Abstract
Megasatellites are DNA tandem arrays made of large motifs; they were discovered in the yeast Candida glabrata. They are widespread in this species (40 copies) but are not found in any other hemiascomycete so far, raising the intriguing question of their origin. They are found mainly in genes encoding cell wall products, suggesting that megasatellites were selected for a function linked to cell–cell adhesion or to pathogenicity. Their putative role in promoting genome rearrangements by interfering with DNA replication will also be discussed.
Keywords: Candida glabrata, Megasatellite, Fragile sites, Cell wall, Genome
Megasatellites are a new class of DNA tandem repeats
In addition to its profound impact on evolutionary genomics and on our understanding of complex genetic networks, the systematic sequencing of whole eukaryotic genomes has also led to the discovery of new genetic elements. One of these discoveries was recently made in the genome of the opportunistic pathogen Candida glabrata. C. glabrata is a hemiascomycetous yeast, often involved in human candidiasis and bloodstream infections, particularly in immunocompromised patients [1, 2]. C. glabrata is more resistant to fluconazole treatments than other pathogenic yeasts [3] and has become the second major causative agent of nosocomial infections due to yeast species. The C. glabrata genome of the reference strain (CBS138) has been completely sequenced [4], revealing that it is phylogenetically closer to Saccharomyces cerevisiae than to the other extensively studied pathogen Candida albicans [5]. We recently investigated the genome of C. glabrata, searching for minisatellites, a family of tandem DNA repeats whose motif size ranges from nine nucleotides to usually fewer than 100 base pairs (reviewed in [6]).
Besides the presence of numerous minisatellites, the C. glabrata genome also contains tandem repeats whose motif size is much longer, ranging from 135 to 417 nucleotides. We called this new family of large tandem repeats, megasatellites [7]. They harbor two remarkable features: they are not found in any other sequenced living species besides C. glabrata and Kluyveromyces delphensis (two Saccharomycetaceae yeasts of the same clade [8]), and they are mainly found in genes proven, or suspected, to encode cell wall proteins, raising the possibility that megasatellites could be directly involved in regulating cell adhesion and pathogenicity. Altogether, 40 megasatellites were found in 33 genes in C. glabrata and classified in families.
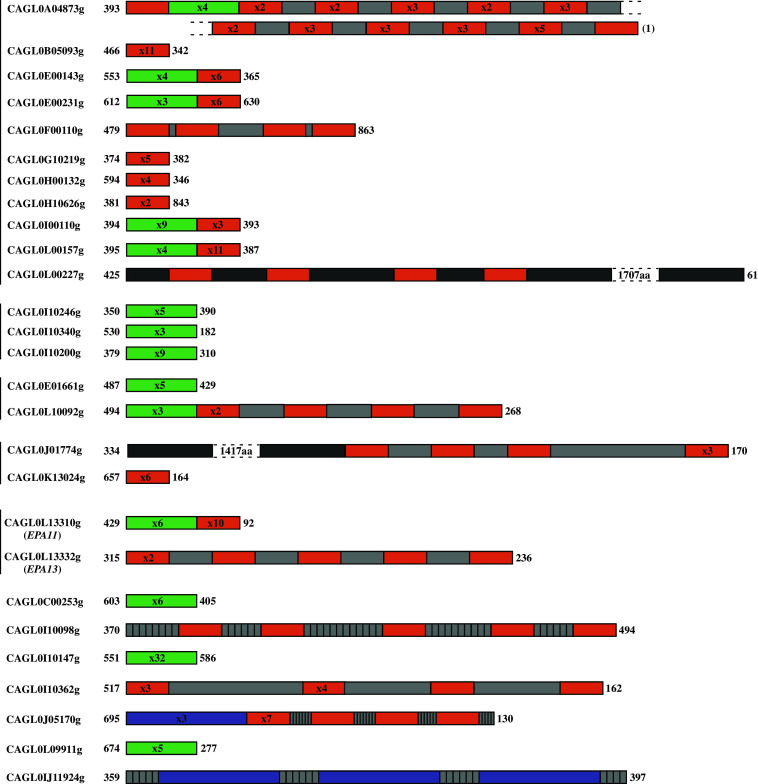
Of these 40 megasatellites, 14 belong to the SFFIT family and 20 belong to the SHITT family (family names come from the conservation of five amino acids in each motif of the megasatellite). Each tandem array contains from 3 to 32 motifs and covers from 405 to 9,600 DNA base pairs (Fig. 1). The remaining six megasatellites contain motifs that do not show obvious similarities with SFFIT or SHITT motifs. Megasatellites are distributed on 11 out of the 13 chromosomes but show some preferential bias toward the subtelomeric regions (Fig. 2), with the right end of chromosome IX carrying seven such elements within 65 kb, a density significantly higher than the genome average (one megasatellite per 224 kb). Subtelomeric regions are highly flexible in Saccharomyces cerevisiae, exhibiting a high level of inter-chromatid and inter-chromosome recombination [9]. It is possible that C. glabrata subtelomeres share similar properties, and that subtelomeric megasatellites recombine with each other, although this remains to be demonstrated.
Fig. 1.
Schematic representation of C. glabrata SHITT and SFFIT megasatellites. Gene names are indicated to the left (http://www.genolevures.org). Vertical lines near gene names indicate paralogous gene families. The region of the gene containing the megasatellite is represented by colored boxes. Orange SHITT motifs. Green SFFIT motifs. The numbers in boxes correspond to the number of tandemly repeated motifs (no number indicates the presence of only one motif). Purple indicates degenerate SFFIT motifs. The degenerate SFFIT motifs found within CAGL0J11924g and CAGL0J05170g are not identical, although they both probably come from a SFFIT motif. Light gray indicates sequences of variable length that are sometimes tandemly repeated and found interspersed within some of the SHITT and SFFIT megasatellites. Dark gray represents glycine- and serine-rich motifs of variable length found only in CAGL0L00227g and CAGL0J01774g. Their size (in amino acids) is indicated when too long to be drawn to scale. Otherwise, boxes are drawn to scale. Numbers shown before and after boxes represent the number of amino acids before and after the repeated motifs. Given the lower sequence coverage and unprecise assembly of subtelomeric regions, it is possible that the number of motifs shown for the 11 subtelomeric megasatellite-containing genes is different from what is represented here (11 genes, from CAGL0A04873g to CAGL0L00227g). (1) The subtelomeric sequence is interrupted within a megasatellite. Among the genes harboring megasatellites, only two have a known function and are involved in cell adhesion (EPA11, EPA13); the function of the other genes is unknown
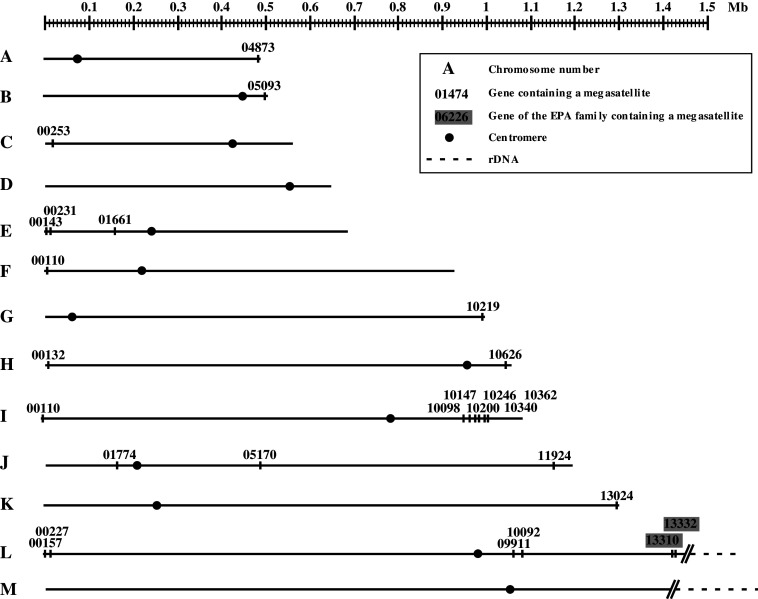
Fig. 2.
Distribution of megasatellites in the C. glabrata genome. The 13 chromosomes are represented in ascending size order and drawn to scale. Genes containing one or more megasatellites are indicated (only the five-digit number is given here, as compared to Fig. 1). The two genes of known function (EPA11, EPA13) are boxed in gray. rDNA tandem repeats are dashed (their precise length is unknown)
Possible involvement of megasatellites in pathogenicity
In S. cerevisiae, several genes involved in cell wall biogenesis contain minisatellites [10–12]. Some of them, called the FLO genes, play a direct role in flocculation and cellular adhesion. It was shown that cell adhesion and flocculation are directly correlated with the length of the minisatellite in FLO1 [12] and that cell–cell adhesion leading to the formation of a biofilm at the surface of sherry wine was directly dependent on the size of a minisatellite in FLO11 [13]. In C. glabrata CBS138 strain, three EPA genes (functional homologues of the FLO genes [14–17]) contain megasatellites (EPA2, EPA11, and EPA13), but three other EPA genes contain simple minisatellites (EPA1, EPA3, and EPA15), and three do not contain any kind of tandem repeat (EPA6, EPA7, and EPA8). In addition, 30 other genes that are not part of the EPA family contain megasatellites. Some of the proteins encoded by these genes exhibit signatures of cell-wall proteins but experimental evidence of their function or their localization is lacking. Interestingly, in S. cerevisiae, three FLO genes (FLO1, FLO5, and FLO9) contain a 135 bp motif, tandemly repeated 7–13 times [11]. This threonine-rich motif shares no obvious similarity with any of the C. glabrata megasatellites. However, it is the same size as the SHITT motif. It is therefore possible that the optimal size for a tandem repeat in these cell-wall embedded proteins is 45 amino acids (135 bp) and that the motif size is therefore under strong selection, whereas the sequence itself is not necessarily conserved.
In budding yeast, telomeric regions are silenced by a multiprotein complex containing the SIR genes, RAP1, ESC1, and the Ku complex [18]. These genes are conserved in C. glabrata, with the exception of SIR1, which is involved in silencing the silent mating-type loci, but not in telomeric silencing [19]. The inactivation of SIR3 and RAP1 was shown to increase the level of expression of several EPA genes, including the megasatellite-containing EPA2 gene [15, 20], suggesting that the mechanism of subtelomeric silencing is probably similar in C. glabrata and in S. cerevisiae. Subtelomeric megasatellite-containing genes are therefore probably also silenced, although this remains to be shown. Therefore, at the present time, the possible role played by megasatellites in C. glabrata pathogenicity is unclear and needs to be clarified in the future.
Megasatellites and genome rearrangements
Given the repeated nature of the large arrays formed by the megasatellites, one may wonder if they could behave like fragile sites and thus induce genome rearrangements. In humans, fragile sites are defined as chromatid constrictions or breaks visible on metaphasic chromosomes when cells are grown in the presence of drugs that impair replication or DNA metabolism [6, 21–23]. Although the precise molecular nature of all fragile sites is not known, some of them have been sequenced. The FRA3B locus contains numerous transposons and LTRs found in direct and inverted orientations; FRA10B and FRA16D loci contain AT-rich minisatellites (42 and 33 bp motif size, respectively); FRAXA, FRAXE, FRAXF, FRA11B, and FRA16A contain CGG trinucleotide repeats.
Some of these fragile site loci are associated with cancer. FRA3B, the most common fragile site in humans, often contains deletions in several gastrointestinal, colon, lung, breast, and cervical cancers [24]. Loss of heterozygosity and a recurrent translocation were also observed at FRA16D in breast and prostate cancers and multiple myelomas [25].
Interestingly, chromosomal translocations and chromosome losses in Candida albicans are often associated with a large DNA tandem repeat, called the major repeat sequence (MRS). It is a complex tandem repeat found at nine different locations in the genome and composed of a 2 kb motif tandemly repeated (RPS), itself including several tandem copies of smaller motifs (16 and 29 bp long). Most chromosome length polymorphisms in this yeast are due to size heterogeneity of the MRS [26].
When Muller and colleagues analyzed chromosomal translocations among different strains of C. glabrata, three major rearrangements, involving chromosomes IV, IX, XII, and XIII were found [27]. The three breakpoints corresponding to these three rearrangements were mapped and sequenced, but they are not located in the proximity of megasatellites, nor do they encompass any kind of repeated element. However, in the same study, it was shown that among 12 deletions ranging in size from 130 to 12 kb detected in the C. glabrata genome, two were located within two megasatellites (one of them being the longest megasatellite of the genome). The probability of this happening by chance is low, suggesting that megasatellites might be involved in the mechanism leading to these two deletions. In a more recent study using a larger number of probes, 11 reciprocal and nonreciprocal translocations involving 11 out of the 13 C. glabrata chromosomes were found [28]. The authors also detected five segmental duplications, including four class III duplications leading to the formation of new chromosomes [29]. In one of these, the duplication breakpoint is located less than 10 kb from a SFFIT megasatellite (MS#208), suggesting that it could be involved in the rearrangement, although this was not formally proven.
In conclusion, observations made on chromosomal plasticity in C. glabrata suggest that some of the rearrangements observed might be triggered by the presence of a megasatellite. However, the majority of megasatellites are not associated with chromosomal rearrangements and frequent rearrangements are observed far from any megasatellite, showing that megasatellites are not systematically involved in rearrangements. Large-scale studies of replication and recombination in C. glabrata are now needed to understand the precise role of megasatellites in chromosomal replication and instability.
Evolution of megasatellites
The last intriguing question concerning megasatellites relates to their mechanism(s) of formation. One simple way to propagate megasatellites is to duplicate the gene(s) that contain them. In C. glabrata, several megasatellites are found in paralogous gene families. The largest of these families encompasses 11 paralogues, each containing a SHITT megasatellite and five of them also containing a SFFIT repeat array (Fig. 1). However, six megasatellites containing either SHITT or SFFIT motifs are found in genes that are present in unique copies in the genome, raising questions about their very origin. If point mutations followed by replication slippage may explain how smaller tandem repeats such as microsatellites are born, it is hard to imagine the same mechanism responsible for the de novo creation of larger tandem arrays. Haber and Louis proposed that minisatellites are formed by replication slippage between two short (5 bp) sequences flanking a 10–20 nucleotide unique sequence [30]. Most of the S. cerevisiae minisatellites may actually be flanked by such short motifs [11], but this does not seem to be the case for C. glabrata megasatellites.
In silico comparisons of megasatellites with others in the same strain (CBS138, the sequenced strain) show that some motifs found in a given gene are actually phylogenetically closer to motifs found in another gene, suggesting that some kind of genetic transfer exists between megasatellites (Rolland, Dujon and Richard, unpublished). This transfer may involve gene conversion, or alternatively one may imagine that SHITT and SFFIT motifs are able to “jump” from one megasatellite to another one, using mechanisms that may be related to transposition or retrotransposition (Fig. 3). There is only one full-size retrotransposon in the C. glabrata genome, a gypsy-like element (Tcg3, gene name CAGL0G07183g, The Génolevures Consortium, http://www.genolevures.org/), two degenerate copies, and two solo LTRs (Cécile Neuvéglise, personal communication). Unless there is another source of reverse transcriptase in this genome, it is difficult to hypothesize that retrotransposition is involved in the spreading of megasatellites.
Fig. 3.
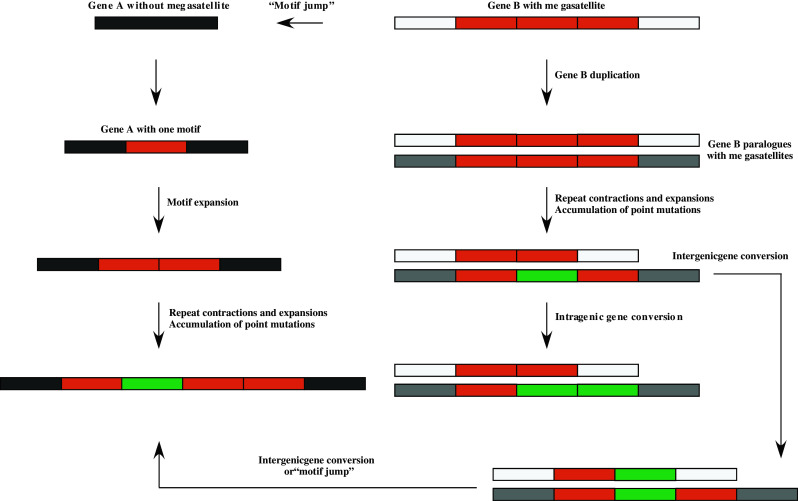
Different mechanisms can lead to megasatellite spreading in the C. glabrata genome. Left Gene A with no megasatellite may acquire a motif by retrotransposition or another mechanism, followed by expansion of the motif into a megasatellite. Right A gene already containing a megasatellite may duplicate itself, leading to the formation of two paralogues, each of them containing an identical megasatellite. Contractions and expansions may now occur, independently in both tandem repeats, and point mutations may accumulate in one or several motifs, leading to slightly different motif sequences (in green). These new motifs may propagate (or disappear) by intergenic (or intragenic) gene conversion. New motifs may also propagate by “jumping” into a megasatellite encoded by a nonparalogous gene (bottom)
In conclusion, the question of the origin of megasatellites is still completely open, but experiments designed specifically to answer this question using molecular tools available in this yeast species should give some answers and may explain why SHITT and SFFIT megasatellites are so widespread in C. glabrata.
Acknowledgments
This work was supported by grant ANR-05-BLAN-0331 from the Agence Nationale de la Recherche. B.D. is a member of the Institut Universitaire de France.
References
- 1.Bodey GP, Mardani M, Hanna HA, Boktour M, Abbas J, Girgawy E, Hachem RY, Kontoyiannis DP, Raad II. The epidemiology of Candida glabrata and Candida albicans fungemia in immunocompromised patients with cancer. Am J Med. 2002;112:380–385. doi: 10.1016/S0002-9343(01)01130-5. [DOI] [PubMed] [Google Scholar]
- 2.Raad I, Hanna H, Boktour M, Girgawy E, Danawi H, Mardani M, Kontoyiannis D, Darouiche R, Hachem R, Bodey GP. Management of central venous catheters in patients with cancer and candidemia. Clin Infect Dis. 2004;38:1119–1127. doi: 10.1086/382874. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates. Clin Microbiol Infect. 2004;10:11–23. doi: 10.1111/j.1470-9465.2004.t01-1-00844.x. [DOI] [PubMed] [Google Scholar]
- 4.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 5.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thierry A, Bouchier C, Dujon B, Richard G-F. Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata . Nucleic Acids Res. 2008;36:5970–5982. doi: 10.1093/nar/gkn594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora . FEMS Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 9.Louis EJ, Naumova ES, Lee A, Naumov G, Haber JE. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowen S, Roberts C, Wheals AE. Patterns of polymorphism and divergence in stress-related yeast proteins. Yeast. 2005;22:659–668. doi: 10.1002/yea.1240. [DOI] [PubMed] [Google Scholar]
- 11.Richard G-F, Dujon B. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol Biol Evol. 2006;23:189–202. doi: 10.1093/molbev/msj022. [DOI] [PubMed] [Google Scholar]
- 12.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci USA. 2006;103:11228–11233. doi: 10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castano I, Pan S-J, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata . Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- 15.De Las Penas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieman MB, McCaffery JM, Cormack BP. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1, 6 glucan-cross-linked cell wall protein. Mol Microbiol. 2002;46:479–492. doi: 10.1046/j.1365-2958.2002.03166.x. [DOI] [PubMed] [Google Scholar]
- 17.Zupancic M, Frieman MB, Smith D, Alvarez RA, Cummings RD, Cormack BP. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol Microbiol. 2008;68:547–559. doi: 10.1111/j.1365-2958.2008.06184.x. [DOI] [PubMed] [Google Scholar]
- 18.Taddei A, Hediger F, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 19.Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 20.Iraqui I, Garcia-Sanchez S, Aubert S, Dromer F, Ghigo J-M, d’Enfert C, Janbon G. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol Microbiol. 2005;55:1259–1271. doi: 10.1111/j.1365-2958.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- 21.Debacker K, Kooy RF (2007). Fragile sites and human disease. Hum Mol Genet 16 (Spec No 2):R150–R158 [DOI] [PubMed]
- 22.Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet. 2005;14:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland GR, Baker E, Richards RI. Fragile sites still breaking. Trends Genet. 1998;14:501–506. doi: 10.1016/S0168-9525(98)01628-X. [DOI] [PubMed] [Google Scholar]
- 24.Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/S0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 26.Magee PT (2007) Genome structure and dynamics in Candida albicans. In: d’Enfert C and Hube B (Eds) Candida: comparative and functional genomics. Caister Academic, Norfolk, UK, pp 7–26
- 27.Muller H, Thierry A, Coppée J-Y, Gouyette C, Hennequin C, Sismeiro O, Talla E, Dujon B, Fairhead C. Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet Biol. 2009;4:264–276. doi: 10.1016/j.fgb.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, Stenderup J, Piskur J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata . Proc Natl Acad Sci USA. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haber JE, Louis EJ. Minisatellite origins in yeast and humans. Genomics. 1998;48:132–135. doi: 10.1006/geno.1997.5153. [DOI] [PubMed] [Google Scholar]