. doi: 10.1074/jbc.271.4.2063. [DOI] [PubMed] [Google Scholar]
- 112.Winshell J, Paulson BA, Buelow BD, Champoux JJ. Requirements for DNA unpairing during displacement synthesis by HIV-1 reverse transcriptase. J Biol Chem. 2004;279:52924–52933. doi: 10.1074/jbc.M409134200. [DOI] [PubMed] [Google Scholar]
- 113.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 114.Wohrl BM, Georgiadis MM, Telesnitsky A, Hendrickson WA, Le Grice SF. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 115.Wohrl BM, Tantillo C, Arnold E, Le Grice SF. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:5343–5356. doi: 10.1021/bi00016a005. [DOI] [PubMed] [Google Scholar]
- 116.Schultz SJ, Champoux JJ. RNase H domain of Moloney murine leukemia virus reverse transcriptase retains activity but requires the polymerase domain for specificity. J Virol. 1996;70:8630–8638. doi: 10.1128/jvi.70.12.8630-8638.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mandal D, Dash C, Le Grice SF, Prasad VR. Analysis of HIV-1 replication block due to substitutions at F61 residue of reverse transcriptase reveals additional defects involving the RNase H function. Nucleic Acids Res. 2006;34:2853–2863. doi: 10.1093/nar/gkl360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Powell MD, Beard WA, Bebenek K, Howard KJ, Le Grice SF, Darden TA, Kunkel TA, Wilson SH, Levin JG. Residues in the alphaH and alphaI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J Biol Chem. 1999;274:19885–19893. doi: 10.1074/jbc.274.28.19885. [DOI] [PubMed] [Google Scholar]
- 119.Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci USA. 2002;99:9515–9520. doi: 10.1073/pnas.142123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bochner R, Duvshani A, Adir N, Hizi A. Mutagenesis of Gln294 of the reverse transcriptase of human immunodeficiency virus type-2 and its effects on the ribonuclease H activity. FEBS Lett. 2008;582:2799–2805. doi: 10.1016/j.febslet.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 121.Hizi A, Tal R, Hughes SH. Mutational analysis of the DNA polymerase and ribonuclease H activities of human immunodeficiency virus type 2 reverse transcriptase expressed in Escherichia coli . Virology. 1991;180:339–346. doi: 10.1016/0042-6822(91)90038-D. [DOI] [PubMed] [Google Scholar]
- 122.Telesnitsky A, Goff SP. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brehm JH, Koontz D, Meteer JD, Pathak V, Sluis-Cremer N, Mellors JW. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J Virol. 2007;81:7852–7859. doi: 10.1128/JVI.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gotte M. Should we include connection domain mutations of HIV-1 reverse transcriptase in HIV resistance testing. PLoS Med. 2007;4:e346. doi: 10.1371/journal.pmed.0040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nikolenko GN, Svarovskaia ES, Delviks KA, Pathak VK. Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J Virol. 2004;78:8761–8770. doi: 10.1128/JVI.78.16.8761-8770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Svarovskaia ES, Delviks KA, Hwang CK, Pathak VK. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J Virol. 2000;74:7171–7178. doi: 10.1128/JVI.74.15.7171-7178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Menendez-Arias L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008;134:124–146. doi: 10.1016/j.virusres.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 128.Nikolenko GN, Palmer S, Maldarelli F, Mellors JW, Coffin JM, Pathak VK. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc Natl Acad Sci USA. 2005;102:2093–2098. doi: 10.1073/pnas.0409823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Delviks-Frankenberry KA, Nikolenko GN, Barr R, Pathak VK. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J Virol. 2007;81:6837–6845. doi: 10.1128/JVI.02820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, Hughes SH, Coffin JM, Jere A, Pathak VK. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci USA. 2008;105:10943–10948. doi: 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 1993;21:4330–4338. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palaniappan C, Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–2070. doi: 10.1074/jbc.271.47.29605. [DOI] [PubMed] [Google Scholar]
- 135.Grobler JA, Dornadula G, Rice MR, Simcoe AL, Hazuda DJ, Miller MD. HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J Biol Chem. 2007;282:8005–8010. doi: 10.1074/jbc.M608274200. [DOI] [PubMed] [Google Scholar]
- 136.Nymark-McMahon MH, Beliakova-Bethell NS, Darlix JL, Le Grice SF, Sandmeyer SB. Ty3 integrase is required for initiation of reverse transcription. J Virol. 2002;76:2804–2816. doi: 10.1128/JVI.76.6.2804-2816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mougel M, Houzet L, Darlix JL. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 139.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000;74:8980–8988. doi: 10.1128/JVI.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roda RH, Balakrishnan M, Hanson MN, Wohrl BM, Le Grice SF, Roques BP, Gorelick RJ, Bambara RA. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J Biol Chem. 2003;278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- 141.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 142.Post K, Kankia B, Gopalakrishnan S, Yang V, Cramer E, Saladores P, Gorelick RJ, Guo J, Musier-Forsyth K, Levin JG. Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009;37:1755–1766. doi: 10.1093/nar/gkn1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramirez BC, Simon-Loriere E, Galetto R, Negroni M. Implications of recombination for HIV diversity. Virus Res. 2008;134:64–73. doi: 10.1016/j.virusres.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 144.Derebail SS, Heath MJ, DeStefano JJ. Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J Biol Chem. 2003;278:15702–15712. doi: 10.1074/jbc.M211701200. [DOI] [PubMed] [Google Scholar]
- 145.Roda RH, Balakrishnan M, Kim JK, Roques BP, Fay PJ, Bambara RA. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J Biol Chem. 2002;277:46900–46911. doi: 10.1074/jbc.M208638200. [DOI] [PubMed] [Google Scholar]
- 146.Fouchier RA, Malim MH. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv Virus Res. 1999;52:275–299. doi: 10.1016/S0065-3527(08)60302-4. [DOI] [PubMed] [Google Scholar]
- 147.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bukrinsky M. A hard way to the nucleus. Mol Med. 2004;10:1–5. [PMC free article] [PubMed] [Google Scholar]
- 149.Oz I, Avidan O, Hizi A. Inhibition of the integrases of human immunodeficiency viruses type 1 and type 2 by reverse transcriptases. Biochem J. 2002;361:557–566. doi: 10.1042/0264-6021:3610557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Herschhorn A, Oz-Gleenberg I, Hizi A. Quantitative analysis of the interactions between HIV-1 integrase and retroviral reverse transcriptases. Biochem J. 2008;412:163–170. doi: 10.1042/BJ20071279. [DOI] [PubMed] [Google Scholar]
- 151.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, Miller JT, Le Grice SF, Clubb RT, Chow SA. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J Biol Chem. 2009;284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Steele SJ, Levin HL. A map of interactions between the proteins of a retrotransposon. J Virol. 1998;72:9318–9322. doi: 10.1128/jvi.72.11.9318-9322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oz Gleenberg I, Avidan O, Goldgur Y, Herschhorn A, Hizi A. Peptides derived from the reverse transcriptase of human immunodeficiency virus type 1 as novel inhibitors of the viral integrase. J Biol Chem. 2005;280:21987–21996. doi: 10.1074/jbc.M414679200. [DOI] [PubMed] [Google Scholar]
- 154.Oz Gleenberg I, Herschhorn A, Goldgur Y, Hizi A. Inhibition of human immunodeficiency virus type-1 reverse transcriptase by a novel peptide derived from the viral integrase. Arch Biochem Biophys. 2007;458:202–212. doi: 10.1016/j.abb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 155.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gleenberg IO, Herschhorn A, Hizi A. Inhibition of the activities of reverse transcriptase and integrase of human immunodeficiency virus type-1 by peptides derived from the homologous viral protein R (Vpr) J Mol Biol. 2007;369:1230–1243. doi: 10.1016/j.jmb.2007.03.073. [DOI] [PubMed] [Google Scholar]
- 157.Sire J, Querat G, Esnault C, Priet S. Uracil within DNA: an actor of antiviral immunity. Retrovirology. 2008;5:45. doi: 10.1186/1742-4690-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Payne SL, Elder JH. The role of retroviral dUTPases in replication and virulence. Curr Protein Pept Sci. 2001;2:381–388. doi: 10.2174/1389203013381008. [DOI] [PubMed] [Google Scholar]
- 159.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J Gen Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 160.Willetts KE, Rey F, Agostini I, Navarro JM, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Priet S, Navarro JM, Querat G, Sire J. Reversion of the lethal phenotype of an HIV-1 integrase mutant virus by overexpression of the same integrase mutant protein. J Biol Chem. 2003;278:20724–20730. doi: 10.1074/jbc.M301768200. [DOI] [PubMed] [Google Scholar]
- 162.Priet S, Navarro JM, Gros N, Querat G, Sire J. Functional role of HIV-1 virion-associated uracil DNA glycosylase 2 in the correction of G:U mispairs to G:C pairs. J Biol Chem. 2003;278:4566–4571. doi: 10.1074/jbc.M209311200. [DOI] [PubMed] [Google Scholar]
- 163.Priet S, Gros N, Navarro JM, Boretto J, Canard B, Querat G, Sire J. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol Cell. 2005;17:479–490. doi: 10.1016/j.molcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 164.Priet S, Navarro JM, Gros N, Querat G, Sire J. Differential incorporation of uracil DNA glycosylase UNG2 into HIV-1, HIV-2, and SIV(MAC) viral particles. Virology. 2003;307:283–289. doi: 10.1016/S0042-6822(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 165.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 168.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 171.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs): past, present, and future. Chem Biodivers. 2004;1:44–64. doi: 10.1002/cbdv.200490012. [DOI] [PubMed] [Google Scholar]
- 173.De Clercq E. The design of drugs for HIV and HCV. Nat Rev Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 174.Ilina T, Parniak MA. Inhibitors of HIV-1 reverse transcriptase. Adv Pharmacol. 2008;56:121–167. doi: 10.1016/S1054-3589(07)56005-9. [DOI] [PubMed] [Google Scholar]
- 175.Gao HQ, Boyer PL, Sarafianos SG, Arnold E, Hughes SH. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J Mol Biol. 2000;300:403–418. doi: 10.1006/jmbi.2000.3823. [DOI] [PubMed] [Google Scholar]
- 176.Sarafianos SG, Das K, Clark AD, Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc Natl Acad Sci USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Halvas EK, Svarovskaia ES, Freed EO, Pathak VK. Wild-type and YMDD mutant murine leukemia virus reverse transcriptases are resistant to 2′,3′-dideoxy-3′-thiacytidine. J Virol. 2000;74:6669–6674. doi: 10.1128/JVI.74.14.6669-6674.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sluis-Cremer N, Arion D, Parikh U, Koontz D, Schinazi RF, Mellors JW, Parniak MA. The 3′-azido group is not the primary determinant of 3′-azido-3′-deoxythymidine (AZT) responsible for the excision phenotype of AZT-resistant HIV-1. J Biol Chem. 2005;280:29047–29052. doi: 10.1074/jbc.M503166200. [DOI] [PubMed] [Google Scholar]
- 180.Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2006;2:e10. doi: 10.1371/journal.ppat.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Deval J, Alvarez K, Selmi B, Bermond M, Boretto J, Guerreiro C, Mulard L, Canard B. Mechanistic insights into the suppression of drug resistance by human immunodeficiency virus type 1 reverse transcriptase using alpha-boranophosphate nucleoside analogs. J Biol Chem. 2005;280:3838–3846. doi: 10.1074/jbc.M411559200. [DOI] [PubMed] [Google Scholar]
- 182.Deval J, Selmi B, Boretto J, Egloff MP, Guerreiro C, Sarfati S, Canard B. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using alpha-boranophosphate nucleotide analogues. J Biol Chem. 2002;277:42097–42104. doi: 10.1074/jbc.M206725200. [DOI] [PubMed] [Google Scholar]
- 183.Selmi B, Boretto J, Sarfati SR, Guerreiro C, Canard B. Mechanism-based suppression of dideoxynucleotide resistance by K65R human immunodeficiency virus reverse transcriptase using an alpha-boranophosphate nucleoside analogue. J Biol Chem. 2001;276:48466–48472. doi: 10.1074/jbc.M107003200. [DOI] [PubMed] [Google Scholar]
- 184.Meyer P, Schneider B, Sarfati S, Deville-Bonne D, Guerreiro C, Boretto J, Janin J, Veron M, Canard B. Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase. EMBO J. 2000;19:3520–3529. doi: 10.1093/emboj/19.14.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schneider B, Meyer P, Sarfati S, Mulard L, Guerreiro C, Boretto J, Janin J, Veron M, Deville-Bonne D, Canard B. Activation of anti-reverse transcriptase nucleotide analogs by nucleoside diphosphate kinase: improvement by alpha-boranophosphate substitution. Nucleosides Nucleotides Nucleic Acids. 2001;20:297–306. doi: 10.1081/NCN-100002300. [DOI] [PubMed] [Google Scholar]
- 186.Boyer PL, Julias JG, Marquez VE, Hughes SH. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J Mol Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 187.Murakami E, Basavapathruni A, Bradley WD, Anderson KS. Mechanism of action of a novel viral mutagenic covert nucleotide: molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antiviral Res. 2005;67:10–17. doi: 10.1016/j.antiviral.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 188.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 189.Ren J, Nichols C, Bird L, Chamberlain P, Weaver K, Short S, Stuart DI, Stammers DK. Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J Mol Biol. 2001;312:795–805. doi: 10.1006/jmbi.2001.4988. [DOI] [PubMed] [Google Scholar]
- 190.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 191.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 192.Das K, Clark AD, Jr, Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. Roles of conformational and positional adaptability in structure-based design of TMC125–R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 193.Vingerhoets J, Azijn H, Fransen E, De Baere I, Smeulders L, Jochmans D, Andries K, Pauwels R, de Bethune MP. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol. 2005;79:12773–12782. doi: 10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ren J, Stammers DK. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 2008;134:157–170. doi: 10.1016/j.virusres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 196.Hsiou Y, Ding J, Das K, Clark AD, Jr, Boyer PL, Lewi P, Janssen PA, Kleim JP, Rosner M, Hughes SH, Arnold E. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J Mol Biol. 2001;309:437–445. doi: 10.1006/jmbi.2001.4648. [DOI] [PubMed] [Google Scholar]
- 197.Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 2008;134:147–156. doi: 10.1016/j.virusres.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ren J, Diprose J, Warren J, Esnouf RM, Bird LE, Ikemizu S, Slater M, Milton J, Balzarini J, Stuart DI, Stammers DK. Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses. J Biol Chem. 2000;275:5633–5639. doi: 10.1074/jbc.275.8.5633. [DOI] [PubMed] [Google Scholar]
- 199.Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark AD, Jr, Knight JL, Julias JG, Clark PK, Krogh-Jespersen K, Levy RM, Hughes SH, Parniak MA, Arnold E. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem Biol. 2006;1:702–712. doi: 10.1021/cb600303y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hachiya A, Kodama EN, Sarafianos SG, Schuckmann MM, Sakagami Y, Matsuoka M, Takiguchi M, Gatanaga H, Oka S. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J Virol. 2008;82:3261–3270. doi: 10.1128/JVI.01154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Herschhorn A, Hizi A. Virtual screening, identification, and biochemical characterization of novel inhibitors of the reverse transcriptase of human immunodeficiency virus type-1. J Med Chem. 2008;51:5702–5713. doi: 10.1021/jm800473d. [DOI] [PubMed] [Google Scholar]