Abstract
Semicarbazide-sensitive amine oxidases (SSAOs) catalyze oxidative deamination of primary amines, but the true physiological function of these enzymes is still poorly understood. Here, we have studied the functional and structural characteristics of a human cell-surface SSAO, AOC2, which is homologous to the better characterized family member, AOC3. The preferred in vitro substrates of AOC2 were found to be 2-phenylethylamine, tryptamine and p-tyramine instead of methylamine and benzylamine, the favored substrates of AOC3. Molecular modeling suggested structural differences between AOC2 and AOC3, which provide AOC2 with the capability to use the larger monoamines as substrates. Even though AOC2 mRNA was expressed in many tissues, the only tissues with detectable AOC2-like enzyme activity were found in the eye. Characterization of AOC2 will help in evaluating the contribution of this enzyme to the pathological processes attributed to the SSAO activity and in designing specific inhibitors for the individual members of the SSAO family.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0076-5) contains supplementary material, which is available to authorized users.
Keywords: SSAO, AOC2, Retina, Monoamine
Introduction
Semicarbazide-sensitive amine oxidases (SSAOs; EC 1.4.3.6) constitute a group of copper-dependent enzymes, which oxidatively deaminate primary endo- and exogenous amines into the corresponding aldehydes in a reaction: R–CH2–NH2 + O2 + H2O → R–CHO + H2O2 + NH3. Although SSAO activity itself has been recognized for several decades, relatively little has been known about the physiological functions of these enzymes. In general, the end-products of the reaction at high concentrations are highly cytotoxic and implicated in playing a role in various processes resulting in cellular damage, like atherosclerosis and vascular complications in diabetes [1–3]. In addition, SSAO activity has been shown to be involved in glucose transport [4–6], adipocyte differentiation [7], and structural organization of vascular smooth muscle [8, 9]. The physiological substrates for the SSAOs are still poorly known, but it is hypothesized that the soluble amines methylamine and/or aminoacetone could be among the naturally occurring substrates for the SSAOs [10–13].
AOC3 (amine oxidase, copper containing; also known as vascular adhesion protein-1, VAP-1) was first described as a 180-kD dimeric endothelial transmembrane glycoprotein mediating leukocyte extravasation from blood to the tissues [14, 15]. Cloning of VAP-1 revealed its homology to the SSAO family, and further studies confirmed that it was indeed able to display SSAO activity [16]. AOC3 is a heart-shaped dimer in which each monomer is composed of three domains, D2–D4 [17, 18]. The two tightly bound D4 domains form the interface of the dimer and contain the deeply buried active sites. The active site is only accessible through a cavity formed by the D3 and D4 domains. The crystal structure of the human AOC3 [18, 19] shows several amino acids with potential key roles in substrate selectivity and binding.
The closely related SSAO, AOC2, was originally cloned from human retina by Imamura et al. Based on northern blot data they designated the clone human retina-specific amine oxidase, hRAO, and showed, with in situ hybridization, that a corresponding RAO mRNA was expressed in the ganglion cell layer of mouse retina [20, 21]. The mouse homologue for AOC2 was recently cloned and the localization of this protein to the retinal ganglion cells was confirmed with immunohistochemistry [22]. Human AOC2 mRNA has also been reported to be present in human adipose tissue and to be up-regulated during in vitro adipocyte differentiation [23, 24]. Apart from the above-mentioned data on the localization of the mRNA and protein, no functional or structural data have been available before for AOC2. We have cloned the AOC2 cDNA from a human lung cDNA library, and studied the functional and structural characteristics of the corresponding protein using enzyme assays, molecular modeling, and co-immunoprecipitations. Here, we report that the mRNA for AOC2 is more widely expressed than described before and that AOC2 can function as a cell-surface SSAO but with different substrate specificity than AOC3. Moreover, we have analyzed the structural differences between these closely-related enzymes by homology modeling, tested the accuracy of the model by site-directed mutagenesis, and shown the possibility of heterodimerization between AOC2 and AOC3 in vitro.
Materials and methods
Antibodies, cells and reagents
The primary antibodies used to detect AOC2 and AOC3 are described in Table 1. The commercial polyclonal AOC2 antibody was from Abnova Corporation (Taipei, Taiwan). Normal rabbit serum, 3G6 (mouse anti-avian T cell antibody, [25]) and MEL-14 (rat anti-mouse L-selectin antibody, a kind gift from E. Butcher) were used as negative controls. The secondary antibody, fluorescent-isothiocyanate (FITC)-conjugated anti-rabbit Ig, was from Sigma (St. Louis, MO) and the tertiary antibody, Alexa Fluor 488-conjugated anti-FITC, was from Molecular Probes (Leiden, The Netherlands). Alexa Fluor 405-conjugated anti-rabbit Ig, Alexa Fluor 546-conjugated anti-mouse Ig (Molecular Probes), and HRP-conjugated anti-rabbit Ig (Dako, Denmark) were used as secondary antibodies. Human embryonic kidney fibroblast 293 cells (HEK293) and HEK293-EBNA were cultured (5% CO2, +37°C) in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco/Invitrogen, Paisley, UK) containing 10% fetal calf serum (FCS), 2 mM l-glutamine, sodium pyruvate, penicillin, and streptomycin. Chinese hamster ovary (CHO) cells were maintained in modified Eagle’s medium (MEM) supplemented with 10% FCS, 2 mM l-glutamine, penicillin, and streptomycin. Ax cells (rat high endothelial venule derived cells, a kind gift from M. Miyasaka) were grown in RPMI-1640 supplemented with 20% FCS, 2 mM l-glutamine, sodium pyruvate, non-essential amino acids, 2-mercapthoethanol, penicillin, and streptomycin. Geneticin was used when appropriate. Methylamine, benzylamine, p-tyramine, tryptamine, 2-phenylethylamine, histamine, spermidine, hydroxylamine, clorgyline, and semicarbazide were from Sigma.
Table 1.
Antibodies recognizing AOC2 and AOC3
| Antibody | Isotype | References | AOC3 | AOC2 |
|---|---|---|---|---|
| poly-VAP | Rabbit IgG | [52] | + | + |
| TK 8-14 | Mouse IgG2a | [53] | + | + |
| 2D10 | Mouse IgG1 | [53] | + | − |
| TK10-79 | Rat IgG | [54] | + | − |
List of antibodies used in immunohistochemistry and how they discriminate between AOC2/AOC3
cDNA cloning and site-directed mutagenesis
A polymerase chain reaction (PCR) fragment of 771 base pairs (bp) covering the nucleotides 38–808 of the AOC3 cDNA (GenBank accession number AF067406) was used as a probe to screen a human lung cDNA library in Lambda EMBL3 SP6/T7 vector (Clontech, Palo Alto, CA). The positive plaques were screened by PCR using AOC3 specific primers from exon 1 (5′-ACTCAGATCTCTACTCGCACT-3′) and exon 4 (5′-ATATGCAGAAAACCAGCTGTC-3′) to exclude the plaques containing the previously isolated AOC3 clone [16]. The amplification was carried out in a Perkin-Elmer GeneAmp PCR Systems 2400, and the protocol was: step 1: 94°C for 5 min; step 2: 94°C for 1 min; step 3: 50°C for 1 min; step 4: 72°C for 1 min; and step 5: 72°C for 5 min. Steps 2–4 were repeated for 29 times. Six plaques produced a band differing from the size of the expected band. The inserts of the isolated phages were subcloned into pUC19 (Invitrogen, CA) for sequencing and subsequently into pcDNA3.1 (Invitrogen) for transfections or pIRES2-EGFP (BD Biosystems Clontech, Palo Alto, CA) for adhesion assays. The clones were sequenced on both strands by the sequencing service facilities of the Department of Medical Genetics, University of Turku (Turku, Finland). Standard molecular biology techniques were used in plaque hybridization, transformation and plasmid and phage DNA purifications. Sequence analyses and alignments were performed using the Wisconsin Package version 10.1-UNIX of the Genetics Computer Group (GCG; Madison, WI), SeqWeb of BioBox and EMBOS at Scientific Computing Ltd (Center for Scientific Computing, CSC; Espoo, Finland) and BLAST at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/).
Point mutations were introduced to the AOC3 cDNA by PCR and using Phusion™ DNA polymerase (Finnzymes, Helsinki, Finland) according to the supplier’s instructions. The following phosphorylated primers (mutations in bold and underlined) were used: 5′-CAACTATGACTATGTGTGGGATACGGTCTTCCA-3′ (f-HE-5) and 5′-GCCAAGGTGGACATAGATCTGACGACCAGCACCGTT-3′ (r-HE-6) for Leu469Gly, 5′-GTGACCACGGCTCCCCGTGGTCTGCAATC-3′ (f-HE-7) and 5′-TGTCACCAGGTTCCGTCCCCGGTGCTTGTA-3′ (r-HE-8) for Met211Val and, 5′-TTGGCATGGGCAAGAACACCACGCC-3′ (f-HE-10) and 5′-AGCCTCCATCCACATAGCGGGTCGTC-3′ (r-HE-11) for Tyr394Asn. The mutations were introduced sequentially to get the triple-mutant Met211Val-Tyr394Asn-Leu469Gly. The PCR products were purified from agarose gel, ethanol precipitated, self-ligated using T4 DNA ligase (New England BioLabs, MA) and electroporated into DH5α Escherichia coli for propagation. The success of the PCR was confirmed by sequencing.
RNA expression
Multiple tissue cDNA (MTC™) panels (BD Biosystems) of adult human tissues were used as templates in PCR. The primers were located at exon 1 and exon 2 in AOC2: the sequence for the forward primer was 5′-AGGTCCTGGGAAAGGAGGACCTGACAG-3′ and that for the reverse 5′-GCCCTTCTCAAAGTAGACACTGCCAGGG-3′. GAPDH primers provided by the manufacturer were used for normalization of the template levels. The amplification was carried out in a Perkin-Elmer GeneAmp PCR Systems 2400, using the method described by the cDNA panel manufacturer. The protocol was: step 1: 94°C for 1 min; step 2: 94°C for 30 s; steps 3–4: 68°C for 2 min; and step 5: 72°C for 5 min. Steps 2–4 were repeated 34 times.
Total RNA was prepared from human fetal skeletal muscle, liver, lung, brain, heart, kidney, skin, cartilage, and tonsil around age 20 weeks of gestation by using the guanidinium isothiocyanate method [26]. The use of adult and fetal tissues was approved by the Institutional Review Board of Medicolegal Affairs (Helsinki and Turku, Finland). Reverse transcription and PCR were performed in a one step reaction by using GeneAmp® Gold RNA PCR Reagent Kit (PE Applied Biosystems, Foster City, CA). The thermal cycles of amplification were carried out in a Perkin-Elmer GeneAmp PCR Systems 2400 and the protocol was according to the manufacturer’s instructions for one step reverse transcriptase-PCR (RT-PCR). One microgram of total RNA and 20 pmol/μl of primers were used in all reactions. The sequences for the specific sense and antisense primers were the same as those used with the MTC-panels (see above). β-actin primers used for normalization were from Stratagene (#302055) (Stratagene, La Jolla, CA). The Southern blot analysis was performed on the RT-PCR reactions, which were separated on agarose gels and blotted onto Hybond N filters. The probe recognizing the amplified sequence of AOC2 was generated from the original AOC2 cDNA by PCR using the same primers as in the RT-PCR reactions. Standard methods in molecular biology were used in Southern blotting, hybridization and autoradiography.
Enzyme assays
HEK293, HEK293-EBNA or CHO cells were transiently transfected using the calcium phosphate and Fugene HD (Roche Applied Science, Indianapolis, IN) procedures. For the calcium phosphate procedure, 5–20 μg of AOC3 or AOC2 cDNA in pcDNA3.1 was used for a 90-mm plate, and the cultures were incubated at 5% CO2, +37°C for 24–48 h. With Fugene transfection reagent, 2 μg of DNA was used for a six-well plate with 3–8 μl of the reagent. After the incubation, the cells were harvested by short trypsination and lysed in a lysis buffer (10 mM Tris–HCl, pH 7.2, +1.5 mM MgCl2 +0.1% Nonidet P-40 (Fluka Chemie, Buchs, Switzerland) in PBS or 0.2% Nonidet P-40 in PBS) for 2 h at +4°C with shaking. The soluble cell lysate was obtained by centrifugation (30 min at +4°C, 12,000g) and collecting the supernatant. The transfection efficiencies were assayed by immunofluorescent staining (see later). SSAO activity of the cell lysates was measured using the Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine; Molecular Probes Europe, Leiden, The Netherlands). In brief, 15–20 μg of cell lysates were preincubated in Krebs–Ringer phosphate glucose (KRPG) 30 min at 37°C with inhibitors (see below). The same amount of non-transfected or mock transfected cell lysate was used as a background control and the values were normalized to the transfection efficiency of AOC2/AOC3. For the initial AOC2 activity screen, the catalytic reactions were initiated by addition of an amine substrate (methylamine, benzylamine, 2-phenylethylamine, p-tyramine, tryptamine, histamine, and spermidine at 1 mM concentration) together with the H2O2-detecting mixture containing horseradish peroxidase and Amplex Red reagent. Fluorescence intensity of the samples was measured with Tecan ULTRA or Tecan Infinite fluoropolarometer (excitation, 545 nm; emission, 590 nm) using the linear range of increase in the fluorescence intensity. H2O2 concentration was calculated from calibration curves generated by serial dilutions of standard H2O2. To evaluate the amount of H2O2 formed in the SSAO mediated reaction, specific SSAO inhibitors semicarbazide (100–1,000 μM) and hydroxylamine (5 μM) were included in the control wells subjected to the same treatment and these values were subtracted from the total amount of H2O2 formed. In order to determine kinetic parameters for AOC2 nine concentrations (0–10,000 μM) of methylamine, benzylamine, 2-phenylethylamine, p-tyramine and tryptamine were used, for AOC3 only methylamine and 2-phenylethylamine were used. Data from 2 to 4 experiments were used for analysis.
The relative activities of the AOC3 and the AOC3 triple mutant (Met211Val-Tyr394Asn-Leu469Gly) transfected HEK293-EBNA cell lysates were measured using 100 μM methylamine, 0.5 mM benzylamine, 0.5 mM p-tyramine and 5 mM phenylethylamine. In addition, the MAO inhibitor clorgyline (1 mM) was included. To determine the kinetic parameters of AOC3 and the AOC3 triple mutant in methylamine and phenylethylamine oxidation, five different concentrations of these substrates were used (0–10,000 μM) as duplicates. Since high concentrations of tyramine and tryptamine reduce the fluorescence in Amplex Red assay, a series of standard curves (fluorescence versus H2O2) were generated with 10–10,000 μM of the two substrates in order to determine the degree to which fluorescence is reduced at each concentration and to correct the values obtained from the assays. The standard curves were prepared from four parallel measurements with five different time points.
The kinetic data were fitted to Michaelis–Menten equation with or without substrate inhibition (f(x) = V max x/(K m + (1 + x/K i)x and f(x) = V max x/(K m + x), respectively; Equation 5.44 in [27]) by non-linear regression with Graph Pad Prism (version 4; GraphPad Software, San Diego, CA, USA). The substrate inhibition equation was used when the parameters converged. Standard errors presented for the kinetic parameters are from the curve fitting.
The enzyme assays of tissue lysates were performed as described above and the semicarbazide concentration used was chosen to be such that no cross-inhibition of MAO-A or MAO-B should happen. The protein concentration of the lysate was measured with Bio-Rad DC Protein Assay (Bio-Rad Laboratories, Hercules, CA).
Adhesion assay
Adhesion assays were performed as described [16]. In brief, Ax cells were transfected with either AOC3 in pIRES2-EGFP or AOC2 in pIRES2-EGFP. Cells were plated on wax-pen circles in gelatin-coated microscope slides and cultured to confluency. Lymphocytes were isolated from healthy volunteers and 2 million cells were applied to each circle in slides. After incubation (30 min, 60 rpm, +7°C), the slides were washed and fixed. The adhesion was analyzed with microscope by counting lymphocytes bound to fluorescent cells in predefined areas.
Immunostainings
Fluorescence stainings of coverslip plated cells
HEK293 cells were grown on coverslips in a 24-well plate and transfected with 0.25 μg of AOC2 in pcDNA3.1 (see “Enzyme assays”). Cells for intracellular stainnings were permeabilized with acetone. All cells were stained with the rabbit anti-VAP polyclonal antibody or normal rabbit serum (negative control). The slides were mounted with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL) and analyzed with a fluorescence microscope (Olympus BX 60).
Immunohistochemistry
Acetone fixed frozen sections (5 μm) of retina, tonsil, appendix, lymph node, liver, skin, and small intestine were incubated with mAb 2D10 (recognizing only AOC3) or polyclonal anti-VAP (recognizing both AOC2 and AOC3) as primary antibodies and Alexa Fluor 405-conjugated anti-rabbit Ig or Alexa Fluor 546-conjugated anti-mouse Ig as secondary antibodies. Fluoromount-G was used as a mounting medium. Analysis was made with either fluorescence microscope (Olympus BX 60) or confocal microscope (Zeiss LSM510 META). Formalin-fixed and paraffin-embedded sections of human retina, optic nerve, and optic tract were dewaxed with UltraClear (J. T. Barker, Deventer, The Netherlands) and rehydrated in a series of decreasing concentrations of alcohol. Antigen retrieval was done by boiling in 0.01 M citric acid for 10 min and endogenous peroxidase activity was removed by incubating in PBS containing 0.3% H2O2. The sections were then stained with mAb TK10-79 (detecting only VAP-1) and polyclonal anti-VAP antibody. Class-matched negative control antibodies and normal rabbit serum were used as negative controls in all stainings. HRP conjugated rabbit anti-rat and swine anti-rabbit immunoglobulins (Dako) were used as secondary antibodies. For developing peroxidase-mediated reaction 3,3-diaminobenzidine in TBS containing 0.03% H2O2 was used. Finally, the slides were counterstained with hematoxylin, dehydrated, cleared in HistoClear (J. T. Barker) and mounted in DePex (VWR International, Poole, England). The sections were analyzed with Olympus BX 60.
Flow cytometry
AOC2 transfected HEK293 cells (see “Enzyme assays”) were harvested, half of the cells were permeabilized with acetone, and all were stained with the rabbit polyclonal anti-VAP antibody or normal rabbit serum (negative control). The acquisition was done with FACScan®, and Win MDI 2.8. was used for the analysis. To confirm the success and the transfection efficiency of the AOC3 or mutant transfections, a portion of the cells were stained with FITC conjugated anti-VAP-1 monoclonal antibody TK 8-14 (2 μg/ml). An equal amount of 3G6-FITC against chicken T cells was used as a negative control.
Homology modeling
The sequences of human AOC3 (VAP-1, Swiss-Prot accession number Q16853) and human AOC2 (hRAO, O75106) were aligned with the program MALIGN [28] in the Bodil visualization and modeling package [29]. The structural models of AOC2 and the heterodimer were constructed with the program MODELLER [30] based on the alignment and the crystal structure of human AOC3 (PDB code 1US1; [18]). The topaquinone residues in both AOC3 and AOC2 were modeled to iminoquinone residues and rotated to a position equivalent to the inhibitor-binding state in the crystal structure of the E. coli amine oxidase in complex with 2-hydrazinopyridine (ECAO; PDB code 1SPU; [31]). Ten different models of AOC2 were generated and, after visual examination, the one with the lowest value of the MODELLER objective function was chosen for further analysis. The program PROSAII [32] was used to evaluate the folding of the model. In the AOC2 model, the side-chains of the amino acids Asn464, His206 and Lys372 were rotated to attain better interactions in the active site. Tyr384 in AOC3 and Tyr378 in AOC2 were adjusted to resemble the rotamer of Tyr381 in the ECAO-2-hydrazinopyridine complex structure (1SPU), in order to give access to the active site. The final AOC2 model was compared to the AOC3 crystal structure after superimposing them with the program VERTAA [33]. Hydrogens were added to the model and the crystal structure with the program REDUCE [34]. The ligands were built with SYBYL® 7.0 (Tripos, St. Louis, Missouri, USA) and docked to the crystal structure of AOC3 and the structural model of AOC2 with GOLD [35, 36]. The ligands were covalently connected to the N5 atom of the iminoquinone to simulate the substrate Schiff base intermediate in the enzymatic reaction. Ten dockings with each ligand were made, and the results with the highest fitness value derived by GOLD were chosen. The figures were made with PyMOL [37].
Co-immunoprecipitations
The full-length AOC3 cDNA was subcloned into a flag-tagged vector pRK5flag, and the corresponding AOC2 cDNA was subcloned into a myc-tagged vector pTB399myc [38] as C-terminal fusion proteins for co-precipitations. HEK293 cells were co-transfected with the two constructs or the corresponding empty vectors (control) using calcium phosphate transfection. After an overnight culture, the cells were washed in PBS and lysed in 700 μl of ice cold lysis buffer [50 mM Tris–HCl, pH 7.2, 150 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, 0.5% NP-40 and an incomplete protease inhibitor cocktail (Roche), 1 mM Na3VO4, 10 mM NaF, 1 mM β-glyserophosphate]. After 5 min, the cells were scraped off and sheared through a 27G needle. The lysates were clarified by centrifugation, and pre-cleared by rocking for 20 min with 17.5 μl Protein G beads. Thereafter, the supernatants were divided into two aliquots and incubated with 17.5 μl Protein G beads together with 5 μg of either the anti-flag mAb M2 (Sigma) or the anti-myc mAb 9E10 (CRL1729 from ATCC) for 1 h at +4°C. After three washings with the lysis buffer, Laemmli’s sample buffer containing 5% 2-ME was added to the beads. After boiling, the immunoprecipitates were resolved in 6-12% SDS-PAGE gels, and subjected to immunoblotting with anti-flag and anti-myc mAbs using ECL.
Statistical analysis
Data are shown as mean values ± SEM. Statistics were performed using Student’s t test. Values of P < 0.05 were chosen as statistically significant.
Results
AOC2 mRNA is expressed in several human tissues
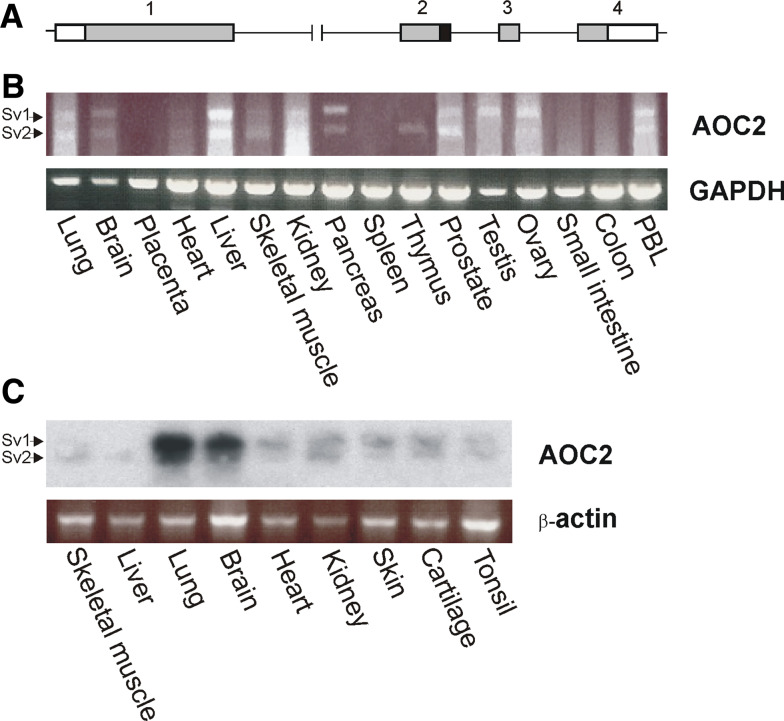
When searching for additional AOC3-related transcripts, a human lung cDNA library was screened with a probe created with PCR corresponding to the sequence between nucleotides 38 and 808 of AOC3 cDNA sequence. Six clones were obtained that represented a cDNA differing from that of AOC3: two of them were found to represent a 130-bp shorter splice variant of AOC3 (manuscript in preparation), and four were similar to the previously documented hRAO [20]. Sequencing of the clones revealed the presence of two alternatively spliced transcripts (as also reported earlier by [21]) differing in size by 81 bps and producing open reading frames (ORFs) of 2,271 bp (splice variant 1, sv1) and 2,190 bp (splice variant 2, sv2). Sv 1 produces the full length AOC2 protein, whereas sv2 gives rise to a 27-amino acid shorter polypeptide. The deleted region in sv2 corresponds to the end of exon 2 of the AOC2 gene (Fig. 1a), which does not contain any of the functionally important conserved sites found in the SSAO genes (Electronic supplementary material, ESM, Fig. 1; for review, see [39]).
Fig. 1.
Expression of AOC2 mRNA in adult and fetal human tissues. a Schematic representation of the exon–intron organization of the human AOC2 gene. The boxes represent the four exons of the gene. 5′- and 3′-untranslated regions (white) and translated regions (grey) as well as the alternatively spliced region in the end of exon 2 (black) are shown. The splicing out of the indicated region in exon 2 produces the shorter isoform Sv2. b Commercial first strand cDNA panels were used to determine the expression of AOC2 and GAPDH (control) in adult human tissues by PCR. c RT-PCR analysis of nine fetal human tissues was performed using AOC2 and beta-actin specific primers. To further confirm the specificity of the resulting amplicons the blot was hybridized with 32P-labeled cDNA probe recognizing AOC2. The two alternatively spliced mRNA species are marked with arrows (sv1 and sv2)
Since we had cloned AOC2 from a human lung cDNA library, it was evident that this molecule was more widely expressed than previously reported. Further characterization of the expression pattern of this transcript was performed with PCR using commercial first strand cDNA panels of adult human tissues and with RT-PCR of human fetal RNAs. The results revealed the presence of AOC2 mRNA in several human tissues: of the adult tissues studied, only placenta, spleen, small intestine, and colon appeared to be devoid of the AOC2 transcripts. In thymus, only the shorter isoform is transcribed and in testis, only the larger one. In most tissues, however, both of the alternatively spliced transcripts are present (Fig. 1b). Because the AOC2 mRNA levels in the fetal tissues were even lower than those in the adult tissues, the RT-PCR gels themselves were further Southern blotted in order to amplify the signal and verify the specificity of the bands produced. All the fetal tissues examined contained AOC2 mRNA, and after normalization with the control beta-actin RT-PCR, only the lung RNA seemed to contain slightly higher amounts of AOC2 mRNA than the other tissues (Fig. 1c). The method used does not, however, allow absolute quantification of the mRNA abundance. Therefore, the expression levels between the different tissues are not directly comparable.
AOC2 is an enzymatically active cell surface SSAO with distinct substrate specificity
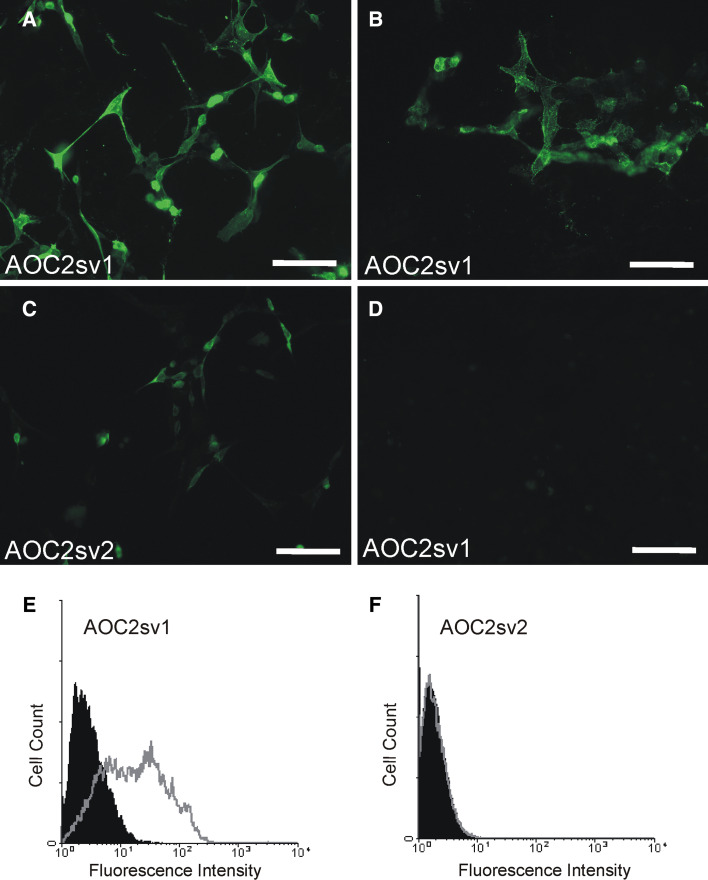
To study the cellular localization of AOC2, both the alternatively spliced variants of AOC2 were cloned into pcDNA3.1 and transfected into HEK293 cells. The expressed polypeptides were detected with a polyclonal anti-VAP antibody, both on coverslip-plated cells using fluorescence microscopy (Fig. 2a–d) and in FACS stainings of harvested cells (Fig. 2e, f). The sv1 isoform was found to be present on the surface of the cells (Fig. 2b, e), but the shorter sv2 isoform, even though detectable in the cytoplasm, was either not translocated to the plasma membrane or the amounts present were below the detection level (Fig. 2c, f). The sv2 isoform was therefore excluded from further studies.
Fig. 2.
Expression of AOC2 in HEK293 cells. Fluorescence microscopy of AOC2 transfected HEK293 cells on coverslips. a AOC2 Sv1 transfected cells (after permeabilization). b AOC2 Sv1 transfected cells (non-permeabilized, surface staining). c AOC2 Sv2 transfected cells (after permeabilization). d AOC2 Sv1 transfected cells (after permeabilization). The cells were stained either with polyclonal anti-VAP antibody (a, b, c) or normal rabbit serum (control, d). The cell surface expression of the AOC2 isoforms was determined also by flow cytometry. e AOC2 Sv1 transfected cells. f AOC2 Sv2 transfected cells. The grey histograms show the staining with the polyclonal anti-VAP antibody and the black histograms the corresponding control staining with normal rabbit serum. Scale bar 100 μm
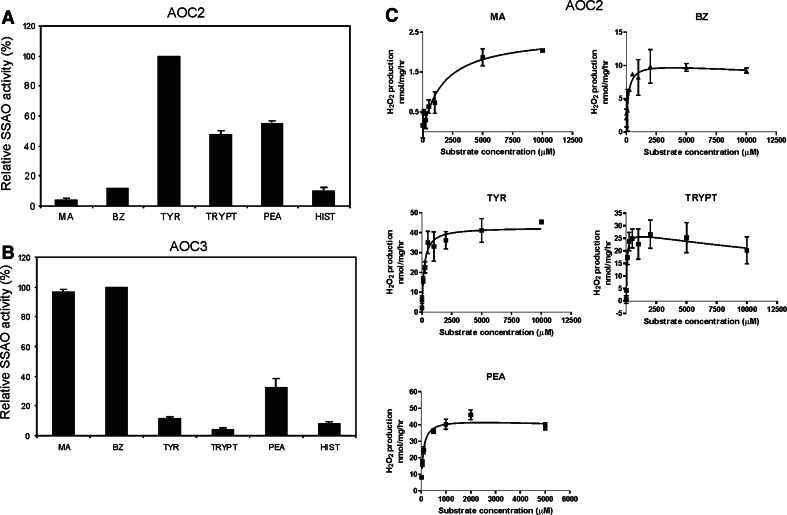
Since the regions important for the monoamine oxidazing properties of AOC3 [39] are conserved in AOC2, the enzymatic activity of AOC2 was studied using the AOC2 sv1 transfected cells and the Amplex Red Assay. For initial screening, several amine substrates were used in the assay to detect the putative SSAO activity, i.e., whether AOC2 is able to catalyze the reaction in which an amine is oxidatively deaminated into the corresponding aldehyde. The highest activities were detected when using 2-phenylethylamine, tryptamine, or p-tyramine as a substrate (Fig. 3a). In addition, since AOC2 could use benzylamine to a lesser degree as a substrate than AOC3 and had practically no enzymatic activity towards methylamine, the substrate preference of AOC2 seemed to be distinctly different from that of the highly homologous SSAO, AOC3 (Fig. 3b). Neither AOC2 nor AOC3, however, could oxidize the polyamine spermidine (data not shown) or histamine, which is one of the substrates for the intracellular SSAO diamine oxidase (DAO) [40, 41].
Fig. 3.
Substrate specificities of AOC2 and AOC3. The substrate specificities were determined in fluorometric assays. The substrates used were: MA methylamine; BZ benzylamine; TYR p-tyramine; TRYPT tryptamine; PEA 2-phenylethylamine; and HIST histamine (all as 1 mM). Results are shown as mean percentage of relative SSAO activities + SEM of four experiments, where 100% is the SSAO activity obtained for the substrate with the highest activity. a The AOC2 transfected cells. b The AOC3 transfected cells. c Kinetic curves of AOC2. The derived kinetic constants are in Table 2
In order to examine the kinetics of the AOC2-based SSAO activity in more detail, we also measured the activities using several concentrations of each substrate (Fig. 3c, Table 2). In addition, to be able to compare the properties of AOC2 to those of AOC3 in cell lysates, we also measured the kinetic constants for AOC3 using methylamine and 2-phenylethylamine, the two substrates that differed significantly between the two proteins in the primary screen, but which do not interfere with the Amplex Red assay as tyramine and tryptamine do (see “Materials and methods”; Fig. 3a, b, Table 2). When the kinetics was compared, AOC2 showed higher activity towards 2-phenylethylamine than AOC3 did (K m 77 and 1,940 μM, respectively), the maximal rate of reaction (V max) for 2-phenylethylamine being 2.5-fold higher in the AOC2 transfectants. The affinity of AOC2 towards methylamine, on the other hand, was low since the K m of AOC2 for this substrate was almost three times higher than that of AOC3 (K m 1,700 and 670 μM, respectively) and the V max was only 6% of the rate seen with the AOC3 lysates. The maximal rates of reaction with the best substrates tested—2-phenylethylamine and tyramine for AOC2 and methylamine for AOC3—were, however, of similar level.
Table 2.
Kinetic constants for AOC2, AOC3 and the AOC3 triple mutant M211V-Y394N-L469G
| Construct/substrate | K m (M) | V max (nmol H2O2/mg per h) | K i (mM) | V max /K m | Percentage of AOC3 V max(MA) | R 2 |
|---|---|---|---|---|---|---|
| CHO transfectants | ||||||
| AOC3 | ||||||
| Methylamine | 670 ± 45 | 39 ± 0.76 | – | 0.06 | 100 | 0.99 |
| 2-Phenylethylamine | 1,940 ± 260 | 17 ± 1.0 | – | 0.009 | 44 | 0.97 |
| AOC2 | ||||||
| Methylamine | 1,700 ± 710 | 2.4 ± 0.4 | – | 0.001 | 6 | 0.84 |
| 2-Phenylethylamine | 77 ± 15 | 44 ± 2.5 | 80 ± 144 | 0.6 | 113 | 0.91 |
| Benzylamine | 167 ± 132 | 11 ± 2.2 | 89 ± 280 | 0.06 | 28 | 0.62 |
| p-Tyramine | 178 ± 51 | 43 ± 2.4 | – | 0.2 | 110 | 0.89 |
| Tryptamine | 56 ± 34 | 28 ± 3.2 | 32 ± 29 | 0.5 | 72 | 0.79 |
| HEK-EBNA transfectants | ||||||
| AOC3 | ||||||
| Methylamine | 1,020 ± 92 | 62 ± 1.6 | – | 0.06 | 100 | 0.99 |
| 2-Phenylethylamine | 8,340 ± 3,550 | 15 ± 3.7 | – | 0.002 | 24 | 0.98 |
| M211V-Y394N-L469G | ||||||
| Methylamine | 1,990 ± 149 | 14 ± 0.3 | – | 0.007 | 23 | 0.99 |
| 2-Phenylethylamine | 478 ± 58 | 18 ± 0.5 | – | 0.04 | 29 | 0.99 |
The K m and V max values were calculated using hyberbolic non-linear regression for Michaelis–Menten equationa and, when the K i value is shown, with a Michaelis–Menten equation with substrate inhibitionb
a f(x) = V max x/(K m + x)
b f(x) = V max x/(K m + (1 + x/K i)x)
As already suggested by the initial substrate screen, the kinetic analysis verified that AOC2 is highly active towards p-tyramine and tryptamine (Table 2). In contrast, the seemingly low activity detected towards benzylamine (Fig. 3a) seems to result from the low V max (11 nmol/mg per h) since the measured K m is as low as 167 μM for this amine.
The calculated V max/K m values, which give an indication of substrate specificity, gave the highest value for AOC2 in 2-phenylethylamine and tryptamine oxidation (0.6 and 0.5, respectively). These values are only 15 and 30% lower than the corresponding value for AOC3 in benzylamine oxidation (0.7; determined with HEK-EBNA transfectants; H. Elovaara, unpublished results) but are both an order of magnitude higher than that of AOC3 methylamine oxidation (0.06). This further demonstrates that AOC2 differs from AOC3 in substrate specificity.
In addition to being a SSAO, AOC3 is known for its ability to support adhesion [15]. To study this possibility with AOC2, we performed adhesion assays using AOC2 transfected endothelial Ax cells and human lymphocytes. The cells were transfected with pIRES-AOC2 and pIRES-AOC3 expression plasmids, which also produce, in addition to the corresponding SSAO protein, green fluorescent protein (GFP), whereby the transfection efficiency of the individual cells could be evaluated using fluorescence microscopy. In these experiments, the cells expressing AOC2 could not support adhesion in contrast to the cells harboring AOC3 (data not shown).
Homology modeling and ligand docking reveal structural differences between AOC2 and AOC3
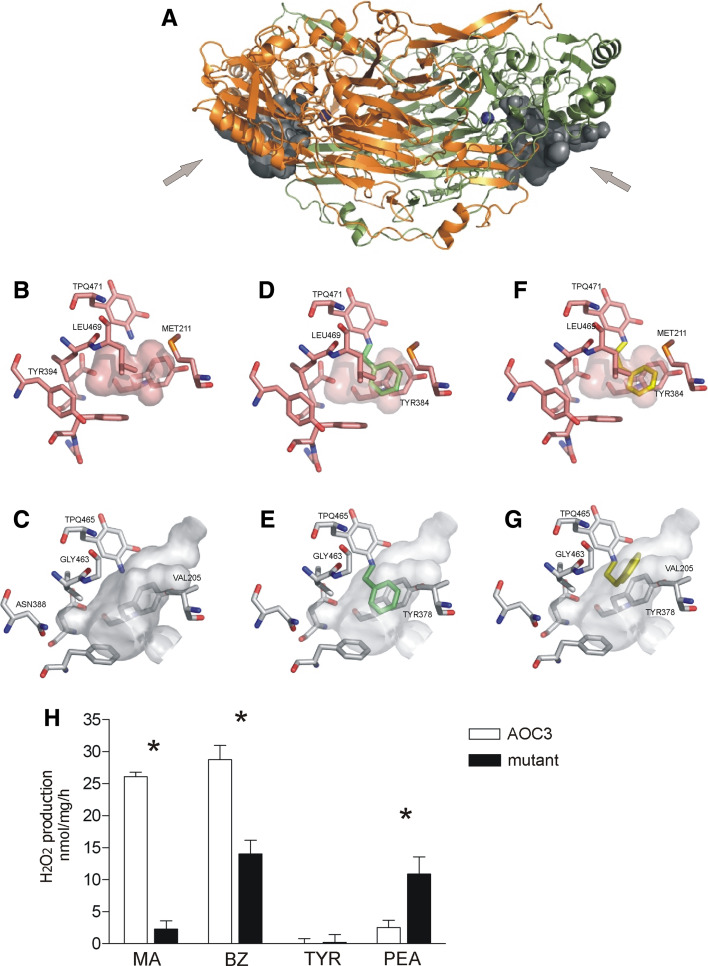
To find a possible structural basis for the differences detected between AOC2 and AOC3 in substrate preference, homology modeling and ligand docking were performed. Based on the model (Fig. 4a), AOC2 forms a heart-shaped dimeric structure similar to that of AOC3 [18, 19]. The AOC2 model has a 68% sequence identity with AOC3, and the monomers superimpose with a root mean square deviation (RMDS) of 0.91 Å. The active site in AOC3 seems to be much smaller than in AOC2 (Fig. 4b, c) and many of the active site residues are not conserved between these SSAOs (ESM, Table). In particular, the large side chains of Met211 and Tyr394 in AOC3, which correspond to Val205 and Asn388 in AOC2, affect the size of the active site cavity. The substrate channel leading to the active site in AOC3 is “guarded” by Leu469 and the side chain of this amino acid has to rotate to allow access for substrates to the active site [18]. In the AOC2 model, the entry is not blocked in a similar way, since Gly463 allows free access to the active site of AOC2 (Fig. 4b, c).
Fig. 4.
Molecular modeling and substrate docking of AOC2. a The model of AOC2. Monomers are colored orange and green and the topaquinone cofactors in both monomers are drawn as blue spheres denoting the active site locations. The active site cavities are shown as surface representations. The access sites of the substrate from the solvent are marked with arrows. The comparison of AOC2 and AOC3 active sites: AOC3 structure b and AOC2 model c. The residues that are described in the text are indicated and the cavity is shown as a surface representation. Ligand docking: in d and e the benzylamine ligand (green) is docked as a reaction intermediate into AOC3 and into AOC2 active site, respectively; in f (AOC3) and g (AOC2), the docked substrate is 2-phenylethylamine (yellow). h Substrate specificities of AOC3 (wild type, white) and AOC3 M211V-Y394N-L469G triple mutant (black). Activity is described as nmol of formed hydrogen peroxide per mg of total protein in cell lysate per hour. The columns represent the average of five experiments, and the error bars SEM. *P < 0.05
The docking experiments show that the aromatic ring of benzylamine is positioned between Tyr384 and Leu469 in AOC3 (Fig. 4d). In the AOC2 model, on the other hand, benzylamine stacks only with Tyr378 and the aromatic ring is not stabilized on the other side, since Leu469 is replaced by Gly463 and the hydrophobic entity is missing (Fig. 4e). 2-phenylethylamine, p-tyramine, and tryptamine cannot be docked in the same position in the AOC3 active site since their hydrocarbon chains are longer with an additional -CH2- group (Fig. 4f) and the aromatic ring crashes with the surroundings. However, 2-phenylethylamine might fit generously into the modeled active site cavity of AOC2, adopting another favorable position in the extra space generated from Leu469 to Gly463 substitution (Fig. 4g). Based on our docking results the aromatic rings in 2-phneylethylamine and tryptamine are positioned in a similar way, but the tyramine ring is flipped 90° in respect to the docked 2-phenylethylamine ring because the hydroxyl group in tyramine forms a hydrogen bond with Lys372 in AOC2 (data not shown). Methylamine, on the other hand, fits well into the active sites of both AOC3 and AOC2. The smaller active site of AOC3, however, makes it possible for methylamine to interact with the hydrophobic side chains of Leu468 and Leu469. In the larger active site of the AOC2 model, the side chains of the corresponding residues, Val462 and Gly463, are smaller and therefore methylamine could not form favorable hydrophobic interactions with them.
To test the validity of the AOC2 model and the results obtained from the modeling and docking studies, we generated an active site mutant for AOC3. To ensure a measurable difference in substrate preference, an AOC3 triple mutant was designed in which the three large non-conserved amino acids—Met211, Tyr394, and Leu469—of AOC3, were mutated to the corresponding ones in AOC2 (Met211Val–Tyr394Asn–Leu469Gly, Fig. 4b, c). The effect of the mutations on the enzymatic activity was tested with four of the substrates (benzylamine, methylamine, 2-phenylethylamine, and p-tyramine). The substrate specificity of the triple mutant was clearly different from that of the wild type AOC3 demonstrating decreased activity towards methylamine and benzylamine (9 and 49% of the wild type activity, respectively), but increased activity towards 2-phenylethylamine (435%) (Fig. 4h). In the case of p-tyramine, the activity of both tested lysates remained close to the background level. To study the activity of the triple mutant in more detail, we measured the activity of the mutant using methylamine and 2-phenylethylamine, the substrates shown to differ significantly between AOC2 and AOC3, in five different concentrations, and determined the kinetic constants for them (Table 2). The values obtained confirm the results from the primary assay with a single concentration and thus verify the importance of the amino acids Met211, Tyr394, and Leu469 in defining the substrate preference.
AOC2 cannot be detected without AOC3 in the tissues studied
Due to the lack of monoclonal antibody that would specifically recognize AOC2 and the fact that the commercial polyclonal AOC2 antibody used failed to recognize AOC2 in our experiments, we used combinations of earlier produced antibodies for AOC3, some of which were able to detect AOC2 as well (Table 1), to study the localization of the protein in vivo. Frozen sections of several human tissues (retina, tonsil, appendix, lymph node, liver, skin, and small intestine) were stained with a polyclonal antibody recognizing both AOC2 and AOC3, and with a monoclonal antibody recognizing only AOC3 (data not shown and Supplemental Fig. 2). Deducing from the fact that all the cell types stained with the polyclonal antibody were also recognized by the monoclonal AOC3-specific antibody, AOC2 was not found to be present independently in any of the tissues studied.
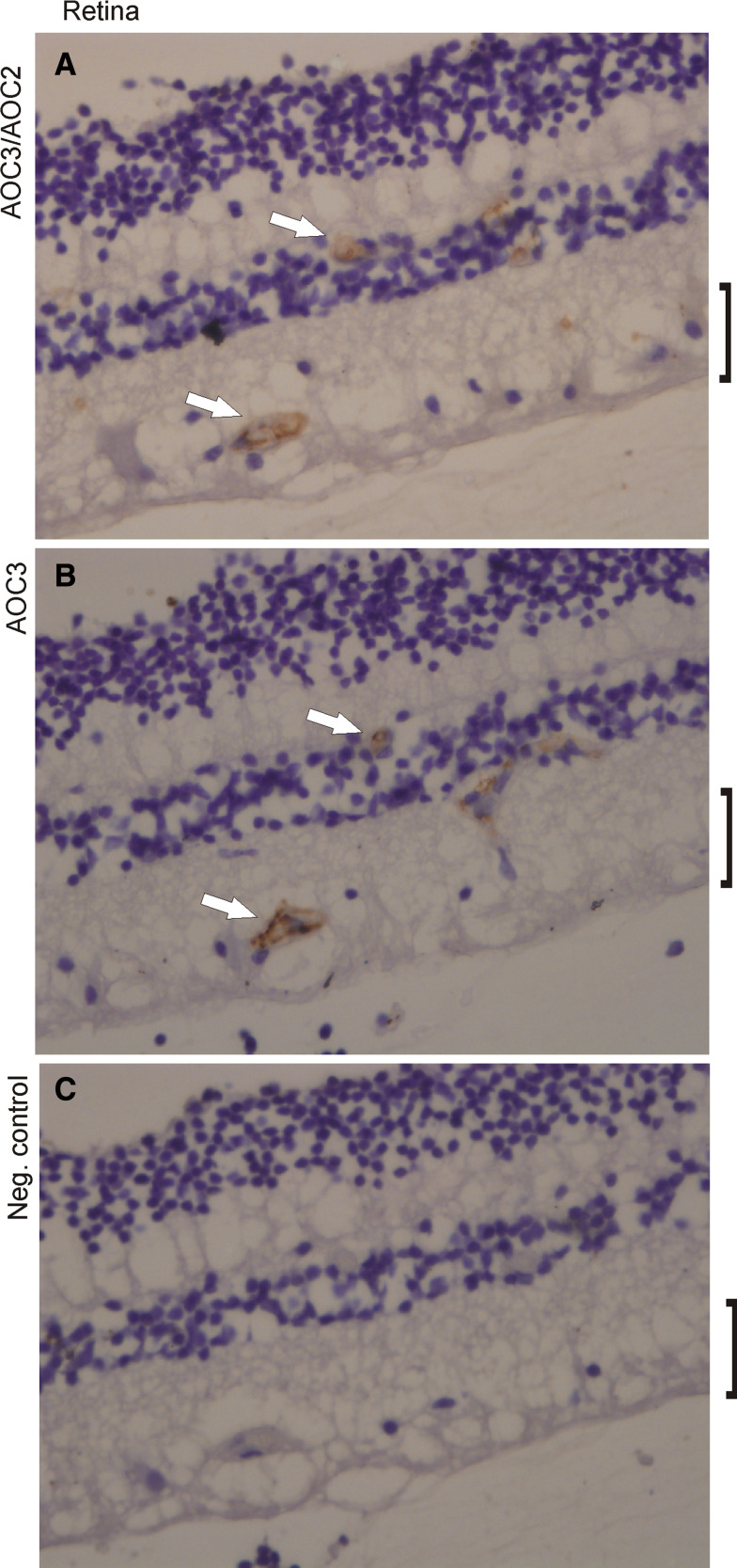
Since the only previously published information on AOC2 at the protein level describes its presence in the ganglion cell layer of mouse retina [22], we stained paraffin-embedded samples of human retina with poly- and monoclonal antibodies for AOC3 as described above (Fig. 5). In contrast to the report on mouse, we could not detect any staining of the human ganglion cell layer with any of the antibodies available to us. The retinal capillaries, on the other hand, were clearly positive with the antibodies used. Since retina shows AOC2-like enzymatic activity (see below) and the polyclonal anti-VAP antibody recognizes AOC2, it strongly suggests that in humans AOC2 in retina is expressed in the same cells as AOC3 in the endothelium of the local vasculature.
Fig. 5.
Expression of AOC2 and AOC3 in human retina. Paraffin sections of human retina were stained with a polyclonal poly-VAP antibody detecting both AOC2 and AOC3 a or TK10-79 antibody which detects AOC3 on paraffin-embedded sections b. c Negative control antibody. The arrows point to the vessels with positive staining. The ganglion cell layer is indicated
AOC2-like SSAO activity is found in human eye
As AOC2 was, however, widely expressed at the mRNA-level and, based on the antibody stainings, there was a possibility that AOC2 is expressed in the same cells as AOC3 in vivo, lysates from human tonsils, adipose tissue, smooth muscle, and retina were used in an initial screen to look for AOC2-like SSAO activity in human tissues. Since p-tyramine is a poor substrate for AOC3 and methylamine is in turn not used by AOC2, these two amine substrates were used in Amplex assays to dissect the individual contribution of the two AOC proteins to the overall SSAO activity. In these assays, the only tissue exhibiting any enzyme activity towards tyramine was retina (data not shown), which suggested that low levels of AOC2 protein are likely to be present in this tissue. Since the AOC2-like SSAO activity in the two retina samples studied differed considerably (835 vs. 85 pmol/mg per h), additional experimentation was carried out. Although difficult to acquire for ethical reasons, we managed to obtain samples of different structures of the eye to repeat the assay with four new retina samples and to study the presence of SSAO activity in the other structures of the eye available to us. In these enzyme assays, the same individual variation in AOC2-like SSAO activity between the retina samples was found (Table 3). Interestingly, of the three ocular structures studied, the highest SSAO activities towards p-tyramine were found in the iris samples. The only structure where the AOC2-like activity was higher than the AOC3-like activity was, however, retina.
Table 3.
SSAO activities in structures of human eye
| Structure | SSAO activity (pmol/mg per h) | |
|---|---|---|
| MA | TYR | |
| Iris (n = 4) | 2,250 ± 770a | 510 ± 190 |
| Choroid (n = 4) | 1,100 ± 310 | 340 ± 140 |
| Retina (n = 4) | 180 ± 20 | 260 ± 260 |
aMean ± SEM
AOC2 is able to form heterodimers with AOC3 in vitro
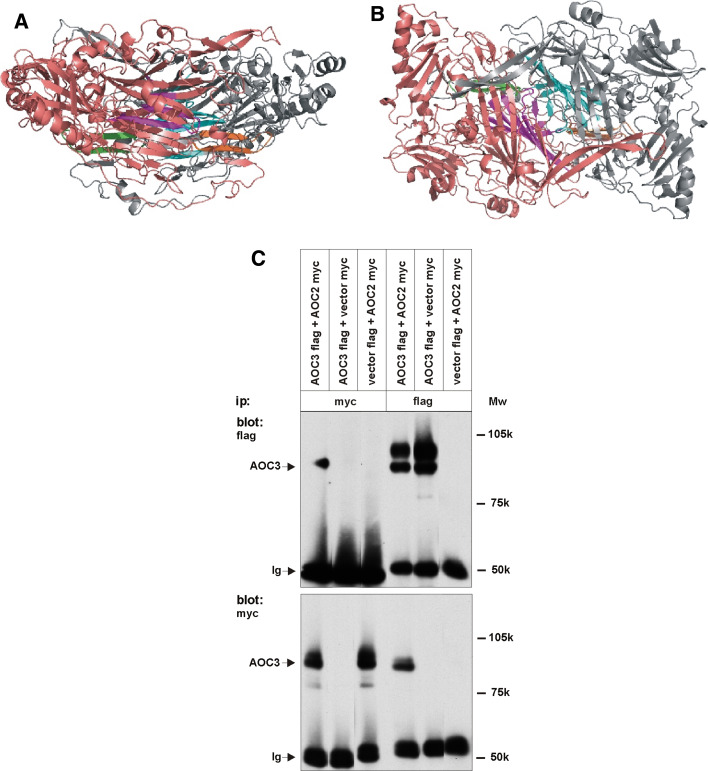
AOC3 is known to be present on the cell surface as a homodimer [42]. If AOC2 is indeed co-expressed with AOC3 in vivo, as the data presented above suggest, heterodimerization of these homologous proteins might occur and even lead to changes in the cell surface expression and/or function of these proteins. To address this possibility, structural modeling of the putative heterodimer was performed. Based on the model (Fig. 6a, b), only 44 residues in the heterodimer interface are different compared with the homodimers, which account for 18% of all substitutions between AOC2 and AOC3. Thus, the majority of the substitutions occur at positions remote from the dimer interface. Most of the substituted residues in the dimer interface are located in two regions: in the large central cavity between the catalytic D4 domains (cyan in AOC2 and purple in AOC3) and in the arm that extends from one monomer to the entrance of the substrate cavity of the other monomer (green in AOC2 and orange in AOC3).
Fig. 6.
Heterodimerization of AOC2 and AOC3. The structural model of the AOC2/AOC3 heterodimer from side a and top b. The AOC2 monomer is colored in grey and the AOC3 monomer in pink. The dimer interface regions containing most of the differences compared to the homodimers are colored as follows: the region lining the central cavity is cyan (AOC2) and purple (AOC3), and the arm close to the entrance of the substrate cavity is green (AOC2) and orange (AOC3). c Lysates of HEK293 cells co-transfected with myc-tagged AOC2 cDNA, flag-tagged AOC3 cDNA or with the corresponding tagged empty vectors in different combinations were immunoprecipitated with either the myc antibody (3 lanes on the left) or the flag-antibody (3 lanes on the right). The samples were separated in SDS-PAGE, blotted to nitrocellulose and the detection was performed with either the flag-antibody (upper panel) or the myc-antibody (lower panel). The size of the AOC monomer and the molecular weight markers are indicated. Ig Immunoglobulin
To explore the possibility of such heterodimerization in vitro, co-immunoprecipitations were performed from lysed HEK293 cells transfected with tagged AOC2 and AOC3 full length cDNAs (Fig. 6c). When myc-tagged AOC2 was immunoprecipitated, the flag-tagged AOC3 was seen to co-immunoprecipitate. Conversely, immunoprecipitation of flag-tagged AOC3 co-precipitated the myc-tagged AOC2. Control co-precipitations with the flag- or myc-tag plasmids without the insert confirmed the specificity of the observed AOC2-AOC3 interaction. The upper AOC3 flag band detected in blotting with the anti-flag antibody may represent some precursor form of the molecule that is not yet able to dimerize. Due to the unavailability of a specific antibody recognizing human AOC2, we are unable to study the heterodimerization with the corresponding endogenous proteins in vivo. Unfortunately, the enzymatic activity of the in vitro formed heterodimers proved to be impossible to test as well, since the harsh protocol used in immunoprecipitation rendered even the homodimers enzymatically inactive.
Discussion
AOC2 was originally cloned from a human retina cDNA library and designated human retina-specific amine oxidase, hRAO [20]. Amongst the tissues studied with northern hybridizations, hRAO mRNA was detected only in retina. The cloning of mouse RAO/AOC2, also from retina, has also been published and its expression pattern studied with quantitative PCR: when the expression levels were compared to that in retina, low levels of mouse RAO/AOC2 mRNA were detected in brain, heart, liver, and testis, and none in lung and spleen [22]. In our study, AOC2 was, however, cloned from a human lung cDNA library—as well as from mouse lung mRNA (data not shown)—and detectable amounts of AOC2 mRNA were found in most of the human tissues studied using RT-PCR. The transcription of AOC2 outside retina has also been reported by Heniquez et al. who showed expression of this gene in human adipocytes [24]. Furthermore, data mining using the expression data set generated by Su et al. in a large-scale microarray analysis of human and mouse transcriptomes reveals that AOC2 mRNA is expressed in certain human tissues and cell lines other than retina [43]. The HEK293 cells used here in the transfection experiments do not, however, contain transcripts of this gene.
The higher expression levels reported for AOC2 in retina suggest that the role of this gene product is especially important in the structures of the eye. We have in this study shown for the first time that AOC2 has SSAO activity, and that it preferably oxidizes aromatic amines. Since this substrate preference of AOC2 differs from that of the more widely and abundantly expressed AOC3, it is conceivable that the different amine substrates present in the eye necessitate the expression of a SSAO species that is specific for these particular amines. It has been reported, for example, that a SSAO in bovine retina and choroid uses dopamine as a substrate, whereas no activity towards this substrate is detected in the iris or the optic nerve, where, however, SSAO activity towards benzylamine is present [44]. This suggests a differential tissue-specific substrate pattern and thereby a specific SSAO expression profile in these anatomically closely located cell types. Which biological amines are the true physiological substrates for each individual SSAO species remains still largely unknown.
Despite the broad mRNA expression profile of AOC2, its expression and function at the protein level was under the detection limit of our methods in other organs apart from the structures of the eye, although we used the sensitive Amplex assay for detecting the enzymatic activity. Moreover, the use of a polyclonal anti-VAP antibody (against AOC2 and AOC3) together with an AOC3-specific monoclonal antibody would have allowed the recognition of AOC2, if it were present separately from AOC3. Based on these results, we may speculate that there is a specific need for AOC2 expression and enzymatic activity in the eye. However, the lack of AOC2 in human ganglion cell layer and its presence in the corresponding retinal layer in the mouse remains an enigma.
We modeled AOC2 and docked the experimentally tested substrates to the active sites of AOC3 and AOC2 in order to recognize the characteristics in the binding mode that account for their substrate preferences. Based on our model, the active site cavity in AOC3 is in general much smaller than in AOC2, which correlates with the preference of AOC2 for larger substrates than AOC3. The preferred substrates for AOC3, in turn, are methylamine and benzylamine, which do fit easily into its small active site. According to the AOC3 structure, Leu469 has an important dual function in AOC3: it limits the access of the substrate to the active site and stabilizes the binding of substrates by hydrophobic interactions. In E. coli amine oxidase (ECAO), there is a glycine instead of a leucine at the entrance of the substrate channel [45] and it prefers 2-phenylethylamine, p-tyramine, and tryptamine like AOC2 does. Since in our model for AOC2 the same amino acid difference was detected, it is conceivable that the change in this gate-keeper amino acid has an important role in defining substrate usage. For example, due to the substitution, methylamine cannot form hydrophobic interactions in the modeled active site of AOC2, and thus it cannot be used as a substrate even though it fits well into the active site itself. Regarding the larger substrates, 2-phenylethylamine, p-tyramine, and tryptamine, AOC2 is able to bind them in a manner not possible in AOC3, where the binding is hindered mainly by Met211. The importance of the size of the active site was further supported by the fact that the Met211Val–Tyr394Asn–Leu469Gly AOC3 mutant with a larger active site than wild type AOC3 was impaired in methylamine oxidation but had gained the ability to use 2-phenylethylamine as a substrate.
In the proposed AOC2/AOC3 heterodimer model, the two active sites would still retain their characteristic substrate preferences; one of them has AOC2 selectivity and the other one AOC3 selectivity. However, the arm that protrudes from the AOC2 subunit to the active site opening of the AOC3 subunit and vice versa has been suggested to have a role in the selective entry of the substrates to the active site [46], and, therefore, the entry of the substrates to the active sites of the possible heterodimer might be affected by this difference compared to the homodimers.
SSAO activity has been implicated in many pathological conditions such as diabetes, chronic liver disease, and congestive heart failure, in which an elevated serum SSAO level has been detected [47, 48]. Recently, it has been shown with knock-out and transgenic mice that AOC3 is responsible for the serum SSAO activity and that the vascular endothelium is the major source of soluble AOC3 in physiological conditions [2]. Since humans—like rodents—lack a functional gene for a secreted amine oxidase that is found in the genome of some other mammals [49], it is reasonable to assume that the source of human serum SSAO activity is also derived from proteolytic cleavage of the cell surface associated SSAO AOC3. Due to the fact that the expression of AOC2 seems to be concentrated in the eye, it is not likely that it contributes to the total serum SSAO activity found in these disease states but has a more local effect whereby the over-expression of this SSAO could result in tissue destruction seen in ocular pathologies like those in diabetic retinopathy.
A recently published study on endotoxin-induced uveitis in rat shows that inhibition of VAP-1 (AOC3) suppresses P-selectin expression in retinal endothelium and leukocyte recruitment to the anterior chamber, vitreous, and retina. [50]. Furthermore, blockade of VAP-1 also suppressed choroidal neovascularization (CNV) through affecting the recruitment of macrophages to the CNV lesion [51]. The existence of another SSAO in the eye with a distinct substrate repertoire may thus have implications on the strategies of inhibiting SSAO activity for therapeutic purposes. The importance of the enzymatic activity of AOC2 in disease processes requires further studies, and the structural characteristics depicted here will help in development of specific inhibitors of SSAO activity discriminating between the AOC3 and AOC2 members of the SSAO family.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1. Sequence alignment of mammalian monoamine oxidazing SSAOs. The deduced amino acid sequences of four monoamine oxidizing mammalian SSAOs from GenBank were compared. The SSAOs shown are human AOC3 (VAP-1, AF067406), mouse AOC3 (mAOC3, mVAP-1, AF054831), human AOC2 (hRAO, D88213) and Bovine Serum Amine Oxidase (BSAO, L27218). Highlighted are: yellow, the hydrophobic N-terminal sequence; red, the conserved signature motif of SSAO active site, where the first tyrosine is post-translationally modified to topaquinone; pink, (putative) catalytic site base; light green, the conserved, Cu(II) binding histidine residues; turquoise, the conserved cysteine residues involved in dimerization; dark blue the putative N-linked glycosylation sites. The RGD sequence of AOC3 is indicated with red letters. The underlined sequence corresponds to the region deleted in the AOC2 splice variant 2 (sv2). For review of the consensus sites important for SSAO activity, see [39]. (PPT 55 kb)
Supplemental Figure 2. Antibodies recognizing AOC3 alone or both AOC3 and AOC2 give identical staining patterns. Frozen sections of human tonsil were stained with the monoclonal antibody 2D10 detecting only AOC3 (left panel) and with the polyclonal antibody poly-VAP detecting both AOC2 and AOC3 (right panel). Scale bar 100 μm. (TIFF 15356 kb)
Acknowledgments
The expert technical assistance of Teija Pöysti, Maritta Pohjansalo, Pirjo Heinilä, Etta-Liisa Väänänen, Riikka Sjöroos, Sari Mäki, and Laila Reunanen is greatly appreciated. Drs. Heikki Irjala and Kalle Alanen are thanked for providing us with the tissue samples. Dr. Gennady Yegutkin is thanked for advice. Professor Mark Johnson is acknowledged for the excellent facilities at the Structural Bioinformatics Laboratory at the Department of Biochemistry and Pharmacy, Åbo Akademi University. This work was supported by the Academy of Finland, the Sigrid Juselius Foundation, the Emil Aaltonen Foundation, and the Varsinais-Suomi Regional Fund of the Finnish Cultural Foundation.
References
- 1.Mathys KC, Ponnampalam SN, Padival S, Nagaraj RH. Semicarbazide-sensitive amine oxidase in aortic smooth muscle cells mediates synthesis of a methylglyoxal-AGE: implications for vascular complications in diabetes. Biochem Biophys Res Commun. 2002;297:863–869. doi: 10.1016/S0006-291X(02)02293-3. [DOI] [PubMed] [Google Scholar]
- 2.Stolen CM, Madanat R, Marti L, Kari S, Yegutkin GG, Sariola H, Zorzano A, Jalkanen S. Semicarbazide sensitive amine oxidase overexpression has dual consequences: insulin mimicry and diabetes-like complications. FASEB J. 2004;18:702–704. doi: 10.1096/fj.03-0562fje. [DOI] [PubMed] [Google Scholar]
- 3.Griendling KK, Sorescu D, Lassegue B, Ushio-Fukai M. Modulation of protein kinase activity and gene expression by reactive oxygen species and their role in vascular physiology and pathophysiology. Arterioscler Thromb Vasc Biol. 2000;20:2175–2183. doi: 10.1161/01.atv.20.10.2175. [DOI] [PubMed] [Google Scholar]
- 4.Enrique-Tarancon G, Castan I, Morin N, Marti L, Abella A, Camps M, Casamitjana R, Palacin M, Testar X, Degerman E, Carpene C, Zorzano A. Substrates of semicarbazide-sensitive amine oxidase co-operate with vanadate to stimulate tyrosine phosphorylation of insulin-receptor-substrate proteins, phosphoinositide 3-kinase activity and GLUT4 translocation in adipose cells. Biochem J. 2000;350(Pt 1):171–180. doi: 10.1042/0264-6021:3500171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zorzano A, Abella A, Marti L, Carpene C, Palacin M, Testar X. Semicarbazide-sensitive amine oxidase activity exerts insulin-like effects on glucose metabolism and insulin-signaling pathways in adipose cells. Biochim Biophys Acta. 2003;1647:3–9. doi: 10.1016/s1570-9639(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 6.Enrique-Tarancon G, Marti L, Morin N, Lizcano JM, Unzeta M, Sevilla L, Camps M, Palacin M, Testar X, Carpene C, Zorzano A. Role of semicarbazide-sensitive amine oxidase on glucose transport and GLUT4 recruitment to the cell surface in adipose cells. J Biol Chem. 1998;273:8025–8032. doi: 10.1074/jbc.273.14.8025. [DOI] [PubMed] [Google Scholar]
- 7.Mercier N, Moldes M, El Hadri K, Feve B. Semicarbazide-sensitive amine oxidase activation promotes adipose conversion of 3T3–L1 cells. Biochem J. 2001;358:335–342. doi: 10.1042/0264-6021:3580335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gokturk C, Nilsson J, Nordquist J, Kristensson M, Svensson K, Soderberg C, Israelson M, Garpenstrand H, Sjoquist M, Oreland L, Forsberg-Nilsson K. Overexpression of semicarbazide-sensitive amine oxidase in smooth muscle cells leads to an abnormal structure of the aortic elastic laminas. Am J Pathol. 2003;163:1921–1928. doi: 10.1016/S0002-9440(10)63550-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langford SD, Trent MB, Boor PJ. Semicarbazide-sensitive amine oxidase and extracellular matrix deposition by smooth-muscle cells. Cardiovasc Toxicol. 2002;2:141–150. doi: 10.1385/CT:2:2:141. [DOI] [PubMed] [Google Scholar]
- 10.Buffoni F. Semicarbazide-sensitive amine oxidases: some biochemical properties and general considerations. Prog Brain Res. 1995;106:323–331. doi: 10.1016/S0079-6123(08)61228-5. [DOI] [PubMed] [Google Scholar]
- 11.Callingham BA, Crosbie AE, Rous BA. Some aspects of the pathophysiology of semicarbazide-sensitive amine oxidase enzymes. Prog Brain Res. 1995;106:305–321. doi: 10.1016/S0079-6123(08)61227-3. [DOI] [PubMed] [Google Scholar]
- 12.O’Sullivan J, Unzeta M, Healy J, O’Sullivan MI, Davey G, Tipton KF. Semicarbazide-sensitive amine oxidases: enzymes with quite a lot to do. Neurotoxicology. 2004;25:303–315. doi: 10.1016/S0161-813X(03)00117-7. [DOI] [PubMed] [Google Scholar]
- 13.Yu PH, Wright S, Fan EH, Lun ZR, Gubisne-Harberle D. Physiological and pathological implications of semicarbazide-sensitive amine oxidase. Biochim Biophys Acta. 2003;1647:193–199. doi: 10.1016/s1570-9639(03)00101-8. [DOI] [PubMed] [Google Scholar]
- 14.Salmi M, Jalkanen S. A 90-kilodalton endothelial cell molecule mediating lymphocyte binding in humans. Science. 1992;257:1407–1409. doi: 10.1126/science.1529341. [DOI] [PubMed] [Google Scholar]
- 15.Salmi M, Jalkanen S. Human vascular adhesion protein 1 (VAP-1) is a unique sialoglycoprotein that mediates carbohydrate-dependent binding of lymphocytes to endothelial cells. J Exp Med. 1996;183:569–579. doi: 10.1084/jem.183.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DJ, Salmi M, Bono P, Hellman J, Leu T, Jalkanen S. Cloning of vascular adhesion protein 1 reveals a novel multifunctional adhesion molecule. J Exp Med. 1998;188:17–27. doi: 10.1084/jem.188.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salminen TA, Smith DJ, Jalkanen S, Johnson MS. Structural model of the catalytic domain of an enzyme with cell adhesion activity: human vascular adhesion protein-1 (HVAP-1) D4 domain is an amine oxidase. Protein Eng. 1998;11:1195–1204. doi: 10.1093/protein/11.12.1195. [DOI] [PubMed] [Google Scholar]
- 18.Airenne TT, Nymalm Y, Kidron H, Smith DJ, Pihlavisto M, Salmi M, Jalkanen S, Johnson MS, Salminen TA. Crystal structure of the human vascular adhesion protein-1: unique structural features with functional implications. Protein Sci. 2005;14:1964–1974. doi: 10.1110/ps.051438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobsson E, Nilsson J, Kallstrom U, Ogg D, Kleywegt GJ. Crystallization of a truncated soluble human semicarbazide-sensitive amine oxidase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:274–278. doi: 10.1107/S1744309105002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura Y, Kubota R, Wang Y, Asakawa S, Kudoh J, Mashima Y, Oguchi Y, Shimizu N. Human retina-specific amine oxidase (RAO): cDNA cloning, tissue expression, and chromosomal mapping. Genomics. 1997;40:277–283. doi: 10.1006/geno.1996.4570. [DOI] [PubMed] [Google Scholar]
- 21.Imamura Y, Noda S, Mashima Y, Kudoh J, Oguchi Y, Shimizu N. Human retina-specific amine oxidase: genomic structure of the gene (AOC2), alternatively spliced variant, and mRNA expression in retina. Genomics. 1998;51:293–298. doi: 10.1006/geno.1998.5357. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Mashima Y, Noda S, Imamura Y, Kudoh J, Shimizu N, Nishiyama T, Umeda S, Oguchi Y, Tanaka Y, Iwata T. Characterization of AOC2 gene encoding a copper-binding amine oxidase expressed specifically in retina. Gene. 2003;318:45–53. doi: 10.1016/S0378-1119(03)00753-4. [DOI] [PubMed] [Google Scholar]
- 23.Bour S, Daviaud D, Gres S, Lefort C, Prevot D, Zorzano A, Wabitsch M, Saulnier-Blache JS, Valet P, Carpene C. Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human adipocytes. Biochimie. 2007;89:916–925. doi: 10.1016/j.biochi.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Heniquez A, Meissonnier G, Visentin V, Prevot D, Carpene C. High expression of semicarbazide-sensitive amine oxidase genes AOC2 and AOC3, but not the diamine oxidase gene AOC1 in human adipocytes. Inflamm Res. 2003;52(suppl 1):S74–S75. doi: 10.1007/s000110300061. [DOI] [PubMed] [Google Scholar]
- 25.Wuorela M, Jalkanen S, Pelliniemi LJ, Toivanen P. Nurse cells of the bursa of Fabricius: do they exist? Eur J Immunol. 1990;20:913–917. doi: 10.1002/eji.1830200429. [DOI] [PubMed] [Google Scholar]
- 26.Chirgwin J, Przybyla A, MacDonald R, Rutter W. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry (Mosc) 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 27.Copeland RA. Enzymes: a practical introduction to structure, mechanism and data analysis. 2. New York: Wiley; 2000. [Google Scholar]
- 28.Johnson MS, Overington JP. A structural basis for sequence comparisons. An evaluation of scoring methodologies. J Mol Biol. 1993;233:716–738. doi: 10.1006/jmbi.1993.1548. [DOI] [PubMed] [Google Scholar]
- 29.Lehtonen JV, Still DJ, Rantanen VV, Ekholm J, Bjorklund D, Iftikhar Z, Huhtala M, Repo S, Jussila A, Jaakkola J, Pentikainen O, Nyronen T, Salminen T, Gyllenberg M, Johnson MS. BODIL: a molecular modeling environment for structure–function analysis and drug design. J Comput Aided Mol Des. 2004;18:401–419. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- 30.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Wilmot CM, Murray JM, Alton G, Parsons MR, Convery MA, Blakeley V, Corner AS, Palcic MM, Knowles PF, McPherson MJ, Phillips SE. Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction. Biochemistry (Mosc) 1997;36:1608–1620. doi: 10.1021/bi962205j. [DOI] [PubMed] [Google Scholar]
- 32.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 33.Johnson M, Lehtonen J (2000) Bioinformatics. In: Higgins D, Taylor W (eds), Oxford University Press, Oxford, United Kingdom, pp 15-50
- 34.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 35.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. doi: 10.1016/S0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 36.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 37.deLano, W (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org
- 38.Tapon N, Nagata K, Lamarche N, Hall A. A new rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kappaB signalling pathways. EMBO J. 1998;17:1395–1404. doi: 10.1093/emboj/17.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jalkanen S, Salmi M. Cell surface monoamine oxidases: enzymes in search of a function. EMBO J. 2001;20:3893–3901. doi: 10.1093/emboj/20.15.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu D, Medzihradszky KF, Adams GW, Mayer P, Hines WM, Burlingame AL, Smith AJ, Cai D, Klinman JP. Primary structures for a mammalian cellular and serum copper amine oxidase. J Biol Chem. 1994;269:9926–9932. [PubMed] [Google Scholar]
- 41.Buffoni F. Histaminase and related amine oxidases. Pharmacol Rev. 1966;18:1163–1199. [PubMed] [Google Scholar]
- 42.Salmi M, Hellman J, Jalkanen S. The role of two distinct endothelial molecules, vascular adhesion protein-1 and peripheral lymph node addressin, in the binding of lymphocyte subsets to human lymph nodes. J Immunol. 1998;160:5629–5636. [PubMed] [Google Scholar]
- 43.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez de Arriba A, Lizcano JM, Balsa D, Unzeta M. Contribution of different amine oxidases to the metabolism of dopamine in bovine retina. Biochem Pharmacol. 1991;42:2355–2361. doi: 10.1016/0006-2952(91)90241-V. [DOI] [PubMed] [Google Scholar]
- 45.Roh JH, Suzuki H, Azakami H, Yamashita M, Murooka Y, Kumagai H. Purification, characterization, and crystallization of monoamine oxidase from Escherichia coli K-12. Biosci Biotechnol Biochem. 1994;58:1652–1656. doi: 10.1271/bbb.58.1652. [DOI] [PubMed] [Google Scholar]
- 46.Wilce MC, Dooley DM, Freeman HC, Guss JM, Matsunami H, McIntire WS, Ruggiero CE, Tanizawa K, Yamaguchi H. Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone. Biochemistry (Mosc) 1997;36:16116–16133. doi: 10.1021/bi971797i. [DOI] [PubMed] [Google Scholar]
- 47.Gronvall-Nordquist JL, Backlund LB, Garpenstrand H, Ekblom J, Landin B, Yu PH, Oreland L, Rosenqvist U. Follow-up of plasma semicarbazide-sensitive amine oxidase activity and retinopathy in Type 2 diabetes mellitus. J Diabetes Complications. 2001;15:250–256. doi: 10.1016/S1056-8727(01)00151-9. [DOI] [PubMed] [Google Scholar]
- 48.Weiss HG, Klocker J, Labeck B, Nehoda H, Aigner F, Klingler A, Ebenbichler C, Foger B, Lechleitner M, Patsch JR, Schwelberger HG. Plasma amine oxidase: a postulated cardiovascular risk factor in nondiabetic obese patients. Metabolism. 2003;52:688–692. doi: 10.1016/S0026-0495(03)00028-3. [DOI] [PubMed] [Google Scholar]
- 49.Schwelberger HG. The origin of mammalian plasma amine oxidases. J Neural Transm. 2007;114:757–762. doi: 10.1007/s00702-007-0684-x. [DOI] [PubMed] [Google Scholar]
- 50.Noda K, Miyahara S, Nakazawa T, Almulki L, Nakao S, Hisatomi T, She H, Thomas KL, Garland RC, Miller JW, Gragoudas ES, Kawai Y, Mashima Y, Hafezi-Moghadam A. Inhibition of vascular adhesion protein-1 suppresses endotoxin-induced uveitis. FASEB J. 2008;22:1094–1103. doi: 10.1096/fj.07-9377com. [DOI] [PubMed] [Google Scholar]
- 51.Noda K, She H, Nakazawa T, Hisatomi T, Nakao S, Almulki L, Zandi S, Miyahara S, Ito Y, Thomas KL, Garland RC, Miller JW, Gragoudas ES, Mashima Y, Hafezi-Moghadam A. Vascular adhesion protein-1 blockade suppresses choroidal neovascularization. FASEB J. 2008;22:2928–2935. doi: 10.1096/fj.07-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maula SM, Salminen T, Kaitaniemi S, Nymalm Y, Smith DJ, Jalkanen S. Carbohydrates located on the top of the “cap” contribute to the adhesive and enzymatic functions of vascular adhesion protein-1. Eur J Immunol. 2005;35:2718–2727. doi: 10.1002/eji.200526351. [DOI] [PubMed] [Google Scholar]
- 53.Kurkijarvi R, Adams DH, Leino R, Mottonen T, Jalkanen S, Salmi M. Circulating form of human vascular adhesion protein-1 (VAP-1): increased serum levels in inflammatory liver diseases. J Immunol. 1998;161:1549–1557. [PubMed] [Google Scholar]
- 54.Bono P, Salmi M, Smith DJ, Jalkanen S. Cloning and characterization of mouse vascular adhesion protein-1 reveals a novel molecule with enzymatic activity. J Immunol. 1998;160:5563–5571. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Sequence alignment of mammalian monoamine oxidazing SSAOs. The deduced amino acid sequences of four monoamine oxidizing mammalian SSAOs from GenBank were compared. The SSAOs shown are human AOC3 (VAP-1, AF067406), mouse AOC3 (mAOC3, mVAP-1, AF054831), human AOC2 (hRAO, D88213) and Bovine Serum Amine Oxidase (BSAO, L27218). Highlighted are: yellow, the hydrophobic N-terminal sequence; red, the conserved signature motif of SSAO active site, where the first tyrosine is post-translationally modified to topaquinone; pink, (putative) catalytic site base; light green, the conserved, Cu(II) binding histidine residues; turquoise, the conserved cysteine residues involved in dimerization; dark blue the putative N-linked glycosylation sites. The RGD sequence of AOC3 is indicated with red letters. The underlined sequence corresponds to the region deleted in the AOC2 splice variant 2 (sv2). For review of the consensus sites important for SSAO activity, see [39]. (PPT 55 kb)
Supplemental Figure 2. Antibodies recognizing AOC3 alone or both AOC3 and AOC2 give identical staining patterns. Frozen sections of human tonsil were stained with the monoclonal antibody 2D10 detecting only AOC3 (left panel) and with the polyclonal antibody poly-VAP detecting both AOC2 and AOC3 (right panel). Scale bar 100 μm. (TIFF 15356 kb)