Abstract
In higher organisms, dietary proteins are broken down into amino acids within the digestive tract but outside the cells, which incorporate the resulting amino acids into their metabolism. However, under certain conditions, an organism loses more nitrogen than is assimilated in the diet. This additional loss was found in the past century to come from intracellular proteins and started an intensive research that produced an enormous expansion of the field and a dispersed literature. Therefore, our purpose is to provide an updated summary of the current knowledge on the proteolytic machinery involved in intracellular protein degradation and its physiological and pathological relevance, especially addressed to newcomers in the field who may find further details in more specialized reviews. However, even providing a general overview, this is an extremely wide field and, therefore, we mainly focus on mammalian cells, while other cells will be mentioned only for comparison purposes.
Keywords: Intracellular protein degradation, Proteases, Proteasomes, Ubiquitin, Lysosomes, Autophagy, Cell injury
Introduction
Intracellular protein degradation is different from digestion of dietary proteins in two main aspects: (1) the proteins that are degraded are the same that were synthesized by the organism, and (2) this degradation occurs within the cells by energy requiring processes. The book of Rudolf Schönheimer (1898–1941) “The Dynamic State of Body Constituents”, published 1 year after his untimely death, led to the important concept that intracellular proteins are continuously being synthesized and degraded within cells, and started an intensive research on this topic. Although this concept was challenged until as late as the mid-1950s, several laboratories firmly established first the existence of an extensive intracellular protein degradation in both prokaryotes and eukaryotes, and later its selectivity and relevance.
Initially, it was assumed that intracellular proteins were degraded within lysosomes, organelles discovered in 1949 by Christian de Duve (Nobel Prize in Physiology or Medicine in 1974, together with Albert Claude and George E. Palade) and co-workers. However, it was soon clear that more selective processes were required for intracellular protein degradation. In particular, the field of ubiquitin research was initiated in the 1980s, and followed when the chemistry of ubiquitin conjugation was described [1, 2]. For these discoveries, Aaron Ciechanover, Avram Hershko and Irwin Rose were awarded with the Nobel Prize of Chemistry in 2004. Research on the mechanistics of this selective degradation of proteins was carried out by these and many other laboratories [3]. On the lysosomal side, a period of quick discoveries on the molecules involved in its main intracellular protein degradation pathway, called autophagy, was initiated in the 1990s using autophagy mutants in Saccharomyces cerevisiae. As a consequence of all these efforts, the field of intracellular protein degradation experienced, at the beginning of this century, an enormous expansion that revolutionized and diversified the initial concept.
General characteristics of intracellular protein degradation
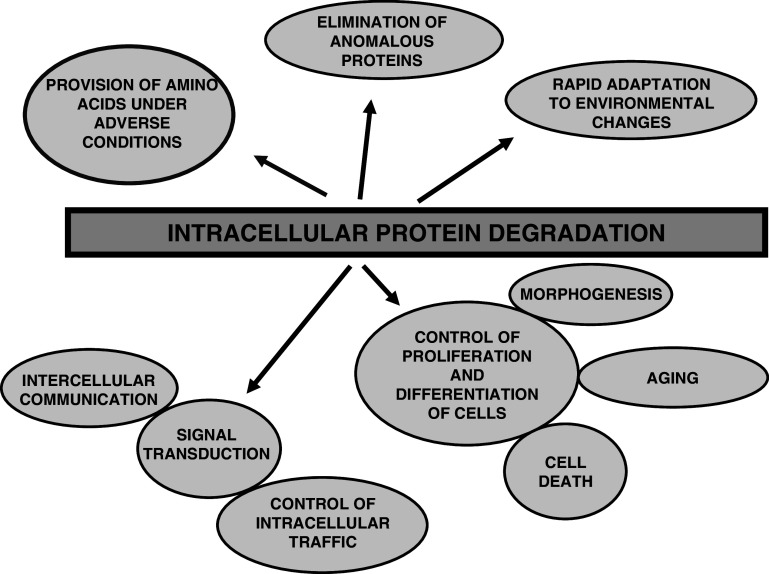
Cell growth and differentiation depend on the rates of protein synthesis and degradation, and both opposite processes are equally important and operate in a coordinated way to maintain cell life. Intracellular protein degradation has several main features. First, it is general, since all intracellular proteins from eukaryotic or prokaryotic organisms so far investigated are continuously being degraded to amino acids. It is also quite extensive, since, for example, in a healthy 70 kg adult human, about 300 g of its proteins are daily degraded and replaced. Therefore, one may ask what evolutionary superiority gives such a process to a cell. Obviously, one important advantage is to degrade defective proteins, whose accumulation could trigger cell aging and/or death. In fact, it has been postulated that many errors occur during protein translation and these defective proteins are quickly eliminated [4]. However, functional proteins are also degraded, and although, at first glance, it may appear inconvenient for the cell to degrade these proteins by mechanisms that, in addition, require energy, a continuous intracellular proteolysis provides several advantages (Fig. 1).
Fig. 1.
Intracellular protein degradation fulfills many functions in mammalian cells. This process, in addition to survey and degrade invasive microorganisms, brings several advantages for the cells and organisms as indicated, including: (1) the provision, under adverse conditions such as starvation, of amino acids to be used as an energy source and/or for the synthesis of proteins essential for survival; (2) the rapid degradation of defective proteins; (3) a better and quick adaptation of cell metabolism to the environment, by modifying the levels of proteins; (4) the control of many important processes such as cell proliferation (division and growth) and differentiation, morphogenesis and regression of retired tissues, cell aging and death (by necrosis and apoptosis), etc.; and (5) signal transduction, including the regulation of intercellular (for example, in the generation of antigenic peptides) and intracellular communication, and the control of protein traffic. All this, taken together, justifies that intracellular protein degradation is universal
A second fundamental and apparently paradoxical feature of intracellular protein degradation is that energy is required, as first observed more than 50 years ago in liver slices. This energy cost is high, despite peptide bond hydrolysis being a catabolic process, and can be explained because the two major sites of intracellular protein degradation in mammalian cells, proteasomes and lysosomes, require energy to accomplish their work.
Finally, a major feature of intracellular protein degradation is that it is both specific and tightly regulated, constantly recognizing the proteins that should be degraded. In contrast to protein synthesis, which follows zero-order kinetics, the degradation rate of a protein is a first order kinetics. This means that: (1) this rate depends on the concentration of the protein, and (2) protein degradation is a random process, and a newly synthesized polypeptide chain has the same probability to be degraded as a molecule from the same protein synthesized long ago. However, this does not exclude that damaged, mutated or otherwise abnormal proteins are selectively degraded. First order kinetics allows one to express the degradation rate of a protein, and thus its stability, in terms of half-life (t 1/2), which is the time required to degrade half of the original molecules. The t 1/2 s of proteins are affected by hormones, nutrients and, in general, by the environmental conditions of the cells, indicating a tight regulation, and it is noticeable that proteins in a same cell, and even in the same localization, have vastly different t 1/2 s (Table 1). In general, housekeeping proteins have t 1/2 s optimized for efficient protein production and, thus, longer than those of regulatory proteins. Because of this specificity, it is possible that certain sequence motifs in a protein can facilitate its degradation. A quite simple example [3] is the presence in some proteins of destabilizing N-terminal residues that define a degradation signal (N-degron) recognized by the ubiquitin-proteasome system (N-end rule). Other known examples are the degradation box of mitotic cyclins and the PEST sequences, both associated to short-lived proteins, and the KFERQ-like sequences, recognized by a specific lysosomal pathway of protein degradation. However, the generality of these observations remains to be proven and, for example, using a proteomic approach, this has been questioned for the PEST hypothesis [6].
Table 1.
Half-lives (t 1/2) (∼) of some rat liver proteins
| Protein/s | Localization | t 1/2 (∼) |
|---|---|---|
| Ornithine decarboxylase | Cytosol | 10 min |
| Fructose 1,6-bisphosphatase | Cytosol | 30 min |
| δ-aminolevulinate synthetase | Mitochondria | 70 min |
| Tyrosine aminotransferase | Cytosol | 90 min |
| Tryptophan oxygenase | Cytosol | 120 min |
| Hydroxymethyl CoA reductase | Endoplasmic reticulum | 150 min |
| Phosphoenol pyruvate carboxykinase | Mitochondria | 5 h |
| Dihydroorotase | Cytosol | 12 h |
| Glucose 6-phosphate dehydrogenase | Cytosol | 15 h |
| Ornithine-oxo-acid aminotransferase | Mitochondria | 19 h |
| Alanine aminotransferase | Mitochondria | 20 h |
| Glutamate dehydrogenase | Mitochondria | 24 h |
| Glucokinase | Cytosol | 33 h |
| Acetyl-CoA carboxylase | Cytosol | 48 h |
| Cytochrome P450 | Endoplasmic reticulum | 50 h |
| Monoamine oxidase | Mitochondria | 55 h |
| Catalase | Peroxisome | 60 h |
| Malate dehydrogenase | Mitochondria | 62 h |
| Cytochrome c reductase | Endoplasmic reticulum | 70 h |
| Arginase | Cytosol | 96 h |
| α-glycerophosphate dehydrogenase | Mitochondria | 96 h |
| Cytochrome b5 | Mitochondria | 122 h |
| Cytochrome c oxidase | Mitochondria | 134 h |
| Carbamoyl phosphate synthetase | Mitochondria | 185 h |
| Proteasomes | Cytosol/nucleus | 199 h |
| Ornithine transcarbamoylase | Mitochondria | 209 h |
| β-glucuronidase | Lysosome | 15 days |
| Lactate dehydrogenase (isozyme 5) | Cytosol | 16 days |
Most of the data are from Ref. [5]
Although individual proteins have quite different t 1/2 s, the global degradation of proteins in a cell can be divided into two main kinetics, separated by a clear discontinuity, that correspond to short-lived (t 1/2 around 1 h) and long-lived (t 1/2 of days) proteins. Short-lived proteins only represent 1% of all proteins but, because of their high turnover, contribute to as much 30% of all intracellular protein degradation, and their nature remains speculative [4, 7]. In addition to regulatory and damaged proteins, they could include newly synthesized proteins that are rapidly degraded to accomplish various functions, such as the generation of a constitutive supply of antigenic peptides [3, 4]. However, the degradation of such a high fraction of newly synthesized proteins is considered by others an experimental artifact [8].
Pathways of intracellular protein degradation
Peptide bonds are hydrolyzed by proteases, to produce peptides and amino acids. In the human genome, the number of identified protease genes is 561 ([9], see also the MEROPS database http://merops.sanger.ac.uk/). Most of these proteases are within cells in almost all compartments, but the majority only catalyze a limited proteolysis. Therefore, in mammalian cells, there are only two main pathways to afford an extensive intracellular proteolysis: proteasomes and lysosomes. Although proteasomes and lysosomes are equally important in the degradation of short-lived and long-lived proteins [7, 10], their relative importance varies with the cell type, its metabolic situation and the specific protein to be degraded.
Proteasomes
Proteasomes [for reviews see 11, 12] are localized in both nucleoplasm and cytosol and represent the main non-lysosomal pathway of intracellular protein degradation. They especially participate in the rapid degradation of defective proteins whose accumulation will have detrimental consequences. For example, proteins that remain partially unfolded in the ER interact with others to prevent their aggregation and to allow their exit, through a channel in the ER membrane, to the cytosol to be degraded by proteasomes. This process, termed ER-associated degradation or ERAD [13], is an important degradation pathway, since about one quarter of the cell’s proteome enters into the ER. However, the presence of defective proteins in the ER also activates lysosomes that contribute to their degradation. In addition to these and other defective proteins, proteasomes preferentially degrade many functional cell proteins with short t 1/2 s, including cyclins and cyclin-dependent kinase inhibitors, transcription factors, tumour suppressors, and proteins catalyzing rate-limiting steps in metabolic routes. In all these cases, proteasomes produce peptides that are quickly hydrolysed by cytosolic peptidases, except some of them with an immunological role. However, proteasomes can also process, by limited proteolysis, some substrates, such as the precursor proteins NF-kappaB1 p105 and NF-kappaB2 p100, to release products with distinct biological activity.
Proteasomes mainly degrade proteins by ubiquitin-dependent (the 26S proteasomes, but not the 20S proteasomes) processes (see below). In addition, 26S proteasomes can also degrade certain proteins independently of ubiquitin. A paradigmatic example is ornithine decarboxylase, although an increasing number of proteins, including p21ras/MAPK, p53, retinoblastoma protein and TCRα, have been shown to be degraded through proteasome-dependent but ubiquitin-independent processes. Also, the 20S proteasome itself can directly degrade unstructured and partially unfolded proteins, produced, for example, by oxidation, without requiring ATP and bypassing ubiquitination. However, the mechanisms by which proteasomes recognize nonubiquitinated substrates are not well understood [14].
Proteasome complexes
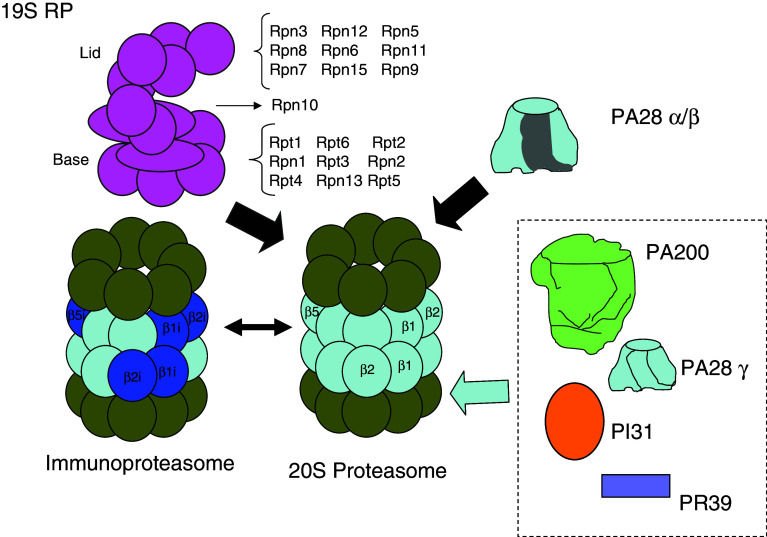
Proteasomes appear in different and widely distributed complexes of high molecular weight [11, 12]. The catalytic core of these complexes (Fig. 2) is the 20S proteasome, which can build larger supramolecular structures. From archaea to eukaryotes, the 20S proteasome is a cylindrical multicatalytic protein complex of 670 kDa, composed of four superimposed rings around a central channel of 17 Å in diameter. In mammalian cells, it is composed of 2 identical copies of 7 distinct α and 7 distinct β subunits (21–35 kDa), arranged in a hollow cylinder composed by 4 heptameric rings organized in an α1–7–β1–7–β1–7–α1–7 structure. Thus, subunits α and β are, respectively, situated in the outer and inner rings, and the two copies of each subunit are located at equivalent positions in the two symmetrical halves of the cylinder, resulting in a structure with a double symmetry and three internal chambers. The two β-rings form the central catalytic chamber, where substrate proteins are cleaved to small peptides by the combined actions of three of the β subunits, with catalytic activities depending on the secondary alcohol of an N-terminal threonine as the nucleophilic species. They are designed as chymotrypsin-like (β5), trypsin-like (β2) and caspase-like (β1, formerly called peptidylglutamyl-peptide hydrolyzing activity) [15]. The beta subunits of the eukaryotic and prokaryotic 20S proteasomes are synthesized in a precursor form with a propeptide at the N-terminus. Although the assembly process of the 20S proteasome is still not fully understood, it has been shown to involve various steps orchestrated by chaperones such as PAC1-4 and Ump1 [16], with the successive formation of: (1) a seven-α-subunits ring, (2) the α ring plus β2, β3 and β4 subunits (13S precursor), (3) a 15S half proteasome with an α and a β ring, and (4) a full, but still inactive, 20S proteasome with the extensions of the propeptides of the β subunits, which are finally autocatalytically removed to expose the N-terminal threonines of the mature 20S proteasome. In the 20S proteasome, the subunits interact tightly and, therefore, access is restricted only to proteins targeted for degradation that enter unfolded through the narrow openings of its outer α-rings. Also, the peptides that are produced within the proteasomes can only emerge through these α-rings openings. Since protein entry and peptide exit can occur through the same α-ring opening, the 20S proteasome is able to process two substrate proteins simultaneously at its six active sites [17]. Certain proteasome subunits can be subjected to N-terminal acetylation (e.g., α2, α5, α7, β3, and β4) and to phosphorylation (e.g., α7). In addition, in mammalian cells, the number of β subunits is higher than 7, since there are three additional inducible catalytic subunits, LMP2, MECL1 and LMP7 (β1i, β2i and β5i). They are non-essential, their levels are increased with γ-interferon or under conditions of an increased immune response, and they are incorporated into the 20S proteasomes at the expenses of the other three β catalytic subunits to form the immunoproteasomes [18]. They do not only differ from constitutive 20S proteasomes in their regulation, but also in their specificities that favor the generation of certain peptides of the appropriate length and C-termination for class I antigen presentation.
Fig. 2.
Different regulatory particles can bind to two ends or to one end of the 20S proteasome. The best known of these complexes is formed when 20S proteasomes bind, in an ATP-dependent process, a 19S regulatory particle (RP), composed of a base and a lid, with the indicated subunits, to produce the 26S proteasome that degrades polyubiquitinated proteins. Upon substrate degradation and ATP hydrolysis, the 26S proteasome separates into four parts, 20S proteasome, lid, base and subunit Rpn10, suggesting a cycle of proteasome assembly and dissociation during proteolysis. Also, the 20S proteasomes can form the PA28-proteasomes when PA28α/β heteroheptamers, whose expression is intensified in the presence of γ-interferon, are bound. These proteasomes and the immunoproteasomes, which are formed from 20S proteasomes when three interferon-inducible subunits (β1i, β2i and β5i) substitute the corresponding three constitutive subunits in the nascent proteasomes, facilitate the generation of antigenic peptides. The functions of other proteasome complexes with PA200, PA28γ, PI31 or PR39 are still poorly known. In addition, hybrid proteasomes are formed when 20S proteasomes bind two different regulatory particles at each end, but this has been firmly established only for proteasomes with the 19S and the PA28α/β RPs
20S proteasomes, or immunoproteasomes, can interact with various regulatory particles to form other proteasomes [11, 12]. One of them, the 26S proteasome (Fig. 2), is the key enzyme of intracellular protein degradation in eukaryotic cells. It is composed of two subcomplexes: (1) a proteolytic core (the 20S proteasome), and (2) a 19S regulatory particle (also called PA700) of more than 900 kDa. The 19S regulatory particle controls the activity of the complex, since it opens the proteolytic chamber by radial displacement of the adjacent subunits of the 20S proteasome [19], and its subunits bind polyubiquitinated proteins, remove and disassemble the polyubiquitin chain, unfold the substrate, and translocate it into the 20S proteasome proteolytic core. In addition, the 19S regulatory particle can also play, independently of the 20S proteasome, non-proteolytic roles, such as in nucleotide excision repair and in the activation of transcription and chromatin remodeling [20]. Up to two of these 19S particles can bind to both ends of the 20S proteasome, thus forming, respectively, 26S and 30S proteasomes. In fact, most 26S proteasomes probably result from the release of one of the 19S regulatory particles of the 30S proteasome during the purification process. In contrast to the 20S proteasome, little is known about the formation of the 26S proteasome, except for its ATP-dependence, that the α rings of the 20S proteasome may function as a template for this assembly and that certain proteins facilitate (the chaperone hsp90) or hinder (PAAF1, proteasomal ATPase-associated factor 1) the process. The 19S regulatory particle has 19 constitutive subunits, most of them with unknown function, distributed in a lid connected to a base. In addition to four non-ATPase subunits (Rpn1, Rpn2, Rpn10, which forms part of the hinge between base and lid, and Rpn13), the base has six homologous ATPase subunits (Rpt1 to Rpt6) that are members of the AAA family of ATPases and which carry out unfolding of the protein to be degraded as well as channel opening. In contrast, and contrary to previously proposed models, ATP hydrolysis is not absolutely required for translocation of the substrate into the cavity of the 20S core particle with the proteolytic active sites [21]. The lid has nine non-ATPase subunits distributed in two subcomplexes (Rpn5, Rpn6, Rpn8, Rpn9 and Rpn11, and Rpn3, Rpn7, Rpn12 and Rpn15). The major activities of the lid are two: interaction with the ubiquitinated substrates (Rpn10/S5a and Rpn13/ADRM1 [22]) and deubiquitination [23]. Rpn11/S13 is probably the major enzyme responsible for coupling deubiquitination with degradation. Uch37, which associates to Rpn13, also has this role, while Ubp6/Usp14, a deubiquitinating enzyme also associated to the base of the 19S regulatory particle, delays Rpn11 deubiquitination [24]. Moreover, occasionally certain E2 and E3 enzymes, or additional non-essential subunits of the 26/30S proteasome (such as Rad23, Dsk2 and Ddi1) which bind to Rpn1, interact transiently with the ubiquitinated proteins. The 19S particle subunits also have different isoforms (e.g., Rpn10a and its alternatively spliced isoform Rpn10b) and can be modified by N-terminal acetylation (e.g., Rpn1, Rpn5, Rpn6, Rpt3 and Rpt6) and myristoylation (e.g., Rpt2).
Another regulatory particle is the 11S regulatory particle (also called PA28α/β or REG), a heteroheptameric complex of two homologous PA28 subunits, α and β (neither of them is an ATPase), inducible by γ-interferon. This particle binds to the immunoproteasome or to the 20S proteasome to form the PA28-proteasome [18], absent in yeast and believed to participate in the generation of antigenic peptides to be presented by the major histocompatibility complex class I. However, the PA28-proteasome should have nonimmunological roles too, and also the 26S proteasomes can generate different sets of peptides that can be class I presented. REGα can also form homoheptamers, and there is a third member of the REG family, REGγ (also called Ki antigen), which resides in the nucleus in the form of homoheptamer caps and is not stimulated by γ-interferon. Although certain intracellular proteins can be degraded by a REGγ-proteasome complex in an ATP- and ubiquitin-independent manner [25], these and other functions need further investigation. Other regulatory complexes either enhance (PA200, which opens the proteasome outer rings) or inhibit (PI31 and PR39, which block the binding of proteasome activators) the activity of the 20S proteasome. Finally, the 11S regulatory complex can bind at one end of the 20S proteasome, while a 19S complex is bound at the other end, to form a hybrid proteasome, but the possible existence of other regulator combinations is still under investigation [12].
Ubiquitination
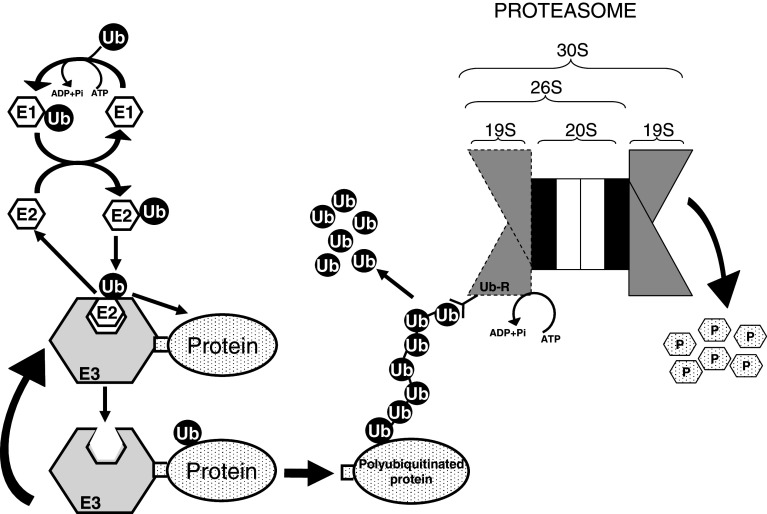
Ubiquitin is a polypeptide of 76 amino acids (8.5 kDa), an invariable diglycine motif at its C-terminus, and a relatively long (28–31 h) t 1/2. Eukaryotic cells have developed a system of conjugation of ubiquitin moieties to substrates [2] that mainly works together with proteasomes to facilitate the temporal and specific regulation of intracellular proteolysis (Fig. 3), but that can also operate independently in non-proteolytic functions. It includes a multi-enzyme cascade, catalyzed by three classes of enzymes, working consecutively to recognize and transfer ubiquitins to substrate proteins. First, E1 (ubiquitin activating enzyme), that was thought to be unique until recently, when a second enzyme was identified [26], catalyzes, in an ATP-dependent reaction, the covalent binding of an ubiquitin molecule to a cysteine residue in the E1 active site, generating a high energy thiol ester bond. Then, the activated ubiquitin is transferred, first to one (or to several in sequence [27]) of the about 20–40 E2 enzymes (ubiquitin conjugating enzymes) present in mammalian cells, via another high energy thiol ester intermediate. Finally, except in some rare instances where an E2, such as RAD6, can directly promote substrate ubiquitination, E2 enzymes transfer ubiquitin to the protein bound to a third enzyme, E3 (ubiquitin-protein ligase). These enzymes participate, either catalytically or non-catalytically, in the formation of an isopeptide bond between the carboxylic group of the C-terminal glycine of the ubiquitin molecule and an ε-amino group of an internal lysine residue (or, less frequently, the amino terminal group, [28]) in the substrate protein. There are about 500–1,000 E3s predicted in the mammalian genome that can be subdivided into different general protein families [29]. More than 90% of E3s belong to the cullin-RING (really interesting new gene) and to the RING finger-like (that include U-box proteins) families that simply facilitate ubiquitin transfer. Other E3 classes are the HECT (homologous to E6-AP C-terminus) domain ligases that form a catalytic ubiquitin intermediate, and multi-subunit complexes such as the SCF (Skp1-Cullin-F-box), SOCS (suppressor of cytokine signalling) and the APC/C (anaphase promoting complex/cyclosome). Combinations of E2 with E3s allow a large flexibility in the recognition of protein substrates and provide specificity.
Fig. 3.
The ubiquitin-proteasome system is the major non-lysosomal pathway of intracellular proteolysis. The process has two parts: (1) substrate polyubiquitination (on the left) and degradation of the tagged protein by the downstream 26S/30S proteasome (on the right). Canonical ubiquitination involves three steps: (1) activation of ubiquitin by E1 enzyme in an ATP-dependent manner, (2) transfer of ubiquitin from E1 to an E2 enzyme, and (3) direct or indirect transfer of ubiquitin to a specific protein substrate recognized by an E3 enzyme. Further incorporation of other activated ubiquitin molecules generates a polyubiquitin chain. Although polyubiquitination is a reversible process, most proteins with polyubiquitin chains of four or more ubiquitins (Ub) attached to an inner Lys of the substrate and each other by their Lys48 are recognized by a subunit (Ub-R) on the 19S regulatory particle of the 26S/30S proteasome, deubiquitinated to generate free ubiquitin, and degraded in the 20S catalytic core to peptides (P) by processes that also consume ATP
To mark proteins for proteasome degradation, ubiquitin forms polyubiquitin chains after an E3 enzyme recognizes specific sequence motifs called degrons in these proteins [30]. Phosphorylation of the substrate proteins is sometimes a pre-requisite for polyubiquitination and, in fact, unstable proteins appear to be rich in amino acids that can be phosphorylated [6]. Once the first ubiquitin has been bound to the proteolytic substrate, the C-terminal glycine of other ubiquitins can be bound to the former ubiquitin by isopeptide linkages, usually through its Lys48 residue, forming a polyubiquitin chain, whose only role is to direct the protein to the proteasome without participating in the beginning of its degradation [31]. Although polyubiquitin chains are mainly formed by adding one unit at a time, preformed ubiquitin chains can also be directly transferred to the proteolytic substrate [32]. Sometimes, enzymes called E4, which are a kind of E3 enzymes, recognize short polyubiquitin chains and catalyze their extension [33].
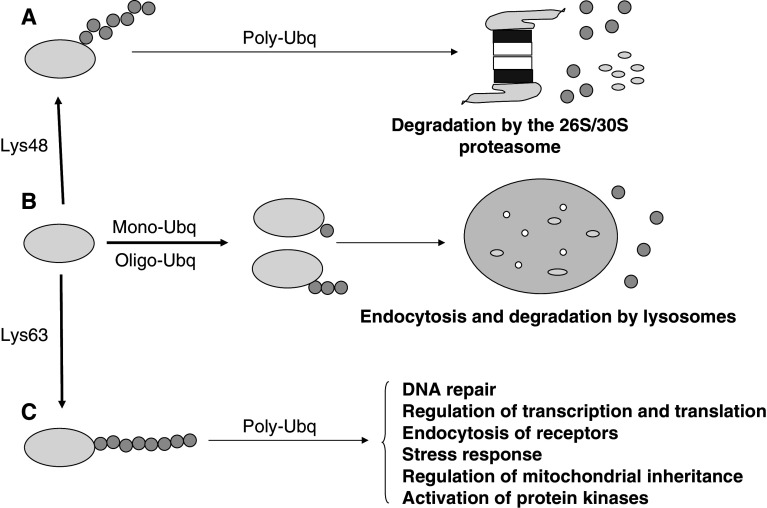
Ubiquitin has other functions (Fig. 4) beyond its crucial proteolytic role with proteasomes. For example, in contrast to the degradation of proteins by proteasomes, which needs chains with four or more ubiquitin molecules, endocytosis of some proteins (ion channels, receptors, proteins of junctional complexes, etc.) requires only monoubiquitination of single or multiple lysine residues [34]. Another well-known example of monoubiquitination is that of histones H2A and H2B, which regulates various nuclear processes related with RNA polymerase II-mediated transcription, and DNA silencing and repair [35]. Even with polyubiquitinated proteins, the topology is important to decide its final fate [30]. Based on the ubiquitin lysine implicated in the isopeptide bond, seven linkages are possible, but they mainly involve Lys48, Lys63 and, to a lesser extent, Lys11. It is usually believed that the globular Lys48-linked polyubiquitin chain is a signal for proteasomal degradation, whereas the more elongated Lys63-linked polyubiquitin chain serves other functions. However, the existence of branched chains from multiple lysines on a single ubiquitin, and of linear polyubiquitin chains with peptide instead of isopeptide bonds, increases the complexity of this simple model and would require further investigation of the fate of proteins with such linkages.
Fig. 4.
Ubiquitination of proteins controls many cellular processes by either proteolytic or non-proteolytic means. The figure illustrates some of the possibilities of ubiquitination (Ubq) of a protein that are important to decide its final fate. a Polyubiquitination at Lys48 in the ubiquitin molecule is the canonical signal for the 26S/30S proteasomal-dependent degradation of cellular proteins. b In contrast, mono or oligoubiquitination (<4 ubiquitins) targets plasma membrane proteins, via endocytosis, to early endosomes, multivesicular bodies and, finally, lysosomes, where they are degraded. In both proteasomal and lysosomal degradation, ubiquitin is first released for further use. Monoubiquitination can also target proteins to different cell compartments or have other roles as mentioned in the text. c Polyubiquitination at Lys63 in the ubiquitin molecule can serve as a non-degradative signal functioning in several cell processes as indicated
The availability of free ubiquitin in the cell is limited, and ubiquitins are released from the chains by deubiquitinating enzymes (DUBs) that maintain the necessary pool [36]. In humans, there are about hundred DUBs, including those associated, constitutively or not, to the proteasome [23]. There are two classes of DUBs, almost all within a family of cysteine proteases called ubiquitin C-terminal hydrolases (UCHs) and ubiquitin specific proteases (UBPs in yeast and USPs in mammals), with low and large molecular masses, respectively. In addition to separate ubiquitins from the protein prior to its proteolysis by the 26S proteasome, DUBs negatively regulate protein degradation by reversing ubiquitination and acting as a proof-reading mechanism in proteins inappropriately targeted for protein degradation.
Ubiquitin is a member of a family of small signaling molecules that use similar enzymatic cascades to mark other proteins by an isopeptide bond or another type of covalent link [37]. They include SUMO-1, -2, -3, Nedd8/Rub1, ISG15, FAT10 and, as we will see later, two proteins implicated in macroautophagy, Atg12 and Atg8, the latter involved not in the classical protein–protein linkage, but in a protein–lipid conjugation. Probably, the best known from these proteins in mammalian cells are SUMO (small ubiquitin-like modifier) proteins, a family with three isoforms (SUMO-1, -2 and -3) of about 12 kDa that, like ubiquitin, can be covalently attached by a serie of enzymatic reactions involving an E1-like activating enzyme (the SAE1/SAE2 heterodimer), an E2-like conjugating enzyme (UBC9), and various SUMO E3 ligases and isopeptidases. However, with rare exceptions, in which sumoylation can precede ubiquitination [38], SUMO is not used to tag proteins for degradation and instead is involved in functions such as regulation of transcription, cell cycle progression, apoptosis, nucleo-cytoplasmic transport, assembly and disassembly of protein complexes, etc.
Other non-lysosomal pathways of protein degradation
In addition to proteasomes, there are other non-lysosomal pathways of protein degradation. For example, tripeptidyl-peptidase II is a high molecular weight oligomeric peptidase that degrades the products of proteasome activity, contributes to the generation of antigenic peptides and can also partially compensate for impaired proteasome function [39]. Calpains 1 and 2 are ubiquitous proteases requiring, respectively, micromolar and milimolar calcium concentrations, that have been implicated in the regulation of important cell processes, such as cell adhesion and migration [40]. Caspases function in cascades to activate other caspases and are mainly implicated in apoptosis, although they could also have other roles (see, for example, [41]). Mitochondria contain various orthologues of Escherichia coli proteases, such as ClpP and Lon, that ensure the complete degradation of excess or damaged mitochondrial synthesized proteins [42], and there are also other proteases in various cell compartments. However, in spite of the importance of these proteases in other processes, their contribution to the total proteolysis in the cells is less relevant than that of proteasomes and lysosomes.
Lysosomes
The lysosome is the final destination for endocytic, autophagic, and secretory molecules targeted for destruction or modification by a variety of acidic hydrolases, including numerous peptidases, called cathepsins. Lysosomes degrade intracellular proteins by quite different mechanisms (Fig. 5), including endocytosis, crinophagy and the various autophagies [43]. Endocytosis (formerly called heterophagy) is the degradative route to lysosomes followed by extracellular material and also by certain plasma membrane proteins that, unlike the low density lipoproteins and the transferrin receptors, do not recycle back to the plasma membrane for reuse. By crinophagy, secretory proteins are degraded by lysosomes when their demand decreases, after fusion of the secretory granules with lysosomes or endosomes instead of with the plasma membrane. Finally, the main lysosomal mechanism for intracellular proteolysis is a “self-eating” autophagy, a general term with three different forms: macroautophagy, microautophagy and chaperone-mediated autophagy. Macroautophagy is the most important and best known and, therefore, it is frequently and simply called autophagy.
Fig. 5.
Proteins can be incorporated into lysosomes for degradation by different mechanisms. Macroautophagy (upper part) is the main lysosomal degradative route and involves the sequestration by a segregating structure of large areas of cytoplasm, typically including whole organelles, to make up autophagosomes. These pre-lysosomes fuse with endosomes and lysosomes to form autolysosomes that degrade its cytoplasmic content. In addition to macroautophagy, other mechanisms (lower part) have been described whereby lysosomes could also participate in intracellular protein degradation, including endocytosis, crinophagy, microautophagy and chaperone-mediated autophagy (for details, see text)
Macroautophagy: sequestration, formation of autophagosomes and maturation into autolysosomes
Macroautophagy (for reviews, see [43, 44]) is mainly a housekeeping pathway to recycle cell components and to supply nutrients for both biosynthetic and energetic processes. It occurs in all nucleated eukaryotic cells and is especially active under nutrient starvation. Although macroautophagy is quite similar in different cells, its roles in mammalian cells are clearly beyond those in yeast, since they include development, programmed cell death and tissue-specific roles. The process requires intermediate filaments and microfilaments to sequestrate, by a double membrane, large areas of cytoplasm, typically including entire organelles. This forms a pre-lysosome, the autophagosome, which is transported, in a dynein-dependent manner, along microtubules and fuses, first with early and//or late endosomes and, then, with lysosomes to produce an autolysosome with hydrolytic enzymes that degrade the autophagosomal inner membrane together with the sequestered content. These processes occur with a t 1/2 of 6–9 min. Finally the degradation products exit from the lysosome for utilization elsewhere. Macroautophagy is mainly non-selective; although an increasing number of reports have shown that, under certain conditions, organelles or even proteins, such as the important adaptor protein for macroautophagy p62/SQSTM1, can be specifically degraded by this lysosomal pathway. Thus, in yeast, but also in mammalian cells, there is a selective degradation of mitochondria, called mitophagy [43, 44], of peroxisomes, called pexophagy [45] and of ribosomes, called ribophagy [46]. Selective macroautophagy is now the focus of much interest, because a better understanding of this mechanism could give some clues as to its role in cell aging and death and in various pathologies.
The origin of the autophagosomal membrane and its lipid composition, which should include lipids that facilitate its expansion and curvature, is still unclear . There are two main models: maturation from a pre-existing compartment (e.g., the ER) and de novo assembly [47]. In yeast, autophagosomes originate from a compartment called preautophagosomal structure (PAS). In mammalian cells, an equivalent structure to PAS has not been found, but a de novo assembled membrane, called phagophore, was proposed years ago to be involved in the formation of the autophagosome [43]. However, there is also evidence which favors a role of ER membranes devoid of ribosomes in this formation, both in yeast and in mammalian cells, where an ER-connected compartment enriched in phosphatidylinositol 3-phosphate co-localizes with some autophagosomal proteins [48]. In addition, alternative sources for autophagosomal membranes exist (for example, the Golgi complex). Therefore, the origin of the segregating membranes in mammalian cells still remains controversial [49].
In yeast, where macroautophagy has been investigated in detail, about 31 different genes, the so-called ATG (AuTophaGy-related [50]) genes, have been identified. They include specific genes and others common to various routes. From those, about 18 appear to be essential for autophagosome formation, many of them with well-identified mammalian orthologues, including those that codify human autophagins, LC3 and beclin 1 (ATG4, ATG6 and ATG8 in yeast, respectively). However, the specific functions of other orthologues remain to be investigated. Table 2 shows the essential autophagy genes, whose products may be classified into five functional groups. The first and second groups correspond to two complexes. The first consists of an Atg1 kinase complex (Atg1–Atg13), which is formed when autophagy is induced by nutrient limitation, which together with an Atg17–Atg29–Atg31 ternary complex recruits to the autophagosomal membrane other Atg proteins [51]. The second group is another complex (with Atg2 and Atg18, and, perhaps, Atg 9) that binds to phosphatidylinositol 3-phosphate at the autophagosomal membrane and has an unknown, but essential, function [52].
Table 2.
Some identified macroautophagy-related genesa
| Yeast name | Mammalian homologue | Function in autophagy |
|---|---|---|
| ATG1 | Ulk1,2 | Ser/Thr kinase, which recruits Atg13 and Atg17, implicated in the nucleation of the autophagosome membrane |
| ATG2 | Atg2a,b | Peripheral membrane protein which forms a complex with Atg9 and Atg18, probably implicated in the nucleation of the autophagosome membrane |
| ATG3 | Atg3 | E2-like enzyme in Atg8-PE conjugation system |
| ATG4 | Atg4a-d b | Cysteine-protease which processes ATG8 before its binding to the lipid in the Atg8-PE conjugation system and releases also PE from some of the Atg8-PE conjugates |
| ATG5 | Atg5 | Forms an Atg12-Atg5 conjugate implicated, with Atg16, in the nucleation of the autophagosome membrane |
| ATG6 (VPS30) | Beclin 1 c | Involved (with Atg14, Vps34/Vps15 and other proteins) in the nucleation of the autophagosome membrane |
| ATG7 | Atg7 | E1-like enzyme in Atg8-PE and Atg12-Atg5 conjugation systems |
| ATG8 | Lc3 d | Ubiquitin-like protein in the Atg8-PE conjugation system (the Atg12-Atg5 conjugation system may act here as an E3-like enzyme) |
| ATG9 | Atg9a,b e | Only known integral membrane protein involved in the formation (with Atg2 and Atg18) of the autophagosome. Cycles between PAS and other cell structures |
| ATG10 | Atg10 | E2-like enzyme in the Atg12-Atg5 conjugation system |
| ATG12 | Atg12 | Ubiquitin-like protein in the Atg12-Atg5 conjugation system |
| ATG13 | Harbi1 | Modifies Atg1 activity and participates in the initiation (with Atg17) of macroautophagy |
| ATG14 | Barkor f | Building of the complex I of class III PI3K (with Atg6, Vps34/Vps15 and other proteins), involved in the formation of the autophagosome and in endosome fusion events |
| ATG16 | Atg16l1,2 g | Associates with the Atg12-Atg5 conjugate and homo-oligomerizes in a high molecular mass structure |
| ATG17 | – | Modifies Atg1 activity and participates (with Atg13) in the initiation of macroautophagy, and as a scaffold in the expansion of the autophagic membrane (with Atg29 and Atg 31) |
| ATG18 | Wipi1 | Probably involved (with Atg2 and Atg9) in the nucleation of the autophagosome membrane |
| ATG29 | – | Appears to function with Atg17 in the expansion of the autophagosome membrane, which leads to the engulfment of the material to be degraded |
| ATG31 | – | Like Atg29 |
PAS Preautophagosomal structure, PE phosphatidylethanolamine, Vps34 class III phosphatidylinositol 3-kinase (PI3K)
aThere other genes which can be considered autophagy-related, such as mTor (TOR1 and TOR2 in yeast, and various components of the class III PI3K complex: Vps34, Vps15 and Uvrag (also present in yeast), and Ambra1 and Bcl2 (only in mammalians)
bIn mammalian cells, the products of Atg4, the autophagins, constitute a 4-members family (a, b, c and d)
c Beclin1, the orthologue of ATG6, codifies a tumor suppressor and its heterozygous deletion increases mice cancer rates
dLC3, microtubule-associated protein 1 light chain 3, is the best characterized product of the orthologues of ATG8 (Gabarap, Gabarapl2, Gate16 and Atg8l) and encodes a protein with three isoforms, A, B and C
eThe product of Atg9a, APG9L1, can be complemented with the product of a second gene, Atg9b (APG9L2)
fBarkor shares in human cells 18% sequence identity and 32% sequence similarity with yeast Atg14 and is probably its human homologue or at least one of them
gIn humans, the Atg16l1 gene is associated with Crohn′s disease
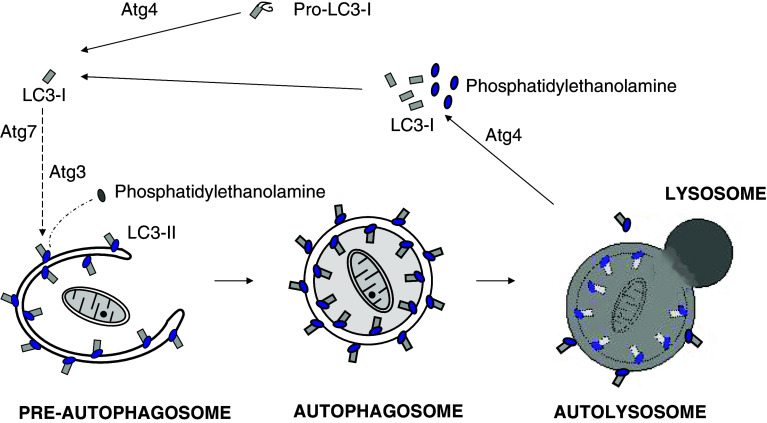
The third and fourth groups are two ubiquitin-like conjugation systems that participate in autophagosome formation [53]. One of those mediates the binding of Atg12 and Atg5, and these two proteins will later form, with Atg16L, a high molecular mass oligomeric structure, of ~800 kDa in mammalian cells, called the Atg16L complex. In Atg12–Atg5 conjugation, Atg7 and Atg10 play, respectively, a role equivalent to the E1 and E2 enzymes of the ubiquitin pathway. In the second conjugation system, Atg8 binds covalently to phosphatidylethanolamine to mediate tethering of the adjacent membranes in the closure of the autophagosomal membrane [54], with Atg7, again, and Atg3 working as E1- and E2-like enzymes, respectively. These two conjugation systems are connected, since the Atg16L complex recognizes the pre-autophagosomal membrane and functions, like an E3 enzyme, as a scaffold for Atg8 lipidation [55]. LC3 is the best characterized of the mammalian orthologues of ATG8. It encodes a protein with three isoforms that follows an important cycle in the formation and maturation of the autophagic vacuole (Fig. 6), and its lipidated form, LC3-II, is frequently used, under certain conditions, as a marker for macroautophagy. This latter system also requires a cysteine-protease (Atg4) both to process Atg8, exposing a C-terminal glycine residue before its binding to the lipid, and also to delipidate it.
Fig. 6.
Covalent binding of LC3 to phosphatidylethanolamine (PE) is essential for autophagosome formation. In mammalian cells, cytosolic LC3-I (Atg8 in yeast) is synthesized as a precursor (Pro-LC3-I). Immediately after its synthesis, a C-terminal fragment is cleaved by Atg4 to produce LC3-I with an exposed glycine residue that binds covalently to PE on the pre-autophagosome membrane to form LC3-II. In this process, the mammalian homologues of Atg7 and Atg3 work, respectively, as the E1- and E2-like enzymes of the ubiquitination system, and the Atg16L complex probably functions as an E3-like enzyme. Once the autophagosome is formed, LC3-II localizes both at the cytosolic and luminal faces of its double membrane. After fusion of the autophagosome with endosomes/lysosomes to form an autolysosome, the luminal LC3-II is degraded by lysosomal cathepsins, while Atg4 recycles LC3-I and PE from LC3-II on the cytosolic face of the autolysosome membrane
The last group is a most relevant complex formed by Vps34 (class III phosphatidylinositol 3-kinase (PI3K)), Vps15 (p150), Atg6 (Beclin 1) and Atg14 (Barkor). Beclin 1 is a tumor suppressor that binds to the anti-apoptotic protein Bcl-2 (B-cell lymphoma 2) under nutrient-rich conditions. Starvation induces JNK1-mediated phosphorylation of Bcl-2 [56], from which Beclin 1 dissociates and interacts with class III PI3K associated to the p150 serine/threonine kinase, to form a macroautophagy activating complex. Other components of this complex that positively regulate macroautophagy are: (1) UVRAG (ultraviolet irradiation resistance-associated gene), which interacts with other proteins, such as Bif1, and stimulates the fusion of lysosomes with late endosomes and, perhaps, with autophagosomes, too [57], and (2) Barkor (Beclin 1-associated autophagy-related key regulator), a homologue of Atg14 that participates in a second complex, without UVRAG and with a clearer role in autophagosome formation [58]. Also, and at least in certain mammalian cells, another protein, Ambra1 (activating molecule in Beclin1-regulated autophagy), binds to Beclin 1 and favors its interaction with class III PI3K [59]. In contrast, Bcl-2 and Bcl-X, which also bind to Beclin1, negatively regulate autophagy. Therefore, all these partners, and probably others, appear to regulate Beclin1 activity and, thus, macroautophagy in mammalian cells.
Other autophagic pathways
Microautophagy is a degradative route to directly internalize portions of cytoplasm into lysosomes by various modifications of the lysosomal membrane that produce, by budding, intralysosomal vesicles that later release their content [43]. Microautophagy degrades nuclear fragments (piecemeal microautophagy of the nucleus or nucleophagy), cell organelles such as peroxisomes (micropexophagy), or single macromolecules. In yeast, some ATG genes are required for micropexophagy and nucleophagy [60], thus suggesting a tight regulation between macro- and microautophagy. However, in mammalian cells, microautophagy is poorly characterized, although it has been reconstructed in vitro.
In mammalian cells, a specific transfer of cytosolic protein molecules through the lysosomal membrane was described. This pathway, called chaperone-mediated autophagy [43], degrades certain proteins, such as ribonuclease A with a KFERQ “signal” sequence (amino acids 7–11). Although this pentapeptide has not been found in other proteins, KFERQ-like sequences are present in other substrates of the pathway, but not in proteins that are not. The pathway is especially active under prolonged starvation, and its requirements, mainly investigated under in vitro conditions, are similar to those described for the biosynthetic transport of proteins to cell organelles: (1) chaperones (hsc73, but also others) in the cytosol and in the lysosomal membrane and lumen, (2) ATP-Mg++, and (3) a receptor on the lysosomal membrane (LAMP-2a, a LAMP-2 isoform), which multimerizes to form the hypothetical translocon.
More recently, a non-canonical autophagy has been described in cancer MCF-7 cells that does not require the entire set of Atg proteins, in particular Beclin1, to form the autophagosome, and that is involved in caspase-independent cell death [61].
Regulation of intracellular protein degradation
It is well known that macroautophagy is physiologically regulated (see [62] for a review). In yeast, nutrient starvation, in particular of nitrogen, is the most potent stimulus of autophagy. In mammalian cells, the main signals controlling macroautophagy are nutritional, but hormones are important regulators too. Thus, early studies with hepatocytes showed that insulin inhibits macroautophagy, whereas glucagon activates it. In addition, in mammalian cells macroautophagy is regulated by glucose, which, in contrast to yeast, increases macroautophagy, vitamins, osmotic stress, various growth factors, and some amino acids, which are the main regulators. Thus, in liver, eight regulatory amino acids of macroautophagy have been identified and, among those, Leu is the most important in many cells. However, regulatory amino acids may also change in some cells [10]. Although the receptors for hormones and growth factors in the cells are, in general, well characterized, this is not the case with the sensors for amino acids. The most accepted point of view [62] postulates that these receptors are intracellular, perhaps close to L-amino acids transporters at the plasma membrane, like the recently described antiporter that imports leucine and other branched amino acids while exporting glutamine [63].
The signals that indicate the availability of nutrients and energy and the presence of growth factors converge at the serine-threonine kinase TOR (target of rapamycin, a lipophilic drug), and several reports indicate the implication of TOR in the transduction of these signals to the macroautophagic machinery (see [62] and [64] and citations therein). Mammalian TOR (mTOR) phosphorylates indirectly Atg1 and directly, among others, two proteins that regulate translation, 4E-BP1 (binding protein 1 of eukaryotic initiation factor 4E) and S6K1 (ribosomal S6 protein kinase 1). There are two mTOR complexes, mTORC1, which is rapamycin-sensitive and phosphorylates 4E-BP1 and S6K1, and mTORC2, which is not inhibited by rapamycin and phosphorylates Akt at S473. Since mTORC1 negatively regulates macroautophagy in hepatocytes, rapamycin activates this proteolytic process [62], but sometimes the results obtained have been different [65]. Therefore, and although it appears clear that amino acids regulate macroautophagy by mTOR, in certain cells alternative signaling routes to mTOR also appear to regulate macroautophagy [10, 66].
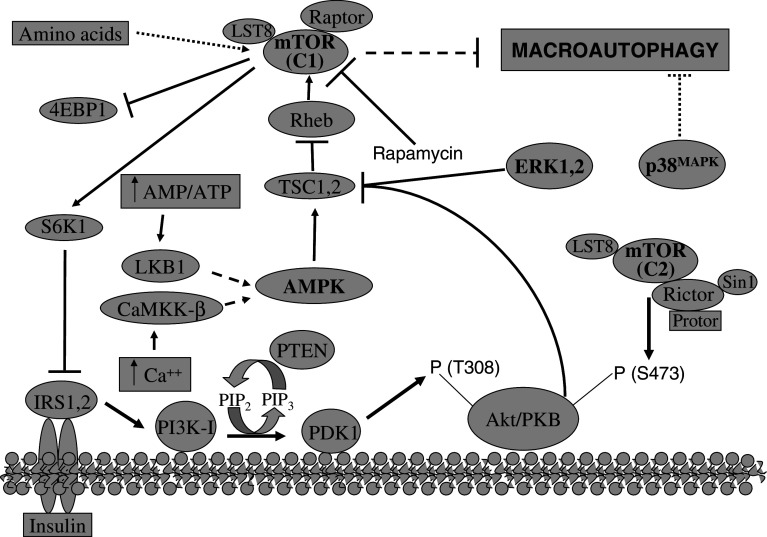
Upstream of mTOR, other molecules (Fig. 7 shows a simplified scheme) participate in this signaling route to the autophagic vacuoles [62, 64, 67]: class IA PI3K (which, contrary to class III PI3K, inhibits sequestration), the phosphatase PTEN (which reverts the effect of class I PI3K), Pdk1 (which is activated by phosphatidylinositol 3,4,5 trisphosphate and binds to the plasma membrane), Akt (which is activated by phosphorylation in T308 by Pdk1 and in S473 by mTORC2), the TSC1/TSC2 complex (which is inactivated by Akt), and a small G protein called Rheb (Ras homology enriched in brain) that activates mTORC1 and is inhibited by the GAP activity of TSC1/TSC2. In addition, AMP-activated protein kinase (AMPK), which is activated by an increase in the AMP to ATP ratio, phosphorylates TSC2 producing the inhibition of Rheb and activating macroautophagy [68]. Also, calcium, which activates AMPK by the Ca++/calmodulin dependent kinase kinase-beta, regulates autophagy [69]. Finally, other molecules have been implicated in the regulation of macroautophagy in different cell types, including ceramides, myo-inositol-1,4,5-trisphosphate, heterotrimeric G proteins, JNK1, ERK 1,2 and p38 MAP kinases, protein kinase A, p19ARF, DRAM, etc. [62, 64].
Fig. 7.
Macroautophagy is mainly regulated by the TOR signalling pathway. The kinase mammalian TOR (mTOR) is the principal negative regulator of macroautophagy. mTOR exists in two distinct complexes: one which is sensitive to the drug rapamycin (C1) and a second, which is not (C2). Regulatory amino acids activate mTORC1 and, thus, inhibit macroautophagy by a still unknown pathway. Insulin and some growth factors inhibit macroautophagy through the insulin receptors (IRS) and class I PI3K (PI3K–I) that produces phosphatidylinositol 3,4,5-trisphosphate (PIP3). The well-known tumor suppressor phosphatase and tensin homologue (PTEN) antagonizes the activity of PI3K–I by dephosphorylating PIP3 to.phosphatidylinositol 4,5-bisphosphate (PIP2). PIP3 recruits the oncoprotein Akt (also called proteinase kinase B or PKB) to the cytosolic face of the plasma membrane where it is phosphorylated at different residues (T308 and S473) by the 3-phosphoinositide-dependent kinase 1 (PDK1) and the mTORC2 complex, respectively. Akt phosphorylates one member (TSC2) of the tuberous sclerosis complex TSC1 (hamartin), TSC2 (tuberin), which is inhibited. TSC1,2 is a negative regulator with GAP activity of the small GTPase Rheb, which in its GTP-bound form activates mTOR-Raptor, inhibiting macroautophagy. On the other hand, a high AMP/ATP ratio or an increase in cytosolic Ca++ produces, through the serine/threonine protein kinase LKB1 or the Ca++/calmodulin-dependent protein kinase kinase beta (CaMKK-β), respectively, the phosphorylation and the activation of the AMP-activated protein kinase (AMPK), which increases macroautophagy, via TSC1,2. mTORC1 phosphorylates its substrates, including 4EBP1 and S6K1, and rapamycin is a strong and quite specific inhibitor of mTORC1. To prevent the overactivation of the mTOR signalling pathway, S6K1 can phosphorylate the insulin receptor substrate 1 (IRS1), inhibiting in this way the proximal part of the insulin signalling pathway. Finally, other signalling pathways are also known to regulate macroautophagy, including those involving the MAP kinases ERK1,2 (Ras-Raf-MEK-ERK pathway) and the p38MAPK, which activate and inhibit, respectively, macroautophagy. ERK1,2, like Akt, also inhibits TSC1,2, while how p38MAPK inhibits macroautophagy is less known
Very little is known about the regulation of other proteolytic systems, probably because macroautophagy was considered until recently the only proteolytic mechanism that was directly activated under extreme situations such as starvation. However, and although the regulation of proteasome degradation was believed to occur only at the level of the substrate proteins, amino acids and insulin reduce, in an additive way, the degradation by proteasomes in human fibroblasts [10] and in C2C12 myotubes [70]. These effects could be due to changes in the activity of proteasomes or of specific E3 ubiquitin ligases. Proteasomes and macroautophagy can be simultaneously regulated, as exemplified by the forkhead transcription factor FOXO3, which can promote skeletal muscle atrophy by activating macroautophagy and the ubiquitin-proteasome pathway, the latter via expression of the muscle-specific ubiquitin ligase atrogin-1 [71].
In summary, in mammalian cells, many signals regulate macroautophagy, but the data are still incomplete and sometimes contradictory, probably because crosstalks occur among various signaling routes that vary depending on cell type. Moreover, most aspects remain unknown, in particular the regulation of the proteolytic pathways different from macroautophagy.
Pathology
Alterations in the proteolytic routes are associated with an increasing number of pathologies, including oncogenesis, neurodegenerative diseases, vacuolar and non-vacuolar myopathies, α-antitrypsin deficiency, infectious disorders, and several autoimmune diseases, such as rheumatoid arthritis and Crohn′s disease, etc. [1, 72]. Therefore, special attention is now devoted to the activity of proteolytic systems in pathological situations, because this information could be important to identify new therapeutic targets.
Dysfunction of programmed cell death leads to various human pathologies, including cancer and several degenerative diseases. Classically, two types of programmed cell death were described. Type I (apoptotic) is mediated by caspases, and, for example, 26S proteasomes become inactivated at an early stage by a caspase-mediated cleavage of three subunits of the 19S regulatory complex [73]. Type II (autophagic or non-apoptotic) is characterized by the apparent destruction of the cell by abundant autophagic vacuoles instead of by phagocytosis of apoptotic bodies. However, it is difficult to decide if macroautophagy has a causal role in cell death or if it simply represents a failed attempt of the cells to promote survival [74]. In fact, starvation-induced macroautophagy is clearly an anti-apoptotic mechanism, and this is probably also the role of macroautophagy in cells with intact apoptotic machinery.
In cancer, there is usually a gain of function of growth factor receptors or protooncogenes such as Akt or PI3K, or loss of functions of tumor suppressors such as PTEN, Beclin1, TSC1,2 or LKB1, and numerous evidence indicates that alterations in the ubiquitin-mediated proteolysis of these and of other proteins that allow the limitless replication of cancer cells contribute to tumorogenesis [1]. Most of the above-mentioned proteins regulate macroautophagy (see Fig. 7), and an inverse correlation between autophagic activity and malignant potential has frequently been observed . For example, when tumor suppresor genes such as Beclin1 or PTEN are overexpressed, macroautophagy is induced [72], suggesting that autophagy is harmful for cancer cells. However, a limited autophagy could represent a benefit to overcome the limitations in nutrients supply.
Neurodegenerative disorders represent another important group of diseases in which proteolysis is relevant, since neurons, with their long axons and dendritic arborization, are particularly vulnerable. However, the exact basis for this susceptibility is unknown. Mutations in the ubiquitin ligase parkin causing autosomal-recessive juvenile Parkinson′s disease represent a classical example of the implication of the ubiquitin system in a neurodegenerative disease, and similar pathologies, such as Huntington′s disease, have been associated with alterations in the normal functioning of the ubiquitin-proteasome system [75]. In these, and in many other neurodegenerative disorders, protein misfolding and increased protein hydrophobicity drive protein aggregation, and the resulting aggresomes may sequester and inactivate beneficial cellular proteins, particularly components of the ubiquitin-proteasome machinery that try to degrade them. However, cells may attempt to compensate for impairments in one form of proteolysis (e.g., proteasomes) by elevating an alternate form (e.g., macroautophagy). Therefore, a common observation in neuronal pathologies, as well as in aging, is the presence of proteolytic alterations and a cross-talk between different proteolytic pathways [76]. Also, a decreased macroautophagy can contribute to the pathogenesis of neurodegenerative diseases, as shown for example by the altered processing of the amyloid precursor protein in a mouse model of Alzheimer’s disease [77]. Therefore, as in other processes, in neurodegenerative diseases, the beneficial or detrimental role of autophagy is context-dependent.
Proteolysis, lysosomal and non-lysosomal, has also been implicated in an increasing number of other pathologies. We will mention only two representative examples. Lysosomal storage diseases are caused by a deficiency in some lysosomal protein that results in lysosomal accumulation of undegraded material, including polyubiquitinated protein aggregates, and usually in neurodegeneration. Common features of most of these diseases are a block in the fusion of autophagosomes with lysosomes/endosomes, and mitochondrial fragmentation and dysfunction [78], thus connecting lysosomal alterations with mitochondrial degenerative processes. The second example refers to the ubiquitin-proteasome system. Although failures of this system have been implicated in the pathogenesis of many diseases, an accelerated breakdown of myofibrils in myocytes by the ubiquitin-proteasome system occurs in muscle atrophy (produced by several diseases, such as cancer, diabetes mellitus, AIDS, sepsis and Cushing′s syndrome, or by muscle denervation or disuse). In addition, the degradative systems are intracellular surveillance systems to eradicate pathogens that reach the cytosol and both ubiquitination [79] and xenophagy (macroautophagic digestion of microbes) [80] play a critical role in protecting the cells from microbial infection. However, some of these microorganisms have evolved mechanisms to circumvent these protective systems. Also, in several viral malignancies, such as those associated to Kaposi′s sarcoma, certain B cell lymphomas and cervical cancer, the associated viruses subvert the ubiquitin-proteasome system [81].
Because intracellular protein degradation alterations are involved in so many diseases, their components have become an attractive target for therapies. The paradigmatic example of this is a potent, specific and reversible proteasome inhibitor, bortezomib (Velcade, formerly known as PS-341) that in 2003 received approval from the USA Food and Drug Administration agency to treat multiple myeloma. Since bortezomib downregulates chemoresistance pathways, it has been successfully used, in combination with other anticancer drugs, to treat cancer cells, specifically in multiple myeloma and in non-Hodgkin′s lymphoma [82]. The exact mode of action of Velcade is not clear, although it inhibits in particular the translocation of NF-κB to the nucleus, triggered by the proteasomal degradation of IκB, thus inactivating the transcription of several genes codifying for anti-apoptotic factors, cytokines, cell adhesion molecules and cell cycle regulators. This results in a variety of general effects, including the induction of apoptosis in cancer cells and the reversal of their resistance to radiation or chemotherapy. Although the activity of proteasomes is crucial for normal cell function, and proteasome inhibitors produce cell death, it appears that rapidly dividing cancer cells are more sensitive to some of these inhibitors than quiescent normal cells. In addition, there are other procedures to inhibit proteasome-dependent degradation that may find clinical application for drug development, such as the inhibition of the formation of polyubiquitin chains by ubistatins, and it is expected that E3 enzymes or deubiquitinases, because of their greater specificity, could be more appropriate therapeutic targets for therapy, since their inhibition will only affect the degradation of certain protein substrates and not all proteasomal activity [82]. Finally, manipulation of autophagy by drugs may soon allow the development of novel therapies with clinical application [83].
Conclusions
In summary, and after the important advances in the knowledge of the complex machinery implicated in the proteolytic pathways using genetic approaches in yeast, and after the recognition of the relevance of proteolytic processes in physiology and pathology, it is expected that research with mammalian systems is also experiencing a considerable progress using various experimental procedures. Thus, for example, transgenic mice models have been developed for the in vivo study of the ubiquitin-proteasome system and of the macroautophagic process. Also, proteomic approaches have been recently introduced [8] which will allow the identification of substrates of the different proteolytic machineries, in particular of specific E3 ligases. These procedures are also relevant to investigate the pathologies associated to alterations in intracellular proteolysis. In fact, and to mention only an example, conditional knock-out mice that do not express in neurons the products of ATG5 or of ATG7 genes develop neurodegeneration [84, 85]. However, there are still many general questions to be answered, in particular those related to the regulation of the different proteolytic pathways in the cells under various conditions, the molecular mechanisms involved in the degradation of intracellular proteins by proteolytic pathways that are different from macroautophagy and proteasomes, or the real implication of alterations in all these pathways in the various pathologies.
Acknowledgments
We thank Asunción Montaner and Nuria Mas for useful comments. Studies of the authors′ laboratories are supported by grant BFU2008-00186BMC from the Spanish Ministerio de Ciencia e Innovación.
References
- 1.Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Grisolía S, Hernández-Yago J, Knecht E. Regulation of mitochondrial protein concentration: a plausible model which may permit assessing protein turnover. Curr Topics Cell Regul. 1985;27:387–396. doi: 10.1016/b978-0-12-152827-0.50040-2. [DOI] [PubMed] [Google Scholar]
- 6.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 7.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:665–675. doi: 10.1016/S1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 8.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 9.Quesada V, Ordoñez GR, Sánchez LM, Puente XS, López-Otín C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucl Acids Res. 2009;37:D239–D243. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteban I, Aguado C, Sánchez M, Knecht E. Regulation of various proteolytic pathways by insulin and amino acids in human fibroblasts. FEBS Lett. 2007;581:3415–3421. doi: 10.1016/j.febslet.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 11.DeMartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 16.Rosenzweig R, Glickman MH. Chaperone-driven proteasome assembly. Biochem Soc Trans. 2008;36:807–812. doi: 10.1042/BST0360807. [DOI] [PubMed] [Google Scholar]
- 17.Hutschenreiter S, Tinazli A, Model K, Tampé R. Two-substrate association with the 20S proteasome at single-molecule level. EMBO J. 2004;23:2488–2497. doi: 10.1038/sj.emboj.7600262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rechsteiner MC, Hill M. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 19.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitinating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 25.Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groettrup M, Pelzer C, Schmidtke G, Hofmann K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem Sci. 2008;33:230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 28.Ciechanover A. N-terminal ubiquitination. Methods Mol Biol. 2005;301:255–270. doi: 10.1385/1-59259-895-1:255. [DOI] [PubMed] [Google Scholar]
- 29.Robinson PA, Ardley HC. Ubiquitin-protein ligases. J Cell Sci. 2004;117:5191–5194. doi: 10.1242/jcs.01539. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. J Biol Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 33.Hoppe T. Multiubiquitination by E4 enzymes: ‘one size’ doesn’t fit all. Trends Biochem Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Seaman MNJ. Endosome protein sorting: motifs and machinery. Cell Mol Life Sci. 2008;65:2842–2858. doi: 10.1007/s00018-008-8354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Millard SM, Wood SA. Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J Cell Biol. 2006;173:463–468. doi: 10.1083/jcb.200602082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 38.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 39.Naujokat C, Fuchs D, Berges C. Adaptive modification and flexibility of the proteasome system in response to proteasome inhibition. Biochim Biophys Acta. 2007;1773:1389–1397. doi: 10.1016/j.bbamcr.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Croall DE, Ersfeld K. The calpains: modular designs and functional diversity. Genome Biol. 2007;28:218. doi: 10.1186/gb-2007-8-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, Cuervo AM, Dunn WA, Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 45.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 46.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 47.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimori T, Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirni A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 51.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 55.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maria Fimia G, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 60.Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, Millen J, Goldfarb DS, Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 62.Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 63.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meléndez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kadowaki M, Razaul Karim M, Carpi A, Miotto G. Nutrient control of macroautophagy in mammalian cells. Mol Aspects Med. 2006;27:426–443. doi: 10.1016/j.mam.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 66.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 67.Abraham RT. Regulation of the mTOR signalling pathway: from laboratory bench to bedside and back again. F1000 Biol Rep. 2009;1:8. doi: 10.3410/B1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 69.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 70.Sadiq F, Hazlerigg DG, Lomax MA. Amino acids and insulin act additively to regulate components of the ubiquitin-proteasome pathway in C2C12 myotubes. BMC Mol Biol. 2007;8:23. doi: 10.1186/1471-2199-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Mol Cell. 2004;14:81–93. doi: 10.1016/S1097-2765(04)00156-X. [DOI] [PubMed] [Google Scholar]
- 74.Levine B, Kroemer G. Autophagy in aging, disease and death: the true identity of a cell death impostor. Cell Death Differ. 2009;16:1–2. doi: 10.1038/cdd.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 76.Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin-proteasome system: collaborators in neuroprotection. Biochim Biophys Acta. 2008;1782:691–699. doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 79.Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 80.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Shackelford J, Pagano JS. Role of the ubiquitin system and tumor viruses in AIDS-related cancer. BMC Biochem. 2007;8(Suppl 1):S8. doi: 10.1186/1471-2091-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9(Suppl 1):S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levine BB, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 85.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]