Abstract
Receptor for advanced glycation end products (RAGE) mediates diverse physiological and pathological effects and is involved in the pathogenesis of Alzheimer’s disease (AD). RAGE is a receptor for amyloid β peptides (Aβ), mediates Aβ neurotoxicity and also promotes Aβ influx into the brain and contributes to Aβ aggregation. Soluble RAGE (sRAGE), a secreted RAGE isoform, acts as a decoy receptor to antagonize RAGE-mediated damages. Accumulating evidence has suggested that sRAGE represents a promising pharmaceutic against RAGE-mediated disorders. Recent studies revealed proteolysis of RAGE as a previously unappreciated means of sRAGE production. In this review we summarize these findings on the proteolytic cleavage of RAGE and discuss the underlying regulatory mechanisms of RAGE shedding. Furthermore, we propose a model in which proteolysis of RAGE could restrain AD development by reducing Aβ transport into the brain and Aβ production via BACE. Thus, the modulation of RAGE proteolysis provides a novel intervention strategy for AD.
Keywords: Receptor for advanced glycation end products, Alzheimer’s disease, Ectodomain shedding, Amyloid β peptide, ADAM10, MMP9
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease and is the most prevalent cause of dementia. AD-affected individuals develop a gradual and progressive decline in cognitive and functional abilities as well as behavioral and psychiatric symptoms leading to a vegetative state and ultimately death [1]. The presence of amyloid plaques and intracellular neurofibrillary tangles are the main neuropathological hallmarks of AD [2]. The primary constituents of the amyloid plaques are heterogeneous, 39–43 amino acid peptides, the amyloid β peptides (Aβ). Aβ peptides are generated by the sequential proteolytic processing of their precursor, the amyloid precursor protein (APP) [3]. Processing of APP occurs in vivo by two competitive pathways: the amyloidogenic pathway is initiated by cleavage at Asp1 of the Aβ sequence mediated by β-secretase and generates soluble APPβ (sAPPβ) and a unique C-terminal membrane-retained fragment, termed CTF-β. Subsequent cleavage of CTF-β by γ-secretase results in the production of Aβ. In contrast, the non-amyloidogenic pathway is mediated by α-secretase, which generates soluble APPα (sAPPα) and the C-terminal fragment-α (CTF-α). Subsequent cleavage of CTF-α by γ-secretase generates a truncated non-toxic peptide. Since α-secretase attacks APP inside the Aβ sequence, the non-amyloidogenic pathway precludes neurotoxic Aβ peptide formation [4].
According to the amyloid cascade hypothesis [5], which states that AD development is due to abnormal accumulation of Aβ in the brains of AD patients causing neurodegeneration and finally the clinical symptoms of dementia, Aβ is considered to play a central role in the pathogenesis of AD. First of all, Aβ is directly toxic to cultured neurons due to its ability to generate reactive oxygen species (ROS) and produce an accumulation of H2O2 and lipid peroxides in cells [6]. Second, Aβ is a potent inducer of the transcription factor nuclear factor-κB (NF-κB) in primary neurons and astrocytes [7]. Third, being chemotactic, Aβ causes migration of microglia, thereby contributing to an increased accumulation of microglial cells surrounding the amyloid plaques [8]. Finally, Aβ potentiates the secretion of the cytokines interleukin (IL)-6 and IL-8 in IL-1β-activated human astrocytoma cells [9]. The predominant forms of Aβ are the Aβ40 and Aβ42 fragments. Soluble Aβ40 is the major form of circulating Aβ and cerebrovascular amyloid, whereas amyloidogenic Aβ42, the major constituent of amyloid plaques, accounts for minor amounts in the circulation [10]. The origin of Aβ deposited in the cerebral vasculature and brain is uncertain. According to the “neuronal theory,” Aβ is produced locally in the brain. In contrast, the “vascular theory” proposes that Aβ originates from the entire body and that circulating soluble Aβ can contribute to neurotoxicity since it crosses the blood brain barrier (BBB) [11]. The “vascular theory” is supported by the compelling evidence that the BBB plays a crucial role in the transport and metabolism of circulating Aβ and the modulation of AD progression [12–14].
The life-long accumulation of Aβ in the brain is determined by the rate of Aβ generation versus Aβ clearance. Strategies to treat AD have focused on both decreasing Aβ production and enhancing its clearance from the brain. Clearance can be accomplished via two major pathways: proteolytic degradation and receptor-mediated export from the brain. Degradation of Aβ in the central nervous system (CNS) could play an important role in clearance [15]. The proteases capable of degrading Aβ include neprilysin, insulin-degrading enzyme (insulysin), plasmin, tissue plasminogen activator, endothelin-converting enzyme and matrix metalloproteinase-9 [16]. An Aβ-lowering strategy based on receptor mediated-Aβ transport has just recently begun to receive more attention. Increasing lines of evidence suggest that the low-density lipoprotein receptor-related protein-1 (LRP-1) and the receptor for advanced glycation end products (RAGE) are involved in receptor-mediated flux of Aβ across the BBB [17]. While LRP-1 appears to mediate the efflux of Aβ from the brain to the periphery, RAGE is implicated in Aβ influx back into the CNS.
Since the subject of this review is on proteolysis of RAGE and AD, we will first briefly describe the biology of RAGE and the interaction of RAGE and Aβ. Then we focus on proteolysis of RAGE and discuss recent exciting findings on ectodomain shedding of RAGE. At the end, we propose an intriguing model in which modulation of RAGE sheddases is a double-edged strategy to alleviate AD by preventing Aβ production and aggregation in the brain.
Biology of RAGE
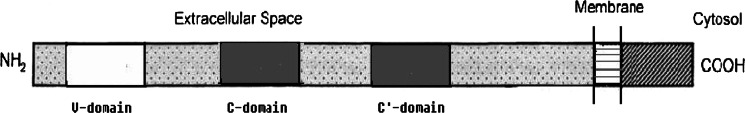
RAGE was first identified as a cell surface receptor for the products of nonenzymatic glycation and oxidation of proteins, the advanced glycation end products (AGEs). The RAGE gene was located on chromosome 6p21.3 in a gene-rich region containing a number of genes involved in inflammation and components of the major histocompatibility complex (MHC) [18]. The human RAGE gene encodes a type-I transmembrane protein of 404 amino acids, composed of an extracellular domain with 344 amino acids, a transmembrane domain with 19 residues and a cytosolic domain with 43 amino acids (Fig. 1) [19]. The large extracellular domain of RAGE contains a N-terminal signal sequence of 22 amino acids and three immunoglobulin (Ig)-like regions, which define the RAGE protein as a member of the immunoglobulin superfamily. These Ig-like regions include one “V”-type domain (IgV sequence from residue 41 to residue 126) followed by two “C”-type domains (IgC sequence from residue 127 to residue 234 and IgC’ from residue 235 to residue 344). The “V”-type domain confers ligand binding and contains two putative N-glycosylation sites. Deglycosylation was shown to affect RAGE binding of certain ligands [20]. The short cytoplasmic domain is critical for signaling downstream of receptor–ligand interaction.
Fig. 1.
Structure of receptor for advanced glycation end products (RAGE). The large extracellular domain of RAGE contains an N-terminal signal sequence and three Ig-like regions including one “V”-type domain followed by two “C”-type domains. The “V”-type domain confers ligand binding and contains two putative N-glycosylation sites. The short cytoplasmic domain is critical for downstream signaling
RAGE transcription is controlled by several transcription factors, including SP-1, AP-2 and NF-κB [21]. During development, RAGE is highly expressed in the nervous system, e.g., in the developing embryonic rat brain [22, 23]. In striking contrast, in mature mammalians there is relatively little expression of RAGE in most tissues except lung and skin. At the cellular level, RAGE is expressed in a variety of cell types, including endothelial, vascular smooth muscle, mononuclear phagocytes, microglial cells, astrocytes and neuronal cells [24]. RAGE expression is highly up-regulated under pathological conditions such as diabetic vascular disease, chronic inflammation, AD and tumors [25].
RAGE is a multi-ligand receptor and in addition to AGEs, its ligands also include amphoterin (also know as high mobility group I DNA-binding protein, HMG-1), Aβ and S100/calgranulins. The engagement of RAGE by these diverse ligands contributes to various pathologic processes ranging from proinflammatory responses, accelerated diabetic atherosclerosis, AD to tumor cell invasion [26]. On the other hand, RAGE also has physiological functions. Judged from the expression patterns of RAGE described above, it is reasonable to assume that RAGE could play roles in development. A few reports have suggested that RAGE might contribute to CNS development. For instance, amphoterin can bind RAGE and contributes to axonal sprouting, which accompanies neuronal development [22]. Recently, RAGE was shown to promote neurite outgrowth in vivo in a unilateral sciatic nerve crush model and the trophic effects could be blocked by soluble RAGE (sRAGE) or antibodies against either RAGE or amphoterin, thus confirming the neurotrophic function of RAGE [27]. These results are in accord with the fact that RAGE is expressed at a high level in the nervous system during development [22]. However, RAGE null mice develop normally, live a natural life span and are fertile [28]; therefore, redundant molecules can compensate for the loss of RAGE. Another recent study revealed the possibility of an involvement of RAGE in the regulation of cell differentiation [29]. In this context, down-regulation of RAGE may be considered as a critical step in tissue reorganization and the formation of lung tumors. Considering that lung is one of the few tissues in which RAGE is constitutively expressed at a high level, we can suppose that in normal lung tissues RAGE maintains cells in the differentiated state and inhibits tumorigenesis. Upon down-regulation of RAGE, cells may become undifferentiated and tumor growth may ensue. Undoubtedly, further experiments are needed to confirm whether RAGE indeed regulates cell differentiation.
As a transmembrane receptor, the engagement of RAGE by its ligands has been reported to trigger intracellular signaling pathways. In most cases, RAGE induces the activation of the immune/inflammatory-associated transcription factor NF-κB. Notably, the human RAGE promoter contains two NF-κB responsive elements that act to form a positive feedback loop such that RAGE is up-regulated where its ligands are present [21]. In addition to NF-κB, RAGE can activate a range of signaling pathways including nuclear factor of activated T-cells (NF-AT) [30], small GTPases Rac1 and Cdc42 [31], mitogen-activated protein kinase (MAPK) and c-Jun N-terminal kinase [32], extracellular signal-regulated kinases 1 and 2 [33], as well as cAMP response element-binding factor [34]. The diversity of signaling cascades identified in RAGE-mediated cellular signaling implies that different RAGE ligands might induce different pathways and thus make the RAGE network more complicated. Although a number of reports suggested that the cytoplasmic domain of RAGE is essential for intracellular signaling, a challenging task will be to elucidate the exact bridging molecules that engage the cytoplasmic domain of RAGE upon activation of the receptor. Dia-1 was recently identified as a direct binding partner of the RAGE cytoplasmic domain and the mediator of downstream Rac1 and Cdc42 activation [31].
RAGE and Aβ interaction
It has been demonstrated that expression of RAGE is increased in AD patients and furthermore, expression of RAGE was colocalized with that of Aβ in human AD brain tissues, in neurons, microglia and vascular elements [35, 36], thus indicating the possibility of a RAGE–Aβ interaction in the pathogenesis of AD. It has been suggested that RAGE may be the nerve cell receptor for Aβ [37–39]. Indeed, RAGE binds soluble Aβ in a dose-dependent manner in the nanomolar range [35]. Studies using an in vitro model of the human BBB showed that RAGE mediated the binding of soluble Aβ1–40 at the apical side of brain capillary endothelium, and RAGE was also involved in soluble Aβ1–40 transcytosis [40]. In addition, RAGE mediates the transport of pathophysiologically relevant concentration of plasma Aβ across the BBB, while deletion of the RAGE gene eliminates transport of free circulating Aβ into the brain [41]. Besides RAGE, gp330/megalin may also transport circulating Aβ in a complex with apoJ [42]. However, gp330/megalin is normally saturated by high levels of plasma apoJ, which precludes a significant influx of Aβ into the CNS under physiological conditions. This leaves RAGE as a probable major influx receptor for Aβ at the BBB. Furthermore, RAGE is involved in Aβ-mediated migration of monocytes across the BBB, which may contribute to Aβ-related vascular disorder [43].
Transport of circulating Aβ into the brain results in expression of proinflammatory cytokines in neurovascular cells and elaboration of endothelin-1 causing decreased cerebral blood flow. Because of the presence of two NF-κB responsive elements in the human RAGE promoter, RAGE–Aβ interaction might induce a possible feedback loop. Thereby, Aβ-induced oxidant stress activates NF-κB, which subsequently binds to the RAGE promoter and upregulates RAGE expression. The interaction of RAGE with Aβ is followed by a series of intracellular activities that trigger inflammatory pathways, which could contribute to the progression of AD [39]. Furthermore, mouse models were developed to assess the impact of RAGE in an Aβ-rich environment. Transgenic mice with targeted neuronal overexpression of either RAGE or dominant negative (DN) RAGE were used for these studies. When compared with mutant APP transgenic mice, double transgenic mice expressing RAGE and mutant APP displayed earlier abnormalities in spatial learning and memory that were accompanied by altered activation of markers of synaptic plasticity and exaggerated neuropathologic changes. In contrast, double transgenic mice bearing DN-RAGE and APP displayed preservation of spatial learning and memory and diminished neuropathologic changes. These data strongly indicate that RAGE is a cofactor for Aβ-induced neuronal pathology in AD models and suggest its potential as a therapeutic target to ameliorate dysfunction associated with AD [44].
Isoforms of RAGE
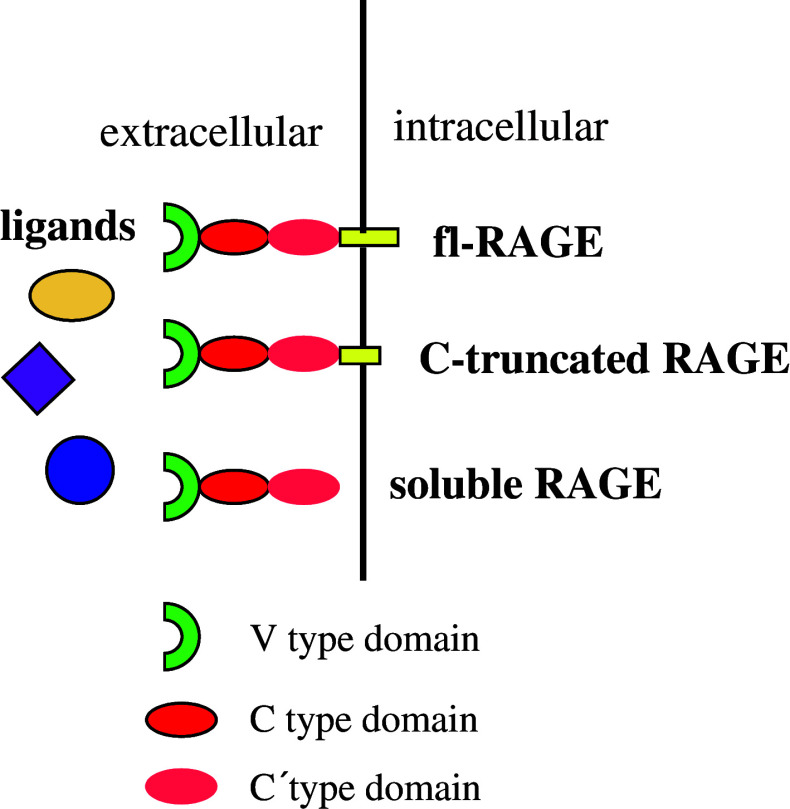
There are three major different forms of RAGE (Fig. 2). These isoforms can be defined as full-length RAGE (fl-RAGE), soluble RAGE (sRAGE) and C-terminal truncated RAGE (C-truncated RAGE) [45]. C-truncated RAGE contains the extracellular and transmembrane domain of RAGE, but lacks the C-terminal intracellular domain important for signal transduction. Therefore, by competing for the binding of RAGE ligands C-truncated RAGE prevents the activation of fl-RAGE. For this reason C-truncated RAGE is also named dominant negative RAGE (DN-RAGE) [35]. Recently, it was shown that the expression level of DN-RAGE in the human brain is similar to that of fl-RAGE [46]. Since a number of RAGE ligands such as AGE and Aβ have devastating effects on cells, binding of these ligands to DN-RAGE would lead to dismissed binding to fl-RAGE and consequently, less deleterious effects mediated by fl-RAGE. But it is important to keep in mind that although trapping of harmful ligands by DN-RAGE may be beneficial during the initial periods of ligand binding, the long-term effects may be less advantageous. The engagement of the ligands at the cell surface by DN-RAGE may lead to the initiation of further ligand recruitment, oxidation, and aggregation, which unfavorably enhances the activation of fl-RAGE [45]. It is undisputed that a detailed understanding of the regulation of DN-RAGE generation is crucial to evaluate DN-RAGE as a target to modulate signaling mediated by fl-RAGE.
Fig. 2.
Three major isoforms of RAGE. Major RAGE isoforms include full-length RAGE (fl-RGAE), soluble RAGE (sRAGE) and C-terminal truncated RAGE (C-truncated RAGE). The existence of multiple RAGE isoforms indicates that the physiopathological effects mediated by RAGE ligands result from the complex interaction of these ligands with different RAGE isoforms
The soluble form of RAGE contains only the extracellular domain of RAGE, while lacking the transmembrane and the cytoplasmic domains. Thereby, sRAGE is released from the cell surface into the extracellular space as a soluble form, which serves as a decoy receptor for the ligands and blocks RAGE signaling by preventing ligands from gaining access to fl-RAGE. Recombinant sRAGE has been used to reduce diabetic late complications [47], to inhibit tumor metastases and invasion [32] and to block transport of Aβ across the BBB [41]. However, it is important to note that sRAGE provides a decoy strategy, i.e., sRAGE sequesters ligands and prevents their interaction with RAGE and, potentially, other receptors as well. So it is imperative to investigate whether sRAGE has any inhibitory effects on normal functions of other receptors before a therapy based on sRAGE can be implemented.
Soluble forms of membrane-associated receptors can be generated via distinct mechanisms. The first involves the alternative splicing of mRNA transcripts that usually encode membrane-associated receptors. Alternative splicing is an important way to generate soluble forms of cell surface receptor; examples include the transforming growth factor (TGF)-β receptor [48], the tumor necrosis factor (TNF) receptor [49] and the IL-17 receptor-like protein [50]. Several alternatively spliced RAGE mRNAs encoding secretory proteins have also been reported to be present in human brain [46].
Ectodomain shedding of membrane proteins
While overwhelming data demonstrated that sRAGE is derived from alternative splicing of the RAGE mRNA, some evidence indicated that sRAGE in mice may be generated as a result of protein cleavage instead of mRNA splicing [51].
The extracellular domains of various cell surface proteins are released through proteolytic cleavage. This type of proteolysis occurs at or near the plasma membrane and is known as ectodomain shedding [52]. The shedding of cell surface proteins can occur either in non-stimulated cells (known as constitutive shedding) or in activated cells (known as inducible shedding) [52]. Stimulants that can induce shedding include phorbol esters [53], calcium ionophores [54] and serum factors [55]. Ectodomain shedding events have been observed for a surprisingly large number of cell surface proteins with distinct functions. Targets of this process include cytokines and cytokine receptors [56–58], growth factors [59, 60], adhesion molecule L1 [61], angiotensin-converting enzyme [62] and proteins associated with neuropathological disorders (APP and prion protein) [63, 64].
It appears increasingly apparent that proteolytic shedding of cell surface proteins is an important cellular post-translational regulatory process. Soluble shedded proteins commonly consist of the extracellular portions or ectodomains of their membrane-bound precursors and thereby retain the ability to bind ligands. They may further serve to affect the nature of cell-signaling events by acting as antagonists, carrier molecules or chaperones to protect the ligands from binding to membrane-bound proteins [65] and in some cases acting as agonists [66]. Cleavage of various membrane proteins contributes to mitogenesis, cell migration, differentiation and various diseased states such as inflammation, tumorigenesis, spongiform encephalopathies and AD [67]. Thus, protein ectodomain shedding can potentially modulate most cellular functions mediated by transmembrane proteins and, therefore, has attracted extensive attention to understand the underlying regulatory mechanisms [68].
Proteases responsible for mediating ectodomain shedding are denoted as “sheddases” or “secretases” and appear to be members of the metzincin superfamily of zinc-dependent proteases, including A disintegrin and metalloproteinases (ADAMs) and matrix metalloproteinases (MMPs). ADAMs are type-I transmembrane proteins that contain a disintegrin-like and a metalloproteinase-like domain. Up to date, more than 40 ADAMs have been identified in the mammalian genome [69]. ADAMs have been implicated in most of the known shedding events [70, 71]. ADAM17, also known as tumor necrosis factor-α converting enzyme (TACE), was the first protease shown to be responsible for shedding events. A number of diverse cell surface proteins, including APP, TNF-α, TNFRs I and II, TGF-α, L-selectin, IL-6R, CD30 and growth factor receptors, undergo proteolysis mediated by TACE. Other members of the ADAM family of proteinases, particularly ADAM9, ADAM10 and ADAM12, have been implicated as sheddases for a wide range of proteins such as Notch and APP [72]. MMPs, also known as matrixins, are a large family of zinc-dependent metalloproteinases that degrade extracellular matrix and basement membrane components [73]. Till now, more than 20 endopeptidases have been classified as MMPs. Based on substrate specificity and the presence of distinct structural domains, MMPs are categorized into six subfamilies: collagenases, gelatinases, stromelysins, matrilysins, membrane type MMPs (MT-MMPs) and other MMPs [74]. The MMPs are homogeneous enzymes and share common structural elements. All members of this family contain a pro-domain and a catalytic domain. The pro-domain is cleaved upon activation while the catalytic domain contains the catalytic machinery including the zinc binding site and a conserved methionine. The metal ions maintain the three dimensional structure of MMPs and are necessary for stability and enzymatic activities. Physiologically, MMPs are thought to be important in wound healing, angiogenesis and bone remodeling [75, 76]. MMPs also have pathological roles in a variety of disease processes exemplified by tumor metastasis and neurodegeneration [77, 78]. In addition, certain MMPs are found to play a role in the shedding of cell surface proteins. Specifically, MT5-MMP has been shown to cleave cadherin [79], MMP7 participates in the constitutive shedding of proTNF-α [80] and MT1-MMP contributes to the shedding of the cell adhesion molecule CD44 [56]. MMP-9 can accomplish cleavage of cell surface proteins including TGF-β [81], IL-2R [82] and intercellular adhesion molecule 1 (ICAM-1) [83].
Shedding of RAGE
Purification of RAGE from mouse lung and protein sequencing revealed that mouse soluble RAGE might be generated by proteolysis [51]. It was also found that in the human lung 80% of RAGE mRNAs encode for the fl-RAGE protein and only 7% encode the endogenous secretory RAGE variant [84]. However, the fl-RAGE protein is difficult to detect in human lung samples where the sRAGE protein is abundant [85]. Collectively these results suggest that mechanisms other than alternative splicing account for the secretion of RAGE. Recently, several independent studies converge to support that proteolysis based ectodomain shedding is the second mechanism that contributes to the production of sRAGE [86–89].
A soluble form of RAGE was detected in supernatant of HEK Flp-In 293 cells transfected with fl-RAGE cDNA, indicating constitutive shedding of RAGE. Several stimuli including phorbol-12-myristat-13-acetate (PMA) and 4-aminophenylmercuric acetate (APMA) enhance the release of sRAGE, representing inducible shedding of RAGE [86]. Furthermore, HMGB1, a RAGE ligand, promoted RAGE shedding [87]. In contrast, another group reported calcium inducible RAGE shedding in HEK 293 cells but failed to observe PMA or RAGE ligands inducible RAGE shedding [89]. Hydrogen peroxide also induced RAGE shedding in neonatal rat oligodendrocytes [88].
By cell surface protein biotinylation experiments, it was demonstrated that sRAGE is biotinylated, providing further evidence that sRAGE is derived from cleavage of fl-RAGE at the cell surface [86]. While two groups identified ADAM10 as the sheddase responsible for RAGE shedding [87, 89], one study revealed both ADAM10 and MMP9 to be involved in RAGE shedding [86]. The contribution of MMP9 may explain the residual shedding of RAGE observed in ADAM10 null cells [87]. Interestingly, sRAGE was detected as doublet bands, and only the low molecular weight protein could be detected after treatment with peptide N-glycosidase F, demonstrating that the two forms of sRAGE result from different glycosylation rather than from different proteolytic cleavage [86]. The fact that sRAGE is glycosylated is consistent with previous observations [20, 51]. The importance of glycosylation for the ability of RAGE to bind its ligands has been demonstrated [20]. Furthermore, it is likely that glycosylation is of the same importance for sRAGE to bind its ligand. As fl-RAGE and shedded RAGE possess the same glycosylation pattern, ligand binding should be indistinguishable between these two isoforms. Ectodomain shedding of RAGE is followed by subsequent proteolytic cleavage by γ-secretase, which releases RAGE intracellular domain into the cytoplasm [86, 89].
Regulation of RAGE shedding
Ectodomain shedding represents a distinguished mechanism to regulate the signaling capacity of cell surface receptors. On one hand, ligands are synthesized as inactive transmembrane proforms and converted to active forms able to bind receptors by shedding. On the other hand, cleaving the ligand binding domain of receptors by shedding can terminate receptor signaling.
The existence of natural sRAGE in biological fluids suggests that sRAGE has important roles in normal physiology as well as in the development of pathological processes. The biological function of sRAGE may be implied by the efficacy of recombinant sRAGE to treat diabetic atherosclerosis [90] and tumor metastases [32] as well as reduce Aβ accumulation in brain [41]. However, the in vivo significance of RAGE shedding to generate sRAGE has not yet been studied. A relatively lower level of plasma sRAGE has been described to be associated with various diseases including mild cognitive impairment [91], AD [92], hypertension [93], rheumatoid arthritis [94] and coronary artery disease [95]. Therefore, the formation of sRAGE by metalloproteinase cleavage might be important to regulate RAGE-mediated cellular functions. The presence of sRAGE in biological fluids could affect the function of RAGE ligands by competing for ligand binding with membrane-bound RAGE. Thus, by preventing interaction of ligands with cell surface RAGE, sRAGE can disrupt ligand-induced RAGE signaling. Given that RAGE is a multi-ligand receptor and a number of disorders have been reported to be associated with activation of the ligand–RAGE axis, modulation of sRAGE generation by regulating proteolysis of RAGE provides a novel and favorable therapeutic way in addition to administration of recombinant sRAGE. Additionally, because RAGE and presumably also sRAGE are considered as pattern recognition receptors [96], sRAGE would be able to prevent ligand binding of other types of pattern recognition receptors by trapping of their ligands.
Since RAGE shedding is inducible by several stimuli, it is worth investigating the underlying mechanisms to exploit this understanding to promote sRAGE production in order to antagonize harmful effects mediated by fl-RAGE. PMA is a potent activator of PKC. In addition to RAGE, several other proteins known to be shed from the cell surface display PMA stimulated shedding. Currently, the mechanism of PMA stimulated ectodomain shedding is poorly understood. Although some reports indicated that PMA stimulated shedding is PKC-dependent, others gave controversial results [97]. Intriguingly RAGE shedding is PKC-dependent [86], suggesting that PKC agonists may be beneficial to alleviate RAGE-mediated disorders. APMA has been extensively used to activate recombinant MMPs in vitro. These metalloproteinases are synthesized as zymogens in which an unpaired cysteine residue of the prodomain binds to the active site zinc ion. In vitro, APMA reacts with the free thiol group of the zymogen and stabilizes an active form of the enzyme that leads to autoproteolytic processing of the propeptide [98]. Although the mechanism by which APMA enhances RAGE shedding was not addressed [86], the known ability of APMA to activate the removal of prodomains from metalloproteinase opens up the possibility that prodomains could play a role in the regulation of RAGE sheddase in cells. Calcium ionophores can activate a range of cell signaling pathways including PKC [99, 100], which may lead to activation of metalloproteinases. This may explain the observed inducibility of RAGE shedding by ionomycin [89] and calcium ionophore A23187 (Zhang L and Postina R, unpublished data). Furthermore, it has been reported that hydrogen peroxide induced RAGE shedding while antioxidant inhibited RAGE shedding [88]. We observed that chelerythrine enhanced RAGE shedding (Zhang L and Postina R, unpublished data). This is in accordance with a recent study reporting chelerythrine stimulated shedding of HB-EGF in a ROS dependent manner [101]. Since chelerythrine was shown to induce intracellular ROS [102], it is important to address the involvement of intracellular peroxides in the regulated ectodomain shedding of RAGE in future studies.
Modulating RAGE shedding for AD therapy
Given the crucial role of RAGE in AD progression as discussed in previous sections, RAGE represents a promising target for AD therapy. RAGE antibodies or sRAGE have proved to be effective means to inhibit RAGE–Aβ interaction, prevent RAGE activation, block Aβ influx across the BBB and alleviate RAGE-mediated cellular perturbation in AD. Furthermore, a set of compounds is being developed as RAGE antagonists for RAGE-related diseases. TTP488 (PF 04494700), an orally available small compound that inhibits RAGE activation, is in phase IIa/IIb clinical trials for AD patients. TTP488 is also in phase II clinical trials for type 2 diabetes patients. Compared to these approaches aiming at the RAGE molecule, manipulation of RAGE shedding may be a better strategy to prevent the progression of AD because RAGE shedding would reduce the amount of harmful membrane-bound RAGE and increase the amount of beneficial sRAGE simultaneously. Indeed, the amount of membrane-bound RAGE is elevated in the brain vasculature of AD patients while the level of sRAGE is significantly reduced in the plasma of AD patients compared with controls [92]. Given the recent identification of ADAM10 and MMP9 as the sheddases for RAGE, it is conceivable that elevated level of membrane-bound RAGE in the brain vasculature as well as the decrease of circulating sRAGE in the plasma may be due to the decrease of either ADAM10/MMP9 expression or ADAM10/MMP9 activity in AD patients. Consistent with this assumption, ADAM10 has been reported to be reduced in the platelets and CSF of AD patients [103, 104]. With regard to MMP9, the level of circulating MMP9 in AD patient’s plasma has been studied by several different groups. Although one group detected an increased level of plasma MMP9 in AD patients [105], others found no significant difference between AD patients and normal controls. On the other hand, increased MMP9 expression levels have been observed in AD brains, but MMP9 was predominantly found in the latent or proenzyme form in the proximity of extracellular amyloid plaques, and it was proposed that the lack of MMP9 activation contributes to the accumulation of insoluble Aβ in plaques [106]. Whether a reduction of MMP9 activation contributes to a decreased level of sRAGE in the plasma and a corresponding elevated amount of membrane-bound RAGE in the brain vasculature in AD needs to be addressed in future studies.
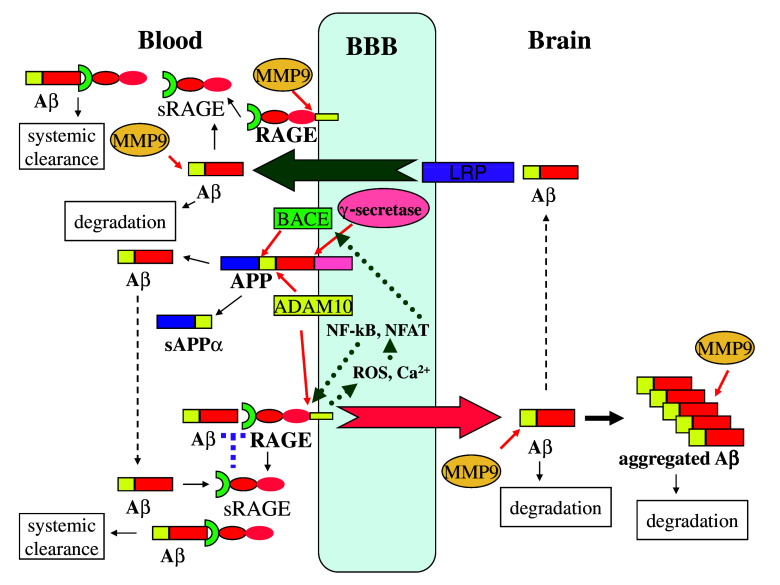
The identification of ADAM10 and MMP9 as the proteases responsible for ectodomain shedding of RAGE provides an unprecedented opportunity to modulate these enzyme activities for AD therapy. As shown in Fig. 3, RAGE/Aβ interaction creates a positive feedback loop to execute and amplify neurotoxic effects mediated by RAGE receptor–Aβ ligand binding. Upon binding Aβ, RAGE initiates several branches of downstream signaling. In one branch, increased ROS activates NF-κB, leading to increased RAGE expression [21]. In another branch, increased calcium activates NFAT, which upregulates BACE expression and enzymatic activity, resulting in elevated Aβ generation by β-secretase [30]. If left uncontrolled, this ligand and receptor interaction will lead to the generation of more and more ligand/receptor and severe pathological consequences. Fortunately, RAGE is subject to ectodomain shedding by ADAM10 and MMP9 and the derived sRAGE can sequester Aβ for systemic clearance and antagonize Aβ binding to cell surface RAGE, on one hand blocking the amplification loop for more Aβ and RAGE generation, on the other hand inhibiting RAGE-mediated transport of Aβ into brain for aggregation. Furthermore, ADAM10 and MMP9 enzymes play other beneficial roles in alleviating AD development. ADAM10 acts as α-secretase for APP to produce neurotrophic and neuroprotective sAPPα [4]. This antagonizes APP processing by β-secretase BACE and reduces Aβ production. MMP9 can degrade both soluble Aβ and tightly aggregated Aβ fibrils, while other Aβ degrading enzymes including endothelin-converting enzyme, insulin-degrading enzyme and neprilysin can only degrade soluble Aβ, implying the significant contribution of MMP9 to clearance of plaques in amyloid-laden brains [107]. In vivo studies further demonstrated that pharmacological inhibition of MMPs increased the brain interstitial fluid Aβ level in an AD mice model, and the Aβ level was significantly increased in MMP9 knockout mice [108].
Fig. 3.
Potential roles of ADAM10 and MMP9 in preventing Aβ aggregation in the brain. Processing of APP by ADAM10 produces neurotrophic sAPPα and precludes Aβ production by BACE and γ-secretase. RAGE promotes influx of circulating Aβ across the BBB, which is antagonized by LRP-1-mediated efflux of Aβ. RAGE initiates ROS/NF-kB and calcium/NFAT axes to promote RAGE and BACE transcription, respectively, which lead to further production of the RAGE receptor and Aβ ligand, forming a positive feedback. Shedding of RAGE by ADAM10/MMP9 decreases the availability of RAGE to bind and transport Aβ. Furthermore, the derived sRAGE can sequester Aβ to inhibit its neurotoxicity and influx into the brain. sRAGE also promotes Aβ systemic clearance. MMP9 degrades both soluble Aβ and aggregated Aβ fibrils, further lessening the Aβ load in the brain. Thus, modulation of ADAM10/MMP9 emerges as a novel strategy for AD therapy
The functional interaction between MMP9 and RAGE is particularly reasonable in view of several aspects. First, both molecules are expressed in human endothelial cells, neurons, astrocytes, and microglia, thus giving the chance to interact with each other. Second, increased expression of both RAGE and MMP9 is found in Aβ rich tissues [35, 106]. Third, overexpression of both molecules is involved in inflammation, tumor growth and migration, atherosclerosis, multiple sclerosis and neurodegenerative disorders such as AD. All these coincidences between MMP9 and RAGE strongly support the possibility of ectodomain shedding of RAGE by MMP9. Although MMP9 has the potential to be utilized for AD therapy by producing sRAGE and degrading Aβ, MMP9 has been shown to play detrimental roles in other diseases such as stroke and metastasis. Thus, whether or not MMP9 is beneficial or harmful may depend on several factors, including the cellular sources, the extracellular environment and the stage of lesion in the development of diseases.
RAGE shedding can be promoted via the increase of ADAM10/MMP9 activity or expression to generate more sRAGE. In contrast, specific ADAM10/MMP9 inhibitors can be used to inhibit RAGE shedding. Given the complex roles of RAGE and other receptors interacting with RAGE ligands, the decision to activate or inhibit RAGE shedding will depend on different physiological or pathological contexts. But considering the overwhelming evidence that supports pathogenic roles of RAGE in AD, it is tempting to promote RAGE shedding for the benefits of AD patients. It has been speculated that sRAGE–ligand complexes are eliminated from the blood via the spleen and/or liver [109]. Considering this, enhancement of RAGE shedding will give rise to clearance of circulating Aβ from blood by sRAGE. On the other hand, by shifting cell surface RAGE into sRAGE, it is possible to reduce the uptake of Aβ into the brain and diminish membrane-bound RAGE-mediated neurotoxicity.
A plethora of molecules have been characterized to stimulate sheddase activities, especially for ADAM10, the α-secretase for APP [4]. Considering the crucial involvement of ADAM10 in RAGE shedding, we are at a great advantage to exploit the current understanding of the modulation of ADAM10 activity in the shedding of other membrane proteins and put it in the context of RAGE shedding. The neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP) was shown to stimulate ADAM10 activity resulting in the α-secretase cleavage of APP and the production of sAPPα [110]. Although the neuroprotective effects of PACAP have been attributed to the activation of the non-amyloidogenic pathway of APP processing, it is possible that PACAP-stimulated ADAM10 activity also enhances RAGE shedding and antagonizes RAGE-mediated neurodegeneration. Further study to examine the sRAGE level in AD animal models treated with PACAP will be important to test this hypothesis and provide valuable information on the promise of PACAP as a small molecule modulator of RAGE shedding for AD therapy. PKC activator bryostatin and a synthetic analog were recently reported to be able to increase α-secretase activity and reverse Aβ-induced abnormality in a cell model [111]. Given the observation that cognitive restorative and antidepressant effects of bryostatin are partly mediated by the activation of PKC isozymes [112] and our demonstration that RAGE shedding is PKC-dependent [86], it will be very attractive to test the effects of bryostatin and its analog on RAGE shedding and the ensuing impact on AD progression. More importantly, bryostatin has been used in dozens of clinical studies for treatment of many kinds of cancer and the first clinical trial for AD patients was initiated in 2008. Therefore, bryostatin and its analog show promise for AD drug development. Abnormal insulin and insulin-like growth factor (IGF)-1 signaling has been implicated in AD besides type 2 diabetes [113]. The IGF-1 signal is transduced via different pathways including MAPK and phosphatidylinositol 3-kinase (PI3K) cascades to stimulate α-secretase activity and increase the shedding of the APP protein family [114]. Insulin can increase the shedding of Klotho by stimulating ADAM10 activity via the PI3K pathway without increasing ADAM10 mRNA and protein levels [115]. Therefore, it is worth investigating the stimulating effects of insulin and IGF-1 on the sheddase responsible for RAGE shedding and correlating insulin, IGF-1, RAGE and sRAGE levels in samples from AD patients. These studies will reveal whether the therapeutic effects of insulin and IGF-1 on AD are partly mediated through modulation of RAGE shedding.
Conclusion
RAGE acts as a multi-ligand receptor that is able to bind a range of ligands and therefore mediates various physiological and pathological effects. RAGE is abnormally upregulated in many situations of disease and consequently contributes to these processes. Although the detailed molecular mechanisms underlying these RAGE-mediated disorders are far from being completely elucidated, extensive research contributed by researchers from different disciplines by employing a variety of strategies has greatly advanced our understanding of RAGE. As a result, insightful views have emerged regarding the development of innovative diagnosis and therapy approaches against RAGE-mediated diseases [116]. The identification and utilization of sRAGE are the most distinguished among these advances. A growing body of evidence suggests the possibility of sRAGE as a biomarker for many RAGE-related diseases, including AD, coronary artery disease, rheumatoid arthritis, diabetes, hypertension and chronic renal failure [117]. On the other hand, recombinant sRAGE has been used to alleviate RAGE-mediated pathological conditions. Therefore, sRAGE represents both a potential biomarker and a promising therapeutic tool for RAGE-mediated disorders. So it is desperately necessary to decipher the generation and pathophysiologic functions of sRAGE. While the predominant view holds that sRAGE is derived from alternative splicing of mRNA, recent studies from independent groups demonstrated clearly that RAGE undergoes ectodomain shedding in both constitutive and regulated manner. Several stimuli including PMA, AMPA and calcium stimulate RAGE shedding and contribute to sRAGE production. Furthermore, ADAM10 and MMP9 are characterized as the sheddases that mediate RAGE shedding. Since ADAM10 is also engaged in reducing Aβ production and MMP9 in promoting Aβ degradation, the exciting findings that sRAGE can be derived from shedding of RAGE by ADAM10/MMP9 open up a fascinating field for modulation of RAGE proteolysis as a new therapeutic approach against AD.
Acknowledgments
This work was supported by the Alzheimer Forschung Initiative e.V. (Düsseldorf, Germany) and the Deutsche Forschungsgemeinschaft (DFG Priority Program SPP1085–Cellular Mechanisms of Alzheimer’s Disease). We thank Dr. Zhijun Zhang (Zhongda Hospital Affiliated to Southeast University, Nanjing, China) for support and helpful discussion.
Contributor Information
Ling Zhang, FAX: +86-25-83272090, Email: zhangling9@hotmail.com.
Yingqun Wang, FAX: +1-215-5732486, Email: yingqunw@mail.med.upenn.edu.
References
- 1.Tanzi RE. A genetic dichotomy model for the inheritance of Alzheimer’s disease and common age-related disorders. J Clin Invest. 1999;104:1175–1179. doi: 10.1172/JCI8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 3.Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postina R. A closer look at alpha-secretase. Curr Alzheimer Res. 2008;5:179–186. doi: 10.2174/156720508783954668. [DOI] [PubMed] [Google Scholar]
- 5.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 6.Behl C, Davis JB, Lesley R, Schubert D. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 7.Kaltschmidt B, Uherek M, Volk B, Baeuerle PA, Kaltschmidt C. Transcription factor NF-kappaB is activated in primary neurons by amyloid beta peptides and in neurons surrounding early plaques from patients with Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:2642–2647. doi: 10.1073/pnas.94.6.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klegeris A, Walker DG, McGeer PL. Activation of macrophages by Alzheimer beta amyloid peptide. Biochem Biophys Res Commun. 1994;199:984–991. doi: 10.1006/bbrc.1994.1326. [DOI] [PubMed] [Google Scholar]
- 9.Gitter BD, Cox LM, Rydel RE, May PC. Amyloid beta peptide potentiates cytokine secretion by interleukin-1 beta-activated human astrocytoma cells. Proc Natl Acad Sci USA. 1995;92:10738–10741. doi: 10.1073/pnas.92.23.10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giri R, Selvaraj S, Miller CA, Hofman F, Yan SD, Stern D, Zlokovic BV, Kalra VK. Effect of endothelial cell polarity on beta-amyloid-induced migration of monocytes across normal and AD endothelium. Am J Physiol Cell Physiol. 2002;283:C895–C904. doi: 10.1152/ajpcell.00293.2001. [DOI] [PubMed] [Google Scholar]
- 11.Zlokovic B. Can blood–brain barrier play a role in the development of cerebral amyloidosis and Alzheimer’s disease pathology. Neurobiol Dis. 1997;4:23–26. doi: 10.1006/nbdi.1997.0134. [DOI] [PubMed] [Google Scholar]
- 12.Zlokovic BV. The blood–brain barrier in health and chronic neurodegenerative disorders. Neuron. 2008;57:178–201. doi: 10.1016/j.neuron.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Zlokovic BV, Yamada S, Holtzman D, Ghiso J, Frangione B. Clearance of amyloid beta-peptide from brain: transport or metabolism? Nat Med. 2000;6:718. doi: 10.1038/77397. [DOI] [PubMed] [Google Scholar]
- 14.Martel CL, Mackic JB, Matsubara E, Governale S, Miguel C, Miao W, McComb JG, Frangione B, Ghiso J, Zlokovic BV. Isoform-specific effects of apolipoproteins E2, E3, and E4 on cerebral capillary sequestration and blood–brain barrier transport of circulating Alzheimer’s amyloid beta. J Neurochem. 1997;69:1995–2004. doi: 10.1046/j.1471-4159.1997.69051995.x. [DOI] [PubMed] [Google Scholar]
- 15.Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, Gerard C, Hama E, Lee HJ, Saido TC. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 16.Selkoe DJ. Clearing the brain’s amyloid cobwebs. Neuron. 2001;32:177–180. doi: 10.1016/S0896-6273(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 17.Zlokovic BV. Clearing amyloid through the blood–brain barrier. J Neurochem. 2004;89:807–811. doi: 10.1111/j.1471-4159.2004.02385.x. [DOI] [PubMed] [Google Scholar]
- 18.Sugaya K, Fukagawa T, Matsumoto K, Mita K, Takahashi E, Ando A, Inoko H, Ikemura T. Three genes in the human MHC class III region near the junction with the class II: gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics. 1994;23:408–419. doi: 10.1006/geno.1994.1517. [DOI] [PubMed] [Google Scholar]
- 19.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 20.Srikrishna G, Huttunen HJ, Johansson L, Weigle B, Yamaguchi Y, Rauvala H, Freeze HH. N-Glycans on the receptor for advanced glycation end products influence amphoterin binding and neurite outgrowth. J Neurochem. 2002;80:998–1008. doi: 10.1046/j.0022-3042.2002.00796.x. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Schmidt AM. Characterization and functional analysis of the promoter of RAGE, the receptor for advanced glycation end products. J Biol Chem. 1997;272:16498–16506. doi: 10.1074/jbc.272.26.16498. [DOI] [PubMed] [Google Scholar]
- 22.Hori O, Brett J, Slattery T, Cao R, Zhang JH, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D, Morser J, Stern D, Schmidt AM. The receptor for advanced glycation end-products (Rage) is a cellular-binding site for amphoterin—mediation of neurite outgrowth and coexpression of rage and amphoterin in the developing nervous-system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 23.Sakaguchi T, Yan SF, Du Yan S, Belov D, Rong LL, Sousa M, Andrassy M, Marso SP, Duda S, Arnold B, Liliensiek B, Nawroth PP, Stern DM, Schmidt AM, Naka Y. Central role of RAGE-dependent neointimal expansion in arterial restenosis. J Clin Invest. 2003;111:959–972. doi: 10.1172/JCI17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brett J, Schmidt AM, Yan SD, Zou YS, Weidman E, Pinsky D, Nowygrod R, Neeper M, Przysiecki C, Shaw A. Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am J Pathol. 1993;143:1699–1712. [PMC free article] [PubMed] [Google Scholar]
- 25.Bucciarelli LG, Wendt T, Rong L, Lalla E, Hofmann MA, Goova MT, Taguchi A, Yan SF, Yan SD, Stern DM, Schmidt AM. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell Mol Life Sci. 2002;59:1117–1128. doi: 10.1007/s00018-002-8491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, Rutledge R, Lin B, Amoscato AA, Zeh HJ, Lotze MT. RAGE (receptor for advanced glycation endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong LL, Trojaborg W, Qu W, Kostov K, Yan SD, Gooch C, Szabolcs M, Hays AP, Schmidt AM. Antagonism of RAGE suppresses peripheral nerve regeneration. FASEB J. 2004;18:1812–1817. doi: 10.1096/fj.04-1899com. [DOI] [PubMed] [Google Scholar]
- 28.Bierhaus A, Haslbeck KM, Humpert PM, Liliensiek B, Dehmer T, Morcos M, Sayed AA, Andrassy M, Schiekofer S, Schneider JG, Schulz JB, Heuss D, Neundorfer B, Dierl S, Huber J, Tritschler H, Schmidt AM, Schwaninger M, Haering HU, Schleicher E, Kasper M, Stern DM, Arnold B, Nawroth PP. Loss of pain perception in diabetes is dependent on a receptor of the immunoglobulin superfamily. J Clin Invest. 2004;114:1741–1751. doi: 10.1172/JCI18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartling B, Hofmann HS, Weigle B, Silber RE, Simm A. Down-regulation of the receptor for advanced glycation end-products (RAGE) supports non-small cell lung carcinoma. Carcinogenesis. 2005;26:293–301. doi: 10.1093/carcin/bgh333. [DOI] [PubMed] [Google Scholar]
- 30.Cho HJ, Son SM, Jin SM, Hong HS, Shin DH, Kim SJ, Huh K, Mook-Jung I. RAGE regulates BACE1 and Aβ generation via NFAT1 activation in Alzheimer’s disease animal model. FASEB J. 2009;23:2639–2649. doi: 10.1096/fj.08-126383. [DOI] [PubMed] [Google Scholar]
- 31.Hudson BI, Kalea AZ, Del Mar AM, Harja E, Boulanger E, D’Agati V, Schmidt AM. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283:34457–34468. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann MA, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern DM, Schmidt AM. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 33.Simm A, Munch G, Seif F, Schenk O, Heidland A, Richter H, Vamvakas S, Schinzel R. Advanced glycation endproducts stimulate the MAP-kinase pathway in tubulus cell line LLC-PK1. FEBS Lett. 1997;410:481–484. doi: 10.1016/S0014-5793(97)00644-3. [DOI] [PubMed] [Google Scholar]
- 34.Huttunen HJ, Kuja-Panula J, Rauvala H. Receptor for advanced glycation end products (RAGE) signaling induces CREB-dependent chromogranin expression during neuronal differentiation. J Biol Chem. 2002;277:38635–38646. doi: 10.1074/jbc.M202515200. [DOI] [PubMed] [Google Scholar]
- 35.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt AM. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 36.Yan SD, Stern D, Kane MD, Kuo YM, Lampert HC, Roher AE. RAGE–Abeta interactions in the pathophysiology of Alzheimer’s disease. Restor Neurol Neurosci. 1998;12:167–173. [PubMed] [Google Scholar]
- 37.Yan SD, Stern D, Schmidt AM. What’s the RAGE? The receptor for advanced glycation end products (RAGE) and the dark side of glucose. Eur J Clin Invest. 1997;27:179–181. doi: 10.1046/j.1365-2362.1996.00072.x. [DOI] [PubMed] [Google Scholar]
- 38.Li JJ, Dickson D, Hof PR, Vlassara H. Receptors for advanced glycosylation endproducts in human brain: role in brain homeostasis. Mol Med. 1998;4:46–60. [PMC free article] [PubMed] [Google Scholar]
- 39.Du YS, Zhu H, Fu J, Yan SF, Roher A, Tourtellotte WW, Rajavashisth T, Chen X, Godman GC, Stern D, Schmidt AM. Amyloid-beta peptide-receptor for advanced glycation endproduct interaction elicits neuronal expression of macrophage-colony stimulating factor: a proinflammatory pathway in Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:5296–5301. doi: 10.1073/pnas.94.21.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackic JB, Stins M, McComb JG, Calero M, Ghiso J, Kim KS, Yan SD, Stern D, Schmidt AM, Frangione B, Zlokovic BV. Human blood–brain barrier receptors for Alzheimer’s amyloid-beta 1–40. Asymmetrical binding, endocytosis, and transcytosis at the apical side of brain microvascular endothelial cell monolayer. J Clin Invest. 1998;102:734–743. doi: 10.1172/JCI2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deane R, Du YS, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, Welch D, Manness L, Lin C, Yu J, Zhu H, Ghiso J, Frangione B, Stern A, Schmidt AM, Armstrong DL, Arnold B, Liliensiek B, Nawroth P, Hofman F, Kindy M, Stern D, Zlokovic B. RAGE mediates amyloid-beta peptide transport across the blood–brain barrier and accumulation in brain. Nat Med. 2003;9:907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 42.Zlokovic BV, Martel CL, Matsubara E, McComb JG, Zheng G, McCluskey RT, Frangione B, Ghiso J. Glycoprotein 330/megalin: probable role in receptor-mediated transport of apolipoprotein J alone and in a complex with Alzheimer disease amyloid beta at the blood–brain and blood–cerebrospinal fluid barriers. Proc Natl Acad Sci USA. 1996;93:4229–4234. doi: 10.1073/pnas.93.9.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giri R, Shen Y, Stins M, Du YS, Schmidt AM, Stern D, Kim KS, Zlokovic B, Kalra VK. beta-amyloid-induced migration of monocytes across human brain endothelial cells involves RAGE and PECAM-1. Am J Physiol Cell Physiol. 2000;279:C1772–C1781. doi: 10.1152/ajpcell.2000.279.6.C1772. [DOI] [PubMed] [Google Scholar]
- 44.Arancio O, Zhang HP, Chen X, Lin C, Trinchese F, Puzzo D, Liu S, Hegde A, Yan SF, Stern A, Luddy JS, Lue LF, Walker DG, Roher A, Buttini M, Mucke L, Li W, Schmidt AM, Kindy M, Hyslop PA, Stern DM, Du Yan SS. RAGE potentiates Abeta-induced perturbation of neuronal function in transgenic mice. EMBO J. 2004;23:4096–4105. doi: 10.1038/sj.emboj.7600415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Q, Keller JN. Evaluation of rage isoforms, ligands, and signaling in the brain. Biochim Biophys Acta. 2005;1746:18–27. doi: 10.1016/j.bbamcr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Ding Q, Keller JN. Splice variants of the receptor for advanced glycosylation end products (RAGE) in human brain. Neurosci Lett. 2005;373:67–72. doi: 10.1016/j.neulet.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 47.Goova MT, Li J, Kislinger T, Qu W, Lu Y, Bucciarelli LG, Nowygrod S, Wolf BM, Caliste X, Yan SF, Stern DM, Schmidt AM. Blockade of receptor for advanced glycation end-products restores effective wound healing in diabetic mice. Am J Pathol. 2001;159:513–525. doi: 10.1016/S0002-9440(10)61723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi ME. Cloning and characterization of a naturally occurring soluble form of TGF-beta type I receptor. Am J Physiol. 1999;276:F88–F95. doi: 10.1152/ajprenal.1999.276.1.F88. [DOI] [PubMed] [Google Scholar]
- 49.Michel J, Langstein J, Hofstadter F, Schwarz H. A soluble form of CD137 (ILA/4–1BB), a member of the TNF receptor family, is released by activated lymphocytes and is detectable in sera of patients with rheumatoid arthritis. Eur J Immunol. 1998;28:290–295. doi: 10.1002/(SICI)1521-4141(199801)28:01<290::AID-IMMU290>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 50.Haudenschild D, Moseley T, Rose L, Reddi AH. Soluble and transmembrane isoforms of novel interleukin-17 receptor-like protein by RNA splicing and expression in prostate cancer. J Biol Chem. 2002;277:4309–4316. doi: 10.1074/jbc.M109372200. [DOI] [PubMed] [Google Scholar]
- 51.Hanford LE, Enghild JJ, Valnickova Z, Petersen SV, Schaefer LM, Schaefer TM, Reinhart TA, Oury TD. Purification and characterization of mouse soluble receptor for advanced glycation end products (sRAGE) J Biol Chem. 2004;279:50019–50024. doi: 10.1074/jbc.M409782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arribas J, Borroto A. Protein ectodomain shedding. Chem Rev. 2002;102:4627–4637. doi: 10.1021/cr010202t. [DOI] [PubMed] [Google Scholar]
- 53.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanderson MP, Erickson SN, Gough PJ, Garton KJ, Wille PT, Raines EW, Dunbar AJ, Dempsey PJ. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J Biol Chem. 2005;280:1826–1837. doi: 10.1074/jbc.M408804200. [DOI] [PubMed] [Google Scholar]
- 55.Hirata M, Umata T, Takahashi T, Ohnuma M, Miura Y, Iwamoto R, Mekada E. Identification of serum factor inducing ectodomain shedding of proHB-EGF and sStudies of noncleavable mutants of proHB-EGF. Biochem Biophys Res Commun. 2001;283:915–922. doi: 10.1006/bbrc.2001.4879. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura H, Suenaga N, Taniwaki K, Matsuki H, Yonezawa K, Fujii M, Okada Y, Seiki M. Constitutive and induced CD44 shedding by ADAM-like proteases and membrane-type 1 matrix metalloproteinase. Cancer Res. 2004;64:876–882. doi: 10.1158/0008-5472.CAN-03-3502. [DOI] [PubMed] [Google Scholar]
- 57.Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol. 2001;153:893–904. doi: 10.1083/jcb.153.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budagian V, Bulanova E, Orinska Z, Ludwig A, Rose-John S, Saftig P, Borden EC, Bulfone-Paus S. Natural soluble interleukin-15Ralpha is generated by cleavage that involves the tumor necrosis factor-alpha-converting enzyme (TACE/ADAM17) J Biol Chem. 2004;279:40368–40375. doi: 10.1074/jbc.M404125200. [DOI] [PubMed] [Google Scholar]
- 59.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 60.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 61.Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, Postina R, Fahrenholz F, Fogel M, Lemmon V, Altevogt P. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parkin ET, Tan F, Skidgel RA, Turner AJ, Hooper NM. The ectodomain shedding of angiotensin-converting enzyme is independent of its localisation in lipid rafts. J Cell Sci. 2003;116:3079–3087. doi: 10.1242/jcs.00626. [DOI] [PubMed] [Google Scholar]
- 63.Allinson TM, Parkin ET, Condon TP, Schwager SL, Sturrock ED, Turner AJ, Hooper NM. The role of ADAM10 and ADAM17 in the ectodomain shedding of angiotensin converting enzyme and the amyloid precursor protein. Eur J Biochem. 2004;271:2539–2547. doi: 10.1111/j.1432-1033.2004.04184.x. [DOI] [PubMed] [Google Scholar]
- 64.Cisse MA, Sunyach C, Lefranc-Jullien S, Postina R, Vincent B, Checler F. The disintegrin ADAM9 indirectly contributes to the physiological processing of cellular prion by modulating ADAM10 activity. J Biol Chem. 2005;280:40624–40631. doi: 10.1074/jbc.M506069200. [DOI] [PubMed] [Google Scholar]
- 65.Mortier E, Bernard J, Plet A, Jacques Y. Natural, proteolytic release of a soluble form of human IL-15 receptor alpha-chain that behaves as a specific, high affinity IL-15 antagonist. J Immunol. 2004;173:1681–1688. doi: 10.4049/jimmunol.173.3.1681. [DOI] [PubMed] [Google Scholar]
- 66.Vollmer P, Peters M, Ehlers M, Yagame H, Matsuba T, Kondo M, Yasukawa K, Buschenfelde KH, Rose-John S. Yeast expression of the cytokine receptor domain of the soluble interleukin-6 receptor. J Immunol Methods. 1996;199:47–54. doi: 10.1016/S0022-1759(96)00163-9. [DOI] [PubMed] [Google Scholar]
- 67.Hooper NM, Karran EH, Turner AJ. Membrane protein secretases. Biochem J. 1997;321(Pt 2):265–279. doi: 10.1042/bj3210265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Gall SM, Bobe P, Reiss K, Horiuchi K, Niu XD, Lundell D, Gibb DR, Conrad D, Saftig P, Blobel CP. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, l-selectin, and tumor necrosis factor alpha. Mol Biol Cell. 2009;20:1785–1794. doi: 10.1091/mbc.E08-11-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reiss K, Saftig P. The “A Disintegrin And Metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin Cell Dev Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Huovila AP, Turner AJ, Pelto-Huikko M, Karkkainen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 71.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deuss M, Reiss K, Hartmann D. Part-time alpha-secretases: the functional biology of ADAM 9, 10 and 17. Curr Alzheimer Res. 2008;5:187–201. doi: 10.2174/156720508783954686. [DOI] [PubMed] [Google Scholar]
- 73.Dzwonek J, Rylski M, Kaczmarek L. Matrix metalloproteinases and their endogenous inhibitors in neuronal physiology of the adult brain. FEBS Lett. 2004;567:129–135. doi: 10.1016/j.febslet.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 74.Hartung HP, Kieseier BC. The role of matrix metalloproteinases in autoimmune damage to the central and peripheral nervous system. J Neuroimmunol. 2000;107:140–147. doi: 10.1016/S0165-5728(00)00225-3. [DOI] [PubMed] [Google Scholar]
- 75.van Hinsbergh VW, Koolwijk P. Endothelial sprouting and angiogenesis: matrix metalloproteinases in the lead. Cardiovasc Res. 2008;78:203–212. doi: 10.1093/cvr/cvm102. [DOI] [PubMed] [Google Scholar]
- 76.Krane SM, Inada M. Matrix metalloproteinases and bone. Bone. 2008;43:7–18. doi: 10.1016/j.bone.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 77.Rydlova M, Holubec L, Jr, Ludvikova M, Jr, Kalfert D, Franekova J, Povysil C, Ludvikova M. Biological activity and clinical implications of the matrix metalloproteinases. Anticancer Res. 2008;28:1389–1397. [PubMed] [Google Scholar]
- 78.Rosenberg GA. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol. 2009;8:205–216. doi: 10.1016/S1474-4422(09)70016-X. [DOI] [PubMed] [Google Scholar]
- 79.Monea S, Jordan BA, Srivastava S, DeSouza S, Ziff EB. Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J Neurosci. 2006;26:2300–2312. doi: 10.1523/JNEUROSCI.3521-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest. 2000;105:143–150. doi: 10.1172/JCI7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 82.Sheu BC, Hsu SM, Ho HN, Lien HC, Huang SC, Lin RH. A novel role of metalloproteinase in cancer-mediated immunosuppression. Cancer Res. 2001;61:237–242. [PubMed] [Google Scholar]
- 83.Fiore E, Fusco C, Romero P, Stamenkovic I. Matrix metalloproteinase 9 (MMP-9/gelatinase B) proteolytically cleaves ICAM-1 and participates in tumor cell resistance to natural killer cell-mediated cytotoxicity. Oncogene. 2002;21:5213–5223. doi: 10.1038/sj.onc.1205684. [DOI] [PubMed] [Google Scholar]
- 84.Hudson BI, Carter AM, Harja E, Kalea AZ, Arriero M, Yang H, Grant PJ, Schmidt AM. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2008;22:1572–1580. doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- 85.Englert JM, Hanford LE, Kaminski N, Tobolewski JM, Tan RJ, Fattman CL, Ramsgaard L, Richards TJ, Loutaev I, Nawroth PP, Kasper M, Bierhaus A, Oury TD. A role for the receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Am J Pathol. 2008;172:583–591. doi: 10.2353/ajpath.2008.070569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang L, Bukulin M, Kojro E, Roth A, Metz VV, Fahrenholz F, Nawroth PP, Bierhaus A, Postina R. Receptor for advanced glycation end products is subjected to protein ectodomain shedding by metalloproteinases. J Biol Chem. 2008;283:35507–35516. doi: 10.1074/jbc.M806948200. [DOI] [PubMed] [Google Scholar]
- 87.Raucci A, Cugusi S, Antonelli A, Barabino SM, Monti L, Bierhaus A, Reiss K, Saftig P, Bianchi ME. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10) FASEB J. 2008;22:3716–3727. doi: 10.1096/fj.08-109033. [DOI] [PubMed] [Google Scholar]
- 88.Qin J, Goswami R, Dawson S, Dawson G. Expression of the receptor for advanced glycation end products in oligodendrocytes in response to oxidative stress. J Neurosci Res. 2008;86:2414–2422. doi: 10.1002/jnr.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Galichet A, Weibel M, Heizmann CW. Calcium-regulated intramembrane proteolysis of the RAGE receptor. Biochem Biophys Res Commun. 2008;370:1–5. doi: 10.1016/j.bbrc.2008.02.163. [DOI] [PubMed] [Google Scholar]
- 90.Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Jr, Chow WS, Stern D, Schmidt AM. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med. 1998;4:1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- 91.Ghidoni R, Benussi L, Glionna M, Franzoni M, Geroldi D, Emanuele E, Binetti G. Decreased plasma levels of soluble receptor for advanced glycation end products in mild cognitive impairment. J Neural Transm. 2008;115:1047–1050. doi: 10.1007/s00702-008-0069-9. [DOI] [PubMed] [Google Scholar]
- 92.Emanuele E, D’Angelo A, Tomaino C, Binetti G, Ghidoni R, Politi P, Bernardi L, Maletta R, Bruni AC, Geroldi D. Circulating levels of soluble receptor for advanced glycation end products in Alzheimer disease and vascular dementia. Arch Neurol. 2005;62:1734–1736. doi: 10.1001/archneur.62.11.1734. [DOI] [PubMed] [Google Scholar]
- 93.Geroldi D, Falcone C, Emanuele E, D’Angelo A, Calcagnino M, Buzzi MP, Scioli GA, Fogari R. Decreased plasma levels of soluble receptor for advanced glycation end-products in patients with essential hypertension. J Hypertens. 2005;23:1725–1729. doi: 10.1097/01.hjh.0000177535.45785.64. [DOI] [PubMed] [Google Scholar]
- 94.Pullerits R, Bokarewa M, Dahlberg L, Tarkowski A. Decreased levels of soluble receptor for advanced glycation end products in patients with rheumatoid arthritis indicating deficient inflammatory control. Arthritis Res Ther. 2005;7:R817–R824. doi: 10.1186/ar1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Falcone C, Emanuele E, D’Angelo A, Buzzi MP, Belvito C, Cuccia M, Geroldi D. Plasma levels of soluble receptor for advanced glycation end products and coronary artery disease in nondiabetic men. Arterioscler Thromb Vasc Biol. 2005;25:1032–1037. doi: 10.1161/01.ATV.0000160342.20342.00. [DOI] [PubMed] [Google Scholar]
- 96.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83:876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 97.Racchi M, Solano DC, Sironi M, Govoni S. Activity of alpha-secretase as the common final effector of protein kinase C-dependent and -independent modulation of amyloid precursor protein metabolism. J Neurochem. 1999;72:2464–2470. doi: 10.1046/j.1471-4159.1999.0722464.x. [DOI] [PubMed] [Google Scholar]
- 98.Birkedal-Hansen H. Proteolytic remodeling of extracellular matrix. Curr Opin Cell Biol. 1995;7:728–735. doi: 10.1016/0955-0674(95)80116-2. [DOI] [PubMed] [Google Scholar]
- 99.Eguchi S, Numaguchi K, Iwasaki H, Matsumoto T, Yamakawa T, Utsunomiya H, Motley ED, Kawakatsu H, Owada KM, Hirata Y, Marumo F, Inagami T. Calcium-dependent epidermal growth factor receptor transactivation mediates the angiotensin II-induced mitogen-activated protein kinase activation in vascular smooth muscle cells. J Biol Chem. 1998;273:8890–8896. doi: 10.1074/jbc.273.15.8890. [DOI] [PubMed] [Google Scholar]
- 100.Franklin RA, Atherfold PA, McCubrey JA. Calcium-induced ERK activation in human T lymphocytes occurs via p56(Lck) and CaM-kinase. Mol Immunol. 2000;37:675–683. doi: 10.1016/S0161-5890(00)00087-0. [DOI] [PubMed] [Google Scholar]
- 101.Kim J, Lin J, Adam RM, Lamb C, Shively SB, Freeman MR. An oxidative stress mechanism mediates chelerythrine-induced heparin-binding EGF-like growth factor ectodomain shedding. J Cell Biochem. 2005;94:39–49. doi: 10.1002/jcb.20276. [DOI] [PubMed] [Google Scholar]
- 102.Yu R, Mandlekar S, Tan TH, Kong AN. Activation of p38 and c-Jun N-terminal kinase pathways and induction of apoptosis by chelerythrine do not require inhibition of protein kinase C. J Biol Chem. 2000;275:9612–9619. doi: 10.1074/jbc.275.13.9612. [DOI] [PubMed] [Google Scholar]
- 103.Colciaghi F, Marcello E, Borroni B, Zimmermann M, Caltagirone C, Cattabeni F, Padovani A, Di Luca M. Platelet APP, ADAM 10 and BACE alterations in the early stages of Alzheimer disease. Neurology. 2004;62:498–501. doi: 10.1212/01.wnl.0000106953.49802.9c. [DOI] [PubMed] [Google Scholar]
- 104.Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med. 2002;8:67–74. [PMC free article] [PubMed] [Google Scholar]
- 105.Lorenzl S, Albers DS, Relkin N, Ngyuen T, Hilgenberg SL, Chirichigno J, Cudkowicz ME, Beal MF. Increased plasma levels of matrix metalloproteinase-9 in patients with Alzheimer’s disease. Neurochem Int. 2003;43:191–196. doi: 10.1016/S0197-0186(03)00004-4. [DOI] [PubMed] [Google Scholar]
- 106.Backstrom JR, Lim GP, Cullen MJ, Tokes ZA. Matrix metalloproteinase-9 (MMP-9) is synthesized in neurons of the human hippocampus and is capable of degrading the amyloid-beta peptide (1–40) J Neurosci. 1996;16:7910–7919. doi: 10.1523/JNEUROSCI.16-24-07910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yan P, Hu X, Song H, Yin K, Bateman RJ, Cirrito JR, Xiao Q, Hsu FF, Turk JW, Xu J, Hsu CY, Holtzman DM, Lee JM. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J Biol Chem. 2006;281:24566–24574. doi: 10.1074/jbc.M602440200. [DOI] [PubMed] [Google Scholar]
- 108.Yin KJ, Cirrito JR, Yan P, Hu X, Xiao Q, Pan X, Bateman R, Song H, Hsu FF, Turk J, Xu J, Hsu CY, Mills JC, Holtzman DM, Lee JM. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006;26:10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Renard C, Chappey O, Wautier MP, Nagashima M, Lundh E, Morser J, Zhao L, Schmidt AM, Scherrmann JM, Wautier JL. Recombinant advanced glycation end product receptor pharmacokinetics in normal and diabetic rats. Mol Pharmacol. 1997;52:54–62. doi: 10.1124/mol.52.1.54. [DOI] [PubMed] [Google Scholar]
- 110.Kojro E, Postina R, Buro C, Meiringer C, Gehrig-Burger K, Fahrenholz F. The neuropeptide PACAP promotes the alpha-secretase pathway for processing the Alzheimer amyloid precursor protein. FASEB J. 2006;20:512–514. doi: 10.1096/fj.05-4812fje. [DOI] [PubMed] [Google Scholar]
- 111.Khan TK, Nelson TJ, Verma VA, Wender PA, Alkon DL. A cellular model of Alzheimer’s disease therapeutic efficacy: PKC activation reverses Abeta-induced biomarker abnormality on cultured fibroblasts. Neurobiol Dis. 2009;34:332–339. doi: 10.1016/j.nbd.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun MK, Alkon DL. Bryostatin-1: pharmacology and therapeutic potential as a CNS drug. CNS Drug Rev. 2006;12:1–8. doi: 10.1111/j.1527-3458.2006.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kojro E, Postina R. Regulated proteolysis of RAGE and AbetaPP as possible link between type 2 diabetes mellitus and Alzheimer’s disease. J Alzheimer’s Dis. 2009;16:865–878. doi: 10.3233/JAD-2009-0998. [DOI] [PubMed] [Google Scholar]
- 114.Adlerz L, Holback S, Multhaup G, Iverfeldt K. IGF-1-induced processing of the amyloid precursor protein family is mediated by different signaling pathways. J Biol Chem. 2007;282:10203–10209. doi: 10.1074/jbc.M611183200. [DOI] [PubMed] [Google Scholar]
- 115.Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA. 2007;104:19796–19801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yan SF, Du YS, Ramasamy R, Schmidt AM. Tempering the wrath of RAGE: an emerging therapeutic strategy against diabetic complications, neurodegeneration, and inflammation. Ann Med. 2009;25:1–15. doi: 10.1080/07853890902806576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Santilli F, Vazzana N, Bucciarelli LG, Davi G. Soluble forms of RAGE in human diseases: clinical and therapeutical implications. Curr Med Chem. 2009;16:940–952. doi: 10.2174/092986709787581888. [DOI] [PubMed] [Google Scholar]