Abstract
Snake venom contains mixture of bioactive proteins and polypeptides. Most of these proteins and polypeptides exist as monomers, but some of them form complexes in the venom. These complexes exhibit much higher levels of pharmacological activity compared to individual components and play an important role in pathophysiological effects during envenomation. They are formed through covalent and/or non-covalent interactions. The subunits of the complexes are either identical (homodimers) or dissimilar (heterodimers; in some cases subunits belong to different families of proteins). The formation of complexes, at times, eliminates the non-specific binding and enhances the binding to the target molecule. On several occasions, it also leads to recognition of new targets as protein-protein interaction in complexes exposes the critical amino acid residues buried in the monomers. Here, we describe the structure and function of various protein complexes of snake venoms and their role in snake venom toxicity.
Keywords: PLA2 complexes, Metalloprotease complexes, Dimeric disintegrin, Serine protease complexes, Covalent and non-covalent three-finger toxin, Synergistic three-finger toxin
Introduction
Snake venom is a mixture of bioactive proteins and polypeptides, produced and stored in a highly specialized venom gland. After their administration through a bite, these proteins and polypeptides function as toxins and attack various physiological systems, often leading to death and debilitation of the prey or victim. These toxins are classified as enzymatic and non-enzymatic proteins. Most snake toxins exhibit their pharmacological activities on their own. However, some proteins form covalent/non-covalent complexes with other proteins to exhibit more potent pharmacological activities. These synergistic interactions between venom proteins enhance the lethal potency of the snake venom.
Since the isolation of crotoxin from the venom of Crotalus durissus terrificus [1], many other snake venom protein complexes have been identified and characterized. Most of the early protein complexes studied are potent neurotoxins and contain either phospholipase A2 (PLA2) or PLA2-like molecules. In the last 2 decades, complexes involving other families of venom proteins, such as metalloproteases, serine proteases, Snaclecs [C-type lectins (CTLs) and C-type lectin-related proteins (CLRPs)] and three-finger toxins (3FTxs), have also been reported. Some of these protein complexes exhibit procoagulant, anticoagulant and antiplatelet activities. Over the last couple of decades, the complex formation and its effects on pharmacological effects of various snake venom protein complexes have been thoroughly characterized. In this review, the structure, biological activities, role of individual subunits and interaction among the subunits of snake venom protein complexes have been discussed.
Phospholipase A2 (PLA2) complexes
PLA2 enzymes are one of the most extensively studied families of snake venom proteins [2]. They mostly exist as monomers. However, several of them interact with other PLA2 (or PLA2-like molecules) or with other proteins to form complexes (Fig. 1) either through covalent or non-covalent interactions. Functionally, these PLA2 complexes exhibit presynaptic neurotoxicity [3] and play an important role during envenomation.
Fig. 1.
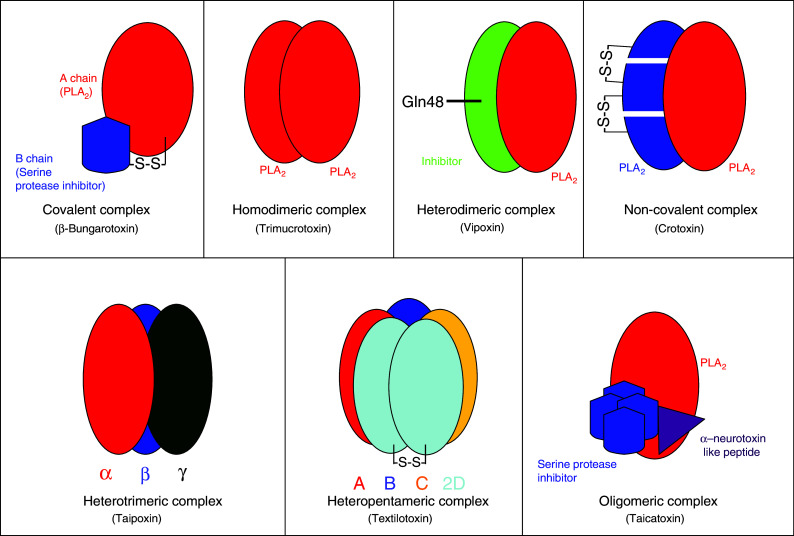
Schematic representation of covalent and non-covalent snake venom PLA2 complexes
Covalent complex
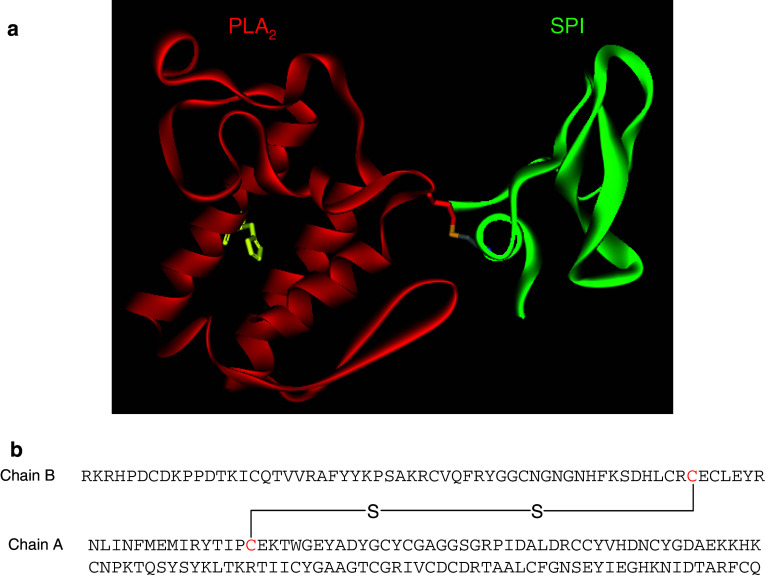
β-Bungarotoxins (β-BuTx) are one of the well-studied PLA2 complexes isolated from the venom of Bungarus multicinctus (Taiwan banded krait) [4]. So far, as many as 16 isoforms have been reported from Bungarus species [5], and 8 of them, namely β1–β5 [6] and SPI–SPIII [7], have been studied in detail. Structurally, they are covalently linked heterodimers (Fig. 2a); chain A is homologous to Group I PLA2 enzyme, while chain B is structurally similar to Kunitz-type serine protease inhibitors and dendrotoxins [8, 9]. As with other PLA2 enzymes, chain A has 122 amino acid residues. It has 13 cysteine residues, of which 12 form six disulphide bridges, but the unique disulphide bridge (Cys11–Cys77) of Group I PLA2 is absent. The extra cysteine residue at the 15th position forms the interchain disulphide bridge with chain B [3]. Chain B is structurally similar to bovine pancreatic trypsin inhibitor (BPTI) and has 61 amino acid residues and 7 cysteine residues. Six of the cysteine residues are conserved, which form three disulphide bridges, but the 7th cysteine residue at the 55th position is involved in the interchain disulphide bond with Cys15 of chain A (Fig. 2b).
Fig. 2.
a Ribbon model of β-bungarotoxin (β-BuTx) (PDB ID: 1BUN) showing the disulphide linkage between chain A and chain B. b Inter-chain disulphide bridge is formed between (highlighted in red) 1st Cys of chain A and 6th Cys of chain B, and the linkage is shown in both the ribbon model and amino sequences
The crystal structure of β-BuTx was determined by Kwong and co-workers [10]. Structurally, the α helices and the Ca2+ binding loop in chain A are similar to other PLA2 molecules. However, there is a conformational change in the region where it interacts with chain B (residue 13–16 and 74–76), as well as in the substrate binding loop (residue 63–65). Similarly, the C-terminal region of chain B shows conformational change due to the interaction with chain A, and this conformational change accounts for the lack of protease inhibitor activity [10]. In addition to the interchain disulphide bridge, ionic interactions between Glu16 of chain A and Lys48 of chain B, and a hydrogen bond between Arg75 of chain A and carbonyl oxygen atom of Leu58 of chain B in the interface play important roles in the interaction and contribute to the overall stability of this covalent complex.
Functionally, β-BuTx binds to the presynaptic site at the neuromuscular junction and disrupts the neurotransmission. In the mid 1980s, Su and Chang [11] studied the effect of β-BuTx on the nerve terminal and summarized the series of events during neurotoxicity as: (1) initial weak reduction in the spontaneous acetylcholine release, (2) subsequent enhancement of the release of acetylcholine and (3) a final decrease in the release of acetylcholine leading to complete failure of neurotransmission. Subsequently, several studies were done to understand the effect of β-BuTx in the nerve terminals. As mentioned earlier, chain B is similar to dendrotoxin from Dendroaspis venoms, which blocks voltage sensitive K+ channels. Hence, when muscle preparation is pretreated with dendrotoxin, the neuromuscular block by β-BuTx slows down. This indicates that both β-BuTx and dendrotoxin target the same receptor/acceptor [12] and that chain B exhibits the blocking activity by binding to the voltage gated K+ channel [12–14]. Chemical modification studies show that the inhibition of PLA2 activity of chain A inhibits the neurotoxic effect, suggesting the involvement of enzymatic activity in the neurotoxicity [15, 16]. Chain B binds to the K+ channels in the target presynaptic site due to its high affinity interaction and in turn helps chain A to bind to the presynaptic site [17–19].
Non-covalent complexes
Homodimeric PLA2 complexes
Trimucrotoxin from the venom of Trimeresurus mucrosquamatus (Taiwan habu) is the only known homodimeric PLA2 complex. Functionally, it exhibits presynaptic neurotoxicity. The subunits are 14 kDa in size, and the amino acid sequence determined by the N-terminal and cDNA sequencing reveals that they belong to the Group II PLA2 enzyme. It is structurally, immunologically and pharmacologically similar to crotoxin (discuss below) and agkistrodotoxin [20]. The crystal structure of this complex has not been determined, and the exact mode of interaction between the subunits is not known.
Heterodimeric complexes
Crotoxin and related toxins: Crotoxin is the major toxic component of C. durissus terrificus (South American rattlesnake) venom and was first reported in 1938 by Slotta and Fraenkel-Conrat. It was the first snake venom protein to be crystallized [1] and was re-crystallized recently [21]. This is the major toxic component of Crotalus venoms, and so far in C. durissus terrificus venom as many as 15 isoforms of crotoxin have been identified and characterized [22, 23].
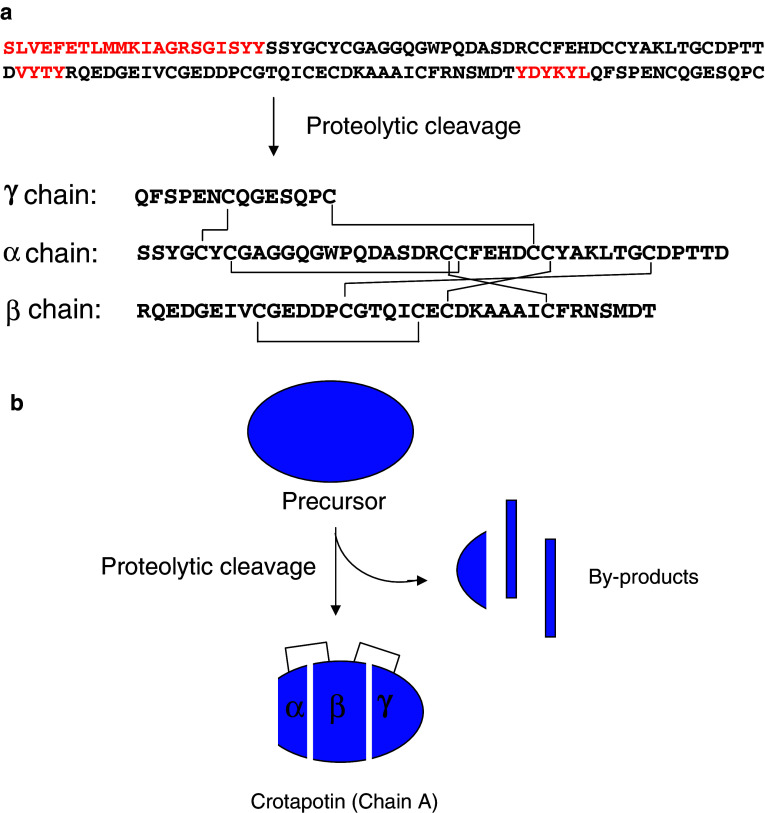
Crotoxin is composed of two subunits, the non-toxic acidic subunit (CA) named crotapotin, which lacks PLA2 activity, and the weakly toxic basic subunit (CB), which exhibits PLA2 activity (Fig. 1) [24]. The CA subunit is made up of three disulfide-linked polypeptide chains. They are generated by proteolytic cleavage of the CA precursor, which is similar to the Group II PLA2 enzyme (Fig. 3). So far, four isoforms (CA1, CA2, CA3 and CA4) of this precursor have been identified in the transcriptome of C. durissus terrificus [25]. The proteases involved in the processing of the CA subunit are yet to be identified. The CB subunit is an intact Group II PLA2 enzyme. Although it was the first toxin to be crystallized, the three-dimensional structure of the complex is not yet available. Only recently, the crystal structure of individual CB subunits has been reported [26].
Fig. 3.
a Proteolytic cleavage of chain A precursor of crotoxin. Chain A precursor undergoes proteolytic cleavage to form three polypeptide α, β and γ chains. These polypeptides are linked by disulphide bridges. The inter-chain disulphide bridge is shown in straight lines. b Schematic representation of proteolysis and inter-chain disulphide bridge
Functionally, crotoxin is a potent neurotoxin [1], which acts at neuromuscular junctions and blocks transmission of nerve signals. Similar to the native crotoxin complex, the CB subunit causes a blockade of neuromuscular transmission and exhibits other pharmacological effects, while CA is pharmacologically inactive [27]. Although the CB subunit exhibits neurotoxic activity, the native crotoxin complex is at least one order of magnitude more potent than CB alone. The CA subunit acts as a chaperone by blocking the CB subunit from binding to non-specific sites and guides the CB subunit to its specific target site, leading to the increased potency. Binding experiments show that the toxin complex dissociates upon binding to the synaptic membranes [9]. If this dissociation is prevented by a covalent linkage between the subunits, the lethal potency of crotoxin is completely abolished [28]. Thus, the dissociation of the complex is important for exhibiting its biological activity. In addition to the neurotoxicity, crotoxin exhibits a number of other biological activities, including local myotoxicity [29], systemic myotoxicity and myoglobinuria [30], inhibition of inflammatory response [31, 32], initiation of platelet aggregation [33, 34], analgesic effect [35], inhibition of macrophage function [31] and cytotoxicity on several tumor cell lines [36–39]. Crotoxin is more cytotoxic to cell lines that express high levels of epidermal growth factor receptors [37]. However, the role of these receptors in cytotoxicity is not yet understood. Crotoxin causes the necrosis of skeletal muscle, and the CB subunit is as potent as the complex in causing skeletal muscle necrosis [29, 40]. This might be due to the basic nature of the CB subunit. Therefore, crotoxin is a multifunctional protein complex that exhibits various biological activities.
Crotoxin-like neurotoxins in other rattlesnake venoms have also been purified and characterized, such as vergrandis toxin from C. vergrandis [41], concolor toxin from the C. viridis concolor and canebrake toxin from C. horridus atricaudatus venoms [41–45]. Other venoms that contain crotoxin-like complexes include Sistrurus catenatus tergeminus, C. mitchelli mitchelli, C. horridus atricaudatus, C. basiliscus and C. durissus cumanensis [46]. Among crotoxin-like complexes, Mojave toxin isolated from the venom of C. scutulatus scutulatus (Mojave rattlesnake) is the second most-studied homolog of crotoxin [42, 47]. The acidic subunit undergoes proteolytic processing similar to crotoxin [24, 48]. Using anti-Mojave toxin antibodies and genomic DNA analysis, Mojave toxin and similar neurotoxins have been found in five subspecies of Crotalus helleri (Southern Pacific rattlesnake) [49]. The presence of Mojave toxin in C. s. scutulatus distributed in Arizona, New Mexico and Texas was analyzed using their DNA samples. The acidic subunit was not amplified from three DNA samples obtained from Arizona, and hence these snakes might lack the Mojave toxin [50].
Functionally, Mojave toxin is presynaptically neurotoxic [43]. This toxin acts as a calcium channel blocker at the presynaptic membrane [51], and dihydropyridines receptors are found to be its molecular target [52]. Similar to crotoxin, the acidic subunit of the Mojave toxin is non-toxic, but it enhances the toxicity of the basic subunit, which has weak PLA2 activity [53].
Vipoxin and related toxins: Vipoxin isolated from the venom of Vipera ammodytes meridionalis (Bulgarian snake) [54] is the only known postsynaptic neurotoxic complex [55] (Fig. 4). Both the subunits of this heterodimeric complex are PLA2 molecules with 60% similarity to each other. However, there is a crucial substitution of His48 to Gln48 at the active site of the acidic subunit, which leads to the loss of enzymatic activity. Interestingly, this acidic subunit (Inh—inhibitory subunit) inhibits the enzymatic activity of the basic subunit [55]. This is a unique example of modulation of the catalytic and toxic functions of PLA2 by an inactive, non-toxic PLA2 [56]. The crystal structure of vipoxin shows that the active PLA2 subunit and its Inh have similar structures that are comparable to other PLA2 structures. The interactions between these two polypeptides are mediated by 12 intermolecular contacts that stabilize the complex. The conformation of the Ca2+ binding loop of the basic subunit in the complex is different from other PLA2 enzymes, resulting in lower enzymatic activity [57]. Functionally, it acts on the postsynaptic membrane to prevent the binding of acetylcholine to its receptor and thus blocks the neuromuscular transmission. Interestingly, the separated basic subunit shows presynaptic neurotoxicity, similar to several other PLA2 enzymes [58].
Fig. 4.
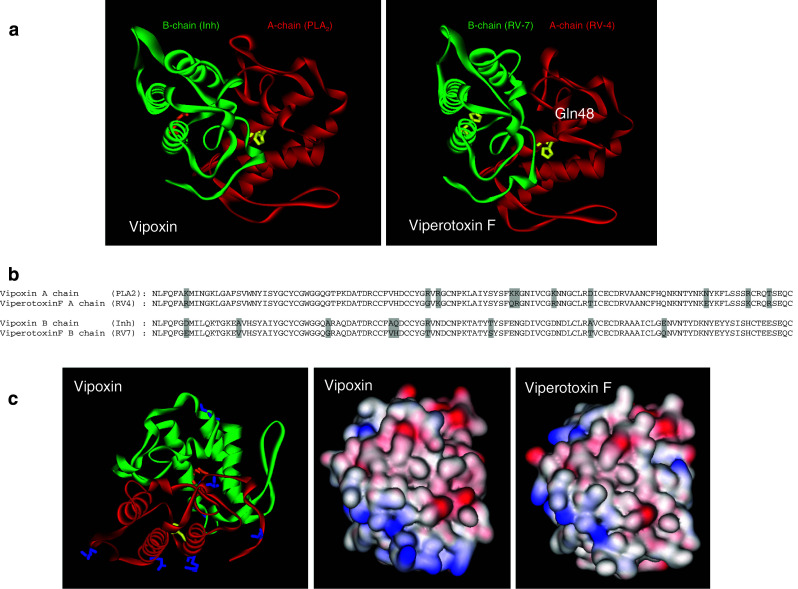
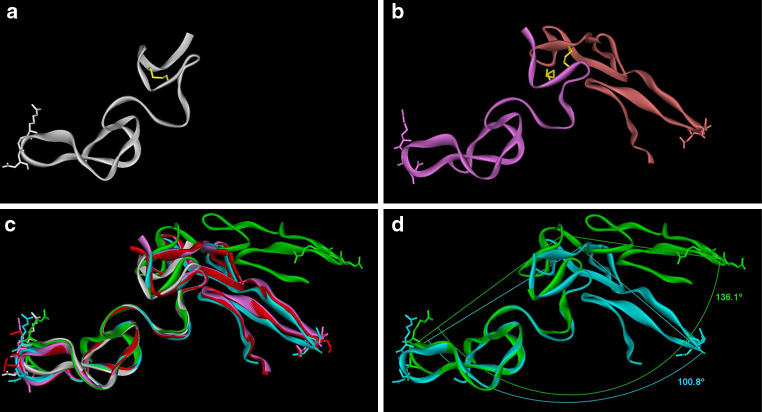
a Ribbon model of vipoxin (PDB ID: 1AOK) and viperotoxin F (PDB ID: 1OQS) showing PLA2 subunit (red) and its inhibitor (green). The active site residue, His48, is replaced with Gln48 in the inhibitor, which is shown in a stick and ball model. b Alignment of amino acid sequence of chain A and B of vipoxin and viperotoxin F. The substituted amino acid residues are highlighted in grey. c The surface residues that are substituted in vipoxin are shown in a stick model (blue color). The surface charge of vipoxin and viperotoxin F is shown
Viperotoxin F, a heterodimeric neurotoxin, is the major lethal component from the venom of Vipera russelli formosensis (Taiwan viper). It is comprised of two subunits: a basic neurotoxic PLA2 component (RV-4) and a non-toxic acidic component with low enzymatic activity (RV-7) [59]. The low enzymatic activity of RV-7 is due to the substitutions in the amino acid residue found at the 7th, 17th, 59th, 114th and 119th positions, which are likely to be involved in binding to the aggregated lipid substrates. RV-7 inhibits the enzymatic activity of RV-4, but enhances its lethal potency and neurotoxic effect similar to acidic subunit of vipoxin. It has been suggested that the acidic component guides the specific binding of RV-4 to the neurotoxic site by preventing its non-specific absorption [59]. The crystal structure of this complex has been solved, and the two components interact through ionic, hydrophobic and hydrogen bonds [57] (Fig. 4). Although viperotoxin F and vipoxin are structurally similar, they differ in their mode of action: (1) the former is a presynaptic neurotoxin, while the latter is a postsynaptic neurotoxin; (2) Inh decreases the toxicity of vipoxin while RV-7 enhances the toxicity of RV-4/RV-7 complex; (3) acidic component of viperatoxin F (RV-7) has low enzymatic activity, whereas vipoxin (Inh) has no activity due to substitution of His48 to Gln48 [59, 60].
A similar heterodimeric neurotoxin, Vipera toxin, has been reported from the venom of Vipera palestinae [61]. Structurally, it is similar to Viperotoxin F [62, 63], and individually, the two subunits are found to be non-lethal, but the reconstituted complex is toxic.
Another heterodimeric neurotoxin reported from snake venom includes Pseudocerastes neurotoxin from the venom of Pseudocerastes fieldi (Field’s horned viper) [64]. It is made of two non-covalently associated subunits, a basic weakly toxic subunit (Cb II) and an acidic subunit. The acidic subunit has two different isoforms, Cb Iα and Cb Iβ [65]. All the subunits are PLA2-like molecules, but the acidic subunits are inactive towards phosphatidylcholine [66], despite having all the conserved putative catalytic residues (His48, Asp49, Ca2+-binding loop, the hydrophobic pocket and interfacial recognition site). Similar to the acidic subunit in viperotoxin, CbI enhances the lethal potency of the basic and weakly toxic subunit, CbII [65].
Heterotrimeric PLA2 complexes
Taipoxin and related toxins: Taipoxin, isolated from Oxyuranus s. scutellatus (Australian taipan snake), is one of the most potent animal toxins. It is a heterotrimeric PLA2 complex that exhibits toxicity through a presynaptic neuromuscular blockade [67]. Structurally, it has three subunits, namely α, β and γ (Fig. 1). Functionally, the α subunit is the most toxic, and the γ subunit is moderately toxic, but the β subunits (β-1 and β-2) are non-toxic [67, 68]. Amino acid sequences reveal that the α and β subunits belong to Group IA and the γ subunit belongs to Group IB PLA2 molecules. The γ subunit is similar to mammalian pancreatic pro-PLA2 enzyme with an additional eight-residue propeptide at the N-terminal. However, the γ subunit retains this propeptide unlike the pancreatic PLA2 in which it is cleaved off during activation. The γ subunit is glycosylated, and the carbohydrate moiety is attached at Asn78 position. In contrast to other subunits that have seven disulphide bridges, γ subunit has eight disulphide bridges, and the extra disulphide bridge is found in between Cys23 and Cys27. The γ subunit is the most important subunit of this complex as it acts as a chaperone to stabilize the complex and protects the α subunit from dissociation [69]. The γ subunit most probably interacts with the basic α subunit at the C-terminal region as it contains a stretch of negatively charged residues that are likely to be involved in this interaction [67]. The three-dimensional structure of this protein complex is not yet known.
Paradoxin isolated from the venom of Parademansia microlepidotus (currently known as Oxyuranus microlepidotus) [70, 71] and Cannitoxin isolated from the venom of Oxyuranus scutellatus canni [72] are homologs of taipoxin. Structurally, they have three subunits, α, β and γ, and functionally they act as presynaptic neurotoxins similar to taipoxin.
Heteropentameric PLA2 complexes
Textilotoxin is structurally the most complex neurotoxin isolated from Pseudonaja textilis venom [73]. It is among the most potent neurotoxins as the LD50 is as low as 1 μg/kg (i.p.) in mice. It has a molecular weight of 70,551 Da and consists of four subunits, namely A, B, C and D, at a 1:1:1:2 ratio [74]. All of the subunits are putative PLA2 molecules (Fig. 1). Subunit A is basic in nature, and it is the most toxic among the subunits. Subunit B is similar to the β subunit of taipoxin in the amino acid sequence and is non-toxic to mice. Subunit C is similar to subunit A, but has low PLA2 activity and lethality. Two identical subunit D polypeptides are linked together by a disulfide bond, and each chain is similar to γ subunit of taipoxin in its amino acid sequence, glycosylation and N-terminal propeptide. It has a single glycosylation site at Asn93 [75]. Similar to subunit C, it shows very low PLA2 activity. The decrease in enzymatic activity is due to the presence of the propeptide at the N-terminal, which blocks the hydrophobic channel [76].
Other PLA2 complexes
Taicatoxin, an oligomeric complex isolated from the venom of Oxyuranus scutellatus scutellatus (Australian taipan snake), is the only known non-neurotoxic PLA2 complex. This complex causes its toxic effects by blocking the Ca2+ channels [77]. It consists of three distinct subunits, an α-neurotoxin-like peptide, a neurotoxic PLA2 and a serine protease inhibitor (Fig. 1). These subunits form the oligomeric complex by non-covalent interactions at an approximate stoichiometry of 1:1:4 [78]. Taicatoxin is reported to retain its Ca2+ channel-blocking activity even after removal of the neurotoxic PLA2 subunit from the complex, but with a decreased potency. This indicates that the neurotoxic PLA2 subunit acts as an enhancer in this complex. However, taicatoxin completely loses its blocking activity when the protease inhibitor is removed from the complex. These results indicate that the α-neurotoxin-like peptide and the protease inhibitor subunit are important in binding of taicatoxin to the Ca2+ channel [78].
Protease complexes
Snake venom, particularly from Viperidae and Crotalidae families, contains proteases that affect the blood coagulation cascade and helps in digestion of prey [79–86]. These proteases are divided into metalloproteases or serine proteases. Although most of them are pharmacologically active by themselves, some require additional components to exhibit optimal activity.
Metalloprotease complexes
On the basis of their domain architecture, snake venom metalloproteases are classified into different types (P-I to P-III) [87, 88]. Only a few members of the class P-III (classified as P-IIId class [89]) metalloproteases are complexes as they have an extra subunit that interacts through covalent or non-covalent interactions. Two of these complexes are well studied and have similar architecture but distinct specificity (described below).
RVV-X is a factor X-activating protein isolated from Russell’s viper venom (Daboia russelli). It is a glycosylated heterotrimeric complex consisting of one heavy and two light chains [90]. The heavy chain consists of a metalloprotease, disintegrin and cysteine-rich domains (Fig. 5). The two light chains are homologous to CLRPs isolated from various snake venoms [91–93]. This protein complex activates factor X to Xa by cleaving the internal Arg51–Ile52 bond of factor X [87, 94]. The two light chains (A and B) are involved in the binding to the Gla domain of factor X. Ca2+ plays an important role in this activation process as Ca2+ induces conformational changes in the Gla domain of factor X, which might be necessary for the RVV-X recognition [92]. The RVV-X crystal structure has been solved recently, and it exhibits a “hook-spanner-wrench” configuration (Fig. 5). The heavy and light chains form the hook and the handle, respectively [95]. The heavy chain is covalently linked to the light chain via a disulphide bridge between the Cys339 of heavy chain and Cys133 of light chain A. The two light chains are linked by disulphide bridges between Cys79 and Cys77 of chain A and B, respectively. The two light chains form a concave structure at the point of contact similar to the ligand-binding site of factor IX/X binding protein. This concave surface functions as an exosite that binds to the Gla domain of factor X. RVV-X is an example of venom complex that has evolved to target specific proteins in the blood coagulation cascade and to cause immediate toxicity to the vertebrate prey by coagulating its blood.
Fig. 5.
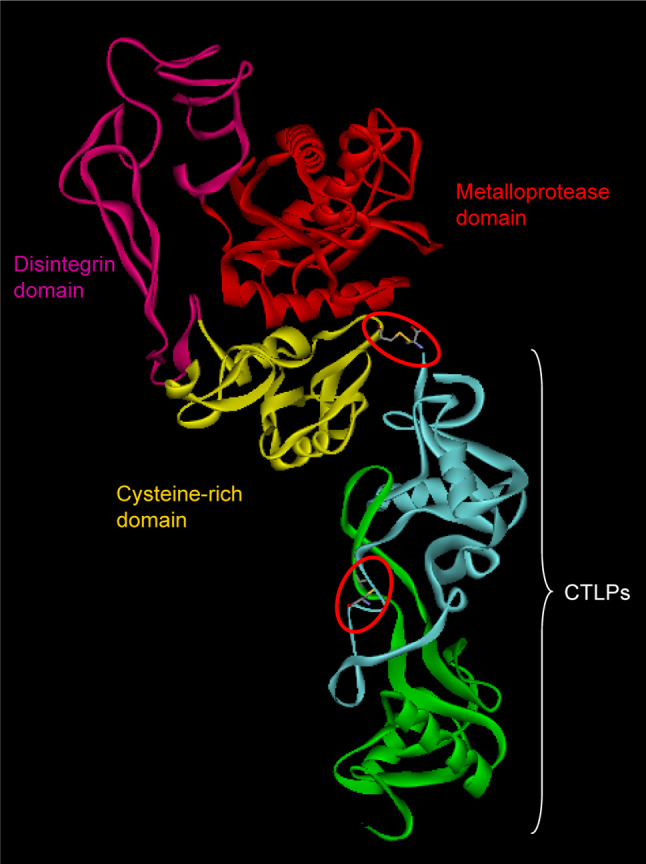
Ribbon model of RVV-X (PDB ID: 2E3X) showing different domains: metalloproteinase domain (red), disintegrin domain (megneta), cysteine rich domain (yellow) and CTLP domain (green and turquoise). The interchain disulphide bridges are encircled (red)
Carinactivase-1 (CA-1) is a heterodimeric protein complex isolated from the venom of Echis carinatus that is a potent prothrombin activator. It is structurally similar to RVV-X, but its heavy and light chains interact non-covalently. The heavy chain is structurally similar to ecarin, and the two regulatory subunits are similar to factor IX/X-binding protein and CLRPs. It primarily recognizes the Ca2+-bound conformation of the Gla domain of prothrombin for activation [96]. The light chains contribute to the specificity as well as calcium dependency. This protein has been used commercially to quantify prothrombin levels in the plasma in newborn infants by a novel method known as the CA-1 method [97]. A prothrombin activator from E. multisquamatus, named multactivase, which is structurally and functionally similar to Carinactivase-1, has also been characterized [98].
Dimeric disintegrins
Disintegrins are a non-enzymatic group of small molecules (4–14 kDa) that binds selectively to integrins molecules. [As disintegrin-like domains form part of metalloproteases (P-II and P-III classes) with the exception of the α chain of acostatin [99], we have included these protein complexes in this section based on their origin.] Although most of them exist as monomers, they are found as homodimers and heterodimers [100]. Dimeric disintegrins are 60–67-amino acid long and have 10 cysteine residues; 8 of them are involved in intrachain disulphide bridge, whereas 2 of them form the interchain disulphide bridge [100–102]. Most disintegrins have a RGD motif at the tip of a loop (Fig. 6), which is responsible for binding to the integrin molecules [103]. However, these functional site residues are replaced by KGD, KTS, VGD, MGD, MLD as well as ECD residues (for details, see reviews [88, 89, 104, 105]. Functionally, disintegrins are known to induce various biological activities, such as platelet aggregation, angiogenesis, metastasis and tumor growth through their interaction with various integrins [85, 106–109].
Fig. 6.
Dimeric disintegrins. (a) Disintegrin monomer (PDB ID: 1J2L) (white ribbon). RGD is shown as a white stick model. Intramolecular disulfide bond (C6:C14) is shown in the yellow stick. b Homodimeric disinegrin (PDB ID: 1RMR) (pink and orange ribbons). Side chains of RGD are shown as pink/orange sticks. Intermolecular disulfide bonds (C7:C12) are shown in the yellow stick. c Superimposition of homodimeric disintegrins (PDB ID: 1RMR in pink ribbon, PDB ID: 1TEJ in cyan ribbon) and heterodimeric integrins (PDB ID: 1Z1X in red ribbon, PDB ID: 3C05 in green ribbon) on monomeric disintegrin (PDB ID: 1J2L in white ribbon). RGDs are shown as the white stick in 1J2L, as the pink stick in 1RMR, as the cyan stick in 1TEJ and as the green stick in 3C05. MLD is shown as the red stick in 1Z1X. d For 1RMR, the angle between chain A Gly43 Cα, chain B Cys6 Cα, and chain B Gly43 Cα is 100.8°; for 1Z1X, the angle between chain A Gly44 Cα, chain B Cys7 Cα and chain B Gly44 Cα is 136.1°
Three-dimensional structures of a number of monomeric (beyond the scope of this review) and dimeric disintegrins have been solved. Schistatin is a homodimeric disintegrin (PDB no. 1RMR) isolated from the venom of E. carinatus [101]. The interchain disulphide bridges are formed between Cys7 of subunit A and Cys12 of subunit B, and between Cys12 of subunit A and Cys7 of subunit B (Fig. 6b). [These cysteine residues form intrachain disulfide in monomeric disintegrins (Fig. 6a).] This structural rearrangement in dimers leads to two integrin-binding sites at opposite ends. In another homodimeric disintegrin (PDB ID: 1Z1X) from E. carinatus venom, the RGD tripeptides are replaced by MLD (Fig. 6c). The effect of this structural change would affect the integrin specificity [104]. In the case of heterodimeric disintegrin (PDB ID: 1TEJ), also from E. carinatus venom, both monomers have a RGD tripeptide sequence, and its neighboring regions are quite similar (QRARGDGNHDY in the A chain compared to QRARGDGNNDY in the B chain). Thus, we speculate that both loops would have similar integrin specificity as schistatin loops. Acostatin, a heterodimeric disintegrin from A. contortrix contortrix venom, has a similar pattern of interchain disulphide bridges. However, the position of the cysteine residues involved in the interchain disulphide bridge are Cys8 and Cys13 [102], and β chains of acostatin have the all conserved cysteine residues and the RGD motif similar to other disintegrins, but the mature protein within the subunits shows only 68.75% identity. Interestingly, the positioning of the RGD loops are distinct compared to all other homodimeric and heterodimeric disintegrins (Fig. 6d). In additions the neighboring segments of A and B chains are also somewhat different. As these surrounding regions contribute to specificity, each of these loops may recognize distinct integrins [110–113]. Hence, the integrin specificity of acostatin would be most likely different compared to other examples described above.
Serine protease complexes
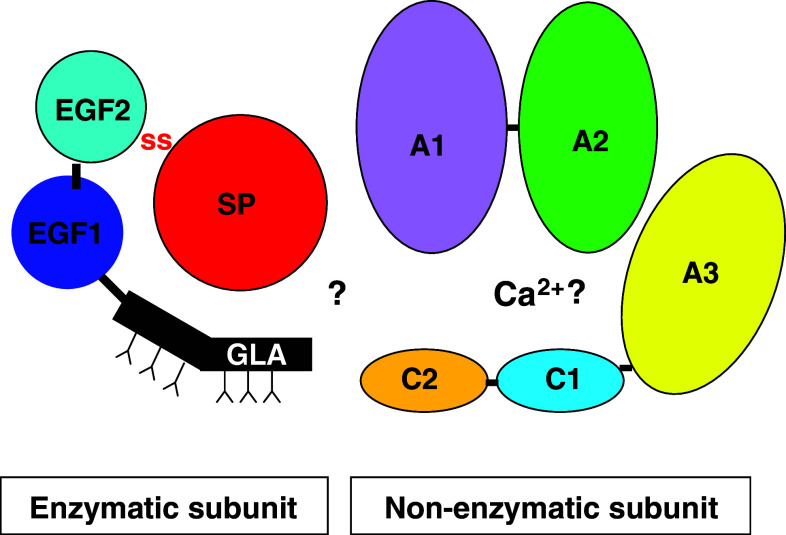
Among snake venom serine proteases, only group C prothrombin activators exist as complexes [114]. Pseutarin C from the P. textilis venom is one of the most well-characterized group C prothrombin activators [115]. This dimeric protein complex is a major toxic component (16–20% of the crude venom) and functions as a toxin. It comprises two subunits that are ~60 and ~220 kDa in size. The smaller catalytic subunit of pseutarin C (PCCS) has serine protease activity. PCCS has light and heavy chains that are held together by an interchain disulphide bridge. The light chain has a Gla domain and two epidermal growth factor-like domains (EGF), while the heavy chain has a serine protease domain (Fig. 7). The larger nonenzymatic subunit (PCNS) is structurally similar to mammalian factor Va; the heavy chain has A1 and A2 domains, whereas the light chain has A3, C1 and C2 domains [115]. It converts prothrombin to thrombin and requires only Ca2+ and phospholipids as co-factors for their optimal activity [115]. PCCS and factor Xa activate prothrombin to thrombin by cleaving the same peptide bonds at Arg274–Thr275 and Arg323–Ile324 [115]. Thus, structurally and functionally, pseutarin C is similar to the blood coagulation factor Xa–Va complex.
Fig. 7.
Diagrammatic representation of Pseutarin C. The enzymatic subunit contains the Gla, EGF1, EGF2 and serine protease domain, whereas the enzymatic subunit contains the A1–A3 and C1–C2 domain. The Gla and EGF domains are linked to the serine protease domain by a disulphide bridge. The glycosylation sites of the Gla domain are shown with “Y” in the Figure. The non-enzymatic subunit contains a heavy chain and a light chain similar to mammalian factor Va. The heavy chain contains A1 and A2 domains, whereas light chain contains the A3, C1 and C2 domains. The interaction between the enzymatic and non-enzymatic subunit is not yet know and hence shown with a “?” symbol
Snake venom l-amino acid oxidase (SV-LAAOs)
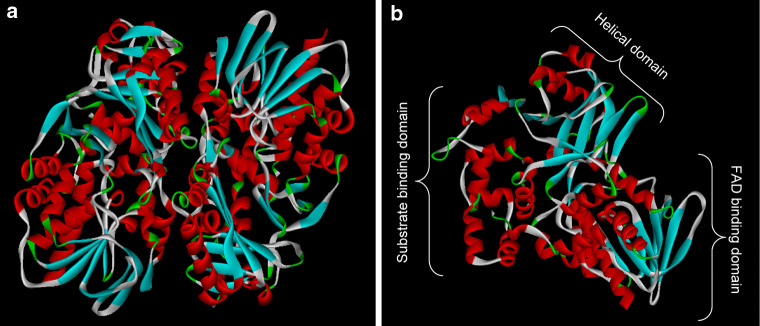
SV-LAAOs are flavoezymes that catalyze the oxidative deamination of l-amino acid to α-keto acids with liberation of hydrogen peroxide and ammonia. The byproduct, hydrogen peroxide, is thought to be responsible for its toxic effects. This group of enzymes is one of the most common components of snake venoms [116–125] and is responsible for the yellow color of the venom. Structurally, they are homodimers with a molecular weight of 110–150 kDa. The subunits are held together either covalently or non-covalently. Each subunit has three domains, the FAD-binding domain, substrate-binding domain and helical domain (Fig. 8) [126]. The FAD-binding domain is similar to the other LAAO found in human, bacteria and fungi and is found to be important for its enzymatic activity [127]. The interface between the substrate-binding domain and the helical domain contributes to the formation of the funnel-like pathway through which the substrate enters the active site [128]. Calloselasma rhodosoma LAAO has two glycosylation sites at Asn172 and Asn361 [129]. The carbohydrate moiety is a bis-sialylated, biantenary core-fucosylated dodecasaccharide. This glycosylation is important for its enzymatic activity [127] and hence its antibacterial activity [129]. The biological effect of this enzyme during envenomation is not clearly understood. However, in vitro studies showed that they display various activities like pro-apoptosis, cytotoxic, antitumoral, hemorraghic, initiation or inhibition of platelet aggregation and antibacterial activity [117, 130–133].
Fig. 8.
Ribbon model of l-amino acid oxidase (PDB ID: 1F8S) showing the homodimer (a) and monomer (b). The monomer has three different domains, the FAD-binding domain, helical domain and substrate-binding domain, which are shown in the ribbon model
Snaclecs (CTLs and CLRPs)
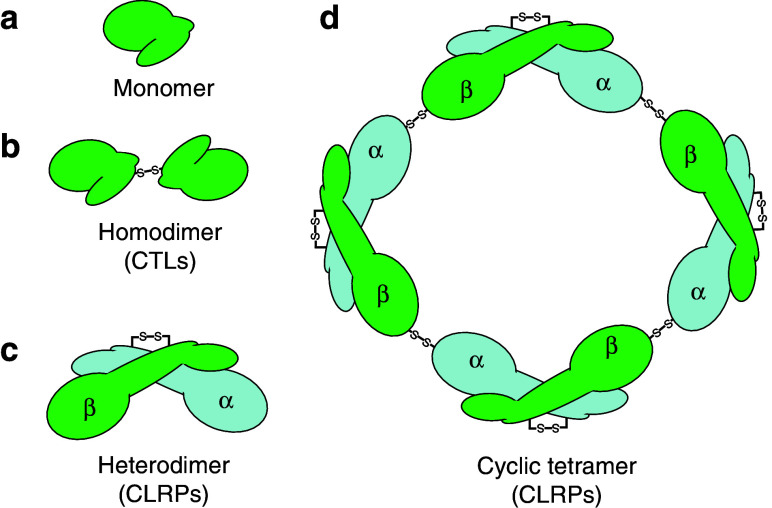
This group of non-enzymatic proteins in snake venom is growing in number and has been renamed as Snaclecs (Snake venom C-type lectins) by the Registry on Exogenous Hemostatic Factors of the Scientific and Standardization Committee of the International Society on Thrombosis and Hemostasis [134]. Snaclecs have two structurally similar subclasses of proteins, namely CTLs and CLRPs. They form a superfamily of non-enzymatic proteins and are commonly found in venoms of all families of snakes. They are either homodimers or heterodimers, and the subunits are mostly held together by a single inter-subunit disulfide bond. At times, four to five dimeric units aggregate to form larger complexes (Figs. 9, 10).
Fig. 9.
Schematic representation of C-type and C-type lectin-like proteins from snake venom
Fig. 10.
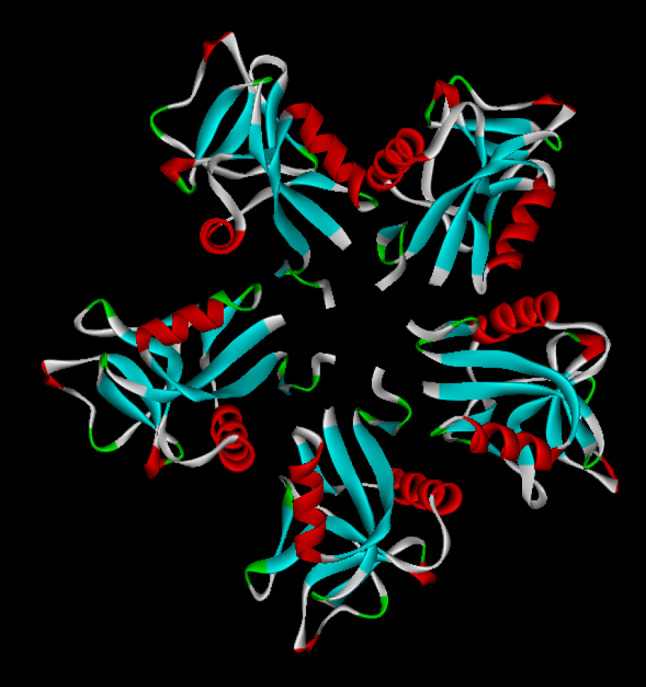
Ribbon model of rattlesnake venom lectins (RSL) (PDB ID: 1JZN) showing pentameric structure
C-type lectins
CTLs exhibit biological activities like adhesion, endocytosis and pathogen neutralization in the presence of Ca2+ [135], hemagglutinating activities [136, 137] and platelet aggregation [138, 139]. They are homodimers of subunits containing 135–141-amino acid residues [140]. These covalent homodimers have been reported from various snakes, such as Crotalus atrox [136], Bothrops jararaca [141], Lacheis muta stenophyrs [142], Protobothrops (formerly, Trimeresurus) stejnegeri [143], Crotalus ruber [144] and B. multicinctus [137]. These proteins are cysteine rich and contain nine half cysteines; eight of them are involved in the intra-subunit disulphide bridge, while the ninth cysteine (Cys86) is involved in inter-subunit interaction [136]. They bind to mono- and oligosaccharides in the presence of Ca2+ via the carbohydrate recognition domain (CRD) [145]. This domain binds to carbohydrates through a Ca2+ ion, which is coordinated by eight conserved amino acid residues, namely Cys31, Gly69, Trp92, Pro97, Cys106, Asp120, Cys123 and Cys131 [136]. In addition, Q97, D98, N119 and E104 are involved in binding to the carbohydrate molecule [146]. Thus, Ca2+ binding [136, 141] and the interchain disulphide bond between the monomers [147] are required for the agglutination activity.
In addition to homodimeric, homooligomeric CTL has been reported from C. atrox venom called rattlesnake venom lectin (RSL) [136, 147]. It induces both RBC (red blood cells) agglutination and platelet aggregation [136, 148]. The crystal structure of this protein [146] shows that each monomer is disulphide linked (C86–C86), and five such dimers interact to form the pentameric structure (Fig. 10). The CRDs remain exposed and simultaneously interact with cell surface glycoconjugates. Thus, complex formation in this molecule results in multiple interaction sites on each surface of the complex [146]. Each subunit of the pentamer can recognize two different carbohydrate molecules and initiate the process of agglutination.
C-type lectin-related proteins (CLRPs)
CLRPs are similar to CTLs, but lack carbohydrate-binding activity and are known to induce various biological activities like anticoagulant, procoagulant and agonist/antagonist of platelet activation [145, 149]. The anticoagulant effect of CLRPs is through binding to coagulation factors like X and IX [150, 151] or to thrombin [152–154]. CLRPs also bind to the platelet membrane receptor and cause platelet aggregation [155]. Structurally, CLRPs have α and β subunits, which may be either heterodimers (αβ) or oligomers of heterodimers (αβ)4 [91, 149] (Fig. 9c, d).
Heterodimeric CLRPs
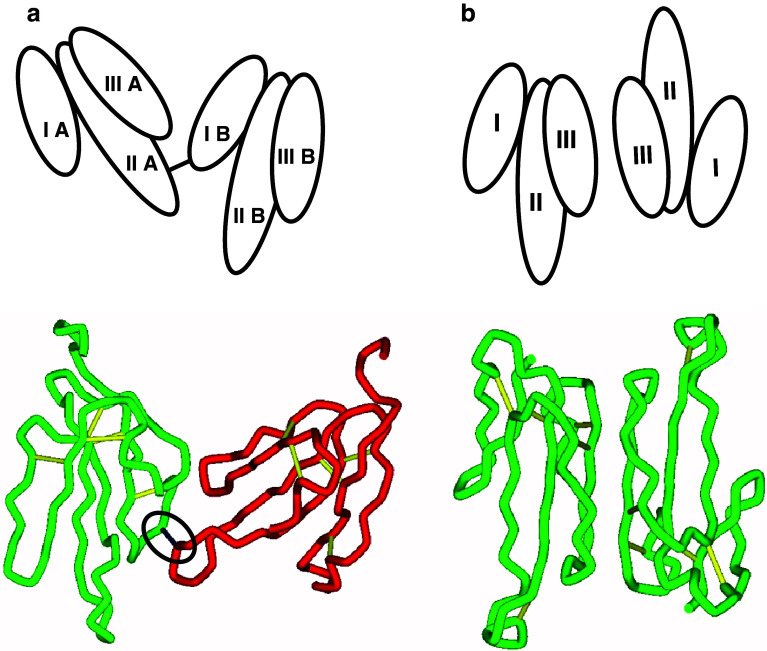
Heterodimeric CLRPs are reported from several snake venoms and are composed of two different α and β subunits (Fig. 11). IX/X binding protein from the venom of Protobothrops (formerly, Trimeresurus) flavoviridis specifically binds to factor IX and X and disturbs the coagulation process [150]. The α and β subunits are homologous to CLRPs and have similar disulphide bonding patterns [156]. The crystal structure of IX/X-bp shows that it is an intertwined dimer, and the central loop of each subunit is swapped within the αβ dimer [157]. The overall structure of the subunit is similar to rat mannose-binding protein. Despite such structural similarities, the Ca2+ binding site in IX/X is different from CTLs.
Fig. 11.
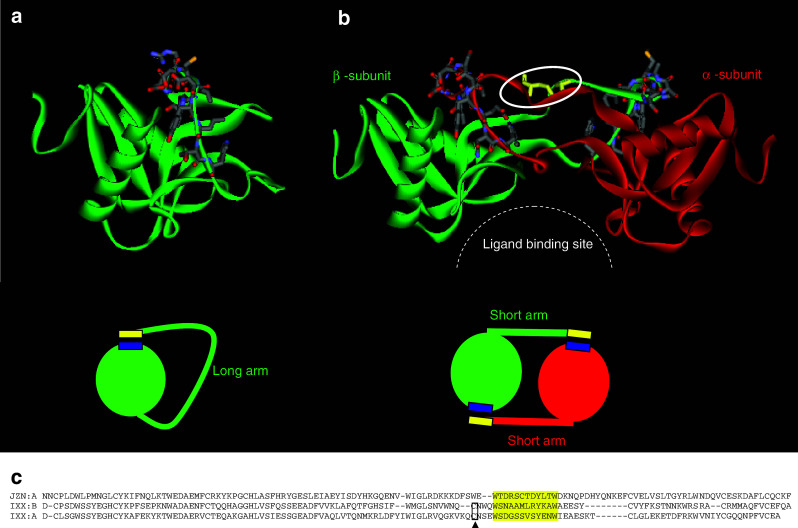
a, b Ribbon and schematic model of rattlesnake venom lectins (RSL) monomer (PDB ID: 1JZN) and factor IX/X-binding protein from Trimeresurus flavoviridis (PDB ID: 1IXX). The amino acid residues involved in domain swapping are shown in the stick model. The ligand-binding site in IX/X-binding protein formed by domain swapping is shown in the dotted arc. c Alignment of amino acid sequence of chain A of RSL, chain A and B of IX/X binding protein. The amino acid residues involved in domain swapping are highlighted in yellow color. The extra cysteine residues that form the interchain disulphide bridge in IX/X binding between chain A and B are boxed and marked (arrowhead)
Bothrojaracin from B. jararaca venom is a potent thrombin inhibitor [152, 154]. It has a molecular weight of 28 kDa with two subunits of 15 and 13 kDa linked together by a disulphide bridge between the seventh cysteine residues of both the subunits [152]. It inhibits thrombin function by interacting non-covalently with exosite I, but it does not interact with the catalytic site [154]. It also interacts with the proexosite I of human prothrombin forming a 1:1 non-covalent complex with the zymogen. Further, it decreases the prothrombin activation by factor Xa in the presence of factor Va [158]. Thus, bothrojaracin is a strong anticoagulant protein complex that inhibits thrombin as well as prothrombin activation.
Rhodocetin, a potent inhibitor of platelet aggregation, is a unique heterodimeric CLRP isolated from the venom of the Malayan Pit Viper (Calloselasma rhobostoma) [159]. Interestingly, the α and β subunits are held together by non-covalent interaction in contrast to other CLRPs where the subunits are covalently held together. It has all the conserved half cysteine residues as other CLRPs, but the seventh cysteine is replaced by serine in the α subunit and arginine in the β subunit. The complex formation at the ratio of 1:1 has been demonstrated by size exclusion chromatography and by SPR studies. The recombined complex exhibits biological activity similar to the native complex [159]. However, the individual subunits were unable to exhibit significant biological activities, indicating that the complex formation is important for its activity. The crystal structure of this protein has been reported [160].
Oligo heterodimeric C-type lectin-related proteins (CLRPs)
Oligo-heterodimeric CLRPs are composed of more than one heterodimeric complex. Convulxin isolated from the venom of C. durissus terrificus is a heterodimeric CLRP that lacks CRD and Ca2+ binding domains [161]. Functionally, it is a potent platelet agonist that acts on glycoprotein VI (GPVI) [162] as well as causes cardiovascular and respiratory disturbances [163]. Crystal structure revealed that it is a heterotetramer of α4β4 subunits [163, 164]. The α and β subunits are linked together by disulphide bridges similar to CLRPs. This heterodimer forms the cyclic heterotetramer complex by additional disulphide bridges between Cys135 of the C-terminal end of α subunit and Cys3 of the N-terminal end of β subunit of the neighboring heterodimer [163] (Fig. 9d).
Flavocetin-A from Protobothrops flavoviridis (formerly, Trimeresurus flavoviridis) venom [165] binds to platelet GP Ibα-subunit with high affinity and strongly inhibits vWF dependent platelet aggregation. The heterotetrameric complex formation in flavocetin-A is similar to convulxin where the heterodimers interact through a disulphide bridge between the C-terminal end of α subunit and N-terminal end of β subunit [166]. The binding sites in the tetramer act cooperatively, resulting in high affinity binding to GP Ibα-subunit [166].
Stejnulxin isolated from Protobothrops (formerly, Trimeresurus) stejnegeri venom is a unique CLRP as it contains one α and two β subunits (α1β2). However, the molecular weight of β1 (20 kDa) and β2 (22 kDa) differs due to the heterogeneity in glycosylation. Stejnulxin is reported to potently induce human platelet aggregation activity in a dose-dependent manner through binding to αIIbβ3 and fibrinogen as anti-GPVI polyclonal antibodies inhibit its activity [167].
Three-finger toxin complexes
The three-finger toxins (3FTxs) are non-enzymatic proteins containing 60–74 amino acid residues, abundantly found in the venoms of elapids (cobras, kraits and mambas) and hydrophiids (sea snakes) [168]. However, their presence in the venoms of colubridae [169–171] and in the transcriptomes of crotalidae snakes (rattlesnakes) [172, 173] has been revealed recently. Structurally, 3FTxs have three β-stranded loops called the “fingers” extending from a small, globular, hydrophobic core that is cross-linked by four conserved disulfide bridges [174, 175]. Although the members of this family of proteins are structurally similar, they exhibit different pharmacological functions, such as neurotoxicity, cardiotoxicity, cytotoxicity, anticoagulant, hypotensive effect, antimicrobial activity and platelet aggregation [174–178]. Thus, it has been proposed that the hydrophobic core from which the three loops emerge has been used to anchor different functional groups as most of the functionally important residues reside in the loops [174]. They exist as mostly monomers in the venom, but covalent and non-covalent dimers have also been reported and are discussed below.
Covalent complexes
Irditoxin is the first covalently linked heterodimeric 3FTx isolated and characterized from the venom of a colubrid snake, Boiga irregularis [179] (Fig. 12a). The three-dimensional structure shows a three-finger fold similar to other non-conventional 3FTxs. In additional to the ten conserved cysteine residues, both subunits have one additional cysteine residue. The eleventh cysteine residues of subunits A and B are present at the tip of the first and second loops at the 17th and 42nd position of the mature proteins respectively, and they form the inter-subunit disulphide bridge. The N-terminal ends of both subunits have additional seven amino acid residues similar to other colubrid three-finger toxins such as denmotoxin and α-colubritoxin [169, 180]. The quaternary structure of this dimer (Fig. 12) is also unique compared to κ-toxins (see below). Functionally, irditoxin is a potent and taxa-specific postsynaptic neurotoxin; it is at least three orders of magnitude more potent in blocking nerve transmission in chick biventer cervicis muscle than in rat hemidiaphragm [179]. However, the role of individual subunits in neurotoxicity is yet to be elucidated.
Fig. 12.
Three-finger toxin (3FTxs) complexes. Schematic and ribbon model of κ-bungarotoxin (non-covalent) (a) and iriditoxin (covalent) (b). The interchain disulphide bridge of irditoxin is encircled
Other covalent dimers of 3FTxs Recently, covalently linked homodimer of α-cobrotoxin (αCT–αCT) and heterodimers of cytotoxin and α-neurotoxin from the venom of Naja kaouthia have been reported [181]. The exact molecular interactions in these dimers are not clearly understood; however, disulphide bonds of the hydrophobic core might be involved in the dimerization. Functionally, the heterodimer, αCT-CX still binds to muscle type and α7 nAChR, but loses its cytotoxic activity. In this case dimerization leads to the loss of activity, whereas the homodimer αCT–αCT recognizes α3β2 neuronal nAChR similar to κ-bungarotoxin and hence is considered as its analogue [181]. Thus, the dimerization leads to a new function. However, these dimeric complexes are found in extremely small amounts (<0.04%) in the venom and hence might not be physiologically relevant.
Non-covalent complexes
κ-Bungarotoxin is a non-covalent homodimeric 3FTx isolated and characterized from the venom of B. multicinctus [182] (Fig. 12b). It exists as a homodimer in solution and exhibits neurotoxicity [183, 184]. Under denaturing conditions the monomers of this non-covalent complex can be separated. The self association of κ-bungarotoxin has been determined by size exclusion chromatography, sedimentation velocity and sedimentation equilibrium [185]. The solution and crystal structures of κ-bungarotoxin have been reported [186–188]. Structurally, the two subunits are oppositely oriented (Fig. 12b), and the interaction between the subunits is mostly through main-chain-main-chain hydrogen bonding. Two more hydrogen bonds between the side chains of Gln48 are involved in the interaction. In addition to this, hydrophobic interactions are also involved in the dimer formation [188]. Structure-function relationship studies have shown that Arg-34 and Pro-36 (second loop) are important for binding to neuronal receptors [189]. Similar κ-neurotoxins, such as κ2-neurotoxin and κ3-neurotoxin from B. multicinctus and κ-flavitoxin from Bungarus flaviceps venoms, have also been reported to form homodimers through self aggregation [190].
Synergistic 3FTxs
Some toxins from mamba venom are known to enhance the low toxicity of certain toxins. For example, S2C4 isolated from the venom of Dendroaspis jamesoni interacts with angusticeps-type toxins and acts synergistically. Individually, these proteins are less toxic, but they act as potent toxins when injected into the mice in combination [191]. The exact molecular interactions of these toxins are not yet known.
Complex formation and toxicity
Snake venom toxicity is due to the cumulative effect of various toxins present in the venom. Although most toxins are active individually, they will exert synergistic effects in combination. For example, neurotoxic components from different families of proteins act in concert and block the neuromuscular transmission. The presynaptic activity of PLA2 toxins and the postsynaptic activity of 3FTxs may readily contribute to immobilization and death of the prey. Similarly, hemorrhagic toxins can induce much more severe damage together with anticoagulant and antiplatelet components of the venom. In Dendroaspis angusticeps venom, dendrotoxin induces the release of acetylcholine by its action on potassium channels, while fasciculins inhibit acetylcholinesterase. Thus, both toxins enhance acetylcholine concentration in the neuromuscular synapse. In these instances, the individual components do not interact with each other, but their individual biological actions in concert enhance the pharmacological efficiency of the venom. In addition to such synergistic actions, complex formation among snake venom proteins contributes significantly towards enhancing the potency of the individual components of the complex. It adds another dimension to the structural and functional diversity of the venom composition. Conformational changes during complex formation lead to modifications to the active site/functional site that result in enhanced activities. Larger size of the complex provides larger binding surfaces and hence increased affinity of interactions with target receptors/ion channels. Sometimes protein complexes achieve entirely new activity because of exposure of newer surface that were buried in individual subunits.
Non-covalent interactions in the complex formation
The process of complex formation is directed through covalent (inter-subunit disulfide bonds) and/or non-covalent interactions between the subunits. Even when covalent linkages occur between the subunits, there are significant non-covalent interactions to keep the partnering subunits together (see descriptions of individual complexes above). Although in CLRPs, domain swapping and interchain disulfide are responsible for the complex formation (Fig. 11), the subunits in rhodocetin interact with each other by domain swapping alone, and both subunits lack the cysteine residues that are involved in the covalent linkage. Therefore, we hypothesize that the subunits indeed interact at first through non-covalent interactions, and subsequently the complex is further stabilized by the disulfide bond. Thus, non-covalent interaction is most likely the primary driving force in the complex formation. It would be interesting to understand the co-evolution of these inter-subunit interfaces.
Complex formations contribute to both structural stability and functional evolution. They are expected to be similar, if not identical, among closely related proteins. Interestingly, the complex formation among CTLs and CLRPs are distinct. In IX/X-binding protein and other CLRPs, complex formation occurs through a phenomenon called “domain swapping.” In domain swapping, exchange of identical structural elements takes place between two or more molecules to form dimers or oligomers [192]. In IX/X bp, the central parts of α and β subunits project out and interact with each other and form a covalent dimer [91]. Due to this domain swapping, the hinge region forms a concave structure between α and β subunits and provides a new functional surface to the heterodimer. In contrast, in CTLs individual subunits are able to bind to carbohydrates, similar to mannose-binding protein, but for the lectin-like function they need at least bivalency, which is achieved through a simple interchain disulfide linkage (Fig. 9b). In these cases domain swapping does not occur. Although dimerization is essential for the expression of their biological activities, it occurs with or without domain swapping, leading to distinct quaternary structures among these closely related proteins. Therefore, it is interesting and intriguing to decipher the molecular mechanism of domain swapping. We analyzed the domain swapping region and its surroundings (Fig. 11c) to understand the domain swapping in CLRPs. The domain swapping region in CLRPs and the corresponding region in CTLs (shown in yellow) are similar, and their interaction with other subunits in CLRPs and within the same subunit in CTLs are also similar (shown in blue); in both cases two Trp residues (one at each end) and a Tyr residue in the middle are buried in the interaction interface. In the case of CTLs, the segment on the C-terminal side of this interaction site (yellow segment) is longer (Fig. 11c), and hence it folds on itself, forming intra-subunit contacts. In contrast, in CLRPs the corresponding segment is six to eight residues shorter and thus unable to fold back and interact within each subunit. This necessitates the interaction of the yellow segment with a second subunit leading to domain swapping (Fig. 11). Thus, although CLRPs and CTLs share significant sequence similarities, the subtle differences in the positioning of interactive segment lead to distinct quaternary structures of these two classes of proteins.
Post-translational modifications in complex formation
Post-translational modifications, including phosphorylation, glycosylation, disulfide bond formation and proteolysis—are known to alter the conformation and function of the proteins. So far, there is no documented evidence for the role of phosphorylation (temporal) and glycosylation in the formation of snake venom protein complexes. However, indeed disulfide bond formation (discussed above) and proteolysis play an important role in the complex formation. The acidic subunit of crotoxin is generated by proteolysis of a PLA2-like precursor protein. It is well documented that this subunit (three disulphide-linked polypeptides) easily forms a complex with the basic subunit. To understand the role of proteolysis in crotoxin complex formation, it is important to determine whether the intact, uncleaved precursor of the acidic subunit interacts with the basic subunit and enhances its presynaptic neurotoxicity.
Evolution of protein complexes
Snake venom contains a handful of superfamilies of proteins that are mostly recruited from the body proteins. Some of these families have evolved from non-toxic to most toxic proteins in the venom gland. They have evolved through gene duplication and accelerated evolution. Group C prothrombin activators of Australian elapid venoms are protein complexes similar to factor Xa and factor Va found in the coagulation cascade. Based on their homology of blood coagulation factors, it is easier to speculate their origin through duplication of genes encoding coagulation factors and recruitment for expression in the venom gland to target prey’s coagulation cascade (for a review see [193]). However, origins of other protein complexes are more difficult to speculate.
It is logical to expect similar protein complexes in the venoms of closely related snake species. Such protein complexes exhibit similar pharmacological properties. For example, heterodimeric crotoxin and related toxins in Crotalus species share similar structure and neurotoxic properties. Similarly, heterotrimeric taipoxin and related toxins in Oxyuranus species share similar structural and functional attributes. Several interesting questions still remain to be answered. They include: (1) how these and other protein complexes in which the subunits are products of independent genes were recruited for their expression in the venom gland; (2) how inter-molecular interfaces evolved or what determined the complementary partners; (2) what were the parameters for initial natural selection—is it structure or function?
Despite the intriguing questions, the similar protein complexes in closely related species are easier to explain through simple molecular phylogeny. In contrast, vipoxin and viperotoxin F isolated from the venoms of Vipera species are unique examples of closely related protein complexes with distinct pharmacological properties. Vipoxin acts as a postsynaptic neurotoxin, whereas viperotoxin F is a presynaptic neurotoxin. Individual basic subunits of both complexes exhibit presynaptic neurotoxicity, while individual acidic subunits are pharmacologically inactive. Although they form similar quaternary complexes (Fig. 4a), vipoxin shows a drastic change in its pharmacology. Therefore, it is interesting to examine the structure of these two closely related complexes. The subunits in both complexes share high structural identity; there are only five and three unconserved changes in amino acids between the basic and acidic subunits (Fig. 4b). Most of these differences appear to be located on the same molecular surface and alter the surface charges (Fig. 4c). It will be interesting to examine the role of these residues in the observed drastic switch in the pharmacology of vipoxin.
Role of subunits
The individual components/subunits contribute significantly to the structure as well as enhanced function of the complex. In the case of crotoxin, the inactive CA subunit acts as a chaperone. The acidic CA subunit blocks the non-specific binding of the basic CB subunit and delivers this enzymatically active subunit to the presynaptic site [9]. Unlike other protein complexes in which the subunits stay together when they interact with their target receptor/ion channel, the acidic subunit dissociates upon binding to the presynaptic site [3]. This dissociation can be explained by two scenarios: (1) the site recognized by the acidic subunit and the receptor at least partially overlap or (2) the acidic subunit and the receptor bind to non-competitive allosteric sites on the basic subunit. In both scenarios, the affinity between the basic subunit and the receptor is expected to be higher than that between the acidic and basic subunits to drive the binding to target receptor and dissociation. In the case of β-BuTx, the B chain determines the affinity to the target potassium channel and delivers the enzymatically active A chain to the presynaptic site. In both these cases, eventual inhibition of neurotransmission is due to the physical destruction and interference by phospholipid hydrolysis at the specific sites [194]. In group C prothrombin activators, the nonenzymatic subunit, similar to factor Va, probably enhances the Vmax of the catalytic subunit. In metalloprotease complexes, RVV-X and carinactivase-1, the respective substrates factor X and prothrombin bind to the light chains through their Gla domains [157, 195]. Thus, their CLRP subunits bind to specific substrates and bring them near the protease domain for activation.
Drawbacks to complex formation
In general, the larger the size of the protein, the slower will be its ability to diffuse and distribute through various tissues. As the protein complexes are larger, it may take more time to diffuse into the prey/victim’s body. At times, such large complexes may become physiologically irrelevant due to their inability to enter the right tissue or organ.
Conclusion and future prospects
Protein complexes are one of the important components in some snake venoms, and they play important roles in venom toxicity. In many snakes they are the major lethal component of the venom. Currently, only a small number of protein complexes have been fully characterized. To date, we understand the role of subunits in a small number of protein complexes. Understanding the structure and function of these complexes will provide a better understanding of their role in venom toxicity. Biophysical interactions and structures of venom complexes will further help us to understand better the protein-protein interactions in snake venom as well as their target receptor/ion channels.
Acknowledgments
This work was supported by grants from the Biomedical Research Council (BMRC) of Singapore. We thank Cho Yeow Koh for preparing Fig. 6. The authors thank the anonymous reviewers for their constructive comments.
References
- 1.Slotta K, Fraenkel-Conrat H. Schlangengifte-III. Mitteilung: Reinigung und Krystallisation des Klapperschlangen-Giftes. Ber Dtsch Chem Ges. 1938;71:1076–1081. doi: 10.1002/cber.19380710527. [DOI] [Google Scholar]
- 2.Kini RM. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Bon C. Multicomponent neurotoxic phospholipases A2. In: Kini RM, editor. Phospholipase A2 enzyme: structure, function and mechanism. Chichester: Wiley; 1997. pp. 269–285. [Google Scholar]
- 4.Chang CC, Lee CY. Isolation of neurotoxins from the venom of Bungarus multicinctus and their modes of neuromuscular blocking action. Arch Int Pharmacodyn Ther. 1963;144:241–257. [PubMed] [Google Scholar]
- 5.Chu CC, Li SH, Chen YH. Resolution of isotoxins in the beta-bungarotoxin family. J Chromatogr A. 1995;694:492–497. doi: 10.1016/0021-9673(94)01173-C. [DOI] [PubMed] [Google Scholar]
- 6.Kondo K, Toda H, Narita K, Lee CY. Amino acid sequences of three beta-bungarotoxins (beta 3-, beta 4-, and beta 5- bungarotoxins) from Bungarus multicinctus venom: amino acid substitutions in the A chains. J Biochem. 1982;91:1531–1548. doi: 10.1093/oxfordjournals.jbchem.a133844. [DOI] [PubMed] [Google Scholar]
- 7.Chu CC, Chu ST, Chen SW, Chen YH. The non-phospholipase A2 subunit of beta-bungarotoxin plays an important role in the phospholipase A2-independent neurotoxic effect: characterization of three isotoxins with a common phospholipase A2 subunit. Biochem J. 1994;303(Pt 1):171–176. doi: 10.1042/bj3030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo K, Toda H, Narita K. Characterization of phospholipase A activity of beta1-bungarotoxin from Bungarus multicinctus venom. I: its enzymatic properties and modification with p-bromophenacyl bromide. J Biochem (Tokyo) 1978;84:1291–1300. doi: 10.1093/oxfordjournals.jbchem.a132248. [DOI] [PubMed] [Google Scholar]
- 9.Bon C, Changeux JP, Jeng TW, Fraenkel-Conrat H. Postsynaptic effects of crotoxin and of its isolated subunits. Eur J Biochem. 1979;99:471–481. doi: 10.1111/j.1432-1033.1979.tb13278.x. [DOI] [PubMed] [Google Scholar]
- 10.Kwong PD, McDonald NQ, Sigler PB, Hendrickson WA. Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action. Structure. 1995;3:1109–1119. doi: 10.1016/S0969-2126(01)00246-5. [DOI] [PubMed] [Google Scholar]
- 11.Su MJ, Chang CC. Presynaptic effects of snake venom toxins which have phospholipase A2 activity (beta-bungarotoxin, taipoxin, crotoxin) Toxicon. 1984;22:631–640. doi: 10.1016/0041-0101(84)90003-5. [DOI] [PubMed] [Google Scholar]
- 12.Harvey AL, Karlsson E. Protease inhibitor homologues from mamba venoms: facilitation of acetylcholine release and interactions with prejunctional blocking toxins. Br J Pharmacol. 1982;77:153–161. doi: 10.1111/j.1476-5381.1982.tb09281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benishin CG. Potassium channel blockade by the B subunit of beta-bungarotoxin. Mol Pharmacol. 1990;38:164–169. [PubMed] [Google Scholar]
- 14.Wu PF, Wu SN, Chang CC, Chang LS. Cloning and functional expression of B chains of beta-bungarotoxins from Bungarus multicinctus (Taiwan banded krait) Biochem J. 1998;334(Pt 1):87–92. doi: 10.1042/bj3340087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu ST, Chu CC, Tseng CC, Chen YH. Met-8 of the beta 1-bungarotoxin phospholipase A2 subunit is essential for the phospholipase A2-independent neurotoxic effect. Biochem J. 1993;295(Pt 3):713–718. doi: 10.1042/bj2950713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang LS, Yang CC. Role of the N-terminal region of the A chain in beta 1-bungarotoxin from the venom of Bungarus multicinctus (Taiwan-banded krait) J Protein Chem. 1988;7:713–727. doi: 10.1007/BF01025579. [DOI] [PubMed] [Google Scholar]
- 17.Petersen M, Penner R, Pierau FK, Dreyer F. Beta-bungarotoxin inhibits a non-inactivating potassium current in guinea pig dorsal root ganglion neurones. Neurosci Lett. 1986;68:141–145. doi: 10.1016/0304-3940(86)90244-2. [DOI] [PubMed] [Google Scholar]
- 18.Rehm H, Betz H. Binding of beta-bungarotoxin to synaptic membrane fractions of chick brain. J Biol Chem. 1982;257:10015–10022. [PubMed] [Google Scholar]
- 19.Rehm H, Betz H. Solubilization and characterization of the beta-bungarotoxin-binding protein of chick brain membranes. J Biol Chem. 1984;259:6865–6869. [PubMed] [Google Scholar]
- 20.Tsai IH, Lu PJ, Wang YM, Ho CL, Liaw LL. Molecular cloning and characterization of a neurotoxic phospholipase A2 from the venom of Taiwan habu (Trimeresurus mucrosquamatus) Biochem J. 1995;311(Pt 3):895–900. doi: 10.1042/bj3110895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos KF, Murakami MT, Cintra AC, Toyama MH, Marangoni S, Forrer VP, Brandao N, Jr, Polikarpov I, Arni RK. Crystallization and preliminary X-ray crystallographic analysis of the heterodimeric crotoxin complex and the isolated subunits crotapotin and phospholipase A2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:287–290. doi: 10.1107/S1744309107006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faure G, Guillaume JL, Camoin L, Saliou B, Bon C. Multiplicity of acidic subunit isoforms of crotoxin, the phospholipase A2 neurotoxin from Crotalus durissus terrificus venom, results from posttranslational modifications. Biochemistry. 1991;30:8074–8083. doi: 10.1021/bi00246a028. [DOI] [PubMed] [Google Scholar]
- 23.Wu SH, Chang FH, Tzeng MC. Separation of the subunits of crotoxin by high-performance liquid chromatography. J Chromatogr. 1983;259:375–377. doi: 10.1016/S0021-9673(01)88024-9. [DOI] [PubMed] [Google Scholar]
- 24.Aird SD, Kaiser II, Lewis RV, Kruggel WG. Rattlesnake presynaptic neurotoxins: primary structure and evolutionary origin of the acidic subunit. Biochemistry. 1985;24:7054–7058. doi: 10.1021/bi00346a005. [DOI] [PubMed] [Google Scholar]
- 25.Faure G, Choumet V, Bouchier C, Camoin L, Guillaume JL, Monegier B, Vuilhorgne M, Bon C. The origin of the diversity of crotoxin isoforms in the venom of Crotalus durissus terrificus . Eur J Biochem. 1994;223:161–164. doi: 10.1111/j.1432-1033.1994.tb18978.x. [DOI] [PubMed] [Google Scholar]
- 26.Marchi-Salvador DP, Correa LC, Magro AJ, Oliveira CZ, Soares AM, Fontes MR. Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom. Proteins. 2008;72:883–891. doi: 10.1002/prot.21980. [DOI] [PubMed] [Google Scholar]
- 27.Habermann E, Breithaupt H. Mini-review: the crotoxin complex—an example of biochemical and pharmacological protein complementation. Toxicon. 1978;16(1):9–30. doi: 10.1016/0041-0101(78)90056-9. [DOI] [PubMed] [Google Scholar]
- 28.Hendon RA, Tu AT. The role of crotoxin subunits in tropical rattlesnake neurotoxic action. Biochim Biophys Acta. 1979;578:243–252. doi: 10.1016/0005-2795(79)90132-6. [DOI] [PubMed] [Google Scholar]
- 29.Gopalakrishnakone P, Hawgood BJ. Morphological changes induced by crotoxin in murine nerve and neuromuscular junction. Toxicon. 1984;22:791–804. doi: 10.1016/0041-0101(84)90162-4. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez JM, Ponce-Soto LA, Marangoni S, Lomonte B. Systemic and local myotoxicity induced by snake venom group II phospholipases A2: comparison between crotoxin, crotoxin B and a Lys49 PLA2 homologue. Toxicon. 2008;51:80–92. doi: 10.1016/j.toxicon.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Sampaio SC, Brigatte P, Sousa-e-Silva MC, dos-Santos EC, Rangel-Santos AC, Curi R, Cury Y. Contribution of crotoxin for the inhibitory effect of Crotalus durissus terrificus snake venom on macrophage function. Toxicon. 2003;41:899–907. doi: 10.1016/S0041-0101(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 32.Sampaio SC, Rangel-Santos AC, Peres CM, Curi R, Cury Y. Inhibitory effect of phospholipase A(2) isolated from Crotalus durissus terrificus venom on macrophage function. Toxicon. 2005;45:671–676. doi: 10.1016/j.toxicon.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Strong PN, Goerke J, Oberg SG, Kelly RB. Beta-Bungarotoxin, a pre-synaptic toxin with enzymatic activity. Proc Natl Acad Sci USA. 1976;73:178–182. doi: 10.1073/pnas.73.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landucci EC, Condino-Neto A, Perez AC, Hyslop S, Corrado AP, Novello JC, Marangoni S, Oliveira B, Antunes E, de Nucci G. Crotoxin induces aggregation of human washed platelets. Toxicon. 1994;32:217–226. doi: 10.1016/0041-0101(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhang HL, Han R, Chen ZX, Chen BW, Gu ZL, Reid PF, Raymond LN, Qin ZH. Opiate and acetylcholine-independent analgesic actions of crotoxin isolated from crotalus durissus terrificus venom. Toxicon. 2006;48:175–182. doi: 10.1016/j.toxicon.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Costa LA, Miles H, Araujo CE, Gonzalez S, Villarrubia VG. Tumor regression of advanced carcinomas following intra- and/or peri-tumoral inoculation with VRCTC-310 in humans: preliminary report of two cases. Immunopharmacol Immunotoxicol. 1998;20:15–25. doi: 10.3109/08923979809034806. [DOI] [PubMed] [Google Scholar]
- 37.Donato NJ, Martin CA, Perez M, Newman RA, Vidal JC, Etcheverry M. Regulation of epidermal growth factor receptor activity by crotoxin, a snake venom phospholipase A2 toxin: a novel growth inhibitory mechanism. Biochem Pharmacol. 1996;51:1535–1543. doi: 10.1016/0006-2952(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 38.Rudd CJ, Viskatis LJ, Vidal JC, Etcheverry MA. In vitro comparison of cytotoxic effects of crotoxin against three human tumors and a normal human epidermal keratinocyte cell line. Invest New Drugs. 1994;12:183–184. doi: 10.1007/BF00873958. [DOI] [PubMed] [Google Scholar]
- 39.Yan CH, Yang YP, Qin ZH, Gu ZL, Reid P, Liang ZQ. Autophagy is involved in cytotoxic effects of crotoxin in human breast cancer cell line MCF-7 cells. Acta Pharmacol Sin. 2007;28:540–548. doi: 10.1111/j.1745-7254.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- 40.Mebs D, Ownby CL. Myotoxic components of snake venoms: their biochemical and biological activities. Pharmacol Ther. 1990;48:223–236. doi: 10.1016/0163-7258(90)90081-C. [DOI] [PubMed] [Google Scholar]
- 41.Kaiser II, Aird SD. A crotoxin homolog from the venom of the Uracoan rattlesnake (Crotalus vegrandis) Toxicon. 1987;25:1113–1120. doi: 10.1016/0041-0101(87)90268-6. [DOI] [PubMed] [Google Scholar]
- 42.Bieber AL, Tu T, Tu AT. Studies of an acidic cardiotoxin isolated from the venom of Mojave rattlesnake (Crotalus scutulatus) Biochim Biophys Acta. 1975;400:178–188. doi: 10.1016/0005-2795(75)90139-7. [DOI] [PubMed] [Google Scholar]
- 43.Ho CL, Lee CY. Presynaptic actions of Mojave toxin isolated from Mojave rattlesnake (Crotalus scutulatus) venom. Toxicon. 1981;19:889–892. doi: 10.1016/0041-0101(81)90086-6. [DOI] [PubMed] [Google Scholar]
- 44.Pool WR, Bieber AL. Fractionation of midget faded rattlesnake (Crotalus viridis concolor) venom: lethal fractions and enzymatic activities. Toxicon. 1981;19:517–527. doi: 10.1016/0041-0101(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 45.Straight RC, Glenn JL. Isolation and characterization of basic phospholipase (PLA2) and acidic subunits of canebrake toxin from Crotalus horridus atricaudatus venom using HPLC. Toxicon. 1988;27:80. [Google Scholar]
- 46.Chen YH, Wang YM, Hseu MJ, Tsai IH. Molecular evolution and structure-function relationships of crotoxin-like and asparagine-6-containing phospholipases A2 in pit viper venoms. Biochem J. 2004;381:25–34. doi: 10.1042/BJ20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aird SD, Kruggel WG, Kaiser II. Amino acid sequence of the basic subunit of Mojave toxin from the venom of the Mojave rattlesnake (Crotalus s. scutulatus) Toxicon. 1990;28:669–673. doi: 10.1016/0041-0101(90)90255-6. [DOI] [PubMed] [Google Scholar]
- 48.Bieber AL, Becker RR, McParland R, Hunt DF, Shabanowitz J, Yates JR, III, Martino PA, Johnson GR. The complete sequence of the acidic subunit from Mojave toxin determined by Edman degradation and mass spectrometry. Biochim Biophys Acta. 1990;1037:413–421. doi: 10.1016/0167-4838(90)90045-h. [DOI] [PubMed] [Google Scholar]
- 49.French WJ, Hayes WK, Bush SP, Cardwell MD, Bader JO, Rael ED. Mojave toxin in venom of Crotalus helleri (Southern Pacific Rattlesnake): molecular and geographic characterization. Toxicon. 2004;44:781–791. doi: 10.1016/j.toxicon.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Wooldridge BJ, Pineda G, Banuelas-Ornelas JJ, Dagda RK, Gasanov SE, Rael ED, Lieb CS. Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:169–179. doi: 10.1016/S1096-4959(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 51.Valdes JJ, Thompson RG, Wolff VL, Menking DE, Rael ED, Chambers JP. Inhibition of calcium channel dihydropyridine receptor binding by purified Mojave toxin. Neurotoxicol Teratol. 1989;11:129–133. doi: 10.1016/0892-0362(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 52.Chambers JP, Wayner MJ, Dungan J, Rael ED, Valdes JJ. The effects of purified Mojave toxin on rat synaptic membrane (Ca2+ + Mg2+)-ATPase and the dihydropyridine receptor. Brain Res Bull. 1986;16:639–643. doi: 10.1016/0361-9230(86)90135-8. [DOI] [PubMed] [Google Scholar]
- 53.Cate RL, Bieber AL. Purification and characterization of Mojave (Crotalus scutulatus scutulatus) toxin and its subunits. Arch Biochem Biophys. 1978;189:397–408. doi: 10.1016/0003-9861(78)90227-8. [DOI] [PubMed] [Google Scholar]
- 54.Tchorbanov B, Grishin E, Aleksiev B, Ovchinnikov Y. A neurotoxic complex from the venom of the Bulgarian viper (Vipera ammodytes ammodytes) and partial amino acid sequence of the toxic phospholipase A2. Toxicon. 1978;16:37–44. doi: 10.1016/0041-0101(78)90058-2. [DOI] [PubMed] [Google Scholar]
- 55.Mancheva I, Kleinschmidt T, Aleksiev B, Braunitzer G. Sequence homology between phospholipase and its inhibitor in snake venom: the primary structure of phospholipase A2 of vipoxin from the venom of the Bulgarian viper (Vipera ammodytes ammodytes, Serpentes) Biol Chem Hoppe Seyler. 1987;368(34):3–352. doi: 10.1515/bchm3.1987.368.1.343. [DOI] [PubMed] [Google Scholar]
- 56.Banumathi S, Rajashankar KR, Notzel C, Aleksiev B, Singh TP, Genov N, Betzel C. Structure of the neurotoxic complex vipoxin at 1.4 A resolution. Acta Crystallogr D Biol Crystallogr. 2001;57:1552–1559. doi: 10.1107/S0907444901013543. [DOI] [PubMed] [Google Scholar]
- 57.Perbandt M, Wilson JC, Eschenburg S, Mancheva I, Aleksiev B, Genov N, Willingmann P, Weber W, Singh TP, Betzel C. Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution. FEBS Lett. 1997;412(57):3–577. doi: 10.1016/s0014-5793(97)00853-3. [DOI] [PubMed] [Google Scholar]
- 58.Tchorbanov B, Aleksiev B, Bukolova-Orlova T, Burstein E, Atanasov B. Subfractionation and recombination of a neurotoxic complex from the venom of the Bulgarian viper (Vipera ammodytes ammodytes) FEBS Lett. 1977;76:266–268. doi: 10.1016/0014-5793(77)80165-8. [DOI] [PubMed] [Google Scholar]
- 59.Wang YM, Lu PJ, Ho CL, Tsai IH. Characterization and molecular cloning of neurotoxic phospholipases A2 from Taiwan viper (Vipera russelli formosensis) Eur J Biochem. 1992;209:635–641. doi: 10.1111/j.1432-1033.1992.tb17330.x. [DOI] [PubMed] [Google Scholar]
- 60.Freedman JE, Snyder SH. Vipoxin: a protein from Russell’s viper venom with high affinity for biogenic amine receptors. J Biol Chem. 1981;256:13172–13179. [PubMed] [Google Scholar]
- 61.Ovadia M, Kochva E, Moav B. Purification and partial characterization of lethal synergistic components from the venom of Vipera palaestinae . Toxicon. 1977;15:549–560. doi: 10.1016/0041-0101(77)90106-4. [DOI] [PubMed] [Google Scholar]
- 62.Simon T, Bdolah A, Kochva E. The two-component toxin of Vipera palaestinae: contribution of phospholipase A to its activity. Toxicon. 1980;18:249–259. doi: 10.1016/0041-0101(80)90003-3. [DOI] [PubMed] [Google Scholar]
- 63.Krizaj I, Bdolah A, Gubensek F, Bencina P, Pungercar J. Protein and cDNA structures of an acidic phospholipase A2, the enzymatic part of an unusual, two-component toxin from Vipera palaestinae . Biochem Biophys Res Commun. 1996;227:374–379. doi: 10.1006/bbrc.1996.1515. [DOI] [PubMed] [Google Scholar]
- 64.Batzri-Izraeli R, Bdolah A. Isolation and characterization of the main toxic fraction from the venom of the false horned viper (Pseudocerastes fieldi) Toxicon. 1982;20:867–875. doi: 10.1016/0041-0101(82)90074-5. [DOI] [PubMed] [Google Scholar]
- 65.Tsai MC, Lee CY, Bdolah A. Mode of neuromuscular blocking action of a toxic phospholipase A2 from Pseudocerastes fieldi (Field’s horned viper) snake venom. Toxicon. 1983;21:527–534. doi: 10.1016/0041-0101(83)90130-7. [DOI] [PubMed] [Google Scholar]
- 66.Francis B, Bdolah A, Kaiser II. Amino acid sequences of a heterodimeric neurotoxin from the venom of the false horned viper (Pseudocerastes fieldi) Toxicon. 1995;33:863–874. doi: 10.1016/0041-0101(95)00034-J. [DOI] [PubMed] [Google Scholar]
- 67.Fohlman J, Eaker D, Karlsoon E, Thesleff S. Taipoxin, an extremely potent presynaptic neurotoxin from the venom of the Australian snake taipan (Oxyuranus s. scutellatus): isolation, characterization, quaternary structure and pharmacological properties. Eur J Biochem. 1976;68:457–469. doi: 10.1111/j.1432-1033.1976.tb10833.x. [DOI] [PubMed] [Google Scholar]
- 68.Lind P, Eaker D. Amino-acid sequence of the alpha-subunit of taipoxin, an extremely potent presynaptic neurotoxin from the Australian snake taipan (Oxyuranus s. scutellatus) Eur J Biochem. 1982;124:441–447. doi: 10.1111/j.1432-1033.1982.tb06612.x. [DOI] [PubMed] [Google Scholar]
- 69.Fohlman J, Lind P, Eaker D. Taipoxin, an extremely potent presynaptic snake venom neurotoxin: elucidation of the primary structure of the acidic carbohydrate-containing taipoxin-subunit, a prophospholipase homolog. FEBS Lett. 1977;84:367–371. doi: 10.1016/0014-5793(77)80726-6. [DOI] [PubMed] [Google Scholar]
- 70.Fohlman J. Comparison of two highly toxic Australian snake venoms: the taipan (Oxyuranus S. scutellatus) and the fierce snake (Parademansia microlepidotus) Toxicon. 1979;17:170–172. doi: 10.1016/0041-0101(79)90296-4. [DOI] [PubMed] [Google Scholar]
- 71.Hodgson WC, Dal Belo CA, Rowan EG. The neuromuscular activity of paradoxin: a presynaptic neurotoxin from the venom of the inland taipan (Oxyuranus microlepidotus) Neuropharmacology. 2007;52:1229–1236. doi: 10.1016/j.neuropharm.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 72.Kuruppu S, Reeve S, Banerjee Y, Kini RM, Smith AI, Hodgson WC. Isolation and pharmacological characterization of cannitoxin, a presynaptic neurotoxin from the venom of the Papuan Taipan (Oxyuranus scutellatus canni) J Pharmacol Exp Ther. 2005;315:1196–1202. doi: 10.1124/jpet.105.093641. [DOI] [PubMed] [Google Scholar]
- 73.Su MJ, Coulter AR, Sutherland SK, Chang CC. The presynaptic neuromuscular blocking effect and phospholipase A2 activity of textilotoxin, a potent toxin isolated from the venom of the Australian brown snake, Pseudonaja textilis . Toxicon. 1983;21:143–151. doi: 10.1016/0041-0101(83)90057-0. [DOI] [PubMed] [Google Scholar]
- 74.Pearson JA, Tyler MI, Retson KV, Howden ME. Studies on the subunit structure of textilotoxin, a potent presynaptic neurotoxin from the venom of the Australian common brown snake (Pseudonaja textilis). 3: the complete amino-acid sequences of all the subunits. Biochim Biophys Acta. 1993;1161(22):3–229. doi: 10.1016/0167-4838(93)90217-f. [DOI] [PubMed] [Google Scholar]
- 75.Pearson JA, Tyler MI, Retson KV, Howden ME. Studies on the subunit structure of textilotoxin, a potent presynaptic neurotoxin from the venom of the Australian common brown snake (Pseudonaja textilis). 2: the amino acid sequence and toxicity studies of subunit D. Biochim Biophys Acta. 1991;1077(14):7–150. doi: 10.1016/0167-4838(91)90051-z. [DOI] [PubMed] [Google Scholar]
- 76.Scott DL, White SP, Otwinowski Z, Yuan W, Gelb MH, Sigler PB. Interfacial catalysis: the mechanism of phospholipase A2. Science. 1990;250:1541–1546. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown AM, Yatani A, Lacerda AE, Gurrola GB, Possani LD. Neurotoxins that act selectively on voltage-dependent cardiac calcium channels. Circ Res. 1987;61:I6–I9. [PubMed] [Google Scholar]
- 78.Possani LD, Martin BM, Yatani A, Mochca-Morales J, Zamudio FZ, Gurrola GB, Brown AM. Isolation and physiological characterization of taicatoxin, a complex toxin with specific effects on calcium channels. Toxicon. 1992;30:1343–1364. doi: 10.1016/0041-0101(92)90511-3. [DOI] [PubMed] [Google Scholar]
- 79.Rucavado A, Lomonte B, Ovadia M, Gutierrez JM. Local tissue damage induced by BaP1, a metalloproteinase isolated from Bothrops asper (Terciopelo) snake venom. Exp Mol Pathol. 1995;63:186–199. doi: 10.1006/exmp.1995.1042. [DOI] [PubMed] [Google Scholar]
- 80.Gutierrez JM, Romero M, Nunez J, Chaves F, Borkow G, Ovadia M. Skeletal muscle necrosis and regeneration after injection of BaH1, a hemorrhagic metalloproteinase isolated from the venom of the snake Bothrops asper (Terciopelo) Exp Mol Pathol. 1995;62:28–41. doi: 10.1006/exmp.1995.1004. [DOI] [PubMed] [Google Scholar]
- 81.Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000;82:841–850. doi: 10.1016/S0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 82.Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 83.Bjarnason JB, Fox JW. Snake venom metalloendopeptidases: reprolysins. Methods Enzymol. 1995;248:345–368. doi: 10.1016/0076-6879(95)48023-4. [DOI] [PubMed] [Google Scholar]
- 84.Castro HC, Zingali RB, Albuquerque MG, Pujol-Luz M, Rodrigues CR. Snake venom thrombin-like enzymes: from reptilase to now. Cell Mol Life Sci. 2004;61:843–856. doi: 10.1007/s00018-003-3325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Markland FS. Snake venoms and the hemostatic system. Toxicon. 1998;36:1749–1800. doi: 10.1016/S0041-0101(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 86.Serrano SM, Maroun RC. Snake venom serine proteinases: sequence homology vs. substrate specificity, a paradox to be solved. Toxicon. 2005;45:1115–1132. doi: 10.1016/j.toxicon.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 87.Hite LA, Jia LG, Bjarnason JB, Fox JW. cDNA sequences for four snake venom metalloproteinases: structure, classification, and their relationship to mammalian reproductive proteins. Arch Biochem Biophys. 1994;308:182–191. doi: 10.1006/abbi.1994.1026. [DOI] [PubMed] [Google Scholar]
- 88.Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 89.Fox JW, Serrano SM. Timeline of key events in snake venom metalloproteinase research. J Proteomics. 2009;72:200–209. doi: 10.1016/j.jprot.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 90.Kisiel W, Hermodson MA, Davie EW. Factor X activating enzyme from Russell’s viper venom: isolation and characterization. Biochemistry. 1976;15:4901–4906. doi: 10.1021/bi00667a023. [DOI] [PubMed] [Google Scholar]
- 91.Morita T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon. 2005;45:1099–1114. doi: 10.1016/j.toxicon.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 92.Takeya H, Nishida S, Miyata T, Kawada S, Saisaka Y, Morita T, Iwanaga S. Coagulation factor X activating enzyme from Russell’s viper venom (RVV-X): a novel metalloproteinase with disintegrin (platelet aggregation inhibitor)-like and C-type lectin-like domains. J Biol Chem. 1992;267:14109–14117. [PubMed] [Google Scholar]
- 93.Gowda DC, Jackson CM, Hensley P, Davidson EA. Factor X-activating glycoprotein of Russell’s viper venom: polypeptide composition and characterization of the carbohydrate moieties. J Biol Chem. 1994;269:10644–10650. [PubMed] [Google Scholar]
- 94.Fujikawa K, Legaz ME, Davie EW. Bovine factor X 1 (Stuart factor): mechanism of activation by protein from Russell’s viper venom. Biochemistry. 1972;11:4892–4899. doi: 10.1021/bi00776a003. [DOI] [PubMed] [Google Scholar]
- 95.Takeda S, Igarashi T, Mori H. Crystal structure of RVV-X: an example of evolutionary gain of specificity by ADAM proteinases. FEBS Lett. 2007;581:5859–5864. doi: 10.1016/j.febslet.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 96.Yamada D, Sekiya F, Morita T. Isolation and characterization of carinactivase, a novel prothrombin activator in Echis carinatus venom with a unique catalytic mechanism. J Biol Chem. 1996;271:5200–5207. doi: 10.1074/jbc.271.9.5200. [DOI] [PubMed] [Google Scholar]
- 97.Yamada D, Morita T. CA-1 method, a novel assay for quantification of normal prothrombin using a Ca2+-dependent prothrombin activator, carinactivase-1. Thromb Res. 1999;94:221–226. doi: 10.1016/S0049-3848(98)00212-6. [DOI] [PubMed] [Google Scholar]
- 98.Yamada D, Morita T. Purification and characterization of a Ca2+-dependent prothrombin activator, multactivase, from the venom of Echis multisquamatus . J Biochem. 1997;122:991–997. doi: 10.1093/oxfordjournals.jbchem.a021862. [DOI] [PubMed] [Google Scholar]
- 99.Okuda D, Koike H, Morita T. A new gene structure of the disintegrin family: a subunit of dimeric disintegrin has a short coding region. Biochemistry. 2002;41:14248–14254. doi: 10.1021/bi025876s. [DOI] [PubMed] [Google Scholar]
- 100.Calvete JJ, Moreno-Murciano MP, Theakston RD, Kisiel DG, Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bilgrami S, Tomar S, Yadav S, Kaur P, Kumar J, Jabeen T, Sharma S, Singh TP. Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 A resolution. J Mol Biol. 2004;341:829–837. doi: 10.1016/j.jmb.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 102.Moiseeva N, Bau R, Swenson SD, Markland FS, Jr, Choe JY, Liu ZJ, Allaire M. Structure of acostatin, a dimeric disintegrin from Southern copperhead (Agkistrodon contortrix contortrix), at 1.7 A resolution. Acta Crystallogr D Biol Crystallogr. 2008;64:466–470. doi: 10.1107/S0907444908002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-X. [DOI] [PubMed] [Google Scholar]
- 104.Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 105.Calvete JJ. Structure-function correlations of snake venom disintegrins. Curr Pharm Des. 2005;11:829–835. doi: 10.2174/1381612053381783. [DOI] [PubMed] [Google Scholar]
- 106.McLane MA, Kuchar MA, Brando C, Santoli D, Paquette-Straub CA, Miele ME. New insights on disintegrin-receptor interactions: eristostatin and melanoma cells. Haemostasis. 2001;31:177–182. doi: 10.1159/000048061. [DOI] [PubMed] [Google Scholar]
- 107.McLane MA, Joerger T, Mahmoud A. Disintegrins in health and disease. Front Biosci. 2008;13:6617–6637. doi: 10.2741/3177. [DOI] [PubMed] [Google Scholar]
- 108.Da Silva M, Lucena S, Aguilar I, Rodriguez-Acosta A, Salazar AM, Sanchez EE, Giron ME, Carvajal Z, Arocha-Pinango CL, Guerrero B. Anti-platelet effect of cumanastatin 1, a disintegrin isolated from venom of South American Crotalus rattlesnake. Thromb Res. 2009;123:731–739. doi: 10.1016/j.thromres.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 109.Sanchez EE, Rodriguez-Acosta A, Palomar R, Lucena SE, Bashir S, Soto JG, Perez JC. Colombistatin: a disintegrin isolated from the venom of the South American snake (Bothrops colombiensis) that effectively inhibits platelet aggregation and SK-Mel-28 cell adhesion. Arch Toxicol. 2009;83:271–279. doi: 10.1007/s00204-008-0358-y. [DOI] [PubMed] [Google Scholar]
- 110.Humphries JD, Askari JA, Zhang XP, Takada Y, Humphries MJ, Mould AP. Molecular basis of ligand recognition by integrin alpha5beta 1. II: specificity of arg-gly-Asp binding is determined by Trp157 OF THE alpha subunit. J Biol Chem. 2000;275:20337–20345. doi: 10.1074/jbc.M000568200. [DOI] [PubMed] [Google Scholar]
- 111.Lazarus RA, McDowell RS. Structural and functional aspects of RGD-containing protein antagonists of glycoprotein IIb-IIIa. Curr Opin Biotechnol. 1993;4:438–445. doi: 10.1016/0958-1669(93)90009-L. [DOI] [PubMed] [Google Scholar]
- 112.Lu X, Davies J, Lu D, Xia M, Wattam B, Shang D, Sun Y, Scully M, Kakkar V. The effect of the single substitution of arginine within the RGD tripeptide motif of a modified neurotoxin dendroaspin on its activity of platelet aggregation and cell adhesion. Cell Commun Adhes. 2006;13:171–183. doi: 10.1080/15419060600726183. [DOI] [PubMed] [Google Scholar]
- 113.Sessions BR, Aston KI, Davis AP, Pate BJ, White KL. Effects of amino acid substitutions in and around the arginine-glycine-aspartic acid (RGD) sequence on fertilization and parthenogenetic development in mature bovine oocytes. Mol Reprod Dev. 2006;73:651–657. doi: 10.1002/mrd.20462. [DOI] [PubMed] [Google Scholar]
- 114.Kini RM, Rao VS, Joseph JS. Procoagulant proteins from snake venoms. Haemostasis. 2001;31:218–224. doi: 10.1159/000048066. [DOI] [PubMed] [Google Scholar]
- 115.Rao VS, Kini RM. Pseutarin C, a prothrombin activator from Pseudonaja textilis venom: its structural and functional similarity to mammalian coagulation factor Xa-Va complex. Thromb Haemost. 2002;88:611–619. [PubMed] [Google Scholar]
- 116.Jin Y, Lee WH, Zeng L, Zhang Y. Molecular characterization of l-amino acid oxidase from king cobra venom. Toxicon. 2007;50:479–489. doi: 10.1016/j.toxicon.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 117.Zhang L, Wu WT. Isolation and characterization of ACTX-6: a cytotoxic l-amino acid oxidase from Agkistrodon acutus snake venom. Nat Prod Res. 2008;22:554–563. doi: 10.1080/14786410701592679. [DOI] [PubMed] [Google Scholar]
- 118.Braga MD, Martins AM, Amora DN, de Menezes DB, Toyama MH, Toyama DO, Marangoni S, Alves CD, Barbosa PS, de Sousa AR, Fonteles MC, Monteiro HS. Purification and biological effects of l-amino acid oxidase isolated from Bothrops insularis venom. Toxicon. 2008;51:199–207. doi: 10.1016/j.toxicon.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 119.Toyama MH, Toyama DO, Passero LF, Laurenti MD, Corbett CE, Tomokane TY, Fonseca FV, Antunes E, Joazeiro PP, Beriam LO, Martins MA, Monteiro HS, Fonteles MC. Isolation of a new l-amino acid oxidase from Crotalus durissus cascavella venom. Toxicon. 2006;47:47–57. doi: 10.1016/j.toxicon.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 120.Samel M, Vija H, Ronnholm G, Siigur J, Kalkkinen N, Siigur E. Isolation and characterization of an apoptotic and platelet aggregation inhibiting l-amino acid oxidase from Vipera berus berus (common viper) venom. Biochim Biophys Acta. 2006;1764:707–714. doi: 10.1016/j.bbapap.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 121.Zhang YJ, Wang JH, Lee WH, Wang Q, Liu H, Zheng YT, Zhang Y. Molecular characterization of Trimeresurus stejnegeri venom l-amino acid oxidase with potential anti-HIV activity. Biochem Biophys Res Commun. 2003;309:598–604. doi: 10.1016/j.bbrc.2003.08.044. [DOI] [PubMed] [Google Scholar]
- 122.Lu QM, Wei Q, Jin Y, Wei JF, Wang WY, Xiong YL. l-amino acid oxidase from Trimeresurus jerdonii snake venom: purification, characterization, platelet aggregation-inducing and antibacterial effects. J Nat Toxins. 2002;11:345–352. [PubMed] [Google Scholar]
- 123.Ali SA, Stoeva S, Abbasi A, Alam JM, Kayed R, Faigle M, Neumeister B, Voelter W. Isolation, structural, and functional characterization of an apoptosis-inducing l-amino acid oxidase from leaf-nosed viper (Eristocophis macmahoni) snake venom. Arch Biochem Biophys. 2000;384:216–226. doi: 10.1006/abbi.2000.2130. [DOI] [PubMed] [Google Scholar]
- 124.Tan NH, Swaminathan S. Purification and properties of the l-amino acid oxidase from monocellate cobra (Naja naja kaouthia) venom. Int J Biochem. 1992;24:967–973. doi: 10.1016/0020-711X(92)90105-A. [DOI] [PubMed] [Google Scholar]
- 125.Rodrigues NH, da Silva JF, Boldrini-Franca J, Fonseca FP, Otaviano AR, Henrique-Silva F, Hamaguchi A, Magro AJ, Braz AS, Dos Santos JI, Homsi-Brandeburgo MI, Fontes MR, Fuly AL, Soares AM, Rodrigues VM. Structural and functional properties of Bp-LAAO, a new l-amino acid oxidase isolated from Bothrops pauloensis snake venom. Biochimie. 1992;91:490–501. doi: 10.1016/j.biochi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 126.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of l-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Torii S, Yamane K, Mashima T, Haga N, Yamamoto K, Fox JW, Naito M, Tsuruo T. Molecular cloning and functional analysis of apoxin I, a snake venom-derived apoptosis-inducing factor with l-amino acid oxidase activity. Biochemistry. 2000;39:3197–3205. doi: 10.1021/bi992416z. [DOI] [PubMed] [Google Scholar]
- 128.Du XY, Clemetson KJ. Snake venom l-amino acid oxidases. Toxicon. 2002;40:659–665. doi: 10.1016/S0041-0101(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 129.Geyer A, Fitzpatrick TB, Pawelek PD, Kitzing K, Vrielink A, Ghisla S, Macheroux P. Structure and characterization of the glycan moiety of l-amino-acid oxidase from the Malayan pit viper Calloselasma rhodostoma . Eur J Biochem. 2001;268:4044–4053. doi: 10.1046/j.1432-1327.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 130.Suhr SM, Kim DS. Identification of the snake venom substance that induces apoptosis. Biochem Biophys Res Commun. 1996;224:134–139. doi: 10.1006/bbrc.1996.0996. [DOI] [PubMed] [Google Scholar]
- 131.Vieira Santos MM, Sant’ana CD, Giglio JR, da Silva RJ, Sampaio SV, Soares AM, Fecchio D. Antitumoral effect of an l-amino acid oxidase isolated from Bothrops jararaca Snake Venom. Pharmacol Toxicol. 2008;102:533–542. doi: 10.1111/j.1742-7843.2008.00229.x. [DOI] [PubMed] [Google Scholar]
- 132.Tonismagi K, Samel M, Trummal K, Ronnholm G, Siigur J, Kalkkinen N, Siigur E. l-amino acid oxidase from Vipera lebetina venom: isolation, characterization, effects on platelets and bacteria. Toxicon. 2006;48:227–237. doi: 10.1016/j.toxicon.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 133.Souza DH, Eugenio LM, Fletcher JE, Jiang MS, Garratt RC, Oliva G, Selistre-de-Araujo HS. Isolation and structural characterization of a cytotoxic l-amino acid oxidase from Agkistrodon contortrix laticinctus snake venom: preliminary crystallographic data. Arch Biochem Biophys. 1999;368:285–290. doi: 10.1006/abbi.1999.1287. [DOI] [PubMed] [Google Scholar]
- 134.Clemetson KJ, Morita T, Kini RM. Scientific and standardization committee communications: classification and nomenclature of snake venom C-type lectins and related proteins. J Thromb Haemost. 2009;7(2):360. doi: 10.1111/j.1538-7836.2008.03233.x. [DOI] [PubMed] [Google Scholar]
- 135.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 136.Hirabayashi J, Kusunoki T, Kasai K. Complete primary structure of a galactose-specific lectin from the venom of the rattlesnake Crotalus atrox. Homologies with Ca2+-dependent-type lectins. J Biol Chem. 1991;266:2320–2326. [PubMed] [Google Scholar]
- 137.Lin LP, Lin Q, Wang YQ. Cloning, expression and characterization of two C-type lectins from the venom gland of Bungarus multicinctus . Toxicon. 2007;50:411–419. doi: 10.1016/j.toxicon.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 138.Clemetson KJ, Navdaev A, Dormann D, Du XY, Clemetson JM. Multifunctional snake C-type lectins affecting platelets. Haemostasis. 2001;31:148–154. doi: 10.1159/000048058. [DOI] [PubMed] [Google Scholar]
- 139.Clemetson KJ, Polgar J, Clemetson JM. Snake venom C-type lectins as tools in platelet research. Platelets. 1998;9:165–169. doi: 10.1080/09537109876636. [DOI] [PubMed] [Google Scholar]
- 140.Ogawa T, Chijiwa T, Oda-Ueda N, Ohno M. Molecular diversity and accelerated evolution of C-type lectin-like proteins from snake venom. Toxicon. 2005;45:1–14. doi: 10.1016/j.toxicon.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 141.Ozeki Y, Matsui T, Hamako J, Suzuki M, Fujimura Y, Yoshida E, Nishida S, Titani K. C-type galactoside-binding lectin from Bothrops jararaca venom: comparison of its structure and function with those of botrocetin. Arch Biochem Biophys. 1994;308:306–310. doi: 10.1006/abbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 142.Aragon-Ortiz F, Mentele R, Auerswald EA. Amino acid sequence of a lectin-like protein from Lachesis muta stenophyrs venom. Toxicon. 1996;34:763–769. doi: 10.1016/0041-0101(96)00011-6. [DOI] [PubMed] [Google Scholar]
- 143.Xu Q, Wu XF, Xia QC, Wang KY. Cloning of a galactose-binding lectin from the venom of Trimeresurus stejnegeri . Biochem J. 1999;341(Pt 3):733–737. doi: 10.1042/0264-6021:3410733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamako J, Suzuki Y, Hayashi N, Kimura M, Ozeki Y, Hashimoto K, Matsui T. Amino acid sequence and characterization of C-type lectin purified from the snake venom of Crotalus ruber . Comp Biochem Physiol B Biochem Mol Biol. 2007;146:299–306. doi: 10.1016/j.cbpb.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 145.Drickamer K. Recognition of complex carbohydrates by Ca2+-dependent animal lectins. Biochem Soc Trans. 1993;21:456–459. doi: 10.1042/bst0210456. [DOI] [PubMed] [Google Scholar]
- 146.Walker JR, Nagar B, Young NM, Hirama T, Rini JM. X-ray crystal structure of a galactose-specific C-type lectin possessing a novel decameric quaternary structure. Biochemistry. 2004;43:3783–3792. doi: 10.1021/bi035871a. [DOI] [PubMed] [Google Scholar]
- 147.Gartner TK, Ogilvie ML. Isolation and characterization of three Ca2+-dependent beta-galactoside-specific lectins from snake venoms. Biochem J. 1984;224:301–307. doi: 10.1042/bj2240301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ogilvie ML, Byl JW, Gartner TK. Platelet-aggregation is stimulated by lactose-inhibitable snake venom lectins. Thromb Haemost. 1989;62:704–707. [PubMed] [Google Scholar]
- 149.Morita T. C-type lectin-related proteins from snake venoms. Curr Drug Targets Cardiovasc Haematol Disord. 2004;4:357–373. doi: 10.2174/1568006043335916. [DOI] [PubMed] [Google Scholar]
- 150.Atoda H, Morita T. A novel blood coagulation factor IX/factor X-binding protein with anticoagulant activity from the venom of Trimeresurus flavoviridis (Habu snake): isolation and characterization. J Biochem (Tokyo) 1989;106:808–813. doi: 10.1093/oxfordjournals.jbchem.a122935. [DOI] [PubMed] [Google Scholar]
- 151.Atoda H, Hyuga M, Morita T. The primary structure of coagulation factor IX/factor X-binding protein isolated from the venom of Trimeresurus flavoviridis. Homology with asialoglycoprotein receptors, proteoglycan core protein, tetranectin, and lymphocyte Fc epsilon receptor for immunoglobulin E. J Biol Chem. 1991;266:14903–14911. [PubMed] [Google Scholar]
- 152.Zingali RB, Jandrot-Perrus M, Guillin MC, Bon C. Bothrojaracin, a new thrombin inhibitor isolated from Bothrops jararaca venom: characterization and mechanism of thrombin inhibition. Biochemistry. 1993;32:10794–10802. doi: 10.1021/bi00091a034. [DOI] [PubMed] [Google Scholar]
- 153.Zingali RB, Ferreira MS, Assafim M, Frattani FS, Monteiro RQ. Bothrojaracin, a Bothrops jararaca snake venom-derived (pro)thrombin inhibitor, as an anti-thrombotic molecule. Pathophysiol Haemost Thromb. 2005;34:160–163. doi: 10.1159/000092416. [DOI] [PubMed] [Google Scholar]
- 154.Arocas V, Zingali RB, Guillin MC, Bon C, Jandrot-Perrus M. Bothrojaracin: a potent two-site-directed thrombin inhibitor. Biochemistry. 1996;35:9083–9089. doi: 10.1021/bi960043l. [DOI] [PubMed] [Google Scholar]
- 155.Li WF, Chen L, Li XM, Liu J. A C-type lectin-like protein from Agkistrodon acutus venom binds to both platelet glycoprotein Ib and coagulation factor IX/factor X. Biochem Biophys Res Commun. 2005;332:904–912. doi: 10.1016/j.bbrc.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 156.Atoda H, Morita T. Arrangement of the disulfide bridges in a blood coagulation factor IX/factor X-binding protein from the venom of Trimeresurus flavoviridis . J Biochem (Tokyo) 1993;113:159–163. doi: 10.1093/oxfordjournals.jbchem.a124020. [DOI] [PubMed] [Google Scholar]
- 157.Mizuno H, Fujimoto Z, Koizumi M, Kano H, Atoda H, Morita T. Structure of coagulation factors IX/X-binding protein, a heterodimer of C-type lectin domains. Nat Struct Biol. 1997;4:438–441. doi: 10.1038/nsb0697-438. [DOI] [PubMed] [Google Scholar]
- 158.Zingali RB, Bianconi ML, Monteiro RQ. Interaction of bothrojaracin with prothrombin. Haemostasis. 2001;31:273–278. doi: 10.1159/000048073. [DOI] [PubMed] [Google Scholar]
- 159.Wang R, Kini RM, Chung MC. Rhodocetin, a novel platelet aggregation inhibitor from the venom of Calloselasma rhodostoma (Malayan pit viper): synergistic and noncovalent interaction between its subunits. Biochemistry. 1999;38:7584–7593. doi: 10.1021/bi982132z. [DOI] [PubMed] [Google Scholar]
- 160.Paaventhan P, Kong C, Joseph JS, Chung MC, Kolatkar PR. Structure of rhodocetin reveals noncovalently bound heterodimer interface. Protein Sci. 2005;14:169–175. doi: 10.1110/ps.04945605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vargaftig BB, Prado-Franceschi J, Chignard M, Lefort J, Marlas G. Activation of guinea-pig platelets induced by convulxin, a substance extracted from the venom of Crotalus durissus cascavella . Eur J Pharmacol. 1980;68:451–464. doi: 10.1016/0014-2999(80)90420-3. [DOI] [PubMed] [Google Scholar]
- 162.Francischetti IM, Saliou B, Leduc M, Carlini CR, Hatmi M, Randon J, Faili A, Bon C. Convulxin, a potent platelet-aggregating protein from Crotalus durissus terrificus venom, specifically binds to platelets. Toxicon. 1997;35:1217–1228. doi: 10.1016/S0041-0101(97)00021-4. [DOI] [PubMed] [Google Scholar]
- 163.Murakami MT, Zela SP, Gava LM, Michelan-Duarte S, Cintra AC, Arni RK. Crystal structure of the platelet activator convulxin, a disulfide-linked alpha4beta4 cyclic tetramer from the venom of Crotalus durissus terrificus . Biochem Biophys Res Commun. 2003;310:478–482. doi: 10.1016/j.bbrc.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 164.Batuwangala T, Leduc M, Gibbins JM, Bon C, Jones EY. Structure of the snake-venom toxin convulxin. Acta Crystallogr D Biol Crystallogr. 2004;60:46–53. doi: 10.1107/S0907444903021620. [DOI] [PubMed] [Google Scholar]
- 165.Taniuchi Y, Kawasaki T, Fujimura Y, Suzuki M, Titani K, Sakai Y, Kaku S, Hisamichi N, Satoh N, Takenaka T. Flavocetin-A and -B, two high molecular mass glycoprotein Ib binding proteins with high affinity purified from. Biochim Biophys Acta. 1995;1244:331–338. doi: 10.1016/0304-4165(95)00052-d. [DOI] [PubMed] [Google Scholar]
- 166.Fukuda K, Mizuno H, Atoda H, Morita T. Crystallization and preliminary X-ray studies of flavocetin-A, a platelet glycoprotein Ib-binding protein from the habu snake venom. Acta Crystallogr D Biol Crystallogr. 1999;55:1911–1913. doi: 10.1107/S0907444999009622. [DOI] [PubMed] [Google Scholar]
- 167.Lee WH, Du XY, Lu QM, Clemetson KJ, Zhang Y. Stejnulxin, a novel snake C-type lectin-like protein from Trimeresurus stejnegeri venom is a potent platelet agonist acting specifically via GPVI. Thromb Haemost. 2003;90:662–671. doi: 10.1160/TH03-05-0269. [DOI] [PubMed] [Google Scholar]
- 168.Fry BG, Wuster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57:110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 169.Fry BG, Lumsden NG, Wuster W, Wickramaratna JC, Hodgson WC, Kini RM. Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: evidence for early origin of venom in snakes. J Mol Evol. 2003;57:446–452. doi: 10.1007/s00239-003-2497-3. [DOI] [PubMed] [Google Scholar]
- 170.Pawlak J, Mackessy SP, Fry BG, Bhatia M, Mourier G, Fruchart-Gaillard C, Servent D, Menez R, Stura E, Menez A, Kini RM. Denmotoxin: a three-finger toxin from colubrid snake Boiga dendrophila (mangrove catsnake)with bird-specific activity. J Biol Chem. 2003;281:29030–29042. doi: 10.1074/jbc.M605850200. [DOI] [PubMed] [Google Scholar]
- 171.Lumsden NG, Fry BG, Ventura S, Kini RM, Hodgson WC. Pharmacological characterisation of a neurotoxin from the venom of Boiga dendrophila (mangrove catsnake) Toxicon. 2005;45:329–334. doi: 10.1016/j.toxicon.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 172.Junqueira-de-Azevedo IL, Ching AT, Carvalho E, Faria F, Nishiyama MY, Jr, Ho PL, Diniz MR. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics. 2006;173:877–889. doi: 10.1534/genetics.106.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pahari S, Bickford D, Fry BG, Kini RM. Expression pattern of three-finger toxin and phospholipase A2 genes in the venom glands of two sea snakes, Lapemis curtus and Acalyptophis peronii: comparison of evolution of these toxins in land snakes, sea kraits and sea snakes. BMC Evol Biol. 2007;7:175. doi: 10.1186/1471-2148-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Menez A. Functional architectures of animal toxins: a clue to drug design? Toxicon. 1998;36:1557–1572. doi: 10.1016/S0041-0101(98)00148-2. [DOI] [PubMed] [Google Scholar]
- 175.Tsetlin V. Snake venom alpha-neurotoxins and other ‘three-finger’ proteins. Eur J Biochem. 1999;264:281–286. doi: 10.1046/j.1432-1327.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 176.Kini RM. Molecular moulds with multiple missions: functional sites in three-finger toxins. Clin Exp Pharmacol Physiol. 2002;29:815–822. doi: 10.1046/j.1440-1681.2002.03725.x. [DOI] [PubMed] [Google Scholar]
- 177.Pawlak J, Mackessy SP, Fry BG, Bhatia M, Mourier G, Fruchart-Gaillard C, Servent D, Menez R, Stura E, Menez A, Kini RM (2006) Denmotoxin: a three-finger toxin from colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J Biol Chem [DOI] [PubMed]
- 178.Rajagopalan N, Pung YF, Zhu YZ, Wong PT, Kumar PP, Kini RM. {beta}-Cardiotoxin: a new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. 2007;21:3685–3695. doi: 10.1096/fj.07-8658com. [DOI] [PubMed] [Google Scholar]
- 179.Pawlak J, Mackessy SP, Sixberry NM, Stura EA, Le Du MH, Menez R, Foo CS, Menez A, Nirthanan S, Kini RM. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2008;23:534–545. doi: 10.1096/fj.08-113555. [DOI] [PubMed] [Google Scholar]
- 180.Pawlak J, Mackessy SP, Fry BG, Bhatia M, Mourier G, Fruchart-Gaillard C, Servent D, Menez R, Stura E, Menez A, Kini RM. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. J Biol Chem. 2006;281:29030–29041. doi: 10.1074/jbc.M605850200. [DOI] [PubMed] [Google Scholar]
- 181.Osipov AV, Kasheverov IE, Makarova YV, Starkov VG, Vorontsova OV, Ziganshin RK, Andreeva TV, Serebryakova MV, Benoit A, Hogg RC, Bertrand D, Tsetlin VI, Utkin YN. Naturally occurring disulfide-bound dimers of three-fingered toxins: a paradigm for biological activity diversification. J Biol Chem. 2008;283:14571–14580. doi: 10.1074/jbc.M802085200. [DOI] [PubMed] [Google Scholar]
- 182.Chiappinelli VA. Kappa-bungarotoxin: a probe for the neuronal nicotinic receptor in the avian ciliary ganglion. Brain Res. 1983;277:9–22. doi: 10.1016/0006-8993(83)90902-2. [DOI] [PubMed] [Google Scholar]
- 183.Dryer SE, Chiappinelli VA. Kappa-bungarotoxin: an intracellular study demonstrating blockade of neuronal nicotinic receptors by a snake neurotoxin. Brain Res. 1983;289:317–321. doi: 10.1016/0006-8993(83)90033-1. [DOI] [PubMed] [Google Scholar]
- 184.Chiappinelli VA, Dryer SE. Nicotinic transmission in sympathetic ganglia: blockade by the snake venom neurotoxin kappa-bungarotoxin. Neurosci Lett. 1984;50:239–244. doi: 10.1016/0304-3940(84)90492-0. [DOI] [PubMed] [Google Scholar]
- 185.Chiappinelli VA, Lee JC. Kappa-Bungarotoxin: self-association of a neuronal nicotinic receptor probe. J Biol Chem. 1985;260:6182–6186. [PubMed] [Google Scholar]
- 186.Oswald RE, Sutcliffe MJ, Bamberger M, Loring RH, Braswell E, Dobson CM. Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: sequence-specific assignments, secondary structure, and dimer formation. Biochemistry. 1991;30:4901–4909. doi: 10.1021/bi00234a010. [DOI] [PubMed] [Google Scholar]
- 187.Sutcliffe MJ, Dobson CM, Oswald RE. Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics. Biochemistry. 1992;31:2962–2970. doi: 10.1021/bi00126a017. [DOI] [PubMed] [Google Scholar]
- 188.Dewan JC, Grant GA, Sacchettini JC. Crystal structure of kappa-bungarotoxin at 2.3-A resolution. Biochemistry. 1994;33:13147–13154. doi: 10.1021/bi00248a026. [DOI] [PubMed] [Google Scholar]
- 189.Chiappinelli VA, Weaver WR, McLane KE, Conti-Fine BM, Fiordalisi JJ, Grant GA. Binding of native kappa-neurotoxins and site-directed mutants to nicotinic acetylcholine receptors. Toxicon. 1996;34:1243–1256. doi: 10.1016/S0041-0101(96)00110-9. [DOI] [PubMed] [Google Scholar]
- 190.Chiappinelli VA, Wolf KM. Kappa-neurotoxins: heterodimer formation between different neuronal nicotinic receptor antagonists. Biochemistry. 1989;28:8543–8547. doi: 10.1021/bi00447a041. [DOI] [PubMed] [Google Scholar]
- 191.Joubert FJ, Taljaard N. Snake venoms: the amino-acid sequence of protein S2C4 from Dendroaspis jamesoni kaimosae (Jameson’s mamba) venom. Hoppe Seylers Z Physiol Chem. 1979;360(57):1–580. doi: 10.1515/bchm2.1979.360.1.571. [DOI] [PubMed] [Google Scholar]
- 192.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kini RM. The intriguing world of prothrombin activators from snake venom. Toxicon. 2005;45:1133–1145. doi: 10.1016/j.toxicon.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 194.Montecucco C, Rossetto O, Caccin P, Rigoni M, Carli L, Morbiato L, Muraro L, Paoli M (2008) Different mechanisms of inhibition of nerve terminals by botulinum and snake presynaptic neurotoxins. Toxicon (in press) [DOI] [PubMed]
- 195.Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. Crystal structure of Mg2+- and Ca2+-bound Gla domain of factor IX complexed with binding protein. J Biol Chem. 2003;278:24090–24094. doi: 10.1074/jbc.M300650200. [DOI] [PubMed] [Google Scholar]