Abstract
The proto-oncogene MYC encodes a nuclear transcription factor that has an important role in a variety of cellular processes, such as cell cycle progression, proliferation, metabolism, adhesion, apoptosis, and therapeutic resistance. MYC amplification is consistently observed in aggressive forms of several solid malignancies and correlates with poor prognosis and distant metastases. While the tumorigenic effects of MYC in patients with head and neck squamous cell carcinoma (HNSCC) are well known, the molecular mechanisms by which the amplification of this gene may confer treatment resistance, especially to immune checkpoint inhibitors, remains under-investigated. Here we present a unique case of a patient with recurrent/metastatic (R/M) HNSCC who, despite initial response to nivolumab-based treatment, developed rapidly progressive metastatic disease after the acquisition of MYC amplification. We conducted comparative transcriptomic analysis of this patient’s tumor at baseline and upon progression to interrogate potential molecular processes through which MYC may confer resistance to immunotherapy and/or chemoradiation and used TCGA-HNSC dataset and an institutional cohort to further explore clinicopathologic features and key molecular networks associated with MYC amplification in HNSCC. This study highlights MYC amplification as a potential mechanism of immune checkpoint inhibitor resistance and suggest its use as a predictive biomarker and potential therapeutic target in R/M HNSCC.
Subject terms: Head and neck cancer, Head and neck cancer
Introduction
Locoregionally advanced head and neck squamous cell carcinoma (HNSCC) is associated with poor 5-year survival of only 40% for non-viral mediated disease, and substantial functional morbidity with combined multimodality therapy1–4. Due to the poor success of systemic cytotoxic chemotherapy in treating recurrent/metastatic (R/M) HNSCC, the recent clinical focus has shifted to immunotherapy with antibodies targeting T cell inhibitory receptors that function as immune checkpoints, such as programmed death 1 (PD-1). Nonetheless, PD-1 inhibitors were reported to unleash anti-tumor immunity and achieve durable clinical responses in only 15–20% of treated patients in front-line recurrent/metastatic (R/M) setting5–8, with 4-year overall survival rate of about 15% overall and up to 22% among cases with a PD-L1 combined positive score (CPS) of 20 or greater5–9. Such disparity in treatment benefits between patients, paired with the absence of any standard effective therapies that target immunotherapy resistance, necessitates the identification of predictive biomarkers to better inform clinicians’ therapeutic decisions3. While it was reported that higher PD-L1 expression, assessed by the CPS, is predictive of a favorable response to immune checkpoint inhibitors (ICI), CPS remains an imperfect biomarker, with the majority of patients ultimately developing therapeutic resistance. Therefore, identification of improved biomarkers of response and resistance is of great importance, as biomarker-directed therapeutics after progression on immunotherapy (such as HRAS) suggest that determining specific mechanisms driving treatment resistance in HNSCC is key to therapeutic development10,11. Furthermore, little is known regarding the mechanisms of resistance to ICIs in head and neck cancer, further limiting the ability to predict non-responders and to investigate novel therapeutics targeting mechanisms of resistance to ICIs in R/M HNSCC12,13.
A high prevalence of alterations in the MYC oncogene is well documented in various solid malignancies, along with its association with aggressive disease and poor clinical outcomes14–16. While recurrent MYC gain-of-function mutations were found in certain human lymphomas17–19, in HNSCC, amplification appears to be the predominant genetic aberration that occurs in the MYC gene20, with an estimated prevalence of 12% in the HNSCC cohort of The Cancer Genome Atlas (TCGA), whereas mutations occur in only a small subset (1.2%) of patients21. MYC amplification is known to have broad influence on the transcriptome, driving upregulation of various mitogenic and survival signaling pathways associated with cell growth, proliferative, anti-apoptotic, and metabolic processes in ovarian, breast, and lung cancers among others22–25. Furthermore, MYC plays an important role in suppressing the host anti-tumor immune response, through various mechanisms involving modifications to the tumor microenvironment via inhibitory cytokines (e.g., TGFβ and immunomodulatory molecules such as PD-L1, CD47, and MHC I)26–29. While the tumorigenic effects of MYC are well known, the molecular mechanisms by which the mutation or amplification of this gene may confer treatment resistance in HNSCC remains under-investigated30. A recent case report of a patient with recurrent/metastatic (R/M) HNSCC highlighted a differential response to nivolumab in metastatic lesions secondary to the acquisition of MYC amplification. The MYC amplified lesion was resistant to the ICI while all other lesions, devoid of MYC amplification, responded to treatment, suggesting MYC amplification may play a role in ICI resistance in R/M HNSCC31. Supporting this suggestion, another study showed that MYC amplification regulates PD-L1 expression and is involved in a decreased response to ICI therapy in esophageal squamous cell carcinoma32.
Here we present a unique case of a patient with R/M HNSCC who, despite initial response to nivolumab-based therapy, developed rapidly progressive, metastatic disease after the acquisition of a MYC amplification. We conducted transcriptomic analysis of this patient’s HNSCC to identify the potential molecular processes through which MYC amplification may confer resistance to immunotherapy. In light of these findings, we searched our in-house genomic sequencing platform and identified seven additional patients with MYC amplified HNSCC along with 48 MYC wild-type matched controls, to further elucidate the broader clinicopathologic characteristics of MYC-driven disease. Finally, we performed gene expression and pathway analysis using the transcriptomic data obtained from TCGA to further explore key molecular networks associated with MYC amplification in HNSCC. Collectively, this work seeks to highlight MYC amplification as a potential mechanism of treatment resistance and suggest its use as a predictive biomarker and a potential therapeutic target in the treatment of R/M HNSCC24,33,34.
Results
Case presentation
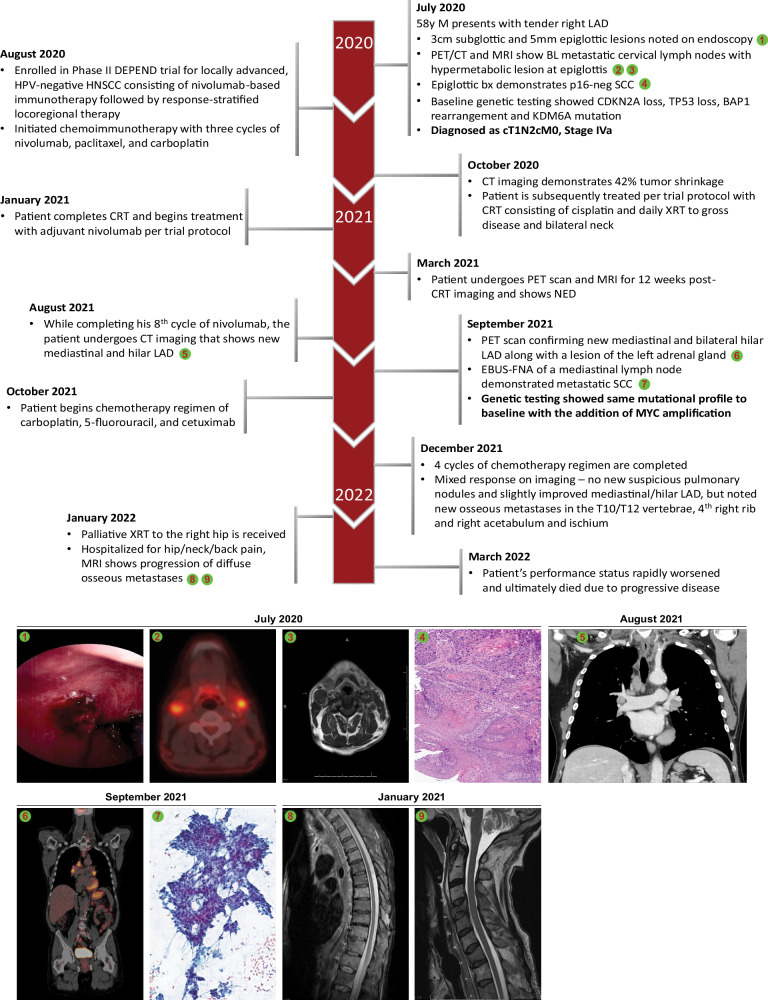
A 58-year-old male with an 80-pack-year tobacco history but quit at diagnosis presented in July 2020 noting a right-sided neck mass (Fig. 1). A CT soft tissue neck with contrast demonstrated abnormally enlarged bilateral lymph nodes with a heterogenous appearance and a soft tissue prominence of the anterior hypopharynx at the level of the glottis. A subsequent ultrasound-guided fine-needle aspiration of a right cervical node demonstrated scant metastatic squamous cell carcinoma involving fibrous and lymphoid tissue, at which time the patient was referred to our institutional multidisciplinary head and neck cancer team. Upon endoscopic evaluation, a 3 cm epiglottic pedunculated lesion was noted. A biopsy of the epiglottic lesion was performed and demonstrated squamous cell carcinoma. Immunohistochemical staining for p16 was negative. Subsequently, a PET/CT scan was conducted, which showed metastatic cervical lymph nodes, most prominent at bilateral level 2, with mild hypermetabolic activity of a lesion along the anterior commissure of the glottis and was staged as (cT1N2cM0, Stage IVa, American Joint Committee on Cancer 8th edition). A baseline Oncoplus molecular analysis was retrospectively conducted on the primary epiglottic specimen, showing CDKN2A loss, TP53 loss, BAP1 rearrangement, and KDM6A mutation. The specimen was found to be microsatellite stable (MSS) and had a tumor mutation burden (TMB) of 18.0 mutations/mb. PD-L1 immunohistochemical staining was negative (CPS < 1%).
Fig. 1. Course of disease.
Timeline depicting the course of disease and treatment, during which the patient acquired a MYC amplification that was not detected on baseline genetic testing and subsequently developed highly aggressive, immunotherapy-resistant disease. Numbered images at the bottom correspond to the indexes indicated across the course of the disease depicted above. BL: bilateral, EBUS-FNA: endobronchial ultrasound-guided fine-needle aspiration.
The patient was enrolled in a phase II clinical trial for locally advanced, HPV-negative HNSCC evaluating nivolumab-based chemoimmunotherapy followed by response-stratified locoregional therapy (NCT03944915). He initiated induction chemoimmunotherapy with three cycles of nivolumab 360 mg day 1, paclitaxel 100 mg/m2 days 1/8/15, and carboplatin AUC 5 day 1 of 21-day cycle followed by imaging demonstrating 42% tumor shrinkage per RECIST 1.1 and was subsequently treated per protocol with chemoradiotherapy (CRT) consisting of cisplatin 100 mg/m2 q21 days with daily fractionated radiation therapy (RT) at 2 Gy per fraction to total of 70 Gy over 35 fractions to gross disease and bilateral neck. Following completion of CRT he initiated adjuvant nivolumab per protocol. Three months after completion of CRT, the patient underwent PET demonstrating a complete metabolic response. MRI demonstrated no residual cervical lymph node metastases or discernible measurable laryngeal tumor consistent with a complete response by RECIST 1.1 criteria, and direct laryngoscopy showed a complete clinical response. However, during adjuvant nivolumab, six months after completion of chemoradiation, the patient underwent CT imaging demonstrating new mediastinal and hilar lymphadenopathy. A subsequent PET scan showed 18F-fluorodeoxyglucose (FDG) avidity of the mediastinal and bilateral hilar adenopathy along with a left adrenal nodule. Endobronchial ultrasound and fine-needle aspiration of a mediastinal lymph node demonstrated p16 negative, metastatic SCC in September 2021. Repeat Oncoplus molecular analysis was performed on the mediastinal lymph node and revealed an acquired MYC amplification in addition to the genetic aberrations detected in the baseline analysis (e.g., CDKN2A and TP53 loss, BAP1 rearrangement as well as KDM6A mutation) supporting a clonal relationship between primary and progressive disease. Of note, this biopsy was also found to be PD-L1 negative on immunohistochemical staining.
The patient started a chemotherapy regimen consisting of carboplatin (AUC 5 day 1), 5-fluorouracil (5-FU, 1000 mg/m2/d days 1–4), and cetuximab (400 mg/m2 loading followed by 250 mg/m2 weekly of 21-day cycle). Repeat imaging demonstrated progressive disease with interval increase in both the intrathoracic lymphadenopathy (LAD) and adrenal lesion as well as new vertebral metastases in T10 and T12 after four cycles of chemotherapy/cetuximab. The patient received palliative XRT to the right hip, however, his performance status rapidly worsened and he ultimately died from progressive disease five months after starting chemotherapy/cetuximab for recurrent/metastatic HNSCC. Given the observation of an acquired MYC amplification upon rapidly progressive disease while receiving anti-PD-1 therapy following an initial partial response, we proceeded to characterize clinical-pathologic features of MYC amplified HNSCC from our internal dataset, performed RNA-Seq of the paired samples with and without MYC amplification from our case study, and analyzed transcriptomic data of MYC amplified cases obtained from TCGA-HNSC dataset.
Clinical-pathologic characterization of recurrent MYC amplified HNSCC
To further interrogate the clinical and molecular profile of tumors with acquired MYC amplification, we performed a retrospective medical records review of patients with HNSCC treated at our institution between 2018 and 2021. We identified 8 cases (including the index patient) who were positive for MYC amplification based on the OncoPlus assay at the time of recurrence, and 48 matched control cases bearing a wild-type MYC (Supplementary Data 1). MYC amplified cases demonstrated a median age of 61, 25% were p16 positive oropharynx, all cases (100%) had the local disease at the time of diagnosis and initially received chemoradiotherapy, while the majority (63%) ultimately received immunotherapy during their treatment course. MYC amplification was significantly associated with enrichment for the laryngeal primary site (Supplementary Table 1) and showed numerical trend toward elevated frequency of TP53 and CDKN2A genetic aberrations, similar to previous reports (Supplementary Table 2)35–37. While median age, gender, treatment modality, p16 status (a surrogate biomarker for HPV-positivity), as well as tumor stage at diagnosis, were not different between the two groups, MYC amplified patients showed non-significant trend toward higher rates of developing recurrent and/or metastatic disease following primary therapy, with 100% of cases among MYC amplified compared to 72.9% among the wild-type MYC counterparts (p = 0.08; Supplementary Table 1). Furthermore, although the overall survival was not statistically significantly different with this small sample size available for analysis, median survival of MYC amplified patients was 40.6 months, compared to 49.1 months across the MYC wild-type individuals (Supplementary Table 1)36.
RNA-Seq analysis of paired patient samples with acquired MYC amplification
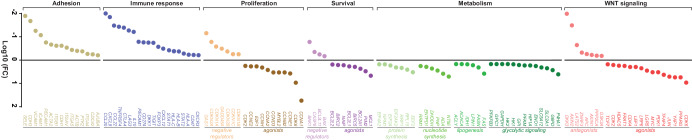
Given that transcriptional changes associated with the acquisition of MYC amplification in setting of immunotherapeutic resistance is poorly understood in HNSCC, we next performed RNA-Seq analysis of the tumor specimens collected from the patient described in the case report at baseline and upon rapid progression of R/M disease while receiving nivolumab (Supplementary Data 2). A purely descriptive comparative transcriptomic analysis of paired samples showed that genes known to be modulated by MYC were upregulated at disease progression, such as WNT/-catenin pathway agonists, genes central to the glycolytic pathway (e.g., encoding phosphofructokinase, hexokinase II and enolase), fatty acid metabolism (e.g., acetyl-CoA carboxylase, fatty acid synthase, and ATP-citrate lyase), regulators of nucleotide (e.g., ribonucleotide reductase and ectonucleoside triphosphate diphosphohydrolase) and protein (e.g., eukaryotic translation and elongation factors) synthesis networks, as well as molecules that play role in cell proliferation and survival (Fig. 2). Conversely, WNT antagonists, tumor suppressors known to negatively regulate cell cycle, apoptosis and cell adhesion were downregulated in the MYC amplified disease. Notably, genes involved in host anti-tumor immune response such as immune activation regulators, class I human leukocyte antigens (HLAs), STAT 1/2, and chemokines associated with recruitment of immunosuppressive cells were almost universally downregulated in the MYC amplified tumor compared to baseline. While these descriptive observations may provide a snapshot of the transcriptomic changes associated with acquired MYC amplification, thousands of genes (nearly 15% of transcriptome) are predicted to be direct targets of MYC, highlighting the complexity of the MYC-regulated signaling network38–40.
Fig. 2. RNA-Seq analysis of rapidly progressive, metastatic disease after the acquisition of a MYC amplification.
RNA sequencing was performed on the matched tumor specimens collected at baseline and upon progression. The reads were aligned against the human reference genome GRCh38/hg38 and gene counts were calculated. The figure indicates fold change between expression levels of the selected genes in tumor biopsy collected upon disease progression (acquired MYC amplification) and baseline specimen (wild-type MYC). Genes with fold change value ± 1.5 were included and grouped by their major roles in WNT signaling, metabolism, survival, and proliferation signaling networks.
Acquisition of MYC amplification is associated with broad transcriptomic changes
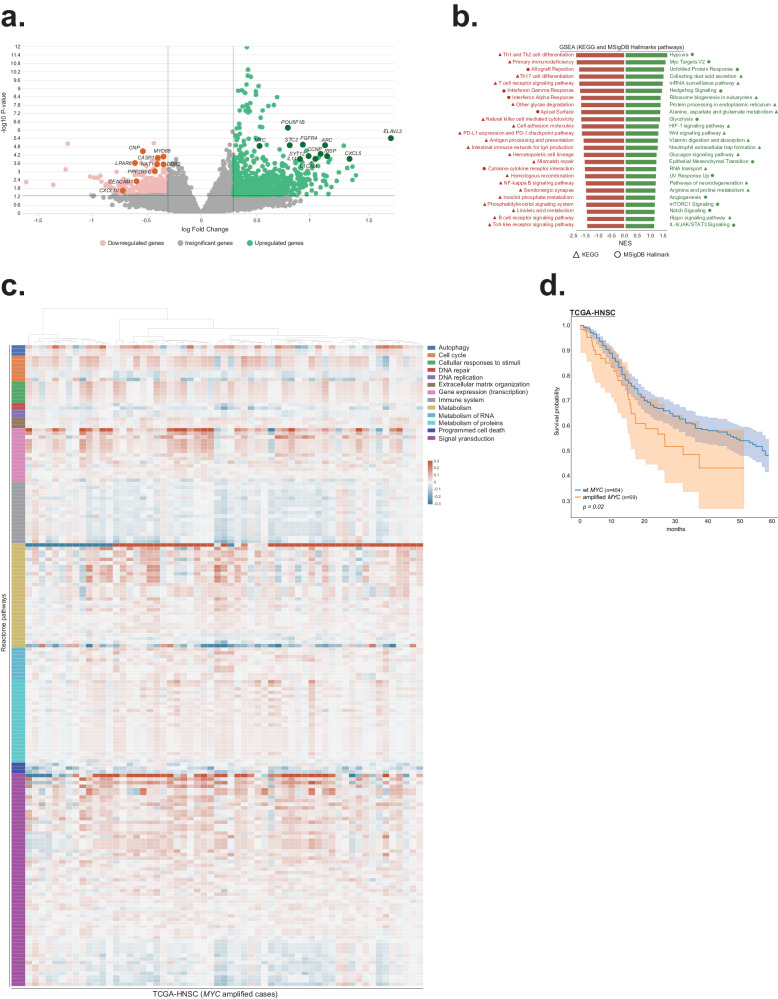
As we could not generate statistically meaningful conclusions based on the analysis of a single case sequencing, we next performed transcriptomic evaluation using samples obtained from the TCGA-HNSC dataset (Supplementary Data 3). Notably, tumors carrying amplified MYC (n = 59) were enriched for laryngeal disease, TP53 and CDKN2A mutations (Supplementary Table 3, Supplementary Fig. 1), and exhibited significant upregulation of numerous driver genes known to be associated with carcinogenesis in several types of cancer, including HNSCC (e.g. ELAVL2, CXCL5, FGFR4, CCNP, STC2, POU5F1B and SYT12)41–50. Significantly down-regulated genes contained known tumor suppressors and regulators of immune response (MYD88, NAT1, DDB2, PPP2R5C, LPAR6, CNP, CASP1, CXCL10, and CEACAM1)51–66 (Fig. 3a, Supplementary Data 4). Interestingly, the relative expression of T cell recruiting chemokines, immune checkpoints, genes associated with antigen presentation, and IFN-γ signaling showed a trend toward downregulation in MYC amplified tumors (Supplementary Fig. 2). As analysis of the expression patterns fails to capture subtle differences between samples that arise from dynamic interactions between genes at the signaling level67, we next applied pathway-based scoring methods that project gene expression data into the biological networks. Gene set enrichment analysis (GSEA)68 using KEGG and MSigDB Hallmark pathway databases revealed that the top up-regulated pathways in MYC amplified tumors were represented by processes that promote tumor growth, metastasis, and chemoresistance such as hypoxia, protein metabolism, glycolysis, epithelial-mesenchymal transition, angiogenesis, WNT (which has been shown to transcriptionally activate MYC expression)69, NOTCH, mTORC1, Hedgehog, Hippo and IL-6/JAK/STAT3 signaling (Fig. 3b, Supplementary Data 5). On the other hand, pathways associated with anti-tumor immune response (e.g., interferon-gamma response, T cell receptor signaling, natural killer cell-mediated cytotoxicity, antigen processing and presentation, Th1/Th2 cell differentiation, and toll-like receptor signaling) were enriched among the top down-regulated signaling axes (Fig. 3b), which may explain, in part, the poor effectiveness of immunotherapies against MYC-driven malignancies70. To query the transcriptomic data in more detail, iPANDA algorithm was pursued to predict differential activation of pathways retrieved from the Reactome database, which provides the hierarchical organization of signaling axes grouped into broader domains of biological functions67,71. Supporting the GSEA analysis, PandaOmics revealed that signaling networks associated with cell cycle progression, gene expression, signal transduction, and metabolism were upregulated in patients carrying the MYC amplification, whereas cellular processes related to the immune system and apoptosis were downregulated (Fig. 3c, Supplementary Data 6). Notably, while HPV frequency, age, gender, and tumor stage at diagnosis were not different between the two groups (Supplementary Table 3), the 5-year overall survival of patients with MYC amplified tumors was lower compared to the MYC wild-type counterparts (Fig. 3d), supporting the relevance of the molecular landscape changes described above to patient’s outcomes.
Fig. 3. Acquisition of MYC amplification is associated with broad transcriptomic changes.
a Volcano plot based on the TCGA-HNSC transcriptomic data comparing patients carrying amplified and wild-type MYC. Logarithmic fold-changes and FDR-corrected q-values were used to build volcano plot for differentially expressed genes (Q-value < 0.05). Green and pink colors depict significantly up-regulated and down-regulated genes respectively. Darker colors highlight genes known to be associated with HNSCC carcinogenesis. b Gene set enrichment analysis based on the TCGA-HNSC transcriptomic data comparing patients carrying amplified and wild-type MYC. Analysis was performed using two collections of gene sets obtained from the Enrichr library: MSigDB Hallmark 2020 and KEGG 2021 Human. Top 25 upregulated (green) and down-regulated (red) pathways from MSigDB Hallmark gene sets (circle) and KEGG database (triangle) are shown. All indicated pathways have FDR q-val < 0.05. c Pathway activation heatmap comparing TCGA-HNSC tumors with amplified and wild-type MYC. Pathway activation (shades of red) or inhibition (shades of blue) is inferred based on the scores obtained from InSilico Pathway Activation Network Decomposition Analysis (iPANDA) algorithm applied to the Reactome pathways database. Pathways are grouped according to the Reactome’s “superpathways” that describe normal cellular functions. d Kaplan–Meier survival probabilities with 95% CIs for patients with MYC amplification and wild-type MYC tumors (two-sided log-rank test p = 0.02).
Discussion
Immune checkpoint inhibition alone or in combination with chemotherapy has an established role in R/M HNSCC and is associated with improved survival, yet mechanisms of acquired resistance are poorly understood5–9. Here we present a unique case of a patient with p16 negative SCC of the epiglottis, staged as T1N2cM0 (IVA), who had a partial response following induction nivolumab/chemotherapy followed by definitive concurrent CRT, demonstrating a complete metabolic response. Interestingly, despite early response to therapy, this patient unfortunately developed rapidly progressive metastatic disease while actively receiving adjuvant nivolumab and was found to have acquired a MYC amplification that was not present on his baseline biopsy. This led to the question of how the acquisition of MYC amplification in this patient’s rapidly progressive metastatic HNSCC may contribute to molecular pathway dysfunction that promoted immune evasion and therapeutic resistance. While MYC amplification is well described in epithelial-type malignancies and associated with a poor prognosis25,72,73, its role in HNSCC therapeutic resistance and immune evasion has not been characterized.
We initially queried our internal database for HNSCC patients with MYC amplification at the time of recurrence along with MYC wild-type controls to characterize the clinicopathologic features of this cohort. Interestingly, we found that MYC amplified HNSCCs were enriched for larynx primary site, and appeared to be associated with higher frequencies of TP53 and CDKN2A genetic aberrations and HPV-negative disease, consistent with previous reports35,74. Yet despite these findings, MYC amplification in our cohort was also identified among other subsites and among patients with HPV-positive HNSCC. Although statistically significant prognostic comparisons were limited by the small sample size in our internal cohort and MYC status was only evaluable at the time of recurrence, there did appear to be a numerical trend towards worse median survival with MYC amplification, as well as higher rates of recurrence, despite the majority of patients ultimately receiving immunotherapy in addition to chemoradiotherapy, conforming with previous observations36.
Although these findings were suggestive of unique clinicopathologic patterns and signal of prognostic implications, the potential role of acquired MYC alterations in therapeutic resistance including immune evasion in HNSCC could not be interrogated. Therefore, we performed RNA-Seq analysis on paired patient samples from our index case to compare in descriptive terms baseline MYC wild-type tumor sample, with acquired MYC amplification at the time of distant metastatic disease while receiving anti-PD-1 therapy. Notably, we found relative upregulation of genes that play role in WNT/-catenin signaling (which is strongly correlated with immune exclusion across multiple human cancer types)75, the glycolytic pathway, fatty acid metabolism, nucleotide regulators, and protein synthesis networks. At the same time, we identified relative downregulation of tumor suppressors known to negatively regulate cell cycle, apoptosis, and cell adhesion, consistent with previous suggestions that MYC amplification in HNSCC is associated with activation of DNA repair pathways and promoting DNA damage checkpoint response, which may contribute to chemotherapeutic and radiotherapeutic resistance76,77. Interestingly, paired RNA-Seq analysis in our patient also identified relative downregulation of genes involved in host anti-tumor immune response including immune activation regulators, class I HLAs, STAT 1/2, and chemokines associated with recruitment of immunosuppressive cells within the tumor microenvironment. Previous studies have corroborated these findings suggesting that MYC activation leads to the enrichment of immunosuppressive tumor microenvironment such as regulatory T-cells, along with the exclusion of CD8+ T-cells and NK cells, in several tumor types including HNSCC, and supported by reports of differential metastasis response to immune checkpoint inhibitors with MYC amplification31,36,70. While formal statistical comparisons cannot be performed in a single patient, these findings suggest potential MYC-mediated mechanisms of immune evasion, that may explain the development of secondary resistance to immune checkpoint inhibitors with the acquisition of a MYC amplification in this patient. Of note, this patient did not have PD-L1 expression in his tumor at baseline, which may also explain immune evasion despite immune checkpoint inhibitor therapy in this patient. However, a very high tumor mutational burden at baseline (18 mut/MB) was noted which would be anticipated to predict response to immune checkpoint inhibition even without PD-L1 expression via generation of higher neoantigen burden facilitating deeper and more durable responses with immune checkpoint inhibition78. This supports our hypothesis that alternative acquired MYC-driven immunosuppressive processes may be playing a role in this patient’s rapid disease progression while receiving nivolumab, even if robust neoantigen is present79,80.
Given our inability to generate statistically meaningful conclusions based on a single case paired sequencing analysis, we interrogated the transcriptomic changes within the publicly available TCGA-HNSC dataset. We again identified enrichment for laryngeal disease and HPV-negative patients, along with increased association with TP53 (as has been characterized previously in the literature and our internal dataset), and a statistically significant worsening of survival compared with MYC wild-type35,74. As negative HPV status and the presence of TP53 mutations are independent predictors of an aggressive disease phenotype in HNSCC81,82, they may also underlie the inferior overall survival seen in MYC amplified patients, requiring mechanistic investigation. Our TCGA-HNSC analysis similarly found upregulation in carcinogenesis driver genes, along with downregulation of tumor suppressor and immune response regulatory pathways. Furthermore, iPANDA pathway activation prediction analysis67,83,84 specifically identified upregulation of signaling networks associated with cell cycle progression, signal transduction, and metabolism, while immune cellular processes and apoptosis were indeed downregulated. These provide additional support for the hypothesis, driven from RNA-Seq analysis of a single case, for MYC-driven mechanisms of immune evasion playing a key role in addition to chemoradiotherapy resistance in the poor outcome for this patient despite initial response to immunotherapy-based multimodality treatment. Our findings highlight the need for more work interrogating MYC-mediated mechanisms of acquired resistance to immunotherapy in HNSCC, as well as potential combinatorial therapeutic strategies to target the cellular processes that drive this therapeutic resistance24,33,34.
Limitations to our study include the single patient case which limits definitive conclusions, as well as the small number of MYC amplified cases in our internal cohort limiting power for statistical comparisons. Additionally, there may be selection bias within a single institution for DNA sequencing and potential confounders due to the retrospective nature of comparisons of MYC amplified with MYC wild-type cohorts which limit definitive conclusions and generalizability. Furthermore, transcriptomic data for TCGA-HNSC patients was obtained from primary tumors, which may not fully reflect the complex network of MYC-induced signaling aberrations that occur upon progression of R/M disease. Finally, while our transcriptomic pathway analysis focused on potential MYC-driven mechanisms of immune evasion, there is likely therapeutic resistance to chemotherapy, radiotherapy, and targeted therapy also at play. Further work is needed to mechanistically differentiate therapeutic resistance to immunotherapy and other therapeutic treatment modalities. As there is no accepted HNSCC cellular or animal model mimicking MYC amplification, we are currently creating a bank of HNSCC organoids carrying amplified MYC. The mechanistic studies using these models will be report in a follow-up publication.
In conclusion, our study suggests that MYC amplification in HNSCC, while a relatively uncommon occurrence, may play a key role not only in chemo and radioresistance but suggests that acquired clonal alteration may drive resistance to immunotherapy through a variety of potential immune suppressive mechanisms, warranting further study. Characterization of potential mechanisms of MYC-driven immune evasion in HNSCC could lead to the development of biomarker-selected targeted combinatorial therapeutic strategies to overcome resistance to immunotherapy.
Methods
Case report/series
A single patient treated at the University of Chicago Medical Center (UCMC) on a prospective clinical trial (NCT03944915) was selected for a case analysis based on the identification of an acquired MYC amplification noted on the UCMC in-house OncoPlus next-generation sequencing-based genomic sequencing platform (https://uchicagomedlabs.testcatalog.org/show/NGPLSF)85. The prospective “DEPEND” trial was a single-arm phase II study of nivolumab, paclitaxel, and carboplatin, followed by response-adaptive chemoradiation and adjuvant nivolumab for patients with locoregionally advanced HPV-negative HNSCC as described previously86.
A patient signed and informed consent under the University of Chicago Institutional Review Board (IRB) approval and provided consent to publish on the basis of anonymized data. Seven additional HNSCC patients with MYC amplification and 48 patients with wild-type MYC to serve as controls were identified from the OncoPlus genetic screen database85. There were no MYC amplified HNSCC patients excluded from the analysis. A retrospective chart review was conducted following IRB-approved protocols to collect demographic and clinical data (including disease course, treatments, and survival). The present study was conducted in accordance with all relevant ethical regulations including the Declaration of Helsinki.
RNA sequencing
Tumor samples were obtained following IRB-approved protocols. Informed written consent was obtained from the patient before sampling. Paraffin-embedded slides were microdissected to obtain >60% neoplastic cells. Neoplastic cellularity was estimated from the sequential slides, which highly reflect cellularity of the section used for RNA sequencing. Total RNA was isolated with AllPrep DNA/RNA Kit (Qiagen) and measured using Agilent 2100 Bioanalyzer. Libraries were prepared using the TruSeq RNA Library Prep Kit and sequencing was performed using the NovaSeq platform (Illumina). The quality control assessment of raw paired-end sequencing reads was performed by FastQC87. Further, the reads were aligned against the human reference genome GRCh38/hg38, and read counts per gene was calculated using STAR (v2.6.1d)88. Visualization for Fig. 2 was generated using GraphPad Prism (version 10.1.1).
Transcriptomic analysis of the data obtained from TCGA-HNSC cohort
Raw counts pre-processing included Upper-quartile normalization and log2-transformation. Patients were segregated into MYC amplified and MYC wild-type subgroups based on GISTIC2 call. Specifically, TCGA-HNSC thresholded gene-level copy number variation (CNV) was estimated using the GISTIC2 method89. Copy number profile was measured experimentally using whole genome microarray at a TCGA genome characterization center. Subsequently, GISTIC2 method was applied using the TCGA FIREHOSE pipeline to produce gene-level copy number estimates. GISTIC2 further thresholded the estimated values to −2,−1,0,1,2, representing homozygous deletion, single copy deletion, diploid normal copy, low-level copy number amplification, or high-level copy number amplification. The high-level thresholds are calculated on a sample-by-sample basis and are based on the maximum median arm-level amplification copy number found in the sample. Only samples with GISTIC2 value of 2 (high-level copy number amplification) were considered MYC amplified (Supplementary Data 3). Gene expression data were uploaded into PandaOmics, an AI-driven target discovery platform, for subsequent analysis. Differential gene expression analysis has been performed using the limma-voom package inside the PandaOmics platform90. Obtained gene-wise p-values were corrected by the Benjamini–Hochberg procedure. Logarithmic fold-changes (LFC) and FDR-corrected Q-values were used to build volcano plot for differentially expressed genes (DEGs) (Q-value < 0.05) (Supplementary Data 4). Then, each of the DEGs was given a status of being oncogene and(or) tumor suppressor gene following the mappings mined from the OncoKB database91,92. To study the dysregulation of cellular processes between MYC amplified and MYC wild-type cases, iPANDA algorithm67 was applied using the Reactome pathways database93. iPANDA calculates the activation or inhibition score for each pathway by combining precalculated gene coexpression data with gene importance factors based on the degree of differential gene expression and pathway topology decomposition. Significantly dysregulated pathways with iPANDA score > 0.01 or < −0.01 were considered as activated and inhibited, respectively (Supplementary Data 6). P-values for the iPANDA pathway activation scores were obtained using weighted Fisher’s combined probability test. Detailed description and statistical credibility of the iPANDA score was previously published67. Unsupervised complete-linkage clustering was performed following Farthest Point algorithm and Euclidean metric94. Gene set enrichment analysis (GSEA) was performed with the GSEAPY python package95 using two collections of gene sets obtained from Enrichr library96 - MSigDB_Hallmark_2020 and KEGG_2021_Human (Supplementary Data 5).
Statistical analysis
Survival analysis was prepared in PandaOmics using the Kaplan–MeierFitter function from the lifelines Python package (two-sided log-rank test). The differences in clinical and disease-specific characteristics i(e.g. age, gender, primary site, HPV/p16 status, TNM stage, gene mutation frequency, etc.) between MYC amplified and wild-type cases (in the internal cohort and TCGA-HNSC dataset) were calculated by two-sample t-tests and chi-square tests of independence using GraphPad Prism software. The significance level was defined as 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the National Institutes of Health grants R01DE027809 and R01DE028674. Thomas Cyberski was partially supported by the Burroughs Wellcome Fellowship.
Author contributions
Conceptualization: A.J.R., E.I. and N.A. Investigation: E.I., A.J.R., T.F.C., A.S., V.M., M.K. and F.P. Resources: E.I., A.J.R., A.Z. and N.A. Data curation: A.S., M.K., L.S., A.T.P., F.P., A.J., W.G., S.K., M.L., E.D., G.C. and A.Z. Writing - original draft preparation: E.I., A.J.R., T.F.C., N.A. and A.S. Writing - review and editing: E.I., N.A., A.J.R., A.T.P., B.B., A.S., C.W., X.C., M.E., M.K., E.D., V.M., M.L., F.P., Y.M., G.C., A.J. and T.F.C. Supervision: E.I., A.J.R. and N.A. Funding acquisition: E.I., N.A., A.J.R., T.F.C. and A.Z. All authors contributed to the article and approved the submitted version.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information files. Source data are provided with this paper. Patients’ de-identified data (such as diagnosis, gender, averaged age, and treatment type) is provided in the manuscript.
Code availability
No code was developed in this study. GSEA software is freely available for download at https://www.gsea-msigdb.org/gsea/index.jsp. PandaOmics is industry-grade commercial software platform used since 2020. The platform is available at https://pandaomics.com. A trial access to the platform is available from InSilico Medicine upon request. A workflow for running the platforms is described in the “Methods” section.
Competing interests
M.K., A.Z. and F.P. are affiliated with InSilico Medicine, a company developing an AI-based end-to-end integrated pipeline for drug discovery and development. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Thomas F. Cyberski, Alka Singh, Michael Korzinkin.
Contributor Information
Evgeny Izumchenko, Email: izumchen@uchicago.edu.
Ari J. Rosenberg, Email: arirosenberg@uchicago.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00606-w.
References
- 1.Johnson DE, et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mody MD, Rocco JW, Yom SS, Haddad RI, Saba NF. Head and neck cancer. Lancet. 2021;398:2289–2299. doi: 10.1016/S0140-6736(21)01550-6. [DOI] [PubMed] [Google Scholar]
- 3.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machtay M, et al. Factors associated with severe late toxicity after concurrent chemoradiation for locally advanced head and neck cancer: an RTOG analysis. J. Clin. Oncol. 2008;26:3582–3589. doi: 10.1200/JCO.2007.14.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burtness B, et al. Pembrolizumab alone or with chemotherapy for recurrent/metastatic head and neck squamous cell carcinoma in KEYNOTE-048: subgroup analysis by programmed death ligand-1 combined positive score. J. Clin. Oncol. 2022;40:2321–2332. doi: 10.1200/JCO.21.02198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtness B, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 7.Cohen EEW, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet. 2019;393:156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 8.Ferris RL, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington KJ, et al. Pembrolizumab with or without chemotherapy in recurrent or metastatic head and neck squamous cell carcinoma: updated results of the phase III KEYNOTE-048 study. J. Clin. Oncol. 2023;41:790–802. doi: 10.1200/JCO.21.02508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavrielatou N, Doumas S, Economopoulou P, Foukas PG, Psyrri A. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat. Rev. 2020;84:101977. doi: 10.1016/j.ctrv.2020.101977. [DOI] [PubMed] [Google Scholar]
- 11.Ho AL, et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J. Clin. Oncol. 2021;39:1856–1864. doi: 10.1200/JCO.20.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MY, Allen CT. Mechanisms of resistance to T cell-based immunotherapy in head and neck cancer. Head Neck. 2020;42:2722–2733. doi: 10.1002/hed.26158. [DOI] [PubMed] [Google Scholar]
- 13.Kok VC. Current understanding of the mechanisms underlying immune evasion from PD-1/PD-L1 immune checkpoint blockade in head and neck cancer. Front. Oncol. 2020;10:268. doi: 10.3389/fonc.2020.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin X, Lin X, Guo L, Wang Y, Zhang G. Distinct clinicopathological characteristics, genomic alteration and prognosis in breast cancer with concurrent TP53 mutation and MYC amplification. Thorac. Cancer. 2022;13:3441–3450. doi: 10.1111/1759-7714.14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastronikolis N, et al. Mechanisms of C-myc oncogenic activity in head and neck squamous cell carcinoma. J. BUON. 2019;24:2242–2244. [PubMed] [Google Scholar]
- 16.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 17.Xu-Monette ZY, et al. Clinical and biologic significance of MYC genetic mutations in De Novo diffuse large B-cell lymphoma. Clin. Cancer Res. 2016;22:3593–3605. doi: 10.1158/1078-0432.CCR-15-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. doi: 10.1182/blood.V95.6.2104. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya N, Roy A, Roy B, Roychoudhury S, Panda CK. MYC gene amplification reveals clinical association with head and neck squamous cell carcinoma in Indian patients. J. Oral. Pathol. Med. 2009;38:759–763. doi: 10.1111/j.1600-0714.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang CV, et al. The c-Myc target gene network. Semin. Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal. Transduct. Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalkat, M. et al. MYC deregulation in primary human cancers. Genes10.3390/genes8060151 (2017). [DOI] [PMC free article] [PubMed]
- 26.Versteeg R, Noordermeer IA, Krüse-Wolters M, Ruiter DJ, Schrier PI. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 1988;7:1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhanasekaran R, et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022;19:23–36. doi: 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 2019;25:301–311. doi: 10.1038/s41591-018-0321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson AM, et al. Cisplatin exposure causes c-Myc-dependent resistance to CDK4/6 inhibition in HPV-negative head and neck squamous cell carcinoma. Cell Death Dis. 2019;10:867. doi: 10.1038/s41419-019-2098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noji R, et al. MYC-PDL1 axis reduces sensitivity to nivolumab in recurrent head and neck squamous cell carcinoma. Oral. Oncol. 2022;124:105666. doi: 10.1016/j.oraloncology.2021.105666. [DOI] [PubMed] [Google Scholar]
- 32.Liang MQ, Yu FQ, Chen C. C-Myc regulates PD-L1 expression in esophageal squamous cell carcinoma. Am. J. Transl. Res. 2020;12:379–388. [PMC free article] [PubMed] [Google Scholar]
- 33.Llombart V, Mansour MR. Therapeutic targeting of “undruggable” MYC. EBioMedicine. 2022;75:103756. doi: 10.1016/j.ebiom.2021.103756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duffy MJ, O’Grady S, Tang M, Crown J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021;94:102154. doi: 10.1016/j.ctrv.2021.102154. [DOI] [PubMed] [Google Scholar]
- 35.Baltaci E, Karaman E, Dalay N, Buyru N. Analysıs of gene copy number changes ın head and neck cancer. Clin. Otolaryngol. 2018;43:1004–1009. doi: 10.1111/coa.12686. [DOI] [PubMed] [Google Scholar]
- 36.Zhao S, et al. Identification and validation of the role of c-Myc in head and neck squamous cell carcinoma. Front. Oncol. 2022;12:820587. doi: 10.3389/fonc.2022.820587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulz P, Heitzer E, Speicher MR. Co-occurrence of MYC amplification and TP53 mutations in human cancer. Nat. Genet. 2016;48:104–106. doi: 10.1038/ng.3468. [DOI] [PubMed] [Google Scholar]
- 38.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmadi SE, Rahimi S, Zarandi B, Chegeni R, Safa M. MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J. Hematol. Oncol. 2021;14:121. doi: 10.1186/s13045-021-01111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao WS, et al. Genome-scale CRISPR activation screening identifies a role of ELAVL2-CDKN1A axis in paclitaxel resistance in esophageal squamous cell carcinoma. Am. J. Cancer Res. 2019;9:1183–1200. [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazaki H, et al. Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006;66:4279–4284. doi: 10.1158/0008-5472.CAN-05-4398. [DOI] [PubMed] [Google Scholar]
- 43.Dutra RL, et al. FGFR4 profile as a prognostic marker in squamous cell carcinoma of the mouth and oropharynx. PLoS ONE. 2012;7:e50747. doi: 10.1371/journal.pone.0050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sánchez-Botet A, et al. The atypical cyclin CNTD2 promotes colon cancer cell proliferation and migration. Sci. Rep. 2018;8:11797. doi: 10.1038/s41598-018-30307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasa L, et al. A systematic analysis of orphan cyclins reveals CNTD2 as a new oncogenic driver in lung cancer. Sci. Rep. 2017;7:10228. doi: 10.1038/s41598-017-10770-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S, et al. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8:5976–5991. doi: 10.18632/oncotarget.13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simó-Riudalbas L, et al. Transposon-activated POU5F1B promotes colorectal cancer growth and metastasis. Nat. Commun. 2022;13:4913. doi: 10.1038/s41467-022-32649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner KW, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J. Clin. Invest. 2013;123:5231–5246. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu K, et al. Synaptotagmin 12 (SYT12) Gene expression promotes cell proliferation and progression of lung adenocarcinoma and involves the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway. Med. Sci. Monit. 2020;26:e920351. doi: 10.12659/MSM.920351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, et al. Targeting KDM2A enhances T-cell infiltration in NSD1-deficient head and neck squamous cell carcinoma. Cancer Res. 2023;83:2645–2655. doi: 10.1158/0008-5472.CAN-22-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu M, Guo S, Stiles JK. The emerging role of CXCL10 in cancer (Review) Oncol. Lett. 2011;2:583–589. doi: 10.3892/ol.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reschke, R. et al. Immune cell and tumor cell-derived CXCL10 is indicative of immunotherapy response in metastatic melanoma. J. Immunother. Cancer10.1136/jitc-2021-003521 (2021). [DOI] [PMC free article] [PubMed]
- 53.Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014;6:97. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi C, et al. Hypermethylation of N-acetyltransferase 1 is a prognostic biomarker in colon adenocarcinoma. Front. Genet. 2019;10:1097. doi: 10.3389/fgene.2019.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C, Zhao Z, Liu H, Yao S, Zhao D. Weighted gene co-expression network analysis identified a novel thirteen-gene signature associated with progression, prognosis, and immune microenvironment of colon adenocarcinoma patients. Front. Genet. 2021;12:657658. doi: 10.3389/fgene.2021.657658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun H, et al. Long non-coding RNA H19 mediates N-acetyltransferase 1 gene methylation in the development of tamoxifen resistance in breast cancer. Exp. Ther. Med. 2022;23:12. doi: 10.3892/etm.2021.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bommi PV, Ravindran S, Raychaudhuri P, Bagchi S. DDB2 regulates epithelial-to-mesenchymal transition (EMT) in oral/head and neck squamous cell carcinoma. Oncotarget. 2018;9:34708–34718. doi: 10.18632/oncotarget.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Wlodarchak N, Xing Y. PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol. 2016;51:162–184. doi: 10.3109/10409238.2016.1143913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B, Liu S, Gao Y, Zheng L, Lu Y. Combined detection of SDC2/ADHFE1/PPP2R5C methylation in stool DNA for colorectal cancer screening. J. Cancer Res. Clin. Oncol. 2023;149:10241–10253. doi: 10.1007/s00432-023-04943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He, J. et al. Lysophosphatidic acid receptor 6 (LPAR6) is a potential biomarker associated with lung adenocarcinoma. Int. J. Environ. Res. Public Health10.3390/ijerph182111038 (2021). [DOI] [PMC free article] [PubMed]
- 62.Tao K, et al. Lysophosphatidic acid receptor 6 (LPAR6) expression and prospective signaling pathway analysis in breast cancer. Mol. Diagn. Ther. 2019;23:127–138. doi: 10.1007/s40291-019-00384-3. [DOI] [PubMed] [Google Scholar]
- 63.Zorniak M, et al. Differential expression of 2’,3’-cyclic-nucleotide 3’-phosphodiesterase and neural lineage markers correlate with glioblastoma xenograft infiltration and patient survival. Clin. Cancer Res. 2012;18:3628–3636. doi: 10.1158/1078-0432.CCR-12-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature10.1038/nature14129 (2015). [DOI] [PMC free article] [PubMed]
- 65.Celardo I, et al. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis. 2013;4:e645. doi: 10.1038/cddis.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sappino AP, et al. The CEACAM1 tumor suppressor is an ATM and p53-regulated gene required for the induction of cellular senescence by DNA damage. Oncogenesis. 2012;1:e7. doi: 10.1038/oncsis.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozerov IV, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat. Commun. 2016;7:13427. doi: 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 70.Li J, Dong T, Wu Z, Zhu D, Gu H. The effects of MYC on tumor immunity and immunotherapy. Cell Death Discov. 2023;9:103. doi: 10.1038/s41420-023-01403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sidiropoulos K, et al. Reactome enhanced pathway visualization. Bioinformatics. 2017;33:3461–3467. doi: 10.1093/bioinformatics/btx441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu J, Chen Y, Olopade OI. MYC and Breast Cancer. Genes Cancer. 2010;1:629–640. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaub FX, et al. Pan-cancer alterations of the MYC oncogene and its proximal network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300.e282. doi: 10.1016/j.cels.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence MS, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582,. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin. Cancer Res. 2019;25:3074–3083. doi: 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ganesan S. MYC, PARP1, and chemoresistance: BIN there, done that? Sci. Signal. 2011;4:pe15. doi: 10.1126/scisignal.2001946. [DOI] [PubMed] [Google Scholar]
- 77.Wang WJ, et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73:1219–1231. doi: 10.1158/0008-5472.CAN-12-1408. [DOI] [PubMed] [Google Scholar]
- 78.Wildsmith S, et al. Tumor mutational burden as a predictor of survival with durvalumab and/or tremelimumab treatment in recurrent or metastatic head and neck squamous cell carcinoma. Clin. Cancer Res. 2023;29:2066–2074. doi: 10.1158/1078-0432.CCR-22-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moroishi T, et al. The hippo pathway kinases LATS1/2 suppress cancer immunity. Cell. 2016;167:1525–1539.e1517. doi: 10.1016/j.cell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ni X, et al. YAP is essential for Treg-mediated suppression of antitumor immunity. Cancer Discov. 2018;8:1026–1043. doi: 10.1158/2159-8290.CD-17-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou G, Liu Z, Myers JN. TP53 mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response. J. Cell Biochem. 2016;117:2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fakhry C, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 83.Makarev E, et al. In silico analysis of pathways activation landscape in oral squamous cell carcinoma and oral leukoplakia. Cell Death Discov. 2017;3:17022. doi: 10.1038/cddiscovery.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pun, F. W. et al. A comprehensive AI-driven analysis of large-scale omic datasets reveals novel dual-purpose targets for the treatment of cancer and aging. Aging Cell10.1111/acel.14017 (2023). [DOI] [PMC free article] [PubMed]
- 85.Kadri S, et al. Clinical validation of a next-generation sequencing genomic oncology panel via cross-platform benchmarking against established amplicon sequencing assays. J. Mol. Diagn. 2017;19:43–56. doi: 10.1016/j.jmoldx.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 86.Rosenberg AJ, et al. A multicenter, open-label, randomized, phase II study of cediranib with or without lenalidomide in iodine 131-refractory differentiated thyroid cancer. Ann. Oncol. 2023;34:714–722. doi: 10.1016/j.annonc.2023.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andrews, S. FastQC a quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 88.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kamya, P. et al. PandaOmics: an AI-driven platform for therapeutic target and biomarker discovery. J. Chem. Inf. Model10.1021/acs.jcim.3c01619 (2024). [DOI] [PMC free article] [PubMed]
- 91.Suehnholz, S. P. et al. Quantifying the expanding landscape of clinical actionability for patients with cancer. Cancer Discov.10.1158/2159-8290.Cd-23-0467 (2023). [DOI] [PMC free article] [PubMed]
- 92.Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol.10.1200/po.17.00011 (2017). [DOI] [PMC free article] [PubMed]
- 93.Gillespie M, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–d692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bar-Joseph, Z., Gifford, D. K. & Jaakkola, T. S. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics17, S22–S29 (2001). [DOI] [PubMed]
- 95.Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics10.1093/bioinformatics/btac757 (2022). [DOI] [PMC free article] [PubMed]
- 96.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information files. Source data are provided with this paper. Patients’ de-identified data (such as diagnosis, gender, averaged age, and treatment type) is provided in the manuscript.
No code was developed in this study. GSEA software is freely available for download at https://www.gsea-msigdb.org/gsea/index.jsp. PandaOmics is industry-grade commercial software platform used since 2020. The platform is available at https://pandaomics.com. A trial access to the platform is available from InSilico Medicine upon request. A workflow for running the platforms is described in the “Methods” section.