Abstract
The specific transport of metal ions, mediated by membrane-localized metal transporters, is of fundamental importance in all eukaryotes. Genome-wide analysis of metal transporters was undertaken, making use of whole genome sequences of the green alga Chlamydomonas reinhardtii, the moss Physcomitrella patens, the lycophyte Selaginella moellendorffii, the monocots rice and sorghum, and the dicots Arabidopsis thaliana, poplar, grapevine, as well as of the yeast Saccharomyces cerevisiae. A repertoire of 430 metal transporters was found in total across eight photosynthetic plants, as well as in S. cerevisiae. Seventy-two full-length metal transporter genes were identified in the Populus genome alone, which is the largest number of metal transporters genes identified in any single species to date. Diversification of some transporter family gene clusters appears to have occurred in a lineage-specific manner. Expression analysis of Populus metal transporters indicates that some family members show tissue-specific transcript abundance. Taken together, the data provide a picture into the diversification of these important gene families.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0445-0) contains supplementary material, which is available to authorized users.
Keywords: Expression evidence, Gene diversification, Metal transporters, Populus trichocarpa
Introduction
Copper, iron, zinc, cobalt, nickel, and manganese are metal cations essential for cellular processes, since they act as important cofactors for many enzymes, are components of transcription factors and other proteins, and are essential for both mitochondrial and chloroplast functions. However, when present at high concentrations, along with non-essential metals such as cadmium, mercury, silver, and lead, essential metals can become extremely toxic, since they can cause oxidative damage or compete with other essential ions. Heavy metals are present in soil as natural components or as a result of human activity. The primary sources of metal pollution are the burning of fossil fuels, mining and smelting of metalliferous ores, downwash from power lines, municipal wastes, fertilizers, pesticides, and sewage. Most existing remediation physicochemical technologies (chemical reduction/oxidation, soil washing, and excavation) are meant primarily for intensive in situ or ex situ treatment of relatively highly polluted sites, and thus are not very suitable for the remediation of vast, diffusely polluted areas where pollutants occur only at relatively low concentrations and superficially [1].
Interestingly enough, in the last few years, the possibility of planting metal hyperaccumulator crops or large biomass producers and then harvesting and incinerating the biomass used for bioenergy production has appeared as an alternative technology [2]. According to some authors [3], trees potentially are the lowest-cost plant type to use for phytoremediation. A number of tree species can grow on land of marginal quality. This allows establishment of trees on sites with low fertility and poor soil structure, keeping costs low for plant establishment. Besides, trees have the most massive root systems of all plants, which penetrate the soil for several meters, farther than most herbaceous plants [3]. In some tree species, such as members of the Salicacea family (Populus and Salix species), above-ground biomass can be harvested, and trees will resprout without disturbance of the site.
Plant growth on metal-polluted soils usually depends on their tolerance level to metals at the cellular level. Tolerance to metals is based on multiple mechanisms such as cell-wall binding, active transport of ions into the vacuole, and formation of complexes with organic acids or peptides. A growing knowledge of these factors important to phytoremediation can provide a basis for genetic modification of plants for improved performance. Trees hardly accumulate metal at the level of hyperaccumulator plants [4]. In order to increase metal accumulation capacities by trees, one way would be to increase the number of uptake proteins for the metal of interest in a model tree. Alternatively, the specificity of uptake proteins could be manipulated to take up only the metal of interest, to the exclusion of otherwise competitive substrates, reducing the overall metal load. Employing these methods predictably requires a thorough knowledge of transport proteins involved in metal uptake and sequestration in trees.
Over the past decade, significant progress has been made in elucidating the molecular basis of metal uptake into plant cells [5]. In addition to the transport of inorganic forms of metals [6], a number of studies have been dedicated to the study of chelate-based transport systems [7, 8], some of which are key determinants in metal homeostasis and tolerance in plants [9, 10]. Metal transporters play crucial roles in many aspects of essential and toxic metal distribution in plants. They are involved in the uptake of metals from the soil to root epidermal and cortical cells [11–14]. Subsequently, they are necessary to load metals to the xylem vessels and therefore for metal transfer from roots to shoots and leaves [15]. Within cells, transporters are important for metal storage or sequestration into the vacuole [16–20] as well as their distribution to organelles [21] such as mitochondria [22] and plastids [23, 24]. However, metal transport proteins in perennial species have only been rarely investigated [16, 21]. A better understanding of the biochemical processes involved in tree heavy metal uptake, transport, accumulation, and tolerance will help to systematically improve phytoremediation using molecular genetic approaches. Systematic screening of plant species and genotypes for metal accumulation and resistance will broaden the spectra of genetic material available for optimization and transfer. Therefore, long-term efforts should be directed toward the development of a “gene repertoire” composed of genes valuable for phytoremediation. The availability of the full-genome sequence of poplar (Populus trichocarpa) provides the opportunity to investigate the metal transporter families in this organism [25]. It is worthwhile because functions for metal transporters not detectable in Arabidopsis thaliana could occur in poplar in view of its different life cycle. Poplar is a perennial species and nutrient remobilization preceding the loss of leaves requires metal transport. Furthermore, comparison of members of one gene family between evolutionary related plants may provide some interesting biological insights. Specific transporters, encoded by multigenic families, are responsible for the uptake and secretion of metal ions, and for their sequestration into organelles [6, 26, 27].
In this context, the present paper describes a survey of metal transporters from the copper transporter (CTR), cation diffusion facilitator (CDF), zinc–iron permease (ZIP), cation exchanger (CAX), natural resistance-associated macrophage protein (NRAMP), and heavy metal ATPases (HM-ATPase) families performed in eight plant genome sequences, broadly sampling the plant evolutionary tree (green alga Chlamydomonas reinhardtii, non-vascular moss Physcomitrella patens, lycophyte Selaginella moellendorffii, monocots rice and sorghum, and dicots Arabidopsis thaliana, poplar and grapevine), as well as with members of the S. cerevisiae genome. Finally, based on a search in available EST and Affymetrix data, we describe poplar expression profiles. Although chelate-based transport systems are of importance in plants for tolerance mechanisms, we have limited the present study to transport mechanisms of inorganic compounds to keep a reasonable size.
Materials and methods
In silico genome automatic annotation and manual curation
A HMM seed alignment of representative protein sequences of diverse organisms was retrieved from http://pfam.sanger.ac.uk/ for each transporter family using selected Pfam domains and used to search a customized database containing the genome annotations of Arabidopsis (Arabidopsis thaliana; TAIR release 7; http://www.arabidopsis.org/), Chlamydomonas (Chlamydomonas reinhardtii; JGI release 4; http://genome.jgi-psf.org/Chlre4/Chlre4.home.html), rice (Oryza sativa; TIGR release 6.1); http://rice.plantbiology.msu.edu/), Physcomitrella (Physcomitrella patens ssp. patens; JGI release 1.1; http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html), poplar (Populus trichocarpa; JGI release 1.1; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), sorgho (Sorghum bicolor); JGI release 1.0; http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html), Selaginella (Selaginella moellendorffii; JGI release 1.0; http://genome.jgi-psf.org/Selmo1/Selmo1.home.html), vine (Vitis vinifera; http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html) and yeast (Saccharomyces cerevisiae; http://www.yeastgenome.org/). The final gene models were chosen based on the criteria of full length (with start and stop codons), longer transcript/coding region, and, most importantly, highly similar with Arabidopsis proteins. Retrieved sequences were corrected when a portion of protein was missing due to a wrong gene model prediction. These corrections were supported by EST sequences when possible or were guided by the alignments with homologous proteins. Sequences showing huge truncations and that could not be completed among others by BLAST search were excluded for phylogenetic analyses.
Phylogenetic tree construction
Full-length amino acid sequences were aligned by CLUSTALW and imported into the Molecular Evolutionary Genetics Analysis (MEGA) package version 4 [28]. Phylogenetic analyses were conducted using the neighbor-joining (NJ) method implemented in MEGA, with the pairwise deletion option for handling alignment gaps, and with the equal input correction model with heterogeneous pattern among lineages for distance computation. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed [29]. The phylogenetic trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances [30]. The evolutionary distances are in the units of the number of amino acid substitutions per site. For the ZIP family, a Poisson correction model for distance computation was also used (homogeneous substitution pattern among lineages), to infer phylogeny on all lineages, since several pairwise distances could not be assigned with the equal input correction model (see below).
EST-based expression analysis
For EST-based expression analysis, BLAST search was performed using the cDNA sequences of poplar transporter genes against the EST database (http://www.ncbi.nlm.nih.gov/dbEST/index.html) available at NCBI (Supplemental Table S1). The expression evidence from EST or full-length cDNA for Populus genes was determined by minimal 97% identity over an alignment of at least 100 bp and at least 80% length of the shorter sequences [31]. Note that the ESTs were identified in libraries from different poplar species (P. trichocarpa, P. nigra, P. tremula) and hybrids (P. trichocarpa × P. deltoides; P. tremula × P. tremuloides).
Populus microarray analysis
Transcript accumulation patterns of all Populus metal transporter genes were analyzed using data from PopGenExpress (http://www.bar.utoronto.ca)––an Affymetrix GeneChip-based resource for poplar transcriptome analysis [32]. The expression data are also available for download from the Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) as accession number GSE13990. As Affymetrix microarray data have been “validated” by qPCR countless times (e.g., [32]) and discrepancies in the order of magnitude of transcript abundance are so small as to be negligible, Affymetrix data should merely be viewed as yet another form of transcript abundance data, akin to Northern blot or qPCR. As the manuscript focuses on trends in difference in transcript abundance (i.e., increase in one tissue, decrease in another), and not on the absolute order of magnitude difference in transcript abundance, a second method of transcript abundance analysis was not undertaken. Details of the plant material and data analysis are described in Wilkins [32]. Briefly, all tissues were collected from P. balsamifera trees and total RNA was hybridized to the Affymetrix poplar genome array. All measurements were performed in biological triplicate. GeneChip analysis was performed using the BioConductor Suite in R [33] using the Affy package [34]. The arrays were pre-processed using GC-robust multiarray analysis [35], which uses the intensity of all probes across all arrays to normalize both for probe-specific and array-specific binding effects. Probe sets corresponding to the putative Populus metal transporter gene models were identified using the Probe Match tool on the NetAffx Analysis Center website (http://www.affymetrix.com/analysis/index.affx). Hierarchical clustering, based on Pearson correlation coefficients of transcript accumulations patterns for all probe sets, was performed to group genes with similar transcript accumulation patterns together. The Heatplus package was used to generate heatmaps to display the transcript accumulation patterns.
Supplemental data
Locus name, protein length, CDS length, strand, chromosome, start and end positions (Supplemental Table S2) were found on specific databases: http://www.arabidopsis.org/, http://genome.jgi-psf.org/Chlre4/Chlre4.home.html, http://rice.plantbiology.msu.edu/, http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html, http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html, http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html, http://genome.jgi-psf.org/Selmo1/Selmo1.home.html, http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html and http://www.yeastgenome.org/.
Results and discussion
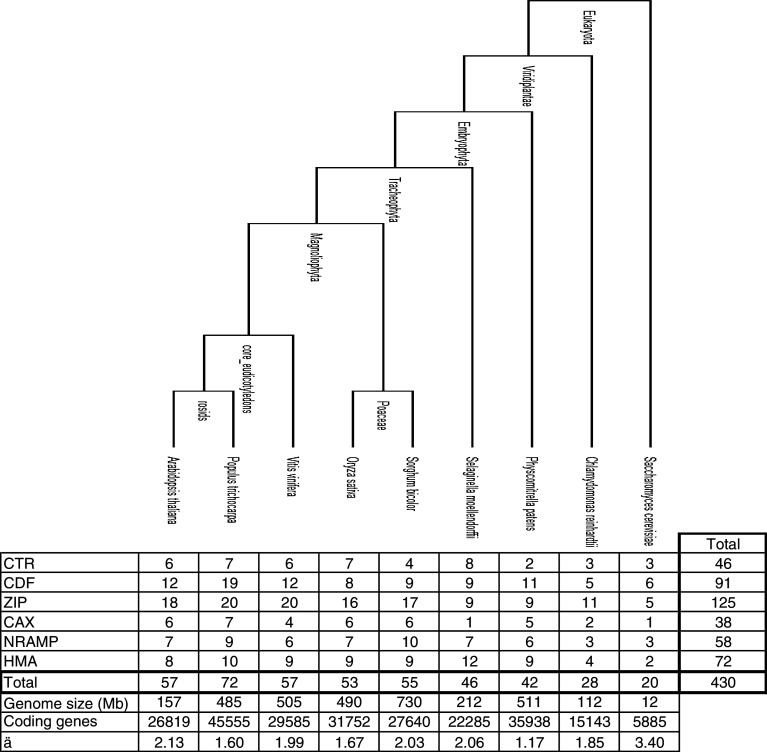
Table 1 lists the six metal transporter families to which belong the 410 metal transporters that we have identified in the eight plants (and the additional 20 genes in S. cerevisiae). These metal transporters represent between 1.17 and 2.1‰ of the total number of coding genes in each of the plant genomes [25, 36–41], which is lower than that found for S. cerevisiae [42]. Here below we describe the metal transporter families. Evolutionary relationships within each transporter family were examined using phylogenetic trees that allow classification in different clusters. Affymetrix Poplar Genome Arrays were used to assess the transcript abundance of metal transporter-encoding genes.
Table 1.
Evolution of the plant metal transporters
On the top side, the phylogenic tree of selected plants is represented. On the bottom side, the number of metal transporter genes from the CTR, CDF, ZIP, CAX, NRAMP, and HMA is given, as well as the genome size and the contribution of metal transporters to the total predicted number of genes for each organism. References for genome data are: At [37], Pt [25], Vv [36], Os [40], Sb [41], Sm (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html), Pp [39], Cr [38], Sc [42]
The copper transporter (CTR) family (TC 1.A.56)
Copper is an essential micronutrient for most organisms, especially in respiring eukaryotes, as it is a cofactor in electron transfer proteins and in enzymes that catalyse redox reactions or oxygen chemistry, such as cytochrome oxidase, ferroxidases, and Cu/Zn superoxide dismutases. Copper deficiency is a challenge encountered by most of the organisms, a challenge they have met through the evolution of sophisticated adaptive mechanisms. Whereas some P-type ATPases function in copper distribution (see below), the CTR family copper transporters function in copper acquisition from the environment or from cellular storage compartments. CTR transporters are constituted by transmembrane polypeptides, containing several copper-binding sequences of functional and/or regulatory value, and assembling as trimers [43]. The CTR family copper transporters were recently included within the channel-type facilitators (http://www.tcdb.org). Copper is transported down a concentration gradient because intracellular copper is immediately sequestered, notably by Cu chaperones.
CTR family copper transporters were first discovered in yeast [44]. Ctr1p and Ctr3p, localized to the plasma membrane, are functionally redundant [45]. A third CTR-type transporter, Ctr2p, mobilizes stored copper from the vacuole under conditions of copper deficiency [17]. CTR transporters are also present in animal and plant cells. Six CTR members (named AtCOPT1-6) were identified in A. thaliana. AtCOPT1, 2, 3, and 5 were functionally characterized [46]. Similarly, three genes were identified and characterized from the alga C. reinhardtii [11]. All these proteins could rescue the mutant phenotype of ctr1 and were described as functional plant Cu transporters.
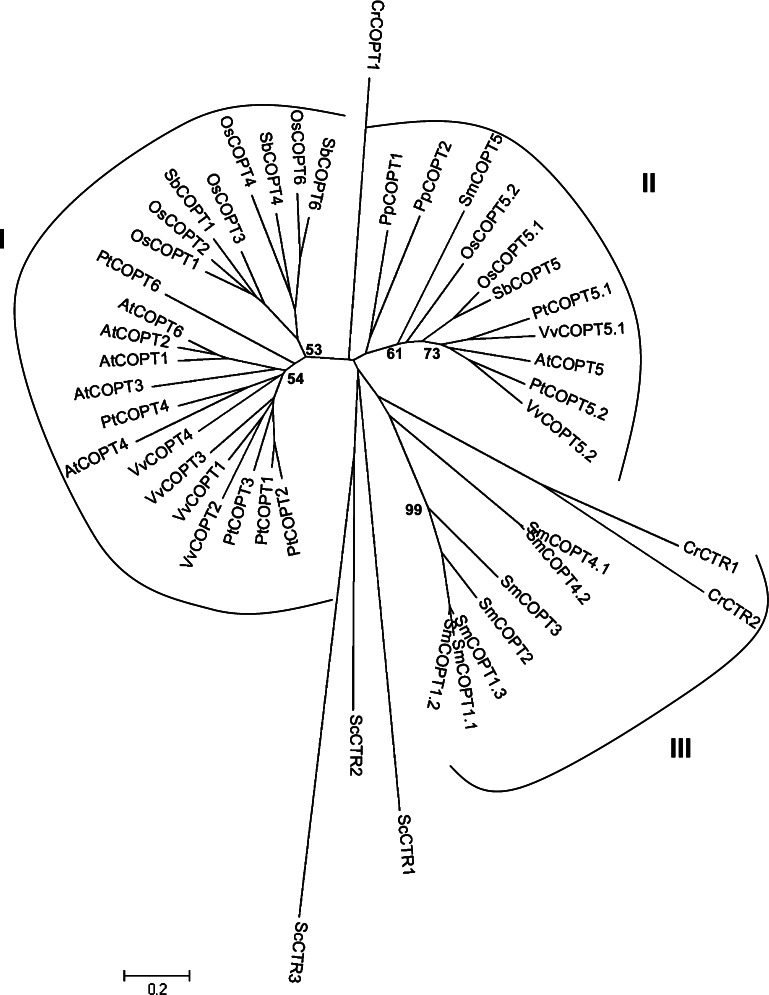
The search for genes encoding CTRs enabled the discovery of a relatively conserved number of genes in the genome of angiosperms (Table 1), with six gene models in A. thaliana and V. vinifera genomes, and seven gene models in P. trichocarpa and O. sativa genomes. However in the genome of S. bicolor, four gene models could only be found. Surprisingly the lycophyte S. moellendorffii genome contains eight gene models. Cluster I includes angiosperm sequences whereas C. reinhardtii, P. patens and S. moellendorffii obviously lacked counterparts in this cluster (Fig. 1). Cluster III includes C. reinhardtii and S. moellendorffii homologues. These C. reinhardtii and S. moellendorffii homologues grouped together with S. cerevisiae CTRs, suggesting a relatively high sequence divergence of these proteins with those from the other photosynthetic organisms. Conversely SmCOPT5 was present in cluster II consisting of either one (A. thaliana and S. bicolor) or two (O. sativa, P. trichocarpa, P. patens, and V. vinifera) CTR members. Group I of CTRs contains between three (for S. bicolor) to five (for O. sativa, P. trichocarpa, and A. thaliana) members.
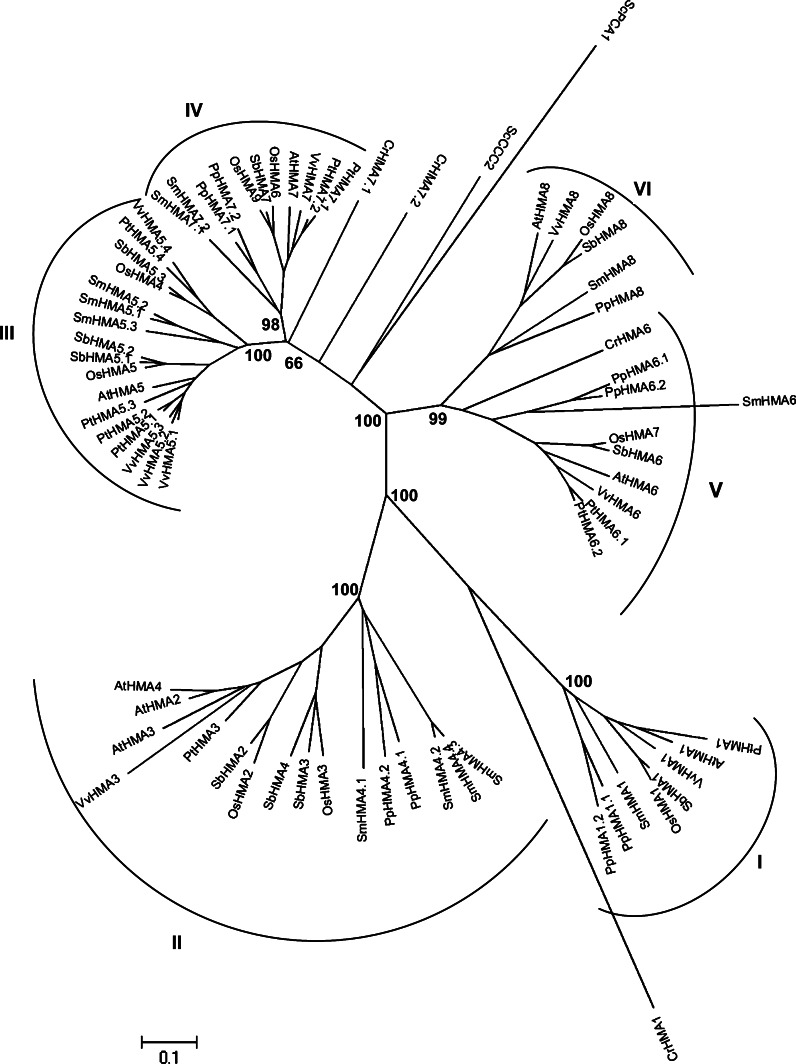
Fig. 1.
An unrooted, neighbor-joining (NJ)-based tree of the copper transporter (Ctr) family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are abbreviated with a two-letter code (At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Os, Oryza sativa; Sb, Sorghum bicolor; Sc, Saccharomyces cerevisiae; Sm, Selaginella moellendorffii; Vv, Vitis vinifera)
Subcellular localization of higher plant CTRs has not yet been reported. Based on localization to the plasma membrane and ability to rescue a yeast mutant defective in high-affinity copper transport, two CTRs from C. reinhardtii were proposed as the assimilatory Cu transporters of this alga [11]. Further studies dealing with the localization of CTR proteins from higher plants are needed to better understand the role of these transporters in Cu homeostasis in model plants.
Each CTR gene is expressed in poplar as revealed by the analysis of EST databases (Fig. 2a). A few ESTs could only be found for PtCOPT3-4,6, whereas PtCOPT1 and PtCOPT5.1 are the CTR transcripts that are the most highly represented in the databases. Interestingly, among the different families of metal transporters, PtCOPT5.1 is the second most highly expressed gene (after PtZIP11.1) with up to 45 ESTs identified. PtCOPT6, PtCOPT4, and PtCOPT3 are the predominant COPT members expressed in xylem, root, and catkins, respectively (Fig. 3). AtCOPT1 functions in soil Cu uptake when Cu becomes limited and it is expressed in both primary and secondary root tips [47]. According to its high level of transcript accumulation in root tissues, it would be tempting to point out a similar role to PtCOPT4. AtCOPT1 is also involved in pollen development [47], a function that may also predominate for its close poplar homologues PtCOPT3 and PtCOPT6, since these genes are highly expressed in poplar male catkins (Fig. 3). The PtCOPT1 transcript is most abundant in leaves, whatever the light treatment.
Fig. 2.
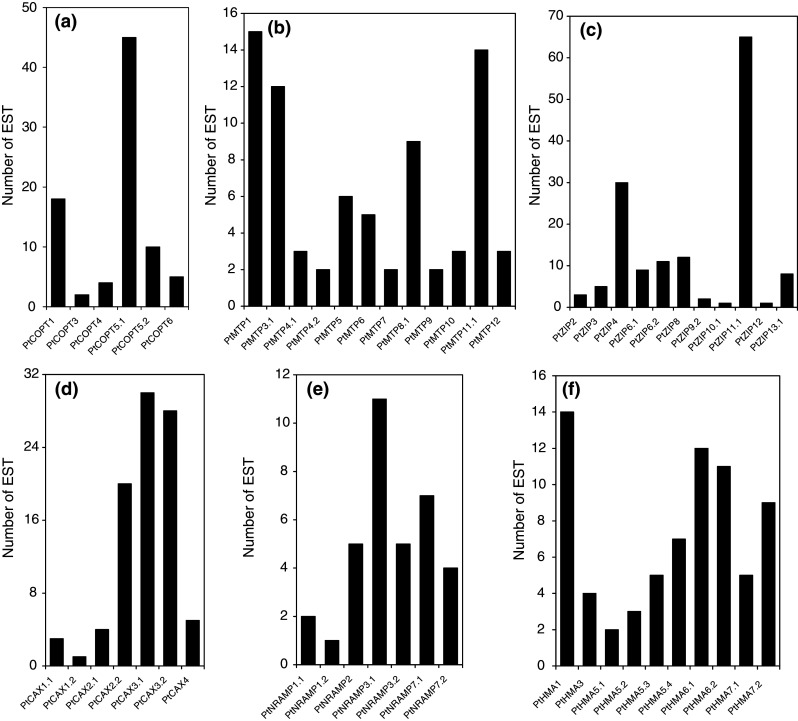
EST-based expression analysis for Populus metal transporter genes. BLAST search was performed using the cDNA sequences of poplar transporter genes against the EST database (http://www.ncbi.nlm.nih.gov/dbEST/index.html) available at NCBI. The expression evidence from ESTs or full-length cDNAs for Populus genes was determined as described in the “Materials and methods” section
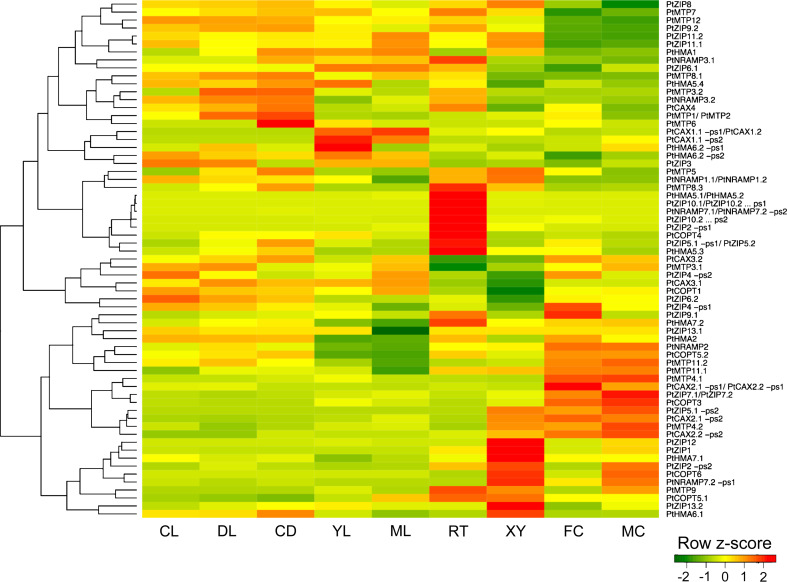
Fig. 3.
Expression of Populus metal transporter genes across a range of tissues, organs, and treatments. The patterns of relative transcript accumulation of each of 65 metal transporter genes as determined by microarray analysis are presented as a heat map, with red indicating higher levels and green indicating lower levels of transcript accumulation. Each column represents the average of three biological replicates. Represented tissues are as follows: CL Seedlings grown in continuous light, DL seedlings grown in continuous darkness and then transferred to light for 3 h, CD seedlings grown in continuous darkness, YL young leaf, ML mature leaf, RT root, XY differentiating xylem, FC female catkins, MC male catkins. Data are normalized within each row
The cation diffusion facilitator (CDF) family (TC 2.A.4)
The CDF transporters first identified by Nies and Silver [27], are ubiquitous, spanning all three kingdoms of life: Archaea, Eubacteria, and Eukaryotes. Studies on CDF transporters may inform strategies for bioremediation and human nutrition and health [6, 27]. CDFs are Me2+/H+ (K+) antiporters that catalyse the efflux of transition metal cations, including Zn2+, Co2+, Fe2+, Cd2+, Ni2+, or Mn2+, from the cytoplasm to the outside of the cell or into subcellular compartments [6, 18, 27, 48]. Phylogenetic analysis classified CDF members into three major groups, which have different selectivity towards the principally transported metal (Zn-, Fe/Zn-, and Mn-CDF), plus one group with unknown metal specificity [49]. The majority of CDF proteins possess six putative transmembrane domains, with cytoplasmic N and C termini, however, some CDF members exhibit different types of secondary structure, like the Msc2 protein of S. cerevisiae, which is predicted to have 12 transmembrane domains. Plant CDF are usually called MTP, for metal tolerance protein.
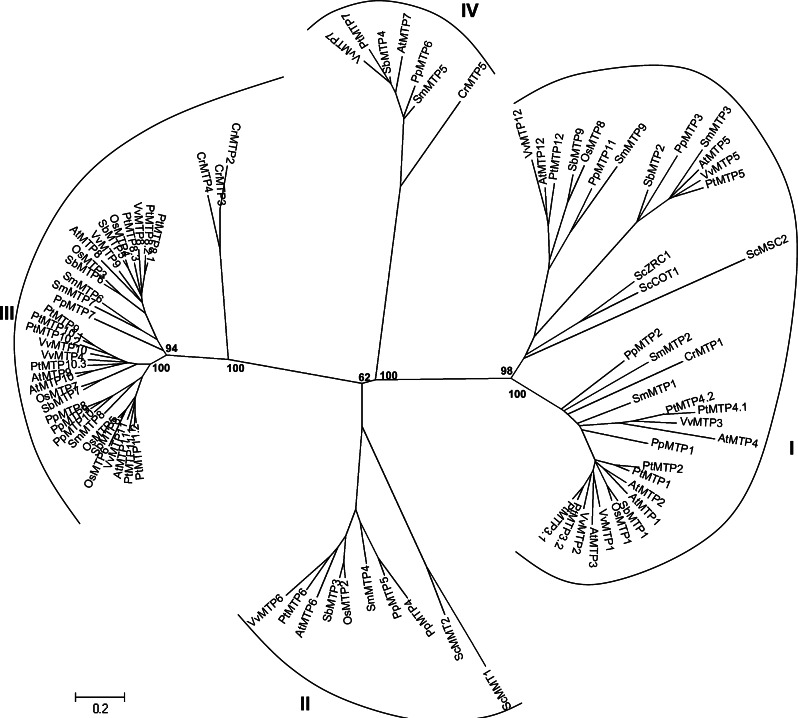
Our phylogenetic analysis, performed on yeast, alga, moss, lycophyte, and several monocotyledons and dicotyledons, confirms the existence of these major groups since all characterized CDF transporters specific for the same cation(s) clustered together (Fig. 4). Cluster I, corresponding to Zn-CDF members, contains the highest number of genes, can be divided into two subclusters typified by the A. thaliana AtMTP1–4 and by AtMTP5 and AtMTP12, respectively. The first subcluster is restricted to photosynthetic organisms since it contains no sequences from fungi as confirmed by previous observations [49]. S. bicolor, O. sativa, and P. patens possess one or two homologues while A. thaliana, V. vinifera, and S. moellendorffii possess three or four homologues and P. trichocarpa possess six homologues. Moreover, we can underline that MTP1, MTP2, and MTP3 proteins of higher plants are very similar and present a sequence identity comprised between 61 and 86%. AtMTP1, AtMTP3, and PtMTP1 have been characterized as Zn transporters and are associated with Zn tolerance by facilitating the Zn transfer into the vacuole [16, 48, 50–52]. All these transporters possess a histidine-rich region between TMD IV and TMD V, which is responsible for Zn binding. With the exception of O. sativa, each plant possesses at least one MTP5 and one MTP12 homologues in the second subcluster. Like ScMSC2, all MTP12 homologues share a 12-transmembrane domain topology. It can be noticed that two MTP5 and two MTP12 members are present in S. moellendorffii, suggesting that a duplication event occurred in this plant during evolution.
Fig. 4.
An unrooted, neighbor-joining (NJ)-based tree of the cation diffusion facilitator (CDF) family in selected plants. Details are given in the legend of Fig. 1. Clusters I, II, and III correspond to Zn-CDF, Zn/Fe-CDF, and Mn-CDF, respectively. For cluster IV no data on metal preference are available
Cluster II includes AtMTP6 homologues from all organisms analyzed. The only exceptions are from P. patens, with two MTP6 homologues and C. reinhardtii with no MTP6 homolog excepted alga. Members of cluster II are Fe and Zn transporters, and correspond to the Fe/Zn-CDF group as described previously [49]. It has been demonstrated that ScMMT1 and ScMMT2 were mitochondrial Fe transporters [22]. To date, no experimental data concerning MTP6 homologues of photosynthetic organisms are available.
Cluster III corresponds to the Mn-CDF group, including homologues from all organisms with the exception of yeasts as previously described [49]. It can be noticed that poplar possesses eight putative Mn transporters whereas the other species possess between four and six Mn-CDF homologues. It may be suggested that duplication events occurred in P. trichocarpa, on MTP8, MTP10, and MTP11 homologues. In particular, MTP8 and MTP10 tandem repeats can be recognized on three linkage groups: LG I (MTP8.2 and MTP8.3), and LG X (MTP10.1, MTP10.2, and MTP10.3), while MTP11 locate on two different LGs on syntenic regions. To date, AtMTP8, AtMTP11, PtMTP11.1, and PtMTP11.2 were demonstrated to be involved in Mn transport, and MTP11 homologues were shown to be localized in Golgi apparatus [18, 21]. The three C. reinhardtii members CrMTP2-4 are included in this group. Another phylogenetic study of the CDF members from alga suggested that they are related to the AtMTP8-11 cluster [53]. The yeast Zn transporter ScZrg17 has homologues only among fungi, so no plant sequence cluster with this ER-localized CDF, and was very distantly related to CDF members [49]. In fact, distances between ScZrg17 and several other members could not be calculated with and it was not included in the tree. AtMTP7 homologues are also present in plants, but to date, there is no biochemical study that could suggest a metal specificity for these members.
We identified 76 ESTs for 12 MTP genes (out of 19) from poplar tissues, revealing that at least two-thirds of the MTP genes are functional in poplar (Fig. 2b). The libraries contained a much higher number of ESTs for PtMTP1 (15 ESTs), PtMTP3.1 (12 ESTs) and PtMTP11.1 (14 ESTs). These data were supported by microarray analysis (Fig. 3). For example, PtMTP8.3 and PtMTP9 had the highest transcript abundance relative to other MTP genes in roots and in xylem, respectively. A subset of five genes (PtMTP1/2, 3.2, 5.4, 6, and 8.1) had the greatest transcript abundance in seedlings relative to other tissues or conditions. Interestingly, PtMTP6 had the highest transcript abundance in leaves under continuous darkness, suggesting a function in metal redistribution under low carbon. Although we could not differentiate PtMTP1 and PtMTP2 on microarrays, the lack of PtMTP2 in EST databases suggest that signals obtained in microarray correspond to PtMTP1, PtMTP4.1, 4.2, 11.1, and 11.2 may be involved in the developmental processes occurring in reproductive organs, as highlighted by their strong expression in these tissues. PtMTP1 and PtMTP11 functions have been well described both in poplar and Arabidopsis [16, 18, 21]. Although genomics has suggested rapid expansion of the CDF family, functional data are lacking for many of the gene family members. The specific expression patterns of some of the poplar CDF members (in xylem, root, or catkins for instance) may help to highlight the function and distribution of these CDF members.
The zinc–iron permeases (ZIP) family (TC 2.A.5)
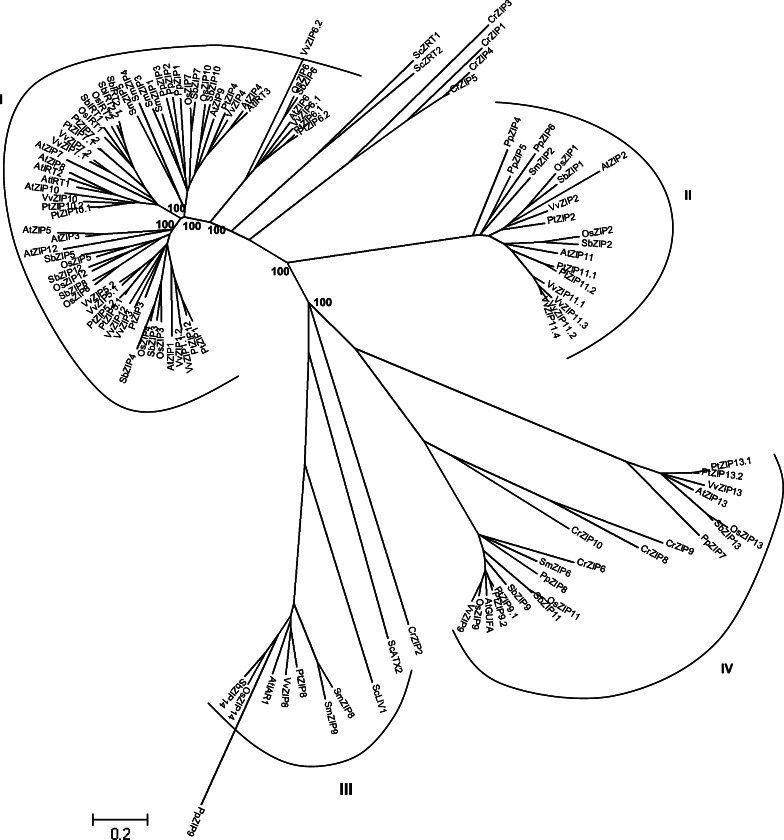
The ZIP family includes the well-characterized members ZRT1 and ZRT2 (zinc-regulated transporter), the high- and the low-affinity zinc transporters from S. cerevisiae and IRT1, the A. thaliana iron-regulated transporter, which is a cation transporter expressed in roots of iron-deficient plants [54]. It has been demonstrated that these proteins transport zinc and/or other metal ion substrates from the extracellular space or organellar lumen into the cytoplasm. ZIP transporters are found in all phylogenetic levels including bacteria, fungi, plants, and mammals [26, 54]. Most ZIP proteins have eight predicted transmembrane domains and similar predicted topologies with the N- and C-termini of the protein located on the extracytoplasmic face of the membrane. ZIP proteins range from 309 to 476 amino acids, this difference is largely due to the length between transmembrane domain III and IV, designated the “variable region”. In most cases, the variable region contains a potential metal-binding domain rich in histidine residues that is predicted to be cytoplasmic. The strong sequence heterogeneity within this loop prevented distance calculations for four proteins (ScZRT3, SmZIP7, CrZIP7, and CrZIP11). Therefore, evolutionary relationships using heterogeneous pattern among lineages were inferred by eliminating these sequences. A tree based on the Poisson correction model for distance computation revealed that these proteins belonged to cluster IV (Fig. S1).
ZIP proteins of eukaryotes have been previously classified into clusters I and II [54]. Cluster I consisted largely of fungal and plant members whereas cluster II was a smaller group of nematodes proteins. Another analysis of over 85 ZIP members identified two additional clusters [26]. The first one, the GUFA cluster (Myxococcus xanthus) includes ZRT3, a zinc transporter from S. cerevisiae. The presence of ZRT3 in the GUFA cluster of proteins clearly links this cluster to potential function in metal ion transport [26]. The LIV-1 subfamily constitutes the second new cluster. As observed in both ZIP and CDF proteins, LIV-1 proteins present a high abundance of histidine residues, suggesting that LIV-1 proteins may play a role in metal ion transport [26]. Several studies have demonstrated that ZIP homologues of cluster II are principally expressed in roots and up-regulated in Fe-deficient roots (i.e., OsIRT1, AtIRT1, and AtIRT2) [12, 55]. Interestingly, AtZIP7 is expressed in both roots and shoots of Arabidopsis under Zn-deficiency treatment [56]. In addition it has been demonstrated that OsIRT2 encodes an Fe transporter of lower affinity than OsIRT1 [13]. Little is known about the cluster III members but it has been shown that AtZIP4 was predominantly expressed in the roots of Zn-deficient [56], OsZIP7.1 was up-regulated in rice shoots by Zn deficiency and OsZIP7.2 was up-regulated in roots under Fe deficiency [57].
As revealed by the phylogenetic analysis (Table 1), the number of ZIP homologues is greater in angiosperms (comprised between 16 and 20) in comparison to P. patens, S. moellendorffii, and C. reinhardtii, which possess nine, nine, and 11 ZIP genes, respectively. The number of ZIP genes in S. cerevisiae is even lower than in plants. In angiosperms, the high number of ZIP transporters would facilitate the absorption of metal ions from the soil and the distribution throughout the plant. Moreover, it is presumed that some of the ZIP proteins will be found to locate to different membranes facilitating the exchange between intracellular components. Our analysis, performed on yeast, alga, moss, lycophytes, and several monocotyledons and dicotyledons confirms the existence of the four clusters described above (Fig. 5).
Fig. 5.
An unrooted, neighbor-joining (NJ)-based tree of the zinc–iron permeases (ZIP) family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are given in the legend of Fig. 1
Cluster I, typified by the A. thaliana ZIP1 and IRT1, includes sequences from most organisms, although the S. cerevisiae and C. reinhardtii ZIP are more distantly related to the other sequences. This cluster corresponds to subfamily I, as previously described [53], and it can be further divided into four subclusters. Phylogenetic analysis reveals that three of these subclusters are restricted to higher plants since they contain no sequences from P. patens, S. moellendorffii, and C. reinhardtii. It can be hypothesized that these clusters appeared later during evolution of the green lineage [53]. The first subgroup is composed of ZIP homologues from monocots and dicots. The number of ZIP homologues is relatively conserved and ranges from four in A. thaliana to six in P. trichocarpa. Few studies on A. thaliana and O. sativa homologues revealed that AtZIP1 and AtZIP3 were predominantly expressed in the roots of Zn-deficient Arabidopsis [56], while OsZIP1, OsZIP3, and OsZIP4 were up-regulated in rice roots and shoots after Zn deprivation. On the contrary, OsZIP2 was up-regulated in rice roots by Zn deficiency and OsZIP5 was only up-regulated in rice shoots by Zn deficiency [55].
The number of subcluster II homologues also is relatively conserved and ranges from two in O. sativa to four in P. trichocarpa. Surprisingly, A. thaliana possesses five ZIP isoforms suggesting that a duplication event occurred in A. thaliana. Several studies have demonstrated that ZIP homologues of this subcluster are principally expressed in roots and up-regulated in Fe-deficient roots (i.e., OsIRT1, AtIRT1, and AtIRT2) [12, 55]. Interestingly, AtZIP7 was expressed in both roots and shoots of Arabidopsis under Zn-deficiency treatment [56]. In addition, it has been demonstrated that OsIRT2 encodes an Fe transporter of lower affinity than OsIRT1 [13].
The third subcluster is restricted to land plants since it does not contain C. reinhardtii ZIP homologues suggesting that this subgroup appeared after the split with green alga. Interestingly, S. moellendorffii and P. patens contain two and three ZIP homologues, respectively, whereas monocots and dicots only possess two and one ZIP homologues, respectively. Except for A. thaliana, which possesses three homologues, it seems that the number of ZIP homologues decreased during evolution and more particular in dicots. Little is known about the cluster III members but it has been shown that AtZIP4 was predominantly expressed in the roots of Zn-deficient [56]. OsZIP7.1 was up-regulated in rice shoots by Zn deficiency and OsZIP7.2 was up-regulated in roots under Fe deficiency [57]. The forth subcluster only contains between 1 and 2 ZIP homologues of each higher plant analyzed. It can be noticed that duplication events occurred in P. trichocarpa and V. vinifera during evolution.
Cluster II, typified by the A. thaliana ZIP2 and ZIP11 proteins also includes representatives from most of the organisms studied, except S. cerevisiae and C. reinhardtii, which lacked obvious counterparts in this cluster. It may be suggested that duplication events occurred in P. trichocarpa and more particular in V. vinifera, which possess four ZIP11 genes that are in tandem on the genomic sequence. Cluster III is made of AtIAR1 and other plant homologues but lacks C. reinhardtii homologues (Fig. 5). The members of this cluster are related to the S. cerevisiae LIV1, a zinc exporter localized to the endoplasmic reticulum, found in all eukaryotes [58] and the S. cerevisiae ATX2, which is involved in manganese homeostasis [59]. Cluster IV contains ZIP homologues from all photosynthetic organisms, including the A. thaliana ZIP13 and GUFA (Fig. 5). All photosynthetic organisms analyzed possess at least one ZIP13 homologue and one GUFA homologues. P. trichocarpa has two ZIP13 and two GUFA homologues derived from a duplication event. Phylogenetic analysis reveals that ScZRT3, which functions in the transport of Zn from the vacuole into the cytoplasm [60], belongs to this cluster (Fig S1). The strong conservation of ZIP13 homologues between the different photosynthetic organisms may reveal an important function of ZIP13 in plant physiology. C. reinhardtii has an unusually high number of ZIP (six) belonging to this cluster (Figs. 5, S1), which was correlated with previous studies [53].
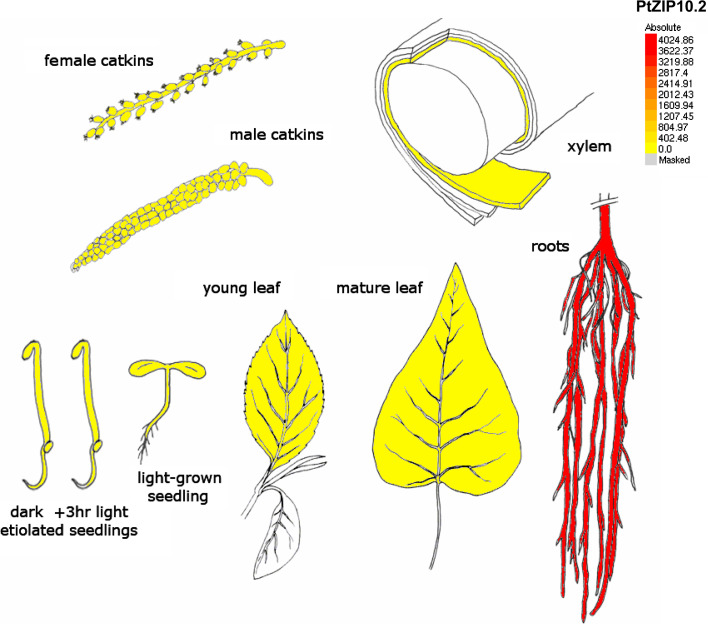
We identified ESTs corresponding to 11 different PtZIPs suggesting that at least half of poplar ZIPs were expressed (Fig. 2c), with PtZIP11.1 having the highest number of EST retrieved for the 72 putative metal transporter genes identified in the Populus genome. Microarray analysis further revealed that ZIP gene transcripts were identified in all of the plant tissues surveyed (Fig. 3). A group of four ZIP genes were highly expressed in the root (PtZIP2, 5.2, 10.1, and 10.2) and another group of genes were characterized by high accumulation in xylem (PtZIP1, 12, and 13.2) tissues, suggesting a key role of PtZIP homologues in the uptake of metal from the soil solution and further transfer to shoots. The Arabidopsis AtZIP2 and AtIRT2, which are close homologues of PtZIP2 and PtZIP10.2 have key function in root metal uptake [12, 61], which is concordant with our data. The specific location of PtZIP10.2 in root tissues is exemplified in Fig. 6. A subset of seven PtZIP genes (PtZIP3, 6.1, 6.2, 8, 9.2, 11.1, 11.2) that were expressed in leaves may indicate a strong involvement of PtZIPs in the control of metal distribution in shoots. A set of 65 ESTs corresponding to PtZIP11.1 retrieved from poplar leaf databases confirms its predominant function in leaf tissues.
Fig. 6.
eFP display of transcript accumulation patterns across a variety of Populus organs and treatments. Poplar eFP browser presents the transcript accumulation pattern of PtZIP10.2 in a variety of tissues and organs. In all cases, red indicates higher levels of transcript accumulation and yellow indicates a lower level of transcript accumulation
The cation exchanger (CAX) subfamily (TC 2.A.19)
CAX proteins belong to the CaCA (for Ca2+: cation antiporter) family. CaCA proteins are integral proteins that transport Ca2+ or other cations using either the H+ or K+ gradient formed by the activity of primary transporters. CAX genes have been found in bacteria, amoeba, lower vertebrates, fungi, and plants. As previously found for the yeast CAX VCX1p, most of the plant CAX proteins (AtCAX1–4, OsCAX1a) are integral proteins of the vacuolar membrane [62]. Two CAXs, namely GmCAX1 and OsCAX3, are present on the plasma membrane [63, 64]. Many plant CAXs are involved in Ca2+ transport and homeostasis, but some of them are also involved in metal homeostasis, especially Mn2+ [62]. Moreover, it has been recently shown that AtCAX4 was required for root development under metal stress [14].
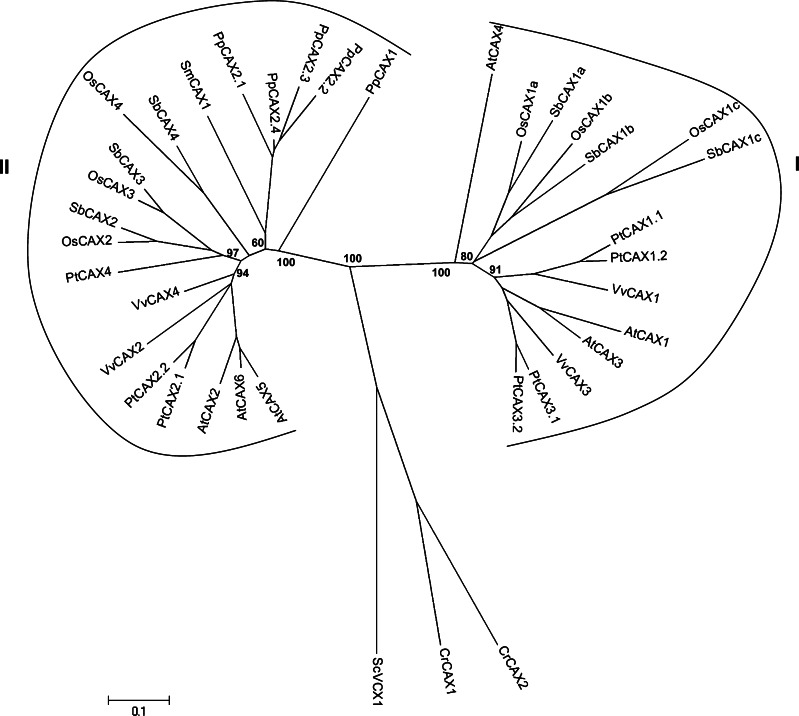
When analyzing 138 full-length CAX polypeptides from species of different kingdoms, three major types (type-I to III) could be identified within this subfamily [62]. The type-I CAXs include members from plants, fungi, and bacteria. Type-II CAXs are found in fungi, Dyctiostelium, and lower vertebrates. Type-III CAXs seem to be more specific to the bacterial kingdom. Within type-I, eight clusters could be identified. Arabidopsis CAXs all clustered either in cluster I (AtCAX1,3,4) or cluster II (AtCAX2,5,6) [62]. In the model plant Arabidopsis, 11 CAX genes were initially named as CAX transporters [52]. However, a recent phylogenetic study revealed that only six of these genes (AtCAX1-6) could be considered as true CAXs [62]. AtCAX7-11 displayed indeed limited primary amino-acid sequence homology with any CAX, suggesting that they likely have different biochemical properties from those of other CAX proteins. Therefore, AtCAX7-11 were renamed as AtCCX1-5 (for Cation Calcium eXchanger). Even if we cannot rule out a possible role of AtCCXs in metal transport, since there is no available information on function for AtCCX1-5, CCX-like genes were not considered in the present study.
A search in the genome of selected plant species revealed a similar number of CAX genes among monocots and dicots (Table 1). O. sativa, A. thaliana, and S. bicolor genomes contain six members, three in each cluster I and II (Fig. 7). Poplar, with seven members, possesses an additional gene in cluster I. According to Tuskan [25], it has to be noted that an event of ancient duplication for PtCAX3.1 and PtCAX1.1 occurred as well as three other recent duplication events for PtCAX1.1 and PtCAX1.2, PtCAX2.1, and PtCAX2.2, PtCAX3.1 and PtCAX3.2. Four members were identified in the V. vinifera genome. A similar number of CAX genes (five) were found in the genome of the moss P. patens whereas a single gene (SmCAX1) was identified in the lycophyte S. moellendorffii (Fig. 7). The CAX members for these two latter organisms are found in cluster II, whereas cluster I lacks sequences from these two organisms. Two members identified in the alga C. reinhardtii are more distantly related to other members of photosynthetic organisms and more closely related to the yeast VCX1. Within cluster II, AtCAX5 and AtCAX6 are located in tandem on chromosome 1, a feature not found for CAXs in the other studied genomes. The close proximity of AtCAX5 and AtCAX6 is therefore likely due to a recent tandem duplication that occurred after the divergence of the Arabidopsis and dicots lineages. AtCAX5 was recently characterized [65] but data are lacking for AtCAX6. Consequently, conclusions about a putative redundancy between these two genes cannot be drawn.
Fig. 7.
An unrooted, neighbor-joining (NJ)-based tree of the CAX (CAtion eXchanger) subfamily in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file and names of the species are given in the legend of Fig. 1
The dichotomy of plant CAXs may be due to different substrate specificities. For instance AtCAX1 (cluster I) is considered to be a specialized calcium transporter, whereas AtCAX2 (cluster II) transports multiple cations including Ca2+, Cd2+, and Mn2+ [66]. Other AtCAX2-like proteins (AtCAX5, HvCAX2, LeCAX2) could also transport Ca2+ and Mn2+ [65]. AtCAX1 is a high-capacity, low-affinity Ca2+/H+ antiporter involved in both signaling events and tolerance to excess Ca2+ [66, 67]. Knockout analysis showed that AtCAX2 (with a lower affinity for Ca2+ transport than AtCAX1) did not have a major physiological role in Ca2+ homeostasis but is important for vacuolar Mn2+ accumulation [68]. However, OsCAX1a (cluster I) could also transport both Ca2+ and Mn2+ [69]. Further exhaustive tests will have to be performed with members of various species to determine whether a broad metal substrate range is a common characteristic to all plant CAXs or solely to CAXs of cluster I.
An alternative hypothesis to explain the dichotomy of plant CAXs emerges from different studies showing different modes of regulation. AtCAX1 is highly expressed in leaf and floral tissues [70] and is induced by elevated Ca2+ [71]. Conversely, AtCAX2 is only expressed at low levels in all tissues and highly induced by any metal [66]. CAXs are also regulated post-translationally. Ca2+ transport activity of AtCAX1 is regulated by a domain on the N-terminal tail through an autoinhibitory process [72]. AtCAX2 is also regulated through a similar autoinhibitory process. However, AtCAX2-like proteins from different plant species including tomato and barley, and also AtCAX5, showed a significant diversity in their functional characteristics, particularly with regard to regulatory mechanisms [65]. Therefore the first hypothesis given above might be more consistent. However, further functional studies are needed to shed light on the origin of this dichotomy in plant CAXs.
In databases, ESTs could be found for all poplar CAX genes suggesting that all of the PtCAX genes identified were expressed (Fig. 2d), which was further confirmed by microarray analysis (Fig. 3). We found a large number of ESTs for PtCAX2.2 (19), PtCAX3.1 (29), and PtCAX3.2 (30). Four sequences belonging to PtCAX2.1 could only be found in databases. A similar finding was found for PtCAX1.1, PtCAX1.2, and PtCAX4, suggesting a low rate of transcript synthesis or expression in specific conditions/cell types. The total number of EST for PtCAX in libraries made from root tissues was relatively low (three EST on a total of 86 retrieved from databases), which was confirmed by the overall low level of expression in roots for these family members (Fig. 3). These observations would suggest that PtCAXs made a minor contribution to Ca2+ or metal homeostasis in poplar roots. However, AtCAX4 was highly upregulated under metal stress conditions and played an important function for root growth under such conditions [14]. PtCAX2.1 and PtCAX2.2 were mostly expressed in reproductive tissues, indicating potential roles of these genes in floral development and/or fruit/seed formation/maturation. PtCAX1.1 and PtCAX1.2 were strongly expressed in leaves, which agreed with the predominant expression of their Arabidopsis closest homolog AtCAX1 in leaves [70].
The natural resistance-associated macrophage protein (NRAMP) family (TC 2.A.55)
The NRAMP family members function as divalent metal transporters in a wide range of organisms, from bacteria to Homo sapiens [73]. Plant NRAMPs also transport a range of divalent metal cations including iron, manganese, cadmium, and zinc [74–80]. Arabidopsis NRAMP1, NRAMP3, and NRAMP4 as well as tomato and apple tree NRAMP1 are up-regulated under iron deficiency [19, 74, 75, 77, 81, 82]. AtNRAMP3 and AtNRAMP4 play a critical role in the mobilization of iron stores during seed germination [83]. They are also involved in Mn export from the vacuoles in mesophyll cells [84]. Arabidopsis plants lacking AtNRAMP3 and AtNRAMP4 are hypersensitive to Cd and Zn [79]. Interestingly, NRAMP3 is constitutively highly expressed in two Zn and Cd hyperaccumulating species, Thlaspi caerulescens and A. halleri [79, 80, 85, 86]. This suggests that NRAMP3 might be involved in increased metal tolerance or accumulation. In addition, AtNRAMP6 was found to modulate Cd sensitivity [78]. Arabidopsis plants lacking AtNRAMP6 are more resistant to Cd than the wild-type, indicating that AtNRAMP6 enhances Cd sensitivity. Recently, it was demonstrated that AtNRAMP1 encodes the high-affinity Mn uptake system in Arabidopsis roots [87]. The decreased Cd sensitivity of nramp1 knockout plant suggests that AtNRAMP1 could also represent a Cd import pathway in Arabidopsis roots.
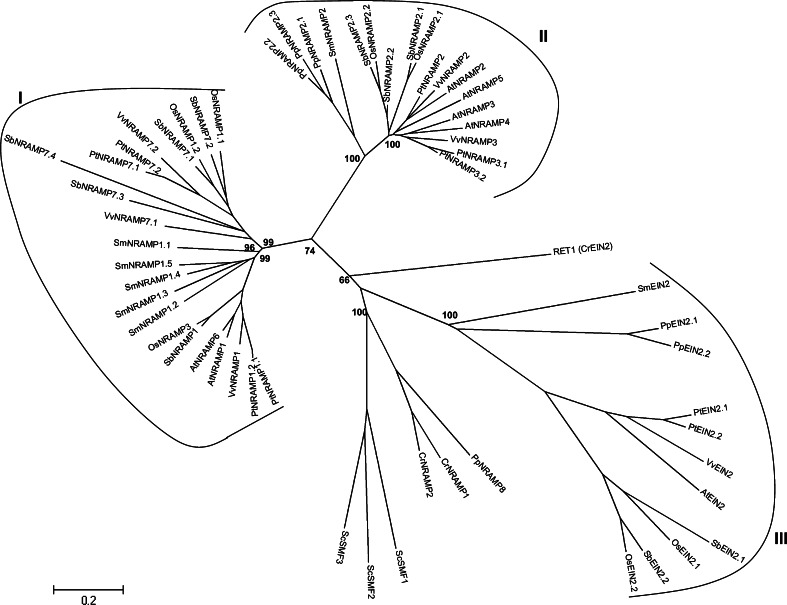
A database search for genes encoding NRAMP proteins in photosynthetic organisms identified between six and 10 gene models in embryophytes, whereas the unicellular organisms C. reinhardtii and S. cerevisiae have only three NRAMP (Table 1). In poplar, six of the nine NRAMP homologues were identified as pairs of closely related genes (NRAMP1.1 and 1.2, 3.1 and 3.2, 7.1 and 7.2), with the members of the pair located about 20 kb apart in the genomic sequence. This genomic organization suggests recent duplications.
Plant NRAMP split in two evolutionary clusters I and II [88]. Cluster I includes the closely related pair AtNRAMP1 and AtNRAMP6 from Arabidopsis thaliana and their homologues from monocotyledonous and dicotyledonous plants (Fig. 8). Cluster II includes the four Arabidopsis genes NRAMP2-5 and their related homologues from the other plants. NRAMP proteins from the unicellular organism C. reinhardtii and S. cerevisiae form separate clusters indicating a diversification of NRAMP transporters after the separation between the embryophytes. Interestingly, the genome of the moss P. patens contains 2 NRAMP sequences that fall within cluster II, close to NRAMP from higher plants and S. mollendorfii and one that clusters with NRAMP from the unicellular alga, C. reinhardtii, in agreement with the intermediate position of mosses in the evolutionary tree. By contrast, all NRAMP sequences from S. mollendorfii, the next node in the evolutionary tree, fall within cluster I or II, together with higher plant NRAMP. Cluster III is defined by sequences related to the Arabidopsis EIN2 (Ethylene Insensitive) gene (Fig. 8). EIN2 homologues have been shown to be necessary for the perception of the plant hormone, ethylene, in several plant species including A. thaliana, rice, petunia, and Medicago truncatula [89]. EIN2 proteins contain a transmembrane moiety fused with a large globular domain. The transmembrane moiety displays overall homology with NRAMP. Within the EIN2 cluster, EIN2 homologues from monocots, dicots, and mosses constitute separate subclusters. The function of moss and S. mollendorfii EIN2 homologues has not yet been reported.
Fig. 8.
An unrooted, neighbor-joining (NJ)-based tree of the NRAMP family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are given in the legend of Fig. 1
For the analysis of the NRAMP metal transporter expression in poplar, we have not considered poplar EIN2 homologues because there is no evidence that they encode metal transporters but they rather encode proteins involved in ethylene signal transduction. A database search identified three ESTs for PtNRAMP1, five ESTs for PtNRAMP2, 16 ESTs for PtNRAMP3, and 11 ESTs for PtNRAMP7 (Fig. 2e). ESTs corresponding to NRAMP genes made the lowest contribution to the whole set of ESTs retrieved for poplar metal transporters. This is confirmed by the overall low level of expression of poplar NRAMP genes (Fig. 3). When the ESTs were sequenced from tissue-specific libraries, their origin (data not shown) was generally in good agreement with the micro-array data. The PtNRAMP3.1 and 3.2 were highly expressed in poplar roots and xylem. This observation partially agrees with the expression pattern of their closest homologues in Arabidopsis AtNRAMP3 and AtNRAMP4, which are expressed in the vascular bundles of roots, stems, and leaves [19, 83]. In addition, the strong expression of PtNRAMP3.2 in seedlings is concordant with the high expression of AtNRAMP3 and AtNRAMP4 in germinating seeds [83]. The predominant expression of PtNRAMP2 in male and female catkins is reminiscent of the expression pattern of their homologue AtNRAMP5, which expressed exclusively in Arabidopsis flowers [88]. AtNRAMP1 expression is localized in roots [74]. Its closest homologues in poplar, PtNRAMP1.1 and 1.2 are expressed at a higher level in roots than in leaves but their highest expression is found in xylem. Moreover, PtNRAM7.1 and 7.2, which have no close homologues in Arabidopsis but belong to the same phylogenetic group as AtNRAMP1, are also highly expressed in roots (1.1) and xylem (1.2). The microarray data indicate a general conservation of NRAMP expression patterns between Arabidopsis and poplar with overlapping but distinct expression patterns for each member of the highly homologous pair of poplar NRAMP genes.
The heavy metal ATPases (HMA) family (TC 3.A.3.5)
Heavy metal ATPases (HMAs) have been classified in the type 1B-ATPase group of the P-ATPase family (TC 3.A3.5) [90]. They can be divided into two subgroups corresponding to the various transported metals, either Cu+/Ag+ or Zn2+/Cd2+/Pb2+/Co2+ [91, 92]. The first subgroup is represented in a wide range of organisms from bacteria to human whereas the presence of HMAs belonging to the second subgroup is restricted to bacteria and plants [92–95]. The present phylogenetic study indicated that the number of HMA homologues is in the same order in all studied organisms except the unicellular ones, photosynthetic or not, C. reinhardtii and S. cerevisiae.
After the annotation of the A. thaliana and rice genomes, based on sequence comparisons, Baxter [95] classified the 16 HMAs (now a ninth HMA has been found in the rice genome) in six clusters. We extend the search for HMAs to the genome of other six plant organisms and to S. cerevisiae. The number of HMA genes is rather conserved among embryophyte plants (between 8 and 11) whereas C. reinhardtii and S. cerevisiae possess a much lower number of HMA genes (Table 1). All HMAs of the various organisms included in the present study get in the six clusters defined by the Baxter classification (Fig. 9), although HMAs from both unicellular organisms are more or less apart, as in the case of the ZIP transporters (Fig. 5). Five of the six clusters present the same distribution of HMAs. Each can be divided into two subclusters. The first subcluster contains monocot and dicot proteins; this subcluster could be separated in two branches. The second subcluster is defined with moss and lycophyte transporters.
Fig. 9.
An unrooted, neighbor-joining (NJ)-based tree of the HMA (heavy metal ATPases) family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are given in the legend of Fig. 1
The members of cluster I, P1B-1-ATPases in the Baxter classification, are related to HMA1 from A. thaliana. AtHMA1 presents two properties relatively apart from all other HMAs. AtHMA1 is expressed at the inner envelope membrane of the chloroplast and is involved in the loading of copper in the stroma. Plants defective for AtHMA1 exhibit a reduced chloroplastic Cu/Zn superoxide dismutase (Cu/Zn-SOD) level and a high light photosensitivity phenotype [23]. By heterologous expression in yeast, it has been demonstrated that AtHMA1 also transported Cd, and more surprisingly calcium [96]. Phylogenetic analysis reveals that this cluster includes one member from each photosynthetic organism (two sequences for P. patens). The five higher plants HMA1 are closely related (Fig. 9). The S. moellendorffii and P. patens homologues are more distant. The C. reinhardtii sequence (it presents the characteristic SPC motif) is largely apart from the other. The only non-photosynthetic organism, the yeast S. cerevisiae, does not present any transporter related to this cluster.
The P1B-2-ATPase cluster II contains the 16 Zn/Cd/Co/Pb divalent cation transporters, in reference to the metal specificity found for the A. thaliana ones. Three A. thaliana proteins, AtHMA2, AtHMA3, and AtHMA4, belong to this second cluster (Fig. 9). AtHMA2 and AtHMA4 are closely related to the prokaryotic Cd/Zn efflux pumps CadA or ZntA. AtHMA4 is expressed at the plasma membrane and is involved in the Zn and Cd xylem loading and in the translocation of these metals from the roots to the shoot [15, 97]. The characterization of AtHMA2 in planta has demonstrated a large functional redundancy with those of AtHMA4 in Zn transport [97]; AtHMA3 likely plays a role in the detoxification of biological (Zn) and non-biological heavy metals (Cd, Co, and Pb) by participating in their vacuolar sequestration [20]. P. trichocarpa possesses only one transporter belonging to this cluster. The protein sequence presents 63.5–72% identity with the three AtHMAs of this cluster. The more related protein seems to be AtHMA2 in respect of size and sequence identity; so we named it PtHMA2. S. moellendorffii, with four proteins, is the most represented organism within this cluster. Although it was demonstrated that ScPCA1 was a Cd transporter, it is situated very far and does not belong to the P1B-2-ATPase cluster.
The other four clusters contain HMAs transporting only monovalent copper (and predicted to transport Ag+). Cluster III (P1B-3-ATPases) is the larger one, comprising 17 transporters from all land plants (Fig. 9). It is the only cluster where the distribution of HMAs in various subclusters is slightly different, presenting a fourth subcluster, containing two HMAs from dicots (PtHMA5.4 and VvHMA5.4) and two from monocots. PtHMA5.1, PtHMA5.2, and PtHMA5.3 proteins are very closely related (sequence identity comprised between 79 and 91%, whereas PtHMA5.4 only presented 57% identity with PtHMA5.1). The genes encoding PtHMA5.1 and PtHMA5.3 are nearly located on the chromosome III, suggesting that a duplication event has occurred. Nevertheless, the gene coding for PtHMA5.2, which is more closely related to PtHMA5.1, is located on another chromosome. The case of HMA5.1 to HMA5.3 from V. vinifera is very similar. The three proteins are closely related and their respective genes are nearly located on the same chromosome (probably a double duplication event). No protein of P. patens is present in this cluster. Only AtHMA5 has been functionally characterized today. AtHMA5 is mostly expressed at the plasma membrane of root cells and is required for Cu compartmentalization and detoxification in roots under conditions of copper excess [98].
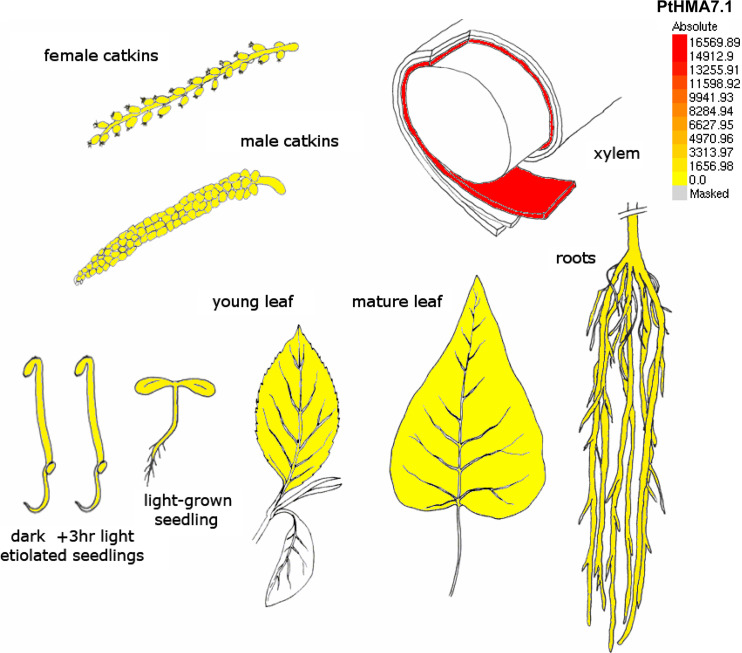
Cluster IV (P1B-4 ATPases in the Baxter classification) contains 11 proteins, including two isoforms from P. trichocarpa (Fig. 9). PtHMA7.1 and PtHMA7.2 protein sequences present 91% identity, but their genes are not nearly located in the genome. Two transporters from C. reinhardtii, CrHMA7.1 and CrHMA7.2, and ScCCC2 from S. cerevisiae are situated on the same branch of the phylogenetic tree, but so far that it is difficult to include them in this cluster. P. trichocarpa presents two HMA proteins, PtHMA6.1 and PtHMA6.2, belonging to P1B-5 ATPase cluster V (Fig. 9). These two transporters are located on different chromosomes; their protein sequences present 87% identity, indicating probably that a duplication event occurred. AtHMA6 is a copper transporter into the chloroplast, localized at the inner envelope membrane and implicated in the Cu+ import to the stroma. AtHMA6 is involved in metal delivery to the Cu/Zn-SOD and is necessary for a correct assembly of plastocyanin [24]. The last cluster VI (P1B-6 ATPases) is the smallest one containing six enzymes, restricted to higher plants except P. trichocarpa, which does not present a protein in this cluster (Fig. 9). As AtHMA1 and AtHMA6, AtHMA8 is a copper transporter into the chloroplast, inserted in the thylakoid membrane. AtHMA8 seems to function in tandem with AtHMA6. It delivers copper into the lumen and is also necessary to obtain a functional plastocyanin [24].
Interestingly, a set of 72 ESTs was identified in EST databases for the P. trichocarpa HMAs evidencing that all poplar HMA gene models are functional genes (Fig. 2f). PtHMA7.1 had the highest expression level of all microarray data and its expression was restricted to xylem tissues (Figs 3, 10). The close homolog PtHMA7.2 was essentially expressed in roots. It could be a case of subfunctionalization. The PtHMA5.1 and 5.3 were strongly expressed in roots, which correlates with the seven corresponding ESTs made root tissues found for these gene models in libraries. This is also in agreement with the function of its closest Arabidopsis homologue (AtHMA5, Fig. 9), which was primarily expressed in roots [98]. The only Zn/Cd/Co/Pb HMA found in P. trichocarpa PtHMA2 was mostly expressed in catkins, and in a lesser extent in roots. The expression profile of its Arabidopsis homolog AtHMA2 is also strongly expressed in these organs. On the contrary, the expression profiles of PtHMA1 and AtHMA1 are not the same. PtHMA1 is strongly expressed is root tissues, which is in contradiction with the well-described function of its closest Arabidopsis AtHMA1 which contributes to Zn(II) detoxification by reducing the Zn content of Arabidopsis thaliana plastids [99].
Fig. 10.
eFP display of transcript accumulation patterns across a variety of Populus organs and treatments. Poplar eFP browser presents the transcript accumulation pattern of PtHMA7.1 in a variety of tissues and organs. In all cases, red indicates higher levels of transcript accumulation and yellow indicates a lower level of transcript accumulation
Conclusions
In this work, we present a repertoire of 430 metal transporters found in eight photosynthetic plants as well as in the yeast model S. cerevisiae. This provides a global estimate of the conservation, amplification, or reduction of these families during evolution. When comparing the present data with those from Tuskan [25], a difference in gene numbers for three of the described families exists. It appears that 20 & 10 gene models are listed in the present study whereas 22 & 8 were reported by Tuskan et al. [25], for the ZIP & NRAMP families, respectively. This discrepancy lies in the different versions of the JGI poplar genome version that were used. Data from the Tuskan paper originated from version 1.0 and the present data from version 1.1. The better assembly and automatic predictions of gene models of the second version allowed us to remove or add gene models for these families. For instance, we found that pseudogenes were present and were removed within the ZIP family. Conversely, gene models, initially suspected to be pseudogenes, could be fully reconstructed and were further kept in the present analysis. The fact that transporter representatives are found in the moss P. patens for the six transporters families examined in this work suggests that metal transporter proteins have evolved early in the transition of plants to the terrestrial environment and were present in the most recent common ancestor of bryophytes and vascular plants. Some data showed further conservation of transporter gene in lineages for some family clusters (CDF cluster II with one to three members, ZIP cluster IV), suggesting possible conservation of function within these lineages.
Conversely, diversification of gene numbers in some transporter clusters has occurred in a lineage-specific manner, suggesting possible diversification of function. The total number of genes in a given species (Table 1) indicates for instance a considerable expansion of CDF, NRAMP, and HMA in land plants as compared to the unicellular alga C. reinhardtii. This is also exemplified by the lack of C. reinhardtii representatives in many clusters (ZIP clusters I, II, and III; CAX cluster II; NRAMP cluster II). C. reinhardtii sequences often clustered apart from other plant species, but very often with S. cerevisiae sequences. Vascular species showed a higher number of CTR sequences (between 4 and 8) than their non-vascular counterparts (2 or 3 sequences). More specifically, the NRAMP cluster I lacks the non-vascular species P. patens and C. reinhardtii, suggesting an important function of NRAMP proteins in metal translocation processes in the vascular system. A strong expansion within the angiosperm lineage is evidenced by the total number of ZIP sequences in angiosperms (between 16 and 20), much higher than the 5–9 sequences found in non-angiosperm species (Table 1). This is further highlighted in the CAX cluster I that lacks C. reinhardtii, P. patens, and S. moellendorffii sequences, and in the ZIP cluster I, in which the number of sequences in much lower for P. patens and S. moellendorffii (three and four sequences, respectively), as compared to angiosperm species (between ten and 13 sequences). The CTR cluster I also indicates a considerable expansion of these transporters in angiosperm species (between four and seven sequences), as compared to non-angiosperm species (one or two sequences). Specific features are also identified within the dicotyledon lineage, for example, in the CDF cluster I where the number of sequences is doubled that all other species. In cluster III of the CDF family, further specialization is observed for the perennial species V. vinifera and P. trichocarpa, where the number of sequences is also twice that identified for non-perennial species. This might be indicative of specialized functions linked to the perennial status of these two species.
On the whole, P. trichocarpa possess a much higher number of metal transporter genes than any other plant species, including the other perennial V. vinifera. Due to whole-genome duplication events, the number of protein-coding genes in Populus is more than that observed in Arabidopsis [25]. In the present study, we have found 72 Populus genes and this is much higher compared to the total of 57 Arabidopsis genes, that is ~1.4 times transporter genes than those found in Arabidopsis. This agrees with the reported observation, based on comparative genomics studies, that for each Arabidopsis gene, 1.4–1.6 putative Populus homologues are found [25]. A specific expansion of gene number was found for the CDF family, especially for cluster III (11 genes for Populus compared to a maximum of five genes in Vitis or Oryza). EST evidence was found for 52 out of the 72 sequences identified. Affymetrix Poplar Genome Arrays confirm transcript abundance for 65 metal transporter-encoding genes. The expression patterns obtained may help to approach the function of some metal transporters, especially in those families (CDF, ZIP), where most members have not yet been characterized in plants. The transcript abundance data described in this manuscript identified differentiating xylem as a cell population in which a subset of poplar metal transport genes were likely expressed. Xylem comprises the conduits that enable the transport of water and solutes from the roots to the aerial tissues. The expression of metal transporters in the cells would facilitate the transport of metals through this important tissue, largely comprised of dead, passively conducting cells, thereby providing soil-derived metals to the aerial portion of the plant body. It is also plausible that metal transporters function in the cells of the other vascular cell types that make up the phloem, where they might participate in the active transport processes that move sugars and amino acids around the plant body. For this particular research, transcript abundance data were unavailable as a first step in testing the hypothesis that metal transporters were active in phloem, but this could be the subject of future research.
The presence of several hundreds of transporter sequences suggests that plants may have rich potential to mobilize and detoxify metals in their environment within their tissues and organs. This work should provide a basis for further experimental molecular and genomic studies of metal homeostasis and tolerance in photosynthetic plants. Genomic and proteomic information gained from these sequenced plant species will greatly accelerate the phytoremediation process in situ. Further developments of phytoremediation will require an integrated multidisciplinary research effort that combines plant biology, genetic engineering, soil chemistry, soil microbiology, ecology, as well as agricultural and environmental engineering.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. An unrooted, neighbor-joining (NJ)-based tree of the zinc–iron permeases (ZIP) family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Distances were calculated with the Poisson correction model for distance computation (homogeneous substitution pattern among lineages). Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are given in the legend of Fig. 1. Supplementary material 3 (PPT 559 kb)
Acknowledgments
This work was supported by an ANR PRECODD project (ANR-06-ECOT-O15-01) and by a PhD grant to A. Migeon from the Agence de l’Environnement et de la Maîtrise de l’Energie (ADEME) and the Conseil Régional de Lorraine. OW was generously supported by a Natural Science and Engineering Research Council of Canada (NSERC) Canadian Graduate Scholarship (CGSD). This work was generously supported in part by funding from NSERC to MMC.
References
- 1.Rulkens WH, Tichy R, Grotenhuis JTC. Remediation of polluted soil and sediment: perspectives and failures. Water Sci Technol. 1998;37:27–35. [Google Scholar]
- 2.Schröder P, Herzig R, Bojinov B, Ruttens A, Nehnevajova E, Stamatiadis S, Memon A, Vassilev A, Caviezel M, Vangronsveld J. Bioenergy to save the world: producing novel energy plants for growth on abandoned land. Environ Sci Pollut Res Int. 2008;15:196–204. doi: 10.1065/espr2008.03.481. [DOI] [PubMed] [Google Scholar]
- 3.Stomp AM, Han KH, Wilbert S, Gordon MP, Cunningham SD. Genetic strategies for enhancing phytoremediation. Ann NY Acad Sci. 1994;721:481–491. doi: 10.1111/j.1749-6632.1994.tb47418.x. [DOI] [PubMed] [Google Scholar]
- 4.Migeon A, Richaud P, Guinet F, Chalot M, Blaudez D. Metal accumulation by woody species on contaminated sites in the north of France. Water Air Soil Pollut. 2009;204:89–101. doi: 10.1007/s11270-009-0029-5. [DOI] [Google Scholar]
- 5.Memon AR, Schröder P. Implications of metal accumulation mechanisms to phytoremediation. Environ Sci Pollut Res Int. 2009;16:162–175. doi: 10.1007/s11356-008-0079-z. [DOI] [PubMed] [Google Scholar]
- 6.Hall JL, Williams LE. Transition metal transporters in plants. J Exp Bot. 2003;54:2601–2613. doi: 10.1093/jxb/erg303. [DOI] [PubMed] [Google Scholar]
- 7.Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot. 2009;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu A, Muller-Rober B, Schulz B. Multifunctionality of plant ABC transporters––more than just detoxifiers. Planta. 2002;214:345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- 9.Kim DY, Bovet L, Kushnir S, Noh EW, Martinoia E, Lee Y. AtATM3 is involved in heavy metal resistance in arabidopsis. Plant Physiol. 2006;140:922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J. 2008;54:141–151. doi: 10.1111/j.1365-313X.2008.03402.x. [DOI] [PubMed] [Google Scholar]
- 11.Page MD, Kropat J, Hamel PP, Merchant SS. Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell. 2009;21:928–943. doi: 10.1105/tpc.108.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vert G, Briat JF, Curie C. Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J. 2001;26:181–189. doi: 10.1046/j.1365-313x.2001.01018.x. [DOI] [PubMed] [Google Scholar]
- 13.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+ . Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 14.Mei H, Cheng NH, Zhao J, Park S, Escareno RA, Pittman JK, Hirschi KD. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 2009;183:95–105. doi: 10.1111/j.1469-8137.2009.02831.x. [DOI] [PubMed] [Google Scholar]
- 15.Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004;576:306–312. doi: 10.1016/j.febslet.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Blaudez D, Kohler A, Martin F, Sanders D, Chalot M. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell. 2003;15:2911–2928. doi: 10.1105/tpc.017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees EM, Lee J, Thiele DJ. Mobilization of intracellular copper stores by the Ctr2 vacuolar copper transporter. J Biol Chem. 2004;279:54221–54229. doi: 10.1074/jbc.M411669200. [DOI] [PubMed] [Google Scholar]
- 18.Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR. Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell. 2003;15:1131–1142. doi: 10.1105/tpc.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 2003;34:685–695. doi: 10.1046/j.1365-313X.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 20.Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJM, Blaudez D, Chalot M, Sanders D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc Natl Acad Sci USA. 2007;104:8532–8537. doi: 10.1073/pnas.0609507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Kaplan J. Characterization of two homologous yeast genes that encode mitochondrial iron transporters. J Biol Chem. 1997;272:28485–28493. doi: 10.1074/jbc.272.45.28485. [DOI] [PubMed] [Google Scholar]
- 23.Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, Finazzi G, Grunwald D, Rappaport F, Vavasseur A, Joyard J, Richaud P, Rolland N. HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J Biol Chem. 2006;281:2882–2892. doi: 10.1074/jbc.M508333200. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam M, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, DePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjärvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leplé JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van De Peer Y, Rokhsar D. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 26.Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–270. doi: 10.1023/A:1012988914300. [DOI] [PubMed] [Google Scholar]
- 27.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 30.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
- 31.Yang X, Kalluri UC, Jawdy S, Gunter LE, Yin T, Tschaplinski TJ, Weston DJ, Ranjan P, Tuskan GA. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008;148:1189–1200. doi: 10.1104/pp.108.121921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team RDC . R: a language and environment for statistical computing. R Foundation for Statistical Computing. Austria: Vienna; 2009. [Google Scholar]
- 34.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy––analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. doi: 10.1198/016214504000000683. [DOI] [Google Scholar]
- 36.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S, Reid J, Malacarne G, Iliev D, Coppola G, Wardell B, Micheletti D, Macalma T, Facci M, Mitchell JT, Perazzolli M, Eldredge G, Gatto P, Oyzerski R, Moretto M, Gutin N, Stefanini M, Chen Y, Segala C, Davenport C, Dematté L, Mraz A, Battilana J, Stormo K, Costa F, Tao Q, Si-Ammour A, Harkins T, Lackey A, Perbost C, Taillon B, Stella A, Solovyev V, Fawcett JA, Sterck L, Vandepoele K, Grando SM, Toppo S, Moser C, Lanchbury J, Bogden R, Skolnick M, Sgaremella V, Bhatnagar SK, Fontana P, Gutin A, Van de Peer Y, Salamini F, Viola R. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin X, Town CD, Venter JC, Fraser CM, Tabata S, Nakamura Y, Kaneko T, Sato S, Asamizu E, Kato T, Kotani H, Sasamoto S, Ecker JR, Theologis A, Federspiel NA, Palm CJ, Osborne BI, Shinn P, Conway AB, Vysotskaia VS, Dewar K, Conn L, Lenz CA, Kim CJ, Hansen NF, Liu SX, Buehler E, Altafi H, Sakano H, Dunn P, Lam B, Pham PK, Chao Q, Nguyen M, Yu G, Chen H, Southwick A, Jeong Mi L, Miranda M, Toriumi MJ, Davis RW, Wambutt R, Murphy G, Dasterhaft A, Stiekema W, Pohl T, Entian KD, Terryn N, Volckaert G, Salanoubat M, Choisne N, Rieger M, Ansorge W, Unseld M, Fartmann B, Valle G, Artiguenave F, Weissenbach J, Quetier F, Wilson RK, De la Bastide M, Sekhon M, Huang E, Spiegel L, Gnoj L, Pepin K, Murray J, Johnson D, Habermann K, Dedhia N, Parnell L, Preston R, Hillier L, Chen E, Marra M, Martienssen R, McCombie WR, Mayer K, White O, Bevan M, Lemcke K, Creasy TH, Bielke C, Haas B, Haase D, Maiti R, Rudd S, Peterson J, Schoof H, Frishman D. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 38.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Grigoriev IV, Rokhsar DS, Grossman AR, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riao-Pachan DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, MartÃnez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin-I T, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto SI, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Sung HC, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van De Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 40.Jacquemin J, Laudié M, Cooke R. A recent duplication revisited: phylogenetic analysis reveals an ancestral duplication highly-conserved throughout the Oryza genus and beyond. BMC Plant Biol. 2009;9:146. doi: 10.1186/1471-2229-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob Ur R, Ware D, Westhoff P, Mayer KFX, Messing J, Rokhsar DS. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 42.Goffeau A, Barrell G, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 43.Peñarrubia L, Andrés-Colás N, Moreno J, Puig S. Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J Biol Inorg Chem. 2010;15:29–36. doi: 10.1007/s00775-009-0591-8. [DOI] [PubMed] [Google Scholar]
- 44.Eide DJ. The molecular biology of metal ion transport in Saccharomyces cerevisiae . Annu Rev Nutr. 1998;18:441–469. doi: 10.1146/annurev.nutr.18.1.441. [DOI] [PubMed] [Google Scholar]
- 45.Puig S, Lee J, Lau M, Thiele DJ. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J Biol Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 46.Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L. Identification of a copper transporter family in Arabidopsis thaliana . Plant Mol Biol. 2003;51:577–587. doi: 10.1023/A:1022345507112. [DOI] [PubMed] [Google Scholar]
- 47.Sancenón V, Puig S, Mateu-Andrés I, Dorcey E, Thiele DJ, Peñarrubia L. The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem. 2004;279:15348–15355. doi: 10.1074/jbc.M313321200. [DOI] [PubMed] [Google Scholar]
- 48.Persans MW, Nieman K, Salt DE. Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense . Proc Natl Acad Sci USA. 2001;98:9995–10000. doi: 10.1073/pnas.171039798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genom. 2007;8:107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arrivault S, Senger T, Krämer U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006;46:861–879. doi: 10.1111/j.1365-313X.2006.02746.x. [DOI] [PubMed] [Google Scholar]
- 51.Becher M, Talke IN, Krall L, Krämer U. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri . Plant J. 2004;37:251–268. doi: 10.1046/j.1365-313x.2003.01959.x. [DOI] [PubMed] [Google Scholar]
- 52.Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanikenne M, Krämer U, Demoulin V, Baurain D. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae . Plant Physiol. 2005;137:428–446. doi: 10.1104/pp.104.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerinot ML. The ZIP family of metal transporters. Biochim Biophys Acta Biomembr. 2000;1465:190–198. doi: 10.1016/S0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 55.Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S. Cloning an iron-regulated metal transporter from rice. J Exp Bot. 2002;53:1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- 56.Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X, Huang J, Jiang Y, Zhang HS. Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.) Mol Biol Rep. 2009;36:281–287. doi: 10.1007/s11033-007-9177-0. [DOI] [PubMed] [Google Scholar]
- 58.Kumánovics A, Poruk KE, Osborn KA, Ward DM, Kaplan J. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae . J Biol Chem. 2006;281:22566–22574. doi: 10.1074/jbc.M604730200. [DOI] [PubMed] [Google Scholar]
- 59.Lin SJ, Culotta VC. Suppression of oxidative damage by Saccharomyces cerevisiae ATX2, which encodes a manganese-trafficking protein that localizes to Golgi-like vesicles. Mol Cell Biol. 1996;16:6303–6312. doi: 10.1128/mcb.16.11.6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae . EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- 62.Shigaki T, Rees I, Nakhleh L, Hirschi KD. Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J Mol Evol. 2006;63:815–825. doi: 10.1007/s00239-006-0048-4. [DOI] [PubMed] [Google Scholar]
- 63.Luo GZ, Wang HW, Huang J, Tian AG, Wang YJ, Zhang JS, Chen SY. A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol Biol. 2005;59:809–820. doi: 10.1007/s11103-005-1386-0. [DOI] [PubMed] [Google Scholar]
- 64.Qi BS, Li CG, Chen YM, Lu PL, Hao FS, Shen GM, Chen J, Wang XC. Functional analysis of rice Ca2+/H+ antiporter OsCAX3 iNYeast and its subcellular localization in plant. Prog Biochem Biophys. 2005;32:876–882. [Google Scholar]
- 65.Edmond C, Shigaki T, Ewert S, Nelson MD, Connorton JM, Chalova V, Noordally Z, Pittman JK. Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochem J. 2009;418:145–154. doi: 10.1042/BJ20081814. [DOI] [PubMed] [Google Scholar]
- 66.Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 2000;124:125–133. doi: 10.1104/pp.124.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mei H, Zhao J, Pittman JK, Lachmansingh J, Park S, Hirschi KD. In planta regulation of the Arabidopsis Ca2+/H+ antiporter CAX1. J Exp Bot. 2007;58:3419–3427. doi: 10.1093/jxb/erm190. [DOI] [PubMed] [Google Scholar]
- 68.Pittman JK, Shigaki T, Marshall JL, Morris JL, Cheng NH, Hirschi KD. Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant Mol Biol. 2004;56:959–971. doi: 10.1007/s11103-004-6446-3. [DOI] [PubMed] [Google Scholar]
- 69.Kamiya T, Maeshima M. Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem. 2004;279:812–819. doi: 10.1074/jbc.M309726200. [DOI] [PubMed] [Google Scholar]
- 70.Cheng NH, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, Lahner B, Salt DE, Hirschi KD. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 2005;138:2048–2060. doi: 10.1104/pp.105.061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirschi KD. Expression of Arabidopsis CAX1 in tobacco: altered calcium homeostasis and increased stress sensitivity. Plant Cell. 1999;11:2113–2122. doi: 10.1105/tpc.11.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pittman JK, Hirschi KD. Regulation of CAX1, an arabidopsis Ca2+/H+ antiporter. Identification of an N-terminal autoinhibitory domain. Plant Physiol. 2001;127:1020–1029. doi: 10.1104/pp.010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–755. doi: 10.1042/0264-6021:3470749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J. 2003;35:295–304. doi: 10.1046/j.1365-313X.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- 77.Xiao H, Yin L, Xu X, Li T, Han Z. The iron-regulated transporter, MbNRAMP1, isolated from Malus baccata is involved in Fe, Mn and Cd trafficking. Ann Bot. 2008;102:881–889. doi: 10.1093/aob/mcn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochem J. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- 79.Oomen RJ, Wu J, Lelievre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MG, Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens . New Phytol. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 80.Wei W, Chai T, Zhang Y, Han L, Xu J, Guan Z. The Thlaspi caerulescens NRAMP homologue TcNRAMP3 is capable of divalent cation transport. Mol Biotechnol. 2009;41:15–21. doi: 10.1007/s12033-008-9088-x. [DOI] [PubMed] [Google Scholar]
- 81.Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem. 2003;278:24697–24704. doi: 10.1074/jbc.M301365200. [DOI] [PubMed] [Google Scholar]
- 82.Lanquar V, Lelièvre F, Barbier-Brygoo H, Thomine S. Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci Plant Nutr. 2004;50:1141–1150. [Google Scholar]
- 83.Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, Barbier-Brygoo H, Thomine S. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lanquar V, Ramos MS, Lelievre F, Barbier-Brygoo H, Krieger-Liszkay A, Kramer U, Thomine S. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weber M, Harada E, Vess C, Roepenack-Lahaye EV, Clemens S. Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J. 2004;37:269–281. doi: 10.1111/j.1365-313X.2003.02013.x. [DOI] [PubMed] [Google Scholar]
- 86.van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Loren Ver, van Themaat E, Koornneef M, Aarts MG. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens . Plant Physiol. 2006;142:1127–1147. doi: 10.1104/pp.106.082073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomine S, Schroeder JI (2004) Plant metal transporters with homology to proteins of the NRAMP family. Eurekahcom The Nramp Family. Landes Biosciences, Georgetown, pp 114–124
- 89.Varma Penmetsa R, Uribe P, Anderson J, Lichtenzveig J, Gish JC, Nam YW, Engstrom E, Xu K, Sckisel G, Pereira M, Baek JM, Lopez-Meyer M, Long SR, Harrison MJ, Singh KB, Kiss GB, Cook DR. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 2008;55:580–595. doi: 10.1111/j.1365-313X.2008.03531.x. [DOI] [PubMed] [Google Scholar]
- 90.Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/PL00006286. [DOI] [PubMed] [Google Scholar]
- 91.Axelsen KB, Palmgren MG. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cobbett CS, Hussain D, Haydon MJ. Structural and functional relationships between type 1B heavy metal-transporting P-type ATPases in Arabidopsis. New Phytol. 2003;159:315–321. doi: 10.1046/j.1469-8137.2003.00785.x. [DOI] [PubMed] [Google Scholar]
- 93.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adle DJ, Sinani D, Kim H, Lee J. A cadmium-transporting P1B-type ATPase iNYeast Saccharomyces cerevisiae . J Biol Chem. 2007;282:947–955. doi: 10.1074/jbc.M609535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003;132:618–628. doi: 10.1104/pp.103.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreno I, Norambuena L, Maturana D, Toro M, Vergara C, Orellana A, Zurita-Silva A, Ordenes VR. AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J Biol Chem. 2008;283:9633–9641. doi: 10.1074/jbc.M800736200. [DOI] [PubMed] [Google Scholar]
- 97.Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in arabidopsis. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, Mayo S, Thiele DJ, Ecker JR, Puig S, Peñarrubia L. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 2006;45:225–236. doi: 10.1111/j.1365-313X.2005.02601.x. [DOI] [PubMed] [Google Scholar]
- 99.Kim YY, Choi H, Segami S, Cho HT, Martinoia E, Maeshima M, Lee Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009;58:737–753. doi: 10.1111/j.1365-313X.2009.03818.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. An unrooted, neighbor-joining (NJ)-based tree of the zinc–iron permeases (ZIP) family in selected plants. The analysis was performed as described in the “Materials and methods” section and the tree was generated using MEGA version 4.1 [28] after sequence alignment. Distances were calculated with the Poisson correction model for distance computation (homogeneous substitution pattern among lineages). Bootstrap values are indicated (1,000 replicates). Branch lengths are proportional to phylogenetic distances. Corresponding gene loci are given in a supplemental file. Names of the species are given in the legend of Fig. 1. Supplementary material 3 (PPT 559 kb)