Abstract
Curcumin, a natural polyphenol, has been described to exhibit effects on signaling pathways, leading to induction of apoptosis. In this study, we observed that curcumin inhibited Hsp90 activity causing depletion of client proteins implicated in survival pathways. Based on this observation, this study was designed to investigate the cellular effects of curcumin combination with the pan-HDAC inhibitors, vorinostat and panobinostat, which induce hyperacetylation of Hsp90, resulting in inhibition of its chaperone function. The results showed that, at subtoxic concentrations, curcumin markedly sensitized tumor cells to vorinostat- and panobinostat-induced growth inhibition and apoptosis. The sensitization was associated with persistent depletion of Hsp90 client proteins (EGFR, Raf-1, Akt, and survivin). In conclusion, our findings document a novel mechanism of action of curcumin and support the therapeutic potential of curcumin/HDAC inhibitors combination, because the synergistic interaction was observed at pharmacologically achievable concentrations, which were ineffective when each drug was used alone.
Keywords: Curcumin, Hsp90, Istone deacetylase inhibitors, Apoptosis, HDAC6
Introduction
Curcumin, a yellow pigment derived from the plant turmeric, Curcuma longa, is known to have anti-inflammatory and anti-oxidant properties [1, 2]. This natural compound has been extensively studied as a chemopreventive agent, in particular for colon carcinoma [3, 4]. The chemopreventive efficacy of curcumin has been ascribed to the modulation of signal-transduction pathways associated with malignant transformation and tumor promotion, through interaction with a variety of proteins implicated in cell proliferation, invasion, and angiogenesis [5]. Indeed, curcumin induces apoptosis in tumor cells, and apoptosis is emerging as a major mechanism by which chemopreventive agents eliminate preneoplastic cells [6]. In spite of the pleiotropic effects reported, the mechanism of action of curcumin remains poorly understood.
Recently, curcumin has been reported to be able to down-regulate some proteins, which are known to be clients of Hsp90, thus suggesting a modulation of Hsp90 function [7, 8]. Hsp90 is an exploitable therapeutic target, because it plays a central role in correct folding and stabilization of a variety of proteins involved in malignant behavior [9–12]. Indeed, heat shock proteins are often over-expressed in tumor cells, thus supporting their ability to survive under stressed conditions [9, 13]. The chaperone function of Hsp90 is regulated by a number of post-translational modifications, including acetylation [14]. The acetylation status of Hsp90 appears to be controlled by the deacetylase HDAC6 [15–17]. Among HDAC enzymes, HDAC6 is unique because of its substrate preferences and cytoplasmic localization [18]. Depletion of HDAC6 or inhibition of its deacetylase activity have been reported to result in reversible Hsp90 hyperacetylation and inhibition of its function [19–21].
Based on these observations, the present study was undertaken to investigate the Hsp90-mediated effects of curcumin and to explore the cellular effects of the combination of curcumin with known HDAC inhibitors, vorinostat, and panobinostat, which are known to inhibit HDAC6, besides other HDAC enzymes [21, 22]. We have found that anti-proliferative concentration of curcumin resulted in down-regulation of a number of Hsp90 client proteins in squamous cell carcinoma A431 and mesothelioma STO cells. The combination of curcumin and HDAC inhibitors at subtoxic concentrations resulted in a marked growth inhibition, enhanced depletion of Hsp90 client proteins, and sensitization to apoptosis.
Materials and methods
Cell lines and culture conditions
The human epidermoid carcinoma A431 and the peritoneal mesothelioma STO cell lines (provided by Dr. N. Zaffaroni, IRCCS Istituto Nazionale Tumori, Milan) were used in this study. A431 and STO cells were routinely grown in RPMI 1640 or in 50/50 DMEM/Ham’s F12 medium (Lonza, Switzerland), respectively, supplemented with 10% (v/v) heat-inactivated foetal bovine serum (Gibco®, Invitrogen, Segrate, Italy) at 37°C in a 5%/95% CO2/air atmosphere.
Drugs and treatment conditions
Panobinostat was synthesized as described in the recent US patent application (6,552,065). Vorinostat was provided by BIOMOL International LP (Plymouth Meeting, PA, USA). Curcumin was provided by Sigma Chemical (St. Louis, MO, USA). Stock solutions of all drugs (10 mg/ml) were prepared in dimethyl sulfoxide (DMSO) (BDH Prolabo, Milan, Italy) and maintained at −20°C. Before each experiment, drugs were diluted in DMSO and used at the concentrations reported in the legends to individual figures. 17-AAG was obtained from InvivoGen (San Diego, USA) and it was provided as a stock solution (5 mM) that we freshly diluted in culture medium before each experiment. Bortezomib was also stocked in 10 mg/ml DMSO solutions, but dilutions were freshly prepared in sterile water. Experiments were performed by incubating cells with drugs for 4, 24, or 72 h at 37°C in culture medium supplemented with 10% (v/v) heat-inactivated foetal bovine serum.
Anti-proliferative assay
Cell sensitivity to drug treatment was determined by growth inhibition assay. Briefly, cells were seeded in 12-well plates (40,000 cells/well), 24 h before experiments. Cells were exposed to the drugs for 72 h and then adherent cells were trypsinized and counted by a cell counter (Beckam Coulter, Fullerton, CA, USA). After 24 h of treatment, cells were incubated in drug-free medium for 72 h and counted by using cell counter. IC50 values, derived from dose–response curves, were defined as drug concentrations required for 50% inhibition of cell growth.
Protein expression analysis
Total cell lysates were prepared rinsing cells twice with ice-cold PBS supplemented with 0.1 mM sodium orthovanadate, then lysing them in hot sample buffer as previously described [23]. After determination of protein concentration (BCA Protein Assay Reagent, Pierce, Thermo Fisher Scientific, Rockford, IL, USA), whole-cell extracts were separated by SDS polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose membranes. Detection of proteins was accomplished using horseradish peroxidase-conjugated secondary antibodies and a chemiluminescence reagent purchased through Amersham Biosciences (Rockford, IL). Primary antibodies used in this study are: anti-Raf-1 (sc-133) and anti-Cdk4 (sc-601) (Santa Cruz Biotechnology, CA, USA), anti-EGFR (Upstate Biotechnology, Millipore, Billerica, MA, USA), anti-Survivin and anti-p23 and anti-Hsc70 (Abcam, Cambridge, UK), anti-Akt (Transduction Laboratories, Lexington, USA), anti-PARP-1 (Calbiochem, Merck Chemicals, Nottingham, UK) and anti-Actin (Sigma).
Quantification of western blot analyses was obtained by Image Quant 5.2 program.
Co-immunoprecipitation and immunoblot analysis
Following the designated treatments, cells were lysed in NET buffer [50 mmol/l Tris (pH 7.4), 150 mmol/l sodium chloride, 0.1% NP40, 1 mmol/l phenylmethylsulfonyl fluoride, 1 mmol/l EDTA, 2.5 μg/ml leupeptin, 5 μg/ml aprotinin] first for 30 min on ice and then for 30 min at 4°C in rotation on a wheel. Nuclear and cellular debris were cleared by centrifugation (10,000g, 10 min, 4°C). Total cellular proteins were then quantified using the BCA protein assay. Cell lysates (500 μg) were incubated with the Raf-1 or Hsp90 polyclonal-specific monoclonal antibodies for 2 h at 4°C. Protein A-Sepharose (Sigma) (90 μl), previously prepared in TNT solution (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton X-100), was added to the lysates and incubated overnight at 4°C. The immunoprecipitates (separated by centrifugation, 15,000g, 2 min, 4°C) were washed twice in NET buffer and twice in PBS with 1% aprotinin and 1 mmol/l PMSF. Proteins were eluted with the SDS sample loading buffer before the immunoblot analyses with specific antibodies against Raf-1, Hsp90, or Hsp70 (Santa Cruz Biotechnology); p23 or Hsc70 (Abcam). Quantification of western blot analyses was obtained by Image Quant 5.2 program.
Determination of apoptosis
Apoptosis was determined by TUNEL assay (Roche) after 72 h exposure or after 24 h of treatment followed by 72 h of drug-free incubation. Treated cells were fixed in 4% paraformaldehyde (RT, 45 min), washed and resuspended in ice-cold PBS. The in situ cell death detection kit (Roche, Germany) was used according to the manufacturer’s instructions, and samples were analyzed by flow cytometry (FACScan; Becton-Dickinson, Franklin Lakes, NJ, USA).
Fluorescence polarization assay
Binding of curcumin to human full length recombinant Hsp90α was determined by a competitive binding fluorescence polarization assay [24], using a fluorescent geldanamycin probe. Recombinant human Hsp90, provided by Assay Designs (Ann Arbor, MI) was cloned from a HeLa cDNA library and expressed in E.coli and purified by multi-step chromatography.
Surface plasmon resonance analyses
SPR analyses were performed using a Biacore 3000 optical biosensor equipped with research-grade CM5 sensor chips (Biacore AB). Using this platform, two separate recombinant Hsp90 surfaces, a BSA surface and an unmodified reference surface, were prepared for simultaneous analyses. Proteins (100 μg/ml in 10 mM sodium acetate, pH 5.0) were immobilized on individual sensor chip surfaces at a flow rate of 5 μl/min using standard amine-coupling protocols [25] to obtain densities of 8–12 kRU. Compound 2 and radicicol were dissolved in 100% DMSO to obtain 4 mM solutions, and diluted 1:200 (v/v) in PBS (10 mM NaH2PO4, 150 mM NaCl, pH 7.4) to a final DMSO concentration of 0.5%. Compounds concentration series were prepared as twofold dilutions into running buffer: for each sample, the complete binding study was performed using a six-point concentration series, typically spanning 0.05–10 μM, and triplicate aliquots of each compound concentration were dispensed into single-use vials. Included in each analysis were multiple blank samples of running buffer alone [26]. Binding experiments were performed at 25°C, using a flow rate of 50 μL/min, with 60 s monitoring of association and 200 s monitoring of dissociation. Simple interactions were adequately fit to a single-site bimolecular interaction model (A + B = AB), yielding a single K D. Sensorgram elaborations were performed using the Biaevaluation software provided by Biacore AB [27].
Limited proteolysis
Limited proteolysis experiments were performed at 37°C, PBS 0.1% DMSO, at a 3-μM recombinant human Hsp90α concentration using trypsin, chymotrypsin and endoprotease V8 as proteolytic agents; 30 μL of solution were used for each experiment. Binary complexes Hsp90/inhibitor were formed by incubating the protein with a 10:1 M excess of the individual inhibitor at 37°C for 15 min prior to proteolytic enzyme addition. Each complex was digested using a 1:100 (w/w) enzyme to substrate ratio. The extent of the reactions was monitored on a time-course basis by sampling the incubation mixture after 5, 15, and 30 min of digestion. Samples were desalted by ziptip C4 (Millipore) and the proteolytic fragments were analyzed by MALDI-TOF/MS using a MALDI-MX micro (Waters). In order to optimize sensitivity and accuracy of the mass measurements, three different m/z ranges were explored for each sample: a first range, from m/z 500 to 3,500 was analyzed in reflector mode; the other two ranges, from m/z 3,500 to 20,000 and from m/z 20,000 to 95,000, were analyzed in linear mode. Each m/z range was calibrated using a suitable peptide or protein mixture. Mass data were elaborated using the Masslynx software (Waters).
Preferential hydrolysis sites on Hsp90 under different conditions were identified on the basis of the fragments released during the enzymatic digestions. When comparative experiments were carried out on Hsp90 in the presence or in the absence of inhibitors, differences in the susceptibility of specific cleavage sites were detected, from which protein regions involved in the conformational changes could be inferred [28].
Statistical analysis
All the reported values represent the mean ± standard deviation (SD) of at least three independent experiments performed in duplicate. Where necessary, data were statistically compared by t test.
Results
Down-regulation of Hsp90 client proteins by curcumin
The analysis of protein expression was performed in two cell lines, the squamous cell carcinoma A431 and mesothelioma STO cells after 24 h-exposure to IC80 of curcumin (5 μM). The two cell lines exhibited a comparable sensitivity to curcumin in the anti-proliferative assay (Fig. 1). As shown in Fig. 2a, EGFR, Raf-1, Survivin, and Cdk4 protein expression were strongly downregulated by curcumin treatment, whereas the effect was less marked for Akt. This effect was consistent with previous observations indicating a delayed depletion of Akt produced by other Hsp90 inhibitors [29]. However, in contrast to the geldanamycin derivative, 17-AAG, curcumin did not cause increased expression of Hsp70 (Fig. 2b).
Fig. 1.
Antiproliferative effect of curcumin on A431 and STO cells. Cells were treated for 72 h with curcumin and cell growth was evaluated 72 h after the treatment by cell counting. Inverted filled triangle A431 cells (IC50, 2.2 μM); open circle STO cells (IC50, 2.1 μM). Data are the mean of three independent experiments
Fig. 2.
a Effect of curcumin on Hsp90 client protein levels in A431 and STO cells. Total cellular extracts were obtained 24 h after treatment with curcumin (5 μM, IC80). Actin is shown as a control for protein loading. C Control, CUR curcumin. b Comparison of the effects of curcumin and 17-AAG on protein levels of Hsp70. Cells were treated as described in (a), with curcumin (5 μM) or 17-AAG (0.1 μM). c Coimmunoprecipitation of Raf-1/Hsp90 in A431 cells treated with 17-AAG or curcumin for 1 h. Cells were treated with equitoxic concentrations of 17-AAG (0.1 μM, IC80) or curcumin (5 μM). Cell lysates were than harvested and immunoprecipitated with anti-Raf-1 rabbit polyclonal antibody. Immunoprecipitates were immunoblotted for Hsp90. Blots were stripped and probed for Raf-1. C Control, AAG 17-AAG, CUR curcumin. d Coimmunoprecipitation of Hsp90/hsc70 and Hsp90/p23 in A431 cells treated with curcumin. Cells were treated with two concentrations of curcumin (2 and 5 μM; i.e., IC50 and IC80) as described above. Immunoprecipitated obtained by anti-Hsp90 antibody were immunoblotted for hsc70 or p23. C Control, CUR2 curcumin 2 μM, CUR5 curcumin 5 μM. e Effect of bortezomib (BOR) on the depletion of Hsp90 client proteins induced by curcumin (CUR) in A431 cells. Total cellular extracts were obtained 24 h after treatment (curcumin 5 μM, bortezomib 0.001 μM). Actin is shown as a control for protein loading. C Control
To investigate the mechanism of down-regulation of Hsp90 clients, co-immunoprecipitation of Raf-1/Hsp90 was performed in A431 cells. After 4 h exposure to curcumin (5 μM) or equitoxic concentration of 17-AAG (0.1 μM), a lower amount of Hsp90 was found in complex with Raf-1 (Fig. 2c). Thus, in spite of a different potency, the effects of the two agents were comparable at equitoxic concentrations. An additional characterization of the effect of curcumin on the association between Hsp90 and co-chaperones was performed in an immunoprecipitation assay with anti-Hsp90 antibody (Fig. 2d). The results showed that anti-proliferative concentrations of curcumin reduced the interaction of Hsc70 (but not of p23) with Hsp90. The inhibition of Hsp90–client interaction is expected to result in client protein degradation mediated by the ubiquitin–proteasome system [12, 30]. The combination of bortezomib, a well-known proteasome inhibitor, with curcumin prevented protein depletion induced by curcumin (Fig. 2e), thus supporting that the observed curcumin-induced protein down-regulation occurred via the ubiquitin–proteasome pathway.
Curcumin-Hsp90 interaction
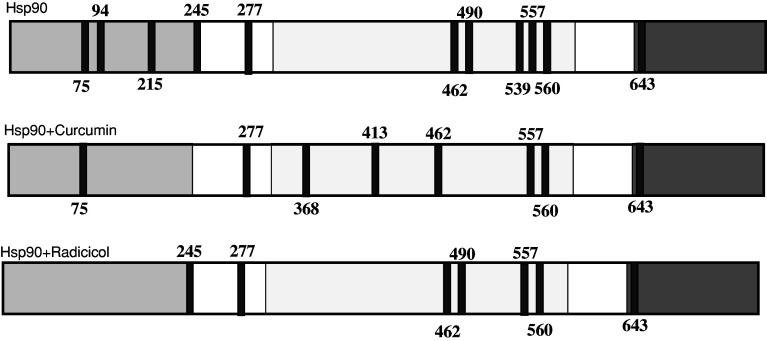
The competitive binding fluorescence polarization assay gave an IC50 of 6.2 ± 0.12 μM for curcumin, 1.09 ± 0.05 μM for 17-AAG, and 0.058 ± 0.001 for radicicol. Thus, the different anti-proliferative potencies of curcumin and 17-AAG (IC50 values, 2.4 and 0.06 μM, respectively) were consistent with their binding affinities to Hsp90. We used the limited proteolysis–mass spectrometry technique for the structural analysis of drug–Hsp90 interaction [28]. This approach is based on the evidence that exposed, weakly structured, and flexible regions of a protein can be recognized by a proteolytic enzyme. The differences in the proteolytic patterns occurring on the protein when it underwent digestion in the presence or in the absence of putative ligands allow identification of the protein regions involved in the molecular interactions [31]. Results achieved in the experiment performed on Hsp90, on the Hsp90/curcumin complex, and on the Hsp90/radicicol complex are summarized in Fig. 3 and in Table 1. A comparison between the proteolysis patterns observed on free Hsp90 and Hsp90/curcumin complex demonstrated that the sites 94, 215, and 245, cleaved in the experiments performed on the isolated proteins, were protected following the complex formation, thus suggesting an interaction involving the Hsp90 N-terminal domain. However, binding of the protein with curcumin also produced an higher susceptibility to enzymatic hydrolysis of same sites located into the middle domain, suggesting a possible long-range effect of the curcumin on the protein tertiary structure. The experiments performed on the Hsp90/radicicol complex indicated a significant protection of the N-terminal domain from enzymatic hydrolysis. This result is in agreement with the radicicol/Hsp90 interaction involving the ATPase portion located into the N-terminal domain of Hsp90 [32].
Fig. 3.
Schematic representation of the results obtained from limited proteolysis experiments on recombinant Hsp90. The preferential cleavage sites detected performing enzymatic digestions on recombinant Hsp90, on the Hsp90/curcumin complex, and on the Hsp90/radicicol complex are in dark grey. The Hsp90 N-terminal domain is highlighted in light grey, the middle domain is boxed and the C-terminal domain is highlighted in grey
Table 1.
Curcumin and radicicol interaction with Hsp90
| Compound | KD (nM)a | Kd (s−1)a | Protected sitesb | Exposed sitesb | Putative interaction region |
|---|---|---|---|---|---|
| Radicicol | 3.04 ± 0.85 | 0.0113 ± 0.0005 | 75, 94, 215 | – | N-terminal domain |
| Curcumin | 6.68 ± 3.28 | 0.0090 ± 0.0008 | 94, 215, 245 | 368, 413 | N-terminal domain |
aDissociation constants obtained by SPR analysis
bProteolytic pattern obtained by limited proteolysis and mass spectrometry analysis
We used surface plasmon resonance to measure kinetic and thermodynamic parameters of ligand–protein complex formation [33]. An SPR-based binding assay as implemented with Biacore technology was, therefore, used to investigate the drug–protein interactions. The values of the constants (Table 1) indicated that curcumin exhibited an high affinity for Hsp90, even if it was lower than that measured for radicicol. However, the difference in the thermodynamic dissociation constants (K D) observed for these two compounds mainly depend on the stability of the respective protein complexes, as inferred by the lower kinetic constant (K d) determined for curcumin in respect of that of radicicol.
Cellular effect of the combination of curcumin with HDAC inhibitors
In combination studies, we used vorinostat and panobinostat, two hydroxamic acid-based compounds, which are well-known pan-histone deacetylase inhibitors including HDAC6. The cell growth inhibition studies were performed using subtoxic concentrations of curcumin (IC10–20) (Fig. 4). Under these conditions, the combined treatment resulted in a marked enhancement of the anti-proliferative activity of either vorinostat or panobinostat as shown by the dose response curves. Based on these observations, further experiments of drug interaction were performed with subtoxic concentrations of all agents (IC20), i.e. curcumin (1.5 μM), panobinostat (11.5 nM), and vorinostat (15 nM). Apoptosis response was examined after 72 h exposure by TUNEL analysis (Fig. 5a). When tested at these concentrations, single drug treatment induced only a marginal extent of apoptosis, whereas the combination treatment markedly enhanced apoptotic response to around 50%. A similar enhancement was observed following a shorter exposure (24 h) and 72 h incubation in drug-free medium. On the contrary, when apoptosis was assessed after 72 h exposure to cytotoxic concentrations of single drug (IC80), a consistent number of apoptotic cells was found; but the shorter exposure followed by drug removal, indicated the reversibility of the effect of these agents, as no significant apoptosis was found (Fig. 5b). These results confirm that, differently from single treatment, the sensitization resulting from the drug interaction did not require prolonged exposure, suggesting an irreversible effect specific of the combined treatment.
Fig. 4.
Antiproliferative effects of HDAC inhibitors, vorinostat (a) or panobinostat (b), alone or in combination with curcumin. Cells were treated for 72 h with panobinostat or vorinostat combined with two subtoxic concentrations of curcumin (1.5 and 0.5 μM); cell growth, after the treatment, was determined by cell counting. Data are the mean of three independent experiments. Filled square single agent (vorinostat or panobinostat), filled circle curcumin 1.5 μM + vorinostat or panobinostat, filled triangle curcumin 0.5 μM + vorinostat or panobinostat. The effect of the subtoxic concentrations of curcumin alone at the tested concentrations is also shown in each cell line. Open triangle curcumin 0.5 μM, open circle curcumin 1.5 μM. Data from three independent assays were analyzed by t test (vorinostat or panobinostat vs combined treatment): *P < 0.05, ** P < 0.005, ***P < 0.0005
Fig. 5.
a Effect of curcumin on apoptosis induced by HDAC inhibitors in A431 cells. Cells were exposed for 72 h to subtoxic concentrations of panobinostat (11.5 nM) or vorinostat (15 nM), alone or in combination with curcumin (1.5 μM). b Apoptotic cell death caused by cytotoxic concentrations of single agent treatment. Cells were exposed for 72 h to cytotoxic concentrations of panobinostat (100 nM), vorinostat (1 μM) and curcumin (5 μM). At the end of the treatment, apoptosis was detected by TUNEL assay and determined by FACS analysis. In a parallel experiment, cells were treated for 24 h and apoptosis was detected after 72 h following drug removal. Data from three independent assays were analyzed by t test (control vs treatment): *P < 0.05, **P < 0.005
Effect of the combination of curcumin and HDAC inibitors on levels of Hsp90 client proteins
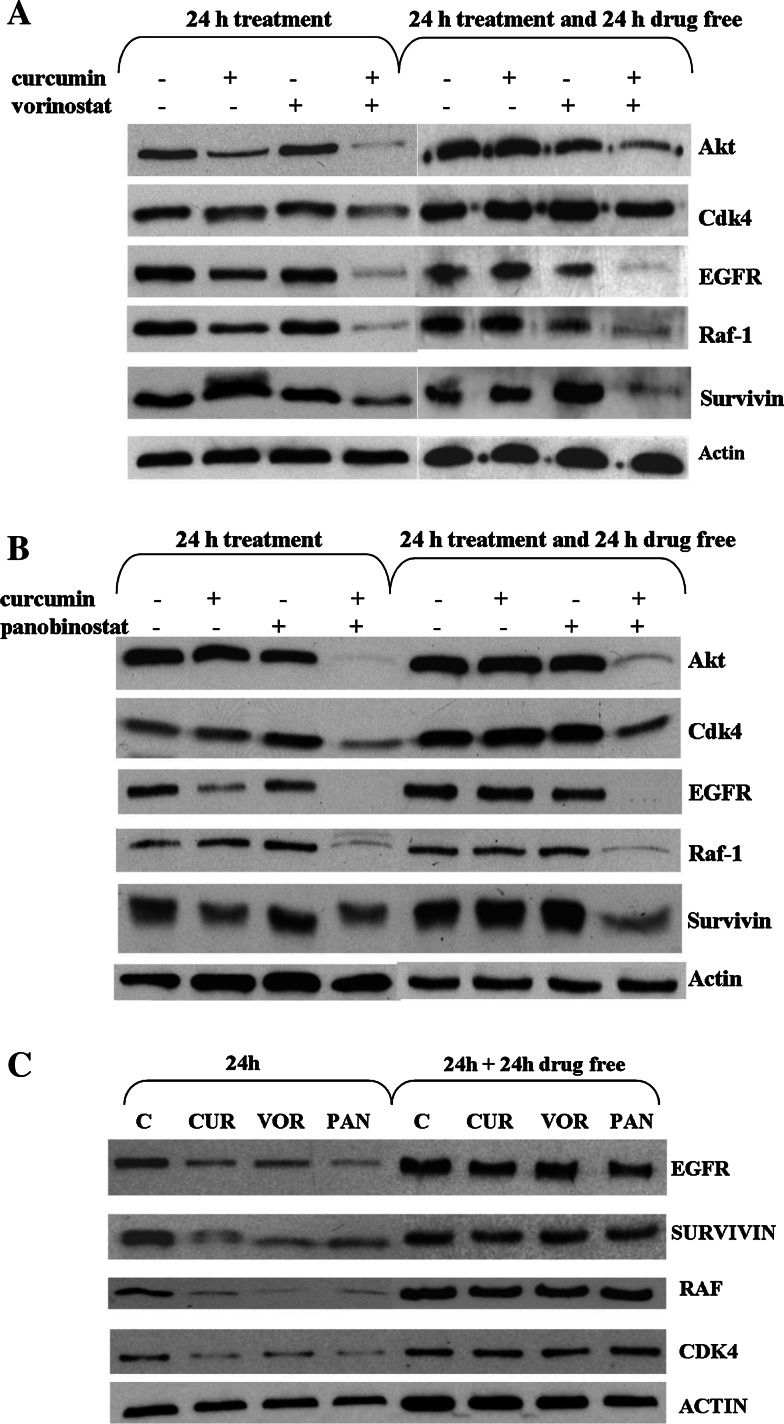
In order to investigate the molecular basis of the synergistic interaction between curcumin and HDAC inhibitors, the effects of single agents and their combination on Hsp90 client proteins were examined by western blot analysis using the same (subtoxic) concentrations of previous experiments. Figure 6 shows that, under these conditions, single agents did not produce appreciable changes of protein expression. In contrast, the combinations of curcumin with either HDAC inhibitors caused depletion of the well-established client proteins, in particular Akt, EGFR, and Raf-1. The down-regulation of Cdk4 and survivin was less marked and may reflect a different kinetics of depletion as suggested by the effect of the combination on survivin 24 h after treatment. The reduction of the Hsp90 client protein levels was persistent even 24 h after drug removal, with the exception of Cdk4. The persistence of the client down-regulation was typical of the combined treatments; indeed, when cells were treated with single agents (IC80) and incubated in drug-free medium for 24 h, the biochemical effects were lost (Fig. 6c). To better document the involvement of Hsp90 inhibition in down-regulation of client proteins, we have performed a co-immunoprecipitation analysis after 4 h exposure, i.e. under conditions where a substantial depletion of client proteins is not expected (Fig. 7). Again in contrast to a negligible effect of single agents, the combination treatment reduced the amount of Hsp90 bound to Raf-1, an effect most evident with the combination of curcumin and vorinostat. This observation was consistent with an enhanced inhibition of Hsp90 function, resulting in a reduced association of Hsp90 with Raf-1. The combined treatment resulted in a concomitant increase in the binding of Raf-1 to Hsp70. The shift of the binding from Hsp90 to Hsp70 has been described as an event preceding proteasome-mediated degradation of the client proteins [30, 34, 35].
Fig. 6.
Effects of combination of curcumin/vorinostat (a) or curcumin/panabinostat (b), or single agent treatment (c) on cellular levels of Hsp90 client proteins. a,b Cells were exposed to each drug alone or in combination for 24 h and processed as indicated in legend to Fig. 2. Subtoxic concentrations of each drug were used (i.e. curcumin 1.5 μM, vorinostat 15 nM, panobinostat 11.5 nM). The analysis was also performed after incubation of the drug-treated cells in drug-free medium for 24 h. c Cells were exposed to cytotoxic concentrations (IC80) of each drug (i.e. curcumin 5 μM, vorinostat 1 μM, panobinostat 0.1 μM) for 24 h and processed as indicated in the legend to Fig. 2. The analysis was also performed after incubation of the drug-treated cells in drug-free medium for 24 h. Actin is shown as a control for protein loading
Fig. 7.
Coimmunoprecipitation of Raf-1/Hsp90 in A431 cells treated with curcumin, vorinostat, panobinostat alone or in combination. A431 cells were exposed to subtoxic concentrations of panobinostat (11.5 nM), vorinostat (15 nM), and curcumin (1.5 μM) alone or in combination, for 4 h. Cell lysates were than harvested and immunoprecipitated with anti-Raf-1 rabbit polyclonal antibody. Immunoprecipitates were immunoblotted for Raf-1. Blots were stripped and probed for Hsp90 and Hsp70
Discussion
Curcumin has been shown to have multiple biological activities. The chemopreventive and anti-tumor effects of curcumin have been related to its ability to interfere with several signal transduction pathways which are involved in cell proliferation and/or apoptosis [6]. However, the underlying molecular mechanisms of these cellular effects are not well defined. In the present study, we have provided evidence that curcumin at cytotoxic/anti-proliferative concentrations was able to directly inhibit Hsp90 function, thereby promoting the chaperone dissociation and the degradation of several Hsp90 client proteins. This finding may account for curcumin’s ability to interfere with multiple signal transduction pathways, because Hsp90 plays a critical role in stabilization of several proteins implicated in the control of cell growth and malignant transformation [9, 10]. As compared with the geldanamycin derivative, 17-AAG, a well-established Hsp90 inhibitor, the potency of curcumin was substantially lower, because the inhibitory effects could be detected in the micromolar range of concentrations. This was consistent with lower anti-proliferative potency and a weaker Hsp90 binding. The mechanism of curcumin-Hsp90 interaction remains to be defined. A lack of up-regulation of Hsp70 suggests a mechanism of interaction different from that of geldanamycin. However, in preliminary limited proteolysis experiments, the proteolysis pattern observed on HSP90α indicated a protection of the N-terminal domain from the enzymatic hydrolysis.
Based on the observations that Hsp90 hyperacetylation by HDAC inhibitors effective against HDAC6 isoform disrupts its chaperone function [15–17], we have investigated the efficacy of curcumin in combination with well-known pan-HDAC inhibitors. Our results provide evidence that the potentiation of the anti-proliferative/cytotoxic activity of both vorinostat and panobinostat by subtoxic concentrations of curcumin (<IC20) might be mediated by inhibition of Hsp90 function. Indeed, the combinations of curcumin and HDAC inhibitors, at concentrations which produced negligible effects when used alone, resulted in a marked induction of apoptosis (Fig. 5). The cellular death could be related to a synergistic inhibition of Hsp90 activity, as under the same conditions, the combined treatment caused an almost complete depletion of several client proteins following 24 h exposure. The synergistic interaction of curcumin and HDAC inhibitor at target level was supported by co-immunoprecipitation experiment indicating an early (4 h) shift of the association of the Raf-1 from Hsp90 to Hsp70 (Fig. 7). Previous reports have shown that the shift in the chaperone association of the client proteins to the Hsp70-containing unstable multichaperone complex induces polyubiquitylation and degradation of the client proteins by the 26 S proteasome [30, 34, 35].
The most striking finding of this study was the long-lasting effect of the combination, which persisted even following removal of the drugs (Fig. 6). As a consequence of the persistent inhibition, the enhancement of apoptosis did not required prolonged exposure. Thus, given the relatively rapid reversibility of cellular effects of HDAC inhibitors [36] and curcumin (Figs. 5b and 6c), these combinations may have therapeutic implications. Since Hsp90 acetylation may induce a modified conformation of the enzyme, a tentative explanation of the synergistic interaction between HDAC inhibitor and curcumin may be that, as a consequence of allosteric effects, changed conformation of the acetylated protein favors the curcumin–Hsp90 interaction. A synergistic interaction between HDAC inhibitors and standard inhibitors of Hsp90 (i.e. geldanamycin analogues) has already been reported in leukemia or sarcoma cells [37–39], and the sensitization has been ascribed to the modulation of Hsp90 chaperone function by its acetylation status [40].
In conclusion, our findings reveal a novel mechanism of action of curcumin, which is consistent with modulation of signaling pathways. The effects of curcumin described in this manuscript support previous observations indicating the ability of curcumin to down-regulate p210bcr/abl, a client protein of Hsp90 [7]. Our obsvervations may be relevant to the therapeutic applications of both HDAC inhibitors and curcumin. Indeed, the therapeutic efficacy of these agents as single-agent therapy is still limited for pharmacological and pharmaco-dynamic reasons. Indeed, besides problems related to bioavailability and metabolism [41], the expected reversibility of target inhibition represents a major drawback of these target-specific agents. Relevant to this point is the observation that the sensitization for drug-induced apoptosis could be produced by very low drug concentrations which are achievable in vivo and expected to be well tolerated. Both curcumin and HDAC inhibitors have been reported to enhance cytotoxicity of various antitumor agents [6, 42–45]. The present study supports the potential interest of a novel combination which can be translated into the design of novel therapeutic approaches.
Acknowledgments
This work was partially supported by the Associazione Italiana per la Ricerca sul Cancro, Milan, by the Fondazione CARIPLO, Milan, and by the Ministero della Salute (Project Alleanza Contro il Cancro), Rome, Italy.
References
- 1.Aggarwal BB, Harikumar KB. Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int J Biochem Cell Biol. 2009;41:40–59. doi: 10.1016/j.biocel.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joe B, Vijaykumar M, Lokesh BR. Biological properties of curcumin-cellular and molecular mechanisms of action. Crit Rev Food Sci Nutr. 2004;44:97–111. doi: 10.1080/10408690490424702. [DOI] [PubMed] [Google Scholar]
- 3.Kawamori T, Lubet R, Steele VE, Kelloff GJ, Kaskey RB, Rao CV, Reddy BS. Chemopreventive effect of curcumin, a naturally occurring anti-inflammatory agent, during the promotion/progression stages of colon cancer. Cancer Res. 1999;59:597–601. [PubMed] [Google Scholar]
- 4.Chauhan DP. Chemotherapeutic potential of curcumin for colorectal cancer. Curr Pharm Des. 2002;8:1695–1706. doi: 10.2174/1381612023394016. [DOI] [PubMed] [Google Scholar]
- 5.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signalling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Reuter S, Eifes S, Dicato M, Aggarwal BB, Diederich M. Modulation of anti-apoptotic and survival pathways by curcumin as a strategy to induce apoptosis in cancer cells. Biochem Pharmacol. 2008;76:1340–1351. doi: 10.1016/j.bcp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Wu LX, Xu JH, Huang XW, Zhang KZ, Wen CX, Chen YZ. Down-regulation of p210(bcr/abl) by curcumin involves disrupting molecular chaperone functions of Hsp90. Acta Pharmacol Sin. 2006;27:694–699. doi: 10.1111/j.1745-7254.2006.00326.x. [DOI] [PubMed] [Google Scholar]
- 8.Jung Y, Xu W, Kim H, Ha N, Neckers L. Curcumin-induced degradation of ErbB2: a role for the E3 ubiquitin legase CHIP and the Michael reaction acceptor activity of curcumin. Biochim Biophys Acta. 2007;1773:383–390. doi: 10.1016/j.bbamcr.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–2918. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- 10.Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581:3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 11.Bagatell R, Whitesell L. Altered Hsp90 function in cancer: a unique therapeutic opportunity. Mol Cancer Ther. 2004;3:1021–1030. doi: 10.4161/cbt.3.10.1142. [DOI] [PubMed] [Google Scholar]
- 12.Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Targeting Hsp90 for cancer therapy. Br J Cancer. 2009;100:1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 14.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 15.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus E, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylase and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 16.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, Moscinski L, Smith C, Wu J, Jove R, Atadja P, Bhalla K. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res. 2003;63:5126–5135. [PubMed] [Google Scholar]
- 20.Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J, Atadja P, Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiskus W, Ren Y, Mohapatra A, Bali P, Mandawat A, Rao R, Herger B, Yang Y, Atadja P, Wu J, Bhalla K. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-alpha levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res. 2007;13:4882–4890. doi: 10.1158/1078-0432.CCR-06-3093. [DOI] [PubMed] [Google Scholar]
- 22.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 23.Zuco V, Zanchi C, Cassinelli G, Lanzi C, Supino R, Pisano C, Zanier R, Giordano V, Garattini E, Zunino F. Induction of apoptosis and stress response in ovarian carcinoma cell lines treated with ST1926, an atipica retinoid. Cell Death Differ. 2004;11:280–289. doi: 10.1038/sj.cdd.4401304. [DOI] [PubMed] [Google Scholar]
- 24.Howes R, Barril X, Dymock BW, Grant K, Northfield CJ, Robertson AG, Surgenor A, Wayne J, Wright L, James K, Matthews T, Cheung KM, McDonald E, Workman P, Drysdale MJ. A fluorescence polarization assay for inhibitors of Hsp90. Anal Biochem. 2006;350:202–213. doi: 10.1016/j.ab.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 25.Johnsson B, Löfås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-R. [DOI] [PubMed] [Google Scholar]
- 26.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Papalia GA, Leavitt S, Bynum MA, Katsamba PS, Wilton R, Qiu H, Steukers M, Wang S, Bindu L, Phogat S, Giannetti AM, Ryan TE, Pudlak VA, Matusiewicz K, Michelson KM, Nowakowski A, Pham-Baginski A, Brooks J, Tieman BC, Bruce BD, Vaughn M, Baksh M, Cho YH, Wit MD, Smets A, Vandersmissen J, Michiels L, Myszka DG. Comparative analysis of 10 small molecules binding to carbonic anhydrase II by different investigators using Biacore technology. Anal Biochem. 2006;359:94–105. doi: 10.1016/j.ab.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Birolo L, Dal Piaz F, Pucci P, Marino G. Structural characterization of the M* partly folded intermediate of wild type and P138A aspartate aminotransferase fron Escherichia coli . J Biol Chem. 2002;277:17428–17437. doi: 10.1074/jbc.M200650200. [DOI] [PubMed] [Google Scholar]
- 29.Sharp SY, Prodromou C, Boxall K, Powers MV, Holmes JL, Box G, Matthews TP, Cheung KM, Kalusa A, James K, Hayes A, Hardcastle A, Dymock B, Brough PA, Barril X, Cansfield JE, Wright L, Surgenor A, Foloppe N, Hubbard RE, Aherne W, Pearl L, Jones K, McDonald E, Raynaud F, Eccles S, Drysdale M, Workman P. Inhibition of the heat showck protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues. Mol Cancer Ther. 2007;6:1198–1211. doi: 10.1158/1535-7163.MCT-07-0149. [DOI] [PubMed] [Google Scholar]
- 30.An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr–abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- 31.Atkinson RA, Joseph C, Dal Piaz F, Birolo L, Stier G, Pucci P, Pastore A. Binding of alpha-actinin to titin: implications for Z-disk assembly. Biochemistry. 2000;39:5255–5264. doi: 10.1021/bi991891u. [DOI] [PubMed] [Google Scholar]
- 32.Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumour antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 33.Cooper MA. Biosensor profiling of molecular interactions in pharmacology. Curr Opin Pharmacol. 2003;3:557–562. doi: 10.1016/j.coph.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Nimmanapalli R, O’Bryan E, Bhalla K. Geldanamycin and its analogue 17-allylamino-17-demethoxygeldanamycin lowers Bcr-Abl levels and induces apoptosis and differentiation of Bcr-Abl-positive human leukemic blasts. Cancer Res. 2001;61:1799–1804. [PubMed] [Google Scholar]
- 35.Stancato LF, Silverstein AM, Owens-Grillo JK, Chow YH, Jove R, Pratt WB. The Hsp90-binding antibiotic geldanamycin decreases Raf levels and epidermal growth factor signalling without disrupting formation of signalling complexes or reducing the specific enzymatic activity of Raf kinase. J Biol Chem. 1997;272:4013–4020. doi: 10.1074/jbc.272.7.4013. [DOI] [PubMed] [Google Scholar]
- 36.Pratt WB, Toft DO. Regulation of signalling protein function and trafficking by the Hsp90/Hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 37.Rahmani M, Reese E, Dai Y, Bauer C, Kramer LB, Huang M, Jove R, Dent P, Grant S. Cotreatment with suberanoylanilide hydroxamic acid and 17-allylamino 17-demethoxygeldanamycin synergistically induces apoptosis in Bcr-Abl+ cells sensitive and resistant to STI571 (Imatinib mesylate) in association with down-regulation of Bcr-Abl, abrogation of signal transducer and activator of transcription 5 activity, and Bax conformational change. Mol Pharmacol. 2005;67:1166–1176. doi: 10.1124/mol.104.007831. [DOI] [PubMed] [Google Scholar]
- 38.George P, Bali P, Annavarapu S, Scuto A, Fiskus W, Guo F, Sigua C, Sondarva G, Moscinski L, Atadja P, Bhalla K. Combination of the histone deacetylase inhibitor LBH589 and the Hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen A, Su L, Campbell B, Poulin NM Nielsen TO (2009) Synergism of heat shock protein 90 and histone deacetylase inhibitors in synovial sarcoma. Sarcoma 794901 [DOI] [PMC free article] [PubMed]
- 40.Rao R, Fiskus W, Yang Y, Lee P, Joshi R. HDAC6 inhibition enhances 17-AAG-mediated abrogation of Hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 41.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 42.Fulda S. Modulation of TRAIL-induced apoptosis by HDAC inhibitors. Curr Cancer Drug Targets. 2008;8:132–140. doi: 10.2174/156800908783769355. [DOI] [PubMed] [Google Scholar]
- 43.Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, antiangiogenesis and inhibition of nuclear factor-kB-regulated gene products. Cancer Res. 2007;67:3853–3861. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 44.Somers-Edgar TJ, Scandlyn MJ, Sturart EC, Le Nedelec MJ, Valentine SP. The combination of epigallocatechin gallate and curcumin suppresses ERα-breast cancer cell growth in vitro and in vivo. Int J Cancer. 2008;122:1966–1971. doi: 10.1002/ijc.23328. [DOI] [PubMed] [Google Scholar]
- 45.Mai A, Altucci L. Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead. Int J Biochem Cell Biol. 2009;41:199–213. doi: 10.1016/j.biocel.2008.08.020. [DOI] [PubMed] [Google Scholar]