Abstract
The pathogenesis of any given human disease is a complex multifactorial process characterized by many biologically significant and interdependent alterations. One of these changes, specific to a wide range of human pathologies, is DNA hypomethylation. DNA hypomethylation signifies one of the major DNA methylation states that refers to a relative decrease from the “normal” methylation level. It is clear that disease by itself can induce hypomethylation of DNA; however, a decrease in DNA methylation can also have an impact on the predisposition to pathological states and disease development. This review presents evidence suggesting the involvement of DNA hypomethylation in the pathogenesis of several major human pathologies, including cancer, atherosclerosis, Alzheimer’s disease, and psychiatric disorders.
Keywords: DNA hypomethylation, G-specific hypomethylation, Cancer, Atherosclerosis, Alzheimer’s disease, Psychiatric disorders
Introduction
“Epigenetics” is defined as heritable changes in gene expression associated with modifications of DNA or chromatin proteins that are not due to any alteration in the DNA sequence [1–3]. Such modifications include the best known and much studied methylation of DNA, a covalent addition of a methyl group (CH3) to the cytosine residue at CpG sequences in mammals [4], and the modifications of the proteins that bind to DNA [5, 6]. These epigenetic modifications are essential for normal development and proper maintenance of cellular functions in adult organisms. Additionally, alterations in DNA methylation, both decreases and increases, are a frequent characteristic of a wide range of human pathologies. Although these alterations are well established and have been studied extensively [2, 3], until recently, most biomedical research has concentrated on the role and mechanisms of hypermethylation under normal physiological conditions, e.g., aging [7], or during pathological conditions [2, 8]. Much less attention has been devoted to the role and place of the disease-linked DNA hypomethylation [9]. This review presents evidence suggesting the involvement of DNA hypomethylation in the pathogenesis of several major human pathologies, including cancer, atherosclerosis, Alzheimer’s disease, and psychiatric disorders.
DNA methylation
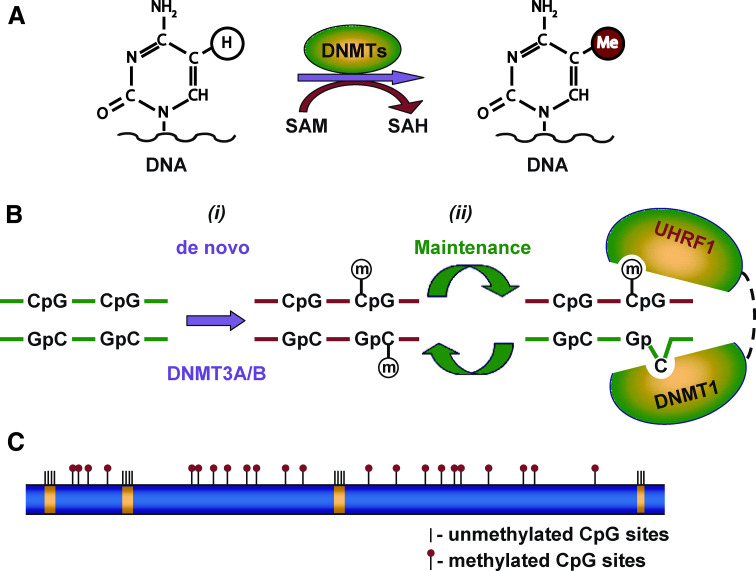
DNA methylation is the addition of a methyl group from the universal methyl donor, S-adenosyl-l-methionine (SAM), to the fifth carbon atom in the cytosine pyridine ring, resulting in the formation of 5-methylcytosine (5meC) [10] (Fig. 1a). This reaction is catalyzed by DNA methyltransferases (DNMTs) [11, 12]. In eukaryotes, this stable post-synthetic epigenetic mark is found exclusively at cytosine residues at CpG sequences [13]. DNA methylation is essential for normal development and the maintenance of cellular homeostasis and functions in adult organisms, particularly for X-chromosome inactivation in females [14], genomic imprinting [15], silencing of repetitive DNA elements [11, 16], regulation of chromatin structure [17], and proper expression of genetic information [18].
Fig. 1.
Schematic model showing cytosine DNA methylation. a Cytosine residues in DNA at CpG sites are converted 5-methylcytosines by the addition of a methyl group from SAM to the fifth carbon atom in the cytosine pyridine ring. This reaction is catalyzed by the enzymatic activity of DNA methyltransferases (DNMTs). b Establishment and maintenance of the DNA methylation pattern during DNA replication. (i) DNA methylation is initiated and established during embryonic development by means of the de novo DNMT3A and DNMT3B DNA methyltransferases. (ii) Maintenance of DNA methylation. During DNA replication, DNA methylation is maintained by a complex coordinated action of the maintenance methyltransferase DNMT1 and UHRF1 [28–30]. The SRA domain of UHRF1 recognizes the hemimethylated CpG site and recruits DNMNT1, which transfers methyl group to the unmethylated cytosine residue on the newly synthesized DNA strand. c DNA methylation landscape in mammalian genomes. Methylation in normal mammalian cells occurs primarily at CpG sites located in repetitive sequences, exons other than first exons, and intergenic DNA (blue). The CpG islands that span the promoter and first exons of the majority of genes are usually unmethylated (yellow) in normal cells and embedded in a matrix of long methylated domains (blue) [33, 34]
DNA methylation is initiated and established by means of the de novo DNA methyltransferase DNMT3 family (DNMT3A and DNMT3B) [11, 19] (Fig. 1b), whose expression is coordinated by DNMT3L [20], Lsh (lymphoid-specific helicase) [21], microRNAs [22], and piRNAs [23]. During DNA replication, DNA methylation is maintained by a complex cooperative interplay of maintenance methyltransferase DNMT1 with the de novo DNA methyltransferases DNMT3A and DNMT3B [24, 25], methyl-CpG-binding protein 2 (MeCP2) [26], histone-modifying enzymes [27], and the UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) protein [28–30] (Fig. 1b).
Total genomic DNA methylation refers to the overall content of 5meC in the genome. Approximately 70–90% of the CpG dinucleotides in the mammalian genome are methylated [31]; however, the CpG sites are not distributed uniformly across the genome [31, 32]. The methylation landscape of mammalian genomes consist of short (<4 kb) unmethylated domains embedded in a matrix of long methylated domains (Fig. 1c) [33, 34]. Promoters and first exons of the majority of genes in the genome are strongly enriched in unmethylated domains and depleted in methylated domains, which are found predominantly in interspersed and tandem repetitive sequences and exons other than first exons [33, 34]. The enrichment of CpG islands, genomic regions that contain the high G + C content and the high frequency of CpG dinucleotides [35], in unmethylated domains is the major difference between the unmethylated and methylated DNA regions [33, 34].
The accurate maintenance of DNA methylation patterns depends on the function and cooperation of several critical factors, including the activity and expression of DNMTs [11, 36], DNA demethylase [37], histone-modifying enzymes [27, 38], the status of one-carbon metabolism [39, 40], DNA integrity [41, 42], and cell proliferation [43]. Disturbances in any or all of these factors may lead to an altered DNA methylation status, including DNA hypomethylation.
Mechanisms of DNA hypomethylation
DNA hypomethylation signifies one of the major DNA methylation states, the other being hypermethylation, and in most cases refers to a relative situation in which there is a decrease from the “normal” methylation level [31]. The mechanism of DNA hypomethylation is still unclear, and, very likely, there is not a single mechanism responsible for demethylation of DNA. However, it is well established that several factors may trigger and contribute to the loss of genomic methylation.
DNA methyltransferases and DNA hypomethylation
A large body of evidence clearly demonstrates that the proper function of DNMTs is crucial in the maintenance of faithful DNA methylation [36]. Reducing the expression of Dnmts through gene-targeting of individual Dnmts, or a combination of Dnmts, is associated with markedly decreased global methylation levels [44–46]. For instance, the reduction of Dnmt1 expression to 10% causes significant hypomethylation of centromeric and endogenous retroviral intracesternal A particle (IAP) repetitive sequences in mice [44]. Likewise, the loss of DNMT function secondary to the inhibition of its activity with demethylating agents, such as 5-aza-2′-deoxycytidine (5-aza-dC) [45], homocysteine, or its metabolite S-adenosyl-l-homocysteine (SAH) [46], results in rapid demethylation of DNA. Additionally, DNA demethylation caused by exposure to a number of environmental chemicals, e.g., arsenic [47], and nutritional and life-style factors, such as dietary bioflavonoids [48], alcohol [49], and cigarettes [50], is associated with an inhibitory effect on the expression and activity of DNA methyltransferases.
The normal status of DNA methylation also depends on cooperation between individual DNMTs [24, 25] and critical regulators of DNMTs function, including DNMT3L [20], Lsh [21, 51], microRNAs [22], and piRNAs [23]. Additionally, the results of a recent study have demonstrated the involvement of lysine-specific demethylase 1 (LSD1) for the maintenance of DNA methylation by regulation of the methylation status of DNMT1 and modulation of its stability [38]. Aberrations in any of these factors may compromise the DNMT function leading to DNA hypomethylation.
One-carbon metabolism and DNA hypomethylation
The methyl groups needed for all cellular biological methylation reactions, including DNA methylation, are acquired from SAM, the primary universal donor of methyl groups, which is derived from methionine through a one-carbon metabolic pathway [10]. This process indispensably connects faithful DNA methylation to the proper functioning of the one-carbon metabolic pathway, which has a great impact on DNA methylation [39, 40]. There are two groups of risk factors that may compromise the normal functioning of the one-carbon metabolic pathway and, subsequently, affect the DNA methylation profile. The first group consists of nonmodifiable genetic risk factors, such as genetic variations in genes encoding enzymes involved in the cellular one-carbon metabolism. Indeed, there are extensive amounts of data showing that single nucleotide polymorphisms in these genes are associated with aberrant DNA methylation [52, 53]. The second group consists of potentially modifiable factors, specifically essential nutrients involved in the metabolism of methyl groups, including methionine, choline, folic acid, and vitamin B12 [39, 40, 54]. Previously, we and others have demonstrated that long-term exposure to an inadequate supply of methionine, choline, folic acid, or vitamin B12 results in a profound loss of cytosine methylation in the livers of male rats and mice [55–57]. Additionally, it is believed that the loss of DNA methylation induced by exposure to arsenic [47, 58], diethanolamine [59], trichloroethylene [60], and alcohol [61], is associated with perturbations in cellular SAM homeostasis.
DNA integrity and DNA hypomethylation
The integrity of the genome is another critical factor that affects the normal status of DNA methylation. Every living organism is exposed to a variety of genomic insults on a daily basis from many endogenous and exogenous sources [62]. The results of several studies have demonstrated that the presence of unrepaired lesions in DNA induced by these factors substantially alters the methylation capacity of DNA methyltransferases, leading to DNA hypomethylation [41, 42, 57]. Specifically, the presence of 8-oxoguanine and 5-hydroxymethylcytosine in DNA, common DNA modifications resulting from oxidative damage to DNA, inhibits the binding of the MeCP2 protein and diminishes the ability of the DNMTs to methylate DNA [41, 63]. Likewise, the presence of pyrimidine photodimers, preferentially induced by sunlight at methylated CpG sites [64], reduces DNA methylation [65]. The significance of these processes in DNA hypomethylation increases progressively with age due to an age-dependent decrease in the proficiency of DNA repair [66].
DNA demethylases and DNA hypomethylation
It is believed that, in addition to a passive loss of DNA methylation through the blocking of methylation of cytosine residues, DNA methyl groups can be removed by active demethylation by means of DNA demethylases. DNA demethylase activity has been attributed to several proteins, including RNA-dependent 5meC glycosylase [67] with the involvement of DNA repair pathway [68], ribozyme-like demethylase [69], and methyl-binding domain 2 (MBD2) [70]. Furthermore, two recent reports claim that DNMT3A and DNMT3B may also act as DNA demethylases [71, 72]. Interestingly, all these proteins are characterized by quite different DNA demethylating mechanisms. However, despite efforts to identify unambiguously a DNA demethylase, the evidence for the existence of an active DNA demethylation in mammals remains inconclusive [37].
DNA hypomethylation and human diseases
DNA hypomethylation and cancer
Classically, the development of cancer in humans has been viewed as a progressive multistep process of transformation of normal cells into malignant cells driven by genetic alterations [73, 74]. However, a wealth of data in the past decade indicating the importance of epigenetic processes has largely changed the view on cancer as being a solely genetic disease [75]. Currently, cancer is recognized as a disease driven by both genetic and epigenetic alterations, and both of these components cooperate and complement each other at every stage of cancer development [75].
The loss of global DNA methylation, the first epigenetic abnormality identified in cancer cells more than a quarter century ago [76–78], continues to be a central feature and one of the most common molecular alterations in human cancers. This is evident by the fact that almost all of major human cancers, including colon [79, 80], gastric [80], lung [81], liver [82], breast [83], bladder [84], ovarian [85, 86], and endometrial [87], are characterized by a profound cancer-linked hypomethylation of the genome. More importantly, the association between the degree of DNA hypomethylation and the grade and stage of cancer gives a firm basis for its use as a biomarker for the diagnosis and prognosis of disease [84–86]. Indeed, the results of several studies have demonstrated that DNA hypomethylation is a more informative prognostic marker than tumor stage or grade [85]. However, a decrease in DNA methylation, by itself, is not sufficient to address precisely the role of DNA hypomethylation in tumorigenesis [88] because it could simply be a secondary consequence of malignant cell transformation reflecting the undifferentiated state of tumors. To provide evidence that hypomethylation has a significant role in cancer development, it is necessary to demonstrate the following: (1) the loss of DNA methylation occurs at a considerable frequency at early stages of carcinogenesis, (2) changes that occur at preneoplastic stages are also present during later stages of cancer, (3) additional changes in methylation are acquired during tumor progression, and (4) a mechanistic link exists between the hypomethylation of DNA and cancer development. The results of numerous studies demonstrating (1) the frequent loss of DNA methylation during premalignant pathological states or during early preneoplastic stages of tumorigenesis [81–84], (2) a greater degree of DNA hypomethylation in tumors compared to preneoplastic tissues [83–87], and (3) cumulative methylation changes during cancer progression from normal to stage IV disease in various cancers [86] provide convincing evidence that loss of DNA methylation in cancer is not a secondary event. Furthermore, a recent large case-control study has furnished solid evidence for an association between DNA hypomethylation and an increased risk of bladder-cancer development [89]. In addition, a decrease in DNA methylation by gene-targeting of Dnmt1 [44, 90, 91] and Lsh [92] results in tumor induction, providing strong evidence for a causative role of DNA hypomethylation in the origin of cancer.
The mechanistic link between the loss of DNA methylation and cancer development, including induction of chromosomal instability, reactivation and transposition of retrotransposable elements, loss of imprinting, and activation of normally silenced genes, is directly related to the DNA methylation landscape of the mammalian genome and the function of DNA methylation in normal cells. As mentioned previously, the mammalian genome consists of relatively short unmethylated domains embedded in a matrix of long stably methylated domains, in which methylation occurs at repetitive elements and within the body of genes [33, 34]. Because of this, loss of DNA methylation largely affects only these areas of the genome. Evidence for this is provided by the strong correlation between the loss of global DNA methylation and the demethylation of repetitive sequences, such as long interspersed nucleotide elements (LINE), short interspersed nucleotide elements (SINE), IAP, and Alu elements in tumors. Furthermore, loss of LINE-1 methylation has been proposed as a surrogate marker for cancer-linked genome demethylation [93].
There are two well-established consequences associated with the loss of DNA methylation at repetitive sequences that may contribute to tumorigenesis. First, demethylation of repetitive sequences located at centromeric, pericentromeric, and subtelomeric chromosomal regions may cause the induction of chromosomal abnormalities. For example, recent findings have demonstrated that DNA hypomethylation at the centromeric region causes permissive transcriptional activity at the centromere [94] and the subsequent accumulation of small minor satellite transcripts that impair centromeric architecture and function [95]. Likewise, hypomethylation of the subtelomeric regions is associated with enhanced transcription of the telomeric region [96, 97]. Second, hypomethylation of LINE-1, SINE, Alu, and IAP retroviral elements causes their activation and transposition [98] that may lead to genomic instability. An integral role of the loss of DNA methylation and the presence of these alterations in the neoplastic process is now commonly accepted.
One of the main functions of DNA methylation in normal cells is genomic imprinting [15], a parent-of-origin-dependent allele-specific expression of a small number of genes (approximately 90). Loss of imprinting (LOI), a loss of monoallelic regulation of imprinted gene expression, is frequently detected in human tumors and currently is considered as one of the most frequent alterations in cancer [99]. The first imprinted gene that exhibited LOI in human cancer was the insulin-like growth factor-II (IGF2) gene [100]. Initially, LOI of IGF2 was linked to increased methylation; however, a number of studies have established that hypomethylation is the reason for LOI of the IGF2 gene in colorectal [101, 102], breast [102], liver [103], and bladder [104] cancers; the H19 gene in colon [102] and lung [105] cancers; and the KCNQ1 gene in breast, liver, and colon cancers [106, 107].
It is well established that more than 70% of the genes in the human genome normally contain unmethylated CpG islands in their promoters [108]. However, a recent analysis of 5,549 autosomal genes with dense CpG island promoters indicates that about 4% of these genes are methylated and silenced under normal conditions [109]. Until recently, the majority of the studies in the field of cancer research have focused on the role of promoter hypermethylation and gene silencing in cancer, which overshadowed the significance of the hypomethylation of normally methylated genes in cancer development. However, mounting evidence indicates that gene-specific hypomethylation also plays an important role in cancer. Table 1 lists selected hypomethylated and overexpressed genes in various human cancers.
Table 1.
Selected list of the hypomethylated genes in human cancers
| Gene | Official gene name | Tumor | References |
|---|---|---|---|
| S100A4 | S100 calcium binding protein A4 | Colon, endometrial, pancreatic | [110–112] |
| CYP2W1 | Cytochrome P450, family 2, subfamily W, polypeptide 1 | Colon | [113] |
| CDH3 | Cadherin 3 (P-cadherin) | Colon, breast | [114, 115] |
| BAGE | B melanoma antigens | Colon | [116] |
| DCN | Decorin | Colon | [117] |
| MAGE-A1 | Melanoma antigen, family A, 1 | Colon, gastric | [118, 119] |
| MAGE-A3 | Melanoma antigen, family A, 3 | Colon, gastric | [118, 119] |
| XAGE-1 | X antigen family | Gastric | [120] |
| CCND2 | Cyclin D2 | Gastric | [121] |
| SERPINB5 | Serpin peptidase inhibitor, clade B, member 5 (Maspin) | Gastric, pancreatic, thyroid | [112, 122–124] |
| MUC2 | Mucin 2 | Gastric | [125] |
| NGALR | Neutrophil gelatinase-associated lipocalin receptor | Esophagus | [126] |
| CD133 | Cell surface protein CD133 | Brain | [127] |
| NAT1 | N-Acetyltransferase | Breast | [128] |
| FEN1 | Flap endonuclease 1 | Breast | [129] |
| SNCG | Synuclein gamma | Breast, ovarian | [130, 131] |
| UPA | Plasminogen activator, urokinase | Breast, prostate | [132] |
| CAV1 | Caveolin 1 | Breast | [133] |
| ZEB2 | Zinc finger E-box binding homeobox 2 | Breast | [134] |
| TFF3 | Trefoil factor 3 | Pancreatic, liver | [112, 135] |
| CLDN4 | Claudin 4 | Pancreatic, ovarian | [112, 136] |
| LCN2 | Lipocalin 2 | Pancreatic | [112] |
| PAX2 | Paired box 2 | Endometrial | [137] |
| DNMT3L | DNA (cytosine-5-)-methyltransferase 3-like | Endometrial | [138] |
| CAGE | Cancer/testis antigen | Endometrial | [139] |
| ER-α | Estrogen receptor-alpha | Endometrial | [140] |
| HNF-1β | Hepatocyte nuclear factor-1 beta | Ovarian | [141] |
| BORIS | Brother of the regulator of imprinted sites | Ovarian | [142] |
| CA9 | Carbonic anhydrase IX | Renal | [143] |
| GLIPR1/RTVP-1 | Glioma pathogenesis-related 1/related to testis-specific, vespid, and pathogenesis proteins 1 | Wilms tumors | [144] |
| HPSE2 | Heparanase 2 | Prostate | [145] |
| PRAME | Preferentialy expressed antigen of melanoma | Myeloid leukemia | [146] |
| DDX43 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 (HAGE) | Myeloid leukemia | [147] |
| PRDM16 | PR domain containing 16 (MEL1) | T-cell leukemia | [148] |
| BCL2 | B-cell CLL/lymphoma 2 | B-cell lymphocytic leukemia | [149] |
| TCL1 | T-cell leukemia/lymphoma 1A | T-cell lymphocytic leukemia | [150] |
| FGFR1 | Fibroblast growth factor receptor 1 | Rhabdomyosarcoma | [151] |
| TNFRSF8 | Tumor necrosis factor receptor superfamily, member 8 (CD30) | Hodgkin lymphoma | [152] |
This list is noticeably shorter than the number of hypermethylated genes in human cancers [153] and even in any specific type of cancer, e.g., breast cancer [154]. This is because the number of genes that can potentially be demethylated (normally methylated) is substantially smaller than the number of genes that can potentially be methylated (normally unmethylated), which is directly predetermined by the methylation landscape of the genome. Despite the different number of cancer-linked hypomethylated and hypermethylated genes, the dynamic of gene-specific methylation changes during tumorigenesis is identical: the progressive accumulation of hypomethylated or/and hypermethylated alterations during tumor development.
DNA hypomethylation, carcinogen exposure, and cancer risk assessment
Environmental exposure to natural and man-made chemical and physical agents is one of the major causes of human cancer [155]. The need for the rapid identification and appropriate regulation of human carcinogens before their dissemination into society is of prime importance for the primary prevention of neoplasia in humans. Until now, research emphasis in cancer risk assessment and cancer epidemiology has focused on the measurement of DNA damage, DNA adduct formation, and mutations induced by specific agents or exposures [156]. The recognition of the role of epigenetic mechanisms in carcinogenesis and results of studies documenting that environmental exposures can alter expression of genetic information not only by genetic but also by epigenetic mechanisms [157] have challenged our current approach to carcinogenicity testing and indicated the need for a new generation of exposure biomarkers [158]. The results obtained in numerous animal studies have demonstrated that early indicators of carcinogenic exposure are epigenetic alterations and the emergence of epigenetically reprogrammed cells with epigenetic alterations similar to those found in malignant cells [155, 159]. Furthermore, it has been proposed that epigenetic alterations, including genomic and repeat-associated hypomethylation, may precede genetic alterations [159, 160]. Additionally, considering the stability and inheritance of epigenetic alterations through transmission of carcinogen-induced aberrant epigenetic patterns from one cell generation to the next, epigenetic alterations may be better biomarkers of carcinogenic exposure. The results of recent human studies have provided strong support for this suggestion [161–164]. For instance, low-level occupational exposure of gas-station attendants and traffic police to benzene has resulted in significant epigenetic alterations, as characterized by a significant reduction of LINE-1 and MAGE1 gene methylation in blood DNA samples, compared to unexposed subjects [161]. Importantly, the aberrant DNA methylation patterns in exposed individuals highly reproduce the aberrant epigenetic patterns found in acute myelogenous leukemia patients. Similar DNA methylation changes in the blood have been found in humans exposed chronically to organic pollutants [162], arsenic [163], and traffic-derived particles [164].
DNA hypomethylation and cardiovascular diseases
Atherosclerosis and its complications are a major cause of mortality, morbidity, and disability in developed Western countries [165]. Atherosclerosis is characterized by the infiltration of lipid particles into the arterial wall, accompanied by the recruitment of inflammatory and immune cells, migration and proliferation of smooth muscle cells, synthesis of extracellular matrix, and development of fibrocellular lesions [166]. In contrast to cancer research, where the role of DNA hypomethylation has been studied for decades, the involvement of DNA hypomethylation in the context of atherosclerosis was first formulated by Newman in 1999 [167]. The hypothesis was based on evidence suggesting that elevated plasma homocysteine is a risk factor for atherosclerosis [168] and that homocysteine and SAH efficiently inhibit DNA methyltransferases, causing hypomethylation of DNA. The significance of the loss of DNA methylation in atherosclerosis is widely documented [169–171]. Substantial global DNA hypomethylation has been found in peripheral white blood cells [172], smooth muscle cells [170, 173], and atherosclerotic lesions [174] in patients with atherosclerosis. These correlative studies, without disputing the underlying role of homocysteine as a risk factor for atherosclerosis, suggest that hypomethylation during atherosclerosis may be a secondary event induced by elevated homocysteine. However, it should be noted that the result of a recent study, in which the occurrence of global DNA hypomethylation prior to the formation of atherosclerotic lesions in genetically atherosclerosis-prone Apoe −/− mice, clearly demonstrated the significance of DNA hypomethylation in the pathogenesis of atherosclerosis and in susceptibility to the disease [175]. Furthermore, transcriptional up-regulation of the 5-lipoxygenase and 15-lipoxygenase genes, key enzymes implicated in the pathogenesis of atherosclerosis [176], is mediated by promoter hypomethylation [177, 178].
DNA hypomethylation and neurodegenerative diseases and psychiatric disorders
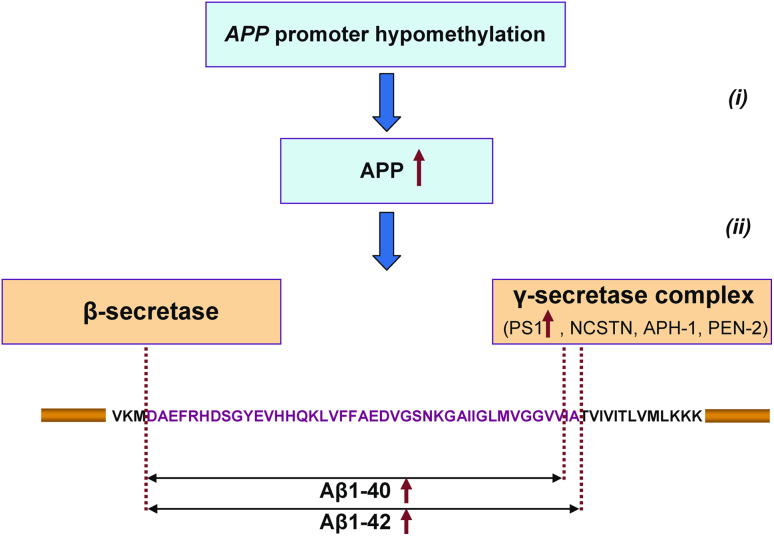
Alzheimer’s disease is an age-related progressive neurodegenerative disorder characterized by the presence of amyloid plaques and intracellular tangles in the brain [179]. The biogenesis and accumulation of amyloid plaques, which consist primarily of 40- to 42-residue β-amyloid peptides (Aβ40 and Aβ42) derived from amyloid precursor protein (APP) as a result of sequential proteolic processing by β-secretase (BACE1) and γ-secretase complex [180], is a key event in Alzheimer’s disease. An association between DNA hypomethylation and Alzheimer’s disease has been noted in several studies. For example, hypomethylation-associated overexpression of the APP gene has been demonstrated in the brain of an Alzheimer’s patient [181] and, in another study, substantial age-dependent APP promoter demethylation has been demonstrated in the cortex from Alzheimer’s patients [182]. Specifically, the frequency of methylation of cytosine residues at -207, -204, -200, and -182 in the APP promoter region in subjects younger than 70 years was substantially greater (55%) compared to subjects older than 70 years (5%) [182]. Additionally, expression of the presenilin 1 (PS1) gene, which encodes a key component of the γ-secretase complex, is regulated by methylation [183]. In light of these considerations, the following hypothetical model is proposed for the pathogenesis of Alzheimer’s disease driven by the DNA hypomethylation events (Fig. 2). First, the age-related hypomethylation of the APP promoter provokes an over-expression of the APP gene, leading to greater levels of APP in brain. Second, hypomethylation and up-regulation of the PS1 gene activates the γ-secretase complex and stimulates the proteolytic cleavage of APP, leading to the accumulation of Aβ40 and Aβ42. Importantly, this model brings together the two most widely accepted hypotheses of Alzheimer’s disease, the amyloid and presenilin hypotheses, into a single mechanism.
Fig. 2.
Hypothetical model of the pathogenesis of Alzheimer’s disease driven by DNA hypomethylation events. Involvement of DNA hypomethylation in the biogenesis and processing of APP in the human brain. First, hypomethylation of the APP promoter provokes an overexpression of the APP gene, leading to greater levels of APP protein in the brain (i). Second, the hypomethylation and up-regulation of the PS1 gene induces the activity of the γ-secretase complex and stimulates the proteolytic cleavage of APP (ii), leading to the accumulation of Aβ40 and Aβ42. Note that genes encoding nicastrin (NCSTN), anterior pharynx defective 1 (APH-1), and presenilin enhancer 2 (PEN-2), three other components of the γ-secretase complex, also contain CpG islands according to http://cpgislands.usc.edu [35] and may be regulated by DNA methylation
Despite the strong evidence that supports a genetic origin of major human psychiatric disorders, no specific gene associated with the development of these disorders has been identified [184]. In contrast, a growing body of evidence suggests the involvement of aberrant epigenetic mechanisms in the pathogenesis of major psychiatric disorders, including schizophrenia and bipolar disorder. For instance, the involvement of promoter hypermethylation of the reelin (RELN) gene in the pathogenesis of schizophrenia is well-established [185]. Another critical gene that has been implicated in the etiology of psychiatric disorders is a catechol-O-methyltransferase (COMT) [184]. The results of recent studies have demonstrated a crucial role of promoter hypomethylation of membrane-bound COMT, a predominant form of COMT that is involved in the degradation of synaptic dopamine in the human brain, in the pathogenesis of schizophrenia and bipolar disorder [186]. Additionally, analysis of leukocyte DNA methylation in 124 male patients with schizophrenia has demonstrated a significant hypomethylation of DNA compared to healthy subjects [187].
DNA hypomethylation and other human pathologies: autoimmune and chronic kidney diseases, and age-related macular degeneration
Similar global and gene-specific hypomethylation changes have been found in patients with uremia [188], systemic lupus erythematosus [189, 190], rheumatoid arthritis [191, 192], and age-related macular degeneration [193]. For example, age-related macular degeneration, the leading cause of irreversible blindness in people 50 years and older [194], is associated with hypomethylation-induced overexpression of the clusterin (CLU) gene [193] that encodes one of the major proteins of drusen, the deposition of which between pigment epithelium and Bruch’s membrane causes blindness.
Concluding remarks
The pathogenesis of any given human disease is a complex multifactorial process characterized by many biologically significant and interdependent alterations. One of these changes, which is specific to many human diseases, is the alteration of DNA methylation, including hypomethylation. It is clear that disease by itself can induce hypomethylation of DNA; however, the loss of DNA methylation can also have an impact on the predisposition to pathological states and disease development. Interestingly, one of the common features of the previously described human chronic pathological states is their association with aging. It is well-established that levels of DNA methylation are markedly decreased upon aging [7]. DNA methylation is a crucial biological process that programs a proper expression of genetic information in mammals. The accurate status of DNA methylation is balanced in mature cells, but with age this balance is strongly shifted in favor of DNA demethylation. Therefore, DNA hypomethylation that occurs during normal aging appears to be a critical risk factor contributing to the development of chronic age-related human pathological states. In addition to age-related hypomethylation, DNA hypomethylation can be caused by various endogenous and exogenous factors, including environmental chemicals and physical agents, lifestyle factors, and infections. This induced DNA hypomethylation may predispose individuals to disease development. However, considering the fact that a remarkable feature of epigenetic abnormalities, including DNA hypomethylation, is their potential reversibility, timely correction and proper maintenance of DNA methylation levels are promising avenues to prevent the development of chronic human diseases.
Footnotes
The views expressed in this paper do not necessarily represent those of the US Food and Drug Administration.
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi HY, Beaudet AL. Epigenetics and human disease. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 435–456. [Google Scholar]
- 3.Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-I. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 191–209. [Google Scholar]
- 7.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 9.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 11.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 12.Li E, Bird A. DNA methylation in mammals. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 341–356. [Google Scholar]
- 13.Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 14.Singer-Sam J, Riggs AD. X-chromosome inactivation and DNA methylation. EXS. 1993;64:358–384. doi: 10.1007/978-3-0348-9118-9_16. [DOI] [PubMed] [Google Scholar]
- 15.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 16.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 19.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 20.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravin AA, Sachidanandam R, Bouc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Osta A. DNMT cooperativity––the developing links between methylation, chromatin structure and cancer. Bioessays. 2003;25:1071–1084. doi: 10.1002/bies.10345. [DOI] [PubMed] [Google Scholar]
- 26.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 27.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 29.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemimethylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn BK. Hypomethylation: one side of a larger picture. Ann NY Acad Sci. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 32.Gama-Sosa MA, Wang RY, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 35.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 37.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 39.Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J Nutr. 2002;132:2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- 40.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet. 2005;14:R139–R147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 41.Weitzman SA, Turk PW, Milkowski DH, Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci USA. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 43.Goodman JI, Counts JL. Hypomethylation of DNA: a possible nongenotoxic mechanism underlying the role of cell proliferation in carcinogenesis. Environ Health Perspect. 1993;101:169–172. doi: 10.2307/3431863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 45.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- 46.Jamaluddin MD, Yang X, Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin Chem Lab Med. 2007;45:1660–1666. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 47.Reichard JF, Schnekenburger M, Puga A. Long-term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 49.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26:5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 51.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeisel SH. Genetic polymorphisms in methyl-group metabolism and epigenetics: lessons from humans and mouse models. Brain Res. 2008;1237:5–11. doi: 10.1016/j.brainres.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castro R, Rivera I, Racasco P, Camilo ME, Jakobs C, Blom HJ, de Almeida IT. 5, 10-methylenetetrahydrofolate reductase (MTHFR) 677C → T and 1298A → C mutations are associated with DNA hypomethylation. J Med Genet. 2004;41:454–458. doi: 10.1136/jmg.2003.017244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oommen AM, Griffin JB, Sarath G, Zempleni J. Roles of nutrients in epigenetic events. J Nutr Biochem. 2005;16:74–77. doi: 10.1016/j.jnutbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52:2071S–2077S. [PubMed] [Google Scholar]
- 56.Christman JK. Lipotrope deficiency and persistent changes in DNA methylation. Adv Exp Med Biol. 1995;375:97–106. doi: 10.1007/978-1-4899-0949-7_9. [DOI] [PubMed] [Google Scholar]
- 57.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehman-McKeeman LD, Gamsky EA, Hicks SM, Vassallo JD, Mar MH, Zeisel SH. Diethanolamine induces hepatic choline deficiency in mice. Toxicol Sci. 2002;67:38–45. doi: 10.1093/toxsci/67.1.38. [DOI] [PubMed] [Google Scholar]
- 59.Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Effect of trichloroethylene and its metabolites, dichloroacetic and trichloroacetic acid, on the methylation and expression of c-Jun and c-Myc protooncogenes in mouse liver: prevention by methionine. Toxicol Sci. 2000;54:399–407. doi: 10.1093/toxsci/54.2.399. [DOI] [PubMed] [Google Scholar]
- 60.Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32:1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 61.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35:7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, Melnyk S. Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J Nutr. 2003;133:3740S–3747S. doi: 10.1093/jn/133.11.3740S. [DOI] [PubMed] [Google Scholar]
- 63.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- 65.Becker FF, Holton P, Ruchirawat M, Lapeyre JN. Perturbation of maintenance and de novo DNA methylation in vitro by UVB (280–340 nm)-induced pyrimidine photodimers. Proc Natl Acad Sci USA. 1985;82:6055–6059. doi: 10.1073/pnas.82.18.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acid Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swisher JF, Rand E, Cedar H, Marie Pyle A. Analysis of putative RNase sensitivity and protease insensitivity of demethylation activity in extracts from rat myoblasts. Nucleic Acids Res. 1998;26:5573–5580. doi: 10.1093/nar/26.24.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 71.Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 72.Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 73.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 74.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 75.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 78.Flatau E, Bogenmann E, Jones PA. Variable 5-methylcytosine levels in human tumor cell lines and fresh pediatric tumor explants. Cancer Res. 1983;43:4901–4905. [PubMed] [Google Scholar]
- 79.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 80.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 81.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin CH, Hsieh SY, Sheen IS, Lee WC, Chen TC, Shyu WC, Liaw YF. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61:4238–4243. [PubMed] [Google Scholar]
- 83.Jackson K, Yu MC, Arakawa K, Fiala E, Youn B, Fiegl H, Müller-Holzner E, Widschwendter M, Ehrlich M. DNA hypomethylation is prevalent in low-grade breast cancers. Cancer Biol Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 84.Nakagawa T, Kanai Y, Ushijima S, Kitamura T, Kakizoe T, Hirohashi S. DNA hypomethylation on pericentromeric satellite regions significantly correlates with loss of heterozygosity on chromosome 9 in urothelial carcinomas. J Urol. 2005;173:243–246. doi: 10.1097/01.ju.0000154632.11824.4d. [DOI] [PubMed] [Google Scholar]
- 85.Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, Marth C, Müller-Holzner E, Zeimet AG, Laird PW, Ehrlich M. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 86.Watts GS, Futscher BW, Holtan N, DeGeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim YI, Giuliano A, Hatch KD, Schneider A, Nour MA, Dallal GE, Selhub J, Mason JB. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::AID-CNCR2820740316>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 88.Christman JK. Diet, DNA methylation and cancer. In: Daniel H, Zempleni J, editors. Molecular nutrition. Oxon: CABI Publishing; 2003. pp. 237–265. [Google Scholar]
- 89.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcίa-Closas R, Chanock S, Tardón A, Serra C, Carrato A, Dosemeci M, Garcίa-Closas M, Esteller M, Fraga M, Rothman N, Malats N. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish bladder cancer study: a case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 91.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan T, Schmidtmann A, Xi S, Briones V, Zhu H, Suh HC, Gooya J, Keller JR, Xu H, Roayaei J, Anver M, Ruscetti S, Muegge K. DNA hypomethylation caused by LSH deletion promotes erythroleukemia development. Epigenetics. 2008;3:134–142. doi: 10.4161/epi.3.3.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong NC, Wong LH, Quach JM, Canham P, Craig JM, Song JZ, Clark SJ, Choo LHA. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet. 2006;2:e17. doi: 10.1371/journal.pgen.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 97.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 98.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 99.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- 100.Cui H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis Markers. 2007;23:105–112. doi: 10.1155/2007/363464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 102.Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S, Woodfine K, Jagodic M, Nativio R, Dunning A, Moore G, Klenova E, Bingham S, Pharoah PD, Brenton JD, Beck S, Sandhu MS, Murrell A. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang SH, Yang DH, Huang W, Zhou HK, Lu XH, Ye G. Hypomethylation of P4 promoter induces expression of the insulin-like growth factor-II gene in hepatocellular carcinoma in a Chinese population. Clin Cancer Res. 2006;12:4171–4177. doi: 10.1158/1078-0432.CCR-05-2261. [DOI] [PubMed] [Google Scholar]
- 104.Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10:2619–2626. doi: 10.1093/hmg/10.23.2619. [DOI] [PubMed] [Google Scholar]
- 105.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–1198. [PubMed] [Google Scholar]
- 106.Scelfo RA, Schwienbacher C, Veronese A, Gramantieri L, Bolondi L, Querzoli P, Nenci I, Calin GA, Angioni A, Barbanti-Brodano G, Negrini M. Loss of methylation at chromosome 11p15.5 is common in human adult tumors. Oncogene. 2002;21:2564–2572. doi: 10.1038/sj.onc.1205336. [DOI] [PubMed] [Google Scholar]
- 107.Ehrich M, Turner J, Gibbs P, Lipton L, Giovanneti M, Cantor C, van den Boom D. Cytosine methylation profiling of cancer cell lines. Proc Natl Acad Sci USA. 2008;105:4844–4849. doi: 10.1073/pnas.0712251105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakamura N, Takenaga K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin Exp Metastasis. 1998;16:471–479. doi: 10.1023/A:1006589626307. [DOI] [PubMed] [Google Scholar]
- 111.Xie R, Loose DS, Shipley GL, Xie S, Bassett RL, Jr, Broaddus RR. Hypomethylation-induced expression of S100A4 in endometrial carcinoma. Mod Pathol. 2007;20:1045–1054. doi: 10.1038/modpathol.3800940. [DOI] [PubMed] [Google Scholar]
- 112.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Coggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 113.Gomez A, Karlgren M, Edler D, Bernal ML, Mkrtchian S, Ingelman-Sundberg M. Expression of CYP2W1 in colon tumors: regulation by gene methylation. Pharmacogenomics. 2007;8:1315–1325. doi: 10.2217/14622416.8.10.1315. [DOI] [PubMed] [Google Scholar]
- 114.Milicic A, Harrison LA, Goodlad RA, Hardy RG, Nicholson AM, Presz M, Sieber O, Santander S, Pringle JH, Mandir N, East P, Obszynska J, Sanders S, Piazuelo E, Shaw J, Harrison R, Tomlinson IP, McDonald SA, Wright NA, Jankowski JA. Ectopic expression of P-cadherin correlates with promoter hypomethylation early in colorectal carcinogenesis and enhanced intestinal crypt fission in vivo. Cancer Res. 2008;68:7760–7768. doi: 10.1158/0008-5472.CAN-08-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paredes J, Albergaria A, Oliveira JT, Jerónimo C, Milanezi F, Schmidt FC. P-cadherin overexpression is an indicator of clinical outcome in invasive breast carcinomas and is associated with CDH3 promoter hypomethylation. Clin Cancer Res. 2005;11:5869–5877. doi: 10.1158/1078-0432.CCR-05-0059. [DOI] [PubMed] [Google Scholar]
- 116.Grunau C, Brun ME, Rivals I, Selves J, Hindermann W, Favre-Mercuret M, Granier G, De Sario A. BAGE hypomethylation, a new epigenetic biomarker for colon cancer detection. Cancer Epidemiol Biomarkers Prev. 2008;17:1374–1379. doi: 10.1158/1055-9965.EPI-07-2656. [DOI] [PubMed] [Google Scholar]
- 117.Adany R, Iozzo RV. Hypomethylation of the decorin proteoglycan gene in human colon cancer. Biochem J. 1991;276:301–306. doi: 10.1042/bj2760301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim KH, Choi JS, Kim IJ, Ku JL, Park JG. Promoter hypomethylation and reactivation of MAGE-A1 and MAGE-A3 genes in colorectal cancer cell lines and cancer tissues. World J Gastroenterol. 2006;12:5651–5657. doi: 10.3748/wjg.v12.i35.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jung EJ, Kim MA, Lee HS, Yang HK, Lee YM, Lee BL, Kim WH. Expression of family A melanoma antigen in human gastric carcinoma. Anticancer Res. 2005;25:2105–2211. [PubMed] [Google Scholar]
- 120.Lim JH, Kim SP, Gabrielson E, Park YB, Park JW, Kwon TK. Activation of human cancer/testis antigen gene, XAGE-1, in tumor cells is correlated with CpG island hypomethylation. Int J Cancer. 2005;116:200–206. doi: 10.1002/ijc.21007. [DOI] [PubMed] [Google Scholar]
- 121.Oshimo Y, Nakayama H, Ito R, Kitadai Y, Yoshida K, Chayama K, Yasui W. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663–1670. [PubMed] [Google Scholar]
- 122.Akiyama Y, Maesawa C, Ogasawara S, Terashima M, Masuda T. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am J Pathol. 2003;163:1911–1919. doi: 10.1016/S0002-9440(10)63549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cao D, Zhang Q, Wu LS, Salaria SN, Winter JW, Hruban RH, Goggins MS, Abbruzzese JL, Maitra A, Ho L. Prognostic significance of maspin in pancreatic ductal adenocarcinoma: tissue microarray analysis of 223 surgically resected cases. Mod Pathol. 2007;20:570–578. doi: 10.1038/modpathol.3800772. [DOI] [PubMed] [Google Scholar]
- 124.Ogasawara S, Maesawa C, Yamamoto M, Akiyama Y, Wada K, Fujisawa K, Higuchi T, Tomisawa Y, Sato N, Endo S, Saito K, Masuda T. Disruption of cell-type-specific methylation at the Maspin gene promoter is frequently involved in undifferentiated thyroid cancers. Oncogene. 2004;23:1117–1124. doi: 10.1038/sj.onc.1207211. [DOI] [PubMed] [Google Scholar]
- 125.Mesquita P, Peixoto AJ, Seruca R, Hanski C, Almeida R, Silva F, Reis C, David L. Role of site-specific promoter hypomethylation in aberrant MUC2 mucin expression in mucinous gastric carcinoma. Cancer Lett. 2003;189:129–136. doi: 10.1016/S0304-3835(02)00549-9. [DOI] [PubMed] [Google Scholar]
- 126.Cui L, Xu LY, Shen ZY, Tao Q, Gao SY, Lv Z, Du ZP, Fang WK, Li EM. NGALR is overexpressed and regulated by hypomethylation in esophageal squamous cell carcinoma. Clin Cancer Res. 2008;14:7674–7681. doi: 10.1158/1078-0432.CCR-08-0420. [DOI] [PubMed] [Google Scholar]
- 127.Tabu K, Sasai K, Kimura T, Wang L, Aoyanagi E, Kohsaka S, Tanino M, Nishihara H, Tanaka S. Promoter hypomethylation regulates CD133 expression in human gliomas. Cell Res. 2008;18:1037–1046. doi: 10.1038/cr.2008.270. [DOI] [PubMed] [Google Scholar]
- 128.Kim SJ, Kang HS, Chang HL, Jung YC, Sim HB, Lee KS, Ro J, Lee ES. Promoter hypomethylation of the N-acetyltransferase 1 gene in breast cancer. Oncol Rep. 2008;19:663–668. [PubMed] [Google Scholar]
- 129.Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, Kernstine KH, Lin D, Shen B. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 131.Czekierdowski A, Czekierdowska S, Wielgos M, Smolen A, Kaminski P, Kotarski J. The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-gamma (SNCG) in ovarian cancer. Neuro Endocrinol Lett. 2006;27:381–386. [PubMed] [Google Scholar]
- 132.Pakneshan P, Szyf M, Rabbani SA. Hypomethylation of urokinase (uPA) promoter in breast and prostate cancer: prognostic and therapeutic implications. Curr Cancer Drug Targets. 2005;5:471–488. doi: 10.2174/156800905774574011. [DOI] [PubMed] [Google Scholar]
- 133.van den Eynden GG, van Laere SJ, van der Auwera I, Merajver SD, van Marck EA, van Dam PB, Vermeulen PB, Dirix LY, van Golen KL. Overexpression of caveolin-1 and -2 in cell lines and in human samples of inflammatory breast cancer. Breast Cancer Res Treat. 2006;95:219–228. doi: 10.1007/s10549-005-9002-1. [DOI] [PubMed] [Google Scholar]
- 134.Rodenhiser DI, Andrews J, Kennette W, Sadicovic B, Mendlowitz A, Tuck AB, Chambers AF. Epigenetic mapping and functional analysis in breast cancer metastasis using whole-genome promoter tiling microarrays. Breast Cancer Res. 2008;10:R62. doi: 10.1186/bcr2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okada H, Kimura MT, Tan D, Fujiwara K, Igarashi J, Makuuchi M, Hui AM, Tsurumaru M, Nagase H. Frequent trefoil factor 3 (TFF3) overexpression and promoter hypomethylation in mouse and human hepatocellular carcinomas. Int J Oncol. 2005;2005(26):369–377. [PMC free article] [PubMed] [Google Scholar]
- 136.Litkouci B, Kwong J, Lo CM, Smedley JG, 3rd, McClane BA, Aponte M, Gao Z, Sarno JL, Hinners J, Welch WR, Berkowitz RS, Mok SC, Garner EI. Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia. 2007;9:304–314. doi: 10.1593/neo.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, Wang D, Li R, Yi X, Zhang H, Sun L, Shang Y. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438:981–987. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- 138.Gokul G, Gautami B, Malathi S, Sowjanya AP, Poli UR, Jain M, Ramarkrishna G, Khosla S. DNA methylation profile at the DNMT3L promoter: a potential biomarker for cervical cancer. Epigenetics. 2007;2:80–85. doi: 10.4161/epi.2.2.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee TS, Kim JW, Kang GH, Park NH, Song YS, Kang SB, Lee HP. DNA hypomethylation of CAGE promotors in squamous cell carcinoma of uterine cervix. Ann NY Acad Sci. 2006;1091:218–224. doi: 10.1196/annals.1378.068. [DOI] [PubMed] [Google Scholar]
- 140.Asada H, Yamagata Y, Taketani T, Matsuoka A, Tamura H, Hattori N, Ohgane J, Hattori N, Shiota K, Sugino N. Potential link between estrogen receptor-alpha gene hypomethylation and uterine fibroid formation. Mol Hum Reprod. 2008;14:539–545. doi: 10.1093/molehr/gan045. [DOI] [PubMed] [Google Scholar]
- 141.Kato N, Tamura G, Motoyama T. Hypomethylation of hepatocyte factor-1beta (HNF-1beta) CpG island in clear cell carcinoma of the ovary. Virchows Arch. 2008;452:175–180. doi: 10.1007/s00428-007-0543-z. [DOI] [PubMed] [Google Scholar]
- 142.Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- 143.Cho M, Uemura H, Kim SC, Kawada Y, Yoshida K, Hirao Y, Konishi N, Saga S, Yoshikawa K. Hypomethylation of the MN/CA9 promoter and upregulated MN/CA9 expression in human renal cell carcinoma. Br J Cancer. 2001;85:563–567. doi: 10.1054/bjoc.2001.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chilukamarri L, Hancock AL, Malik S, Zabkiewicz J, Baker JA, Greenhough A, Dallosso AR, Huang THM, Royer-Pokora B, Brown KW, Malik K. Hypomethylation and aberrant expression of the glioma pathogenesis-related gene in Wilms tumors. Neoplasia. 2007;9:970–978. doi: 10.1593/neo.07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S, Ribeiro-Filho LA, Terashima M, Fujime M, Igawa M, Dahiya R. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 146.Schenk T, Stengel S, Goellner S, Steinbach D, Saluz HP. Hypomethylation of PRAME is responsible for its aberrant overexpression in human malignancies. Genes Chromosomes Cancer. 2007;46:796–804. doi: 10.1002/gcc.20465. [DOI] [PubMed] [Google Scholar]
- 147.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, Garate L, Cordeu L, Cervantes F, Prosper F, Heiniger A, Torres A. Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica. 2007;92:153–162. doi: 10.3324/haematol.10782. [DOI] [PubMed] [Google Scholar]
- 148.Yoshida M, Nosaka K, Yasunaga J, Nishikata I, Morishita K, Matsuoka M. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood. 2004;103:2753–2760. doi: 10.1182/blood-2003-07-2482. [DOI] [PubMed] [Google Scholar]
- 149.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. Bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 150.Yuille MR, Condie A, Stone EM, Wilsher J, Bradshaw PS, Brooks L, Catovsky D. TCL1 is activated by chromosomal rearrangement or by hypomethylation. Genes Chromosomes Cancer. 2001;30:336–341. doi: 10.1002/gcc.1099. [DOI] [PubMed] [Google Scholar]
- 151.Goldstein M, Meller I, Orr-Urtreger A. FGFR1 over-expression in primary rhabdomyosarcoma tumors is associated with hypomethylation of a 5′ CpG island and abnormal expression of the AKT1, NOG, and BMP4 genes. Genes Chromosomes Cancer. 2007;46:1028–1038. doi: 10.1002/gcc.20489. [DOI] [PubMed] [Google Scholar]
- 152.Watanabe M, Ogawa Y, Itoh K, Koiwa T, Kadin ME, Watanabe T, Okayasu I, Higashihara M, Horie R. Hypomethylation of CD30 CpG islands with aberrant JunB expression drives CD30 induction in Hodgkin lymphoma and anaplastic large cell lymphoma. Lab Invest. 2008;88:48–57. doi: 10.1038/labinvest.3700696. [DOI] [PubMed] [Google Scholar]
- 153.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 154.Hinshelwood RA, Clark SJ. Breast cancer epigenetics: normal human mammary epithelial cells as a model system. J Mol Med. 2008;86:1315–1328. doi: 10.1007/s00109-008-0386-3. [DOI] [PubMed] [Google Scholar]
- 155.Wild CP. Environmental exposure measurement in cancer epidemiology. Mutagenesis. 2009;24:117–125. doi: 10.1093/mutage/gen061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat Rev Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 157.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:53–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moggs JG, Goodman JI, Trosko JE, Roberts RA. Epigenetics and cancer: implications for drug discovery and safety assessment. Toxicol Appl Pharmacol. 2004;196:422–430. doi: 10.1016/j.taap.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 159.Karpinets TV, Foy BD. Tumorigenesis: the adaptation of mammalian cells to sustained stress environment by epigenetic alterations and succeeding matched mutations. Carcinogenesis. 2005;26:1323–1334. doi: 10.1093/carcin/bgi079. [DOI] [PubMed] [Google Scholar]
- 160.Sawan C, Vaissière T, Murr R, Herceg Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutat Res. 2008;642:1–13. doi: 10.1016/j.mrfmmm.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 161.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 162.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pisner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Banglageshi adults. Am J Clin Nutr. 2007;86:1179–1186. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 164.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J (2009) Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. doi:10.1164/rccm.200807-1097OC [DOI] [PMC free article] [PubMed]
- 165.Mensah GA, Brown DW. An overview of cardiovascular disease burden in the United States. Health Aff (Millwood) 2007;26:38–48. doi: 10.1377/hlthaff.26.1.38. [DOI] [PubMed] [Google Scholar]
- 166.Raines EW, Ross R. Biology of atherosclerotic plaque formation: possible role of growth factor lesion development and the potential impact of soy. J Nutr. 1995;125:624S–630S. doi: 10.1093/jn/125.3_Suppl.624S. [DOI] [PubMed] [Google Scholar]
- 167.Newman PE. Can reduced folic acid and vitamin B12 levels cause deficient DNA methylation producing mutations which initiate atherosclerosis? Med Hypothesis. 1999;53:421–424. doi: 10.1054/mehy.1998.0794. [DOI] [PubMed] [Google Scholar]
- 168.Nehler MR, Taylor LM, Jr, Porter JM. Homocysteinemia as a risk factor for atherosclerosis: a review. Cardiovasc Surg. 1997;5:559–567. doi: 10.1016/S0967-2109(97)00062-8. [DOI] [PubMed] [Google Scholar]
- 169.Dong C, Yoon W, Goldschmidt-Clermont J. DNA methylation and atherosclerosis. J Nutr. 2002;132:2406S–2409S. doi: 10.1093/jn/132.8.2406S. [DOI] [PubMed] [Google Scholar]
- 170.Hiltunen MO, Ylä-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler Thromb Vasc Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 171.Zaina S, Lindholm MW, Lund G. Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? J Nutr. 2005;135:5–8. doi: 10.1093/jn/135.1.5. [DOI] [PubMed] [Google Scholar]
- 172.Castro R, Rivera I, Struys EA, Jansen EEW, Ravasco P, Camilo ME, Blom HJ, Jakobs C, de Almeida IT. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 173.Yideng J, Jianzhong Z, Ying H, Juan S, Jinge Z, Shenglan W, Xiaoqun H, Shuren W. Homocysteine-mediated expression of SAHH, DMNTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol. 2007;26:603–611. doi: 10.1089/dna.2007.0584. [DOI] [PubMed] [Google Scholar]
- 174.Hiltunen MO, Turunen MP, Häkkinen TP, Rutanen J, Hedman M, Mäkinen K, Turunen AM, Aalto-Setälä K, Ylä-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 175.Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 176.Zhao L, Funk CD. Lipoxygenase pathways in atherosclerosis. Trends Cardiovasc Med. 2004;14:191–195. doi: 10.1016/j.tcm.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 177.Uhl J, Klan N, Rose M, Entian KD, Werz O, Steinhilber D. The 5-lipoxygenase promoter is regulated by DNA methylation. J Biol Chem. 2002;277:4374–4379. doi: 10.1074/jbc.M107665200. [DOI] [PubMed] [Google Scholar]
- 178.Liu C, Xu D, Sjöberg J, Forsell P, Björkholm M, Claesson HE. Transcriptional regulation of 15-lipoxygenase expression by promoter methylation. Exp Cell Res. 2004;297:61–67. doi: 10.1016/j.yexcr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 179.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 180.Wolfe MS. When loss is gain: reduced presenilin proteolytic function leads to increased Aβ42/Aβ40. EMBO Rep. 2007;8:136–140. doi: 10.1038/sj.embor.7400896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- 182.Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region -224 ~ -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999;70:288–292. doi: 10.1016/S0169-328X(99)00163-1. [DOI] [PubMed] [Google Scholar]
- 183.Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 184.Abdolmaleky HM, Thiagalingam S, Wilcox M. Genetics and epigenetics in major psychiatric disorders: dilemmas, achievements, applications, and future scope. Am J Pharmacogenomics. 2005;5:149–160. doi: 10.2165/00129785-200505030-00002. [DOI] [PubMed] [Google Scholar]
- 185.Grayson DR, Jia X, Chen Y, Sharma RP, Michell CP, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shimabukuro M, Sasaki T, Imamura A, Tsujita T, Fuke C, Umekage T, Tochigi M, Hiramatsu K, Miyazaki T, Oda T, Sugimoto J, Jinno Y, Okazaki Y. Global hypomethylation of peripheral leukocyte DNA in male patients with schizophrenia: a potential link between epigenetics and schizophrenia. J Psychiatr Res. 2007;41:1042–1046. doi: 10.1016/j.jpsychires.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 188.Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, Vacca M, D’Esposito M, D’Urso M, Galetti P, Zappia V. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinemia in patients with uraemia. Lancet. 2003;361:1693–1699. doi: 10.1016/S0140-6736(03)13372-7. [DOI] [PubMed] [Google Scholar]
- 189.Richardson B. DNA methylation and autoimmune disease. Clin Immunol. 2003;109:72–79. doi: 10.1016/S1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 190.Sekigawa I, Kawasaki M, Ogasawara H, Kaneda K, Kaneko H, Takasaki Y, Ogawa H. DNA methylation: its contribution to systemic lupus erythematosus. Clin Exp Med. 2006;6:99–106. doi: 10.1007/s10238-006-0103-x. [DOI] [PubMed] [Google Scholar]
- 191.Neidhart M, Rethage J, Kuchen S, Künzler P, Crowl RM, Billingham ME, Gay RE, Gay S. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 2000;43:2634–2647. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 192.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 193.Suuronen T, Nuutinen T, Ryhänen T, Kaarniranta K, Salminen A. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357:397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 194.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]