Abstract
Brassinosteroids (BRs) are steroidal plant hormones that are essential for growth and development. It has been proposed that BRs are synthesized via two parallel pathways, the early and late C-6 oxidation pathways according to the C-6 oxidation status. The tomato (Lycopersicon esculentum) Dwarf gene encodes a cytochrome P450 that has been shown to catalyze the C-6 oxidation of 6-deoxocastasterone to castasterone. We isolated an Arabidopsis ortholog (AtBR6ox gene) of the tomato Dwarf gene. The encoded polypeptide has characteristics of P450s and is classified into the CYP85 family. The AtBR6ox and tomato Dwarf gene were expressed in yeast and the ability of the transformed yeast cells to metabolize 6-deoxo-BRs was tested. Metabolites were analyzed by gas chromatography-mass spectrometry. Both enzymes catalyze multiple steps in BR biosynthesis: 6-deoxoteasterone to teasterone, 3-dehydro-6-deoxoteasterone to 3-dehydroteasterone, 6-deoxotyphasterol to typhasterol, and 6-deoxocastasterone to castasterone. Our results indicate that the AtBR6ox gene and the tomato Dwarf gene encode steroid-6-oxidases and that these enzymes have a broad substrate specificity. This suggests that the BR biosynthetic pathway consists of a metabolic grid rather than two separate parallel pathways.
Since the discovery of brassinolide (BL; Grove et al., 1979), more than 40 natural analogs, collectively called brassinosteroids (BRs), have been isolated and characterized (Fujioka and Sakurai, 1997a, 1997b; Fujioka, 1999). Exogenous application of BRs to plants between nanomolar and micromolar concentrations causes a wide spectrum of physiological effects, including promotion of cell elongation and division, enhancement of tracheary element differentiation, retardation of abscission, enhancement of gravitropic-induced bending, promotion of ethylene biosynthesis, and enhancement of stress resistance as reviewed by Clouse and Sasse (1998) and Sasse (1999). A number of BR-deficient mutants have been discovered in Arabidopsis, pea (Pisum sativum), and tomato (Lycopersicon esculentum; for reviews, see Clouse and Feldmann, 1999; Schumacher and Chory, 2000). These mutants exhibit dwarfism under both light and dark conditions. Many of these mutants also have dark-green leaves, reduced fertility, a prolonged lifespan, and display abnormal skotomorphogenesis. BR-insensitive mutants have been identified in Arabidopsis, pea, tomato, and rice (Oryza sativa; Clouse et al., 1996; Kauschmann et al., 1996; Li and Chory 1997; Nomura et al., 1999; Koka et al., 2000; Yamamuro et al., 2000).
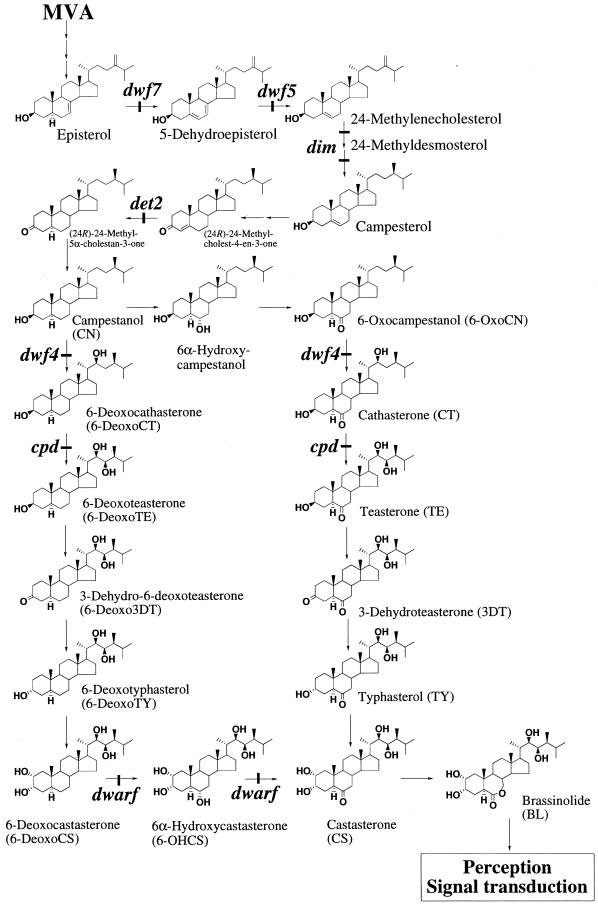
We have studied the biosynthetic pathways leading to BL (the most active BR) using cultured cells of Catharanthus roseus and proposed the two alternative biosynthetic pathways shown in Figure 1 (Fujioka and Sakurai, 1997a, 1997b). One is the early C-6 oxidation pathway, in which oxidation at C-6 occurs before the introduction of vicinal hydroxyls at C22 and C23 of the side chain (Fig. 1). The other is the late C-6 oxidation pathway in which C-6 is oxidized after the introduction of hydroxyls on the side chain and the A ring (Fig. 1). The natural occurrence of intermediates from both the early and late pathways have been shown in a variety of plants. Most of the steps in both the early and late C-6 oxidation pathways have been defined by feeding plants deuterium-labeled substrates and then identifying the metabolites using gas chromatography-mass spectrometry (GC-MS). In contrast, the physiological or biochemical relevance of two alternative pathways remained unclear.
Figure 1.
BR biosynthesis pathway. The proposed biosynthetic pathway for BL from mevalonate (MVA) is shown with the steps blocked in reported mutants.
A number of studies have reported BR biosynthesis and signal transduction mutants in Arabidopsis. For example, det2 (Li et al., 1996; Noguchi et al., 1999b), cpd (Szekeres et al., 1996), dwf4 (Choe et al., 1998), dwf1/dim (Klahre et al., 1998; Choe et al., 1999a), ste1/dwf7 (Choe et al., 1999b), sax1 (Ephritikhine et al., 1999), dwf5 (Choe et al., 2000), fackel (Jang et al., 2000; Schrick et al., 2000), and bri1 (Clouse et al., 1996; Li and Chory, 1997). From the analysis of these mutants, we identified BL, castasterone (CS), typhasterol (TY), teasterone (TE), 6-deoxocastasterone (6-DeoxoCS), 6-deoxotyphasterol (6-DeoxoTY), 6-deoxoteasterone (6-DeoxoTE), and 6-deoxocathasterone (6-DeoxoCT) as endogenous BRs in various Arabidopsis tissues, such as shoots, siliques, and seeds (Fujioka et al., 1996, 1998; Noguchi et al., 1999a, 2000). All BRs identified in Arabidopsis are important components of either the early or late C-6-oxidation pathways, indicating that both pathways are functional in this species. We studied the metabolism of deuterium-labeled BR intermediates in Arabidopsis very recently and demonstrated the operation of the biosynthetic sequence: campestanol (CN) → 6-DeoxoCT → 6-DeoxoTE → 3-dehydro-6-deoxoteasterone (6-Deoxo3DT) → 6-DeoxoTY → 6-DeoxoCS → 6α-hydroxyCS (6-OHCS) → CS → BL (Noguchi et al., 2000). We also showed the operation of the biosynthetic sequence: TE → 3-dehydroteasterone (3DT) → TY → CS → BL. These studies established that the previously determined biosynthetic pathway is also present in Arabidopsis.
The tomato Dwarf gene was isolated by transposon tagging (Bishop et al., 1996) and shown to encode a cytochrome P450 enzyme that converts 6-DeoxoCS to CS via 6-OHCS, a step where the early C-6 oxidation and the late C-6 oxidation pathways are connected (Bishop et al., 1999). This step is the furthest downstream step in BL biosynthesis among those known for mutations and enzymes. A defect in the Dwarf gene results in deficiency of CS and causes dwarfism with stem elongation and leaf expansion being suppressed (Bishop et al., 1996; Bishop et al., 1999). These studies demonstrated that 6-deoxo BRs exhibit very weak biological activity and it remains to be determined whether any or all of the 6-oxo BRs, e.g. TE, 3DT, TY, or CS, are active per se or become active after being converted to BL. Therefore, it is physiologically important to understand the biosynthesis of 6-oxo BRs including BL with the C-6 oxidation step potentially being one of the key regulatory steps in BR biosynthesis. Furthermore, intermediates of the late C-6 oxidation pathway are predominant over those of the early C-6 oxidation pathway in many species including Arabidopsis, tomato, and pea (Choi et al., 1997; Yokota et al., 1997; Bishop et al., 1999; Noguchi et al., 1999a; Koka et al., 2000; Nomura et al., 2000).
Especially high accumulation of 6-DeoxoCS has often been recorded for many plants, suggesting that the C-6 oxidation is a rate-limiting step. Such regulation seems to be cancelled in BR-insensitive mutants because CS accumulates at aberrant levels in pea lka (Nomura et al., 1997, 1999) and tomato curl-3 (T. Nomura, T. Yokota, and G.J. Bishop, unpublished data). In Arabidopsis, both CS and BL accumulate in the bri1 mutant (Noguchi et al., 1999a), indicating that C-6 oxidation is controlled by a feedback mechanism in steady-state conditions of wild-type plants. It is interesting that in tomato, the presence of BL has not been demonstrated, suggesting that CS is a biologically active BR. Therefore, the Dwarf enzyme may be a key enzyme governing the physiological role of BRs in tomato.
Dwarf enzyme functionally expressed in yeast cells can convert 6-DeoxoCS to CS but cannot oxidize CN to 6-oxocampestanol (6-oxoCN; Bishop et al., 1999). However, there is no information on whether the Dwarf enzyme catalyzes the conversion of other intermediates of the late C-6 oxidation pathway. Arabidopsis seedlings recently were found to oxidize the C-6 of both 6-DeoxoTY and 6-DeoxoCS (Noguchi et al., 2000). This finding has raised a question: In Arabidopsis, is such C-6 oxidation regulated by a single Dwarf ortholog or by multiple genes? In the present work, we report the isolation of a Dwarf gene ortholog termed AtBR6ox from Arabidopsis and the functional analysis of both AtBR6ox and Dwarf.
RESULTS
Cloning of an Arabidopsis Homologue for the Tomato Dwarf Gene
Databases were searched for homologs of the tomato Dwarf gene (Bishop et al., 1996) and three expressed sequence tag (EST) clones were found to have the highest sequence similarities; they are F5F3, 224F7, and 313B11 (the respective GenBank/EMBL/DNA Data Bank of Japan [DDBJ] accession nos. are AA713019, N65267, and AA394869). Based on partial DNA sequencing and restriction mapping, it was found that the clones corresponded to the same gene, but only F5F3 had sufficient length to contain the full-length cDNA (data not shown). When the entire nucleotide sequence of the cDNA insert of F5F3 was determined and compared with the Dwarf gene, it was apparent that the first intron remained un-spliced with the consensus splice sites at the putative exon-intron boundaries in the cDNA. Therefore, we re-isolated cDNA clones by reverse transcriptase (RT)-PCR. The isolated clones contained the same open reading frame (ORF) sequence, except that each of these clones had base substitutions in independent positions that are likely to be derived from PCR errors (data not shown).
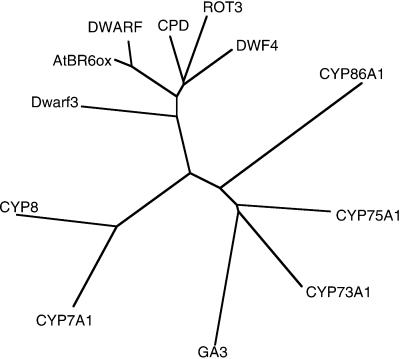
The complete ORF cDNA sequence encodes a polypeptide of 466 amino acid residues. The deduced amino acid sequence of the cDNA is shown in Figure 2A. We designated the corresponding gene AtBR6ox. The deduced amino acid sequence had characteristics of P450s. For example, the heme-binding consensus sequence FxxGxxxCxG (lowercase x indicates variable amino acid residues; Nelson et al., 1996) is conserved (Fig. 2A). The N terminus retains a hydrophobic region that likely functions as an anchor to the endoplasmic reticulum membrane (data not shown). AtBR6ox nucleotide sequence has 71% homology with that of the tomato Dwarf gene and the deduced amino acid sequence has 81% similarity and 68% identity (Fig. 2A). It also has high sequence similarity to members of CYP90 from Arabidopsis, namely with CPD, DWF4, and ROT3 at the amino acid level (Fig. 2B). A phylogenic relationship was calculated and is shown in Figure 3. This indicates that AtBR6ox is nearer to the tomato Dwarf than to Arabidopsis P450 genes in the CYP90 family, some of which are thought to be involved in BR biosynthesis. AtBR6ox is also designated CYP85A1 according to the nomenclature of the P450 superfamily (Nelson et al., 1996).
Figure 2.
Sequence alignment of AtBR6ox and related genes. A, Sequence alignment of AtBR6ox and the tomato Dwarf gene. Reverse contrast characters highlight identical amino acid residues and conserved ones are indicated by hatched characters. Gaps introduced to improve the alignment are shown by hyphens. The GenBank/EMBL/DDBJ accession nos. of the AtBR6ox gene (CYP85) and the tomato Dwarf gene (CYP85) are AB035868 and U54770, respectively. The heme-binding signature sequence is underlined. B, Sequence alignment of Arabidopsis P450s that are related to BR biosynthesis. In the consensus line, capital letters indicate identical amino acid residues in all genes and small letters indicate the most commonly conserved amino acid residues. The GenBank/EMBL/DDBJ accession nos. of ROT3 (CYP90C), DWF4 (CYP90B), and CPD (CYP90A) are AB008097, AF044216, and X87363, respectively. The heme-binding signature sequence is underlined.
Figure 3.
Phylogenetic relationship between AtBR6ox and selected P450 genes. AtBR6ox belongs to the group consisting of Dwarf (Tomato, CYP85), CPD (Arabidopsis, CYP90A), ROT3 (Arabidopsis, CYP90C), and DWF4 (Arabidopsis, CYP90B). All of these genes are thought to be involved in BR biosynthesis, except ROT3. The GenBank/EMBL/DDBJ accession nos. of Dwarf3 (Maize, CYP88A) and GA3 (Arabidopsis, CYP701A) are U32579 and AF047720, respectively; both are suggested to be involved in gibberellin (GA) biosynthesis. The first P450 genes functionally identified from higher plants are CYP73A (Helianthus cinnamate 4-hydroxylase, Z17369) and CYP75A (Petunia flavonoid-3′, 5′-hydroxylase, D14588); both belong to the higher plant-specific Group A of P450 genes. The accession nos. of the other members are D30718 (CYP8), M93133 (CYP7A), and X90458 (CYP86A).
Genome Analysis
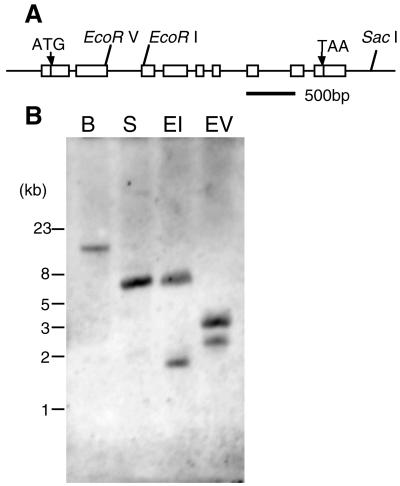
We also searched for genomic clones of the AtBR6ox gene in the databases. The nucleotide sequence of the cDNA clone, F5F3 completely matched to sequences from an Arabidopsis genomic clone, K15E6, sequenced by the Arabidopsis genome project (Sato et al., 1998; the GenBank, EMBL, and DDBJ accession no. is AB009048). Comparison of the cDNA and the genomic clone revealed that the AtBR6ox gene consists of nine exons (Fig. 4A). All the exon-intron boundaries consist of the conserved sequences of consensus splice sites. Clone K15E6 maps to chromosome 5 (Sato et al., 1998). To investigate the number of related genes in Arabidopsis, a genomic DNA gel blot was hybridized with an RNA probe from cDNA clone F5F3. The result after low-stringency washes is shown in Figure 4B. The result indicates that there are no other genes that cross-hybridized to the AtBR6ox gene according to the restriction map of the genomic sequence. The band pattern was the same after high-stringency washes (not shown).
Figure 4.
Analysis of the AtBR6ox gene. A, The physical structure of the AtBR6ox gene in Arabidopsis. White boxes represent exons. Slanting bars indicate restriction sites. There are no BamHI sites in the genome region presented. Start and stop codons are indicated by arrows. B, Genomic Southern analysis. The blot contains 2 μg genomic DNA that was digested with BamHI (B), SacI (S), EcoRI (EI), or EcoRV (EV), and was hybridized with a digoxigenin-labeled RNA probe derived from the F5F3 clone. The blot was washed under low-stringency condition.
Functional Analysis
The sequence similarity between AtBR6ox and Dwarf does not necessarily imply that the AtBR6ox enzyme catalyzes the conversion of the same substrates in Arabidopsis as the Dwarf enzyme does in tomato. To establish the biochemical function of the AtBR6ox product, the gene was functionally expressed in yeast. The protein-coding region of the complete sequence was synthesized by fusing the 5′-coding region of PCR clone pCRCSS12 and the 3′ region of the EST clone, F5F3. A cDNA sequence of the protein-coding region was then sub-cloned into a yeast expression vector, pYeDP60, and was expressed in the yeast strain, WAT11, which carries Arabidopsis NADPH-P450-reductase (Urban et al., 1997). In this strain, both AtBR6ox and P450 reductase were over expressed in the presence of Gal (Pompon et al., 1996). An induced culture of the yeast transformant was incubated with 5 μg of deuterated [2H6]BRs. Products from the incubation were analyzed by GC-MS. Confirmation of the identity of the products was provided by a direct comparison of the relative abundance of characteristic ions of the metabolites and standard compounds (Table I). Conversion rates obtained by GC-selected ion monitoring for AtBR6ox and Dwarf are summarized in Tables II and III, respectively.
Table I.
GC-MS identification of [2H6]-labeled BRs in yeast cultures expressing AtBR6ox
| Compound | Retention Time on GC | Characteristic Ions (Relative Intensity Percentage) |
|---|---|---|
| min | m/z | |
| [2H6]CSa | ||
| Standard | 11.82 | 518 [M+] (33), 399 (5), 358 (11), 287 (23), 161 (100) |
| Metabolite | 11.83 | 518 [M+] (33), 399 (5), 358 (12), 287 (22), 161 (100) |
| [2H6]6-OHCSb | ||
| Standard | 11.40 | 592 [M+] (2), 577 (22), 502 (64), 271 (66), 161 (100) |
| Metabolite | 11.42 | 592 [M+] (2), 577 (23), 502 (72), 271 (63), 161 (100) |
| [2H6]TYb | ||
| Standard | 11.23 | 550 [M+] (81), 535 (50), 521 (100), 460 (38), 161 (58) |
| Metabolite | 11.23 | 550 [M+] (71), 535 (46), 521 (100), 460 (46), 161 (66) |
| [2H6]3DTa | ||
| Standard | 11.92 | 476 [M+] (12), 316 (9), 287 (7), 245 (13), 161 (100) |
| Metabolite | 11.92 | 476 [M+] (11), 316 (8), 287 (6), 245 (13), 161 (100) |
| [2H6]TEb | ||
| Standard | 11.95 | 550 [M+] (25), 535 (66), 521 (100), 460 (2), 161 (25) |
| Metabolite | 11.97 | 550 [M+] (25), 535 (65), 521 (100), 460 (3), 161 (21) |
M+, Molecular ion; m/z, mass-charge ratio.
Methaneboronate derivative.
Methaneboronate-trimethylsilyl derivative.
Table II.
C6 oxidation of BRs by AtBR6ox expressed in yeast
| Substrate | Product | Conversion Ratea
|
||
|---|---|---|---|---|
| First experiment | Second experiment | Vector | ||
| % | ||||
| [2H6]6-DeoxoCS | [2H6]CS | 35 | 42 | N.D.b |
| [2H6]6-DeoxoCS | [2H6]6-OHCS | 16 | 13 | N.D. |
| [2H6]6-DeoxoTY | [2H6]TY | 23 | – | N.D. |
| [2H6]6-Deoxo3DT | [2H6]3DT | 36 | 38 | N.D. |
| [2H6]6-DeoxoTE | [2H6]TE | 13 | 21 | N.D. |
| [2H6]6-DeoxoCT | [2H6]CT | N.D. | N.D. | N.D. |
| [2H6]CN | [2H6]6-OxoCN | N.D. | N.D. | N.D. |
The conversion rate was calculated roughly as a percentage of the amount of each metabolite versus the amount of substrate added to the culture.
N.D., Not detected.
Table III.
C6 oxidation of BRs by Dwarf expressed in yeast
| Substrate | Product | Conversion Ratea
|
|
|---|---|---|---|
| First experiment | Vector | ||
| % | |||
| [2H6]6-DeoxoCS | [2H6]CS | 62 | N.D.b |
| [2H6]6-DeoxoCS | [2H6]6-OHCS | 30 | N.D. |
| [2H6]6-DeoxoTY | [2H6]TY | 66 | N.D. |
| [2H6]6-Deoxo3DT | [2H6]3DT | 96 | N.D. |
| [2H6]6-DeoxoTE | [2H6]TE | 50 | N.D. |
| [2H6]6-DeoxoCT | [2H6]CT | N.D. | N.D. |
| [2H6]CN | [2H6]6-OxoCN | N.D. | N.D. |
The conversion rate was calculated as a percentage of the amount of each metabolite versus the amount of substrate added to the culture. Non-labeled BRs were added to the media before extraction and used as internal standards.
N.D., Not detected.
When a yeast strain expressing AtBR6ox was incubated with [2H6]6-DeoxoCS, both [2H6]CS and [2H6]6-OHCS were identified as metabolites (Tables I and II), whereas BL was not identified in this incubation. From this result, we concluded that AtBR6ox encodes a steroid-6-oxidase. Other 6-deoxo compounds were also fed to the yeast to know whether these can be substrates for the AtBR6ox enzyme. It was found that [2H6]6-DeoxoTY, [2H6]6-Deoxo3DT, and [2H6]6-DeoxoTE were converted to [2H6]TY, [2H6]3DT, and [2H6]TE, respectively. However, [2H6]6-DeoxoCT and [2H6]CN were not converted to [2H6]cathasterone (CT) and [2H6]6-OxoCN, respectively (Tables I and II). No conversion was detected in a yeast strain that was transformed only with the vector, pYeDP60.
A yeast strain expressing the tomato Dwarf gene was also examined for its substrate specificity using the same 6-deoxo BRs, revealing that the transformant had the same substrate specificity as the AtBR6ox-expressing yeast (Table III).
DISCUSSION
We isolated the AtBR6ox gene, an Arabidopsis ortholog of the tomato Dwarf gene. The primary structure of the isolated AtBR6ox gene suggests that it belongs to the P450 superfamily. Phylogenic sequence comparison in Figure 3 revealed that it is most similar to the tomato Dwarf gene, among known P450s. AtBR6ox and its homologs form a family with CPD and DWF4, BR-related P450 genes, in a phylogenic tree. The only exception is the maize (Zea mays) Dwarf3 gene that was reported to be involved in gibberellin biosynthesis (Winkler and Helentjaris 1995). The present work demonstrated that AtBR6ox and tomato Dwarf are involved in C-6 oxidation, and hence the corresponding proteins were designated BR-6-oxidases. Sequence comparison in Figure 2A revealed the existence of consensus amino acid sequences for BR-6-oxidases. On the other hand, sequence comparison in Figure 2B revealed consensus sequences in Arabidopsis P450 mono-oxygenases in CYP85 and CYP90 families suggesting specified protein structures. Most members of these families are involved in BR biosynthesis.
This study revealed that both Arabidopsis AtBR6ox and tomato Dwarf enzymes catalyze identical multiple reactions in which the C-6 position of 6-DeoxoCS, 6-DeoxoTY, 6-Deoxo3DT, and 6-DeoxoTE are oxidized. In a number of plants including Arabidopsis, both of these substrates and products seem to occur naturally, suggesting that these reactions may be operative in planta. In Arabidopsis seedlings, TY has been recovered as a metabolite of 6-DeoxoTY (Noguchi et al., 2000), supporting the presence of the pathway from 6-DeoxoTY to TY in planta. In contrast, neither 3DT nor TE was recovered from 6-Deoxo3DT and 6-DeoxoTE (Noguchi et al., 2000). A number of possibilities can account for this discrepancy between in yeast and in planta metabolism so far. For example, in planta metabolic flux from 6-DeoxoTE to TE and 6-Deoxo3DT to3DT cannot be detected because the 6-oxo BRs are turned over very rapidly to regulate their endogenous levels at low levels. Another possibility is that in planta metabolic flows from 6-DeoxoTE to TE and 6-Deoxo3DT to3DT are minor or even absent; flows from 6-DeoxoTY to TY and flows from 6-DeoxoCS to CS are major.
Although tomato Dwarf oxidizes the same substrates as AtBR6ox, no 6-oxo BRs such as 6-OHCN, 6-oxoCN, CT, TE, 3DT, and TY have been identified as endogenous BRs (Yokota et al., 1997, Bishop et al., 1999, Koka et al., 2000). Therefore, conversion of 6-DeoxoCS to CS seems to be the only major C-6 oxidation pathway in tomato tissues. Further biochemical and metabolic studies of BR-6-oxidases will be needed to clarify the C-6 oxidation mechanism and its difference between Arabidopsis and tomato.
Yeasts transformed with AtBR6ox and Dwarf could not catalyze C-6 oxidation of [2H6]6-DeoxoCT and [2H6]CN (Tables I and II). The possibility is not likely that these substrates cannot be incorporated into yeast cells because [2H6]6-DeoxoCT and [2H6]CT were metabolized to [2H6]6-DeoxoTE and [2H6]6-OxoCN when fed to C. roseus cells and Arabidopsis seedlings (Noguchi et al., 2000; Suzuki et al., 1995). We recently found the natural occurrence of 6-OxoCN in Arabidopsis (S. Fujioka, unpublished data), which may suggest that an unknown BR-6-oxidase catalyzing CN to 6-OxoCN exists in Arabidopsis. However, genomic DNA gel-blot analysis did not support the existence of such an Arabidopsis gene that can hybridize with the AtBR6ox gene (Fig. 4). Further efforts to isolate a putative unknown BR-6-oxidase are in progress. In an RNA gel-blot analysis using the N-terminal region of the AtBR6ox gene as a probe, we observed no signal (data not shown). When using full-length AtBR6ox cDNA, a single band was observed, but it was significantly smaller than the expected mature mRNA (data not shown) and also than the ORF sizes of the five cDNA clones derived from RT-PCR. We concluded that the in vivo level of AtBR6ox transcription is extremely low and further approaches, such as RT-PCR, are needed to study the expression and the regulatory function of the AtBR6ox gene.
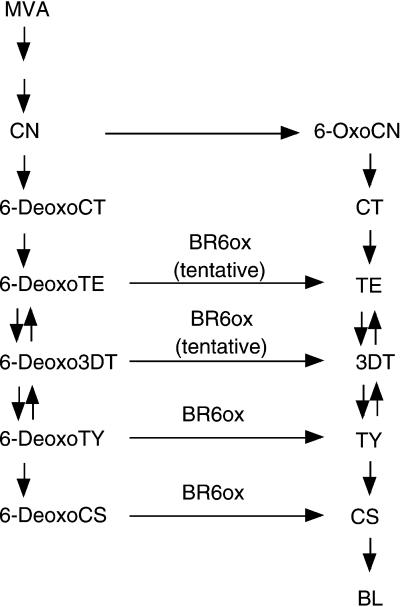
Altogether, pathways from 6-DeoxoTY to TY, from 6-Deoxo3DT to 3DT, and from 6-DeoxoTE to TE are postulated to be present in Arabidopsis and possibly other plants, although the latter two remain tentative (Fig. 5). However, occurrence of these pathways in tomato seems less likely. It is also indicated that these C-6 oxidation steps as well as the conversion of 6-DeoxoCS to CS are catalyzed by a single BR-6-oxidase. The BR biosynthetic pathway seems to consist of a metabolic grid similar to that in GA biosynthesis, rather than of two independent parallel pathways, the early and late C-6-oxidation pathways.
Figure 5.
BR biosynthesis pathway. BR-6-oxidase (BR6ox) converts 6-DeoxoTE to TE, 6-Deoxo3DT to 3DT, 6-DeoxoTY to TY, and 6-DeoxoCS to CS.
MATERIALS AND METHODS
Plant Materials
Arabidopsis ecotype Colombia (Col-0) was used as wild type in this study. Seedlings were grown aseptically on one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1.5% (w/v) Suc and 0.8% (w/v) agar in a growth cabinet (at 22°C, continuous illumination of 100 μmol mm−2 s−1).
Isolation of cDNA Clones
Arabidopsis EST clones were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Total RNAs were extracted from 10-d-old Arabidopsis seedlings grown on one-half-strength Murashige and Skoog agar medium in light by the guanidine-hydrochloride method (Kawakami and Watanabe 1988). cDNA was synthesized with a Ready To Go T-Primed First Strand Kit (Amersham Pharmacia Biotech Inc., Buckinghamshire, UK) according to the manufacturer's instructions. The full-length cDNA fragment of AtBR6ox was amplified by PCR with the cDNA, LA Taq Polymerase (Takara Shuzo, Kyoto), and primers complementary to regions of the putative start and stop codons: CSS-F2 (5′ CAG AGC AGA AAA CAG AGT GAG ATG G 3′) and CSS-R1 (5′ TAC GTC TTC TGT ATC CTC TGC GTG C 3′). The fragments were cloned into the pCR2 vector using a TOPO TA Cloning Kit (Invitrogen, Groningen, The Netherlands) according to the manufacturer's instructions. They were designated pCRCSS12, 19, 93, 94, 98, and 99. The nucleotide sequences of these clones were determined and it was found that the first intron was properly spliced out, except for clone pCRCSS99.
DNA Sequence Analyses
DNA sequences were determined using an automated DNA sequencer (model 373A and model 310A DNA Sequencing System, PE Biosystems, Foster City, CA) according to the manufacturer's instructions. The nucleotide sequence was compiled and analyzed with GENETYX-Mac (Software Development Co., Ltd., Tokyo). The BLAST (Altschul et al., 1990) program was used to search for entries of homologous sequences in the databases at DDBJ. The ClustalW program on the server at DDBJ was used to align the amino acid sequences and to draw phylogenic relationships using the Neighbor-Joining method (Saitou and Nei, 1987). The aligned sequences were shaded using the Boxshade program, available on the server at the European Molecular Biology Network.
Southern Analysis
Genomic DNA was isolated from Arabidopsis using Nucleon PhytoPure Plant and Fungal DNA Extraction Kits (Amersham International PLC) according to the manufacturer's instructions. The DNA (2 μg) was digested, fractionated in 0.8% (w/v) agarose-Tris-EDTA gel, and transferred to positively charged nylon membranes (Roche, Mannheim, Germany) according to Sambrook et al. (1989). For probe preparation, the cDNA clone F5F3 was digested with BamHI. The full-length antisense RNA was labeled with digoxigenin using a DIG RNA Labeling Kit (Roche) according to the manufacturer's instructions. The probe was hybridized to the blot at 50°C overnight and washed four times with 1 × SSPE (Sambrook et al., 1989), 0.1% (w/v) SDS for low-stringency washes, or washed two times with 1 × SSPE, 0.1% (w/v) SDS and then an additional two times with 0.1 × SSPE, 0.1% (w/v) SDS at 50°C for high-stringency washes. Washed blots were reacted with Anti-Digoxigenin-AP and CPD-Star (Roche) according to the manufacturer's instructions, and then detected with a luminescent image analyzer (LAS 1000, Fujifilm, Tokyo).
Yeast Expression Vector
An EST clone of the AtBR6ox gene, F5F3, contained the first intron, which was not spliced out properly. Each of the full-length cDNA clones isolated by RT-PCR had base substitutions, derived from PCR artifacts, in different positions. Therefore, the full-length cDNA clone corresponding to the complete sequence was made by fusing the 5′-coding region of clone pCRCSS12 and the 3′ region of the EST clone, F5F3, as follows. F5F3 was completely digested with BamHI then digested partially with ClaI. The band consisting of the 3′ region of the cDNA and the vector, pBluescriptII SK(-), was purified from a gel. The 5′ region of pCRCSS12 was amplified by PCR with a cDNA clone, pCRCSS12, and primers: CSS-F-bam primer (5′ GGG GAT CCA TGG GAG CAA TGA TGG TG 3′) and CSS-R2-cla primer (5′ AAG CAT CGA TTG TGG GTA ACC AG 3′). The amplified DNA was digested with BamHI and ClaI, and then ligated to the 3′ region of the cDNA derived from F5F3. The clones were sequenced to verify the sequence accuracy. A cDNA clone of the full-length sequence was identified and designated pBCSS4. The full-length cDNA was excised from pBCSS4 with BamHI and KpnI, and then sub-cloned into a yeast expression vector, pYeDP60 (Pompon et al., 1996). The resulting plasmid was designated pYCS41.
Yeast Functional Assay
Yeast expression of the tomato (Lycopersicon esculentum) Dwarf gene was performed as descried previously (Bishop et al., 1999). For the Arabidopsis gene, pYCS41 and pYeDP60 were transformed to yeast strain WAT11 (Pompon et al., 1996) using a Frozen-EZ Yeast Transformation Kit (ZYMO Research, Orange, CA). Transformants were selected on SGI-agar medium (Pompon et al., 1996). Isolated colonies were cultured in 3 mL of SGI medium overnight at 30°C. Yeast cells were collected by centrifugation, and then washed twice with sterile distilled water. They were then inoculated into 30 mL of YPL medium (Pompon et al., 1996) to induce expression of the AtBR6ox gene and an Arabidopsis NADPH-P450-reductase gene (Urban et al., 1997). After 6 h of cultivation, substrate solutions (5 μg/5 μL ethanol) of [2H6]6-DeoxoCS, [2H6]6-DeoxoTY, [2H6]6-Deoxo3DT, [2H6]6-DeoxoTE, [2H6]6-DeoxoCT, and [2H6]CN were added to the culture as substrates for the AtBR6ox protein. After 16 h of incubation, the BRs were extracted and analyzed as described previously (Bishop et al., 1999; Noguchi et al., 1999a). Products from the incubations were analyzed by GC-MS/selected ion monitoring. Confirmation of the identity of the products was provided by their full-scan mass spectra. Quantification was based on calibration curves constructed using [2H6]-labeled and non-labeled BRs. The conversion rate was roughly calculated as a percentage of the amount of each metabolite versus the amount of substrate added to the culture for Arabidopsis. For the tomato experiment, non-labeled BRs were added to the media before extraction and used as internal standards to calculate the conversion rate.
ACKNOWLEDGMENTS
We thank Drs. P. Urban and D. Pompon for providing pYeDP60 and yeast strain WAT11.
Footnotes
This work was supported by the RIKEN Special Postdoctoral Researchers Program and by Grants-in-Aid for Scientific Research (no. 11640661 to Y.S. and no. 10460050 to S.F.) from the Ministry of Education, Science, Culture and Sports of Japan. Y.S. was a Special Postdoctoral Researcher at RIKEN. T.N. was supported by the Japan Society for the Promotion of Science. G.J.B. was supported as an STA (Royal Society) Research Fellow.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones J. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;8:959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y. The tomato DWARF enzyme catalyzes C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Choi Y, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A. An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry. 1997;44:609–613. [Google Scholar]
- Clouse SD, Feldmann KA. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse S, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 163–190. [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H. The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 1999;18:315–320. doi: 10.1046/j.1365-313x.1999.00455.x. [DOI] [PubMed] [Google Scholar]
- Fujioka S. Natural occurrence of brassinosteroids in the plant kingdom. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 21–45. [Google Scholar]
- Fujioka S, Choi YH, Takatsuto S, Yokota T, Li J, Chory J, Sakurai A. Identification of castasterone, 6-deoxocastasterone, typhasterol and 6-deoxotyphasterol from the shoots of Arabidopsis thaliana. Plant Cell Physiol. 1996;37:1201–1203. doi: 10.1093/oxfordjournals.pcp.a029074. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Yokota T, Takatsuto S, Yoshida S. Brassinosteroids in Arabidopsis thaliana. Phytochemistry. 1998;48:595–599. doi: 10.1016/s0031-9422(98)00065-x. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Biosynthesis and metabolism of brassinosteroids. Physiol Plant. 1997a;100:710–715. [Google Scholar]
- Fujioka S, Sakurai A. Brassinosteroids. Nat Prod Rep. 1997b;14:1–10. doi: 10.1039/np9971400001. [DOI] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffen GL, Flippen-Anderson JL, Cook JC. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature. 1979;281:216–217. [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000;14:1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Kawakami N, Watanabe A. Change in gene expression in radish cotyledons during dark-induced senescence. Plant Cell Physiol. 1988;29:33–42. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–90. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000;122:85–98. doi: 10.1104/pp.122.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–38. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–498. [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Tax FE, Feldmann KA. Biosynthetic pahways of brassinolide in Arabidopsis. Plant Physiol. 2000;124:201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999a;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999b;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kitasaka Y, Takatsuto S, Reid JB, Fukami M, Yokota T. Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 1999;119:1517–1526. doi: 10.1104/pp.119.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T. Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 1997;113:31–37. doi: 10.1104/pp.113.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T (2000) Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry (in press) [DOI] [PubMed]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasse J. Physiological actions of brassinosteroids. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 137–161. [Google Scholar]
- Sato S, Kaneko T, Kotani H, Nakamura Y, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5: IV. Sequence features of the regions of 1,456,315 bp covered by nineteen physically assigned P1 and TAC clones. DNA Res. 1998;5:41–54. doi: 10.1093/dnares/5.1.41. [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Chory J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2000;3:79–84. doi: 10.1016/s1369-5266(99)00038-2. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Inoue T, Fujioka S, Saito T, Takatsuto S, Yokota T, Murofushi N, Yanagisawa T, Sakurai A. Conversion of 24-methylcholesterol to 6-oxo-24-methylcholestanol, a putative intermediate of the biosynthesis of brassinosteroids, in cultured cells of Catharanthus roseus. Phytochemistry. 1995;40:1391–1397. [Google Scholar]
- Szekeres M, Nemeth K, Koncz Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Winkler R, Helentjaris T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell. 1995;7:1307–1317. doi: 10.1105/tpc.7.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1605. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Nomura T, Nakayama M. Identification of brassinosteroids that appear to be derived from campesterol and cholesterol in tomato shoots. Plant Cell Physiol. 1997;38:1291–1294. [Google Scholar]