Abstract
The human α2-plasmin inhibitor (A2PI) possesses unique N- and C-terminal extensions that significantly influence its biological activities. The C-terminal segment, A2PIC (Asn398-Lys452), contains six lysines thought to be involved in the binding to lysine-binding sites in the kringle domains of human plasminogen, of which four (Lys422, Lys429, Lys436, Lys452) are completely and two (Lys406, Lys415) are partially conserved. Multiple Lys to Ala mutants of A2PIC were expressed in Escherichia coli and used in intrinsic fluorescence titrations with kringle domains K1, K4, K4 + 5, and K1 + 2 + 3 of human plasminogen. We were able to identify the C-terminal Lys452 as the main binding partner in recombinant A2PIC (rA2PIC) constructs with isolated kringles. We could show a cooperative, zipper-like enhancement of the interaction between C-terminal Lys452 and internal Lys436 of rA2PIC and isolated K1 + 2 + 3, whereas the other internal lysine residues contribute only to a minor extent to the binding process. Sulfated Tyr445 in the unique C-terminal segment revealed no influence on the binding affinity to kringle domains.
Keywords: α2-Plasmin inhibitor, Plasminogen-binding region, Kringle, Lysine-binding site, Fluorescence titration
Introduction
The human α2-plasmin inhibitor (A2PI), a member of the serine protease inhibitor (serpin) super family [1], was discovered in 1976 [2–4]. A2PI is a single-chain plasma glycoprotein [3, 5] synthesized in the liver [6, 7] with a carbohydrate content of 14% [5]. It has been shown that A2PI contains four fully glycosylated N-glycosylation sites (Asn87, Asn250, Asn270, and Asn277) [8, 9]. Two different N-terminal forms of A2PI circulate in human plasma: Met-A2PI, a 464-residue protein with Met as N-terminus, and Asn-A2PI, a 452-residue form with Asn as N-terminus [10, 11]. The numbering in this publication is according to Asn-A2PI. Mature human A2PI, which corresponds to Asn-A2PI [10–13], contains four cysteine residues (Cys31, Cys64, Cys104, and Cys113), two of which form a single disulfide bridge (Cys31-Cys104) [14] and a sulfated tyrosine residue (Tyr445) of unknown function [15]. A2PI possesses unique N- and C-terminal extensions that significantly influence its biological activities. The N-terminal portion at Gln2 is cross-linked to Lys303 in the Aα chain of fibrin(ogen) by activated coagulation factor XIIIa [16, 17]. The C-terminal moiety of A2PI (A2PIC) extends approximately 55 amino acids beyond the consensus serpin sequence [12] and is highly conserved among species, which indicates a distinct and specific function for the C-terminal Asn398-Lys452 segment [18] (Scheme 1). It has been shown that A2PIC mediates the binding to plasmin(ogen) kringles [19–21]. The X-ray crystal structure of N-terminally truncated murine A2PI, which shares 71% sequence identity with human A2PI, shows that A2PI exhibits the typical serpin fold [22]. Unfortunately, the structure of the unique N- and C-terminal extensions still remains unsolved.
Scheme 1.
Multiple sequence alignment of human, bovine, murine, rabbit, and rat A2PIC. The numbering is according to the Asn form of human A2PI. The Pro399Ala mutation is shaded in gray, sulfated Tyr445 is underlined, Lys residues are in bold type and the symbols asterisk, colon, dot, and blank space stand for identity, high, average, and poor similarity, respectively
A2PI is the principal physiological inhibitor of plasmin (Plm) [2–4], the key enzyme of the fibrinolytic cascade. Plasminogen (Pgn), the zymogen of Plm, is a 90-kDa multidomain plasma glycoprotein synthesized in the liver [23]. It is composed of an N-terminal PAN module [24], five kringle domains [25], and a trypsin-like serine protease unit [26]. All kringle domains contain a lysine-binding site (LBS), which is responsible for the interaction of Pgn with, e.g., fibrin, A2PI, and small molecules with positively charged group(s) such as lysine and lysine analogous, like 6-aminohexanoic acid (6-AHA) or trans-4-(aminomethyl)cyclohexanecarboxylic acid (t-AMCHA) (reviewed in [27]). Plm reacts with A2PI to form an enzymatically inactive 1:1 equimolar complex [3, 5, 28, 29] upon cleavage of the scissile peptide bond (Arg365-Met366) in the reactive center loop (RCL) of A2PI [30]. Kinetic data suggest a two-step reaction for the rapid inhibition of Plm: a fast reversible second-order reaction (k 1, 3.8 × 107 M−1 s−1) followed by a slower irreversible first-order transition (k 2, 4.2 × 107 M−3 s−1) [19, 28, 29, 31, 32]. In the second step, the scissile peptide bond in the RCL of A2PI is cleaved by the protease subunit of Plm. The first step is rate limiting and results in the formation of a non-covalent complex between A2PIC and kringle domains of Plm. The complex formation between Plm and A2PI is competitively inhibited by plasminogen kringles 1–3 (K1–3) and kringle 4 (K4) or by 6-AHA [19]. A2PI inhibits miniplasmin, a Plm fragment possessing only kringle 5 (K5) and the fully active protease domain, approximately 60-fold slower than intact Plm [31]. 30% of mature A2PI in human plasma is partially degraded lacking at least 26 residues (Gly427-Lys452) of the C-terminal segment [20]. This truncated form also inhibits Plm less rapidly than native A2PI [33, 34]. These observations emphasize the physiological relevance of this non-covalent interaction between the unique A2PIC and Plm kringle domains.
Human A2PIC contains six lysine residues, of which four (Lys452, Lys436, Lys429, and Lys422) are completely and two (Lys406 and Lys415) are partially conserved (Scheme 1). It is well known that internal as well as C-terminal lysine residues often exhibit a distinct affinity for LBS in kringles. Initial studies gave evidence for the participation of the C-terminal Lys452 residue in the binding to the Plm kringle domains [18]. Furthermore, a synthetic C-terminal peptide of A2PI (Gly427-Lys452) has been shown to competitively block the reversible association of A2PI to Plm kringle domains [20, 21]. Substitution of Lys452 with arginine or its removal with carboxypeptidase B was found to reduce the interaction between these peptides and Plm [21, 35]. These data led to the suggestion that the multiple lysine residues within A2PIC contribute, probably in a zipper-like manner, to the binding to the ‘five-kringle array’ in Plm [18, 35]. In contrast to these findings, kinetic data by Wang et al. [36, 37] indicate that Lys436 might play an important role in the interaction between Plm and A2PI.
To elucidate the exact role of the four completely conserved lysine residues (Lys452, Lys436, Lys429, and Lys422) and the partially conserved Lys415 and Lys406 in the binding to the LBS of Pgn kringle domains, we have expressed wild-type, single, double, triple, quadruple, quintuple, and sextuple Lys mutants of recombinant A2PIC(Pro399Ala) (rA2PIC) in Escherichia coli. Association constant (K a) values of the different mutant rA2PIC constructs to human recombinant kringle 1 (rK1) and kringles 4–5 (rK4–5) and human K4 and K1–3, which were obtained by limited proteolysis of Pgn with porcine elastase, have been determined applying intrinsic fluorescence ligand titrations. To investigate the influence of sulfated Tyr445 in the interaction with human kringles rK3 and K4, the K a values of synthetic peptides Glu442-Lys452 of the C-terminal region of A2PI, A2PIC(Glu442-Lys452), with and without sulfated Tyr445 have also been obtained by intrinsic fluorescence ligand titrations.
Materials and methods
Materials
Restriction endonucleases, Pfu DNA Polymerase, T4 DNA Ligase, activated coagulation factor X (FXa) and trypsin (porcine, seq. grade modified) came from Promega (Madison, WI, USA). Porcine elastase was purchased from Serva (Heidelberg, Germany). UltraPure™ Agarose was obtained from Invitrogen (Carlsbad, CA, USA). E.Z.N.A.® MicroSpin Gel Extraction Kit was purchased from peQLab (Erlangen, Germany). GenElute™ HP Plasmid Miniprep Kit and Luria–Bertani Broth (LB Broth) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Primers were synthesized either by Microsynth (Balgach, Switzerland) or Sigma-Aldrich. Sephadex G-50m and G-50sf were purchased from GE Healthcare (Waukesha, WI, USA). Acrylamide, Bio-Gel P-6DG and Bio-Gel P-300 were from Bio-Rad (Hercules, CA, USA). Lysine Bio-Gel P-300 was prepared according to Brunisholz et al. [38] and Ni2+-NTA Agarose was purchased from QIAGEN (Venlo, The Netherlands).
Bacterial strains and plasmids
Escherichia coli strain XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proABlacIqZ∆M15 Tn10 (Tetr)]} was obtained from Stratagene (La Jolla, CA, USA) and used for routine transformations and plasmid preparations. Rosetta™ [F–ompT hsdSB(r–Bm–B) gal dcm (DE3) pRARE [39] (CamR)] E. coli strain was purchased from Novagen (Madison, WI, USA) and used for the expression of recombinant A2PIC (rA2PIC) and rA2PIC mutants. The pColdII plasmid was obtained from Takara (Kyoto, Japan) and used for the expression of rA2PIC. The plasmid contains a lac operator, a cspA 5′-untranslated region, a translation enhancing element, a hexaHis Tag sequence (His-tag), a multiple cloning site, and a cspA 3′-untranslated region immediately upstream of the cspA transcription initiation site and carries an ampicillin resistance [40]. The pET100/D-TOPO® plasmid (Invitrogen) containing the rA2PIC peptide was prepared as described previously [8].
Methods
DNA manipulations
Isolation of plasmid DNA was done as described by the manufacturer. DNA fragments generated by restriction endonuclease cleavage were purified on 1.2 or 0.5% (depending on their size) agarose gels, stained with ethidium bromide and visualized by UV light. DNA fragments were recovered from the agarose gels by the E.Z.N.A.® MicroSpin Gel Extraction Kit according to the supplier’s instructions.
Construction of expression vectors for wild-type and mutant rA2PIC peptides
The C-terminal region of A2PI (Asn398-Lys452) was amplified in a PCR using the pET100/D-TOPO® plasmid containing rA2PIC or rA2PIC(Lys452Ala) as a template. The 5′ primer F/rA2PIC, which binds to the FXa cleavage site of the sense strand of the pET100/D-TOPO® plasmid and the first four bases of rA2PIC, introduced a KpnI and a FXa cleavage site upstream of the codon for Asn398. The 3′ primer R/rA2PIC, complementary to the coding strand of rA2PIC, was used to introduce two stop codons and a BamHI cleavage site downstream of the codon for Lys452. Primers used are listed in Table 1. The KpnI and BamHI cleaved PCR product was cloned into the KpnI-/BamHI-digested pColdII plasmid and transformed into XL1-Blue E. coli cells. Clones were selected and the insert was verified by DNA sequence analysis at Microsynth (Balgach, Switzerland).
Table 1.
Primers used for the generation of rA2PIC mutants, pColdIImut vector, rK5, and rK4–5
| Primer name | DNA sequence |
|---|---|
| F/pelB | 5′-GGCGACTAGTATGAAATACCTGCTGCCGACCG-3′ |
| R/pelB | 5′-CGGCACTAGTCGAATTAATTCCGATATCCATGGCC-3′ |
| F/pColdIImut | 5′-GGCGACTAGTCATCATCATCATCATCATATGGAGCTCGG-3′ |
| R/pColdIImut | 5′-CGGCACTAGTCACTTTGTGATTCATGGTGTATTACCTC-3′ |
| F/rA2PIC | 5′-CACCGGTACCATCGAGGGTAGAAACC-3′ |
| R/rA2PIC | 5′-GCGCGGATCCCTATTACTTGGG-3′ |
| F/rK4–5 | 5′-CACCGGTACCATCGAGGGTAGAGTACAGGACTGCTACCAT-3′ |
| R/rK4–5 | 5′-CGCGGGATCCCTATTAAAATGAAGGGGCCGCACA-3′ |
| F/rK5 | 5′-CACCGGTACCATCGAGGGTAGAACTGCATCCGAAGAAGACTGTATG-3′ |
| R/rK5 | 5′-CGCGGGATCCCTATTAGGCCGCACACTGAGGGACAT-3′ |
| Lys406Ala | 5′-GCGGGAGCTCGCGGAACAGCAGGATTC-3′ |
| Lys415Ala | 5′-GATTCCCCGGGCAACGCGGACTTCCTCCAGAG-3′ |
| Lys422Ala | 5′-TTCCTCCAGAGCCTGGCAGGCTTCCCCCG-3′ |
| Lys429Ala | 5′-GGCTTCCCCCGCGGAGACGCGCTTTTCGGCCCTGACTT-3′ |
| Lys436Ala | 5′-GCTTTTCGGCCCTGACTTAGCACTTGTGCCCCCCATGGAG-3′ |
| Lys452Ala | 5′-GCGCGGATCCCTATTAAGCGGG-3′ |
| Lys452Ser | 5′-CGCGGGATCCCTATTAAGAGGGGCTGCCAAACTGGGGG-3′ |
| Pro399Ala | 5′-CGAGGGTAGAAACGCCAGTGCACCGC-3′ |
Mismatched bases are shown in bold type
For the construction of the expression vectors rA2PIC(Lys452Ala) and rA2PIC(Lys452Ser) the 3′-mismatch primers Lys452Ala and Lys452Ser (Table 1) were used, respectively. The point mutation at the position Lys452 to Ser was introduced to improve the solubility of rA2PIC(Lys452Ala).
Construction of expression vectors for single, double, triple, quadruple, quintuple, and sextuple Lys to Ala mutant rA2PIC peptides by site-directed mutagenesis
The method used is based on the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). In brief, the appropriate pColdII expression vector (0.5 μg/μL) was selected as template and used in a PCR with the suitable 5′ primer in a concentration of 1.25 μg/μL. Primers used are listed in Table 1. After amplification, the template DNA was digested by adding 1 μL DpnI (8 h at 37°C). 4% of DpnI-digested PCR product was then transformed into XL1-Blue E. coli cells. Clones were selected and the insert was verified by DNA sequence analysis.
Substitution of Pro399 to Ala in all rA2PIC peptides
A point mutation at position Pro399 was introduced in all rA2PIC peptides using site-directed mutagenesis as described above, to improve the cleavage efficiency of FXa [41, 42]. The Pro399Ala primer is shown in Table 1. All mentioned rA2PIC peptides in this publication are hereafter referred to as carriers of the Pro399Ala mutation.
Construction of the pColdIImut(pelB) vector
To overcome the poor expression yields with the pColdII expression system for certain constructs, we have modified the pColdII vector by introducing the pelB leader sequence, which can also be used as enhancer for the total protein expression [43], between the TEE and His-tag sequence of the pColdII vector. pColdIImut was amplified in a PCR using the pColdII vector as template. The 5′ primer F/pColdIImut and 3′ primer R/pColdIImut were used to introduce a SpeI cleaving site (ACTAGT) between the TEE and the His-tag sequences. Cleavage with SpeI and ligation, followed by transformation into XL1-Blue E. coli cells and selection of appropriate clones, yielded the mutated pColdIImut vector. The pelB leader sequence was amplified in a PCR using the pET22b(+) vector as a template. The 5′ primer F/pelB and 3′ primer R/pelB were used to introduce a SpeI cleaving site up- and downstream of the pelB leader sequence. The SpeI cleaved PCR product was cloned into the SpeI-digested pColdIImut plasmid and transformed into XL1-Blue E. coli cells. Clones were selected and the insert was verified by DNA sequence analysis. The primers used are listed in Table 1.
Construction of the expression vector for rK5 and rK4–5
The rK5 construct (Thr456-Ala543) was amplified in a PCR using the pPLGKG plasmid (contains the cDNA of human Pgn) as a template. The 5′-mismatch primer F/rK5, which binds to the sense strand of Pgn, introduced a KpnI and a FXa cleavage site upstream of the codon for Thr456. This 5′-mismatch primer was also used to introduce a point mutation at position Pro457 to Ala to enhance the cleavage efficiency of FXa [41, 42]. The 3′ primer R/rK5, complementary to the coding strand of Pgn, was used to introduce two stop codons and a BamHI cleavage site downstream of the codon for Ala543. The primers used are compiled in Table 1. The cloning, transformation, and verification of clones was carried out according to the above-described procedure for the construction of expression vectors for wild-type and mutant rA2PIC peptides except that the pColdIImut(pelB) vector was used. The mentioned rK5 in this article is hereafter referred to as rK5 construct (Thr456-Ala543) and carrier of the Pro457Ala mutation.
The rK4–5 construct (Val355-Phe546) was generated as described above except that 5′ primer F/rK4–5, 3′ primer R/rK4–5 and pColdII were used. The mentioned rK4–5 in this publication is hereafter referred to as rK4–5 construct (Val355-Phe546).
Expression and isolation of wild-type and mutant rA2PIC peptides
The expression was carried out according to a slightly modified protocol of the manufacturer’s instructions. The agar plates and LB Broth culture media always contained 100 μg ampicillin/mL and 34 μg chloramphenicol/mL. Cells were grown at 37°C in LB Broth medium in 2-L Erlenmeyer flasks to an OD600 of about 0.4–0.6 absorbance units (AU). The cell cultures were refrigerated at 15°C for 35 min before isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 2 mM. The cultures were thereafter incubated at 15°C and 220rpm for 21–24 h and finally harvested by centrifugation (Sorvall RC 3C Plus, 4,000g, 65 min, 4°C). The cell pastes were stored at −80°C until further use. The protein expression was verified by SDS-PAGE (15%) and the proteins were stained with Coomassie brilliant blue G-250.
For the isolation of wild-type and mutant rA2PIC peptides, the cell pastes were thawed and suspended in lysis buffer (6 M guanidine hydrochloride and 300 mM NaCl in 50 mM Tris, pH 8.0; 10 mL/g of wet cell paste), stirred overnight at 4°C and centrifuged (Sorvall RC 5B Plus, 12,000g, 45 min, 4°C). The supernatants were loaded onto a Ni2+-NTA Agarose column (2.2 cm × 9 cm; 1 mL Ni2+-NTA Agarose/1,000 mL cell culture) equilibrated with the lysis buffer, pH 8.0. The column was thereafter washed with lysis buffer, pH 8.0 and 6.3. The recombinant peptides were eluted from the columns with lysis buffer, pH 4.5. The peptides were desalted by dialysis against 20 mM NH4HCO3, lyophilized and stored at 4°C until further use. The peptides were analyzed by RP-HPLC, amino acid analysis, ESI-MS, and Edman sequence analysis.
Expression, isolation, and characterization of recombinant kringle constructs rK1, rK3, rK5, and rK4–5
rK1 and rK3 were expressed, isolated, refolded and characterized according to Marti et al. [44, 45].
rK4–5 and rK5 were expressed and isolated according to the above-described protocol for wild-type and mutant rA2PIC peptides except that the proteins were refolded instead of desalting by dialysis. rK4–5 was refolded and isolated according to Douglas et al. [46]. rK5 was refolded according to the same protocol as rK4–5 [46]. In addition, isolated rK5 was lyophilized and further purified by size exclusion chromatography on a Sephadex G-50sf column (1.5 cm × 100 cm; 0.13 M formic acid). The constructs were analyzed as mentioned above.
Isolation of human plasminogen and generation of the K1–3 and K4 domains
Pgn was isolated from citrated plasma by affinity chromatography on Lysine Bio-Gel P-300 as previously described [38]. K1–3 and K4 were prepared via limited proteolysis of Pgn with porcine elastase, isolated and purified as previously described [25].
Purification and characterization of the synthetic peptides A2PIC(Glu442-Lys452) with and without sulfated Tyr445
The synthetic peptides A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) were purified by preparative RP-HPLC on a butyl column (Wide-Pore™, 5 μm, 4.6 mm × 250 mm; J.T. Baker) with a Hewlett-Packard liquid chromatograph 1090. 50 mM ammonium acetate in water, pH 7.0, was used as solution A and 25 mM ammonium acetate, 80% (v/v) acetonitrile in water, pH 7.0, as solution B. A linear gradient (0–60% solution B in 60 min) was used at a flow rate of 1 mL/min.
RP-HPLC
RP-HPLC separations were carried out on an Aquapore butyl column (BU-300, 7 μm, 300 Å, 2.1 mm × 100 mm; Perkin Elmer) or on a Nucleosil 300 phenyl column (7 μm, 2 mm × 100 mm; Dr. Maisch HPLC, Ammerbuch, Germany) with a Hewlett-Packard liquid chromatograph 1090. 0.1% (v/v) trifluoroacetic acid (TFA) in water was used as solution A and 0.1% (v/v) TFA, 80% (v/v) acetonitrile in water as solution B. A linear gradient (0–100% solution B in 60 min) was used at a flow rate of 0.3 mL/min.
Amino acid analysis
Samples were hydrolyzed in the gas phase with 6 M HCl containing 0.1% (v/v) phenol for 22 h at 115°C under N2 vacuum according to Chang and Knecht [47]. The liberated amino acids were reacted with phenylisothiocyanate, and the resulting phenylthiocarbamoyl amino acids were analyzed by RP-HPLC on a Nova Pack C18 column (4 μm, 3.9 mm × 150 mm; Waters) with a Dionex Summit® HPLC system (Dionex, Olten, Switzerland) equipped with an automatic injection system according to Bidlingmeyer et al. [48]. The corresponding ammonium acetate buffer replaced the 0.14 M sodium acetate buffer, pH 6.3.
Mass spectrometry
The mass of the peptides was determined by ESI-MS (VG Platform mass spectrometer; Micromass Instruments, UK). The samples were dissolved in 0.5% (v/v) formic acid, 50% (v/v) acetonitrile in water.
Removal of the His-tag sequence
To remove the His-Tag, constructs were incubated with FXa at 37°C for 16 h in 50 mM Tris, pH 8.0, containing 100 mM NaCl and 5 mM CaCl2 at an enzyme/substrate ratio 1:800 (w/w). Cleaved peptides were separated from His-tag and undigested constructs on a Ni2+-NTA Agarose column (2.2 cm × 9 cm). rA2PIC and rK5, both without His-tag, were finally desalted either on a Sephadex G-50m or on a Bio-Gel P-6DG column (1.5 cm × 100 cm) (50 mM NH4HCO3, pH 8.0).
Tryptic peptide mass mapping
For an explicit identification of the diverse rA2PIC mutants, the peptides were digested with trypsin at 37°C overnight in 0.1 M NH4HCO3 at an enzyme/substrate ratio 1:100 (w/w). The peptide mass mapping was carried out by ESI-MS.
Intrinsic fluorescence titration
The influence of wild-type and mutant rA2PIC peptides on the intrinsic fluorescence of different Pgn kringles was measured in 50 mM sodium phosphate, pH 8.0, at 25°C with a Cary Eclipse fluorescence spectrophotometer (Varian) as reported by Menhart et al. [49]. The ligand (wild-type and mutant rA2PIC) was added to a 5-μM kringle solution at intervals of 1.66, 3.3, 5, 10, 20, 33, or 40 μM depending on the binding affinity of the investigated kringle until the intrinsic fluorescence was constant. Fluorescence was measured at 340 or 344 nm, depending on the investigated kringle, with excitation at 296 nm. K a values were determined according to Scatchard [50].
A threefold determination of the peptide content in the ligand solutions by amino acid analysis allowed an accurate evaluation of the K a values.
The K a values for the interaction of A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) with Pgn rK3 and K4 were obtained as described above.
Results
Recombinant expression of rA2PIC and rA2PIC mutants
We have successfully applied the pColdII expression system to improve the expression of rA2PIC and rA2PIC mutants. The identification of rA2PIC peptides by SDS-PAGE showed a main band indicating a relatively high level of expression (data not shown). The expression yield of the rA2PIC mutants was in the range of 5–35 mg of pure peptide/2 L of cell culture due to variable expression levels of the rA2PIC constructs. The introduction of the point mutation at position Pro399 to Ala dramatically enhanced the cleavage efficiency of FXa, since it has been shown that the P2′ position at the FXa cleavage site (IEGR) is important for the enzymatic activity of FXa [41, 42]. We could overcome the poor solubility of rA2PIC(Lys452Ala) by introducing a Lys452 to Ser mutation. All rA2PIC peptides eluted as sharp, symmetrical peaks on RP-HPLC columns. After removal of the His-tag, the measured masses agree well with the calculated values (Table 2). Amino acid analyses and Edman sequence analyses yielded the expected compositions and N-terminal sequences (data not shown), respectively, and both methods were applied for the characterization of all following constructs.
Table 2.
Characterization of rA2PIC peptides by ESI-MS (experimental and theoretical values)
| Construct | Mass (exp.) (Da) | Mass (theor.) (Da) |
|---|---|---|
| rA2PIC(wt) | 6,145.0 ± 0.4 | 6,145.9 |
| rA2PIC/K406A | 6,087.4 ± 0.5 | 6,088.8 |
| rA2PIC/K415A | 6,087.5 ± 0.8 | 6,088.8 |
| rA2PIC/K422A | 6,087.9 ± 0.6 | 6,088.8 |
| rA2PIC/K429A | 6,088.0 ± 0.6 | 6,088.8 |
| rA2PIC/K436A | 6,088.8 ± 1.2 | 6,088.8 |
| rA2PIC/K452A | 6,088.0 ± 0.1 | 6,088.8 |
| rA2PIC/K452S | 6,104.3 ± 0.2 | 6,104.8 |
| rA2PIC/K436/452A | 6,031.1 ± 0.3 | 6,031.7 |
| rA2PIC/K429/452A | 6,031.4 ± 0.2 | 6,031.7 |
| rA2PIC/K429/436A | 6,031.8 ± 0.2 | 6,031.7 |
| rA2PIC/K422/429/436A | 5,974.3 ± 0.4 | 5,974.6 |
| rA2PIC/K415/422/429/436A | 5,917.3 ± 0.7 | 5,917.5 |
| rA2PIC/K406/415/422/429/436A | 5,860.4 ± 0.4 | 5,860.4 |
Recombinant expression, purification, and refolding of Pgn rK1, rK3, rK5, and rK4–5
rK1 and rK3 were expressed in E. coli, purified, refolded, and characterized as previously described [44, 45]. The isolation over Lysine Bio-Gel P-300 ensured that the disulfide bridges were folded correctly. The measured masses of 10,167.3 ± 0.6 Da for rK1 and 9,503.8 ± 1.0 Da for rK3 correspond well with the calculated masses of 10,167.2 and 9,505.4 Da, respectively.
rK5 was successfully expressed in E. coli using the pColdIImut(pelB) vector with which we achieved a significantly higher expression yield compared to the unmodified pColdII plasmid. A point mutation at position Pro457 to Ala was introduced to enhance the cleavage efficiency of FXa as mentioned above. The identification of rK5 by SDS-PAGE showed a main band indicating a relatively high level of expression (data not shown). The expression yield was about 19 mg of pure protein/2 L of cell culture. rK5 was isolated as described above and refolded according to Marti et al. [44, 45]. The measured mass of 9,769.0 ± 0.6 Da corresponds well with the calculated value of 9,768.7 Da.
rK4–5 was successfully cloned into the pColdII vector and expressed in E. coli [51]. Approximately 5–10 mg of pure protein/2 L of cell culture was obtained. rK4–5 was isolated and refolded as described for rK5 and the measured mass of 23,734.1 ± 4.9 Da corresponds well with the calculated mass of 23,735.3 Da.
Characterization of the purified synthetic peptides A2PIC(Glu442-Lys452/Tyr445[SO4]) and A2PIC(Glu442-Lys452)
The measured masses of 1,295.8 ± 0.3 Da for A2PIC(Glu442-Lys452) and 1,376.1 ± 0.3 Da for A2PIC(Glu442-Lys452/Tyr445[SO4]) correspond well with the calculated masses of 1,296.4 and 1,376.4 Da, respectively.
Binding of wild-type and mutant rA2PIC to human Pgn K4
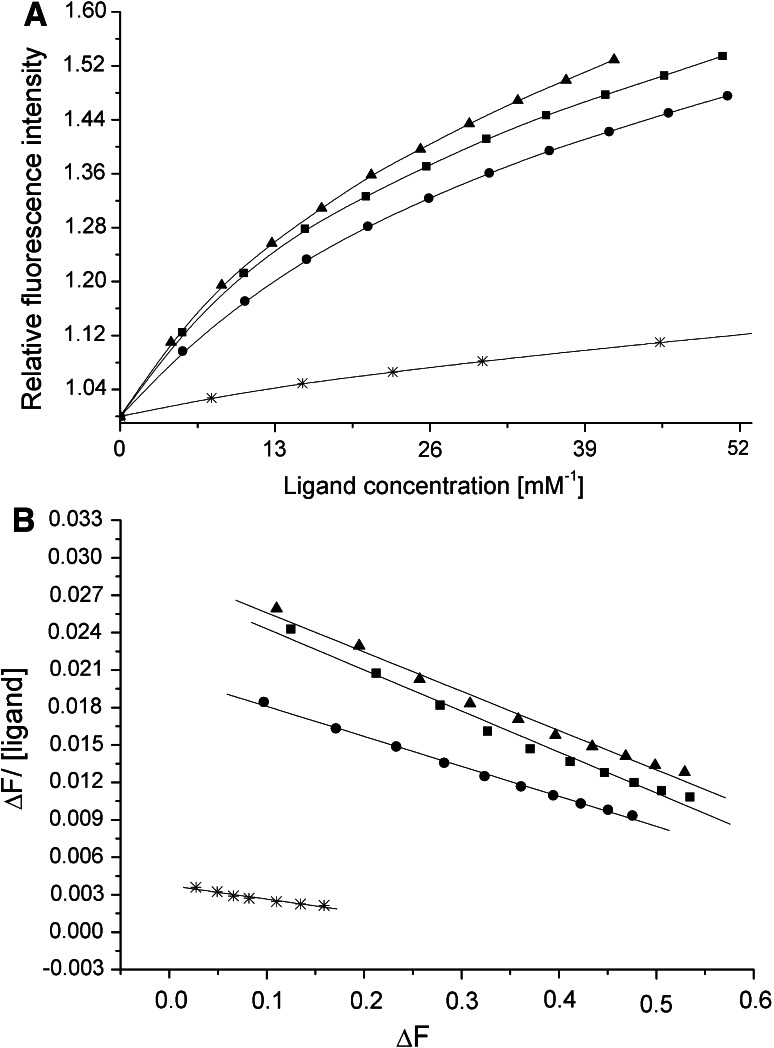
Wild-type rA2PIC exhibits a K a value of 34 ± 2 mM−1 for Pgn K4, which is in the range of previously published data [18]. Replacing the C-terminal Lys452 residue by either Ser or Ala substantially decreases the affinity (rA2PIC(K452S), K a = 11 ± 2 mM−1) by a factor of ~3. Our data confirm that Lys452 is the main binding partner in the interaction with the isolated K4 domain in solution, which is in good agreement with published data [18]. The single Lys to Ala mutants of the totally conserved internal Lys residues Lys436 (K a = 32 ± 2 mM−1), Lys429 (K a = 33 ± 2 mM−1), and Lys422 (K a = 33 ± 3 mM−1), and the partially conserved Lys415 (K a = 44 ± 6 mM−1) and Lys406 (K a = 42 ± 6 mM−1) contribute only to a minor extent to the total binding affinity to the LBS of K4. The double rA2PIC(Lys429Ala/Lys436Ala) (K a = 29 ± 0.5 mM−1), triple rA2PIC(Lys422Ala/Lys429Ala/Lys436Ala) (K a = 28 ± 1 mM−1), quadruple rA2PIC(Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 24 ± 1 mM−1), and quintuple mutants rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 23 ± 2 mM−1) exhibit a steady decrease of the K a values with a decreasing number of Lys residues. The data confirm the weak affinity for internal Lys residues to the LBS of K4 [18, 52, 53]. The sextuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala/Lys452Ala) mutant exhibits a very weak affinity (K a = ~3 mM−1). A comparison of the fluorescence titration and Scatchard plot of wild-type rA2PIC with mutants rA2PIC(Lys436A), rA2PIC(Lys452Ser), and rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) is given in Fig. 1 and all K a values are summarized in Table 3. The determined K a value for 6-AHA of 46 ± 4 mM−1 was in good agreement with published data [18].
Fig. 1.
a Averaged fluorescence titration of K4 with wild-type rA2PIC (filled squares), single Lys to Ala mutants rA2PIC(Lys436Ala) (filled triangles), and rA2PIC(Lys452Ser) (star), and quintuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (filled circles). The fluorescence intensity is normalized to 1 and the x-axis is enlarged. b Corresponding Scatchard plots and calculated regression curves
Table 3.
Association constants K a (mM−1) of wild-type and mutant rA2PIC peptides to various kringle constructs of Pgn
| Ligand | rK1 | K1–3 | K4 | rK4–5 |
|---|---|---|---|---|
| 6-AHA | 75 ± 11a | 107 ± 11 | 46 ± 4 | ~52 |
| Wild type | 77 ± 4 | 176 ± 30 | 34 ± 2 | 18 ± 1 |
| K406A | 42 ± 6 | |||
| K415A | 44 ± 6 | |||
| K422A | 177 ± 22 | 33 ± 3 | ||
| K429A | 170 ± 9 | 33 ± 3 | ||
| K436A | 70 ± 1 | 26 ± 10 | 32 ± 2 | 16 ± 2 |
| K452S | 8 ± 0.5 | 17 ± 4 | 11 ± 2 | 1.4 ± 0.5 |
| K452A | 11.6 ± 0.5a | 9.1 ± 0.2a | ||
| K429/36A | 17 ± 6 | 29 ± 0.5 | 16 ± 5 | |
| K429/52A | 8 ± 0.5 | 30 ± 5 | ||
| K422/29/36A | 22 ± 7 | 28 ± 1 | 18 ± 5 | |
| K415/22/29/36A | 12 ± 2 | 24 ± 1 | ~17 | |
| K406/15/22/29/36A | 53 ± 7 | 46 ± 12 | 23 ± 2 | 17 ± 4 |
| K406/15/22/29/36/52A | ~3 | ~4 | ~3 | |
| A2PIC(E442-K452/Y445[SO4]) | 29 ± 4 | |||
| A2PIC(E442-K452) | 24 ± 4 |
aFrom [18]
Binding of wild-type and mutant rA2PIC to Pgn rK1
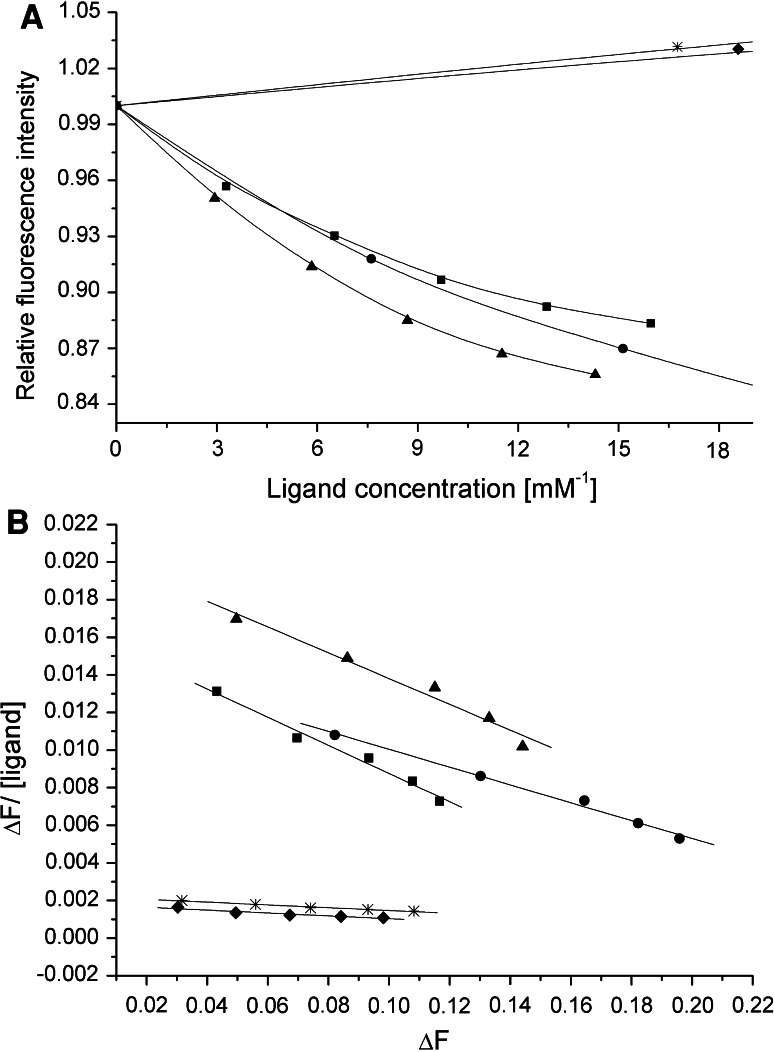
The wild-type rA2PIC exhibits a K a value of 77 ± 4 mM−1 for Pgn rK1 and replacement of the C-terminal Lys452 residue by Ser or Ala decreases the affinity (rA2PIC(Lys452Ser), K a = 8 ± 0.5 mM−1) by a factor of ~10, which is in good agreement with published data [18]. The double mutant rA2PIC(Lys429Ala/Lys452Ala) (K a = 8 ± 0.5 mM−1) exhibits the same K a value as the single mutant rA2PIC(Lys452Ser), while the single mutant of internal Lys436 rA2PIC(Lys436Ala) (K a = 70 ± 1 mM−1) shows no significant difference compared to the wild-type rA2PIC. The quintuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 53 ± 7 mM−1) exhibits a partially decreased affinity compared to the wild-type rA2PIC. The sextuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala/Lys452Ala) shows a weak K a value of ~3 mM−1. Our data confirm that Lys452 is the main binding partner in the interaction with the isolated rK1 domain in solution, which is in good agreement with published data [18], and that rK1 exhibits only a weak but distinct affinity for internal Lys residues [18, 52, 53]. A selection of the fluorescence titrations and Scatchard plots of wild-type rA2PIC, single rA2PIC(Lys436A) and rA2PIC(Lys452Ser), double rA2PIC(Lys429Ala/Lys452Ala), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) mutants is shown in Fig. 2 and all K a values are summarized in Table 3. The K a value of 75 ± 11 mM−1 for 6-AHA was determined by Frank et al. [18] and is in the same range as wild-type rA2PIC.
Fig. 2.
a Averaged fluorescence titration of rK1 with wild-type rA2PIC (filled squares), single Lys to Ala mutants rA2PIC(Lys436Ala) (filled triangles), and rA2PIC(Lys452Ser) (star), double rA2PIC(Lys429Ala/Lys452Ala) (filled diamonds), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (filled circles) mutants. The fluorescence intensity is normalized to 1 and the x-axis is enlarged. b Corresponding Scatchard plots and calculated regression curves
Binding of wild-type and mutant rA2PIC to human Pgn K1–3
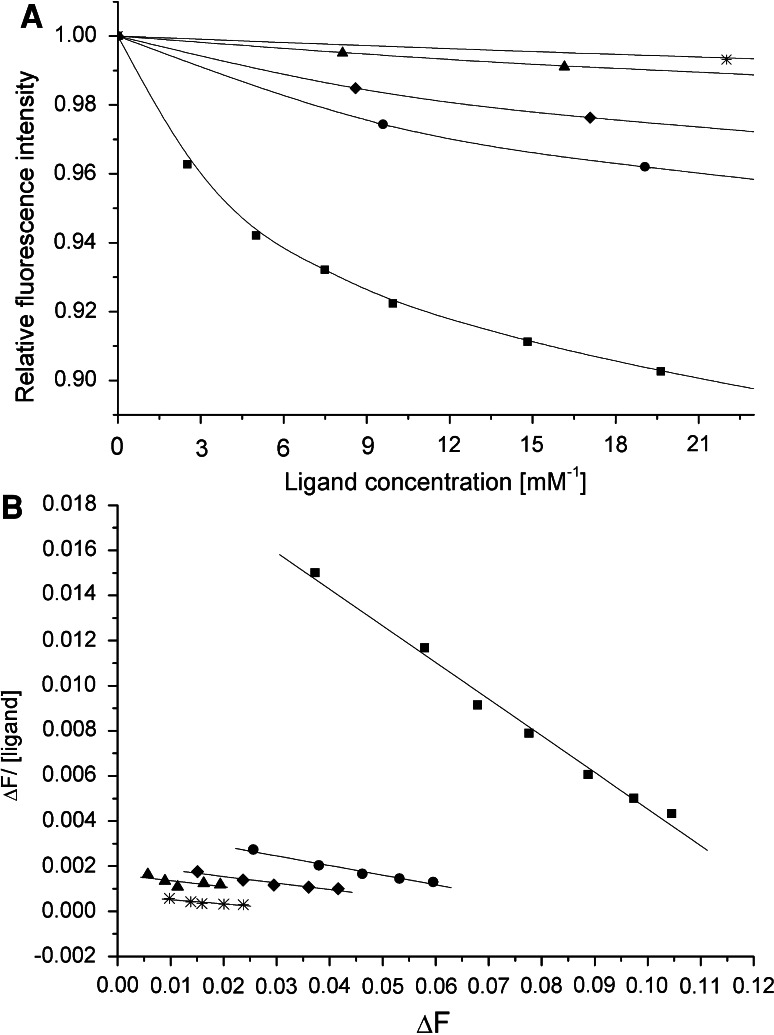
The K a values of wild-type rA2PIC (K a = 176 ± 30 mM−1) and internal single mutants of the totally conserved Lys429 (K a = 170 ± 9 mM−1) and Lys422 (K a = 177 ± 22 mM−1) for Pgn K1–3 are in a similar range. The K a values drop dramatically after replacement of the C-terminal Lys452 (rA2PIC(Lys452Ser), K a = 17 ± 4 mM−1) or the internal Lys436 (rA2PIC(Lys436Ala), K a = 26 ± 10 mM−1) residue by Ser or Ala, respectively. The double rA2PIC(Lys429Ala/Lys436Ala) (K a = 17 ± 6 mM−1) and rA2PIC(Lys429Ala/Lys452Ala) (K a = 30 ± 5 mM−1), triple rA2PIC(Lys422Ala/Lys429Ala/Lys436Ala) (K a = 22 ± 7 mM−1), quadruple rA2PIC(Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 12 ± 2 mM−1), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 46 ± 12 mM−1) mutants exhibit a highly reduced affinity, thus confirming the importance of the Lys452 and Lys436 residues in the interaction with the isolated triple-kringle domain K1–3 in solution. The fluorescence titrations and Scatchard plots of wild-type rA2PIC, single rA2PIC(Lys436A) and rA2PIC(Lys452Ser), double rA2PIC(Lys429Ala/Lys452Ala), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) mutants are shown in Fig. 3. The sextuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala/Lys452Ala) exhibits a very weak affinity (K a = ~4 mM−1) and the Lys analog 6-AHA exhibits a K a value of 107 ± 11 mM−1, which is considerably smaller compared to that of wild-type rA2PIC. All K a values are summarized in Table 3.
Fig. 3.
a Averaged fluorescence titration of K1–3 with wild-type rA2PIC (filled squares), single Lys to Ala mutants rA2PIC(Lys436Ala) (filled triangles) and rA2PIC(Lys452Ser) (star), double rA2PIC(Lys429Ala/Lys452Ala) (filled diamonds), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (filled circles) mutants. The fluorescence intensity is normalized to 1 and the x-axis is enlarged. b Corresponding Scatchard plots and calculated regression curves
Binding of wild-type and mutant rA2PIC to Pgn rK4–5 and rK5
The K a value of wild-type rA2PIC (K a = 18 ± 1 mM−1) for the rK4–5 double-kringle domain is smaller than for K4 alone. As in K4 and rK1, the C-terminal Lys452 (rA2PIC(Lys452Ser), K a = 1.4 ± 0.5 mM−1) is the main binding partner for the interaction with the LBS of isolated rK4–5 in solution. The single rA2PIC(Lys436Ala) (K a = 16 ± 2 mM−1), double rA2PIC(Lys429Ala/Lys436Ala) (K a = 16 ± 5 mM−1), triple rA2PIC(Lys422Ala/Lys429Ala/Lys436Ala) (K a = 18 ± 5 mM−1), quadruple rA2PIC(Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = ~17 mM−1), and quintuple rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 17 ± 4 mM−1) mutants exhibit comparable affinities to the LBS of rK4–5 as wild-type rA2PIC. The sextuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala/Lys452Ala) shows no detectable change in intrinsic fluorescence and an accurate K a value could not be determined. 6-AHA exhibits a K a value of ~52 mM−1, which is significantly higher than the K a value of wild-type rA2PIC. All K a values are summarized in Table 3.
Due to the low affinity of K5 for Lys residues, no significant fluorescence change could be detected. This is consistent with earlier studies, which showed the very weak affinity of K5 for C-terminal and internal Lys residues [18, 54].
Binding of A2PIC(Glu442-Lys452/Tyr445[SO4]) and A2PIC(Glu442-Lys452) to Pgn rK3 and K4
rK3 was used for intrinsic fluorescence titrations with the short synthetic C-terminal segments A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) to investigate the influence of the sulfated Tyr445 residue. The A2PIC(Glu442-Lys452/Tyr445[SO4]) exhibits a very weak affinity with a K a value of ~3 ± 1 mM−1 and is in a comparable range to the non-sulfated peptide A2PIC(Glu442-Lys452) (K a = ~5 ± 1 mM−1).
K4 was used to confirm the ability of A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) to bind to the LBS of Pgn kringles. The K a values of A2PIC(Glu442-Lys452/Tyr445[SO4]) (K a = 29 ± 4 mM−1) and A2PIC(Glu442-Lys452) (K a = 24 ± 4 mM−1) are in a similar range (see Table 3). The quintuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) (K a = 23 ± 2 mM−1) and the synthetic C-terminal peptide A2PIC(Glu442-Lys452) (K a = 24 ± 4 mM−1), both containing only the C-terminal Lys452, show very similar binding affinities.
Discussion
Our investigations of wild-type and single, double, triple, quadruple, quintuple, and sextuple Lys to Ala mutants of the Pgn-binding region of A2PI (Asn398-Lys452), rA2PIC, via intrinsic fluorescence titrations, identify the C-terminal Lys452 as the main binding partner in the interaction with the LBS of isolated K4 in solution, which is consistent with published data [8, 18]. We could show that all internal Lys residues (Lys436, Lys429, Lys422, Lys415 and Lys406) contribute only to a minor but distinct extent to the binding to the LBS of K4, which is demonstrated by a steady decrease of the K a values with a decreasing number of Lys residues. We find similar results for isolated rK1, which is also consistent with published data [8, 18], and the double-kringle domain rK4–5 and the triple-kringle domain K1–3 in solution. The K a value for wild-type rA2PIC and rK4–5 is smaller than for wild-type rA2PIC and K4, which could be due to a shielding effect of the rK5 domain in the flexible rK4–5 double-kringle domain or due to the N-terminal His-tag, which was still present. Due to the weak binding affinity of K5 to internal and C-terminal Lys residues [18, 54], we could not detect a significant change in the intrinsic fluorescence of isolated rK5 in solution. Frank et al. [18] found that the K a values for wild-type rA2PIC with isolated rK2, rK3mut (contains an active LBS in contrast to wild-type rK3, which is devoid of a functionally active LBS [45]) and K5 in solution are in the range of 3–4 mM−1, which is indicative for a minor importance of these Pgn kringles in the interaction with wild-type rA2PIC. In contrast to these findings with isolated kringle domains, we identified Lys452 and Lys436 as important binding partners for the interaction with the isolated triple-kringle domain K1–3 in solution. The association constant of K1–3 for wild-type rA2PIC is two- to threefold increased compared to rK1, and replacing of the conserved internal Lys422 and Lys429 with Ala does not affect this enhanced binding affinity for K1–3. In addition, we could also demonstrate that the other internal Lys residues (Lys415 and Lys406) do not contribute significantly in the binding to the LBS of K1–3 and that the quintuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala) exhibits a K a value which is in the same range (K a = ~50 mM−1) as for the interaction with isolated rK1. This enhanced binding affinity of K1–3 for wild-type rA2PIC points towards a cooperative effect of Lys452 and Lys436, since it has been shown that the binding affinity of isolated rK2 in solution for Lys residues is very low [18, 44]. Our data underscore the fact that the Lys residues in the unique A2PIC mediate the binding to Pgn kringle domains [19–21, 31–34]. Wang et al. [36, 37] found in their kinetic studies with A2PI mutants that Lys436 is important for the interaction with plasmin in solution, immobilized plasminogen, or immobilized K1–3. They showed, in contrast to our findings, that Lys452 is not involved in this non-covalent interaction. The question arises, why the C-terminal Lys452 should be of no importance since it has been shown that the LBS of rK1 and K4 exhibit a much higher affinity for N α-acetyllysine, a ligand that mimics C-terminal lysines, than for N α-acetyllysine methyl ester, a mimic of internal Lys residues [18, 44, 52, 53]. Furthermore, another interesting fact supports the importance of Lys452. As Frank et al. [18] have already discussed, the distance between the last two lysines (Lys436 and Lys452) in fully extended linear A2PIC is ~30 Å, which would match the distance required to bridge the LBSs of K1 and K2 as estimated from the K1–3 crystal structure [55], where the distance from Trp144 (K1 LBS) to Trp225 (K2 LBS) is ~30 Å. This bridging seems to point towards an induced-fit model, where the binding of Lys452 to the LBS of K1 is reinforced by the binding of Lys436 to the LBS of K2, which results in a conformational change of the K1–3 triple-kringle construct as a whole, rather than structural changes within domains, resulting in an enhanced binding compared to isolated rK1 or rK2 in solution. This hypothesis is supported by the fact that the multi-kringle domains are very flexible structures, since the conformational change upon activation of Pgn to Plm is quite dramatic (reviewed in [27]). Another fact derived from the X-ray crystal structure, supporting the importance of the Lys452 and Lys436 residues, is that the C-terminal extension lies close to the RCL in such a way that the more distal conserved Lys residues (Lys452 and Lys436) could readily engage the kringles in binding [22]. In addition, Sofian et al. [56] prepared three peptides, peptide 1 (Asn398-Leu421), peptide 2(Asp416-Leu435), and peptide 3(Leu430-Lys452), and the corresponding antibodies to these different regions of A2PIC, and investigated their effects on plasmin–antiplasmin kinetics and fibrinolysis. They could show that peptide 3 and antibody 3 exhibit a ~10-fold decrease of the rate of association between A2PI and Plm, which underscores the importance of Lys452 and Lys436, whereas peptides 1 and 2 and antibodies 1 and 2, corresponding to the remaining internal Lys residues (Lys429/Lys422 and Lys415/Lys406, respectively), seemed to have no significant effect on the serpin inhibition reaction. Nevertheless, it might be possible that the C-terminal segment adopts different conformations in the whole A2PI or as peptide in solution. These conformational differences could result in different binding affinities for kringle domains. However, the structure of the unique A2PIC remains unsolved [22], leaving the question of different conformations open. In addition, we have expressed recombinant A2PI and Lys to Ala mutants in the C-terminal segment of A2PI at high levels in E. coli [51]. No binding affinities could be determined due to extremely low solubility of the recombinant A2PI constructs at physiological conditions (pH and salts). Furthermore, Frank et al. [18] demonstrated by NMR and CD measurements that rA2PIC is a rather flexible polypeptide of low β-structure supporting the discussed induced-fit model.
We also investigated the effect of sulfated Tyr445, which is present in the unique C-terminal extension of native human A2PI [15]. A possible function of this posttranslational modification could be an interaction with Pgn K3, which contains no active LBS [45] but a heparin-binding site (Prof. Dr. M. Llinás, personal communication). It is therefore possible that sulfated Thyr445 in A2PIC might interact with the heparin-binding site in K3. We applied intrinsic fluorescence titrations to investigate the effect of two synthetic peptides A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) on the interaction with isolated Pgn rK3 in solution. The K a values of ~3 ± 1 mM−1 for the sulfated and ~5 ± 1 mM−1 for the non-sulfated A2PIC(Glu442-Lys452) revealed that sulfated Tyr445 has no detectable effect on the interaction of rK3 and A2PIC(Glu442-Lys452/Tyr445[SO4]) in solution. To ensure that the two synthetic peptides are capable of binding to the LBS in kringle domains, we investigated the binding of A2PIC(Glu442-Lys452) and A2PIC(Glu442-Lys452/Tyr445[SO4]) to isolated Pgn K4 in solution. The achieved K a values of 29 ± 4 mM−1 for the sulfated and 24 ± 4 mM−1 for the non-sulfated A2PIC(Glu442-Lys452) are both in a similar range. An interesting finding is that the K a value of A2PIC(Glu442-Lys452) is in the same range as the quintuple mutant rA2PIC(Lys406Ala/Lys415Ala/Lys422Ala/Lys429Ala/Lys436Ala), which also contains only the C-terminal Lys452.
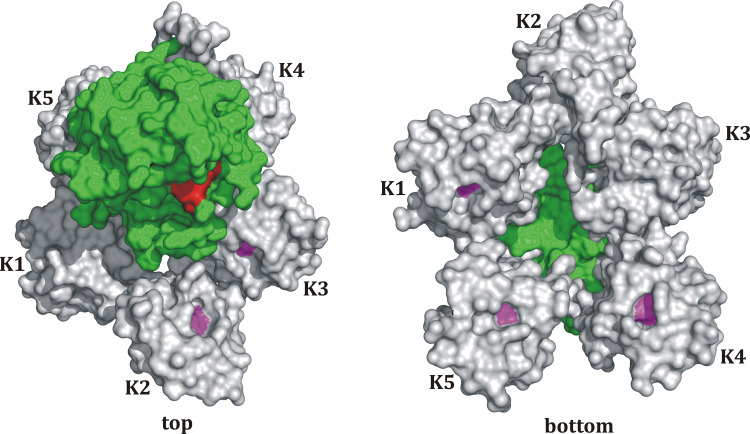
We further attempted to model the 3D structure of the whole Pgn. The model is based on the amino acid sequence of Pgn, which was used as a template for the automated structure modeling using SWISS-MODEL [57–59]. Connected and overlapping 3D structures of Pgn fragments (K1–K2; Leu81-Thr245, parts of K3–K4; Thr291-Ser441, parts of K4–K5; Leu402-Ala543, K1–K2–K3; Leu81-Cys333, and K5; Asp461-Pro544) were selected and thereafter assembled and attached to the 3D structure of the catalytic domain of Plm (1BML) using the visualization and rendering software PyMOL. The obtained model of the whole plasmin(ogen) molecule is shown in Fig. 4. The 3D structure model exhibits a spiral-type shape, which resembles the known shape of Glu1-Pgn visualized by electron microscopy [60, 61]. An interesting finding is that the LBS of K1, K4, and K5 are orientated opposite the active site of the protease domain whereas the LBS of K2 and K3 are orientated towards it. The opposite orientation of the LBS of K1 and K4 would allow Plm to bind to the target surface and leave the active site in the protease domain freely accessible. The model supports the findings that the K1- and K4-LBS act as independent and complementary domains both able to support Pgn binding and activation, and both seem to be required to ensure Pgn anchorage and subsequent proteolytic activity [62]. The remaining kringles seem to be involved in the modulation of Plm activity rather than anchorage of Pgn to the target surface since it has been shown that K5 interacts with long-chain fatty acids [63] as well as various cell surface proteins [64–66] and that K3 exhibits a heparin-binding site (Prof. Dr. M. Llinás, personal communication). The model is also consistent with our findings, that isolated rK1 and K4 exhibit a strong affinity for C-terminal Lys452, which supports the independence of both domains [62], and that the reinforced interaction between C-terminal Lys452 and internal Lys436 in the interaction with the isolated K1–3 domain (two- to threefold increase compared to rK1) underscores the high flexibility of multi-kringle domains (reviewed in [27]) allowing Lys436 to interact with the LBS of K2.
Fig. 4.
Model structure of the whole Pgn molecule. The model is based on the amino acid sequence of Pgn, which was used as a template for the automated structure modeling using SWISS-MODEL [57–59]. Connected and overlapping 3D structures of Pgn fragments (K1–K2; Leu81-Thr245, parts of K3–K4; Thr291-Ser441, parts of K4–K5; Leu402-Ala543, K1–K2–K3; Leu81-Cys333, and K5; Asp461-Pro544) were selected and thereafter assembled and attached to the 3D structure of the catalytic domain of Plm (1BML) using the visualization and rendering software PyMOL. Each kringle domain (K) is shown in gray, the central Trp of each LBS is presented in pink, the protease domain is colored in green, and the active site residues are marked in red
Conclusions
We could identify the C-terminal Lys452 as the main binding partner in the interaction of rA2PIC with isolated rK1, K4, and rK4–5 in solution. We could show a cooperative, zipper-like enhancement of the interaction between Lys452 and internal Lys436 of the rA2PIC and the isolated K1–3 triple-kringle domain in solution. The data allow the assumption that the C-terminal Lys452 is the primary binding partner whereas Lys436 functions as enhancer rather than as the primary binding partner. A co-crystallization of rA2PIC and Pgn or the K1–5 multi-kringle domain would definitively solve the interaction mechanism between the kringle domains and A2PIC. Our findings point towards a major interaction of the C-terminal Lys452 of rA2PIC with both LBS in K1 and K4, which act as independent and complementary domains both required to ensure Pgn anchorage and subsequent proteolytic activity, and that the remaining kringles (K2, K3, and K5) seem to be involved in the modulation of Plm activity rather than anchorage of Pgn to the target surface. The function of sulfated Tyr445 still remains to be solved.
Acknowledgments
We would like to thank PD Dr. A. Walz (Theodor Kocher Institute, University of Bern, Switzerland) for the generation of the synthetic peptides A2PIC(Glu442-Lys452) with and without sulfated Tyr445, Prof. Dr. U. Baumann (University of Bern, Switzerland) for providing the pET22b(+) vector and Prof. L.-O. Hedén (University of Lund, Sweden) for supplying the pPLGKG plasmid. We also thank Mr. U. Kämpfer for expert technical assistance, Mr. Christian Trachsel for his support building the plasminogen model, and Mr. S. Halbherr and Mrs. A. Mori for performing fluorescence measurements during their bachelor theses.
Abbreviations
- 6-AHA
6-Aminohexanoic acid
- A2PI
Human α2-plasmin inhibitor
- A2PIC
C-terminal moiety of A2PI (Asn398-Lys452)
- rA2PIC
Recombinant A2PIC (A2PIC/Pro399Ala)
- FXa
Activated coagulation factor X
- His-tag
Peptide MNHKVH6MELGTIEGR
- K1
Kringle 1 of Pgn (Cys84-Cys162), generated as rK1 (Lys78-Glu164)
- K2
Kringle 2 of Pgn (Cys166-Cys243), generated as rK2 (Cys162Thr/Glu163Ser/Glu164-Thr244/Cys169Gly)
- K3
Kringle 3 of Pgn (Cys256-Cys333), generated as rK3 (Thr253-Ser335/Cys297Ser)
- rK3mut
Mutated K3 domain of Pgn (rK3/Lys311Asp)
- K4
Kringle 4 of Pgn (Cys358-Cys435), generated as fragment Val355-Ala440 of Pgn
- K5
Kringle 5 of Pgn (Cys462-Cys541), generated as rK5 (Thr456-Ala543, Pro457Ala)
- K1–3
Kringles 1–3 of Pgn (Cys84-Cys333), generated as fragment Tyr80-Val338 of Pgn
- K4–5
Kringles 4–5 of Pgn (Cys358-Cys541), generated as rK4–5 (Val355-Phe546)
- LB Broth
Luria–Bertani Broth
- LBS
Lysine-binding site
- PAN
Plasminogen N-terminal domain
- Pgn
Human plasminogen
- Plm
Human plasmin
- RCL
Reactive center loop
- Serpin
Serine protease inhibitor
- t-AMCHA
trans-4-(Aminomethyl)cyclohexanecarboxylic acid
References
- 1.Lijnen HR, Wiman B, Collen D. Partial primary structure of human alpha 2-antiplasmin-homology with other plasma protease inhibitors. Thromb Haemost. 1982;48:311–314. [PubMed] [Google Scholar]
- 2.Collen D. Identification and some properties of a new fast-reacting plasmin inhibitor in human plasma. Eur J Biochem. 1976;69:209–216. doi: 10.1111/j.1432-1033.1976.tb10875.x. [DOI] [PubMed] [Google Scholar]
- 3.Moroi M, Aoki N. Isolation and characterization of α2-plasmin inhibitor from human plasma. J Biol Chem. 1976;251:5956–5965. [PubMed] [Google Scholar]
- 4.Müllertz S, Clemmensen I. The primary inhibitor of plasmin in human plasma. Biochem J. 1976;159:545–553. doi: 10.1042/bj1590545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiman B, Collen D. Purification and characterization of human antiplasmin, the fast-acting plasmin inhibitor in plasma. Eur J Biochem. 1977;78:19–26. doi: 10.1111/j.1432-1033.1977.tb11709.x. [DOI] [PubMed] [Google Scholar]
- 6.Sumi Y, Ichikawa Y, Nakamura Y, Miura O, Aoki N. Expression and characterization of Pro α2-plasmin inhibitor. J Biochem. 1989;106:703–707. doi: 10.1093/oxfordjournals.jbchem.a122920. [DOI] [PubMed] [Google Scholar]
- 7.Saito H, Goodnough LT, Knowles BB, Aden DP. Synthesis and secretion of α2-plasmin inhibitor by established human liver cell lines. Proc Natl Acad Sci. 1982;79:5684–5687. doi: 10.1073/pnas.79.18.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locher M (2004) Strukturelle und funktionelle Untersuchungen am α2-Plasmininhibitor. Inauguraldissertation, Doctoral Dissertation, University of Bern, Switzerland
- 9.Ries M, Easton RL, Longstaff C, Zenker M, Morris HR, Dell A, Gaffney PJ. Differences between neonates and adults in carbohydrate sequences and reaction kinetics of plasmin and α2-antiplasmin. Thromb Res. 2002;105:247–256. doi: 10.1016/S0049-3848(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 10.Lee KN, Jackson KW, Christiansen VJ, Chung KH, McKee PA. A novel plasma proteinase potentiates α2-antiplasmin inhibition of fibrin digestion. Blood. 2004;103:3783–3788. doi: 10.1182/blood-2003-12-4240. [DOI] [PubMed] [Google Scholar]
- 11.Koyama T, Koike Y, Toyota S, Miyagi F, Suzuki N, Aoki N. Different NH2-terminal form with 12 additional residues of α2-plasmin inhibitor from human plasma and culture media of Hep G2 cells. Biochem Biophys Res Commun. 1994;200:417–422. doi: 10.1006/bbrc.1994.1465. [DOI] [PubMed] [Google Scholar]
- 12.Tone M, Kikuno R, Kume-Iwaki A, Hashimoto-Gotoh T. Structure of human α2-plasmin inhibitor deduced from the cDNA sequence. J Biochem. 1987;102:1033–1041. doi: 10.1093/oxfordjournals.jbchem.a122141. [DOI] [PubMed] [Google Scholar]
- 13.Holmes WE, Nelles L, Lijnen HR, Collen D. Primary structure of human α2-antiplasmin, a serine protease inhibitor (serpin) J Biol Chem. 1987;262:1659–1664. [PubMed] [Google Scholar]
- 14.Christensen S, Valnickova Z, Thøgersen IB, Olsen EH, Enghild JJ. Assignment of a single disulphide bridge in human alpha2-antiplasmin: implications for the structural and functional properties. Biochem J. 1997;323:847–852. doi: 10.1042/bj3230847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hortin G, Fok KF, Toren PC, Strauss AW. Sulfation of a tyrosine residue in the plasmin-binding domain of α2-antiplasmin. J Biol Chem. 1987;262:3082–3085. [PubMed] [Google Scholar]
- 16.Kimura S, Aoki N. Cross-linking site in fibrinogen for α2-plasmin inhibitor. J Biol Chem. 1986;261:15591–15595. [PubMed] [Google Scholar]
- 17.Sakata Y, Aoki N. Significance of cross-linking of α2-plasmin inhibitor to fibrin in inhibition of fibrinolysis and in hemostasis. J Clin Invest. 1982;69:536–542. doi: 10.1172/JCI110479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank PS, Douglas JT, Locher M, Llinás M, Schaller J. Structural/functional characterization of the α2-plasmin inhibitor C-terminal peptide. Biochemistry. 2003;42:1078–1085. doi: 10.1021/bi026917n. [DOI] [PubMed] [Google Scholar]
- 19.Wiman B, Lijnen HR, Collen D. On the specific interaction between the lysine-binding sites in plasmin and complementary sites in alpha2-antiplasmin and in fibrinogen. Biochim Biophys Acta. 1979;579:142–154. doi: 10.1016/0005-2795(79)90094-1. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki T, Morita T, Iwanaga S. Identification of the plasminogen-binding site of human alpha 2-plasmin inhibitor. J Biochem. 1986;99:1699–1705. doi: 10.1093/oxfordjournals.jbchem.a135645. [DOI] [PubMed] [Google Scholar]
- 21.Hortin GL, Gibson BL, Fok KF. Alpha 2-antiplasmin’s carboxy-terminal lysine residue is a major site of interaction with plasmin. Biochem Biophys Res Commun. 1988;155:591–596. doi: 10.1016/S0006-291X(88)80535-7. [DOI] [PubMed] [Google Scholar]
- 22.Law RH, Sofian T, Kan WT, Horvath AJ, Hitchen CR, Langendorf CG, Buckle AM, Whisstock JC, Coughlin PB. X-ray crystal structure of the fibrinolysis inhibitor α2-antiplasmin. Blood. 2008;111:2049–2052. doi: 10.1182/blood-2007-09-114215. [DOI] [PubMed] [Google Scholar]
- 23.Raum D, Marcus D, Alper CA, Levey R, Taylor PD, Starzl TE. Synthesis of human plasminogen by the liver. Science. 1980;208:1036–1037. doi: 10.1126/science.6990488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tordai H, Bányai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. doi: 10.1016/S0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 25.Sottrup-Jensen L, Claeys H, Zajdel M, Petersen TE, Magnusson S (1978) In: Davidson JF, Rowan RM, Samama MM, Desnoyers PC (eds) Progress in chemical fibrinolysis and thrombolysis, vol 3. Raven Press, New York, pp 191-209
- 26.Wang X, Lin X, Loy JA, Tang J, Zhang XC. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;281:1662–1665. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]
- 27.Ponting CP, Marshall JM, Cederholm-Williams SA. Plasminogen: a structural review. Blood Coagul Fibrinolysis. 1992;3:605–614. doi: 10.1097/00001721-199210000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Christensen U, Clemmensen I. Kinetic properties of the primary inhibitor of plasmin from human plasma. Biochem J. 1977;163:389–391. doi: 10.1042/bj1630389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiman B, Collen D. On the kinetics of the reaction between human antiplasmin and plasmin. Eur J Biochem. 1978;84:573–578. doi: 10.1111/j.1432-1033.1978.tb12200.x. [DOI] [PubMed] [Google Scholar]
- 30.Shieh B-H, Travis J. The reactive site of human α2-antiplasmin. J Biol Chem. 1987;262:6055–6059. [PubMed] [Google Scholar]
- 31.Wiman B, Boman L, Collen D. On the kinetics of the reaction between human antiplasmin and a low-molecular-weight form of plasmin. Eur J Biochem. 1978;87:143–146. doi: 10.1111/j.1432-1033.1978.tb12360.x. [DOI] [PubMed] [Google Scholar]
- 32.Wiman B, Collen D. On the mechanism of the reaction between human alpha 2-antiplasmin and plasmin. J Biol Chem. 1979;254:9291–9297. [PubMed] [Google Scholar]
- 33.Kluft C, Los P, Jie AF, van Hinsbergh VW, Vellenga E, Jespersen J, Henny CP. The mutual relationship between the two molecular forms of the major fibrinolysis inhibitor alpha-2-antiplasmin in blood. Blood. 1986;67:616–622. [PubMed] [Google Scholar]
- 34.Clemmensen I, Thorsen S, Müllertz S, Petersen LC. Properties of three different molecular forms of the α2plasmin inhibitor. Eur J Biochem. 1981;120:105–112. doi: 10.1111/j.1432-1033.1981.tb05675.x. [DOI] [PubMed] [Google Scholar]
- 35.Hortin GL, Trimpe BL, Fok KF. Plasmin’s peptide-binding specificity: characterization of ligand sites in α2-antiplasmin. Thromb Res. 1989;54:621–632. doi: 10.1016/0049-3848(89)90128-X. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Yu A, Wiman B, Pap S. Identification of amino acids in antiplasmin involved in its noncovalent ‘lysine-binding-site’-dependent interaction with plasmin. Eur J Biochem. 2003;270:2023–2029. doi: 10.1046/j.1432-1033.2003.03578.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Karlsson A, Sjöström I, Wiman B. The interaction between plasminogen and antiplasmin variants as studied by surface plasmon resonance. Biochim Biophys Acta. 2006;1764:1730–1734. doi: 10.1016/j.bbapap.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Brunisholz R, Lerch P, Rickli EE. Structural comparison between human, porcine and bovine plasminogen. In: Neri Serneri GG, Prentice CRM, editors. Haemostasis and thrombosis. London: Academic press; 1979. pp. 757–761. [Google Scholar]
- 39.Novy R, Drott D, Yaeger K, Mierendorf R. Overcoming the codon bias of E. coli for enhanced protein expression. inNovations. 2001;12:1–3. [Google Scholar]
- 40.Qing G, Ma L-C, Khorchid A, Swapna GVT, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T, Montelione GT, Ikura M, Inouye M. Cold-shock induced high-yield protein production in Escherichia coli . Nat Biotechnol. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 41.Bromfield KM, Quinsey NS, Duggan PJ, Pike RN. Approaches to selective peptidic inhibitors of factor Xa. Chem Biol Drug Des. 2006;68:11–19. doi: 10.1111/j.1747-0285.2006.00404.x. [DOI] [PubMed] [Google Scholar]
- 42.Ludeman JP, Pike RN, Bromfield KM, Duggan PJ, Cianci J, Le Bonniec B, Whisstock JC, Bottomley SP. Determination of the P1′, P2′ and P3′ subsite-specificity of factor Xa. Int J Biochem Cell Biol. 2003;35:221–225. doi: 10.1016/S1357-2725(02)00128-0. [DOI] [PubMed] [Google Scholar]
- 43.Sletta H, Tøndervik A, Hakvåg S, Vee Aune TE, Nedal A, Aune R, Evensen G, Valla S, Ellingsen TE, Brautaset T. The presence of N-terminal secretion signal sequences leads to strong stimulation of the total expression levels of three tested medically important proteins during high-cell-density cultivations of Escherichia coli . Appl Environ Microbiol. 2007;73:906–912. doi: 10.1128/AEM.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marti DN, Hu C-K, An SSA, von Haller P, Schaller J, Llinás M. Ligand preferences of kringle 2 and homologous domains of human plasminogen: canvassing weak, intermediate, and high-affinity binding sites by 1H-NMR. Biochemistry. 1997;36:11591–11604. doi: 10.1021/bi971316v. [DOI] [PubMed] [Google Scholar]
- 45.Marti D, Schaller J, Ochensberger B, Rickli E. Expression, purification and characterization of the recombinant kringle 2 and kringle 3 domains of human plasminogen and analysis of their binding affinity for ω-aminocarboxylic acids. Eur J Biochem. 1994;219:455–462. doi: 10.1111/j.1432-1033.1994.tb19959.x. [DOI] [PubMed] [Google Scholar]
- 46.Douglas JT, von Haller PD, Gehrmann M, Llinás M, Schaller J. The two-domain NK1 fragment of plasminogen: folding, ligand binding, and thermal stability profile. Biochemistry. 2002;41:3302–3310. doi: 10.1021/bi016018j. [DOI] [PubMed] [Google Scholar]
- 47.Chang J-Y, Knecht R. Direct analysis of the disulfide content of proteins: methods for monitoring the stability and refolding process of cystine-containing proteins. Anal Biochem. 1991;197:52–58. doi: 10.1016/0003-2697(91)90354-V. [DOI] [PubMed] [Google Scholar]
- 48.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984;336:93–104. doi: 10.1016/S0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 49.Menhart N, Sehl LC, Kelley RF, Castellino FJ. Construction, expression, and purification of recombinant kringle 1 of human plasminogen and analysis of its interaction with ω-amino acids. Biochemistry. 1991;30:1948–1957. doi: 10.1021/bi00221a031. [DOI] [PubMed] [Google Scholar]
- 50.Scatchard G. The attractions of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660–672. doi: 10.1111/j.1749-6632.1949.tb27297.x. [DOI] [Google Scholar]
- 51.Lejon S (2008) Human α2-plasmin inhibitor, a serpin with unique structural and functional properties. Doctoral Dissertation, University of Bern, Switzerland
- 52.Rejante MR (1992) Proton NMR studies on the structure and ligand-binding properties of human plasminogen kringles 1 and 4. Doctoral Dissertation, Carnegie Mellon University, Pittsburgh, PA
- 53.Petros AM, Ramesh V, Llinás M. 1H NMR studies of aliphatic ligand binding to human plasminogen kringle 4. Biochemistry. 1989;28:1368–1376. doi: 10.1021/bi00429a064. [DOI] [PubMed] [Google Scholar]
- 54.Thewes T, Constantine K, Byeon I-JL, Llinás M. Ligand interactions with the kringle 5 domain of plasminogen. JBC. 1990;265:3906–3915. [PubMed] [Google Scholar]
- 55.Abad MC, Arni RK, Grella DK, Castellino FJ, Tulinsky A, Geiger JH. The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin. J Mol Biol. 2002;318:1009–1017. doi: 10.1016/S0022-2836(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 56.Sofian T, Horvath A, Hitchen C, Forsyth S, Coughlin P (2008) Antibody to the C-terminal extension of antiplasmin enhances fibrinolysis. In: Proceedings of the 5th international symposium on serpin biology, structure and function, serpins2008, Leuven, Belgium
- 57.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 58.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 60.Tranqui L, Prandini M-H, Chapel A. The structure of plasminogen studied by electron Microscopy. Biol Cell. 1979;34:39–42. [Google Scholar]
- 61.Weisel JW, Nagaswami C, Korsholm B, Petersen LC, Suenson E. Interactions of plasminogen with polymerizing fibrin and its derivatives, monitored with a photoaffinity cross-linker and electron microscopy. J Mol Biol. 1994;235:1117–1135. doi: 10.1006/jmbi.1994.1061. [DOI] [PubMed] [Google Scholar]
- 62.Ho-Tin-Noé B, Rojas G, Vranckx R, Lijnen HR, Anglés-Cano E. Functional hierarchy of plasminogen kringles 1 and 4 in fibrinolysis and plasmin-induced cell detachment and apoptosis. FEBS J. 2005;272:3387–3400. doi: 10.1111/j.1742-4658.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 63.Huet E, Cauchard JH, Berton A, Robinet A, Decarme M, Hornebeck W, Bellon G. Inhibition of plasmin-mediated prostromelysin-1 activation by interaction of long chain unsaturated fatty acids with kringle 5. Biochem Pharmacol. 2004;67:643–654. doi: 10.1016/j.bcp.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 64.Cao Y, Chen A, An SS, Ji RW, Davidson D, Llinás M. Kringle 5 of plasminogen is a novel inhibitor of endothelial cell growth. J Biol Chem. 1997;272:22924–22928. doi: 10.1074/jbc.272.36.22924. [DOI] [PubMed] [Google Scholar]
- 65.Gonzalez-Gronow M, Kalfa T, Johnson CE, Gawdi G, Pizzo SV. The voltage-dependent anion channel is a receptor for plasminogen kringle 5 on human endothelial cells. J Biol Chem. 2003;278:27312–27318. doi: 10.1074/jbc.M303172200. [DOI] [PubMed] [Google Scholar]
- 66.Davidson DJ, Haskell C, Majest S, Kherzai A, Egan DA, Walter KA, Schneider A, Gubbins EF, Solomon L, Chen Z, Lesniewski R, Henkin J. Kringle 5 of human plasminogen induces apoptosis of endothelial and tumor cells through surface-expressed glucose-regulated protein 78. Cancer Res. 2005;65:4663–4672. doi: 10.1158/0008-5472.CAN-04-3426. [DOI] [PubMed] [Google Scholar]