Abstract
Bet3, a transport protein particle component involved in vesicular trafficking, contains a hydrophobic tunnel occupied by a fatty acid linked to cysteine 68. We reported that Bet3 has a unique self-palmitoylating activity. Here we show that mutation of arginine 67 reduced self-palmitoylation of Bet3, but the effect was compensated by increasing the pH. Thus, arginine helps to deprotonate cysteine such that it could function as a nucleophile in the acylation reaction which is supported by the structural analysis of non-acylated Bet3. Using fluorescence spectroscopy we show that long-chain acyl-CoAs bind with micromolar affinity to Bet3, whereas shorter-chain acyl-CoAs do not interact. Mutants with a deleted acylation site or a blocked tunnel bind to Pal-CoA, only the latter with slightly reduced affinity. Bet3 contains three binding sites for Pal-CoA, but their number was reduced to two in the mutant with an obstructed tunnel, indicating that Bet3 contains binding sites on its surface.
Keywords: Golgi, Vesicular transport, Tethering, TRAPP, Bet3, Palmitoylation, Acyl-CoA
Introduction
Bet3 was identified to be a component of the multi-subunit transport protein particle (TRAPP) trafficking complex that plays an important role for the recruitment of ER-derived vesicles to the Golgi apparatus [1–5]. Two TRAPP complexes have been characterized that differ in size and localization within the Golgi and function [2]. Both TRAPP complexes facilitate nucleotide exchange on the small GTPase Ypt1 [6–8]. X-ray crystallography with human and mouse Bet3 revealed a novel protein fold and, surprisingly, a long-chain fatty acid covalently bound via a thioester linkage to the conserved residue cysteine 68. The carbon chain is not exposed at the surface of the molecule, but is completely buried inside a hydrophobic tunnel [9, 10]. Acylated Bet3 forms homodimers and also heterodimers with structurally related, but non-acylated TRAPP subunits, such as TPC6/Trs33 [11–14].
A variety of fatty acids may be attached to cysteine residues of proteins through an acylation reaction. S-acylation (also termed palmitoylation) may be dynamic with rapid cycles of acylation and deacylation, in contrast to other hydrophobic modifications, i.e., myristoylation or isoprenylation. Palmitoylation controls targeting of the modified protein to membranes or membrane subdomains, affects protein–protein interactions, and influences the stability of proteins [15]. Palmitoylation of proteins is also involved in intracellular transport. Pal-CoA, the lipid donor in the palmitoylation reaction, stimulates transport through the Golgi in vitro, supporting budding as well as fusion of vesicles. It was suggested that this stimulatory effect might be due to palmitoylation of a protein, which thereby gets activated to function in the transport reaction [16, 17].
Palmitoyl-CoA is capable of spontaneously S-acylating cysteine residues of several proteins and especially of peptides at their natural palmitoylation sites in vitro [18–25]. Most of these reactions are unlikely to operate inside cells. They are either too slow or require unphysiologically high pH values or Pal-CoA concentrations to account for palmitoylation in vivo.
The family of DHHC–CRD proteins (polytopic membrane proteins with the sequence DHHC and a cysteine-rich domain) has been identified as acyltransferases for a variety of substrate proteins in yeast and mammals [26]. However, Bet3 is unique in this respect since it has strong self-palmitoylating activity also in vivo. Incubation of purified Bet3 with [3H]-palmitoyl-CoA leads to a rapid and complete attachment of fatty acids to cysteine 68 [27]. This conclusion was supported by a study on the palmitoyl proteome of S. cerevisiae. Various DHHC proteins were deleted, and the effect on palmitoylation of individual proteins was determined. Only Bet3 showed a palmitoylation that was undiminished in any of the DHHC-mutant strains, strongly arguing in favor of an autocatalytic mode for its acylation [28].
The only mechanistic understanding of acylation reactions of proteins has been derived from structural studies on the N-myristoyl transferase (NMT), which attaches the rare C:14 carbon chain via an amide linkage to an N-terminal glycine residue. NMT binds almost exclusively myristoyl-CoA with nanomolar affinity, the binding site contains all the elements responsible for acyl chain-length measurement. Acyl transfer occurs through the nucleophilic addition–elimination reaction: an oxyanion hole formed by main-chain atoms of NMT polarizes the thioester carbonyl of Myr-CoA and stabilizes the transition state while deprotonation of the ammonium of the Gly acceptor is mediated by NMT’s C-terminal carboxylate [29].
Here we have characterized the self-palmitoylation reaction of Bet3 in more detail. We found that Bet3 has an inherent propensity to be autoacylated, which is fine-tuned by amino acids located in the vicinity of the acylation site. Especially the conserved arginine 67 enhances the acylation reaction, probably by facilitating deprotonation of cysteine 68 such that it could function as a nucleophile attacking the carbonyl of the C–S bond in Pal-CoA. Acyl-CoAs containing long-chain fatty acids bind with micromolar affinity to Bet3, whereas shorter-chain acyl-CoAs do not interact. Bet3 mutants with a blocked tunnel bind to Pal-CoA with only slightly reduced affinity and revealed two acyl-CoA binding sites instead of three calculated for wild-type Bet3. We conclude that Bet3 contains acyl-CoA binding sites on its surface which might be required to extract Pal-CoA from intracellular membranes. Our findings might be helpful to understand palmitoylation performed by DHHC proteins which are promising, but poorly characterized drug targets [26].
Materials and methods
Recombinant protein expression
For expression of GST fusion protein in E. coli, human Bet3 cDNA was cloned into the bacterial expression vector pGEX-4T1 (GE Healthcare, Munich, Germany). Site-directed mutagenesis was performed by overlap extension PCR [30]. E. coli BL21 (DE3) cells were transformed with the resulting plasmids. An overnight culture was inoculated in YT medium containing 100 μg/ml ampicillin. The preculture was diluted 20-fold in the same medium, grown to OD600 0.7 at 37°C, and induced with 1 μM isopropyl-β-D-thiogalactopyranoside. Growth was continued for 3 h, cells were harvested by centrifugation, and pellets were stored at −80°C. To obtain GST-Bet3, cells were lysed using a French press (SLM-AMINCO) in PBS (phosphate buffered saline) supplemented with 1 mM PMSF, 1 mM DTT and DNAse I (Roche). A cleared lysate obtained by centrifugation was loaded on 2 ml glutathione–Sepharose (Amersham) in a polypropylene column. The column was washed with PBS, protein was eluted with elution buffer (50 mM Tris pH 8, 10 mM GSH), and elution fractions were dialyzed against 25 mM Tris, pH 8.4, 120 mM NaCl.
To obtain Bet3 without GST-tag, the following procedure was used: In some experiments cells expressing GST-Bet3 were lysed by sonication in PBS supplemented with 1 mM PMSF, 1 mM DTT, and DNase I (Boehringer, Mannheim, Germany). A cleared lysate obtained by centrifugation was loaded on 5 ml of glutathione–Sepharose (GE Healthcare) in a polypropylene column. The column was washed with PBS. GST was cleaved by on-column incubation with 100 U thrombin overnight at room temperature. Protein was eluted with elution buffer (50 mM Tris, 1 M NaCl, pH 7.4) over a 1-ml HiTrap benzamidine column (GE Healthcare), to retain thrombin. Protein solutions were concentrated and the buffer exchanged to PBS with Amicon Ultra Centrifugal Filter devices (10,000 MWCO) (Millipore).
Palmitoylation assay
[3H]-Pal-CoA, prepared as described [22], was dried in a SpeedVac and dissolved in 20 mM Tris–HCl (pH 7.4), 120 mM NaCl, 0.05% Triton X-100 to yield a final concentration of 100,000 CPM/μl. The palmitoylation reaction included purified protein, GST-Bet3 wild-type and mutants (0.5–8 μl, 10 μg, 4 μM), 5 μl of [3H]-Pal-CoA (1 μM, 500,000 CPM), and 1 mM DTT in a final volume of 100 μl of PBS buffer and was incubated for 20 min at 30°C. One ml of chloroform/methanol (1:1) was added to the samples, and precipitated proteins were pelleted (15 min, 14,000 × g), washed, resuspended in 20 μl of non-reducing SDS-PAGE sample buffer, and boiled for 5 min. Samples were subjected to SDS-PAGE, staining with Coomassie, and fluorography for 5–48 h. The palmitoylation assays shown in Figs. 1, 2, 3, 5, 6, 7 were performed at least two times with similar results.
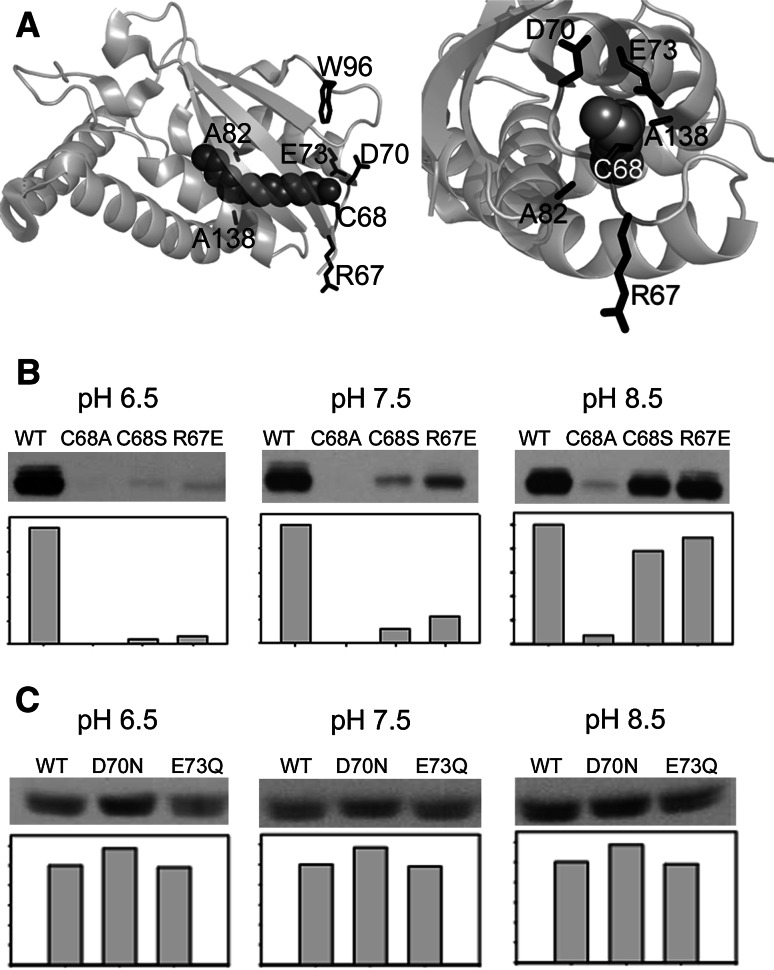
Fig. 1.
Testing the pH dependence of palmitoylation of Bet3. a Orthogonal views of the crystal structure of human Bet3: the covalently bound fatty acid inserted into the hydrophobic tunnel, amino acids mutated in this study and the single tryptophan (W 96) used for fluorescence spectroscopy are highlighted. b, c Comparison of the self-palmitoylating activity of Bet3 mutants relative to wild-type Bet3 at different pH values. b Equal amounts of purified wild-type GST-Bet3 (WT) and GST-Bet3 mutants containing amino-acid substitutions at cysteine 68 (C68S, C68A) or in the vicinity of the palmitoylation site (R67E) were incubated with [3H]-Pal-CoA in PIPES/NaCl at pH 6.5 or Tris/NaCl buffer at pH 7.5 or 8.5 for 20 min at 30°C. Samples were then subjected to SDS-PAGE and fluorography. Shown are the resulting fluorograms and the quantification of individual bands (wt = 100%) by densitometry. c Similar amounts of purified wild-type GST-Bet3 and GST-Bet3 mutants containing amino-acid substitutions at aspartate 70 (D70N) and glutamate 73 (E73Q) were incubated with Pal-CoA as described in b. Shown are the resulting fluorograms and the quantification of individual bands (D70N = 100%) by densitometry
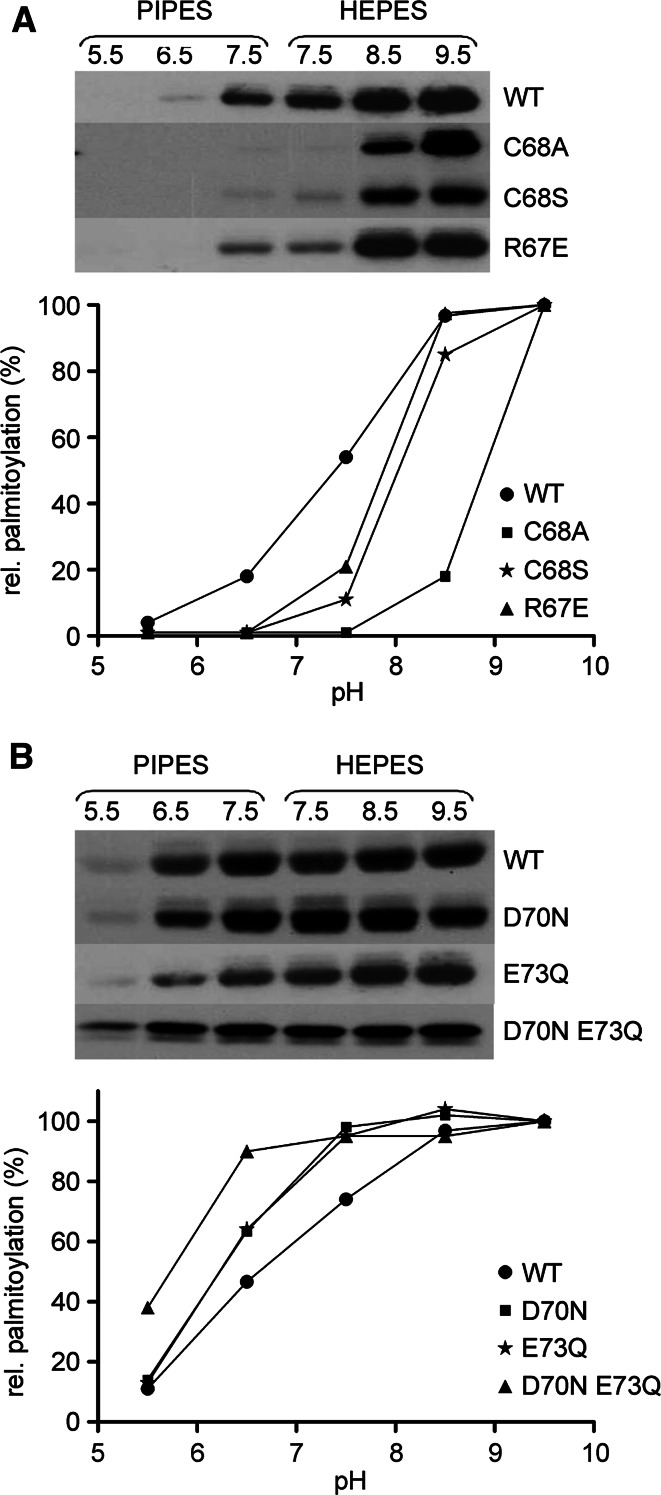
Fig. 2.
Comparison of the self-palmitoylating activity of Bet3 mutants at different pH values. a Wild-type GST-Bet3 (WT) and R67E, C68S, C68A mutants were incubated with [3H]-Pal-CoA in PIPES/NaCl buffer at pH 5.5, 6.5, or 7.5 or in Tris/NaCl buffer at pH 7.5, 8.5, or 9.5 for 20 min at 30°C. b Comparison of the self-palmitoylating activity of wild-type GST-Bet3 and GST-Bet3 E73Q, D70N, and the double mutant E73Q–D70N at different pH values as described in a. The fluorograms were quantified by densitometry of bands, and relative palmitoylation (absorbance of bands at pH 9.5 = 100%) is plotted against pH. Note that the strength of the self-palmitoylation reaction (intensity of bands) cannot be compared between the wild-type and mutant Bet3 proteins, since individual fluorograms were exposed for different time periods
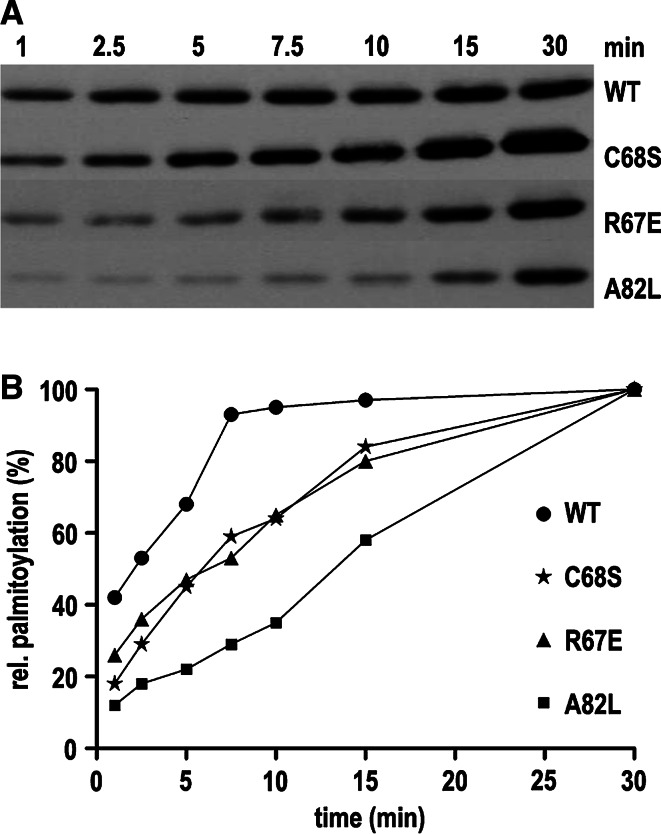
Fig. 3.
Self-palmitoylation of Bet3 is retarded in mutants that are less efficiently palmitoylated at neutral pH. Purified GST-Bet3 proteins, wild-type, and mutants, as described in Fig. 1, were incubated at pH 7.5 with [3H]-Pal-CoA for 1, 2.5, 5, 7.5, 10, 15, or 30 min at 30°C prior to SDS-PAGE and fluorography. The fluorograms were quantified by densitometry of bands, and relative palmitoylation (absorbance of bands after 30 min of incubation = 100%) is plotted against the time of incubation. Note that individual fluorograms were exposed for different time periods, and, thus, intensity of bands cannot be compared between wild-type and mutant Bet3 proteins
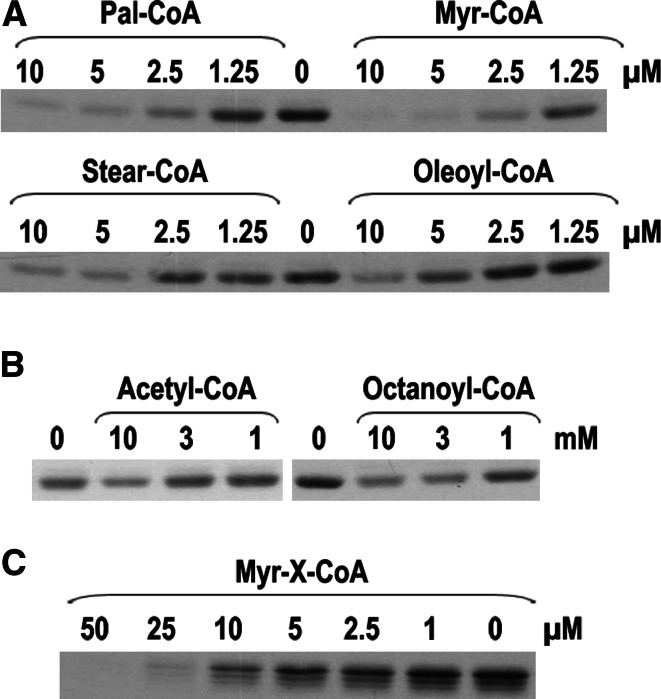
Fig. 5.
Long-chain acyl-CoAs inhibit self-palmitoylation of Bet3 at micromolar concentration. a Myristoyl-CoA, palmitoyl-CoA, stearoyl-CoA and 9-cis-oleoyl-CoA at the indicated final concentrations were incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C. The resulting fluorograms are shown. b Acetyl-CoA and octanoyl-CoA at the indicated final concentrations were incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C. Note that millimolar concentrations of these CoA derivatives were required to reveal a slight reduction in palmitoylation. c Myr-X-CoA (a non-hydrolyzable Myr-CoA analog with a thioether instead of a thioester linking myristate and CoA) at the indicated final concentration was incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C
Fig. 6.
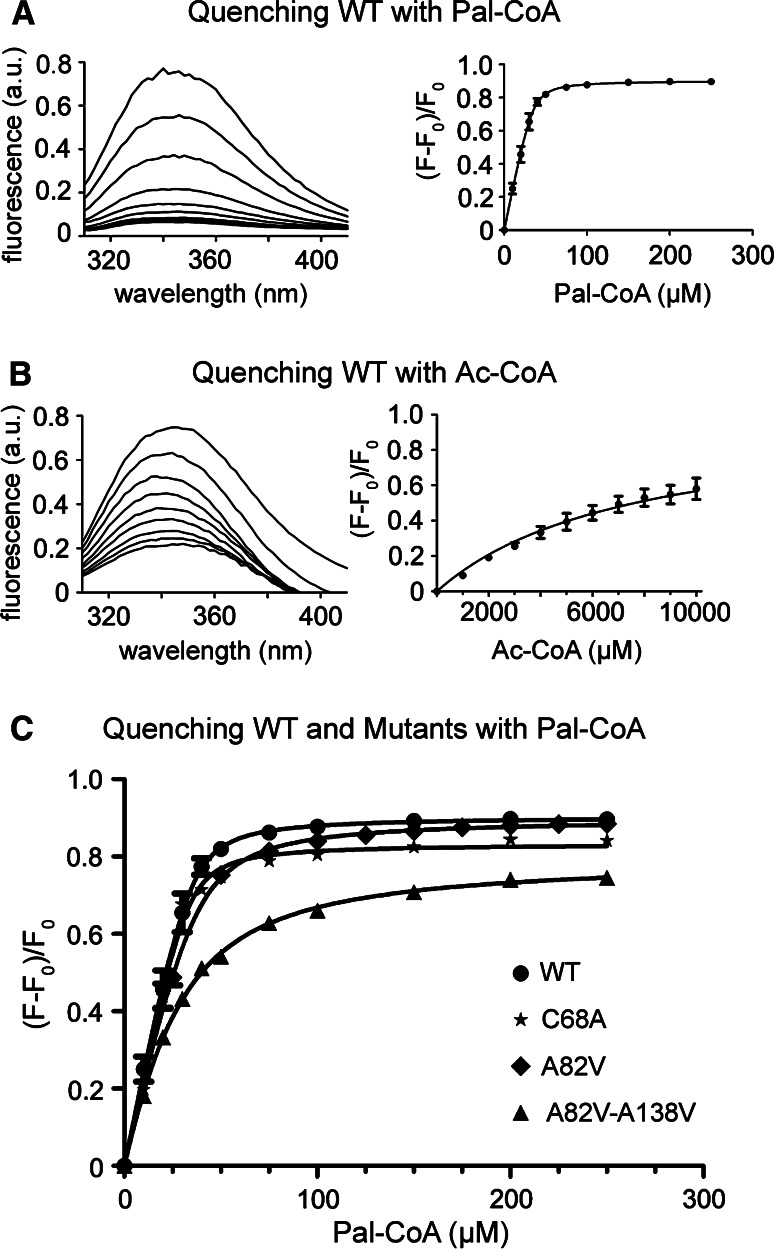
Fluorescence spectroscopy to analyze binding of Acyl-CoAs to Bet3. a Left panel Emission spectra of purified wild-type Bet3 between 310 and 410 nm. Excitation at 295 nm. The uppermost trace represents the intrinsic tryptophan fluorescence of Bet3 in the absence of acyl-CoA. It is successively quenched upon addition of palmitoyl-CoA which was added in steps of 10 μM (final concentration) to Bet3 (subsequent traces). No further reduction of the tryptophan fluorescence occurs at a final Pal-CoA concentration of 75 μM (undermost traces). Right panel Quantitative analysis of binding of Pal-CoA to wild-type Bet3 (described in c). Error bars indicate the standard deviation calculated from three experiments. b Left panel Emission spectra of purified Bet3: Similar to A, but acetyl-CoA was added to Bet3 in steps of 1 mM (final concentration). Note that the intrinsic tryptophan fluorescence is not completely quenched, even at a final concentration of 10 mM acetyl-CoA. Right panel Quantitative analysis of acetyl-CoA binding to wild-type Bet3 (described in c). Error bars indicate the standard deviation calculated from three experiments. c Quantitative analysis of binding of Pal-CoA to wild-type and mutant Bet3. Fitting of fluorescence intensities at the emission maximum (344 nm) were normalized [(F–F 0)/F 0, F 0 fluorescence intensity without ligand] and plotted against the Pal-CoA concentration. The data points were fitted and the number of acyl-CoA bindings sites listed in Tables 2 and 3 were calculated as described in the Materials and methods section. Error bars indicate the standard deviation calculated from three experiments. Where not indicated, error bars are not larger than the symbols
Fig. 7.
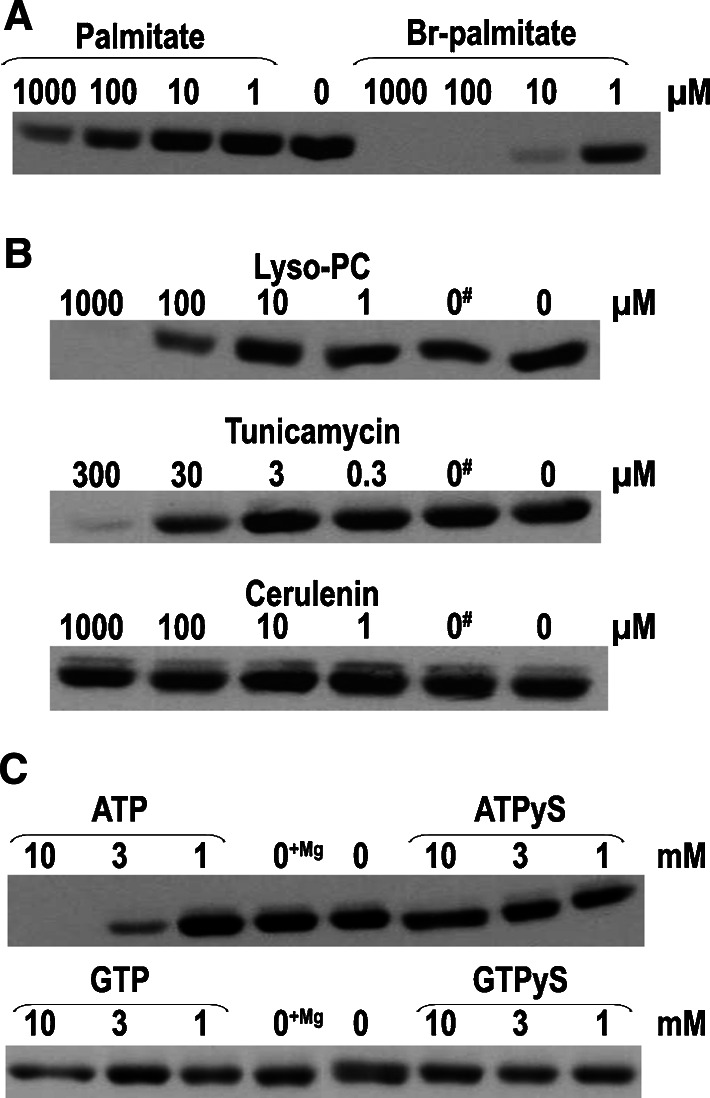
Testing inhibitors of the self-palmitoylation reaction. a Palmitate or 2-bromo-palmitate at the indicated final concentrations were incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C. b Lyso-phosphatidylcholine, cerulenin or tunicamycin at the indicated final concentrations were incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C. #: 1% chloroform, the solvent for the three inhibitors, was added to the sample. c The designated nucleotides at the indicated final concentrations and MgCl2 (1 mM) were incubated with wild-type GST-Bet3 and [3H]-Pal-CoA for 15 min at 30°C
Structure determination
The Trs33B:Bet3C68A complex was purified essentially as described before [13]. The protein was concentrated to 12 mg/ml in buffer containing 20 mM Tris, 200 mM NaCl, 2 mM DTT, pH 7.5 and 1 mM Myr-X-CoA, a non-hydrolysable myristoyl-CoA thioether. Crystals were obtained at 20°C by the sitting-drop method using a semi-automated dispensing system [31] in conditions containing 22% PEG3350, 0.2 M Li2SO4, 0.1 M Bis–Tris pH 6.5. Crystals were flash-frozen in liquid nitrogen in cryo solution containing 20% PEG3350, 15% glycerol, 0.2 M Li2SO4, 0.1 M BisTris pH 5.6, 250 μM Myr-X-CoA, and data were collected at ESRF BM14. Initial phases were obtained using Phaser [32] with the Trs33B:Bet3 complex (PDB: 2CFH) as a search model. After model bias removal (http://tuna.tamu.edu/), the final structure was obtained in several rounds of model building with Coot [33] and refinement using REFMAC5 [34]. The coordinates and diffraction amplitudes were deposited in the Protein Data Bank with accession code 3KXC.
Fluorescence spectroscopy
Purified Bet3 was diluted with Tris buffer (5 mM Tris, 100 mM NaCl, pH 7.4) to a concentration of 10 μM. Measurements were carried out with a Cary Eclipse Fluorescence spectrophotometer (Varian) at room temperature using a quartz glass cuvette (100 μl volume). Samples were excited at 295 nm, and the emission scanned in the range of 310–450 nm. Each scan was repeated five times, and scanned graphs were averaged. Emission maxima were read out at 344 nm. Emission of the protein was recorded and then the acyl-CoA concentration increased stepwise by adding 1 μl of stock solution (in Tris buffer, concentration 1–100 mM). The reduction of the fluorescence intensity was normalized (F 0–F/F 0; F 0 fluorescence intensity without ligand, F fluorescence intensity at a certain acyl-CoA concentration) and plotted against the acyl-CoA concentration. Taking substrate depletion into account, the following equation can be derived for acyl-CoA binding to Bet3 [35]:
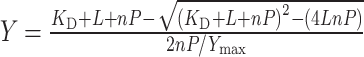 |
where K D denotes the apparent dissociation constant for acyl-CoA, L and P denote the ligand and protein concentration, respectively, n is the number of ligand binding sites per protein, and Y max is the final level of the relative fluorescence change. In all fits using Prism (GraphPad), protein concentration was fixed to 10 μM and K D, n and Y max were allowed to vary. The results shown were calculated from at least three different experiments.
Circular dichroism
For circular dichroism spectroscopy, protein samples of human Bet3 purified from E. coli were diluted to 0.16 mg/ml in PBS buffer. Measurements were carried out with a Jasco J720 spectropolarimeter at 20°C from 200 to 260 nm with a 0.2 cm path-length cuvette.
Results
Characterization of the mechanism of the Bet3 self-palmitoylation reaction
Insertion of the fatty acid of Pal-CoA into the hydrophobic tunnel, leaving the CoA moiety outside, would be a plausible mechanism to initiate self-palmitoylation of Bet3. This would position the thioester bond of the Pal-CoA molecule at the entrance of the tunnel where the fatty acid acceptor, cysteine 68, is located. Acyl-chain transfer then might occur through nucleophilic attack of the sulphur of cysteine 68 on the carbonyl-C of the C–S bond in Pal-CoA to catalyze the generation of acylated Bet3. It has been postulated that, besides availability of an activated fatty acid (acyl-CoA), the formation of a deprotonated cysteine (a thiolate) controls S-acylation [36]. Free cysteine has a pK a value of 8.5 so that deprotonation occurs infrequently at the neutral pH of the cell. However, the apparent pK a values may differ widely for individual cysteines present inside a peptide chain because the local environment strongly influences deprotonation of the SH group [37].
We first analyzed the effect of thiolates on self-palmitoylation of Bet3 by facilitating their formation using buffers with decreasing proton concentration. We compared wild-type GST-Bet3 and mutants with modifications of the acylation site. Cysteine 68 was changed to alanine (C68A), or to serine (C68S) which, due to its hydroxyl group retains a potential acylation site. The palmitoylation assays were performed with GST-Bet3 fusion proteins, since the GST tag provided stability and protection against proteolytical degradation. However, performing the assays with Bet3 alone produced identical results (not shown). Another mutant contained glutamic acid instead of the strictly conserved arginine in the vicinity of the acylation site (R67E). Figure 1a depicts the structure of Bet3 with the location of mutated amino acids highlighted.
Purified proteins were incubated with [3H]-Pal-CoA in buffer adjusted to pH 6.5, 7.5 or 8.5 for 20 min, and samples were subjected to SDS-PAGE and fluorography. At pH 6.5, only palmitoylation of wild-type Bet3 was readily detectable. The R67E mutant showed reduced acylation at neutral pH, probably by affecting deprotonation of cysteine 68. With increasing pH acylation of the mutants C68S and R67E becomes obvious, and at pH 8.5 it is almost as strong as the acylation of Bet3 wt (Fig. 1b). Even faint acylation of C68A becomes apparent at pH 8.5, probably representing background acylation occurring at other cysteine residues. Two additional cysteines are found in human Bet3, residue 31, which is partially surface-exposed, and residue 132, which is in the center of the protein so that unfolding at unphysiological pH would be a prerequisite for its palmitoylation.
Deprotonation of Cys 68 might be facilitated when amino acids in its vicinity serve as acceptor of the proton. Such a function has been proposed for the carboxylate present at the C-terminus of the NMT which accepts a proton from the designated acylation site in the protein, in this case the ammonium of an N-terminal glycine residue [29]. Likely candidates for a role as proton acceptors apparent from the structure of Bet3 are His 69, and the acidic amino acids Asp 70 and Glu 73 (Fig. 1a). In contrast to His 69, which is not present in yeast Bet3, Glu 73 (and especially Asp 70) are partially conserved within Bet3 orthologs and were therefore mutated to their respective amides. However, these Bet3 mutants were acylated at neutral pH to a similar (E73Q), or even slightly higher (D70N) extent as compared to Bet3 wt (Fig. 1c).
In the next set of experiments the self-palmitoylation activity of each Bet3 mutant was compared at various pH values between 5.5 and 9.5 (at more basic values the self-palmitoylation activity of Bet3 is inhibited [27]). Palmitoylation of wild-type Bet3 increased gradually up to pH 8.5, where it reached a plateau, indicating an apparent pKa of the reaction in the neutral range. The pH profiles of Bet3 mutants R67E, C68S and C68A are shifted toward basic pH, which is most evident for the C68A mutant (Fig. 2a). In contrast, the pH profiles of D70N and E73Q mutants are shifted toward acidic pH (Fig. 2b), an observation that was even more striking when both amino acids were exchanged simultaneously (D70N–E73Q). The results suggest a specific influence of the mutations on the apparent pK a value of the palmitoylation reaction.
Self-palmitoylation of Bet3 proceeds rapidly, i.e., incorporation of [3H]-palmitate saturates in less than 10 min (Fig. 3; [27]). In contrast, acylation of Bet3 mutants R67E, C68S and A82L, a mutant with an obstructed hydrophobic tunnel, advances much more slowly (Fig. 3), which suggests a reduced enzymatic activity.
Structure of the Trs33B:Bet3-C68A dimer crystallized in the presence of a non-hydrolysable acyl-CoA analogue
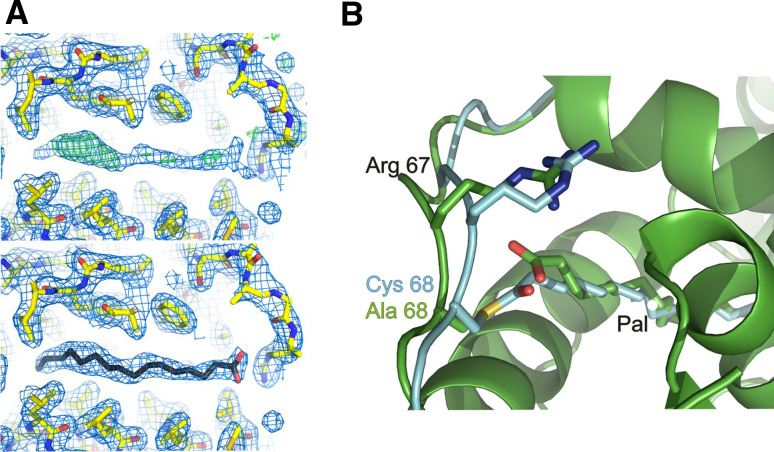
To provide a structural basis for the mechanism of Bet3 acylation we attempted the co-crystallization of the mutant in complex with non-hydrolysable myristoyl-CoA thioether (Myr-X-CoA) which binds to Bet3 as described below (see Fig. 5c). This protein–ligand complex should represent a reaction intermediate of Bet3 acylation. We determined the structure at 2-Å resolution using molecular replacement with the Trs33B:Bet3 complex (Table 1). To reduce model bias, the palmitoyl chain occupying the hydrophobic tunnel and the α2-α3 loop containing the palmitoylation site were excluded from the initial refinement, and the model was submitted to the model bias removal server (http://tuna.tamu.edu/). Surprisingly, Myr-X-CoA was not present in the crystal, which was confirmed by generating an anomalous difference map from a data set collected at low energy (1.7 Å): no sulphur signal from the ligand could be detected in the map (not shown). Nevertheless, the hydrophobic tunnel of Bet3 is occupied by an elongated density, which we found to be best accounted for by modeling a palmitate molecule (Fig. 4a). The palmitate was probably co-purified during the isolation of the protein from E. coli to stabilize the molecule [27]. Myr-X-CoA binds to Bet3 with higher affinity than palmitate (see Figs. 5, 7, below), but the additional CoA moiety might have interfered with the formation of crystal contacts, which may have led to the formation of crystals containing palmitate instead. Although we did not achieve our goal to crystallize a Bet3-ligand complex, the first structure of the Bet3 C68A mutant has peculiar properties that might be interesting with regard to the mechanism of the acylation reaction.
Table 1.
Data collection and refinement statistics for the structure of the Trs33B:Bet3 C68A complex
| X-ray diffraction data | |
|---|---|
| Space group | P212121 |
| Unit cell | |
| a [Å] | 47.17 |
| b [Å] | 69.26 |
| c [Å] | 121.38 |
| Resolution [Å] | 30–2.0 (2.1–2.0) |
| Unique reflections | 26,752 (3,311) |
| Redundancy | 4.05 (4.15) |
| <I/σ(I)> | 16.8 (3.0) |
| Completeness [%] | 94.9 (87.6) |
| R (sym) [%] | 6.3 (50.1) |
| Refinement | |
| Resolution [Å] | 25–2.0 |
| R(work)/R(free) [%] | 19.7/23.4 |
| RMSD bond distances [Å] | 0.009 |
| RMSD bond angles [°] | 1.117 |
| Mean B value [Å2] protein | 42.2 |
| Water (100) | 41.5 |
| Palmitate (1) | 56.4 |
| Ramachandran statistics | |
| Residues in most favored regions [%] | 97.3 |
| Residues in additionally allowed regions [%] | 2.7 |
Values in parentheses refer to the outer shell of reflections
Fig. 4.
X-ray structural analysis of the Trs33B:Bet3C68A mutant. a The hydrophobic tunnel of Bet3 C68A is occupied by an elongated molecule. A prominent, unaccounted electron density is visible after building the peptide chain of Bet3 (upper panel 2F 0–F c map, blue at 0.9 σ contour level and F 0–F c map, green at 3 σ contour level). Palmitate could be modeled into the hydrophobic channel (lower panel 2F 0–F c map, blue at 1.0 σ contour level). b Superposition of the α2-α3 loop region of mutant Bet3 C68A (green complex presented here) and wild-type Bet3 (cyan in the Trs33A:Bet3 complex, PDB: 2C0J) which were crystallized in the same space group
Compared to the structure of wild-type Bet3, the α2-α3 loop containing the palmitoylated residue Cys 68 as well as Arg 67 adopts a similar conformation in Bet3 C68A (Fig. 4b). Because there is no covalent bond to palmitate in the mutant structure, the entrance to the hydrophobic tunnel is left slightly wider open. In addition, the guanidino group of Arg 67 has rotated towards the carboxyl group of palmitate and towards the Cβ of Ala 68, which would represent the active center during the acylation reaction of wild-type Bet3. Because Cys 68 and Arg 67 are adjacent residues, their side chains are 7.2 Å apart, too far for a direct deprotonation of Cys 68. Thus, Arg 67 probably lowers the pK a of Cys 68 through electrostatic interactions and stabilizes the negative charge developing during thiolate formation and the acylation reaction.
Characterization of the substrate requirements of the palmitoylation reaction
Next we analyzed the chemical composition required for a lipid molecule to function as ligand of Bet3. Increasing concentrations of unlabeled acyl-CoAs were added to the in vitro palmitoylation reaction. Myr- (C 14:0), Pal- (C 16:0), Stear- (C 18:0) and Oleoyl-CoA (C 18:1) almost completely inhibited [3H]-palmitoylation of Bet3 at concentrations of around 10 μM (Fig. 5a), indicating that they compete with [3H]-Pal-CoA for a binding site. In contrast, only a slight inhibition of [3H]-palmitoylation of Bet3 was observed when 1,000-fold higher concentrations of acetyl- or octanoyl-CoA were used (Fig. 5b). It cannot be inferred from these experiments that all long-chain acyl-CoAs are actually transferred to Bet3. Since myristate was found as the prevailing fatty acid covalently linked to Bet3 purified from E. coli [9], it is likely that the other acyl-CoAs can also be utilized by Bet3, but their ability to react with Bet3 is not a prerequisite for binding of acyl-CoAs to Bet3: The Myr-CoA analogue Myr-X-CoA inhibited self-palmitoylation of Bet3 at only marginally diminished concentration compared to Myr-CoA (Fig. 5c).
For a quantitative assessment of Bet3 palmitoylation, we employed fluorescence spectroscopy to analyze binding of the substrate to the protein. Bet3 contains only one tryptophan residue at position 96, which is exposed at the surface of the molecule near the end of the hydrophobic tunnel (Fig. 1a). When excited at 295 nm, purified Bet3 (without GST) displayed a broad emission peak between 330 and 360 nm (uppermost trace in Fig. 6a). Adding increasing amounts of CoAs with long chain fatty acids, i.e., Myr-CoA, Pal-CoA, Stear-CoA, and Oleoyl-CoA to Bet3 quenched its intrinsic fluorescence gradually and yielded complete quenching at a final concentration in the lower micromolar range (shown exemplarily for Pal-CoA in Fig. 6a). In contrast, CoAs esterified to short carbon chains, i.e., acetyl-CoA (C 2:0, Fig. 6b) and octanoyl-CoA (C 8:0, not shown), did not completely quench the intrinsic fluorescence of Bet3, even at a final concentration of 10 mM. The reduction in fluorescence intensity was normalized and plotted against the acyl-CoA concentration (shown for palmitoyl- and acetyl-CoA in Fig. 6a, b, right panel). The data were fitted as described in “Materials and methods” with an equation taking into account ligand depletion upon substrate binding. From the fitting, the number of acyl-CoA-binding sites per protein and a value for the apparent dissociation constant for acyl-CoA (K D) (comprised of the individual K Ds of each acyl-CoA-binding site) were obtained (Tables 2, 3).
Table 2.
KD [μM] values including standard deviation for binding of various acyl-CoAs to wild-type Bet3 and to the non-acylated C68A mutant as calculated from at least three different fluorescence spectroscopy measurements
| Acyl-CoA | Bet3 wt | Bet3 C68A |
|---|---|---|
| Stearoyl-CoA | 1.3 ± 0.8 | 2.9 ± 0.8 |
| Oleoyl-CoA | 2.1 ± 0.6 | 1.1 ± 0.3 |
| Palmitoyl-CoA | 1.8 ± 0.7 | 1.6 ± 0.6 |
| Myristoyl-CoA | 2.0 ± 0.8 | 2.3 ± 1.6 |
| Octanoyl-CoA | 7,252 ± 1,412 | 11,256 ± 3,055 |
| Acetyl-CoA | 7,624 ± 2,110 | 7,321 ± 1,327 |
| 2-Br-Palmitate | 1.3 ± 0.5 | 1.9 ± 0.3 |
Table 3.
KD values [μM] including standard deviation and number (no.) of interaction sites for binding of Pal-CoA to wild-type and mutant Bet3 as calculated from at least three different fluorescence spectroscopy measurements
| Bet3 mutant | K D | No. of interaction sites |
|---|---|---|
| Wild type | 1.8 ± 0.7 | 3.5 ± 0.2 |
| C68A | 1.6 ± 0.6 | 3.3 ± 0.2 |
| A82 V | 4.0 ± 0.5 | 3.7 ± 0.1 |
| A82 V-A138 V | 15.8 ± 2.3 | 2.1 ± 0.4 |
When analyzing the most common acyl-CoAs-containing long-chain, saturated fatty acids, i.e., Myr-CoA, Pal-CoA, and Stear-CoA, the intrinsic fluorescence of Bet3 was almost completely quenched at concentrations of around 50 μM. K D values of 2.0 μM (Myr-CoA), 1.8 μM (Pal-CoA), and 1.3 μM (Stear-CoA) were calculated, indicating that they bound with similar affinity to Bet3 (see Table 2). Almost identical results (K D 2.1 μM) were observed for an unsaturated acyl-CoA (oleoyl-CoA), indicating that a kink in the fatty acid does not diminish binding to Bet3. In contrast, K D values in the millimolar range were calculated for CoAs containing short acyl chains (Fig. 6b; Table 2).
Does self-acylation of Bet3 require activation of the fatty acid substrate? Addition of [3H]-palmitate did not label Bet3, even when tenfold higher radioactivity compared to [3H]-Pal-CoA was used in the assay and when CoA and ATP were also present to possibly activate the fatty acid (data not shown). Free palmitate inhibited the palmitoylation reaction only at millimolar concentrations (Fig. 7a, left part). The other constituent of activated fatty acids, CoA, did not inhibit the self-palmitoylating activity of Bet3 either, even at concentrations of 10 mM (data not shown).
Thus, the requirements for a substrate to react with Bet3 are a long-chain fatty acid linked to CoA, neither the carbon chain nor CoA alone interacts with micromolar affinity. We also asked whether other fatty acid-containing compounds, especially those used as inhibitors of protein palmitoylation, interact with Bet3. Lyso-phosphatidylcholine and tunicamycin both contain a long acyl chain, either bound to glycerol or (via sugar moieties) to adenine, and both inhibit self-palmitoylation of Bet3, with a half maximal inhibition at a concentration of around 100 μM (Fig. 7b). In contrast, cerulenin, which contains an unsaturated, C8 carbon chain, does not inhibit the reaction, even at millimolar concentration. Cerulenin and tunicamycin are established inhibitors of fatty acid synthesis and N-linked glycosylation, respectively, but have also been used by some as inhibitors of protein palmitoylation [38]. Presently, the most popular inhibitor of protein palmitoylation is 2-bromo-palmitate [38], which also very efficiently blocks palmitoylation of Bet3 at low μM concentration (Fig. 7a) and binds to wild-type Bet3 with a K D value of 1.3 μM (Table 2).
The palmitoylation reaction catalyzed by the yeast DHHC protein Akr1p is stimulated by ATP [39]. Surprisingly, however, ATP at concentrations of 10 mM completely inhibited self-palmitoylation of Bet3. The phenomenon was specific for the adenine trinucleotide and required a hydrolyzable ATP, since GTP and ATPγs did not block the reaction (Fig. 7c).
Determination of the number of Pal-CoA interaction sites in Bet3 and molecular requirements for binding
We next investigated whether Bet3 mutants with blocked or diminished self-palmitoylation activity also have a reduced binding affinity for the substrate. First we asked whether covalent transfer of the fatty acids to Cys 68 is a prerequisite for binding of acyl-CoAs to Bet3. In the fluorescence spectroscopy assay, Bet3 C68A revealed very similar K D values for binding of various acyl-CoAs compared to wild-type Bet3 (Fig. 6c; Table 2). Furthermore, binding of the other long-chain acyl-CoAs was not altered either, and short-chain acyl-CoAs did not bind (Table 2). Thus, wild-type Bet3 and Bet3 C68A have identical affinities for the lipid substrate, indicating that binding and fatty acid transfer are distinct events.
Next we tested our initial assumption that insertion of the fatty acid moiety into the hydrophobic tunnel is a prerequisite for binding of Pal-CoA to Bet3. Surprisingly, measuring the affinity for Pal-CoA of the Bet3 mutant containing a narrowed tunnel (A82V) revealed a K D value of 4.0 μM, close to that of wild-type Bet3 (Fig. 6c; Table 3).
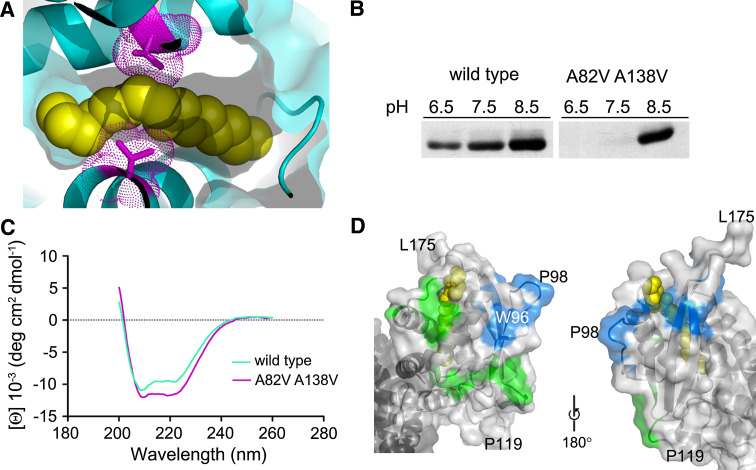
Since Bet3 A82V or A82L still shows residual self-palmitoylating activity at neutral pH [27], it is likely that the mutation does not sufficiently restrict the tunnel to completely exclude insertion of a fatty acid. In the crystal structure of the Bet3 A82V mutant, the protein indeed was found to be palmitoylated at cysteine 68, with an acyl chain occupying the hydrophobic tunnel (Kümmel/Heinemann, unpublished). We therefore engineered an additional mutation into Bet3 A82V. Alanine 138, which is located in the middle of the hydrophobic tunnel opposite to alanine 82, was replaced by the bulky amino acid valine (Fig. 8a) to completely bock the tunnel. In accordance with this consideration, self-palmitoylation of Bet3 A82V-A138V is completely blocked at neutral pH (Fig. 8b).
Fig. 8.
Testing a Bet3 mutant with a completely blocked hydrophobic tunnel. a Detail of the Bet3 crystal structure showing the hydrophobic tunnel. The fatty acid is depicted as yellow space-filling model. The amino acids mutated in A82V-A138V to block the hydrophobic tunnel are modeled purple with their electron densities indicated. The tunnel is narrowed by the two opposing valines (V138 top, V82 bottom). b Similar amounts of purified wild-type GST-Bet3 and GST-Bet3 A82V-A138V were incubated with [3H]-Pal-CoA in PIPES/NaCl at pH 6.5 or Tris/NaCl buffer at pH 7.5 or 8.5 for 20 min at 30°C. c CD spectra of wild-type Bet3 and Bet3 A82V-A138V indicate that the secondary structure of Bet3 was not changed significantly by the mutation. d Patches of hydrophobic amino acids at the surface of Bet3 that might bind the fatty acid of Acyl-CoAs. The green area contains the amino acids L60, A61, V65, G66, V77, V81, L87, G88, I89, I106, L107, P111, G159, V160, the blue area consists of F71, I93, W96, P98, A99, G100, A145, F167, I168, I171
Since lack of acylation increases the inherent instability of the Bet3 molecule [27], we compared the overall structure of Bet3 A82V-A138V with wild-type Bet3 using circular dichroism (CD) spectroscopy. However, the CD spectra for both molecules were similar, no indications for a random coil formation were detected (Fig. 8c), which indicates that the secondary structure of Bet3 was not changed significantly by the mutation.
Determination of the K D of the A82V-A138V mutant for Pal-CoA revealed a reduction in its affinity to values of 15.8 μM, indicating that it still binds to Pal-CoA with only marginally reduced affinity (Fig. 6c; Table 3). From the fluorescence spectroscopy assay and the model we used to fit the data, the concentration of bound Pal-CoA and the number of acyl-CoA binding sites per Bet3 monomer was calculated (Table 3). Whereas wild-type Bet3, non-acylated Bet3 C68A and Bet3 A82V apparently contain three binding sites per Bet3 monomer, their number was found to be reduced to two in the Bet3 mutant containing a completely blocked tunnel. This indicates that the hydrophobic tunnel is indeed a binding site for acyl-CoA, but that other sites with similar affinity exist as well, probably at the surface of the molecule.
Discussion
In this study we have analyzed binding of acyl-CoAs to Bet3 and investigated the chemical mechanism that allows transfer of the fatty acid from CoA to the acylation site of the protein, cysteine 68. The self-palmitoylating activity of wild-type Bet3 is enhanced by increasing the pH to slightly basic values (Figs. 1, 2) suggesting that palmitoylation requires the formation of a deprotonated cysteine. At unphysiologically high pH values (9.5), even the Bet3 mutant containing alanine at the original acylation site is palmitoylated (Fig. 2), indicating that Bet3 has an inherent propensity to be acylated. One factor that controls S-acylation of Bet3 is the pK a value at which thiolate formation of individual putative acylation sites occurs. The authentic acylation site of Bet3, cysteine 68, is apparently in an environment that allows deprotonation at neutral pH. Even a serine, that has a very high pK a value of 14 as free amino acid, is acylated at only slightly basic values (pH 8.5) when introduced at this position (Figs. 1, 2). Thus, amino acids in the vicinity of the acylation site facilitate deprotonation of Cys 68 and even of Ser 68. A candidate for such an activity is the strictly conserved arginine 67, which is required for efficient acylation of Bet3 at neutral pH (Fig. 1). The functional defect of Bet3 R67E can be complemented by increasing the pH to 8.5. Furthermore, the crystal structure of Bet3 C68A shows that the guanidino group of Arg 67 has rotated towards the acylation site (Fig. 4) as compared to a previously published structure of wild-type Bet3 crystallized in the same space group [11]. The subtle structural change can be attributed to the fact that the palmitate ligand is negatively charged. This probably resembles charged reaction intermediates, in contrast to uncharged, covalently linked palmitate in the wild-type structure. These observations are also consistent with the assumption that Arg 67 stabilizes deprotonation of Cys 68. Alternatively, arginine 67 may polarize the carbonyl bond in the thioester linkage of Pal-CoA or stabilize the negative charge of a putative tetrahedral reaction intermediate.
The moderately conserved amino acids Asp 70 and Glu 73 apparently do not act as acceptor for protons released by thiolate formation of Cys 68. In contrast to Arg 67, their exchange did not diminish self-palmitoylation of Bet3 and did not cause a shift in the pH optimum of the reaction to basic values (Figs. 1, 2). Instead, we observed increased acylation at acidic pH, indicating that both amino acids might prevent acylation of Bet3 at unphysiologically low pH values. Thus, the inherent propensity of Bet3 to be autoacylated is fine-tuned by the amino acids Asp 70, Glu 73, and especially by Arg 67 such that it occurs more rapidly (Fig. 3) and at neutral pH, and therefore at conditions which allow self-palmitoylation of Bet3 inside cells.
The second factor that controls efficient self-palmitoylation of Bet3 is enrichment of acyl-CoAs in the vicinity of putative acylation sites. The affinity of Bet3 for binding of Pal-CoA is relatively modest (micromolar, Fig. 6; Table 2) compared to e.g., the NMT, which binds myristoyl-CoA with nanomolar affinity. However, the affinity of Bet3 for acyl-CoAs is approximately one order of magnitude higher compared to Pal-CoA concentrations inside cells (5–150 μM) [40]. Bet3 binds the most common long-chain acyl-CoAs (and also palmitoylation inhibitors with long carbon chains), but CoA alone or CoAs esterified to short carbon chains do not interact with Bet3 (Figs. 5, 6, 7). The binding properties of Bet3 thus reflect the fatty acid composition of most S-acylated proteins, which contain a variety of long-chain fatty acids [15]. Since wild-type Bet3 and Bet3 C68A have identical affinities for their lipid substrate (Table 2), stable binding of acyl-CoAs does not require fatty acid transfer to the acylation site.
Contrary to our initial assumption, Bet3 mutants with a blocked tunnel bound to Pal-CoA with only slightly reduced affinity and revealed two acyl-CoA binding sites instead of three calculated for wild-type Bet3 (Table 3). This indicates that the hydrophobic tunnel is indeed a binding site for acyl-CoAs, but that other sites with similar affinity exist as well, probably at the surface of the molecule. Displaying the hydrophobic interaction potential of Bet3 revealed two continuous regions on its surface, which might interact with acyl chains (depicted green and blue in Fig. 8d).
Do these presumed acyl-CoA binding sites at the surface of the molecule play a role for self-palmitoylation of Bet3 inside cells? Since the cytosolic acyl-CoA-binding protein (ACBP) binds its ligand with nanomolar affinity, virtually every cytosolic acyl-CoA molecule exists in a complex with ACBP [40]. Thus, free acyl-CoAs are available for Bet3 only at intracellular membranes, such as the ER and the Golgi apparatus, where they are needed for the synthesis of phospholipids and also for DHHC-catalyzed S-acylation of proteins. Bet3 has an intrinsic membrane binding activity, which targets the protein to the same intracellular location [9, 10]. Bet3 probably does not penetrate deeply into the bilayer but resides at the membrane surface. If the interacting capacity of Bet3 for acyl-CoAs were confined to the hydrophobic tunnel, it would be difficult for the molecule to interact with acyl-CoAs fatty acid, which is embedded in the hydrophobic interior of the bilayer. In contrast, a high-affinity acyl-CoA-binding site on the surface of Bet3 might be more suitable to facilitate interaction with and extraction of acyl-CoAs from the membrane.
How is it conceivable that an acyl-CoA molecule bound to the surface of Bet3 is inserted into the tunnel and its fatty acid transferred to cysteine 68? We have previously proposed a molecular model for the extrusion of the fatty acid from the hydrophobic tunnel of Bet3 [10]. The mechanism involves a rotation of approximately 30° in the kinked helix α2 about a hinge region close to residue Gly 52, coupled with a maximal displacement of approximately 9 Å that would open the hydrophobic tunnel. Although purely speculative, it is possible that non-acylated Bet3 contains an open tunnel. This state will be energetically highly unfavorable. Binding of acyl-CoAs to the surface of Bet3 might trigger a conformational change that allows helix α2 to rotate back to its original position whereby the fatty acid of an acyl-CoA molecule would be inserted into the hydrophobic tunnel. Narrowing or blocking the tunnel by introduction of bulky amino acids reduces acylation of Bet3, probably because the conformational change that causes fatty acid insertion is hindered. The conformational change might also be sensitive to the pH, since an extended hydrogen-bonding pattern seen in the Bet3 structure within the putative hinge region must be modulated to allow movement of helix α2. This assumption would explain how a partial restriction of the tunnel can be complemented by a slightly basic pH during the palmitoylation reaction. Insertion of acyl-CoA into the tunnel is a prerequisite for acylation, since Bet3 containing a completely blocked tunnel is not acylated (Fig. 8).
As we have already pointed out [10], there is some structural similarity between the palmitate-binding α-helical face of Bet3 and the non-specific lipid transfer proteins (LTPs) from plants and mammals [41–43]. LTPs bind a variety of lipids, including Pal-CoA, with dissociation constants in the micromolar range. A further similarity to Bet3 is their lipid-binding site, a hydrophobic tunnel extending through the protein and bending slightly in the middle. The hydrocarbon chain of each fatty acid is buried in the cavity, and the carboxyl group is localized at the aqueous interface in contact with surface residues and water molecules. In barley LTP, the binding pocket can contract or expand to accommodate fatty acids of various sizes. These conformational changes are accompanied by a bending motion in helix αA and structural adjustments at the C-terminus and at helix αC, which are similar to those proposed above for Bet3. It is thus conceivable that Bet3 is evolutionarily related to an LTP. By mutational acquisition of a potential acylation site at the entrance of the tunnel and of amino acids that facilitate its deprotonation, Bet3 has gained a self-palmitoylation activity that is used to stabilize the structure of the molecule. Other subunits of TRAPP exhibiting the same fold, such as TPC5/Trs31p and TPC6/Trs33p, do not contain an acylation site. Self-palmitoylation is lost, and the collapse of the molecule onto an empty cavity is prevented by bulky hydrophobic side chains that occupy the tunnel accommodating palmitate in Bet3.
In accordance with this assumption is our observation that the hydrophobic tunnel of the Trs33B:Bet3-C68A dimer is occupied by non covalently bound palmitate (Fig. 4). Apparently, Bet3, which can not stabilize its structure by self-palmitoylation of cysteine 68, nevertheless inserts a fatty acid into the tunnel. Non-covalently bound palmitate might also be present in purified wild-type Bet3, but this moiety can be exchanged by Pal-CoA, for which Bet3 has a much higher affinity, in order for self-palmitoylation to occur.
In summary, our results suggest the following mechanism mediating self-palmitoylation of Bet3. Acyl-CoA binds to the surface of the molecule, and its fatty acid is subsequently inserted into the hydrophobic tunnel. The thioester bond of the Pal-CoA molecule is now situated at the entrance of the tunnel where the fatty acid acceptor, cysteine 68, is located. Facilitated by amino acids in its vicinity, especially by arginine 67, cysteine 68 is deprotonated. The resulting thiolate functions as a nucleophile attacking the carbonyl-C of the C–S bond in Pal-CoA. The fatty acid is then attached to cysteine 68, and CoA is released from Bet3. Since an acylated Bet3 molecule is no longer available for a second round of catalysis, this mechanism is different from a classical enzymatic reaction. However, the speed of the reaction in vitro, its pH and Pal-CoA requirements and the observation that Bet3 does not require functional DHHC proteins for its acylation inside cells [28], distinguishes self-palmitoylation of Bet3 from purely chemical acylation reactions described previously [19–25].
Acknowledgments
This work was funded by the DFG (SFB 740, TP C1) and supported by the Fonds der Chemischen Industrie. We thank Ingrid Poese for experimental assistance and the staff at ESRF BM14 for excellent beamline support.
Footnotes
D. Kümmel and J. Walter contributed equally to this work.
References
- 1.Barrowman J, Sacher M, Ferro-Novick S. TRAPP stably associates with the Golgi and is required for vesicle docking. EMBO J. 2000;19:862–869. doi: 10.1093/emboj/19.5.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/S1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 3.Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, III, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 5.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. The TRAPP complex: insights into its architecture and function. Traffic. 2008;9:2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S, Reinisch KM. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YG, Sohn EJ, Seo J, Lee KJ, Lee HS, Hwang I, Whiteway M, Sacher M, Oh BH. Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP. Nat Struct Mol Biol. 2005;12:38–45. doi: 10.1038/nsmb871. [DOI] [PubMed] [Google Scholar]
- 10.Turnbull AP, Kummel D, Prinz B, Holz C, Schultchen J, Lang C, Niesen FH, Hofmann KP, Delbruck H, Behlke J, Muller EC, Jarosch E, Sommer T, Heinemann U. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. EMBO J. 2005;24:875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MS, Yi MJ, Lee KH, Wagner J, Munger C, Kim YG, Whiteway M, Cygler M, Oh BH, Sacher M. Biochemical and crystallographic studies reveal a specific interaction between TRAPP subunits Trs33p and Bet3p. Traffic. 2005;6:1183–1195. doi: 10.1111/j.1600-0854.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Kummel D, Muller JJ, Roske Y, Henke N, Heinemann U. Structure of the Bet3-Tpc6B core of TRAPP: two Tpc6 paralogs form trimeric complexes with Bet3 and Mum2. J Mol Biol. 2006;361:22–32. doi: 10.1016/j.jmb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Kummel D, Muller JJ, Roske Y, Misselwitz R, Bussow K, Heinemann U. The structure of the TRAPP subunit TPC6 suggests a model for a TRAPP subcomplex. EMBO Rep. 2005;6:787–793. doi: 10.1038/sj.embor.7400463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 16.Pfanner N, Glick BS, Arden SR, Rothman JE. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. J Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfanner N, Orci L, Glick BS, Amherdt M, Arden SR, Malhotra V, Rothman JE. Fatty acyl-coenzyme A is required for budding of transport vesicles from Golgi cisternae. Cell. 1989;59:95–102. doi: 10.1016/0092-8674(89)90872-6. [DOI] [PubMed] [Google Scholar]
- 18.Quesnel S, Silvius JR. Cysteine-containing peptide sequences exhibit facile uncatalyzed transacylation and acyl-CoA-dependent acylation at the lipid bilayer interface. Biochemistry. 1994;33:13340–13348. doi: 10.1021/bi00249a021. [DOI] [PubMed] [Google Scholar]
- 19.Belanger C, Ansanay H, Qanbar R, Bouvier M. Primary sequence requirements for S-acylation of beta(2)-adrenergic receptor peptides. FEBS Lett. 2001;499:59–64. doi: 10.1016/S0014-5793(01)02513-3. [DOI] [PubMed] [Google Scholar]
- 20.Bizzozero OA, Bixler HA, Pastuszyn A. Structural determinants influencing the reaction of cysteine-containing peptides with palmitoyl-coenzyme A and other thioesters. Biochim Biophys Acta. 2001;1545:278–288. doi: 10.1016/s0167-4838(00)00291-0. [DOI] [PubMed] [Google Scholar]
- 21.Duncan JA, Gilman AG. Autoacylation of G protein alpha subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 22.Veit M. Palmitoylation of the 25-kDa synaptosomal protein (SNAP-25) in vitro occurs in the absence of an enzyme, but is stimulated by binding to syntaxin. Biochem J. 2000;345(Pt 1):145–151. doi: 10.1042/0264-6021:3450145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veit M, Sachs K, Heckelmann M, Maretzki D, Hofmann KP, Schmidt MF. Palmitoylation of rhodopsin with S-protein acyltransferase: enzyme catalyzed reaction versus autocatalytic acylation. Biochim Biophys Acta. 1998;1394:90–98. doi: 10.1016/s0005-2760(98)00097-6. [DOI] [PubMed] [Google Scholar]
- 24.Bano MC, Jackson CS, Magee AI. Pseudo-enzymatic S-acylation of a myristoylated yes protein tyrosine kinase peptide in vitro may reflect non-enzymatic S-acylation in vivo. Biochem J. 1998;330(Pt 2):723–731. doi: 10.1042/bj3300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bizzozero OA, McGarry JF, Lees MB. Autoacylation of myelin proteolipid protein with acyl coenzyme A. J Biol Chem. 1987;262:13550–13557. [PubMed] [Google Scholar]
- 26.Tsutsumi R, Fukata Y, Fukata M. Discovery of protein-palmitoylating enzymes. Pflugers Arch. 2008;456:1199–1206. doi: 10.1007/s00424-008-0465-x. [DOI] [PubMed] [Google Scholar]
- 27.Kummel D, Heinemann U, Veit M. Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc Natl Acad Sci USA. 2006;103:12701–12706. doi: 10.1073/pnas.0603513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, III, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatnagar RS, Futterer K, Waksman G, Gordon JI. The structure of myristoyl-CoA:protein N-myristoyl transferase. Biochim Biophys Acta. 1999;1441:162–172. doi: 10.1016/s1388-1981(99)00155-9. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 31.Heinemann U, Bussow K, Mueller U, Umbach P. Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc Chem Res. 2003;36:157–163. doi: 10.1021/ar010129t. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Heck M, Hofmann KP. G-protein-effector coupling: a real-time light-scattering assay for transducin-phosphodiesterase interaction. Biochemistry. 1993;32:8220–8227. doi: 10.1021/bi00083a024. [DOI] [PubMed] [Google Scholar]
- 36.Dietrich LE, Ungermann C. On the mechanism of protein palmitoylation. EMBO Rep. 2004;5:1053–1057. doi: 10.1038/sj.embor.7400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resh MD. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton JA. Fatty acid interactions with proteins: what X-ray crystal and NMR solution structures tell us. Prog Lipid Res. 2004;43:177–199. doi: 10.1016/j.plipres.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Lerche MH, Kragelund BB, Bech LM, Poulsen FM. Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure. 1997;5:291–306. doi: 10.1016/S0969-2126(97)00186-X. [DOI] [PubMed] [Google Scholar]
- 43.Lerche MH, Poulsen FM. Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins. Protein Sci. 1998;7:2490–2498. doi: 10.1002/pro.5560071202. [DOI] [PMC free article] [PubMed] [Google Scholar]