Abstract
Lysine acetylation is a post-translational modification that critically regulates gene transcription by targeting histones as well as a variety of transcription factors in the nucleus. More recent reports have also demonstrated that numerous proteins located outside the nucleus are also acetylated and that this modification has profound consequences on their functions. This review describes the latest findings on the substrates acetylated outside the nucleus and on the acetylases and deacetylates that catalyse these modifications. Protein acetylation is emerging as a major mechanism by which key proteins are regulated in many physiological processes such as migration, metabolism and aging as well as in pathological circumstances such as cancer and neurodegenerative disorders.
Keywords: Acetylation, Metabolism, Migration, Neurodegenerative disorders, Cancer, HDAC, Sirtuins
Introduction
Post-translational modifications of many if not all proteins critically regulate their biological functions. It is now well accepted that these modifications are required to face challenges and stress from the environment, to trigger a variety of processes ranging from cell proliferation, differentiation, autophagy to apoptosis. These modifications, when not properly regulated, also contribute to multiple pathologies such as cancer and auto-immune diseases. Therefore, several disease-related research programs are currently ongoing in order to better understand the pathways that involve these post-translational modifications and to subsequently define new therapeutic targets.
Protein phosphorylation is the most widely studied modification but early evidence suggested that many other protein modifications such as methylation, ubiquitination, sumoylation, O-GlcNacylation and lysine acetylation also occur in vivo. Lysine acetylation, defined as the addition of an acetyl moiety to the ε-amino group of a lysine residue, has been linked for many years to gene transcription. Indeed, nuclear core histones were initially identified as substrates of lysine acetyltransferases (KAT)/lysine deacetyltransferases (KDAC) which catalyses the adding or removal of an acetyl moiety to/from protein substrates in order to promote or repress gene transcription, respectively [1, 2]. The nomenclature for these modifying enzymes was recently updated [3]. Other proteins such as transcription factors, cytoskeletal proteins and metabolic enzymes were subsequently identified as substrates of (de)acetylases, and recent studies dedicated to identifying the so-called “acetylome” demonstrated that hundreds of proteins are indeed acetylated in vivo [4, 5]. Remarkably, a number of non-nuclear proteins were found to be acetylated and many of them are surprisingly located in the mitochondria [4]. Therefore, these recent studies strongly suggest that the biological contexts that involve protein acetylation and their physiological relevance are most likely severely underestimated today.
This review focuses on the physiological and pathological processes that rely on lysine acetylation, with a special emphasis on the substrates located outside the nucleus. We discuss the significant advances that have recently been made on the identity of the substrates and highlight the poverty of information available regarding the identity of the enzymes that catalyse these post-translational modifications.
Acetylation and deacetylation outside of the nucleus: who are the actors?
Since histones were identified as first substrates of proteins that catalyse the removal of an acetyl moiety, these enzymes were termed histone deacetylases (HDAC) even though some of them do not actually target histones. HDACs can be classified into two distinct families. The “classical” family includes 11 members that are structurally related to the yeast Hda1/Rpd3 proteins and require Zn2+ to efficiently promote protein deacetylation [6]. The second family includes the NAD+-dependent yeast sir2 homologues [7]. Histone deacetylases can be further divided into four classes based on their phylogenetic conservation. In this classification, classes I, II (a and b) and IV are the classical HDAC proteins that include 11 members (HDAC1–11), whereas homologues of yeast sir2, referred to as “sirtuins” (SIRTs), represent the class III deacetylases and include 7 members (SIRT1–7).
The subcellular localization of these enzymes supported the notion that protein deacetylation does not exclusively occur in the nucleus. Indeed, whereas human SIRT6 and SIRT7 are nuclear proteins, SIRT3, SIRT4 and SIRT5 are located in the mitochondria while SIRT1 and SIRT2 can be found both in the nucleus and in the cytoplasm, in a cell- and tissue-dependent manner [7]. Similarly, class IIa members (HDAC4, 5, 7 and 9) shuttle between the nucleus and the cytoplasm through the association with partners such as MEF2 and 14-3-3 proteins which promote their nuclear translocation or their cytoplasmic retention, respectively [8]. Although HDAC6 has been shown to be enriched in active but not silent genes [9], this acetylase is mainly cytoplasmic and promotes the deacetylation of multiple substrates such as α-tubulin, Hsp90, cortactin and peroxiredoxins in this cell compartment [10–15]. Surprisingly, very little information is available regarding the acetylases that promote the post-translational modifications of these cytoplasmic proteins. Yet, recent reports that we will now describe strongly indicate that protein acetylation outside the nucleus play critical roles in multiple processes ranging from cytoskeleton remodelling and cell migration to metabolism and aging, and consequently in diseases such as cancer and neurological disorders.
(De)acetylation in aging and age-related diseases
Caloric restriction has been known for decades to extend life span, but the molecular mechanisms underlying these effects were only recently identified and involve protein deacetylation by sirtuins. This post-translational modification is now seen as a major mechanism by which the activity of multiple substrates is regulated, with key consequences on aging and diseases associated with aging [16]. Indeed, sirtuins have been defined as major players linked to longevity and stress tolerance in budding yeast and other lower eukaryotic organisms [17, 18]. Yeast Sir2 (silent information regulator 2) and orthologs act as anti-aging genes, and increasing their activity extends life span in Saccharomyces cerevisiae (yeast), Caenorhabditis elegans (worms) and Drosophila melanogaster (flies) [19–21]. This key property of sirtuins results from their capacity to act as NAD+-dependent protein deacetylases, and the use of the energic intermediate NAD+ as co-factor appears to be crucial for their role in preventing diseases associated with aging [22, 23]. As a proof for sirtuins acting as key mediators of the beneficial effects of caloric restriction, the increased yeast replicative life span seen upon reduction of glucose is abrogated upon invalidation of the SIR2 gene [24]. Although this life-extending effect of sirtuins partially relies on their ability to deacetylate nuclear histones at various loci including ribosomal DNA, a pathway that reduces the rate of formation of toxic ribosomal circles [25], these deacetylases also target a variety of substrates in the mitochondria which have been strongly linked with aging and age-related diseases. Aging is at least the result of oxidation of macromolecules in cells because of the generation of reactive oxygen species (ROS) by mitochondria. SIRT3, a member synthesized by brown adipose tissue in response to cold, allows reduction of ROS when overexpressed [26].
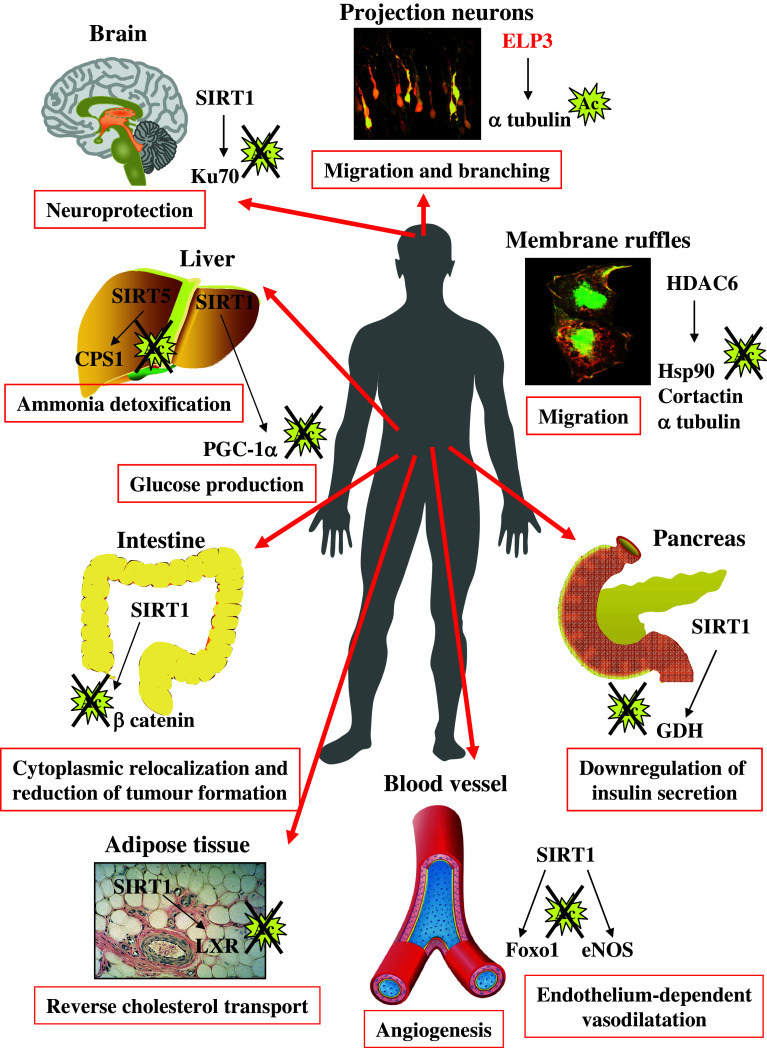
The age-dependent atherosclerosis and more specifically the regulation of lipid and cholesterol metabolism involve SIRT1. Indeed, whereas this sirtuin regulates the activity of the nuclear receptor LXR by targeting its deacetylation, a pathway that promotes the reverse cholesterol transport [27], a conditional deletion of SIRT1 in mouse endothelial cells impairs sprouting angiogenesis as well as branching morphogenesis. Genes involved in blood vessel development and vascular remodelling are consequently downregulated (Fig. 1). Moreover, SIRT1 deficiency in endothelial cells impairs the angiogenic response following an ischemic insult, at least in part because of defective forkhead transcription factor Foxo1 deacetylation [28]. Likewise, endothelial nitric oxide synthase (eNOS) has also been described as a substrate of SIRT1 (Fig. 1). In this case, SIRT1 actually promotes endothelium-dependent vasodilatation by deacetylating eNOS [29].
Fig. 1.
Physiological and physiopathological roles of acetylation or deacetylation of non-nuclear substrates. Deacetylases (sirtuins and HDAC6, in black) and the acetylase ELP3 (in red) are represented with their respective substrates. The biological significance of acetylation or deacetylation of these substrates is indicated. Updated from [102]
Whereas the ability of sirtuins to regulate cellular senescence remains unclear as conflicting reports have been published on this issue [30–32], recent reports illustrated a role for these deacetylases as modulators of autophagy. When over-expressed, SIRT1 induces basal levels of autophagy by forming a complex with autophagy components (Atg5, -7 and -8) whose deacetylations activate the catabolic process [33]. The relevance of this mechanism is supported by overlapping phenotypes of Sirt1- and Atg5-deficient mice as both of them show an accumulation of damages organelles, disruption of energy homeostasis and early perinatal mortality [33].
The positive effects of sirtuins on aging define them as strong targets for therapeutic purposes in neurodegenerative disorders. This was indeed supported by the beneficial effects of caloric restriction and sirtuin activators such as resveratrol in rodent models of neurodegenerative disorders [21, 34–36]. More specifically, one of the first studies that identified sirtuin activation as a protective mechanism in neurodegenerative disorders took advantage of the Wallerian degeneration slow (wilds) mouse strain. A transacted axon is subjected to a self-destructive degeneration process at its distal portion referred to as “Wallerian degeneration”. The wilds mouse line shows delayed axonal degeneration in response to axonal injury due to SIRT1 activation [37]. SIRT1 levels are actually increased as a protective response to neurodegenerative circumstances seen in rodent models of Alzheimer disease or amyotrophic lateral sclerosis [38]. Increased SIRT1 levels are correlated with a decrease in the acetylation state of p53 and PGC-1α, two substrates of SIRT1. Additional substrates such as Ku70, a protein that sequesters the pro-apoptotic Bax protein away from mitochondria, may also contribute to the neuroprotective effect of SIRT1 (Fig. 1) [38]. In conclusion, protein acetylation is emerging as a key mechanism that regulates aging and aging-related diseases, but there also, a lot of work still has to be done in order to identify the acetylases specifically involved in these processes.
(De)acetylation and metabolism
The fact that sirtuins require NAD+ to promote protein deacetylation logically suggested a functional link between sirtuin activity and intracellular energetics. Indeed, acetyl-CoA and NAD+, two co-substrates for (de)acetylations, are key indicators of the cellular energy state. Moreover, the initial link between caloric restriction and sirtuin activation implied that these deacetylases had to play critical roles in metabolism. Caloric restriction may have multiple effects on metabolism, and primary targeted sites are the liver, pancreas, muscle and adipose tissue. Blood glucose levels are maintained during starvation at least in part through hepatic gluconeogenesis, and the sirtuins appear to play critical roles in this physiological adaptation. In this context, a strong link between sirtuin biology and metabolism was established by demonstrating that the peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1 α) was deacetylated by SIRT1 [39–41]. This pathway triggers gluconeogenic gene induction and increases hepatic glucose output [41]. Moreover, PGC-1 α deacetylation by SIRT1 stimulates mitochondrial biogenesis and induces oxidative phosphorylation gene in muscles [42]. As a result, this pathway increases exercise tolerance and thermogenesis, which protects against obesity and associated metabolic dysfunction such as insulin resistance [42]. These conclusions were further supported by the ability of small molecule activators of SIRT1 to improve symptomatic conditions in diet and genetically induced obese mice and in a rat model of diabetes [43].
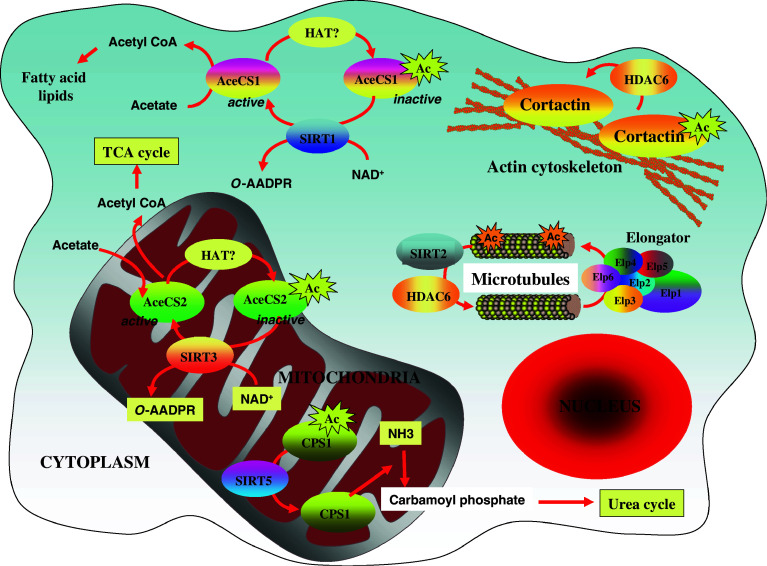
SIRT3 activity has also been linked to metabolism as this enzyme deacetylates and activates the mitochondria form of AceCS2, an enzyme that catalyses the formation of acetyl CoA, an intermediate of the tricarboxylic acid (TCA) cycle, from acetate [44, 45] (Fig. 2). Therefore, SIRT3 improves the metabolic use of acetate, a pathway that ensures energy production under conditions when ATP is scarce [44, 45]. Of note, SIRT1 deacetylates and activates the cytoplasmic AceCS1 to provide acetyl CoA. This latter molecule will then acts as a source for fatty acid and cholesterol synthesis [44] (Fig. 2).
Fig. 2.
Pathways regulated by some sirtuins and HDAC6 in the mitochondria and in the cytoplasm. Substrates of SIRT1, 2, 3 and 5 as well of HDAC6 are illustrated. These pathways regulate the actin cytoskeleton, microtubule (de)acetylations, TCA and urea cycles as well as levels of fatty acids (see text for details)
The functional link between metabolic pathways and the sirtuin-dependent deacetylations in the mitochondria recently got stronger by the demonstration that SIRT5, which is located in the mitochondrial matrix, deacetylates and activates carbamoyl phosphate synthetase 1 (CPS1), the enzyme required for the first step in the urea cycle [46] (Fig. 2). NAD levels increase in liver mitochondria upon fasting and this pathway triggers SIRT5-mediated CPS1 deacetylation and subsequent redirection to amino acid catabolism. Therefore, SIRT5 has been defined as a key actor for ammonia detoxification and disposal [46].
Sirtuins also regulates some metabolic pathways through deacetylase-independent functions. Indeed, pancreatic insulin secretion and thus serum glucose levels are also modulated by SIRT4. This mitochondrial enzyme represses the activity of glutamate dehydrogenase (GDH) through mono-ADP-ribosylation in pancreatic β cells. As a result, both ATP production and insulin secretion by β cells are downregulated in response to amino acids [47]. ADP-ribosylation is a post-translational modification that involves a N- or S-glycolidic linkage between a specific amino acid on the acceptor protein and the ADP-ribose residue of NAD+. GDH catalyses the conversion of glutamate to the TCA-intermediate α-ketoglutarate and this reaction ultimately allows the generation of ATP and stimulates insulin secretion.
These numerous studies strongly indicate that sirtuins, as actors for the regulation of glucose homeostasis and adipogenesis, may be elegant targets for treatments of diabetes and obesity.
(De)acetylation and cell migration
The first experimental link between protein deacetylation and cell migration came from the identification of HDAC6 as an α tubulin deacetylase [12–14]. These studies demonstrated that HDAC6-overexpressing NIH3T3 cells moved faster than control NIH3T3 cells in response to serum and that such process required the deacetylase activity of HDAC6 [13]. These data suggested that HDAC6-mediated α tubulin deacetylation regulates microtubule-dependent cell motility (Fig. 1). However, a subsequent study suggested that HDAC6-mediated microtubule deacetylation was not sufficient to promote cell migration [48]. HDAC6 actually translocates to actin-enriched membrane ruffles in response to growth factor stimulation and becomes associated with macropinosomes where this deacetylase targets Hsp90 [11, 48]. This pathway is required for Rac1 activation, ruffle formation, macropinocytosis and cell motility, suggesting that the actin cytoskeleton was the main target of HDAC6 for the regulation of cell migration, at least in fibroblasts. As far as cell motility is concerned, Hsp90 is not the only substrate whose deacetylation is regulated by HDAC6. Indeed, the F-actin binding protein cortactin was also defined as a substrate of this cytoplasmic deacetylase (Fig. 1) [12]. Cortactin, a protein that is found at the leading edge of migrating cells, is deacetylated by HDAC6, a post-translational modification that triggers its translocation to the cell periphery and enhances its binding to F-actin [12]. Cortactin is also deacetylated by SIRT1 and an inverse correlation was found between SIRT1 levels and acetylated cortactin in breast tissues [49]. All these studies confirmed the critical role of the actin cytoskeleton in the link between protein acetylation and cell migration.
Lysine acetylation in the cytoplasm: which are the enzymes?
Whereas multiple studies identified the enzymes required for the deacetylation of several cytoplasmic substrates, the acetylases that target these proteins remained uncharacterized for many years. It is currently unclear why these acetylases remain unidentified in most cases today. Considering the large number of acetylated proteins in the mitochondria for example, it is worth saying that high levels of acetyl-CoA found in this cell compartment may actually result in lysine acetylation without the need of any enzyme to catalyse this modification. Alternatively, several cytoplasmic proteins are acetylated by KAT which are also located in the nucleus. For example, β-catenin is acetylated by PCAF, a KAT also known to mediate histone acetylation in the nucleus [50]. This latter study thus demonstrates that acetylation of non-nuclear proteins does not necessarily mean that the enzymes involved in these modifications have to be exclusively located in the cytoplasm. It is also worth mentioning that cytoplasmic acetylated proteins are not first modified in the nucleus, as demonstrated for acetylated α-tubulin which is exclusively cytoplasmic.
Elongator, a mainly cytoplasmic KAT
A recent study dedicated to the elucidation of the roles played by a KAT, named Elongator, brought new insights on the mechanisms by which an acetyltransferase is involved in lysine acetylation in the cytoplasm. Elongator is composed of six subunits that include ELP1, the scaffold protein, and ELP3 (also named KAT9 [3]), the catalytic subunit and regulates cell motility, at least by promoting the transcriptional elongation of selected genes coding for proteins involved in cell migration [51–54]. Histone H3 was the first substrate of elp3 to be identified in yeast, yet the mainly cytoplasmic localization of Elongator indicated that other proteins may be targeted by this complex in this cell compartment. This hypothesis was validated by the demonstration that ELP1 or ELP3-depleted colon cancer-derived cells showed decreased levels of acetylated α tubulin [55]. Interestingly, elp1 or elp3-depleted projection neurons also showed defects in cell migration and branching in the developing cortex and those defects were also observed upon expression of a non-acetylable α tubulin mutant in the developing cortex (Fig. 1) [55]. The acetylation of α tubulin serves as a recognition signal for molecular motor anchoring and underlies microtubule-dependent transport of various proteins and organelles [56, 57]. It is thus tempting to suggest that part of the microtubule-dependent intracellular transport may be defective upon Elongator invalidation. Interestingly, this acetylase complex also regulates cell migration via a direct interaction with filamin A [58]. The localization of filamin A in membrane ruffles and the organization of the actin cytoskeleton were defective in Elongator-deficient cells [58]. Therefore, these data indicated that Elongator regulates cell migration by targeting multiple substrates in distinct cell compartments. Although these studies undoubtedly brought some insights into the role played by Elongator, some key questions remained unanswered. Among them, cell migration is equally defective upon HDAC6 or Elongator invalidation, yet those complexes have opposite functions on the levels of acetylated α tubulin. Those data imply that these complexes may not exclusively target the same substrates. In other words, the (de)acetylation of HDAC6 or Elongator-specific substrates may actually critically regulate cell motility.
(De)acetylation and cancer
Numerous clinical trials that involve HDAC inhibitors (SAHA, MS-275, CI-994) have been successfully conducted for the treatment of both haematological and solid tumours [59]. Although initially unexpected because of the wide role of HDAC proteins in gene regulation, the positive effects of HDAC inhibitors appears to be specific for cancerous cells. Indeed, HDAC inhibitors trigger cell cycle arrest, apoptosis, autophagy, differentiation and also inhibit metastasis and angiogenesis of cancerous cells through multiple mechanisms [60]. HDAC inhibitors alter the levels of cell cycle regulatory proteins such as p21WAF1/CIP1 and p27KIP1 and cause Rb hyperphosphorylation [61–67]. They actually trigger cell apoptosis by inducing pro-apoptotic members such as Bax, Bak, Nova, Bim and Puma and also by decreasing the expression of anti-apoptotic proteins such as Bcl-2, Bcl-XL and Mcl-1 [68–72]. As HDAC inhibitors can also induce TRAIL expression, it is now accepted that these drugs trigger apoptosis by linking both death receptor and mitochondrial pathways [70]. In agreement with a pro-apoptotic role of HDAC inhibitors, these drugs also activate p53 by promoting its acetylation on key residues [73]. Importantly, HDAC inhibitors also downregulate positive regulators of cell proliferation such as cyclins D1 and D2, c-Myc and c-Src [74–79]. HDAC inhibitors prevent tumour progression by downregulating angiogenesis-related gene expression [80–88].
Surprisingly, a small percentage of genes are actually influenced by HDAC inhibitors [89–91]. This observation indicates that the mechanisms by which HDAC inhibitors trigger cell death in cancerous cells most likely do not exclusively rely on altered gene transcription. This notion is supported by the ability of HDAC members to target many additional proteins beside histones and transcription factors in the nucleus. Numerous cytoplasmic substrates are indeed targeted by HDAC proteins and the acetylation level of some of them appears to underlie the beneficial effects of HDAC inhibitors for the therapy of solid and haematological cancers.
HDAC6 is probably the best characterized HDAC that targets a variety of cytoplasmic substrates. α-tubulin, Hsp90 and cortactin have indeed all been defined as HDAC6 substrates, and these modifications are relevant in cancer as they critically regulate actin remodelling, dynamics of cell adhesion and motility [11–13, 48, 92, 93]. β-catenin is also deacetylated by HDAC6 and this is required for its nuclear translocation of this substrate through the EGF-dependent pathway [94]. Interestingly, HDAC6 expression is induced upon oncogenic Ras transformation, and fibroblasts deficient for this HDAC member are more resistant to both oncogenic Ras and ErbB2-mediated cell transformations. Several HDAC6-deficient cancer cell lines show a reduced anchorage-independent growth and a decreased ability to form tumours in mice. HDAC6-deficient mice are more resistant to chemical carcinogen-induced skin tumours [95]. Therefore, those data strongly indicate that HDAC6 is a key target that underlies the anti-tumour effects of HDAC inhibitors through deacetylation of non-nuclear proteins. Loss or inhibition of HDAC6 may also interfere with tumour growth by causing accumulation of toxic and misfolded proteins. Indeed, as HDAC6 is involved in the clearance of toxic misfolded protein aggregates through the aggresome-autophagy pathway, tumour cells that produce excessive amounts of misfolded proteins may be efficiently treated with HDAC6 inhibitors [96, 97].
SIRT1 is another deacetylase that critically regulates tumour development. Indeed, SIRT1 deacetylates β-catenin and, in contrast to the modification triggered by HDAC6, this deacetylation appears to promote cytoplasmic localization of β-catenin (Fig. 1). As a result, induction of SIRT1 in a β-catenin-driven mouse model of colon cancer reduces tumour formation [98]. A tumour suppressor function of SIRT1 is also supported by its high levels of expression in normal colon mucosa and benign adenomas compared to lower levels in advanced tumours [99]. Thus, drugs that activate SIRT1 may be of interest, at least for the treatment of β-catenin-driven malignancies. Of note, this conclusion may not apply to all kinds of cancers as another report concluded that Sirt1 deficiency in mice had no effect on the incidence of skin papillomas induced by the classical two-stage carcinogenesis protocol [100].
Conclusions and perspectives
An understanding of the biological relevance of protein acetylation in health and diseases is emerging, yet we are only at the beginning of the story. Despite significant progress, especially in the identity of the acetylated proteins in vivo, key issues remain unanswered. First, we still have to learn more about the enzymes required for this post-translational modification. Significant progress has been recently made on protein deacetylation in the cytoplasm by HDACs or sirtuins, but much less is known on the acetylases involved. Indeed, while it is generally accepted that acetylases such as p300 or CBP target multiple substrates in the nucleus (histones, transcription factors), the hundreds of non-nuclear proteins acetylated are targeted by acetylases whose identity remains unknown in most cases. Very few acetylases besides ELP3 are actually mainly localized in the cytoplasm and in the mitochondria, yet it is assumed that acetylation in those cell compartments requires these enzymes. Given the unexpectedly high number of substrates, it is likely that protein acetylation is as crucial as protein phosphorylation in life. New mice models where these enzymes are specifically deleted are clearly required to exhaustively study their roles in biology. Progress in mice genetics as well as in mass spectrometry-based proteomic analysis will undoubtedly shed more light on these important issues. The activity of these acetylases or deacetylases is also regulated through some post-translational modifications (“regulation of the regulators”) [101]. Here also, key processes were revealed which demonstrated how complex the network might be for the regulation of protein acetylation in distinct cell compartments.
Given the key roles of protein acetylation and deacetylation in biology, it is not surprising to see that multiple clinical trials are currently ongoing in order to develop therapeutic drugs that specifically target the enzymes involved in these reactions. The pharmacological modulation of sirtuin activity that was initiated through the use of resveratrol represents a very promising approach for the treatment of diverse human diseases such as diet-induced metabolic and neurodegenerative disorders or tumourigenesis. Extending our life span by targeting these enzymes may also be conceivable today. Future studies dedicated to the elucidation of sirtuins and HDAC-dependent pathways and to the extensive identification of their substrates should undoubtedly help to design very specific drugs for the treatment of unexpectedly numerous and diverse human pathologies.
Acknowledgments
P.C., L.N. and A.C. are Senior Research Assistant, Research Associate and Senior Research Associate, respectively, of the Belgian National Funds for Scientific Research (F.R.S.-F.N.R.S.). This work was supported by grants from the F.R.S.-F.N.R.S., TELEVIE, the Belgian Federation against cancer, the Concerted Research Action Program (04/09-323, University of Liege), the Inter-University Attraction Pole 6/12 (Federal Ministry of Science), the “Centre Anti-Cancéreux”, the Fonds Léon Fredericq and the Fondation Médicale Reine Elisabeth.
References
- 1.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 3.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 4.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 7.Haigis MC, Guarente LP. Mammalian sirtuins––emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 8.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parmigiani RB, Xu WS, Venta-Perez G, Erdjument-Bromage H, Yaneva M, Tempst P, Marks PA. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, Yao TP, Lane WS, Seto E. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westphal CH, Dipp MA, Guarente L. A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci. 2007;32:555–560. doi: 10.1016/j.tibs.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 18.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 19.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans . Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 21.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 22.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 23.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae . Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 25.Sinclair DA, Guarente L. Extrachromosomal rDNA circles––a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 26.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 28.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Gan Q, Han L, Li J, Zhang H, Sun Y, Zhang Z, Tong T. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS One. 2008;3:e1710. doi: 10.1371/journal.pone.0001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, Murphy MM, Appella E, Alt FW. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 35.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 36.Maswood N, Young J, Tilmont E, Zhang Z, Gash DM, Gerhardt GA, Grondin R, Roth GS, Mattison J, Lane MA, Carson RE, Cohen RM, Mouton PR, Quigley C, Mattson MP, Ingram DK. Caloric restriction increases neurotrophic factor levels and attenuates neurochemical and behavioral deficits in a primate model of Parkinson’s disease. Proc Natl Acad Sci USA. 2004;101:18171–18176. doi: 10.1073/pnas.0405831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 38.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 41.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 48.Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol. 2007;27:8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, Nicosia SV, Zhang X. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 50.Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell. 2009;20:419–427. doi: 10.1091/mbc.E08-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, Siebenlist U, Merville MP, Slaugenhaupt SA, Bours V, Svejstrup JQ, Chariot A. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 53.Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 55.Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 56.Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 58.Johansen LD, Naumanen T, Knudsen A, Westerlund N, Gromova I, Junttila M, Nielsen C, Bottzauw T, Tolkovsky A, Westermarck J, Coffey ET, Jaattela M, Kallunki T. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J Cell Sci. 2008;121:854–864. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 59.Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–1302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- 60.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 61.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan H, Heckman CA, Boxer LM. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol Cell Biol. 2005;25:1608–1619. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lavelle D, Chen YH, Hankewych M, DeSimone J. Histone deacetylase inhibitors increase p21(WAF1) and induce apoptosis of human myeloma cell lines independent of decreased IL-6 receptor expression. Am J Hematol. 2001;68:170–178. doi: 10.1002/ajh.1174. [DOI] [PubMed] [Google Scholar]
- 64.Nome RV, Bratland A, Harman G, Fodstad O, Andersson Y, Ree AH. Cell cycle checkpoint signaling involved in histone deacetylase inhibition and radiation-induced cell death. Mol Cancer Ther. 2005;4:1231–1238. doi: 10.1158/1535-7163.MCT-04-0304. [DOI] [PubMed] [Google Scholar]
- 65.Rosato RR, Wang Z, Gopalkrishnan RV, Fisher PB, Grant S. Evidence of a functional role for the cyclin-dependent kinase-inhibitor p21WAF1/CIP1/MDA6 in promoting differentiation and preventing mitochondrial dysfunction and apoptosis induced by sodium butyrate in human myelomonocytic leukemia cells (U937) Int J Oncol. 2001;19:181–191. doi: 10.3892/ijo.19.1.181. [DOI] [PubMed] [Google Scholar]
- 66.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res. 2003;63:3637–3645. [PubMed] [Google Scholar]
- 67.Strait KA, Warnick CT, Ford CD, Dabbas B, Hammond EH, Ilstrup SJ. Histone deacetylase inhibitors induce G2-checkpoint arrest and apoptosis in cisplatinum-resistant ovarian cancer cells associated with overexpression of the Bcl-2-related protein Bad. Mol Cancer Ther. 2005;4:603–611. doi: 10.1158/1535-7163.MCT-04-0107. [DOI] [PubMed] [Google Scholar]
- 68.Mitsiades N, Mitsiades CS, Richardson PG, McMullan C, Poulaki V, Fanourakis G, Schlossman R, Chauhan D, Munshi NC, Hideshima T, Richon VM, Marks PA, Anderson KC. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- 69.Neuzil J, Swettenham E, Gellert N. Sensitization of mesothelioma to TRAIL apoptosis by inhibition of histone deacetylase: role of Bcl-xL down-regulation. Biochem Biophys Res Commun. 2004;314:186–191. doi: 10.1016/j.bbrc.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 70.Shankar S, Singh TR, Fandy TE, Luetrakul T, Ross DD, Srivastava RK. Interactive effects of histone deacetylase inhibitors and TRAIL on apoptosis in human leukemia cells: involvement of both death receptor and mitochondrial pathways. Int J Mol Med. 2005;16:1125–1138. [PubMed] [Google Scholar]
- 71.Singh TR, Shankar S, Srivastava RK. HDAC inhibitors enhance the apoptosis-inducing potential of TRAIL in breast carcinoma. Oncogene. 2005;24:4609–4623. doi: 10.1038/sj.onc.1208585. [DOI] [PubMed] [Google Scholar]
- 72.Zhang XD, Gillespie SK, Borrow JM, Hersey P. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther. 2004;3:425–435. [PubMed] [Google Scholar]
- 73.Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, Murakami T, Minami S, Matsunaga T, Takayama T, Kato J, Niitsu Y. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res. 2003;63:8948–8954. [PubMed] [Google Scholar]
- 74.Takai N, Kawamata N, Gui D, Said JW, Miyakawa I, Koeffler HP. Human ovarian carcinoma cells: histone deacetylase inhibitors exhibit antiproliferative activity and potently induce apoptosis. Cancer. 2004;101:2760–2770. doi: 10.1002/cncr.20709. [DOI] [PubMed] [Google Scholar]
- 75.Dehm SM, Bonham K. SRC gene expression in human cancer: the role of transcriptional activation. Biochem Cell Biol. 2004;82:263–274. doi: 10.1139/o03-077. [DOI] [PubMed] [Google Scholar]
- 76.Heruth DP, Zirnstein GW, Bradley JF, Rothberg PG. Sodium butyrate causes an increase in the block to transcriptional elongation in the c-myc gene in SW837 rectal carcinoma cells. J Biol Chem. 1993;268:20466–20472. [PubMed] [Google Scholar]
- 77.Lallemand F, Courilleau D, Sabbah M, Redeuilh G, Mester J. Direct inhibition of the expression of cyclin D1 gene by sodium butyrate. Biochem Biophys Res Commun. 1996;229:163–169. doi: 10.1006/bbrc.1996.1774. [DOI] [PubMed] [Google Scholar]
- 78.Souleimani A, Asselin C. Regulation of c-myc expression by sodium butyrate in the colon carcinoma cell line Caco-2. FEBS Lett. 1993;326:45–50. doi: 10.1016/0014-5793(93)81758-r. [DOI] [PubMed] [Google Scholar]
- 79.Takai N, Desmond JC, Kumagai T, Gui D, Said JW, Whittaker S, Miyakawa I, Koeffler HP. Histone deacetylase inhibitors have a profound antigrowth activity in endometrial cancer cells. Clin Cancer Res. 2004;10:1141–1149. doi: 10.1158/1078-0432.ccr-03-0100. [DOI] [PubMed] [Google Scholar]
- 80.Deroanne CF, Bonjean K, Servotte S, Devy L, Colige A, Clausse N, Blacher S, Verdin E, Foidart JM, Nusgens BV, Castronovo V. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 81.Kwon HJ, Kim MS, Kim MJ, Nakajima H, Kim KW. Histone deacetylase inhibitor FK228 inhibits tumor angiogenesis. Int J Cancer. 2002;97:290–296. doi: 10.1002/ijc.1602. [DOI] [PubMed] [Google Scholar]
- 82.Michaelis M, Michaelis UR, Fleming I, Suhan T, Cinatl J, Blaheta RA, Hoffmann K, Kotchetkov R, Busse R, Nau H, Cinatl J., Jr Valproic acid inhibits angiogenesis in vitro and in vivo. Mol Pharmacol. 2004;65:520–527. doi: 10.1124/mol.65.3.520. [DOI] [PubMed] [Google Scholar]
- 83.Mie Lee Y, Kim SH, Kim HS, Jin Son M, Nakajima H, Jeong Kwon H, Kim KW. Inhibition of hypoxia-induced angiogenesis by FK228, a specific histone deacetylase inhibitor, via suppression of HIF-1alpha activity. Biochem Biophys Res Commun. 2003;300:241–246. doi: 10.1016/s0006-291x(02)02787-0. [DOI] [PubMed] [Google Scholar]
- 84.Pili R, Kruszewski MP, Hager BW, Lantz J, Carducci MA. Combination of phenylbutyrate and 13-cis retinoic acid inhibits prostate tumor growth and angiogenesis. Cancer Res. 2001;61:1477–1485. [PubMed] [Google Scholar]
- 85.Qian DZ, Wang X, Kachhap SK, Kato Y, Wei Y, Zhang L, Atadja P, Pili R. The histone deacetylase inhibitor NVP-LAQ824 inhibits angiogenesis and has a greater antitumor effect in combination with the vascular endothelial growth factor receptor tyrosine kinase inhibitor PTK787/ZK222584. Cancer Res. 2004;64:6626–6634. doi: 10.1158/0008-5472.CAN-04-0540. [DOI] [PubMed] [Google Scholar]
- 86.Rossig L, Li H, Fisslthaler B, Urbich C, Fleming I, Forstermann U, Zeiher AM, Dimmeler S. Inhibitors of histone deacetylation downregulate the expression of endothelial nitric oxide synthase and compromise endothelial cell function in vasorelaxation and angiogenesis. Circ Res. 2002;91:837–844. doi: 10.1161/01.res.0000037983.07158.b1. [DOI] [PubMed] [Google Scholar]
- 87.Sawa H, Murakami H, Ohshima Y, Murakami M, Yamazaki I, Tamura Y, Mima T, Satone A, Ide W, Hashimoto I, Kamada H. Histone deacetylase inhibitors such as sodium butyrate and trichostatin A inhibit vascular endothelial growth factor (VEGF) secretion from human glioblastoma cells. Brain Tumor Pathol. 2002;19:77–81. doi: 10.1007/BF02478931. [DOI] [PubMed] [Google Scholar]
- 88.Williams RJ. Trichostatin A, an inhibitor of histone deacetylase, inhibits hypoxia-induced angiogenesis. Expert Opin Investig Drugs. 2001;10:1571–1573. doi: 10.1517/13543784.10.8.1571. [DOI] [PubMed] [Google Scholar]
- 89.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 90.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 92.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 93.Tran AD, Marmo TP, Salam AA, Che S, Finkelstein E, Kabarriti R, Xenias HS, Mazitschek R, Hubbert C, Kawaguchi Y, Sheetz MP, Yao TP, Bulinski JC. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J Cell Sci. 2007;120:1469–1479. doi: 10.1242/jcs.03431. [DOI] [PubMed] [Google Scholar]
- 94.Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ. HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization. J Biol Chem. 2008;283:12686–12690. doi: 10.1074/jbc.C700185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee YS, Lim KH, Guo X, Kawaguchi Y, Gao Y, Barrientos T, Ordentlich P, Wang XF, Counter CM, Yao TP. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 97.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 98.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, Yeatman T, Coppola D, Chen J. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 101.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 102.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]