Abstract
Three novel glycine-rich peptides, named ctenidin 1–3, with activity against the Gram-negative bacterium E. coli, were isolated and characterized from hemocytes of the spider Cupiennius salei. Ctenidins have a high glycine content (>70%), similarly to other glycine-rich peptides, the acanthoscurrins, from another spider, Acanthoscurria gomesiana. A combination of mass spectrometry, Edman degradation, and cDNA cloning revealed the presence of three isoforms of ctenidin, at least two of them originating from simple, intronless genes. The full-length sequences of the ctenidins consist of a 19 amino acid residues signal peptide followed by the mature peptides of 109, 119, or 120 amino acid residues. The mature peptides are post-translationally modified by the cleavage of one or two C-terminal cationic amino acid residue(s) and amidation of the newly created mature C-terminus. Tissue expression analysis revealed that ctenidins are constitutively expressed in hemocytes and to a small extent also in the subesophageal nerve mass.
Keywords: Ctenidin, Cupiennius salei, Glycine-rich peptides, Antimicrobial, Tissue expression
Introduction
Arthropods, lacking the highly sophisticated adaptive immune system of vertebrates, rely on their innate immune system to survive in environments that also harbor many potentially pathogenic microorganisms. This innate immune system consists of cellular as well as humoral responses to invaders. Cellular responses include phagocytosis, nodulation, and encapsulation. Humoral responses include, e.g., proteolytic cascades leading to melanization of invaders or hemolymph coagulation systems, as well as the production of several different killing factors. These killing factors can be reactive oxygen species, reactive nitrogen intermediates, or antimicrobial peptides (AMPs) [1, 2].
AMPs play a key role in innate immunity and are produced throughout the phylogenetic tree. Despite their diversity in sequence and structure, they share some common features, such as small size (generally ≤10 kDa), as well as cationic and amphipathic properties [3–6]. Based on structural features and amino acid composition, AMPs are usually grouped into three main classes: (1) linear amphipathic, α-helical AMPs, (2) cyclic or open ended cyclic AMPs containing cysteine, and (3) AMPs with an overrepresentation of particular amino acids, such as proline, histidine or glycine. Besides this classical grouping, there is also another class of AMPs comprised of peptides that are generated by partial hydrolysis of large precursors with no antimicrobial properties [7].
Activation of AMP production differs depending on species. In holometabolous insects, AMPs are produced predominantly in the fat body, their synthesis is induced upon infection and they are then released into the hemolymph. In hemimetabolous insects, like termites [8], and other invertebrates, like mussels [9], shrimps [10], horseshoe crabs [11], scorpions [12] or spiders [13, 14], AMPs are produced constitutively and stored in hemocyte granules, and released into the hemolymph [10, 11, 13, 15] and/or fused with phagocytic vesicles following an infection [9].
One class of AMPs with an overrepresentation of one particular amino acid are the glycine-rich peptides. They are able to inhibit the growth of fungi and have been isolated and characterized from different taxonomic groups, including plants [16, 17], amphibians [18], and arthropods [13, 19–21]. The percentage of glycine residues in these peptides varies considerably, from 10–30% in some species to more than 60% in the shepherins [17] and acanthoscurrins [13].
Until now, the immune system of only one spider species, the mygalomorph spider Acanthoscurria gomesiana, has been investigated. From naive hemocytes of this spider, two different AMPs were isolated, gomesin and acanthoscurrins. Both are produced constitutively and stored in granules of hemocytes, from which they can be released into the hemolymph upon infection. Gomesin is a small, 18-residue AMP forming two disulphide bridges and adopting a β-hairpin-like structure active against Gram-positive and Gram-negative bacteria and fungi [14]. Acanthoscurrins are glycine-rich AMPs with a unique amino acid composition active against Gram-negative bacteria and yeast. They are 130 and 132 amino acids long, but are composed of only seven different amino acids, 72–73% of which are glycines [13].
In this study, we present the isolation and characterization of ctenidins, a glycine-rich peptide family, from naive hemocytes of the spider Cupiennius salei. Three isoforms of the peptide were identified, as well as two of the three corresponding mRNAs. Mass spectrometry and cDNA sequence analysis showed that ctenidins result from the processing of a precursor containing a signal peptide immediately followed by the mature peptide. Furthermore, ctenidins were found to be C-terminally amidated. The genomic organization is simple, containing no introns. We also show that the ctenidins are mainly expressed in hemocytes and to a lesser extent in the subesophageal nerve mass.
Materials and methods
Animals
Mature females of the wandering spider Cupiennius salei Keyserling were used for the experiments. The spiders were obtained from a permanent breeding line that has been maintained for several years. Each C. salei was housed separately in a 2-l glass jar at a room temperature of 23°C and with a light:dark regime of 12:12 h.
Hemolymph collection
The spiders were anesthetized with CO2 and the hemolymph (~300 μl/animal) was collected by cardiac puncture with a 27G needle on a 1-ml syringe. The hemolymph was collected in the presence of sodium citrate buffer, pH 4.6, to prevent coagulation [22]. The hemocytes were separated from the plasma by centrifugation at 800g at 4°C for 10 min. They were then washed once with sodium citrate buffer and again centrifuged at 800g at 4°C for 10 min, yielding ~1.3 × 107 hemocytes/spider, with an average weight of 1 ng per hemocyte.
Peptide extraction and purification
The hemocytes collected from the hemolymph of 160 spiders were resuspended in doubly distilled water (ddH2O) containing 0.1% trifluoroacetic acid (TFA), and homogenized by ultrasonication (Sonoplus, Bandelin, Switzerland), which was carried out twice for 1 min at 30% power, pulse level 5, on ice. Lysed hemocytes were then centrifuged at 20,800g for 30 min to remove cell debris. Supernatants were pooled and applied to reversed phase HPLC (RP-HPLC) on an Atlantis® Prep T3 column (10 × 100 mm, 5 μm; Waters, USA) equilibrated with ddH2O, 0.1% (TFA). Elution was performed with a stepwise gradient of 0% acetonitrile (ACN) for 3 column volumes (CV), 0–22% ACN, 0.1% TFA, in 1.1 CV, 22–60% ACN, 0.1% TFA, in 15.5 CV, at a flow rate of 2 ml/min (Fig. 1a).
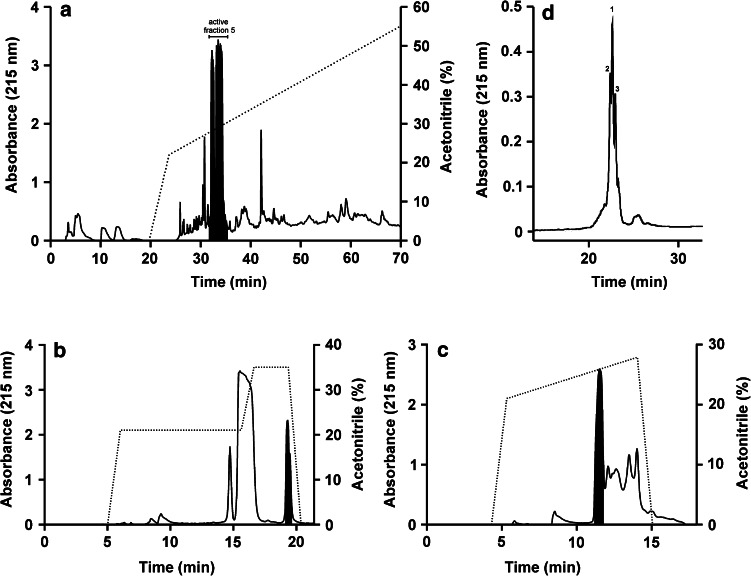
Fig. 1.
Purification of ctenidins from C. salei hemocytes. a Preparative RP-HPLC chromatogram of hemocyte lysates. The area in black corresponds to fraction number 5, which showed activity against E. coli and S. aureus. b, c Further separation of the bactericidal active fraction (black areas) was done by two subsequent preparative RP-HPLC purification steps. d Small scale RP-HPLC chromatogram of the active purified sub-fraction containing the ctenidins. The numbers of the peaks correspond to the numbering of the different ctenidins
The active fraction identified by bactericidal assays was further purified using a nucleosil 100-5 C18 Nautilus column (4.6 × 250 mm; Macherey & Nagel, Switzerland), equilibrated with ddH2O, 0.1% TFA, under stepwise isocratic conditions of 0% ACN, 21% ACN, 0.1% TFA, and 35% ACN, 0.1% TFA, at a flow rate of 1 ml/min (Fig. 1b). A final purification using the nucleosil 100-5 C18 Nautilus column (4.6 × 250 mm) was performed with a stepwise gradient of 0–21% ACN, 0.1% TFA in 0.25 CV and 21–45% ACN, 0.1% TFA, in 7.2 CV, at a flow rate of 1 ml/min, yielding the ctenidins (Fig. 1c).
HPLC purification was carried out at room temperature on an Aekta purifier 900 HPLC system (Pharmacia Biotech, Sweden). The column effluent was monitored for absorbance at 215 nm. All fractions were concentrated under vacuum, and reconstituted in ddH2O.
RP-HPLC of the ctenidin 1–3 containing fraction
The above marked black bactericidal fraction (Fig. 1c) was further separated by RP-HPLC on a Reprosil-Pur 300 Phenyl column (2.0 × 100 mm, 5 μm; Dr. Maisch HPLC, Switzerland) in a Hewlett Packard liquid chromatograph 1090 (Hewlett Packard) applying a gradient of 10–60% ACN (80% v/v), 0.1% TFA in 60 min at a flow rate of 0.4 ml/min (Fig. 1d). All peaks were collected as indicated and characterized by MS.
The samples thus obtained were dissolved in 100 mM Tris–HCl buffer, pH 7.6 containing 10 mM CaCl2 and were cleaved with endoproteinase Arg-C (sequencing grade; Roche Diagnostics) at an enzyme:substrate ratio of 1:50 (w/w) during 18 h at 37°C. The peptides were separated by RP-HPLC.
Amino acid analysis
Samples were hydrolyzed in the gas phase with 6 M hydrochloric acid containing 0.1% phenol (by vol) for 22 h at 115°C under N2 vacuum according to Chang and Knecht [23]. The liberated amino acids were labeled with phenylisothiocyanate and the resulting phenylthiocarbamoyl amino acids were analyzed by RP-HPLC on a Nova Pak ODS column (3.9 × 150 mm, 4 μm; Waters) in a Dionex Summit liquid chromatograph (Dionex) with an automatic injection system [24].
Amino acid sequence analysis
N-terminal sequence analysis was carried out in a Procise cLC 492 protein sequencer from Applied Biosystems. The released PTH amino acids were analyzed on-line by RP-HPLC.
Mass spectrometry
Electrospray ionization-mass spectrometric (ESI–MS) analyses were performed on a LTQ Orbitrap XL mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray ion source, resulting in accurate monoisotopic masses. Samples were dissolved in ACN/ddH2O (1:1, vol/vol) containing 1% formic acid. All analyses were carried out in the positive ion mode. Analysis of digested ctenidins was performed by matrix assisted laser desorption/ionisation-time of flight mass spectrometry (MALDI-TOF–MS) on a VoyagerTM Elite BioSpectrometryTM Research Station (Applied Biosystems).
Antibacterial assays
The presence of antibacterial activity was determined by a liquid growth inhibition assay. The bacteria strains used were Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213, and the yeast strain was Candida albicans. Single colonies of bacteria or yeast were picked and grown in Mueller–Hinton broth (bacteria) or Sabouraud-2% dextrose broth (yeast) at 36°C overnight. These cultures were then diluted and grown to the exponential growth phase, where they were again diluted to a density of 1,000 cells/ml. Then, 90 μl of these suspensions were transferred to the wells of a 96-well U-bottom microtiter plate, and 10 μl of the substance to be tested or ddH2O were added. The plate was incubated at 36°C while shaking for 1 h (bacteria) or 2 h (yeast), typically resulting in a two- to fourfold amplification in cell numbers in the controls. After incubation, the whole 100 μl of each well were plated on Mueller–Hinton agar plates (bacteria), or Sabouraud-2% dextrose agar plates (yeast) supplemented with 50 μg/ml chloramphenicol and 12.5 μg/ml tetracycline. After incubation at 36°C overnight, the colonies were counted. MICs were determined and are expressed as (a)–(b) intervals of concentrations, where (a) is the highest concentration tested at which the microorganisms are still growing, and (b) is the lowest concentration at which the growth is 100% inhibited.
To differentiate between bactericidal or bacteriostatic activity of the ctenidins, E. coli was incubated in the presence of ctenidins at its MIC for 16 h at 25°C. The cultures were then plated on Mueller–Hinton agar plates and colony-forming units counted after an overnight incubation at 36°C.
RNA isolation
Total RNA was isolated from hemocytes using the Absolutely RNA® Miniprep Kit (Stratagene, Switzerland) or the RNeasy® Mini Kit (Qiagen, Switzerland). RNA was isolated following the instructions of the manufacturer in the manual of the kit, including the optional on-column DNA digestion.
cDNA synthesis
3′- and 5′-RACE ready cDNAs were synthesized from 1 μg of the isolated total RNA using the BD SMART™ RACE cDNA Amplification Kit (BD Biosciences Clontech). This system incorporates an anchor sequence (“BD SMART sequence”) either at the 3′- or the 5′- end of the generated cDNA which can be used as primer binding sites for subsequent RACE PCRs.
3′- and 5′-RACE PCR
A set of five primers for 3′-RACE and five primers for 5′-RACE PCR was designed based on the sequence of acanthoscurrin [13]: Acantho fwd beg 1, Acantho fwd beg d, Acantho fwd mid 1, Acantho fwd mid 2, Acantho fwd end 2, Acantho rev mid 1, Acantho rev mid 2, Acantho rev end 1, Acantho rev end 2, Acantho rev end 3 (Table 1). These primers were used for PCR with 1/100th of the synthesized cDNA and the universal primer UPM from the BD SMART™ RACE cDNA Amplification Kit, taking advantage of the BD SMART sequence incorporated into the cDNAs. PCR was performed for 5 min at 94°C, followed by 5 cycles of 94°C for 30 s, 65°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 61°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 59°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 1.5 min, 15 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1.5 min, and a final elongation at 72°C for 7 min. PCR products were run on a 0.7% agarose gel containing 0.5 ng ethidium bromide/ml. Single bands were cut out using a scalpel, and DNA was extracted from the gel using a MinElute® Gel Extraction Kit (Qiagen). Then, 1/10th of the extracted DNA was used as template in another round of PCR, using the same primers as before and the following PCR program: 1 min at 95°C, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, 68°C for 1.5 min, and a final elongation at 68°C for 5 min. PCR products were then purified using the MinElute® PCR Purification Kit (Qiagen).
Table 1.
Primers used in this study
| 3′-RACE primers | |
| Acantho fwd beg 1 | 5′-TATGGCGGTGGAAGGTATGG-3′ |
| Acantho fwd beg d | 5′-TAYGGHGGHGGHAGRTAYGG-3′ |
| Acantho fwd mid 1 | 5′-GGTGGACTCGGAGGCGGC-3′ |
| Acantho fwd mid 2 | 5′-GGACTGGGAGGAGGAAAAGGACT-3′ |
| Acantho fwd end 2 | 5′-GGATACGGAGGTGGATACGG-3′ |
| 5′-RACE primers | |
| Acantho rev mid 1 | 5′-GCCGCCTCCGAGTCCACC-3′ |
| Acantho rev mid 2 | 5′-AGTCCTTTTCCTCCTCCCAGTCC-3′ |
| Acantho rev end 1 | 5′-CCGTATCCACCACCACGACC-3′ |
| Acantho rev end 2 | 5′-CCGTATCCACCTCCGTATCC-3′ |
| Acantho rev end 3 | 5′-GCCTTTGTATTTTCCGCCTCC-3′ |
| Ctenidin 5′RACE 1 | 5′-TTTTCCAACATATCAAATTATTTT-3′ |
| Ctenidin mid rev: | 5′-CCTACGTCGTCTTTGCCGTCTATTAC-3′ |
| Ctenidin Nterm re: | 5′-GCCGTATCCACCTCCTCTTCTACC-3′ |
| Universal primers | |
| UPM | 5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ |
| NUP | 5′-AAGCAGTGGTATCAACGCAGAGT-3′ |
| Genomic primers | |
| Ctenidin sig fwd | 5′-ATGAAGCATTTGATTCCACTAATTGTAATG-3′ |
| Ctenidin gen rev | 5′-TGTCCTATTACAAATATTAAGTGATGACAG-3′ |
| Tissue expression primers | |
| Lycosa S3A fwd | 5′-CGTAAGGATTGGTACGATGT-3′ |
| Lycosa S3A rev | 5′-TCCATTCCGTGAAAGTTGGT-3′ |
From the sequencing results of these PCR products, another primer for 5′-RACE PCR was designed: Ctenidin 5′RACE 1 (Table 1). 5′-RACE PCR was performed with this primer and the nested universal primer (NUP), using 1/100th of the synthesized cDNA as template and the following PCR program: 5 min at 94°C, followed by 5 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 48°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 46°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 44°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 43°C for 30 s, 72°C for 1.5 min, 10 cycles of 94°C for 30 s, 42°C for 30 s, 72°C for 1.5 min, and a final elongation at 72°C for 7 min. The PCR products were purified using the MinElute® PCR Purification Kit.
Another primer for 5′-RACE PCR was designed based on the sequencing results from the products of the above PCR, Ctenidin mid rev (Table 1). 5′-RACE PCR was performed with this primer and the nested universal primer NUP, using 1/100th of the synthesized cDNA as template and the following PCR program: 5 min at 94°C, followed by 5 cycles of 94°C for 30 s, 53°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 1.5 min, 20 cycles of 94°C for 30 s, 51°C for 30 s, 72°C for 1.5 min, and a final elongation at 72°C for 7 min. PCR products were run on a 1% low-melt agarose gel containing 0.5 ng ethidium bromide/ml. Single bands were cut out using a scalpel, and DNA was extracted from the gel using a MinElute® Gel Extraction Kit (Qiagen). Half of the extracted DNA was then used as template in another round of PCR, using the same primers and PCR program as before. PCR products were then purified using the MinElute® PCR Purification Kit (Qiagen).
To completely solve the sequence, yet another primer for 5′-RACE PCR had to be designed based on the sequencing results from the products of the above PCR, Ctenidin Nterm re (Table 1). This primer was used together with the universal primer UPM for 5′-RACE PCR with 1/100th of the synthesized cDNA as template. The following PCR program was used: 5 min at 94°C, followed by 5 cycles of 94°C for 30 s, 68°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 65°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 61°C for 30 s, 72°C for 1.5 min, 5 cycles of 94°C for 30 s, 59°C for 30 s, 72°C for 1.5 min, 10 cycles of 94°C for 30 s, 57°C for 30 s, and a final elongation at 72°C for 7 min. The PCR products were purified using the MinElute® PCR Purification Kit (Qiagen).
Cloning and sequencing
Purified PCR products were cloned into pDrive vector using the QIAGEN PCR Cloning Kit. The products in the pDrive vector were sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit. Sequences were separated on an ABI3130XL automated sequencer using POP7 polymer on a 50-cm array and acquired using sequence detection software v.3.0 (all from Applied Biosystems). Sequences were analyzed with BioEdit v7.0.8 software and putative signal peptides were analyzed using the prediction server SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP).
DNA isolation
Genomic DNA was isolated from hemocytes of two adult female C. salei, using the DNeasy® Blood & Tissue Kit (Qiagen). Isolation was carried out following the manufacturer’s protocol for cultured cells.
Genomic PCR
PCR was carried out with 1/200th of the isolated genomic DNA as template. The primers used were Ctenidin sig fwd and Ctenidin gen rev. The PCR program was 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 42°C for 30 s, 72°C for 1 min and a final elongation at 72°C for 2 min. PCR products were analyzed on a 1.5% agarose gel containing 0.5 ng ethidium bromide/ml.
Tissue expression analysis
Various tissues (ovaries, subesophageal nerve mass, hepatopancreas, venom glands, heart, silk glands, hemocytes, and muscle) were extracted from CO2 anesthetized, unchallenged female C. salei. Total RNA was extracted from these tissues using the RNeasy® Mini Kit (Qiagen). 1 μg of total RNA was reverse-transcribed into cDNA using the BD SMART™ RACE cDNA Amplification Kit. The amount of template used for expression analysis was normalized according to the band intensity values obtained with the ribosomal protein S3A transcript as control. Primers used for this control PCR were Lycosa S3A fwd and Lycosa S3A rev (Table 1), based on the sequence of Lycosa singoriensis S3A transcript (GenBank EU 247153.1). The primers used for the expression analysis of ctenidin were Ctenidin sig fwd and Ctenidin mid rev (Table 1). The PCR program was 3 min at 94°C, followed by 33 cycles of 94°C for 30 s, 44°C for 30 s, 72°C for 30 s, and a final elongation at 72°C for 2 min. PCR products were run on a 2% agarose gel containing 0.5 ng ethidium bromide/ml. Band intensities were quantified using the ImageMaster® VDS 3.0 software (Pharmacia Biotech, Sweden).
Results
Purification of antimicrobial peptides from hemolymph
In searching for antimicrobially active peptides produced by the spider C. salei as immune defense, we isolated hemocytes from the hemolymph of mature female spiders by centrifugation. The peptides from the hemocytes were extracted under acidic conditions after disrupting the cells by sonication. The acidic extract was directly submitted to RP-HPLC, resulting in 15 fractions that were tested for antimicrobial activity in antibacterial assays. Fraction number 5 showed activity against both the Gram-negative E. coli and the Gram-positive S. aureus (Fig. 1a, black area). Since the fraction contained several peptides, it was further separated by two RP-HPLC steps as described in material and methods (Figs. 1b and c), observing activities of the sub-fractions (black areas) by liquid growth inhibition assays, finally yielding an active fraction containing three peptides which could not be separated further on a large scale (Fig. 1d).
Characterization
The three peptides were analyzed by high resolution MS, resulting in the monoisotopic molecular masses of 8,810.022, 9,507.372 and 9,564.373 Da. However, sequencing of the three combined peptides by Edman-degradation revealed only a single N-terminal sequence, DRGYGGGRRGGGYGGGGYGG. The sequence had a high glycine residue content, and the assignment of the sequence became ambiguous after 20 amino acid residues due to the increased lag. Amino acid analysis revealed a unique composition consisting of more than 71% Gly, and only eight other amino acids, Asx (3.5%), Glx (1.4%), Arg (5.2%), Tyr (8.6%), Val (2.6%), Ile (0.9%), Leu (4.0%), and Lys (2.6%).
Cloning and sequencing
Based on the first 20 amino acids, a set of degenerate primers was designed for 3′-RACE PCR of hemocyte cDNA. Another set of primers for 3′- as well as 5′-RACE PCRs was designed based on the sequence of acanthoscurrin, a glycine-rich AMP isolated from the hemocytes of the mygalomorph spider A. gomesiana [13]. This second set of primers was designed because of similarities in the amino acid sequences and the amino acid compositions between the peptides and acanthoscurrin. With these primers, hemocyte cDNA was screened for glycine-rich sequences. While the degenerate primers did not yield any sequences, one of the 3′-RACE primers from the acanthoscurrin sequence yielded the C-terminus of one of the glycine-rich peptides. By primer walking sequencing, the whole cDNA sequences of two of the three peptides were identified. We named these peptides ctenidin 1 and 2, adapted from Ctenidae, the family name of the spider C. salei. The ctenidin 1 cDNA consists of 771 bp, starting with a 3′-UTR of 53 bp, followed by an open reading frame (ORF) of 423 bp, a 5′-UTR of 270 bp, and the poly-A tail. The ctenidin 2 cDNA is identical, except that 30 bp are missing in the ORF of ctenidin 2 compared to ctenidin 1 (Fig. 2). The deduced amino acid sequences of the two peptides start with a putative signal peptide of 19 amino acids, the cleavage site for the signal peptidase most likely being located after the alanine residue preceding the aspartic acid in position 20, as predicted by SignalP 3.0 software (data not shown). The sequence following the signal peptide overlapped with the sequence obtained by Edman-degradation of the peptides. The calculated monoisotopic masses of 8,996.099 and 9,693.414 Da for the two peptides differed from the measured masses of 8,810.022 and 9,507.372 Da by 186.077 or 186.042 Da, respectively. This mass difference corresponds to the lack of the two C-terminal amino acids, a glycine and a lysine, due to the posttranslational amidation process of the C-terminal glycine of the mature peptides (differences between measured and calculated masses after processing: 0.023 or 0.059 Da, respectively). Mature ctenidin 1 is composed of 119 amino acid residues and ctenidin 2 comprises 109 amino acid residues.
Fig. 2.
Ctenidin 1 and 2 cDNA sequences. The deduced amino acid sequence is presented under the nucleotide sequence. The mature peptide sequence is underlined; the part missing in ctenidin 2 is double underlined. The nucleotide sequence of the mature peptide is in bold. The dotted line marks the post-translationally cleaved off lysine and the glycine residue involved in C-terminal amidation of the other glycine. Asterisks mark the stop codon. The polyadenylation signal is shown in bold italics
Characterization of full-length peptides
To confirm this, the three peptides were separated by RP-HPLC on a small scale (Fig. 1b) and each was incompletely digested with the endoproteinase Arg-C. The resulting fragments were separated by RP-HPLC, and MALDI-TOF–MS of all fragments was performed. The resulting masses of the C-terminal fragments (1,896.8 Da, average mass) confirmed the hypothesis that the C-terminal lysine and glycine are cleaved off post-translationally. The glycine likely contributes to the amidation of the preceding glycine in both ctenidins, the amidation being shown by the monoisotopic masses of the mature peptides. By the same method, the amino acid sequence of the third peptide, which we named ctenidin 3, was determined. The fragments generated by Arg-C digestion revealed two additional glycines in the N-terminal region of ctenidin 3 compared to ctenidins 1 and 2, resulting in a fragment of 2,353.8 Da (average mass) instead of 2,239.2 Da (average mass), and a slightly different C-terminus (Fig. 3a). The C-terminal glycine of ctenidin 1 and 2 is missing in ctenidin 3; instead, it ends with a tyrosine, the resulting mass of the C-terminal fragment being 1,840.1 Da (average mass). Like ctenidin 1 and 2, ctenidin 3 is C-terminally amidated, as evidenced by the monoisotopic mass of the mature peptide. The position of the additional glycines in the N-terminal region was determined by Edman-degradation of the corresponding fragment. The rest of the amino acid sequence is identical to that of the other ctenidins.
Fig. 3.
Comparisons of the three mature ctenidins and of ctenidin 1 to acanthoscurrin 1. a Alignment of the three mature ctenidins. The fragment of ctenidin 3 that was sequenced by Edman-degradation to determine the position of the additional glycines is underlined. The fragments used to prove the C-terminal processing are underlined by a dotted line. b Comparison of ctenidin 1 to acanthoscurrin 1. All glycine-repeats are highlighted in gray
Tissue expression
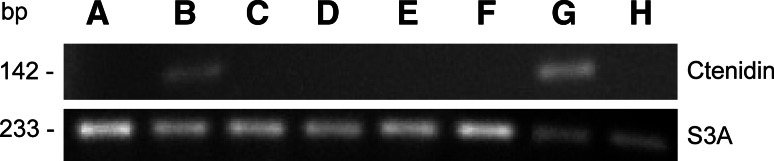
Various spider tissues were isolated and the expression of ctenidin mRNA in them was determined using semi-quantitative RT–PCR. The tissue cDNAs were normalized against the transcript of the ribosomal protein S3A, and a primer pair detecting all three ctenidins was used. Only hemocytes showed an abundant expression of ctenidin mRNA, while subesophageal nerve mass showed little expression. No expression was detected in the remaining tissues (Fig. 4).
Fig. 4.
RT-PCR analysis of ctenidin-gene expression in different tissues. Expression was assayed in ovaries (A), subesophageal nerve mass (B), hepatopancreas (C), venom glands (D), heart (E), silk glands (F), hemocytes (G), and muscle (H)
Gene structure
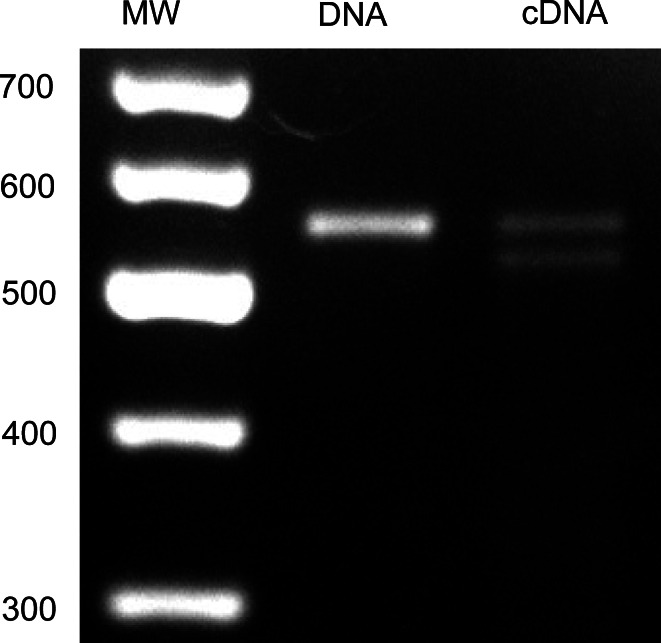
Genomic PCR was performed to solve gene structure of the ctenidins. The resulting PCR product had the same size as the ctenidin 1 PCR product obtained with cDNA as template, suggesting that there are no introns in the ORF of ctenidin 1 (Fig. 5). This was confirmed by sequencing, which showed the same sequences for products obtained from DNA and from mRNA. Interestingly, genomic PCR yielded the sequence of ctenidin 1, but also of ctenidin 3, of which the cDNA sequence had not been solved (Fig. 6). Similarly to ctenidin 1, the gene sequence of ctenidin 3 did not contain any introns. Sequencing revealed a slightly different C-terminus for ctenidin 3. Compared to ctenidin 1 and 2, it contained a point mutation resulting in the mutation of the last glycine in the mRNA sequence to an arginine. The deduced amino acid sequence showed a difference of 342.20 Da to the measured mass of ctenidin 3, which is explained by the post-translational processing of the C-terminus. In the case of ctenidin 3, the two C-terminal cationic residues Arg and Lys are removed, with the remaining Gly used to amidate the preceding Tyr, resulting in the C-terminus of the mature peptide. This also explains the slightly different C-terminus of ctenidin 3 compared to ctenidin 1 and 2. On the other hand, genomic PCR did not yield the ctenidin 2 sequence, which was clearly observed in PCR with cDNA (Fig. 5).
Fig. 5.
Ctenidin gene structure. PCR products of hemocyte cDNA and genomic DNA with ctenidin-specific primers. The 100-bp molecular weight marker is marked with MW; band sizes are in bp
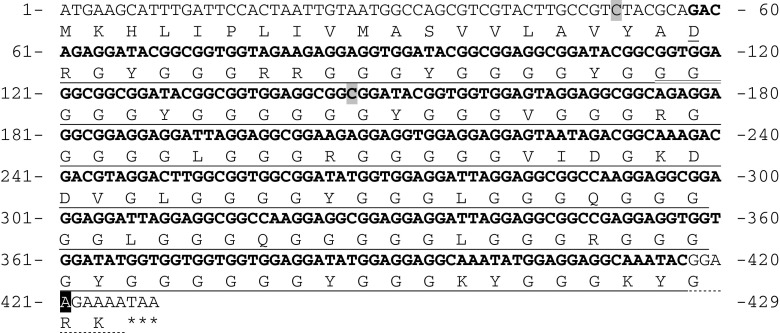
Fig. 6.
Ctenidin 3 ORF gene sequence. The deduced amino acid sequence is presented under the nucleotide sequence. The mature peptide sequence is underlined. The nucleotide sequence of the mature peptide is in bold. The dotted line marks the post-translationally cleaved off lysine and the glycine residue involved in C-terminal amidation of the other glycine. The additional glycines compared to ctenidin 1 and 2 are double underlined. Silent mutations are highlighted in light gray, the mutation causing a Gly-Arg exchange is highlighted in black. Asterisks mark the stop codon
Antimicrobial activity
To investigate the antimicrobial activity of the ctenidins, the mixture of the three peptides isolated from hemocytes was quantified by Edman degradation. From a total of 150 spiders, 261.5 μg ctenidins were isolated. The antimicrobial activity of the ctenidins was tested against the following three test organisms: E. coli (Gram-negative bacterium), S. aureus (Gram-positive bacterium), and C. albicans (yeast). The ctenidins showed activity against E. coli with a MIC of 2.5–5 μM. The growth of S. aureus was clearly reduced in the presence of ctenidins compared to the control group, but not inhibited in the range of the concentrations tested (up to 10 μM). Due to lack of material, no higher concentrations could be tested. No activity was detected in the range of the concentrations tested (up to 5 μM) against C. albicans.
When E. coli was incubated in the presence of ctenidins at its MIC of 5 μM for 16 h, a full growth recovery was observed, hinting at bacteriostatic rather than bactericidal activity.
Discussion
We report here for the first time the purification and characterization of an AMP from hemocytes of an araneomorph spider, C. salei. So far, investigations on the immune system of spiders have been limited to the mygalomorph species A. gomesiana [13–15, 25]. Three isoforms of the AMP named ctenidin were identified. Ctenidins are linear, cationic peptides, possessing a net charge of +5 (not taking into account the C-terminal amidation). With more than 70% glycine residues, ctenidins clearly represent new members of the family of glycine-rich AMPs. As seen in other glycine-rich AMPs, only a few amino acids make up the peptide. In the case of the ctenidins, these are Gly (71.2%), Asp (3.5%), Gln (1.4%), Arg (5.2%), Tyr (8.6%), Val (2.6%), Ile (0.9%), Leu (4.0%), and Lys (2.6%). This composition resembles the composition of acanthoscurrins isolated from A. gomesiana. Interestingly, acanthoscurrins exhibit 1.7 times more hydrophobic amino acid residues (mainly Leu) than the ctenidins, but 4 times less Asp. While related to acanthoscurrins, ctenidins show considerable structural differences to other glycine-rich peptides (Table 2), as has been reported before for the glycine-rich AMPs [13, 19].
Table 2.
Structural comparison among glycine-rich antimicrobial peptides
| Peptides | Molecular masses (Da) | pI | G (%) | K + R (%) | H (%) | Hydrophobic residues (%) |
|---|---|---|---|---|---|---|
| Ctenidin 1 | 9,514 | 9.6 | 71.4 | 7.5 | 0.0 | 7.5 |
| Ctenidin 2 | 8,816 | 9.6 | 70.6 | 8.3 | 0.0 | 7.6 |
| Ctenidin 3 | 9,571 | 9.6 | 71.6 | 7.5 | 0.0 | 7.5 |
| Acantho 1 | 10,226 | 9.9 | 72.9 | 6.8 | 0.0 | 12.8 |
| Acantho 2 | 10,112 | 9.9 | 72.5 | 6.9 | 0.0 | 13.0 |
| Armadillidin | 5,260 | 12.0 | 47.2 | 11.1 | 11.1 | 18.9 |
| AFP | 7,118 | 6.7 | 31.3 | 4.5 | 20.9 | 4.5 |
| Holotricin 3 | 6,838 | 7.4 | 63.9 | 1.2 | 15.7 | 6.0 |
| Tenecin 3 | 7,323 | 7.5 | 44.2 | 0.0 | 19.5 | 9.1 |
| Shepherin 1 | 2,362 | 7.3 | 67.9 | 0.0 | 28.6 | 0.0 |
| Shepherin 2 | 3,259 | 7.3 | 65.8 | 0.0 | 21.1 | 0.0 |
Modified from Herbinière et al. [19]
Antibacterial peptides from insects: AFP, holotricin 3 and tenecin 3 [47, 48, 51]; from spider: acanthoscurrins 1 and 2 [13]; from woodlice: armadillidin [19]; and from plants: shepherin 1 and 2 [17]. Molecular masses (average) and pI were calculated using http://www.expasy.org/tools/protparam.html
While a complete sequencing of the ctenidins by Edman degradation was not possible, the full sequences of the ctenidins were identified by screening hemocyte cDNA for glycine-rich sequences. Within this context, it is worth mentioning that the sequence of the ctenidins with its many glycine repeats led to problems in PCR, yielding many truncated products, probably due to a ‘sliding’ of the polymerase caused by the glycine repeats. Two of the ctenidins identified, ctenidin 1 and 2, are almost identical on the peptide and nucleotide level, except for a sequence of ten amino acids missing in ctenidin 2. The third isoform, ctenidin 3, differs by two additional glycines inserted in the N-terminal part of the peptide, a slightly different C-terminus, and two additional silent point mutations on the nucleotide level (Fig. 6), as well as some point mutations in the 3′-UTR (not shown). The full-length peptides overall resemble the acanthoscurrins, mainly due to the repeats of 3–6 glycines, mostly interrupted by a single different amino acid (Fig. 3b). But despite this resemblance, the peptides show only a few sequence similarities. This leaves the question open as to whether the peptides originate from a common ancestor or have evolved independently, although the latter case seems rather unlikely when comparing the peptides. An interesting feature that distinguishes the ctenidins from the acanthoscurrins is the presence of a ten amino acid residues-long sequence in the middle of the peptide, VIDGKDDVGL, which interrupts the glycine repeats. The function of this sequence is not clear, as it gives the ctenidins a local negative charge, flanked on either side by hydrophobic amino acid residues.
Comparisons of the calculated and the measured masses of the mature peptides suggest the cleavages of the last two (ctenidin 1 and 2) or three (ctenidin 3) C-terminal amino acids, the glycine being involved in the amidation of the preceding amino acid (another glycine or tyrosine). This is a mechanism also identified in spider venom toxins [26, 27]. It is probable that the C-terminal basic arginine or lysine residue could be removed in a first processing step by a basic residue-specific carboxypeptidase. In a second step, the remaining glycine-extended peptide would be converted into a des-Gly peptide amine by α-amidation [28]. A similar amidation signal of X-Gly-Arg/Lys-Arg/Lys is also found in scorpion toxins [29, 30]. Post-translational amidation of the C-terminus is a common modification of AMPs of chelicerates and crustaceans [13, 14, 19, 31, 32], making the peptides more stable and raising their positive net charge, an important feature of most AMPs.
Ctenidins are synthesized as prepeptides containing a signal peptide of 19 amino acid residues, directly followed by the mature peptide (Figs. 2 and 6). The presence of the signal peptides makes it likely that the ctenidins are directed to and stored in hemocyte granules. This is a common process in invertebrates, as has been identified in other spider AMPs [15, 33], as well as in shrimps [10], limulids [34], mussels [9], and termites [8]. The content of these granules can then be released into the hemolymph upon infection or fuse with phagocytic vesicles inside the hemocytes.
Tissue expression of ctenidins determined by RT–PCR revealed that they are constitutively expressed in hemocytes, which is in agreement with the above-mentioned mechanism. Of the remaining tissues, only the subesophageal nerve mass showed a weak expression, and no signal was detectable in the other tissues (Fig. 4). This contrasts somewhat with the other spider AMPs, which are exclusively expressed in hemocytes [13, 33]. Tachyplesin, an AMP from horseshoe crabs, is also expressed mainly in hemocytes, but to a lesser extent also in the brain [34], like the ctenidins. Also, other AMPs of invertebrates can be expressed in nervous tissues [35].
The gene structure of the ctenidin genes is very simple, showing no introns in the so far identified genes of the long isoforms ctenidin 1 and 3 (Fig. 5). Since ctenidin 1 and 2 are identical except for a missing piece in ctenidin 2, alternative splicing of mRNA from the same gene could have been the cause of the two isoforms. This mechanism was disproved since there are no introns. Either ctenidin 2 originates from its own gene, which we were not able to amplify, or it is generated by a so far unknown RNA processing mechanism other than splicing from the ctenidin 1 gene. The two identified genes have likely evolved from gene duplication events during the evolution of C. salei, as it is also described for a duplicated Ultrabithorax (Ubx) gene [36] or three Hox genes (Dfd, Scr, and Ubx) [37] of this spider.
Ctenidins showed activity against E. coli at a MIC of 2.5–5 μM, similar to that of acanthoscurrins. When incubated for 16 h, the growth of E. coli had recovered. This indicates that ctenidins act in a bacteriostatic rather than bactericidal manner, although the lack of material did not allow us to confirm this observation by raising the ctenidin concentration. The loss of activity after a given time might be due to degradation of the peptides. The targets of glycine-rich peptides in bacteria and fungi are not clear, but can be diverse. Besides the classical membrane disruption or its depolarization [38], ctenidins could inhibit intracellular components as mentioned for proline-rich peptides [39] or could hamper bacterial cell division as suggested for the glycine- and proline-rich coleoptericins [40]. Another glycine-rich peptide family, the attacins, exerts its activity against Gram-negative bacteria by inhibiting the synthesis of an outer membrane protein by interfering with the transcription of its genes [41]. Finally, the proline-rich pyrrhocoricin inhibits the ATPase activity of the bacterial heat shock protein DnaK, thereby preventing chaperone-assisted protein folding [42]. No structures of truly glycine-rich peptides are available, making it difficult to predict a target or functional mechanism of such peptides. While a recent report suggests a β-sheet structure for the C-terminal part of acanthoscurrin [43], the cationic plasticins, glycine-leucine-rich peptides from frog skin, were shown to go through multiple conformational transitions, including helix states, β-structures, and disordered states [44]. We had to measure the combined MIC of the three ctenidin isoforms as not enough material was present to separate the peptides on a larger scale to obtain enough material for separate tests. The three isoforms were approximately present in a 1:1:1 ratio, as judged by the chromatogram of the analytical separation (Fig. 1d). Since their sequences are so similar, we expect the MICs of the single isoforms to differ only marginally, if at all. The growth of the Gram-positive bacterium S. aureus was slowed down in the presence of ctenidins to about 30% of the normal growth (data not shown), but was not completely inhibited. This selective activity against Gram-negative bacteria is in agreement with other glycine-rich AMPs [13, 45]. The specificity for Gram-negative bacteria might be due to the composition of their outer membrane. Positively charged amino acid residues might bind specifically to components of the outer membrane of Gram-negative bacteria, like the glycolipid lipopolysaccharide, as has been shown for the bactericidal/permeability-increasing (BPI)/lipopolysaccharide-binding protein (LBP) protein family [46]. Specificity might also be mediated by the inhibition of the synthesis of Gram-negative-specific proteins [41]. In contrast, armadillidin, a glycine-rich AMP isolated from the woodlouse Armadillidium vulgare, showed activity against the Gram-positive bacterium B. megaterium but not against Gram-negative bacteria [19]. Rather surprisingly, no activity of ctenidins was detected against the yeast C. albicans up to the tested concentration of 5 μM, whereas the acanthoscurrins exhibit fungicidal activities at a MIC of 1.3–2.6 μM. The 22 amino acid residues peptide leptoglycin, isolated from skin secretions of the frog Leptodactylus pentadactylus, exhibits high sequence similarities to the ctenidins and is also only active against Gram-negative bacteria, but not against either Gram-positive bacteria or fungi [45]. Most other glycine-rich AMPs are active against yeast [13, 17, 47, 48], with few exceptions [49], and several plant glycine-rich peptides are induced by wounding or fungal infection and are therefore considered to be defense-related proteins [50].
Conclusions
In conclusion, we have shown that the spider C. salei expresses three isoforms of a glycine-rich AMP that we named ctenidins. Two of the isoforms originate from their own genes, while a gene for ctenidin 2 was not detected. While RNA splicing can be ruled out, it is possible that its mRNA is generated by an unknown processing mechanism from the ctenidin 1 mRNA. Ctenidins are constitutively expressed in hemocytes and nervous tissue, thus their expression is independent of immune challenges. They are active against E. coli and are therefore assumed to be generally active against Gram-negative bacteria. Future research will focus on the identification of further antimicrobial compounds of the spider immune system and possible synergistic interactions among the ctenidins themselves and with other compounds to shed light on how the innate immune system of spiders deals efficiently with different kinds of invaders.
Acknowledgment
We thank Dr. D. Destoumieux-Garzón and Dr. H. Murray for critical comments on the manuscript, and the Swiss National Science Foundation for funding.
References
- 1.Jiravanichpaisal P, Lee BL, Söderhäll K. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology. 2006;211:213–236. doi: 10.1016/j.imbio.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 2.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/S0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 3.Hwang PM, Vogel HJ. Structure-function relationships of antimicrobial peptides. Biochem Cell Biol. 1998;76:235–246. doi: 10.1139/bcb-76-2-3-235. [DOI] [PubMed] [Google Scholar]
- 4.Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 5.Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 6.van ‘t Hof W, Veerman EC, Helmerhorst EJ, Amerongen AV. Antimicrobial peptides: properties and applicability. Biol Chem. 2001;382:597–619. doi: 10.1515/BC.2001.072. [DOI] [PubMed] [Google Scholar]
- 7.Sonenshine DE, Hynes WL, Ceraul SM, Mitchell R, Benzine T. Host blood proteins and peptides in the midgut of the tick Dermacentor variabilis contribute to bacterial control. Exp Appl Acarol. 2005;36:207–223. doi: 10.1007/s10493-005-2564-0. [DOI] [PubMed] [Google Scholar]
- 8.Lamberty M, Zachary D, Lanot R, Bordereau C, Robert A, Hoffmann JA, Bulet P. Insect immunity: constitutive expression of a cysteine-rich antifungal and a linear antibacterial peptide in a termite insect. J Biol Chem. 2001;276:4085–4092. doi: 10.1074/jbc.M002998200. [DOI] [PubMed] [Google Scholar]
- 9.Mitta G, Vandenbulcke F, Roch P. Original involvement of antimicrobial peptides in mussel innate immunity. FEBS Lett. 2000;486:185–190. doi: 10.1016/S0014-5793(00)02192-X. [DOI] [PubMed] [Google Scholar]
- 10.Destoumieux D, Munoz M, Cosseau C, Rodriguez J, Bulet P, Comps M, Bachère E. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J Cell Sci. 2000;113:461–469. doi: 10.1242/jcs.113.3.461. [DOI] [PubMed] [Google Scholar]
- 11.Iwanaga S, Lee BL. Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol. 2005;38:128–150. doi: 10.5483/bmbrep.2005.38.2.128. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez de la Vega RC, García BI, D’Ambrosio C, Diego-García E, Scaloni A, Possani LD. Antimicrobial peptide induction in the haemolymph of the Mexican scorpion Centruroides limpidus limpidus in response to septic injury. Cell Mol Life Sci. 2004;61:1507–1519. doi: 10.1007/s00018-004-4096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzini DM, da Silva PI, Jr, Fogaça AC, Bulet P, Daffre S. Acanthoscurrin: a novel glycine-rich antimicrobial peptide constitutively expressed in the hemocytes of the spider Acanthoscurria gomesiana . Dev Comp Immunol. 2003;27:781–791. doi: 10.1016/S0145-305X(03)00058-2. [DOI] [PubMed] [Google Scholar]
- 14.Silva PI, Jr, Daffre S, Bulet P. Isolation and characterization of gomesin, an 18-residue cysteine-rich defense peptide from the spider Acanthoscurria gomesiana hemocytes with sequence similarities to horseshoe crab antimicrobial peptides of the tachyplesin family. J Biol Chem. 2000;275:33464–33470. doi: 10.1074/jbc.M001491200. [DOI] [PubMed] [Google Scholar]
- 15.Fukuzawa AH, Vellutini BC, Lorenzini DM, Silva PI, Jr, Mortara RA, da Silva JM, Daffre S. The role of hemocytes in the immunity of the spider Acanthoscurria gomesiana . Dev Comp Immunol. 2008;32:716–725. doi: 10.1016/j.dci.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Egorov TA, Odintsova TI, Pukhalsky VA, Grishin EV. Diversity of wheat anti-microbial peptides. Peptides. 2005;26:2064–2073. doi: 10.1016/j.peptides.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Park CJ, Park CB, Hong SS, Lee HS, Lee SY, Kim SC. Characterization and cDNA cloning of two glycine- and histidine-rich antimicrobial peptides from the roots of shepherd’s purse, Capsella bursa-pastoris . Plant Mol Biol. 2000;44:187–197. doi: 10.1023/A:1006431320677. [DOI] [PubMed] [Google Scholar]
- 18.El Amri C, Nicolas P. Plasticins: membrane-damaging peptides with ‘chameleon-like’ properties. Cell Mol Life Sci. 2008;65:895–909. doi: 10.1007/s00018-007-7445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbinière J, Braquart-Varnier C, Grève P, Strub JM, Frère J, Van Dorsselaer A, Martin G. Armadillidin: a novel glycine-rich antibacterial peptide directed against gram-positive bacteria in the woodlouse Armadillidium vulgare (terrestrial isopod, Crustacean) Dev Comp Immunol. 2005;29:489–499. doi: 10.1016/j.dci.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Otvos L., Jr Antibacterial peptides isolated from insects. J Pept Sci. 2000;6:497–511. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 21.Sperstad SV, Haug T, Vasskog T, Stensvag K. Hyastatin, a glycine-rich multi-domain antimicrobial peptide isolated from the spider crab (Hyas araneus) hemocytes. Mol Immunol. 2009;46:2604–2612. doi: 10.1016/j.molimm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Söderhäll K, Smith VJ. Separation of the haemocyte populations of Carcinus maenas and other marine decapods, and prophenoloxidase distribution. Dev Comp Immunol. 1983;7:229–239. doi: 10.1016/0145-305X(83)90004-6. [DOI] [PubMed] [Google Scholar]
- 23.Chang JY, Knecht R. Direct analysis of the disulfide content of proteins: methods for monitoring the stability and refolding process of cystine-containing proteins. Anal Biochem. 1991;197:52–58. doi: 10.1016/0003-2697(91)90354-V. [DOI] [PubMed] [Google Scholar]
- 24.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984;336:93–104. doi: 10.1016/S0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 25.Pereira LS, Silva PI, Jr, Miranda MT, Almeida IC, Naoki H, Konno K, Daffre S. Structural and biological characterization of one antibacterial acylpolyamine isolated from the hemocytes of the spider Acanthocurria gomesiana . Biochem Biophys Res Commun. 2007;352:953–959. doi: 10.1016/j.bbrc.2006.11.128. [DOI] [PubMed] [Google Scholar]
- 26.Choi SJ, Parent R, Guillaume C, Deregnaucourt C, Delarbre C, Ojcius DM, Montagne JJ, Celerier ML, Phelipot A, Amiche M, Molgo J, Camadro JM, Guette C. Isolation and characterization of Psalmopeotoxin I and II: two novel antimalarial peptides from the venom of the tarantula Psalmopoeus cambridgei . FEBS Lett. 2004;572:109–117. doi: 10.1016/j.febslet.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Ostrow KL, Mammoser A, Suchyna T, Sachs F, Oswald R, Kubo S, Chino N, Gottlieb PA. cDNA sequence and in vitro folding of GsMTx4, a specific peptide inhibitor of mechanosensitive channels. Toxicon. 2003;42:263–274. doi: 10.1016/S0041-0101(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 28.Kapuscinski M, Green M, Sinha SN, Shepherd JJ, Shulkes A. Peptide alpha-amidation activity in human plasma: relationship to gastrin processing. Clin Endocrinol. 1993;39:51–58. doi: 10.1111/j.1365-2265.1993.tb01750.x. [DOI] [PubMed] [Google Scholar]
- 29.Dai L, Corzo G, Naoki H, Andriantsiferana M, Nakajima T. Purification, structure-function analysis, and molecular characterization of novel linear peptides from scorpion Opisthacanthus madagascariensis . Biochem Biophys Res Commun. 2002;293:1514–1522. doi: 10.1016/S0006-291X(02)00423-0. [DOI] [PubMed] [Google Scholar]
- 30.Zeng XC, Wang SX, Zhu Y, Zhu SY, Li WX. Identification and functional characterization of novel scorpion venom peptides with no disulfide bridge from Buthus martensii Karsch. Peptides. 2004;25:143–150. doi: 10.1016/j.peptides.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Destoumieux D, Bulet P, Loew D, Van Dorsselaer A, Rodriguez J, Bachère E. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei (Decapoda) J Biol Chem. 1997;272:28398–28406. doi: 10.1074/jbc.272.45.28398. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura T, Furunaka H, Miyata T, Tokunaga F, Muta T, Iwanaga S, Niwa M, Takao T, Shimonishi Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus): isolation and chemical structure. J Biol Chem. 1988;263:16709–16713. [PubMed] [Google Scholar]
- 33.Lorenzini DM, Fukuzawa AH, da Silva PI, Jr, Machado-Santelli G, Bijovsky AT, Daffre S. Molecular cloning, expression analysis and cellular localization of gomesin, an anti-microbial peptide from hemocytes of the spider Acanthoscurria gomesiana . Insect Biochem Mol Biol. 2003;33:1011–1016. doi: 10.1016/S0965-1748(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 34.Shigenaga T, Muta T, Toh Y, Tokunaga F, Iwanaga S. Antimicrobial tachyplesin peptide precursor: cDNA cloning and cellular localization in the horseshoe crab (Tachypleus tridentatus) J Biol Chem. 1990;265:21350–21354. [PubMed] [Google Scholar]
- 35.Schikorski D, Cuvillier-Hot V, Leippe M, Boidin-Wichlacz C, Slomianny C, Macagno E, Salzet M, Tasiemski A. Microbial challenge promotes the regenerative process of the injured central nervous system of the medicinal leech by inducing the synthesis of antimicrobial peptides in neurons and microglia. J Immunol. 2008;181:1083–1095. doi: 10.4049/jimmunol.181.2.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damen WG, Hausdorf M, Seyfarth EA, Tautz D. A conserved mode of head segmentation in arthropods revealed by the expression pattern of Hox genes in a spider. Proc Natl Acad Sci USA. 1998;95:10665–10670. doi: 10.1073/pnas.95.18.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwager EE, Schoppmeier M, Pechmann M, Damen WG. Duplicated Hox genes in the spider Cupiennius salei . Front Zool. 2007;4:10. doi: 10.1186/1742-9994-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winans KA, King DS, Rao VR, Bertozzi CR. A chemically synthesized version of the insect antibacterial glycopeptide, diptericin, disrupts bacterial membrane integrity. Biochemistry. 1999;38:11700–11710. doi: 10.1021/bi991247f. [DOI] [PubMed] [Google Scholar]
- 39.Park KH, Park Y, Park IS, Hahm KS, Shin SY. Bacterial selectivity and plausible mode of antibacterial action of designed Pro-rich short model antimicrobial peptides. J Pept Sci. 2008;14:876–882. doi: 10.1002/psc.1019. [DOI] [PubMed] [Google Scholar]
- 40.Sagisaka A, Miyanoshita A, Ishibashi J, Yamakawa M. Purification, characterization and gene expression of a glycine and proline-rich antibacterial protein family from larvae of a beetle, Allomyrina Dichotoma . Insect Mol Biol. 2001;10:293–302. doi: 10.1046/j.0962-1075.2001.00261.x. [DOI] [PubMed] [Google Scholar]
- 41.Carlsson A, Engström P, Palva ET, Bennich H. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with omp gene transcription. Infect Immun. 1991;59:3040–3045. doi: 10.1128/iai.59.9.3040-3045.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of dnak and prevents chaperone-assisted protein folding. Biochemistry. 2001;40:3016–3026. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 43.Remuzgo C, Andrade GF, Temperini ML, Miranda MT. Acanthoscurrin fragment 101–132: total synthesis at 60°C of a novel difficult sequence. Biopolymers (Pep Sci) 2009;92:65–75. doi: 10.1002/bip.21110. [DOI] [PubMed] [Google Scholar]
- 44.El Amri C, Lacombe C, Zimmerman K, Ladram A, Amiche M, Nicolas P, Bruston F. The plasticins: membrane adsorption, lipid disorders, and biological activity. Biochemistry. 2006;45:14285–14297. doi: 10.1021/bi060999o. [DOI] [PubMed] [Google Scholar]
- 45.Sousa JC, Berto RF, Gois EA, Fontenele-Cardi NC, Honório JE, Jr, Konno K, Richardson M, Rocha MF, Camargo AA, Pimenta DC, Cardi BA, Carvalho KM. Leptoglycin: a new Glycine/Leucine-rich antimicrobial peptide isolated from the skin secretion of the South American frog Leptodactylus pentadactylus (Leptodactylidae) Toxicon. 2009;54:23–32. doi: 10.1016/j.toxicon.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iijima R, Kurata S, Natori S. Purification, characterization, and cDNA cloning of an antifungal protein from the hemolymph of Sarcophaga peregrina (flesh fly) larvae. J Biol Chem. 1993;268:12055–12061. [PubMed] [Google Scholar]
- 48.Lee SY, Moon HJ, Kurata S, Natori S, Lee BL. Purification and cDNA cloning of an antifungal protein from the hemolymph of Holotrichia diomphalia larvae. Biol Pharm Bull. 1995;18:1049–1052. doi: 10.1248/bpb.18.1049. [DOI] [PubMed] [Google Scholar]
- 49.Pelegrini PB, Murad AM, Silva LP, Dos Santos RC, Costa FT, Tagliari PD, Bloch C, Jr, Noronha EF, Miller RN, Franco OL. Identification of a novel storage glycine-rich peptide from guava (Psidium guajava) seeds with activity against Gram-negative bacteria. Peptides. 2008;29:1271–1279. doi: 10.1016/j.peptides.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Sachetto-Martins G, Franco LO, de Oliveira DE. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta. 2000;1492:1–14. doi: 10.1016/s0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 51.Lee YJ, Chung TJ, Park CW, Hahn Y, Chung JH, Lee BL, Han DM, Jung YH, Kim S, Lee Y. Structure and expression of the tenecin 3 gene in Tenebrio molitor . Biochem Biophys Res Commun. 1996;218:6–11. doi: 10.1006/bbrc.1996.0002. [DOI] [PubMed] [Google Scholar]