Abstract
β-Glucosidases (3.2.1.21) are found in all domains of living organisms, where they play essential roles in the removal of nonreducing terminal glucosyl residues from saccharides and glycosides. β-Glucosidases function in glycolipid and exogenous glycoside metabolism in animals, defense, cell wall lignification, cell wall β-glucan turnover, phytohormone activation, and release of aromatic compounds in plants, and biomass conversion in microorganisms. These functions lead to many agricultural and industrial applications. β-Glucosidases have been classified into glycoside hydrolase (GH) families GH1, GH3, GH5, GH9, and GH30, based on their amino acid sequences, while other β-glucosidases remain to be classified. The GH1, GH5, and GH30 β-glucosidases fall in GH Clan A, which consists of proteins with (β/α)8-barrel structures. In contrast, the active site of GH3 enzymes comprises two domains, while GH9 enzymes have (α/α)6 barrel structures. The mechanism by which GH1 enzymes recognize and hydrolyze substrates with different specificities remains an area of intense study.
Keywords: Biological function, Structure, Substrate-specificity, Glycoside hydrolase, Glycosides, Structure–function relationships
Introduction
Beta-glucosidases (β-d-glucopyranoside glucohydrolases, E.C. 3.2.1.21) are enzymes that hydrolyze glycosidic bonds to release nonreducing terminal glucosyl residues from glycosides and oligosaccharides. These enzymes are found universally, in all domains of living organisms, Archaea, Eubacteria, and Eukaryotes, in which they play a variety of functions. These include biomass conversion in microorganisms, breakdown of glycolipids and exogenous glucosides in animals, and lignification, catabolism of cell wall oligosaccharides, defense, phytohormone conjugate activation, and scent release in plants, as well as both sides of plant–microbe and plant–insect interactions.
Although the definition of β-glucosidases is straightforward, the abundance of nonreducing terminal β-linked d-glucosyl residues in nature, some examples of which are shown in Fig. 1, has led to the assignment of many E.C. numbers for enzymes that hydrolyze their glycosidic bond. Among these enzymes are glucosyl ceramidases or glucocerebrosidases (3.2.1.45), glucan 1,4-β-glucosidases (3.2.1.58), glucan 1,3-β-glucosidases (3.2.1.74), steryl-β-glucosidase (3.2.1.104), strictosidine β-glucosidase (3.2.1.105), amygdalin hydrolase (3.2.1.117), prunasin hydrolase (3.2.1.118), vicianin β-glucosidase (3.2.1.119), raucaffricine β-glucosidase (3.2.1.125), and coniferin β-glucosidase (3.2.1.126). In addition, β-glucosidases often exhibit additional activities, such as β-d-fucosidase (3.2.1.38), β-d-galactosidase (3.2.1.23), β-d-mannosidase (3.2.1.25), and β-disaccharidase activities, such as β-apiosyl-β-d-glucosidase (3.2.1.161).
Fig. 1.
Structures of example β-glucosidase substrates. The plant cyanogenic glucosides linamarin, dhurrin, prunasin, and its precursor amygdalin. Other defense-related glycosides include 2-O-β-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOAGlc) and the flavonoids apigenin 7-O-β-d-glucoside, the isoflavonoids diadzin and genistin, and phloridzin. Coniferin and coumaryl alcohol represent monolignol β-glucosides, while abscissic acid glucosyl ester is a phytohormone glucoconjugate and salicin and indoxyl β-glucoside are other plant glycosides with similarity to phytohormones. Strictosidine is the metabolic precursor to a wide array of monoterpene alkaloids. Cellobiose and laminaribiose represent plant cell-wall-derived oligosaccharides and can be extended with the same linkage to give the corresponding triose, tetraose, etc. In the lower right is an example of a glucosyl ceramide, one of the substrates for human acid β-glucosidase (GBA1) and other mammalian β-glucosidases
Since the name and the corresponding E.C. number(s) tell us little about mechanism of action, structure, and relationship to other glycoside hydrolases, and one enzyme may catalyze hydrolysis of several related substrates, Henrissat developed an alternative classification system for glycoside hydrolases based on amino acid sequence and structural similarity [1–3]. In this system, those enzymes with overall amino acid sequence similarity and well-conserved sequence motifs are grouped into the same family. At this writing, 115 glycoside hydrolase families are listed in the frequently updated Carbohydrate Active enZYme (CAZY) Web site (http://www.cazy.org) [3]. The β-glucosidases that have been described in the literature fall in glycoside hydrolase families GH1, GH3, GH5, GH9, and GH30, [1, 3–5]. In addition, the human bile acid β-glucosidase/GBA2 glucocerebrosidase and its relatives are yet to be assigned to a family. The families that have similar catalytic domain structures and conserved catalytic amino acids, suggestive of a common ancestry and catalytic mechanism, are grouped into clans [2, 3]. Of these, clan GH-A has the largest number of families, and it includes the β-glucosidase-containing families GH1, GH5, and GH30.
GH1 includes the largest number of characterized β-glucosidases; therefore, it will be the emphasis of this review, with the other β-glucosidase families receiving brief mention. We will consider the roles of β-glucosidases and related enzymes in animals, in plants, and in microorganisms, followed by the application of these enzymes. Then, we will describe the structure, mechanism, and the resulting general properties of β-glucosidases that have been purified, and end with a perspective on what is known and needs for further study.
β-Glucosidases and their functional roles
Roles of β-glucosidases and their relatives in mammals
Mammals contain several β-glucosidases, including the family GH1 lactase-phloridzin hydrolase and cytoplasmic β-glucosidase, the GH30 human acid β-glucosidase (GBA1) and the bile acid β-glucosidase or GBA2. These enzymes are thought to play roles in metabolism of glycolipids and dietary glucosides. In addition, a group of related family GH1 proteins is thought to play signaling functions.
Perhaps the best-studied mammalian β-glucosidase is the human acid β-glucosidase, which is generally considered a glucosyl ceramidase. Defects in the function of this enzyme and its transport to the lysosome lead to Gaucher disease, in which glycoceramides accumulate in the lysosomes of tissue leukocytes leading to their engorgement and buildup in the tissues [6]. Since enzyme replacement therapy is one way of alleviating the symptoms for this disease, the enzyme has been produced in recombinant mammalian and insect cells, and gene-activated human cells, and the structures of the enzymes determined [7–9]. Other means of treatment for Gaucher disease include use of imino sugars like deoxynojirimycin and isofagomine and their hydrophobic derivatives, which may inhibit synthesis of glycoceramides, but also bind to mutant GBA1 in the ER and help it to fold properly for transport to the lysosome [6]. However, these inhibitors also inhibit other β-glucosidases, such as GBA2 [10, 11].
A human bile acid β-glucosidase (GBA2) was found associated with liver microsomes [12]. Immunofluorescence showed a perinuclear reticulated localization [10], consistent with its earlier assignment to the endoplasmic reticulum (ER), while over-expressed green fluorescent protein tagged enzyme was localized near the plasma membrane [11]. When the bile acid β-glucosidase gene was knocked out in mice, little effect was seen on bile acid metabolism, but there was an accumulation of glucoceramides in the Seritoli cells of the testes, leading to round-headed sperm and decreased fertility [10]. Cells transfected with the gene showed an increased ability to hydrolyze fluorescent glucoceramides, confirming the protein's identity as GBA2, the nonlysosomal glycoceramidase [10, 11].
Currently, five GH1 proteins are known in humans: lactase-phloridzin hydrolase (LPH), cytoplasmic β-glucosidase, Klotho (α-Klotho, KL) β-Klotho (β-KL), and Klotho-LPH-related protein (KLPH) [13]. LPH, an intestinal hydrolase involved in food digestion, has both β-glucosidase activity toward exogenous glucosides, such as phloridzin, and β-galactosidase activity toward lactose. The precursor protein consists of four GH1 domains, the first two of which are removed during maturation, leaving a type I membrane protein with the LPH3 and LPH4 domains bound to the intestinal epithelial cells by a C-terminal transmembrane segment [14]. Flavonoid glucosides appear to be hydrolyzed at both the LPH3 active site and the lactase active site in the LPH4 domain [15, 16]. Deficiency of this enzyme leads to lactose intolerance, one of the most common enzyme deficiencies in humans.
The broad specificity cytoplasmic β-glucosidase has been studied for 30 years [17], and it has recently been given the additional name Klotho-related protein [18]. The cytoplasmic β-glucosidase is found in high levels in hepatocytes and brush border epithelial cells, and it has been shown to hydrolyze plant-derived flavonoid glucosides with high efficiency [19, 20], as shown in Table 1. Recently, the cytoplasmic β-glucosidase was shown to partially account for residual hydrolysis of glucoceramides and galactoceramides in fibroblasts treated with conduritol β-epoxide (CBE), a potent inhibitor of human GBA1 [18]. Although the recombinant enzyme produced in Escherichia coli showed extremely slow hydrolysis of natural glycoceramides (Table 1), its structure was solved and found to include lipids in positions suggestive of a glycoceramide binding site and its k cat /K m values for fluorescently labeled glycoceramides were comparable to those of flavonoid glycosides. The structure of the human cytoplasmic β-glucosidase expressed in Pichia pastoris was solved independently, and the residues likely to be involved in its binding of quercetin 4′-O-glucoside were identified by molecular docking and site-directed mutagenesis [13]. No lipids were observed in the active site of the enzyme produced in P. pastoris.
Table 1.
Substrate specificity of human cytoplasmic β-glucosidase: substrate K m and apparent k cat values
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Reference |
|---|---|---|---|---|
| Artificial aryl glycosides | ||||
| pNP-β-d-Glc | 1,800 | 12 | 6.9 | Berrin [20] |
| pNP-β-d-Fuc | 370 | 11 | 29 | Berrin [20] |
| pNP-α-l-Ara | 570 | 6.0 | 10 | Berrin [20] |
| pNP-β-d-Gal | 3,100 | 18 | 5.6 | Berrin [20] |
| Flavonoids and isoflavonoids | ||||
| Quercitin 4′-glucoside | 34 | ND | Day [19] | |
| Quercetin 4′-glucoside | 32 | 1.1 | 34 | Berrin [20] |
| Quercetin 7-glucoside | 42 | 0.7 | 16 | Berrin [20] |
| Apigenin 7-glucoside | 22 | 1.5 | 71 | Berrin [20] |
| Luteolin 4′-glucoside | 10 | 1.2 | 117 | Berrin [20] |
| Luteolin 7-glucoside | 50 | 3.0 | 61 | Berrin [20] |
| Genistin | 13 | ND | Day [19] | |
| Genistin | 35 | 1.5 | 44 | Berrin [20] |
| Glycosphingolipids | ||||
| C6-NBD-GlcCer | 4.6 | 0.121 | 26 | Hayashi [18] |
| C6-NBD-Gal-Cer | 2.0 | 0.255 | 128 | Hayashi [18] |
| C18-Glc-Cer | 14 | 0.0072 | 0.51 | Hayashi [18] |
| C18-Gal-Cer | 9.2 | <0.0002 | <0.02 | Hayashi [18] |
The Klotho subfamily of mammalian GH1 proteins (i.e., KL, β-KL, and KLPH) lack a complete active site with both the catalytic acid/base and nucleophile and thus have no β-glucosidase activity [12]. However, KL has been shown to have weak glucuronidase activity and to modify glycosylation of the transient receptor potential ion channel TRPV5, suggesting they may act as glycoside hydrolases [21]. KL was identified by its induction of rapid aging-like symptoms in knock-out mice [22], and it plays regulatory roles in calcium and phosphate homeostasis [21, 23]. All Klotho subfamily members have N-terminal secretory signal sequences and C-terminal transmembrane domains, and KL has a secretory form as well [24, 25]. The KLPH has a single GH1 domain, while KL and β-KL have two GH1 domains, all of which lack essential catalytic amino acids [24].
Insect β-glucosidases and myrosinases
Although the Drosophila melanogaster genome contains only one GH1 gene, suggesting that insects may not have expanded this gene family at an early stage, other insects have adapted glycosides and glycoside hydrolases from the plants on which they feed for protection and digestive purposes [26]. Digestive β-glycosidases from GH1 have been isolated from insect larvae that feed on plants [27, 28]. Similarly, myrosinases have been isolated from specialist insects that feed on crucifers, such as the cabbage aphid, Brevicoryne brassicae [29]. The larval β-glycosidases mentioned above can hydrolyze gluco-oligosaccharides and plant glycosides, such as cellobiose, gentiobiose, and amygdalin [27], in line with their digestive functions. These insect β-glycosidases and myrosinases have sequences most similar to each other, then to vertebrate LPH, suggesting they diverged from the same animal GH1 gene ancestor, after its divergence from plants. These genes have since evolved to meet the unique needs of the herbivorous insects in their battle with plant defenses to exploit the plant nutrients.
Roles of GH1 β-glucosidases in plants
Functional diversity and multiplicity
It is in plants that β-glucosidases have been found to play the widest array of biological functions, which include roles in defense, symbiosis, cell wall catabolism and lignification, signalling, and plant secondary metabolism. Several putative β-glucosidase genes have been shown either to be induced by biotic or abiotic stress or to be necessary for successful response to the stress [30–34]. With the advent of genomics, it became clear that about 40 GH1 β-glucosidases are expressed in a typical plant, many in the same tissues [35, 36]. The roles of these enzymes are presumed to be determined by their substrate-specificities, their tissue and subcellular localization, and the conditions under which they come into contact with their physiological substrates.
To match this enormous functional diversity, plants have the largest number of GH1 family proteins. For example, 48 GH1 genes for putative β-glucosidases and thioglucosidases are found in Arabidopsis thaliana [35] and 40 GH1 genes are found in rice genome sequences [36]. A number of these represent pseudogenes, and, in the case of rice, two appear to be endophyte genes, but nonetheless both plants appear to express over 30 putative GH1 β-glucosidases. Sequence-based phylogenetic analysis grouped these proteins into eight clusters that include both rice and Arabidopsis representatives and two clusters found only in Arabidopsis and other plants of the family Capparales, including a cluster of classical thioglucosidases (myrosinases) and a cluster of ER and peroxisomal β-glucosidases and myrosinases. In addition, several groups of enzymes from other plants do not fall into the Arabidopsis and rice phylogenetic clusters, including the well-studied chloroplastic β-glucosidases of maize, sorghum, wheat and other cereals, which are not found in rice. Most of these plant GH1 enzymes are closely related to one another, but the lineage of SFR2 [33] shows higher similarity to enzymes from thermophilic bacteria and Archaea than other plant enzymes, and this lineage is thought to be distinct within GH1 [37]. A few of the Arabidopsis and rice enzymes have been shown to be primarily β-d-mannosidases [35, 38], so it is possible that some of the others will have different glycone specificities as well, but most are likely to be β-d-glucosidases. Given that plants also contain GH3 and GH5 β-glycosidases with β-glucosidase activity [5, 39], the precise number of β-glucosidase isoenzymes in a particular plant species has yet to be determined.
Defense and microbial interaction
Plants have long been known to contain glycosides that release toxic compounds, such as cyanide and hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones) [40, 41], and the use of β-glucosidases as “detonators” of these chemical “bombs” has recently been reviewed [42]. In general, the defense glycosides are stored in a different cell or a different cellular compartment from the β-glucosidases that hydrolyze them to release toxic compounds. The defense compounds tend to be stored in the vacuole, while their corresponding β-glucosidases are often found in the apoplast or plastid. Both β-glucosidases and thioglucosidases have been found to play these roles, and specialist insects that feed on these plants have adapted these enzymes to diffuse the glycoside bombs or use them for their own defense [26]. β-Glucosidase-mediated defenses are also required for endophytic fungi to develop symbiotic relationships with plants, evidently by modulating the growth of these microorganisms [43].
Plants have developed a wide range of compounds for defense, examples of which can be seen in Fig. 1. Cyanogenic β-glucosides, including linamarin from clover, cassava and various other plants, dhurrin from sorghum, and prunasin from cherry and other stone fruits, are hydrolyzed to release an α-hydroxynitrile, which then breaks down either enzymatically or spontaneously to release cyanide and an aldehyde [41, 42]. Noncyanogenic defense compounds, such as γ- and β-hydroxynitriles and isoflavones in legumes, other flavonoids, coumarins, hydroxaminic acids, such as 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) in maize and wheat, and saponins are also stored as β-d-glucosides, which are hydrolyzed by specific β-glucosidases [44–54].
Aside from sequestration of the enzyme in the chloroplast or apoplast, several GH1 hydrolases are found in other compartments. The AtBGLU26 (PEN2) myrosinase is found in the pyroxisome [34], while AtBGLU23 (PYK10) is the most abundant protein in an ER-derived compartment called the ER body, which is only found in crucifers [55, 56]. AtBGLU26 has been shown to be critical to the Arabidopsis defense against nonspecialist fungi [34, 57]. AtBGLU23 is a β-glucosidase that has been found to be critical for establishment of symbiosis with the endophytic fungus Piriformospora indica by preventing it from overgrowing the roots and triggering a defense response [43, 58]. AtBGLU23 and the closely related isoenzymes AtBGLU21 and AtBGLU22 have recently been shown to be specific for scopolin, the most abundant coumarin glucoside in Arabidopsis roots, thereby explaining the antifungal role of AtBGLU23 [54].
Upon cell disruption, plant defensive β-glucosidases and β-thioglucosidases often bind to cytoplasmic aggregating factors, which are thought to help localize the otherwise soluble β-glucosidases and β-thioglucosidases at the site of injury, ensuring a maximal release of defense compounds [53, 59–63]. The functional significance of the interactions of the various defensive β-glucosidases from different cellular compartments and their multiple aggregating factors is an area of active investigation.
Cell wall metabolism
The cell wall of plants is the largest repository of carbohydrates in nature, much of which are β-linked glucosyl residues, so it is not surprising that β-glucosidases should play important roles in cell wall development. β-Glucosidases, in fact, appear to play roles in both the degradation of oligosaccharides generated in cell wall turnover and release of monolignols from their glycosides to allow lignification to stabilize secondary cell walls.
Several β-glucosidases that hydrolyze cell-wall-derived oligosaccharides have been identified over the years and have been studied primarily in monocots. For example, a β-glucosidase in germinating barley seedlings showed activity toward β-1,3- and β-1,4-linked oligosaccharides [64–66]. More recently, it has been shown that this enzyme displays greater preference for mannooligosaccharides, which are also found in barley endosperm cell walls [67]. Rice seedling β-glucosidases have also been shown to hydrolyze oligosaccharides, with varying preferences [36, 38, 68–71]. Rice BGlu1 (Os3BGlu7), Os3BGlu8, and Os7BGlu26 hydrolyzed cellooligosaccharides with increasing efficiency as the degree of polymerization (DP) increased from 2 to 6, while Os4BGlu12 showed little increase in activity with DP and Os3BGlu6 hydrolyzed disaccharides best. The Os3BGlu7 and Os3BGlu8 isoenzymes are widely expressed in rice tissues, so they may be needed for release of glucose from oligosaccharides generated in cell wall remodeling at various stages of plant development. Since the rice β-glucosidase isoenzymes mentioned above also hydrolyze plant-derived glycosides, they may play other roles in the plant as well.
The lignification of secondary cell walls is thought to involve the activation of monolignols by removal of β-glucosyl residues from monolignol glycosides, like cinnamyl alcohol β-glucosides [72]. A coniferin β-glucosidase was identified from lodgepole pine tree xylem [73]. Immunological analysis indicated that this protein was localized to the differentiating region of the xylem, consistent with a role in lignification. More recently, two Arabidopsis β-glucosidases (AtBGLU45 and AtBGLU46) that cluster with lodgepole pine coniferin β-glucosidase in phylogenetic analysis were shown to hydrolyze coniferin, syringin and coumaryl alcohol glucoside [74].
Phytohormone activation
Many phytohormone glucosides are found in plants, and there has been some debate as to whether these are terminal inactivation products or storage forms that can be readily activated by specific β-glucosidases. Partially purified rice β-glucosidases were shown to hydrolyze gibberellin glucosides [75], while maize β-glucosidase (Zm-p60.1, an isoform of ZmGlu1, which hydrolyzes the defense compound DIMBOAGlc) was shown to hydrolyze and activate cytokinin β-glucosides in vitro, as well as in vivo after infusion of the exogeneous substrate [76]. Abscissic acid (ABA) glucosyl ester (ABA-GE) was shown to be transported from roots to leaves and be hydrolyzed by extracellular β-glucosidase in the leaves, although free ABA is transported in larger amounts [77]. An enzyme hydrolyzing the auxin glucosyl ester 6-O-(4-O)-indole-3-ylacetyl-β-d-glucose has also been identified, though the nature of this enzyme remains to be determined [78].
Recently, it has been shown that a specific Arabidopsis ER β-glucosidase (AtBGLU18, AtBG1) is activated to hydrolyze ABA-GE in response to drought stress [79]. Mutation of the gene for this enzyme caused early germination and defective stomata closing, which could be rescued by transgenic expression of the gene, but not by a gene encoding an inactive mutant, thereby verifying its role in increasing ABA levels. This is perhaps the clearest demonstration of a physiological role for a β-glucosidase in phytohormone activation and suggests that other phytohormone glucoconjugates also serve as β-glucosidase-activated storage forms.
Secondary metabolism
The monoterpene alkaloid intermediate strictosidine is hydrolyzed by a specific cytoplasmic β-glucosidase to allow metabolism to various monoterpene alkaloids, depending on the plant [80]. This enzyme has been characterized from several plants, and that of Rauvolfia serpentina has been structurally characterized [81]. One of the downstream products from strictosidine is raucaffricine, a glucoside that can be deglucosylated by raucaffricine β-glucosidase for further metabolization to ajmaline [82]. Recently, another β-glucosidase hydrolyzing alkaloid glucosides was isolated from Psychotria ipecacuanha, further expanding on this theme [83]. As such, β-glucosidases can play metabolic roles to release glucosyl blocking groups from metabolic intermediates and allow their metabolism to various natural products, many of which are of medicinal importance.
Other functions
Plant β-glucosidases may play a variety of other roles. For instance, they appear to play roles in releasing volatiles from glycoside storage forms. This includes flower scents [84] as well as attractants for parasitic wasps that can attack herbivores feeding on the plant [85]. With the wide variety of glucosides found in plants, it is likely many roles remain to be discovered.
Roles of GH1 β-glucosidases in microorganisms
Although much research has been done on β-glucosidases from microorganisms, most of it has focused on their application rather than their endogenous function. As such, most of the enzymes that have been studied in the context of their natural function are those involved in bioconversion to produce glucose from plant biomass, or in breaking through plant cell walls to establish pathogenic or symbiotic relationships [86]. Bacterial β-glucosidases are often components of large complexes called cellosomes, contain polysaccharide degrading endoglucanases and carbohydrate binding proteins to localize the complex and to the cellulose surface and the cell membrane [87, 88]. Alternatively, some microorganisms secrete soluble endoglycosidases and exoglycosidases for this function, including exoglucanases/β-glucosidases. Fungi, such as the white rot fungus Phanerochaete chrysosporium, may contain cytoplasmic β-glucosidases and extracellular exoglucanases, some of which may act in metabolism of the organism’s own cell wall, in addition to plant cell wall biomass metabolism [89–91].
Applications of β-glucosidases
As noted above, β-glucosidases are of interest for biomass conversion, since conversion of β-glucans, the largest source of biomass in the world, is generally accomplished by three enzymes, endo-β-glucanases (e.g., cellulases), exo-β-glucanases (e.g., cellobiosidases) and β-glucosidases [86]. Limiting factors in conversion of cellulose to glucose for fermentation to alcohol include the inhibition of cellulases by oligosaccharides and the lack of adequate β-glucosidase production by certain microorganisms used for biomass breakdown. Thus, the identification and production of β-glucosidases, especially those with high glucose tolerance, has been of interest, and applicable β-glucosidases have been isolated from bacteria and fungi [86, 89–91].
There are hundreds of different β-glucosidic flavor precursors in plants, and their hydrolysis often enhances the quality of the beverages and foods produced from them [92, 93]. Generally, there are native β-glucosidases in source-plant tissues that hydrolyze these flavor precursors to produce the desired aglycone moiety. Enzymes from the source plants or other sources may be added to foods and beverages before, during, or after processing to enhance food quality. Enzymes with desirable properties may be targeted for breeding programs to increase their abundance in the plants or for overproduction in transgenic microbial or plant hosts, and for engineering to improve their catalytic properties for flavor enhancement and stability.
Aside from flavor enhancement, foods, feeds, and beverages may be improved nutritionally by release of vitamins, antioxidants, and other beneficial compounds from their glycosides. For instance, vitamin B6 (pyridoxine) can be released from its glucoside by enzymes in rice, in which pyridoxine glucoside is its predominant form [70, 94]. Other vitamins are also found as glucosides in plant sources, and release of their aglycones may improve their nutritional availability, despite the presence of animal and microbial β-glucosidases in the small intestine to aid in this process. Therefore, animal feeds are often treated with crude β-glucosidases. Much work has been done to identify β-glucosidases that can hydrolyze soy isoflavone glycosides, the aglycones of which have antioxidant properties [45–47, 95, 96]. Similarly, the pungent taste of food made from cruciferous vegetables (e.g., broccoli, cabbage, cauliflower, horseradish, kale, mustard, watercress, etc.) is derived from the products of the myrosinase/glucosinolate system, which may also have anticarcinogenic effects, although they may cause endemic goiter in large amounts [93, 97].
The compartmentalized β-glucosidase-β-glucoside defense systems found in such food and feed-plant tissues as cassava roots and leaves, lima beans, flax seeds, and clover leaves produce HCN when the tissue is macerated during preparation or by chewing [93]. The bitterness in almonds is caused by the presence of cyanogenic glucosides [98]. Cassava is highly consumed in parts of Asia, Africa, and South America, and contains the cyanogenic β-glucoside linamarin and its β-glucosidase linamarase. When consumed raw, cyanide poisoning can occur with symptoms of difficulty in breathing, paralysis, convulsion, coma, and even death. Similar symptoms can arise when bitter almonds are eaten raw. Processing of cassava roots by maceration results in the enzyme releasing the HCN, and subsequent aeration and washing removes the products of cyanogenesis. Alternatively, thorough cooking destroys the linamarase enzyme, preventing cyanide release.
In addition to their catalysis of hydrolysis, β-glucosidases also catalyze reverse hydrolysis and transglycosylation reactions, which can be used to synthesize oligosaccharides and glycosides of interest [99]. Various mutations have been developed to maximize the products of these transglycosylation reactions by manipulating the catalytic mechanism, as described below.
Biochemistry of β-glucosidases
Structures of β-glucosidases
β-Glucosidases have various structures, but the overall fold of the catalytic domain is similar in each GH family. The families GH1, GH5, and GH30 belong to the Clan GH-A, and they all have similar (β/α)8-barrel domains that contain their active site. In contrast, GH3 enzymes have two domains contributing to their active site. GH9 enzymes have (α/α)6-barrel structures, while the GBA2 family shows weak homology to proteins with this (α/α)6 structure as well (Fig. 2). We will consider these in turn, followed by a more in-depth look at GH1 enzymes, which serve as an excellent model for studying the structural basis for diverse substrate specificities.
Fig. 2.
Structures of β-glucosidases from different GH families. These include β-glucosidases or related enzymes from GH1 (Zea mays ZmGlu1, PDB code 1E1E), GH3 (Hordeum vulgare Exo I β-glucan glucohydrolase, PDB code 1EX1), GH5 (Candida albicans exo-β-(1,3)-glucanase Exg exoglucanase, PDB code 1CZ1), GH30 (Homo sapiens, acid β-glucosidase/glucocerebrosidase GBA1, PDB code 2V3D), and GH9 (Vibrio parahaemolyticus, putative exoglucanase, PDB code 3H7L). The structural cartoons are colored in a spectrum from blue to red from their N- to C-termini, with the catalytic nucleophile and acid–base residues shown in stick for those enzymes in which they are known. The ligands shown are glucose in the GH3 barley ExoI and N-butyl-deoxynojirimycin in the GH30 human GBA1, both of which are shown with carbons in pink. The human GBA2 (bile acid β-glucosidase) shows low levels of sequence similarity to (α/α)6 enzymes, suggesting its catalytic domain may be similar to the GH9 structure. Drawn with Pymol (DeLano)
The clan GH-A enzymes of families GH1, GH5, and GH30 all have a common (β/α)8-barrel structure and their active sites contain two conserved carboxylic acid residues on β-strands 4 and 7, serving as the catalytic acid/base and nucleophile, respectively [100, 101]. Although structures are available for all three of these families, our focus will be on GH1 here, since the relatively closely related GH1 plant β-glycosidases show a high diversity of substrate specificities, the basis of which will be considered later. The lengths and subunit masses of these GH1 enzymes vary considerably, depending on the presence of auxiliary domains and redundant GH1 domains (as in human LPH), but the catalytic domain itself ranges from around 440 to 550 residues, depending on the lengths of the variable loops at the C-terminal ends of the β-strands of the (β/α)8-barrel [102]. These monomers form a wide range of quaternary structures, including monomeric enzymes, dimers, tetramers, hexamers, octamers, and large aggregates.
The GH3 β-glucosidases and exoglucanases have a two-domain structure, a (β/α)8-barrel followed by an α/β sandwich comprising a 6-stranded β-sheet sandwiched between three α-helices on either side [103]. The active site of GH3 enzymes is situated between the (β/α)8 and (α/β)6 sandwich domains, each of which contributes one catalytic carboxylate residue (Fig. 2). The catalytic nucleophile for barley Exo I is an aspartate at residue D285, which resides on the loop after β-strand 7 of the (β/α)8 barrel, while the catalytic acid/base is glutamate E491, which is on a long loop extending from the (α/β)6 sandwich domain [104].
Only a few GH9 proteins have been verified to be β-glucosidases [105, 106], as most proteins in this family are endoglycosidases. This family consists of (α/α)6 barrels. Recently, the structure of a Vibrio parahaemolyticus protein with 69% amino acid sequence identity over 567 residues with the Vibrio cholera β-glucosidase, was determined (PDB accession 3H7L, Fig. 2). These family GH9 enzymes are the first β-glucosidases shown to act through an inverting mechanism (the prevailing mechanism in family GH9), which is unusual, since all other β-glucosidases described so far act through a retaining mechanism [106].
The human GBA2 and its relatives are not related to other β-glycosidases, but show weak similarities to certain (α/α)6 enzymes in homology searches. The GBA2 sequence contains no secretory pathway signal sequence and a single putative transmembrane domain, but was predicted to have its N-terminus in the endoplasmic reticulum and C-terminus in the cytoplasm [11]. The position of this putative transmembrane α-helix falls in the middle of a sequence homologous to soluble (α/α)6 amylohydrolase, chitobiose phosphorylase and α-l-rhamnosidase (3.2.1.40) enzymes, and the low confidence of the transmembrane topology prediction call this topology into question, but the enzyme is clearly associated with membranes by some means [10, 11].
Catalytic mechanisms
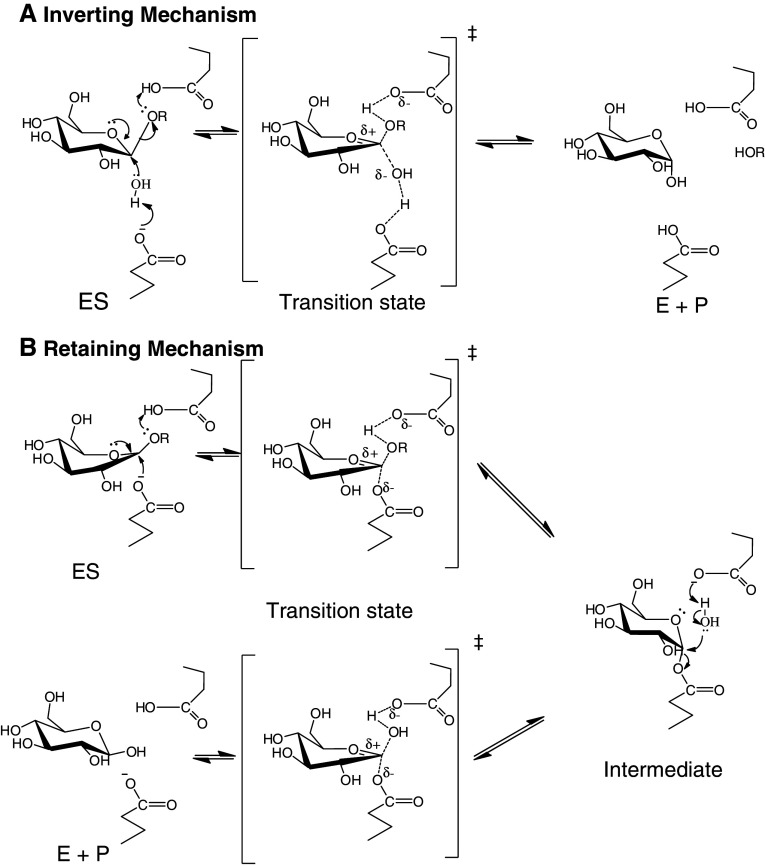
Glycoside hydrolases perform catalysis using two mechanisms, one with inversion and one with retention of chirality at the anomeric carbon [105]. Both of these mechanisms use a pair of acidic and nucleophilic residues, usually carboxylic acids, on either side of the sugar, approximately 5 Å apart in the retaining mechanism, and 10 Å apart in the inverting mechanism, in which a water molecule must fit between the catalytic base and the substrate. The GH9 β-glucosidases use an inverting mechanism, in which an activated water molecule makes a direct nucleophilic attack on the anomeric carbon to displace the aglycone in a single step, as shown in Fig. 3a [106]. The catalytic base extracts a proton from the incoming water molecule while the catalytic acid protonates the leaving group aglycone. In contrast, most β-glucosidases that have been characterized (i.e., GH1, GH3, and GH30 enzymes) are retaining enzymes, and they perform catalysis in two steps, glycosylation and deglycosylation (Fig. 3b). In glycosylation, the aglycone departs with the donation of a proton from the catalytic acid/base and nucleophilic attack of the catalytic nucleophile on the anomeric carbon to yield an α-linked covalent enzyme-glycone intermediate. In the deglycosylation step, the process is reversed, as a water molecule attacks with basic assistance from the catalytic acid/base to displace the catalytic nucleophile from the glucose.
Fig. 3.
Retaining catalytic mechanisms of inverting and retaining β-glucosidases. a The inverting mechanism that is seen in family GH9 glycoside hydrolases, including β-glucosidases. A single displacement of the aglycone by the water leads to an anomeric carbon with inverted chirality. b The commonly accepted mechanism for hydrolysis with retention of anomeric configuration as seen GH Clan A and family GH3 β-glucosidases. The glucosyl moiety is distorted into an 1S3 skew boat upon binding to the enzyme in preparation to form the 4H3 half chair conformation of the proposed transition state [107, 108]. The first step is glycosylation, in which the catalytic acid donates a proton to the leaving group, while the catalytic nucleophile attacks from the opposite side to form an α-linked intermediate. In the second, deglycosylation step, the catalytic base (the same carboxylate as the catalytic acid) extracts a proton from a water molecule, improving its nucleophilic power to attack at the anomeric carbon and displace the enzyme. Hydrolysis by either mechanism is equivalent in the organism, since mutarotation of the released glucose will lead to a racemic mixture of glucose in solution after a short time
Both the glycosylation and deglycosylation steps are thought to pass through oxocarbenium-ion-like transition states. The glucose of the incoming substrate has sometimes been observed to be distorted into a 1S3 skew boat as it moves toward the 4H3 half-chair shape in the first transition state, although in other structures it is poorly defined by the electron density, apparently due to high mobility [108–112]. The structures of certain putative transition state mimics have also been solved in the active site and shown to have a structure close to the 4H3 half-chair, although others appeared to inhibit by mechanisms other than transition state mimicry [108, 113–118].
The presence of the covalent intermediate was first demonstrated with the GH1 Agrobacterium sp. β-glucosidase by covalent labeling with 2,4-dinitrophenyl-2-deoxy-2-fluoroglucoside [119, 120]. In this inhibitor, the electronegative fluoride atom destabilizes the transition state for both half reactions, while the 2,4-dinitrophenylate provides an excellent leaving group to allow the glycosylation step to proceed rapidly. This traps the enzyme in the covalent intermediate and allows the catalytic nucleophile to be identified by tryptic digest and mass spectrometry. This covalent intermediate has also been observed in crystal structures for both the 2-F-glucoside and, in some cases, in the nonfluorinated glucosyl residue in certain acid/base catalyst mutants [104, 112, 121, 122]. The covalent inhibitor CBE has also been used to identify the catalytic nucleophile in some cases [104], but it is less specific and sometimes labels nearby amino acids. The acid/base catalyst of cassava β-glucosidase was also identified with a mechanism-based inhibitor, N-bromoacetyl-β-d-glucopyranosylamine [123], but most acid/base residues have been identified through homology, proximity to the glycosidic bond oxygen in crystal structures or site-directed mutagenesis [124].
The double-displacement mechanism for retaining β-glucosidases leads to predictions that mutants of these enzymes in which the acid/base or nucleophile is removed can be rescued by small nucleophiles and utilized for transglycosylation [107, 124–127]. When the acid/base of Agrobacterium sp. β-glucosidase was mutated to glycine (E170G), the hydrolysis of 2,4-dinitrophenyl β-d-glucoside (dNPGlc), which has a leaving group that does not require protonation (pKa = 3.96), lost its pH dependence from 7 to 9 and could be rescued by various small nucleophiles, such as azide, which produced β-d-glucosyl azide [124]. This verified E170 as the catalytic acid/base and was consistent with the double displacement mechanism in which the role of the catalytic acid has been circumvented. Similarly, conversion of the Abg catalytic nucleophile to a small nonnucleophilic amino acid, i.e., Ala, Ser, or Gly, resulted in an inactive enzyme that could be rescued by azide or fluoride to form α-d-glucosides, thereby converting a retaining enzyme to an inverting enzyme [125, 126]. Alternatively, the use of α-fluoroglucoside, in which the fluoride replaces the enzyme nucleophile in the covalent intermediate, allowed transfer of a β-linked glucosyl moiety onto a sugar or other alcohol. Since these nucleophile mutants have low hydrolytic activity, but relatively high transferase activity, they were designated glycosynthases [127]. Both the acid/base and the nucleophile mutants have potential uses in glycoconjugate synthesis.
Mechanism of substrate binding and specificity
Although the residues responsible for the hydrolytic mechanism are well characterized, how β-glycosidases recognize and interact with their substrates, which in large part determines their diverse functions, is less clear. GH1 enzymes are a prime model for these studies and the structures of 23 GH1 enzymes and their variants are currently available, including three from archaea, nine from bacteria, two from animals, one from a fungus, and eight from plants (CAZY website, [5]). The complexes of several of these enzymes with substrates, inhibitors and covalent intermediates are available, allowing in-depth analysis of residues likely to be critical to substrate and transition state binding. Although many of the prokaryotic enzymes show rather similar and broad substrate specificities, the complexes of β-glycosidase S from the archaeon Sulfolobus solfataricus and β-glucosidase A from the eubacteria Thermotoga maritima with a range of inhibitors has provided a wealth of information on catalytic and inhibitory mechanisms [113–118]. In addition, site-directed mutagenesis of GH1 enzymes with and without experimentally determined structures has been done to test the roles of various residues.
The GH1 enzymes may have rather broad glycone specificity, for instance one enzyme may hydrolyze β-d-glucosides, β-d-fucosides, β-d-mannosides, β-d-galactosides and α-l-arabinosides, or may be specific for one or a few glycone sugars. Marana [128] analyzed GH1 specificity and concluded that a conserved glutamate, which bridges the glycone hydroxyl groups 4 and 6 in enzymes with β-glucosidase and β-galactosidase activities but is replaced in 6-phosphoglycosidases, is critical for the distinction between enzymes. However, it still remains to be determined how GH1 enzymes can be primarily β-glucosidases or β-mannosidases or show different ranges of allowed glycones, even though they bind the sugar with the same conserved residues [38, 65, 109, 112]. It is worth noting that binding of the aglycone has also been observed to affect the sugar binding position [109, 129], so residues more distant in the substrate binding pocket cannot be excluded from playing roles in glycone specificity.
The basis of the tremendous diversity in function of β-glucosidases, especially in plants, is the substrate aglycone specificity differences that determine their natural substrates. Structures of complexes of enzymes with inhibitors and mutant enzymes with substrates, along with mutagenesis and chimera studies comparing similar enzymes with divergent specificities, have suggested that the basis of aglycone specificity is complex. Although this includes mutagenesis and structural studies of human cytoplasmic β-glucosidase [13, 130], the plant GH1 enzymes have served as the primary model, due to their high diversity in aglycone specificity.
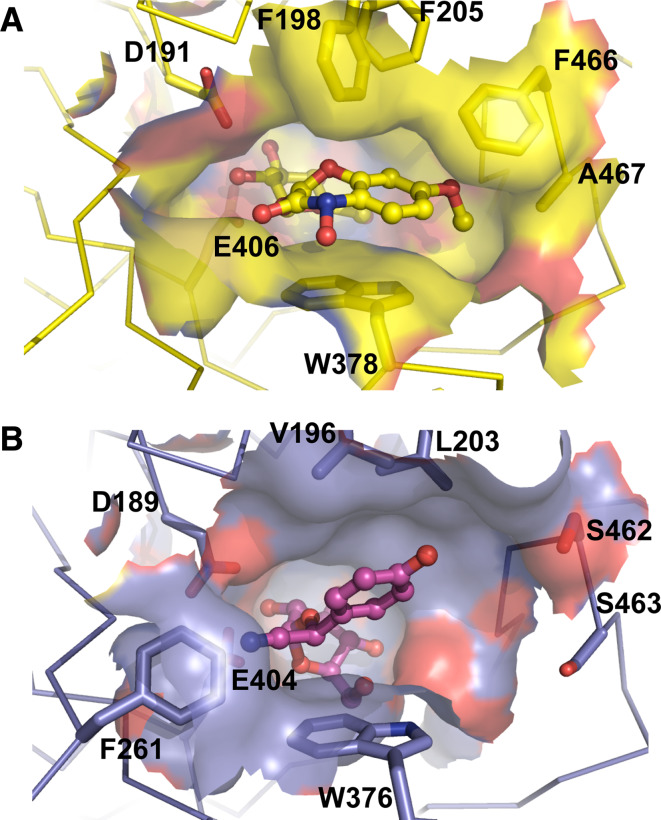
Maize ZmGlu1 and sorghum dhurrinase 1 (SbDhr1) are closely related, displaying 70% amino acid sequence identity, but have distinct specificities. ZmGlu1 has broad specificity, but cannot hydrolyze dhurrin, the natural substrate of SbDhr1, while SbDhr1 hydrolyzes only dhurrin. Studies of reciprocal ZmGlu1/SbDhr1 chimeric enzymes [131] and subsequent structural and site-directed mutagenesis studies [109–111, 129] indicated that the aglycone specificity determining sites are different in ZmGlu1 and SbDhr1. The structures of a catalytically inactive ZmGlu1 mutant (Glu1E191D) in complex with the natural substrate DIMBOAGlc (Fig. 4a), its free aglycone DIMBOA, and the unhydrolyzed competitive inhibitor dhurrin showed that the aglycone moiety of the substrate is sandwiched between four aromatic residues, W378 on one side and F198, F205, and F466 on the other [109]. The 7-methoxy group of DIMBOA also has a hydrophobic contact with A467. All of these residues, except W378, are variable among β-glucosidases that differ in substrate specificity, which led to the conclusion that these sites and the active-site shape are the basis of aglycone binding specificity in β-glucosidases. In the case of Dhr1, the three phenylalanines are replaced with V196, L203, and S462, and the active site is smaller (Fig. 4b). A water-mediated H-bond between the dhurrin phenolic hydroxyl and Dhr1 S462 provides a more polar and stronger binding interaction than seen in ZmGlu1 [109–111]. This apparently led to a more stable 1S3 skew boat conformation of the glucosyl residue, whereas in ZmGlu1 the conformation appeared to be variable, leading to poor electron density around the anomeric carbon. Mutagenesis of these aglycone-binding residues in the Zm60.8 isoform of ZmGlu1 confirmed their importance to hydrolysis of synthetic substrates [132].
Fig. 4.
Active site configurations of maize β-glucosidase 1 (Glu1, a) and sorghum dhurrinase 1 (Dhr1, b). The active sites of maize Glu1 and sorghum Dhr1 enzymes are shown for the structures of the Glu1 E189D mutant in complex with DIMBOA glucoside (PDB entry 1E56) and Dhr1 E191D mutant in complex with dhurrin (PDB entry 1VO3) [109, 111]. The sidechains of residues noted to interact with the aglycone are shown in stick representation behind the active site surface, which is colored as the underlying residues, which are colored with carbon in yellow for Glu1 and purple for Dhr1, nitrogen in blue, and oxygen in red. The ligands are shown in ball and stick representation with similar coloration. The Phe 261 residue, which narrows the active site in Dhr1, is also shown in front of the catalytic nucleophile Glu 404. Figure created with Pymol
However, the structural investigation of ZmGlu1 to SbDhr1 mutants and the subsequent structure of SbDhr1 and other GH1 hydrolases showed that the above-mentioned variable residues alone cannot designate substrate specificity [111, 131]. Although the Trp corresponding to ZmGlu1 W378 is nearly invariable in other plant GH1 enzymes, its positional variation was found to be critical for binding of substrates like dhurrin and strictosidine [81, 111, 131]. Even the closely related wheat β-glucosidase (TaGlu) was found to have different amino acids at the other aglycone-binding residues found in ZmGlu1, despite the fact that it also hydrolyzes DIMBOAGlc [52]. The oligosaccharide binding site in rice BGlu1 runs out of the active site in a different direction from that of DIMBOAGlc binding in ZmGlu1, so that L442, corresponding to ZmGlu F466, makes no interaction with the substrate, while N190, corresponding to ZmGlu F205, interacts only indirectly ([112], and PDB code 3F5K). Instead, N245 plays a key role in binding to the third glucosyl residue in cellooligosaccharides, while the corresponding residues in SbDhr1 (F261) and rice Os3BGlu6 (M251) appear to block off their active sites, which do not bind to such long substrates [71, 111]. Thus, a different, though overlapping, set of active site residues has been recruited to interact with the aglycone in each GH1 β-glucosidase that has been investigated, in contrast to the highly conserved glycone binding site.
Kinetic parameters for substrate hydrolysis
β-Glucosidases have variable kinetic parameters toward their substrates. The K m values for natural substrates and other good substrates are typically 1 mM or less, but these values vary roughly 1,000-fold. Similarly, β-glucosidases have relatively low k cat values (~300 s−1 or lower), which may be physiologically beneficial in some roles, but one must suspect the physiological relevance or the enzyme preparation if these values are too low. Comparison of the k cat /K m (efficiency coefficient) values is generally used to evaluate potential natural substrates, as two substrates with similar K m values may have vastly different catalytic efficiencies. It is instructive to look at the published values for the hydrolysis of putative natural substrates by natural and recombinant preparations of human cytoplasmic β-glucosidases (Table 1). The plant-derived flavonoid luteolin 4′-glucoside and a synthetic fluorescent glycoceramide, C6-NBD-GalCer, give similar efficiency coefficients, although the former is hydrolyzed approximately five-fold faster than the latter. The natural glycoceramides that have longer acyl chains had much slower hydrolysis rates. Nonetheless, it was shown that RNAi knockdown of the cytoplasmic β-glucosidase resulted in an increase in glycoceramide concentrations, suggesting they may serve as natural substrates in the cell [18].
When β-glucosides with different efficiencies differ in the leaving-group ability of their aglycones, the rate-limiting step will be the glycosylation reaction, while either the glycosylation or deglycosylation step, or both, might be rate-limiting if substrates differ in the glycone. For example, many β-glucosidases hydrolyze p-nitrophenyl-β-d-fucoside (pNPFuc) with higher efficiency than p-nitrophenyl-β-d-glucoside (pNPGlc). In the case of maize β-glucosidases [50] and rice Os3BGlu7 [70], the K m values are similar, and the V max values for pNPFuc are clearly higher, however, in Dalbergia isoflavonoid β-glucosidases [46, 47, 133] and rice Os3BGlu6 [71], the V max is similar or higher for pNPGlc and the K m is ten-fold lower for pNPFuc.
Inhibitors and cofactors
β-Glucosidases are inhibited by transition-state sugar analogues, substrate analogue glycosides, and free aglycones of their substrates, as well as slowly hydrolyzed substrates, such as the fluoroglucosides mentioned earlier. Structural and thermodynamic analysis of 18 putative substrate analogues suggested many may not act as true transition-state analogues, but may nonetheless bind and inhibit T. maritima β-glucosidase [118]. Since the aglycone and glycone-binding pockets in the active site are distinct, sugar analogs shaped similar to the half-chair conformation of the transition state can bind to the glycone-binding site and inhibit the enzyme, whereas free aglycones may bind to the aglycone-binding site. Free glucose is a poor inhibitor (typically K i = 100–200 mM) because glucose must be distorted toward the half-chair conformation for binding to the glycone-binding site, which is thought to require a portion of the energy of aglycone binding. In contrast, free aglycones can be potent competitive inhibitors because they bind to the aglycone-binding site without energetically unfavorable distortion.
Although most metal ions do not inhibit β-glucosidases, Ag+ and Hg2+, are potent β-glucosidase inhibitors, and inhibition by Cu2+ and Fe3+ has also been reported [134]. Although β-O-glucosidases are not known to require any cofactors, ascorbate is known to enhance the activity of β-S-glucosidases (myrosinases) by acting as a surrogate catalytic base [135]. The chelation of Zn2+ between the monomers in the biological dimer of myrosinase suggests that metal ions could act in stabilization of some GH1 enzymes.
pH and temperature optima and stability
β-Glucosidases show a range of pH optima and stabilities, depending on their source and amino acid sequence. The pH optima of most β-glucosidases range between pH 4 and 7.5, depending on their source and cellular location, and they tend to be stable over a range of pH from 4 to 9. It is usually safe to store these enzymes at 0–4°C at pH 7–8, once major protease contaminants have been removed, but this should be tested with each enzyme. As with other proteins, pH extremes, copurifying proteases, and microbial contamination may result in degradation, although many β-glucosidases are resistant to proteases due to their tightly folded core structure. Nonetheless, proteolysis can result in recombinant proteins losing their purification tags and in purified active enzymes appearing to have two subunits on SDS-PAGE due to an internal cleavage that leaves the fold intact, in the authors’ experience. Some β-glucosidases are resistant to denaturation by ionic detergents such as SDS, which allows extraction with buffers containing up to 3% SDS and zymogram development after SDS-PAGE when samples are applied without heating.
Thermostability and temperature optima vary greatly among enzymes. Mesophilic β-glucosidases may show highest activity at 30–65°C, but are generally inactivated at and above 55–70°C [46, 53, 58, 73]. High activity at temperatures above the extremes of the enzyme’s natural environment is not physiologically relevant and may result in rapid heat denaturation, so assays are often run at 30–40°C. On the other hand, β-glucosidases from thermophilic bacteria, such as T. maritima BglA, may have temperature optima of over 100°C [136]. Engineering of a bacterial β-glucosidase to have the same N-terminal and C-terminal residues as T. maritima BglA, allowed hydrogen bonding between these termini and stabilized the enzyme, suggesting such interactions may be important for high stability [137]. This thermostability is also thought to be due to an increased number of proline residues, electrostatic bridges, and internal water molecules, and binding of more subunits in the quaternary structure compared to many mesophilic enzymes [138].
Summary and future prospects
The description of β-glucosidases in this review is limited in detail, due to the vast amounts of data that have been generated in the last several years. Nonetheless, we hope the reader will appreciate the wide variety of functions that β-glucosidases play in nature, from biomass degradation by microorganisms, to glycolipid and xenogenic β-glucoside breakdown in animals, to roles in defense, phytohormone regulation, cell wall metabolism, and secondary metabolism in plants, where the β-glucosidases have attained their greatest multiplicity and diversity of function. Although β-glucosidases like sorghum SbDhr1 can be very specific, others show overlapping ranges of activities for multiple substrates, such as the glucocerebrosidase and flavonoid β-glucosidase activities of human β-glucosidases. It seems likely that β-glucosidases play many as yet undiscovered roles, as well as potential for many applications.
Although the catalytic mechanism is well understood for GH Clan A and GH3 β-glucosidases, the means by which their exact substrate specificity is established has proven to be divergent for even closely related β-glucosidases. This and the high multiplicity of putative β-glucosidase in plants limit the conclusions that can be drawn from genomic sequences as to the putative specificities and functions of new β-glucosidase homologs. Nonetheless, the insights gained from structural and mutagenic studies provide a starting point from which to investigate the functions of new β-glucosidases. As more substrate specificities and structures are determined, it should become more feasible to predict substrate specificity from the sequences of as yet uninvestigated β-glucosidases.
Acknowledgments
Rodjana Opassiri and three anonymous reviewers are thanked for useful comments on the manuscript. JRKC was supported by Suranaree University of Technology and the Thailand Research Fund.
References
- 1.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrissat B, Davies G. Structural and sequence-based classification of glycosyl hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 3.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunner R, Wirtz W, Rose JKC, Darbill AG, Govers F, Scheel D, Nurnberger T. A β-glucosidase/xylosidase from the phytopathogenic oomycete, Phytophthora infestans . Phytochemistry. 2002;59:689–696. doi: 10.1016/S0031-9422(02)00045-6. [DOI] [PubMed] [Google Scholar]
- 5.Opassiri R, Pomthong B, Akiyama T, Nakphaichit M, Onkoksoong T, Ketudat-Cairns M, Ketudat Cairns JR. A stress-induced rice β-glucosidase represents a new subfamily of glycosyl hydrolase family 5 containing a fascin-like domain. Biochem J. 2007;408:241–249. doi: 10.1042/BJ20070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butters TD. Gaucher disease. Curr Opin Chem Biol. 2007;11:412–418. doi: 10.1016/j.cbpa.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 7.Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. X-ray structure of human acid-β-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hedge RS, Grabowski GA. Analyses of variant acid β-glucosidases: effects of Gaucher disease mutations. J Biol Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman RL, Wustman BA, Huertas P, Powe AC, Pine CW, Khanna R, Schlossmacher MG, Ringe D, Petsko GA. Structure of acid β-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat Chem Biol. 2007;3:101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 10.Yildiz Y, Matern H, Thompson B, Allegood JC, Warren RL, Ramirez DMO, Hammer RE, Hamra FK, Matern S, Russell DW. Mutation of β-glucosidase 2 causes glycolipid storage disease and impaired male fertility. J Clin Invest. 2006;116:2985–2994. doi: 10.1172/JCI29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boot RG, Verhoek M, Donker-Koopman W, Strijland A, van Marle J, Overkleeft HS, Wennekes T, Aerts JM. Identification of the non-lysosomal glucosylceramidase as beta-glucosidase 2. J Biol Chem. 2007;282:1305–1312. doi: 10.1074/jbc.M610544200. [DOI] [PubMed] [Google Scholar]
- 12.Matern H, Boermans H, Lottspeich F, Matern S. Molecular cloning and expression of human bile acid beta-glucosidase. J Biol Chem. 2001;276:37929–37933. doi: 10.1074/jbc.M104290200. [DOI] [PubMed] [Google Scholar]
- 13.Tribolo S, Berrin J-G, Kroon PA, Czjzek M, Juge N. The structure of human cytoplasmic β-glucosidase unravels substrate aglycone specificity of a family 1 glycoside hydrolase. J Mol Biol. 2007;370:964–975. doi: 10.1016/j.jmb.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 14.Mantei N, Villa M, Enzler T, Wacker H, Boll W, James P, Hunziker W, Semenza G. Complete primary structure of human and rabbit lactase-phlorizin hydrolase: implications for biosynthesis, membrane anchoring and evolution of the enzyme. EMBO J. 1988;7:2705–2713. doi: 10.1002/j.1460-2075.1988.tb03124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arribas JC, Herrero AG, Martín-Lomas M, Cañada FJ, He S, Withers SG. Differential mechanism-based labeling and unequivocal activity assignment of the two active sites of intestinal lactase/phlorizin hydrolase. Eur J Biochem. 2000;267:6996–7005. doi: 10.1046/j.1432-1327.2000.01784.x. [DOI] [PubMed] [Google Scholar]
- 16.Day AJ, Canada FJ, Diaz JC, Kroon PA, Mclauchlan R, Faulds CB, Plumb GW, Morgan MR, Williamson G. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Lett. 2000;468:166–170. doi: 10.1016/S0014-5793(00)01211-4. [DOI] [PubMed] [Google Scholar]
- 17.Daniels LB, Coyle PJ, Chiao YB, Glew RH, Labow RS. Purification and characterization of a cytosolic broad specificity beta-glucosidase from human liver. J Biol Chem. 1981;256:13004–13013. [PubMed] [Google Scholar]
- 18.Hayashi Y, Okino N, Kakuta Y, Shikanai T, Tani M, Narimatsu H, Ito M. Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase. J Biol Chem. 2007;282:30889–30900. doi: 10.1074/jbc.M700832200. [DOI] [PubMed] [Google Scholar]
- 19.Day AJ, DuPont MS, Ridley S, Rhodes M, Rhodes MJ, Morgan MR, Williamson G. Deglycosylation of flavonoid and isoflavonoid glycosides by human small intestine and liver beta-glucosidase activity. FEBS Lett. 1998;436:71–75. doi: 10.1016/S0014-5793(98)01101-6. [DOI] [PubMed] [Google Scholar]
- 20.Berrin J-G, McLauchlan WR, Needs P, Williamson G, Puigserver A, Kroon PA, Juge N. Functional expression of human liver cytosolic β-glucosidase in Pichia pastoris. Insights into its role in the metabolism of dietary glucosides. Eur J Biochem. 2002;269:249–258. doi: 10.1046/j.0014-2956.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 21.Chang Q, Hoefs S, van der Kemp AW, Topala CN, Bindels RJ, Hoenderop JG. The beta-glucuronidase klotho hydrolyzes and activates the TRPV5 channel. Science. 2005;310:490–493. doi: 10.1126/science.1114245. [DOI] [PubMed] [Google Scholar]
- 22.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsuki T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima YI. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 23.Nabeshima Y, Imura H. α-Klotho: a regulator that integrates calcium homostasis. Am J Nephrol. 2008;28:455–464. doi: 10.1159/000112824. [DOI] [PubMed] [Google Scholar]
- 24.Ito S, Fujimori T, Hayashizaki Y, Nabeshima Y. Identification of a novel mouse membrane-bound family 1 glycosidase-like protein, which carries an atypical active site structure. Biochim Biophys Acta. 2002;1576:341–345. doi: 10.1016/s0167-4781(02)00281-6. [DOI] [PubMed] [Google Scholar]
- 25.Shiraki-Iida T, Aizawa H, Matsumura Y, Sekine S, Iida A, Anazawa H, Nagai R, Kuro-o M, Nabeshima Y. Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Lett. 1998;424:6–10. doi: 10.1016/S0014-5793(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 26.Zagrobelny M, Bak S, Møller BL. Cyanogenesis in plants and arthropods. Phytochemistry. 2008;69:1457–1468. doi: 10.1016/j.phytochem.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Marana SR, Jacobs-Lorena M, Terra WR, Ferrieira C. Amino acid residues involved in substrate binding and catalysis in an insect digestive β-glycosidase. Biochim Biophys Acta. 2001;1545:41–52. doi: 10.1016/s0167-4838(00)00260-0. [DOI] [PubMed] [Google Scholar]
- 28.Ferrieira AHP, Marana SR, Terra WR, Ferreira C. Purification, molecular cloning, and properties of a β-glycosidase isolated from midgut lumen of Tenebrio molitor (Coleoptera) larvae. Insect Biochem Mol Biol. 2001;31:1065–1076. doi: 10.1016/S0965-1748(01)00054-6. [DOI] [PubMed] [Google Scholar]
- 29.Jones AME, Bridges M, Bones AM, Cole R, Rossiter JT. Purification and characterisation of a non-plant myrosinase from the cabbage aphid Brevicoryne brassicae . Insect Biochem Mol Biol. 2001;31:1–5. doi: 10.1016/S0965-1748(00)00157-0. [DOI] [PubMed] [Google Scholar]
- 30.Malboobi MA, Lefebvre DD. A phosphate-starvation inducible β-glucosidase gene (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol. 1997;34:57–68. doi: 10.1023/A:1005865406382. [DOI] [PubMed] [Google Scholar]
- 31.van de Ven WT, LeVesque CS, Perring TM, Walling LL. Local and systemic changes in squash gene expression in response to silver winged whitefly feeding. Plant Cell. 2000;12:1409–1423. doi: 10.1105/tpc.12.8.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorlby G, Fourier N, Warren G. The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a beta-glucosidase. Plant Cell. 2004;16:2192–2203. doi: 10.1105/tpc.104.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, Llorente F, Molina A, Parker J, Somerville S, Schulze-Lefert P. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Escamilla-Treviño LL, Zeng L, Lalgondar M, Bevan DR, Winkel BSJ, Mohamed A, Cheng C, Shih M, Poulton JE, Esen A. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol Biol. 2004;55:343–367. doi: 10.1007/s11103-004-0790-1. [DOI] [PubMed] [Google Scholar]
- 36.Opassiri R, Pomthong B, Okoksoong T, Akiyama T, Esen A, Ketudat Cairns JR. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 2006;6:33. doi: 10.1186/1471-2229-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques AR, Coutinho PM, Videira P, Fialho AM, S-Correia I. Sphingomonas paucimobilis beta-glucosidase Bgl1: a member of a new bacterial subfamily in glycoside hydrolase family 1. Biochem J. 2003;370:793–804. doi: 10.1042/BJ20021249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuntothom T, Luang S, Harvey AJ, Fincher GB, Opassiri R, Hrmova M, Ketudat Cairns JR. Rice family GH1 glycosyl hydrolases with β-d-glucosidase and β-d-mannosidase activities. Arch Biochem Biophys. 2009;491:84–95. doi: 10.1016/j.abb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Arthan D, Kittakoop P, Esen A, Svasti J. Furostanol glycoside 26-O-β-glucosidase from the leaves of Solanum torvum . Phytochemistry. 2006;67:27–33. doi: 10.1016/j.phytochem.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 40.Niemeyer HM. Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defense chemicals in the Gramineae. Phytochemistry. 1988;27:3349–3358. doi: 10.1016/0031-9422(88)80731-3. [DOI] [Google Scholar]
- 41.Poulton JE. Cyanogenesis in plants. Plant Physiol. 1990;94:401–405. doi: 10.1104/pp.94.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchéz-Perez R, Møller BL, Bak S. β-Glucosidases as detonators of plant chemical defense. Phytochemistry. 2008;69:1795–1813. doi: 10.1016/j.phytochem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Sherameti I, Venus Y, Drzewiecki C, Tripathi W, Dan VM, Nitz I, Varma A, Grundler F, Oelmüller R. PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thanliana and the endophytic fungus Piriformospora indica . Plant J. 2008;54:428–439. doi: 10.1111/j.1365-313X.2008.03424.x. [DOI] [PubMed] [Google Scholar]
- 44.Morant AV, Bjarnholt N, Kragh ME, Kjaergaard CH, Jørgensen K, Paquette SM, Piotrowski M, Imberty A, Olsen CE, Møller BL, Bak S. The beta-glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus . Plant Physiol. 2008;147:1072–1091. doi: 10.1104/pp.107.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki H, Takahasi S, Watanabe R, Fukushima Y, Fujita N, Noguchi A, Yokoyama R, Nishitani K, Nishino T, Nakayama T. An isoflavone conjugate-hydrolyzing β-glucosidase from the roots of soybean (Glycine max) seedlings. J Biol Chem. 2006;281:30251–30259. doi: 10.1074/jbc.M605726200. [DOI] [PubMed] [Google Scholar]
- 46.Chuankhayan P, Hua Y, Svasti J, Sakdarat S, Sullivan PA, Ketudat Cairns JR. Purification of an isoflavonoid 7-O-β-apiosyl-glucoside β-glycosidase and its substrates from Dalbergia nigrescens Kurz. Phytochemistry. 2005;66:1880–1889. doi: 10.1016/j.phytochem.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 47.Chuankhayan P, Rimlumduan T, Tantanuch W, Mothong N, Kongsaeree PT, Metheenukul P, Svasti J, Jensen ON, Ketudat Cairns JR. Functional and structural differences between isoflavonoid β-glycosidases from Dalbergia sp. Arch Biochem Biophys. 2007;468:205–216. doi: 10.1016/j.abb.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA. Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula . Proc Natl Acad Sci USA. 2007;104:17909–17915. doi: 10.1073/pnas.0708697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esen A. Purification and partial characterization of maize (Zea mays L.) β-glucosidase. Plant Physiol. 1992;98:174–182. doi: 10.1104/pp.98.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babcock GD, Esen A. Substrate specificity of maize β-glucosidase. Plant Sci. 1994;101:31–39. doi: 10.1016/0168-9452(94)90162-7. [DOI] [Google Scholar]
- 51.Nikus J, Daniel G, Jonsson LM. Subcellular localization of beta-glucosidase in rye, maize and wheat seedlings. Plant Physiol. 2001;111:466–472. doi: 10.1034/j.1399-3054.2001.1110406.x. [DOI] [PubMed] [Google Scholar]
- 52.Sue M, Yamazaki K, Yajima S, Nomura T, Matsukawa T, Iwamura H, Miyamoto T. Molecular and structural characterization of hexameric beta-d-glucosidases in wheat and rye. Plant Physiol. 2006;141:1237–1247. doi: 10.1104/pp.106.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nisius A. The stroma centre in Avena plastids: an aggregation of β-glucosidase responsible for the activation of oat-leaf saponins. Planta. 1988;173:474–481. doi: 10.1007/BF00958960. [DOI] [PubMed] [Google Scholar]
- 54.Ahn YO, Shimizu B, Sakata K, Gantulga D, Zhou Z, Bevan DR, Esen A. Scopulin-hydrolyzing β-glucosidases in the roots of Arabidopsis. Plant Cell Physiol. 2010;51:131–143. doi: 10.1093/pcp/pcp174. [DOI] [PubMed] [Google Scholar]
- 55.Hara-Nishimura I, Matsushima R. A wound-inducible organelle derived from endoplasmic reticulum: a plant strategy against environmental stress? Curr Opin Plant Biol. 2003;6:538–588. doi: 10.1016/j.pbi.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I. A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER retention signal in Arabidopsis . Plant J. 2003;33:493–502. doi: 10.1046/j.1365-313X.2003.01636.x. [DOI] [PubMed] [Google Scholar]
- 57.Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 58.Nagano AJ, Matsushima R, Hara-Nishimura I. Activation of an ER-body-localized β-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana . Plant Cell Physiol. 2005;46:1140–1148. doi: 10.1093/pcp/pci126. [DOI] [PubMed] [Google Scholar]
- 59.Falk A, Taipalensuu J, Ek B, Lenman M, Rask L. Characterization of rapeseed myrosinase-binding protein. Planta. 1995;195:387–395. doi: 10.1007/BF00202596. [DOI] [PubMed] [Google Scholar]
- 60.Esen A, Blanchard DJ. A specific β-glucosidase-aggregating factor (BGAF) is responsible for the β-glucosidase null phenotype in maize. Plant Physiol. 2000;122:563–572. doi: 10.1104/pp.122.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanchard DJ, Cicek M, Chen J, Esen A. Identification of beta-glucosidase aggregating factor (BGAF) and mapping of BGAF binding regions on maize beta-glucosidase. J Biol Chem. 2001;276:11895–11901. doi: 10.1074/jbc.M008872200. [DOI] [PubMed] [Google Scholar]
- 62.Kittur FS, Lalgondar M, Yu HY, Bevan DR, Esen A. Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J Biol Chem. 2007;282:7299–7311. doi: 10.1074/jbc.M607417200. [DOI] [PubMed] [Google Scholar]
- 63.Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I. Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana . Plant Cell Physiol. 2008;49:969–980. doi: 10.1093/pcp/pcn075. [DOI] [PubMed] [Google Scholar]
- 64.Leah R, Kigel J, Svedsen I, Mundy J. Biochemical and molecular characterization of a barley seed β-glucosidase. J Biol Chem. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- 65.Hrmova M, Harvey AJ, Wang J, Shirley NJ, Jones GP, Stone BA, Hoj PB, Fincher GB. Barley β-d-glucan exohydrolases with β-d-glucosidase activity. J Biol Chem. 1996;271:5277–5286. doi: 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- 66.Hrmova M, MacGregor EA, Biely P, Stewart RJ, Fincher GB. Substrate binding and catalytic mechanism of a barley β-d-glucosidase/(1, 4)-β-d-glucan exohydrolase . J Biol Chem. 1998;273:11134–11143. doi: 10.1074/jbc.273.18.11134. [DOI] [PubMed] [Google Scholar]
- 67.Hrmova M, Burton RA, Biely P, Lahnstein J, Fincher GB. Hydrolysis of (1,4)-β-d-mannans in barley (Hordeum vulgare L.) is mediated by the concerted action of (1,4)-β-d-mannan endohydrolase and β-d-mannosidase. Biochem J. 2006;399:77–90. doi: 10.1042/BJ20060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akiyama T, Kaku H, Shibuya N. A cell wall-bound β-glucosidase from germinated rice: purification and properties. Phytochemistry. 1998;48:49–54. doi: 10.1016/S0031-9422(97)01099-6. [DOI] [PubMed] [Google Scholar]
- 69.Opassiri R, Ketudat Cairns JR, Akiyama T, Wara-Aswapati O, Svasti J, Esen A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003;165:627–638. doi: 10.1016/S0168-9452(03)00235-8. [DOI] [Google Scholar]
- 70.Opassiri R, Hua Y, Wara-Aswapati O, Akiyama T, Svasti J, Esen A, Ketudat Cairns JR. β-Glucosidase, exo-β-glucanase and pyridoxine transglucosylase activities of rice BGlu1. Biochem J. 2004;379:125–131. doi: 10.1042/BJ20031485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seshadri S, Akiyama T, Opassiri R, Kuaprasert B, Ketudat Cairns J. Structural and enzymatic characterization of Os3BGlu6, a rice β-glucosidase hydrolyzing hydrophobic glycosides and (1 → 3)- and (1 → 2)-linked disaccharides. Plant Physiol. 2009;151:47–58. doi: 10.1104/pp.109.139436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hösel W, Surholt E, Borgmann E. Characterization of β-glucosidase isoenzymes possibly involved in lignification from chick pea (Cicer arietinum L.) cell suspension culture. Eur J Biochem. 1978;84:487–492. doi: 10.1111/j.1432-1033.1978.tb12190.x. [DOI] [PubMed] [Google Scholar]
- 73.Dhamawardhana DP, Ellis BE, Carlson JE. A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 1995;107:331–339. doi: 10.1104/pp.107.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Escamilla-Treviño LL, Chen W, Card ML, Shih MC, Cheng CL, Poulton JE. Arabidopsis thaliana β-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry. 2006;67:1651–1660. doi: 10.1016/j.phytochem.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 75.Schliemann W. Hydrolysis of conjugated gibberellins by β-glucosidases from dwarf rice (Oryza sativa L. cv. Tan-ginbozu) J Plant Physiol. 1984;116:123–132. doi: 10.1016/S0176-1617(84)80069-3. [DOI] [PubMed] [Google Scholar]
- 76.Brzobohatý B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- 77.Dietz K-J, Sauter A, Wichert K, Messdaghi D, Hartung W. Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J Exp Bot. 2000;51:937–944. doi: 10.1093/jexbot/51.346.937. [DOI] [PubMed] [Google Scholar]
- 78.Jakubowska A, Kawalczyk S. A specific enzyme hydrolyzing 6-O(4-O)-indole-3-ylacetyl-β-d-glucose in immature kernels of Zea mays . J Plant Physiol. 2005;162:207–213. doi: 10.1016/j.jplph.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 79.Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–1120. doi: 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 80.Stöckigt J, Zenk MH. Strictosidine (isovincoside): the key intermediate in the biosynthesis of monoterpenoid indole alkaloids. J Chem Soc Chem Commun. 1977;1977:646–648. doi: 10.1039/c39770000646. [DOI] [Google Scholar]
- 81.Barleben L, Panjikar S, Ruppert M, Koepke J, Stöckigt J. Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell. 2007;19:2886–2897. doi: 10.1105/tpc.106.045682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Warzecha H, Gerasimenko I, Kutchan TM, Stöckigt J. Molecular cloning and functional bacterial expression of a plant β-glucosidase specifically involved in alkaloid biosynthesis. Phytochemistry. 2000;54:657–666. doi: 10.1016/S0031-9422(00)00175-8. [DOI] [PubMed] [Google Scholar]
- 83.Nomura T, Quesada AL, Kutchan TM. The new beta-d-glucosidase in terpenoid-isoquinoline alkaloid biosynthesis in Psychotria ipecacuanha . J Biol Chem. 2008;283:34650–34659. doi: 10.1074/jbc.M806953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reuveni M, Sagi Z, Evnor D, Hetzroni A. β-Glucosidase activity is involved in scent production in Narcissus flowers. Plant Sci. 1999;147:19–24. doi: 10.1016/S0168-9452(99)00097-7. [DOI] [Google Scholar]
- 85.Mattiacci L, Dicke M, Posthumus MA. Beta-glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc Natl Acad Sci USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilbert HJ, Stålbrand H, Brumer H. How the walls come tumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11:338–348. doi: 10.1016/j.pbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 87.Doi RH, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 88.Carvalho AL, Dias FM, Nagy T, Prates JA, Proctor MR, Smith N, Bayer EA, Davies GJ, Ferreira LM, Romaño MJ, Fontes CM, Gilbert HJ. Evidence for a dual binding mode of dockerin modules to cohesins. Proc Natl Acad Sci USA. 2007;2007(104):3089–3094. doi: 10.1073/pnas.0611173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lymar ES, Li B, Renganathan V. Purification and characterization of a cellulose-binding β-glucosidase from cellulose-degrading cultures of Phanerochaete chrysosporium . Appl Environ Microbiol. 1995;61:2976–2980. doi: 10.1128/aem.61.8.2976-2980.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Igarashi K, Tani T, Kawal R, Samejima M. Family 3 β-glucosidase from cellulose-degrading culture of the white-rot fungus Phanerochaete chrysosporium . J Biosci Bioeng. 2003;95:572–576. doi: 10.1016/s1389-1723(03)80164-0. [DOI] [PubMed] [Google Scholar]
- 91.Tsukada T, Igarashi K, Yoshida M, Samejima M. Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium . Appl Microbiol Biotechnol. 2006;73:807–814. doi: 10.1007/s00253-006-0526-z. [DOI] [PubMed] [Google Scholar]
- 92.Günata Z. Flavor enhancement in fruit juices and derived beverages by exogenous glycosidases and consequences of the use of enzyme preparations. In: Whitaker JR, Voragen AGJ, Wong DWS, editors. Handbook of food enzymology. New York: Marcel Dekker Inc.; 2003. pp. 303–330. [Google Scholar]
- 93.Esen A. β-Glucosidases. In: Whitaker JR, Voragen AGJ, Wong DWS, editors. Handbook of food enzymology. New York: Marcel Dekker Inc.; 2003. pp. 791–804. [Google Scholar]
- 94.Yasumoto K, Tsuji H, Iwami K, Mitsuda H. Isolation from rice bran of a bound form of vitamin B6 and its identification as 5′-O-β-d-glucopyranosyl-pyridoxine. Agric Biol Chem. 1977;41:1061–1067. [Google Scholar]
- 95.Chuankhayan P, Rimlumduan T, Svasti J, Ketudat Cairns JR. Hydrolysis of soybean isoflavonoid glycosides by Dalbergia β-glucosidases. J Agric Food Chem. 2007;55:2407–2412. doi: 10.1021/jf062885p. [DOI] [PubMed] [Google Scholar]
- 96.Ismail B, Hayes K. β-Glycosidase activity toward different glycosidic forms of isoflavones. J Agric Food Chem. 2005;53:4918–4924. doi: 10.1021/jf0404694. [DOI] [PubMed] [Google Scholar]
- 97.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sánchez-Pérez R, Jørgensen K, Olsen CE, Dicenta F, Møller BL. Bitterness in almonds. Plant Physiol. 2008;146:1040–1052. doi: 10.1104/pp.107.112979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crout DH, Vic G. Glycosidases and glycosynthetases in glycoside and oligosaccharide synthesis. Curr Opin Chem Biol. 1998;2:98–111. doi: 10.1016/S1367-5931(98)80041-0. [DOI] [PubMed] [Google Scholar]
- 100.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jenkins J, Lo Leggio L, Harris G, Pickersgill R. Beta-glucosidase, beta-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold beta/alpha architecture and with two conserved glutamates near the carboxy-terminal ends of beta-strands four and seven. FEBS Lett. 1995;362:281–285. doi: 10.1016/0014-5793(95)00252-5. [DOI] [PubMed] [Google Scholar]
- 102.Sanz-Aparicio J, Hermoso JA, Martinez-Ripoll M, Lequerica JL, Polaina J. Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases. J Mol Biol. 1998;275:491–502. doi: 10.1006/jmbi.1997.1467. [DOI] [PubMed] [Google Scholar]
- 103.Varghese JN, Hrmova M, Fincher GB. Three-dimensional structure of a barley β-d-glucan exohydrolase; a family 3 glycosyl hydrolase. Structure. 1999;7:179–190. doi: 10.1016/S0969-2126(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 104.Hrmova M, Varghese JN, De Gori R, Smith BJ, Driguez H, Fincher GB. Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-d-glucan glucohydrolase. Structure. 2001;9:1005–1016. doi: 10.1016/S0969-2126(01)00673-6. [DOI] [PubMed] [Google Scholar]
- 105.Park JK, Wang L-X, Patel HV, Roseman S. Molecular cloning and characterization of a unique β-glucosidase from Vibrio cholorae . J Biol Chem. 2002;277:29555–29560. doi: 10.1074/jbc.M202978200. [DOI] [PubMed] [Google Scholar]
- 106.Qi M, Jun H-S, Forsbert CW. Cel9D, an atypical 1, 4-β-d-glucan glucohydrolase from Fibrobacter succinogenes: characteristics, catalytic residues, and synergistic interactions with other cellulases. J Bacteriol. 2008;109:1976–1984. doi: 10.1128/JB.01667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rye CS, Withers SG. Glycosidase mechanisms. Curr Opin Chem Biol. 2000;4:573–580. doi: 10.1016/S1367-5931(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 108.Davies GJ, Ducros VM-A, Varrot A, Zechel DL. Mapping the conformational itinerary of β-glucosidases by X-ray crystallography. Biochem Soc Trans. 2003;31:523–527. doi: 10.1042/BST0310523. [DOI] [PubMed] [Google Scholar]
- 109.Czjzek M, Cicek M, Zamboni V, Bevan DR, Henrissat B, Esen A. The mechanism of substrate (aglycone) specificity in β-glucosidases is revealed by crystal structures of mutant maize β-glucosidase-DIMBOA, -DIMBOAGlc, and dhurrin complexes. Proc Natl Acad Sci USA. 2000;97:13555–13560. doi: 10.1073/pnas.97.25.13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Czjzek M, Cicek M, Zamboni V, Burmeister WP, Bevan DR, Henrissat B, Esen A. Crystal structure of a monocotyledon (maize ZMGlu1) β-glucosidase and a model of its complex with p-nitrophenyl β-d-thioglucoside. Biochem J. 2001;354:37–46. doi: 10.1042/0264-6021:3540037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verdoucq L, Morinière J, Bevan DR, Esen A, Vasella A, Henrissat B, Czjzek M. Structural determinants of substrate specificity in family 1 β-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant β-glucosidase with strict specificity, in complex with its natural substrate. J Biol Chem. 2004;279:31796–31803. doi: 10.1074/jbc.M402918200. [DOI] [PubMed] [Google Scholar]
- 112.Chuenchor W, Pengthaisong S, Robinson RC, Yuvaniyama J, Oonanant W, Bevan DR, Esen A, Chen C-J, Opassiri R, Svasti J, Ketudat Cairns JR. Structural insights into rice BGlu1 β-glucosidase oligosaccharide hydrolysis and transglycosylation. J Mol Biol. 2008;377:1200–1215. doi: 10.1016/j.jmb.2008.01.076. [DOI] [PubMed] [Google Scholar]
- 113.Zechel DL, Boraston AB, Gloster TM, Boraston CM, Macdonald JM, Tilbrook DMG, Stick RV, Davies GJ. Iminosugar glycosidase inhibitors: structural and thermodynamic dissection of the binding of isofagomine and 1-deoxynojirimycin to β-glucosidases. J Am Chem Soc. 2003;125:14313–14323. doi: 10.1021/ja036833h. [DOI] [PubMed] [Google Scholar]
- 114.Vincent F, Gloster TM, Macdonald J, Morland C, Stick RV, Dias FM, Prates JA, Fontes CM, Gilbert HJ, Davies GJ. Common inhibition of both beta-glucosidases and beta-mannosidases by isofagomine lactam reflects different conformational itineraries for pyranoside hydrolysis. Chembiochem. 2004;5:1596–1599. doi: 10.1002/cbic.200400169. [DOI] [PubMed] [Google Scholar]
- 115.Gloster TM, Macdonald JM, Tarling CA, Stick RV, Withers SG, Davies GJ. Structural, thermodynamic, and kinetic analyses of tetrahydroozazine-derived inhibitors bound to β-glucosidases. J Biol Chem. 2004;279:29236–49242. doi: 10.1074/jbc.M407195200. [DOI] [PubMed] [Google Scholar]
- 116.Gloster TM, Madsen R, Davies GJ. Dissection of conformationally restricted inhibitors binding to a beta-glucosidase. Chembiochem. 2006;7:738–742. doi: 10.1002/cbic.200600005. [DOI] [PubMed] [Google Scholar]
- 117.Gloster TM, Roberts S, Perugino G, Rossi M, Moracci M, Panday N, Terinek M, Vasella A, Davies GJ. Structural, kinetic, and thermodynamic analysis of glucoimidazole-derived glycosidase inhibitors. Biochemistry. 2006;45:11879–11884. doi: 10.1021/bi060973x. [DOI] [PubMed] [Google Scholar]
- 118.Gloster TM, Meloncelli P, Stick RV, Zechel D, Vasella A, Davies GJ. Glycosidase inhibition: an assessment of the binding of 18 putative transition-state mimics. J Am Chem Soc. 2007;129:2345–2354. doi: 10.1021/ja066961g. [DOI] [PubMed] [Google Scholar]
- 119.Withers SG, Street IP, Bird P, Dolphin DH. 2-Deoxy-2-fluoroglucosides: a novel class of mechanism based inhibitors. J Am Chem Soc. 1987;109:7530–7531. doi: 10.1021/ja00258a047. [DOI] [Google Scholar]
- 120.Withers SG, Warren RAJ, Street IP, Rupitz K, Kempton JB, Aebersold R. Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. J Am Chem Soc. 1990;112:5887–5889. doi: 10.1021/ja00171a043. [DOI] [Google Scholar]
- 121.Burmeister WP, Cottaz S, Driguez H, Iori R, Palmieri S, Henrissat B. The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure. 1997;5:663–675. doi: 10.1016/S0969-2126(97)00221-9. [DOI] [PubMed] [Google Scholar]
- 122.Noguchi J, Hayashi Y, Baba Y, Okino N, Kimura M, Ito M, Kakuta Y. Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase. Biochem Biophys Res Commun. 2008;374:549–552. doi: 10.1016/j.bbrc.2008.07.089. [DOI] [PubMed] [Google Scholar]
- 123.Keresztessy Z, Kiss L, Hughes MA. Investigation of the active site of the cyanogenic beta-d-glucosidase (linamarase) from Manihot esculenta Crantz (cassava). II. Identification of Glu-198 as an active site carboxylate group with acid catalytic function. Arch Biochem Biophys. 1994;315:323–330. doi: 10.1006/abbi.1994.1507. [DOI] [PubMed] [Google Scholar]
- 124.Wang Q, Trimbur D, Graham R, Warren RA, Withers SG. Identification of the acid/base catalyst in Agrobacterium faecalis beta-glucosidase by kinetic analysis of mutants. Biochemistry. 1995;34:14554–14562. doi: 10.1021/bi00044a034. [DOI] [PubMed] [Google Scholar]
- 125.Wang Q, Graham RW, Trimbur D, Warren RAJ, Withers SG. Changing enzymic reaction mechanisms by mutagenesis: conversion of a retaining glucosidase to an inverting enzyme. J Am Chem Soc. 1994;116:11594–11595. doi: 10.1021/ja00104a060. [DOI] [Google Scholar]
- 126.Mackenzie LF, Wang Q, Warren RAJ, Withers SG. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J Am Chem Soc. 1998;120:5583–5584. doi: 10.1021/ja980833d. [DOI] [Google Scholar]
- 127.Ly HD, Withers SG. Mutagenesis of glycosidases. Annu Rev Biochem. 1999;68:487–522. doi: 10.1146/annurev.biochem.68.1.487. [DOI] [PubMed] [Google Scholar]
- 128.Marana SR. Molecular basis of substrate specificity in family 1 glycoside hydrolases. IUBMB Life. 2006;58:63–73. doi: 10.1080/15216540600617156. [DOI] [PubMed] [Google Scholar]
- 129.Verdoucq L, Czjzek M, Moriniere J, Bevan DR, Esen A. Mutational and structural analysis of aglycone specificity in maize and sorghum β-glucosidases. J Biol Chem. 2003;278:25055–25062. doi: 10.1074/jbc.M301978200. [DOI] [PubMed] [Google Scholar]
- 130.Berrin J-G, Czjzek M, Kroon PA, McLauchlan WR, Puigserver A, Williamson G, Juge N. Substrate (aglycone) specificity of human cytosolic β-glucosidase. Biochem J. 2003;373:41–48. doi: 10.1042/BJ20021876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cicek M, Blanchard D, Bevan DR, Esen A. The aglycone specificity-determining sites are different in 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)-glucosidase (maize beta-glucosidase) and dhurrinase (sorghum beta-glucosidase) J Biol Chem. 2000;275:20002–20011. doi: 10.1074/jbc.M001609200. [DOI] [PubMed] [Google Scholar]
- 132.Zouhar J, Vévodová J, Marek J, Damborský J, Su X-D, Bryzobohatý B. Insights into the functional architecture of the catalytic center of a maize β-glucosidase Zm-p60.1. Plant Physiol. 2001;127:973–985. doi: 10.1104/pp.010712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Srisomsap C, Svasti J, Surarit R, Champattanachai V, Sawangareetrakul P, Boonpuan K, Subhasitanont P, Chokchaichamnankit D. Isolation and characterization of an enzyme with beta-glucosidase and beta-fucosidase activities from Dalbergia cochinchinensis Pierre. J Biochem. 1996;119:585–590. doi: 10.1093/oxfordjournals.jbchem.a021282. [DOI] [PubMed] [Google Scholar]
- 134.Zollner H. Handbook of enzyme inhibitors. Weinheim: VCH; 1989. pp. 94–95. [Google Scholar]
- 135.Burmeister WP, Cottaz S, Rollin P, Vasella A, Henrissat B. High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. J Biol Chem. 2000;275:39385–39393. doi: 10.1074/jbc.M006796200. [DOI] [PubMed] [Google Scholar]
- 136.Kengen SW, Luesink EJ, Stams AJ, Zehnder AJ. Purification and characterization of an extremely thermostable beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus . Eur J Biochem. 1993;213:305–312. doi: 10.1111/j.1432-1033.1993.tb17763.x. [DOI] [PubMed] [Google Scholar]
- 137.Nam KH, Kim S-J, Kim M-Y, Kim JH, Yeo Y-S, Lee C-M, Jun H-K, Hwang KY. Crystal structure of engineered beta-glucosidase from soil metagenome. Proteins. 2008;73:788–793. doi: 10.1002/prot.22199. [DOI] [PubMed] [Google Scholar]
- 138.Chi YI, Martinez-Cruz LA, Jancarik J, Swanson RV, Robertson DE, Kim SH. Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Lett. 1999;445:375–383. doi: 10.1016/S0014-5793(99)00090-3. [DOI] [PubMed] [Google Scholar]