Abstract
Accumulation of abnormal proteins and endoplasmic reticulum stress accompany neurodegenerative diseases including Huntington’s disease. We show that the expression of mutant huntingtin proteins with extended polyglutamine repeats differentially affected endoplasmic reticulum signaling cascades linked to the inositol-requiring enzyme-1 (IRE1) pathway. Thus, the p38 and c-Jun N-terminal kinase pathways were activated, while the levels of the nuclear factor-κB-p65 (NF-κB-p65) protein decreased. Downregulation of NF-κB signaling was linked to decreased antioxidant levels, increased oxidative stress, and enhanced cell death. Concomitantly, calpain was activated, and treatment with calpain inhibitors restored NF-κB-p65 levels and increased cell viability. The calpain regulator, calpastatin, was low in cells expressing mutant huntingtin, and overexpression of calpastatin counteracted the deleterious effects caused by N-terminal mutant huntingtin proteins. These results show that calpastatin and an altered NF-κB-p65 signaling are crucial factors involved in oxidative stress and cell death mediated by mutant huntingtin proteins.
Keywords: Nuclear factor κappa B, Oxidative stress, Calpastatin, Endoplasmic reticulum stress, Cell death
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease that causes severe motor and cognitive impairment and marked brain atrophy with loss of neurons, especially in caudate-putamen and the frontal lobes [1]. HD is caused by an expansion of CAG-repeats in the first exon of the IT15 gene [2]. Wild-type IT15 encodes for a large 350-kDa huntingtin protein that is expressed in most tissues, including the nervous system. In neurons, huntingtin expression is localized to neuronal cell bodies, axons, and dendrites [3, 4]. The biological functions of HD are not fully understood but both loss-of-function and gain-of-function following mutant huntingtin expression have been described [3]. Alterations in mitochondria and in other organelles with increased oxidative stress are linked to disease progress in HD [5, 6].
Mitochondria are the major source of reactive oxygen species (ROS) that in large amounts damage cell membranes and interfere with various cellular functions [7]. Increased levels of ROS are counteracted by antioxidant systems including the antioxidants superoxide dismutase (SOD) and thioredoxins (Trx) [7, 8]. Apart from mitochondria, the endoplasmic reticulum (ER) produces ROS during normal cell metabolism and protein folding [7, 8]. ER dysfunctions trigger the unfolded protein response (UPR) that is mediated by three proximal ER sensors: PRK-like ER protein kinase/pancreatic eIF2α kinase (PERK/PEK), the activating transcription factor-6 (ATF6), and the inositol-requiring enzyme-1 (IRE1) [9–12]. UPR is a cellular adaptation mechanism, but prolonged or severe UPR can also activate signaling pathways leading to cell death [13, 14], and the final outcome of the UPR activation is cell context-dependent.
We have recently shown that the expression of full-length and N-terminal huntingtin proteins with extended polyglutamine (polyQ) repeats cause ER stress [15]. Here we have analyzed the ER stress signaling further and studied the IRE1α pathway that is linked to the regulation of various protein kinases. We observed that the NF-κB-65-kDa protein, p65 (also called RelA) [16], is selectively downregulated in cells expressing N-terminal mutants and the disease-inducing variant of full-length huntingtin, while the p38-mitogen-activated protein kinase (MAPK) and c-jun-N-terminal kinase (JNK) are activated. NF-κB is an important transcription factor for cell survival and protection against cell stress [16]. We observed a decrease in antioxidant proteins with an increase in ROS production in cells having reduced NF-κB-p65 caused by mutant huntingtin proteins.
Materials and methods
Cell culture
PC6.3 neuronal cells were cultured in RPMI-1600 (Biochrom) medium supplemented with 5% fetal calf serum and 10% horse serum. Cells were transfected with expression vectors encoding different polyQ repeats of huntingtin exon-1 fused to enhanced green fluorescent protein (EGFP), or using full-length huntingtin constructs with 17Q and 75Q repeats (a kind gift from Dr. Saudou) using the Transfectin reagent (BioRad) as described before [15]. Controls cells were similarly transfected with EGFP plasmid (Clontech). In some experiments, the full-length huntingtin constructs were co-transfected with this plasmid (4:1). Expression plasmid for human calpastatin was from Toyobo Company, Japan; 1 μM MG132 (Calbiochem), 50 μM Boc-Asp-(Ome)-fluoromethylketone (BAF, Calbiochem), brefeldin A (Sigma), 1 mM dithiothreitol (DTT; Sigma), 5 mM N-acetyl cysteine (NAC; Calbiochem), 20 μM N-acetyl-Leu-Leu-Nle-CHO (ALLN; Calbiochem), 20 μM Z-l-Abu-CONH(CH2)3-morpholine (referred as CX295, Calbiochem) were added 2 h after transfection. ER stress was also induced by treating cells with 1 μg/ml tunicamycin (Sigma) or 2 μM thapsigargin (Sigma) for different times (4–16 h). Staurosporine was used in a concentration of 1 μM for 20 h.
Hoescht 33342 (Sigma) was employed to stain dying cells showing condensed and fragmented DNA [15]. More than 300 fluorescent cells in each well were analyzed and experiments were repeated five times. Statistical analyses were done using Student’s t-test.
Immunocytochemistry
PC6.3 cells plated on polylysine/laminine-coated coverslips were fixed for 20 min using 4% paraformaldehyde. Cells were incubated for 1 h in phosphate-buffered saline (PBS) containing 0.1% Triton-X-100 and 5% bovine serum albumin (Sigma), followed by incubation overnight with primary NF-κB-p65 (1:200, Santa Cruz Biotechnology) and nitrotyrosine antibodies (1:500, Upstate Biotechnology) as instructed by the vendor. Cells were washed using PBS, incubated for 1 h using Alexa594-conjugated secondary antibodies (1:500, Molecular Probes), counterstained for 1 min using Hoechst 33342 Blue (4 μg/ml; Sigma), and mounted in gel-mounting medium (Sigma). Controls without primary antibodies showed no staining.
Immunoblots
Cells were lysed in RIPA buffer (150 mM NaCl, 1% Triton-X-100, 0.5% sodium deoxycholate, 50 mM Tris–HCl and 0.1% SDS pH 8.0) and supplemented with protease inhibitors (Roche). Protein concentrations were determined (BioRad BC assay), and equal amounts (40 μg) were separated using SDS-PAGE, and blotted onto nitrocellulose filters (Amersham). Filters were blocked for 1 h in 5% milk-TBS followed by an overnight incubation at +4°C using primary antibodies. These included antibodies against p-JNK (diluted 1:250, Cell Signaling), p–c-jun (1:1,000, Cell Signaling), c-jun (1:2,000, Santa Cruz), p38 MAPK (1:1,000, Cell Signaling), p-p38 MAPK (1:2,000, Promega), extracellular signal-regulated kinase (ERK) 1:1,000, Cell Signaling), p-ERK (1:1,000, Cell Signaling), NF-κB-p65 (1:250, Santa Cruz), p100/p52 (1:1,000, Cell Signaling), IκB-α (1:1,000, Cell Signaling), Sod1 (1:1,000, Calbiochem), Sod2 (1:15,000, LabFrontier), Trx2 (1:1,000, LabFrontier), catalase (1:500, Abcam), α-spectrin (1:1,000, Chemicon), calpastatin (1:1,000, Santa Cruz), anti-human calpastatin (1:1,000, Sigma), huntingtin (1:1,000, Millipore), p-eIF2α (1:1,000, Cell Signaling), GFP (1:5,000, Roche), active caspase-3 p17 (1:1,000, Cell Signaling) and β-actin (1:1,000, Sigma). After washing, the filters were incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,500, Pierce), followed by detection using the enhanced chemiluminescent method (Pierce). Filters were stripped for 30 min at 60°C (62.5 mM Tris–HCl, pH 6.8, 100 mM ß-mercaptoethanol, 2% SDS) and reprobed with antibodies for β-actin (1:2,000; Sigma). Poly-ADP-ribose polymerase (PARP) Western blotting was done as previously described [15].
Polymerase chain reaction
RNA was prepared using the mammalian RNA purification kit (Sigma), and cDNA made by reverse transcription using 50 U SuperScript II reverse transcriptase as described earlier. PCR were started with a 5-min denaturation step at 95°C followed by 30 cycles carried out at 95°C for 60 s, 52° (Trx2)/58° (Calpastatin)/60° (Sod2, Sod1) for 60 s and 72°C for 90 s. The following primers were used:
Sod2: 5′-GCCTGCACTGAAGTTCAATG-3′ (forward primer)
5′-ATCTGTAAGCGACCTTGCTC-3′ (reverse primer)
Sod1: 5′-CAAGCGGTGAACCAGTTGTG-3′ (forward primer)
5′-TGAGGTCCTGCAGTGGTAC-3′ (reverse primer)
Calpastatin: 5′-GCCCCAAACTCTTCCTAAGC-3′ (forward primer)
5′-AGCATGGTCGATTCCCATAG-3′ (reverse primer)
ß-actin: 5′-CTTCAACACCCCAGCCATG-3′ (forward primer);
5′-GTGGTACGACCAGAGGCATAC-3′ (reverse primer)
ROS measurements
The ROS levels were measured by loading cells for 15 min with 10 μM dihydroethidium (DHE; Molecular Probes) followed by examination of cells using the fluorescence-activated cell sorter FACSAria (BD Biosciences) as described earlier [17]. The number of DHE-positive cells was measured by gating GFP-expressing cells using excitation at 488 nm and emission at 502 nm for GFP and 595 nm for DHE.
NF-κB reporter assay
PC6.3 cells in six-well plates were transfected with 0.5 μg of full-length huntingtin expression plasmids in conjunction with 0.5 μg of the NF-κB reporter plasmid, containing multiple NF-κB sites linked to firefly luciferase gene [17]; 0.02 μg Renilla luciferase pRL-TK was used as a control for transfection efficiency. Cells were harvested after 48 h using Passive Lysis Buffer, and the renilla and firefly luciferase activities were measured using the dual luciferase substrate and a luminometer (GloMax® 20/20, Promega, Biofellows, Helsinki, Finland) [17]. Results are shown as fold increase in firefly luciferase activity normalized to the Renilla luciferase activity.
Calcium measurements
Cells plated on polyornithine-laminin-coated glass coverslips were loaded with 4 μM Fura-2 for 20 min at 37°C in TBM (137 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1.2 mM MgCl2, 0.44 mM KH2PO4, 4.2 mM NaHCO3, 10 mM glucose, 1 mM probenecid ([p-(dipropylsulfamoyl)benzoic acid]) buffered with 20 mM Hepes, pH 7.4, rinsed once, and used immediately. Ca2+ measurements were performed at 35°C using Nikon TE2000 fluorescence microscope (20×/0.75 air objective) and Andor iXon 885 EM-CCD camera under the control of Nikon NIS Elements software with 4D extension. For Ca2+ imaging, the cells were excited with alternating 340- and 380-nm light and the emitted light collected through a 400-nm dichroic mirror and a 450-nm long-pass filter. Transfected cells were selected based upon GFP-fluorescence. For Mn2+ quench, weighted sum of fluorescence at 340 and 380 nm was used as described [18]. Additions were made by constant perfusion.
Quantifications and statistics
Immunoblots and nitrotyrosine stainings were quantified using the ImageJ software. Results are expressed as percentage of controls (mean ± SD, n = 3–5). Statistical analyses were done using Student’s t-test with Bonferroni for multiple comparisons. p < 0.05 was considered as significant.
Results
Expression of mutant huntingtin proteins selectively downregulates NF-κB-p65
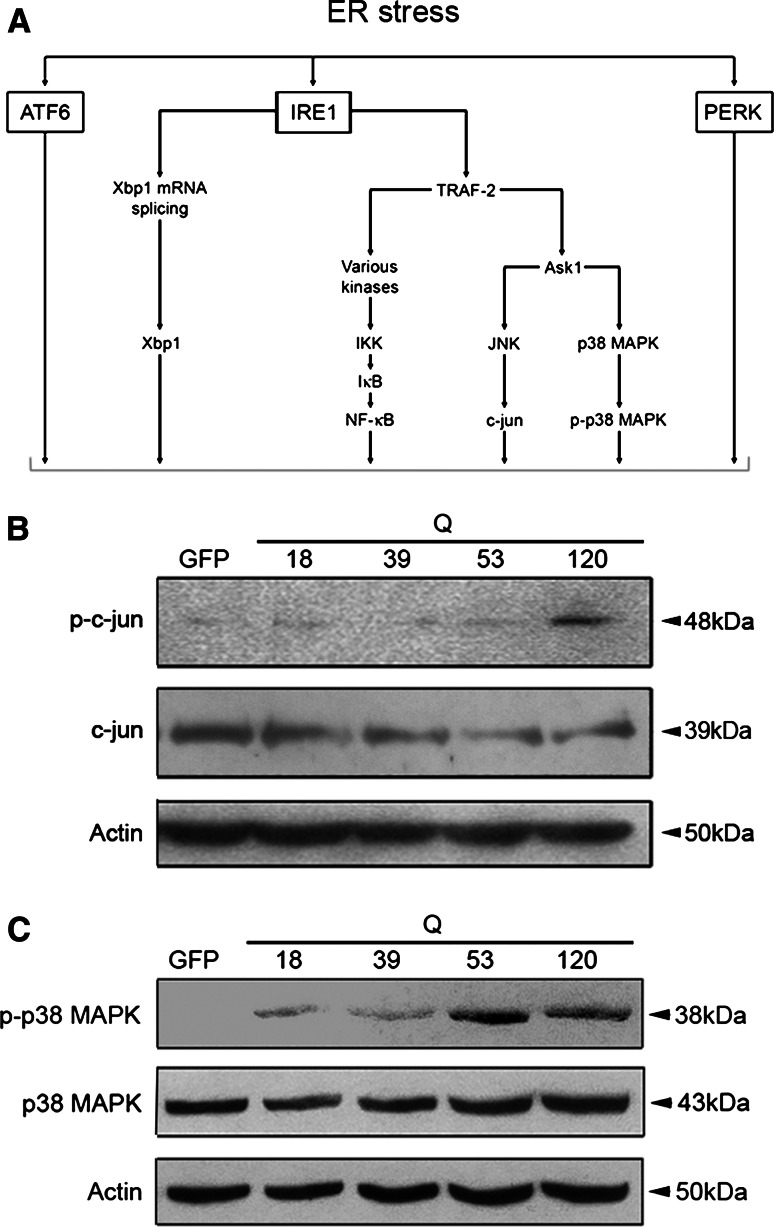
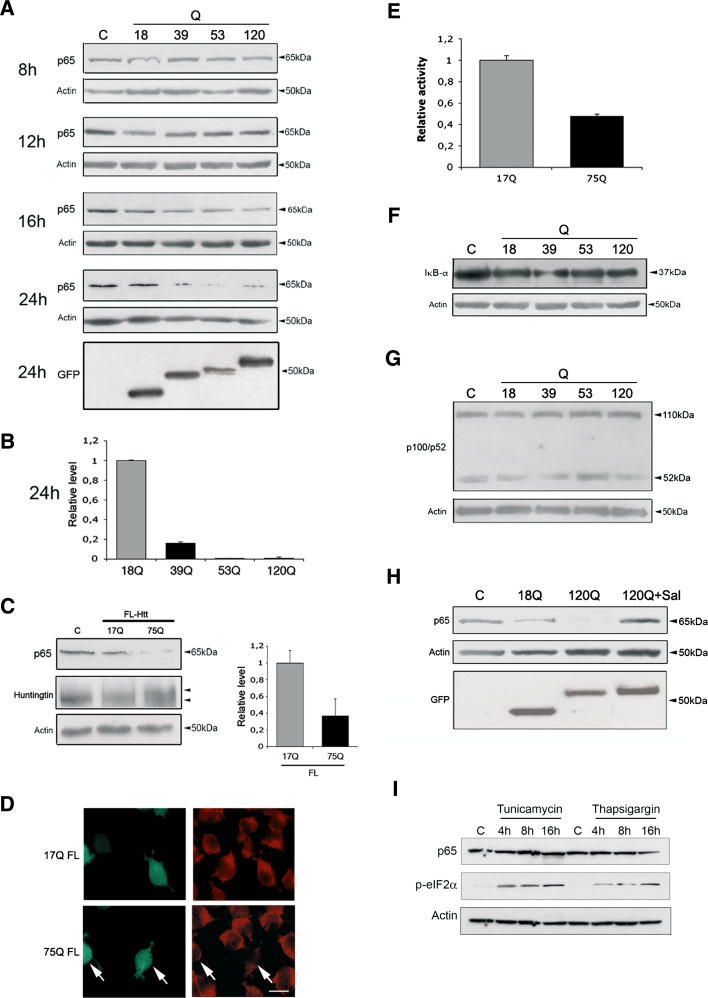
ER stress is known to activate distinct signaling pathways in the cell [9–12]. In this study, we focused on the IRE1α pathway that is involved in cell death and adaptation to cell stress (Fig. 1a). Data showed that expression of mutant N-terminal huntingtin fragment proteins activated the p38-MAPK and the c-jun-N-terminal kinase (JNK) pathways, as shown by increased protein phosphorylation (Fig. 1b, c). In contrast, at the same time the level of the NF-κB-p65 protein was decreased in the PC6.3 cells (Fig. 2a). The reduction in p65 levels was correlated with the length of the polyQ repeat in the huntingtin fragment proteins, and became evident after 16 h (Fig. 2a, b). To substantiate findings obtained with the N-terminal huntingtin fragment proteins, we expressed full-length huntingtin containing either a 17Q- or 75Q- repeat. Data showed that the disease-causing 75Q-repeat huntingtin reduced NF-κB-p65 levels, while the control protein did not (Fig. 2c). Immunostaining revealed that NF-κB-p65 immunoreactivity was preferentially reduced in cells overexpressing the full-length mutant huntingtin (Fig. 2d). Experiments using a NF-κB-luciferase reporter further showed a decrease in NF-κB activity in cells expressing this full-length mutant huntingtin, whereas the control protein had no effect (Fig. 2e). These data show that NF-κB signaling is affected in cells expressing mutant huntingtin proteins.
Fig. 1.
Analysis of the IRE1 pathway. Neuronal PC6.3 cells were transfected with expression constructs encoding different N-terminal huntingtin fragment proteins, and the activation of different kinases was analyzed after 24 h by immunoblotting as described in “Materials and methods”. a Schema of the IRE1-signaling pathways. b–c Immunoblots. Phosphorylation of c-jun and of p38 is increased in cells expressing the mutant huntingtin fragment proteins. ß-actin was used as control
Fig. 2.
NF-κB pathway is downregulated by mutant huntingtin proteins. a Cells were treated as above followed by immunoblotting. NF-κB-p65 levels decreased after 16 h in cells expressing the mutant N-terminal huntingtin proteins. C = EGFP-transfected cells. Lowermost panel shows expression levels for GFP-huntingtin constructs. b Quantification was done as described in “Materials and methods”. **p < 0.01 for proteins containing 39Q, 53Q and 120Q versus 18Q repeats, n = 4. ß-actin was used as control. c Cells were transfected for 48 h with full-length huntingtin constructs having either 17 (17Q) or 75 polyglutamine (75Q) repeats. NF-κB-p65 levels decreased in cells expressing the disease-causing 75Q-huntingtin. Left immunoblotting. Right quantification. **p < 0.01 for 75Q versus 17Q, n = 3. d Immunostaining. Cells were co-transfected with full-length huntingtin constructs and EGFP (green fluorescence) followed by immunostaining. NF-κB-p65 immunoreactivity (red) decreased in cells expressing the 75Q-huntingtin but not in those expressing the 17Q-huntingtin. Scale bar 10 μm. e The NF-κB reporter assay was performed as described in “Materials and methods”. Data show relative luciferase activity (firefly normalized to Renilla). Quantification showed a decrease in NF-κB gene activity upon expression of 75Q-huntingtin. **p < 0.001 for 75Q versus 17Q, n = 3. f–g Immunoblot. No changes in IκB levels or in the p100/p52 NF-κB pathway upon expression of N-terminal huntingtin proteins. h Immunoblot. Salubrinal increases NF-κB-p65 levels reduced by the 120Q-huntingtin fragment protein. **p < 0.01 for salubrinal + 120Q versus 120Q, n = 3. GFP-N-terminal huntingtin expression levels are shown with GFP Western blotting. i Immunoblot. Neither tunicamycin nor thapsigargin affects NF-κB-p65 levels. ER stress is induced as shown by increases in phosphorylation of eIF2alfa
NF-κB is a signaling factor that can be activated in cells either via the classical inhibitor of κB-p65- (IκB-) dependent pathway or by the non-classical NF-κB2 pathway, involving cleavage of the 100-kDa protein (p100) [16]. IκB levels were not altered by the mutant huntingtin protein (Fig. 2f) nor was the NF-κB2 pathway affected, as shown by unchanged levels of p100 and p52 (Fig. 2g). This demonstrates that the p65 protein is selectively decreased in cells expressing mutant huntingtin proteins leading to a reduced NF-κB activity.
We then studied the link between ER stress and NF-κB-p65 protein using salubrinal that is an inhibitor of ER stress [19]. Salubrinal was shown to counteract cell degeneration produced by N-terminal mutant huntingtin proteins [15]. We observed that salubrinal also inhibited the decrease in NF-κB-p65 induced by mutant huntingtin fragment proteins (Fig. 2h), indicating an involvement of ER stress in this process. Treatment of cells with either tunicamycin or thapsigargin to chemically induce ER stress did not downregulate NF-κB-p65 (Fig. 2i). This shows that the decline in NF-κB-p65 protein was specifically caused by ER stress induced by the expression of the mutant huntingtin proteins.
Lack of NF-κB-p65 sensitizes cells to mutant huntingtin-induced cell death
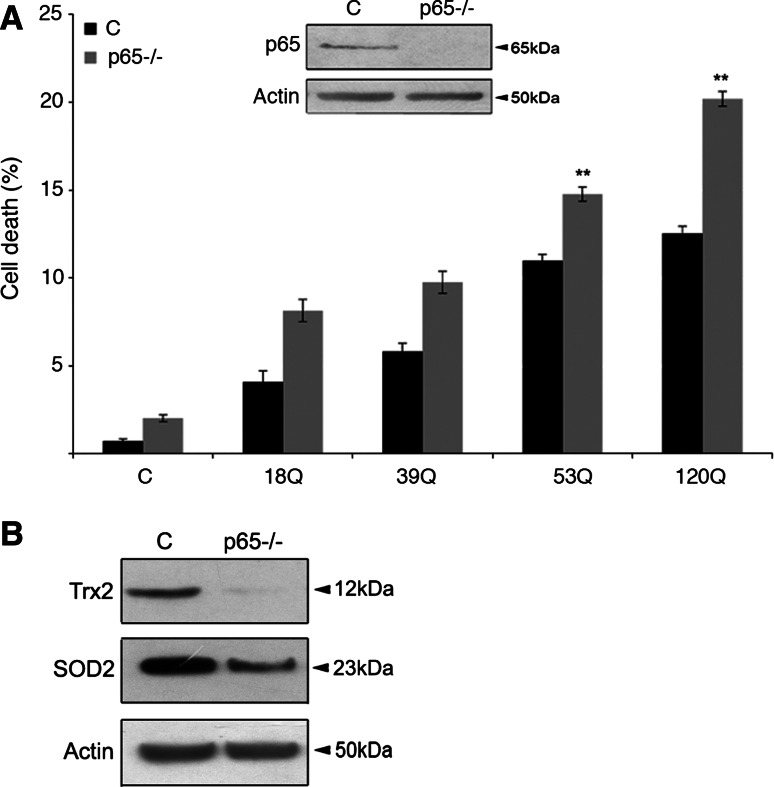
To study the involvement of NF-κB-p65 in mutant huntingtin-mediated cell death further, we analyzed p65-deficient mouse embryonic fibroblasts (MEFs) [17, 20]. Expression of N-terminal mutant huntingtin proteins induced cell death in wild-type MEFs, which increased with the length of the polyQ repeat (Fig. 3a). However, cell death was significantly enhanced in p65-deficient MEFs and the largest increase was observed with the 120Q-huntingtin fragment protein. Quantification showed that in this case cell death occurred in about 20 ± 2% of the p65-deficient cells compared to 11 ± 2% in controls (p < 0.05; Fig. 3a). In the p65-deficient fibroblasts, the levels of the antioxidants Trx2 and Sod2 were lower compared with control cells (Fig. 3b).
Fig. 3.
Lack of NF-κB-p65 sensitizes cells to mutant huntingtin protein-induced cell death. a Cell death was assessed by counting the number of apoptotic nuclei in control (C) and p65-deficient (p65−/−) fibroblasts as described in “Materials and methods”. Cell death increased in p65−/− cells transfected with the 53Q- and 120Q-huntingtin fragment proteins. **p < 0.01 for p65−/− versus c, n = 5. Insert shows immunoblots for NF-κB-p65. b Immunoblot. Note that the antioxidants Trx2, and Sod2 are decreased in p65−/− cells. Immunoblotting was repeated three times and typical experiment is shown
Downregulation of NF-κB-p65 results in decreased levels of antioxidants and in increased oxidative stress
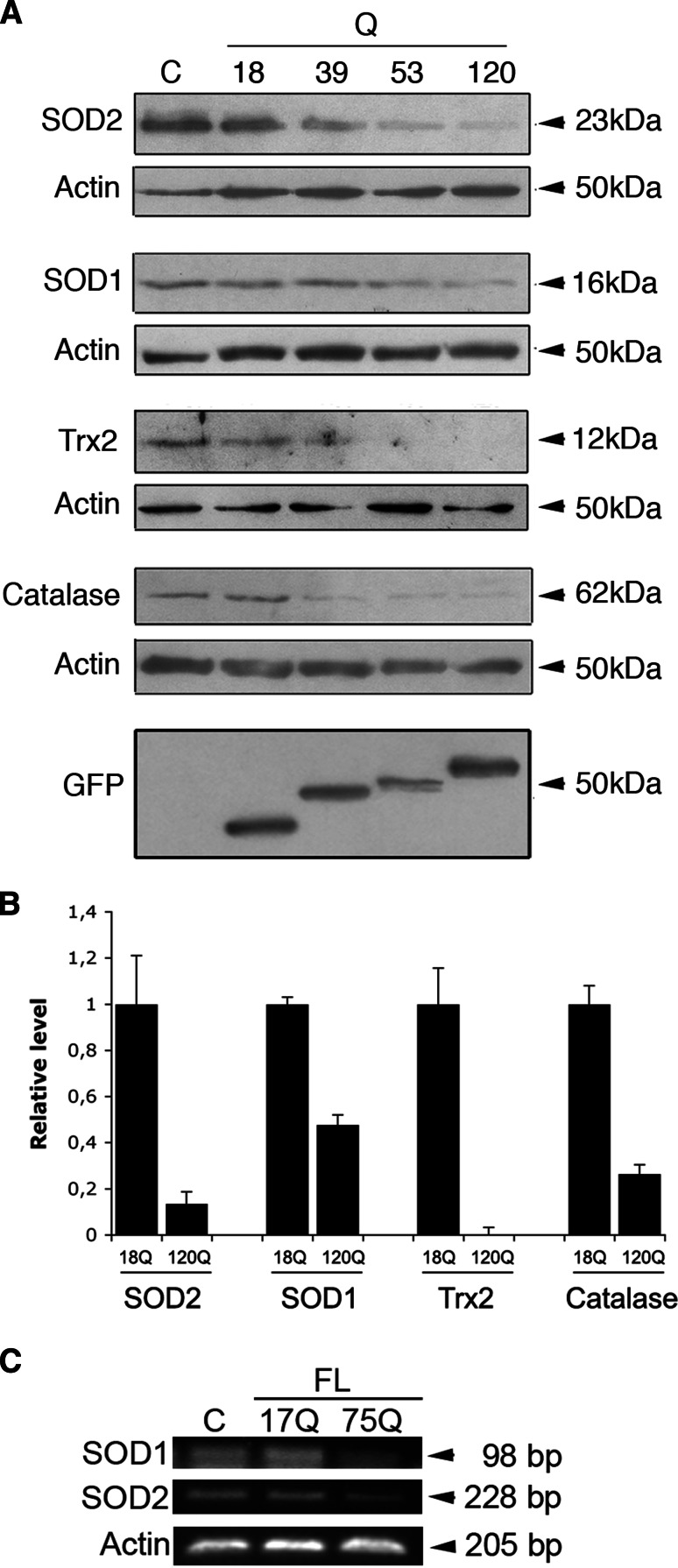
NF-κB is an important transcription factor that regulates different genes in protection against cell stress [16]. We have previously shown that particularly the mitochondrial antioxidants, Sod2 and Trx2, are regulated by NF-κB activation in the PC6.3 cells [17]. We therefore studied whether the antioxidant protein levels are altered subsequent to decrease in NF-κB-p65 induced by mutant huntingtin proteins. Data showed that Sod2 and Trx2 as well as the cytosolic antioxidants Sod1 and catalase were all significantly downregulated by the N-terminal mutant huntingtin proteins (Fig. 4a, b). Particularly, in cells expressing the 120Q-containing huntingtin fragment protein the levels of Sod2 and Trx2 were dramatically decreased (Fig. 4a, b). In addition, the mRNA levels decreased after expression of the disease-causing 75Q-huntingtin, as studied here for Sod1 and Sod2 (Fig. 4c). This shows that both mRNA and protein levels of important cellular antioxidants are decreased by mutant huntingtin proteins concomitant with the reduction in NF-κB-p65 levels.
Fig. 4.
Decreased cellular antioxidants after expression of mutant huntingtin proteins. a Immunoblots. The antioxidants Sod2, Sod1, Trx2, and catalase decreased in cells expressing mutant N-terminal fragment huntingtin proteins. ß-actin was used as control. GFP-N-terminal huntingtin expression is shown in the lowermost panel. b Quantification. Expression levels are normalized to that in cells expressing the 18Q-huntingtin fragment protein. Values are mean ± SD. p < 0.05 for 18Q- versus 120Q-protein-expressing cells, n = 3. c Cells were transfected with full-length huntingtin constructs (FL) for 24 h, and mRNA levels were analyzed by using semiquantitative RT-PCR as described in “Materials and methods”. Note a decrease in Sod2 and Sod1 mRNA levels by the disease-inducing 75Q-huntingtin. Immunoblotting was repeated three times and the typical experiment is shown
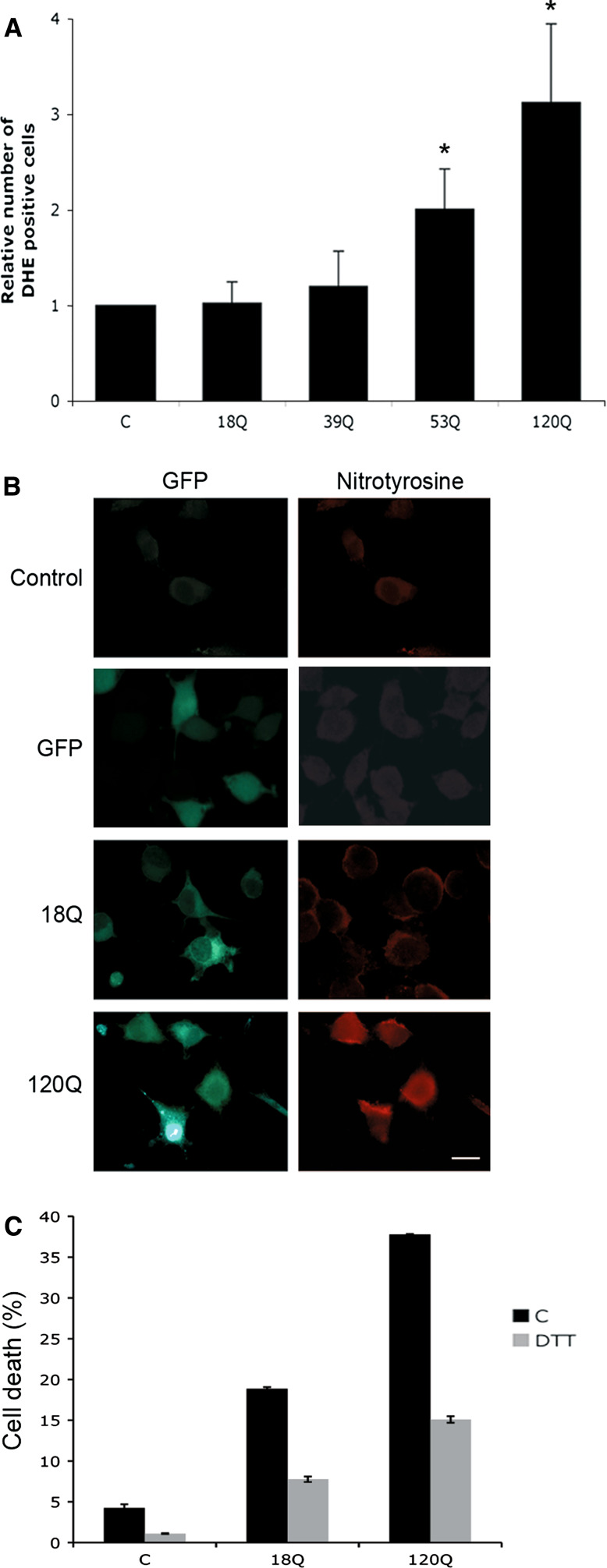
The decreased levels of antioxidants were further associated with an increased production of ROS in cells expressing the 53Q- and 120Q-huntingtin fragment proteins as shown using the compound dihydroethidium (DHE) in living cells (Fig. 5a), and using anti-nitrotyrosine antibodies for immunostaining (Fig. 5b). Increased production of ROS caused cell degeneration, and antioxidants, including DTT (Fig. 5c) and NAC (data not shown), significantly decreased cell death induced by the 120Q-huntingtin fragment protein.
Fig. 5.
Increased ROS production in cells expressing mutant huntingtin proteins. a Cells were transfected for 24 h and levels of ROS were measured using FACS as described in “Materials and methods”. *p < 0.05 for 53Q and 120Q-huntingtin fragment protein versus 18Q-protein, n = 3. b Immunostaining. Nitrotyrosine immunoreactivity (red) increased in cells expressing 120Q-huntingtin protein (green) compared with non-transfected control cells or those expressing 18Q N-terminal huntingtin or GFP protein. Scale bar 10 μm. c Treatment of cells with the antioxidant DTT increased cell survival. p < 0.01 for DTT + 120Q versus 120Q alone, n = 5
Role of calpain activity in NF-κB-p65 downregulation
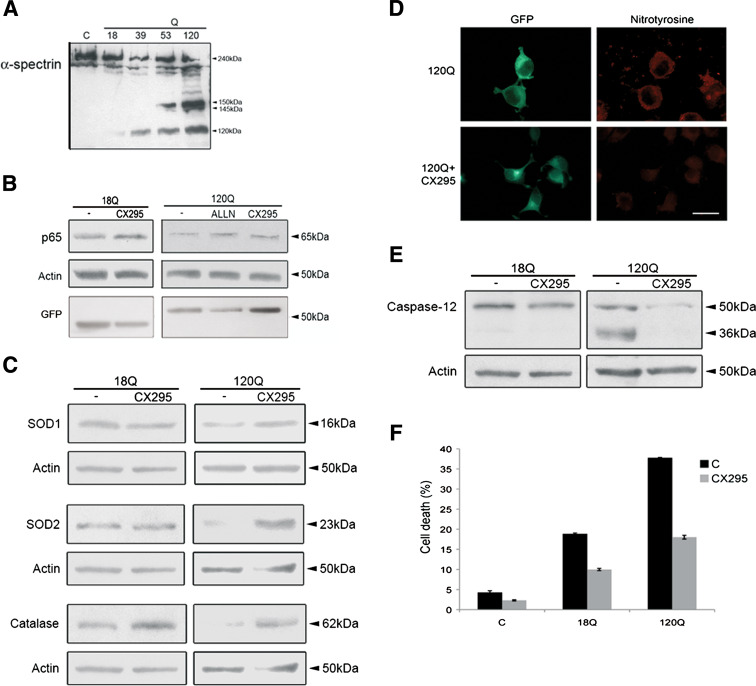
To study the mechanisms underlying the observed decrease in NF-κB-p65, we analyzed the effects of different compounds. The protease calpain is activated during ER stress [21] and could potentially influence NF-κB-p65. Immunoblotting showed the presence of the 145-kDa fragment of α-spectrin that is cleaved by calpain in cells expressing the 53Q- and 120Q- huntingtin fragment proteins (Fig. 6a). Treatment of cells with the calpain inhibitors, ALLN and CX295 [22], counteracted this decrease in NF-κB-p65 (Fig. 6b). The levels of the antioxidants Sod1, Sod2 and catalase also increased in cells treated with CX295 (Fig. 6c). As shown using anti-nitrotyrosine antibodies, CX295 reduced oxidative stress induced by the 120Q-containing huntingtin fragment protein (Fig. 6d). CX295 also counteracted the cleavage of pro-caspase-12 to the 36-kDa fragment, which occurs during ER stress (Fig. 6e), and increased cell survival (Fig. 6f). These results suggest that increased calpain activity is involved in the regulation of NF-κB-p65 and in subsequent oxidative stress produced by mutant huntingtin proteins.
Fig. 6.
Increased calpain activity regulates NF-κB-p65 levels and oxidative stress in cells expressing mutant huntingtin proteins. a Immunoblot for α-spectrin. Note the appearance of the calpain-specific 145-kDa cleavage product of α-spectrin in cells expressing 53Q- and 120Q-huntingtin fragment proteins. b Immunoblot. The calpain inhibitors, ALLN and CX295, restored NF-κB-p65 levels. ß-actin was used as control. p < 0.01 for 120Q + ALLN and for 120Q + CX295 versus 120Q, n = 3. Expression of GFP-N-terminal huntingtin is shown in the lowermost panel. c Immunoblots. CX295 prevented restored antioxidant levels. p < 0.05 for 120Q + CX295 versus 120Q for Sod1, Sod2 and catalase, n = 3. d Immunostaining. Nitrotyrosine immunoreactivity (red) is decreased by CX295 in cells expressing the 120Q-containing huntingtin protein (green). Scale bar 10 μm. e Immunoblots. Cleavage of caspase-12 (36-kDa) caused by the 120Q-huntingtin fragment protein is reduced using CX295. f Cell death induced by the 120Q-huntingtin fragment protein was decreased using CX295. p < 0.01 for 120Q + CX295 versus 120Q, n = 5
Calpastatin downregulation and the role of calcium in mutant huntingtin protein-expressing cells
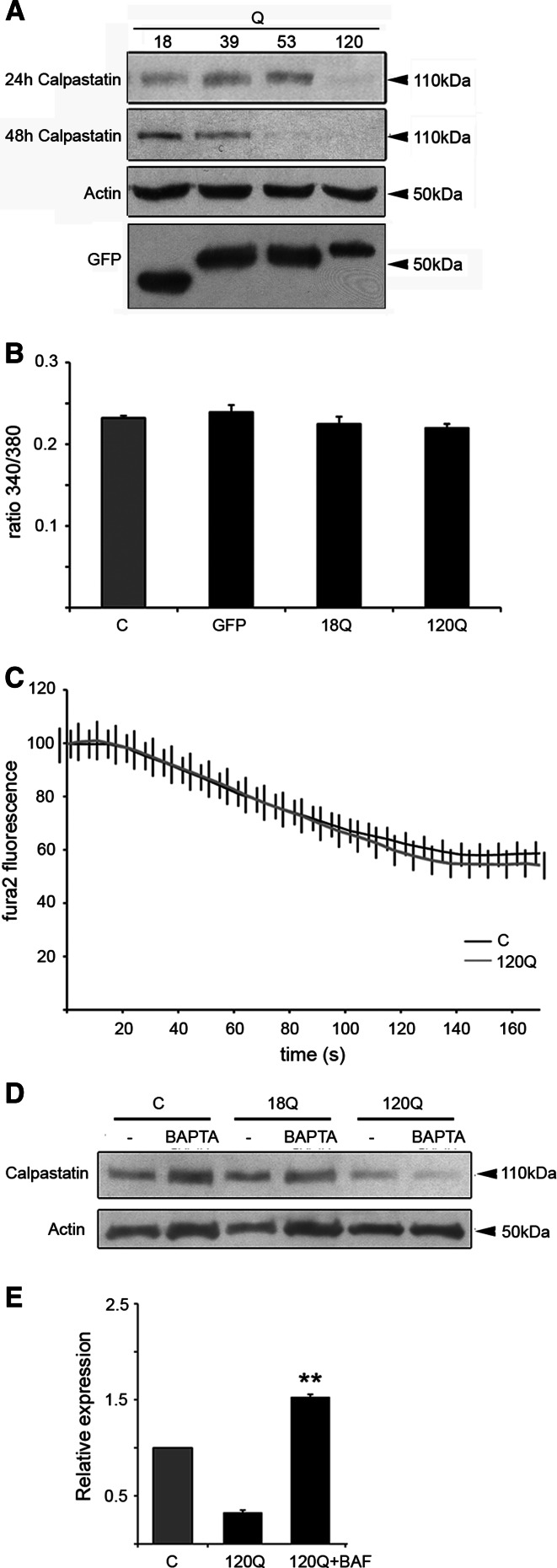
We then studied whether the endogenous calpain inhibitor calpastatin plays a role in the effect of the mutant huntingtin on calpain activity. Data showed that the levels of calpastatin decreased in cells expressing the 120Q-huntingtin fragment protein at 24 h, and at 48 h also in cells expressing 53Q (Fig. 7a).
Fig. 7.
Calpastatin levels decreased in cells expressing mutant huntingtin proteins. a Immunoblot. The levels of calpastatin decreased in cells expressing 120Q-huntingtin fragment protein at 24 h. Decrease in calpastatin levels was also detected in 53Q-expressing cells at 48 h. ß-actin was used as the control. Immunoblotting was repeated three times and a typical experiment is shown. Expression of GFP-N-terminal huntingtin is shown in the lowermost panel. b Ca2+ measurements in transfected cells were done as described in “Materials and methods”. Basal Ca2+ levels are given as the ratio of the fura-2 fluorescence at the excitation wavelengths 340 and 380 nm (“ratio 340/380”). c Mn2+ influx measurements were done as described in “Materials and methods”. The data are given as weighted sum of fluorescence at 340 and 380 nm. Mn2+ significantly quenched the fluorescence but there was no difference in the quenching rate or degree between control and 120Q- huntingtin fragment protein-expressing cells. d Immunoblot. Treatment with BAPTA-AM to reduce calcium levels does not inhibit the reduction in calpastatin levels. e Cells were treated with 50 μM BAF to inhibit cellular caspases. BAF restored calpastatin levels reduced by the 120Q-containing huntingtin protein. **p < 0.01 for 120Q + BAF versus 120Q, n = 3
Apart from calpastatin, calpain can be regulated by increased intracellular calcium [22–24]. Particularly μ-calpain is sensitive to increased Ca2+, whereas activation of m-calpain is also mediated by ERK activation [23, 24]. We observed a transient ERK phosphorylation after expression of mutant huntingtin proteins (data not shown). In contrast, there was no significant difference between control and mutant huntingtin fragment protein-expressing cells with regard to intracellular Ca2 (Fig. 7b). However, intracellular Ca2+ levels are tightly regulated by various extrusion and sequestration mechanisms, and local changes can be hard to detect. The Mn2+ quench technique is useful to detect subtle Ca2+ influxes, as Mn2+ is not removed from the cytosol by means of Ca2+ pumps [25]. In our experiments, essentially all cells showed a basal Mn2+ influx with no difference between control and 120Q-huntingtin fragment protein-expressing cells (Fig. 7c). These data indicate that under the present conditions, mutant huntingtin fragment proteins do not significantly elevate cytosolic Ca2+ nor do they influence the Ca2+ influx. In line with this, the compound BAPTA-AM that chelates intracellular Ca2 did not restore the calpastatin level reduced by the 120Q-huntingtin fragment protein (Fig. 7d). In studying the underlying mechanisms, we observed that calpastatin mRNA did not change (data not shown), indicating an effect on the degradation of the protein. However, the pan-caspase inhibitor BAF was able to restore the levels of calpastatin in cells expressing the mutant huntingtin protein (Fig. 7e), which suggests that cellular caspases are involved in the regulation of calpastatin under these conditions.
Calpastatin counteracts effects produced by N-terminal mutant huntingtin proteins
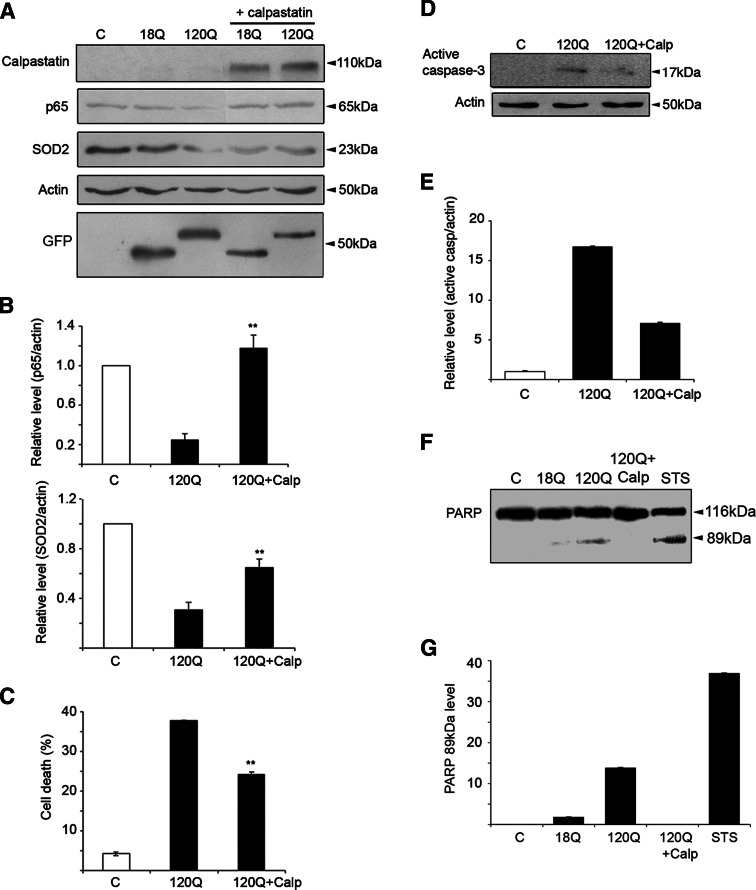
To study whether calpastatin may protect against the adverse effects of the mutant huntingtin fragment proteins, we transfected cells with the cDNA encoding calpastatin (Fig. 8a). Overexpression of calpastatin increased NF-κB-p65 and the antioxidants levels reduced by the 120Q-containing huntingtin fragment protein (Fig. 8b, c). Calpastatin also increased cell viability compromised by the mutant huntingtin fragment protein (Fig. 8d). Activation of different caspases including caspase-3 has been linked to cell death caused by mutant huntingtin proteins [26]. The presence of the cleaved caspase-3 p17 band was significantly reduced in calpastatin-expressing cells (Fig. 8d–e). Furthermore, we analyzed the caspase-3 substrate PARP using Western blotting as an indication of active caspase-3. Figure 8f and g show that the caspase-3-mediated PARP-processing (89-kDa band) is significantly reduced in calpastatin-expressing cells.
Fig. 8.
Calpastatin overexpression counteracts the downregulation of NF-κB-p65 and of antioxidants and increases cell viability. a Immunoblots. Human calpastatin was overexpressed for 24 h together with the 120Q-huntingtin fragment protein followed by immunoblotting. The level of exogenous calpastatin was a shown using a human-specific antibody (lanes 4, 5). Calpastatin increased NF-κB-p65 and Sod2 levels. ß-actin was used as control. Expression of GFP-N-terminal huntingtin is shown in the lowermost panel. b Quantification. **p < 0.01 for calpastatin + 120Q versus 120Q, n = 3. c Calpastatin-reduced cell death induced by the mutant huntingtin protein. **p < 0.01 for calpastatin + 120Q versus 120Q, n = 4. d Immunoblot. Calpastatin reduces caspase-3 activation as evident from levels of cleaved caspase-3 (17-kDa band). e Quantification of the 17-kDa band. **p < 0.01 for 120Q versus calpastatin + 120Q, n = 3. f Immunoblots of PARP cleavage. Calpastatin reduces the caspase-3 mediated cleavage of PARP (89-kDa band). Staurosporine (STS) was used as a positive control. g Quantification of the 89-kDa band. p < 0.01 for 120Q versus 120Q + calpastatin
Discussion
The results in this paper show that there is a selective downregulation of NF-κB-p65 signaling in neuronal PC6.3 cells expressing mutant N-terminal fragment and full-length mutant huntingtin proteins. The reduced levels of NF-κB-p65 were linked to a decreased NF-κB activity, shown by the NF-κB-luciferase reporter, and a decreased NF-κB-p65 signaling with reduced levels of antioxidants in cells expressing mutant huntingtin proteins (Fig. 9). The antioxidants decreased in these cells by mutant huntingtin included the mitochondrial proteins, Sod2 and Trx2, as well as the cytosolic ones, Sod1 and catalase. These antioxidants play an important role in protecting cells against the damage caused by increased levels of ROS. Decreased levels of antioxidants may therefore render cells more vulnerable to oxidative stress-induced damage and increased cell death. We observed increased oxidative damage and cell death and an increased production of ROS in cells expressing the mutant huntingtin proteins. Together, these results indicate that the reduced NF-κB-p65 signaling contributes to the increase in oxidative stress and cell degeneration observed in mutant huntingtin protein-expressing cells.
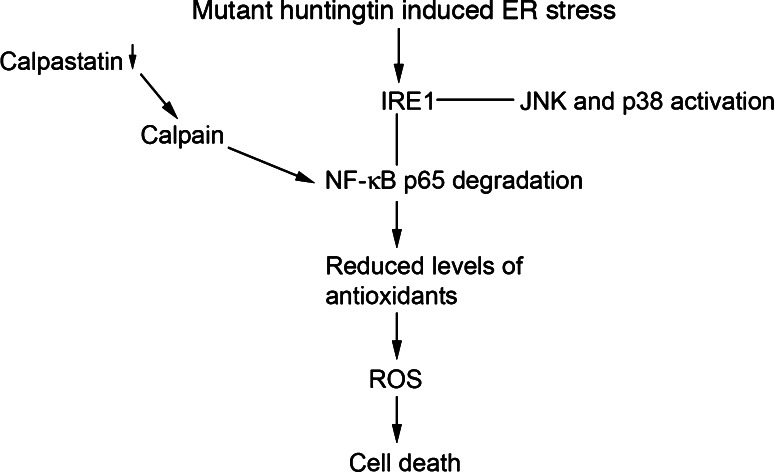
Fig. 9.
Summary of the results. Expression of mutant huntingtin induces a calpain-mediated NF-κB-p65 degradation that is followed by decreased antioxidant levels and increased oxidative stress
Previous studies on HD patients and in animal models of the disease have indicated a role for oxidative stress in HD. Thus, postmortem HD brain specimens show increased levels of lipofuscin, lipid peroxidation, and an accumulation of the oxidated DNA metabolite, 8-hydroxydeoxyguanosine, as well as increased heme oxygenase and 3-nitrotyrosine levels that represent markers for oxidative stress [26, 27]. Biochemical analyses have further revealed changes in complex I–IV and aconitase activities in the mitochondria, along with increases in lactate and decreased ATP generation, indicative of mitochondrial dysfunctions in HD [28, 29]. Moreover, in the R6/1 and R6/2 mice, which represent animal models for HD, the expression of nitric oxide synthase was shown to change with age while the levels of Sod2 decreased [30, 31]. Expression of mutant huntingtin also decreases the levels of the transcriptional coactivator, peroxisome proliferator-activated receptor-gamma coactivator-1 alpha (PGC-1α) that is involved in the regulation of cellular antioxidants [32, 33]. PGC-1α gene-deficient mice show an increased neurodegeneration mainly in the striatum, whereas the overexpression of PGC-1α in the R6/2 mice is neuroprotective [33]. In this work, we show that an unperturbed NF-κB-p65 signaling is crucial in regulating antioxidants levels and oxidative stress in the neuronal PC6.3 cells with lower levels producing oxidative stress and an increased vulnerability of cells to undergo cell death. PGC-1α and NF-κB-p65 signaling are both known to increase antioxidants and enhance cellular defense reactions against oxidative stress [16, 33]. It is therefore likely that different cell-signaling systems are affected in HD, which leads to changes in gene expression with increased cell degeneration. The network of genes affected in HD may include different antioxidants, anti-apoptotic proteins, as well as various growth factors and their signaling molecules.
To study the mechanisms for the reduced NF-κB-p65 in mutant huntingtin-expressing cells we focused on the calpain/calpastatin system. Previously, an increased calpain activity was observed in brain in HD [34]. In a recent study with the YAC transgenic HD model, the increased calpain activity in striatum was linked to alterations in activity and toxicity of NMDA receptors in neurons [35]. Calpain has also been shown to cleave mutant huntingtin following calcium-induced neuronal damage [34, 36]. Recently, calpain was shown to also cleave the p35 protein that is involved in the phosphorylation of the cyclin-dependent kinase-5 and which may influence HD pathology, as shown in other neurodegenerative diseases [37]. In the present study, we identified the NF-κB-p65 protein as a target for the increased calpain activity induced by mutant huntingtin proteins. The calpain inhibitors ALLN and CX295 inhibited the degradation of NF-κB-p65 and decreased cell degeneration induced by the expression of the mutant huntingtin fragment proteins. In line with our cellular data, previous biochemical analyses have shown that calpains are able to cleave the NF-κB-p65 in vitro [38].
In cells, calpastatin is the major regulator of calpain activity [21], which, however, can be regulated by high intracellular calcium levels. We observed reduced calpastatin levels in cells expressing mutant huntingtin proteins. This decrease in calpastatin was restored using the pan-caspase inhibitor BAF (Fig. 7b), indicating a role for cellular caspases in the control of calpastatin under these conditions. Previous studies have shown that mutant huntingtin can activate different cellular caspases with increased cell death [15, 39]. However, other proteases also probably become activated in the mutant huntingtin-expressing cells and may contribute to the degradation of calpastatin observed in this study. The identity of such proteases regulating the calpastatin/calpain system remains to be studied further.
The importance of calpastatin for the regulation of NF-κB-p65 and antioxidants level was studied using overexpression of calpastatin in conjunction with N-terminal mutant huntingtin proteins. Results showed that overexpression of calpastatin counteracted the decrease in NF-κB-p65 and Sod2 levels caused by the mutant proteins. Calpastatin also enhanced cell viability and reduced caspase-3 cleavage in cells expressing the 120-Q huntingtin fragment protein. These results identify calpastatin as an important upstream regulator in the NF-κB-p65 pathway and oxidative stress induced by mutant huntingtin proteins.
Increased ER stress is observed in many human disorders including neurodegenerative diseases [14]. In line with this work, recent studies on HD and other polyQ diseases have provided evidence for the involvement of ER stress in disease pathogenesis [40–43]. There are also indications that ER stress and oxidative damage are linked through the close communication between the ER and mitochondria. Significant levels of ROS are produced in ER during protein folding and in oxidation reactions [9]. In this study, we showed that inhibition of ER stress by salubrinal restored NF-κB-p65 levels that were decreased in cells expressing the mutant huntingtin proteins (Fig. 2i). In addition, recent data indicate that the ER-signaling pathway may be activated differently during ER stress [44]. We observed that within the IRE1-signaling pathway, the JNK and p38 kinases were activated, whereas NF-κB-p65 was downregulated in cells expressing the mutant huntingtin proteins. This shows that a careful analysis of signaling systems linked to ER stress is necessary in order to understand their roles in the pathogenesis of different diseases.
It has recently been shown that the NF-κB-signaling pathway is involved in cell toxicity mediated by huntingtin [45, 46]. Thus it was shown that DNA damage can trigger proteolysis of huntingtin in a caspase-dependent manner, which was promoted by the activation of I kappa B kinase beta (IKKβ) and inhibited by I kappa B kinase alpha (IKKα) [46]. The IKKs act by phosphorylating the IκB protein as well as other proteins leading to activation of different c-Rel proteins and the NF-κB pathway. We show here that mutant huntingtin proteins reduce the levels of p65 and the NF-κB pathway associated with ER stress and the activation of calpain. These results together with those by Khoshnan et al. [45, 46] show that NF-κB signaling is modulated by mutant huntingtin contributing to increased cell degeneration.
Taken together, the present study shows that the N-terminal mutant huntingtin proteins as well as the disease-inducing 75Q-containing huntingtin decreased NF-κB signaling in PC6.3 neuronal cells leading to increases in oxidative stress and cell death. The underlying mechanism for this involved a decrease in calpastatin, which activated calpain and decreased the NF-κB-p65 level. These results may be useful in defining novel targets for cell protection and for counteracting some of the adverse cellular changes observed in HD.
Acknowledgments
We thank A. Nørremølle and L. Hasholt for the N-terminal huntingtin, F. Saudou for the full-length huntingtin expression vectors, U. Arumäe for PC6.3 cells, T. Gilmore for Rel-A-deficient cells, and E. Lehto and J. Mäkelä for skillful technical assistance. Supported by the Academy of Finland, Finska Läkaresällskapet and University of Helsinki Research Funds as well as the Sigrid Juselius, Arvo and Lea Ylppö, Magnus Ehrnrooth, Maud Kuistila, Liv och Hälsa, Minerva, von Frenckell, Emil Aaltonen and Oskar Öflund Foundations.
References
- 1.van Dellen A, Grote HE, Hannan AJ. Gene–environment interactions, neuronal dysfunction and pathological plasticity in Huntington’s disease. Clin Exp Pharmacol Physiol. 2005;32:1007–1019. doi: 10.1111/j.1440-1681.2005.04313.x. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 3.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, Boyce F, Aronin N. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 4.Sharp AH, Loev SJ, Schilling G, Li SH, Li XJ, Bao J, Wagster MV, Kotzuk JA, Steiner JP, Lo A, Hedreen J, Sisodia S, Snyder SH, Dawson TM, Ryugo DK, Ross CA. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 5.Gil J, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 6.Lipinski MM, Yuan J. Mechanisms of cell death in polyglutamine expansion diseases. Curr Opin Pharmacol. 2004;4:85–90. doi: 10.1016/j.coph.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 8.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 11.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;174:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 13.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 14.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 15.Reijonen S, Putkonen N, Nørremølle A, Lindholm D, Korhonen L. Inhibition of endoplasmic reticulum stress counteracts neuronal cell death and protein aggregation caused by N-terminal mutant huntingtin proteins. Exp Cell Res. 2008;314:950–960. doi: 10.1016/j.yexcr.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 17.Kairisalo M, Korhonen L, Blomgren K, Lindholm D. X-linked inhibitor of apoptosis protein increases mitochondrial antioxidants through NF-kappaB activation. Biochem Biophys Res Commun. 2007;364:138–144. doi: 10.1016/j.bbrc.2007.09.115. [DOI] [PubMed] [Google Scholar]
- 18.Moore ED, Becker PL, Fogarty KE, Williams DA, Fay FS. Ca2+ imaging in single living cells: theoretical and practical issues. Cell Calcium. 1990;11:157–179. doi: 10.1016/0143-4160(90)90068-6. [DOI] [PubMed] [Google Scholar]
- 19.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. Selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 20.Gapuzan ME, Schmah O, Pollock AD, Hoffmann A, Gilmore TD. Immortalized fibroblasts from NF-kappaB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor alpha after transformation by v-Ras. Oncogene. 2005;24:6574–6583. doi: 10.1038/sj.onc.1208809. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomgren K, Hallin U, Andersson AL, Puka-Sundvall M, Bahr BA, McRae A, Saido TC, Kawashima S, Hagberg H. Calpastatin is up-regulated in response to hypoxia and is a suicide substrate to calpain after neonatal cerebral hypoxia–ischemia. J Biol Chem. 1999;274:14046–14052. doi: 10.1074/jbc.274.20.14046. [DOI] [PubMed] [Google Scholar]
- 23.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 24.Glading A, Chang P, Lauffenburger DA, Wells A. Epidermal growth factor receptor activation of calpain is required for fibroblast motility and occurs via an ERK/MAP kinase signaling pathway. J Biol Chem. 2000;275:2390–2398. doi: 10.1074/jbc.275.4.2390. [DOI] [PubMed] [Google Scholar]
- 25.Åkerman KE, Shariatmadari R, Krjukova J, Larsson KP, Courtney MJ, Kukkonen JP. Ca2+ -dependent potentiation of muscarinic receptor-mediated Ca2+ elevation. Cell Calcium. 2004;36:397–408. doi: 10.1016/j.ceca.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington’s disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polidori MC, Mecocci P, Browne SE, Senin U, Beal MF. Oxidative damage to mitochondrial DNA in Huntington’s disease parietal cortex. Neurosci Lett. 1999;272:53–56. doi: 10.1016/S0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- 28.Grünewald T, Beal MF. Bioenergetics in Huntington’s disease. Ann NY Acad Sci. 1999;893:203–213. doi: 10.1111/j.1749-6632.1999.tb07827.x. [DOI] [PubMed] [Google Scholar]
- 29.Milakovic T, Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem. 2005;280:30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 30.Santamaría A, Pérez-Severiano F, Rodríguez-Martínez E, Maldonado PD, Pedraza-Chaverri J, Ríos C, Segovia J. Comparative analysis of superoxide dismutase activity between acute pharmacological models and a transgenic mouse model of Huntington’s disease. Neurochem Res. 2001;26:419–424. doi: 10.1023/A:1010911417383. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Severiano F, Escalante B, Vergara P, Ríos C, Segovia J. Age-dependent changes in nitric oxide synthase activity and protein expression in striata of mice transgenic for the Huntington’s disease mutation. Brain Res. 2002;951:36–42. doi: 10.1016/S0006-8993(02)03102-5. [DOI] [PubMed] [Google Scholar]
- 32.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Gafni J, Ellerby LM. Calpain activation in Huntington’s disease. J Neurosci. 2002;22:4842–4849. doi: 10.1523/JNEUROSCI.22-12-04842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowan CM, Fan MM, Fan J, Shehadeh J, Zhang LY, Graham RK, Hayden MR, Raymond LA. Polyglutamine-modulated striatal calpain activity in YAC transgenic Huntington disease mouse model: impact on NMDA receptor function and toxicity. J Neurosci. 2008;28:12725–12735. doi: 10.1523/JNEUROSCI.4619-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun B, Fan W, Balciunas A, Cooper JK, Bitan G, Steavenson S, Denis PE, Young Y, Adler B, Daugherty L, Manoukian R, Elliott G, Shen W, Talvenheimo J, Teplow DB, Haniu M, Haldankar R, Wypych J, Ross CA, Citron M, Richards WG. Polyglutamine repeat length-dependent proteolysis of huntingtin. Neurobiol Dis. 2002;11:111–122. doi: 10.1006/nbdi.2002.0539. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti P, Vila I, Rifé M, Lizcano JM, Alberch J, Ginés S. Dopaminergic and glutamatergic signaling crosstalk in Huntington’s disease neurodegeneration: the role of p25/cyclin-dependent kinase 5. J Neurosci. 2008;28:10090–10101. doi: 10.1523/JNEUROSCI.3237-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu ZQ, Kunimatsu M, Yang JP, Ozaki Y, Sasaki M, Okamoto T. Proteolytic processing of nuclear factor kappa B by calpain in vitro. FEBS Lett. 1996;385:109–113. doi: 10.1016/0014-5793(96)00360-2. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez Mejia RO, Friedlander RM. Caspases in Huntington’s disease. Neuroscientist. 2001;7:480–489. doi: 10.1177/107385840100700604. [DOI] [PubMed] [Google Scholar]
- 40.Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noh JY, Lee H, Song S, Kim NS, Im W, Kim M, Seo H, Chung CW, Chang JW, Ferrante RJ, Yoo YJ, Ryu H, Jung YK. SCAMP5 links endoplasmic reticulum stress to the accumulation of expanded polyglutamine protein aggregates via endocytosis inhibition. J Biol Chem. 2009;284:11318–11325. doi: 10.1074/jbc.M807620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho KJ, In Lee B, Cheon SY, Kim HW, Kim HJ, Kim GW. Inhibition of Ask1 reduces ER stress and nuclear huntingtin fragments in a mouse model of Huntington disease. Neuroscience. 2009;163:1128–1134. doi: 10.1016/j.neuroscience.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 43.Carnemolla A, Fossale E, Agostoni E, Michelazzi S, Calligaris R, De Maso L, Del Sal G, MacDonald ME, Persichetti F. Rrs1 is involved in endoplasmic reticulum stress response in Huntington disease. J Biol Chem. 2009;284:18167–18173. doi: 10.1074/jbc.M109.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khoshnan A, Ko J, Watkin EE, Paige LA, Reinhart PH, Patterson PH. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khoshnan A, Ko J, Tescu S, Brundin P, Patterson PH. IKKalpha and IKKbeta regulation of DNA damage-induced cleavage of huntingtin. PLoS One. 2009;4:e5768. doi: 10.1371/journal.pone.0005768. [DOI] [PMC free article] [PubMed] [Google Scholar]