Abstract
G proteins are universal molecular switches in eukaryotic signal transduction. The Arabidopsis genome sequence reveals no RAS small GTPase and only one or a few heterotrimeric G proteins, two predominant classes of signaling G proteins found in animals. In contrast, Arabidopsis possesses a unique family of 11 Rop GTPases that belong to the Rho family of small GTPases. Previous studies indicate that Rop controls actin-dependent pollen tube growth and H2O2-dependent defense responses. In this study, we tested the hypothesis that the Rop GTPase acts as a versatile molecular switch in signaling to multiple developmental processes in Arabidopsis. Immunolocalization using a general antibody against the Rop family proteins revealed a ubiquitous distribution of Rop proteins in all vegetative and reproductive tissues and cells in Arabidopsis. The cauliflower mosaic virus 35S promoter-directed expression of constitutively active GTP-bound rop2 (CA-rop2) and dominant negative GDP-bound rop2 (DN-rop2) mutant genes impacted many aspects of plant growth and development, including embryo development, seed dormancy, seedling development, lateral root initiation, morphogenesis of lateral organs in the shoot, shoot apical dominance and growth, phyllotaxis, and lateral organ orientation. The rop2 transgenic plants also displayed altered responses to the exogenous application of several hormones, such as abscisic acid-mediated seed dormancy, auxin-dependent lateral shoot initiation, and brassinolide-mediated hypocotyl elongation. CA-rop2 and DN-rop2 expression had opposite effects on most of the affected processes, supporting a direct signaling role for Rop in regulating these processes. Based on these observations and previous results, we propose that Rop2 and other members of the Rop family participate in multiple distinct signaling pathways that control plant growth, development, and responses to the environment.
G proteins are pivotal molecular switches in eukaryotic signal transduction that controls a wide spectrum of processes ranging from odorant perception to cell cycle control. To generate functional diversity and specificity, G protein structural variants have evolved in different organisms. Two major classes of signaling G proteins are known: heterotrimeric G proteins and the Ras superfamily of monomeric GTPases. Among the five families of the Ras superfamily (RAS, RHO, RAB/YPT, RAN, and ARF), only RAS and RHO are considered bona fide signaling switches; the others are primarily involved in the regulation of trafficking of vesicles or large molecules (Moore and Blobel, 1993; Stamnes and Rothman, 1993; Lazar et al., 1997). Mammals possess a large number of heterotrimeric G proteins that are formed from the combinations of 20 α, 5 β, and 7 γ subunits (Glick et al., 2000; Mumby, 2000). Thus, more than one-third of mammalian signaling pathways are dependent on heterotrimeric G proteins (Glick et al., 2000; Mumby, 2000). Furthermore, RAS and RAS-like signaling switches have also been shown to control a large number of signaling pathways in animals (Bos, 2000). In contrast, only a single prototype for each of the Gα, Gβ, and Gγ subunits and no RAS orthologs have been identified in plants (Ma et al., 1990; Weiss et al., 1994; Arabidopsis Genome Initiative, 2000; Mason and Botella, 2000). Pharmacological studies have implicated heterotrimeric G proteins in the gibberellin (GA), phytochrome, and abscisic acid signaling pathways (for review, see Ma, 1994; Yang, 1996). Genetic studies suggest that Gα has a role in GA signaling and seed development in rice (Ashikari et al., 1999; Ueguchi-Tanaka et al., 2000). However, it remains to be seen whether heterotrimeric G proteins has a widespread role in signaling in plants as in animals. Consistent with lack of RAS GTPases, Arabidopsis apparently does not possess receptor Tyr kinases that typically regulate RAS in animals (Arabidopsis Genome Initiative, 2000). Instead, receptor-like Ser/Thr kinases appear to be a major class of membrane receptors in plants (Braun and Walker, 1996; Arabidopsis Genome Initiative, 2000; Chory and Wu, 2001).
Do plants use a novel type of G proteins as a predominant molecular switch? Plants possess a large family of genes encoding the Rop (Rho-related GTPase from plants) GTPase, which belong to a distinct subfamily of the RHO family (Yang and Watson, 1993; Delmer et al., 1995; Winge et al., 1997; Li et al., 1998; Zheng and Yang, 2000). RHO was initially identified as a regulator of actin organization, but has now been shown to control other processes including gene expression, cell wall synthesis, and cell cycle progression in yeast and animals (Hall, 1998; Mackay and Hall, 1998). In those phyla, the RHO family can be subdivided into three major subfamilies, Rho, Cdc42, and Rac, each having distinct roles in controlling specific forms of F-actin and related cellular processes (Mackay and Hall, 1998). The Arabidopsis genome sequence reveals no orthologs of Cdc42, Rac, and Rho, and Rop appears to be unique to plants and to have evolved from the same ancestor as Cdc42 and Rac (Li et al., 1998; Zheng and Yang, 2000).
Recent studies have revealed an important role for Rop in plant signaling. First of all, the pollen-specific Rop1 (Arabidopsis Rop1 and its pea ortholog) has been shown to control polar growth in pollen tubes. Studies using an anti-Rop1 antibodies indicate that Rop1 is localized to the plasma membrane at the tip of pollen tubes and is essential for pollen tube growth (Lin et al., 1996; Lin and Yang, 1997). Transgenic expression of constitutively active (CA) and dominant negative (DN) rop1 mutants in Arabidopsis reveals a role for Rop1 in the control of cell polarity development in pollen tubes (Li et al., 1999). A similar role for Rop5 has also been shown using transient expression of rop5 mutants in tobacco pollen tubes (Kost et al., 1999). Genes encoding three nearly identical Rops (Rop1, Rop3, and Rop5) are expressed in pollen, suggesting that these Rop genes may be functionally redundant in pollen tubes (Li et al., 1999). Rop1 controls pollen tube polar growth via modulating the dynamics of tip actin and the formation of tip-focused calcium gradients (Li et al., 1999; Fu et al., 2001).
Rop has also been shown to control the production of H2O2 and defense responses. Transgenic expression of CA and DN mutants for a rice Rop (OsRac1) and a cotton Rop (Rac13) altered H2O2 production (Kawasaki et al., 1999; Potikha et al., 1999). The former is linked to cell death and disease resistance in rice leaves (Ono et al., 2001), and the latter has been implicated in the synthesis of secondary cell wall in cotton fiber cells (Potikha et al., 1999). Tobacco transgenic plants expressing the alfalfa MsRac1 antisense gene exhibit reduced responses to elicitor treatments (Schiene et al., 2000). It is interesting that a Rop-like protein is specifically associated with the active CLAVATA1 receptor kinase complex, suggesting that a Rop GTPase may participate in the signaling to meristem maintenance (Trotochaud et al., 1999).
We have shown that a Rop protein is localized to the tonoplast of developing vacuoles in the pea tapetum, supporting a role for Rop in the regulation of vacuole development (Lin et al., 2001). Different Rops exhibit differential subcellular localization to the plasma membrane, a perinuclear organelle, and the cytosol (Bischoff et al., 2000; Ivanchenko et al., 2000). Together these results provide evidence that different Rops may have distinct cellular functions. Analyses by reverse transcription-polymerase chain reactions show that several Rop genes are expressed in all parts of Arabidopsis plants, supporting a broad role for Rop signaling in the control of plant processes.
In this study, we tested the hypothesis that Rop acts as a versatile switch in multiple signaling pathways in Arabidopsis. We have demonstrated that proteins of the Rop family are distributed in various tissues and cell types in Arabidopsis. It is important that we have shown that CA-rop2 and DN-rop2 mutants induce pleiotropic developmental phenotypes. In most cases, CA-rop2 and DN-rop2 cause opposite effects. These results strongly suggest that the Rop GTPase switch participates in the signaling to multiple distinct developmental processes in Arabidopsis.
RESULTS
Rop Proteins Are Distributed in All Tissues But Preferentially Accumulate in Meristems and Rapidly Growing Cells
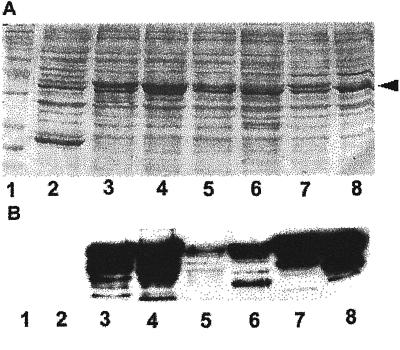
To assess how widespread Rop signaling occurs during plant growth and development, we first investigated the distribution of Rop proteins using immunolocalization involving anti-Rop1 antibodies (Lin et al., 1996). Because of the structural conservation, the anti-Rop1 polyclonal antibodies reacted with all Rop recombinant proteins representing various Rop groups (Zheng and Yang, 2000), although those less related to Rop1 (e.g. Rop7 and Rop8) produced weaker signals (Fig. 1). These results suggest that these anti-Rop1 antibodies are reactive with all Arabidopsis Rops. Thus, immunolocalization using these antibodies will largely reveal protein distribution patterns for the whole Rop family in Arabidopsis.
Figure 1.
Western-blotting analysis of GST-Rop fusions. Different Rop-coding sequences were fused to the C terminus of GST and expressed in Escherichia coli as described (Wu et al., 2000). E. coli extracts were separated on a 12% (w/v) SDS PAGE gel, blotted onto a nitrocellulose membrane, and reacted with anti-Rop1Ps antibodies as described (Lin et al., 1996). A, Shows the membrane stained with 0.1% (w/v) Poceau S for loading control of total E. coli proteins. Arrowhead indicates the position of GST-Rop fusion proteins. B, Shows Rop-specific signals detected by the anti-Rop1Ps antibodies. For some lanes, multiple bands were detected as a result of protein degradation. Negative control lane (GST) did not show any signal even after an extended exposure. Lane 1, protein Mr markers; lane 2, GST; lane 3, Arac10; lane 4, Arac8; lane 5, Rop8; lane 6, Rop7; lane 7, Rop6; lane 8, Rop2.
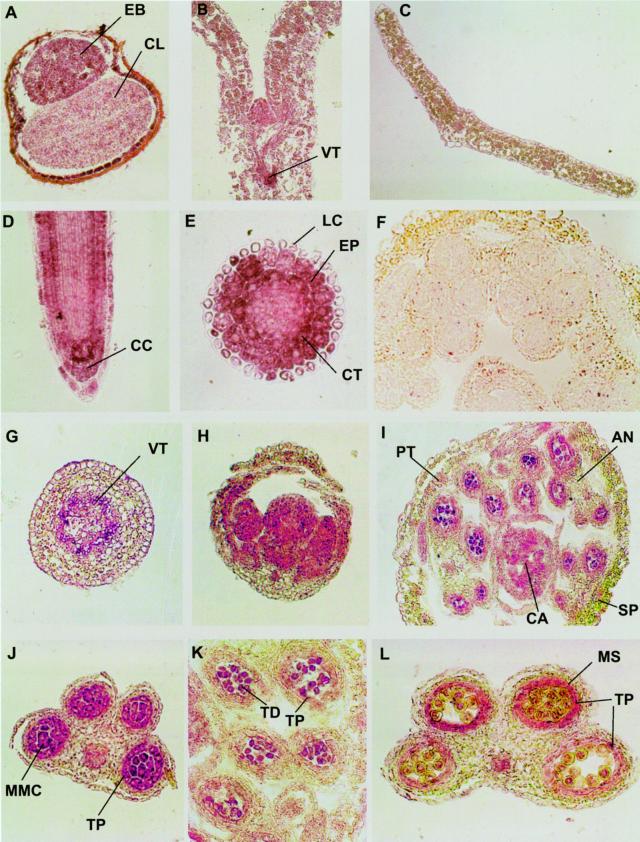
In imbibed seeds, Rop staining was stronger in the embryo than cotyledons, but the strongest staining was found in the inner layer of seed coats (Fig. 2A). The inner layer cells may be equivalent to the aleurone layer in cereal seeds, although their physiological role is unknown. During vegetative growth, Rop proteins are consistently abundant in various tissues with active cell division, such as meristems and leaf primordia (Fig. 2, B and D). In the root tip, Rop signals are also very strong in columella cells and certain lateral root cap cells (Fig. 2D). In the elongation zone of roots, levels of Rop proteins are high in rapidly expanding cells in the epidermis and the cortex, but low in the endodermis, and barely detectable in the stele (Fig. 2, D and E). Moderate levels of Rop proteins are also found in the epidermal and mesophyll cells of expanding cotyledons (Fig. 2B). In addition, high levels of Rop are found in differentiating vascular tissues in all organs (Fig. 2, B, G, and J). Rop proteins appear to be less abundant in epidermal and parenchyma tissues of mature leaves and stems (Fig. 2, C and G).
Figure 2.
Immunolicalization of Rop proteins in various Arabidopsis tissues. Ten-micrometer cryosections of Arabidopsis tissues were incubated with anti-Rop1Ps antibodies and alkaline phosphatase-conjugated secondary antibodies as described in text. A, Cross section of an imbibed seed. B, Longitudinal section of a shoot apex and cotyledons of seedling. C, Cross section of a rosette leaf. D, Longitudinal section of a root tip. E, Cross section of a root tip near the elongation zone of the root. F, Cross section of a closed flower bud stained with preimmune control. G, A cross section of a stem. H, Longitudinal section of a young flower bud. I, Cross section of a closed flower. J, K, and L, Cross sections of anthers at the microspore mother cell, tetrad, and early microspore stages, respectively. Purple color indicates Rop staining, whereas yellow and green colors indicate anthocyanin and chlorophyll pigments from sections of frozen tissues.
In floral buds and flowers, high levels of Rop proteins are also consistently found in inflorescence meristems, organ primordia, developing ovules, and anthers (Fig. 2, H and I). In anthers, Rop is primarily localized to the tapetum, dividing microsporogenic cells, tetrads, microspores, and mature pollen (Fig. 2, I–L). In the tapetum, Rop accumulation exhibits dynamic changes throughout the development of the male gametophyte as shown in pea (Lin et al., 2001). Rop signals are strongest at the microspore mother cell stage, decrease to barely detectable levels at the tetrad stage, increase to a high level during early mitotic stages of microspores, and again decreased to minimal levels during pollen maturation. Staining with preimmune sera did not produce any signals in anthers (Fig. 2F) and various meristematic tissues (data not shown), indicating that the anti-Rop1 antibody staining was specific. These analyses show that Rop proteins are ubiquitously distributed in various tissues and cell types throughout the life cycle of Arabidopsis plants.
Transgenic Expression of rop2 Mutant Genes in Arabidopsis
The ubiquitous distribution of Rop proteins in Arabidopsis plants supports the notion that Rop may serve as a common switch in plant signaling. To further test this hypothesis, we chose Rop2 for functional analyses using transgenic expression of dominant mutant genes for the following reasons. First, we have shown that Rop2 is constitutively expressed in different vegetative parts in Arabidopsis by using reverse transcriptase (RT)-PCR (Li et al., 1998). Furthermore, Rop2 belongs to the largest Rop group (group IV) that includes Rop1 through Rop6 (Zheng and Yang, 2000). Because high sequence identity among members of this group (86%–95%), phenotypes induced by rop2 dominant mutants may provide a useful indication of most if not all physiological functions for this group of Rop proteins. At least some members of the Rop family may be functionally redundant, and thus this gain-of-function approach is expected to be important for our initial functional studies of the Rop family before loss-of-function mutations are available for all Rop members.
We generated two opposite mutations for Rop2: CA and DN. CA or DN mutant proteins permanently bind GTP or GDP and thus are expected to constitutively activate or block Rop2-dependent signaling, respectively (Zheng and Yang, 2000). We expect CA-rop2 and DN-rop2 mutant genes to induce opposite phenotypes if Rop2 acts as a switch in specific signaling pathways.
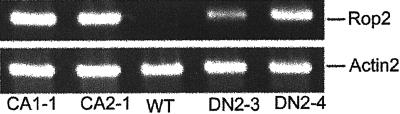
The mutant genes under the control of the cauliflower mosaic virus (CaMV) 35S promoter were stably expressed in Arabidopsis Columbia ecotype. Multiple independent transgenic lines with similar or identical morphological phenotypes were obtained for each mutant gene, and T2 or T3 generations of two independent lines for each construct were used for detailed analyses of phenotypes throughout the life cycle of these plants. RT-PCR analysis confirmed that these transgenic lines expressed the corresponding mutant genes (Fig. 3). We were unable to obtain homozygous lines for CA-rop2 due to embryo lethality (see below). Because CA-rop2 causes a distinct cotyledon phenotype, wild-type siblings that segregated out from the heterozygous line were easily identified and excluded from most of our phenotype analyses. For DN-rop2 transgenic plants, single T-DNA insertions only caused weak phenotypes, and thus a homozygous line was only used for the analyses of some phenotypes described below. Most of the DN-rop2 phenotypes were characterized using two independent lines, each containing at least two T-DNA insertions.
Figure 3.
RT-PCR analysis of rop2 transgene expression. Total RNAs were isolated from wild-type (WT) plants and from four rop2 mutant lines used in this study (CA1-1 and CA2-1, CA-rop2 mutants; DN2-3 and DN2-4, DN-rop2 mutants). CA-rop2 and DN-rop2 cDNAs were amplified using the transgene-specific primers as described in text. The Actin2 primers were included in the PCR reactions as an internal control (Li et al., 1998). A 20-μL PCR product was analyzed on an agarose gel.
CA-rop2 and DN-rop2 Affected Seed Dormancy
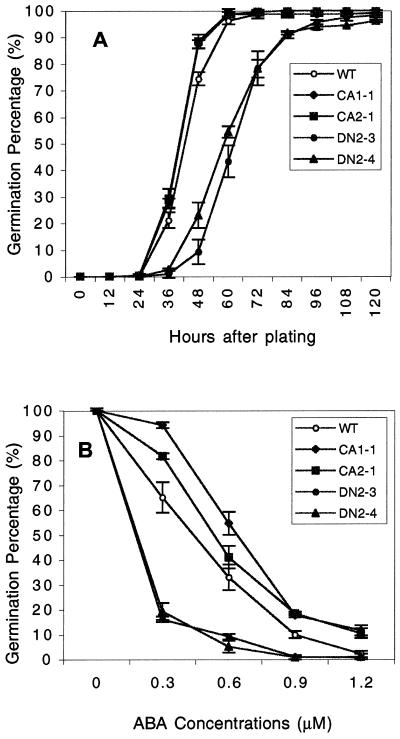
We first determined the effect of CA-rop2 and DN-rop2 on seed germination. Newly harvested seeds were germinated on Murashige and Skoog agar medium, and a time course of germination was determined. As shown in Figure 4A, DN-rop2 seeds showed a dramatic delay in seed germination, whereas CA-rop2 seeds germinated faster than wild-type seeds. At 48 h, germination rates are 16.2%, 74%, and 87% for DN-rop2, WT, and CA-rop2 seeds, respectively. The actual differences in germination rates between WT and transgenic plants were expected to be greater than those shown, because the germination rates for both CA-rop2 and DN-rop2 seed populations were skewed by the presence of WT seeds in the progeny of T2 heterozygous plants. The germination rate for all genotypes reached 100% or nearly 100% after 5 d, suggesting that the expression of Rop2 mutant genes does not affect the viability of seeds. Furthermore, 100% of the seeds germinated with identical kinetics for all genotypes when seeds were cold treated for 4 d before germination (data not shown), suggesting that Rop is involved in the regulation of seed dormancy.
Figure 4.
Effect of CA-rop2 and DN-rop2 expression on seed dormancy. A, Seed dormancy analysis. Newly harvested T4 seeds were plated on agar medium and germination was scored as emergence of radicles at various times after plating (n = 70). B, Effect of ABA on seed germination. Seeds were plated on agar plates containing different concentrations of ABA and cold treated for 4 d prior to incubation at 22 C. Germination was scored 72 h later (n = 50). Error bars show se.
Because abscisic acid (ABA) is a well-known hormone that controls seed dormancy, we next tested the effect of ABA on the germination of cold-treated rop2 transgenic seeds (Fig. 4B). Compared to wild type, DN-rop2 seeds were hypersensitive to ABA inhibition of germination. In the presence of 0.3 μm ABA, greater than 70% of wild-type seeds germinated, whereas only approximately 10% of DN2-4 seeds germinated at 48 h (Fig. 4B). In contrast, CA-rop2 seeds were less sensitive to ABA than wild-type seeds. For example, approximately 88% CA2-1 seeds germinated compared with 73% for wild-type seeds at 48 h. These results suggest that Rop is involved in the negative regulation of ABA-mediated seed dormancy.
CA-rop2 and DN-rop2 Affect Seedling Development
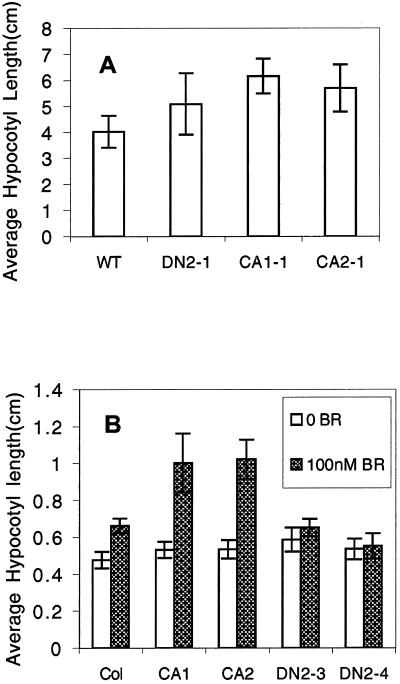
We next investigated the effect of rop2 expression on Arabidopsis seedling development. Cold-treated seeds were germinated on Murashige and Skoog agar medium in dark or in light. Dark-grown CA-rop2 seedlings exhibited a phenotype similar to constitutive photomorphogenesis, including cotyledon expansion and inhibition of hypocotyl elongation; whereas the majority of DN-rop2 seedlings had longer hypocotyls in dark (Fig. 5A), supporting a potential role for Rop in photomorphogenesis. However, responses of transgenic seedlings to light were complex. Under higher light intensity (33 μmol m−2 s−1), CA-rop2 seedlings had longer hypocotyl lengths than either WT or DN-rop2 seedlings, whereas no significant differences in hypocotyl elongation were found between WT and DN-rop2 seedlings (Fig. 6A). Under lower light intensity (20 μmol m−2 s −1), hypocotyl lengths of both CA-rop2 and DN-rop2 seedlings were not significantly different from wild type. Similarly light-grown seedlings in liquid cultures did not shown significant differences in hypocotyl lengths between the three genotypes (Fig. 6B).
Figure 5.
Phenotypes of rop2 transgenic plants at different developmental stages. A, Ten-day-old seedlings grown in an agar medium. B, Ten-day-old dark-grown seedling grown in an agar medium containing 100 nm BR. C, Rosette stage. D, Five-week-old plants. E, Silique oreitation changes in rop2 transgenic plants. F, Leaf morphology of rop2 transgenic plants. G, Fully expanded cotyledons. H, Aborted embryos in CA-rop2 plants indicated by arrowheads. Scale bar = 1 cm (A–F); = 1 mm (G and H).
Figure 6.
Hypocotyl elongation of rop2 transgenic plants. A, Hypocotyl lengths of 10-d-old seedlings grown on agar plates under higher light intensity (8-h dark/16-h light, 33 μmol m−2 s−1). B, Hypocotyl lengths of seedlings grown in liquid culture in the presence of 100 nm BR under lower light intensity (8-h dark/16-h light, 20 μmol m−2 s−1). Filled columns represent 100 nm BR treatment. Blank columns represent hypocotyl elongation without BR treatment. Seedlings were grown in liquid one-half Murashige and Skoog medium plus 1% (w/v) Suc with shaking for 5 d. Error bars stand for sd (n = 20).
Because light control of seedling development is thought to act at least in part through the regulation of the level of hormones such as brassinolides (BRs), we assessed whether CA-rop2 and DN-rop2 expression altered BR responses in seedlings grown in liquid medium (Azpiroz et al., 1998). As shown in Figure 6B, light-grown CA-rop2 seedlings were more sensitive to the stimulation of hypocotyl elongation by 100 nm BR, but DN-rop2 seedlings were less sensitive than WT seedlings. In addition, we found that 100 nm BR dramatically exaggerated CA-rop2 induction of hypocotyl expansion and petiole elongation in dark-grown seedlings (Fig. 6B). These results support a role for Rop in the regulation of BR-mediated Arabidopsis seedling development. Furthermore, CA-rop2 or DN-rop2 expression did not alter seedling responses to GA (data not shown).
CA-rop2 and DN-rop2 Alter the Initiation of Lateral Roots
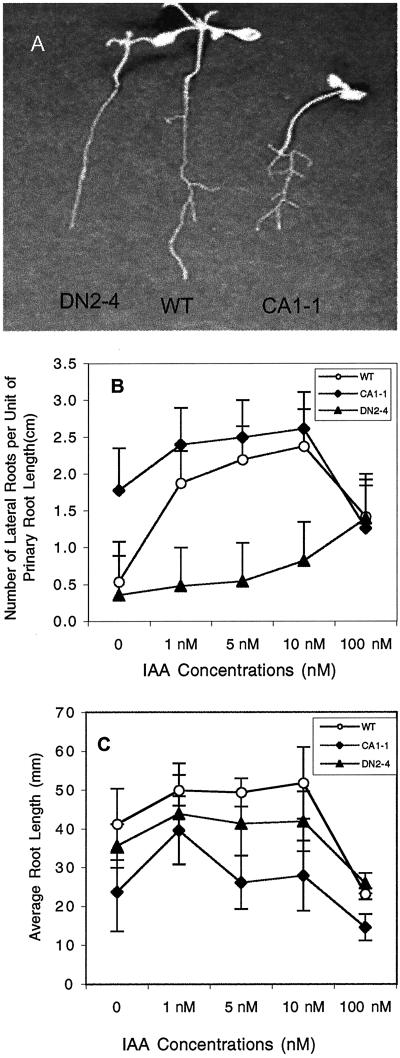
We then examined the effect of CA-rop2 and DN-rop2 expression on adult phenotypes. Kanamycin-resistant T2 or T3 transgenic plants were selected on an agar medium before being transferred to a new agar plate or soil for morphological analyses. To determine the effect of rop2 on root growth and development, kanamycin-resistant seedlings were grown on an agar plate placed vertically and incubated under light for 10 d. As shown in Figure 7A, CA-rop2 and DN-rop2 expression affected both the elongation of primary roots and the formation of lateral roots but apparently did not significantly alter radial expansion of roots. CA-rop2 seedlings have reduced length of primary roots but increased number of lateral roots. Ten-day-old wild-type plants produced an average of 3.0 lateral roots per primary roots, whereas CA-rop2 plants possessed an average of 5.9 lateral roots. In contrast, DN-rop2 plants had reduced lateral roots, averaging 2.0 per primary root. DN-rop2 expression also caused reduction in the length of the primary root (Fig. 7A), suggesting that the increased lateral root initiation in CA-rop2 plants is likely the direct result from CA-rop2 expression but not indirect effect of CA-rop2 via the inhibition of primary root elongation. The average leaf number was not affected by CA-rop2 or DN-rop2 expression (data not shown), suggesting that the alteration in primary root formation was not due to general growth inhibition.
Figure 7.
Effects of CA-rop2 and DN-rop2 expression on the auxin induction of lateral root initiation in rop2 transgenic plants. A, Lateral root initiation in 10-d-old seedlings; CA-rop2 seedlings had more lateral roots than wild type, whereas DN-rop2 seedlings had fewer. B, Effects of IAA on the initiation of lateral roots. C, Effects of IAA on the elongation of primary roots. Error bars show sd (n = 20).
The alteration of lateral root formation by CA-rop2 and DN-rop2 expression is consistent with a role for Rop in auxin regulation of lateral root formation. Thus, we determined the effect of CA-rop2 and DN-rop2 expression on auxin-stimulation of lateral root formation. Because both CA-rop2 and DN-rop2 inhibited primary root elongation, we measured the lateral root forming capacity using the average number of lateral roots per unit of the primary root. As shown in Figure 7B, treatment with 1 nm indole-3-acetic acid (IAA) increased lateral formation by 3 folds in wild-type plants, and maximum responses occurred at 10 nm IAA. CA-rop2 plants produced lateral roots dramatically better than WT in the absence of exogenous IAA and reached the maximum lateral root forming capacity at 1 nm IAA, the lowest concentration tested. In contrast, DN-rop2 plants exhibited drastically reduced sensitivity to IAA stimulation of lateral root formation; little stimulation occurred even in the presence of 10 nm of IAA. However, DN-rop2 and CA-rop2 expression did not affect IAA inhibition of primary root elongation (Fig. 7C). These results suggest that Rop either potentiates IAA control of lateral root formation or participates in an auxin signaling pathway that controls lateral root formation.
CA-rop2 and DN-rop2 Alter Shoot Apical Dominance
We next sought to investigate the effect of DN-rop2 and CA-rop2 expression on aerial phenotypes of adult plants. When grown in soil, both types of transgenic plants display pleiotropic phenotypes with altered plant architecture (Fig. 5, C and D). At the rosette stage (Fig. 5C), DN-rop2 expression dramatically reduced the plant stature, and CA-rop2 expression also slightly reduced the plant size. Rop2 overexpression did not significantly alter plant morphology. Furthermore, CA-rop2- and Rop2-expressing leaves are greener than WT, whereas DN-rop2 leaves had reduced greening. At the mature stage, the height of DN-rop2 plants was dramatically reduced, whereas CA-rop2 expression only slightly reduced the height (Fig. 5D).
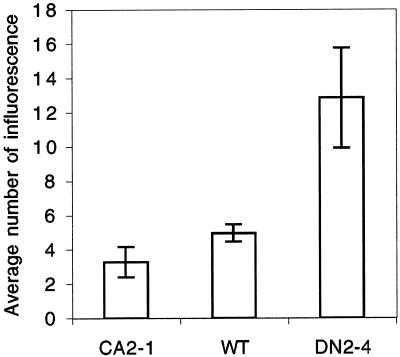
The most striking phenotype is the alteration in shoot apical dominance. CA-rop2 plants displayed enhanced shoot apical dominance. On the average, 7-week-old WT plants had 5.0 inflorescence shoots, whereas CA-rop2 plants had 3.2 inflorescence shoots (Figs. 5D and 8). Thus, the architecture of these plants resembles transgenic plants over-accumulating IAA due to the expression of bacterial iaaM gene (Romano et al., 1995). In contrast, DN-rop2 plants show reduced shoot apical dominance; a strong DN-rop2 line (DN2-4) produced an average of approximately 12 inflorescence shoots per plant (Fig. 8). The number of shoots per plant was variable among DN2-4 T3 plants, although most of them had more shoots than wild-type plants. Furthermore, DN-rop2 plants exhibited different degrees of dwarfism. This phenotypic variation was probably caused by different copy numbers of the DN-rop2 gene present in the heterogeneous siblings.
Figure 8.
Effects of CA-rop2 and DN-rop2 expression on shoot apical dominance. The number of inflorescence produced from each rosette was scored from 40-d-old plants grown at 22°C in growth room (33 μmol m−2 s−1 light, 16-h light/8-h dark; n = 20). Error bars show sd.
CA-rop2 and DN-rop2 Alter Organ Morphogenesis
As shown in Figure 5, F and G, CA-rop2 and DN-rop2 expression also drastically altered leaf morphology. WT Col-0 rosette leaves are oval-shaped in both early and late stages. CA-rop2 leaves, especially in early stage, became more or less diamond-shaped. The widest portion of CA-rop2 leaves is near the tip of the leaf, whereas the middle portion of the WT leaf is widest. In contrast, DN-rop2 leaves are rather irregularly shaped, the base of the leaf usually becomes widened. CA-rop2 leaves are longer both in leaf blades and petioles, whereas DN-rop2 leaves are much shorter than WT. The ratio of the long to wide axis in CA-rop2 leaves is greater (3.21 ± 0.32) compared with WT leaves (2.31 ± 0.20), but smaller in DN-rop2 leaves (1.91 ± 0.16), suggesting that rop2 mutants altered leaf polarity.
Both CA-rop2 and DN-rop2 leaves are more curly than WT. It is interesting that CA-rop2 leaves curl along the long axis, similar to Arabidopsis transgenic plants overproducing IAA (Romano et al., 1995), whereas DN-rop2 leaves tend to curl along the wide axis. Although the overall leaf size is not significantly affected in CA-rop2 plants, DN-rop2 leaves are generally much smaller than WT leaves. Hence, DN-rop2 leaves resemble rosette leaves of auxin-resistant or BR-insensitive mutants. Finally, CA-rop2 leaves are highly serrated, and the waxy appearance of the leaf abaxial surface disappears so that the texture of the abaxial side appears similar to that of the adaxial side (data not shown).
In contrast to near round-shaped WT Col-0 cotyledons, cotyledons in CA-rop2 seedlings are elongated, whereas DN-rop2 cotyledons are slightly smaller and rounder (Fig. 5G). Floral organ morphology was also affected in rop2 transgenic plants. In general, CA-rop2 plants have larger floral organs resulting in larger flowers, whereas DN-rop2 flowers are slightly smaller (data not shown).
CA-rop2 and DN-rop2 Alter the Spatial Control of Organ Development
Mechanisms that determine the orientation of plant organs are poorly understood. We found that the expression of rop2 altered organ orientation in the shoot. CA-rop2 plants have increased angles for lateral branches and siliques, whereas DN-rop2 plants have reduced angles (Fig. 5E). For example, WT Col-0 plants normally bear siliques at an angle of approximately 60 degree from the shoot axis, but CA-rop2 plants frequently show a silique angle of nearly 90 degrees, and occasionally greater than 90 degrees. DN-rop2 siliques are normally formed at an angle of much smaller than 60 degrees. Phyllotaxis changes of siliques are also very common in CA-rop2 plants. WT siliques are positioned on the stem in a spiral pattern with an angle approximately 137 degrees between adjacent siliques. In CA-rop2 plants, adjacent siliques form an angle much smaller or greater than 137 degree. Similar phyllotaxis alterations were also observed for lateral inflorescence (data not shown). These observations suggest that Rop is involved in the control of the orientation and the positioning of lateral organs of the shoot.
CA-rop2 Affects Embryo Development
We found that CA-rop2 plants had reduced seed setting and that siliques from these plants are wrinkled and deformed, suggesting a likely defect in embryo development for a portion of ovules. Siliques from heterozygous CA-rop2 plants contain aborted embryos (Fig. 5H), although embryo development up to the torpedo stage appears to be normal. Because we were unable to obtain homozygous CA-rop2 plants, these observations suggest that two copies of the CA-rop2 gene cause defect in late embryogenesis or embryo maturation. To further confirm that CA-rop2 mutants did not cause defect in gamete development or maternal effects on embryo development, we conducted a reciprocal cross between heterozygous CA-rop2 plants and WT plants. The rate of transmission of the mutant gene was estimated based on percentage of plants with kanamycin resistance and CA-rop2 cotyledon phenotypes. The ratio of CA-rop2 and WT plants was 1:1 in the F1 progeny from either cross (i.e. either WT or CA-rop2 plants as pollen donor) (data not shown). Taken together, these results indicate that high levels of CA-rop2 expression are lethal to the embryo probably during late stages of embryo development or embryo maturation.
Association of Rop2 Expression Patterns with the Phenotypes of rop2 Transgenic Plants
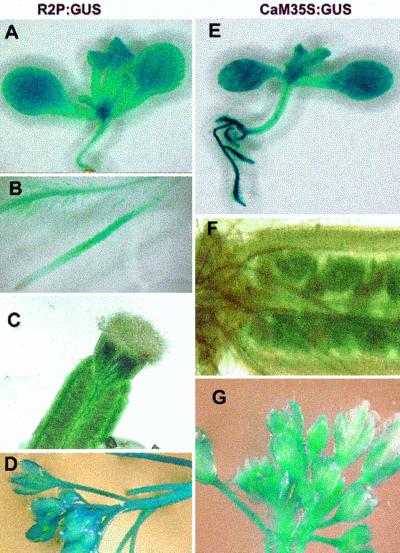
We expect that at least some Rops may have redundant cellular functions, although they may have distinct developmental function due to distinct developmental expression patterns. Thus, some of the phenotypes of rop2 transgenic plants described above may be indicative of the function of other members of the Rop gene family. To assess which aspects of the phenotypes do not reflect the developmental function of Rop2 as a result of the 35S promoter-mediated ectopic expression of rop2, we compared spatial Rop2 expression pattern with the 35S promoter expression using promoter:GUS fusion analysis. As shown in Figure 9, consistent with constitutive accumulation of Rop2 transcripts in different organs, the 0.9-kb Rop2 5′-flanking sequence directs GUS expression in all organs. In leaves, cotyledon, sepals, and petals, Rop2:GUS is constitutively expressed in all cells, although GUS expression is somewhat stronger in vascular bundles (Fig. 9, A and D). This expression pattern is very similar to that of 35S:GUS (Fig. 9, E and G). Thus, the cotyledon and leaf phenotypes observed in rop2 transgenic plants most likely reflect the physiological function of Rop2.
Figure 9.
Expression patterns for Rop2:GUS (R2P:GUS) in comparison with CaMV 35S:GUS expression. A through D, R2P:GUS; E through G, 35S:GUS. A and E, GUS expression patterns in 10-d-old seedlings. B, R2P:GUS expression patterns in roots. C and F, GUS in young pollinated siliques. D and G, GUS expression in floral buds.
In hypocotyls and stems, Rop2:GUS expression is primarily restricted to vascular bundles, little expression is found in parenchyma and epidermal tissues. In contrast, 35S:GUS expression occurs in all cell types in these organs. Furthermore, Rop2:GUS expression is mainly found in the elongation and differentiation zones of the root but not in root tips. This expression pattern is different from the 35S:GUS expression, which is ubiquitous in roots. Finally, Rop2:GUS is primarily expressed in the walls of carpels (Fig. 9C), but 35S:GUS is only expressed in the ovules but not in the carpel walls. Thus, the embryo phenotype induced by CA-rop2 expression most likely reflects the function of a Rop, which is different from Rop2. The rop2 transgenic phenotypes that are inconsistent with Rop2 expression likely reflect the function of other Rops closely related to Rop2, including Rop3, Rop4, Rop5, and Rop6 (Li et al., 1998; Zheng and Yang, 2000).
DISCUSSION
Our studies using transgenic expression of rop2 strongly suggest that the Rop-family GTPases control many distinct developmental processes in plants. Processes affected by the mutant genes include embryo development, seed dormancy, seedling development, shoot apical dominance, lateral root initiation, morphogenesis and orientation of shoot lateral organs, and phyllotaxis. Furthermore, we have shown that CA-rop2 and DN-rop2 expression generally causes opposite effects on the transgenic phenotypes. In addition, overexpression of the WT Rop2 gene did not result in obvious phenotypic changes observed in plants expressing CA-rop2 mutants (data not shown). This observation suggests that CA-rop2-induced phenotypes are due to the activation of specific Rop-dependent pathway(s) and that signal-mediated Rop regulation plays a critical role for the function of Rop. Taken together, our results provide evidence that Rop acts as a molecular switch in multiple signaling pathways that control a wide spectrum of plant growth and developmental processes in Arabidopsis.
Rop Signaling and Plant Organ Morphogenesis
The results described in this report indicate that Rop modulates morphogenesis of aerial organs including cotyledons, leaves, and floral organs. Because of the complex nature of leaf development, mechanisms underlying leaf morphogenesis remain poorly understood. Both hormonal and biophysical cues have been implicated in the modulation of leaf morphogenesis (Van Volkenburgh, 1999). However, to what extent and how each of these cues determines organ morphology are unclear. Rop may provide an important marker for investigating signaling mechanisms underlying organ morphogenesis in plants.
One possible mechanism by which Rop modulates organ morphogenesis is its control of cell morphogenesis. Our studies suggest that Rop2 has a role in the modulation of cell morphogenesis (Fu, Li, and Yang, unpublished results). However, the observed effects of CA-rop2 and DN-rop2 expression on cell morphogenesis seem unlikely to fully account for the dramatic alteration in leaf shapes induced CA-rop2 and DN-rop2. Rop may also modulate leaf morphogenesis via a hormone-dependent mechanism. The vertical curling in CA-rop2 leaves resembles IAA-overproducing transgenic plants (Romano et al., 1995). Likewise, the leaf DN-rop2 morphology is similar to auxin-resistant mutants (Timpte et al., 1994; Hobbie and Estelle, 1995). Furthermore, DN-rop2 leaves are similar to rounded leaves caused by mutations in the ROT3 gene, which encodes a cytochrome P450 involved in BR biosynthesis (Kim et al., 1998). Finally, CA-rop2 induces leaf serration, a phenotype resembling Arabidopsis leaves ectopically expressing the KNAT1 gene associated with increased levels of cytokinin (Chuck et al., 1996). The similarities in leaf morphology between transgenic plants expressing rop2 and hormone-response mutants are interesting, given that Rop may regulate hormone responses as discussed below.
A Potential Rop Involvement in the Regulation of Auxin and/or BR Responses
Our results suggest that one or more Rops may be involved in the regulation of plant responses to auxin and/or BRs. First of all, many of the phenotypes induced by CA-rop2 and DN-rop2 expression are reminiscent of mutants or transgenic plants altered in responses to these hormones or their accumulation. CA-rop2 adult plant phenotypes, e.g. increased shoot apical dominance, later root formation, and vertical leaf curling, are similar to the phenotypes caused by iaaM gene overexpression in Arabidopsis (Romano et al., 1995). On the contrary, DN-rop2 phenotypes, including reduced shoot apical dominance and lateral root formation, resemble several auxin-resistant mutants (Hobbie and Estelle, 1995; Timpte et al., 1995; Ruegger et al., 1997). More importantly, our results show that CA-rop2 and DN-rop2 expression causes alteration in responses to treatments with exogenous IAA and BR. CA-rop2 caused enhanced responses to IAA stimulation of lateral root formation, and the response in CA-rop2 plants reached a plateau at a much lower IAA concentration compared with WT plants. In agreement with these CA-rop2 effects, DN-rop2 dramatically reduced IAA stimulation of lateral root formation. However, IAA inhibition of primary root growth was not affected by CA-rop2 and DN-rop2 expression. Hence, Rop2 may control lateral root formation by increasing sensitivity of IAA stimulation of lateral root formation, promoting the accumulation of auxin at the site of lateral root initiation, or participating in specific auxin signaling pathways that control lateral root formation. The auxin response-related aerial phenotypes in CA-rop2 and DN-rop2 plants support a role for Rop in specific auxin-signaling pathways or auxin transport. Further studies including the use of specific Rop knockout mutants and various auxin-resistant mutants should help to clarify the potential role for Rop in mediating auxin responses.
Some DN-rop2 adult phenotypes, including dwarfism, decreased shoot apical dominance, and reduced leaf size and length/width ratio, are also similar to BR-insensitive or -synthetic mutants (Clouse et al., 1996; Fujioka et al., 1997; Schumacher and Chory, 2000). BR is known to stimulate hypocotyl elongation of light-grown seedlings. It is interesting that CA-rop2 expression also promotes hypocotyl elongation under relatively high light intensity. It is important that light-grown CA-rop2 seedlings are more sensitive to BR stimulation of hypocotyl elongation, whereas DN-rop2 seedlings are less sensitive. These results are consistent with the involvement of Rop in the positive regulation of BR responses. However, this notion appears to contradict the effect of CA-rop2 expression in dark-grown seedlings, i.e. cotyledon opening and reduction in hypocotyl elongation, a phenotype associated with BR-insensitive or -biosynthetic mutants. One explanation for this apparent contradiction is that CA-rop2 expression exaggerated the negative feedback regulation of BR, because high BR levels accumulate in dark and exogenous BR application inhibits hypocotyl elongation in dark-grown seedlings. Rop alternatively might be a positive regulator of photomorphogenesis that is independent of BR regulation.
It is interesting that Rop is associated with the responses to both auxin and BR, the two hormones triggering many parallel responses. Several possibilities could explain this observation. First, Rop could integrate distinct BR and auxin signaling pathways to produce the observed overlapping effects. Second, a single Rop or different Rops could participate in the regulation of respective auxin and BR responses or biosynthesis. Last but not the least, Rop signaling could act to cross-talk between the regulatory pathways leading to the auxin- and BR-dependent processes. Clearly further studies are needed to understand how Rop is involved in the regulatory function of the two important plant hormones.
Rop Negatively Regulates ABA-Mediated Seed Dormancy
Our results indicate that Rop is a negative regulator of seed dormancy. We have shown that CA-rop2 or DN-rop2 expression respectively promotes or inhibits the germination of freshly harvested seeds. Cold treatments eliminate the effect of rop2 on seed germination, indicating that Rop specifically affects seed dormancy. More importantly, germination of DN-rop2 seeds is hypersensitive to the inhibition of seed germination by ABA, whereas CA-rop2 expression reduces the sensitivity of ABA inhibition of germination. These results suggest that Rop may be a negative regulator of ABA responses. It is interesting that protein farnesylation has been shown to participate in the negative regulation of ABA responses (Qian et al., 1996; Pei et al., 1998). Farnesylated proteins contain a C-terminal CAAX motif (C, Cys; A, an aliphatic amino acid; and X, any amino acid except for Leu). Although Rop2 contains the C-terminal CAFL (a geranylgeranylation motif), two other Rops (Arac7 and Arac8) contain a farnesylation motif (Li et al., 1998; Zheng and Yang, 2000). Thus, it is possible that these Rops are farnesylation targets involved in the negative regulation of ABA responses.
An alternative explanation for the negative effect of Rop on ABA-mediated seed dormancy is that Rop signaling antagonizes the ABA effect on seed dormancy. It is interesting that a recent study suggests that BRs may also antagonize ABA-mediated seed dormancy (Steber and McCourt, 2001). This is consistent with our finding that Rop signaling promotes BR-dependent hypocotyl elongation. Nonetheless it remains to be determined whether Rop acts as a negative regulator in the ABA signaling pathway and/or as a positive regulator in a pathway antagonizing ABA-mediated seed dormancy.
Rop Is Involved in Embryo Development
Embryo lethality in homozygous CA-rop2 plants suggests Rop involvement in embryo development. Because Rop is known to control cell polarity development in pollen tubes (Li et al., 1999) Rop might participate in the control of zygote polarity or proper spatial pattern of cell division critical for early embryogenesis (Laux and Juergens, 1997; Souter and Lindsey, 2000). However, our preliminary analyses suggest that all embryos in CA-rop2 plants appear normal up to the torpedo-stage. Thus, CA-rop2-induced embryo abortion is apparently not due to its effect on cell polarity development. Furthermore, alteration in cell polarity and division patterns during early embryogenesis generally causes defects in body patterning but not embryo abortion. However, it is probable that a different Rop is involved in the control of cell polarity and division patterns that is not significantly affected by CA-rop2 or DN-rop2 expression. This is consistent with our observation that seedlings with one cotyledon or three cotyledons are occasionally found in the severe DN-rop2 (DN2-4) siblings (H. Li, J.-J. Shen, Z.-L. Zheng, Y. Lin, and Z. Yang, unpublished data). The abortion phenotype in CA-rop2 embryos supports a role for Rop in the regulation of embryo maturation or viability maintenance.
Rop and Organ Orientation and Phyllotaxis
We have shown that the expression of rop2 alters both phyllotaxis and orientation of lateral organs in the shoot. CA-rop2 expression altered the spiral arrangement of lateral branches and siliques on the stem. Although the mechanism for phyllotaxis control is not well understood, it likely involves a supracellular patterning mechanism (Bowman et al., 1989; Egea-Cortines et al., 1999). Thus, Rop could be involved in the regulation of pattern formation, analogous to Ras GTPase signaling in the control of photoreceptor cell patterning in Drosophila (Yamamoto, 1994). Because phyllotaxis is determined by the patterning of organ primordium formation in the shoot apical meristem (SAM), Rop could regulate spatio-temporal patterns of cell differentiation or proliferation in SAM. In this regard, it is interesting to note that a Rop-like GTPase interacts with CLAVATA1, a receptor-like kinase known to control SAM maintenance (Trotochaud et al., 1999).
CA-rop2 and DN-rop2 mutants also affect the angle of lateral branches and pedicel in relation to the primary growth axis. The mechanism for the control of this angle is again unclear, but presumably involves signal-mediated differential cell elongation and/or division on adaxial and abaxial sides of the primordia for lateral shoots or flowers. Loss of function mutations in BREVIPEDICELLUS (BP) (Koornneef et al., 1983) and ERECTA1 that encodes a receptor-like Ser/Thr kinase (Torri et al., 1996) causes similar changes in pedicel orientation as CA-rop2 expression. However, Rop is the only intracellular signaling protein known to control the orientation of lateral organs in the shoot. The opposite effects of CA-rop2 and DN-rop2 mutants on the pedicel orientation suggest that Rop signaling controls the orientation of lateral organs in the shoot.
Concluding Remarks
We have shown that the expression of CA-rop2 and DN-rop2 mutants impacts a variety of distinct growth and developmental processes. Rop2:GUS expression patterns suggest that many phenotypes are consistent with the function of Rop2, whereas some phenotypes may be due to the function of other Rop genes. It is probable that the Rop GTPase controls additional developmental processes not revealed by the rop2 mutants for the following reasons. First, the Rop-family proteins are distributed in several tissues (e.g. root apices and anthers) where the expression of these mutants did not cause any obvious phenotypes. Second, the rop2 dominant mutants may not interfere with pathways controlled by distantly related Rops including Rop7, Rop8, Arac7, Arac8, and Arac10.
Our results are consistent with the regulation of Rop by hormonal and developmental signals. However, our current study did not identify specific signals that activate Rop2-dependent pathways. Moreover, the observed phenotypes induced by the rop2 mutants may also be due to the modulation of the Rop switch by environmental cues, given the known relationship between hormones and external cues and the effects of the environment on plant development. Nonetheless, our study provides strong evidence that the Rop GTPase acts as a versatile molecular switch in controlling plant growth and development. Loss-of-function Rop mutants will facilitate testing this hypothesis and defining which pathways are controlled by each Rop or each subset of Rops.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia was used in all experiments described in this paper. To characterize adult phenotypes, WT or transgenic plants were grown at 22°C in growth rooms with a light regime of 8-h darkness and 16-h light (33 μmol m−2 s−1). For shoot apical dominance analysis, the total number of inflorescence from 35-d-old plants was counted. For embryo development analysis, young siliques were peeled and photographed under a dissecting microscope. To characterize seedling or root phenotypes, seeds were plated on a agar medium (one-half Murashige and Skoog salt, 10 g L−1 Suc, and 8 g L−1 phytoagar) after surface sterilization (1 min in 70% [v/v] ethanol and 5 min in 50% [v/v] bleach and 0.05% [v/v] Tween 20) and incubated in darkness or in light (33 μmol m−2 s−1). For lateral root initiation analysis, agar plates were placed vertically for 10 d (8-h dark, 16-h light, 20 μmol m−2 s−1) before lateral roots were counted under a dissecting scope. To determine leaf initiation, plants were grown in tissue culture room (8-h dark, 16-h light, 20 μmol m−2 s−1) for 12 d before visible true leaves were counted. For the measurement of hypocotyl lengths, 10-d-old seedlings grown on agar plates vertically placed in a growth room (8-h dark, 16-h light, 33 μmol m−2 s−1) were used.
Transgenic Expression of the Rop2 Promoter:β-Glucuronidase Fusion Gene
To study the expression pattern for Rop2, a 3.5-kb EcoRI/BamHI genomic fragment flanking the Rop2-coding sequence was subcloned into pBluescript II/SK (Stratagene, La Jolla, CA) to allow the use of a HindIII site at the 5′ end of the genomic sequence. To introduce a SalI site 20 bp downstream of the Rop2 ATG codon, the sense T7 primer and the antisense primer containing a SalI site were used for PCR amplification of the putative Rop2 promoter. The amplified fragment (900 bp upstream of ATG) was digested with HindIII and SalI and then translationally fused with the GUS gene in pBI101.2 Vector (CLONTECH Laboratories, Palo Alto, CA). The resulting plasmid, designated as pBR2P:GUS, was introduced into Arabidopsis by Agrobacterium-mediated transformation as described below.
Transgenic plants were examined for GUS expression using a histochemical GUS activity assays as described (Jefferson et al., 1987). All plants showed similar GUS staining patterns and GUS expression, and a representative line was examined in details.
Generation of rop2 Mutants
To create CA and DN mutations for Rop2, we amplified the Rop2-coding sequence (Li et al., 1998) using PCR and subcloned it into the HindIII and XbaI sites of pSELECT (Promega, Madison, WI). Site-directed mutagenesis was conducted by using ALTERED Sites in vitro mutagenesis system (Promega). Four oligonucleotides containing the desired changes (underlined) were used: G15V, 5′-GTCGGAGATGTTGCCGTCGG-3′; T20N, 5′-GTCGGAAAAAATTGCATGCTC-3′; Q64L, 5′-CTGCTGGTCTGGAGGACTAC-3′ and D121A, 5′-ACAAAACTCGCTCTTCGAGA-3′. The sites of mutations were confirmed by sequencing. G15V and Q64L mutations are predicted to cause constitutive activation of Rop2 and are designated as CA1 and CA2, respectively; whereas T20 N and D121A mutations are expected to produce DN effects on Rop2 and are designated as DN1 and DN2, respectively. The mutant genes were then cloned into HindIII and XbaI sites of pKYLX vector behind the 35S CaMV promoter with enhancer (Schardl et al., 1987). For overexpression of the wild-type Rop2 gene, the coding sequence was subcloned in the same vector.
Arabidopsis Transformation
The above constructs were introduced into the Agrobacterium tumefaciens GV3101 by electroporation and transformed into Arabidopsis ecotype Columbia wild-type plants by using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transgenic plants were selected on Murashige and Skoog medium (Life Technologies/Gibco-BRL, Rockville, MD) containing kanamycin. Transgenic seedlings were transferred to soil and grown at 22°C in growth room with 16-h-light and -dark cycles. Lines that showed consistent phenotypes in T2 transgenic plants were selected and used for this study. Copy number of the transgene was estimated by the ratio of kanamycin-resiatant plants to kanamycin-sensitive plants in T2 generations. Homozygous lines were selected from T3 generations.
Hormone Treatments
All plant hormones used in this study were purchased from Sigma (St. Louis). For auxin and GA treatments, seedlings were grown in Murashige and Skoog agar plates in the tissue culture room as described above. After surface sterilization, approximately 50 seeds were plated on agar plates with or without IAA or GA. GA amd IAA dissolved in ethanol were added to autoclaved medium immediately prior to plating. Agar plates were incubated vertically at 22°C in the tissue culture room (8-h dark/16-h light, 20 μmol m−2 s−1 light) before growth measurements. For BR treatment, seeds were germinated and cultured in a Murashige and Skoog liquid medium with shaking for 5 d prior to the measurement of hypocotyl lengths, and other conditions were the same as used for auxin and GA treatments.
Seed Dormancy Assays and ABA Treatment
Newly harvested seeds were used in seed dormancy assays. Seeds were plated on agar plates after surface sterilization and incubated in the tissue culture room (8-h dark/16-h light, 20 μmol m−2 s−1 light). Germination was scored as breakage of radicles from seed coat. For ABA treatments, seeds were plated on Murashige and Skoog agar medium containing ABA (Sigma) and cold-treated at 4°C for 4 d prior to incubation in the tissue culture room. Germination was scored at various times after incubation in the tissue culture room.
RT-PCR Analysis of Transgene Expression
To confirm that the CA-rop2 and DN-rop2 transgenic plants express the transgenes, total RNA was isolated from 10-d-old seedlings using the Trizol Reagent (Life Technologies/Gibco-BRL). Two micrograms of total RNAs were used in a 20-μL reverse transcription reaction as described (Li et al., 1998). Four microliters of reaction products were used in a 100-μL PCR reaction. A 5′-Rop2 gene-specific primer (5′GGCCATGGGCATGGCGTCAAGGTTTA) and a 3′-primer (5′CGAACTCAGTAGGATTCTGGTGTG) covering the pKYLX vector sequence in front of the transcription terminator were used to specifically amplified the CA-rop2 and DN-rop2 transgenes. As an internal control, primers for the Arabidopsis Actin 2 gene (5′-CTAGGATCCAAAATGGCCGATGGTGAGG, 5′-GAAACTCACCACCACGAACCAG) were included in the PCR reactions as described (Li et al., 1998).
Western-Blot Analysis of Recombinant Rops and Immunolocalization of Rop Proteins in Arabidopsis Tissues
To investigate tissue distribution of Rop proteins, we used immunolocalization and affinity-purified anti-Rop1Ps antibodies (Lin et al., 1996). To assess whether the antibodies react with all Rops from Arabidopsis, representative Rops from each of the four Rop groups (Rop2, Rop6, Rop7, Rop8, Arac8, and Arac10) were expressed as glutathione-S transferase fusion proteins and used for western blotting analysis using the anti-Rop1Ps antibodies. For immunolocalization, tissues from seedlings and adult plants (Columbia ecotype) were embedded, sectioned, reacted with the anti-Rop1Ps antibodies as described previously (Lin et al., 2001). Preimmune serum was used as a negative control.
ACKNOWLEDGMENTS
We thank members in the Yang laboratory for their helpful discussions and technical assistance.
Footnotes
This work was supported by the U.S. Department of Agriculture and Department of Energy grants (to Z.Y.).
LITERATURE CITED
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, Locascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bischoff F, Vahlkamp L, Molendijk A, Palme K. Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol Biol. 2000;42:515–30. doi: 10.1023/a:1006341210147. [DOI] [PubMed] [Google Scholar]
- Bos J. Ras. In: Hall A, editor. GTPases. Oxford: Oxford University Press; 2000. pp. 67–88. [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Walker JC. Plant transmembrane receptors: new pieces in the signaling puzzle. Trends Biochem Sci. 1996;21:70–73. [PubMed] [Google Scholar]
- Chory J, Wu D. Weaving the complex web of signal transduction. Plant Physiol. 2001;125:77–80. doi: 10.1104/pp.125.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Pear JR, Andrawis A, Stalker DM. Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol Gen Genet. 1995;248:43–51. doi: 10.1007/BF02456612. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang Z. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol. 2001;152:1019–1032. doi: 10.1083/jcb.152.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick JL, Meigs TE, Casey PJ. G proteins II: Gq, G12, and Gz. In: Hall A, editor. GTPases. Oxford: Oxford University Press; 2000. pp. 35–66. [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Vejlupkova Z, Quatrano RS, Fowler JE. Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J. 2000;24:79–90. doi: 10.1046/j.1365-313x.2000.00855.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. Linkage map of Arabidopsis thaliana. J Heredity. 1983;74:265–272. [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH. Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Juergens G. Embryogenesis: a new start in life. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar T, Gotte M, Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 1999;11:1731–1742. doi: 10.1105/tpc.11.9.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wu G, Ware D, Davis KR, Yang Z. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Seals DF, Randall SK, Yang Z. Dynamic localization of Rop GTPases to the tonoplast during vacuole development. Plant Physiol. 2001;125:241–251. doi: 10.1104/pp.125.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wang Y, Zhu J-K, Yang Z. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yang Z. Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell. 1997;9:1647–1659. doi: 10.1105/tpc.9.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. GTP-binding proteins in plants: new members of an old family. Plant Mol Biol. 1994;26:1611–1636. doi: 10.1007/BF00016493. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc Natl Acad Sci USA. 2000;97:14784–14788. doi: 10.1073/pnas.97.26.14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Mumby SM. G proteins I: Gs and Gi. In: Hall A, editor. GTPases. Oxford: Oxford University Press; 2000. pp. 1–34. [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA. 2001;98:759–764. doi: 10.1073/pnas.021273498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science. 1998;282:287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 1999;119:849–858. doi: 10.1104/pp.119.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Zhou D, Ju R, Cramer CL, Yang Z. Protein farnesyltransferase in plants: molecular characterization and involvement in cell cycle control. Plant Cell. 1996;8:2381–2394. doi: 10.1105/tpc.8.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Robson PRH, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6–1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Schiene K, Puhler A, Niehaus K. Transgenic tobacco plants that express an antisense construct derived from a Medicago sativa cDNA encoding a Rac-related small GTP-binding protein fail to develop necrotic lesions upon elicitor infiltration. Mol Gen Genet. 2000;263:761–770. doi: 10.1007/s004380000248. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Chory J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2000;3:79–84. doi: 10.1016/s1369-5266(99)00038-2. [DOI] [PubMed] [Google Scholar]
- Souter M, Lindsey K. Polarity and signalling in plant embryogenesis. J Exp Bot. 2000;51:971–983. doi: 10.1093/jexbot/51.347.971. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Steber CM, McCourt P. A role for Brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. The axr2–1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torri KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;11:393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA. 2000;97:11638–11643. doi: 10.1073/pnas.97.21.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Volkenburgh E. Commissioned review: leaf expansion: an integrating plant behavior. Plant Cell Environ. 1999;22:1463–1473. [Google Scholar]
- Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H. Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1) Proc Natl Acad Sci USA. 1994;91:9554–9558. doi: 10.1073/pnas.91.20.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM. Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- Wu G, Li H, Yang Z. Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop- specific GTPase stimulation. Plant Physiol. 2000;124:1625–1636. doi: 10.1104/pp.124.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D. Signaling mechanisms in induction of the R7 photoreceptor in the developing Drosophila retina. Bioessays. 1994;16:237–244. doi: 10.1002/bies.950160406. [DOI] [PubMed] [Google Scholar]
- Yang Z. Signal transducing proteins in plants: an overview. In: Verma DPS, editor. Signal Transduction in Plant Growth and Development. New York: Springer Wien; 1996. pp. 1–37. [Google Scholar]
- Yang Z, Watson JC. Molecular cloning and characterization of rho, a ras-related small GTP- binding protein from the garden pea. Proc Natl Acad Sci USA. 1993;90:8732–8736. doi: 10.1073/pnas.90.18.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-L, Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol. 2000;44:1–9. doi: 10.1023/a:1006402628948. [DOI] [PubMed] [Google Scholar]