Abstract
One of the most extraordinary events in the lifetime of a cell is the coordinated separation of sister chromatids during cell division. This is truly the essence of the entire mitotic process and the reason for the most profound morphological changes in cytoskeleton and nuclear organization that a cell may ever experience. It all occurs within a very short time window known as “anaphase”, as if the cell had spent the rest of its existence getting ready for this moment in an ultimate act of survival. And there is a good reason for this: no space for mistakes. Problems in the distribution of chromosomes during cell division have been correlated with aneuploidy, a common feature observed in cancers and several birth defects, and the main cause of spontaneous abortion in humans. In this paper, we critically review the mechanisms of anaphase chromosome motion that resisted the scrutiny of more than 100 years of research, as part of a tribute to the pioneering work of Miguel Mota.
Keywords: Pac-man, Flux, Dynein, Anaphase, Force
Introduction
When the English mountaineer George Mallory1 was asked why he wanted to climb Mount Everest, he replied simply “because it is there”. This can be viewed as a sign of admiration and respect for one of Nature’s most impressive beauties and challenge for mankind. In this sense, anaphase is the cell's own Mount Everest, justifying the interest of many generations of cell biologists in the mechanisms behind chromosome movement. A kernel difference, however, is that the latter remain to be conquered after more than a century of research. Part of the problem towards a universal theory for “the mitotic mechanism” has been related to the fact that different organisms do it differently, and a standing point of debate concerns the mechanisms of force production behind anaphase chromosome motion, which constitute the main subject and purpose of this review.
Anaphase: historical note
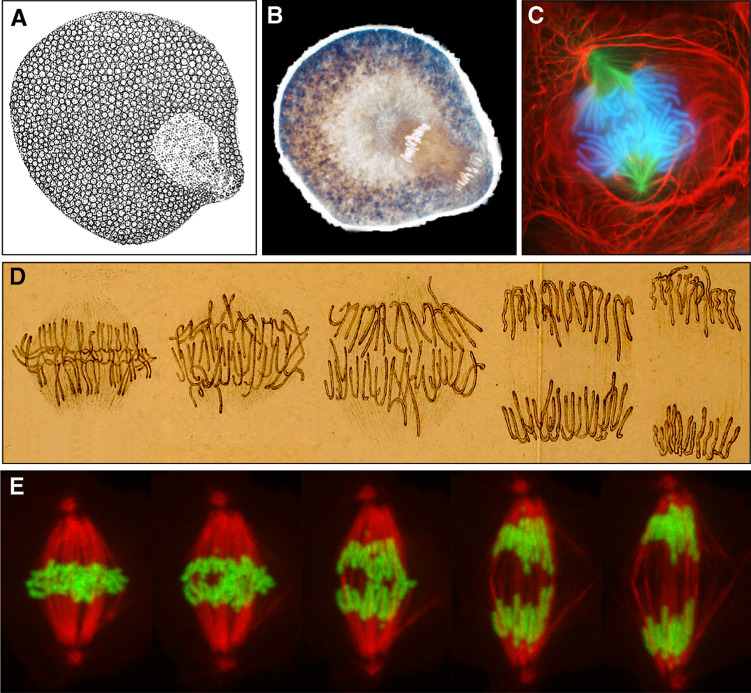
The first representation of a cell in anaphase dates from 1871 when the Russian embryologist Alexander Kowalevski captured the moment of the formation of the first micromeres in the worm Rhynchelmis [1, 2] (Fig. 1). At the time, he believed to be seeing division-products of the nucleolus. It was only after the work of Flemming that it was realized that what was being observed represented the longitudinal splitting of the chromosomes [3]. The term “anaphases” (from the Greek ana, meaning going back) was introduced by Strasburger to account for “the phases passed through [by the nucleus] from the complete separation of the daughter-segments to the final establishment of the daughter-nuclei” [4, 5]. As originally used by Strasburger, “anaphases” included the modern “telophase”, which was subsequently classified as a separate stage [6]. Curiously, E.B. Wilson in his 1925 edition of “The Cell in Development and Heredity”, still refers to “anaphases” in the plural but with the connotation of earlier and later events related to the longitudinal separation of sister chromatids [7]. The first clear demonstration that the anaphase movement consists of separate processes or steps was provided by Hans Ris [8, 9]. These steps, later classified as “anaphase A” and “anaphase B” [10], correspond, respectively, to the shortening of kinetochore fibers (k-fibers) resulting in chromosome approximation to the poles and to the elongation of the whole spindle resulting in further separation of poles and daughter groups of chromosomes. Importantly, Ris noted that in some systems these two steps appear to be temporally distinct events, while in others they may overlap (Fig. 1). Therefore, the use of a temporal connotation for the different “anaphases” may in some cases be inadequate.
Fig. 1.
a First illustration of anaphase during the division of the first micromeres in the worm Rhynchelmis by A. Kowalevski (adapted from [2]). b Contemporary view of micromere formation in the sand dollar embryo (courtesy of George von Dassow, University of Washington, WA, USA). c Newt lung cell in anaphase as viewed by fluorescence microscopy (courtesy of Conly Rieder, Wadsworth Center, NY, USA). d Sequence of the anaphase movement in a plant cell as originally depicted by Strasburger (adapted from [4]). e Sequence of the anaphase movement from a time-lapse movie of a Drosophila S2 cell stably expressing mCherry-α-tubulin (red) and GFP-H2B-Histone (green) (courtesy of Sara Moutinho-Pereira, IBMC, University of Porto, Portugal). Note the simultaneity of anaphase A and B
Anaphase forces
Chromosome velocity during anaphase is so slow (it varies from 0.2 to 5 μm/min) that it has been anecdotally compared with the speed of continental drift or fingernail growth. However, as noted by Nicklas, the main goal of mitosis is the correct distribution of chromosomes to daughter cells. Therefore, selection must favor precision rather than speed [11]. It has been suggested that chromosome velocity is slow in order to allow the decatenation of entangled DNA strands on the sister chromatids through the action of Topoisomerase II [12]. The recent discovery of PICH, a protein that associates with persistent DNA threads during anaphase, whose resolution is dependent on the activity of Topoisomerase II, strongly supports this view [13, 14]. The slow movement of chromosomes during anaphase may also provide the necessary time to correct and accurately segregate chromosomes with merotelic attachments, i.e., chromosomes with individual kinetochores bound to microtubules from both poles [15, 16].
Because anaphase chromosome movement is slow and chromosomes are relatively small, inertia and mass are negligible [17], and the viscous resistance (drag) is the only force that must be overcome. The force required to move a chromosome at typical anaphase velocities against viscous resistance has been estimated to be approximately 0.1 pN [18, 19]. On the other hand, the viscosity of the cytoplasm is not rate limiting for chromosome poleward velocity, since long ones seem to move just as fast as shorter ones that should offer less resistance [18, 20]. In a landmark study, Nicklas was able to measure the maximal force generated by the spindle during anaphase A in grasshopper spermatocytes, which he found to be around 700 pN, a value that is several orders of magnitude greater than the one previously estimated to be required for a chromosome to move against viscous drag [21]. Therefore, the velocity of chromosome movement to the poles must be governed by something other than viscous resistance, which can exert the measured amounts of force.
Anaphase A models
Amongst many rather ingenuous theories put forward until the mid-1950s, which attempted to explain anaphase chromosome movement by evoking currents, diffusion, tactoids, oscillating bodies, colloidal phenomena, or electrical and magnetic forces (reviewed by [22, 23]), the one based on fibrillar contractility [24, 25], the classic “traction fiber” model, appeared the most appealing. According to this theory, chromosomes could approach the poles by contraction of k-fibers anchored to the pole. However, the main objections were the observations that k-fibers do not increase in diameter between metaphase and telophase, as an elastic fiber would, and the lack of polar deformation due to anchoring. An alternative model based on “centromere repulsion” was proposed by Watase [26] and he found his most enthusiastic supporter in Darlington [27]. Yet this model was challenged because it could not explain the partitioning of maternal from paternal chromosomes during the monopolar division of primary spermatocytes in Sciara flies [28, 29]. These observations together with the fact that in some plant hybrids chromosomes move asynchronously during mitosis [30, 31], provided the foundations for the “chromosome autonomy theory”, which argues that chromosomes are moved individually and control their own movement. On the other hand, if centromeres are not repelled and k-fibers do not contract, their shortening during anaphase must be accompanied by gradual disassembly, which could occur from their polar or chromosomal ends.
It is now well established that the mitotic spindle is composed of dynamic microtubules. However, the most controversial issue has been whether changes in k-fiber length by loss of microtubule subunits simultaneously determine the velocity and provide the force for chromosome movement [10, 32–34] or if the rate of change in k-fiber length simply limits/governs the velocity of chromosome movement caused by a mechanically separate force producer [35, 36]. We will next detail and expand on current models of force production during anaphase A, which represent variations from the two main lines of thought that have just been depicted (Fig. 2).
Fig. 2.
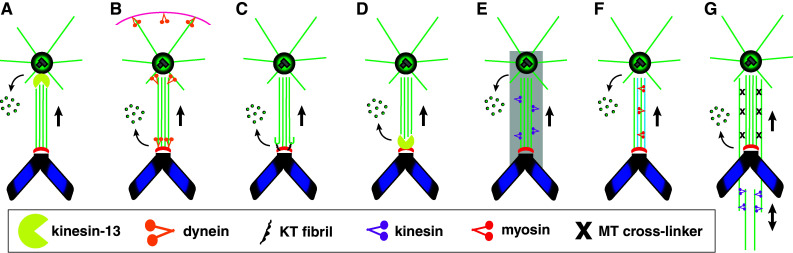
Schematic representation of anaphase models. a Traction-fiber model where active microtubule depolymerization of k-fibers by kinesin-13 proteins occurs at the minus ends. b Kinetochore dynein model where dynein ATPase activity drives anaphase A; dynein at the poles and cortex provides a pulling force that prevents spindle collapse and may drive anaphase B. c Conformational wave model where the bending of protofilaments at microtubule plus ends coupled with kinetochore fibrils drives anaphase A independently of ATP hydrolysis. d Kinetochore pac-man model where active microtubule plus end depolymerization by kinesin-13 proteins occurs at kinetochores. e Spindle matrix model where immobilized kinesin motors exert a poleward force on k-fibers. f Actin/myosin model (blue lines) where actin is a structural component of k-fibers which contracts by the action of myosin. g Coupled sliding model where motors of the kinesin family slide interpolar microtubules leading to spindle elongation and whose poleward force is transmitted along k-fibers by microtubule cross-linking molecules. Combinations between different models are possible
Force generation by microtubule depolymerization at minus-ends
In 1949, Östergren postulated that spindle fibers were composed of “aggregates” in a dynamic equilibrium with their subunit molecules, and that subtraction or addition of subunits results in shortening or lengthening of spindle fibers thus moving chromosomes [37]. Bungo Wada working with living Tradescantia stamen hair cells proposed that “the disintegration of each fiber from its distal end to the kinetochore […] takes place in the spindle poles, and the traction fibers are drawn continuously into the spindle poles […] and bring their kinetochore to the spindle poles” [38]. In Haemanthus endosperm, granules in front of a kinetochore at the start of anaphase move at the same speed of chromosomes in anaphase, and the distance between granules and kinetochores does not change for a considerable period, supporting the theory that k-fibers shorten essentially from their pole-associated end [39]. These ideas were substantiated by the seminal works of Shinya Inoue with the polarization microscope [10, 32, 33], who proposed that the poleward chromosome movement of anaphase is governed by the energy derived from the microtubule disassembly reaction. In agreement, Dietz proposed the “stable-state equilibrium” model based on continuous formation of k-fibers at kinetochores which are continuously disorganized at the spindle pole, predicting a permanent flux of microtubule subunits from the equatorial plate towards the poles [34]. After anaphase onset, polymerization at the kinetochore stops and depolymerization takes place predominantly at the poles pulling the kinetochore polewards [10]. This constitutes the modern vision of a traction fiber model.
Although the transport properties of the spindle as a possible mechanism for chromosome movement have been known since Schaede, who refers to a “streaming of spindle substance toward the poles” [40], it was only after the works of Inoue and Forer that it was demonstrated that kinetochore microtubules are dynamic structures. They used U.V. microbeam irradiation of k-fibers to create areas of reduced birefringence, which move poleward at approximately the same rates of anaphase chromosomes [41–43]. Subsequently, Hamaguchi and collaborators reported a poleward movement of spindle microtubules by tracking bleached marks in sand dollar egg spindles injected with fluorescently labeled tubulin [44]. However, from this study, it was not clear whether the bleached marks corresponded to kinetochore microtubules. A similar phenomenon was observed on kinetochore microtubules in photobleaching studies by Gorbsky and Borisy [45], as well as photoactivation of a fluorescent tubulin derivative by Mitchison [46]. These experiments extended a previous work aimed at determining the sites of microtubule assembly and disassembly in the mitotic spindle, including k-fibers [47], and provided definitive proof for the existence of a “polewards microtubule flux”. Importantly, “flux” was subsequently shown to require ATP hydrolysis but was insensitive to vanadate (which at micromolar concentrations inhibits dynein but not kinesin ATPase activity in vitro), suggesting the involvement of motors of the kinesin family [48]. As we know more about “flux”, we begin to realize that this process may actually involve two independent components: the active depolymerization of spindle microtubules at their minus ends, which may occur in the absence of any detectable poleward sliding of microtubules [49, 50] and the poleward sliding of microtubules, including k-fibers, which may occur in the absence of any detectable depolymerization at their minus ends [51–53]. Several works in many organisms and cell types have shown that, with few known exceptions, the velocity at which microtubules depolymerize at their minus ends is equal to or slower than that of chromosome movement to the poles [54]. Therefore, in some cases, microtubule minus end depolymerization may account for the entire shortening of k-fibers during anaphase A, whereas in other cases this would be insufficient, revealing the existence of alternative, possibly cooperative, mechanisms. The classic examples of the former include plants [55] and insect spermatocytes where microtubule minus-end depolymerization is even faster than chromosome motion to the poles [56]. Drosophila embryos, S2 cells, Xenopus oocyte extract spindles, newt lung cells, mouse, pig, and human cells are all examples of the latter [49, 50, 57–65]. Indeed, in these systems, microtubule minus end depolymerization is turned off or significantly attenuated during anaphase [54, 65, 66]. A possible explanation can be brought up by evoking anaphase B. Brust-Mascher and colleagues [58, 67] proposed a model for anaphase B in which the velocity of spindle elongation is governed by the extent that microtubule minus end depolymerization is suppressed. As pointed out before, anaphase B in many systems cannot be temporally distinguished from anaphase A, which means that spindle elongation in those systems occurs within the very first instant upon the onset of anaphase. In agreement, plant or insect spermatocyte spindles, which maintain high depolymerization activity at microtubule minus ends, elongate little [9, 68]. Finally, if microtubule minus end depolymerization generates a force capable of moving chromosomes, it implies that k-fibers must be anchored at the poles (and these anchored to the cell cortex) or along the length of the fiber, otherwise spindles would be predicted to collapse.
The microtubule depolymerizing activity at the poles has been attributed to the kinesin-13 protein Kif2a in Xenopus and mammals, and to Klp10A in Drosophila [51, 61, 62, 69, 70]. However, the fact that newly created kinetochore microtubule minus ends by laser microsurgery are stable, but are still able to slide poleward at flux rates in metaphase [52, 65], suggests that the microtubule depolymerizing activity at the poles might be a cellular response to regulate spindle length and not necessarily the flux-driving force. Indeed, recent studies showed that spindle lengthening as result of mechanical compression in mitotic Ptk1 cells is associated with the downregulation of microtubule minus end depolymerization, without affecting poleward microtubule sliding [53]. Taken together, these results suggest that microtubule depolymerization at the poles acts as a governor that limits and regulates spindle length in response to external force producers that slide microtubules poleward [65], similar to what has been proposed for spindle elongation during anaphase B [67]. According to this model, one would predict that inhibition of Klp10A or Kif2a attenuates “flux” even if the force sliding microtubules poleward is still present, consistent with what has been experimentally observed [61, 64, 65, 70].
Force generation by kinetochore motors
Kuwada and Belar [71, 72] introduced the kinetochores as important agents in the mechanics of chromosome movement. However, it was only after the classic experiments of Carlson with acentric chromosome fragments in grasshopper neuroblasts [73], together with the observation that kinetochores often lead the way during anaphase, that it became established that the active element of the chromosome responsible for anaphase movement is the kinetochore. The key question was how? In 1936, Eleanor Carothers remarked that spindle fibers “apparently, are reincorporated into the anaphase chromosomes as they move towards the poles” and proposed that “chromatids move apart through some inner mechanism and that the [spindle fibers] act more as guides than as traction fibers” [74]. These ideas were firmly structured into a new hypothesis of the anaphase movement by Miguel Mota in 1956 on the occasion of the International Genetics Symposia in Japan [75]. Accordingly, Mota proposed that “the chromosome has in itself the energy for the anaphase movement and that the centromere [i.e. the kinetochore] is not only the point through which the chromosome is moved but also its own engine”. In a rather unorthodox analogy, Mota compared the action of the kinetochore to that of a jet engine, which generates a poleward reaction that pushes the chromosome during anaphase, while releasing a non-birefringent “substance” that results from the transformation of the spindle material (Fig. 3). At that time, this possible “jet effect” was even thought to account for the still unexplained deformation of the spindle at the equator observed at the onset of anaphase I in time lapse movies of grasshopper spermatocytes [9]. With the inherited limitations of what was known by then about the properties of the spindle material, it is truly remarkable that this new anaphase hypothesis was formulated 9 years before the discovery of the first microtubule motor [76], and more than 30 years before the identification of microtubule motors at kinetochores [77, 78]. This vision of anaphase can thus be considered the first premonition of the mechanism later baptized as “pac-man” [79] in analogy to the Arcade videogame of the same name.
Fig. 3.

Hans Ris (right) chairs the session where Miguel Mota (left) presented his view of the kinetochore as an “engine” in the International Genetics Symposia held in Japan in 1956. Mota used to fly his own Piper Cub from Lisbon to Porto to use the Electron Microscope. His passion for aeroplanes evidently had a strong influence in the conceptualization of his anaphase hypothesis
The direct demonstration of kinetochores as active elements in the anaphase movement came only 30 years after Mota’s original hypothesis. First, it was observed that cells microinjected with biotin-labeled tubulin in late metaphase and fixed at various times showed no label proximal to the kinetochores by mid-anaphase, suggesting that tubulin dissociates from kinetochores (the released “substance” of Mota) as they move poleward [47]. An explanation advanced was that a kinetochore “motor” could be turned on in anaphase but is inactive or less active in metaphase. Subsequently, through photobleaching studies on spindle microtubules, Gorbsky and collaborators [66, 80] showed that chromosomes move to and through a persistent bleach mark, with little change of position of the mark relative to the poles. In a different set of experiments, Nicklas cut microtubules across the middle of the spindle between chromosomes and the pole, and upon anaphase onset in partially lysed preparations of grasshopper spermatocytes, chromosomes moved to the ends of microtubules at the edge of the cut [81, 82]. Overall, this work provided compelling evidence that at least some force producers for chromosome movement to the poles during anaphase were located within or near the kinetochore attachment site.
Kinetochore dynein
The microtubule minus end-directed motor dynein is enriched at the fibrous corona of unattached kinetochores [77, 78, 83], representing an obvious candidate for a kinetochore motor that could drive chromosomes towards the poles. Experimental evidence in support of this view, however, came many years before dynein was localized to kinetochores, but the results were often contradictory. In lysed Ptk1 cells, chromosome motion could be stopped by KCl extraction and re-initiated after the addition of a flagellar dynein-containing extract [84]. Addition of antisera against fragment A of flagellar dynein to isolated mitotic apparatuses completely blocked both chromosome-to-pole movement and spindle elongation in an ATP-dependent manner [85]. However, in this case, it seemed that the cell did not even enter anaphase, which can now be interpreted in the light of the role of kinetochore dynein in the inactivation of the spindle-assembly checkpoint (SAC) [86] and not necessarily due to a direct requirement for chromosome motion. The addition of vanadate into lysed mitotic cells in the presence of EGTA to buffer the concentration of free Ca2+, caused only an impact on anaphase B [87, 88], which was subsequently confirmed after inhibition of dynein ATPase activity with erythro-9-[3-(2-hydroxynonyl)] adenine [89]. Curiously, millimolar concentrations of vanadate or anti-dynein antibody injections did not affect anaphase movements in unlysed Ptk1 cells [90, 91].
Subsequent genetic analysis in budding and fission yeast further revealed that spindle dynamics and chromosome movement in these systems do not require dynein function [92–95]. On the other hand, injection of dynein antibodies or overexpression of human p50 dynamitin in Drosophila syncytial embryos was reported to affect chromosome movement by impairing both anaphase A and B [96, 97]. However, from these studies and due to the rapid syncytial divisions, it was not clear whether the observed effects on anaphase A were truly due to a requirement for kinetochore dynein in chromosome movement or to an indirect effect caused by “forced” mitotic exit. Additionally, the contribution of dynein motor activity at the cell cortex or at the poles could not be ruled out. Indeed, hypomorphic dynein mutant neuroblasts in Drosophila are known to accumulate in prometaphase/metaphase with a high frequency of spindle defects, and p50 dynamitin has been identified in the cell cortex of syncytial embryos [98]. In another study, Savoian and colleagues [99, 100] used zw10 and rod Drosophila mutant spermatocytes to specifically perturb dynein localization at kinetochores, and showed that anaphase chromosome motion to the poles was severely affected in those mutants. However, some potential caveats with this experimental approach are related to the fact that zw10/rod are also required to recruit the SAC protein Mad2 to unattached kinetochores, thereby compromising SAC function [101–103]. Kinetochore dynein is also involved in the initial MT capture at the entry of mitosis, which could indirectly compromise chromosome motility if anaphase is triggered with normal kinetics [100]. Subsequent work in Ptk1 cells overexpressing p50 dynamitin reported a 30% reduction in chromosome-to-pole velocity after abrogation of the SAC [86]. More recently, Yang and colleagues [104] selectively decreased the kinetochore pool of dynein by knocking down ZW10 in human U2OS cells. The authors reported a 40% reduction in chromosome-to-pole velocity during anaphase, suggesting that kinetochore dynein is important in powering chromosome motion. It is somewhat surprising that the observed reduction only corresponds to about half the reported “pac-man” activity in U2OS cells [61], suggesting that other mechanisms or factors are involved. Moreover, it remains to be elucidated whether the observed reduction is a direct result of lack of dynein processive motion along k-fiber microtubules, or whether it is due to defective/unstable microtubule attachments, possibly associated with a defective SAC [103–105]. The recent discovery of spindly, a protein that targets dynein to kinetochores in Drosophila, C. elegans, and humans, but which does not impair the recruitment of Mad2 to kinetochores [106–108], may prove to be an important tool for the clarification of the above-mentioned issues.
In a very interesting study by Vorozhko and colleagues [109], injection of Ndc80 antibodies into mitotic Xenopus S3 cells prevented microtubule end-on attachments, did not affect the recruitment of dynein/dynactin to kinetochores, and chromosomes were still able to undergo rapid dynein-mediated poleward movements typical of initial capture of microtubules during prometaphase, but at anaphase onset chromosomes did not move poleward. Noteworthy, at this stage, not even the fast dynein-mediated poleward movements were observed, suggesting that kinetochore dynein motor activity is switched off at the beginning of anaphase. These results support that microtubule end-on attachment, rather than dynein motor activity at kinetochores, is essential for chromosome movement to the poles. It should be emphasized that dynein levels at kinetochores become reduced as microtubules attach and are barely detectable as cells enter anaphase [98, 110]. Recently, it was shown that phosphorylation of Threonine 89 on dynein intermediate chain directs binding to ZW10 and that microtubule attachment induces dynein dephosphorylation to undetectable levels after metaphase chromosome alignment [111]. Still, dynein could be attached to the kinetochore in very low numbers and contribute significantly to poleward force production. Nicklas calculated that hydrolysis of about 20 ATP molecules is enough to move one chromosome against viscous drag, with a net power output of 3 × 10−2 ATP units/s/chromosome [18, 112]. For a single dynein molecule in flagella, the total power expended is ~20 ATP units/s [113, 114]. In a system working at 20% efficiency, the hydrolysis of 4 ATP units/s/dynein molecule is a reasonable estimate, which means that, in theory, a single (flagellar) dynein motor produces a net power output that is 100× greater than the one required to move a chromosome. However, this argument loses weight by the fact that knocking-down ~85% of ZW10, which should reduce but not fully deplete kinetochore dynein, causes a significant reduction of chromosome movement to the poles [104]. Taken together, these results indicate that, if directly involved, dynein motor activity at kinetochores plays a minor role in powering anaphase chromosome motion.
Force generation by microtubule depolymerization from plus ends
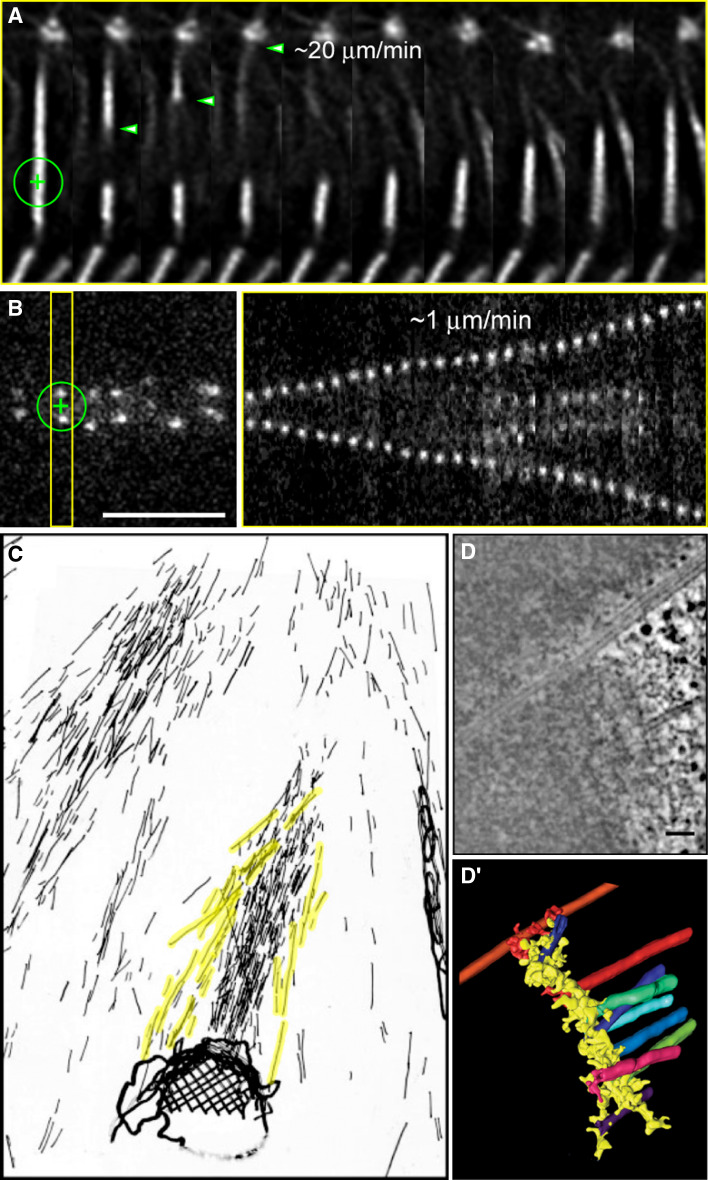
That microtubule depolymerization induced by colchicine, cold, or high pressure in living cells can generate a force that is sufficient to move chromosomes has been known since the works of Inoue, Salmon, and colleagues [10, 43, 115–117]. These works have also shown that the faster the depolymerization of microtubules, the faster the poleward movement of chromosomes, which means that microtubule depolymerization either directly provides the motile force or acts as a rate-limiting step that defines the speed of chromosome-to-pole motion. However, it was not known where the actual depolymerization of the microtubule was occurring, with the dynamic equilibrium model favoring depolymerization at the microtubule minus ends [10]. In 1988, Koshland and colleagues showed that microtubule shortening was able to move chromosomes in vitro by tubulin dissociation from the kinetochore [118]. Importantly, the rate of microtubule depolymerization following dilution of tubulin was the same in the absence of ATP, suggesting that the poleward force at the kinetochore may be generated by some form of energy stored in the microtubule lattice [119], such as that provided by a conformational change associated with GTP-hydrolysis and/or curling of protofilaments [118]. This became known as the “conformational-wave” model. Similarly, microtubule depolymerization induced by a constant stream of buffer that lacked tubulin, ATP, and GTP was found to move chromosomes in vitro, with one or more depolymerizing microtubule producing a force that was at least 1 pN [120]. This is significantly less than the maximum force a spindle can produce during anaphase A, but more than sufficient to move a chromosome in a viscous cytoplasm at the observed velocities [121]. Curiously, the maximum force produced by a single depolymerizing microtubule has been estimated to be approximately 40 pN [122], which is essentially the same maximum force estimated per kinetochore microtubule from real measurements in grasshopper spindles [121]. It should be noted that the velocity of chromosome movement by depolymerizing microtubules (~16 μm/min; [120]) is significantly faster than normal anaphase chromosome movement and somewhat slower than the observed rates of free microtubule plus end depolymerization in vitro [123], in Xenopus extract spindles [124] and of an entire k-fiber in living Drosophila culture cells [52] (Fig. 4). This suggests that kinetochores on chromosomes are resistive couplers against microtubule depolymerization in vivo (Fig. 4). Indeed, subsequent in vitro studies have shown that the velocity of microtubule depolymerization-coupled movement when chromosomes are attached is about fivefold slower than the rate of shortening for microtubules free in solution ([125]; see also [126]).
Fig. 4.
a Laser-mediated severing of a k-fiber in a Drosophila S2 cell stably expressing GFP-α-tubulin during metaphase. Note the fast depolymerization of the pole-proximal fragment and that the chromosome which remains attached to the severed k-fiber maintains its equatorial position (adapted from [52]). b Laser-mediated severing of the centromeric region in a Drosophila S2 cell stably expressing CID-GFP during metaphase. Note the slow poleward migration of each daughter kinetochore after surgery. Scale bar (a,b) 5 μm. c 3D-electron microscope reconstruction of a severed k-fiber from crane flies after irradiation with a UV microbeam (adapted from [177]). Non-kinetochore microtubules in the vicinity of the resulting k-fiber stub were pseudocolored in yellow. d Single slice from a tomographic reconstruction of a Ptk1 kinetochore showing both end-on and lateral MT binding (courtesy from Yimin Dong and Bruce McEwen, Wadsworth Center, NY, USA). Scale bar 100 nm. d’ 3D surface rendering of the 3D volume of the same kinetochore
In a search for potential coupling molecules at the kinetochore, it was found that the kinesin-7 motor CENP-E, but not dynein or MCAK, was required for microtubule depolymerization-dependent motion of chromosomes (but not attachment!) in vitro [127]. Importantly, this was found to be independent of the ATPase activity of CENP-E. Recently, it was shown that a motor-independent function of CENP-E is to promote kinetochore microtubule turnover, which may potentiate depolymerizing microtubules to do work on chromosomes [128]. Another important candidate found in budding yeast was the Dam1 complex, which can form rings around microtubules in vitro [129]. However, while such rings may efficiently couple microtubule depolymerization to chromosome movement in a system where kinetochores bind only to a single microtubule [130, 131], they are not essential in fission yeast [132], and orthologues have never been found outside fungi. Additionally, it was subsequently shown that ring formation is not required for microtubule depolymerization-driven motion of the Dam1 complex [133, 134].
Another question that remains to be addressed is how the energy stored within the microtubule lattice could be converted into sufficient mechanical work to move a chromosome poleward during anaphase. In 2005, McIntosh and co-workers provided theoretical and experimental evidence in support of the conformational-wave model [135, 136]. By conjugating streptavidin-coated glass microbeads to biotinylated microtubules, they observed that, upon depolymerization, microtubules exert a brief pull on the beads before being released. This was interpreted as if the force exerted on the beads was generated by the bending of protofilaments as microtubules depolymerize, and the authors were successful in measuring the resulting forces. Accordingly, it was determined that the maximal force generated was ~46 pN, which might correspond to the bending of 1 or 2 protofilaments. This is about 10 times the force developed by kinesin molecules [137], and it is expected that at kinetochores, with a proper coupler, all protofilaments would act in concert, increasing the force generated by a depolymerizing microtubule to at least 30–65 pN [136]. In agreement, the force produced by this system was shown to be dependent on the curvature of protofilaments [138]. Finally, it has also been suggested that electrostatic interactions between kinetochores and the depolymerizing microtubule plus ends could provide additional force for chromosome translocation during anaphase A [139].
The fact that microtubule depolymerization from kinetochores can generate a force that is sufficient to drive chromosome motion in vitro does not mean that it does so in vivo. Early works on nocodazole-induced microtubule depolymerization during prometaphase/metaphase in vertebrate cells have demonstrated that tubulin loss occurred at kinetochores, leading to chromosome approximation to the poles [140, 141]. Moreover, it has been demonstrated that, like depolymerizing microtubules in vitro, kinetochore microtubule plus ends in yeast, worms, Drosophila, and vertebrate somatic cells have bent protofilaments [142–146]. Definitive work by Grishchuk and McIntosh [94] showed that deletion of all the three minus end-directed motors found in the fission yeast genome did not abolish the poleward movement of chromosomes during anaphase. Because fission yeast lacks any detectable flux and associated depolymerization at the microtubule minus ends during anaphase [147], the most plausible explanation for these results is that anaphase A in this system is driven by depolymerization at microtubule plus ends. What about the structural and molecular nature of the coupler at kinetochores? For the skeptics about rings and collars at kinetochores, McIntosh and colleagues [148] used electron tomography to show that kinetochore fibrils of about 50 nm are connected to curved protofilaments at microtubule plus ends, defining a structural entity at kinetochores that could effectively work as a coupler for chromosome motion by microtubule plus end depolymerization. By measuring the curvature of bending protofilaments associated with kinetochore fibrils, they estimated that each depolymerizing microtubule could produce a force of ~40 pN, in agreement with previous estimations [122]. They further showed that, in some successful cases, glass beads coated with reconstituted Ndc80 complex, a 57-nm-long component of the core attachment site at kinetochores [149], could couple microtubule depolymerization to movement towards the minus ends. This data suggest that the Ndc80 complex may correspond to the kinetochore fibrils observed by electron tomography, but also that additional factors may be involved in microtubule depolymerization-coupled movement [150]. The use of “bonsai” forms of the Ndc80 complex that retain microtubule binding capacity but are only 15 nm long [151] should prove useful in determining the molecular nature of the kinetochore fibrils.
It has been noted that birefringence of k-fibers increases at anaphase onset [152], which, together with a firmer attachment of chromosomes to k-fibers during anaphase [45, 153], suggests increased microtubule stabilization. At the entry of anaphase, CDK1 activity drops abruptly, which is known to promote microtubule stabilization in vitro and in vivo [154, 155], whereas prevention of CDK1 inactivation by expression of non-degradable cyclin B caused a 20% reduction in the velocity of anaphase A in Xenopus egg extracts [156]. Moreover, in some systems, like insect spermatocytes, kinetochore microtubules polymerize rather then depolymerize at their plus ends during anaphase [56, 157]. Curiously, a similar phenomenon has been reported in Ptk1 cells injected with tubulin but only during early anaphase, without affecting normal chromosome-to-pole velocity [158]. All these observations suggest that, at least for some systems, an active mechanism counteracts kinetochore microtubule stabilization/polymerization at anaphase onset leading to their disassembly from plus ends.
Force generation by microtubule depolymerases at kinetochores
It has been difficult to determine whether anaphase A requires ATP. In this regard, the work of Spurck and Pickett-Heaps may have shed light on the problem [119]. By using permeabilized Ptk1 and newt lung cells, these authors noted that anaphase A could be resumed either by the addition of ATP or by treatments that promote microtubule disassembly, such as cold or calcium. These observations suggest that ATP is required for kinetochore microtubule disassembly but not to power a processive kinetochore motor. Three broad protein families with ATPase activity have been shown to promote microtubule disassembly in vitro and/or in vivo: kinesin-8, kinesin-13, and class II AAA ATPases [159, 160]. Kinesin-8 proteins additionally show plus end-directed movement in microtubule-based motility assays, localize to kinetochores in several systems, and have been found to couple microtubule depolymerization to the movement of plastic beads [161]. However, they appear to be dispensable for anaphase A in fission yeast [162]. Curiously, chromosomes in Drosophila kinesin-8 mutants showed reduced poleward velocities during anaphase in male meiosis I [163]. Because RNAi-mediated depletion of kinesin-8 proteins in living Drosophila and human somatic cells accelerates chromosome movement during anaphase A [64, 164], the observations in Drosophila primary spermatocytes may be explained by the presence of merotelic attachments.
Kinesin-13 proteins, such as KLP10A and KLP59C in Drosophila, have been localized to inner kinetochores and inner centromeres, respectively [70]. Specifically, KLP59C has been proposed to actively depolymerize microtubules at kinetochores in Drosophila embryos, as antibody injections completely block kinetochore pac-man activity and reduce chromosome-to-pole motion by 60% in this system [70]. However, KLP59C depletion by RNAi in Drosophila culture cells does not affect anaphase A [64, 165]. Indeed, KLP59C location to inner centromeres does not permit access to most microtubule plus ends that terminate at the kinetochore outer plate in Drosophila somatic cells [145]. The specific requirement of KLP59C in Drosophila embryos may result from an adaptation to a rapid mitosis where KLP59C could depolymerize microtubules which are “fed” and made accessible by kinetochore dynein [97, 166]. In this regard, the third Drosophila kinesin-13, KLP59D, localizes to kinetochores and was recently shown to promote kinetochore microtubule plus end depolymerization [167]. Among the kinesin-13 proteins in human cells, only Kif2b and MCAK (Kif2c) localize to centromeres/kinetochores [69, 168, 169], but MCAK is not required for anaphase A [61]. Interestingly, chromosome velocities in monopolar spindles that form after Kif2b RNAi are severely reduced [168]. Spindle bipolarity after Kif2b RNAi can be restored by simultaneous knock-down of MCAK, with chromosomes moving towards the poles in anaphase at ~0.6 μm/min, which represents a two-thirds reduction in their normal velocity (Duane Compton, personal communication). This observation is intriguing as Kif2b is no longer detectable at kinetochores during anaphase, possibly due to its low abundance [168]. Finally, it has been reported that Katanin, an AAA ATPase that severs microtubules, is also important for the pac-man activity at kinetochores in Drosophila S2 cells [170]. Depletion of this protein reduces chromosome-to-pole movement by 50%, but it is believed that Katanin does not directly depolymerize microtubule plus ends and might work in concert with some kinesin-13 proteins at kinetochores. We conclude that some kinetochore proteins of the kinesin-13 family may directly promote the active depolymerization of microtubule plus ends during anaphase.
Force generation by non-microtubule elements
One intriguing aspect related to the velocity of poleward chromosome movement during anaphase is the apparent independency from the number of microtubules attached to kinetochores [116, 171–173], which can be interpreted as if kinetochore microtubules or kinetochores themselves are not force-generating elements. This idea has been vigorously defended over the last 40 years by Forer, Pickett-Heaps and colleagues, who believe that, at least for some cell types, the force for chromosome-to-pole movement is generated by non-microtubule elements in the spindle. Two main ideas have been proposed for the nature of this force-generating element: a sliding mechanism based on actin and myosin similar to the one responsible for muscle contraction [174], or the action of motors anchored to a spindle matrix [175, 176]. In both models, the force is transmitted along the length of the k-fibers, which might act as “governors” to limit the velocity of poleward movement.
Actin/myosin
There is no shortage of literature reporting the presence of actin and myosin forms in the mitotic spindle in a wide variety of cell types and organisms, including humans (summarized in [177]). The question is whether their presence reflects any functional role in force generation during chromosome movement to the poles. Crane fly spermatocytes treated before or during anaphase with high doses of cytochalasin D and latrunculin B, two drugs that disrupt actin filaments and/or inhibit actin polymerization, caused chromosome movement to stop or slow down [178]. The washout of the drugs usually reverted normal chromosome movement. In a subsequent study, somewhat contradicting findings were reported in the same system where addition of latrunculin B before anaphase did not cause an effect on chromosome movement during anaphase [179]. Under these experimental conditions, inhibition of myosin with two different drugs, 2,3-butanedione 2-monoxime (BDM), an inhibitor of myosin ATPase activity, and Y-27632, an inhibitor of Rho kinase which phosphorylates Myosin II, but not the addition of cytochalasin D, blocked chromosome movement. These observations led the authors to propose that crane fly spermatocytes have redundant mechanisms for anaphase chromosome movements and that myosin might work on actin filaments, or interact with kinetochore microtubules, either directly or through an intermediate component. It should be noted, however, that treatment of crane fly spermatocytes at anaphase onset with cytochalasins at 10-fold lower concentrations inhibit cytokinesis with no apparent effect on chromosome velocity [180]. Increasing cytochalasins to concentrations exceeding those needed to inhibit cytokinesis in this or other systems caused alterations in kinetochore structure and lagging chromosomes during anaphase [180, 181]. Nevertheless, in a different set of studies, treatment of crane fly spermatocytes with calyculin A, which hyperactivates myosin, was shown to accelerate poleward chromosome movement [182]. Finally, microinjection of phalloidin, a fungal toxin that prevents actin depolymerization, into Haemanthus endosperm slows down anaphase chromosome movement by 50% [183].
As usual in science, and typically in the mitosis field, several works on different model systems have led to opposite conclusions to what concerns the involvement of actin/myosin in force production for chromosome movement. First, anti-myosin sera had little or no effect on chromosome motion in isolated mitotic apparatus from equinoderms [85], or when microinjected into living eggs [184], while inhibiting cytokinesis in the latter case. Quantitatively, neither k-fiber shortening nor spindle elongation were affected by doses of antibody up to eightfold higher than those required to inhibit cytokinesis in injected cells [185]. Similarly, inhibition of actin/myosin by several means, including treatments with phalloidin and cytochalasins, did not have an effect on anaphase chromosome movements, while inhibiting cytokinesis in lysed or unlysed Ptk1 cells [186, 187]. Finally, treatment of living LLC-PK1 cells stably expressing GFP-α-tubulin with 20 μM cytochalasin D, the same concentration shown to inhibit chromosome motion in insect spermatocytes [178, 188], was sufficient to inhibit cytokinesis but had no significant effect on k-fiber shortening or spindle elongation when added right after anaphase onset (Fig. 5). Overall, these data pose strong challenges for a role of actin/myosin in the anaphase movement, but exceptions such as some insect spermatocytes and eventually in plants might exist in nature.
Fig. 5.
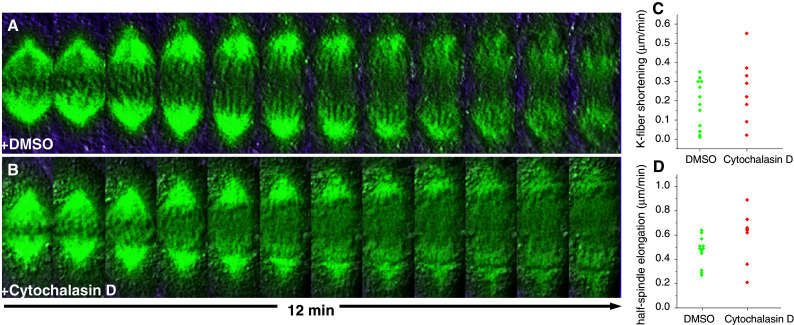
a, b Time-lapse sequences of LLC-PK cells stably expressing GFP-α-tubulin after treatment with DMSO or 20 μM Cytochalasin D, respectively. Chromosomes can be visualized by differential interference contrast in the superimposed image. Time between frames = 1 min. c, d Distributions of measurements of k-fiber shortening and half-spindle elongation velocities after treatment with DMSO or 20 μM Cytochalasin D. The mean k-fiber shortening velocities in DMSO and Cytochalasin D were, respectively, 0.19 ± 0.13 (n = 6 cells) and 0.26 ± 0.17 (n = 4 cells) for the represented period. Error intervals represent standard deviation. The differences are not statistically significant (P = 0.296; t test). The median half-spindle elongation velocities in the same cells after DMSO or Cytochalasin D were, respectively, 0.47 ± 0.12 and 0.60 ± 0.21. Error intervals represent standard deviation. The differences are not statistically significant (P = 0.058; Mann–Whitney rank sum test)
The spindle matrix
Several candidates have been proposed to form a non-microtubular spindle matrix but evidence for a direct role in anaphase remains controversial [189]. In Drosophila, a widely conserved protein, Megator, has been recently implicated in anaphase chromosome movement [190], through a specific role in spindle elongation. Surprisingly, this protein and its human counterpart Tpr were shown to be required for an efficient SAC response, and their depletion accelerated anaphase entry [190, 191]. Curiously, the observed defects in spindle elongation were significantly rescued by increasing the duration of mitosis prior to anaphase onset, suggesting that they were due to the formation of an immature spindle [190], but further experiments are necessary to completely rule out any direct involvement of a “spindle matrix” in the force-production system that drives anaphase movements.
Force generated by microtubule sliding
A “pump hypothesis” for chromosome movement during anaphase A has been envisioned, in which k-fibers anchored along their length to other spindle microtubules slide poleward like other cellular bodies not attached to the spindle [192]. Experimental evidence for this hypothesis came from experiments with UV microbeam irradiation of k-fibers by Forer, who showed that less than half of the k-fiber (not anchored at the pole!) is needed to move a chromosome [41]. Subsequently, McIntosh and colleagues proposed a sliding filament mechanism where a “motor” permanently attached to one microtubule is transitorily associated with another microtubule or filament (e.g., actin) [193]. This original hypothesis applied to anaphase A required non-kinetochore and kinetochore microtubules to be anti-parallel. However, it was later discovered that 90–95% of microtubules in each half-spindle have identical polarities [194–196]. After the discovery of microtubule treadmilling in vitro, a model based on poleward-sliding of anti-parallel microtubules coordinated with the opposite end assembly/disassembly of all spindle microtubules was proposed [197]. In this model, the movement of chromosomes during anaphase is driven by sliding of anti-parallel interpolar microtubules that overlap in the equatorial region, which produce an additional poleward force on kinetochore microtubules through linkages at the region of convergence near the poles. Similar ideas were proposed by Goode, but in this case lateral interactions along the entire k-fiber were envisioned to cross-link parallel non-kinetochore microtubules [198]. Kinetochore and non-kinetochore microtubules in close proximity have been widely reported in the literature, with the best evidence provided by 3D electron microscopy reconstructions of spindles and kinetochore microtubules [199, 200] (Fig. 4). Indeed, it has been estimated that every vertebrate kinetochore is tangentially associated with 3–8 non-kinetochore microtubules [200]. Additionally, several proteins, such as dynein and its co-factor NuMA, kinesin-14 members or the Drosophila microtubule-associated protein ASP have also been implicated in spindle pole focusing by mediating microtubule cross-linking [91, 201, 202]. In this regard, we favor a general distribution of microtubule cross-linkers along the length of the k-fiber, as this would explain why chromosomes lie at the equator after UV or laser microbeam irradiation of the respective k-fibers during metaphase [52, 203]. This behavior has been explained by evoking the participation of actin/myosin and/or a spindle-matrix in insect spermatocytes [204]. An alternative possibility would be that adjacent microtubules that do not terminate at the kinetochore could in principle transmit force to kinetochore microtubules via lateral cross-linking (Fig. 4). Accordingly, we and our collaborators have recently proposed a mechanical model of the spindle in which the flux-driving force is generated by motor proteins, such as kinesin-5 [205, 206], that could slide anti-parallel interpolar microtubules coupled to k-fibers by cross-linking molecules so that a poleward pushing force is transmitted along the entire k-fiber but not generated within it [65]. In this model, microtubule slippage from their kinetochore attachment sites in response to poleward pushing forces allows these same forces to redistribute and converge, while microtubule depolymerases at the poles regulate spindle length. In this way, the metaphase state reflects the uniformity of spindle forces, which subsequently ensure the synchronous poleward movement of chromosomes during anaphase.
Anaphase A versus anaphase B
In all the classic models of anaphase chromosome movement, a lot of attention is given to anaphase A and the importance of anaphase B appears to be largely depreciated. We do believe that, with few exceptions, anaphase B plays equal or even a most critical role than anaphase A ensuring accurate chromosome segregation in animals as this would intrinsically contribute to the uniformity of forces acting on individual chromosomes. In yeasts, spindle elongation is by far the main mechanism contributing to separate chromosomes during anaphase (spindle elongation is 5–7 times the metaphase spindle length, which also implies microtubule polymerization and growth). This may be related to the fact that, in these cases, there is no true metaphase plate and chromosomes enter anaphase from distinct positions relative to the spindle equator [207–209], which could compromise mitotic fidelity if anaphase A was the predominant mechanism.
Spindle elongation during anaphase B is a well-established motor-dependent process with increased sensitivity to the capacity to hydrolyze ATP than in anaphase A [85, 87, 210, 211], suggesting that the molecular motors and/or energetic requirements involved in each process are different. The elongation of the spindle may be driven by forces that are intrinsic to the spindle (e.g., pushing forces by antiparallel interpolar microtubule sliding) and/or by forces outside the spindle (e.g., separation of spindle poles by cortical-mediated pulling forces on astral microtubules), also known as anaphase C [212]. The microtubule sliding mechanism proposed by McIntosh and colleagues [193] explains at least part of the spindle elongation in many systems, including diatoms, yeasts, and Drosophila embryos [96, 213–216]. At the molecular level, the players involved in each system might be different, although their modus operandi must be essentially the same: cross-linking, sliding, and eventually growth/stabilization of interpolar microtubules of opposite polarity. These functions are provided by MAPs that localize to the spindle midzone, such as members of the PRC1, XMAP215, and CLASP families, as well as by force-producing motors of the kinesin-5 or -6 families [96, 217–225].
The elongation of the spindle based on pulling forces acting on astral microtubules has been primarily suggested by Boveri [226], and was largely supported by microsurgery experiments in some fungi (but not yeasts!), echinoderms, worms, insect spermatocytes, and vertebrates about a century later [82, 227–232]. In these experiments, either the asters or spindle midzone were cut with a microneedle or destroyed with a laser microbeam. While aster removal or destruction slowed down or halted spindle elongation, cuts through the spindle midzone consistently accelerated this movement several-fold. These data indicated that, in some systems, pulling forces acting on astral microtubules are the main drivers of spindle elongation, whereas spindle midzone microtubules resist or govern the rate of pole separation. Cytoplasmic dynein located at the cell cortex has been implicated in this astral MT pulling mechanism [89, 90, 96, 233–235]. On the other hand, proteins required for microtubule polymerization and growth/stabilization of interpolar microtubules [236], such as CLASPs [219], coordinated with the activity of kinesin-5 at the spindle midzone, might work as governors [237].
Conclusion
Here, we attempted to provide the different views of the anaphase movement that have been put forward over more than 100 years, and to identify the weaknesses and strengths of each prevailing model in the present days. In this sense, we deliberately adopted a “neutral” position by confronting apparently contradicting findings, leaving the decision to the readers on their favorite model without going for the mainstream. It seems clear, however, from this brief discussion that the “mitotic mechanisms” are not universal, and each organism uses one or a combination of several processes relying at least partially on microtubule properties. One should bear in mind that the ultimate goal of anaphase is the effective separation of the two sets of chromatids, from the shorter to the longer, far enough so that none get caught midway by the cleavage of the cell during cytokinesis. In this regard, recent works on how chromosome segregation is coordinated with cytokinesis provided important mechanistic insight towards a full picture of the anaphase mechanism [238–240]. For the future, it is expected that anaphase aficionados and newcomers to the field will be able to quantify the biophysical relevance and molecular nature of each of the discussed mechanisms in order to establish direct comparisons between evolutionary different systems towards a better understanding of life. As for the first complete ascent of Mount Everest by Sir Edmund Hillary, we wholeheartedly believe that “the final solution of the mitotic problem will almost certainly make use of parts of several different hypothesis, and none of these can be considered as having failed if it has contributed to the final answer” [22].
Acknowledgments
The authors would like to dedicate this review to Miguel Mota as part of a recent tribute to his seminal but largely ignored conceptual contributions to the mechanism of anaphase. We are indebted to Arthur Forer, Conly Rieder, David Sharp, Duane Compton, Gary Gorbsky, Jonathan Scholey, Rebecca Heald, Richard McIntosh, and Ted Salmon for discussing and sharing their views on anaphase movements. We would also like to thank Sara Moutinho-Pereira, Yimin Dong, Bruce McEwen, Conly Rieder, George von Dassow, and Miguel Mota for providing pictures, Irina Matos for library research, Gohta Goshima, Patrick Heun, and Pat Wadsworth for the gift of cell lines, and all the members of the Maiato laboratory for their critical reading of the manuscript. Finally, we apologize to all those whose work contributed for our current understanding of the mechanisms of chromosome movement, but which could not be directly cited due to space limitations. Work in the Maiato laboratory is funded by grants PTDC/BIA-BCM/66106/2006 and PTDC/SAU-OBD/66113/2006 from Fundação para a Ciência e a Tecnologia of Portugal, and the Gulbenkian Programme in the Frontiers of Life Sciences. The title of this review is inspired in the song “Perpetual movement” by the Master of Portuguese guitar, Carlos Paredes (http://www.youtube.com/watch?v=k9cqXIk2B04).
Footnotes
English mountaineer who took part in the first British expeditions to Mount Everest in the early 1920s. In June 1924, Mallory disappeared somewhere high on the North-East ridge during or after completing the final stage of his attempt to make the first ascent of the world’s highest mountain.
References
- 1.Baker J. The cell-theory: a restatement, history, and critique: Part V. The multiplication of nuclei. Q J Microscop Sci. 1955;3–96:449–481. [Google Scholar]
- 2.Kowalevski A. Embryologische Studien an Wurmerm und Arthropoden. Mem Acad Imper Sci St. Petersburg. 1871;16:1–70. [Google Scholar]
- 3.Flemming W. Beiträge zur Kenntniss der Zelle und ihrer Lebenserscheinungen. Arch Mikr Anat. 1879;16:302–436. doi: 10.1007/BF02956386. [DOI] [Google Scholar]
- 4.Strasburger E. Die Controversen der indirecten Kerntheilung. Arch Mikr Anat. 1884;23:246–304. doi: 10.1007/BF02952312. [DOI] [Google Scholar]
- 5.Carter LA. The somatic mitosis of Stegomyia fasciata . Q J Microscop Sci. 1918;2–63:375–386. [Google Scholar]
- 6.Heidenhain M. Neue Untersuchungen über die Centralkörper und ihre Beziehungen zum Kern-und Zellenprotoplasma. Arch Mikr Anat. 1894;43:423–758. doi: 10.1007/BF02933880. [DOI] [Google Scholar]
- 7.Wilson EB (1925) The cell in development and heredity. Macmillan, New York
- 8.Ris H. A quantitative study of anaphase movement in the aphid Tamalia . Biol Bull. 1943;96:90–106. doi: 10.2307/1538001. [DOI] [PubMed] [Google Scholar]
- 9.Ris H. The anaphase movement of chromosomes in the spermatocytes of the grasshopper. Biol Bull. 1949;96:90–106. doi: 10.2307/1538001. [DOI] [PubMed] [Google Scholar]
- 10.Inoue S, Ritter H., Jr Dynamics of mitotic spindle organization and function. Soc Gen Physiol Ser. 1975;30:3–30. [PubMed] [Google Scholar]
- 11.Nicklas RB. Chromosome movement: current models and experiments on living cells. Soc Gen Physiol Ser. 1975;30:97–117. [PubMed] [Google Scholar]
- 12.Gorbsky GJ. Chromosome motion in mitosis. Bioessays. 1992;14:73–80. doi: 10.1002/bies.950140202. [DOI] [PubMed] [Google Scholar]
- 13.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 14.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pidoux AL, Uzawa S, Perry PE, Cande WZ, Allshire RC. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J Cell Sci. 2000;113(Pt 23):4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- 16.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 17.Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. doi: 10.1119/1.10903. [DOI] [Google Scholar]
- 18.Nicklas RB. Chromosome velocity during mitosis as a function of chromosome size and position. J Cell Biol. 1965;25(Suppl):119–135. doi: 10.1083/jcb.25.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor EW (1965) Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. In: Copley AL (ed) Proceedings of the fourth international congress of rheology (1963). Interscience, New York, Part 4, Symposium on Biorheology, pp 175–191
- 20.Barber HN. The rate of movement of chromosomes on the spindle. Chromosoma. 1939;1:33–50. doi: 10.1007/BF01271622. [DOI] [Google Scholar]
- 21.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrader F (1953) Mitosis—the movements of chromosomes in cell division, Columbia University Press, New York
- 23.Mazia D (1961) Mitosis and the physiology of cell division. In: Brachet J, Mirsky AE (eds) The cell-biochemistry, physiology, morphology. Academic, New York, pp 77–412
- 24.Klein E. Observations on the structure of cells and nuclei. Q J Microscop Sci. 1878;18:315–339. [Google Scholar]
- 25.Van Beneden E. Recherches sur la maturation de l’oeuf, la fécondation et la division cellulaire. Arch Biol. 1883;4:265–641. [Google Scholar]
- 26.Watase S. Studies on cephalopods. I. J Morph. 1891;4:247–303. doi: 10.1002/jmor.1050040302. [DOI] [Google Scholar]
- 27.Darlington CD (1937) Recent advances in cytology. Blakiston, Philadelphia
- 28.Metz CW. Monocentric mitosis with segregation of chromosomes in Sciara and its bearing on the mechanism of mitosis. Biol Bull. 1933;64:333–347. doi: 10.2307/1537202. [DOI] [Google Scholar]
- 29.Metz CW. Factors influencing chromosome movements in mitosis. Cytologia. 1936;7:219–231. [Google Scholar]
- 30.Bleier H. Experimentell-cytologische Untersuchungen. I. Einfluss abnormaler Temperatur auf die Reduktionsteilung. Z Zellf u mikr Anat. 1930;11:218–236. doi: 10.1007/BF00534161. [DOI] [Google Scholar]
- 31.Bleier H (1930) Untersuchungen über das Verhalten der verschiedenen Kernkomponenten bei der Reduktionsteilung von Bastarden La Cellule 40
- 32.Inoue S. Motility of cilia and the mechanism of mitosis. Rev Mod Phys. 1959;31:402–408. doi: 10.1103/RevModPhys.31.402. [DOI] [Google Scholar]
- 33.Inoue S, Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol. 1967;50(Suppl):259–292. doi: 10.1085/jgp.50.6.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietz R. An assembly hypothesis of chromosome movement and the changes of the spindle length during anaphase I in spermatocytes of Pales ferruginea. Chromosoma. 1972;38:11–76. doi: 10.1007/BF00319955. [DOI] [PubMed] [Google Scholar]
- 35.Luykx P. Cellular mechanisms of chromosome distribution. Int Rev Cytol Suppl. 1970;2:1–173. [PubMed] [Google Scholar]
- 36.Forer A. Possible roles of microtubules and actin-like filaments during cell division. In: Padilla GM, Cameron IL, Zimmermann AM, editors. Cell cycle controls. New York.: Academic; 1974. pp. 319–336. [Google Scholar]
- 37.Östergren G. Luzula and the mechanism of chromosome movements. Hereditas. 1949;35:445–468. doi: 10.1111/j.1601-5223.1949.tb02884.x. [DOI] [PubMed] [Google Scholar]
- 38.Wada B. The mechanism of mitosis based on studies of the submicroscopic structure and of the living state of the Tradescantia cell. Cytologia. 1950;16:1–26. [Google Scholar]
- 39.Bajer AS, Molè-Bajer J. Cine-analysis of some aspects of mitosis in endosperm. In: Rose GG, editor. Cinemicrography in cell biology. New York.: Academic; 1963. pp. 357–409. [Google Scholar]
- 40.Schaede R. Kritische Untersuchungen über die mechanik der karyokinese. Planta. 1929;8:383–397. doi: 10.1007/BF01916754. [DOI] [Google Scholar]
- 41.Forer A. Local reduction of spindle fiber birefringence in living Nephrotoma suturalis (Loew) spermatocytes induced by ultraviolet microbeam irradiation. J Cell Biol. 1965;25(SUPPL):95–117. doi: 10.1083/jcb.25.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forer A. Characterization of the mitotic traction system, and evidence that birefringent spindle fibers neither produce nor transmit force for chromosome movement. Chromosoma. 1966;19:44–98. doi: 10.1007/BF00332793. [DOI] [PubMed] [Google Scholar]
- 43.Inoue S. Organization and function of the mitotic spindle. In: Allen RD, Kamiya N, editors. Primitive motile systems in cell biology. New York: Academic; 1964. pp. 549–598. [Google Scholar]
- 44.Hamaguchi Y, Toriyama M, Sakai H, Hiramoto Y. Redistribution of fluorescently labeled tubulin in the mitotic apparatus of sand dollar eggs and the effects of taxol. Cell Struct Funct. 1987;12:43–52. doi: 10.1247/csf.12.43. [DOI] [PubMed] [Google Scholar]
- 45.Gorbsky GJ, Borisy GG. Microtubules of the kinetochore fiber turn over in metaphase but not in anaphase. J Cell Biol. 1989;109:653–662. doi: 10.1083/jcb.109.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- 48.Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumont S, Mitchison TJ. Compression regulates mitotic spindle length by a mechanochemical switch at the poles. Curr Biol. 2009;19:1086–1095. doi: 10.1016/j.cub.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- 55.Dhonukshe P, Vischer N, Gadella TW., Jr Contribution of microtubule growth polarity and flux to spindle assembly and functioning in plant cells. J Cell Sci. 2006;119:3193–3205. doi: 10.1242/jcs.03048. [DOI] [PubMed] [Google Scholar]
- 56.LaFountain JR, Jr, Cohan CS, Siegel AJ, LaFountain DJ. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell. 2004;15:5724–5732. doi: 10.1091/mbc.E04-08-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr Biol. 2002;12:1670–1674. doi: 10.1016/S0960-9822(02)01183-1. [DOI] [PubMed] [Google Scholar]
- 58.Brust-Mascher I, Scholey JM. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol Biol Cell. 2002;13:3967–3975. doi: 10.1091/mbc.02-05-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 62.Ferenz NP, Wadsworth P. Prophase microtubule arrays undergo flux-like behavior in mammalian cells. Mol Biol Cell. 2007;18:3993–4002. doi: 10.1091/mbc.E07-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J Cell Biol. 2009;186:11–26. doi: 10.1083/jcb.200904153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorbsky GJ, Sammak PJ, Borisy GG. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J Cell Biol. 1987;104:9–18. doi: 10.1083/jcb.104.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci USA. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bajer AS, Molè-Bajer J. Spindle dynamics and chromosome movements. Int Rev Cytol Suppl. 1972;3:1–271. [Google Scholar]
- 69.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 71.Kuwada Y. Chromosome arrangement.I. Mem Coll Sci, Kyoto Univ. 1929;4:199–264. [Google Scholar]
- 72.Belar K. Beitrage zur kausalanalyse der mitose. II. Arch Entwicklungsmech. 1929;118:359–480. doi: 10.1007/BF02108879. [DOI] [PubMed] [Google Scholar]
- 73.Carlson JG. Mitotic behavior of induced chromosomal fragments lacking spindle attachments in the neuroblasts of the grasshopper. Proc Natl Acad Sci USA. 1938;24:500–507. doi: 10.1073/pnas.24.11.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carothers EE. Components of the mitotic spindle with special reference to the chromosomal and interzonal fibers in the Acrididae. Biol Bull. 1936;71:469–491. doi: 10.2307/1537374. [DOI] [Google Scholar]
- 75.Mota M (1957) A new hypothesis of the anaphase movement. In: Proceedings of the international genetics symposia (1956), Cytologia, Supplement, pp 113–116
- 76.Gibbons IR, Rowe AJ. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 77.Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- 78.Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- 79.Cassimeris LU, Walker RA, Pryer NK, Salmon ED. Dynamic instability of microtubules. Bioessays. 1987;7:149–154. doi: 10.1002/bies.950070403. [DOI] [PubMed] [Google Scholar]
- 80.Gorbsky GJ, Sammak PJ, Borisy GG. Microtubule dynamics and chromosome motion visualized in living anaphase cells. J Cell Biol. 1988;106:1185–1192. doi: 10.1083/jcb.106.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nicklas RB, Lee GM, Rieder CL, Rupp G. Mechanically cut mitotic spindles: clean cuts and stable microtubules. J Cell Sci. 1989;94(Pt 3):415–423. doi: 10.1242/jcs.94.3.415. [DOI] [PubMed] [Google Scholar]
- 82.Nicklas RB. The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J Cell Biol. 1989;109:2245–2255. doi: 10.1083/jcb.109.5.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wordeman L, Steuer ER, Sheetz MP, Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McIntosh JR, Cande WZ, Snyder JA. Structure and physiology of the mammalian mitotic spindle. Soc Gen Physiol Ser. 1975;30:31–76. [PubMed] [Google Scholar]
- 85.Sakai H, Mabuchi I, Shimoda S, Kuriyama R, Ogawa K, Mohri H. Induction of chromosome motion in the glycerol-isolated mitotic apparatus: nucleotide specificity and effects of anti-dynein and myosin sera on the motion. Dev Growth Differ. 1976;18:211–219. doi: 10.1111/j.1440-169X.1976.00211.x. [DOI] [PubMed] [Google Scholar]
- 86.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cande WZ. Nucleotide requirements for anaphase chromosome movements in permeabilized mitotic cells: anaphase B but not anaphase A requires ATP. Cell. 1982;28:15–22. doi: 10.1016/0092-8674(82)90370-1. [DOI] [PubMed] [Google Scholar]
- 88.Cande WZ (1989) Mitosis in vitro. In: Hyams JS, Brinkley BR (eds) Mitosis—molecules and mechanisms. Academic, London, pp 303–326
- 89.Cande WZ. Inhibition of spindle elongation in permeabilized mitotic cells by erythro-9-[3-(2-hydroxynonyl)] adenine. Nature. 1982;295:700–701. doi: 10.1038/295700a0. [DOI] [PubMed] [Google Scholar]
- 90.Cande WZ, Wolniak SM. Chromosome movement in lysed mitotic cells is inhibited by vanadate. J Cell Biol. 1978;79:573–580. doi: 10.1083/jcb.79.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Courtheoux T, Gay G, Reyes C, Goldstone S, Gachet Y, Tournier S. Dynein participates in chromosome segregation in fission yeast. Biol Cell. 2007;99:627–637. doi: 10.1042/BC20070047. [DOI] [PubMed] [Google Scholar]
- 96.Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- 98.Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 99.Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Savoian MS, Goldberg ML, Rieder CL. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat Cell Biol. 2000;2:948–952. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- 101.Basto R, Gomes R, Karess RE. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila . Nat Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- 102.Buffin E, Lefebvre C, Huang J, Gagou ME, Karess RE. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol. 2005;15:856–861. doi: 10.1016/j.cub.2005.03.052. [DOI] [PubMed] [Google Scholar]
- 103.Chan GK, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- 104.Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gassmann R, Essex A, Hu JS, Maddox PS, Motegi F, Sugimoto A, O’Rourke SM, Bowerman B, McLeod I, Yates JR, 3rd, Oegema K, Cheeseman IM, Desai A. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan YW, Fava LL, Uldschmid A, Schmitz MH, Gerlich DW, Nigg EA, Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vorozhko VV, Emanuele MJ, Kallio MJ, Stukenberg PT, Gorbsky GJ. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 2008;117:169–179. doi: 10.1007/s00412-007-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.King JM, Hays TS, Nicklas RB. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J Cell Biol. 2000;151:739–748. doi: 10.1083/jcb.151.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Whyte J, Bader JR, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, Vaughan KT. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nicklas RB. Mitosis. Adv Cell Biol. 1971;2:225–297. doi: 10.1007/978-1-4615-9588-5_5. [DOI] [PubMed] [Google Scholar]
- 113.Gibbons IR. Studies on the adenosine triphosphatase activity of 14 S and 30 S dynein from cilia of Tetrahymena . J Biol Chem. 1966;241:5590–5596. [PubMed] [Google Scholar]
- 114.Brokaw CJ. Adenosine triphosphate usage by flagella. Science. 1967;156:76–78. doi: 10.1126/science.156.3771.76. [DOI] [PubMed] [Google Scholar]
- 115.Inoue S. The effect of colchicine on the microscopic and submicroscopic structure of the mitotic spindle. Exp Cell Res (Suppl) 1952;2:305–318. [Google Scholar]
- 116.Salmon ED. Spindle microtubules: thermodynamics of in vivo assembly and role in chromosome movement. Ann N Y Acad Sci. 1975;253:383–406. doi: 10.1111/j.1749-6632.1975.tb19216.x. [DOI] [PubMed] [Google Scholar]
- 117.Salmon ED, Begg DA. Functional implications of cold-stable microtubules in kinetochore fibers of insect spermatocytes during anaphase. J Cell Biol. 1980;85:853–865. doi: 10.1083/jcb.85.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 119.Spurck TP, Pickett-Heaps JD. On the mechanism of anaphase A: evidence that ATP is needed for microtubule disassembly and not generation of polewards force. J Cell Biol. 1987;105:1691–1705. doi: 10.1083/jcb.105.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 122.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tirnauer JS, Salmon ED, Mitchison TJ. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol Biol Cell. 2004;15:1776–1784. doi: 10.1091/mbc.E03-11-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hunt AJ, McIntosh JR. The dynamic behavior of individual microtubules associated with chromosomes in vitro. Mol Biol Cell. 1998;9:2857–2871. doi: 10.1091/mbc.9.10.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maffini S, Maia AR, Manning AL, Maliga Z, Pereira AL, Junqueira M, Shevchenko A, Hyman A, Yates JR, 3rd, Galjart N, Compton DA, Maiato H. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore–microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 130.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 131.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Force production by depolymerizing microtubules: a theoretical study. Proc Natl Acad Sci USA. 2005;102:4353–4358. doi: 10.1073/pnas.0501142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 137.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 138.Molodtsov MI, Grishchuk EL, McIntosh JR, Ataullakhanov FI. Measurement of the force developed by disassembling microtubule during calcium-induced depolymerization. Dokl Biochem Biophys. 2007;412:18–21. doi: 10.1134/S1607672907010061. [DOI] [PubMed] [Google Scholar]
- 139.Liu J, Onuchic JN. A driving and coupling “Pac-Man” mechanism for chromosome poleward translocation in anaphase A. Proc Natl Acad Sci USA. 2006;103:18432–18437. doi: 10.1073/pnas.0608962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Centonze VE, Borisy GG. Pole-to-chromosome movements induced at metaphase: sites of microtubule disassembly. J Cell Sci. 1991;100(Pt 1):205–211. doi: 10.1242/jcs.100.1.205. [DOI] [PubMed] [Google Scholar]
- 141.Cassimeris L, Salmon ED. Kinetochore microtubules shorten by loss of subunits at the kinetochores of prometaphase chromosomes. J Cell Sci. 1991;98(Pt 2):151–158. doi: 10.1242/jcs.98.2.151. [DOI] [PubMed] [Google Scholar]
- 142.O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae . Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O’Toole ET, McDonald KL, Mantler J, McIntosh JR, Hyman AA, Muller-Reichert T. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans . J Cell Biol. 2003;163:451–456. doi: 10.1083/jcb.200304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Maiato H, Hergert PJ, Moutinho-Pereira S, Dong Y, Vandenbeldt KJ, Rieder CL, McEwen BF. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mastronarde DN, Morphew MK, McIntosh JR. HVEM tomography of PtK cells shows that the plus ends of kinetochore microtubules flare outward in prometaphase, metaphase and anaphase. Mol Biol Cell (Suppl) 1997;8:171a. [Google Scholar]
- 147.Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe . Curr Biol. 1999;9:1423–1426. doi: 10.1016/S0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- 148.McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Inoue S. Polarization optical studies of the mitotic spindle. I. The demonstration of spindle fibers in living cells. Chromosoma. 1953;5:487–500. doi: 10.1007/BF01271498. [DOI] [PubMed] [Google Scholar]
- 153.Nicklas RB, Staehly CA. Chromosome micromanipulation I. The mechanics of chromosome attachment to the spindle. Chromosoma. 1967;21:1–16. doi: 10.1007/BF00330544. [DOI] [PubMed] [Google Scholar]
- 154.Verde F, Labbe JC, Doree M, Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- 155.Moutinho-Pereira S, Debec A, Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Murray AW, Desai AB, Salmon ED. Real time observation of anaphase in vitro. Proc Natl Acad Sci USA. 1996;93:12327–12332. doi: 10.1073/pnas.93.22.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen W, Zhang D. Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat Cell Biol. 2004;6:227–231. doi: 10.1038/ncb1104. [DOI] [PubMed] [Google Scholar]
- 158.Wadsworth P, Shelden E, Rupp G, Rieder CL. Biotin-tubulin incorporates into kinetochore fiber microtubules during early but not late anaphase. J Cell Biol. 1989;109:2257–2265. doi: 10.1083/jcb.109.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 160.White SR, Lauring B. AAA + ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 161.Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- 163.Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- 164.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rath U, Rogers GC, Tan D, Gomez-Ferreria MA, Buster DW, Sosa HJ, Sharp DJ. The Drosophila kinesin-13, KLP59D, impacts Pacman- and Flux-based chromosome movement. Mol Biol Cell. 2009;20:4696–4705. doi: 10.1091/mbc.E09-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hays TS, Salmon ED. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J Cell Biol. 1990;110:391–404. doi: 10.1083/jcb.110.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moens PB. Kinetochore microtubule numbers of different sized chromosomes. J Cell Biol. 1979;83:556–561. doi: 10.1083/jcb.83.3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fuseler JW. Temperature dependence of anaphase chromosome velocity and microtubule depolymerization. J Cell Biol. 1975;67:789–800. doi: 10.1083/jcb.67.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Forer A. Do anaphase chromosomes chew their way to the pole or are they pulled by actin? J Cell Sci. 1988;91(Pt 4):449–453. doi: 10.1242/jcs.91.4.449. [DOI] [PubMed] [Google Scholar]
- 175.Pickett-Heaps J, Spurck T, Tippit D. Chromosome motion and the spindle matrix. J Cell Biol. 1984;99:137s–143s. doi: 10.1083/jcb.99.1.137s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pickett-Heaps JD, Forer A, Spurck T. Rethinking anaphase: where “Pac-Man” fails and why a role for the spindle matrix is likely. Protoplasma. 1996;192:1–10. doi: 10.1007/BF01273239. [DOI] [Google Scholar]
- 177.Forer A, Spurck T, Pickett-Heaps JD, Wilson PJ. Structure of kinetochore fibres in crane-fly spermatocytes after irradiation with an ultraviolet microbeam: neither microtubules nor actin filaments remain in the irradiated region. Cell Motil Cytoskeleton. 2003;56:173–192. doi: 10.1002/cm.10144. [DOI] [PubMed] [Google Scholar]
- 178.Forer A, Pickett-Heaps JD. Cytochalasin D and latrunculin affect chromosome behaviour during meiosis in crane-fly spermatocytes. Chromosome Res. 1998;6:533–549. doi: 10.1023/A:1009224322399. [DOI] [PubMed] [Google Scholar]
- 179.Fabian L, Forer A. Redundant mechanisms for anaphase chromosome movements: crane-fly spermatocyte spindles normally use actin filaments but also can function without them. Protoplasma. 2005;225:169–184. doi: 10.1007/s00709-005-0094-6. [DOI] [PubMed] [Google Scholar]
- 180.LaFountain JR, Jr, Janicke MA, Balczon R, Rickards GK. Cytochalasin induces abnormal anaphase in crane-fly spermatocytes and causes altered distribution of actin and centromere antigens. Chromosoma. 1992;101:425–441. doi: 10.1007/BF00582837. [DOI] [Google Scholar]
- 181.Wrench GA, Snyder JA. Cytochalasin J treatment significantly alters mitotic spindle microtubule organization and kinetochore structure in PtK1 cells. Cell Motil Cytoskeleton. 1997;36:112–124. doi: 10.1002/(SICI)1097-0169(1997)36:2<112::AID-CM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 182.Fabian L, Troscianczuk J, Forer A. Calyculin A, an enhancer of myosin, speeds up anaphase chromosome movement. Cell Chromosome. 2007;6:1. doi: 10.1186/1475-9268-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schmit AC, Lambert AM. Microinjected fluorescent phalloidin in vivo reveals the F-actin dynamics and assembly in higher plant mitotic cells. Plant Cell. 1990;2:129–138. doi: 10.1105/tpc.2.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kiehart DP, Mabuchi I, Inoue S. Evidence that myosin does not contribute to force production in chromosome movement. J Cell Biol. 1982;94:165–178. doi: 10.1083/jcb.94.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cande WZ, McDonald K, Meeusen RL. A permeabilized cell model for studying cell division: a comparison of anaphase chromosome movement and cleavage furrow constriction in lysed PtK1 cells. J Cell Biol. 1981;88:618–629. doi: 10.1083/jcb.88.3.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Snyder JA, Cohen L. Cytochalasin J affects chromosome congression and spindle microtubule organization in PtK1 cells. Cell Motil Cytoskeleton. 1995;32:245–257. doi: 10.1002/cm.970320402. [DOI] [PubMed] [Google Scholar]
- 188.Fabian L, Forer A. Possible roles of actin and myosin during anaphase chromosome movements in locust spermatocytes. Protoplasma. 2007;231:201–213. doi: 10.1007/s00709-007-0262-y. [DOI] [PubMed] [Google Scholar]
- 189.Johansen KM, Johansen J. Cell and molecular biology of the spindle matrix. Int Rev Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- 190.Lince-Faria M, Maffini S, Orr B, Ding Y, Claudia F, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ostergren G, Mole-Bajer J, Bajer A. An interpretation of transport phenomena at mitosis. Ann N Y Acad Sci. 1960;90:381–408. doi: 10.1111/j.1749-6632.1960.tb23258.x. [DOI] [PubMed] [Google Scholar]
- 193.McIntosh JR, Hepler PK, Van Wie DG. Model for mitosis. Nature. 1969;224:659–663. doi: 10.1038/224659a0. [DOI] [Google Scholar]
- 194.Euteneuer U, McIntosh JR. Polarity of midbody and phragmoplast microtubules. J Cell Biol. 1980;87:509–515. doi: 10.1083/jcb.87.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Telzer BR, Haimo LT. Decoration of spindle microtubules with Dynein: evidence for uniform polarity. J Cell Biol. 1981;89:373–378. doi: 10.1083/jcb.89.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Margolis RL, Wilson L, Keifer BI. Mitotic mechanism based on intrinsic microtubule behaviour. Nature. 1978;272:450–452. doi: 10.1038/272450a0. [DOI] [PubMed] [Google Scholar]
- 198.Goode D. Microtubule turnover as a mechanism of mitosis and its possible evolution. Biosystems. 1981;14:271–287. doi: 10.1016/0303-2647(81)90034-4. [DOI] [PubMed] [Google Scholar]
- 199.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Spurck T, Forer A, Pickett-Heaps J. Ultraviolet microbeam irradiations of epithelial and spermatocyte spindles suggest that forces act on the kinetochore fibre and are not generated by its disassembly. Cell Motil Cytoskeleton. 1997;36:136–148. doi: 10.1002/(SICI)1097-0169(1997)36:2<136::AID-CM4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 204.Forer A, Spurck T, Pickett-Heaps JD. Actin and myosin inhibitors block elongation of kinetochore fibre stubs in metaphase crane-fly spermatocytes. Protoplasma. 2007;232:79–85. doi: 10.1007/s00709-007-0265-8. [DOI] [PubMed] [Google Scholar]
- 205.Mitchison TJ. Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos Trans R Soc Lond B. 2005;360:623–629. doi: 10.1098/rstb.2004.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aist JR, Williams PH. Ultrastructure and time course of mitosis in the fungus Fusarium oxysporum . J Cell Biol. 1972;55:368–389. doi: 10.1083/jcb.55.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 209.Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hoffmann-Berling H. The significance of adenosinetriphosphate in cell division and nuclear division in anaphase. Biochim Biophys Acta. 1954;15:226–236. doi: 10.1016/0006-3002(54)90063-3. [DOI] [PubMed] [Google Scholar]
- 211.Lee GM. Characterization of mitotic motors by their relative sensitivity to AMP-PNP. J Cell Sci. 1989;94(Pt 3):425–441. doi: 10.1242/jcs.94.3.425. [DOI] [PubMed] [Google Scholar]
- 212.McIntosh JR (1994) The roles of microtubules in chromosome movement. In: Hyams JS, Lloyd CW (eds) Microtubules, Wiley, New York, pp 413–434
- 213.Pickett-Heaps JD, Tippit DH. The diatom spindle in perspective. Cell. 1978;14:455–467. doi: 10.1016/0092-8674(78)90232-5. [DOI] [PubMed] [Google Scholar]
- 214.Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 216.Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 217.Severin F, Habermann B, Huffaker T, Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J Cell Biol. 2001;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pereira AL, Pereira AJ, Maia AR, Drabek K, Sayas CL, Hergert PJ, Lince-Faria M, Matos I, Duque C, Stepanova T, Rieder CL, Earnshaw WC, Galjart N, Maiato H. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol Biol Cell. 2006;17:4526–4542. doi: 10.1091/mbc.E06-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mollinari C, Kleman JP, Saoudi Y, Jablonski SA, Perard J, Yen TJ, Margolis RL. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell. 2005;16:1043–1055. doi: 10.1091/mbc.E04-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pellman D, Bagget M, Tu YH, Fink GR, Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae . J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Boveri T. Zellenstudien. Jena: II Fischer; 1888. [Google Scholar]
- 227.Aist JR, Berns MW. Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol. 1981;91:446–458. doi: 10.1083/jcb.91.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Aist JR, Liang H, Berns MW. Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J Cell Sci. 1993;104(Pt 4):1207–1216. doi: 10.1242/jcs.104.4.1207. [DOI] [PubMed] [Google Scholar]
- 229.Hiramoto Y, Nakano Y. Micromanipulation studies of the mitotic apparatus in sand dollar eggs. Cell Motil Cytoskeleton. 1988;10:172–184. doi: 10.1002/cm.970100122. [DOI] [PubMed] [Google Scholar]
- 230.Waters JC, Cole RW, Rieder CL. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J Cell Biol. 1993;122:361–372. doi: 10.1083/jcb.122.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 232.Kronebusch PJ, Borisy GG. Mechanics of anaphase B movement. In: Sakai H, Mohri H, Borisy GG, editors. Biological functions of microtubules and related structures. Tokyo: Academic; 1982. pp. 233–245. [Google Scholar]
- 233.Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 235.Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans . Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 236.Shelden E, Wadsworth P. Interzonal microtubules are dynamic during spindle elongation. J Cell Sci. 1990;97(Pt 2):273–281. doi: 10.1242/jcs.97.2.273. [DOI] [PubMed] [Google Scholar]
- 237.Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mendoza M, Norden C, Durrer K, Rauter H, Uhlmann F, Barral Y. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol. 2009;11:477–483. doi: 10.1038/ncb1855. [DOI] [PubMed] [Google Scholar]
- 239.Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 240.Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]