Abstract
Aurora kinase A (AURKA) is an essential mitotic serine/threonine kinase and its abnormal expression is observed in many malignancies, yet the exact role for AURKA in tumorigenesis still remains elusive. Here, through a transcription factor array, we show that the transcription activity of E2F1 was increased by AURKA overexpression. Meanwhile, the E2F1 protein level was found to be upregulated and a correlation between AURKA and E2F1 expression was observed in cancer specimens. Further analysis revealed that AURKA increased E2F1 protein stability by inhibiting proteasome-dependent degradation of this protein. Additionally, a microRNA cluster, miR-17-92, was found to be upregulated upon AURKA overexpression, and this stimulation was largely repressed by E2F1 knockdown. Chromatin immunoprecipitation further demonstrated that AURKA enhanced E2F1 occupancy to the promoter of the miR-17-92 cluster. These data reveal a novel link between AURKA and microRNAs via the regulation of E2F1, providing new clues for understanding the role of AURKA in tumorigenesis.
Keywords: AURKA, miR-17-92, E2F1, Transcription factor, Tumorigenesis
Introduction
AURKA encodes an important member of the Aurora kinase family, an evolutionally conserved serine/threonine kinase that plays a crucial role in centrosome cycling during mitosis. Located on 20q13, a region frequently found to be amplified, AURKA amplification and overexpression have been observed in various types of tumor, including breast, colon, ovarian, bladder, hepatocellular, and pancreatic cancers [1–6]. The oncogenic activity of AURKA was first demonstrated by the ectopic expression of AURKA in rodent fibroblast cells, which led to full transformation of these cells [6]. Further investigations revealed that AURKA is involved in the regulation of a series of signaling molecules that have key roles in modulation of cell proliferation and apoptosis, such as p53 [7], BRCA1 [8], c-Myc [9], AKT [10], as well as NFκB pathways [11], indicating a complex network by which AURKA promotes cell transformation and tumorigenesis. However, despite the piles of literature, the exact mechanism by which AURKA contribute to tumor remains to be elucidated.
To further explore downstream events taking place after AURKA overexpression, we take advantage of a transcription factor array capable of monitoring the transcription activities of general transcription factors in the cells, and by this method, we identified a key transcription factor, E2F1, whose transactivity is upregulated in response to AURKA overexpression. We then further demonstrated that AURKA induces expression of a microRNA (miRNA) cluster, the miR-17-92 cluster, through the regulation of the E2F1 transcription factor.
Materials and methods
Tissue specimens
All specimens were collected from surgically excised tissues of breast cancer patients treated at the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China. The protocol was approved by the Institutional Review Board of the hospital.
Plasmids, cell culture, siRNA, and transfection
The AURKA expression plasmid was constructed by cloning AURKA cDNA into the pcDNA4™/TO/myc-His-B vector (Invitrogen) using the following primers: forward: 5′CGGGATCCATGGACCGATTAAAGAAAAC3′, reverse: 5′GCCTCGAGTAAGACTGTTTGCTAGC3′. AURKA kinase-dead (KD) mutant was created through K162M site-directed mutagenesis as described previously [12]. The 293TREX cell line was kindly provided by Professor Chen Quan and was cultured in DMEM, and the breast cancer cell lines, SK-BR-3 and MCF-7, as well as the esophageal cancer cell line EC9706, were cultured in RPMI 1640, supplemented with 10% fetal bovine serum, respectively. AURKA siRNA sequence: sense5′AUGCCCUGUCUUACUGUCATT3′, antisense5′ UGACAGUAAGACAGGGCAUTT3′; E2F1 siRNA sequence: sense5′GCAUCCAGCUCAUUGCCAATT3′, antisense5′UUGGCAAUGAGCUGGAUGCTT3′; the luciferase gene (GL2) siRNA sequence: sense5′ CGUACGCGGAAUACUUCGATT3′, antisense 5′UCGAAGUAUUCCGCGUACGTT3′. All cell transfections were performed with Lipofectamine2000 (Invitrogen) according to the manufacturer’s instruction. For stable cell-line establishment, cells were selected with Zeocin 24 h after transfection at a concentration of 100 μg/ml for 2 weeks.
Transcription factor array
Cells were grown to 70–80% confluence and induced with doxycycline for 24 h in medium with 0.5% serum. Nuclear extract was isolated using the Nuclear Extraction Kit (Panomics), and the array hybridization was performed using TranSignal™ protein/DNA arrays (Panomics) according to the manufacturer’s instructions as described previously [13].
Electrophoresis mobility shift assay (EMSA)
EMSA was performed using the DIG Gel Shift Kit (Roche Applied Science) according to the manufacturer’s protocol. Briefly, nuclear extracts were prepared from 293TREX–AURKA and 293TREX-Vector cells treated with or without doxycycline for 24 h. Two micrograms of nuclear extracts was incubated with the previously reported digoxigenin (DIG)-labeled E2F-binding oligonucleotide 5′ATTTAAGTTTCGCGCCCTTTCTCAA3′ [14]. The DNA/protein complex was then separated on a nondenaturing 6% polyacrylamide gel and then blotted by electro-blotting to a positively charged nylon membrane. The DIG-labeled DNA fragments were visualized by an enzyme immunoassay using anti-DIG-AP and the chemiluminescent substrates.
Reporter assays
Cells were cotransfected in 24-well plates with pGL3-6 × E2F-luciferase and pRL-SV40 as well as the indicated plasmids. Firefly and Renilla luciferase activities were measured consecutively by using the Dual Luciferase Assay (Promega) 24 h after transfection.
RNA extraction and real-time PCR
Total RNA was extracted with TRIZOL (Invitrogen) and reverse-transcribed to cDNA using MMLV Reverse Transcriptase (Promega). The quantitative real-time PCR of each sample was performed in triplicate on an ABI Prism 7000 (Applied Biosystems, CA) with SYBR Green PCR core reagents (Applied Biosystems, CA) according to the manufacturer’s protocol. β-actin was used as a reference gene. The relative transcript levels in the treated sample compared to the control sample were determined as fold changes. Primers: miR-17-92 cluster: Forward: 5′CTGTCGCCCAATCAAACTG3′, Reverse: 5′GTCACAATCCCCACCAAAC3′; β-actin: Forward: 5′GGCGGCACCACCATGTACCCT3′, Reverse: 5′AGGGGCCGGACTCGTCATACT3′.
Chromatin immunoprecipitation (ChIP)
ChIP assay was conducted with ChIP-IT™ Chromatin Immunoprecipitation Kits & Shearing Kits (Active Motif) according to the manufacturer’s protocol. The antibodies for immunoprecipitation include an anti-E2F1 (sc-193x, Santa Cruz) antibody or mouse IgG (Active Motif). Primers specific for putative E2F-binding sites upstream of the miR-17-92 cluster were reported previously [15]: P1 Forward: 5′GAAGAAGGAGGTGCTCCTGA3′; Reverse: 5′GCCCAGCTGCATTTAGTAAGA3′; P2 Forward: 5′ATTCACCCACATGGTCCTTC3′; Reverse: 5′GCCTGCGCTTTACTACGAC3′.
Immunoblotting and immunohistochemistry
Standard procedures were used for protein preparation and immunoblotting. The antibodies used are listed as below: anti-AURKA (IAK1, BD Biosciences), anti-ACTIN (Sigma-Aldrich), anti-laminB (Santa Cruz, sc-6217), anti-TFIID (Santa Cruz, sc-204). Immunohistochemistry was done as described previously. Specimens were incubated with anti-AURKA (Abcam, Ab1287; 1:700) and anti-E2F1 (Santa Cruz, sc-251; 1:150) at 4°C overnight following the antigen retrieval in pH9.0 EDTA buffer in an autoclave for 2 min. All immunostaining experiments were assessed by an experienced pathologist. The protein expression within the cancer tissue was evaluated and categorized according to the percentage and intensity of positive cells (−, +, ++, and +++). Tumors were then further grouped into weak (− and +) and strong (++ and +++) expression of each protein.
Statistical analysis
SPSS 15.0 for Windows (SPSS Inc.) was used for statistical analysis. Correlation between the AURKA expression levels and E2F1 expression profiles on a per-case basis was analyzed by Spearman’s rank test. Values of p < 0.01 were considered statistically significant.
Results
The profiling of transcription factors that responded to AURKA overexpression
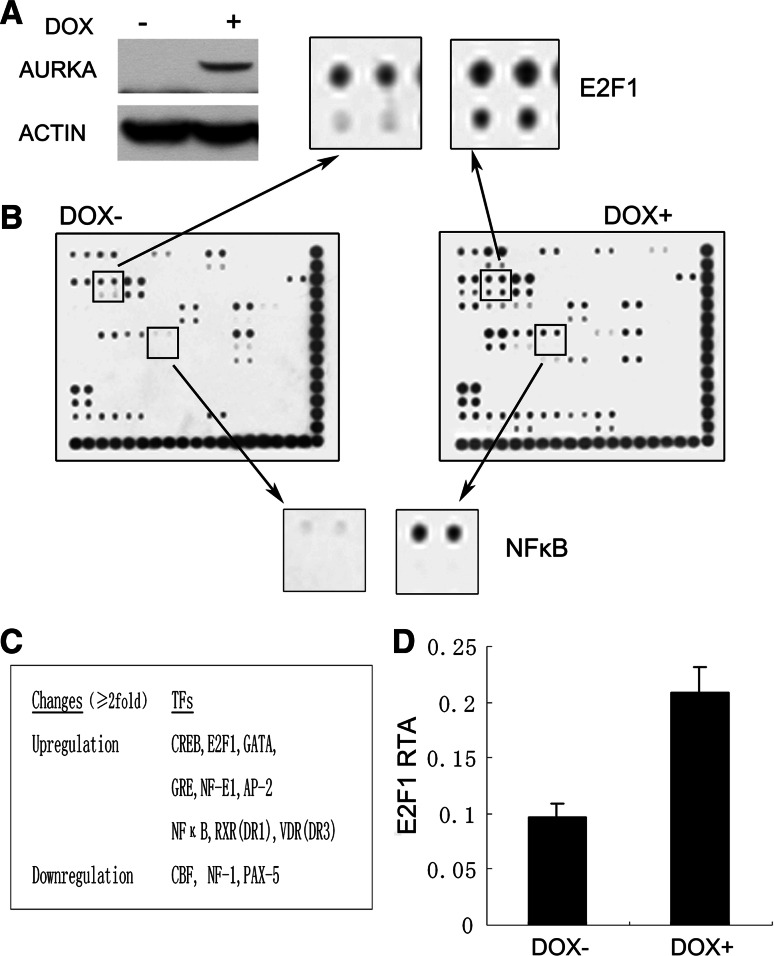
To get a better understanding of downstream events of abnormal AURKA expression in tumorigenesis, a transcription factor array that simultaneously profiles the activities of 54 transcription factors was employed to screen for the potential transcription factors that respond to AURKA induction and thus may be involved in AURKA-elicited tumorigenesis. An inducible cell line 293TREX–AURKA in which exogenous AURKA was efficiently induced by doxycycline (DOX) was generated (Fig. 1a). The nuclear extracts of the cells cultured in the presence or absence of DOX were subjected to the transcription factor array assay. As shown in Fig. 1b, c, several transcription factors significantly responded to AURKA expression (≥two-fold). The activities of nine transcription factors, including some general factors like CREB, GRE, and GATA were elevated, while three out of 54 had their transcription activities decreased. The changes of these global transcription factors might, to a certain extent, reflect the intensive impact of AURKA overexpression on the cell transcriptome and the cellular activities. A key transcription factor, NFκB was also shown to be upregulated after AURKA induction, consistent with a previous report in which NFκB pathway was discovered to be activated by AURKA [11], demonstrating the reliability of the transcription array.
Fig. 1.
Transcription factor activity profiling of 293TREX cells with high and low AURKA expression. a Immunoblotting analysis of AURKA expression in untreated and doxycycline(DOX)-treated 293TREX–AURKA cells. Actin was also probed to act as a loading control. b Transcription factor arrays hybridized with nuclear extracts from untreated (DOX−) or DOX-treated (DOX+) 293TREX–AURKA cells. Magnified panels show the spots representing E2F1 and NFκB transcription factors that were upregulated upon AURKA overexpression. c The summary of the transcription factors with significant changes (either upregulation or downregulation ≥two-fold) in their transcription activities upon AURKA induction. d Relative transcription activities (RTA) of E2F1 in DOX− and DOX+ cells after being normalized to respective internal controls. The data were depicted as mean ± SD (n = 4)
To our surprise, after being normalized to respective internal control, the spots representing the E2F1 transcription factor in DOX-treated cells also showed about a two-fold increase when compared to those of the DOX-untreated samples (Fig. 1b, d), suggesting an elevation of E2F1 transcription activity. A number of studies have shown that E2F1 plays an important role in the regulation of both cell proliferation and apoptosis [16, 17]. The active form of E2F1 controls expression of various genes required for DNA replication, as well as further cell-cycle progression [18, 19]. The particular status of E2F1 prompted us to investigate the potential role of AURKA in the regulation of E2F1.
AURKA increased E2F1 transcription activity
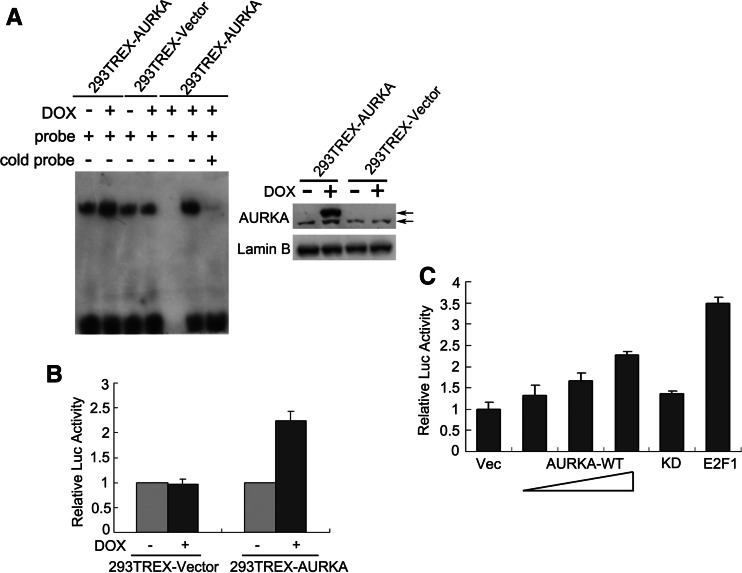
Electrophoresis mobility shift assay (EMSA) and reporter assays were then performed to confirm the array results. As shown in Fig. 2a, it is demonstrated by EMSA that induction of exogenous AURKA greatly enhanced the DNA-binding ability of E2F1. In contrast, there was little change in 293TREX-Vector cells in which the endogenous AURKA expression was unaffected by DOX treatment. AURKA overexpression in 293TREX–AURKA cells also led to increased transcription of luciferase reporter genes linked to the E2F1-binding sites (Fig. 2b), further suggesting a role of AURKA in the elevation of E2F1 transcription activity. To rule out the possibility of cell type-specific effects, EC9706, an esophageal cancer cell line, was also used to perform the transient transfection with AURKA expression plasmids. As shown in Fig. 2c, the reporter activity was upregulated by ectopic AURKA in a dose-dependent manner, while the kinase-dead (KD) mutant form of AURKA exhibited little effects, confirming that AURKA regulates E2F1 transcription activity, as well as suggesting that the kinase activity is critical for this regulation.
Fig. 2.
AURKA increased E2F1 transcription activity. a E2F1 DNA-binding ability was examined by electrophoresis mobility shift assay (EMSA) in nuclear extracts of 293TREX–AURKA cells. The lysates of 293TREX-Vector cells were examined as well to exclude the unspecific effects of DOX. The extracts of DOX-treated 293TREX–AURKA cells were also incubated either without probes or with probes in combination with excess cold probes to indicate the specificity of the probes. Right panel shows equal amounts of nuclear extracts were used in EMSA for the two cell lines, respectively. The upper and the lower arrows indicate exogenous and endogenous AURKA, respectively. b, c Reporter gene analysis with the pGL3-6 × E2F-luciferase plasmids. 293TREX–AURKA cells treated with or without DOX (b) and EC9706 cells transfected with 50 ng Vec, 50/100/200 ng AURKA-WT, 50 ng AURKA-KD and 50 ng E2F1 expression plasmids (c) were assayed for E2F-dependent luciferase activities. Firefly/Renilla luciferase ratios were used to calculate fold induction. Vec, pcDNA4 vector; KD, the kinase-dead mutant form of AURKA. All experiments were performed in triplicate and are depicted as mean ± SD
AURKA upregulated E2F1 protein expression
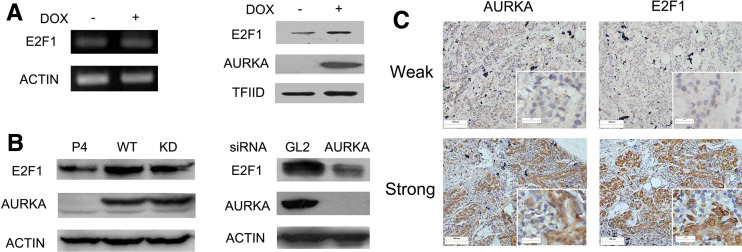
To reason how AURKA increased transactivity of E2F1, we examined E2F1 mRNA and protein levels. Surprisingly, the E2F1 mRNA level was not affected by AURKA overexpression (Fig. 3a). However, both the induction of AURKA in 293TREX–AURKA cells and the transient transfection of AURKA in SK-BR-3 breast cancer cells resulted in a similar increase of E2F1 on the protein level. Besides, in MCF-7 cells previously reported to harbor AURKA overexpression [20], knockdown of AURKA through RNAi efficiently reduced E2F1 protein (Fig. 3b), further confirming the modulation of E2F1 protein by AURKA. To verify the clinical relevance of the in vitro results regarding AURKA regulation over E2F1 protein, 100 samples from patients with primary breast tumors were examined by immunohistochemical staining. Examples of weak and strong staining for both AURKA and E2F1 are shown in Fig. 3c and the correlation between each paired immunohistochemical scores was analyzed with Spearman’s rank test. The result showed that there was a positive correlation between AURKA and E2F1 expression (p = 0.003; Table 1), further supporting a role for AURKA in the regulation of E2F1.
Fig. 3.
AURKA upregulated E2F1 protein expression. a E2F1 mRNA and protein levels in 293TREX–AURKA cells treated with/without DOX for 24 h. b E2F1 protein levels in SK-BR-3 cells transiently transfected with AURKA plasmids (left) and in MCF7 cells treated with AURKA siRNA for 48 h (right). P4, pcDNA4 vector; WT and KD, the wild-type and kinase-dead mutant form of AURKA, respectively; GL2 siRNA was used as a negative control for AURKA siRNA. c Immunohistochemical analysis of AURKA and E2F1 expression in breast cancer samples. Representative photographs of weak and strong staining for AURKA and E2F1 are shown. The original magnification: ×100 (pictures, size bars 100 μm), and ×400 (insets, size bars 20 μm)
Table 1.
Summary of AURKA and E2F1 expression in breast cancers
| AURKA staining | E2F1 staining | Total (%) | |
|---|---|---|---|
| Weak (%) | Strong (%) | ||
| Weak | 50 (50) | 18 (18) | 68 (68) |
| Strong | 14 (14) | 18 (18) | 32 (32) |
| Total | 64 (64) | 36 (36) | 100 (100) |
The level of AURKA and E2F1 expression were determined in 100 surgical specimens of primary breast cancer, as shown in Fig. 3c. The correlation was analyzed using the Spearman’s rank correlation test, p = 0.003
AURKA alleviated E2F1 proteasome-dependent degradation
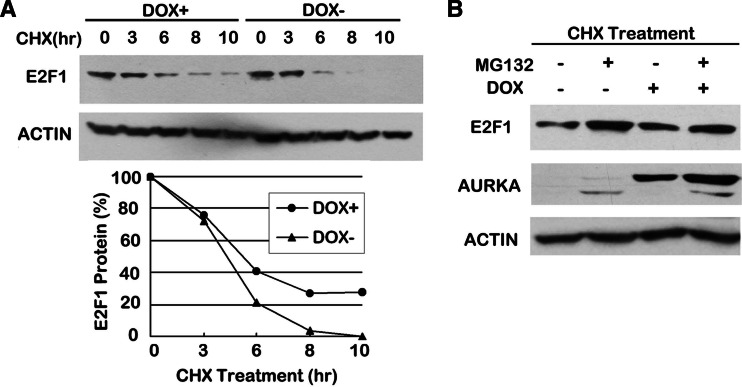
Since E2F1 was unchanged at the mRNA level, we next consider whether the degradation rate of E2F1 was altered by AURKA overexpression. To address this question, 293TREX–AURKA cells were exposed to cycloheximide (CHX), an inhibitor of protein synthesis, in time-course experiments. We estimated the relative amounts of E2F1 in cells treated with CHX for 3, 6, 8, and 10 h and found consistently higher levels of E2F1 in cells with AURKA induction than in cells without AURKA overexpression (Fig. 4a). Treatment with CHX for 6 h decreased E2F1 level by approximately 80% in the DOX– cells but only less than 60% in the DOX+ cells; after 10 h treatment, E2F1 expression was almost undetectable in DOX− cells while there were still about 20% of the original E2F1 level remaining in DOX+ cells, suggesting an impaired protein degradation rate of E2F1 when AURKA is overexpressed.
Fig. 4.
AURKA alleviated E2F1 proteasome-dependent degradation. a Lysates of 293TREX–AURKA cells treated with or without DOX were collected at the indicated time points after addition of CHX and subjected to immunoblotting. The amounts of E2F1 were quantified by densitometry and normalized to the corresponding actin expression. The degradation rate was shown by comparing the amounts of normalized E2F1 at each time point with the original levels (0 h) in DOX+ and DOX− cells, respectively. b Lysates of 293TREX–AURKA cells treated with or without DOX were collected 8 h after addition of CHX alone or in combination with MG132 and subjected to immunoblotting
As it has long been recognized that the turnover of E2F1 protein is through the ubiquitin-mediated proteosome pathway [21], we thus assess the effect of proteosome inhibition on E2F1 degradation in cells with/without DOX treatment. As shown in Fig. 4b, in the presence of CHX, treatment with proteosome inhibitor MG132 unsurprisingly resulted in an accumulation of E2F1 in both DOX− and DOX+ cells; however, the degree of restoration between the two samples varied significantly: cells overexpressing AURKA alleviated E2F1 degradation when exposed to CHX alone, thus generating only a little increase of E2F1 protein level when MG132 was added, while a more abrupt elevation of E2F1 was observed upon proteosome inhibition in the cells without AURKA induction. These data combined together indicate a role for AURKA in increasing the stability of E2F1 by inhibition of proteosome-dependent degradation of this protein.
AURKA induced miR-17-92 cluster expression
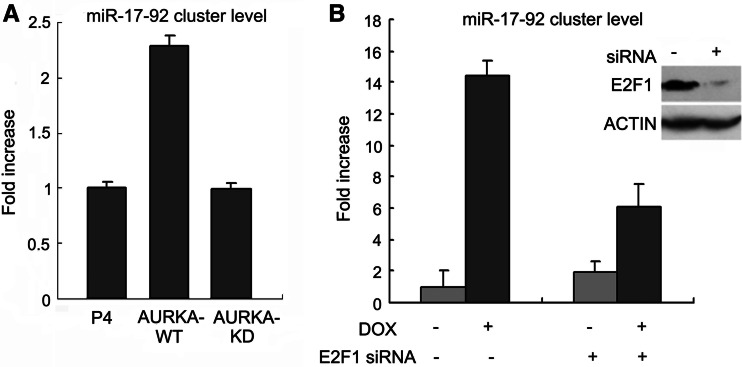
To study the functional consequences of E2F1 transcription activity elevation induced by AURKA, we examined the expression of the validated E2F1 target genes. The miRNA cluster miR-17-92 is an oncogenic polycistronic cluster of miRNAs that were previously identified to be regulated by E2F transcription factors [15, 22]. The oncogenic activity of the miR-17-92 cluster has been extensively studied recently [23]. We then tried to figure out whether AURKA is functionally linked to miR-17-92 cluster. As shown in Fig. 5a, transient expression of AURKA plasmids increased the expression of miR-17-92 cluster in SK-BR-3 breast cancer cells, while the KD mutant form of AURKA failed to induce the miR-17-92 cluster level, indicating the importance of kinase activity of AURKA in the regulation of miR-17-92 cluster. Furthermore, in 293TREX–AURKA cells, AURKA expression induced a more dramatic elevation of the miR-17-92 expression level, reaching to almost 15-fold of that in the DOX-untreated cells (Fig. 5b). However, when E2F1 was knocked down by siRNA, the induction of miR-17-92 by AURKA was greatly repressed to an extent as low as threefold. These results taken together demonstrated that AURKA was able to induce miR-17-92 cluster expression and E2F1 might contribute a large part to this modulation. The remaining induction caused by AURKA independent of E2F1 might probably be mediated by additional factors. Another oncoprotein, c-Myc, is likely to be the player, as Yang et al. [9] showed that c-Myc was upregulated by AURKA while O’Donnell et al. [24] reported that miR-17-92 cluster was also regulated by c-Myc. Further work is obviously required to validate this hypothesis.
Fig. 5.
AURKA-induced miR-17-92 cluster expression. a Real-time PCR analysis of the miR-17-92 cluster expression in SK-BR-3 cells transiently transfected with the indicated plasmids. All experiments were performed in triplicate and are expressed as mean ± SD. b Real-time PCR analysis of the miR-17-92 cluster expression in 293TREX–AURKA cells treated with E2F1 siRNA for 48 h in the absence or presence of DOX treatment. All experiments were performed in triplicate and are expressed as mean ± SD. The efficiency of E2F1 siRNA was tested by immunoblotting with GL2 siRNA as a negative control
AURKA enhanced in vivo binding of E2F1 to the promoter of the miR-17-92 cluster
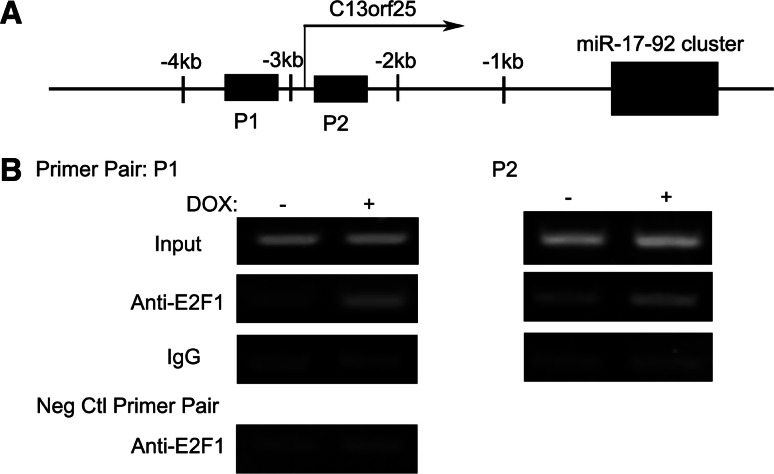
To confirm that AURKA-mediated regulation of the miR-17-92 cluster is indeed directly through E2F1, chromatin immunoprecipitation assay was performed using two amplicons, which were previously reported to overlap two different E2F-binding sites at miR-17-92 cluster promoter [15, shown in Fig. 6a]. As shown in Fig. 6b, with the DNA templates pulled down by an anti-E2F1 antibody, much stronger PCR amplifications were observed for both of the two amplicons in the DOX-treated cells than those in the control samples, while the unspecific immunoprecipitation with mouse IgG generated no obvious difference between the two samples, demonstrating that the in vivo-enhanced association of the endogenous E2F1 with the miR-17-92 promoter occurred upon AURKA overexpression. Therefore, the induction of the miR-17-92 cluster by AURKA may at least be in part through increasing the binding of E2F1 to the promoter.
Fig. 6.
AURKA-enhanced in vivo binding of E2F1 to the promoter of the miR-17-92 cluster. a Schematic representation of the promoter of the miR-17-92 cluster. The transcription start site of C13orf25 and the primer pairs (P1 and P2) flanking the putative E2F1-binding sites are indicated. b PCR analysis of E2F1 chromatin immunoprecipitates. Chromatins prepared from 293TREX–AURKA cells treated with DOX or not were subjected to immunoprecipitation with either an anti-E2F1 antibody or mouse IgG. Associated DNA was detected by PCR with two primer pairs: P1 (flanking E2F-binding site 1) and P2 (flanking E2F-binding site 2). Input DNA of the two samples were subjected to PCR to ensure that equal amounts of total DNA were used in the immunoprecipitation. A negative control primer pair flanking a genomic DNA sequence that should not be bound by E2F1 was also used to exclude the possibility that the variances are due to contamination of different amounts of genomic DNA through unspecific immunoprecipitation
Discussion
Recent studies unveiling substrates of AURKA have helped to elucidate the physiological functions of AURKA in centrosome cycling and cell mitosis, yet how this kinase promotes tumorigenesis still needs to be further explored. Concerning the importance of AURKA in chromosome segregation, it’s often tempting to speculate that centrosome aberrancy arising from abnormal AURKA might provide a route to aneuploidy and thereby contribute to tumor development. However, recent findings showed that some mitosis-related proteins are also involved in regulation of transcription either directly or through transcription factors so as to aid the pathogenesis of cancer. Securin, a mitotic protein that keeps separase from promoting sister chromatids separation before the onset of anaphase, is found to be a global transcription factor which is able to activate c-Myc, FGF2, and cyclinD3 in different cellular processes [25]. PLK1 is another important kinase that controls centrosome maturation. Interestingly, its expression is reported to repress p53 transcription activity but to activate the heat shock factor 1 (HSF1) transcription factor [26]. Similarly, AURKA has been reported to regulate two important transcription pathways in different cancer cells. Briassouli et al. [11] demonstrated that AURKA overexpression in HeLa cells resulted in phosphorylation of IκBα, which is critical for activation of the NFκB transcription pathway. Darr et al. [27] found that AURKA deregulation relieved β-catenin/TCF4 transcription pathway by increasing the Ser-9 phosphorylation of GSK3β in gastric cancer cells. Here, through transcription array profiles, we show that another general transcription factor E2F1 is also under the regulation of AURKA. AURKA overexpression inhibited the turnover of the E2F1 protein, and the accumulation further resulted in upregulation of E2F1 transcription activity. It is of note that the induction of E2F1 is not that dramatic (Figs. 1b, 3b, 4b), yet this may not be unexpected, as the overexpression and hyper-activation of E2F1 can lead to cell death rather than proliferation through p53-dependent apoptosis [17]. Importantly, the moderate accumulation of E2F1 we show here is sufficient to upregulate its transcription activity and elevate the expression of the downstream target miR-17-92 cluster, and therefore E2F1 may function in this manner to play a role in AURKA-induced tumorigenesis. This discrepancy may be explained by the “fine-tuning” theory that only the precisely appropriate oncogenic signals, no more and no less, are required for a cell on its way to transformation. There is currently no evidence for us to show how AURKA exactly functions to inhibit E2F1 degradation. However, there are several possibilities. Possibly, AURKA overexpression might induce phosphorylation and inactivation of some ligase in ubiquitin system, thus leading to impairment of E2F1 degradation. Alternatively, E2F1 stability was reported to be increased through phosphorylation at the ser-31 site by ataxia telangiectasia mutated (ATM) or ATM- and Rad3-related (ATR) kinase [28]. It is therefore interesting to explore whether AURKA has a role in this modulation.
Our data for the first time provide evidence that miRNAs are involved in the network of AURKA. Recent studies have uncovered the involvement of some miRNAs in the development of cancer. The amplification of the miR-17-92 cluster is seen in large-B cell lymphoma and in other malignant lymphomas, while miRNAs from this cluster are discovered to be overexpressed in several human solid tumors [29]. As a result, miRNAs from the miR-17-92 cluster are thought to be oncogenic miRNAs or “oncomirs”. Our discovery of AURKA regulation over the miR-17-92 cluster might therefore provide an additional mechanism for the role of AURKA in malignant transformation besides mitotic control. As miRNAs function through the RNA interference to regulate gene expression, it would be of great interest to explore which genes are targeted by miRNAs from the miR-17-92 cluster in this cascade. This knowledge would nevertheless have implications for further understanding the role of AURKA in tumorigenesis.
Acknowledgments
We thank Prof. Ashok R. Venkitaraman for kindly providing the pIRES-AURKA_KD-EGFP plasmid and Prof. Yu Zhang for the pGL3-6 × E2F-luciferase plasmid and E2F1 expression plasmid. This work was supported by National Natural Science Foundation (39925020, 30721001), National Basic Research Program (2004CB518701).
References
- 1.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin Cancer Res. 2004;10:2065–2071. doi: 10.1158/1078-0432.CCR-1057-03. [DOI] [PubMed] [Google Scholar]
- 3.Rojanala S, Han H, Munoz RM, Browne W, Nagle R, Von Hoff DD, Bearss DJ. The mitotic serine threonine kinase, Aurora-2, is a potential target for drug development in human pancreatic cancer. Mol Cancer Ther. 2004;3:451–457. [PubMed] [Google Scholar]
- 4.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12–q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 5.Watanabe T, Imoto I, Katahira T, Hirasawa A, Ishiwata I, Emi M, Takayama M, Sato A, Inazawa J. Differentially regulated genes as putative targets of amplifications at 20q in ovarian cancers. Jpn J Cancer Res. 2002;93:1114–1122. doi: 10.1111/j.1349-7006.2002.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumor amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 7.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by Aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 8.Sankaran S, Crone DE, Palazzo RE, Parvin JD. Aurora-A kinase regulates breast cancer associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 2007;67:11186–11194. doi: 10.1158/0008-5472.CAN-07-2578. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Ou CC, Feldman RI, Nicosia SV, Kruk PA, Cheng JQ. Aurora-A kinase regulates telomerase activity through c-Myc in human ovarian and breast epithelial cells. Cancer Res. 2004;64:463–467. doi: 10.1158/0008-5472.CAN-03-2907. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- 11.Briassouli P, Chan F, Savage K, Reis-Filho JS, Linardopoulos S. Aurora-A regulation of nuclear factor-kappaB signaling by phosphorylation of IkappaBalpha. Cancer Res. 2007;67:1689–1695. doi: 10.1158/0008-5472.CAN-06-2272. [DOI] [PubMed] [Google Scholar]
- 12.Anand S, Penrhyn-Lowe S, Venkitaraman AR. Aurora-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 13.Govindarajan B, Bai X, Cohen C, Zhong H, Kilroy S, Louis G, Moses M, Arbiser JL. Malignant transformation of melanocytes to melanoma by constitutive activation of mitogen-activated protein kinase kinase (MAPKK) signaling. J Biol Chem. 2003;278:9790–9795. doi: 10.1074/jbc.M212929200. [DOI] [PubMed] [Google Scholar]
- 14.Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 15.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 16.Cam H, Dynlacht BD. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell. 2003;3:311–316. doi: 10.1016/S1535-6108(03)00080-1. [DOI] [PubMed] [Google Scholar]
- 17.Pan H, Yin C, Dyson NJ, Harlow E, Yamasaki L, Van Dyke T. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/S1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 18.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 19.Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 20.Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 21.Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, DePinho RA, Livingston DM, Grossman SR. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci USA. 2001;98:4455–4460. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 23.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 25.Tong Y, Eigler T. Transcriptional targets for pituitary tumor-transforming gene-1. J Mol Endocrinol. 2009;43:179–185. doi: 10.1677/JME-08-0176. [DOI] [PubMed] [Google Scholar]
- 26.Martin BT, Strebhardt K. Polo-like kinase 1: target and regulator of transcriptional control. Cell Cycle. 2006;5:2881–2885. doi: 10.4161/cc.5.24.3538. [DOI] [PubMed] [Google Scholar]
- 27.Dar AA, Belkhiri A, El-Rifai W. The Aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–875. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 29.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]