Abstract
The optic atrophy 3 (OPA3) gene, which has no known homolog or biological function, is mutated in patients with hereditary optic neuropathies. Here, we identified OPA3 as an integral protein of the mitochondrial outer membrane (MOM), with a C-terminus exposed to the cytosol and an N-terminal mitochondrial targeting domain. By quantitative analysis, we demonstrated that overexpression of OPA3 significantly induced mitochondrial fragmentation, whereas OPA3 knockdown resulted in highly elongated mitochondria. Cells with mitochondria fragmented by OPA3 did not undergo spontaneous apoptotic cell death, but were significantly sensitized to staurosporine- and TRAIL-induced apoptosis. In contrast, overexpression of a familial OPA3 mutant (G93S) induced mitochondrial fragmentation and spontaneous apoptosis, suggesting that OPA3 mutations may cause optic atrophy via a gain-of-function mechanism. Together, these results indicate that OPA3, as an integral MOM protein, has a crucial role in mitochondrial fission, and provides a direct link between mitochondrial morphology and optic atrophy.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0365-z) contains supplementary material, which is available to authorized users.
Keywords: Fission, Mitochondria, MOM, OPA3, Optic atrophy
Introduction
Mitochondria are dynamic organelles maintained by frequent fission and fusion processes that influence their morphology [1, 2]. The dynamic behavior of mitochondria plays a central role in a variety of cellular processes, including cell survival, development, proliferation, and apoptosis [3–5]. Mitochondrial dysfunction is emerging as a central event in the pathogenesis of several neurodegenerative diseases such as Charcot-Marie-Tooth (CMT) 2A [6], Parkinson’s disease [7], and hereditary optic neuropathies [8]. CMT2A is characterized by muscle weakness and axonal degeneration in sensory and motor neurons; it is caused by a mutation in mitofusin 2 (Mfn2), which mediates mitochondrial fusion. Inherited autosomal dominant optic atrophy (ADOA) involves the loss of visual acuity due to the degeneration of optic nerve and retinal ganglion cells. The major form of ADOA is caused by a mutation in optic atrophy 1 (OPA1), which is a protein essential for mitochondrial fusion.
The OPA3 gene has been found to be mutated in patients with autosomal, dominantly inherited optic atrophy and cataracts, as well as Costeff optic atrophy syndrome, also known as 3-methylglutaconic aciduria (MGA) type III [9, 10]. Mitochondrial inner membrane (MIM) proteomics and sequence analysis of the OPA3 protein predicted that OPA3 localizes to the inner membrane [9, 11]. Homozygous ENU-induced OPA3 mutant mice (OPA3L122P) display the features of MGA type III: reduced visual function, loss of retinal ganglion cells, and degeneration of the optic nerve [12]. Although OPA3 was proposed to play an important role in optic atrophy and neuronal degeneration, OPA3 protein function and regulation that underlie OPA3-linked pathogenic processes remain poorly understood. Here, we show that OPA3 is an integral protein of the mitochondrial outer membrane (MOM) and has an important role in regulating mitochondrial dynamics and sensitivity to apoptotic cell death.
Materials and methods
Reagents
OPA3-specific polyclonal antibody was raised against a GST-fused partial OPA3 protein (residues 61–179). Purified OPA3 antibody was used at a 1:500 dilution for Western-blot analysis (Supplementary Fig. 1D in ESM). Antibodies against FLAG, GFP, actin, and myc were purchased from Sigma (St. Louis, MO, USA). Antibodies against Tom 20, Tim 23, OPA1, cytochrome c, and DRP1 were from BD Bioscience. Antibodies against caspase-3 and PARP were purchased from Cell Signaling (Beverly, MA, USA). The polyclonal Bax antibody was from Upstate Biotechnology (Bellerica, MA, USA). Fluor 594-conjugated goat anti-mouse and goat anti-rabbit IgG and Fluor 488-conjugated goat anti-mouse and goat anti-rabbit IgG were purchased from Molecular Probes (Eugene, OR, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Proteinase K was from Sigma, and digitonin was purchased from Calbiochem (La Jolla, CA, USA).
Expression constructs, cell culture, and transfection
OPA3 cDNA (NP_079412) was amplified by PCR, digested with EcoR1 and Sal1, and ligated into the pFLAG-CMV-6A plasmid (Sigma; indicated as FLAG-OPA3) and the pCMV 3Tag4B plasmid (Stratagene, La Jolla, CA, USA; indicated as OPA3-myc). Wild-type OPA3, OPA3 deletion mutants, OPA3 variant 1 (NP_001017989, indicated as OPA3-v1) and Drosophila OPA3 [NP_651192, indicated as Drosophila OPA3 (dOPA3)] were amplified using specific primers, digested with restriction endonucleases, and ligated into the pEYFP-N1 plasmid (Clontech, Mountain View, CA, USA; indicated as OPA3-YFP and OPA3-YFP mutants). The nucleotide sequences of the wild-type and mutant OPA3 inserts were confirmed by DNA sequencing. Mito-YFP and mito-DsRed (Clontech) were used as mitochondrial controls.
HeLa cells and COS-7 cells were grown in Dulbecco’s complete medium supplemented with 10% heat-treated fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). The cells were transfected using Effectene reagent (Qiagen, Hilden, Germany) and FuGene 6 (Roche, Basel, Switzerland).
Fluorescence recovery after photobleaching
To measure mitochondrial fusion, fluorescence recovery after photobleaching (FRAP) was performed as described previously [13]. Briefly, cells in two-well chamber slides (Nunc, Rochester, NY, USA) were transfected with mito-YFP, together with myc-fused vector or OPA3-myc (1:3 ratio), respectively. After 15 h, the cells were imaged using a LSM510 Meta imaging station (Carl Zeiss MicroImaging Inc.). A small region of identical size (white circle, ROI) was photobleached in mito-YFP-expressing cells, using a 30.0-mV argon laser set to 488 nm with 30% laser power output and 100% transmission, until the fluorescence intensity of the region reached zero. The region was then monitored for YFP fluorescence recovery. Fluorescence intensity was normalized to the intensity of the ROI in the first image of the series, and fluorescence intensity recovery rates were plotted. Data represent the mean ± SD of experiments with 70 cells per treatment.
Assessment of apoptotic cell death
To measure staurosporine (STS)- and TRAIL-induced apoptosis, cells were incubated in the absence or presence of 5 μg/ml cyclohexamide (CHX) for 3 h and then treated with 300 ng/ml TRAIL or 1 μM STS. The cells were immunostained with anti-cytochrome c or anti-Bax antibodies and observed under a confocal microscope. For caspase-3 activation and poly(ADP-ribose) polymerase (PARP) cleavage, cells were harvested and analyzed by Western blotting with anti-PARP and anti-active caspase-3 antibodies.
Subcellular fractionation and proteinase K assay
For subcellular fractionation, cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), and resuspended in five volumes of buffer containing 10 mM HEPES–KOH, pH 7.4, 38 mM NaCl, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM phenylmethyl sulfonyl fluoride (PMSF) with 250 mM sucrose. The cells were homogenized with 50 strokes in a Dounce homogenizer, and the homogenates were cleared by centrifugation (750 × g, 10 min, 4°C). The mitochondria were collected by centrifugation (10,000 × g, 10 min, 4°C) and resuspended in mitochondria extraction buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.2% Triton X-100, 0.3% NP-40, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). The supernatant was used to prepare the cytosolic fraction by ultracentrifugation (100,000 × g, 30 min, 4°C). For the proteinase assay, the mitochondrial fraction was washed with IB buffer (220 mM mannitol, 70 mM sucrose, 10 mM HEPES/KOH, pH 7.6, 1 mM MgCl2, 1 mM DTT, and 1 mM EDTA), proteinase K (10 μg/ml) was added to the samples in IB buffer or swelling buffer (20 mM HEPES/KOH, pH 7.4), and the samples were incubated on ice for 30 min. Protease activity was terminated by the addition of SDS-PAGE sample buffer.
In vitro mitochondrial import
Mitochondria (1 mg/ml) were isolated from healthy cells and incubated in import buffer (220 mM mannitol, 70 mM sucrose, 10 mM HEPES/KOH, pH 7.2, 1 mM MgCl2, 1 mM EDTA and 1 mM dithiothreitol) containing 1 mM ATP, 0.1 mM GTP, and 5 mM NADH. Import reactions were started by addition of [35S]-labeled OPA3 synthesized by in vitro transcription and translation in rabbit reticulocyte lysate (TNT coupled system, Promega). After incubation, mitochondria were recovered by centrifugation at 10,000 × g for 10 min at 4°C. Isolated mitochondria were incubated with trypsin for 20 min at 4°C after import.
Western-blot analysis
Cell lysates, mitochondrial extracts, and cytosolic fractions were analyzed by Western blotting. SDS-PAGE was carried out in 4–12% polyacrylamide gradient gels (NuPAGE, Invitrogen, Carlsbad, CA, USA). The separated proteins were electroblotted onto nitrocellulose or polyvinylidene fluoride (PVDF) membranes (Invitrogen). Anti-FLAG, anti-myc, anti-Tom 20, anti-OPA1, anti-OPA3, anti-actin, anti-GFP, anti-PLC-γ, and anti-DRP1 antibodies were used.
RNA interference
The experiments using OPA3 siRNAs and RNAi negative control duplexes were performed using an Invitrogen Stealth RNAi system. The target sequences for OPA3 were: 5′-AGCAAGCCGCTTGCCAACCGTATTA-3′ (#156), 5′-GCCGAAGCGAGTTCTTCAAGACCTA-3′ (#157) and 5′-TCACTGGGTGGAGATGCGGACCAAG-3′ (#158). DRP1 shRNA and FIS1 shRNA were cloned as described previously [14]. One day after the cells were transfected with these siRNAs, the medium was changed, and the cells were grown for an additional 2 days prior to further experiments.
Immunofluorescence assay
Cells grown in two-well chamber slides were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.15% Triton X-100 in PBS for 15 min at room temperature, and then blocked with 3% bovine serum albumin in PBS for 45 min at room temperature. The slides were incubated with monoclonal anti-Tom 20, monoclonal anti-cytochrome c, polyclonal anti-Bax, polyclonal anti-FLAG, or polyclonal anti-myc antibodies as the primary antibody. After washing with PBS, the slides were incubated with goat anti-mouse IgG conjugated with Alexa Fluor 488 or 594 or goat anti-rabbit IgG conjugated with Alexa Fluor 488 as the secondary antibody. The slides were observed under a Zeiss LSM 510 confocal microscope using 63× and 40× Apochromat objectives (Zeiss). The excitation wavelengths for YFP, Alexa Fluor 594, Alexa Fluor 488, and DsRed were 514, 594, 488, and 543 nm, respectively.
Digitonin differential permeabilization
For topology experiments, differential permeabilization was performed as described previously [15]. Briefly, HeLa cells were transfected with the indicated plasmids. After 15 h, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with increasing concentrations of digitonin in PBS, and immunofluorescently labeled using anti-FLAG, anti-myc, anti-Tom 20, or anti-cytochrome c antibodies.
Results
OPA3 affects mitochondrial morphology
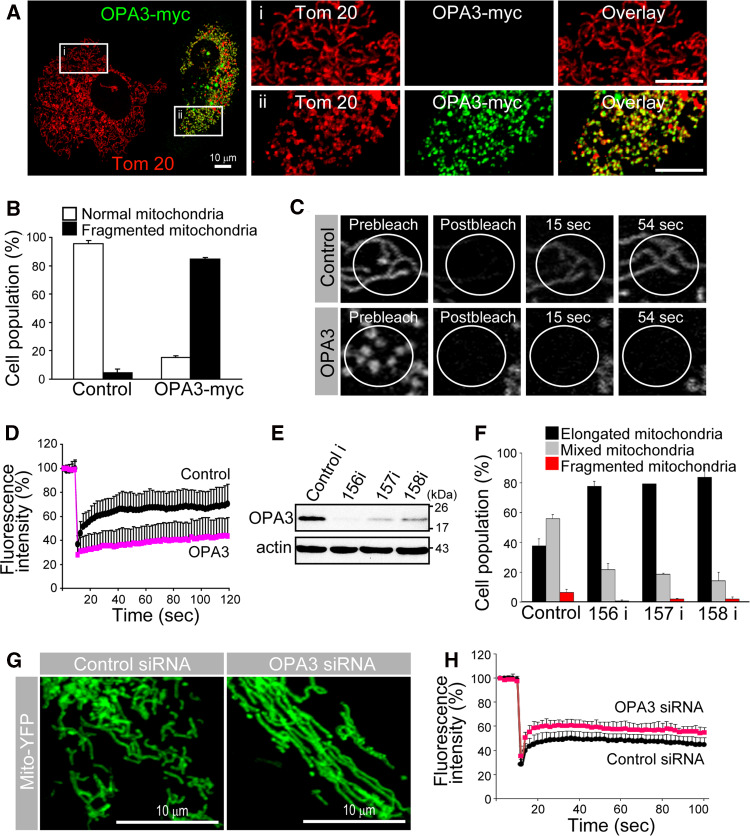
To determine the function of OPA3, we first investigated whether it had an impact on mitochondrial morphology in HeLa, COS 7, and 293 cells. The C-terminally myc-tagged OPA3 (OPA3-myc) expressed in HeLa cells co-localized with the mitochondrial marker protein Tom 20 (Fig. 1a). Interestingly, HeLa cells transfected with OPA3-myc frequently showed abnormal mitochondrial morphology, which led to extensive mitochondrial fragmentation (Fig. 1a(ii)). In contrast, most of the cells transfected with control myc-vector had mitochondria with normal morphology, and short- and long-form mitochondria were present in each cell (Fig. 1b). Less than 5% of the cells transfected with control myc-vector had fragmented mitochondria, whereas most (84 ± 1%) of the cells transfected with OPA3-myc had fragmented mitochondria. To quantify the OPA3-induced mitochondrial fragmentation in HeLa cells, we analyzed mitochondrial volume and connectivity using the FRAP technique. In the control cells, the fluorescence of the mitochondrial matrix-targeted yellow fluorescent protein (mito-YFP) recovered rapidly (within ~10 s) into the bleached area of the mitochondria; however, the recovery of fluorescence into the bleached area was significantly blocked in OPA3-myc-transfected cells (Fig. 1c, d). These results clearly indicate that OPA3-myc overexpression induced mitochondrial fragmentation.
Fig. 1.
Effect of OPA3 on mitochondrial morphology. a, b Cells were stained with fluorescently labeled anti-myc (green) and anti-Tom 20 (red) antibodies. Higher magnification of the white square is presented in the right panel: top without OPA3-myc (i); and bottom with OPA3-myc (ii). Data are the mean ± SD of three experiments, each with 100 cells per treatment. Bar 10 μm. c, d The white circle (ROI) is the photobleached area monitored for recovery of mito-YFP fluorescence. Each line represents the mean of >70 measurements (d). e Total extracts from cells transfected with OPA3 RNAi (156i, 157i, and 158i) were analyzed by Western blotting with anti-OPA3 antibody. Actin was used as a loading control. f, g Mitochondria with OPA3 siRNA or control siRNA, together with mito-YFP, were analyzed by confocal microscopy. The number of cells displaying normal mitochondria (mixed), elongated mitochondria, and fragmented mitochondria were counted. Data are the mean ± SD of three experiments, each with 100 cells per treatment. h The fusion activity of individual mitochondria measured by fluorescence recovery after photobleaching in living OPA3-depleted HeLa cells
To confirm the effect of OPA3 on mitochondrial morphology, we designed three specific siRNAs (156i, 157i, and 158i) for knockdown of endogenous OPA3 protein. The decreased level of OPA3 protein was validated using a rabbit anti-OPA3 polyclonal antibody generated against truncated human OPA3 (62–179) with GST (Supplementary Fig. 1A). After transient transfection with OPA3 siRNA, the levels of OPA3 protein were significantly reduced compared to those in control siRNA transfectants (Fig. 1e). In HeLa cells transfected with control siRNA, less than ~40% of the mitochondria showed elongated mitochondrial morphology, and a small fraction (6.5 ± 2.2%) exhibited the short form (Fig. 1f, g). By contrast, most OPA3-knockdown HeLa cells had more elongated and tubular mitochondria. Fluorescence recovery in bleached areas was significantly more prominent and rapid in OPA3 RNAi cells than in control siRNA transfectants (Fig. 1h).
Knockdown of DRP1 and FIS1 does not affect mitochondrial fragmentation induced by OPA3
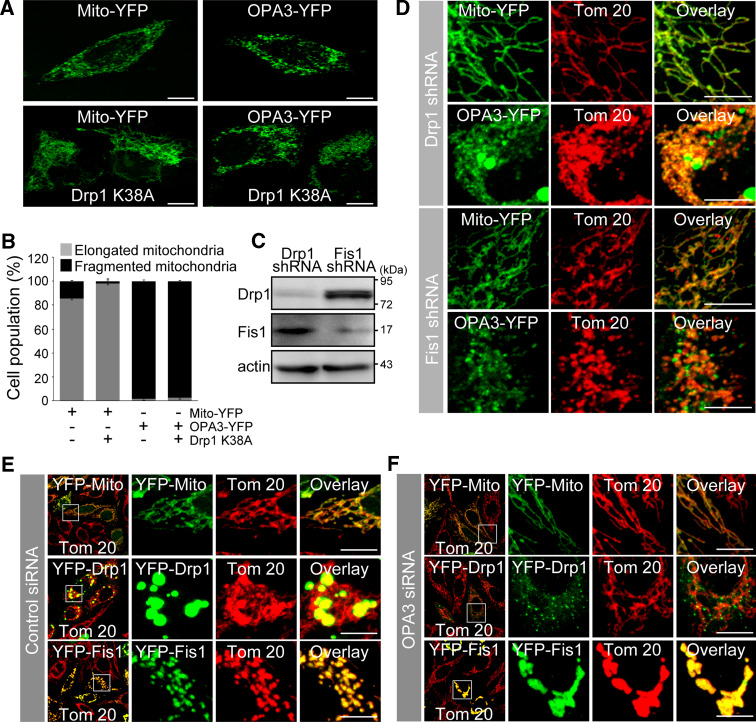
To test whether OPA3-induced mitochondrial fission was dependent on the activity of dynamin-related protein (DRP) 1, we cotransfected C-terminally YFP-tagged OPA3 (OPA3-YFP) along with N-terminally myc-tagged DRP1 K38A (myc-DRP1 K38A), a dominant-negative mutant. As expected, 98% of the transfected cells with myc-DRP1 K38A showed highly interconnected mitochondria, while there was no effect of myc-DRP1 K38A on the OPA3-mediated mitochondrial fragmentation (Fig. 2a, b). Consistent with this, knockdown of DRP1 or fission (FIS) 1 using RNA interference also did not affect mitochondrial fragmentation induced by OPA3 (Fig. 2c, d). To examine the effects of OPA3 knockdown on DRP1/FIS1-mediated mitochondrial fission, HeLa cells were transfected with OPA3 siRNA along with YFP-DRP1 or YFP-FIS1. We observed increased recruitment of large punctate YFP-DRP1 at the fission sites of the mitochondria and extensive mitochondrial fragmentation by overexpression of YFP-FIS1 (Fig. 2e). Conversely, OPA3 knockdown significantly induced a cytosolic distribution of YFP-DRP1 in a more diffuse manner, with smaller punctate foci and reduced FIS1-induced mitochondrial fragmentation (Fig. 2f). These results clearly indicate that OPA3 can induce mitochondrial fragmentation independently on DRP1/FIS1-mediated mitochondrial fission.
Fig. 2.
Effect of OPA3 depletion on DRP1/FIS1-mediated mitochondrial fission. a, b HeLa cells were transfected with the indicated constructs (1:3 ratios). After 15 h, the cells were fixed and analyzed by confocal microscopy. Bar 10 μm. c HeLa cells stably transfected with DRP1 shRNA or FIS1 shRNA were harvested and analyzed by Western blotting with anti-DRP1, anti-FIS1, and anti-actin antibodies. d HeLa cells stably expressing DRP1 shRNA or FIS1 shRNA were transfected with the indicated constructs. Cells were fixed, stained with anti-Tom 20 antibody (red) and analyzed by confocal microscopy. Bar 10 μm. e–g HeLa cells were transfected with control siRNA (e) or OPA3 siRNA (f), along with mito-YFP, YFP-DRP1, or YFP-FIS1. Cells were fixed, stained with anti-Tom 20 (red) and analyzed by confocal microscopy. Higher magnification of the white square is presented in the right panel of the merged image. Bar 10 μm
OPA3 increases sensitivity to apoptotic signals
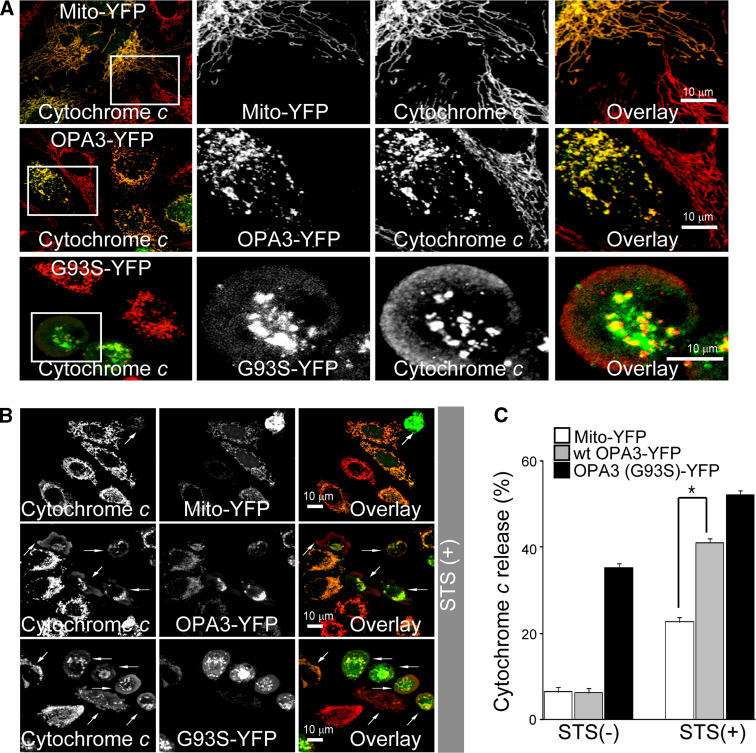
It has been established that altered mitochondrial morphology is closely related with apoptotic sensitivity [5]. We examined whether extrinsic expression of OPA3 could lead to cell death in the absence or presence of apoptotic stimuli. HeLa cells were transiently transfected with OPA3-YFP or mito-YFP and further examined the cytochrome c release and mitochondrial intermembrane potential. Cells with fragmented mitochondria attributable to OPA3 overexpression showed normal cytochrome c distribution (Fig. 3a) and concentrated TMRE fluorescence intensity (data not shown), indicating that OPA3 overexpression alone is not sufficient to induce spontaneous apoptotic cell death. However, treatment with STS significantly increased the redistribution of cytochrome c from the mitochondria to the cytosol in cells with OPA3-YFP (41.0 ± 1.2%), compared with cells with mito-YFP (22.6 ± 2.1%; Fig. 3b, c), showing that OPA3-induced mitochondrial fragmentation augments the sensitivity to STS-stimulated apoptosis. On the other hand, cytochrome c appeared in the cytosol of HeLa cells transfected with a patient OPA3 mutant, OPA3 (G93S) in the absence of external apoptotic stimuli (Fig. 3a–c). Cells with OPA3 (G93S)-YFP showed extensive mitochondrial fragmentation (Supplementary Fig. 2B), comparable to that induced by the overexpression of wild-type OPA3.
Fig. 3.
Overexpression of OPA3 increases the release of cytochrome c with apoptotic stimuli. a, b HeLa cells were transfected with the indicated constructs. After 15 h, the cells were treated without (a) or with (b) 1 μM STS for 30 min, stained with fluorescently labeled anti-cytochrome c antibody (red), and analyzed by confocal microscopy. Higher magnification of the white square a is presented in the right panel of the merged image. Bar 10 μm. White arrows b indicate cells with YFP and cytosolic cytochrome c. c The number of cells displaying cytosolic and mitochondrial cytochrome c were counted and are shown as a percentage of the total cells counted in each green image cell population. Data are the mean ± SD of three experiments, each with 100 cells per condition. *p < 0.05
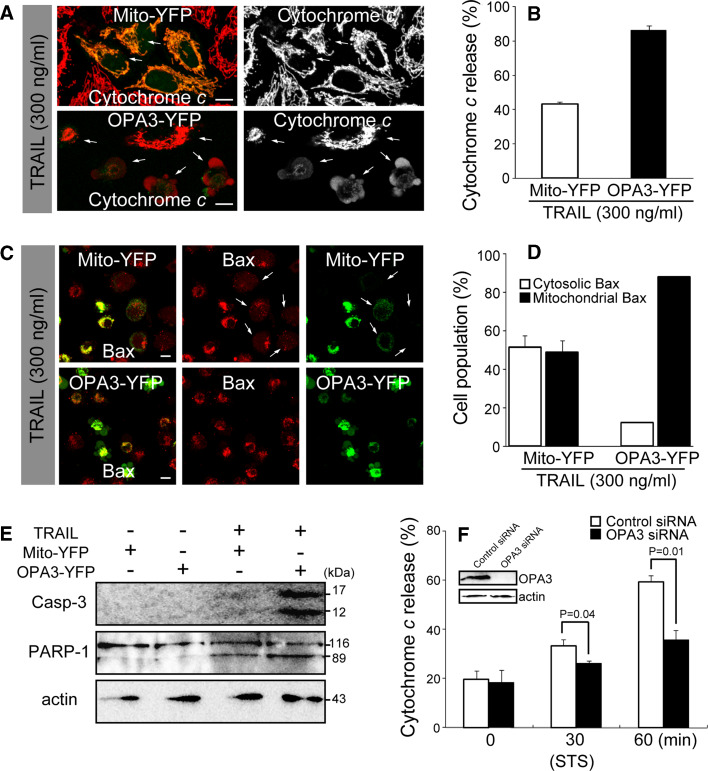
OPA3 expression also increased the sensitivity to TRAIL-induced cell apoptosis. Treatment of the cells with TRAIL induced redistribution of cytochrome c from the mitochondria to the cytosol in cells with OPA3-YFP (85.8 ± 2.8%), compared with cells with mito-YFP (43.2 ± 1.2%; Fig. 4a, b). Consistent results were obtained for the translocation of Bax to mitochondria (Fig. 4c, d), caspase-3 activation, and PARP cleavage (Fig. 4e). To validate the apparent role of OPA3 in apoptotic sensitivity, we examined OPA3-knockdown HeLa cells and measured cytochrome c release following STS treatment. As shown in Fig. 4f, OPA3 knockdown significantly decreased the release of cytochrome c by STS, indicating that depletion of OPA3 could protect against apoptotic cell death.
Fig. 4.
Overexpression of OPA3 leads to apoptotic sensitivity of cells. a–e HeLa cells were transfected with the indicated constructs. Cells were treated with TRAIL for 6 h, stained with fluorescently labeled anti-cytochrome c antibody (a) or anti-Bax antibody (c), and analyzed by confocal microscopy. The number of cells displaying cytosolic cytochrome c (b) and Bax translocation (d) were counted, and are shown as a percentage of the total cells counted in each green image cell population. Data are the mean ± SD of three experiments, each with 100 cells per condition. Bar 10 μm. White arrows indicate cells with YFP and cytosolic cytochrome c (a), or with YFP and cytosolic Bax (c). e Caspase-3 activation and PARP cleavage were assessed by Western blotting with the indicated antibodies. f HeLa cells were transfected with the control siRNA or OPA3 siRNA. Cells were treated with 1 μM STS for the indicated time. Cells were stained with anti-cytochrome c antibody, and then analyzed by confocal microscopy. The numbers of cells displaying cytosolic cytochrome c were counted, and are shown as a percentage of the total cells counted. Data are the mean ± SD of three experiments, each with 100 cells per condition. Knockdown of OPA3 was assessed by Western blotting using anti-OPA3 and anti-actin antibodies (f)
OPA3 is in the mitochondrial outer membrane
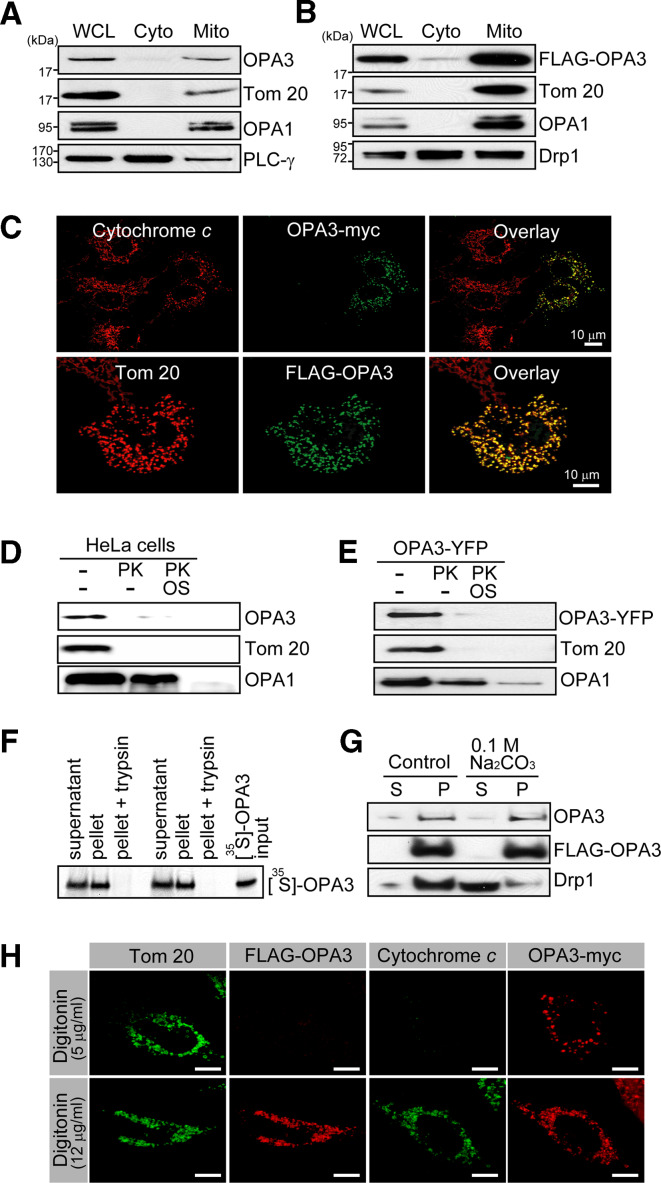
To determine the localization of OPA3, we performed subcellular fractionations. Both endogenous and extrinsically introduced OPA3 were detected in the mitochondrial fraction, but not in the cytosolic fraction (Fig. 5a, b). Consistent with this, OPA3-myc and FLAG-OPA3 colocalized with mitochondrial proteins such as cytochrome c and Tom 20 in HeLa cells (Fig. 5c, upper panel) and COS7 cells (Fig. 5c, lower panel). After confirming that OPA3 was located in the mitochondria, we investigated whether it is localized in the MOM or the MIM by treating mitochondrial fractions with proteinase K. The MIM protein OPA1 was degraded by proteinase K only after osmotic shock (Fig. 5d). In contrast, endogenous OPA3 and the MOM protein Tom 20 were digested by proteinase K even in the absence of osmotic shock, indicating that OPA3 is localized in the MOM (Fig. 5d). Similar results were obtained using OPA3-YFP (Fig. 5e), FLAG-OPA3, and OPA3-myc (Supplementary Fig. 1B, C). Consistent with this, we did not detect [35S]-labeled OPA3 upon trypsin treatment of mitochondria that had been incubated with [35S]-labeled OPA3 in an ATP regeneration system (Fig. 5f). As OPA3 might have been degraded by proteinase K whether it is peripherally or integrally associated with the MOM, we used high-pH sodium carbonate (Na2CO3) to extract the peripherally attached proteins from the MOM. DRP1 was extracted with this alkali treatment, consistent with its known peripheral attachment to the MOM (Fig. 5g). In contrast, endogenous OPA3 and FLAG-OPA3 were precipitated with the membrane fraction in both control buffer and 0.1 M Na2CO3 (Fig. 5g). These results collectively indicate that OPA3 is an integral protein, localized in the MOM.
Fig. 5.
Localization of OPA3 on the mitochondrial outer membrane (MOM). a, b HeLa cells or FLAG-OPA3-expressing HeLa cells were harvested. Two subcellular fractions were prepared, cytosol (Cyto) and mitochondria (Mito), and analyzed by Western blotting with the indicated antibodies. WCL Whole-cell lysate. c HeLa cells (upper panel) and COS7 cells (low panel) were transfected with OPA3-myc and FLAG-OPA3, respectively. Cells were fixed, and stained with fluorescently labeled anti-cytochrome c and anti-myc antibodies, or anti-Tom 20 and anti-FLAG antibodies, followed by confocal microscopy. Mitochondrial fractions (d) and mitochondrial fraction with OPA3-YFP (e) in control buffer or swelling buffer (OS) were digested by proteinase K (PK). Western blots of the separated proteins were probed with anti-OPA3, anti-GFP, anti-Tom 20, and anti-OPA1 antibodies. f Mitochondria labeled with in vitro translated [35S]-OPA3 were incubated with and without 200 μg/ml trypsin. Proteins were separated by SDS-PAGE. [35S]-OPA3 was detected using Kodak X-ray film. g Alkali (Na2CO3)-treated mitochondria were extracted and subjected to further ultracentrifugation to separate soluble (S) and pelleted proteins (P). Western blots of the separated proteins were probed with anti-OPA3, anti-FLAG, and anti-DRP1 antibodies. h Cells with FLAG-OPA3 or OPA3-myc were permeabilized with the indicated concentrations of digitonin and were co-immunostained with fluorescently labeled anti-FLAG and anti-Tom 20 antibodies, or anti-myc, and anti-cytochrome c antibodies. Bar 10 μm
To understand the role of OPA3 in mitochondrial dynamics and function, it is important to understand its topology in the MOM. To this end, we employed a differential permeabilization technique, using different concentrations of digitonin to selectively break the plasma membrane and MOM. In HeLa cells treated with 5 μg/ml digitonin (Fig. 5h), Tom 20 was immunostained by an antibody against its TPR motif, which is exposed to the cytosol, but cytochrome c was not immunostained by an antibody against cytochrome c, which is localized in the mitochondrial intermembrane space. Both cytochrome c and Tom 20 were immunostained in HeLa cells treated with 12 μg/ml digitonin. As shown in Fig. 5h, an antibody against the C-terminally fused myc tag immunostained OPA3-myc in mitochondria after treatment with 5 μg/ml digitonin. However, cytochrome c was not detected by immunostaining under the same conditions, confirming that the C-terminus of OPA3 is exposed to the cytosol. Immunostaining of FLAG-OPA3 by an antibody against the N-terminally fused FLAG tag was positive only in the presence of 12 μg/ml digitonin, which also resulted in positive immunostaining of cytochrome c. Together, these results demonstrate that OPA3 is anchored in the MOM with its N-terminal region exposed to the mitochondrial intermembrane space and its C-terminal region exposed to the cytosol.
Its N-terminus targets OPA3 to mitochondria
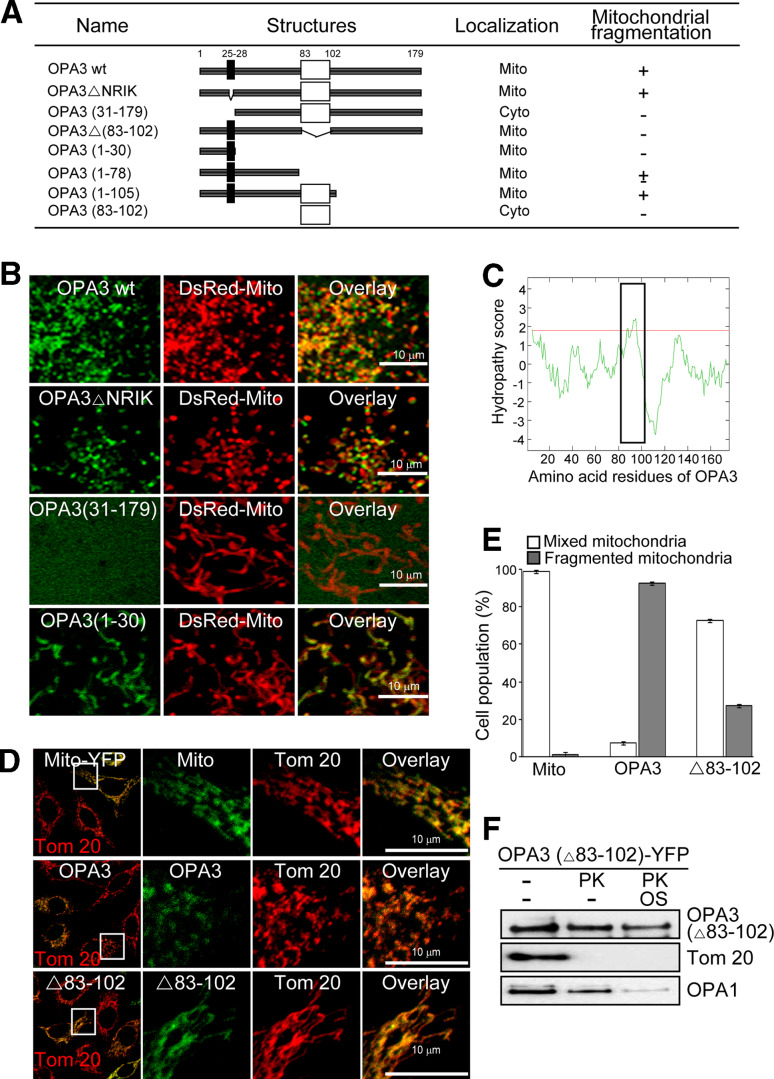
OPA3 amino acid residues 25–29 (NRIKE) have been proposed to be a mitochondrial targeting peptide that is imported into the mitochondrial matrix [9]. To examine this mitochondrial targeting sequence, we tested the localization of an NRIK deletion mutant of OPA3 fused N-terminally to YFP. Consistent with our earlier results, full-length OPA3-YFP was localized on the mitochondria in HeLa cells using DsRed-Mito as the mitochondrial control (Fig. 6a). Contrary to our expectation, OPA3 (ΔNRIK)-YFP was also localized on the mitochondria, suggesting that the N-terminal NRIKE sequence is not crucial for mitochondrial targeting of OPA3 (Fig. 6a, b). To determine the mitochondrial-targeting domain of OPA3, we made several deletion mutants, including a mutant lacking the putative hydrophobic region, corresponding to amino acids 83–102, which represents a predicted membrane-spanning segment according to the TMHMM server (http://www.cbs.dtu.dk), the DAS transmembrane prediction server (http://www.sbc.su.se), and the Kyte-Doolittle hydropathy plot server (Fig. 6a, c). The C-terminally truncated OPA3-YFP mutants, including OPA3 (Δ83–102), OPA3 (1–30), and OPA3 (1–105), were localized to the mitochondria, whereas the N-terminal deletion mutants of OPA3, OPA3 (20–179) (data not shown) and OPA3 (31–179) were found in the cytoplasm (Fig. 6a, b). These results suggest that the N-terminal 30 residues of OPA3 are required for mitochondrial targeting.
Fig. 6.
Hydrophobic region of OPA3 in the MOM is sufficient to trigger mitochondrial fragmentation. a Diagram of C-terminally YFP-fused OPA3 deletion mutants; +, fragmented mitochondria; Mito, mitochondria; Cyto, cytoplasm. b HeLa cells were transfected with the indicated constructs and analyzed by confocal microscopy. DsRed-Mito was used as a mitochondrial marker. c The amino acid sequence of OPA3 was analyzed by Kyte-Doolittle hydropathy plot server. The putative membrane-spanning segment is shown as a black rectangle. d, e Cells were co-transfected with the indicated constructs. Mitochondrial morphology was analyzed by confocal microscopy. Data are the mean ± SD of three experiments, each with 100 cells per treatment. f Mitochondrial fractions with OPA3 (Δ83–102)-YFP in control buffer or swelling buffer (OS) were digested by proteinase K (PK). Western blots of the separated proteins were probed with anti-GFP, anti-Tom 20, and anti-OPA1 antibodies
Hydrophobic region of OPA3 is required for mitochondrial fragmentation
We next examined the effect of the functional domain of OPA3 on mitochondrial morphology. As shown in Fig. 6d, e, overexpression of OPA3-YFP induced significant mitochondrial fragmentation, despite the low expression level of OPA3-YFP. Cells with the C-terminal deletion mutant of OPA3 that retained the hydrophobic region (residues 83–102) also displayed excessively fragmented mitochondria. In contrast, cells transfected with OPA3 (Δ83–102), the OPA3 deletion mutant lacking the hydrophobic region, did not exhibit mitochondrial morphological changes, with >70% of the cells showing mitochondrial networks consisting of tubules (Fig. 6d, e). Although OPA3 (Δ83–102)-YFP localized to the mitochondria, the proteinase K assay showed that the construct failed to localize to the MOM (Fig. 6f). These results suggest that OPA3 residues 83–102 are required for mitochondrial fragmentation and MOM localization of OPA3.
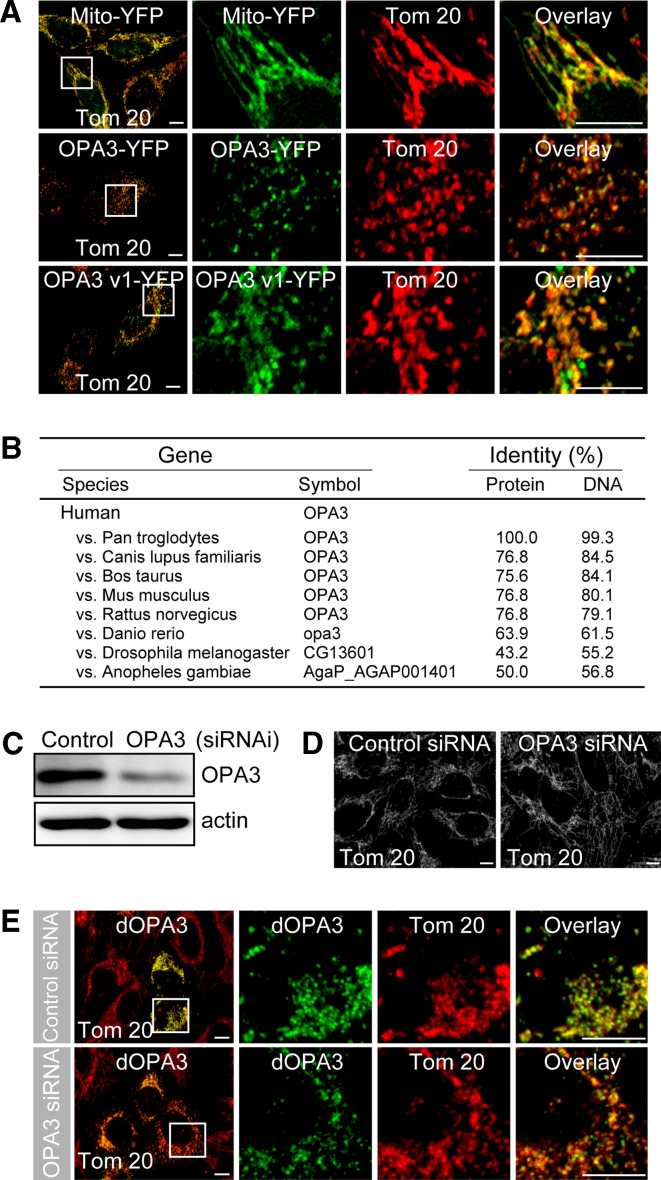
Human OPA3 has two splicing variants (v1, NP_001017989 and v2, NP_079412), which show 81.7% amino acid identity (data not shown). Interestingly, residue 93 (G93) of OPA3 variant 2 (NP_079412, as used in this study) is mutated in the OPA3 of an ADOA patient and is absent from OPA3 variant 1 (NP_001017989). Thus, we investigated whether the expression of OPA3 variant 1 (OPA3-v1) resulted in the same phenotype as the expression of variant 2 (OPA3-v2). As shown in Fig. 7a, OPA3-v1-YFP showed colocalization with Tom 20 in mitochondria and exhibited extensive mitochondrial fragmentation, indicating that mutation of residue 93 of OPA3 is not necessary for mitochondrial fragmentation.
Fig. 7.
OPA3 orthologs in several species can induce mitochondrial fragmentation. a HeLa cells were transfected with the indicated constructs. Cells were fixed and stained with anti-Tom 20 antibody (red). Mitochondrial morphology was analyzed by confocal microscopy. Bar 10 μm. b Diagram showing high amino acid sequence identity with that of human OPA3. c–e HeLa cells were transfected with OPA3 siRNA or control siRNA, together with Drosophila OPA3 (dOPA3). Knockdown of OPA3 was assessed by Western blotting using anti-OPA3 and anti-actin antibodies (c). Cells were fixed, stained with anti-Tom 20 antibody (red) and analyzed by confocal microscopy. Higher magnification of the white square is presented in the right panel of the merged image. Bar 10 μm
Drosophila OPA3, CG13601, rescued the mitochondrial function of human OPA3
To assess the evolutionary relationships of OPA3, we performed a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Pfam analysis (http://pfam.sanger.ac.uk). Pfam analysis revealed 179 homologs containing OPA3 domains in eukaryotes (data not shown). Twenty-one orthologs formed monophyletic subgroups. As shown in Fig. 7b, eight of these orthologs showed high amino acid sequence identity with human OPA3. These results suggest that proteins with an OPA3 domain also have an important role in mitochondrial fragmentation in other species. To this end, we investigated whether the elongated mitochondrial morphology observed in OPA3-downregulated cells could be rescued by expression of an OPA3 ortholog from Drosophila (dOPA3). HeLa cells were transfected with OPA3 siRNA, and we assessed the OPA3 knockdown by Western blotting (Fig. 7c). Because the target sequence we used for OPA3 siRNA is unique to the human OPA3 sequence, dOPA3 constructs could be used to express dOPA3-YFP in the OPA3 siRNA-transfected cells. Consistent with previous results, most OPA3-knockdown HeLa cells had more elongated tubular mitochondria (Fig. 7d). As expected, dOPA3-YFP expressed in cells transfected with either control siRNA or OPA3 siRNA colocalized with Tom 20 on the mitochondria (Fig. 7e). The OPA3 siRNA cells expressing dOPA3 showed extensive mitochondrial fragmentation (Fig. 7e), indicating that expression of Drosophila OPA3 could rescue the mitochondrial morphology induced by human OPA3 knockdown. These results also show that OPA3 orthologs in several species function in mitochondrial fragmentation.
Discussion
In this study, we identified OPA3 as an integral protein of the MOM with effects on mitochondrial dynamics involving mitochondrial fission. We observed a change in mitochondrial morphology to fragmented, ring-like mitochondria after the introduction of either wild-type or a mutant OPA3. The OPA3-induced alteration in mitochondrial morphology increased cellular sensitivity to apoptotic stimuli, supporting a close connection between mitochondrial fragmentation and the apoptotic process. Recent studies have shown that the dysfunction in mitochondrial dynamics owing to mutations in MIM proteins such as OPA1, which is essential for mitochondrial fusion, is closely associated with optic neuropathies [8, 16]. Dysfunction in mitochondrial dynamics resulting from mutations in MOM proteins such as Mfn2, which is essential for mitochondrial fusion, has been associated with the pathogenesis of CMT type 2 with optic atrophy [17], and a mutation of OPA3 has been linked to Costeff optic atrophy syndrome [9].
Ring-like mitochondrial fragmentation has been reported in studies of the overexpression of major MOM fission mediators such as DRP1 and FIS1 [18, 19]. Loss of fusion activity in ring-like mitochondria has been reported with the downregulation of MOM fusion mediators such as Mfn1 and Mfn2 [20]. Mitochondrial morphology significantly changes in various OPA3-overexpressing mammalian cells (e.g., ring-like mitochondrial fragmentation). OPA3 depletion by RNA interference induced an elongated mitochondrial morphology, suggesting a role of OPA3 as a mitochondrial fission factor or an inhibitor of fusion. We showed that expression of dominant-negative DRP1 (DRP1 K38A) had no appreciable effect on OPA3-mediated mitochondrial fragmentation. Consistent with this, OPA3 was able to induce mitochondrial fragmentation following the knockdown of DRP1 or FIS1 via RNA interference. These results suggest that OPA3 might serve as a fission factor for DRP1/FIS1-independent mitochondrial fission. Recently, the Rho-family GTPase Miro was shown to induce mitochondrial fission without an obvious connection to DRP1 [21]. Preliminary data from our laboratory have demonstrated that OPA3 interacts with endogenous Mfn1 (unpublished data). Thus, it is possible that the activity of Mfn1 could increase in the absence of OPA3, such that hyperfusion by Mfn1 activity is sufficient to inhibit mitochondrial fragmentation induced by overexpression of DRP1 or FIS1.
In the present study, C-terminal deletion mutants of OPA3 seemed not to affect the ability of OPA3 to induce mitochondrial fragmentation, as long as the hydrophobic region (83–102) was preserved. The OPA3 mutant lacking the hydrophobic region (Δ83–102) showed a mitochondrial distribution, but failed to induce mitochondrial fragmentation. A plausible explanation is the missorting of the OPA3 mutant (Δ83–102) to the mitochondrial matrix. Thus, our data show that OPA3 in the MOM was sufficient to trigger complete mitochondrial fission, although additional work is required to clarify the mechanism of OPA3-mediated mitochondrial fragmentation.
Our results confirm the sub-mitochondrial localization of OPA3 and its function in mitochondrial morphology. OPA3 was predicted to be an MIM protein [9, 11] and was listed as a MIM protein in a proteomic profile of MIM proteins extracted by alkali treatment of mitochondria [11]. However, it is possible that the alkali-extracted MIM proteins might have been contaminated with outer membrane proteins, as Da Cruz et al. [11] showed that 3.5 and 5% of the proteins in the MIM proteomic profile are known to reside outside the mitochondria and to localize on the MOM, respectively. Moreover, by a differential permeabilization technique, OPA3 was shown to be anchored to the MOM, with its N- and C-terminal regions exposed to the intermembrane space and cytoplasm, respectively.
A link between mitochondrial fission and apoptosis has been indicated by experiments in which, with the overexpression of a DRP1 dominant-negative mutant (DRP1K38A), Mfn1 and Mfn2 were able to prevent apoptosis triggered by many stimuli [22, 23]. Furthermore, the mitochondrial fission machinery has been shown to accelerate cell death induced by many stimuli [14]. In our experiments, OPA3 overexpression did not induce cytochrome c release, caspase-3 activation, or Bax translocation; nevertheless, as indicated by assessment of apoptotic cell death, OPA3-overexpressing cells were sensitized to apoptotic stimuli, compared with control cells. Thus OPA3 itself had no pro-apoptotic effect, but rather potentiated cell death, via mitochondrial fragmentation, after apoptotic stimulation.
Hereditary optic atrophy is characterized by progressive loss of visual acuity and bilateral loss of retinal ganglion cells [16]. Fibroblasts of an ADOAC patient harboring an OPA3 mutation (G93S) were more susceptible to apoptotic stimuli [10], and a mouse model of human Costeff syndrome induced by an OPA3 mutation (L122P) is also characterized by loss of retinal ganglion cells and degeneration of optic nerve axons [12]. In the present study, mutant OPA3 (G93S) increased cell death and mitochondrial fragmentation in the absence of apoptotic stimuli. Our findings substantiate the participation of OPA3 in both mitochondrial dynamics and the pathogenesis of optic atrophy. Further studies are necessary to determine the detailed mechanism of OPA3 action.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. The mitochondrial localization of OPA3. (A) The 20-kDa endogenous OPA3 was identified using anti-OPA3 antibody. HeLa cells were transfected with the indicated constructs, harvested, and analyzed by Western blotting using anti-OPA3 antibody (diluted 1:500). (B) Mitochondrial fraction with FLAG-OPA3 in control buffer or swelling buffer (OS) were digested by proteinase K (PK). Western blots of the separated proteins were probed with anti-FLAG, anti-Tom 20, and anti-OPA1 antibodies. (C) Mitochondrial fraction with OPA3-myc in control buffer or swelling buffer (OS) was digested by trypsin. Alkali (Na2CO3)-treated mitochondria were extracted, after ultracentrifugation to separate soluble and pelleted proteins. Western blots of the separated proteins were probed with anti-myc antibody (TIFF 20491 kb)
Supplementary Fig. 2. Effect of the patient OPA3 mutant (G93S) on mitochondrial morphology (A) HeLa cells were transfected with OPA3-YFP or OPA3(G93S)-YFP. After 15 h, the cells were fixed and stained with fluorescently labeled Tom 20 antibody, and analyzed by confocal microscopy. Bar, 10 μm. (B) HeLa cells were transfected with the indicated constructs. Cells were treated with TRAIL for 6 h, stained with fluorescently labeled anti-cytochrome c antibody and analyzed by confocal microscopy. The number of cells displaying cytosolic cytochrome c was counted, and are shown as a percentage of the total cells counted in cell population (TIFF 19932 kb)
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (KRF-2008-359-C00026). This research was also supported in part by a grant (2009K001282) from the Brain Research Center of the twenty-first Century Frontier Research Program funded by the Ministry of Education, Science, and Technology, Republic of Korea (to C. C.). We acknowledge Dr. Richard J. Youle’s valuable comments on the manuscript and support in some aspects of the work. We also thank Soojay Banerjee for technical support.
Contributor Information
Seung-Wook Ryu, Phone: +82-42-3504381, FAX: +82-42-3504380, Email: ryus@kaist.ac.kr.
Chulhee Choi, Phone: +82-42-3504321, FAX: +82-42-3504380, Email: cchoi@kaist.ac.kr.
References
- 1.Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaffe MP. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 3.Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12:178–184. doi: 10.1016/S0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda S, Hirose S. Stage-specific enhanced expression of mitochondrial fusion and fission factors during spermatogenesis in rat testis. Biochem Biophys Res Commun. 2003;311:424–432. doi: 10.1016/j.bbrc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 6.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 9.Anikster Y, Kleta R, Shaag A, Gahl WA, Elpeleg O. Type III 3-methylglutaconic aciduria (optic atrophy plus syndrome, or Costeff optic atrophy syndrome): identification of the OPA3 gene and its founder mutation in Iraqi Jews. Am J Hum Genet. 2001;69:1218–1224. doi: 10.1086/324651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynier P, Amati-Bonneau P, Verny C, Olichon A, Simard G, Guichet A, Bonnemains C, Malecaze F, Malinge MC, Pelletier JB, Calvas P, Dollfus H, Belenguer P, Malthiery Y, Lenaers G, Bonneau D. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J Med Genet. 2004;41:e110. doi: 10.1136/jmg.2003.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, Martinou JC. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem. 2003;278:41566–41571. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- 12.Davies VJ, Powell KA, White KE, Yip W, Hogan V, Hollins AJ, Davies JR, Piechota M, Brownstein DG, Moat SJ, Nichols PP, Wride MA, Boulton ME, Votruba M. A missense mutation in the murine Opa3 gene models human Costeff syndrome. Brain. 2008;131:368–380. doi: 10.1093/brain/awm333. [DOI] [PubMed] [Google Scholar]
- 13.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 14.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parcellier A, Tintignac LA, Zhuravleva E, Dummler B, Brazil DP, Hynx D, Cron P, Schenk S, Olivieri V, Hemmings BA. The carboxy-terminal modulator protein (CTMP) regulates mitochondrial dynamics. PLoS One. 2009;4:e5471. doi: 10.1371/journal.pone.0005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Votruba M, Aijaz S, Moore AT. A review of primary hereditary optic neuropathies. J Inherit Metab Dis. 2003;26:209–227. doi: 10.1023/A:1024441302074. [DOI] [PubMed] [Google Scholar]
- 17.Chung KW, Kim SB, Park KD, Choi KG, Lee JH, Eun HW, Suh JS, Hwang JH, Kim WK, Seo BC, Kim SH, Son IH, Kim SM, Sunwoo IN, Choi BO. Early onset severe and late-onset mild Charcot-Marie-Tooth disease with mitofusin 2 (MFN2) mutations. Brain. 2006;129:2103–2118. doi: 10.1093/brain/awl174. [DOI] [PubMed] [Google Scholar]
- 18.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 19.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J Cell Biol. 2004;167:87–98. doi: 10.1083/jcb.200405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugioka R, Shimizu S, Tsujimoto Y. Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J Biol Chem. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. The mitochondrial localization of OPA3. (A) The 20-kDa endogenous OPA3 was identified using anti-OPA3 antibody. HeLa cells were transfected with the indicated constructs, harvested, and analyzed by Western blotting using anti-OPA3 antibody (diluted 1:500). (B) Mitochondrial fraction with FLAG-OPA3 in control buffer or swelling buffer (OS) were digested by proteinase K (PK). Western blots of the separated proteins were probed with anti-FLAG, anti-Tom 20, and anti-OPA1 antibodies. (C) Mitochondrial fraction with OPA3-myc in control buffer or swelling buffer (OS) was digested by trypsin. Alkali (Na2CO3)-treated mitochondria were extracted, after ultracentrifugation to separate soluble and pelleted proteins. Western blots of the separated proteins were probed with anti-myc antibody (TIFF 20491 kb)
Supplementary Fig. 2. Effect of the patient OPA3 mutant (G93S) on mitochondrial morphology (A) HeLa cells were transfected with OPA3-YFP or OPA3(G93S)-YFP. After 15 h, the cells were fixed and stained with fluorescently labeled Tom 20 antibody, and analyzed by confocal microscopy. Bar, 10 μm. (B) HeLa cells were transfected with the indicated constructs. Cells were treated with TRAIL for 6 h, stained with fluorescently labeled anti-cytochrome c antibody and analyzed by confocal microscopy. The number of cells displaying cytosolic cytochrome c was counted, and are shown as a percentage of the total cells counted in cell population (TIFF 19932 kb)