Abstract
The last phase of leaf development, generally referred to as leaf senescence, is an integral part of plant development that involves massive programmed cell death. Due to a sharp decline of photosynthetic capacity in a leaf, senescence limits crop yield and forest plant biomass production. However, the biochemical components and regulatory mechanisms underlying leaf senescence are poorly characterized. Although several approaches such as differential cDNA screening, differential display, and cDNA subtraction have been employed to isolate senescence-associated genes (SAGs), only a limited number of SAGs have been identified, and information regarding the regulation of these genes is fragmentary. Here we report on the utilization of enhancer trap approach toward the identification and analysis of SAGs. We have developed a sensitive large-scale screening method and have screened 1,300 Arabidopsis enhancer trap lines and have identified 147 lines in which the reporter gene GUS (β-glucuronidase) is expressed in senescing leaves but not in non-senescing ones. We have systematically analyzed the regulation of β-glucuronidase expression in 125 lines (genetically, each contains single T-DNA insertion) by six senescence-promoting factors, namely abscisic acid, ethylene, jasmonic acid, brassinosteroid, darkness, and dehydration. This analysis not only reveals the complexity of the regulatory circuitry but also allows us to postulate the existence of a network of senescence-promoting pathways. We have also cloned three SAGs from randomly selected enhancer trap lines, demonstrating that reporter expression pattern reflects the expression pattern of the endogenous gene.
The final developmental phase of many plant organs is senescence. Plants exhibit two types of senescence: mitotic senescence and post-mitotic senescence (Gan and Amasino, 1999). A shoot apical meristem cell can undergo a certain number of mitotic divisions to produce organs such as leaves and flowers. Cessation of the cell division in the meristem is called mitotic senescence or proliferative senescence (Hensel et al., 1994). This type of senescence is also observed in yeast and mammalian cells and is often referred to as replicative senescence. Telomere shortening has been implicated in controlling replicative senescence in mammals (Bodnar et al., 1998). In contrast, post-mitotic senescence occurs in organs such as leaves and petals. Once formed, cells in these organs rarely undergo cell division (Colón-Carmona et al., 1999) but these cells undergo cell growth and ultimately cell degeneration or senescence; telomere length in these cells remains stable during leaf growth and senescence (Riha et al., 1998; Zentgraf et al., 2000). We have been interested in understanding mechanisms that control leaf senescence, a type of post-mitotic senescence.
Leaf senescence is a structurally, physiologically, and genetically orchestrated process, whereby cellular organelles and their constituents are sequentially broken down, and the released nutrients are recycled to actively growing organs such as young leaves, developing seeds, and fruits (Noodén, 1988). Leaf senescence is accompanied, and perhaps driven, by changes in gene expression; the majority of genes that are active in non-senescing leaves such as those involved in photosynthesis are down-regulated while a subset of genes are up-regulated during senescence. Studies involving inhibitors of RNA and protein biosynthesis have shown that activation of new genes is required for leaf cells to undergo senescence (for review, see Noodén, 1988; Gan and Amasino, 1997). Therefore, efforts have been made to isolate genes whose transcript abundance increases during leaf senescence, but only a limited number of senescence-associated genes (SAGs) have been isolated (e.g. Davies and Grierson, 1989; Becker and Apel, 1993; Hensel et al., 1993; Taylor et al., 1993; Lohman et al., 1994; Drake et al., 1996; Buchanan-Wollaston and Ainsworth, 1997; Hajouj et al., 2000), most of which possess moderate basal levels of expression prior to leaf senescence. The molecular mechanisms underlying leaf senescence remain poorly understood.
Leaf senescence is a complex process that may be controlled by an array of internal and environmental factors (Noodén, 1988; Smart 1994; Dai et al., 1999) through a regulatory network (Gan and Amasino, 1997). To fully understand the molecular basis of leaf senescence, it is necessary to identify genes whose products are components of the biochemical and regulatory pathways of leaf senescence. To accomplish this we have utilized an enhancer trap/detection strategy to identify SAGs from Arabidopsis. Enhancer detection is a very powerful molecular genetic tool that has been successfully used to isolate novel genes from a variety of organisms including Drosophila), the nematode Caenorhabditis elegans, mouse, and Arabidopsis (for review, see Bellen, 1999). A typical enhancer trap construct carries a reporter gene fused to a minimal promoter. This minimal promoter alone has no transcriptional activity, but when the construct inserts in the proximity of a chromosomal gene, the cis regulatory elements of the chromosomal gene promoter direct expression of the reporter gene. Therefore, the reporter gene expression patterns (e.g. the senescence-associated pattern that we are interested in) represent the chromosomal gene expression patterns. Genes associated with enhancer traps can be readily cloned using the T-DNA sequence tag.
Here, we report that by screening 1,300 Arabidopsis enhancer trap lines we have identified 147 lines in which the reporter gene GUS (β-glucuronidase) is expressed in senescing leaves but not in non-senescing leaves. 68% of the lines segregated for one T-DNA insertion, whereas the rest of the lines possessed multiple T-DNA insertions. In addition to senescing leaves, a subset of these enhancer traps are expressed in senescing flowers, siliques, and/or stems. Analysis of 125 lines reveals that they are differently regulated by senescence-promoting factors such as abscisic acid (ABA), ethylene, jasmonic acid (JA), brassinosteroids, dehydration, and darkness. In addition, we report the cloning and expression of three SAGs from randomly selected enhancer trap lines.
RESULTS AND DISCUSSION
Identification of 147 Senescence-Associated Enhancer Trap Lines in Arabidopsis
A senescence-associated enhancer trap line (Sel) is referred to as an enhancer trap line in which the reporter gene expression is detected in senescing leaves but not in non-senescing ones. The reporter gene in the Arabidopsis enhancer trap lines that we have screened encodes the enzyme β-glucuronidase (GUS; Campisi et al., 1999).
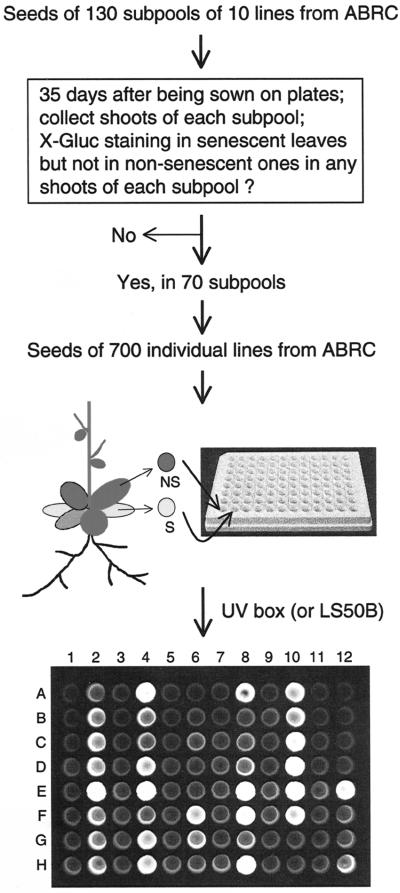
The strategy used to screen for Sels is shown in Figure 1. To identify senescence-associated genes, we plated enhancer trap seeds on kanamycin (Kan)-containing plates. When the first two to three rosette leaves of the plants become senescent, plants were harvested and subjected to standard histochemical GUS staining using X-Gluc as a substrate (Jefferson, 1987). Approximately 10% of the plants exhibited GUS expression in senescing leaves but not in non-senescing ones. We also used a second assay to monitor GUS activity in putative senescence lines; specifically, one non-senescing leaf and one senescing leaf were collected from each plant and incubated in separate wells of 96-well plates with opaque walls and optically clear bottoms in the presence of the fluorogenic substrate MUG. An example of the MUG screening results is shown in Figure 1. If only the senescing leaf showed GUS activity, the original plant was transplanted to soil to produce seeds. Using this method, we have identified 147 independent putative Sels.
Figure 1.
Flow chart indicating the series of steps used to screen for leaf senescence-associated enhancer trap lines. The photograph at bottom shows an example of using a 96-well plate for screening. Each well contains 70 μL of 4-methylumbelliferyl-d-glucuronide (MUG) solution. Individual enhancer trap lines were screened by placing a non-senescing leaf in a single well (odd-numbered column: 1, 3, 5, 7, 9, and 11) and a senescing leaf in the well of even-numbered column (2, 4, 6, 8, 10, and 12) from the same plant. For example, wells A1 (non-senescent) and A2 (senescing) leaf samples are from the same plant, as are A3 (non-senescing) and A4 (senescing) samples, etc. Samples in wells A11/12 through D11/12 are from wild-type Arabidopsis (for control). ABRC, Arabidopsis Biological Resource Center at the Ohio State University. LS50B is a model of Perkin-Elmer's luminescence spectrometers.
As described above, 147 out of 1,300 lines displayed senescence-associated GUS expression in leaves. This frequency (11.3%) is slightly higher than the frequency found in fruitfly, where 5% to 10% of enhancer trap strains expressed a reporter gene in very specific tissues and cells (Bellen, 1999), and is a little lower than the frequency (16%) of enhancer trap lines that exhibited GUS expression during floral abscission/senescence (Campisi et al., 1999).
Genetic Analysis and Segregation of T-DNA Insertions in Each of the 147 Sels
Although the majority of enhancer trap lines contain only a single T-DNA insertion, some lines contain multiple T-DNA insertions (Azpiroz-Leehan and Feldmann, 1997; Campisi et al., 1999). Multiple T-DNA insertions complicate analysis, cloning, and characterization of the tagged gene of interest. Thus, we performed a genetic analysis of all 147 Sels to determine the number of T-DNA insertions in each line. In brief, each line was backcrossed to the wild-type Arabidopsis (ecotype Columbia glabrous1 or Col-gl1) to produce backcross1 (BC1) seeds. The BC1 progeny were allowed to self-pollinate to produce BC1-F2 seeds. Both BC1 and BC1-F2 seeds were sown on plates containing kanamycin to determine the segregation ratio (kanamycinR:kanamycinS). The plants were also subjected to the MUG assay to assess if the senescence-related GUS expression cosegregated with the kanamycin-resistant marker gene. We found that 100 lines (or 68%) segregated a single T-DNA insertion, and that 47 lines possessed more than one insertion; this observation is similar to those of previous analyses (Azpiroz-Leehan and Feldmann et al., 1997; Campisi et al., 1999).
The BC1-F2 progeny of the lines with multiple insertions were selfed to produce BC1-F3 and BC1-F4 progeny. In some cases the BC1-F3 and BC1-F4 plants were backcrossed to wild-type plants a second time to allow segregation of T-DNA insertions not related to the senescence pattern. By doing so we obtained additional 25 lines in which a senescence-associated GUS expression cosegregated with a single T-DNA insertion. Thus, we obtained a total of 125 Sels, each containing a single T-DNA insertion.
Several lines of evidence lead us to believe that these 125 Sels represent 125 SAGs. First, as discussed previously, the enhancer trap vector (http://www.dartmout.edu/t̃jack/#pD991) carries the GUS reporter under the control of the 35S minimal promoter. This minimal promoter alone has no transcriptional activity (Benfey and Chua, 1990), but when the construct inserts in the proximity of a chromosomal gene, the cis elements of the chromosomal gene direct expression of the reporter. Therefore, what is actually “trapped” is the regulatory cis element(s) on a promoter that confers specificity to a gene, which is different from the original meaning of an “enhancer,” which is to intensify or increase quantitatively the expression level of a gene. Second, the likelihood that the enhancer trap reflects the expression of a nearby endogenous gene is very high; in fruitfly, it is rare that reporter expression is detected in cells or tissues in which the endogenous gene is not normally expressed (Bellen, 1999). Third, several genes have been isolated starting with an enhancer trap pattern in Arabidopsis, and in each case the expression of the endogenous gene mimics the expression pattern of the enhancer trap (Springer et al., 1995; Grossniklaus et al., 1998; Gu et al., 1998; Campisi et al., 1999; Swaminathan et al., 2000).
Expression Patterns of the Reporter Gene GUS in the Sels
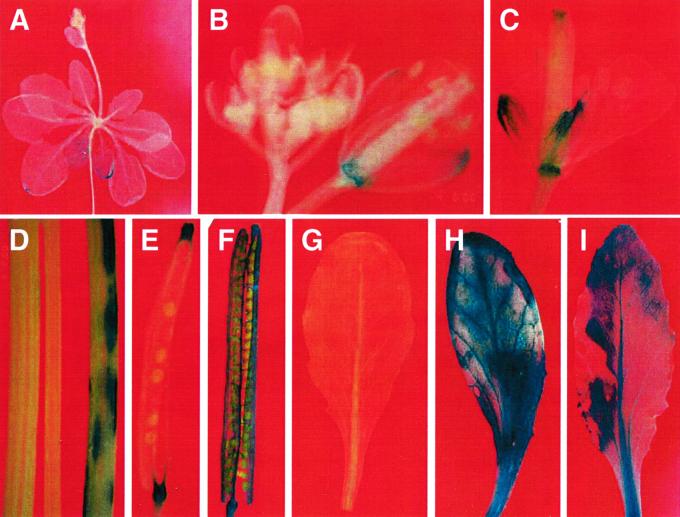
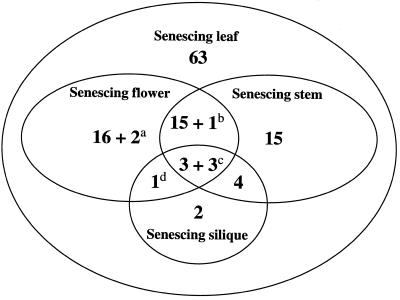
SAG12 and SAG13, two senescence-specific genes from Arabidopsis, are expressed in senescing flowers, siliques, and stems in addition to senescing leaves (Gan, 1995). It is possible that some of the 125 tagged genes are also expressed in other senescing organs in addition to senescing leaves. Therefore, we surveyed the GUS expression patterns in senescing flowers, siliques, and stems in each Sel. Some examples of this analysis are shown in Figure 2. In 63 lines, GUS was expressed in senescing leaves but not in senescing flowers, siliques, and stems, whereas in the other 62 lines, the reporter gene was also expressed in senescing flowers, siliques, and/or stems in addition to senescing leaves (Fig. 3, and Table I). It is interesting that in four lines (Sel95, Sel96, Sel98, and Sel142) GUS expression was detected in young siliques and/or stems (Fig. 3) in addition to senescing ones. The fact that many genes are expressed in both senescing leaves and senescing flowers suggests that there are common components in these senescence pathways. The identification of many leaf-specific senescence patterns suggests that some components are specific for leaf senescence.
Figure 2.
Examples of the GUS expression patterns in Arabidopsis senescence-associated enhancer trap lines (Sels). A, GUS expression in senescing leaves of a 5-week-old plate-grown Sel61 plant. B, GUS activities in senescing sepals and receptacle but not in young flower buds of Sel74. C, GUS expression in senescing sepals, stigma, and receptacle of Sel6. D, GUS expression in a senescing stem (right) of Sel25 but not in its young stem (middle). On the left is a senescing stem from a wild-type plant. E, GUS staining in the receptacle, and remains of stigma and style of a senescent silique of Sel74. F, GUS activity in a senescent silique of Sel142. G through I, Induction of GUS expression in Sel142 leaves by ABA (H) and senescence (I) compared with a non-senescing leaf (G). The leaf shown in H is a non-senescing leaf of Sel142 treated with 0.1 mm ABA for 6 h.
Figure 3.
Venn diagram of the numbers of Sels that display overlapping and nonoverlapping senescence-specific GUS expression among leaves, flowers, stems, and siliques. All Sels displayed senescence-specific reporter expression in leaves, whereas a subset of them also exhibited senescence-specific GUS expression in flowers, stems, and/or siliques with the following exceptions. a, In these two lines (Sel16 and Sel65) GUS staining was observed in both young and senescing flowers; b, this line (Sel96) showed constitutive GUS expression in stems; c, constitutive GUS staining was visible in siliques (Sel142), in siliques and stems (Sel95), or in siliques and flowers (Sel139); and d, this line (Sel98) showed constitutive reporter expression in siliques. A complete list of GUS staining patterns of these Sels can be seen in Table I.
Table I.
The spatial patterns of GUS expression in senescence-associated Arabidopsis enhancer trap lines
| Linea | Leaf | Flower | Silique | Stem |
|---|---|---|---|---|
| Sel36 | sb | s | s | s |
| Sel74 | s | s | s | s |
| Sel93 | s | s | s | s |
| Sel142 | s | s | cc | s |
| Sel95 | s | s | c | c |
| Sel139 | s | c | c | s |
| Sel9 | s | s | – | s |
| Sel40 | s | s | – | s |
| Sel43 | s | s | – | s |
| Sel59 | s | s | – | s |
| Sel61 | s | s | – | s |
| Sel66 | s | s | – | s |
| Sel88 | s | s | – | s |
| Sel94 | s | s | – | s |
| Sel103 | s | s | – | s |
| Sel110 | s | s | – | s |
| Sel111 | s | s | – | s |
| Sel121 | s | s | – | s |
| Sel130 | s | s | – | s |
| Sel135 | s | s | – | s |
| Sel146 | s | s | – | s |
| Sel96 | s | s | – | c |
| Sel98 | s | s | c | – |
| Sel20 | s | – | s | s |
| Sel55 | s | – | s | s |
| Sel64 | s | – | s | s |
| Sel68 | s | – | s | s |
| Sel2 | s | s | – | – |
| Sel6 | s | s | – | – |
| Sel15 | s | s | – | – |
| Sel16 | s | c | – | – |
| Sel28 | s | s | – | – |
| Sel29 | s | s | – | – |
| Sel30 | s | s | – | – |
| Sel32 | s | s | – | – |
| Sel37 | s | s | – | – |
| Sel47 | s | s | – | – |
| Sel49 | s | s | – | – |
| Sel63 | s | s | – | – |
| Sel65 | s | c | – | – |
| Sel67 | s | s | – | – |
| Sel92 | s | s | – | – |
| Sel100 | s | s | – | – |
| Sel117 | s | s | – | – |
| Sel127 | s | s | – | – |
| Sel1 | s | – | s | – |
| Sel113 | s | – | s | – |
| Sel7 | s | – | – | s |
| Sel25 | s | – | – | s |
| Sel26 | s | – | – | s |
| Sel44 | s | – | – | s |
| Sel48 | s | – | – | s |
| Sel50 | s | – | – | s |
| Sel53 | s | – | – | s |
| Sel62 | s | – | – | s |
| Sel69 | s | – | – | s |
| Sel72 | s | – | – | s |
| Sel76 | s | – | – | s |
| Sel102 | s | – | – | s |
| Sel115 | s | – | – | s |
| Sel125 | s | – | – | s |
| Sel136 | s | – | – | s |
The following lines in which GUS is expressed in senescing leaves but not in flowers, siliques, and/or stems, are not listed: Sel3, Sel4, Sel5, Sel8, Sel10, Sel13, Sel14, Sel18, Sel19, Sel22, Sel23, Sel24, Sel27, Sel31, Sel34, Sel35, Sel38, Sel39, Sel41, Sel42, Sel45, Sel46, Sel54, Sel57, Sel58, Sel60, Sel70, Sel71, Sel73, Sel75, Sel78, Sel80, Sel82, Sel84, Sel85, Sel86, Sel87, Sel89, Sel90, Sel91, Sel97, Sel99, Sel101, Sel104, Sel105, Sel106, Sel108, Sel109, Sel112, Sel114, Sel119, Sel120, Sel122, Sel123, Sel124, Sel128, Sel132, Sel134, Sel140, Sel141, Sel143, Sel145, and Sel147.
s, GUS staining is detected in senescing organs but not in non-senescing ones.
c, Constitutive; i.e. GUS staining is detected in both senescing and non-senescing organs.
Regulation of the Reporter Gene GUS in the Sels by Senescence-Promoting Factors
We systematically analyzed the regulation of the GUS reporter gene expression in all 125 Sels by senescence-promoting factors such as ABA, ethylene, JA, brassinosteroids, dehydration, and darkness treatments (see Table II). Similar to many other promoter-reporter gene studies, GUS expression provides insightful information about how the expression of SAGs is regulated in each of the Sels.
Table II.
Induction of GUS expression in non-senescent leaves of Arabidopsis senescence-associated enhancer trap lines
| Linea | Plant Growth Regulators
|
Environmental Stresses
|
||||
|---|---|---|---|---|---|---|
| Ethylene | eBR | JA | ABA | Darkness | Dehydration | |
| Sel20 | +b(3.2)c | +(2.0) | +(2.3) | – | +(2.6) | – |
| Sel6 | +(3.0) | +(3.0) | – | – | – | +(3.9) |
| Sel26 | +(2.9) | +(2.6) | – | – | – | – |
| Sel88 | +(2.6) | +(3.0) | – | – | – | – |
| Sel62 | +(2.1) | – | – | – | +(2.2) | – |
| Sel32 | +(4.3) | – | – | – | – | – |
| Sel36 | +(3.2) | – | – | – | – | – |
| Sel42 | +(3.0) | – | – | – | – | – |
| Sel46 | +(3.1) | – | – | – | – | – |
| Sel61 | +(2.0) | – | – | – | – | – |
| Sel71 | +(5.4) | – | – | – | – | – |
| Sel97 | +(5.3) | – | – | – | – | – |
| Sel120 | +(3.7) | – | – | – | – | – |
| Sel130 | +(3.2) | – | – | – | – | – |
| Sel135 | +(3.1) | – | – | – | – | – |
| Sel121 | – | – | +(3.8) | – | +(2.7) | – |
| Sel2 | – | – | +(2.5) | – | – | – |
| Sel4 | – | – | +(2.9) | – | – | – |
| Sel9 | – | – | +(3.3) | – | – | – |
| Sel10 | – | – | +(2.7) | – | – | – |
| Sel25 | – | – | +(3.4) | – | – | – |
| Sel29 | – | – | +(2.5) | – | – | – |
| Sel47 | – | – | +(5.3) | – | – | – |
| Sel55 | – | – | +(3.0) | – | – | – |
| Sel58 | – | – | +(3.1) | – | – | – |
| Sel69 | – | – | +(3.1) | – | – | – |
| Sel115 | – | – | +(4.4) | – | – | – |
| Sel117 | – | – | +(2.0) | – | – | – |
| Sel68 | – | – | – | +(16.5) | – | – |
| Sel142 | – | – | – | +(16.8) | – | – |
| Sel1 | – | – | – | – | +(2.4) | – |
| Sel31 | – | – | – | – | +(2.7) | – |
| Sel91 | – | – | – | – | +(3.7) | – |
| Sel111 | – | – | – | – | +(3.2) | – |
| Sel114 | – | – | – | – | +(3.5) | – |
| Sel124 | – | – | – | – | +(13.7) | – |
| Sel37 | – | – | – | – | – | +(4.8) |
| Sel72 | – | – | – | – | – | +(5.1) |
| Sel74 | – | – | – | – | – | +(17.0) |
| Sel95 | – | – | – | – | – | +(2.2) |
GUS expression in the following lines is not induced by any of these six factors: Sel3, Sel5, Sel7, Sel8, Sel13, Sel14, Sel15, Sel16, Sel18, Sel19, Sel22, Sel23, Sel24, Sel27, Sel28, Sel30, Sel30, Sel34, Sel35, Sel38, Sel39, Sel40, Sel41, Sel43, Sel44, Sel45, Sel48, Sel49, Sel50, Sel53, Sel54, Sel57, Sel59, Sel60, Sel63, Sel64, Sel65, Sel66, Sel67, Sel70, Sel73, Sel75, Sel76, Sel78, Sel80, Sel82, Sel84, Sel85, Sel86, Sel87, Sel89, Sel90, Sel92, Sel93, Sel94, Sel96, Sel98, Sel99, Sel100, Sel101, Sel102, Sel103, Sel104, Sel105, Sel106, Sel108, Sel109, Sel110, Sel112, Sel113, Sel119, Sel122, Sel123, Sel125, Sel127, Sel128, Sel132, Sel134, Sel136, Sel139, Sel140, Sel141, Sel143, Sel145, Sel146, and Sel147.
+, Indicates that GUS expression is induced.
The nos. in parentheses are the induction folds.
Many physiological and biochemical studies have shown that leaf senescence is regulated by a complex array of endogenous and environmental factors. ABA, ethylene, dehydration, and darkness are the most commonly studied senescence-inducing factors/treatments (for review, see Noodén, 1988; Smart, 1994). However, the mechanisms by which these factors induce leaf senescence are unknown. One model is that each of these factors may induce a subset of SAGs. This study involving the use of a large population of putative Sels has provided molecular evidence for this model. For example, the ethylene and JA treatments induce the reporter expression in 15 and 14 Sels, respectively; these two sets of Sels are different from each other except for one line (Sel20). In a similar manner, the darkness and dehydration treatments induce different sets of Sels.
The ability of JA to induce senescence has been demonstrated in previous studies (Ueda and Kato, 1980; for review, see Smart, 1994). We have found recently that several genes involved in JA biosynthesis are up-regulated during leaf senescence in Arabidopsis, and that the JA level in senescing Arabidopsis leaves increases (Y. He, H. Fukushige, D. Hildebrand, and S. Gan, unpublished data). In this study, we found that JA induces GUS expression in 14 of the 125 senescence enhancer-trap lines; only ethylene induces GUS expression in more lines. Based on this, JA appears to be an important senescence-promoting factor.
Brassinosteroids are a class of plant growth regulators that play an essential role in diverse developmental programs including senescence (for review, see Clouse and Sasse, 1998). Two lines of evidence suggest a leaf senescence-promoting role of brassinosteroids. First, external application of epibrassinolide (eBR; a commonly used brassinosteroid species in studies involving brassinosteroids) induces leaf senescence in mung bean plants (He et al., 1996). Second, several Arabidopsis brassinosteroid mutants that are deficient in either brassinosteroid biosynthesis (e.g. det2) or in the brassinosteroid signal transduction pathway (e.g. bri1) display a delayed leaf senescence phenotype (in addition to other characteristic changes; for review, see Clouse and Sasse, 1998). The molecular mechanism of brassinosteroid action on leaf senescence is unknown. Our data shows that external application of eBR does induce reproducibly the reporter gene expression in four enhancer trap lines, suggesting that brassinosteroids play a role in activating the senescence process.
Regulatory Network of Leaf Senescence in Arabidopsis
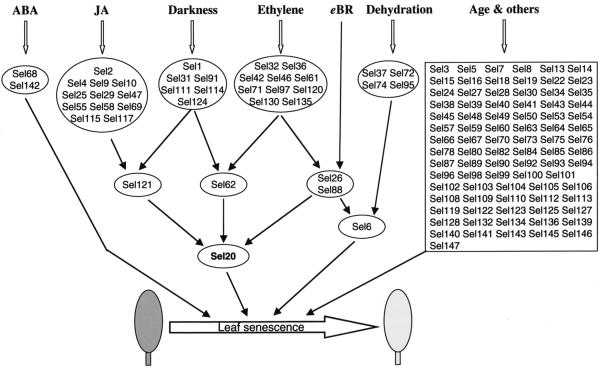
It has been postulated that there may be multiple pathways that respond to various autonomous and environmental factors, and that these pathways are possibly interconnected to form a regulatory network to control leaf senescence (Gan and Amasino, 1997). The above data not only demonstrate that each senescence-promoting factor up-regulates a subset of potential SAGs, but also shows that the GUS expression in certain lines is induced by one or more senescence-promoting factors, which enables us to place potential tagged SAGs into a regulatory network of leaf senescence (Fig. 4). The underlying rationale of this network is as follows. If a SAG is induced by only one stimulus, then this gene is likely in the upstream portion of the regulatory network. For example, the reporter gene GUS in Sel2 is induced by JA only; thus, we place it as an upstream component that presumably is responsive only to JA signaling (Fig. 4). If a gene is regulated by multiple stimuli, then this gene may function in the downstream portion of the proposed regulatory network. For example, the GUS expression in Sel20 is up-regulated by JA, ethylene, eBR, and darkness; therefore, we placed Sel20 in the downstream portion of the proposed network (Fig. 4).
Figure 4.
A putative leaf senescence regulatory network in Arabidopsis. GUS expression in Sels of the top-level circles is regulated by only one of the senescence-promoting factors tested, whereas the reporter gene in Sels of the second-level circles (e.g. Sel121, Sel62) is induced by two of the factors. GUS in Sel6 (third-level circle) is induced by three factors (ethylene, eBR, and dehydration), whereas GUS in Sel20 (fourth-level circle) is induced by four factors (JA, darkness, ethylene, and eBR). GUS expression in Sels in the rectangle does not obviously respond to any of the six factors tested; it may be regulated by age or other factors. It should be noted that this is a very simplified model, as discussed in the text.
There are 85 lines in which the GUS expression is not induced by any of the six senescence-promoting factors. In accordance, we put those genes in the related Sels under the control of age and other factors (“age and others” in Fig. 4). It has been shown that leaf age is a major factor that controls leaf senescence in many plant species such as Arabidopsis (Hensel et al., 1993) and soybean (Jiang et al., 1993). Therefore, it is not surprising that some of the SAGs in these 85 lines are regulated by leaf age. Other factors, not examined in this study, such as sugars (Dai et al., 1999), salicylic acid, extreme temperatures, flooding, pathogen infection, and reproductive growth (for review, see Noodén, 1988; Smart, 1994) also induce leaf senescence.
It is predictable that blocking a particular gene of the network may not have a significant effect on the progression of leaf senescence; this feature has been referred to as the plasticity of leaf senescence (Gan and Amasino, 1997). In homozygous Sel lines, some of the tagged SAGs should have been knocked out due to the T-DNA insertion. However, we did not observe a significantly delayed leaf senescence phenotype in any of these Sels, which is consistent with the existence of a senescence regulatory network.
It should be noted that the postulated leaf senescence regulatory network shown in Figure 4 is a very simplified model, and that the regulation of leaf senescence may be much more complex. For example, JA could regulate the GUS expression in Sel20 directly instead of via Sel2 and/or Sel121. Sels that are included in a circle may be regulated differently. The circuitry will begin to be unraveled as we clone and analyze the tagged genes in these lines. The following represents our first effort in this regard.
Cloning and Expression of SAGs from Sels
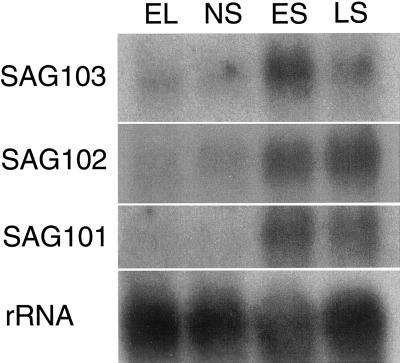
To demonstrate that the enhancer trap expression patterns reflect the expression of the endogenous genes, we first cloned flanking sequences of T-DNA in three randomly chosen Sels, namely Sel25, Sel139, and Sel142. A 1-kb genomic fragment flanking the right border of T-DNA in Sel139 was cloned using the thermal asymmetric interlaced PCR (Liu et al., 1995). Part of this fragment is identical to an Arabidopsis expressed sequence tag (EST; accession no. AI995772). RNA gel-blot analysis showed that this gene is expressed in senescing leaves but not in young, expanding leaves nor in fully expanded but non-senescing leaves (Fig. 5). The gene associated with the Sel139 enhancer trap has been named SAG101. We have found recently that SAG101 encodes an acyl hydrolase (Y. He and S. Gan, unpublished data).
Figure 5.
RNA gel-blot analysis of the steady-state mRNA levels of three newly cloned SAGs during leaf development in Arabidopsis. These SAGs were cloned from randomly selected Sels (SAG101 from Sel139, SAG102 from Sel142, and SAG103 from Sel25). Total RNA was isolated from expanding leaves (EL), fully expanded but non-senescing leaves (NS), early-stage senescing leaves (ES; showing up to 25% yellowing), or late-stage senescing leaves (LS; showing more than 50% yellowing). Approximately 10 μg of total RNA was loaded in each lane. The blot was hybridized with 18S rRNA probe to show relative loading in each lane.
Analysis of flanking DNA from Sel142 revealed that it was identical to an Arabidopsis EST (accession no. T46688) that encodes a protein of unknown function. The gene associated with the Sel142 enhancer trap has been named SAG102. SAG102 is expressed at very low levels during early stages of leaf development, but is up-regulated during leaf senescence (Fig. 5).
DNA flanking the T-DNA insertion in Sel25 was cloned by inverse PCR and shown to be similar to an Arabidopsis EST (accession no. AA598098) that encodes a protein of unknown function. The gene associated with the Sel25 enhancer trap has been named SAG103. Northern-blot analysis shows that SAG103, like SAG102, displays a very low level of expression in non-senescing leaves. The steady-state mRNA level of SAG103 peaks in leaves at an early senescence stage (Fig. 5).
CONCLUSIONS
We have developed a large-scale method for screening enhancer trap lines for genes up-regulated during leaf senescence. By using this method, we have identified 147 Arabidopsis enhancer trap lines in which the GUS reporter is expressed in senescing leaves but not in non-senescing ones. In these lines, we analyzed the effects of six senescence-promoting factors: ethylene, JA, ABA, brassinosteroids, dehydration, and darkness. Based on this analysis, we have constructed a regulatory network of leaf senescence in Arabidopsis. We have also cloned three genes from randomly selected enhancer trap lines that display senescence-associated up-regulation. The fact that mRNAs for these genes are up-regulated during leaf senescence validates our approach of cloning SAGs starting with a reporter expression pattern.
MATERIALS AND METHODS
Arabidopsis Enhancer Trap Lines
Seeds of 1, 300 independent Arabidopsis enhancer trap lines (ecotype Col-gl1; Campisi et al., 1999) in pools of 100, 10, or in some cases individual lines were obtained from the Arabidopsis Biological Resource Center at the Ohio State University (Columbus). The related stock numbers are as follows: cs19651A (13 pools of 100), cs19653, cs19655, cs19656, cs19658, cs19659, cs19660, cs19663, cs19664, cs19666, cs19669, cs19670 through cs19673, cs19675, cs19678, cs19685, cs19686, cs19689, cs19692, cs19698, cs19701 through cs19704, cs19707 through cs19710, cs19713, cs19714, cs19717 through cs19720, cs19722 through cs19724, cs19726, cs19730, cs19732, cs19733, cs19737 through cs19744, cs19746, cs19747, cs19751, cs19752, cs19754, cs19756, cs19758 through cs19760, cs19762, cs19767 through cs19770, cs19773, cs19774, and cs19777 through cs19780 (pools of 10). Seeds of individual Sels will be supplied upon request.
Seed Germination and Growth Conditions
The seeds were sterilized two to three times in 70% (v/v) ethanol containing 0.1% (v/v) Triton X-100 for 2 min, rinsed with two changes of 95% (v/v) ethanol, and then poured and dried on a sterile Whatman No. 1 filter paper (Whatman, Maidstone, UK) in a hood. Sterile seeds were sown on petri plates (100 × 20 mm) containing one-half strength of Murashige and Skoog salts, 40 μg mL−1 Kan, and 0.8% (w/v) phytoagar (hereafter Murashige and Skoog/Kan plates). The enhancer trap construct carries a selective marker gene that renders the transgenic plants resistant to kanamycin (Campisi et al., 1999). After imbibition at 4°C overnight, seeds were germinated in an Arabidopsis growth chamber (Percival Scientific, Boone, IA) at 23°C with 65% relative humidity under approximately 150 μmol m−2 s−1continuous light from a mixture of cool-white fluorescent (60%) and incandescent (40%) bulbs. In some cases, 2-week-old seedlings were transplanted to Fafard super-fine mix soils (Conrad Fafard, Inc., Agawam, MA) and grown under conditions similar to those in the Arabidopsis growth chamber.
Screening for Sels
First, 13 pools of 100 enhancer trap lines were grown in Murashige and Skoog/Kan plates. There were 400 to 500 plants in each pool. After 35 d of growth in the Arabidopsis growth chamber under the conditions as described above, the first two to three rosette leaves (from the bottom) of the miniature plants become senescent. Plants of each pool were then harvested and subjected to standard histochemical GUS staining using X-Gluc as a substrate (Jefferson, 1987). Approximately 10% of the plants from each of the 13 pools showed GUS expression in senescing leaves but not in non-senescing ones. All these 1,300 lines in sub-pools of 10 lines were similarly screened, of which 70 sub-pools displayed GUS staining in senescing leaves only. There were 50 to 70 plants in each sub-pool. Seeds of individual lines of these 70 sub-pools (total 70 × 10 = 700 lines) subsequently were sown on Murashige and Skoog/Kan plates separately, and at least five miniature plants from each line were tested using 1 mm MUG as a substrate. To be specific, one non-senescing leaf and one senescing leaf were collected from each plant and were placed in separate wells of Costar 96-well plates with opaque walls and optically clear bottoms as shown in Figure 1. The well contained 70 μL of MUG solution. After 12 h of incubation in a 37°C oven, the plate was put on a UV box to visualize GUS activity in each well. The GUS enzyme converts MUG to methylumbelliferone that fluoresces upon UV irradiation (Jefferson, 1987). If the senescing leaf (but not the non-senescing one) exhibited GUS activity, the original plant was transplanted to soil to produce seeds.
Treatments of Senescence-Promoting Factors (ABA, Ethylene, JA, eBR, Darkness, and Dehydration)
Rosette leaf 6 (counted from bottom) was harvested from a 23-d-old Arabidopsis plant of each line. Under our growth conditions (see above), leaf 6 at the time of sampling is fully expanded; these fully expanded leaves do not start senescing until 3 d later, in planta. Five leaves from each line were used for each treatment. Ethylene treatment was performed according to Chen and Bleecker (1995). In brief, leaves were placed in two Plexiglas boxes, one filled with air (control) and the other with air containing 80 μL L−1 ethylene. The ethylene concentration was measured using an HP 5890 Series II gas chromatograph (Hewlett-Packard, Palo Alto, CA). The leaves were incubated for 18 h in ethylene or in air. ABA and JA treatments were performed as described by Park et al. (1998). To be specific, leaves were floated on 3 mm MES 2-([N-morpholino]-ethanesulfonic acid) buffer (pH 5.8) containing 0.1 mm ABA (mixed isomers from Sigma) or 50 μm (−)−JA (naturally occurring form from Sigma) for 18 h in our Arabidopsis growth chambers. Leaves on MES buffer only were used as controls. In a similar manner, leaves were floated on MES buffer containing 0 (control) or 1 μm eBR (gift from Prof. Yuju Zhao, Shanghai Institute of Plant Physiology, The Chinese Academy of Sciences, China; see He et al., 1996) for 18 h. Eighteen-hour treatment with 1 μm eBR is sufficient to induce gene expression (Clouse et al., 1992). For dehydration treatment, leaves were put on filter paper to dry for 1 h under dim light (30 μmol m−2 s−1 at 35% relative humidity; Weaver et al., 1998). Leaves put under the same conditions except for saturated humidity (to prevent leaves from losing water) were used as controls. For darkness treatment, leaves were floated on 3 mm MES buffer (pH 5.8) and incubated for 36 h in Arabidopsis growth chambers with all lights off (darkness) or on (control). After various treatments, a small leaf disc (with diameter of 4 mm) from each leaf was put immediately into a well of a 96-well plate containing 70 μL of 1 mm MUG solution (Jefferson, 1987). After 12 h of incubation in a 37°C oven, 60 μL of reaction solution from each well was transferred into a fresh Costar 96-well plate (with opaque walls and optically clear bottoms; Corning Inc., Corning, NY), and the plate was scanned using the Luminescence Spectrometer LS50B (Perkin-Elmer, Beaconsfield, Buckinghamshire, UK) to measure the GUS activity in each well. The excitation and emission wavelength were set at 365 and 455 nm, respectively (Jefferson, 1987). The ratio of fluorescence intensities in treated leaves compared to respective controls was calculated, and in this study we arbitrarily set a ratio of 2 as a cutoff line, i.e. if a treatment results in a ratio of ≥2, the treatment is regarded as “inducible.” Results of this rapid GUS assay method are comparable with those of Jefferson's method.
Cloning, Sequencing, and RNA Gel-Blot Analysis of Tagged Genes
Thermal asymmetric interlaced PCR (Liu et al., 1995) was performed to clone DNA fragments flanking the right border of T-DNA in Sel139 and Sel142. The primers (AD3 and oligo 86 for Sel139 and AD1 and oligo 86 for Sel142) and PCR conditions are essentially the same as described by Campisi et al. (1999). Inverse PCR was used to clone the DNA fragment that flanks the left border of T-DNA insert in Sel25. In brief, 1 μg of genomic DNA of Sel25 was digested using XbaI and self-ligated. This ligation was used as a template to PCR amplify a 0.4-kb DNA with the following pair of primers that anneal to the left border of T-DNA and GUS coding region, respectively: 5′CTACAGGACGGACCATGGTC3′ and 5′GATTTCCCGGACATGAAG3′. All PCR products were cloned into pGEM-T vector (Promega, Madison, WI). DNA cycle sequencing reactions were performed using SP6 or T7 or custom-made primers on the GeneAmp PCR System 2400 according to the manufacturer's protocol (Perkin-Elmer, Foster City, CA). The sequencing was run on an ABI Prism 310 Genetic Analyzer (Perkin-Elmer).
Total RNA extraction from Arabidopsis leaves and northern-blot analysis were carried out as previously described (Gan, 1995).
ACKNOWLEDGMENTS
We thank Drs. George Wagner, Arthur Hunt, and David Hildebrand (University of Kentucky, Lexington) and Dr. Donald Hunter (University of California, Davis) for critically reading this manuscript; Drs. Richard Amasino and Anthony Bleecker (University of Wisconsin, Madison) and Michael Reid (University of California, Davis) for useful discussions; and the Arabidopsis Biological Resources Center (Ohio State University, Columbus) for providing enhancer trap seeds.
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 2001–35304–09994 to S.G.) and by the Tobacco and Health Research Institute's Biotechnology Program at the University of Kentucky (grant to S.G.). Y.H. was supported in part by the University of Kentucky's Research Challenge Trust Fund (Plant Sciences). J.D.S. was supported in part by the University of Kentucky's Science Outreach Center. This is publication no. 01–06–25 of the Kentucky Agricultural Experiment Station.
LITERATURE CITED
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Becker W, Apel K. Differences in gene expression between natural and artificially induced leaf senescence. Planta. 1993;189:74–79. [Google Scholar]
- Bellen HJ. Ten years of enhancer detection: lessons from the fly. Plant Cell. 1999;11:2271–2281. doi: 10.1105/tpc.11.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Chua N-H. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science. 1990;250:959–966. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridization. Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Chen QG, Bleecker AB. Analysis of ethylene signal transduction kinetics associated with seedling growth responses and chitinase induction in Arabidopsis. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME. Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol. 1992;100:1377–1383. doi: 10.1104/pp.100.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell. 1999;11:1253–1266. doi: 10.1105/tpc.11.7.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Grierson D. Identification of cDNA clones for tomato (Lycopersicon esculentum Mill.) mRNAs that accumulate during fruit ripening and leaf senescence in response to ethylene. Planta. 1989;179:73–80. doi: 10.1007/BF00395773. [DOI] [PubMed] [Google Scholar]
- Drake R, John I, Farrell A, Cooper W, Schuch W, Grierson D. Isolation and analysis of cDNAs encoding tomato cysteine proteases expressed during leaf senescence. Plant Mol Biol. 1996;30:755–767. doi: 10.1007/BF00019009. [DOI] [PubMed] [Google Scholar]
- Gan S. Molecular characterization and genetic manipulation of plant senescence. Ph.D. thesis. Madison: University of Wisconsin; 1995. [Google Scholar]
- Gan S, Amasino RM. Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Developmental targeting of gene expression by the use of a senescence-specific promoter. In: Reynolds P, editor. Inducible Gene Expression in Plants. New York: CAB International; 1999. pp. 169–186. [Google Scholar]
- Grossniklaus U, Vielle-Calzada J-P, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;125:1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- Hajouj T, Michelis R, Gepstein S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 2000;124:1305–1314. doi: 10.1104/pp.124.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xu R, Zhao Y. Enhancement of senescence by epibrassinolide in leaves of mung bean seedling. Acta Phytophysiol Sin. 1996;22:58–62. [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol. 1994;106:863–876. doi: 10.1104/pp.106.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jiang CZ, Rodermel SR, Shibles RM. Photosynthesis, Rubisco activity and amount, and their regulation by transcription in senescing soybean leaves. Plant Physiol. 1993;101:105–112. doi: 10.1104/pp.101.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant. 1994;92:322–328. [Google Scholar]
- Noodén LD. The phenomenon of senescence and aging. In: Noodén LN, Leopold AC, editors. Senescence and Aging in Plants. San Diego: Academic Press; 1988. pp. 1–50. [Google Scholar]
- Park J-H, Oh SA, Kim YH, Woo HR, Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Riha K, Fajkus J, Siroky J, Vyskot B. Developmental control of telomere length and telomerase activity in plants. Plant Cell. 1998;10:1691–1698. doi: 10.1105/tpc.10.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CM. Gene expression during leaf senescence. New Phytol. 1994;126:419–448. doi: 10.1111/j.1469-8137.1994.tb04243.x. [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. Gene trap tagging of PROLIFERA, an essential MCM2–3-5-like gene in Arabidopsis. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T. An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol. 2000;124:1658–1667. doi: 10.1104/pp.124.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, Delcardayre SB, Raines RT, Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiol. 1980;66:246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Hinderhofer K, Kolb D. Specific association of a small protein with the telomeric DNA-protein complex during the onset of leaf senescence in Arabidopsis thaliana. Plant Mol Biol. 2000;42:429–438. doi: 10.1023/a:1006324008600. [DOI] [PubMed] [Google Scholar]