Abstract
SoxB1 factors, which include Sox1, 2, and 3, share more than 90% amino acid identity in their DNA binding HMG box and participate in diverse developmental events. They are known to exert cell-type-specific functions in concert with other transcription factors on Sox factor-dependent regulatory enhancers. Due to the high degree of sequence similarity both within and outside the HMG box, SoxB1 members show almost identical biological activities. As a result, they exhibit strong functional redundancy in regions where SoxB1 members are coexpressed, such as neural stem/progenitor cells in the developing central nervous system.
Keywords: Sox factor, SoxB1 member, ES cells, iPS cells, Neural stem cells, Human eye disorder
Introduction
The male sex determination gene Sry (sex-determining region of Y chromosome) was the first Sox (SRY-related HMG box) gene family member identified [1]. Sox proteins contain a 79-amino acid HMG box domain responsible for sequence-specific DNA binding [2, 3]. About 30 Sox genes have been identified, including 20 that are present in both the mouse and the human genome [4]. Mammalian Sox genes have been divided into eight groups, designated A–H, based on phylogenetic analysis of HMG box regions. The B group is further divided into two subgroups: SoxB1 proteins (Sox1, 2, 3) containing transcriptional activation domains and SoxB2 proteins (Sox14, 21) containing repressor domains [5]. SoxB1 subfamily transcription factors are predominantly expressed in the early embryo, developing testis, and nervous system, and are important for cell fate determination and cell differentiation during mouse development (Table 1), as shown by dominant negative and knockout mouse studies [6–9].
Table 1.
Expression and biological functions of SoxB1 members
| Gene | Expressed domains | Functions |
|---|---|---|
| Sox1 | Embryonic nerve system (CNS), lens, urogenital ridge | Lens development (induction and maintenance of gamma-crystallin gene expression) [81] |
| Deletion in KO mice leads to microphthalmia, cataracts, and spontaneous seizures [81, 82] | ||
| Sox2 | Inner cell mass, primitive ectoderm, trophoblast stem cells, embryonic nerve system (CNS), lens, neurogenic regions in adult brain | Deletion in KO mice is embryonic lethal at implantation stage (involved in gene expression in FGF4 [13–15], Oct4 [50, 51], Nanog [47], UTF1 [21], etc., together with Oct-3/4) |
| Induction of gamma- and delta-crystallin gene expression [24, 25] | ||
| Induction of Nestin gene expression in neural stem/progenitor cells [27, 28] | ||
| Involved in its own expression in ES cells and neural stem cells [29–31, 34] | ||
| Required for maintaining neural stem cell state and neurogenic potential [78–80, 86, 90] | ||
| Sox3 | Epiblast, embryonic CNS, lens, urogenital ridge | Required for early embryogenesis, gonadal function, and hypothalamo-pituitary axis formation [89, 96] |
| Candidate gene for human X-linked mental retardation syndromes [97] |
SoxB1 members control the expression of distinct sets of genes in a cell-type-specific manner by interacting with specific partners [10]. In this review, we give an overview of the functions of SoxB1 family genes in embryonic development and stem cells, focusing on the role of Sox2.
Cell-specific function and partnerships of SoxB1 transcription factors
Sox proteins bind to DNA in a sequence-specific manner (5′-A/TA/TCAAA/TG-3′) through their HMG domain [3, 11, 12] and either activate or repress transcription [4, 10]. The HMG domain is highly conserved among Sox factors; accordingly, in vitro DNA binding studies reveal no significant differences in recognition sequence specificity among family members. However, each Sox factor recognizes distinct sets of regulatory regions containing Sox binding sites in vivo [10]. How do Sox factors recognize their appropriate target genes and regulate their transcriptional levels in a cell-type-specific manner?
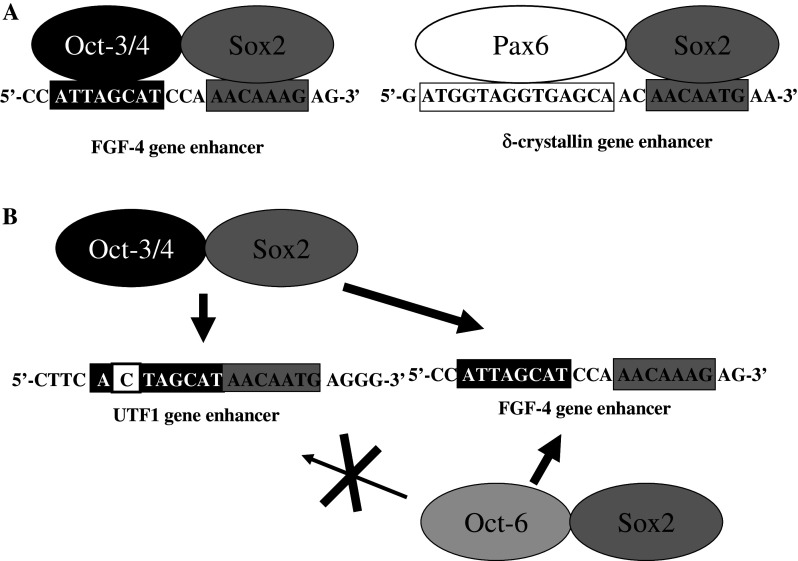
An important clue came from analysis of fibroblast growth factor-4 (FGF-4) expression in embryonic carcinoma (EC) and embryonic stem (ES) cells [13–15]. The regulatory region controlling FGF-4 expression in EC and ES cells contains Octamer factor and Sox binding sites, which serve as binding sites for embryonic Octamer factor 3/4 (Oct-3/4) [16–18] and Sox2, respectively (Fig. 1a). Binding of both factors to their recognition sites is essential for activating transcription, since mutation of either regulatory element results in complete loss of transcription. Subsequently, we demonstrated that the expression of undifferentiated embryonic cell transcription factor 1 (UTF1) gene [19, 20] in ES cells and EC cells is regulated in a similar manner [21], suggesting that the combined action of Oct-3/4 and Sox2 is a general mechanism that regulates gene expression in ES and EC cells. Indeed, a number of ES and EC cell genes, including Oct-3/4 and Sox2 themselves, are now known to be regulated by the same mechanism (for details, see below). However, unlike other regulatory elements containing Octamer factor and Sox binding sites, the UTF1 regulatory region can selectively recruit Oct-3/4–Sox2 complexes and prevent the binding of similar protein complexes, such as Oct-1–Sox2 and Oct-6–Sox2 (Fig. 1b). This specificity is dictated by the unique ability of the Oct-3/4 POU-homeodomain to recognize a variant of the Octamer motif in the UTF1 regulatory element. It is noteworthy that both FGF-4 and UTF1 regulatory regions are not located in the proximity of their promoter regions, but a couple of kilobases away from them. Therefore, Nowling et al. [22, 23] were interested in examining whether Oct-3/4–Sox2 complex directly interacts with pre-initiation complex on the basal FGF-4 gene promoter or some additional factor(s) are involved in the cooperation. They found that the co-activator p300 physically associates with Sox2 and mediates the transcriptional stimulating activity of Oct-3/4–Sox2 protein complex to the promoter.
Fig. 1.
Combinatorial action of Sox2 and its partner proteins on regulatory enhancers. a Sequences of Sox-dependent regulatory regions of FGF-4 and δ-crystallin genes. b Selective recruitment of the Oct-3/4–Sox2 complex by the UTF1 regulatory region. The UTF1 regulatory enhancer serves as a specific binding site for Oct-3/4–Sox2 complex, but prevents similar protein complexes, such as Oct-1–Sox2, from binding to the variant Octamer sequence (for details, see reference [21])
Sox2 cooperates with other transcription factors in other cell types as well [10]. For instance, transcription of δ-crystallin in lens cells is activated by the DC5 enhancer only when both Sox2 and Pax6 are bound [24, 25]. Cooperation with Pax6 appears to be restricted to SoxB1 group members, since other types of Sox factors, such as Sox9, fail to activate transcription through DC5, even though Sox9 binds to DC5 in vitro as strongly as do Sox1, 2, or 3. Moreover, when Sox2 and Pax6 were co-expressed in vivo, both proteins were found by chromatin immunoprecipitation (ChIP) to bind to DC5, whereas neither was able to bind to DC5 when expressed alone [24]. These data show that Pax6 and SoxB1 proteins are unable on their own to form stable protein–DNA complexes with the DC5 enhancer in vivo, even though they are able to do so in vitro. Thus, interactions between SoxB1 proteins and their partners are important not only for selecting target sequences, but also for stable binding to regulatory regions in vivo.
Similarly, the N-3 regulatory region, which controls Sox2 expression in the diencephalon and optic vesicle, activates transcription by the combined action of Sox2 and Pax6 [26]. The core sequence of N-3 comprises a Sox binding sequence and a noncanonical Pax6 binding sequence arranged in the same orientation and spacing as found in the DC5 enhancer, suggesting that as for Oct-3/4 and Sox2 in ES and EC cells, functional cooperation between Sox factors and Pax6 is a widespread mechanism of gene regulation in regions where Pax6 and Sox factors are coexpressed.
The regulatory region of the Nestin gene provides another example of SoxB1 factors activating cell-type-specific transcription [27, 28]. Like the UTF1 and FGF-4 enhancers, the Nestin regulatory region contains Octamer and Sox binding sites but is not active in EC and ES cells. Rather, it functions specifically in neural stem/progenitor cells, in which Sox2 cooperates with Octamer factors Brn 1/2/4 or Oct-6 to activate transcription. Unlike other regulatory regions containing Octamer factor and Sox binding sites, the Sox2 regulatory region 2 (SRR2) is able to support transcription in both ES cells and neural stem/progenitor cells [29, 30]. Oct-3/4 binds to this site to stimulate transcription in ES cells, whereas Brn1/2/4 and Oct-6 cooperate with Sox2 at SRR2 to drive expression of genes in neural stem/progenitor cells.
Thus, regulatory regions containing Octamer factor and Sox binding sites can be divided into three groups based on their specificity: those that function in ES cells, those that function in neural stem cells, and those that function in both. However, apart from the UTF1 regulatory region, which contains an Octamer binding sequence variant that is only recognized by the Oct-3/4 and Sox2 complex (Fig. 1b) [21], it is not clear at present why regulatory regions with similar organizations display such distinct cell-type specificity.
Sox2 regulatory region 2 shows strong regional specificity in CNS
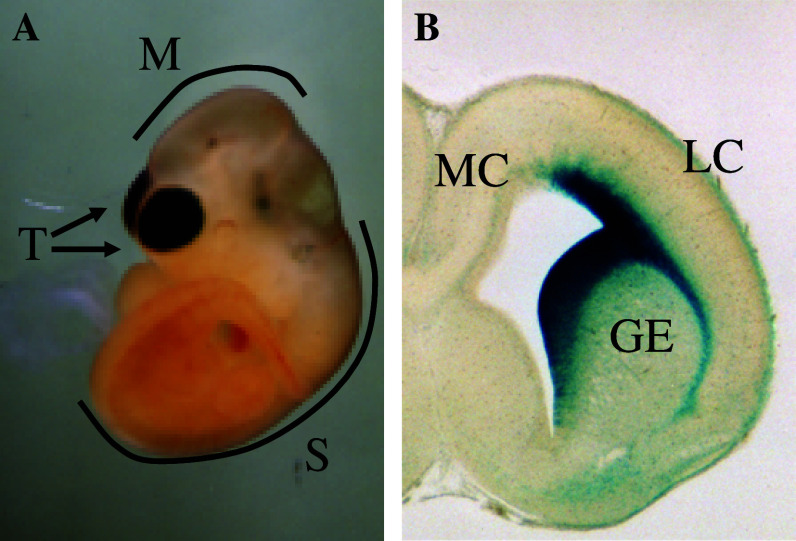
SRR2 and the Nestin enhancer have distinct regional specificities in neural stem/progenitor cells of the developing central nervous system (CNS), as shown by transgenic analyses [31]. The Nestin enhancer is active in neural stem/progenitor cells throughout the CNS [27, 28], whereas SRR2 functions only in the ventricular and subventricular zone of restricted telencephalic areas (Fig. 2). In the rostral telencephalon, SRR2 is active in the ventricular and subventricular zones of the ganglionic eminence and lateral cortex, but not the medial cortex. In the adult mouse brain, SRR2 activity was detected in the subventricular zone of the lateral ventricle and along the rostral migrating stream, where actively dividing cells reside. At least some of these cells display the hallmark properties of neural stem cells, as shown by an in vitro clonogenic assay. However, the molecular mechanisms governing the regional activity of SRR2 are not known at present, since Brn1, Brn2, and Sox2 are expressed in the entire CNS. A likely possibility is that the chromatin structure around SRR2 allows access to Brn-Sox2 complexes specifically in the lateral cortex and ganglionic eminence, but not in the medial telencephalon and other regions of the CNS. This hypothesis is supported by our ChIP analysis demonstrating that Sox and Brn factors interact with SRR2 specifically in the telencephalon [31]. Thus, the specific recruitment of these proteins to SRR2 in the telencephalon appears to define the spatiotemporal activity of the enhancer in the developing nervous system.
Fig. 2.
Strong regional specificity of Sox2 regulatory region 2 (SRR2) in midgestation mouse embryos. a β-galactosidase activity of SRR2-β-geo transgenic mouse embryo at 10.5 days postcoitum (dpc). b Section of β-gal-stained embryonic brain at 16.5 dpc. These data were originally published in [30]. T Telencephalon, M midbrain, S spinal cord, GE ganglionic eminence, LC lateral cortex, MC medial cortex
However, this brings up another question: how is it that Sox2 is expressed in neural stem/progenitor cells along the entire CNS, when SRR2 only functions in a restricted area of the telencephalon? The Sox2 locus contains an additional regulatory element that drives expression in the dorsal telencephalon [32, 33], in a pattern largely complementary to that of SRR2, suggesting that endogenous Sox2 expression in the telencephalon reflects the combined actions of these two regulatory elements. Moreover, regulatory regions controlling expression in other areas of the CNS, such as the midbrain and spinal cord, have been identified [34–36]. Thus, the endogenous Sox2 expression pattern results from a piecemeal compilation of regional expression domains controlled by different regulatory elements. The expression patterns of a number of other genes, including Otx2 and Pax6, are similarly patched together by numerous regulatory regions that each control expression within a specific subregion [37–41]. This is likely the best way to ensure expression within similar but molecularly distinct populations throughout the CNS.
Sox2 is a key regulator of ES cell pluripotency
At early embryonic stages, Sox2 expression is mainly restricted to cells with stem cell characteristics, such as the inner cell mass, epiblast, trophoblast stem cells, and germ cells [8, 42, 43], and is downregulated in cells with restricted developmental potential. Sox2 null embryos die immediately after implantation, and knockdown or conditional knockout of Sox2 in mouse ES cells leads to spontaneous differentiation [8, 44, 45], underscoring its importance in the maintenance of pluripotency. As described above, Sox2 forms a complex with Oct-3/4 to activate transcription of numerous genes in ES and EC cells whose regulatory regions contain Octamer factor and Sox binding sites. In addition to FGF-4 and UTF1, Fbxo15 [46], Nanog [47, 48], and Lefty1 [49] are transcriptionally regulated by the Oct-3/4–Sox2 complex. Interestingly, the Oct-3/4 and Sox2 loci themselves possess enhancers that can be activated by the Oct-3/4–Sox2 complex in a stem-cell-specific manner [29, 50, 51], creating an autoregulatory transcriptional loop that maintains pluripotency of ES cells (Fig. 3). Impairment of this regulatory circuit likely leads to ES cell differentiation; indeed, Cdx2 abrogates the circuit by forming a reciprocal inhibitory loop with Oct-3/4, thereby promoting trophectodermal differentiation [52]. Although inducible Sox2 null ES cells fail to maintain pluripotency and differentiate primarily into trophectoderm-like cells, Sox2 is unexpectedly not required for activation of most regulatory enhancers containing Octamer factor and Sox binding sites (Utf1, Fgf4, Pou5f1, Sox2, Nanog, and Lefty1) [45]. This is likely due to functional redundancy, since Sox4 [5], Sox11 [53], and Sox15 [54] are also present in ES cells. Nonetheless, DNA microarray analysis with Sox2 null ES cells revealed that expression of Nr5a2 [55], a positive regulator of Oct-3/4 expression, was downregulated. Conversely, Nr2f2 [56], a negative regulator of Oct-3/4 expression, was upregulated, suggesting that Sox2 is required for ES cell pluripotency because it maintains the requisite level of Oct-3/4 expression. Consistent with this hypothesis, forced expression of Oct-3/4 renders Sox2 expression dispensable for the maintenance of ES cell pluripotency [45]. Although Sox2 is indispensable for the maintenance of ES cell pluripotency, it has been shown that a moderate increase (twofold or less) in the level of Sox2 expression in ES cells has been shown to reduce their self-renewal activity and promote differentiation into neuroectoderm, mesoderm, and trophectoderm, but not endoderm, while a high level of Sox2 expression affects the ES cell viability [57, 58]. Thus, like Oct-3/4 [59], Sox2 also functions as a molecular rheostat to control the self-renewal and differentiation of ES cells.
Fig. 3.
Autoregulatory circuit of Oct-3/4 and Sox2 gene expression in ES cells. Expression of Oct-3/4 and Sox2 are controlled by the Oct-3/4-Sox2 complex through the DE and SRR2 enhancers, respectively. A–E represent downstream genes, including Nanog and UTF1, that are under the control of this pluripotency transcription factor complex. Oct-3/4 also functions to repress Cdx2 gene expression in which Sox2 is probably not involved. Although not depicted in the figure, Cdx2 blocks Oct-3/4 gene expression and commits differentiation to trophoblast cell lineage. Thus, reciprocal repression between Oct-3/4 and Cdx2 appears to be involved in cell fate determination between inner cell mass cells, which are the in vivo equivalent of ES cells, and trophectoderm within the blastocyst. DE Distal enhancer, PE proximal enhancer
A recent ChIP-on-Chip analysis revealed that more than half of the promoter regions occupied by Oct-3/4 are also bound by Sox2 in human ES cells [60]. Interestingly, the transcription factor Nanog, which also plays a crucial role in maintaining ES cell pluripotency, binds to most of these same promoters in ES cells, suggesting that these three transcription factors form a core transcriptional regulatory unit. Moreover, a substantial number of transcriptionally inactive genes in ES cells are also co-occupied by these pluripotency regulators. Notably, many of these encode transcription factors, such as HoxB1, Pax6, and Myf5, that regulate various developmental processes. Thus, together with Oct3/4 and Nanog, Sox2 maintains the stem cell state in ES cells by repressing expression of genes encoding key developmental regulators and by activating genes involved in self-renewal and pluripotency. It is not clear how these three pluripotency factors can activate some genes and repress others, but polycomb repressive complexes are known to bind to a substantial subset of inactive genes occupied by pluripotency regulators [61].
iPS cells
Induced pluripotent stem (iPS) cells have similar pluripotency to ES cells but can be generated from somatic cells, such as mouse embryonic fibroblast cells or human skin cells. This can be accomplished by forced expression of four transcription factors, Oct-3/4, Sox2, Klf4, and c-Myc [62, 63], though c-Myc does not appear to be essential for iPS cell generation [64]. Either Sox1 or Sox3 can substitute for Sox2 to produce iPS cells, providing further evidence of the functional equivalence among SoxB1 members. Recently, forced expression of Sox2 was shown to be dispensable for generation of iPS cells when neural stem/progenitor cells are used as the starting somatic cells [65–67]. Obviously, since neural stem/progenitor cells endogenously express Sox2, these results do not mean that Sox2 is not required for generation of iPS cells. However, iPS cells have successfully been generated from somatic cells that do not express any of the genes deemed essential for reprogramming by treating Oct-3/4-Klf4-transfected mouse embryonic fibroblast cells with BIX-01294, a G9a histone methyltransferase inhibitor, and BayK8644, an L-channel calcium agonist [68]. It is likely that BIX-01294 facilitates shifting of the epigenetic balance from a silenced to an active transcriptional state at the pluripotency genes necessary for reprogramming. However, it is unclear how and why BayK8644 impacts the reprogramming process, since activation of L-type calcium channels had never been linked to reprogramming before. Elucidating the molecular mechanisms by which these small molecules exert their function will uncover the real role of Sox2 in iPS cell production.
Role of SoxB1 members in CNS and lens development
Besides being expressed in early embryos, Sox2 is also found in the developing CNS [9, 32, 42, 69–73]. Sox2 is first uniformly expressed in the neural plate, in which most cells are multipotent. However, once the columnar epithelium of the neural plate acquires a more complex, stratified structure, expression becomes restricted to the germinal layer, where multipotent neural stem cells are enriched. Expression of other SoxB1 group members, such as Sox1 and Sox3, overlaps extensively with that of Sox2 in the CNS [9, 42, 69, 74]. Since SoxB1 factors are functionally redundant [7, 9, 75–79], loss of any individual SoxB1 gene may lead to defects in the specific regions where the mutated gene is uniquely expressed. For example, loss of Sox2 expression in mouse retina causes the complete loss of proliferative and differentiation capacity of neural retinal progenitor cells, because although other SoxB1 members are initially expressed in the anterior neural plate and invaginating optic vesicle, only Sox2 expression is maintained throughout neural retinal cell development [80]. Likewise, Sox1 mutant mice are viable and show no overt CNS abnormalities, probably due to the expression of other SoxB1 members [81, 82]. Interestingly, however, these mutant mice show defects in lens cells. This is presumably because Sox3 is never expressed in the lens, while Sox2 expression is restricted to an early stage of lens cell development. As Sox2 is abruptly downregulated in the lens around 12.5 days post coitum (dpc), Sox1 gene expression takes over and persists thereafter.
However, retinal and lens cells are rather exceptional cases, since expression of SoxB1 members overlaps in most regions of the CNS [9, 42, 69, 74]. This makes it difficult to assess the function of individual SoxB1 factors by gene-targeting analysis. Inhibiting SoxB1 function with the dominant negative form of Sox2 causes delamination of neural stem/progenitor cells from the ventricular zone and exit from the cell cycle [9]. Conversely, constitutive expression of Sox2 inhibits neuronal differentiation, resulting in the maintenance of neural stem/progenitor character and retention in the ventricular zone. Moreover, the defects caused by the dominant negative form of Sox2 can be rescued by expression of Sox1, further underscoring the functional redundancy among SoxB1 group members.
SoxB1 members play a central role in maintaining the undifferentiated state of neural stem/progenitor cells by counteracting the activity of proneural basic helix-loop-helix (bHLH) proteins, such as Ngn2 [83]. In turn, the capacity of proneural bHLH proteins to promote neuronal differentiation depends on their ability to suppress the expression of SoxB1 members. However, SoxB1 members do not directly inhibit expression of bHLH genes; instead, Notch signaling, which is also involved in maintaining neural stem/progenitor cell identity [80, 84–86], is thought to directly repress expression of bHLH genes [87]. Interestingly, forced expression of SoxB1 factors allows cells to maintain stem/progenitor character even in the absence of Notch signaling. How then do the SoxB1 proteins counteract the neurogenic activity of proneural bHLH proteins, such as Ngn2 and E47, to maintain neural stem/progenitor cell identity? One possibility is that they directly block E47/Ngn2 complexes. Alternatively, SoxB1 members may produce a molecular environment in which proneural bHLH protein complexes are unable to promote neuronal differentiation. Notably, SoxB1 members fail to inhibit transcriptional activation by the E47/Ngn2 protein complex at the E box in the NeuroD promoter. Moreover, high levels of Ngn2 and E47 do not abrogate the neural stem/progenitor cell-promoting activity of SoxB1 members. These findings support the latter model [87], but more work is necessary to elucidate the mechanism by which SoxB1 proteins function.
Interestingly, in vitro differentiation of embryonic neural stem/progenitor cells into mature neurons, but not into astroglia, is profoundly affected in Sox2 hypomorphic mutant neurospheres [88]. Neurospheres in which Sox2 was knocked down are able to differentiate into β-tubulin-positive immature neurons with abnormally poor arborization but fail to progress into more mature MAP-2-positive neurons. Moreover, overexpression of Sox2 in neural cells at an early but not late stage of differentiation can rescue the neuronal maturation defect. Some of the β-tubulin-positive immature neurons from Sox2-deficient neural stem/progenitor cells coexpress a glial marker (GFAP), suggesting that Sox2 functions to suppress GFAP gene expression in wild-type neuronal cells. Forced expression of Sox2 in astroglia also suppresses GFAP expression but does not affect astroglial morphology or expression of other glial marker genes, such as S100 and connexin-43. These data suggest that Sox2 acts as a direct transcriptional repressor of GFAP. Indeed, the genomic region upstream of the GFAP promoter contains potential Sox2 binding sites. This regulatory region has been shown to amplify the level of GFAP transcription, and its ability to activate transcription is impaired by the forced expression of Sox2. In addition, Sox2 deficiency results in a decrease in the number of GABAergic neurons in the newborn mouse cortex and adult olfactory bulb, with some showing abnormal morphology and migration. This loss of GABAergic neurons may be the primary cause of the epilepsy occasionally observed in Sox2 knockdown mice and Sox2-deficient individuals.
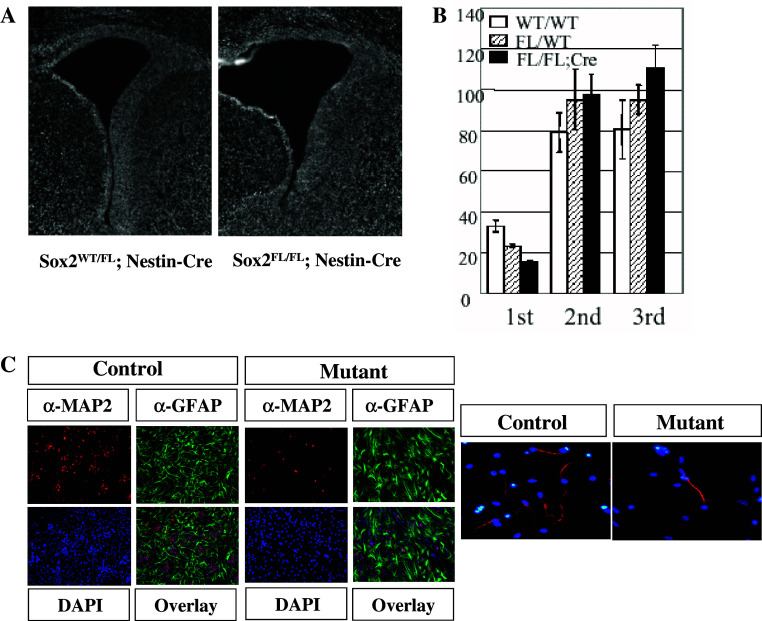
Notwithstanding the functional redundancy issues, each SoxB1 member has been knocked out and examined. As described above, Sox1 null mutant mice are viable and show no obvious CNS defects [81, 82], although they usually have spontaneous seizures between 4 and 6 weeks of age. Loss of Sox3 results in early lethality in chimeras due to a gastrulation defect. When Sox3 was deleted using a conditional knockout strategy, X-linked Sox3 null mutant males had a variable phenotype, with one-third appearing normal and most dying before weaning stage [89]. Sox3 is required in the presumptive hypothalamus and infundibulum for proper morphogenesis of Rathke’s pouch and later for hypothalamo-pituitary axis formation. Since Sox2 null mice also show early embryonic lethality [8], we have disrupted Sox2 specifically in neural stem/progenitor cells in the developing brain using the Nestin-Cre deleter strain [90]. These mice have enlarged lateral ventricles, a phenotype that was not observed in either Sox1 or Sox3 mutant embryos (Fig. 4a). Moreover, mutant mice possess significantly fewer neurosphere-forming cells in the developing mouse brain (Fig. 4b). Interestingly, Sox2-deficient primary neurospheres are able to produce secondary and tertiary neurospheres as efficiently as wild-type neurosphere cells. However, although Sox2 null neurospheres are able to differentiate into both neurons and glia, the efficiency of neuronal but not glial production is reduced (Fig. 4c, left panel). Impaired neurogenic potential of the mutant was evident also when neuronal differentiation was induced with forskolin and retinoic acid for 3 days [91] to neural stem/progenitor cells cultured as adherent cells according to Nakashima et al. [92] (Fig. 4c, right panel). These results indicate that Sox2 null neural stem cells are able to self-renew quite well, probably with the help of Sox1 and Sox3 expression, but fail to maintain normal neurogenic potential. Notably, the level of Sox3 expression was elevated in the mutant embryos, suggesting functional compensation by Sox3 upon loss of Sox2. We and others have previously demonstrated that SoxB1 autoregulatory loops direct the expression of Sox1 and Sox2 in neural stem/progenitor cells [9, 30]. However, our data suggest that expression of Sox3 is controlled at least in part by a SoxB1-independent mechanism, since Sox3 expression was upregulated in Sox2-deficient cells. Although the molecular basis of this upregulation is not known at present, one intriguing possibility is that Sox3 is regulated differently from other SoxB1 members, so that its expression is assured even when SoxB1 factor-dependent Sox expression is impaired. Though disruption of different SoxB1 members has variable effects on CNS development, these differences likely do not reflect functional differences among SoxB1 members, but rather differences in expression level, since many lines of evidence indicate that SoxB1 members exert equivalent biological functions in neural stem/progenitor cells [9, 80, 83].
Fig. 4.
Morphological and functional analyses of mouse embryonic brains in which Sox2 has been conditionally knocked out. a Coronal sections of 19.5 dpc mutant (Sox2Fl/FL; Nestin-Cre) and littermate control (Sox2WT/FL; Nestin-Cre) embryos. b Primary, secondary, and tertiary neurosphere assay of 14.5 dpc mutant (Sox2FL/FL; Nestin-Cre) and littermate control (Sox2WT/FL) brains. c Attenuated neurogenic potential of Sox2-deficient neurosphere cells. Neurospheres are induced to differentiate using fetal bovine serum and retinoic acid (left panel). Impaired neurogenic potential is also evident in neural stem/progenitor cells cultured as adherent cells. Two panels in a and left panel in c were originally published in [90]. Right panel in c represents original data of S.M. and A.O.
Human eye disorders with Sox2 mutations
Bilateral anaphthalmia/microphthalmia is the most severe type of eye malformation. It is known that in humans, 3% of unilateral and ~10% of bilateral eye disorders are linked to haploid insufficiency of Sox2 [93, 94]. Most mutations in the Sox2 gene identified to date are nonsense mutations leading to truncations of the Sox2 protein. No individuals diagnosed with a Sox2-related eye disorder had a parent with the same mutation in the Sox2 gene, suggesting that in most cases, this genetic disorder occurs as the result of a fully penetrant de novo mutation. Therefore, the risk of this disease to siblings is usually very small. However, in one case, a phenotypically normal mother was found to have germline mosaicism of a loss-of-function mutation. In such cases, the risk of Sox2-related eye disorders to siblings may be up to 50% [95]. Sox2-related eye disorders are often associated with other abnormalities, such as myopathy, esophageal atresia, learning disabilities, and defects in male genital organs, although it is not clear how these phenotypes are associated with Sox2 function. It is also of note that heterozygous Sox2 mice do not usually exhibit obvious phenotypes, apart from male fertility defects. However, such differences in haploinsufficiency between mice and humans are not rare.
Concluding remarks
Like other Sox factors, SoxB1 proteins function mainly by regulating transcription. Their functions are context-dependent, as they are modified by the action of partner proteins that interact with SoxB1 members. For example, Oct-3/4 works together with Sox2 to support ES and EC cell-specific gene expression. Numerous studies have demonstrated the importance of SoxB1 members in initial specification and maintenance of stem cell state in neural stem/progenitor cells and in the maintenance of ES cell pluripotency. It was undoubtedly a bit of a surprise for researchers in the field of neural development, especially those studying Xenopus, to find that one of their favorite molecules, Sox2, was important not only for neural stem cells but also for embryonic stem cells in mice. It is still somewhat of an enigma that among all the SoxB1 family members, only Sox2 was assigned this task. Understanding the molecular mechanisms governing the partner switch from Oct3/4 in ES cells to other partners in neural stem/progenitor cells remains a compelling challenge. Interestingly, from a molecular phylogenetic point of view, the Sox2 proteins of egg-laying animals (e.g., fish and birds, in which Sox2 is not expressed in early embryonic cells) segregate sharply from the orthologs of placental mammals, in which Sox2 is expressed at early stages. Perhaps as a consequence of this difference, Sox2 expression in the mouse and probably in other mammals marks the neural stem/progenitor lineage throughout its entire developmental time. As mentioned earlier, this may facilitate the reprogramming of neural stem/progenitor cells into pluripotent stem cells. It is tempting to speculate that the developmental default pathway of ES cells follows the neural differentiation program under the direction of Sox2. This idea could be tested by Sox2 ChIP-on-Chip experiments with ES cells undergoing neural differentiation.
Acknowledgements
The authors are indebted to the members of Okuda’s laboratory for many stimulating discussions. We gratefully acknowledge financial support from the Ministry of Education, Science, Sports, Science and Technology for our previous work on the expression and function of Sox2 in mouse ES cells and neural stem/progenitor cells.
References
- 1.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 2.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 3.Wright EM, Snopek B, Koopman P. Seven new members of the Sox gene family expressed during mouse development. Nucleic Acids Res. 1993;21:744. doi: 10.1093/nar/21.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 5.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 6.Pevny LH, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/S0959-437X(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 7.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multiple cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham V, Khudyakou J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/S0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 10.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/S0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 11.Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992;11:3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 14.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specifc spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in early embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- 17.Rosner MB, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PWJ, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- 18.Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 19.Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Nabeshima Y, Boon K, Keaveney M, Stunnenberg HG, Muramatsu M. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukushima A, Okuda A, Nishimoto M, Seki N, Hori T, Muramatsu M. Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. J Biol Chem. 1998;273:25840–25849. doi: 10.1074/jbc.273.40.25840. [DOI] [PubMed] [Google Scholar]
- 21.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowling TK, Johnson LR, Wiebe MS, Rizzino A. Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J Biol Chem. 2000;275:3810–3818. doi: 10.1074/jbc.275.6.3810. [DOI] [PubMed] [Google Scholar]
- 23.Nowling T, Bernadt C, Johnson L, Desler M, Rizzino A. The co-activator p300 associates physically with and can mediate the action of the distal enhancer of the FGF-4 gene. J Biol Chem. 2003;278:13696–13705. doi: 10.1074/jbc.M207567200. [DOI] [PubMed] [Google Scholar]
- 24.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondoh H, Uchikawa M, Kamachi Y. Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int J Dev Biol. 2004;48:819–827. doi: 10.1387/ijdb.041868hk. [DOI] [PubMed] [Google Scholar]
- 26.Inoue M, Kamachi Y, Matsunami H, Imada K, Uchikawa M, Kondoh H. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells. 2007;12:1049–1061. doi: 10.1111/j.1365-2443.2007.01114.x. [DOI] [PubMed] [Google Scholar]
- 27.Josephson R, Muller T, Picket J, Okabe S, Reynolds K, Turner PA, Zimmer A, McKay RD. POU transcription factors control expression of CNS stem cell-specific gene. Development. 1998;125:3087–3100. doi: 10.1242/dev.125.16.3087. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka S, Kamachi Y, Tanouchi A, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyagi S, Saito T, Mizutani K, Masuyama N, Gotoh Y, Iwama A, Nakauchi H, Masui S, Niwa H, Nishimoto M, Muramatsu M, Okuda A. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol Cell Biol. 2004;24:4207–4220. doi: 10.1128/MCB.24.10.4207-4220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyagi S, Nishimoto M, Saito T, Ninomiya M, Sawamoto K, Okano H, Muramatsu M, Oguro H, Iwama A, Okuda A. The Sox2 regulatory region 2 functions as neural stem cell-specific enhancer in the telencephalon. J Biol Chem. 2006;281:12281–13374. doi: 10.1074/jbc.M512669200. [DOI] [PubMed] [Google Scholar]
- 32.Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a β-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- 33.Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R, Scholer H, Nicolis SK. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- 34.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;5:509–519. doi: 10.1016/S1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 35.Kondoh H, Uchikawa M. Dissection of chick genomic regulatory regions. Methods Cell Biol. 2008;87:313–316. doi: 10.1016/S0091-679X(08)00217-3. [DOI] [PubMed] [Google Scholar]
- 36.Kamachi Y, Iwafuchi M, Okuda Y, Takemoto T, Uchikawa M, Kondoh H. Evolution of non-coding regulatory sequences involved in the developmental process: reflection of differential employment of paralogous genes as highlighted by Sox2 and group B1 Sox genes. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:55–68. doi: 10.2183/pjab.85.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin C, Kleinjan DA, Doe B, van Heyningen V. New 3’ elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech Dev. 2002;112:89–100. doi: 10.1016/S0925-4773(01)00646-3. [DOI] [PubMed] [Google Scholar]
- 38.Kleinjan DA, Seawright A, Childs AJ, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev Biol. 2004;265:462–477. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 40.Kurokawa D, Takasaki N, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development. 2004;131:3307–3317. doi: 10.1242/dev.01219. [DOI] [PubMed] [Google Scholar]
- 41.Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- 42.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/S0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- 43.D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci USA. 2003;100:11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanova N, Dobrin R, Lu R, Kotenko L, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 45.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MSH, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 46.Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol. 2003;23:2699–2708. doi: 10.1128/MCB.23.8.2699-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of Nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 49.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Miyazaki J, Matoba R, Ko MS, Niwa H. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chew J-L, Loh Y-H, Zhang W, Chen X, Tam W-L, Yeap L-S, Li P, Ang Y-S, Lim B, Robson P, Ng H-H. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 53.Wiebe MS, Nowling TK, Rizzino A. Identification of novel domains within Sox-2 and Sox-11 invovled in autoinhibition of DNA binding and partnership specificity. J Biol Chem. 2003;278:17901–17911. doi: 10.1074/jbc.M212211200. [DOI] [PubMed] [Google Scholar]
- 54.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential roles of Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem. 2005;280:24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- 55.Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Palas C, Kruijer W. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol Cell Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boer B, Kopp J, Mallanna S, Desler M, Chakravarthy H, Wilder PJ, Bernadt C, Rizzino A. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 2007;35:1773–1786. doi: 10.1093/nar/gkm059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kopp J, Oremsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 59.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 60.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch DK, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee TI, Jenner R, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:1–12. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 64.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 65.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 66.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 67.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Ortmeier CO, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 68.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Uwanogho DA, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- 70.Collignon J, Sockanathan S, Hacker A, Cohentannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow P, Lovell-Badge R. A comparison of the properties of Sox3 with Sry and two related genes Sox1 and Sox2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 71.Rex M, Orme A, Uwanogho D, Tointon K, Wigmore PM, Sharpe PT, Scotting PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997;209:323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 72.Sasai Y. Roles of Sox factors in neural determination: conserved signaling in evolution? Int J Dev Biol. 2001;45:321–326. [PubMed] [Google Scholar]
- 73.Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 75.Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- 76.Sanchez-Soriano N, Russell S. The Drosophila Sox-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- 77.Buescher M, Hing FS, Chia W. Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- 78.Overto PM, Meadows LA, Urban J, Russell S. Evidence for differential and redundant function of Sox genes Dichaete and SoxN during CNS development in Drosophila. Development. 2002;129:4219–4228. doi: 10.1242/dev.129.18.4219. [DOI] [PubMed] [Google Scholar]
- 79.Ferri ALM, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 80.Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a does-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the γ-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malas S, Postlethwate M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, Meldrum B, Constanti A, Episkopou V. SOX1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexitability. Neuroscience. 2003;119:421–432. doi: 10.1016/S0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- 83.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activty. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 84.Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 85.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 86.Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 87.Holmberg J, Hansson E, Malewicz M, Sanberg M, Perlmann T, Lendahl U, Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135:1843–1851. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- 88.Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, Wanke E, Brunelli S, Favaro R, Ottolenghi S, Nicolis S. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135:541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- 89.Rissoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- 90.Miyagi S, Masui S, Niwa H, Saito T, Shimazaki T, Okano H, Nishimoto M, Muramatsu M, Iwama A, Okuda A. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 91.Nakashima K, Takizawa T, Ochiai W, Yanagisawa M, Hisatsune M, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci USA. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/S0092-8674(04)00248-X. [DOI] [PubMed] [Google Scholar]
- 93.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 94.Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, Salt A, Collin JR, Vivian AJ, Free SL, Thompson P, Williamson KA, Sisodiya SM, van Heyningen V, Fitzpatrick DR. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. [DOI] [PubMed] [Google Scholar]
- 95.Faivre L, Williamson KA, Faber V, Laurent N, Grimaldi M, Thauvin-Robinet C, Durand C, Mugneret F, Gouyon JB, Bron A, Huet F, Hayward C, Heyningen VV, Fitzpatrick DR. Recurrence of SOX2 anophthalmia syndrome with gonosomal mosaicism in a phenotypically normal mother. Am J Med Genet A. 2006;140A:636–639. doi: 10.1002/ajmg.a.31114. [DOI] [PubMed] [Google Scholar]
- 96.Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–8091. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Laumonnier F, Ronce N, Hamel BC, Thomas P, Lespinasse J, Raynaud M, Paringaux C, Van Bokhoven H, Kalscheuer V, Fryns JP, Chelly J, Moraine C, Briault S. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet. 2002;71:1450–1455. doi: 10.1086/344661. [DOI] [PMC free article] [PubMed] [Google Scholar]