Abstract
Cells have evolved to develop molecules and control mechanisms that guarantee correct chromosome segregation and ensure the proper distribution of genetic material to daughter cells. In this sense, the establishment, maintenance, and removal of sister chromatid cohesion is one of the most fascinating and dangerous processes in the life of a cell because errors in the control of these processes frequently lead to cell death or aneuploidy. The main protagonist in this mechanism is a four-protein complex denominated the cohesin complex. In the last 10 years, we have improved our understanding of the key players in the regulation of sister chromatid cohesion during cell division in mitosis and meiosis. The last 2 years have seen an increase in evidence showing that cohesins have important functions in non-dividing cells, revealing new, unexplored roles for these proteins in the control of gene expression, development, and other essential cell functions in mammals.
Keywords: Centrosomes, Chromosome segregation, Cohesin, Cohesinopathies, Gene expression control, Insulators, Neuron development, Sister chromatid cohesion
Introduction
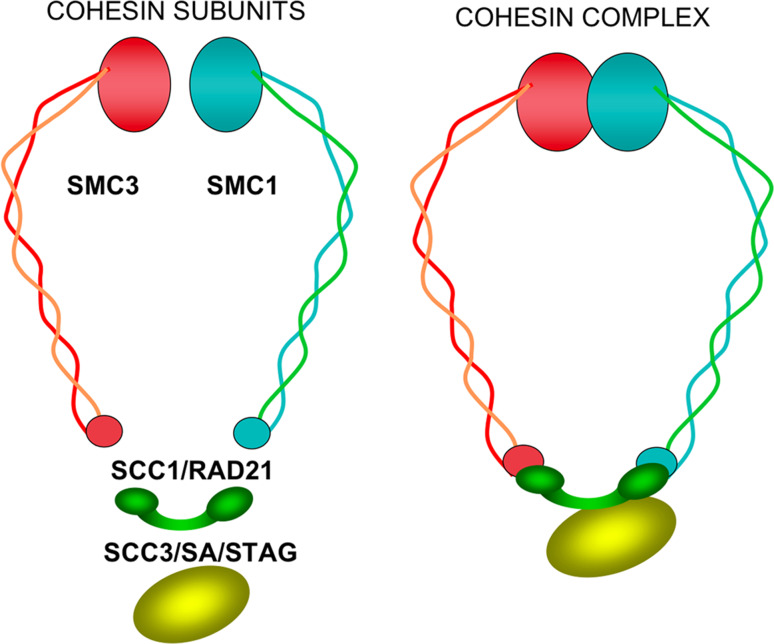
During cell division, the pair of recently synthesized DNA molecules, now called sister chromatids, remain in close proximity until the precise moment at which they must be segregated during the metaphase/anaphase transition. Sister chromatid cohesion is mediated by a multiprotein cohesin complex, which was first characterized in Saccharomyces and Xenopus [1–3]. This complex is essentially composed of four evolutionarily conserved core subunits: two that belong to the structural maintenance of chromosomes (SMC1 and SMC3) family of proteins, one kleisin α (from the Greek word for closure) subunit SCC1/RAD21, and stromalin SCC3/SA/STAG (Fig. 1). Vertebrates have two mitotic SCC3/SA/STAG members, SA1/STAG1 and SA2/STAG2 [4], which do not coexist and are present in different cohesin complexes in Xenopus and human [5, 6]. Based on the structural characteristics of SMC and electronic microscopy results, it has been proposed that these complexes form a ring-like structure (Fig. 1) and mediate cohesion by embracing chromatin fibers from the two sister chromatids [7, 8].
Fig. 1.
Cohesin subunits and ring-like structure proposed for the cohesin complex. Structural maintenance of chromosomes SMC1 and SMC3 form a heterodimer, interacting through their hinge regions. The SMC1 and SMC3 head domains, which contain the ATPase motifs of these proteins, interact with the C- and N-termini of the SCC1/RAD21 kleisin subunit, respectively, closing the ring. The SCC3/SA/STAG subunit interacts with SCC1/RAD21, contributing to maintenance of the ring structure
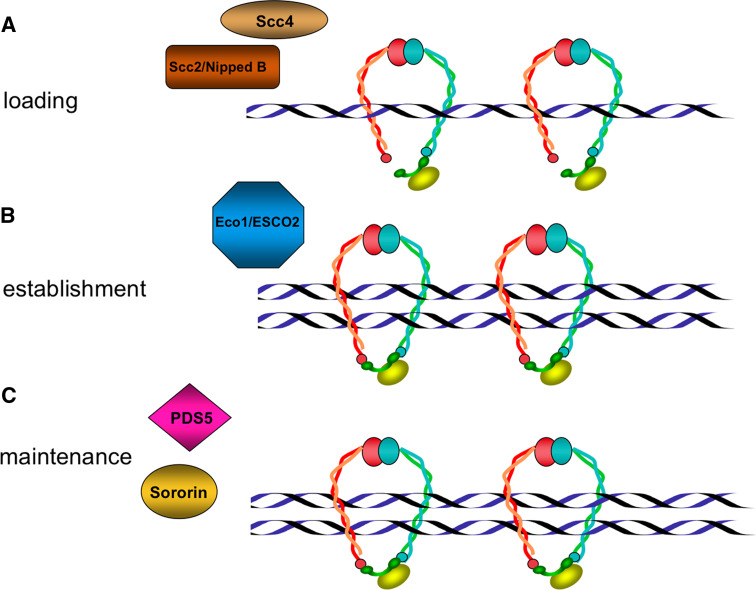
During mitotic cell cycles in Saccharomyces cerevisiae, cohesin complexes are loaded near the G1-to-S phase, but in most organisms studied, cohesins bind to chromatin in telophase [9]. Cohesin complex loading to chromatin depends on the Scc2/Scc4 adherin complex [10, 11], while the establishment and maintenance of cohesion depend on the function of an ever-increasing number of proteins. Eco1/Ctf7p acetyltransferase [12–15], Ctf4, a protein associated with DNA polymerase, and the clamp-loader RFC complex proteins Ctf8 and Ctf18 are reported to be necessary for the establishment of correct cohesion [16, 17]. Another four cohesion-regulator proteins, PDS5 [6, 18, 19], WAPL [20, 21], sororin [22], and haspin [23], are involved in the control of the association/dissociation of cohesin complexes to chromatin (Figs. 2, 3).
Fig. 2.
Representation of some factors that contribute to cohesion regulation. a Adherins Scc2/Scc4, also denoted NippedB/Mau-2, form the cohesin loading complex. b The replication fork-associated Eco1/ESCO2 acetyltransferase is required to establish cohesion. c Precocious dissociation of sister (PDS5) and sororin are the best-characterized factors implicated in the maintenance of sister chromatid cohesion
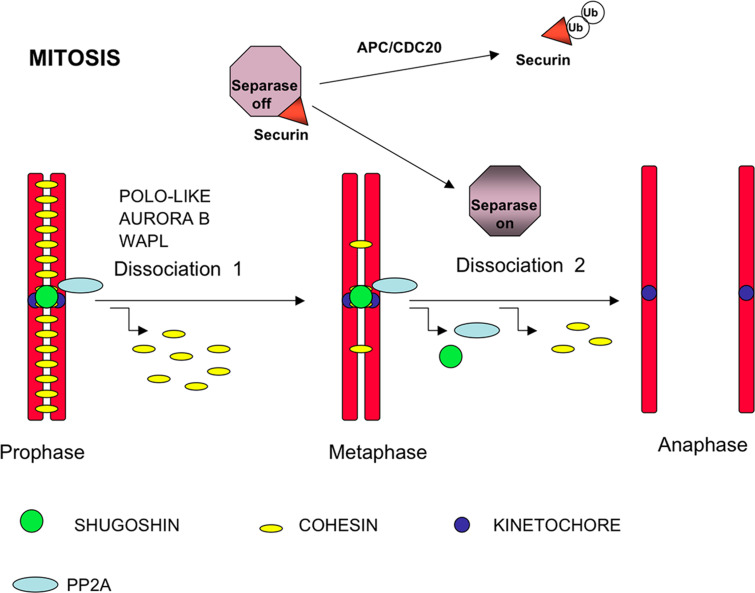
Fig. 3.
Chromosome segregation in fly and vertebrate mitosis. In prophase chromosomes, the cohesin complexes are located along the middle region of the arms and at the inner centromere domain. During the prophase/metaphase transition, the kinases Aurora-B and/or Polo-like kinase 1 (PLK1) phosphorylate the SA2 subunit of most cohesin complexes at the arms and promote their removal from chromosomes. Wing apart-like (WAPL) is a regulator of sister chromatid resolution in the mitotic prophase. Centromere cohesins are protected from phosphorylation by phosphatase PP2A, which is recruited to the centromere by shugoshin. Once all chromosomes are bi-oriented at the metaphase plate in the metaphase/anaphase transition, shugoshin and PP2A are delocalized from the centromeres. At this time, the APC/CDC20 complex ubiquitinizes the separase inhibitor securin, which is dissociated from separase, allowing cleavage of the few remaining cohesin complexes at the arms and centromere cohesins
In addition to their canonical role as “chromosome glue” during cell division, cohesins are involved in additional cellular mechanisms, including centromeric heterochromatin formation, post-replicative double-strand break repair, and gene expression in interphase. A number of recent studies report cohesin expression and function in post-mitotic cells as well as the involvement of cohesin and cohesin-interacting protein gene mutations in human pathologies. These findings are changing our concept of the cohesin complexes, indicating that the name “cohesin” is probably a partial reflection of the many functions of these proteins in the cell.
This review illustrates the general features of the cohesins involved in the regulation of sister chromatid cohesion in chromosome arms, centromeres, and telomeres during mitosis and meiosis and focuses essentially on the role of cohesins in functions other than chromosome segregation.
Cohesins and chromosome cohesion
Sister chromatid arm cohesion
In mitosis in the budding yeast S. cerevisiae, cohesins are loaded into the chromatin near the G1-to-S phase, probably joining with the replication process [9], and the cohesion of both arms and the centromere is lost during the metaphase/anaphase transition. The action of a specific protease called separin/separase is needed to remove cohesin complexes from chromatin; this protease cleaves specific residue bonds of the cohesin complex Scc1 subunit and promotes the dissociation of cohesin complexes from DNA [24]. Until it is activated, separase remains inactive by binding to its specific inhibitor securin [25–28]. In metazoa, however, dissociation of the cohesin complexes from chromatin takes place in two distinct processes (Fig. 3). Most cohesin complexes are removed from chromosome arms during prophase by a separase-independent pathway [29], in a step triggered by phosphorylation of the SA2/STAG2 subunit by Aurora B and Polo-like kinases [30]. Cohesin complexes subsequently remain at the centromeres, maintaining centromeric cohesion until the chromosomes are correctly bioriented and the spindle assembly checkpoint is satisfied in metaphase. Securin is ubiquitinized, mediated by activation of the anaphase-promoting complex/cyclosome (APC/C). It is then released from the securin/separase complex, allowing the activation of separase, which cleaves the RAD21 subunit from centromeric cohesin complexes and triggers the onset of anaphase [31]. This is the common mechanism; in cells lacking separase or expressing a non-cleavable SCC1 subunit, cohesin complexes from arms do not dissociate completely during nocodazole arrest, suggesting that the complete removal of arm cohesion depends on separase [32]. Cohesin cofactor WAPL is the product of the previously identified Drosophila wings apart-like (WAPL) gene involved in heterochromatin organization [33]. Two recent reports have shown that human WAPL regulates the resolution of sister chromatid cohesion and promotes cohesin complex dissociation by direct interaction with the RAD21 and SA/STAG cohesin subunits [20, 21].
Meiosis is the specific division process in germ cells by which diploid cells yield haploid gametes through a single round of DNA replication and two successive steps of chromosome segregation. During meiosis I in most organisms studied to date, the sister chromatid arm cohesion is removed by a separase-dependent mechanism throughout the metaphase I/anaphase I transition [9]. In the budding yeast S. cerevisiae, the meiotic cohesin complex contains Smc1, Smc3, Scc3, and Rec8 instead of Rad21. In fission yeast there is a second Scc3 paralogue, termed Rec11, and different cohesin complexes are described in S. pombe meiosis [34]. In mammals, the meiotic-specific paralogues of SMC1, SCC1/RAD21, and SA/STAG1/2 are thus SMC1β [35], REC8 [36, 37], and STAG3 [38], respectively. Cytological and biochemical analyses have revealed the contribution of different cohesin complexes to synaptonemal complex (SC) formation and meiosis I arm cohesion during mammalian meiosis. Current knowledge of the diverse cohesin complexes and their contribution to chromosome segregation during mammalian meiosis has been extensively reviewed by Suja and Barbero [39].
Sister chromatid cohesion at centromeres
This cohesion is needed to prevent premature chromatid separation before all chromosomes are correctly bi-oriented into the metaphase plate during mitosis and for correct transit through meiosis I and homologue segregation in meiosis. Phosphorylation by Polo-like and Aurora B kinases also participates in the removal of centromeric cohesins, but unlike arm cohesion, this is a separase-dependent mechanism in all organisms studied to date [9]. Phosphorylation of centromeric cohesin by Polo-like and Aurora B kinases enhances separase cleavage of these complexes [40, 41] (Fig. 4).
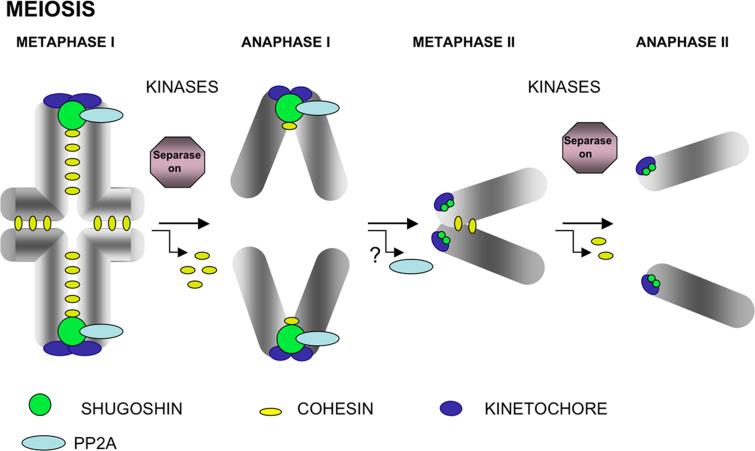
Fig. 4.
Chromosome segregation in meiosis. In metaphase I bivalents, cohesin complexes are located at the interchromatid domain along the arms and at the inner centromere domain below the closely associated sister kinetochores. At the onset of anaphase I, only the cohesin complexes at the arms are cleaved by separase to allow segregation of recombined homologues to opposite poles. Centromere cohesins are protected by SGO/PP2A. In metaphase II chromosomes, sister kinetochores attach to microtubules from opposite poles, and cohesin complexes are found at the inner centromere domain. In late prometaphase II chromosomes in mammals, the interaction of microtubules from opposite poles with sister kinetochores generates tension across the centromere and triggers the redistribution of SGO2 and probably PP2A delocalization from centromeres. During the metaphase II/anaphase II transition, separase is now able to cleave remaining centromeric cohesin complexes to trigger chromatid segregation. The chromosomes depicted are telocentric
An important question is “What is the mechanism protecting centromere cohesin during the loss of arm cohesion in mitosis prometaphase and meiosis I”? Some years ago, Kitajima et al. [42] identified a protein family involved in the protection of centromeric cohesion in fission yeast, which they denoted shugoshins (from the Japanese “guardian of the spirit”). A single shugoshin (Sgo1) is found in the budding yeast S. cerevisiae, but two (Sgo1 and Sgo2) have been characterized in fission yeast and mammals; Sgo1 is essential in yeast meiosis, whereas Sgo2 has a major involvement in mitosis. In contrast, SGOL2, the mammalian orthologue of yeast Sgo2, is essential for the meiotic cell cycle but not for mitosis in vivo [43]. Shugoshin recruits and forms a complex with phosphatase PP2A at the centromeres in mitosis, suggesting that blockade of the centromeric cohesin phosphorylation by specialized kinases is the mechanism by which these proteins avoid premature loss of centromere cohesion [44, 45] (Fig. 3). Recent results from mammalian studies suggest that shugoshins are also components of the tension-sensing machinery during mitosis [46] and meiosis II [47–49].
During the metaphaseI/anaphaseI transition in mammalian meiosis, shugoshin (essentially SGOL2) protects centromeric cohesins from separase by targeting phosphatase PP2A at the centromeres, suggesting a putative blockade of kinase action on cohesins. During metaphase II, SGOL2 relocalizes in a tension-dependent manner in mouse spermatocytes [47] and oocytes [49]. This relocation could cause PP2A to delocalize from centromeres, allowing the action of activated separase and removal of centromere cohesion and sister chromatid segregation (Fig. 4).
Two new cohesion-regulator molecules, sororin and haspin have been implicated in centromere cohesion. Sororin was first identified in a screen for substrates of the anaphase promoting complex (APC) in vertebrates, and no homologues have been described in others organisms [22]. Different results in somatic cells suggest that sororin interacts with the cohesin complex and that it is essential for the maintenance of sister chromatid cohesion. Sororin is ubiquitinized and degraded after sister chromatid cohesion is dissolved [22, 50]. Studies on sororin-depleted and shugoshin-depleted cells indicate that sororin and shugoshin may act in concert in the protection of centromeric cohesion [51]. More recently, sororin has been found to be needed for the efficient repair of DNA double-strand breaks in G2 and for maintaining the stably chromatin-bound cohesin in G2, suggesting a crucial cohesin regulator role for this protein [52]. Haspin/Gsg2 is a histone H3 threonine-3 kinase that colocalizes with the cohesin complex at inner centromeres during vertebrate mitosis. The depletion of haspin in human cells results in premature separation of sister chromatids, and its overexpression rescues the phenotype of SGO1 deficiency, suggesting a role for haspin in the maintenance of centromeric cohesion prior to anaphase [23]. These findings reveal that these two proteins have important functions in the control of sister chromatid cohesion, but the precise molecular mechanisms by which sororin and haspin regulate the cohesin association to chromatin remain to be elucidated.
Sister chromatid cohesion in telomeres
Studies in yeast have provided evidence that cohesins are also responsible for sister chromatid telomere cohesion [53, 54]. An initial study in HeLa cells showed that depletion of tankyrase-I, a telomeric poly (ADP-ribose) polymerase involved in TRF1 location, yields mitotic cells with sister chromatid separated at the arms and centromeres but still associated at the telomeres [55]. This result indicates that, in human cells, telomeres have specific requirements for separation and suggests a unique tankyrase-I dependent mechanism for their resolution in mitosis. One year later, however, Chang et al. [56], using RNAi, showed that tankyrase-I depletion activates the spindle check point, resulting in pre-anaphase arrest with intact sister chromatid cohesion, thereby bringing the role of tankyrase-I in telomere cohesion into question.
More recently, Canudas et al. [57], in a study to clarify this discrepancy, identified the SA1/STAG1 subunit of cohesin complex as the protein that binds to TRF1 (a tankyrase-I substrate) and TIN2 (a TRF1-binding partner). Depletion of cohesin SA1/STAG1 or of the telomeric proteins TRF1 and TIN2 restores normal separation of sister chromatid telomeres in tankyrase-I depleted cells, providing additional evidence of a role for tankyrase-I in telomere cohesion. These data suggest that tankyrase-I could be involved in different pathways and that telomere cohesion may be regulated in mitotic human cells by both cohesins and components of telomere chromatin.
SA2/STAG2-containing complexes do not associate with TRF1 in HeLa cells [57]. The ratio of SA1 to SA2 complexes is variable, depending on cell type [5], but there is currently no reasonable explanation for the presence of two different Scc3 orthologues (SA1 and SA2) in vertebrate cells. Results showing telomeric cohesion involving SA1 but not SA2 are possibly the first functional difference reported for the two kinds of vertebrate mitotic cohesin complexes.
There are also differing results on the involvement of cohesin function in telomere dynamics during meiosis. The absence of cohesin subunit Rec8 induces the persistence of telomere clustering in yeast meiosis [58], and mice lacking the specific meiotic cohesin SMC1β show defective telomere/nuclear envelope attachment [59]. For a recent review of further aspects of telomere dynamics in meiosis, see Scherthan [60].
Cohesins and regulation of gene expression
Cohesins and human developmental disorders
Although most studies of cohesins have focused on chromosome cohesion during cell division, there is evidence that cohesins participate in other important cell processes, such as DNA damage repair [61] and heterochromatin formation [62]. Nonetheless, one of the most surprising findings has been the discovery of a link between cohesin mutations and human diseases. Two groups, Krantz et al. [63] and Tonkin et al. [64], found that the NIPBL gene, the homologue to Scc2 adherin gene involved in cohesin loading to chromatin in yeast, is mutated in the human Cornelia de Lange syndrome (CdLS; OMIM: 122470, 300590, 610759). This pathology is a multiple neuro-developmental disorder characterized by facial dysmorphisms, mental retardation, growth delay, and upper limb abnormalities. Subsequent studies of several cases of this syndrome showed that mutations in SMC1α [65, 66] and SMC3 [66] cause a mild variant of CdLS.
Roberts syndrome/SC phocomelia (RBS; OMIM: 268300) is an autosomal recessive disorder related phenotypically to CdLS; patients present craniofacial abnormalities, growth retardation, and limb reduction. Cells from RBS patients show a lack of cohesion at the heterochromatic regions around the centromeres and at the Y chromosome long arm [67]. Vega et al. [68] reported that RBS is caused by mutations in ESCO2, the homologue to yeast Eco1p, which encodes a protein required for the establishment of cohesion between sister chromatids during S phase (Fig. 2). Analyses of different mutations in RBS patients point to the loss of ESCO2 acetyltransferase activity as a molecular mechanism involved in RBS pathology [69].
Mice that lack PDS5B die shortly after birth and have multiple developmental anomalies that resemble those found in humans with CdLS [70]. PDS5A and PDS5B are large HEAT-repeat proteins that associate with chromatin in a cohesin-dependent manner in both human cells and Xenopus egg extracts [71]. They are not required for cohesin association to chromosomes, but they are needed to maintain cohesion (Fig. 2). The study of chromosomes from Pds5B −/− mouse cells showed no defects in sister chromatid cohesion, indicating a PDS5B function beyond chromosome segregation [70]. Two excellent reviews by McNairn and Gerton [72] and Liu and Krantz [73], respectively, provide extensive information on the relationships between cohesins and human disease.
Cohesins and insulators
The phenotypes and molecular causes of these diseases and the expression of cohesins in post-mitotic cells suggest novel activities for cohesins that are distinct from that of chromatin glue. The first results linking cohesin and the control of gene expression came from research in yeast, flies, and mammalian cells. Mutational analysis in S. cerevisiae implicated Smc proteins in boundary element function [74, 75]. In Drosophila nipped-B, an orthologue of the yeast Scc2 protein facilitates long-range activation of the cut gene [76], and in human cells, SA2/STAG2 may be able to act as a transcriptional co-activator [77]. Although the results of these studies indicate that cohesins are also implicated in gene expression regulation and development, the mechanism(s) by which they carry out these functions remain unknown. Several groups have recently studied cohesin localization in mammalian chromosomes and found that numerous cohesin-binding sites overlap the CCCTC-binding factor (CTCF), an insulator protein that participates in blocking enhancer–promoter interactions [78–80]. The studies also demonstrate that cohesins are required for the CTCF insulation function and for control of H19/IGF2 locus imprinting in G2 and G1 phases in mice. Because there is no sister chromatid cohesion in the G1 phase, the function of cohesins in the control of H19/IGF2 transcription is independent of their role in sister chromatid cohesion.
Using a quantitative proteomic approach to identify cofactors recruited by CTCF, Rubio et al. [81] found that Scc3/SA1/STAG1 associates preferentially with DNA containing the CTCF target sequence of the c-myc insulator element. Their results suggest an interaction between cohesin and CTCF and that this interaction occurs via the Scc3/SA1 subunit, as the other cohesin subunits RAD21, SMC1, and SMC3 showed no significant enrichment in proteomic analysis. This is an interesting observation since the Scc3/SA/STAG subunit had remained up to this time a relative mystery within the structure/activity of the cohesin complex. SMC1/SMC3 heterodimers are the ring-forming subunits with ATPase activity, SMC3 is an Eco1 acetylase substrate, and acetylation of SMC3 is required for sister chromatid cohesion in S phase [82–84]. Kleisin Scc1/Rad21 clearly contributes to the maintenance of the ring, as the N- and C-terminal domains of this subunit interact with head SMC3 and SMC1 domains, respectively [85], and is the principal cleavage substrate for separase [31]. The Scc3/SA/STAG subunit emerges with important roles in several cohesin functions: as the substrate of Polo-like kinase to remove cohesin from the arms during mitosis [30], as a co-activator of transcription based on the luciferase-reporter assay in transfected cells [77], interaction with the CTCF in the insulation effect [81], and as being essential for axon pruning in neuron morphogenesis [86].
Cohesins and neuron development
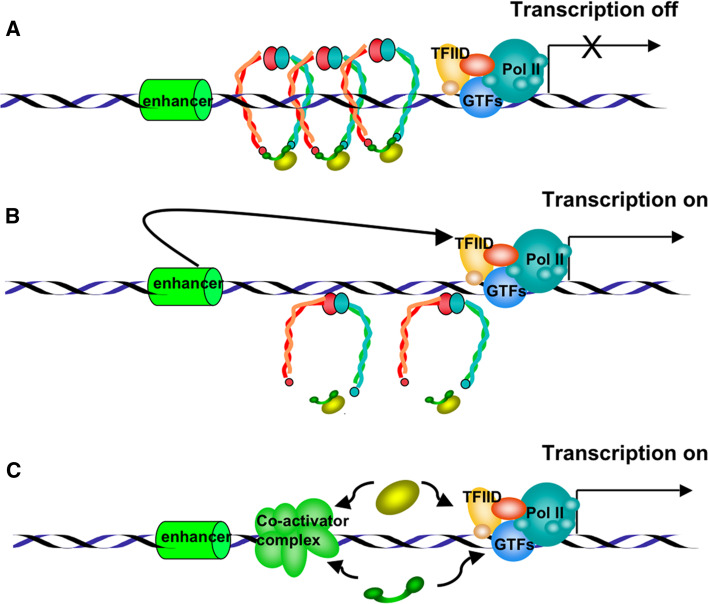
Initial results linking sister chromatid cohesion and neuronal morphogenesis were reported by Takagi et al. [87] and Seitan et al. [88], who demonstrated that the Caenorhabditis elegans Scc4, which is the homologue of Mau-2 [11], the partner of Scc2 in the cohesin-loading complex, is implicated in neuron migration. Two groups have provided evidence that the cohesin complex subunits are needed for morphogenesis of non-dividing neurons in Drosophila. In a genetic screening using a modified piggyBac vector for insertional mutagenesis, Schuldiner et al. [86] identified the two cohesin subunits, SMC1 and SA/STAG, as essential for axon pruning. Similar conclusions were reached in a study by Pauli et al. [89], who generated flies expressing a modified version of cohesin subunit RAD21, RAD21TEV, a substrate of tobacco etch virus (TEV) protease. Through tissue-specific expression of TEV protease and RAD21 cleavage, these researchers showed that axon pruning is specifically blocked in mushroom body neurons. Seeking the putative role of cohesins in these neuron effects, Schuldiner et al. [86] analyzed the expression of the steroid hormone receptor EcR, which encodes EcR-B1 protein, an essential regulator of axon pruning in the mushroom body. They observed that EcR-B1 expression is reduced in SMC1-depleted γ-neurons and that the pruning defect is reversed by EcR-B1 overexpression, suggesting that in this case cohesins facilitate EcR transcription. This result is the opposite of the effect previously described for cohesin function in insulators, but it is not the only evidence of a positive regulation of gene transcription by cohesins. In human cells, SA2/STAG2 co-activates a multimeric NF-κB reporter construct and enhances the activity of the p65/RelA transactivation domain [77]. Zebrafish embryos lacking cohesin subunit RAD21 or SMC3 do not express runx3 and lose runx1 expression during early embryonic development [90]. These findings suggest that the role of cohesin in transcription regulation may be gene-dependent (Fig. 5).
Fig. 5.
Representation of putative models by which cohesins may regulate transcription. a Chromosome-bound cohesin complexes block the transcriptional activation signals from a distal enhancer element, repressing gene expression. b Dissociation of cohesin complexes from chromatin allows the transmission of positive signals from distal elements, activating transcription. c Isolated cohesin subunits may act as transcriptional co-activators by interacting with the transcription initiation complex or with other co-activators
These results on neural development as well as the role of cohesins in insulation function have important connotations in CdLS and RBS, which are characterized by mental retardation and structural limb defects. Revenkova et al. [91] recently generated mutations in the SMC1α and SMC3 hinge domain, where several CdLS mutations cluster, and studied in vitro binding to DNA. They found that mutated hinge dimers bind DNA with higher affinity than wild-type proteins and that SMC-mutated cell lines showed a higher frequency of spontaneous chromosome aberrations than SMC-wild type lines; this was the first suggestion of a molecular link for CdLS.
Other cohesin functions
In addition to the cohesin function in chromosome dynamics in sister chromatid cohesion, DNA repair, and the control of gene expression, new roles for cohesin roles are now emerging in other cell machineries.
Immunodepletion of HeLa cells using anti-hSMC1, -hSMC3, -SA1/SA2, and -hRAD21 antibodies have been found to inhibit microtubule aster assembly in HeLa cells [92]. The authors of this study also described cohesin localization in spindle poles and an interaction between cohesin and NuMA, a spindle pole-associated factor required for mitotic spindle organization, suggesting a role for cohesins in mitotic spindle aster assembly. In a recent study of SMC1 in the spindle pole, Wong and Blobel [93] reported that SMC1 is recruited at the spindle pole by microtubule-bound RNA export factor Rae1; SMC1 binding to Rae1 was stimulated by the phosphorylation of two SMC1 serine residues by spindle-pole-anchored ataxia-telangiectasia mutant (ATM) or a related kinase. These authors proposed that ATM phosphorylation of SMC1/SMC3 heterodimers takes place exclusively at the spindle pole, preventing the spread of cohesin/Rae1 interactions away from the spindle pole. They also speculated that SMC1/SMC3 heterodimers would recruit other cohesin complex subunits, completing the ring structure and embracing microtubules at the spindle pole.
In addition, phosphorylation of SMC-subunits of the cohesin complex have also been found to be involved in cellular responses to DNA damage. The phosphorylation of Ser-957 and/or Ser-966 of human SMC1 by ATM is required for the activation of the S-phase checkpoint in response to ionizing radiation [94]. To address the functional importance of SMC1 phosphorylation, Kitagawa et al. [95] generated murine cells in which these two SMC1 phosphorylation sites were mutated. These mutated cells presented normal focus of ATM, NBS1 and BRCA1 proteins after ionizing radiation, but they exhibited a defective S-phase checkpoint, decreased survival, and chromosomal aberrations after DNA damage. The same positions in SMC1 are phosphorylated after treatment with hydroxyurea or UV irradiation in an ATM-independent manner, indicating that other kinase(s) must be implicated in SMC1 phsophorylation in response to other cellular stresses [96]. Luo et al. [97] recently reported that SMC3 is phosphorylated at two distinct serine residues by two different kinases: Ser-1083 phosphorylation of human SMC3 is inducible by ionizing radiation and is ATM-dependent, and Ser-1067 is phosphorylated constitutively by CK2 kinase and is not enhanced by ionizing radiation. However, both Ser-1067 and Ser-1083 phosphorylation were required for S-phase checkpoint. These results show an important function for the SMC subunits of the cohesin complex in DNA repair, but how phosphorylated SMC1 and SMC3 work in the DNA damage response and whether they perform this role as a subunit of the cohesin complex need to be addressed.
The kinesin-related protein KIF3A and their associated protein SMAP have been reported to interact with SMC3 [98], again supporting a cohesin function in microtubule organization. It has also been suggested that SMC1 is associated with both centrioles of the centrosome at the G0/G1 cell cycle stage in various mammalian cell lines, including HeLa [99]. In this context, recent studies of human SGO1 isoforms have shed new light on centriole cohesion. In a first study, Wang et al. [100] described a new splice variant of human SGO1, sSGO1, which lacks SGO1 exon 6 and has a distinct subcellular localizations in human cells. In a more recent work, these authors reported that human sSGO1 localizes at the centrosome in interphase and at spindle poles in mitosis. Their studies using RNAi to generate SGO1 knockdown in HeLa cells provides compelling evidence that sSGO1 has a important function in centriole cohesion that is regulated by Polo-like kinase 1 [101]. Although the relationship of sSGO1 to putative centriole cohesins was not addressed in this work, the results on cohesin localization in centrioles suggest that the sSGO1 function in maintaining centriole cohesion would be cohesin-dependent.
Concluding remarks
From the first identification of cohesins in yeast and Xenopus a decade ago, cohesin functions have expanded from sister chromatid cohesion to different aspects of chromosome dynamics, gene expression, development, and centrosome/microtubule organization. These roles reflect the flexibility of the cohesin complex in interacting with various factors, including cohesion regulators (Plk1, PDS5, WAPL, SCC2/SCC4), insulation factors (CTCF), spindle pole-associated proteins (NuMA, Rae1, ATM kinase) and, probably, other still-unknown proteins that regulate the many tasks of the cohesins. Although the interactions among the different cohesin complex subunits that maintain the ring structure and are involved in separase cleavage have been studied extensively (for two recent extensive reviews, see 102 and 103), little is known about the cohesin domains involved in interactions with the diverse regulators. Are there specific cohesin complexes depending on the function? What are the molecular signals that drive cohesins to repress or activate transcription? Are all cohesin complex components required for all the cohesin roles discussed here, or can some of these functions be performed by specific isolated subunits (Fig. 5c)? These are some of the questions that remain to be addressed in future studies, to unravel the molecular networks in which cohesins are essential architects.
Acknowledgments
I apologize to all colleagues whose contributions have not been referenced due to space restrictions. We thank C. Mark for editorial assistance. This work was supported by the Spanish Ministerio de Educación y Ciencia (grants BFU2006-04406/BMC) and the Comunidad de Madrid (P-BIO-0189-2006).
References
- 1.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/S0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 2.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S cerevisiae . Cell. 1997;91:47–57. doi: 10.1016/S0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carramolino L, Lee BC, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero JL, Zipori D. SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. doi: 10.1016/S0378-1119(97)00121-2. [DOI] [PubMed] [Google Scholar]
- 5.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3 subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/S0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 10.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/S1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 11.Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Cft7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–323. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/MCB.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/S0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 16.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Cell Biol. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Yoder J, Antoshechkin I, Han M. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol Cell Biol. 2003;23:7698–7707. doi: 10.1128/MCB.23.21.7698-7707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Ren Q, Yang H, Conrad MN, Guacci V, Kateneva A, Dresser ME. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol Microbiol. 2005;56:670–680. doi: 10.1111/j.1365-2958.2005.04582.x. [DOI] [PubMed] [Google Scholar]
- 20.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 22.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 25.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 26.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 27.Ciosk R, Zacharie W, Michaelis C, Shevchenco A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/S0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 28.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 29.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/S0092-8674(00)00132-X. [DOI] [PubMed] [Google Scholar]
- 30.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesions from chromosome arms and loss of arm cohesion during early mitosis depends on phsophorylation of SA2. PLoS Biol. 2005;3:419–432. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Biol. 2007;120:4188–4196. doi: 10.1242/jcs.011528. [DOI] [PubMed] [Google Scholar]
- 33.Vernì F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster . Genetics. 2000;154:1693–1710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- 35.Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parisi S, McKay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 38.Prieto I, Suja JA, Pezzi N, Kremer L, Martínez-A C, Rufas JS, Barbero JL. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- 39.Suja JA, Barbero JL (2009) Cohesin complexes and sister chromatid cohesion in mammalian meiosis In: Benavente R, Volff JN (eds). Genome dynamics, vol 5. Karger, Basel, pp 94-116 [DOI] [PubMed]
- 40.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/S0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 41.Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157:219–229. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 43.Llano E, Gómez R, Gutiérrez-Caballero C, Herrán Y, Sánchez-Martín M, Vázquez-Quiñones L, Hernández T, de Alava E, Cuadrado A, Barbero JL, Suja JA, Pendás AM. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008;22:2400–2413. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 45.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 46.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Gómez R, Valdeolmillos A, Parra MT, Viera A, Carreiro C, Roncal F, Rufas JS, Barbero JL, Suja JA. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007;8:173–180. doi: 10.1038/sj.embor.7400877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:24–413. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 50.Rankin S. Sororin, the cell cycle and sister chromatid cohesion. Cell Cycle. 2005;4:1039–1042. doi: 10.4161/cc.4.8.1926. [DOI] [PubMed] [Google Scholar]
- 51.Díaz-Martínez LA, Giménez-Abián JF, Clarke DJ. Regulation of centromeric cohesion by sororin independently of the APC/C. Cell Cycle. 2007;6:714–724. doi: 10.4161/cc.6.6.3935. [DOI] [PubMed] [Google Scholar]
- 52.Schmitz J, Watrin E, Lénárt P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 53.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae . PLoS Biol. 2004;2:1325–1339. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antoniacci LM, Skibbens RV. Sister-chromatid telomere cohesion is non-redundant and resists both spindle forces and telomere motility. Curr Biol. 2006;16:902–906. doi: 10.1016/j.cub.2006.03.060. [DOI] [PubMed] [Google Scholar]
- 55.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 56.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 57.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol. 2005;170:213–223. doi: 10.1083/jcb.200501042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 60.Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64:117–124. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watrin E, Peters JM. Cohesin and DNA damage repair. Exp Cell Res. 2006;312:2687–2693. doi: 10.1016/j.yexcr.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 62.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe . Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 63.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 65.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 66.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Den Berg DJ, Francke U. Roberts syndrome: a review of 100 cases and a new rating system for severity. Am J Med Genet. 1993;47:1104–1123. doi: 10.1002/ajmg.1320470735. [DOI] [PubMed] [Google Scholar]
- 68.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 69.Gordillo M, Vega H, Trainer AH, Hou F, Sakai N, Luque R, Kayserili H, Basaran S, Skovby F, Hennekam RC, Uzielli ML, Schnur RE, Manouvrier S, Chang S, Blair E, Hurst JA, Forzano F, Meins M, Simola KO, Raas-Rothschild A, Schultz RA, McDaniel LD, Ozono K, Inui K, Zou H, Jabs EW. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet. 2008;17:2172–2180. doi: 10.1093/hmg/ddn116. [DOI] [PubMed] [Google Scholar]
- 70.Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–3201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]
- 71.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 72.McNairn AJ, Gerton JL. Cohesinopathies: one ring, many obligations. Mutat Res. 2008;647:103–111. doi: 10.1016/j.mrfmmm.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 73.Liu J, Krantz ID. Cohesin and human disease. Annu Rev Genomics Hum Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae . Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 76.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lara-Pezzi E, Pezzi N, Prieto I, Barthelemy I, Carreiro C, Martínez A, Maldonado-Rodríguez A, López-Cabrera M, Barbero JL. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem. 2004;279:6553–6559. doi: 10.1074/jbc.M307663200. [DOI] [PubMed] [Google Scholar]
- 78.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 80.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 81.Rubio E-D, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 83.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 86.Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takagi S, Benard C, Pak J, Livingstone D, Hekimi S. Cellular and axonal migrations are misguided along both body axes in the maternal-effect mau-2 mutants of Caenorhabditis elegans . Development. 1997;124:5115–5126. doi: 10.1242/dev.124.24.5115. [DOI] [PubMed] [Google Scholar]
- 88.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:1411–1425. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horsfield JA, Anagnostou SH, Hu JK, Cho KHY, Geister R, Lieschke G, Crosier KE, Crosier PS. Cohesin dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 91.Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2008;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gregson CH, Schmiesing JA, Kim JS, Kobayashi SZ, Yokomori K. Potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- 93.Wong W, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc Natl Acad Sci USA. 2008;105:15441–15445. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY-HP, In Y (2002) SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev 16: 571-582 [DOI] [PMC free article] [PubMed]
- 95.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J Biol Chem. 2008;283:19176–19182. doi: 10.1074/jbc.M802299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shimizu K, Shirataki H, Honda T, Minami S, Takai Y. Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J Biol Chem. 1998;273:6591–6594. doi: 10.1074/jbc.273.12.6591. [DOI] [PubMed] [Google Scholar]
- 99.Guan J, Etwurtzel E, Kvist U, Yuan L. Cohesin protein SMC1 is a centrosomal protein. Biochem Biophys Res Commun. 2008;372:761–764. doi: 10.1016/j.bbrc.2008.05.120. [DOI] [PubMed] [Google Scholar]
- 100.Wang X, Yang Y, Dai W. Different subcellular localizations of two human Sgo1 isoforms. Cell Cycle. 2006;5:635–640. [PubMed] [Google Scholar]
- 101.Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skibbens RV. Mechanisms of sister chromatid pairing. Int Rev Cell Mol Biol. 2008;269:283–339. doi: 10.1016/S1937-6448(08)01005-8. [DOI] [PubMed] [Google Scholar]
- 103.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]