Abstract
Poly(ADP-ribose) polymerase-1 (Parp-1) and the protein deacetylase SirT1 are two of the most effective NAD+-consuming enzymes in the cell with key functions in genome integrity and chromatin-based pathways. Here, we examined the in vivo crosstalk between both proteins. We observed that the double disruption of both genes in mice tends to increase late post-natal lethality before weaning consistent with important roles of both proteins in genome integrity during mouse development. We identified increased spontaneous telomeric abnormalities associated with decreased cell growth in the absence of either SirT1 or SirT1 and Parp-1 in mouse cells. In contrast, the additional disruption of Parp-1 rescued the abnormal pericentric heterochromatin, the nucleolar disorganization and the mitotic defects observed in SirT1-deficient cells. Together, these findings are in favor of key functions of both proteins in cellular response to DNA damage and in the modulation of histone modifications associated with constitutive heterochromatin integrity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0105-4) contains supplementary material, which is available to authorized users.
Keywords: Poly(ADP-ribosyl)ation, Acetylation, Genome integrity, Chromatin modifications, Sirtuins
Introduction
Regulation of higher-order chromosome structure and function is a critical process that underlies many aspects of chromatin-based tasks such as transcription, differentiation, DNA repair processes, and mitotic chromosome segregation.
Between the regulatory mechanisms that control chromatin structure and integrity in response to DNA damage is the modification of histones and other nuclear proteins by poly(ADP-ribose) polymers catalyzed by poly(ADP-ribose) polymerases (Parps) [1]. Among the 17 members of the Parps family, Parp-1 and Parp-2 heterodimerize share common binding partners and have been described as active players of the single-strand break/base excision repair process. Parp-1- and Parp-2-deficient mice and cells are very sensitive to both ionizing radiations and alkylating agents, thus supporting a role of both Parps in the cellular response to DNA damage [2]. Moreover, Parp-1 −/−;Parp-2 −/− embryos die at gastrulation demonstrating the crucial role of poly(ADP-ribosyl)ation during embryonic development [2].
Accumulating recent data support the emerging view that poly(ADP-ribosyl)ation might play a key role in the epigenetic regulation of chromatin dynamics [3–8]. More specifically, several lines of evidence support the view that Parp-1 and Parp-2 play prominent roles in the maintenance of constitutive and facultative heterochromatin integrity with, however, the emergence of specific functions for Parp-2 [9]. Both proteins localize to telomeres [10–13], centromeres [14, 15], and rDNA [16] where they interact with and regulate specific partners.
Parp-2−/− cells exhibit DNA damage induced kinetochore defects whereas the Parp-1 +/−;Parp-2 −/− background displays specific female embryonic lethality associated with X chromosome instability [17]. More recently, we identified the pericentric heterochromatin proteins HP1α and HP1β as binding partners of Parp-1 and Parp-2. Both proteins specifically poly(ADP-ribosyl)ate HP1α [18]. These data introduce both Parps as new actors of the HP1-mediated subcode histone underlying the histone code. Moreover, although little is known about the crosstalk between poly(ADP-ribosyl)ation and other epigenetic marks, recent studies point to a functional dynamic interplay between poly(ADP-ribosyl)ation and histone acetylation. Indeed, Parp-2−/− germ cells display defective meiotic sex chromosome inactivation linked with derailed histone H4 acetylation [19]. In cortical neurons treated with nerve-growth factors or in stimulated cardiomyocytes, the DNA-independent activation of Parp-1 results in ERK2-catalyzed Elk1 phosphorylation and acetylation of histone H3 and H4 [4].
Another enzyme responsible for chromatin modulations associated with heterochromatin formation is the NAD+-dependent histone deacetylase SirT1. SirT1 is the founding member of the sirtuin family and the mammalian ortholog of yeast Sir2 that is implicated in chromatin silencing, establishment and maintenance of telomere heterochromatin, longevity, and genome stability in lower eukaryotes [20]. The in vivo function of mammalian SirT1 has been investigated by the generation and characterization of three different SirT1-deficient mouse models displaying slightly distinct phenotypes most likely associated with different genetic backgrounds. Whereas early post-natal lethality was reported for two SirT1 mutant mice, in one case associated with developmental defects of the retina and the heart, a more severe phenotype of embryonic lethality associated with impaired histone modifications and defective DNA damage response was recently described in a third model [21–23].
It has also been shown that SirT1 promotes heterochromatin formation through the coordination of several events that includes deacetylation of histone H4 lysine 16 (H4K16) and histone H3 lysine 9 (H3K9), and the recruitment and deacetylation of histone H1 and the histone methyl-transferase Suv39h1 that both contribute to the establishment of marks associated with repressive heterochromatin [24, 25].
We have previously identified an interplay of Parp-1 and SirT1 in cellular response to DNA damage [26]. However, the precise relationship of Parp-1 and SirT1 in the regulation of chromatin-based processes is not known and remains a central issue for our understanding of epigenetic crosstalks. In this work, we generated and characterized an animal and cellular model deficient in both Parp-1 and SirT1 and studied the coordinated functions of both enzymes in the formation and function of constitutive heterochromatin. We observe that the double disruption of both Parp-1 and SirT1 genes tends to increased late postnatal lethality before weaning in mice. The characterization of the cellular model reveals that SirT1-deficient cells evidence increased spontaneous genome instability and telomeric aberrations that contribute to decreased cell growth, regardless of the Parp-1 status. In contrast, our results predict an essential contribution of both enzymes in the modulation of histone modifications associated with pericentric heterochromatin integrity, a prerequisite for nucleolus integrity and faithful chromosome segregation.
Materials and methods
Mice
Parp-1-deficient mice (C57Bl/6 background) and SirT1-deficient mice (129Sv-CD1background) have been described [22, 27]. Parp-1+/− females were crossed with SirT1+/− males producing double heterozygous Parp-1+/−;SirT1+/− offspring that were subsequently backcrossed. The breeding cages from Parp-1+/−;SirT1+/− matings were examined daily, and the first day that pups were found in the cage was considered day 1 of postnatal life. Mice were genotyped by PCR as described [22, 27]. Animals were maintained in a conventional animal house facility under 12-h dark-light cycles and given standard diet and water ad libidum.
Isolation and immortalization of MEFs
Primary mouse embryonic fibroblasts were harvested from 13.5-day-old embryos originating from Parp-1+/−;SirT1+/− matings according to standard procedures. Cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 1 mg/ml D-glucose (Invitrogen), 10% fetal calf serum (Eurobio), and 1% gentamicin (Invitrogen) until spontaneous immortalization. Two independent immortalized 3T3 clones of each genotype giving similar results were used throughout the study. For transfection experiments, Parp-1+/+;SirT1−/− cells (2 × 104/24-well plates) were transfected using the JetPEI method (Polyplus Transfections, Illkirch, France) with 0.2 μg of the pCruz-HA-SirT1 vector (Addgene, Cambridge) and 3.8 μg of pcDNA3 (Invitrogen) as carrier and processed 48 h later for either immunofluorescence or ultrastructural analysis.
Western-blot analysis
Exponentially growing 3T3 cells (5 × 106) were lysed by three cycles of freezing and thawing in 50 μl of lysis buffer [20 mM Tris–HCl, pH 7.5, 400 mM KCl, 5 mM DTT, 20% glycerol, 0.5% Nonidet P-40, 0.5 mM Pefabloc (Roche) and protease inhibitor cocktail (PIC, Roche)]. Cleared lysates were quantified by Bradford protein assay. Equivalent amounts of total protein extracts (100 µg) were resuspended in Laemmi buffer, and analyzed by 10% SDS/PAGE and immunoblotting. Blots were probed with an anti-Parp-1 (EGT-69) mouse antibody (1:10 000; [16]), an anti-SirT1 rabbit antibody (1:2500, Millipore 07-131) or an anti-actin rabbit antibody (1:500, Sigma A2066) followed by peroxidase-conjugated secondary antibodies, and developed using the ECL+ detection kit (Amersham, Little Chalsfont, UK).
Colony-forming assay (CFA) and cell growth
For CFA, 1,500 exponentially growing 3T3 cells were seeded in 100-mm culture dishes in triplicates. Eight to ten days later, cells were fixed for 30 min in formaldehyde (3.7%), stained with crystal violet (0.1%), and colonies were photographed and scored. For analysis of cell growth, exponentially growing 3T3 cells (4 × 104) were seeded in 60-mm dishes in triplicates. On days 1, 2, 3, and 4, cells were harvested by trypsinization and counted.
Immunofluorescence
Immunofluorescence was performed essentially as described previously [10]. Briefly, exponentially growing 3T3 cells (5 × 104 cells/12-well plates) were grown on glass coverslips, washed twice with PBS 1X, fixed for 10 min at 25°C in 4% formaldehyde, diluted in PBS 1X-0.1% Triton X-100, and washed again 3 times for 5 min at 25°C with PBS 1X-0.1% Triton X-100-0.1% skimmed milk. Cells were then incubated overnight at 4°C with the appropriate antibodies as following: mouse anti-B23 (1:2000, kindly provided by P. K. Chan, Houston, TX), mouse α-tubulin antibody (1:500, Sigma T9026), human CREST antibody (1:400, kindly provided by K.H. Andy Choo, Australia), rabbit H3K9me3 antibody (1:2000, Abcam ab8898), mouse HP1α antibody (1:500, Upstate 05-689), rabbit anti-phosphoH3-S10 (1:500, Upstate 06-570) and rabbit SirT1 antibody (1:500, Millipore 07-131). After three washes for 5 min at 25°C with PBS-0.1% Triton X-100-0.1% skimmed milk, cells were incubated with the appropriate Alexa-labeled secondary antibodies for 2 h at 25°C as following: Alexa Fluor (488 or 568) goat anti-rabbit IgG (1:1500, Molecular Probes), Alexa Fluor (488 or 568) goat anti-mouse IgG (1:1500, Molecular Probes), and Alexa Fluor 488 goat anti-human IgG (1:400, Molecular Probes, Invitrogen). DNA was counterstained with 4′,6-diamidino-2-phenylindol DAPI (25 ng/ml in PBS 1X) and slides were mounted in Mowiol. Images were captured using a Leica microscope (Leica Microsystems) equiped with an ORCA-ER chilled CCD camera (Hammamatsu) and the capture software Openlab (Improvision).
Electronic microscopy
Exponentially growing 3T3 cells were seeded on glass coverslips (5 × 104 cells/24-well plates) for 24 h, washed twice in 0.1 M Cacodylate buffer, fixed in Karnovsky’s solution, and post-fixed in 1% osmium tetroxide in 0.1 M Cacodylate buffer for 1 h at 4°C. Following samples were dehydrated with increasing concentrations of ethanol and embedded in Epon 812. Ultrathin sections (50–70 nm thick) were contrasted with uranyl actetate and lead citrate and examined with a Morgagni 268D Philips electron microscope.
Cell cycle analysis
Exponentially growing 3T3 cells (5 × 105) were seeded in 100 mm Petri dishes. After 2 days, cells were trypsinized, washed in PGE buffer (PBS 1X, 1% glucose, 1 mM EDTA) and fixed for at least 1 h on ice in EtOH 70% in PGE. Samples were centrifuged at 1,500g for 5 min and cells were rehydrated in 1 ml PGE O/N at 4°C. For the identification of cells in G2/M, samples were prepared and fixed as above, washed twice in PTB buffer (PBS 1X, 0.5% Tween-20, 0.5% BSA) and incubated with rabbit anti-phosphoH3-S10 antibody (1:500, Upstate 06-570) for 90 min at room temperature. After two washes in PTB buffer, cells were incubated in a dark chamber with Alexa Fluor (488) goat anti-rabbit IgG (1:1500, Molecular Probes) for 90 min at room temperature. Cells were finally washed with PTB and rehydrated in 1 ml PGE O/N at 4°C. Before analysis, samples were treated for 30 min with 100 µg/ml RNAse I, filtered and stained with propidium iodide at a final concentration of 50 µg/ml. Flow cytometry analysis was carried out using a fluorescence-activated cell sorter (FACS calibur) and the Cell Quest Software (Becton Dickinson).
Metaphase spreads
Exponentially growing 3T3 cells (1 × 106) were synchronized using 0.5 µg/ml colcemid for 5 h at 37°C, or O/N for the Parp-1−/−;SirT1−/− cells. After hypotonic treatment (KCl 75 mM) for 30 min at 25°C, cells were washed once with Methanol:acetic acid fixing solution (3:1), centrifuged and incubated again O/N at 4°C in fixing solution. Next, samples were centrifuged at 2,500g at 4°C and cells were resuspended in 500 µl of fixation solution until the cell suspension looks milky. Metaphases were dropped on clean microscopic slides and left to air dry. For chromosome counting, slides were washed twice with PBS 1X, DNA-stained using DAPI for 10 min at 25°C and mounted in Mowiol. At least 30 well-spread metaphase chromosomes were captured and scored per cell line using a Leica microscope (Leica Microsystems) as above.
Fluorescent in situ hybridization experiments (FISH)
Metaphase spreads and analysis of telomere aberrations were performed as previously described [28]. Briefly, cells were treated with colcemid and exposed to hypotonic treatment as above. Cells were fixed and stored in Methanol/acetic acid (3:1). Before hybridization, cells were dropped on slides and air-dried overnight. After fixation in 4% PFA, slides were pre-treated with pepsin (0.5 mg/ml for 10 min at 37°C), then dehydrated in ethanol and air-dried. Cells were denaturated (3 min at 80°C on a heat block) with hybridization solution [70% deionized Formamide, 1% blocking reagent (Sigma) and 10 mM Tris–HCl, pH 7.2) containing PNA probe coupled with Cy3 fluorochrome (Cy3-OO-(CCCTAA)3; Applied Biosystems]. After hybridization for 2 h at room temperature, slides were rehybridized overnight with denaturated (10 min at 85°C) FITC-pan-centromeric probe (Cambio). Hybridized spreads were then washed twice with 70% deionized formamide (10 mM Tris-HCl, pH 7.2) and then three times with Tris 50 mM, NaCl 150 mM, Tween 20, 0.05%. Finally, spreads were stained with DAPI before mounting. Slides were observed under a fluorescent microscope (Olympus IX81). Image acquisition by coolsnap HQ camera (Princetown Instruments) was controlled by MetaMorph software (Universal Imaging) Chromosome numbers and telomeric aberrations were quantified per metaphases on three gray-scale 16-bit merged images (Dapi, FITC, and Cy3). All statistical analyses (ANOVA and Fisher’s test) were done with StatView software.
Results
Increased late post-natal lethality in Parp-1−/−;SirT1−/− double knockout mice
To examine the combined effect of the Parp-1 and SirT1 null mutations, we created and bred Parp-1+/−;SirT1+/− double heterozygotes to generate the various possible combinations of Parp-1 and SirT1 targeted alleles. Results are summarized in Table 1. Genotyping of a total of 399 offspring 1 week after birth (days 7–10) revealed only half the proportion of Parp-1+/+;SirT1−/− pups (3% compared to 6.25% expected) consistent with previously described early postnatal loss [21, 22]. Interestingly, we also observed half the proportion of Parp-1+/−;SirT1−/− (5% compared to 12.5% expected) and Parp-1−/−;SirT1−/− (3% compared to 6.25% expected) mice. Thus, the additional disruption of Parp-1 did not increase early post-natal death. Mice of the other genotypes were obtained at approximately Mendelian frequency. However, when survival of the pups was monitored at weaning (days 30–35), we observed an additional loss of 5 out of 11 (45%) Parp-1+/+;SirT1−/− pups and 9 out of 21 (43%) Parp-1+/−;SirT1−/− pups that is further increased by the complete disruption of Parp-1 (80% loss of Parp-1−/−;SirT1−/− pups, 8 out of 10). Together, these data are in line with a determinant role of SirT1 in post-natal development as reported [21, 22] and suggest an additional contribution of Parp-1 in mid to late post-natal development. Interestingly, both remaining Parp-1−/−;SirT1−/− mice (independently of sex) survived into adulthood, appeared healthy, and lagged behind in growth (Supplementary electronic material, ESM, Fig. 1). In addition, according to previous reports [22, 29] we observed sterility for Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− mice (box sexes) when caged with wild-type animals (unpublished observation).
Table 1.
Increased post-natal lethality in Parp−/−;SirT1−/− double knockout mice
| Parp-1; SirT1 | Expected (%) | Observed (days 7–10) | Lethality (<1 week) | Observed (days 30–35) | Lethality (1–5 weeks) (%) (n = 399) | |
|---|---|---|---|---|---|---|
| n | % | % | n | |||
| +/+; +/+ | 6.25 | 31 | 8 | 0 | 27 | 13 |
| +/+; ± | 12.5 | 73 | 18 | 0 | 65 | 10 |
| ±; +/+ | 12.5 | 60 | 15 | 0 | 54 | 10 |
| ±; ± | 25 | 113 | 28 | 0 | 102 | 10 |
| −/−; +/+ | 6.25 | 27 | 7 | 0 | 26 | 3 |
| −/−; ± | 12.5 | 52 | 13 | 0 | 44 | 15 |
| +/+; −/− | 6.25 | 11 | 3 | 56 | 6 | 45 |
| ±; −/− | 12.5 | 21 | 5 | 58 | 12 | 43 |
| −/−; −/− | 6.25 | 10 | 3 | 60 | 2 | 80 |
Genetic analysis of 399 offspring derived from Parp-1+/−;SirT1+/− double heterozygous matings. Genotyping was performed at approximately 1 week (days 7–10) by PCR analysis of tail DNA. Early (1 week) and mid to late (1–5 weeks) post-natal lethality was monitored. All animals viable at weaning survived into adulthood without obvious health and/or behavior abnormalities
Decreased cell growth and increased genome instability in Parp-1+/+;SirT1−/− and Parp-1−/−;SirT1−/− 3T3 cells
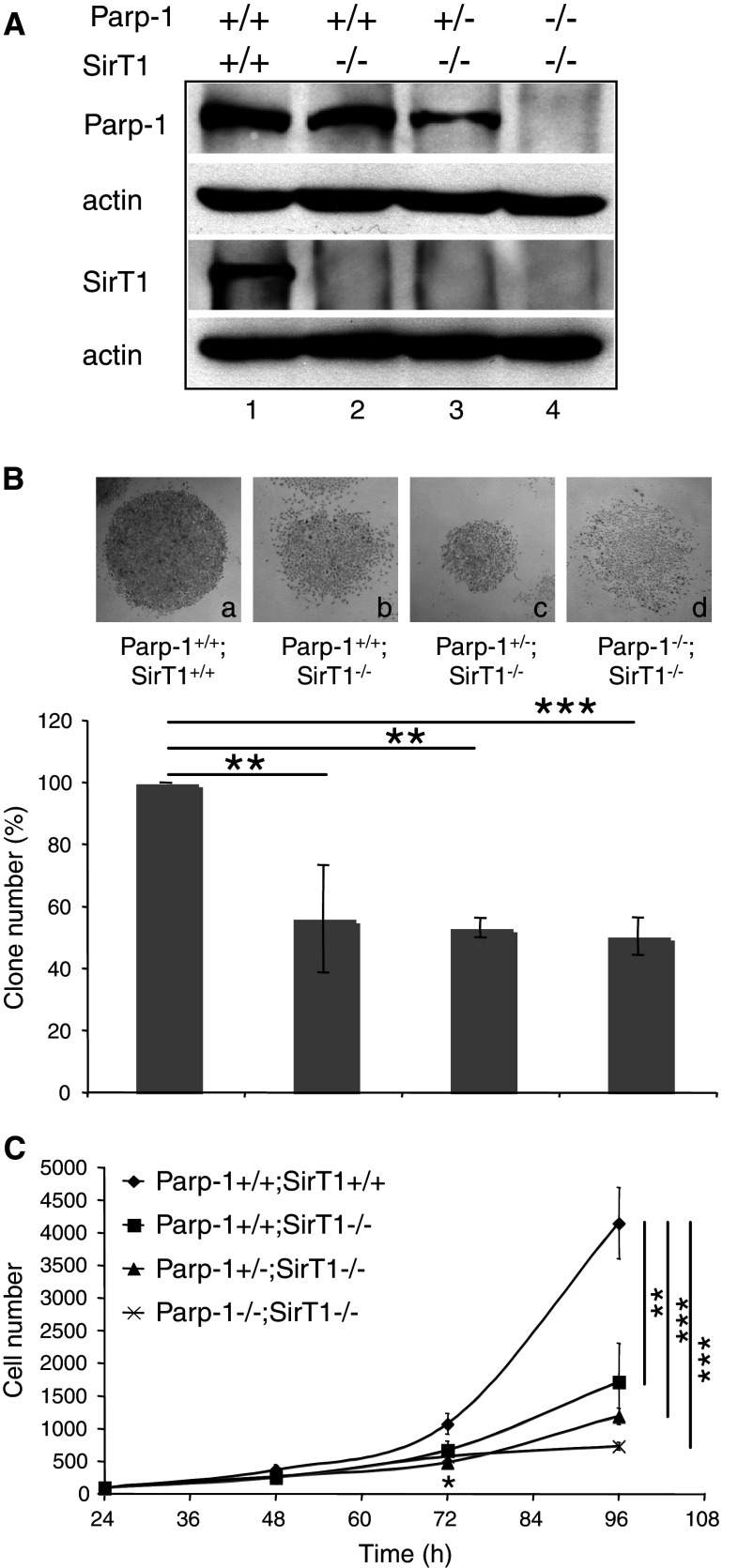
To further investigate the in vivo functional link between Parp-1 and SirT1 and overcome the reduced number of Parp-1−/−;SirT1−/− mice available, we generated spontaneously immortalized mouse embryonic fibroblasts (3T3) (Fig. 1A). Western blot analysis of whole cell extracts using, respectively, anti-Parp-1 and anti-SirT1 antibodies revealed the presence of Parp-1 in Parp-1+/+;SirT1+/+ (lane 1), Parp-1+/+;SirT1−/− (lane 2), and to a lesser extent in Parp-1+/−;SirT1−/− (lane 3), whereas SirT1 was detected in only Parp-1+/+;SirT1+/+ cells (lane 1). None of the proteins were detected in the Parp-1−/−;SirT1−/− cells (lane 4). As an initial experiment, we examined whether the absence of both Parp-1 and SirT1 affected cell growth. Clonogenic assays revealed a comparable marked reduction in colony formation in Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells compared to Parp-1+/+;SirT1+/+ cells (Fig. 1B). To further evaluate a defect on cell proliferation, the four different cell lines were seeded at equal densities, and cell numbers were determined at the indicated times. As shown in Fig. 1C, we observed a significant and similar retarded cell growth in Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/−cells compared to the Parp-1+/+;SirT1+/+ control cells. In contrast, even though the growth curve of Parp-1−/−;SirT1−/− cells looked linearly shaped, the growth rate was not significantly different from that of Parp-1+/+;SirT1−/− cells. We next assessed whether spontaneous cell death could contribute to the decreased growth rate by Tunel assays, but we could not detect increased spontaneous apoptosis (data not shown).
Fig. 1.
Establishment, and growth of Parp-1+/+;SirT1+/+, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− 3T3 cell lines. A Western blot analysis for the expression of Parp-1, SirT1 and β-actin in spontaneously immortalized MEFs (3T3). Equivalent amounts of total protein extracts from wild-type Parp-1+/+;SirT1+/+ (lane 1), Parp-1+/+;SirT1−/− (lane 2), Parp-1+/−;SirT1−/− (lane 3), and Parp-1−/−;SirT1−/− (lane 4) 3T3 cells were separated by SDS-PAGE and analyzed by western blotting with the appropriate antibodies. B Clonogenic assay. Colonies were stained with 0.1% crystal violet in ethanol and counted by visual inspection. Insets represent a microphotography of one representative colony for each genotype. Data are presented as the percent of control colony number. Each point represents the mean of triplicate samples. Data shown are representative of results obtained in three different experiments. P value was calculated by ANOVA test: **P < 0.01, ***P < 0.001. C Growth curves. Equal cell numbers of Parp-1+/+;SirT1+/+ (filled diamond), Parp-1+/+;SirT1−/− (filled circle), Parp-1+/−;SirT1−/− (filled triangle), and Parp-1−/−;SirT1−/− (×—×) were plated in six-well plates, and for each experiment, triplicate wells were counted over a period of 4 days. Points represent the mean of three independent experiments. P value was calculated by ANOVA test comparing the different genotypes to the control cell line: *P < 0.05, **P < 0.01, ***P < 0.001
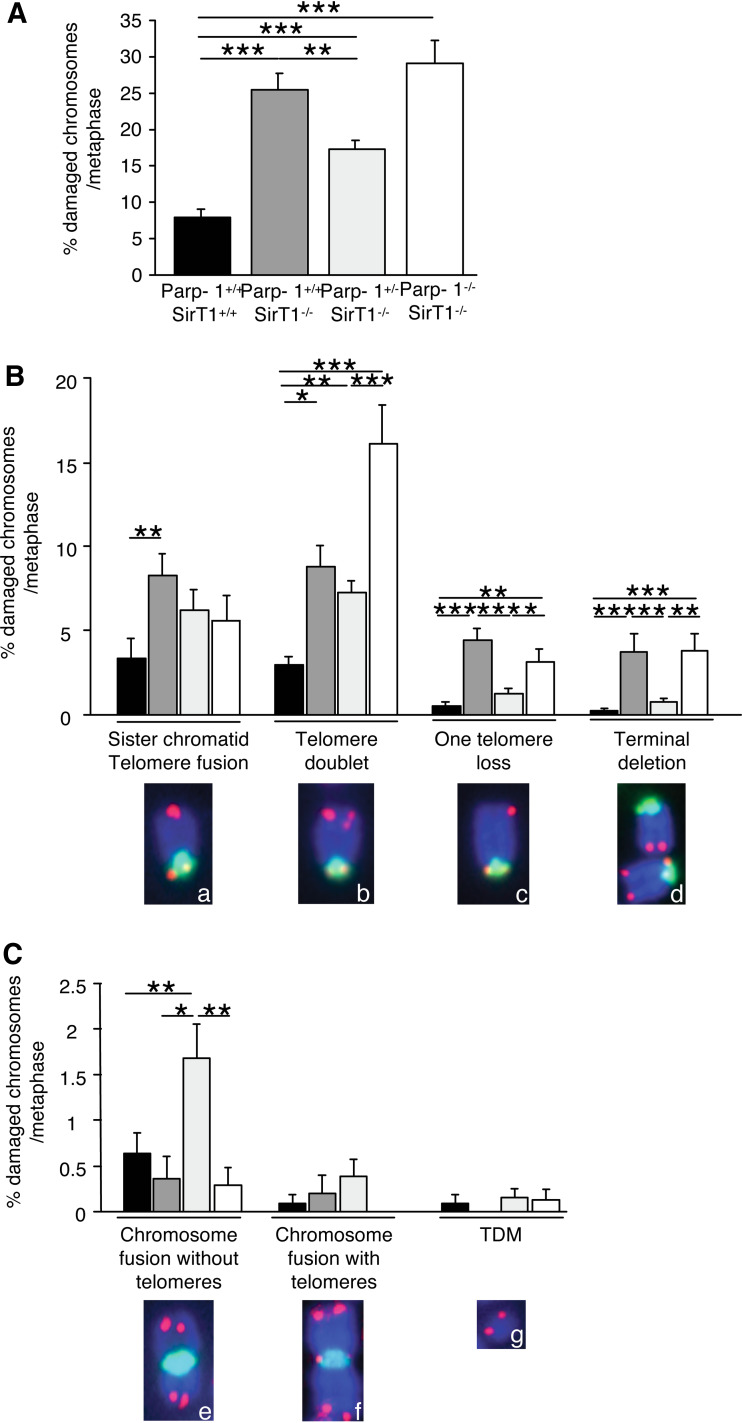
We then investigated whether this slower cell growth was caused by increased genome instability as previously reported [23]. We therefore monitored spontaneous telomeric aberrations using telomeric FISH analysis in the different cell lines. As shown in Fig. 2A, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells displayed a significant and specific increase in spontaneous telomeric aberrations (25.5 ± 2.25, 17.2 ± 1.43, and 28.5 ± 2.8% of chromosomes with telomere aberrations per metaphase, respectively) compared to the Parp-1+/+;SirT1+/+ control cells (7.8 ± 1.25%). More particularly, we found an increase in terminal deletions and/or chromosome fusions without telomeres at the fusion points, evidencing an increased genome instability in the absence of either SirT1 or SirT1 and Parp-1 (Fig. 2B, C). Interestingly, most of the telomere aberrations that were increased in Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells compared to Parp-1+/+;SirT1+/+ control cells resulted from fusion or recombination occurring during or after telomere replication in S or G2 phases (i.e., telomere doublets, one telomere losses and sister chromatid telomere fusions [28]; Fig. 2B). Other chromosome abnormalities including chromosome fusions with telomeres at the fusion points and telomeric DNA-containing double minute chromosomes (TDM), the latter resulting from recombination in G1, were not affected (Fig. 2C).
Fig. 2.
Telomere instability in Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− 3T3 cell lines. Increased spontaneous telomere aberrations in Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− 3T3 cells compared to Parp-1+/+;SirT1+/+ control cells. Telomere aberrations were detected by FISH on metaphase spreads as described in Materials and Methods. Total (A) or each type (B,C) of telomere aberrations are expressed as percentages of damaged chromosomes per metaphasis. The different types of telomere aberrations given in (B) are: chromosomes with sister chromatid telomere fusion (insert a), chromosomes with one extra-telomere signal referred to as telomere doublets (insert b), chromosomes with one telomere loss (insert c), chromosomes with terminal deletion (insert d), fused chromosomes without (insert e) or with telomeres (insert f) at the fusion points, and telomeric DNA-containing double minutes chromosomes (TDM, insert g). Inserts illustrate the different telomere aberrations identified in Parp-1+/+;SirT1−/− cells except for TDM identified in Parp-1−/−;SirT1−/− cells (green, pan-centromeric probe; red, PNA probe). Results were obtained from n = 18, 21, 20, and 15 metaphases for Parp-1+/+;SirT1+/+, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells, respectively. Ordinary ANOVA tests revealed significant differences between genotypes intelomere aberrations (ANOVA, F 3,67 = 23.42, P < 0.0001) (A), telomere doublet (F 3,67 = 9.11, P < 0.0001), one telomere loss (F 3,67 = 13.29, P < 0.0001), terminal deletion (F 3,67 = 8.098, P < 0.001) (B), and chromosome fusion with telomeres (F 3,67 = 5.32, P = 0.024) (C). P value was calculated by Fischer’s test: P *< 0.05, **P < 0.01, ***P < 0.001
Together, these results indicate that cells deficient in SirT1 evidence increased spontaneous genome instability and telomeric aberrations that contribute to decreased cell growth, regardless of the Parp-1 status.
Impaired pericentric heterochromatin organization in SirT1-deficient cells is suppressed by additional disruption of Parp-1
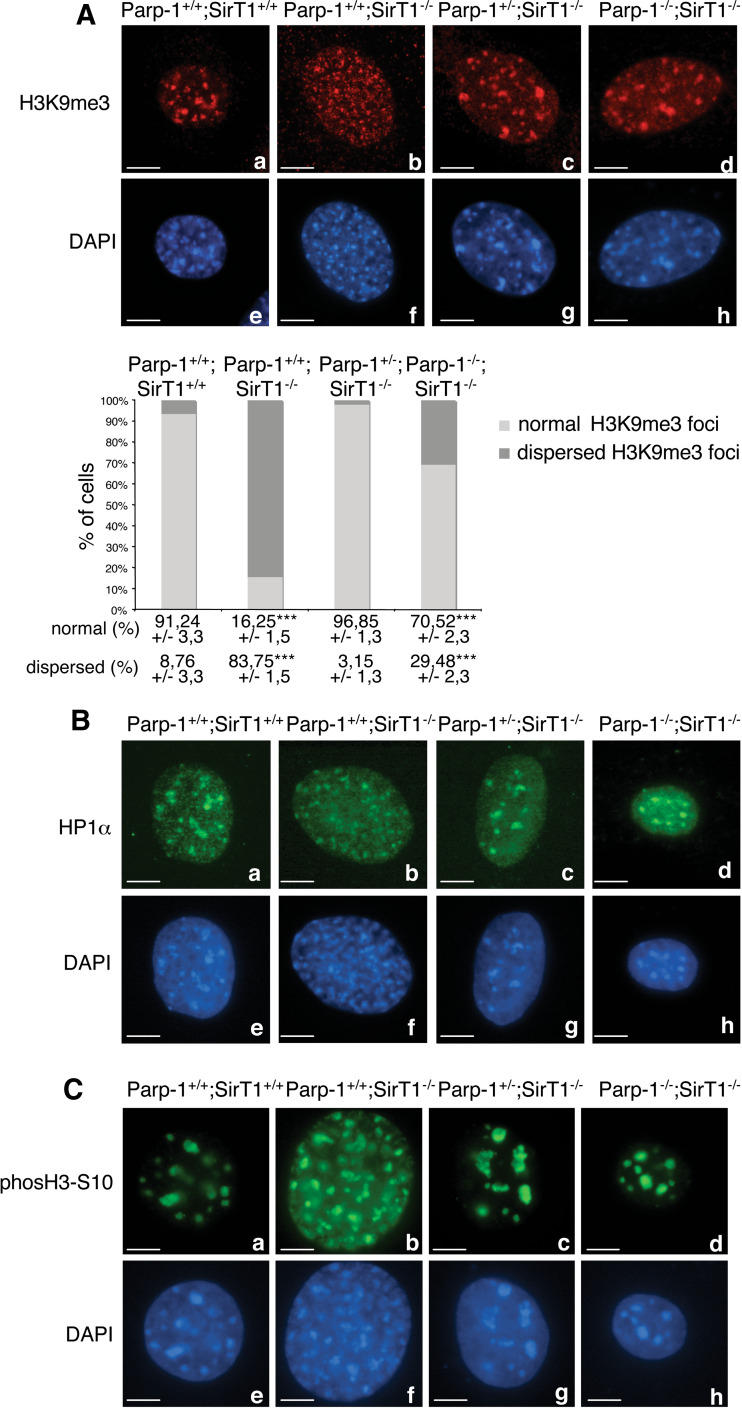
Loss of SirT1 affects Suv39h1-dependent methylation of histone H3 (H3K9me3) and impairs localization of heterochromatin protein 1 (HP1) to heterochromatic foci [24, 25]. To understand the functional link between both Parp-1 and SirT1 in the chromatin context, we monitored H3K9me3 and HP1 localization by immunofluorescence in the different cell lines (Fig. 3). Consistent with published results [23, 24], Parp-1+/+;SirT1−/− cells showed a significant reduction of Suv39h1-mediated H3K9me3 staining in heterochromatic regions in more than 80% of the cells examined, in comparison to the wild-type cells (Fig. 3A, compare b with a). In contrast, the global levels of euchromatic H3K9me3 seemed unaffected as indicated by similar levels of integrated fluorescence intensity in these regions (analyzed by ImageJ software, unpublished observation). Moreover, the heterochromatic localization of HP1 was also affected in most of the Parp-1+/+;SirT1−/− cells (Fig. 3B, compare b with a). This phenotype was accompanied by a pattern of dispersed and fragmented DAPI-stained heterochromatic foci in Parp-1+/+;SirT1−/− cells as compared to normal clustered heterochromatic foci in the control Parp-1+/+;SirT1+/+ cells (Fig. 3A–C, compare f with e). Of note, no heterochromatic-linked hyperacetylation of H4K16 could be detected in the Parp-1+/+;SirT1−/− cells (data not shown) probably due to a compensating histone H4-deacetylase activity of SirT2 [30]. Under similar conditions, a wild-type-like accumulation of H3K9me3 and HP1 to heterochromatic foci was detected in Parp-1−/−;SirT1+/+ cells ([18]; ESM, Fig. 2A).
Fig. 3.
Interplay of Parp-1 and SirT1 in the epigenetic modification of chromatin. A Upper panel Representative immunofluorescence images for the comparative distribution of H3K9me3 in Parp-1+/+;SirT1+/+ (a), Parp-1+/+;SirT1−/− (b), Parp-1+/−;SirT1−/− (c), and Parp-1−/−;SirT1−/− (d) interphase cells. DNA and heterochromatic foci are counterstained with DAPI (e–h). Whereas H3K9me3 marks heterochromatic regions in Parp-1+/+;SirT1+/+ cells (a), a dispersed speckled pattern is observed in a majority of Parp-1+/+;SirT1−/− cells (b). The additional disruption of Parp-1 induces a wild-type-like relocation of H3K9me3 to pericentric heterochromatin (c–d). Lower panel Quantification of the percentage of cells displaying normal or dispersed heterochromatic staining of H3K9me3. An average of 500 cells per cell line were scored in >20 randomly selected immunofluorescence fields. Results are averages from at least three independent experiments. Mean values ± SD are indicated. P value was calculated by ANOVA test: ***P < 0.001. B Representative immunofluorescence images for the comparative distribution of HP1 in Parp-1+/+;SirT1+/+ (a), Parp-1+/+;SirT1−/− (b), Parp-1+/−;SirT1−/− (c), and Parp-1−/−;SirT1−/− (d) interphase cells. DNA and heterochromatic foci are counterstained with DAPI (e–h). Note that the localization/mislocalization of HP1 to pericentric heterochromatin in the different cell lines follows the pattern described for H3K9me3 in (A). C Upper panel, representative immunofluorescence images for the comparative distribution of phosH3-S10 in Parp-1+/+;SirT1+/+ (a), Parp-1+/+;SirT1−/− (b), Parp-1+/−;SirT1−/− (c), and Parp-1−/−;SirT1−/− (d) interphase cells. Lower panel, DNA and heterochromatic foci are counterstained with DAPI (e–h). Scale bars 7 µm
Interestingly, the additional disruption of Parp-1 rescued the localization of both H3K9me3 and HP1 to heterochromatic regions in more than 90% of Parp-1+/−;SirT1−/− and 70% of Parp-1−/−;SirT1−/− cells (Fig. 3A, B, compare c and d with b). In contrast, the re-introduction of Parp-1 into Parp-1−/−;SirT1−/− cells induced a dispersion of H3K9me3 staining in the cells examined, similar to that observed in Parp-1+/+;SirT1−/− cells whereas a wild-type-like accumulation of H3K9me3 onto heterochromatic foci was found in the untransfected cells (ESM, Fig. 2B). To corroborate our result, we also examined the Suv39h1-dependent G2-specific concentration of phosH3-S10 at heterochromatic foci [31] (Fig. 3C). We found a dispersion of phosH3-S10 into many small dots in a majority of Parp-1+/+;SirT1−/− cells whereas a normal heterochromatic distribution similar to control Parp-1+/+;SirT1+/+ cells was observed in most of the Parp-1+/−;SirT1−/− and Parp-1−/−; SirT1−/− cells (Fig. 3C, compare b with a, c and d).
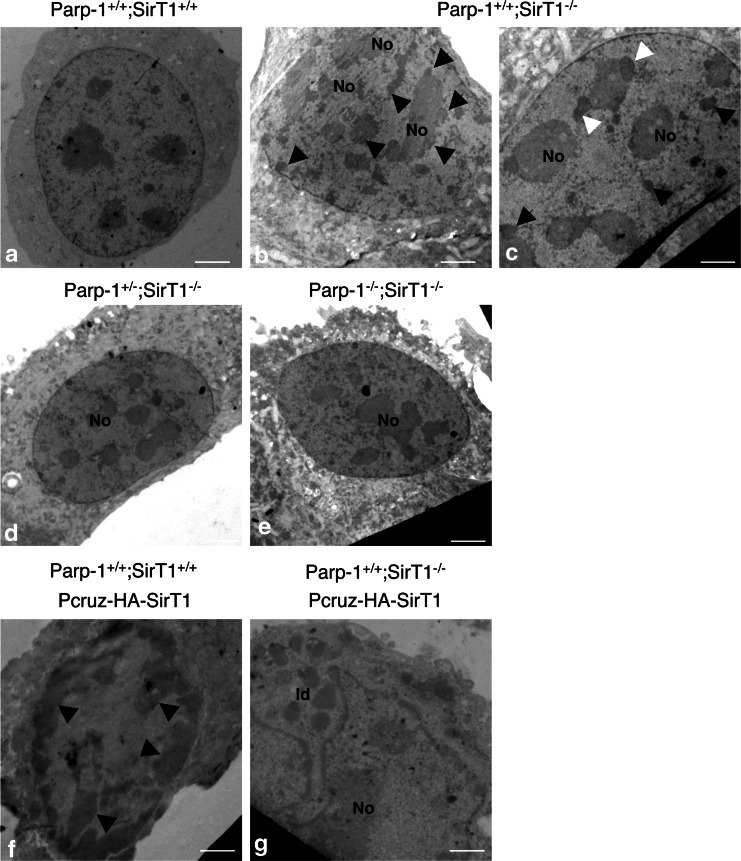
To further corroborate these results and examine the chromatin state, we performed an ultrastructural analysis of the different cell lines by transmission electronic microscopy (Fig. 4). The analysis revealed the accumulation of partially fragmented heterochromatin aggregating around the nucleoli and at the nuclear periphery in Parp-1+/+;SirT1−/− cells (Fig. 4b). We also observed in some cases the formation of nucleolar caps surrounding the central nucleolar body of Parp-1+/+;SirT1−/− cells (Fig. 4c), a phenomenon normally occuring during physiological or drug-induced transcriptional arrest [32]. These alterations were not seen in Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells rather displaying normal condensed heterochromatin uniformely distributed in the nucleus as in Parp-1+/+;SirT1+/+ cells (Fig. 4a, d, e). In contrast, we detected an aberrant hypercondensation of heterochromatin in Parp-1+/+;SirT1+/+ cells overexpressing SirT1 (Fig. 4f), consistent with a major role of SirT1-dependent histone deacetylation in chromosome condensation [23]. Finally, a wild-type-like homogeneous distribution of normal condensed chromatin was observed in SirT1-reconstituted Parp-1+/+;SirT1−/− cells (Fig. 4g) indicating that the abnormal chromatin state observed in Parp-1+/+;SirT1−/− is a direct consequence of SirT1 loss.
Fig. 4.
Interplay of Parp-1 and SirT1 in the organization of pericentric heterochromatin. Electronic microscopy examination of chromatin. Heterochromatin appears uniformly distributed in Parp-1+/+;SirT1+/+ cells (a) but aberrantly fragmented in Parp-1+/+;SirT1−/− cells as indicated by black arrows (b). Some Parp-1+/+;SirT1−/− cells also display nucleolar caps (white arrows) surrounding a central nucleolus (c). A wild-type like organization of heterochromatin is observed again in Parp-1+/−;SirT1−/− (d) and Parp-1−/−;SirT1−/− cells (e). Parp-1+/+;SirT1+/+ cells transfected with SirT1 (f) show an hypercondensation of peripheral heterochromatin (black arrows) whereas Parp-1+/+;SirT1−/− cells transfected with SirT1 (g) display a normal distribution of chromatin but evidence for some cells, the accumulation of cytoplasmic lipid droplets (ld). No Nucleolus, ld lipid droplets. Scale bars 3.25 µm. The transfection efficiency of pCruz-HA-SirT1 was verified by western blot analysis for the absolute levels of SirT1 and actin (ESM, Fig. 3)
Altogether, these data reveal that the impaired pericentric heterochromatin assembly and the associated perturbed histone tail modifications observed in Parp-1+/+;SirT1−/− cells (this study and [23, 24]) can be rescued by the additional disruption of Parp-1 suggesting a functional relationship between Parp-1 and SirT1 in regulating pericentric heterochromatin organization.
Abnormal nucleolus architecture in SirT1-deficient cells is rescued by additional disruption of Parp-1
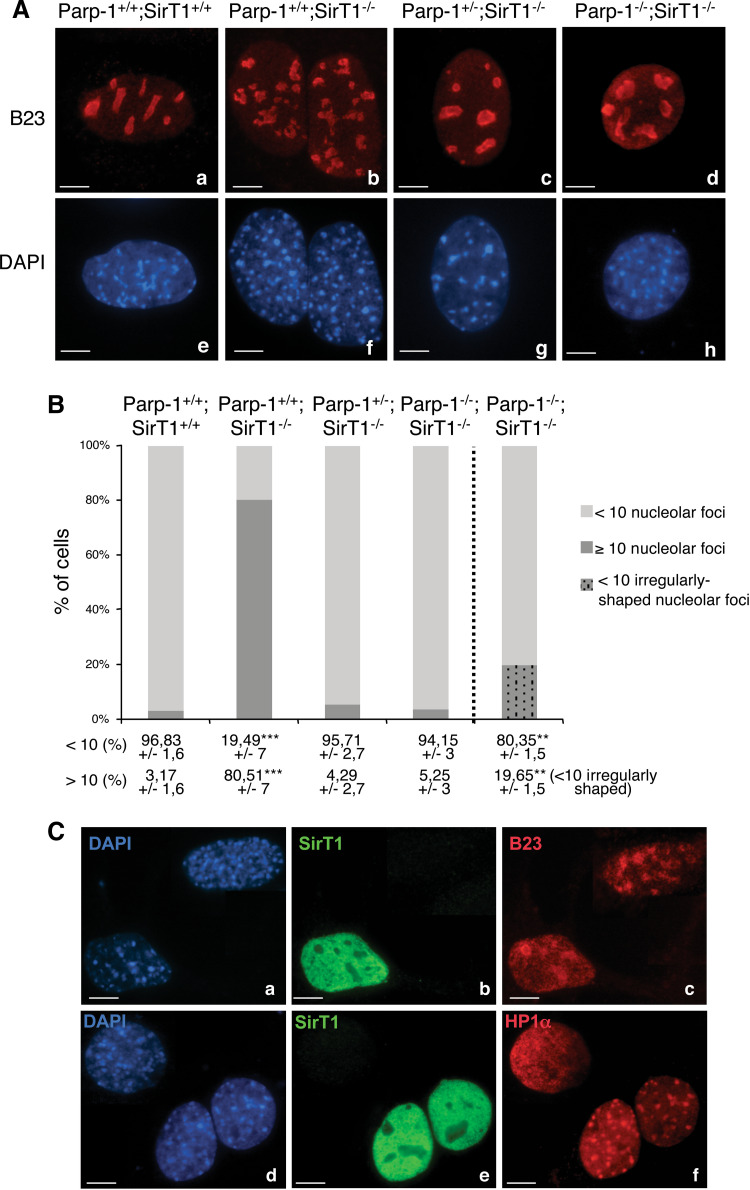
Recent studies have shown that heterochromatin and the associated proteins Suv39h1, HP1, and SirT1 regulate the organization of nucleoli [33, 34]. Interestingly, when we stained the cells for H4K16 acetylation, we observed a particular fragmented-like nucleolar exclusion of acetylH4K16 in SirT1-deficient cells reminiscent of nucleolar fragmentation (data not shown). Therefore, to evaluate the crosstalk of Parp-1 and SirT1 in this process, we characterized in detail the morphology of the nucleolar compartment in the different cell lines by staining for the nucleolar component B23 (Fig. 5). Whereas Parp-1+/+;SirT1+/+ cells all contained less than 10 nucleoli, up to 80% of the Parp-1+/+;SirT1−/− cells contained a significantly higher number of rather irregularly shaped and multilobed nucleoli (Fig. 5A, compare b with a, and B). Of note, the additional disruption of Parp-1 suppressed this phenotype since more than 90% of Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells contained less than 10 nucleoli (Fig. 5A c, d, B). However, these nucleoli were irregularly shaped in about 20% of Parp-1−/−;SirT1−/− cells. Similar results were obtained by staining for the nucleolar phosphoprotein C23 (data not shown). Of note, a wild-type-like number of nucleolar foci was seen in Parp-1−/−;SirT1+/+ cells ([16] and data not shown).
Fig. 5.
Interplay of Parp-1 and SirT1 in the architecture of the nucleolus. A Representative immunofluorescence images for the immunodetection of the nucleolar marker B23 (red) in Parp-1+/+;SirT1+/+ (a), Parp-1+/+;SirT1−/− (b), Parp-1+/−;SirT1−/− (c) and Parp-1−/−;SirT1−/− (d) cells counterstained with DAPI (blue, e–h). The absence of SirT1 results in nucleoli fragmentation as shown by the numerous B23 marked subnuclear structures (b). In contrast only few nucleoli (<10) are counted in Parp-1+/+;SirT1+/+ (a), Parp-1+/−;SirT1−/− (c) and Parp-1−/−;SirT1−/− (d) cells. B Histogram showing the percentage of cells with a normal number of nucleolar foci (<10) or multiple nucleolar foci (>10). Mean values (±SD) are indicated. Of note, around 20% of the Parp-1−/−;SirT1−/− cells displayed a normal number but irregularly-shaped nucleoli. A total average of 500 cells were scored per cell line in >20 randomly selected immunofluorescence fields. P value was calculated by ANOVA test: **P < 0.01, ***P < 0.001. C Transient transfection of Parp-1+/+;SirT1−/− cells with wild-type SirT1 (green, b, e) restores nucleolus integrity marked by B23 (red, c) and relocalization of HP1α onto pericentric heterochromatin (red, f) when compared to an untransfected cell. DNA and heterochromatic foci are counterstained with DAPI (a, d). Scale bars 7 µm
Again to ascertain whether nucleolus disorganization and impaired pericentric heterochromatin composition are direct consequences of the absence of SirT1, we examined the pattern of B23 and HP1α in Parp-1+/+;SirT1−/− cells transiently transfected with the SirT1 expression vector (Fig. 5C). Our data show that the SirT1-reconstituted Parp-1+/+;SirT1−/− cells contain less than 10 nucleoli and display a wild-type-like accumulation of HP1α onto heterochromatic foci when compared to the untransfected Parp-1+/+;SirT1−/− cells.
Mitotic defects in SirT1-deficient cells is suppressed by the additional disruption of Parp-1
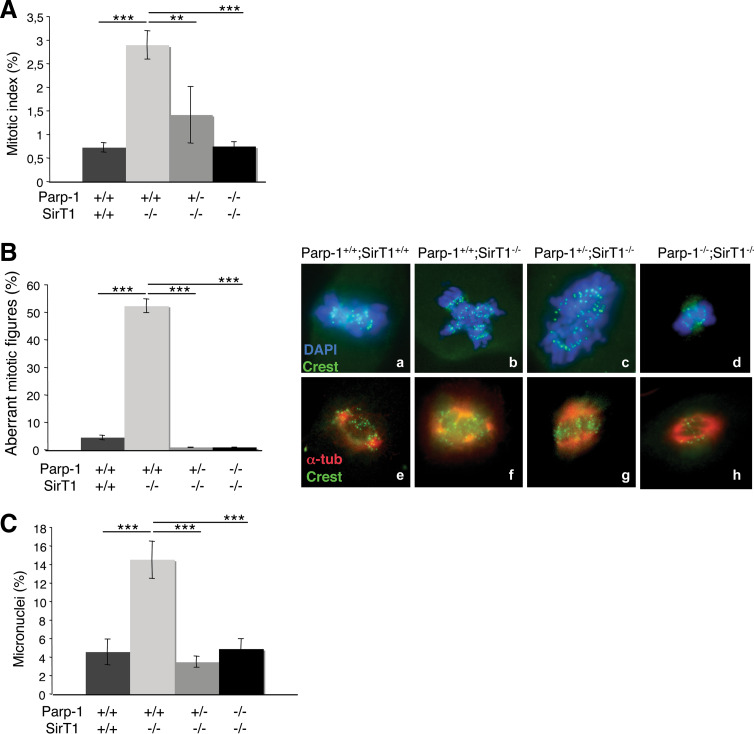
Disorganization of the nucleoli at interphase and perturbed heterochromatin structure have previously been associated with the accumulation of abnormal mitotic cells [31, 33–35]. To determine if the absence of SirT1 or both Parp-1 and SirT1 resulted in mitotic arrest, we first compared the mitotic index of each mutant cell line to the wild-type controls (Fig. 6A). We identified mitotic figures using DAPI staining to mark condensed chromosomes and α-tubulin staining relating to spindle-like structures. As shown in Fig. 6A, we observed a fourfold increase of the spontaneous mitotic index in Parp-1+/+;SirT1−/− cells compared to the Parp-1+/+;SirT1+/+ controls. In contrast, the additional disruption of either one or both alleles of Parp-1 restored the mitotic index near wild-type levels. Further quantification of the mitotic population revealed a significant accumulation of abnormal mitotic figures displaying an aberrant metaphase configuration with condensed DNA but failure of chromosome alignment on the metaphase plate and extra-α-tubulin containing microtubule organizing centres in Parp-1+/+;SirT1−/− cells (12-fold increase) compared to Parp-1+/+;SirT1+/+ cells (Fig. 6B). Again, the additional disruption of Parp-1 efficiently rescued the abnormal mitotic phenotype.
Fig. 6.
Interplay of Parp-1 and SirT1 in the regulation of mitotic division. A Histogram showing the percentage of Parp-1+/+;SirT1+/+, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells in mitosis. Cells (prometaphase–metaphase–anaphase–telophase) were scored by immunofluorescence (DAPI staining). Values represent the mean (±SD) of three independent experiments (n = 500). P value was calculated by ANOVA test: ** P < 0.01 and *** P < 0.001. B Left panel Histogram showing the percentage of aberrant mitosis for each genotype indicated. Cells were scored by DAPI staining. Values represent the mean (±SD) of three independent experiments (n = 500). Right panel Immunofluorescence of Parp-1+/+;SirT1+/+ (a,e), Parp-1+/+;SirT1−/− (b,f), Parp-1+/−;SirT1−/− (c,g), and Parp-1−/−;SirT1−/− (d,h) paraformaldehyde-fixed cells showing measurements of centromere disposition (upper panel, a–d) by the staining of centromeric proteins (Crest, green) and DNA (DAPI, blue) and measurements of spindle structure (lower panel, e–h) by the staining of the spindle (α-tubulin, red) and DNA (DAPI, blue). P value was calculated by ANOVA test: ***P < 0.001. C Histogram showing the percentage of Parp-1+/+;SirT1+/+, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells with micronuclei. Cells were scored by DAPI staining. Values represent the mean (±SD) of three independent experiments (n = 500). In Parp-1+/+;SirT1−/− cells, the number of aberrant mitotic figures (chromosome missegregation) and the number of micronuclei (genomic instability) increased considerably compared with that in Parp-1+/+;SirT1+/+ as well as Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells. P value was calculated by ANOVA test: ***P < 0.001
Both impaired heterochromatin formation and mitotic defects could be a cause of genomic instability. To investigate this, we analyzed the frequency of spontaneous micronucleus formation in each cell line (Fig. 6C). The Parp-1+/+;SirT1−/− cells showed a spontaneous threefold increase in the number of cells displaying micronuclei relative to the control Parp-1+/+;SirT1+/+ cells. In contrast, a wild-type-like level of micronuclei was observed in Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells.
Disruption of mouse SirT1 results in increased chromosome number that is counterbalanced by an additional disruption of Parp-1
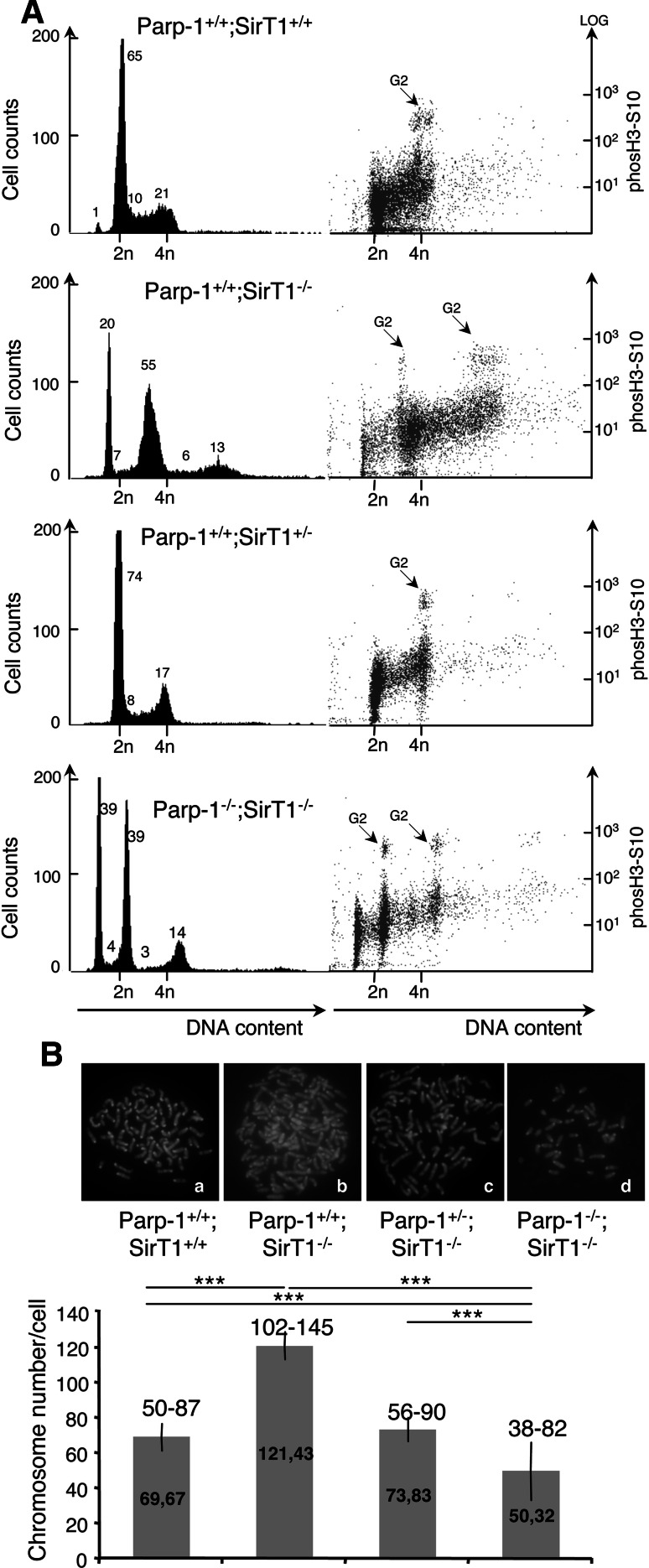
Next, we examined the cell cycle distribution of each cell line by FACS analysis using phospho-H3 staining as a marker of the G2/M phase (Fig. 7A). Compared to the unique normal cell cycle profile of the Parp-1+/+;SirT1+/+ cells, we observed two populations in both Parp-1+/+;SirT1−/− cells and Parp-1−/−;SirT1−/− cells. Whereas a majority of the Parp-1+/+;SirT1−/− cells evidenced subtantial increase in DNA content as shown by a shift of the cell cycle profile towards increased propidium iodide staining, the Parp-1−/−;SirT1−/− cells rather displayed normal to lower DNA content. A wild-type-like cell cycle profile was detected in Parp-1+/−;SirT1−/− cells.
Fig. 7.
Increased DNA content in SirT1-deficient cells suppressed by the additional disruption of Parp-1. A FACS analysis of Parp-1+/+; SirT1+/+, Parp-1+/+;SirT1−/−, Parp-1+/−;SirT1−/−, and Parp-1−/−;SirT1−/− cells. The right hand part shows the identification of cells in G2/M by phosH3S10 staining versus DNA content. The majority of Parp-1+/+;SirT1−/− cells display an increased DNA content compared to the control Parp-1+/+;SirT1+/+ or the Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells. Some Parp-1−/−;SirT1−/− cells also evidence a lower DNA content. The profiles shown are representative of three independent experiments. B Karyotype analysis showing a significantly increased chromosome number in Parp-1+/+;SirT1−/− cells (b) compared to the control Parp-1+/+;SirT1+/+ (a) or the Parp-1+/−;SirT1−/− (c) and Parp-1−/−;SirT1−/− cells (d). At least 30 individual metaphases were counted per cell line. The mean value (±SD) of three independent experiments is shown on the histogram bars. The range of chromosome numbers is indicated on the top. Note, the identification of Parp-1−/−;SirT1−/− cells with a reduced number of chromosomes (n = 38) compared to Parp-1+/+;SirT1+/+ cells (n = 50). P value was calculated by ANOVA test: ***P < 0.001
To further verify this observation, we looked for chromosome number in each cell line by karyotype analysis (Fig. 7B). Consistent with the FACS data, the majority of the Parp-1+/+;SirT1−/− cells were near pentaploid (102–145 chromosomes), whereas the control SirT1+/+;Parp-1+/+ cells were hypotetraploid (50–87 chromosomes), a usual state of spontaneously immortalized MEFs. Interestingly, with the additional disruption of Parp-1, the number of chromosomes decreased again to control levels (range (56–90) for Parp-1+/−;SirT1−/− and range (38–82) for Parp-1−/−;SirT1−/−). Of note, these results are in agreement with the immunofluorescence studies showing that most of the Parp-1+/+;SirT1−/− cells displayed a significant greater nuclei size compared to the Parp-1+/+;SirT1+/+ or Parp-1+/−;SirT1−/− cells whereas a smaller nuclear size was observed for some Parp-1−/−;SirT1−/− cells (Fig. 3).
In addition, no spontaneous aneuploidy was detected in our models of Parp-1 single-deficient cell lines ([27, 36], and data not shown). This is in contrast to Kanai et al. [37] describing spontaneous hypoploidy (1N and 3N) and hyperploidy (8N) associated with centrosomes hyperamplification in the absence of Parp-1. This difference might be related to the different mouse models and backgrounds used to generate the cell lines.
Discussion
In a previous report, we established a tight functional link between the two NAD+ consuming enzymes Parp-1 and SirT1 that determine cell survival and response to genotoxic stress [26]. More recently, both proteins also emerged as key regulators of chromatin structure and function that drive various cellular processes in which both proteins are involved [3, 18, 24, 25].
In this study, we establish an important role of Parp-1 and SirT1 in mouse post-natal development and provide in vivo evidence for a possible functional interplay between both proteins in the definition of constitutive heterochromatin that implement nucleoli organization, mitotic division, and genomic stability.
Contribution of Parp-1 and SirT1 in postnatal development, cell growth and telomere protection
According to previous investigations [21, 22], we show here that approximately 50% of the mice carrying a deletion of SirT1 died at early post-natal stages revealing an essential role of SirT1 in mammalian development. Although the cause of the death remains uncertain, in the model of Cheng et al. [21] this lethality phenotype was associated with cardiac defects. Consistent with this observation, the inhibition of SirT1 deacetylase activity in stressed cardiomyocytes, or its reduction in conditions of robust Parp activation by oxidative stress and concomitant NAD+ depletion both contribute to cell death during heart failure [38–40]. In contrast, Parp-deficient mice are protected from angiotensin II-mediated cardiomyocyte cell death [41]. In addition, overactivation of Parp-1 in SirT1-deficient cells exposed to high levels of oxidative damage leads to AIF-mediated cell death by chromatinolysis [26]. Therefore, we reasoned that, if the postnatal lethality in SirT1 mutant mice can be attributed to Parp-1 overactivation, the simultaneous disruption of Parp-1 should provide a protective effect. Of note, no increased post-natal lethality is observed in Parp-1 mutant mice (this study and [27]). In contrast, our present data suggest that the combined disruption of Parp-1 and SirT1 in mice tends to increase the late post-natal lethality with only 2 out of 10 (20%) surviving into adulthood but with a smaller stature then that of the Parp-1+/+;SirT1−/− mice. What is the cause of the exacerbated lethality in the Parp-1−/−;SirT1−/− mice? In contrast to SirT1-deficient 3T3 cells, in which chromatin-associated abnormalities are suppressed by the additional disruption of Parp-1 (see below), this rescue event if occurring in Parp-1−/−;SirT1−/− mice is not enough to restore genome integrity in the whole organism. Interestingly, SirT1 has recently been shown to play an important role in cellular response to double-strand breaks as shown by a marked reduction in γH2AX, Rad51, BRCA1, and NBS1 foci formation and a reduced repair efficiency in SirT1-deficient cells exposed to γ-irradiation or oxidative stress [23, 42]. Given the key role of Parp-1 in single-strand break repair, it is tempting to speculate that the combined disruption of both DNA repair factors in mice is one of the major reason of post-natal lethality because of increased genomic instability. As such, the viability of the remaining Parp-1−/−;SirT1−/− mice could at least in part be assigned to the compensating activities of the DNA repair factor Parp-2 and the histone H4 deacetylase SirT2 being expressed to varying degrees in the mixed genetic background.
To counteract the problem of limited numbers of Parp-1−/−;SirT1−/− available mice and further investigate the functional interplay between both enzymes, we next decided to derive and characterize 3T3 cells of the different genotypes.
By analogy to the significantly reduced weight growth of the mutant mice, we observed marked decreased cell growth in SirT1-deficient cells that again was not reverted by the additional disruption of Parp-1. Even though different possible reasons could account for this observation, our data suggest a contribution of telomere instability in the slower growth rate observed and, as such, provide evidence for a possible participation of both proteins in telomere maintenance. Accordingly, in Saccharomyces cerevisiae, Sir2 is required for the establishment and maintenance of telomeric heterochromatin [43]. In addition, Parp-1 has previously been shown to favor telomere protection and synthesis [11, 12]. Alternatively, interesting explanations also come from the present characterization of the four cell lines as discussed below.
Impaired pericentric heterochromatin and nucleolar architecture associated with mitotic defects in the absence of SirT1
Previous reports have identified an important role of SirT1 in promoting heterochromatin formation through the coordination of several events, including the recruitment and deacetylation of histone H1 resulting in chromatin silencing and the interaction with and deacetylation of the HMTase Suv39h1 that facilitates the establishment of the repression mark 3HK9me3 onto pericentric heterochromatin [24, 25]. Accordingly, we show here that the absence of SirT1 results in dispersed 3HK9m3 localization in more than 80% of the Parp-1+/+;SirT1−/− cells resulting in perturbed enrichment of the heterochromatin protein HP1α and phosH3-S10 to these heterochromatic subdomains, thus indicating impaired pericentric heterochromatin organization, further confirmed by the electronic microscopy studies. Together, these results reveal that the absence of SirT1 causes dramatic disorganization of chromatin in parallel with changes in its epigenetic modifications.
Altered pattern of histone modifications at pericentric heterochromatin and the reduced accumulation of HP1α have recently been associated with abnormal nucleolar organization. The absence of the heterochromatin regulators Su (var) 3-9 HMTase, HP1, and the RNAi pathway in Drosophila resulted in the formation of multiple nucleoli [34]. According to these results and previous findings, we show here that the disruption of SirT1 in mouse cells induces the accumulation of multiple irregularly shaped nucleoli.
Accurate organization of pericentric heterochromatin is also of fundamental importance for mitotic fidelity and correct chromosome segregation. For example, Suv39 h-deficient mice display impaired heterochromatin integrity and chromosome missegregation in both somatic and germ cells [31]. Perturbed enrichment of HP1 to these chromatin regions has been associated to defective chromosome segregation in mitosis [44]. Finally, the disruption of SirT1 in mice or its inhibition by histone deacetylase inhibitors in tumor cells was shown to induce chromatin relaxation that consequently leads to aberrant mitosis [23, 45]. In line with these data, we show here that the combined deregulation of histone modifications (3HK9me3, phosH3-S10) and defective recruitment of the histone code mediator HP1 in SirT1-deficient cells result in increased chromosome missegregation in mitosis.
Altogether, the alterations in histone modifications combined with the desorganization of the nucleolus and the mitotic abnormalities that together contribute to genomic instability in SirT1-deficient cells, could all account for their significant reduced cell growth as observed.
In addition, these results suggest that SirT1 participates in the maintenance of higher-order pericentric heterochromatin structure, which is required to preserve the structural integrity of the nucleolus and chromosome segregation.
Rescue of SirT1 phenotype by the additional disruption of Parp-1
At the animal level, the combined disruption of Parp-1 and SirT1 favors late post-natal lethality and reduces weight growth. In contrast, at the cellular level, the additional disruption of Parp-1 reverts the chromatin-associated phenotypes of SirT1 and the resulting abnormalities in nucleoli integrity and chromosome segregation. This not only supports recent evidence that Parp-1 and its activity play pivotal roles in epigenetic events [3] but also suggest a subtle regulated crosstalk between Parp-1 and SirT1 that implement chromatin structure and function.
Further characterization of the functional link between both proteins and their activities represents an important step in understanding how they co-operate. The possibility exists that the Parp-1-dependent-poly(ADP-ribosyl)ation of specific histones at specific sites communicates with histone H4 acetylation by SirT1. In line with this hypothesis, recent studies point to a functional interplay between poly(ADP-ribosyl)ation and histone acetylation that regulate facultative X chromosome inactivation in male germ cells [19] or transcriptional activity in stimulated cortical neurons or cardiomyocytes [4]. Another intriguing question arising from this work is whether SirT1 deacetylates Parp-1 to regulate its chromatin-related functions. Indeed, recent work by Hassa et al. [46] has provided evidence for an in vivo acetylation of Parp-1 by the chromatin-modifying factor p300/CREB-binding protein that plays an important role on NF-kB-dependent gene activation. One can envisage that the maintenance of higher-order pericentric heterochromatin structure requires deacetylation of Parp-1 as a way to regulate the protein and/or inhibit poly(ADP-ribosyl)ation and favor chromatin condensation. Consistent with this, Parp-1 was found to be acetylated and activated under stress conditions whereas SirT1-mediated deacetylation inhibits the enzymatic activity of Parp-1 [47]. In addition, SirT1-deficient cells display increased DNA-damage-induced Parp-1 activity and heterochromatin disorganization characterized by deregulated histone modifications [23, 26]. As such, the additional disruption of Parp-1 would rescue both phenotypes to ensure nucleoli integrity and faithful mitosis as seen in the Parp-1+/−;SirT1−/− and Parp-1−/−;SirT1−/− cells. In contrast, no reversion of the retarded cell growth is observed in these cells possibly owing to the simultaneous absence of Parp-1-dependent processing of single-strand breaks [48] and SirT1-dependent repair of double-strand breaks [23, 42].
In conclusion, our findings provide insights into the in vivo functional link between the two NAD+-dependent enzymes Parp-1 and SirT1. They are in favor of important roles of both proteins (1) in cellular response to DNA damage and the maintenance of telomere integrity, as well as (2) in the modulation of histone modifications associated with heterochromatin formation and structure, a crucial process for nucleolus integrity and faithful chromosome segregation. Future studies are needed to figure out how the interplay between both molecules serves in shaping epigenetic programs and genome integrity.
Electronic Supplementary Material
Sup.Fig.1. Retarded growth of Parp-1;SirT1 double knockout mice. Average weight curves of Parp-1+/+;SirT1+/+ (◆), Parp-1+/−;SirT1+/− (◊), Parp-1+/+;SirT1−/− (●), Parp-1+/−;SirT1−/− (▲) and Parp-1−/−;SirT1−/− (×) females (left panel) and males (right panel) ; n, number of mice included in the weigth curves. Parp-1−/−;SirT1−/− mice were 30-40% smaller compared to controls. For the female, this represents an exacerbation of the 20-30% reduction seen in Parp-1+/+; SirT1−/− mice (this study and (McBurney, M.W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb, J.R., Lansdorp, P.M. and Lemieux, M. (2003). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23, 38-54.)). (PPT 206 kb)
(A) Normal patterns of H3K9me3 and HP1α in Parp-1-deficient cells. Representative immunofluorescence images for the comparative distribution of H3K9me3 (b,c) and HP1α (f,h) in Parp-1+/+;SirT1+/+ (a,b,e,f) and Parp-1−/−;SirT1+/+ (c,d,g,h) interphase cells. DNA and heterochromatic foci are counterstained with DAPI (a,c,e,g). Scale bars, 4,45 μm. (B) Dispersed distribution of H3K9me3 in Parp-1−/−;SirT1−/− cells reconstituted with Parp-1. Parp-1−/− ;SirT1−/− cells were transfected with either EGFP-Parp-1 or GST-Parp-1 and processed for immunofluorescence 36h later. a,d: DAPI-stained DNA. b: EGFP fluorescence. e: green labeled anti-GST antibody. c,f: red labeled anti anti-H3K9me3 antibody. Scale bars, 11 μm. (PPT 4483 kb)
Western-blot analysis for the expression of SirT1 and β-actin in SirT1-transfected cells. Equivalent amounts of total protein extracts from mock-transfected (lanes 1,3) or pCruz-HA-SirT1 transfected (lanes 2,4) Parp-1+/+;SirT1+/+ (lanes 1-2) and Parp-1+/+;SirT1−/− (lanes 3-4) 3T3 cells were separated by SDS-PAGE and analyzed by Western blotting with the appropriate antibodies. (PPT 80 kb)
Acknowledgments
We thank G. de Murcia and J.C. Amé for helpful discussions. This work was supported by funds from Centre National de la Recherche Scientifique, Université de Strasbourg, Agence Nationale de la Recherche and Ligue Nationale Contre le Cancer, Comité du Bas-Rhin.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle. 2005;4:96–101. doi: 10.4161/cc.4.1.1398. [DOI] [PubMed] [Google Scholar]
- 6.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouleau M, Aubin RA, Poirier GG. Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Sci. 2004;117:815–825. doi: 10.1242/jcs.01080. [DOI] [PubMed] [Google Scholar]
- 8.Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/S0070-2153(03)01007-X. [DOI] [PubMed] [Google Scholar]
- 9.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, de Murcia G, Schreiber V. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beneke S, Cohausz O, Malanga M, Boukamp P, Althaus F, Burkle A. Rapid regulation of telomere length is mediated by poly(ADP-ribose) polymerase-1. Nucleic Acids Res. 2008;36:6309–6317. doi: 10.1093/nar/gkn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol Biol Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 14.Saxena A, Wong LH, Kalitsis P, Earle E, Shaffer LG, Choo KH. Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum Mol Genet. 2002;11:2319–2329. doi: 10.1093/hmg/11.19.2319. [DOI] [PubMed] [Google Scholar]
- 15.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J Biol Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 16.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 17.Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, De Murcia G. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 18.Quenet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F. The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1alpha. FASEB J. 2008;22:3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- 19.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci USA. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 21.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 25.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 27.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennarun G, Granotier C, Hoffschir F, Mandine E, Biard D, Gauthier LR, Boussin FD. Role of ATM in the telomere response to the G-quadruplex ligand 360A. Nucleic Acids Res. 2008;36:1741–1754. doi: 10.1093/nar/gkn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolthur-Seetharam U, Teerds K, de Rooij DG, Wendling O, McBurney M, Sassone-Corsi P, Davidson I. The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod. 2009;80:384–391. doi: 10.1095/biolreprod.108.070193. [DOI] [PubMed] [Google Scholar]
- 30.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 32.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, Moreno V, Juarranz A, Stockert JC, Langst G, Grummt I, Bickmore W, Esteller M. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 36.Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol Cell Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 39.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 40.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 41.Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291:H1545–1553. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- 42.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/S0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R, Liu ST, Chen W, Bonner M, Pehrson J, Yen TJ, Adams PD. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol Cell Biol. 2007;27:949–962. doi: 10.1128/MCB.01639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens FE, Beamish H, Warrener R, Gabrielli B. Histone deacetylase inhibitors induce mitotic slippage. Oncogene. 2008;27:1345–1354. doi: 10.1038/sj.onc.1210779. [DOI] [PubMed] [Google Scholar]
- 46.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 47.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Konstatin B, Samant S, Hottiger MO, Gupta MP (2009) SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly (ADP-ribose) polymerase 1. Mol Cell Biol. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 48.Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup.Fig.1. Retarded growth of Parp-1;SirT1 double knockout mice. Average weight curves of Parp-1+/+;SirT1+/+ (◆), Parp-1+/−;SirT1+/− (◊), Parp-1+/+;SirT1−/− (●), Parp-1+/−;SirT1−/− (▲) and Parp-1−/−;SirT1−/− (×) females (left panel) and males (right panel) ; n, number of mice included in the weigth curves. Parp-1−/−;SirT1−/− mice were 30-40% smaller compared to controls. For the female, this represents an exacerbation of the 20-30% reduction seen in Parp-1+/+; SirT1−/− mice (this study and (McBurney, M.W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb, J.R., Lansdorp, P.M. and Lemieux, M. (2003). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23, 38-54.)). (PPT 206 kb)
(A) Normal patterns of H3K9me3 and HP1α in Parp-1-deficient cells. Representative immunofluorescence images for the comparative distribution of H3K9me3 (b,c) and HP1α (f,h) in Parp-1+/+;SirT1+/+ (a,b,e,f) and Parp-1−/−;SirT1+/+ (c,d,g,h) interphase cells. DNA and heterochromatic foci are counterstained with DAPI (a,c,e,g). Scale bars, 4,45 μm. (B) Dispersed distribution of H3K9me3 in Parp-1−/−;SirT1−/− cells reconstituted with Parp-1. Parp-1−/− ;SirT1−/− cells were transfected with either EGFP-Parp-1 or GST-Parp-1 and processed for immunofluorescence 36h later. a,d: DAPI-stained DNA. b: EGFP fluorescence. e: green labeled anti-GST antibody. c,f: red labeled anti anti-H3K9me3 antibody. Scale bars, 11 μm. (PPT 4483 kb)
Western-blot analysis for the expression of SirT1 and β-actin in SirT1-transfected cells. Equivalent amounts of total protein extracts from mock-transfected (lanes 1,3) or pCruz-HA-SirT1 transfected (lanes 2,4) Parp-1+/+;SirT1+/+ (lanes 1-2) and Parp-1+/+;SirT1−/− (lanes 3-4) 3T3 cells were separated by SDS-PAGE and analyzed by Western blotting with the appropriate antibodies. (PPT 80 kb)