Abstract
Although INO80 chromatin remodeling enzyme has been shown in yeast to play roles in non-transcriptional nuclear processes such as DNA replication, its cellular functions in higher eukaryotes have remained largely unexplored. Here, we provide evidence that human INO80 (hINO80) participates in both DNA replication and chromosome segregation during the normal cell division cycle. hINO80 binds to chromatin localizing at replication forks during the S-phase, and is required for efficient DNA synthesis and S-phase progression. Unexpectedly, hINO80 associates with spindle microtubule during mitosis, and its deficiency leads to defective microtubule assembly and abnormal chromosome segregation. Consistent with these results, hINO80 is critical for suppressing aneuploidy and structural chromosome abnormalities. This work therefore not only emphasizes the evolutionary importance of INO80 in DNA replication but also reveals a new role for this remodeler in chromosome segregation, both of which likely come into play in maintaining the genome integrity.
Keywords: Human INO80 chromatin remodeling enzyme, DNA replication, Spindle assembly, Chromosome segregation, Genome stability
Introduction
The central task of eukaryotic cell division cycle is to accurately replicate chromosomal DNA and equally distribute identical chromosome copies to two daughter cells. In addition, the epigenetic code also must be precisely copied during replication and the chromosomes should be reconfigured into highly ordered and compacted forms prior to segregation. Further, chromatin structure is expected to be coordinately changed locally and globally for the accurate and efficient execution of these chromosomal processes. Defect or dysregulation in any of those processes not only brings about failure in faithful transmission of genomic information but also results in disruption of genome integrity, which potentially can lead to development of diseases such as cancer [1–4].
Two major types of enzymes are responsible for the modulation of chromatin structure; histone-modifying enzymes and ATP-dependent chromatin remodelers. While the former controls the accessibility of chromatin by adding or removing chemical modifications on histone proteins, the latter do by disrupting the histone–DNA interactions with the energy of ATP hydrolysis, which usually drives sliding of nucleosomes along DNA, entire removal of histones from DNA, or exchange of canonical histones in nucleosomes with histone variants. Thus far, characterized ATP-dependent chromatin remodelers all exist in the form of a multiprotein complex containing a catalytic core subunit that belongs to the large Snf2 ATPase family, and are involved in the various chromosomal processes such as transcription and DNA repair [5–7].
INO80 complex is a recent member of the ATP-dependent chromatin remodeling complex family. The yeast INO80 (yINO80) complex contains at least 15 polypeptides including the INO80 core subunit with significant similarity to the Swi2/Snf2 member of the Snf2 ATPase family [8]. The human counterpart is comprised of eight orthologs of the subunits of the yINO80 complex, including human INO80 (hINO80) ATPase, actin, actin-related proteins and the Rvb AAA+ ATPases, and additional metazoan-specific subunits [9]. Both yINO80 and hINO80 complexes have DNA-dependent ATPase activity and ATP-dependent nucleosome sliding activity in vitro [8–10]. Although the INO80 complex contains multiple ATPases, the INO80 subunit is responsible for in vitro remodeling activity and in vivo cellular functions, and the other associating factors are thought to have more specialized and species-specific roles [11].
A number of recent studies show that the yINO80 complex is implicated in a wide range of chromosomal processes [12, 13]. As a typical ATP-dependent chromatin remodeler, the yINO80 complex plays an important role in transcription [14, 15]. The yINO80 complex has been shown to directly participate in the repair of DNA double strand breaks (DSBs) [15–19] and in the control of cell-cycle checkpoint in response to DSBs [20, 21]. Very recently, studies have shown that the yINO80 complex also has a role in DNA replication such that it helps to stabilize and restart collapsed replication forks induced by replication stress [22–24]. Since most of the INO80 studies thus far have been focused on yeast, whether and to what degrees the functions conferred by the yINO80 complex are conserved in higher eukaryotes has yet to be investigated.
In the present study, we investigated the cellular functions of the hINO80 chromatin remodeling enzyme. We found that hINO80 is important for normal cell-cycle progression, particularly in S-phase progression and mitosis. Our data suggest that hINO80 is directly implicated not only in DNA replication but also in microtubule assembly and thereby chromosome segregation, the latter of which has not been identified from the studies with yeast, and that hINO80 appears to exert these dual functions by dynamically changing its subcellular localization during cell cycle progression. In keeping with its roles in DNA replication and chromosome segregation, hINO80 turns out to be critical for maintaining the chromosome integrity during normal cell proliferation.
Methods and materials
Cells and antibodies
HeLa-S3 cells were transfected with the vector expressing the hINO80-specific siRNA or corresponding empty vector (see below) by calcium phosphate, and selected against puromycin at the concentrations of 300 ng/ml. Two independent stable lines expressing hINO80-specific siRNA were named si-hINO80-2 and si-hINO80-3 cells, and the stable cells containing the empty vector are referred to as vector cells. Polyclonal anti-hINO80 antibodies were raised in rabbits against a synthetic peptide, RKQGKGTNPSGGR, corresponding to 1549–1561 amino acids of hINO80 (GenBank accession no. NM_017533). The sources of other antibodies used in this study were as follows: γ-H2AX, p-H3-Ser10 from Upstate; actin, α-tubulin, GFP, cyclin B1, Cyclin A, and Cdc2 from Santa Cruz; PCNA from Abcam; Flag from Sigma; GAPDH from AbFrontier; p-Cdc2-Y15 from Cell Signaling; BrdU from Molecular Probes. Rabbit IgG serum (I8140) was purchased from Sigma.
Plasmid constructions
The vector expressing hINO80-specific siRNA was constructed by inserting annealed oligonucleotides, 5′-GAT CCC CCT TGG TCT CCA TTT CAT ATT CAA GAG ATA TGA AAT GGA GAC CAA GTT TTT A-3′ and 5′-AGC TTA AAC TTG GTC TCC ATT TCA TAT TCT CTT GAA ATA TGA AAT GGA GAC CAA GGG G-3′, into the BglII–HindIII sites of pSuperior.puro vector (Oligoengine). The full-length hINO80 cDNA sequence was amplified by PCR from the clone KIAA1259 (Kazusa DNA Research Institute, Japan), and ligated into the NotI–EcoRI sites of pcDNA3.1/myc-His(-) to generate pcDNA-hINO80. The expression vector for f-hINO80 was constructed by cloning the Kpn1–NotI fragment of pcDNA-hINO80 into p3XFlag-CMV-14 vector (Sigma). The f-hINO80-K549A expression vector was generated on the basis of the f-hINO80 vector by site-directed mutagenesis according to the QuickChange Site-Directed Mutagenesis protocol (Stratagene) using oligonucleotides 5′-GAA ATG GGC CTT GGT GCA ACA GTA CAG AGC ATT-3′ and 5′-AAT GCT CTG TAC TGT TGC ACC AAG GCC CAT TTC-3′. The RFP-hINO80 expression vector was constructed by cloning the PCR-amplified full-length hINO80 cDNA sequences into the FseI–AscI sites of pCS2mRFP using primers 5′-GCC GGG CCG GCC CAT GGC CTC GGA GTT GGG TGC C-3′ and 5′-GCC GGG CGC GCC TTA CCG TCC TCC AGA GGG GTT-3′.
Synchronization and cell-cycle analysis
HeLa cells at 30–40% confluency were synchronized at G0 by serum starvation for 38–48 h using the culture medium with 0.1% serum. For synchronization at G1/S, cells were subjected to double thymidine block in which cells were grown in the medium containing 2 mM of thymidine for 18 h, released in the normal medium for 8 h, and again grown in the thymidine medium for 18 h. For synchronization at G2/M, cells were treated with nocodazole at 100 ng/ml for 18 h, and round-shaped mitotic cells were collected by shake-off. FACS analysis for cell-cycle profiles and mitotic cells were performed as previously described [25].
Live cell image analysis
GFP-H2B-expressing HeLa cells were transfected with indicated plasmids using Fugene 6 (Roche). Cells were arrested at G1/S by double thymidine block and released in the normal medium, and live cell images were captured using time-lapse UltraView RS-5 (Perkin Elmer) at indicated time or time intervals, with Z-stacks of 12 optical sections (z-increment 2 μm) acquired when necessary. The live images were also converted into videos, which can be provided upon request.
BrdU incorporation assays
Cells were pulse-labeled with 20 μM BrdU for 15 min, and after washing with PBS, the cells were fixed with 70% cold ethanol for overnight at −20°C, and then incubated with 10 μg/ml RNase A for 30 min at 37°C. DNA was denatured in the solution containing 2 M HCl and 0.5% Triton X-100 for 30 min at RT, and neutralized with 0.1 M boric acid for 2 min. After washing with PBS, the cells were incubated with the solution containing 0.1% Tween-20, 1% BSA, and Alexa Fluor 488-conjugated anti-BrdU antibodies (Molecular Probes) for 1 h in the dark. Cells were washed again and stained with 20 μg/ml of PI for 20 min before being subjected to FACS analysis.
Chromatin fractionation
Chromatin fractionation experiments were performed as previously described [26]. In brief, cells were suspended in the extraction buffer containing 50 mM Hepes pH 7.5, 150 mM NaCl, 1 mM EDTA, protease inhibitors, and 0.2% NP-40, and incubated for 5 min on ice. Fraction I was collected by centrifugation at 1,000 × g for 5 min. Pellet was re-suspended in the same buffer and centrifuged to collect Fraction II. The pellet was re-suspended in the fractionation buffer containing 0.5% NP-40 and further incubated for 40 min on ice, and centrifuged at 16,000 × g for 15 min to collect Fraction III (supernatant) and Fraction IV (pellet).
Immunofluorescence microscopy
Cells were placed on poly-l-lysine-coated slides, fixed with 100% methanol for 10 min at −20°C, made permeable by incubation in 0.5% Triton X-100 for 10 min, and blocked with 2% BSA in PBS for 1 h at RT. Cells were then washed for 7 min in PBS-T three times and incubated with primary antibodies overnight at 4°C followed by incubation with secondary antibodies for 30 min at 30°C. Cells were washed for 5 min with PBS-T four times and mounted using Vectashield medium with DAPI to visualize nuclei. Confocal fluorescence images were captured by using LSM 510 Meta (Carl Zeiss).
Immunoprecipitation experiments
Cells were lysed with RIPA or α-tubulin lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl, 0.5% NP40, 1 mM PMSF, leupeptin, aprotinin, 0.5 mM DTT, and 50 mM NaF) for 30 min on ice. The cell lysates were sonicated ten times each for 10 s at 30% amplitude using Cole-Parmer 400-W homogenizer and clarified by centrifugation. The supernatant was pre-cleared by filtering through Protein G Sepharose 4 fast flow for 1 h, and incubated with M2 affinity Gel (Sigma) for f-hINO80 or with indicated antibodies for other proteins overnight at 4°C. In case of using antibodies, protein G Sepharose was added and incubated for 3 h at 4°C. Precipitated beads were intensively washed with lysis buffer before being subjected to SDS-PAGE and immunoblot analysis.
Transfection
Transfections with plasmids or synthetic siRNA were performed using Lipofectamine 2000 (Invitrogen) typically for 48 h. The sequences of siRNA are as follows: hINO80, 5′-UUA AGA GUG UGA UUU CUC AUU-3′ and 5′-UGA GAA AUC ACA CUC UUA AUU-3′; non-specific control, 5′-CCU ACG CCA CCA AUU UCG UUU-3′ and 5′-ACG AAA UUG GUG GCG UAG GUU-3′.
Karyotype analysis
Cells were treated with 0.1 μg/ml colcemid (Biological Industries, Israel) for 1 h, incubated with 75 mM KCl for 30 min at 37°C, and then fixed with three changes of methanol/acetic acid (3:1). Fixed cells were subjected to slide spreading and Giemsa staining. Twenty metaphase spreads for each sample were analyzed with phase contrast microscopy (Zeiss) and Power Gene 810 (Applied Imaging, UK). Chromosomal abnormalities were determined according to the instruction of ISCN [27].
Results
hINO80 knockdown results in slow cell growth
To investigate the cellular functions of hINO80, we generated two independent HeLa cells stably expressing hINO80-specific siRNA, named si-hINO80-2 and si-hINO80-3, which showed approximately 70% knockdown of hINO80 protein expression (Fig. 1a). Both of these cells exhibited rather enlarged and elongated morphology (data not shown), and slow growth rate compared to parental cells or the cells containing empty vector (Fig. 1b). Transient transfection of the parental HeLa and other types of cells with synthetic si-hINO80 confirmed these results, and also indicated that the slow growth rate of hINO80-deficient cells is owing to slow cell growth rather than cell death (Fig. 1c).
Fig. 1.
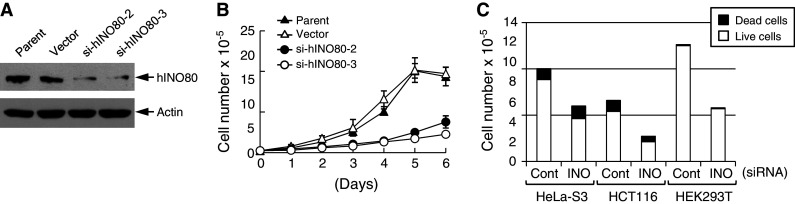
hINO80 knockdown results in slow cell growth. a Whole-cell lysates of parental HeLa-S3, vector, si-hINO80-2 and si-hINO80-3 cells were analyzed for the expression of hINO80 by immunoblottings. The expression of actin was also analyzed as loading control. b The growth rates of parent, vector, and si-hINO80-2/3 cells were determined by counting viable cells using the Tryphan Blue exclusion method. The data are presented as mean ± standard deviation (SD) from three separate experiments. c Indicated cells were transfected with control or synthetic hINO80-specific siRNAs for 24 h, and further cultured for 48 h. Live and dead cells were determined by the Tryphan Blue exclusion method
hINO80-deficient cells show delayed cell-cycle progression from G1/S to mitosis
To further investigate the slow growth of hINO80-deficient cells, we performed live-cell image analysis. HeLa cells, engineered to express histone H2B fused to green fluorescence protein (GFP-H2B), were transfected with empty or the si-hINO80 vectors. Simultaneously, the expression vectors for red fluorescence protein (RFP) were also transfected as a means to identify transfected cells. Cells were then arrested at G1/S and released for 24 h, with live cell images captured every hour using a time-lapse fluorescence microscope. While most of the vector-transfected cells (97%) entered mitosis before 14 h following the release from G1/S arrest, an average of 44% of the si-hINO80-transfected cells did not enter mitosis within this period of time (Fig. 2a–c). Notably, nearly half of such cells did not divide until 24 h post-release. The comparison of cell-cycle progression indicated that si-hINO80-2/3 cells are delayed for mitotic entry by several hours on average (Fig. 2d). Further confirming these results, analysis of cell-cycle profiles by flow cytometry with anti-phospho-H3 antibodies showed that si-hINO80-2/3 cells have significantly decreased mitotic cells (Fig. 2e).
Fig. 2.
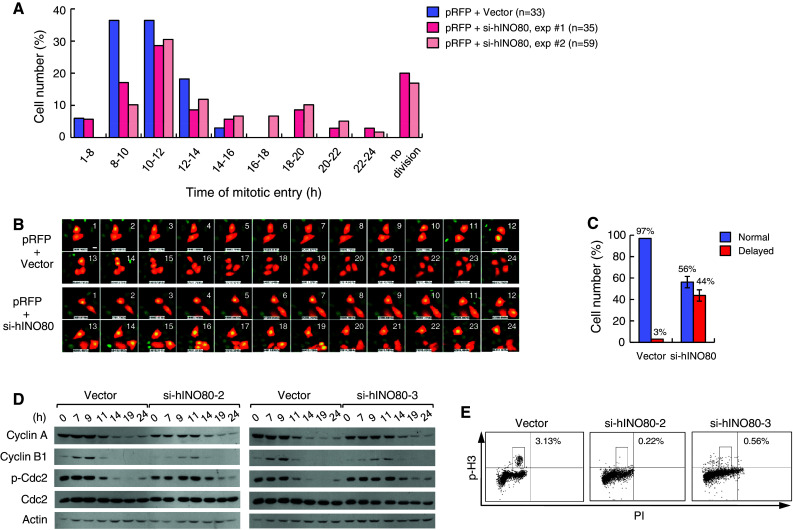
hINO80-deficient cells show a delayed cell-cycle progression from G1/S to mitosis. a GFP-H2B-expressing HeLa cells were cotransfected with the RFP expression vector plus either empty or si-hINO80 vectors at the molar ratios of 1:10. Live cell images were captured every hour after release from G1/S arrest using a time-lapse fluorescence microscope. RFP-positive cells were evaluated for mitotic entry time, whose distribution for three independent transfections was depicted as a graph. The number of RFP-positive cells analyzed for each sample is shown in the parenthesis. b Representative live cell images are shown with the hours after release from G1/S arrest. c A summary of the data in (b) is shown. ‘Normal’ indicates entering mitosis no later than 14 h after release from G1/S arrest, and ‘delayed’ later than 14 h. The data for the transfection with si-hINO80 vectors are presented as mean ± SD from two independent experiments. d G1/S-arrested cells were released for indicated times, and whole-cell lysates were analyzed for the expression of indicated proteins by immunoblottings (actin is for loading control). e Percentages of mitotic cells were determined by subjecting cells to FACS analysis after staining with anti-phospho-H3 antibodies and PI. The scale bars on all the microscopic cell images in this paper indicate 5 μM
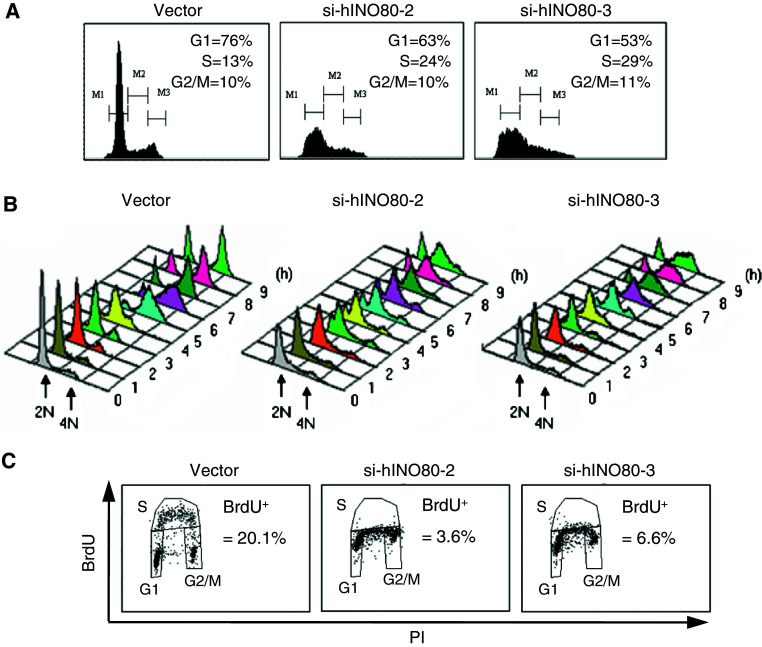
hINO80-deficient cells exhibit S-phase prolongation and impaired DNA synthesis
To dissect the cell-cycle phases in which hINO80 would have a role, we analyzed cell-cycle profiles and found that si-hINO80-2/3 cells have an approximately two-fold increase of S-phase population with no typical G2/M peak compared to vector cells (Fig. 3a), suggesting that hINO80 deficiency has an effect on the progression of the S-phase. Next, we monitored the S-phase progression by releasing cells from arrest at the border of G1/S-phases. Both of the si-hINO80-2/3 cells normally entered the S-phase but hardly traversed the S-phase and had difficulty in proceeding to G2/M (Fig. 3b). To test the possibility that the S-phase prolongation in si-hINO80-2/3 cells is due to DNA replication defect, we performed BrdU incorporation assays and found that si-hINO80-2/3 cells showed a low rate of DNA synthesis compared to vector cells (Fig. 3c). These results show that hINO80 is important for S-phase progression and DNA replication.
Fig. 3.
hINO80-deficient cells exhibit S-phase prolongation and impaired DNA synthesis. a Cell-cycle profiles were determined by subjecting indicated cells to FACS analysis after staining with PI. b Indicated cells were collected every hour after release from G1/S arrest and analyzed for cell-cycle profiles by FACS. c Indicated cells were pulse-labeled with BrdU for 15 min, stained with anti-BrdU antibodies and PI before being subjected to FACS analysis. Percentages of BrdU-positive cells are shown. All the experiments in this figure were repeated several times and the same results were obtained
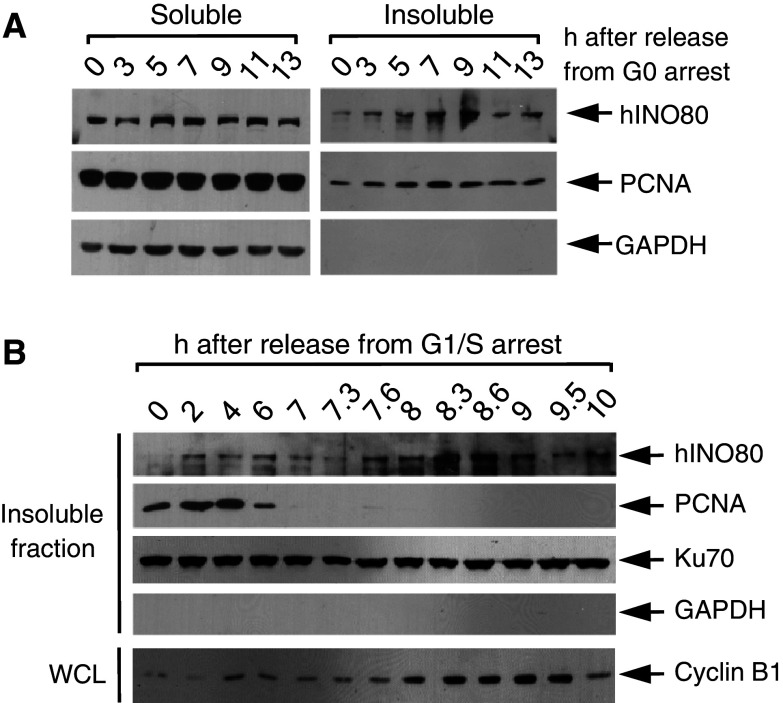
hINO80 binds to chromatin during the S-phase
Next, we asked whether hINO80 is directly involved in DNA replication. If hINO80 participates in DNA replication, it should bind to chromatin during this process. We therefore carried out biochemical fractionation experiments. Sub-confluent HeLa cells were arrested at G0 by serum starvation and subjected to fractionation various times after release. As shown in Fig. 4a, as cell cycle advanced away from G0, the hINO80 proteins found in insoluble fraction proportionally increased, reaching a peak around the 9-h time point, which corresponds to the S-phase. Similar experiments with cells released from G1/S arrest showed that insoluble hINO80 increased during 2–6 h post-release and decreased thereafter (Fig. 4b), confirming that hINO80 associates with chromatin during the S-phase. In both of these experiments, the kinetics of chromatin association of hINO80 was very similar to that of proliferating cell nuclear antigen (PCNA), a protein that highly localizes at replication forks and acts as a processivity factor for DNA polymerases, raising the possibility that hINO80 may target replication forks. In the meantime, we observed an intriguing result that a large amount of hINO80 is found in insoluble fraction peaking at the time between 8 and 9 h after release from G1/S arrest, corresponding to late G2 and mitotic phases as judged by cyclin B levels (Fig. 4b and see below).
Fig. 4.
hINO80 binds to chromatin during the S-phase. a HeLa cells were released from G0 arrest for indicated times and subjected to detergent fractionation. Soluble and insoluble chromatin fractions were analyzed for the levels of hINO80 and PCNA. GAPDH was analyzed as a cytoplasmic marker protein. b HeLa cells were released from G1/S arrest for the indicated time and subjected to detergent fractionation as in (a). Only the blots for the insoluble fractions are shown. Ku70 was used as the protein-loading control. The whole-cell lysates prepared from aliquots of the cells harvested at each time point were analyzed for the expression of cyclin B1
hINO80 is co-localized with PCNA at replication forks during the S-phase
The results showing that hINO80 associates with chromatin at the similar kinetics as PCNA raised a possibility that hINO80 may target the DNA replication forks. To test this possibility, we determined the subcellular localization of hINO80 by immunofluorescence microscopy. First, cells were released from arrest at G0. hINO80 existed primarily within the cytoplasm at 3 and 5 h, started to appear as spots in the nucleus at 7 h, and clearly formed large and distinct foci overlapping with PCNA foci at 9 h after release (Fig. 5a). To make sure that the hINO80 focus formation occurs during the S-phase, the cells were released from arrest at G1/S. We found that the hINO80 foci overlapping with PCNA were detected at 4 and 5 h, and disappeared at 7 h post-release (Fig. 5b and data not shown). These results, in keeping with the chromatin fractionation data, suggest that hINO80 binds to chromatin localizing at replication forks during DNA synthesis. Importantly, the PCNA-overlapping hINO80 foci were detected from some cells (likely S-phase cells) of asynchronized normal cultures (Fig. 5c), indicating that hINO80 localization at replication forks occurs during DNA synthesis under normal conditions. Interestingly, the formation of PCNA foci during the S-phase was not significantly affected by hINO80 deficiency (Fig. 5d), suggesting that hINO80 is not essential for the assembly and/or maintenance of replication forks.
Fig. 5.
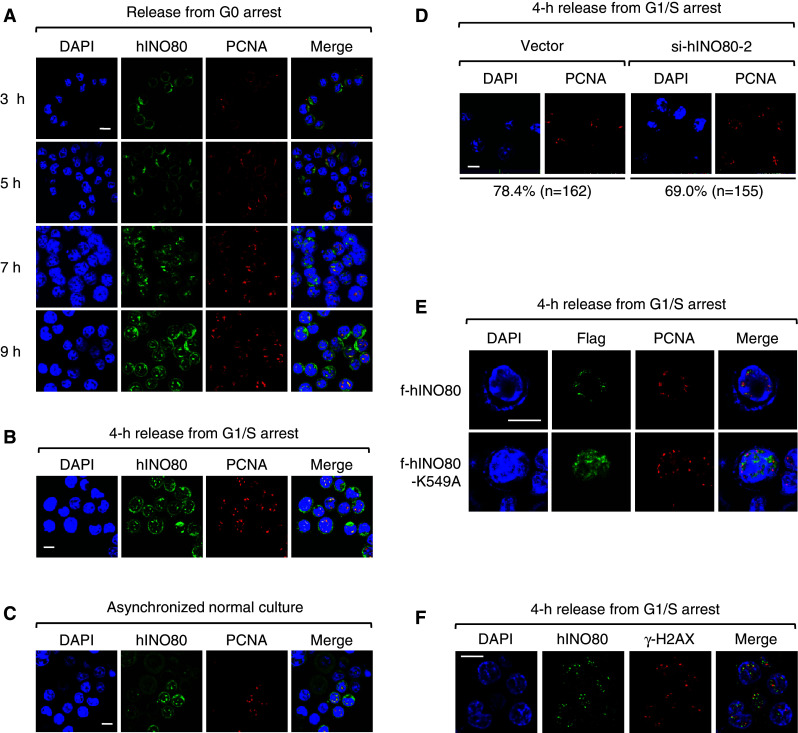
hINO80 is co-localized with PCNA at replication forks during the S-phase. a Sub-confluent HeLa cells were arrested at G0 by serum starvation, and after release for various times, the cells were fixed and dually stained with the antibodies for hINO80 and PCNA. Representative confocal images are shown. Quantitation of colocalization only within the nucleus using the LSM510 META-3.2 software indicates that hINO80 and PCNA overlaps by 90% at the 9-h time point. Note that the cells gradually become larger in size as the cell cycle moves forward into the S-phase. b HeLa cells were released from G1/S arrest for 4 h and fixed for staining with the antibodies for hINO80 and PCNA before confocal images were captured. Quantitation of colocalization between hINO80 and PCNA done as in (a) indicates that they overlap by 90%. c Asynchronized normal HeLa cells were fixed and dually stained with the antibodies for hINO80 and PCNA. Confocal images of the population containing S-phase cells were captured. d Indicated cells were released from G1/S arrest for 4 h and fixed for staining with the PCNA antibodies. The percentages of PCNA focus forming cells were determined by counting the indicated number of cells. e 293T cells were transfected with the expression vectors for f-hINO80 or f-hINO80-K549A and arrested at G1/S. After release for 4 h, cells were fixed for staining with the antibodies for PCNA and Flag. f HeLa cells were arrested at G1/S and released for 4 h before cells were fixed and dually stained with the antibodies for hINO80 and γ-H2AX. Representative confocal images of each experiment are shown
To determine whether the ATPase activity is required for replication fork localization of hINO80, we transfected 293T cells with the expression vectors for Flag-tagged hINO80 (f-hINO80) or the ATPase mutant version of f-hINO80 in which Ala was substituted for Lys at the 549 position (f-hINO80-K549A); this Lys corresponds to the one of yINO80 that is known to be critical for ATPase activity, DNA helicase activity and the ability of nucleosome sliding [8, 10]. We found that f-hINO80-K549A did not form nuclear foci whereas f-hINO80 normally formed the nuclear foci overlapping PCNA during the S-phase just as seen for the endogenous hINO80 (Fig. 5e). Thus, the ATPase activity of hINO80 appears to be important for localization of this protein to the replication forks.
Since the yINO80 complex has been shown to interact with the phosphorylated H2AX (γ-H2AX) at the sites of DSBs [15–17], we tested a possibility that hINO80 localizes at replication forks by interacting with γ-H2AX that can be induced by DSBs spontaneously occurring around replication forks. Immunofluorescence microscopy showed that hINO80 foci do not overlap with γ-H2AX, although they exist close to each other (Fig. 5f). In addition, despite intensive immunoprecipitation experiments, we did not see evidence for the interaction between hINO80 and γ-H2AX (data not shown). Therefore, it is unlikely that hINO80 targets the replication forks by means of interacting with γ-H2AX.
hINO80 associates with microtubules during mitosis and interacts with α-tubulins
The results, showing that hINO80 strongly associates with insoluble fraction around mitotic phase (Fig. 4b), prompted us to determine where the proteins localize during mitosis by immunofluorescence microscopy. HeLa cells were arrested at G2/M by nocodazole, and after release for various times cells were dually stained with the antibodies for hINO80 and α-tubulin. To our great surprise, the staining patterns of hINO80 were very similar to those of α-tubulin during the entire mitotic phase; hINO80 overlapped with the asteric microtubules at prophase, the mitotic spindles at metaphase, the central spindles at anaphase, and the midbody at telophase and cytokinesis (Fig. 6a). To exclude the possibility that the hINO80 staining patterns are due to a non-specific cross activity of the hINO80 antibodies towards α-tubulins, we transfected a vector expressing myc-tagged hINO80 (m-hINO80) into 293T cells and subjected these cells to immunofluorescence microscopy at various times after release from G2/M arrest. As shown in Fig. 6b, m-hINO80 exactly overlaps with the mitotic spindle, excluding the possible non-specific effects of the hINO80 antibodies. To further confirm these results, we performed similar experiments with 293T cells transfected by f-hINO80, and observed that staining with the Flag antibodies gave very similar patterns to those with the α-tubulin antibodies (Fig. 6c). Immunoblot analysis of the lysates from 293T cells transfected with empty, m-hINO80, or f-hINO80 vectors verified that the antibodies for hINO80, myc or Flag have little, if any, non-specific reactions towards the other cellular proteins (Fig. 6d). Interestingly, f-hINO80-K549A also could associate with the mitotic microtubules, suggesting that the ATPase activity is not necessary for the microtubule association of hINO80 during mitosis. The hINO80 association with spindles was also observed from mitotic cells in asynchronized normal culture (data not shown), indicating that these phenomena are not due to perturbation of the cell cycle by nocodazole treatment.
Fig. 6.
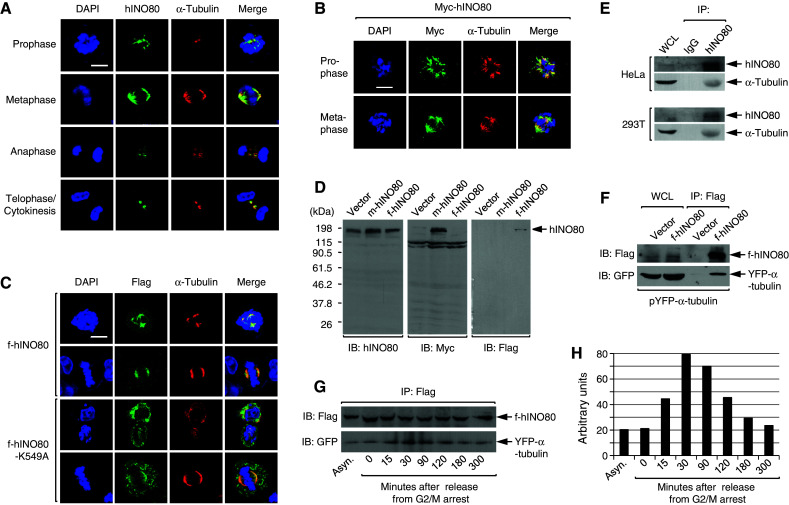
hINO80 is associated with spindle microtubules during the entire mitosis. a HeLa cells were released from G2/M arrest for various times and fixed for staining with the antibodies for hINO80 and α-tubulin. Representative confocal images are shown. b 293T cells were transfected with the myc-hINO80 vector, and after release for various times from G2/M arrest cells were fixed for staining with the myc antibodies. Representative confocal images are shown. c 293T cells were transfected with the f-hINO80 or f-hINO80-K549A vectors and arrested at G2/M. After release for various times, the cells were fixed for staining with the antibodies for Flag and α-tubulin. Representative confocal images are shown. d 293T cells were transfected with empty, myc-hINO80, or flag-hINO80 vectors, and whole-cell lysates were analyzed by immunoblotting using the hINO80 antibodies. The same blot was re-probed sequentially by myc and flag antibodies. e HeLa and 293T cells were released from G2/M arrest for 30 min, and whole-cell lysates were prepared for IP with the hINO80 antibodies and IgG as a control. The IP samples were subjected to immunoblot analysis to examine the interaction between the endogenous hINO80 and α-tubulin proteins. f 293T cells were cotransfected with pYFP-α-tubulin plus either empty or the f-hINO80 vectors, and after 30-min release from G2/M arrest, whole-cell lysates were prepared for IP with the Flag antibodies to determine the interaction between f-hINO80 and YFP-α-tubulin. g 293T cells transfected as in (f) were cultured asynchronously or released for various times from G2/M arrest before harvest, and whole-cell lysates were prepared and subjected to IP with the Flag antibodies followed by immunoblot analysis as indicated. A representative gel of three independent experiments is shown. h The gel in (g) was quantitated by densitometer and the relative intensity of YFP-α-tubulin bands after normalization to f-hINO80 bands was depicted as a graph
Next, we performed immunoprecipitation (IP) experiments to check whether hINO80 interacts with tubulins during mitosis. When hINO80 was precipitated from the lysates prepared from either HeLa or 293T cells in mitosis, α-tubulins were co-precipitated (Fig. 6e), indicating that hINO80 interacts with α-tubulins directly or indirectly. The interaction of hINO80 and α-tubulins was further confirmed by similar IP experiments with 293T cells co-transfected with the expression vectors for f-hINO80 and α-tubulin fused to yellow fluorescence protein (YFP-α-tubulin) (Fig. 6f). To determine whether the interaction between hINO80 and α-tubulin is specific to mitosis, we co-transfected 293T cells with the f-hINO80 and YFP-α-tubulin vectors and performed IP with lysates from these cells either cultured asynchronously or released for various times from G2/M arrest. We found that the interaction between f-hINO80 and YFP-α-tubulin largely increases during mitosis compared to asynchronous cells and the cells in G1 (3-h and longer release from G2/M arrest) (Fig. 6g, h). Thus, it appears that hINO80 highly interacts with α-tubulins during mitosis and this interaction occurs much less during interphase.
hINO80 is required for microtubule assembly during mitosis
Having found that hINO80 associates with spindle microtubules, we tested the possibility that hINO80 is involved in microtubule assembly during mitosis. To this end, we transfected cells with either empty or si-hINO80 vectors along with the RFP vector (to identify transfected cells), and evaluated RFP-positive cells for the formation of mitotic microtubules after release from G2/M arrest. Strikingly, approximately 85% of the si-hINO80 vector-transfected cells displaying condensed mitotic chromosomes failed to assemble microtubules, whereas only a few of the empty vector-transfected mitotic cells showed such defects (Fig. 7a, b), indicating that hINO80 is important for the formation of spindle microtubules during mitosis. To further confirm these results, we transfected cells with three different plasmids; the YFP-α-tubulin and the RFP vectors plus either empty or si-hINO80 vectors. Among the si-hINO80 vector-transfected cells whose chromosomes were obviously in condensed mitotic forms, about 84% were defective in microtubule assembly, whereas most of the empty vector-transfected cells (89.7%) were able to form asteric microtubules or mitotic spindles (Fig. 7c, d), further verifying the importance of hINO80 in the microtubule assembly during mitosis.
Fig. 7.
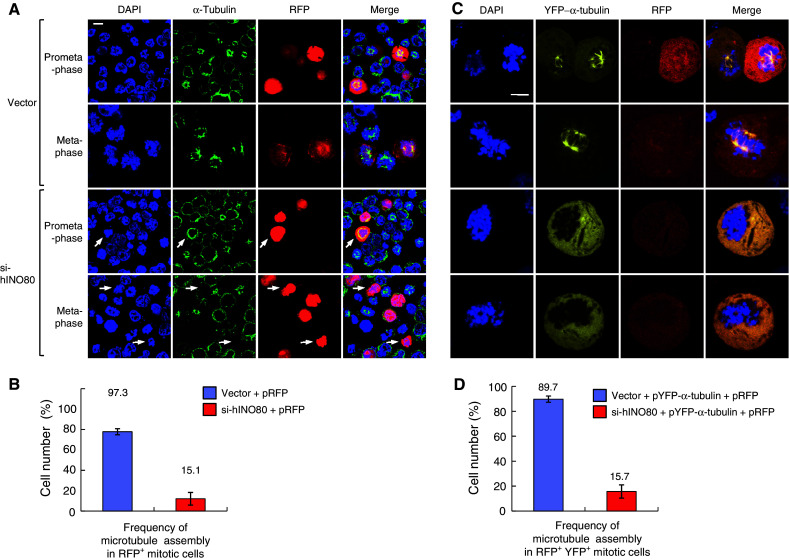
hINO80 is required for microtubule assembly during mitosis. a 293T cells were cotransfected with the RFP vectors plus either empty or si-hINO80 vectors at the molar ratios of 1:10. After release from G2/M arrest for 30 min (prometaphase) or 60 min (metaphase), cells were fixed for staining with the α-tubulin antibodies and DAPI. Representative confocal images are shown. The arrows indicate the RFP-positive cells defective in microtubule assembly during mitosis. b Data from the experiments in (a) were depicted as a graph. The frequency of microtubule assembly in RFP-positive mitotic cells was presented as mean ± SD from two independent experiments. Approximately 100 RFP-positive cells were counted for each sample. c 293T cells were cotransfected with the three different plasmids, the RFP and pYFP-α-tubulin vectors plus either empty or si-hINO80 vectors at the ratios of 1:1:10, and arrested at G2/M and released as in (a). Representative confocal images are shown. d Data from the experiments in (c) were depicted as a graph. The frequency of microtubule assembly in RFP/YFP double-positive mitotic cells was presented as mean ± SD from two independent experiments. Approximately 60 RFP/YFP double-positive mitotic cells were counted for each sample
hINO80 deficiency causes abnormal chromosome segregation and multinucleation
Given the importance of hINO80 for microtubule assembly, we examined whether hINO80 deficiency leads to related mitotic aberrations by the similar live-cell image analysis as previously described. The HeLa cells expressing GFP-H2B were co-transfected with the RFP vector plus either empty or si-hINO80 vectors, and released from G1/S arrest with live-cell images captured every 5 min by a time-lapse fluorescence microscope. The cells transfected with empty vector typically completed mitosis within 40–50 min with little apparent defects (Fig. 8a, b, top). In contrast, the cells transfected with si-hINO80 vector exhibited various types of mitotic defects, which can be categorized into three major groups. The first group of such cells was not able to form prophase chromosomes and hence failed to undergo subsequent mitotic stages (Fig. 8a). This type of defect, occurring most frequently among the three groups, is probably owing to delayed mitotic entry as described before (Fig. 2). The second group of cells showed the anaphase bridge which is characterized by separating sister chromatids being connected to each other (Fig. 8a, b, middle). Some of these cells underwent chromosome fragmentation and cell death following anaphase bridge (for example, Fig. 8b, middle). The third group included the cells showing abortive anaphase, in which chromatid separation does not occur completely, and the cells containing multinuclei; abortive anaphase and multinucleation are often detected in the same cells (Fig. 8a, b, bottom). The induction of multinucleation by hINO80 deficiency was more evident in stable knockdown cells, si-hINO80-2/3 (Fig. 8c, d), which is probably owing to accumulation of such cells through multiple rounds of cell division. These results show that hINO80 is important for chromosome segregation, in keeping with its association with mitotic microtubules.
Fig. 8.
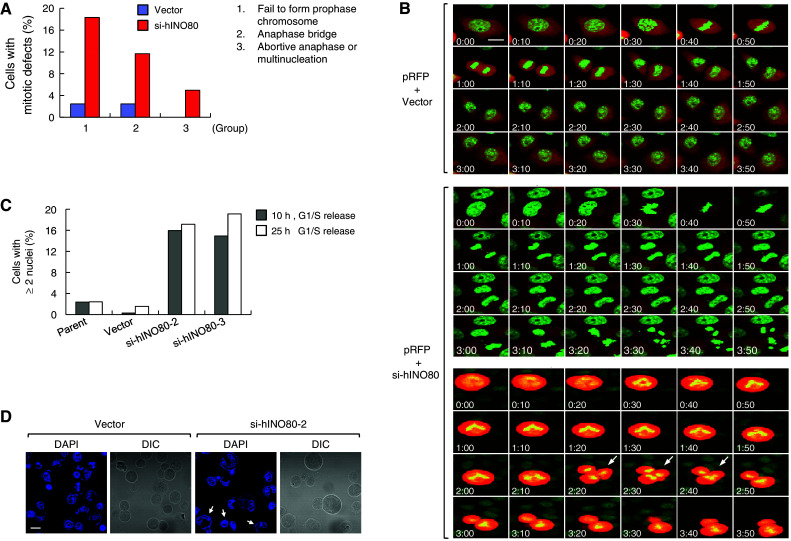
hINO80 deficiency causes abnormal chromosome segregation and multinucleation. a The GFP-H2B-expressing HeLa cells were transfected with the RFP vectors together with either empty or si-hINO80 vectors at the molar ratio of 1:10. Confocal live-cell images were captured every 5 min after release from G1/S arrest. Approximately 80 RFP-positive cells were evaluated for mitotic defects and the results were presented as a graph. b Representative confocal images are shown for the cells transfected with the RFP vector plus either empty (top) or si-hINO80 vectors (middle and bottom). The middle panel is a representative of the cells showing anaphase bridge (followed by chromosome fragmentation and cell death in this particular case), and the bottom is a representative of the cells showing abortive anaphase (indicated by arrows); accompanied by abnormal chromosome segregation into three in this particular case. The images at 10-min intervals were arranged so as to span the entire mitosis (the cells presumably starting to enter prophase were labeled by 0:00 time). c G1/S-arrested cells were released for 10 or 25 h, fixed and stained with DAPI. Percentages of the cells containing two or more nuclei were determined by counting 500–600 cells and depicted as a graph. d Representative confocal images used for the quantitation in (c) are shown. The cells containing two or more nuclei are indicated by arrows
hINO80-deficient cells exhibit aneuploidy and structural chromosome abnormalities
Since our data thus far implicated hINO80 in the chromosomal processes that are intimately associated with chromosome integrity, we asked whether hINO80 deficiency would result in chromosome abnormalities. First, we analyzed si-hINO80-2 cells for their karyotypes using a standard mitotic spreading method. All the parental (data not shown) and vector cells that were analyzed had an identical karyotype of the characteristic triploid chromosome complement of HeLa cells with a certain degree of deviation (Fig. 9a, b). Considering this karyotype as a background, we found that si-hINO80-2 cells exhibited a large degree of additional chromosome abnormalities. The most remarkable aberrations were the changes of chromosome number or aneuploidy; 100% of analyzed cells exhibit chromosome loss and 50% gain. hINO80 deficiency also induced various types of structural chromosome abnormalities, including translocations, deletions, telomeric association, dicentric chromosomes, chromatin/chromatid breaks as well as fragmented chromosomes whose origin is ambiguous, termed as markers (Fig. 9a, b).
Fig. 9.
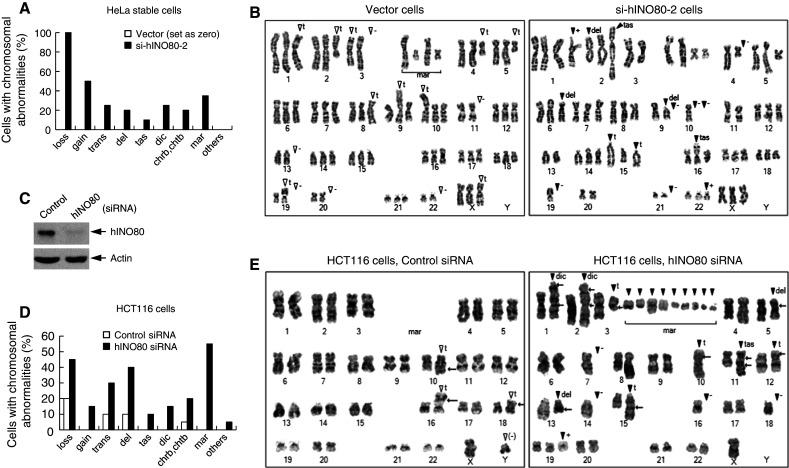
hINO80 is critical for the maintenance of chromosome integrity. a Twenty metaphase spreads for each of the indicated cells were subjected to karyotype analysis. By setting the karyotype of vector cells (all identical in 20 spreads) as a background, the chromosomal aberrations that were additionally generated in si-hINO80-2 cells were evaluated. The frequency of various types of chromosomal aberrations was depicted as a graph. Abbreviations are as follows: loss chromosome loss, gain chromosome gain, trans translocation, del deletion, tas telomeric association, dic dicentromeric chromosomes, chrb chromosome breaks, chtb chromatid break, mar marker chromosomes. b Representative images of the karyotypes analyzed in (a) are shown. Symbols are same as in (a) and as follows: plus (+) sign chromosome gain, minus sign (−) chromosome loss, t translocation. An open arrow head indicates the chromosomal aberrations already present in control cells and the closed arrow head indicates those newly generated in h-pSuper-hINO80 cells. c HCT116 cells were transfected with either non-specific control siRNA or hINO80-specific siRNA. Aliquots of transfected cells were analyzed for the expression of hINO80 and actin (loading control) by immunoblottings. d The remaining cells were subjected to karyotype analysis. The frequency of various types of chromosomal aberrations detected from 20 mitotic spreads was depicted as a graph. e Representative images of the karyotypes analyzed in (d) are shown. The cells transfected with control siRNA show the karyotype identical to the typical one of parental cells, which only has translocations in chromosomes 10, 16, and 18, with approximately half of the cell population missing Y chromosome. Abbreviations and symbols are same as in (a) and (b)
To further confirm the above results, we carried out similar experiments with HCT116 human colon cancer cells, known to have a near diploid karyotype. We transfected the cells with synthetic si-hINO80 or non-specific control siRNA, and verified that the cellular levels of hINO80 were efficiently reduced by more than 80% (Fig. 9c). The cells transfected with control siRNA had all identical diploid karyotype with minor deviations as reported for HCT116 cells; translocations in chromosomes 10, 16, and 18, with approximately half of the population missing Y chromosome (Fig. 9d, e). In contrast, the si-hINO80-transfected cells exhibited a large degree of newly generated chromosome aberrations, including aneuploidy as well as the structural abnormalities similar to those observed for si-hINO80-2 cells (Fig. 9d, e). It should be noted that the karyotypes of the analyzed parental and vector HeLa cells were all identical and all the analyzed control HCT116 cells had the same karyotypes as reported in the literature, indicating that there was no spontaneous karyotype change in those cells at least for the time period during which the experiments were performed. Thus, the increase of chromosomal abnormalities in si-hINO80-2 and hINO80 siRNA-transfected HCT116 cells is a result of the depletion of hINO80. These data clearly demonstrate that hINO80 is critical for maintaining the genome integrity under the conditions of normal cell proliferation.
Discussion
In the present study, we have investigated the cellular functions of the hINO80 chromatin remodeling enzyme. We find that hINO80 is important for normal cell cycle progression, particularly in S-phase progression and mitosis. hINO80 dynamically changes its subcellular localization during the cell-cycle progression such that it exists in the cytoplasm at G1, binds to chromatin localizing at replication forks in the nucleus during S-phase, and then associates with spindle microtubules during mitosis. hINO80 deficiency leads to slow cell growth with a prolonged S-phase and impaired DNA replication as well as with various mitotic defects such as defective microtubule assembly, abnormal chromosome segregation, and multinucleation. Thus, hINO80 appears to have direct roles both in DNA replication and in chromosome segregation, the key chromosomal processes that regulate genome integrity. Consistent with these results, hINO80-deficient cells exhibit aneuploidy as well as various structural chromosome abnormalities under normal culture conditions, suggesting that hINO80 functions as a suppressor of genome instability during normal cell division cycle.
The yINO80 complex has recently been shown to be directly implicated in stabilizing and restarting the collapsed forks induced by replication stress [22–24]. A very recent work has provided evidence that the yINO80 complex has a role in the DNA damage tolerance during replication [28]. Since the studies provide some conflicting data, whether the yINO80 complex functions in unperturbed normal DNA replication remains unclear [22–24, 28]. Our data, showing that hINO80 is required for efficient DNA synthesis and binds to chromatin localizing at PCNA foci under the conditions without replication stress, suggest that hINO80 is involved in normal DNA replication. Whether hINO80 also functions in the restart of collapsed forks and the damage tolerance during replication remains to be investigated. Further studies will be necessary to address whether and to what degree the role of INO80 complex in replication is conserved between yeasts and higher eukaryotes.
How does hINO80 target the replication forks? In an effort to answer this question, we tested some replication fork-associated proteins for possible interaction with hINO80. As previously mentioned, hINO80 foci do not overlap with the spontaneously formed γ-H2AX during the S-phase although they always exist very close to each other (Fig. 5f). In addition, we could not detect the interaction between hINO80 and γ-H2AX under the same chromatin IP conditions that permitted the interaction between the SWI/SNF complex and γ-H2AX [26]. In spite of intensive IP experiments, we did not see evidence for an interaction between hINO80 and PCNA or for the interaction of hINO80 with MCM2 which is known to be involved in the initiation of genome replication. Thus, hINO80 is not likely to target replication forks via interaction with γ-H2AX or the components of the replication machinery such as PCNA and MCM2. Interestingly, our preliminary data suggest that hINO80 becomes modified by phosphorylation when binding to chromatin during the S-phase (J.E.H and J.K., unpublished observations), which may provide an important clue to understanding how hINO80 targets the replication forks.
Although not necessarily expected as a chromatin remodeler, our data clearly show that hINO80 associates with microtubules and is required for efficient microtubule assembly during mitosis. Consistent with these results, one of the most frequently detected chromosomal abnormalities in hINO80-deficient cells is aneuploidy, which is known to be mainly caused by malfunctions of the mitotic apparatus and/or mitotic checkpoint defects [29]. The induction of multinucleation by hINO80 deficiency also supports the importance of hINO80 in microtubule assembly. Since the central spindles and midbody in the late stages of mitosis are thought to serve as the sites at which numerous regulatory proteins are concentrated and have multiple functions in cytokinesis [30], hINO80 may function in stabilizing such late-stage microtubules to ensure cytokinesis and prevent multinucleation. Interestingly, the INO80 associating factor, Arp4, has been reported to associate with centromeric regions and function in the assembly of kinetochore in yeast [31]. Also, a recent work showed that Tip49a, the AAA+ ATPase subunit of INO80 complex, is involved in mitotic spindle assembly in fly and mammalian cells [32]. Thus, the importance of INO80 complex in microtubule assembly appears to be evolutionarily conserved, although the exact role of hINO80 in this process and the underlying mechanisms remain to be further investigated.
DNA replication, together with the chromosome segregation during mitosis, provides the major sources of genome instability during normal cell proliferation. DSBs can be generated during DNA synthesis when replication forks encounter naturally occurring obstacles such as chemical adducts or single-strand DNA nicks. If not properly managed, such DSBs can lead to the gross structural chromosome aberrations such as translocations, interstitial or terminal deletions, and telomeric associations [1]. All of these types of structural abnormalities are clearly evident in hINO80-deficient cells, supporting the importance of hINO80 in DNA replication. Further emphasizing this point, hINO80-deficient cells frequently exhibit an anaphase bridge, which is thought to occur from telomeric associations that lead to the formation of dicentric chromosomes through the breakage–bridge fusion cycle [33]. Although a recent study reported that mammalian INO80 complex plays a role in DNA repair by interacting with transcription factor YY1, loss of which induces aneuploidy and structural chromosome abnormalities [34], our work directly shows for the first time the importance of INO80 in genome stability.
Our data show that whereas f-hINO80 forms the nuclear foci overlapping with PCNA during the S-phase, the ATPase mutant version of f-hINO80 lacking nucleosome remodeling activity does not (Fig. 5e). In contrast, both f-hINO80 and ATPase mutant f-hINO80 can normally associate with spindle microtubules during mitosis (Fig. 6b). These results lead us to speculate that the operating mechanisms of hINO80 in DNA replication and microtubule assembly may differ. While hINO80 could facilitate DNA replication by reconfiguring nucleosomes around the replication forks via its remodeling activity, it might help microtubule assembly using some other mechanisms not involving the remodeling activity.
Another study in our laboratory shows that hINO80 also contributes to DSB repair via the expression of Rad54B and XRCC3 genes. Therefore, our results, revealing the multiple roles for the hINO80 complex in DNA repair, replication and chromosome segregation, all of which are critically responsible for genome integrity, suggest that this chromatin remodeler may have evolved toward maintaining genome integrity, possibly by coordinating such chromosomal processes. Given the intimate association of genome instability with tumorigenesis, it will be of great interest to see if the INO80 complex can function as a tumor suppressor.
Acknowledgments
We thank Jae-Ho Lee (Ajou University, Korea), Dae-Sik Lim (KAIST, Korea), and the Kazusa DNA Research Institute (Japan) for kindly providing GFP-H2B HeLa cells, pYFP-α-tubulin, and the full-length hINO80 cDNA clone, respectively. This work was supported by the Molecular and Cellular BioDiscovery Research Program (M10748000334-08N4800-33410) grant to J.K. from the Korea Science and Engineering Foundation (KOSEF) funded by the Korea Ministry of Education, Science and Technology (MEST), and also supported in part by the grant to J.K. (R01-2007-000-10571-0) from KOSEF funded by MEST, and by grant No. R15-2006-020 from the National Core Research Center (NCRC) program of MEST and KOSEF through the Center for Cell Signaling and Drug Discovery Research at Ewha Womans University.
Footnotes
S.-K. Hur and E.-J. Park contributed equally to this work.
References
- 1.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev. 2004;14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Swedlow JR, Hirano T. The making of the mitotic chromosome: modern insights into classical questions. Mol Cell. 2003;11:557–569. doi: 10.1016/S1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 5.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 8.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 9.Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutiérrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 10.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 11.Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 15.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 16.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 17.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae . Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukuda T, Lo YC, Krishna S, Sterk R, Osley MA, Nickoloff JA. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst) 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, Weissman JS, Delrow J, Krogan NJ, Haber JE, Shen X. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 23.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 24.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer LG, Tommerrup N. An international system for human cytogenetic nomenclature. Basel: S. Karger Publishers, Inc.; 2005. [Google Scholar]
- 28.Falbo KB, Alabert C, Katou Y, Wu S, Han J, Wehr T, Xiao J, He X, Zhang Z, Shi Y, Shirahige K, Pasero P, Shen X. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Struct Mol Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 30.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 31.Ogiwara H, Ui A, Kawashima S, Kugou K, Onoda F, Iwahashi H, Harata M, Ohta K, Enomoto T, Seki M. Actin-related protein Arp4 functions in kinetochore assembly. Nucleic Acids Res. 2007;35:3109–3117. doi: 10.1093/nar/gkm161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ducat D, Kawaguchi S, Liu H, Yates JR, 3rd, Zheng Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol Biol Cell. 2008;19:3097–3110. doi: 10.1091/mbc.E07-11-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, Wu C, Shi Y. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]