Abstract
Expression of the prion protein is necessary for infection with prion diseases. Altered expression levels may play an important role in susceptibility to infection. Therefore, understanding the mechanisms that regulate prion protein expression is of great importance. It was previously shown that expression of the prion protein is to some degree regulated by an alternative promoter within intron 1. Studies using GFP and luciferase reporter systems were undertaken to determine key sites for the repression and activation of expression of the prion protein driven by intron 1. We identified a region within intron 1 sufficient to drive prion protein expression. Our findings highlight two potential repressor regions. Both regions have binding sites for the known repressor Hes-1. Hes-1 overexpression caused a dramatic decrease in PrP protein expression. Additionally, we have identified Atox-1 as a transcription factor that upregulates prion protein expression. These findings clearly indicate that intron 1 plays a key role in regulation of prion protein expression levels.
Keywords: Prion, CJD, BSE, Scrapie, Atox-1, Hes-1, Transcription factor
Introduction
Prion diseases or transmissible spongiform encephalopathies are unique among neurodegenerative disease in being transmissible between individuals and even species [1]. Prion diseases include bovine spongiform encephalopathy, scrapie and Creutzfeldt-Jakob disease. The agent that is associated with disease transmission is a misfolded form of a normal cellular metalloprotein known as the prion protein (PrP). Expression of PrP is necessary for transmission, as PrP-knockout mice are resistant to infection [2] and conditional knockout during disease progress results in cessation of observed behavioural changes and neuronal loss [3]. In contrast, increased expression in animal models is associated with increased susceptibility and shortened incubation time [4]. As with most neurodegenerative diseases, prion diseases remain untreatable and diagnosis in humans remains difficult prior to the onset of irreversible symptoms.
The requirement for neuronal expression of PrP for both infection and disease progression implies that understanding regulation of PrP expression is key to determining factor that may influence onset and susceptibility. As both knockout and decreased expression of PrP effectively reduce or abolish susceptibility to infection, establishing the inherent mechanisms by which the expression of PrP is regulated might be beneficial to the development of a treatment for prion diseases.
The Prnp gene encodes PrP. Prnp has a three exon structure in all characterised species. However, in hamsters and humans, the second exon may not be spliced into the final mRNA sequence [5, 6]. The third and final exon encodes the entire open reading frame (ORF) of the protein. There is also evidence for a splice variant in both cattle and mice that includes exon 2 and 3 but not exon 1 [7]. Various motifs involved in promoter control have been identified within the promoter region, and intron 1 has been identified as essential for full promoter activity [7, 8]. Intron 1 possibly plays a greater role in the expression of the protein than just a regulator element. Intron 1 in isolation of the promoter has been shown to be sufficient to drive expression [7]. In the absence of intron 1, exon 1 inhibits promoter activity in most cell types studied suggesting that Prnp has a unique regulatory structure in which sequences in intron 1 are the dominant elements in controlling expression.
Currently, there is limited information on the key transcription factors that regulate PrP expression. There have been suggestions that Prnp expression is enhanced by metal-activated transcription factor-1 (MTF-1) [9] and repressed by Ying-Yang-1 (YY1) [10]. All other information is indirect, implied by increased PrP expression in cells in response to exogenous factors. The presence of heat shock elements allows the Prnp promoter to become activated in response to forms of cellular stress [11, 12], indicating that total levels of PrP within the cell would be up-regulated as the cell attempted to combat the stress insult. Up-regulation of the Prnp promoter activity has been observed as a result of stress induced by nitric oxide (NO) radicals [13] and oxidative stress induced by hyperbaric oxygen [14], while hypoglycaemia [15] also provides evidence of a role for PrPc in dealing with stress insults. Copper-induced oxidative stress also up-regulates PrP indirectly via binding of p53 following phosphorylation by ataxia-telangiectasia mutated (ATM) [16]. In addition to this up-regulation, down-regulation of the Prnp promoter has been observed in response to ATRA [17].
In the current study, we have concentrated on determining the key regulatory domains of intron 1. In particular, we have identified two significant inhibitory domains. Analysis of the binding sites in these domains suggests that the main repressor of PrP expression is Hes-1. In addition, we have identified Atox-1 as an important regulator of PrP expression. MTF-1 and NRF-2 play no role in the regulation of PrP through intron 1.
Materials and methods
Reagents were purchased from Sigma–Aldrich (Poole, UK) unless otherwise stated.
Cell culture
All cell lines were maintained at 37°C and 5% CO2 in a humidified incubator. Cell lines used for these studies were predominantly mouse N2A cells (Neuro-2A, ATCC Number: CCL-131). For some experiments, the mouse cerebellar neuron-neuroblastoma fusion line F21 were used [18]. Cell lines were cultured in DMEM or EMEM with 10% fetal bovine serum (Lonza) and 1% penicillin–streptomycin.
Plasmid constructs
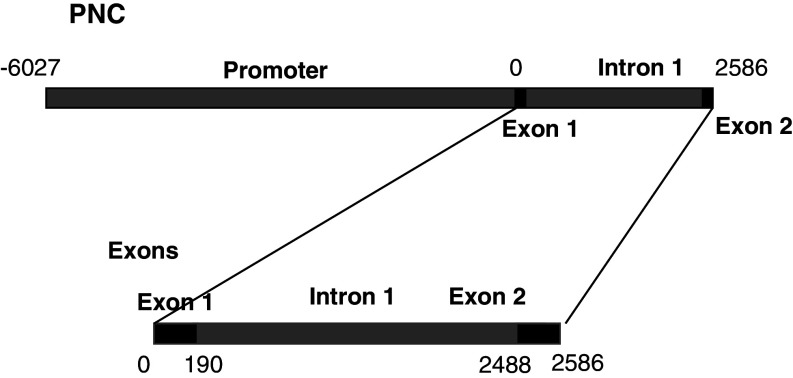
PrP promoter sequences were as described previously [7]. Previously, cloned constructs using the GFP-reporter system were the PNC construct which contains the whole promoter and Exon 1, Intron 1 and Exon 2, and the Exons construct which contains Exon 1, Intron and Exon 2 only (Fig. 1). Both fragments were clones into pd2-EGFP-1 (Invitrogen). Modifications of these constructs were made to create three new plasmids. PNC-xT and Exons-xT were modified from the parent construct by digestion with BspE1 which excises 1,220 pb of DNA (865–2,085) from either construct. The plasmids were then religated. A third construct was generated by subcloning the excised fragment (865–2,085), after treatment with the Klenow polymerase fragment, into the Sma 1 site of pd2-EGFP-1. This construct was named Intron-TATA.
Fig. 1.
A schematic diagram showing the structure of the two main Prnp gene constructs used in this study. The PNC (promoter and non-coding regions) covered the whole of the main promoter, exon 1, intron 1 and exon 2, while the Exons construct covered only Exon 1, intron 1 and exon 2. The numbers indicate distance in base pairs from the transcription initiation site (0)
Constructs using the luciferase reporter system were prepared by cloning previously generated fragments and mutants into pGL3-Basic (Invitrogen). The inserts from Exons, exon-xT (∆865–2085) and Intron-TATA (865–2085) were digested from the pd2-EGFP-1 and cloned into pGL3-Basic. Further constructs were prepared by modification of the pGL3-Basic-Exons construct as follows.
Digestion with HindIII removed a fragment. This allowed generation of both constructs 1–2,036 and 2,036–2,586.
Cleaving with Nde1 cut at 1,072 bp. In combination with Xho1 this allowed generation of 1,072–2,586. Digestion with Nde1 and Spe1 allowed release of 1,072–1,650. Religation of this created ∆1072–1659. Digestion with Spe1 and Xho1 generated 1,650–2,586.
Mutagenesis was carried out to insert an Spe1 site at 1,327. Digestion of this mutant with Spe1 released a fragment and allowed generation of ∆1,327–1,650.
PCR-based mutagenesis was used to insert a Nhe1 site at position 1,525 using split oligonucleotides to mutate the site (forward 5′-AGTAGAGCTAGCGTATCTGATGCAAATTTT). Digestion with Xho1 and Nhe1 with subsequent ligation produced the 1,525–2,586 construct.
The insertion of an Nhe1 site at 1,950 using splint oligonucleotides and PCR mutagenesis (forward 5′-AGTAGAGCTAGCAGTCAGGTGATCCT) and subsequent digestion of the product with Xho1 and Nhe1 generated the 1,950–2,586 construct.
A further mutant with an Nhe1 site at 2312 was also created similarly (forward 5′-AATATAGCTAGCGAGTTCAGTGTCACG). Digestion with Xho1 and Nhe1 resulted in the 2,312–2,586 construct.
Both the 2,312–2,586 and 2,036–2,586 constructs were mutated to include a HindIII site at bp 2,488. Subsequent digestion with HindIII results in the excision of DNA encoding Exon 2 and allowed generation of the 2,036–2,488 and the 2,312–2,488 constructs.
All constructs were checked by DNA sequencing.
The full open reading frame of three transcription factors were amplified from cDNA generated from mRNA extract from SH-SY5Y or N2A cells. Specific primer pairs were used to amplify the products which were then cloned into pCDNA3.1 The primer pairs used were 5′-ATTTATGCTAGCATGATGGACTTGGAGCTG and 5′-TCACGCGGATCCCTAGTTTTTCTTAACATC for NRF2, 5′-GCTCATCTAAGCTTATGGGGGAACACAGTCCAG and 5′-TCTAGACTCGAGTCATCACTTGGAGAAGCTGCT for MTF1, 5′-CCAAGCTTATGCCGAAGCACGAGTTCTCC and 5′-CCGGATCCCTACTTGGGGCCAAGGTAGGA for Atox-1 and 5′-TAGGGATCCATGCCAGCTGATATAATGGAG and 5′-TAGCTCGAGTCAGTTCCGCCACGGCC for Hes-1. The products were confirmed by DNA sequencing.
Transfection
Cells were plated allowing 2.5 × 104 cells/well of a 24-well tray. These were then incubated at 37°C for 2 days until the cells had reached around 75% confluence. Each well was transfected with 300 ng of one of the pGL3-Basic experimental reporters and 10 ng of pRLSV40 control vector. The activity of each experimental reporter was analysed in triplicate. A master mix for each transfection was produced as follows. DNA concentration was determined using a spectrophotometer (Carey) and the volume of DNA required to provide 1.05 μg of DNA was calculated. The concentration of pRLSV40 was also determined and the volume required to give 35 ng/μL was calculated. These volumes of DNA and pRLSV40 were mixed in a thin-walled PCR tube and 175 μL serum free medium was added to the master mix under sterile conditions in the safety cabinet. Fugene transfection reagent was then added at a ratio of 3 μL Fugene to 1 μg DNA. The tubes were inverted to mix the reagents and then incubated at room temperature for 15 min. Then, 50 μL was added to each well. Cells were then incubated for 18–37 h at 37°C in an atmosphere of 5% CO2 prior to measuring luciferase activity. Single transfections with pEGFP-based reporter constructs were similarly performed.
Stable lines were generated for the cell lines transfected with pCDNA3 containing the ORF for NRF2, MTF-1, Hes-1 and Atox-1 or just the empty vector by growing the cells continuously in the presence of 0.4 μg/mL of the antibiotic G418.
Confocal microscopy
Cells were cultured in chambered coverslips (Nunclon–Fisher, Loughborough, UK), at approximately 50% confluency. Images were captured using a Zeiss LSM 510 confocal microscope. A minimum of ten cells were studied per experiment and experiments were repeated a minimum of four times.
Luciferase assays
Media was removed and the 24-well plates were tapped onto paper towel to remove excess liquid. Then, 1 mL passive lysis buffer (PLB) (Promega) was added to 4 mL dH2O and 100 μL was added per well. The plates were then stored at −80°C. The 24-well plates were defrosted and 50 μL from each well was transferred into wells on a white 96-well plate. The assay was then performed as per manufacturers instructions (Promega Dual-luciferase reporter assay) using a FLUOstar Omega (BMG Labtech) plate reader.
Western blot
Protein extracts from cells were prepared in a buffer containing phosphate buffered saline (PBS) pH 7.3, 1.5% Triton X-100 and 1% Igepal. Extracts were cleared of debris by low speed centrifugation and the protein concentration quantified using a BCA (bicinchoninic acid) assay according to the manufacturer’s instructions. Proteins were separated using polyacrylamide gel electrophoresis. The gel was run at 35 mA, 100 W for 50 min. Proteins were then transferred onto an immobilon membrane (Millipore) using a semi-dry transfer unit (Biorad). Membranes were blocked in 5% non-fat milk powder in PBS-T (0.1% Tween 20 in PBS). The membrane was incubated with primary antibody anti-PrP-antibody 8B4 [41] of the appropriate concentration for 1 h. The membrane was then washed three times for 10 min each on a rocker with PBS, incubated with an appropriate horse radish peroxidise conjugated secondary antibody (Dako) for 1 h and then sequentially washed 2 × 2 min, 1 × 15 min and finally 3 × 5 min with PBS-T. The membrane was then incubated with ECL Plus reagent (Amersham Biosciences) for 5 min, with signal detected on X-ray film (FujiFilm). The membrane was stripped by washing in methanol for 10 s, followed by stripping buffer (25 mM Glycine, 1% w/v SDS, pH 2.0) for 1 h and then TBS-T for 5 min. The membranes were then reprobed with other antibodies (rabbit anti-tubulin; Sigma). Signal intensities were quantified using ImageJ or Scion Image software.
Native gel analysis of GFP expression
Cells were extracted by first washing in Hanks’ Balanced Salt Solution, then incubated in extraction buffer (20 mM Tris acetate, 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 10 mM sodium-B-glycerophosphate, and 1% Triton X-100, 1 mM sodium orthovanadate, 2 mM phenylmethylsulphonylfluoride (PMSF), and 1 mM benzamidine) for 20 min at 37°C. Cell lysates were collected and briefly spun to separate the cell debris from the soluble protein. Protein concentrations were determined using a BCA assay. Gels were prepared and run using an ATTO (US) gel tank system. Then 12% native gels were mixed using 3.39 mL H2O, 4 mL 30% acrylamide, 2.5 mL 1.5 M Tris (pH 8.8), 100 μL 10% (w/v) ammon,ium persulphate, and 10 μL TEMED per gel, added in the order the reagents are listed. Next, 50 μg of protein was loaded in each well and gels were run at 30 mA per gel for approximately 1 h. Gels were removed from the chamber and analysed using a Fujifilm FLA-5000 phosphoimager using excitation and emission wavelengths of 488 and 530, respectively (optimum wavelengths for GFP). Bands were quantitated using the phosphoimager’s inbuilt analysis software.
Bioinformatics
Conserved transcription factor binding sites were determined using ConSite [19] which is freely available online (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite). The intron 1 DNA sequence of three species (Homo sapiens, Mus musculus and Rattus norvegicus) was searched pair-wise against intron 1 of Bos taurus with a 15-base region at both 5′ and 3′ ends removed from H. sapiens, M. musculus and R. norvegicus to prevent the detection of conserved splice sites at intron–exon boundaries. TESS (Transcription Element Search System, http://www.cbil.upenn.edu/cgi-bin/tess/tess) was used to determine transcription factor binding sites within intron 1 of the B. taurus Prnp gene.
Statistics
Statistical analysis was performed for non-parametric data using Microsoft Excel. The tests used were either the Mann–Whitney test or ANOVA. In all cases, p values less that 0.05 were considered to indicate a significant difference.
Results
TATA box in Prnp intron 1
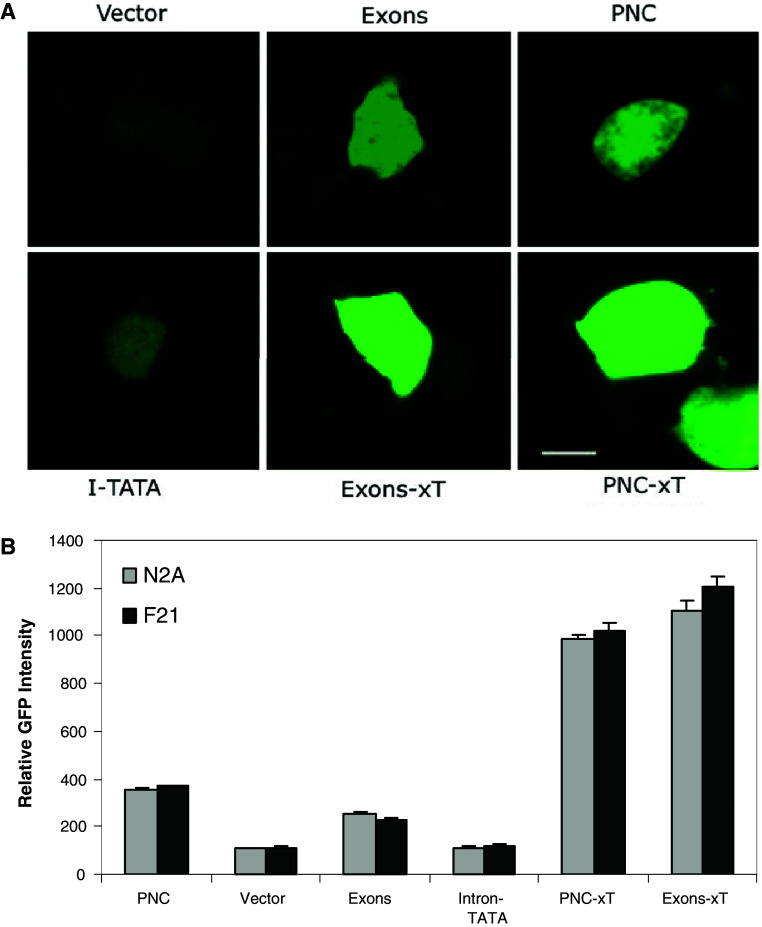
Our previous studies highlighted the importance of regulatory regions within intron 1 of the Prnp gene to PrP expression [7]. In that work, we suggested that the promoter-like activity of intron 1 could be related to a TATA sequence present in the intron. In order to assess the importance of this region to expression of PrP, we used reporter-based systems to compare promoter activity with and without this region of the intron, We used a GFP-based system in which either the whole promoter region including the non-coding regions (PNC: composed of the promoter, exon 1, intron 1 and exon 2) was cloned or the non-coding regions alone (Exons). Mutants were made of both constructs in which 1,220 bp were deleted from intron 1 between 864 and 2,085 bp from the start of exon 1. This produced the constructs PNC-xT and Exons-xT. Lastly, the region deleted (865–2,085) from these constructs was cloned into pd2EGFP-1 to determine any activity of the deleted region (Intron-TATA). Each construct and the empty vector (pd2EGFP-1) were transfected into two neuronal cell lines (N2A, F21) and the level of GFP fluorescence of individual cells determined quantitatively with confocal microscopy (Fig. 2).
Fig. 2.
Deletion of a region in Intron 1 increased GFP-reporter expression in single N2A or F21 cells. a Confocal images were captured for single cells that had been transfected with GFP-reporter plasmids carrying either the PNC, or Exons inserts or mutants of these. Cells were also transfected with plasmids without insert or with the Intron-TATA fragment (I-TATA). Images of N2A cells with these reporter plasmids are shown. b The level of GFP fluorescence was determined digitally and compared to the untransfected cells to determine a value of relative GFP intensity. The graph shows the data for both N2A and F21 cells. Shown are the mean and SE of 40–50 cells per construct: both the PNC-xT and Exons-xT constructs were significantly different to both the parental PNC and Exons constructs (p < 0.05, ANOVA); the Intron-TATA construct was not significantly different to the vector; there was no significant difference between N2A and F21 cells for any of the constructs (p > 0.05)
The GFP expression driven by both the PNC and Exons constructs was high when compared to the empty vector and in keeping with previous results [7]. In both cases, deletion of the 865–2,085 region of intron 1 resulted in a significant and high increase in GFP expression (Fig. 2). In contrast, the Intron-TATA construct showed no significant activity when compared to the empty vector. This implies that the region 865–2,085 does not contain an active TATA box but is more likely to contain repressor regions that when inactive result in increased PrP expression.
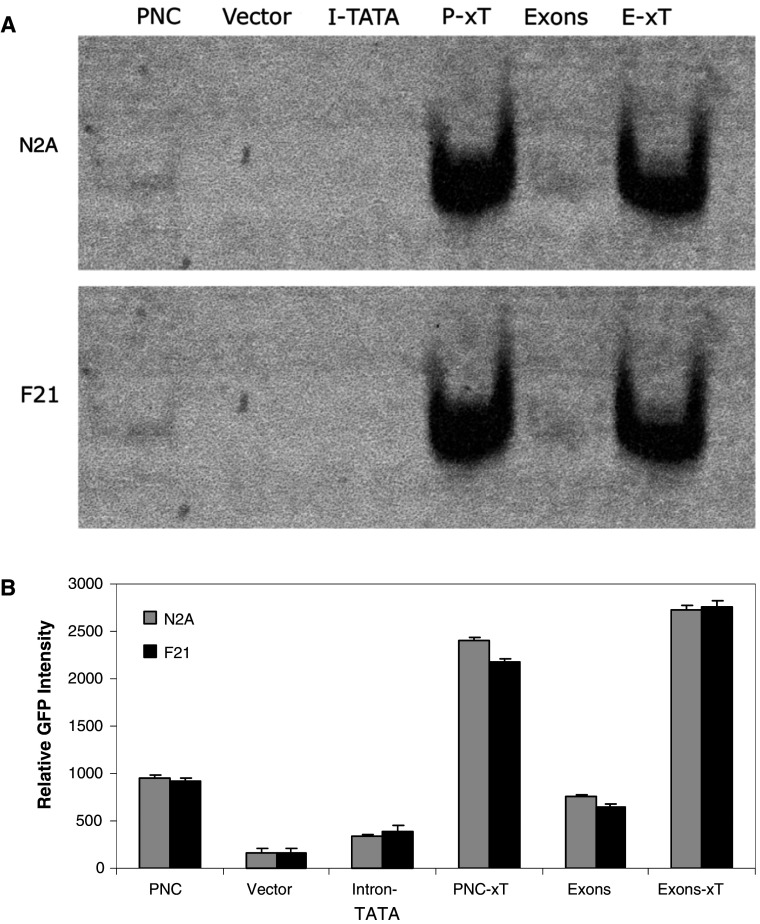
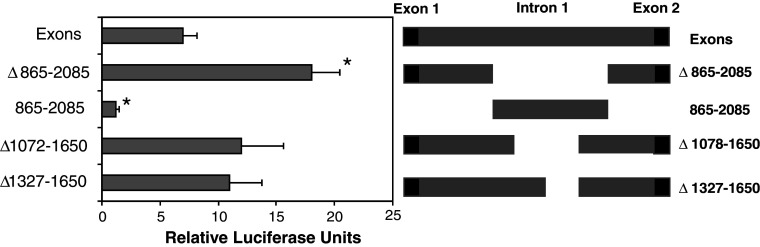
This result was confirmed in two ways. First, GFP expression in cell populations was analysed by extraction of protein from transfected cell lines, native gel electrophoresis and phosophoimager based quantitation of GFP bands. The results confirm that the region 865–2,085 has no promoter-like activity while deletion of this region either from the Exons construct or the PNC construct resulted in a significant increase in GFP expression (Fig. 3). Second, we used an alternative reporter system using luciferase activity as the indicator of promoter activity in transfected N2A cells. In this case, the Exon 1, Intron 1 and Exon 2 (1–2,586 bp) were cloned into the luciferase reporter construct pGL3-Basic (firefly). Three mutants were made from this construct with deletions of either 865–2,085, 1,072–1,650 or 1,327–1,650 bp. These deletions were made to more specifically target the TATA region (at 1,610) and any potential basal transcription machinery recognition sites. The region 865–2,085 was also cloned into the luciferase reporter as before. The level of firefly luciferase activity for each construct was compared after normalising to the renilla luciferase expression from the co-transfected control plasmid. In each case, the deletions resulted in higher activity of the reporter than the original Exons construct (Fig. 4). The ∆865–2,085 mutant showed the highest activity suggesting that this mutant had significantly more repressor elements deleted than ∆1,327–1,650 or ∆1,072–1,650. The 865–2,085 fragment again showed no activity suggesting that the region has no site for the initiation of transcription. The results suggest that the region 1,327–1,650 contains a major repressor binding site. There is possibly a second repressor site either in the region 1,650–2,085 or 865–1,072.
Fig. 3.
GFP-reporter expression in cell population confirmed role of Intron 1. a Cell lines stable transfected with the GFP-reporter plasmids were harvested and protein extracts prepared. The extracts were electrophoresed on a native acrylamide gel and GFP fluorescence detected in the gels with a phosphoimager. Examples of the gels for both N2A and F21 cells are shown. b The densitometric software of the phosphoimager was used to determine the intensity of the bands which were compared to the intensity of bands for cells that were not transfected. Shown are the mean and SE for six experiments: both the PNC-xT and Exons-xT constructs were significantly different (p < 0.05) ANOVA to both the parental PNC and Exons constructs; the Intron-TATA construct was not significantly different to the vector (p > 0.05); there was no significant difference between N2A and F21 cells for any of the constructs
Fig. 4.
Luciferase reporter assays indicated a repressor region is present in Intron 1. N2A cells were transfected with pGL3-Basic carrying the Exons fragment of Prnp or a series of mutants. The diagram on the right shows the Exons construct and the different mutants used. Luciferase reporter assays were performed and values were compared to those of cells transfected with the vector only. The data is presented as relative luciferase units. Shown are the mean and SE for at least three experiments each: significant difference was seen between Exons and ∆865–2,085 (p < 0.05, ANOVA); the value for the smaller fragment, 865–2,085, was significantly less than those of all others (p < 0.05)
Defining key regulatory regions in Intron 1
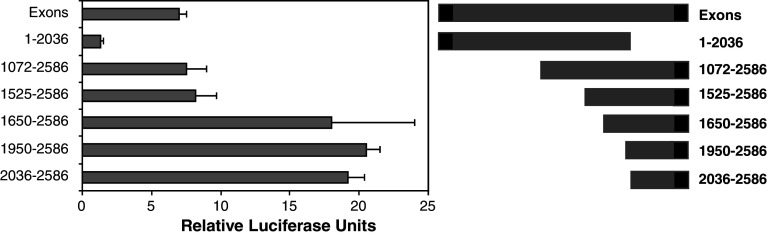
We analysed further mutants of the Exons construct to determine key regulatory regions of intron 1. The activity of these constructs in terms of promoting expression was assessed using the luciferase system as described above. A mutant deletion of the 3′ end intron 1 and all of exon 2 of the Exons construct (1–2,036) showed no activity in the luciferase activity when compared to controls (Fig. 5). This indicated that the transcription machinery binding site lies in the 3′ end of intron 1 or Exon 2. Further mutants were made with stepwise deletion of 5′ regions of intron 1 to determine how the deletions alter activity of the promoter in intron 1. Two mutants, 1,072–2,586 and 1,525–2,586, showed no significant difference from the parent Exons construct in terms of luciferase activity detected in transfected N2A cells. This implies that Exon 1 does not play an important role in the activity of the construct and also that the region up to 1,525 does not include important regulatory sites including repressor regulatory sites (Fig. 5). Three further truncated mutants were also analysed and included 1,650–2,586, 1,950–2,586 and 2,036–2,586. The activity of the three constructs resulted in significantly higher reporter activity than the Exons construct or the mutant 1,525–2,586 (Fig. 5). However, there was no significant difference between the three mutants. The results imply that deletion of the region 1,525–1,650 resulted in the loss of a repressor binding site. This is supported by further deletions that did not increase the reporter activity observed. It also implies that there is no significant repressor elements between 1 and 1,525 in the Exons construct.
Fig. 5.
Mutation analysis of Intron 1 promoter-like activity. N2A cells were transfected with pGL3-Basic carrying the Exons fragment of Prnp or a series of mutants. The diagram on the right shows the Exons construct and the different mutants used. Luciferase reporter assays were performed and values were compared to those of cells transfected with the vector only. The data is presented as relative luciferase unit. Shown are the mean and SE for at least three experiments each: 1–2,036 was significantly less than all other values (p < 0.05); 1,650–2,586, 1,950–2,586 and 2,036–2,586 were all significantly higher than other the other values but were not significantly different from each other (p > 0.05); and 1,072–2,586 and 1,525–2,586 were not significantly different to the Exons value (p > 0.05)
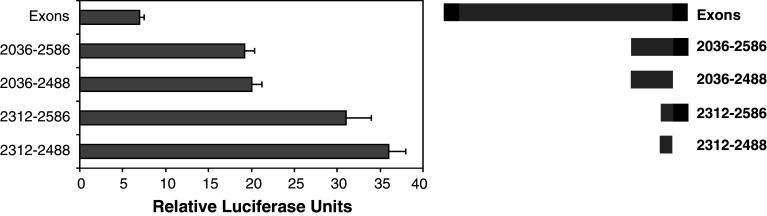
An additional mutant lacking Exon 2 (2,036–2,488) was also assessed in the luciferase reporter system. The activity of this construct in promoting luciferase expression was not significantly different to the 2,036–2,586 construct containing exon 2. This verifies that exon 2 does not play a significant role in the promoter activity measure for intron 1 and does not appear to contain repressor elements (Fig. 6). Two further mutants of the Exons construct were studied. These were 2,312–2,586 and 2,312–2,488. The mutants showed significantly higher promoter activity than any of the other mutants tested in this study (Fig. 6). The activity of both constructs was equivalent and around five times that of the parent Exons construct. This information suggests that the region 2,312–2,488 contains the transcription machinery binding site. It also suggests that there is a potent repressor element between 2,036 and 2,312 in intron 1.
Fig. 6.
Repressor elements defined for Intron 1 of Prnp. N2A cells were transfected with pGL3-Basic carrying the Exons fragment of Prnp or a series of mutants. The diagram on the right shows the Exons construct and the different mutants used. Luciferase reporter assays were performed and values were compared to those of cells transfected with the vector only. The data is presented as relative luciferase units. Shown are the mean and SE for at least three experiments each. Values for the Exons construct were significantly less than all others (p < 0.05); 2,312–2,586 and 2,312–2,488 were significantly higher than all others but were not significantly different from each other; 2,036–2,586 and 2,036–2,586 were not significantly different (p > 0.05)
Analysis of the transcription factor binding sites for the Exons construct was carried out using a range of available programs including TESS and ConSite. These programs allow identification of binding site for candidate repressor or activator transcription factors. Analysis to identify repressors that align with regions we have shown to have repressor activity indicated that YY1 (Ying-Yang-1) and Hes-1 are the two best candidates for repressors acting in this system. Most importantly, these sites are present intron 1 of Prnp from four species (mouse, rat, human and cattle). YY1 has already been proposed as a repressor for PrP expression [10]. In its repressor role, YY1 binds to the consensus sequence CCWTNTTNNNW. Three sites of this kind exist in intron 1 at 1,032, 1,118 and 1,738 bp from the start of exon 1. As indicated above, deletion up to 1,525 in the Exons construct did not alter the level of reporter activity in the luciferase assays indicating that these regions do not have repressor activity. While deletion of the region 1,525–1,650 resulted in loss of a repressor site, there was no significant change on the loss of the region 1,650–1,950. This suggests that YY1 sites possibly do not play any role. In contrast the Hes-1 recognition site CTTGTG was identified at three positions (1,326, 1,548 and 2,175 bp) within intron 1. Deletion of the site at 1,548 was associated with an increased reporter activity and deletion of the site at 2,175 resulted in the largest change in activity we observed. This indicated that Hes-1 is the best possible candidate for a repressor in this system.
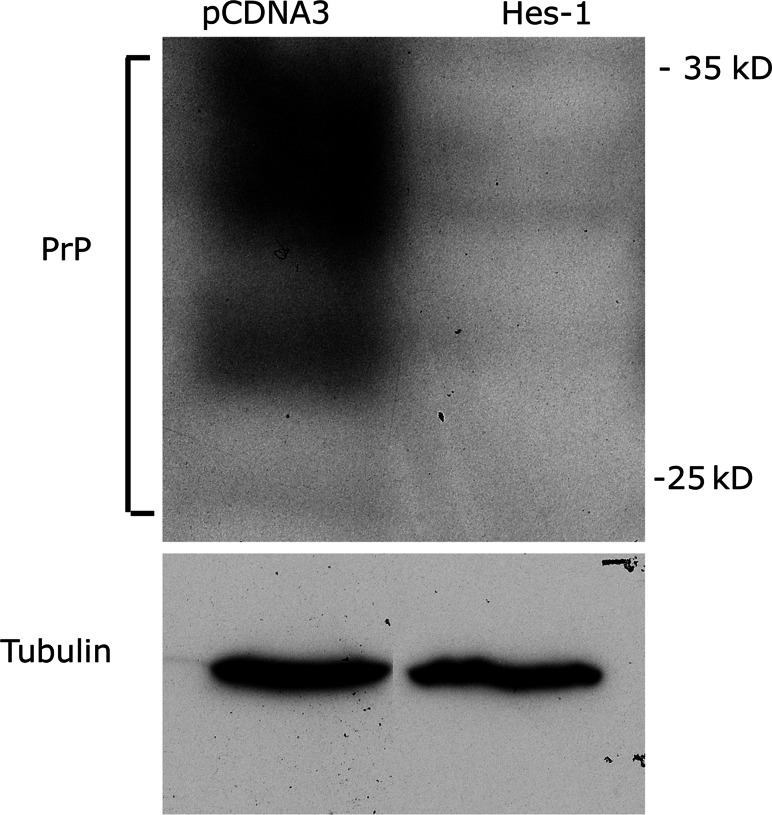
The open reading frame for Hes-1 was cloned into pCDNA3 and N2A cells transfected with the plasmid or the empty vector. Three separate stably expressing cell lines were generated. The level of PrP protein expression was then assessed using western blot of protein extracts and immunodetection with a specific antibody. Hes-1 overexpression caused a dramatic decrease in PrP expression (Fig. 7). Densitometric analysis indicated a reduction in PrP expression of 81 ± 8%. This finding confirms that Hes-1 is a major repressor of PrP expression.
Fig. 7.
Inhibition of PrP protein expression by Hes-1 overexpression. A series of stable N2A cell lines were generated that had been transfected with a plasmid to overexpress Hes-1 or the empty vector (pCDNA3). Protein extracts were prepared from the cell lines and western blots performed. PrP specific bands were identified by immunodetection. As a control, immunodetection of tubulin was carried out. An example is shown Hes-1 caused a dramatic decrease in PrP expression with no change in tubulin
Activation of PrP expression by Intron 1
The potential role of intron 1 to increase PrP expression through the binding of transcription factors was examined by co-transfection of cells with the Exons luciferase reporter and plasmids for the expression of selected transcription factors. PrP expression is known to increase in response to both oxidative stress and metals [20]. Using the TESS program we identified a number of potential transcription factors that could be playing a role in the regulation of PrP expression through intron 1. Nrf2 is a transcription factor associated with resistance to oxidative stress [21]. One Nrf2 binding site was identified (TGAATCCACA) 2,282 bp from the start of exon 1. Two transcription factors are associated with a response to increased copper concentrations. These are MTF-1 and Atox-1 [22, 23]. The Exons construct has one MTF-1 binding site (agGGCACCCAGCcg 125 bp from Exon 1 start) and 2 Atox-1 binding sites (GAAAGA, 1,552 and 2,023 bp from Exon 1 start).
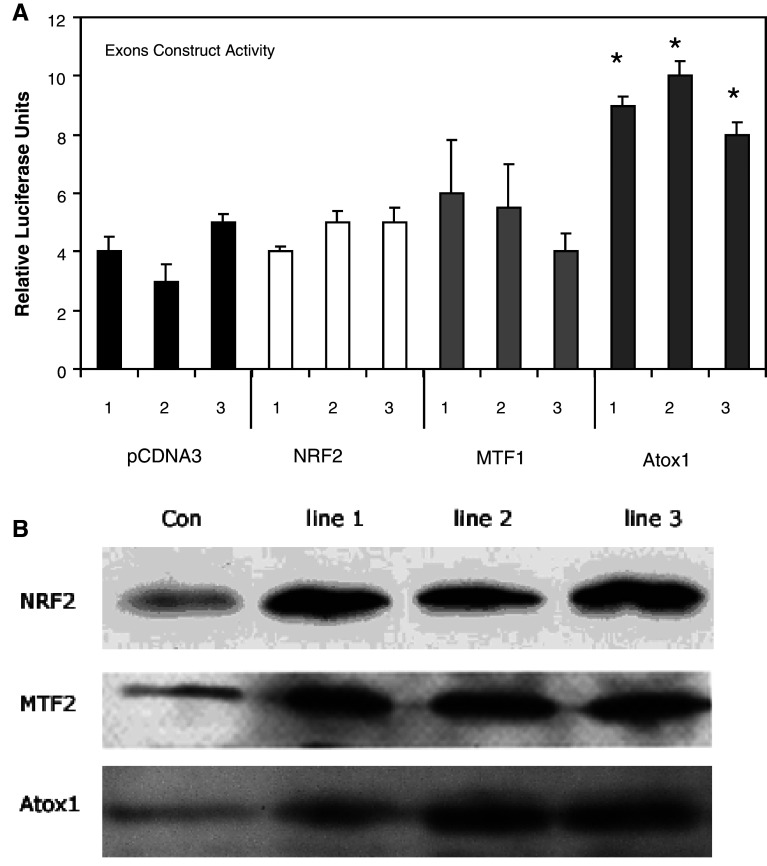
We carried out experiments to assess whether increased expression of these transcription factors would elevate the activity of the Exons construct. The open reading frame (ORF) for MTF-1, NRF-2 and Atox-1 were amplified by PCR and cloned into the pCDNA3 expression vector. Both stable and/or transient cell lines were created by transfecting with the plasmids carrying the ORF for each transcription factor as well as for the empty vector. In each case, three independent stable cell lines were created and analysed. Overexpression of the proteins was confirmed by western blot and immunodetection with a specific antibody (Fig. 8). All the cell lines created were co-transfected with the Exons construct and the level of luciferase activity compared to the control. Neither NRF2 nor MTF-1 overexpression resulted in an increase in the expression of the luciferase reporter (Fig. 8). In contrast, all cell lines transfected with Atox-1 showed an approximate doubling of luciferase activity from the Exons report construct. Confirmation that Atox-1 overexpression increases the expression of PrP protein in cells was obtained. Protein extracts were prepared from cells either overexpressing Atox-1 or transfected with the pCDNA3 vector. The extracts were electrophoresed, western blotted and the level of PrP protein assessed by immunodetection with a specific antibody. The analysis indicated an approximate 2.5-fold higher PrP expression (Fig. 9). This suggests that Atox-1 is a transcription factor that regulates PrP expression through binding to intron 1.
Fig. 8.
Luciferase reporter assays indicated that Atox-1 is an activator of PrP expression. Stable cell lines were generated using N2A cells that carried pCDNA3 with either no insert or the ORF for MTF-2, NRF2 or Atox-1. Three different stable cell lines for each construct was prepared and transiently transfected with the pGL3-Basic-Exons construct. a Luciferase reporter assays were performed and values were compared to those of N2A cells transfected with the pGL3-Basic-Exons construct only. Shown are the mean and SE for at least three experiments: asignificant difference in the level of luciferase activity was seen when comparing the empty vector values (pCDNA3) and Atox-1 values only (p > 0.05, ANOVA). b Western blots of protein extracts from each of the three cell lines for each transcription factor studied. Immunodetection with specific antibodies for each transcription factor verified overexpression of the relevant transcription factor occurred in each cell line
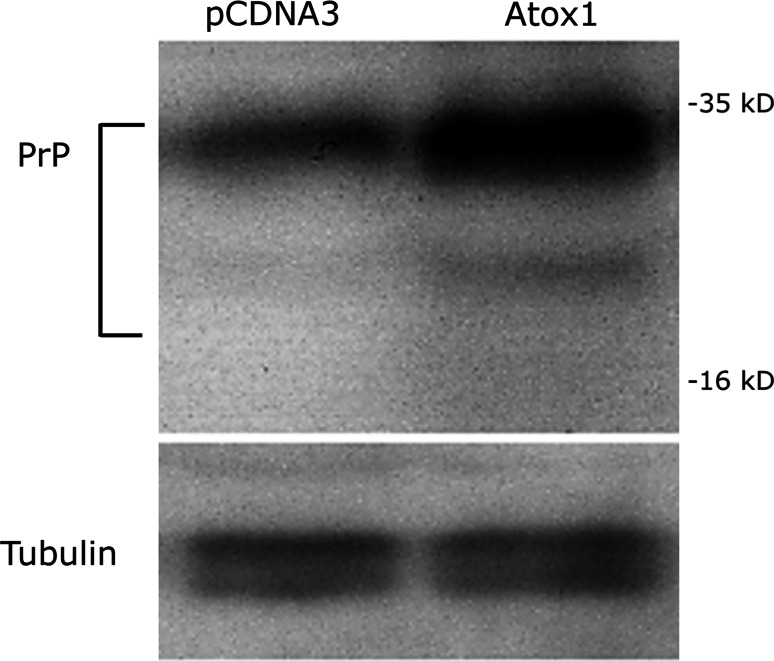
Fig. 9.
Western blot to detect PrP in Atox-1 overexpressing cells. N2A cells were transfected with pCDNA3 with no insert or pCDNA3 carrying the ORF for Atox-1. Stable cell lines were established and protein extracts prepared and electrophoresed on an SDS-PAGE gel. Following transfer to a membrane by western blotting a specific antibody was used to detect bands for PrP expressed by the cells. Following stripping of the blot, the membrane was reprobed with an anti-tubulin antibody to confirm equal protein loading. PrP expression was greater in the Atox-1 expressing cell line. The experiment was repeated four times. Intensity of PrP bands for Atox-1 were 2.6 ± 0.5-fold higher than for pCDNA3
Discussion
We previously suggested that intron 1 of Prnp had promoter-like activity [7]. This activity is present in numerous different cell lines (neuronal, glial, muscle cell) and is conserved between species [7]. In the current work, we have studied intron 1 in isolation to map potential regulator regions and identify transcription factors that might act through recognition of sites in the intron. Another study looking at the interaction between intron 1 and the promoter also suggested a role for intron 1 in Prnp expression but did not show any independent promoter activity [8]. However, this could be due to the different, non-neuronal cell lines used in their study. This study also suggested a strong role of 5′ region of intron 1 while our study did not suggest that this region has a role in the promoter activity driven by intron 1 independently of the promoter. In our previous work [7], we showed that exon 1 inhibits the activity of the main promoter in the absence of intron 1. It is quite possible that the 5′ region of intron 1 plays a role in regulating the inhibitory role of exon 1.
While the 5′ region of intron 1 may have regulatory effects on other regions of the Prnp gene, the 3′ end of intron has domains that regulate its promoter-like activity. The critical region for expression is that directly 5′ to exon 2. The region 2,312–2,488 is likely to be the site of binding for the transcriptional machinery. In agreement with our work, the region of bovine Prnp intron 1 between 114 and 892 bases upstream of the translational start site has been shown to drive considerable reporter activity in a bovine fibroblast cell line [8]. Previously, we suggested that transcriptional initiation would be associated with an apparent TATA site around 1,359–1,609 [7]. However, deletion of this site had no effect on reported promoter activity. Similarly, this region of the gene on its own showed no promoter activity.
This region of the intron also has significant repressor elements as their deletion resulted in step wise increase in the observed reporter activity. The two repressor regions identified lie within 1,525–1,650 and 2,036–2,312 bp from the start of Exon 1. Utilising the Transcriptional Element Search Software (TESS), we identified binding sites for HES-1, YY1, ELP, C/EBPalpha and AP-2 within intron 1, which repress promoter activity in other systems [24–27]. Importantly, these sites are also present within the same region of intron 1 in other species. However, the deletion analysis suggests that of these only YY1 and Hes-1 could potentially act as repressors. YY1 has already been suggested as a repressor for PrP expression [10]. While deletion of one YY1 site is possibly associated with increased reporter activity, deletion of a further site had no effect. This suggests that YY1 might not affect expression driven by intron 1. The previously described YY1 repressor site was located in the main promoter [10]. In contrast to YY1, deletion of both Hes-1 sites resulted in significant increases in reporter activity, making it a more likely candidate for the natural repressor of PrP expression as driven by intron 1. This was confirmed by overexpression of Hes-1 which caused massive decrease in PrP protein expression. The implication of this finding is that regulation of Hes-1 activity could be of great importance in terms of suppressing PrP expression. Our data clearly support a role of Hes-1 in the regulation of PrP via intron 1 and, while Hes-1 may directly bind to intron 1, but until this is established by further analysis, there could still be indirect mechanisms by which Hes-1 reduces PrP expression as well.
There have been a number of suggestions for transcription factors that activate PrP expression. Factors that have been suggested include MTF-1, SP1 and p53 [9, 16]. While most such studies consider the traditional promoter [28], they often overlook intron 1 as a major regulatory region. Additionally, many of these studies suggest binding sites without analysis of which regions of the promoter are active [29]. The one exogenous factor that has been repeatedly shown to increase the levels of PrP protein in cells is copper [30–32]. It is therefore logical to assume that a transcription factor upregulated in response to increased metal levels would increase PrP expression. While MTF-1 is a logical choice, we found no clear evidence that it would increase PrP expression via intron 1. In contrast, Atox-1 not only has the appropriate recognition site, which is located in regions of the intron that show the highest reporter activity, but overexpression of Atox-1 causes elevation of PrP protein expression. Atox-1 was originally identified as a copper chaperone [33] but has recently been shown to act as a copper-dependent transcription factor [23]. It is proposed that Atox-1 forms a homodimer on binding copper and is then able to enter the nucleus and bind DNA [34]. Currently, the number of genes linked to transcriptional regulation by Atox-1 is small, but includes extracellular superoxide dismutase (EC-SOD) [35]. It should be note that knockout of EC-SOD results in an increase in PrP expression and visa versa [36].
While we and others [32] showed no role for MTF-1 in regulating the expression of PrP, other researchers have indicated an important role for this transcription factor [9]. First, our results are restricted to responses mediated through sequences in intron 1. Second, the findings of Bellingham et al. [9] were observed under conditions in which intracellular copper concentrations were modified due to altered expression of the MNK-ATPase. Altering intracellular copper concentrations could have the effect of recruiting MTF-1 as a regulator of PrP while under normal physiological parameters its involvement in PrP expression is minimal.
The importance of PrP expression mediated through intron 1 is highlighted by studies looking at polymorphisms and indels in intron 1 and their link to BSE and other prion diseases [37–40]. In particular, alterations to intron 1 have been studied as a possible risk factor for susceptibility. As intron 1 has now been established as both a promoter-like region and a binding region for active transcriptional regulators, the alterations in intron 1 are now even more likely to play an important role in disease susceptibility.
In summary, we have carried out a detailed study of intron 1 in terms of its potential to play a major role in regulating prion protein expression. While a putative TATA box was shown not to play any significant role, we have shown that a small fragment of the intron can act as a promoter on its own and that the intron contains sites associated with both repression and activation of PrP expression. We suggest Hes-1 is a repressor of PrP expression while we have shown that Atox-1 is a transcription factor able to significantly increase PrP expression. The dependence of Atox-1 activity on copper binding further cements the link between PrP and copper metabolism.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 3.Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 4.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Bolton D. A novel hamster prion protein mRNA contains an extra exon: increased expression in scrapie. Brain Res. 1997;751:265–274. doi: 10.1016/S0006-8993(96)01407-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee IY, Westaway D, Smit AFA, Wang K, Seto J, Chen L, Acharya C, Ankener M, Baskin D, Cooper C, Yao H, Prusiner SB, Hood LE. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res. 1998;8:1022–1037. doi: 10.1101/gr.8.10.1022. [DOI] [PubMed] [Google Scholar]
- 7.Haigh CL, Wright JA, Brown DR. Regulation of prion protein expression by non coding regions of the Prnp gene. J Mol Biol. 2007;368:915–927. doi: 10.1016/j.jmb.2007.02.086. [DOI] [PubMed] [Google Scholar]
- 8.Inoue S, Tanaka M, Horiuchi M, Ishiguro N, Shinagawa M. Characterisation of the bovine prion protein gene: the expression requires interaction between the promoter and intron. J Vet Med Sci. 1997;59:175–183. doi: 10.1292/jvms.59.175. [DOI] [PubMed] [Google Scholar]
- 9.Bellingham SA, Coleman LA, Masters CL, Camakaris J, Hill AF. Regulation of prion gene expression by transcription factors SP1 and metal transcription factor-1. J Biol Chem. 2009;284:1291–1301. doi: 10.1074/jbc.M804755200. [DOI] [PubMed] [Google Scholar]
- 10.Burgess ST, Shen C, Ferguson LA, O’Neill GT, Docherty K, Hunter N, Goldmann W. Identification of adjacent binding sites for the YY1 and E4BP4 transcription factors in the ovine PrP (Prion) gene promoter. J Biol Chem. 2009;284:6716–6724. doi: 10.1074/jbc.M807065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shyu WC, Kao MC, Chou WY, Hsu YD, Soong BW. Heat shock modulates prion protein expression in human NT-2 cells. Mol Neurosci. 2000;11:771–774. doi: 10.1097/00001756-200003200-00023. [DOI] [PubMed] [Google Scholar]
- 12.Shyu WC, Harn HJ, Saeki K, Kubosaki A, Matsumoto Y, Onodera T, Chen CJ, Hsu YD, Chiang YH. Molecular modulation of expression of prion protein by heat shock. Mol Neurobiol. 2002;26:1–12. doi: 10.1385/MN:26:1:001. [DOI] [PubMed] [Google Scholar]
- 13.Wang V, Chuang TC, Hsu YD, Chou WY, Kao MC. Nitric oxide induces prion protein via MEK and p38 MAPK signalling. Biochem Biophys Res Comm. 2005;333:95–100. doi: 10.1016/j.bbrc.2005.05.091. [DOI] [PubMed] [Google Scholar]
- 14.Shyu WC, Lin SZ, Saeki K, Kubosaki A, Matsumoto Y, Onodera T, Chiang MF, Thajeb P, Li H. Hyperbaric oxygen enhances the expression of prion protein and heat shock protein 70 in a mouse neuroblastoma cell line. Cell Mol Neurobiol. 2004;24:257–268. doi: 10.1023/B:CEMN.0000018620.41913.d2. [DOI] [PubMed] [Google Scholar]
- 15.Shyu WC, Chen CP, Saeki K, Kubosaki A, Matsumoto Y, Onodera T, Ding DC, Chiang MF, Lee YJ, Lin SZ, Li H. Hypoglycaemia enhances the expression of prion protein and heat-shock protein 70 in a mouse neuroblastoma cell line. J Neurosci Res. 2005;80:887–894. doi: 10.1002/jnr.20509. [DOI] [PubMed] [Google Scholar]
- 16.Qin K, Zhao L, Ash RD, McDonough WF, Zhao RY. ATM-mediated transcriptional elevation of prion in response to copper-induced oxidative stress. J Biol Chem. 2009;284:4582–4593. doi: 10.1074/jbc.M808410200. [DOI] [PubMed] [Google Scholar]
- 17.Rybner C, Hilion J, Sahraoui T, Lanotte M, Botti J. All-trans retinoic acid down-regulates prion protein expression independently of granulocyte maturation. Leukaemia. 2002;16:940–948. doi: 10.1038/sj.leu.2402443. [DOI] [PubMed] [Google Scholar]
- 18.Holme A, Daniels M, Sassoon J, Brown DR. A novel method of generating neuronal cell lines from gene-knockout mice to study prion protein membrane orientation. Eur J Neurosci. 2003;18:571–579. doi: 10.1046/j.1460-9568.2003.02780.x. [DOI] [PubMed] [Google Scholar]
- 19.Lenhard B, Sandelin A, Mendoza L, Engström P, Jareborg N, Wasserman WW. Identification of conserved regulatory elements by comparative genome analysis. J Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DR, Schmidt B, Kretzschmar HA. Expression of prion protein in PC12 is enhanced by exposure to oxidative stress. Int J Dev Neurosci. 1997;15:961–972. doi: 10.1016/S0736-5748(97)00042-7. [DOI] [PubMed] [Google Scholar]
- 21.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 22.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. Novel role of antioxidant-1 (Atox1) as a copper-dependent transcription factor involved in cell proliferation. J Biol Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotomura N, Ninomiya Y, Umesono K, Niwa O. Transcriptional regulation by competition between ELP isoforms and nuclear receptors. Biochem Biophys Res Comm. 1997;230:407–412. doi: 10.1006/bbrc.1996.5972. [DOI] [PubMed] [Google Scholar]
- 25.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 28.Funke-Kaiser H, Theis S, Behrouzi T, Thomas A, Scheuch K, Zollmann FS, Paterka M, Paul M, Orzechowski HD. Functional characterization of the human prion protein promoter in neuronal and endothelial cells. J Mol Med. 2001;79:529–535. doi: 10.1007/s001090100270. [DOI] [PubMed] [Google Scholar]
- 29.Mahal SP, Asante EA, Antoniou M, Collinge J. Isolation and functional characterisation of the promoter region of the human prion protein gene. Gene. 2001;268:105–114. doi: 10.1016/S0378-1119(01)00424-3. [DOI] [PubMed] [Google Scholar]
- 30.Brown DR. Role of the prion protein in copper turnover in astrocytes. Neurobiol Dis. 2004;15:534–543. doi: 10.1016/j.nbd.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Armendariz AD, Gonzalez M, Loguinov AV, Vulpe CD. Gene expression profiling in chronic copper overload reveals upregulation of Prnp and APP. Physiol Genomics. 2004;20:45–54. doi: 10.1152/physiolgenomics.00196.2003. [DOI] [PubMed] [Google Scholar]
- 32.Varela-Nallar L, Toledo EM, Larrondo LF, Cabral AL, Martins VR, Inestrosa NC. Induction of cellular prion protein gene expression by copper in neurons. Am J Physiol Cell Physiol. 2006;290:C271–C281. doi: 10.1152/ajpcell.00160.2005. [DOI] [PubMed] [Google Scholar]
- 33.Naeve GS, Vana AM, Eggold JR, Kelner GS, Maki R, Desouza EB, Foster AC. Expression profile of the copper homeostasis gene, rAtox1, in the rat brain. Neuroscience. 1999;93:1179–1187. doi: 10.1016/S0306-4522(99)00175-X. [DOI] [PubMed] [Google Scholar]
- 34.Muller PA, Klomp LW. ATOX1: a novel copper-responsive transcription factor in mammals? Int J Biochem Cell Biol. 2009;41:1233–1236. doi: 10.1016/j.biocel.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Itoh S, Ozumi K, Kim HW, Nakagawa O, McKinney RD, Folz RJ, Zelko IN, Ushio-Fukai M, Fukai T. Novel mechanism for regulation of extracellular SOD transcription and activity by copper: role of antioxidant-1. Free Radic Biol Med. 2009;46:95–104. doi: 10.1016/j.freeradbiomed.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kralovicova S, Fontaine SN, Alderton A, Alderman J, Ragnarsdottir KV, Collins SJ, Brown DR. The effects of prion protein expression on metal metabolism. Mol Cell Neurosci. 2009;41:135–147. doi: 10.1016/j.mcn.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 37.McCormack JE, Baybutt HN, Everington D, Will RG, Ironside JW, Manson JC. PRNP contains both intronic and upstream regulatory regions that may influence susceptibility to Creutzfeldt-Jakob disease. Gene. 2002;288:139–146. doi: 10.1016/S0378-1119(02)00466-3. [DOI] [PubMed] [Google Scholar]
- 38.Seabury CM, Womack JE, Piedrahita J, Derr JN. Comparative PRNP genotyping of US cattle sires for potential association with BSE. Mamm Genome. 2004;15:828–833. doi: 10.1007/s00335-004-2400-6. [DOI] [PubMed] [Google Scholar]
- 39.Nakamitsu S, Miyazawa T, Horiuchi M, Onoe S, Ohoba Y, Kitagawa H, Ishiguro N. Sequence variation of bovine prion protein gene in Japanese cattle (Holstein and Japanese Black) J Vet Med Sci. 2006;68:27–33. doi: 10.1292/jvms.68.27. [DOI] [PubMed] [Google Scholar]
- 40.Kerber AR, Hepp D, Passos DT, de Azevedo Weimer T. Polymorphisms of two indels at the PRNP gene in three beef cattle herds. Biochem Genet. 2008;46:1–7. doi: 10.1007/s10528-007-9113-y. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Liu T, Wong B-S, Pan T, Morillas M, Swietnicki W, O'Rourke K, et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J Mol Biol. 2000;301:567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]