Casler et al. identify that the ER-localized protein Scs2 interacts with a FFAT motif in the C-terminus of the mitochondria–PM tether Num1 to form a tripartite mitochondria–ER–PM contact site. Loss of the Num1–Scs2 interaction severely perturbs mitochondrial division rates. Unexpectedly, they also identify a novel role of mitochondria–ER–PM contact sites in regulating PM PI(4)P metabolism.
Abstract
The mitochondria–ER–cortex anchor (MECA) forms a tripartite membrane contact site between mitochondria, the endoplasmic reticulum (ER), and the plasma membrane (PM). The core component of MECA, Num1, interacts with the PM and mitochondria via two distinct lipid-binding domains; however, the molecular mechanism by which Num1 interacts with the ER is unclear. Here, we demonstrate that Num1 contains a FFAT motif in its C-terminus that interacts with the integral ER membrane protein Scs2. While dispensable for Num1’s functions in mitochondrial tethering and dynein anchoring, the FFAT motif is required for Num1’s role in promoting mitochondrial division. Unexpectedly, we also reveal a novel function of MECA in regulating the distribution of phosphatidylinositol-4-phosphate (PI(4)P). Breaking Num1 association with any of the three membranes it tethers results in an accumulation of PI(4)P on the PM, likely via disrupting Sac1-mediated PI(4)P turnover. This work establishes MECA as an important regulatory hub that spatially organizes mitochondria, ER, and PM to coordinate crucial cellular functions.
Introduction
Membrane contact sites (MCSs) play critical roles in spatially organizing cells and facilitating the transfer of biological materials between organelles. MCSs are defined as sites of close apposition between two membranes that are physically tethered by protein–protein or protein–lipid interactions and perform specific biological functions (Scorrano et al., 2019). Recently, genetic screens using synthetic reporters have identified MCSs between every organelle pair tested (Kakimoto et al., 2018; Shai et al., 2018). Thus, MCSs are a ubiquitous mechanism used by cells to facilitate communication between organelles. Despite their prevalence, few MCSs have been characterized at the molecular level, and, for those that have, their precise functions remain elusive.
To understand MCS biology, the exquisite molecular details that mediate the formation and function of MCSs must be elucidated. The simple organelle architecture of budding yeast has proven to be an excellent model for studying MCS form and function. A breakthrough study used a synthetic tethering system in yeast to identify the ER–mitochondria encounter structure (ERMES) that tethers mitochondria to the ER (Kornmann et al., 2009). Subsequent studies found that ER–mitochondria contact sites are not only involved in non-vesicular lipid transport between the organelles but also mark sites of mitochondrial division (Friedman et al., 2011; Kawano et al., 2018; Murley et al., 2013). In addition to contacts with mitochondria, the ER makes extensive contact with the plasma membrane (PM) in yeast, and the proteins that mediate this contact have been thoroughly studied. Scs2 and Scs22, homologs of the mammalian VAP proteins, are integral membrane proteins that function as two of seven known ER–PM tethers in yeast (Loewen et al., 2007; Manford et al., 2012; Quon et al., 2018; Stefan et al., 2011). A major function of Scs2 and Scs22 is to recruit proteins to the ER. For Scs2, its major sperm protein (MSP) domain binds to two phenylalanines in an acidic tract (FFAT) motifs with high affinity, providing a mechanism for protein recruitment (Loewen and Levine, 2005; Murphy and Levine, 2016). The recruitment of FFAT motif-containing proteins to Scs2-mediated ER–PM contact sites plays a critical role in the regulation of phosphatidylinositol-4-phosphate (PI(4)P) synthesis and transport (Stefan et al., 2011). PI(4)P is enriched on the PM and Golgi membranes and has been implicated in maintaining PM identity and the recruitment of polybasic proteins (Hammond et al., 2012). Stt4, the yeast homolog of phosphatidylinositol-4-kinase type IIIα (PI4KIIIα), localizes to stable assemblies on the PM, termed phosphoinositide kinase (PIK) patches, where it synthesizes PI(4)P from the precursor PI. Stt4 is recruited to PIK patches by Efr3, a FFAT motif-containing PIK patch component that requires Scs2 for proper localization to sites of ER–PM contact (Baird et al., 2008; Omnus et al., 2020). Therefore, both ER–mitochondria and ER–PM contact sites have emerged as critical regulators of lipid trafficking between organelles (Lackner, 2019; Zaman et al., 2020).
While most described MCSs are between two organelles, tripartite MCSs have also been identified. In yeast, the mitochondria–ER–cortex anchor (MECA) tethers the PM, ER, and mitochondria (Fig. 1 A). The core component of MECA, Num1, is a 313-kD protein that consists of an N-terminal coiled-coil (CC) domain, a calcium-binding EF hand-like motif (EFLM), 12 64-AA repeats, a C-terminal PH domain, and several regions of unknown function (Anderson et al., 2022; Farkasovsky and Küntzel, 1995; Kormanec et al., 1991; Lackner et al., 2013; Tang et al., 2009; Yu et al., 2004; Fig. 1 B). With the aid of Mdm36, Num1 self-associates to form cortical foci that tether the mitochondrial network to the PM (Hammermeister et al., 2010; Ping et al., 2016; Tang et al., 2012). Mitochondrial tethering is accomplished through two distinct lipid-binding domains in Num1—an N-terminal CC domain, which interacts with cardiolipin on the mitochondrial outer membrane, and a C-terminal PH domain, which binds phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) on the PM (Ping et al., 2016; Yu et al., 2004). Loss of either lipid binding domain results in a collapse of the mitochondrial network into the center of the cell. Num1 also plays a role in controlling the rate of mitochondrial division. Loss of Num1 substantially reduces the number of mitochondrial division events but does not completely prevent mitochondrial division from occurring (Cerveny et al., 2007; Harper et al., 2023; Klecker et al., 2013; Lackner et al., 2013). In a recent study using synthetic tethering systems to recapitulate mitochondria–PM tethering in the absence of Num1, we discovered that, unexpectedly, the role of Num1 in regulating mitochondrial division is not directly related to its function in controlling the cortical distribution of mitochondria (Harper et al., 2023). Whether the Num1–ER interaction plays a role in regulating mitochondrial division is unknown.
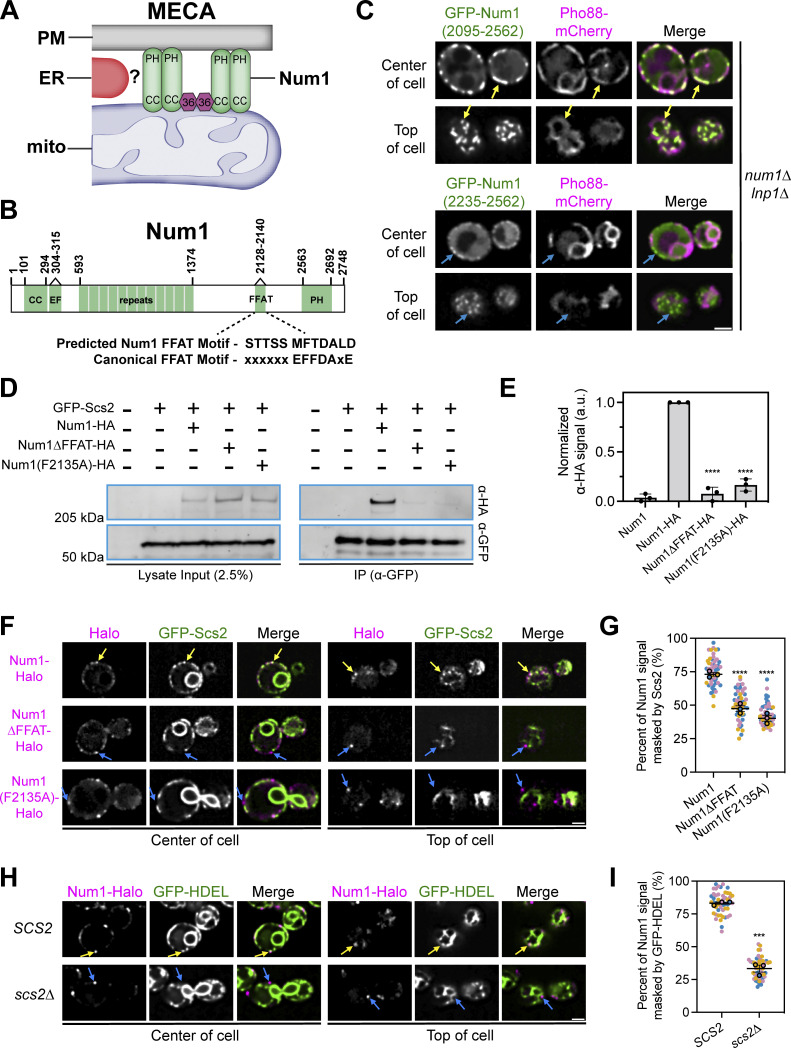
Figure 1.
Num1 interacts with the integral ER protein Scs2 via a C-terminal FFAT motif. (A) Cartoon representation of the mitochondria–ER–cortex anchor (MECA). Num1 (green) interacts directly with mitochondrial membranes and the PM via its coiled-coil (CC) and pleckstrin homology (PH) domains, respectively. Mdm36 (purple hexagon) enhances Num1 self-association. Num1 interacts with the ER (red) via an unknown mechanism. (B) Cartoon domain schematic of Num1 comparing the predicted FFAT motif to the canonical FFAT motif. EF, EF hand-like motif. FFAT, two phenylalanines in an acidic tract. (C) Truncation analysis to identify the minimal region required to localize Num1 to the ER. The indicated Num1 truncations were expressed from the strong GPD promoter in a num1∆ lnp1∆ background. The ER was marked with Pho88-mCherry. Yellow arrows point to cortical, punctate accumulations of Num1(2095-2562) that colocalize with the ER. Blue arrows point to cortical, punctate accumulations of Num1(2235–2562) that do not colocalize with the ER. Images are single slices from the center and top of the same cell. Scale bar, 2 µm. (D) Coimmunoprecipitation of Num1, but not Num1∆FFAT or Num1(F2135A), with Scs2. Cells expressing GFP-Scs2 with the indicated Num1 alleles were lysed and subjected to affinity purification with an α-GFP antibody followed by SDS-PAGE and Western blot analysis. A representative blot of three independent experiments is shown. (E) Quantification of the coimmunoprecipitation experiments in D. For each lane, the α-HA signal at the expected Num1 MW (313 kD) was quantified and normalized to the intensity of the band from the Num1-HA lysate. Each dot represents one experimental replicate and the error bars represent SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001). All statistical analyses are in comparison to the Num1-HA condition. (F) Fluorescence micrographs of cells expressing genomically tagged Num1-Halo, Num1(F2135A)-Halo or Num1∆FFAT-Halo, and GFP-Scs2. Images are single slices from the center or top of the same cell. Individual channels are shown in grayscale. Yellow arrows indicate Num1 foci that contain Scs2 while blue arrows indicate foci that do not contain Scs2. Scale bar, 2 µm. (G) Quantification of the amount of Num1 signal that is masked by Scs2. See Materials and methods for a detailed description of the quantification methodology. Each dot represents one cell. Imaging replicates are depicted as different colors and the average of each replicate is shown as a circle of the appropriate color with a black outline. At least 50 cells per condition were measured. The horizontal line indicates the mean of the three imaging replicates with error bars representing SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used. All statistics are in comparison to Num1 (**** = P < 0.0001). (H and I) The same as F and G except the cells expressed Num1-Halo and the ER marker GFP-HDEL in a wild type or scs2∆ background. Scale bar, 2 µm. To determine statistical significance, an unpaired t test was used (*** = P < 0.001). Source data are available for this figure: SourceData F1.
In addition to controlling mitochondrial distribution and regulating mitochondrial division, the multifunctional Num1 protein also helps position the mitotic spindle during nuclear inheritance by serving as a cortical anchor for dynein (Farkasovsky and Küntzel, 2001; Tang et al., 2012). Interestingly, despite orchestrating distinct cellular functions, the mitochondrial tethering and dynein anchoring functions of Num1 are interconnected. The disruption of dynein anchoring at cortical Num1 clusters results in the impairment of mitochondrial function via an unknown mechanism (White et al., 2022). Additionally, dynein is preferentially anchored at mitochondria-associated Num1 clusters, and this preferential anchoring may contribute to the fidelity of organelle inheritance by biasing mitochondrial inheritance prior to nuclear inheritance (Anderson et al., 2022; Schmit et al., 2018). Precisely how the diverse functions of Num1 are coordinated, however, is unknown.
While the mechanism by which Num1 contacts mitochondria and the PM is known, the molecular basis for the Num1–ER interaction has not been clearly defined. Previous work has implicated Scs2 as a likely Num1 binding partner. IP-MS experiments indicate an association between Num1 and Scs2, and the loss of Scs2 alters the distribution of Num1 foci (Chao et al., 2014; Lackner et al., 2013; Omer et al., 2018). The mechanism by which Num1 interacts with Scs2, however, is unclear. One study suggests that AAs 306–330 of Num1 interact with Scs2 (Chao et al., 2014). However, this region contains the majority of the Num1 EFLM, and mutants lacking the EFLM still show robust ER localization (Anderson et al., 2022; Lackner et al., 2013). Therefore, additional work is required to clarify how Num1 is recruited to the ER.
In this study, we sought to clarify the mechanism by which Num1 contacts the ER and identify novel functions of this tripartite mitochondria–ER–PM contact site. We found that Num1 contains a FFAT motif in its C-terminus that interacts with Scs2 and is required for Num1 to localize to the cortical ER. The Num1–ER interaction is not required for Num1 to function as a mitochondrial tether and dynein anchor, but it is required for Num1’s role in maintaining mitochondrial division rates. Unexpectedly, we also identified a novel role of MECA in maintaining PI(4)P distribution on the PM. Specifically, we found that the interaction between Num1 and each of the three membranes it tethers is required to maintain the enrichment of PI(4)P on the daughter cell PM. Our work supports a model in which MECA functions as a critical organizational hub that facilitates communication between mitochondria, the ER, and the PM to regulate lipid transport and mitochondrial dynamics.
Results
Num1 interacts with Scs2 via a FFAT motif in its C-terminus
The core component of the MECA, Num1, has been shown to localize to the cortical ER via fluorescence microscopy and biochemical fractionation experiments; however, the precise mechanism by which Num1 localizes to the ER is unknown (Lackner et al., 2013; Omer et al., 2018; Fig. 1 A). First, we sought to identify the minimal region of Num1 required for ER localization. Previous truncation experiments indicated that the minimal Num1-ER binding domain is likely present in the C-terminus of the protein (Lackner et al., 2013). Thus, we expressed Num1 truncations lacking the majority of the N-terminus of the protein as well as the C-terminal PH domain and assessed their ability to localize to cortical ER (Fig. 1 C). Determining localization to the cortical ER through microscopy is complicated by the fact that cortical ER occupies up to 20–45% of the PM in budding yeast, making true localization to the ER difficult to differentiate from chance (West et al., 2011). Thus, to clarify cortical ER localization, we performed the analysis in an LNP1 deletion strain. Loss of Lnp1 disrupts cortical ER morphology and creates large regions of the cortex that lack ER (Chen et al., 2012). GFP-Num1(2095–2562) showed robust colocalization with an ER marker while further truncations did not (Fig. 1 C, yellow arrows). Interestingly, GFP-Num1(2235–2562) displayed weak cortical localization that was not biased toward the ER, indicating that another uncharacterized protein–protein interaction between Num1 and a cortical protein may exist (Fig. 1 C, blue arrows). Based on these results, we conclude that Num1 contains an ER-interacting motif within AAs 2095–2235 of its C-terminus.
Previous work identified Scs2 as a likely ER-localized Num1-interacting protein (Chao et al., 2014; Lackner et al., 2013; Omer et al., 2018). Chao et al. (2014) identified Num1 in IP-MS pulldown experiments of Scs2 and mapped a binding interaction to AAs 306–330, a region that includes the EFLM. In contrast to these results, however, we have previously shown that alleles of Num1 lacking the EFLM are still robustly associated with the ER (Anderson et al., 2022; Lackner et al., 2013). Interestingly, a recent, elegant computational study predicted that Num1 contains a strong Scs2-interacting FFAT motif at AAs 2128–2140, but not 306–330 (Slee and Levine, 2019; Fig. 1 B). The predicted Num1 FFAT motif resides in the region we identified by truncation analysis to contain the minimal Num1 ER-interacting motif (Fig. 1 C). To test the hypothesis that Num1 interacts with Scs2 via a C-terminal FFAT motif, we generated two alleles of Num1 containing either an internal deletion of AAs 2128–2140 (hereafter referred to as Num1∆FFAT) or a point mutant of a predicted key aromatic residue in the FFAT motif (hereafter referred to as Num1(F2135A)) and assessed their ability to interact with Scs2 via coimmunoprecipitation analysis. We found that full-length Num1, but not Num1∆FFAT or Num1(F2135A), coimmunoprecipitated with Scs2 (Fig. 1, D and E).
We next examined the localization of Num1 and Scs2 in vivo using fluorescence microscopy. To do so, we expressed Num1-Halo and GFP-Scs2 from their endogenous loci. Previous studies have established that N-terminal tagged versions of Scs2 are functional (Loewen et al., 2007; Manford et al., 2012). We observed a substantial overlap of the Num1 signal with Scs2, while Num1∆FFAT and Num1(F2135A) clearly decorated regions of the cortex that were devoid of Scs2 (Fig. 1, F and G). The Num1 FFAT motif mutants retain the C-terminal PH domain, so cortical localization is expected even in the absence of an interaction with the cortical ER. Next, to see if loss of Scs2 affected the ER localization of Num1, we analyzed the localization of full-length Num1 compared with a general ER marker, GFP-HDEL, in an scs2∆ strain. Loss of Scs2 resulted in a substantial decrease in the overlap between the Num1 and GFP-HDEL signals (Fig. 1, H and I).
Based on these results, we conclude that Scs2 recruits Num1 to the ER by interacting with a FFAT motif present at AAs 2128–2140. Our results can potentially be reconciled with those of Chao et al. (2014), in which they suggest Num1 contains a FFAT motif at AAs 306–330. In our recent work, we demonstrate that the Num1 EFLM (AAs 304–315) binds calcium and likely plays a role in regulating the function of Num1 as a dynein anchor (Anderson et al., 2022). While loss of the EFLM alone does not appreciably alter Num1 recruitment to the ER, loss of AAs 306–330 may alter Num1 conformation in a manner that masks the C-terminal FFAT motif or otherwise disrupts the Num1–Scs2 interaction. Regardless, based on the results presented here and previously published computational work, we conclude that AAs 2128–2140 of Num1 comprise a bona fide Scs2-interacting FFAT motif (Slee and Levine, 2019).
The Num1–Scs2 interaction is sufficient to tether ER and eisosomes
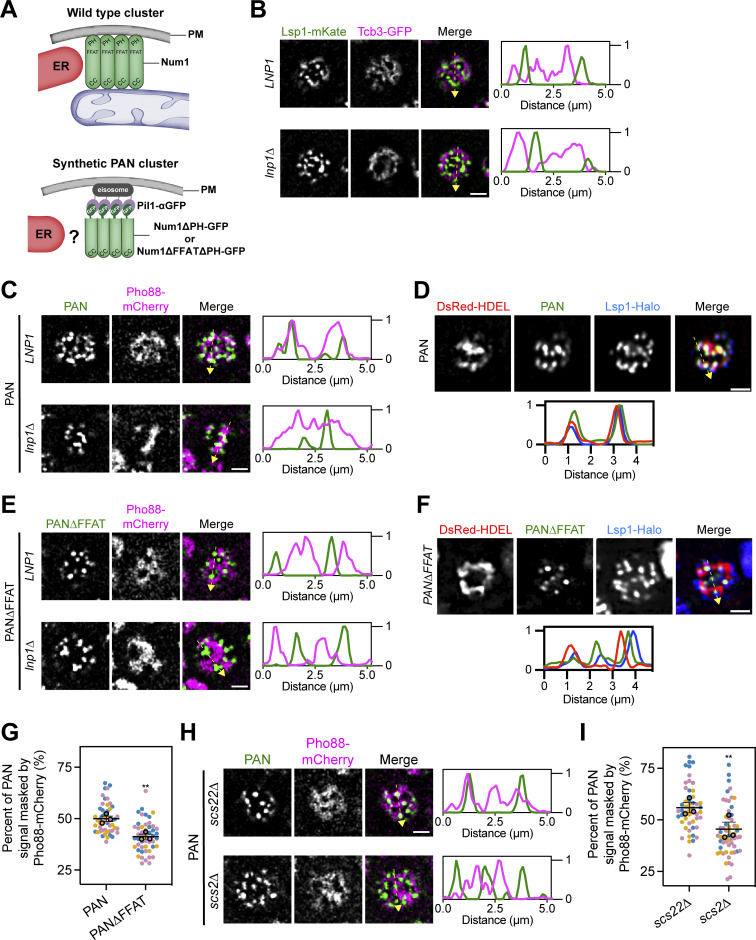
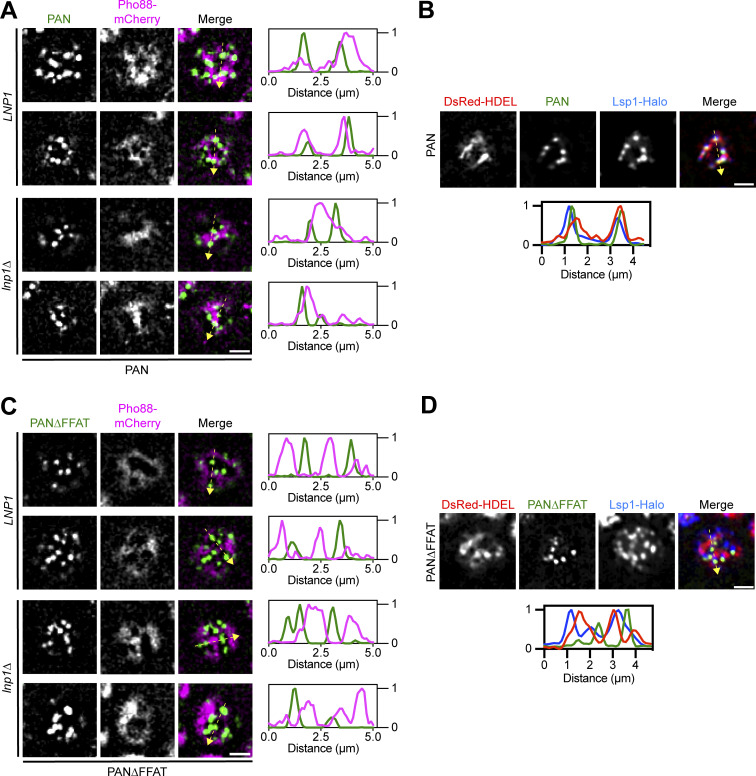
We next wanted to probe the strength of the Num1–Scs2 interaction by testing if it was sufficient to tether typically unassociated structures. Previously, we developed an α-GFP nanobody-based system to ectopically localize Num1 to eisosomes to study differences between mitochondria-associated and mitochondria-free clusters of Num1 (Schmit et al., 2018). Eisosomes are discrete, stable compartments of the PM with poorly understood functions (Walther et al., 2006). In this system, a GFP-tagged version of Num1 lacking the PH domain is recruited to eisosomes via an interaction with an α-GFP nanobody fused to the eisosome component Pil1 (Fig. 2 A). These artificial clusters of Num1 are referred to as PAN, for Pil1-associated Num1, and clusters and strains expressing PAN are referred to as PAN cells. Our previous study focused on how PAN clusters tether mitochondria and anchor dynein but did not examine whether PAN clusters were also ER-associated (Fig. 2 A). Eisosomes are not thought to be ER-associated (Stradalova et al., 2012), which we confirmed using fluorescence microscopy to examine the localization of the cortical ER, marked by Tcb3-GFP, relative to eisosomes, marked by Lsp1-mKate, in wild type and lnp1∆ mutants (Hoffmann et al., 2019; Walther et al., 2006). As expected, eisosomes were found at regions of the cell cortex that lack ER (Fig. 2 B). If the Num1–Scs2 interaction is of sufficient strength, we hypothesized that the PAN system may alter ER–eisosome associations. To test this, we expressed an ER marker, Pho88-mCherry, in PAN cells and examined the relationship between the ER and PAN clusters via fluorescence microscopy. Remarkably, in PAN cells, ER membranes were tightly associated with nearly all PAN clusters (Fig. 2 C and Fig. S1 A). While we have previously demonstrated that all PAN clusters are at eisosomes, we wanted to ensure that our artificial systems capture a true change in eisosome–ER association (Schmit et al., 2018). Thus, we also examined the localization of an eisosome component, Lsp1, in PAN cells. As expected, Lsp1 showed increased ER association in PAN cells (Fig. 2 D and Fig. S1 B). To determine whether this phenomenon was dependent on the Num1–Scs2 interaction, we generated a version of PAN in which Num1 lacked the FFAT motif as well as the PH domain (hereafter referred to as PAN∆FFAT) (Fig. 2 A). In PAN∆FFAT cells, the ER exhibited a near mutually exclusive localization with PAN∆FFAT clusters as well as eisosomes, similar to that seen between the ER and eisosomes in wild type cells (Fig. 2, B, E, and F; and Fig. S1, C and D). Quantification revealed that significantly more PAN signal was overlapped by ER signal when the Num1 FFAT motif was present (Fig. 2 G). Finally, we also examined the association of the ER and eisosomes in strains lacking Scs2 or its paralog Scs22. In PAN cells lacking Scs2, we observed clear separation of the ER and eisosome signals, similar to PAN∆FFAT cells, whereas cells lacking Scs22 looked similar to PAN cells (Fig. 2, H and I). Our results indicate that the Num1–Scs2 interaction is sufficiently strong to tether the ER and eisosomes, which do not normally associate. Because either loss of Num1’s FFAT motif or loss of Scs2 alone is sufficient to break the PAN-ER association, any other potential Num1–ER interactions, including with Scs22, are unlikely to be particularly strong. These experiments also serve as proof of the principle that the association of the ER and eisosomes can be altered, which could be used as a tool to study interactions between these compartments. Taken together, our results reveal that Scs2 is a novel component of MECA that mediates mitochondria–ER tethering.
Figure 2.
The Num1–Scs2 interaction is sufficient to tether ER and eisosomes. (A) Cartoon representation of wild type Num1 clusters and Num1 clusters generated using the PAN system. Wild type Num1 clusters require mitochondria for formation and are ER-associated (Kraft and Lackner, 2017). PAN clusters are formed by targeting GFP-tagged Num1 alleles lacking the PH domain to Pil1 fused to an α-GFP nanobody (Schmit et al., 2018). Whether PAN clusters are associated with ER is unknown. PAN; Pil1-associated-Num1. (B) Fluorescence micrographs of cells expressing an eisosome marker, Lsp1-mKate, and a cortical ER marker, Tcb3-GFP, in both an LNP1 and lnp1∆ background. Individual channels are shown in grayscale. Images are a single slice from the top of a cell. The dashed yellow arrow marks the location analyzed in the accompanying linescans to the right of the micrographs. Scale bar, 2 µm. (C) Fluorescence micrographs of PAN cells expressing the ER marker Pho88-mCherry. Images and linescans are arrayed in the same manner as B. (D) Images of PAN cells expressing the ER marker DsRed-HDEL and an eisosome marker Lsp1-Halo. Images are arrayed in the same manner as B except the linescan is depicted below. (E and F) Identical to C and D except cells expressed Num1∆FFAT∆PH-GFP. (G) Quantification of the percentage of Num1∆PH-GFP or Num1∆FFAT∆PH-GFP signal masked by Pho88-mCherry from the data sets from C and E. The quantification was performed and presented in (Fig. 1 G). To determine statistical significance, an unpaired t test was used (** = P < 0.01). (H) Fluorescence micrographs of PAN cells expressing the ER marker Pho88-mCherry in a scs22∆ or scs2∆ background. Images and linescans are arrayed in the same manner as B. (I) Quantification of the data in H, performed and presented the same as in Fig. 1 G.
Figure S1.
Additional images of the ER and eisosomes in PAN and PAN∆FFAT cells. (A) Additional examples of PAN and the ER marker Pho88-mCherry in LNP1 and lnp1∆ cells (related to Fig. 2, C and D). Images are fluorescence micrographs from the top of the cell. Individual channels are shown in grayscale. Two examples from an LNP1 and lnp1∆ strain are shown. The dashed yellow arrow marks the location analyzed in the accompanying linescans to the right of the micrographs. Scale bar, 2 µm. (B) Fluorescence micrographs of cells expressing Lsp1-Halo and the ER marker DsRed-HDEL in PAN cells. Images are from the top of the cell. Individual channels are shown in grayscale. The dashed yellow arrow marks the location analyzed in the accompanying linescans. Scale bar, 2 µm. (C and D) Identical to A and B except PAN∆FFAT cells were imaged (related to Fig. 4, D and E).
The Num1–Scs2 interaction is not required for Num1 to function as a mitochondrial tether or dynein anchor
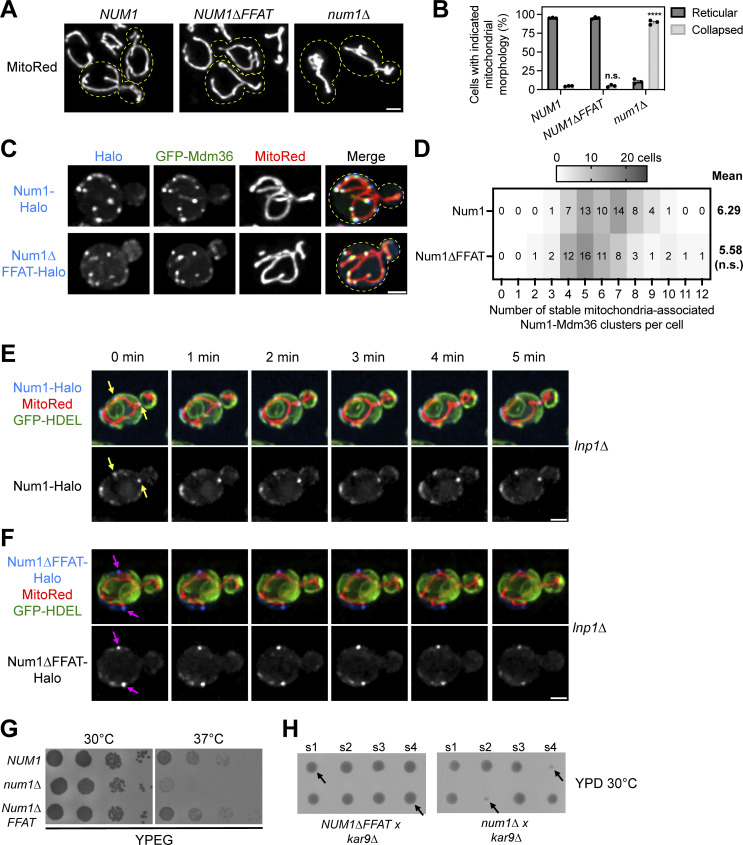
Next, we tested whether the Num1–Scs2 interaction is involved in regulating known Num1 functions. First, we examined whether the Num1–Scs2 interaction is required for Num1 to tether mitochondria. Loss of Num1 results in a collapse of the mitochondrial network into the center of the cell (Cerveny et al., 2007; Lackner et al., 2013; Fig. 3, A and B). Expression of the Num1∆FFAT allele, however, caused no appreciable change in the number of cells displaying collapsed mitochondrial networks compared with full-length Num1 (Fig. 3, A and B). We next expressed genomically tagged versions of Num1-Halo or Num1∆FFAT-Halo with GFP-Mdm36 and a mitochondrial marker, matrix-targeted DsRed (MitoRed), and used 4D confocal microscopy to examine the number of stable mitochondrial tethering points. Both Num1 and Num1∆FFAT formed cortically localized punctate structures that colocalized with Mdm36 and tethered mitochondria over time (Fig. 3 C). Quantification of these data revealed that both Num1 and Num1∆FFAT form a similar number of mitochondrial tethering points per cell (Fig. 3 D). Thus, the Num1–Scs2 interaction is not required for Num1-mediated mitochondria–PM tethering.
Figure 3.
The Num1–Scs2 interaction is not required for Num1 to function as a mitochondrial tether or dynein anchor. (A) Representative micrographs of cells expressing the mitochondrial matrix marker MitoRed in the indicated genetic backgrounds. Yellow dashed lines indicate cell outlines. Scale bar, 2 µm. (B) Quantification of the percentage of cells from A that display either reticular or collapsed mitochondrial networks. Each dot represents one imaging replicate containing 100 cells. The error bars represent the SEM between the three replicates. A detailed description of the quantification methodology is presented in the methods section. To determine the statistical significance between the percentage of collapsed mitochondrial networks seen per condition, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to NUM1 cells. (C) Fluorescence micrographs of cells expressing genomically tagged Num1-Halo or Num1∆FFAT-Halo with GFP-Mdm36 and the mitochondrial matrix marker MitoRed. Images are max projections of a full Z-stack. Individual channels are shown in grayscale. Yellow dashed lines indicate the cell outlines. Scale bar, 2 µm. (D) Quantification of the number of stable mitochondria-associated Num1-Mdm36 clusters per cell from the data shown in C. See Materials and methods for a detailed description of the quantification methodology. The mean number of tethering points is indicated to the right of the heat map. 58 cells were counted per condition. Statistical significance was determined by an unpaired t test (n.s. = not significant). (E) Images from Video 1 of cells expressing genomically tagged Num1-Halo, MitoRed, and GFP-HDEL in an lnp1∆ background. Images are max projections of a full Z-stack. The Num1-Halo channel is shown in grayscale. The yellow arrows indicate ER-associated, Num1-mediated mitochondrial tethering points that persist throughout the time course. Scale bar, 2 μm. (F) Images from Video 2 which is identical to E except the cells expressed Num1∆FFAT-Halo. The magenta arrows indicate Num1∆FFAT-mediated mitochondrial tethering points that are not ER-associated and persist throughout the time course. (G) 10-fold serial dilutions of the indicated strains were spotted on YPEG medium and grown at 30°C or 37°C for 2 days. The image is a representative example of three biological replicates. (H) Diploid NUM1∆FFAT kar9∆ and num1∆ kar9∆ cells were sporulated, and the spores from individual tetrads were arranged in a column on YPD medium. The s1-4 label denotes spores 1-4 from an individual tetrad. Two tetrads are shown per background and black arrows point to double mutants. The images are representative of at least 12 tetrad dissections.
We next probed whether the mitochondrial tethering points formed by Num1∆FFAT were still ER-associated. To do so, we expressed an ER marker, GFP-HDEL, and a mitochondrial matrix marker, MitoRed, in cells expressing either full-length Num1-Halo or Num1∆FFAT-Halo and examined individual mitochondrial tethering points by 4D confocal microscopy. These experiments were performed in an lnp1∆ background to more easily assess ER localization via microscopy. Our results revealed that, while essentially all Num1 foci were ER associated (Video 1 and Fig. 3 E, yellow arrows), many Num1∆FFAT foci were clearly spatially separated from ER membranes while still actively tethering mitochondria (Video 2 and Fig. 3 F, magenta arrows). In 98% of cells expressing full-length Num1, all Num1 foci were associated with both the ER and mitochondria (n = 100 cells from three movies). In contrast, in cells expressing Num1∆FFAT, the percentage of cells in which all Num1 foci were associated with both the ER and mitochondria decreased to 21%; the other 79% of cells contained at least one Num1∆FFAT foci that was clearly spatially separated from the ER (n = 100 cells from three movies). Thus, ER association is not required for Num1 to function as a mitochondria–PM tether.
Video 1.
Num1 tethers mitochondrial and ER membranes over time. 4D confocal microscopy video of cells expressing Num1-Halo, GFP-HDEL, and MitoRed in an lnp1∆ background. The video is a max projection of a full Z-stack. The individual Num1-Halo channel is shown in grayscale beneath the merged video. Time is indicated in min:s format. Still images from this video are depicted in Fig. 3 E. Scale bar, 2 µm. The frame rate is 10 fps.
Video 2.
Num1∆FFAT tethers mitochondrial but not ER membranes over time. 4D confocal microscopy video of cells expressing Num1∆FFAT-Halo, GFP-HDEL, and MitoRed in an lnp1∆ background. The video is a max of a full Z-stack. The individual Num1∆FFAT-Halo channel is shown in grayscale beneath the merged video. Time is indicated in min:s format. Still images from this video are depicted in Fig. 3 F. Scale bar, 2 µm. The frame rate is 10 fps. Num1∆FFAT-Halo is annotated as “Num1dFFAT-Halo” in the video.
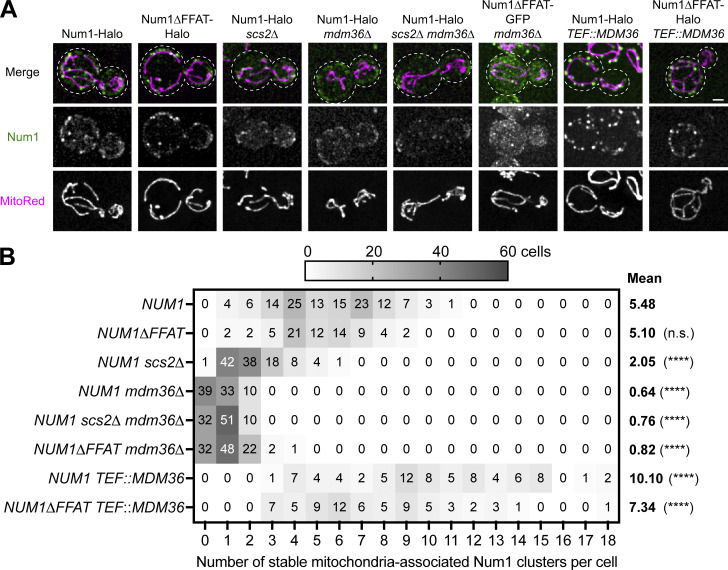
We next sought to better understand how the known components of MECA, Mdm36 and Scs2, regulate the formation of Num1 clusters. Previous reports indicated that the loss of Scs2 reduces the number and alters the localization of Num1 foci (Chao et al., 2014; Omer et al., 2018). A complicating factor in interpreting those experiments, however, is that Scs2 regulates the recruitment of many proteins to the ER and its loss results in pleiotropic effects that alter membrane properties (Murphy and Levine, 2016). To circumvent this, we used the Num1∆FFAT allele, which cleanly separates the Num1–Scs2 interaction from the pleiotropic effects of an Scs2 deletion. We expressed either full-length Num1 or Num1∆FFAT in cells lacking Mdm36 or Scs2 alone or in combination and examined the number of stable Num1-mediated mitochondrial tethering points. While deleting Scs2 resulted in a decrease in the number of Num1 foci, as previously reported (Omer et al., 2018) (∼5.48 compared to ∼2.05 foci per cell in wild type and scs2∆ cells, respectively), the number of Num1 foci in cells expressing Num1∆FFAT (∼5.10 foci per cell) was comparable with wild type (Fig. S2, A and B). Thus, the reduction in the number of Num1 foci and the change in localization is likely due to pleiotropic effects from the loss of Scs2, rather than the loss of the Num1–Scs2 interaction. On the other hand, either loss or overexpression of Mdm36 drastically decreased or increased the number of Num1 foci, respectively, as previously reported (Lackner et al., 2013; Omer et al., 2020; Fig. S2, A and B). Our previous work demonstrates that Mdm36 directly interacts with Num1 to promote cluster formation, potentially through aiding Num1 self-association, and that Num1 clusters are required to stably anchor mitochondria (Lackner et al., 2013; Ping et al., 2016). Taken together, these results indicate that the amount of Mdm36 is the primary determinant regulating the number of Num1 foci and suggest that cells could utilize the expression of Mdm36 as a means to regulate the extent of Num1-mediated mitochondria–ER–PM tethering.
Figure S2.
Mdm36 is the primary determinant of the number and size of Num1 clusters. (A) Fluorescence micrographs of cells expressing the indicated Num1 fusion proteins with the mitochondrial matrix marker MitoRed in the indicated genetic backgrounds. Images are max projections of a full Z-stack. Individual channels are shown in grayscale. Dashed white lines represent cell outlines. The Num1 channels’ brightness/contrast settings were individually set to maximize the visualization of diffuse signals in the Mdm36 deletion strains, and the intensity between images should not be directly compared. Scale bar, 2 µm. (B) A heat map depicting the number of stable Num1-mediated mitochondrial tethering points per cell from the data set shown in A. A stable tethering point is defined as an accumulation of Num1 above background that persists for at least 1.5 min. See Materials and methods for a further description of the quantification methodology. The mean number of tethering points is indicated to the right of the heat map. Between 70 and 120 cells were counted per condition. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001, n.s. = not significant). All statistical analyses are in comparison to NUM1.
In addition to its role in positioning mitochondria, Num1 also functions as an anchor for dynein during nuclear inheritance (Kormanec et al., 1991; Tang et al., 2012). Our previous work established that, unexpectedly, loss of Num1-mediated dynein anchoring results in a respiratory growth defect (White et al., 2022). To see if loss of the Num1–Scs2 interaction impaired the ability of Num1 to function as a dynein anchor, we performed growth assays on media that forced cells to respire. As previously reported, the loss of Num1 resulted in a severe growth defect on respiratory media at elevated temperatures (Fig. 3 G). By contrast, cells expressing Num1∆FFAT grew at a rate indistinguishable from wild type, indicating that dynein was properly anchored (Fig. 3 G). To further test if the Num1–Scs2 interaction is required for dynein anchoring, we performed tetrad analysis by crossing strains expressing Num1∆FFAT or lacking Num1 to a strain harboring a KAR9 deletion. Budding yeast contains two partially redundant nuclear positioning pathways, mediated by Num1/dynein and Kar9, and the loss of both results in a significant growth defect (Miller and Rose, 1998). Tetrad analysis demonstrated that, unlike num1∆ kar9∆ spores, Num1∆FFAT kar9∆ spores grow at rates similar to wild type, indicating that the dynein pathway of nuclear positioning is functional in the absence of the Num1–Scs2 interaction (Fig. 3 H).
Loss of the Num1–Scs2 interaction reduces the rate of mitochondrial division
Num1 was previously proposed to be required for mitochondrial division (Cerveny et al., 2007). We recently demonstrated, however, that Num1 enhances, but is not required for, mitochondrial division, and loss of Num1 severely reduces the rate of mitochondrial division (Harper et al., 2023; Lackner et al., 2013). The function of Num1 in mitochondrial division extends beyond cortically tethering the mitochondrial network, as artificial mitochondria–PM tethers cannot rescue mitochondrial division defects in the absence of Num1 (Harper et al., 2023). Mitochondria–ER MCSs are known to influence mitochondrial division (Friedman et al., 2011). Thus, we sought to test whether loss of the Num1–ER interaction impacts mitochondrial division.
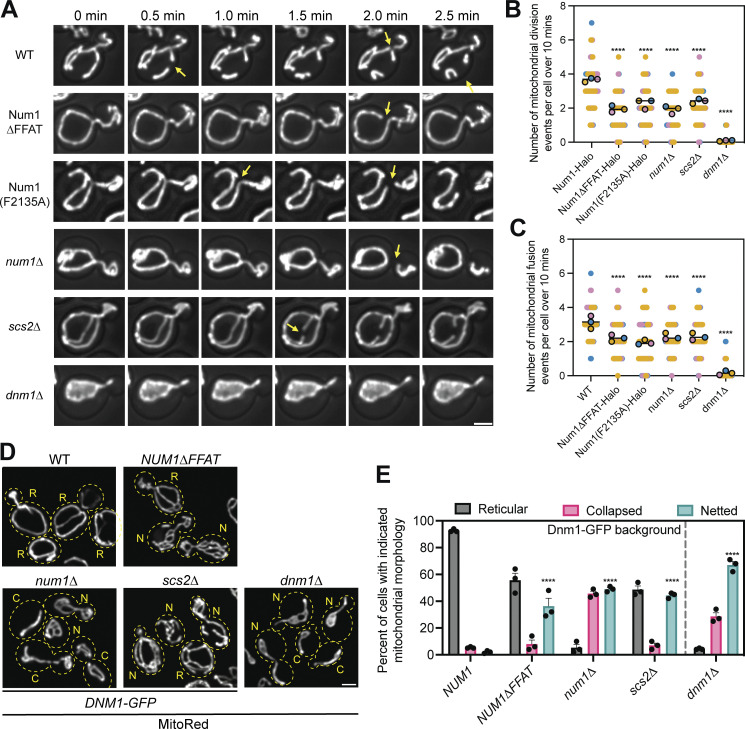
Our previous analysis demonstrated that loss of the Num1–Scs2 interaction did not grossly perturb mitochondrial morphology (Fig. 3, A and B). Mutations that severely compromise mitochondrial fission or fusion lead to hyperfused/netted or fragmented mitochondrial networks, respectively (Bleazard et al., 1999; Sesaki and Jensen, 1999). Mitochondrial morphology is preserved, however, if both mitochondrial fusion and fission processes are equally inhibited (Sesaki and Jensen, 1999). Therefore, mitochondrial morphology is not always a reliable readout for subtle defects in mitochondrial fusion or fission dynamics. To determine if Num1-mediated mitochondria–ER tethering plays a role in regulating mitochondrial division, we used 4D confocal microscopy to measure the rate of mitochondrial division. Remarkably, despite maintaining a cortically distributed mitochondrial network, cells expressing Num1∆FFAT or Num1(F2135A) showed a decrease in the rate of mitochondrial division similar to that observed in cells lacking Num1 (Fig. 4, A and B; and Video 3). Importantly, the observed phenotype was not due to the presence of the epitope tag on Num1 or Num1∆FFAT, as multiple different tagged versions of these proteins showed a similar mitochondrial division rate (Fig. S3 A). The defect in the mitochondrial division was not as severe as the complete loss of the mitochondrial division machinery, as deletion of Dnm1, a GTPase required for mitochondrial division, reduced division rates to nearly zero (Bleazard et al., 1999; Fig. 4, A and B). The mitochondrial division rate in cells lacking Scs2 was also similar to that observed in cells lacking Num1 or expressing Num1∆FFAT (Fig. 4, A and B). Interestingly, similar defects in mitochondrial division rates have been reported on the loss of VAP proteins in mammalian cells (Boutry and Kim, 2021). In line with previous reports demonstrating that mitochondrial fusion and fission rates are balanced, a comparable decrease in mitochondrial fusion rates was also observed for all genotypes with reduced mitochondrial fission rates (Nunnari et al., 1997; Sesaki and Jensen, 1999; Fig. 4 C).
Figure 4.
Loss of the Num1–Scs2 interaction reduces the rate of mitochondrial division. (A) Selected micrographs from Video 3 of cells expressing the mitochondrial matrix marker MitoRed in the indicated genetic backgrounds. Z-stacks were collected every 30 s for 10 min. Images are max projections of the full Z-stack. Yellow arrows indicate mitochondrial division events. The fluorescence signal is shown in grayscale merged with a BF image. Scale bar, 2 µm. Images are taken from various points throughout the supplemental video to highlight mitochondrial division events. The time above the images references the time elapsed between images, not the specific time from Video 3. (B) Quantification of the number of mitochondrial division events observed per cell over 10 min. Mitochondrial division events were manually scored by examining full Z-stacks to determine when two mitochondrial tubules separated. Imaging replicates are depicted as different colors and the average of each replicate is shown as a circle of the appropriate color with a black outline. Each replicate contains twenty cells for a total of 60 cells measured per condition. The horizontal line indicates the mean of the three imaging replicates with error bars representing SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001). All statistical analyses are in comparison to Num1-Halo. (C) The same as B except mitochondrial fusion events were scored. (D) Representative micrographs of cells expressing the mitochondrial matrix marker MitoRed and a C-terminally tagged version of Dnm1-GFP from the endogenous DNM1 locus in the indicated genetic background. Dashed yellow lines indicate cell outlines. The accompanying letter indicates the category of mitochondrial morphology that particular cell was scored as: R = reticular, N = netted, C = collapsed. See Materials and methods section for a detailed explanation of how each category was determined. Scale bar, 2 μm. (E) Quantification of the percentage of cells displaying the indicated mitochondrial morphology for each genetic background. Each dot represents one imaging replicate containing 100 cells. The bars represent the average of the three imaging replicates and the error bars represent SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001). All statistical analyses are in comparison to NUM1 and are comparing the netted category.
Video 3.
Loss of the Num1–Scs2 interaction reduces the rate of mitochondrial division. 4D confocal microscopy video of cells expressing MitoRed (shown in grayscale) in the indicated genetic backgrounds. Fluorescence channels are merged with a bright field image. Each cell comes from a unique movie and are tiled to aid visual comparison. The video is a max projection of full Z-stacks. Time is indicated in min:s format. Still images from this video are depicted in Fig. 4 A. Scale bar, 2 µm. The frame rate is 10 fps. In the labels, the lowercase “d” is used to represent the typical ∆ symbol to indicate gene deletions.
Figure S3.
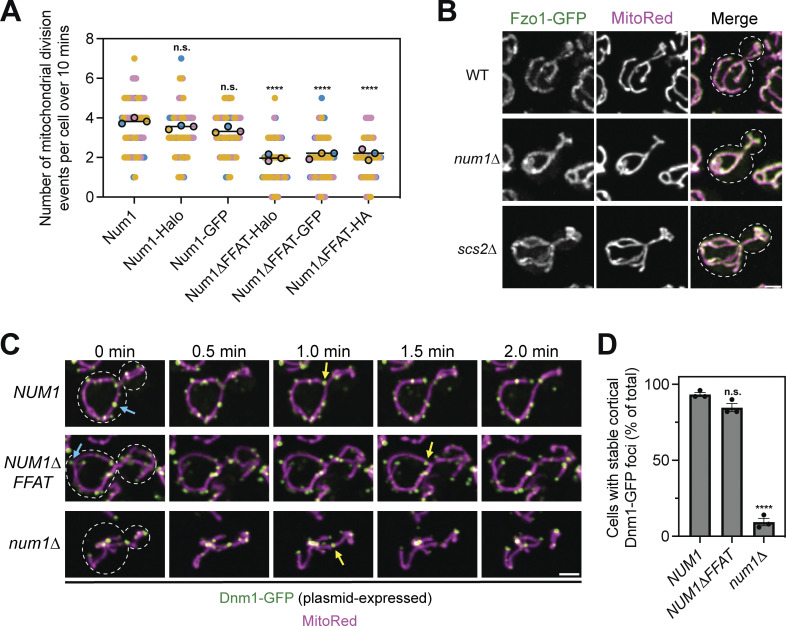
Loss of Num1 does not alter the localization of the mitochondrial division or fusion machinery. (A) The presence of a fluorescent or epitope tag on Num1 does not influence the rate of mitochondrial division. Imaging and quantification of mitochondrial division was performed identically to (Fig. 4 B), except cells were expressing the indicated Num1 or Num1∆FFAT fusion proteins. The data from Num1-Halo and Num1∆FFAT-Halo are reproduced from (Fig. 4 B) to aid visual comparison. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to Num1-Halo. (B) Fluorescent micrographs of cells expressing MitoRed and genomically tagged Fzo1-GFP in wild type, num1∆, and scs2∆ backgrounds. Individual channels are shown in grayscale. Dashed white lines indicate cell outlines. Scale bar, 2 µm. (C) Time-lapse images of cells expressing MitoRed and Dnm1-GFP, expressed from a plasmid, in the indicated genetic backgrounds. The blue arrows point to stable Dnm1-GFP foci that remain cortically localized throughout the time-lapse. The yellow arrows indicate accumulations of Dnm1-GFP on the mitochondria immediately prior to a division event. Images are max projections of full Z-stacks. Scale bar, 2 µm. (D) Quantification of the percent of cells containing at least one cortically localized Dnm1-GFP focus. Cells expressed Dnm1-GFP as an extra copy from a plasmid under control of the native promoter. To be counted, Dnm1-GFP foci needed to remain cortically localized for at least 2 min. Each dot represents one imaging replicate containing 50 cells. Error bars represent SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to NUM1.
To further characterize the mitochondrial division defect, we analyzed mitochondrial morphology in cells expressing a hypomorphic allele of Dnm1, Dnm1-GFP expressed from the endogenous locus. This Dnm1 allele displays reduced mitochondrial division activity and, when coupled with mutations that further compromise mitochondrial division, results in the formation of mitochondrial nets, a phenotype strongly associated with perturbations to mitochondrial division (Harper et al., 2023). In agreement with our analysis of mitochondrial division rates, a significantly higher fraction of cells containing mitochondrial nets was observed in num1∆, NUM1∆FFAT, and scs2∆ cells expressing Dnm1-GFP in comparison with NUM1 cells expressing Dnm1-GFP (Bleazard et al., 1999; Fig. 4, D and E).
Finally, we observed no obvious changes in the behavior of key components of the mitochondrial division or fusion machinery that could explain the observed mitochondrial division defect. Fzo1, a protein involved in outer mitochondrial membrane fusion, displayed typical mitochondrial outer membrane localization in both Num1 and Scs2 mutants (Hermann et al., 1998; Fig. S3 B, 100% of cells, n = 100 cells per replicate over three imaging replicates). Num1 has previously been shown to be required to maintain a stable population of cortical Dnm1 foci that may influence mitochondrial division events (Lackner et al., 2013). Cortical Dnm1 foci were still present in cells expressing Num1∆FFAT, however, making it unlikely that the defect in mitochondrial division is due to the presence or absence of cortical Dnm1 foci (Fig. S3, C and D). Additionally, Dnm1 could readily be detected at sites of mitochondrial division in both num1∆ and NUM1∆FFAT cells (Fig. S3 C).
Taken together, our results demonstrate that, unexpectedly, the defect in mitochondrial division in the absence of Num1 can be explained entirely by the loss of Num1-mediated mitochondria–ER tethering rather than mitochondria–PM tethering. Thus, a specific aspect of the mitochondria–ER tethering mediated by Num1 and Scs2 regulates the rate of mitochondrial division.
Num1 influences the distribution of PI(4)P on the PM
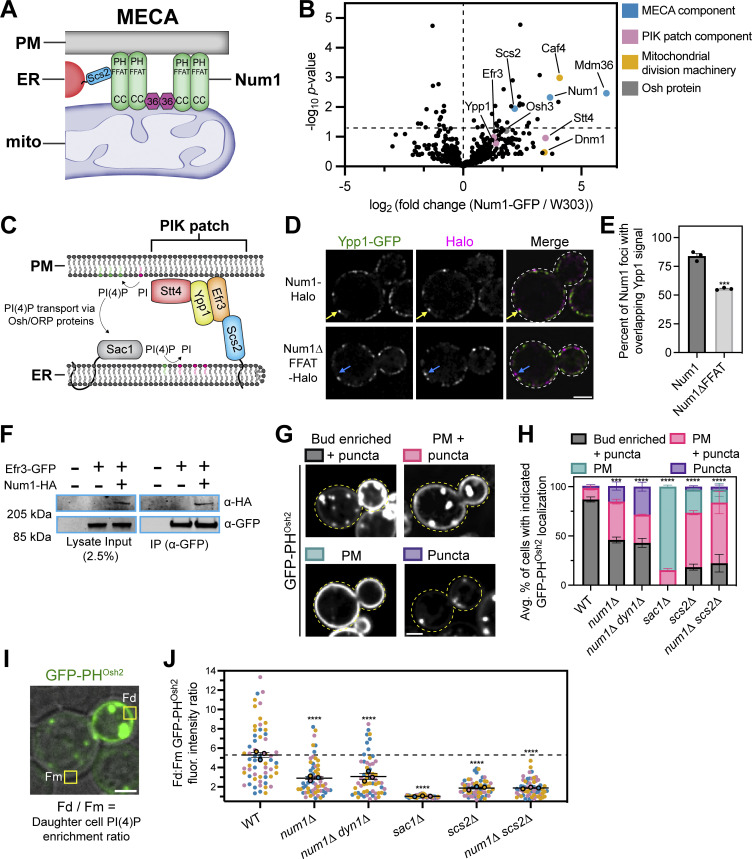
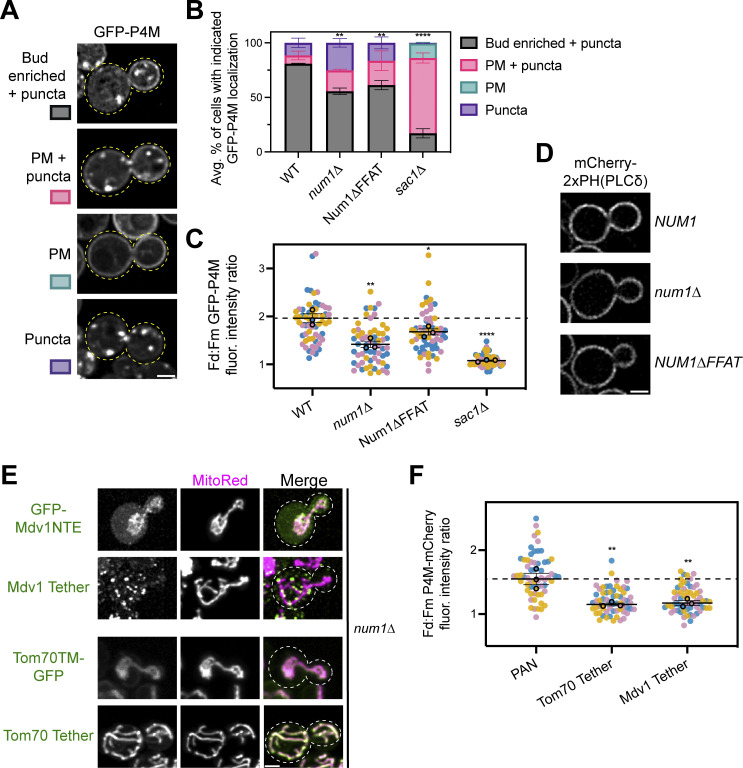
Our work demonstrates that Num1 regulates the distribution of the mitochondrial network via mitochondria–PM tethering and regulates the rate of mitochondrial division via mitochondria–ER tethering; however, the functional purpose of maintaining a tripartite MCS has remained elusive. Therefore, we sought to identify novel functions of the tripartite mitochondria–ER–PM contact site mediated by MECA. To do so, we performed a coIP-MS analysis of Num1 to search for novel protein interactors (data presented in Data S1). In agreement with previous results, two of the proteins identified were Mdm36 and Scs2, the other components of MECA (Chao et al., 2014; Lackner et al., 2013; Fig. 5, A and B). Interestingly, three of the known components of the PM-localized PIK patches, Ypp1, Efr3, and Stt4, were consistently enriched in our Num1 pulldowns, albeit not to a degree that was deemed statistically significant (P = 0.17, 0.1, and 0.11, respectively) (Baird et al., 2008; Fig. 5 B). These proteins form a complex on the PM that synthesizes PI(4)P from the precursor phosphatidylinositol (PI) (Balla, 2013; Fig. 5 C). Osh3, a member of the ORP/Osh family of lipid binding proteins that has been implicated in PI(4)P transport, was also weakly enriched (P = 0.062; Fig. 5 B) (Omnus et al., 2020). Previous studies have demonstrated that PI(4)P synthesis in yeast is regulated by ER–PM contacts mediated by Scs2, Efr3, and ORP/Osh proteins (Baird et al., 2008; Omnus et al., 2020; Schulz et al., 2009; Stefan et al., 2011). Additionally, several recent studies have proposed a role for PI(4)P in regulating mitochondrial dynamics (Boutry and Kim, 2021; Nagashima et al., 2020; Terriente-Felix et al., 2020). Thus, we sought to determine if there was any functional crosstalk between MECA and PIK patches.
Figure 5.
Num1 interacts with PIK patches and is involved in PI(4)P homeostasis. (A) Scs2 is a novel component of MECA. A version of the MECA cartoon shown in Fig. 1 A depicting Scs2 as the molecular link between Num1 and the ER. (B) IP-MS pulldown analysis of Num1-GFP. Each dot represents one identified protein and proteins of interest are highlighted by various colors. The vertical dashed line is centered at 0 to enable easy visual comparison of enriched hits while the horizontal dashed line represents a significance threshold of P = 0.05. (C) A cartoon representation of phosphoinositide kinase (PIK) patches. PIK patches consist of Efr3, Ypp1, and Stt4 (the PI4Kinase), and catalyze the formation of PI(4)P (depicted as green lipids) from precursor PI (depicted as magenta lipids) on the PM. Efr3 interacts with Scs2 via a FFAT motif which helps control the localization of PIK patches to ER–PM contact sites. PI(4)P generated at the PM is presented to Sac1, the ER-localized PI(4)P phosphatase, via members of the Osh/ORP protein family and is converted back to PI. This cartoon was based on models and work presented in Omnus et al. (2020). (D) Num1 localizes adjacent to the PIK patch component Ypp1. Super resolution fluorescence micrographs of cells expressing Ypp1-GFP and either Num1-Halo or Num1∆FFAT-Halo. Individual channels are shown in grayscale. Images are single slices from the center of a cell. Yellow arrows point to Num1 foci that are adjacent to Ypp1 foci. Blue arrows point to Num1∆FFAT foci that are not adjacent to a Ypp1 focus. Scale bar, 2 µm. (E) Quantification of the percentage of Num1 or Num1∆FFAT foci that contain overlapping Ypp1 signal. The Ypp1 signal was thresholded so that only punctate fluorescence was visible and Num1 foci were manually scored for the presence or absence of Ypp1 signal. Each dot represents the percentage of Num1 or Num1∆FFAT foci containing Ypp1 signal per imaging replicate with each replicate containing at least 50 foci. The bars represent the mean and the error bars represent the SEM of the three replicates. To determine statistical significance, an unpaired t test was used (*** = P < 0.001). (F) Coimmunoprecipitation of Num1 with Efr3. Cells expressing genomically tagged Efr3-GFP with or without Num1-HA were lysed and subjected to affinity purification with an α-GFP antibody followed by SDS-PAGE and Western blot analysis. A representative blot of three independent experiments is shown. (G) Array of representative fluorescence micrographs showing the observed localization patterns for the PI(4)P biosensor GFP-PHOsh2. The Bud enriched + puncta image is from a wild type strain, the PM + puncta and Puncta images are from a num1∆ strain, and the PM image is from a sac1∆ strain. Images are single slices from the center of the cell. Dashed yellow lines indicate cell outlines. Scale bar, 2 µm. (H) Quantification of the percentage of cells in the indicated genetic backgrounds showing the GFP-PHOsh2 localization patterns depicted in G. Each portion of the stacked bar graph represents the average percentage of cells displaying the indicated GFP-PHOsh2 localization pattern of three biological replicates. Error bars represent SEM. Imaging replicates consisted of at least 38 cells for a total of at least 130 cells per condition. See Materials and methods for a complete description of the quantification methodology. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (*** = P < 0.001, **** = P < 0.0001). All statistical analyses are comparing the Bud enriched + puncta category to the WT condition. (I) Quantification strategy to measure the fold enrichment of PI(4)P on the daughter cell PM compared to the mother cell PM. The image is a single slice from the center of a wild type cell expressing GFP-PHOsh2. The fluorescence channel is merged with a bright field image to show the cell boundaries. “Fd” and “Fm” refer to the measured GFP-PHOsh2 PM signal intensity in the daughter and mother cell, respectively. See Materials and methods for a complete description of the quantification methodology. Scale bar, 2 µm. (J) Quantification of the ratio of GFP-PHOsh2 enrichment in daughter cells compared to mother cells using the strategy depicted in I. Each dot represents the PM GFP-PHOsh2 ratio measured from a single cell. Imaging replicates are depicted as different colors and the average of each replicate is shown as a circle of the appropriate color with a black outline. Each replicate contains 20 cells for a total of 60 cells measured per condition. The horizontal line indicates the mean of the three imaging replicates with error bars representing SEM. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001). All statistical analyses are in comparison to the WT condition. The dashed black line depicts the average GFP-PHOsh2 enrichment in WT cells. Source data are available for this figure: SourceData F5.
Based on our IP-MS experiments, we predicted that MECA foci would localize near PIK patches. To test this, we expressed fluorescently tagged versions of Num1 and the PIK patch component Ypp1 and examined their localization using super-resolution confocal microscopy. As expected, nearly all Num1 foci were adjacent to Ypp1 foci (Fig. 5, D and E). This is consistent with a previous report that demonstrated PIK patches localize to ER–PM MCSs (Omnus et al., 2020). Notably, there are many Ypp1 foci per cell and the majority of them did not have adjacent Num1 foci. To test if the observed localization was dependent on the Num1–Scs2 interaction, we performed the same analysis on cells expressing Num1∆FFAT. In Num1∆FFAT cells, we saw a significant decrease in the number of Num1 foci adjacent to Ypp1 foci (Fig. 5, D and E). Therefore, the interaction of Num1 with Scs2 is potentially required to facilitate the localization of MECA to PIK patches. Finally, in agreement with our IP-MS data, Num1 was detected in Efr3 coimmunoprecipitation experiments (Fig. 5 F). Based on this evidence, we conclude that MECA associates with PIK patches in vivo.
We next examined if MECA influences the functionality of PIK patches. To assess PIK patch function, we used a genetically encoded fluorescent PI(4)P biosensor, the PH domain of Osh2, which has been shown to recognize PI(4)P with high affinity (Roy and Levine, 2004; Stefan et al., 2011). In a wild type background, GFP-PHOsh2 shows a strong enrichment on the growing daughter cell PM and very little accumulation on the mother cell PM (Stefan et al., 2011; Fig. 5 G). Additionally, a significant portion of GFP-PHOsh2 localizes to intracellular punctate structures that have previously been shown to be Golgi membranes (Roy and Levine, 2004; Fig. 5 G). Due to large cell-to-cell variation in the cellular distribution of the PI(4)P biosensor, we used two separate quantification strategies to rigorously document changes in the localization of GFP-PHOsh2 in different genetic backgrounds. To quantify the overall cellular localization of the probe, we defined four discrete localization categories, “bud enriched + puncta,” “PM + puncta,” “PM,” and “puncta,” that captured the range of observed phenotypes (Fig. 5 G). Detailed descriptions of how each category was defined can be found in the methods section. To document the change in the enrichment of PI(4)P on the daughter cell PM compared with the mother cell PM, we measured the fluorescence intensity of the probe over defined regions of the PM and plotted the ratio of daughter cell PM fluorescence to mother cell PM fluorescence (Fig. 5 I). Remarkably, in num1∆ cells, we saw a significant change in the distribution of GFP-PHOsh2. Many cells lost the strongly polarized distribution of GFP-PHOsh2 and displayed a more uniform distribution along both the mother and daughter cell PM (Fig. 5, H and J). In addition, some cells showed a severe loss of GFP-PHOsh2 from the PM and only displayed the punctate population (Fig. 5 H). Importantly, the altered distribution of the biosensor was not due to disrupting the function of Num1 as a dynein anchor, as the same effect was observed in a num1∆ dyn1∆ strain (Fig. 5, H and J). To validate that the probe was reflecting changes in PI(4)P distribution, we also expressed GFP-PHOsh2 in a sac1∆ strain. Sac1 is the ER-localized PI(4)P phosphatase that hydrolyzes PI(4)P into PI, and loss of Sac1 results in a large increase in PI(4)P levels on the PM (Del Bel and Brill, 2018; Stefan et al., 2011; Fig. 5 C). As previously reported, loss of Sac1 resulted in the majority of GFP-PHOsh2 being uniformly distributed on the PM with less observable punctate signal (Stefan et al., 2011; Fig. 5, H and J). Scs2 has also been reported to control the distribution of PI(4)P, potentially through organizing PIK patches via recruitment of Efr3 to the ER or by helping maintain ER–PM contact sites (Omnus et al., 2020; Stefan et al., 2011). As expected, loss of Scs2 resulted in a significant decrease in the daughter cell enrichment of PI(4)P (Fig. 5, H and J). Interestingly, in the absence of both Num1 and Scs2, the distribution of PI(4)P was similar to that of the single Scs2 deletion (Fig. 5, H and J). Thus, the loss of Num1 potentially alters PI(4)P distribution via the same mechanism as the loss of Scs2.
While GFP-PHOsh2 has been shown to preferentially bind PI(4)P, it also has a weak affinity for PI(4,5)P2 (Lenoir et al., 2015; Levine and Munro, 2002). Since PI(4,5)P2 is enriched on the PM, interpreting our results could be complicated if PI(4,5)P2 localization is significantly different in the Num1 deletion strain (Balla, 2013). The PH domain of Num1 has been shown to bind PI(4,5)P2 with very high affinity, raising the possibility that loss of Num1 could result in changes in PI(4,5)P2 localization (Yu et al., 2004). To validate that the changes in GFP-PHOsh2 distribution are due to changes in PI(4)P distribution, we repeated the previous experiments using another PI(4)P biosensor, the PI(4)P binding domain of the secreted effector protein SidM from Legionella pneumophila (referred to as P4M), which does not have affinity for PI(4,5)P2 (Hammond et al., 2014). GFP-P4M showed a similar distribution pattern to GFP-PHOsh2, with most cells showing an enrichment of the biosensor on the PM of the daughter cell compared with the mother cell, as well as punctate intracellular accumulations (Fig. S4, A–C). Loss of Sac1 resulted in the majority of cells showing uniform GFP-P4M signal on the PM of both mother and daughter cells (Fig. S4, A–C). One noticeable difference between the two biosensors, however, was that, under most genetic conditions, the intracellular punctate signal of GFP-P4M was still highly prevalent (Fig. S4, A–C). Importantly, loss of Num1 resulted in changes in the distribution of GFP-P4M that were similar to those observed for GFP-PHOsh2, thus confirming that loss of Num1 resulted in a change in PI(4)P distribution (Fig. S4, A–C). Finally, to test whether the distribution of PI(4,5)P2 was also affected in these genetic backgrounds, we used the PI(4,5)P2 biosensor mCherry-2xPH-PLCδ (Lemmon et al., 1995). In all conditions tested, 100% of cells showed a similar distribution of mCherry-2xPH-PLCδ along the PM (Fig. S4 D; at least 150 cells analyzed per genetic condition over three imaging replicates). Based on these results, we conclude that the loss of Num1 alters the distribution of PI(4)P on the PM.
Figure S4.
PI(4)P, but not PI(4,5)P2, localization is altered upon loss of Num1, and synthetic mitochondria–PM tethers are not sufficient to restore PI(4)P polarization in the absence of Num1. (A) An array of representative fluorescence micrographs showing the different observed localization patterns for the PI(4)P biosensor GFP-P4M. The Bud enriched + puncta image is from a wild type strain, the PM + puncta and Puncta images are from a num1∆ strain, and the PM image is from a sac1∆ strain. Images are single slices from the center of the cell. Dashed yellow lines indicate cell outlines. Scale bar, 2 µm. (B) Quantification of the percentage of cells in the indicated genetic backgrounds showing the GFP-P4M localization patterns depicted in A. Quantification was performed identically to that done for the GFP-PHOsh2 biosensor in Fig. 5 H. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (** = P < 0.01, **** = P < 0.0001). All statistical analyses are comparing the Bud enriched + puncta category to the WT condition. (C) Quantification of the ratio of GFP-P4M enrichment in daughter cells compared to mother cells using the strategy depicted in (Fig. 5 I). Quantification was performed and is presented identically to (Fig. 5 J). To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (* = P < 0.05, ** = P < 0.01, **** = P < 0.01). All statistical analyses are in comparison to the WT condition. The dashed line depicts the average GFP-P4M enrichment in wild type cells. (D) Representative fluorescence micrographs of cells of the indicated genetic backgrounds expressing the PI(4,5)P2 biosensor mCherry2xPH(PLCδ). Images are single slices from the center of a Z-stack. Scale bar, 2 μm. (E) Representative fluorescence micrographs showing the localization of GFP-Mdv1NTE, the Mdv1 Tether, Tom70TM-GFP, or the Tom70 Tether with the mitochondrial marker MitoRed. Individual channels are shown in grayscale. Images are max projections of a full Z-stack. Dashed white lines indicate cell outlines. Scale bar, 2 µm. (F) Quantification of the ratio of mCherry-P4M enrichment in daughter cells compared to mother cells using the strategy depicted in Fig. 5 G. Quantification was performed identically to that done for the GFP-PHOsh2 biosensor in Fig. 5 J except the dashed line depicts the average mCherry-P4M enrichment in PAN cells. The quantification for PAN cells is duplicated from Fig. 6 D to aid visual comparison. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (** = P < 0.01). All statistical analyses are in comparison to the PAN condition.
Mitochondria–ER–PM tethering by Num1 is required to maintain the polarized distribution of PI(4)P on the PM
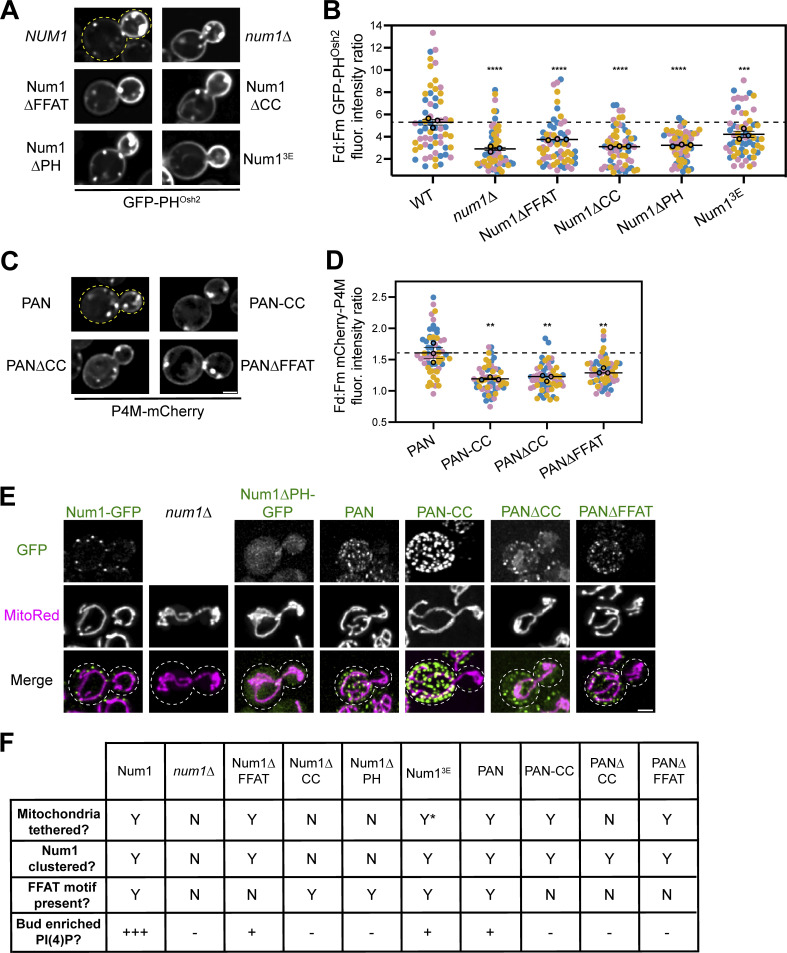
To further understand how Num1 regulates the distribution of PI(4)P, we sought to identify whether its function as a molecular tether was required. First, we examined the distribution of PI(4)P when expressing Num1 mutants that alter the ability of Num1 to function as a mitochondria–PM or mitochondria–ER tether. Disrupting the Num1–ER interaction by expressing Num1∆FFAT resulted in a modest reduction in daughter cell PM PI(4)P enrichment (Fig. 6, A and B). Expression of either Num1∆CC or Num1∆PH, both of which disrupt Num1-mediated mitochondria–PM tethering, significantly reduced the enrichment of PI(4)P on the daughter cell PM, but did not have as strong of an effect as the complete loss of Num1 (Lackner et al., 2013; Tang et al., 2012; Fig. 6, A and B). The reduction, but not complete disruption, of mitochondria–PM tethering in cells expressing the Num13E mutant, which weakens the Num1–mitochondria interaction, resulted in a mild decrease in daughter cell PM PI(4)P enrichment (Ping et al., 2016; Fig. 6, A and B). Taken together, these results indicate that the functions of Num1 as both a mitochondrial and ER tether are important for maintaining the distribution of PI(4)P.
Figure 6.
The function of Num1 as a mitochondria–ER–PM tether is required to maintain polarized PI(4)P distribution. (A) Representative fluorescence micrographs of cells expressing the indicated Num1 alleles and GFP-PHOsh2. Yellow dashed lines represent cell outlines. Scale bar, 2 µm. (B) Quantification of the ratio of GFP-PHOsh2 enrichment in daughter cells compared to mother cells in the indicated genetic backgrounds using the strategy depicted in Fig. 5 I. Quantification was performed and is presented identically to Fig. 5 J. The dashed black line depicts the average GFP-PHOsh2 enrichment in WT cells. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (*** = P < 0.001, **** = P < 0.0001). All statistical analyses are in comparison to the WT condition. The WT and num1∆ data are duplicated from Fig. 5 J to aid visual comparison. (C) Representative fluorescence micrographs of cells expressing the indicated PAN variants and P4M-mCherry. Scale bar, 2 µm. (D) Quantification of the ratio of mCherry-P4M enrichment in daughter cells compared to mother cells expressing the indicated PAN alleles using the strategy depicted in Fig. 5 I. Quantification was performed and is presented identically to Fig. 5 J. The dashed black line depicts the average mCherry-P4M enrichment in PAN cells. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (** = P < 0.01). All statistical analyses are in comparison to the PAN condition. (E) Array of representative fluorescence micrographs showing the indicated Num1 or PAN alleles with the mitochondrial matrix marker MitoRed. Individual channels are shown in grayscale. Images are max projections of a full Z-stack. Scale bar, 2 µm. Dashed white lines represent cell outlines. Due to variations in construct expression, the green channels are set to different brightness/contrast settings to enhance visualization, and the intensity between images should not be directly compared. (F) Table summarizing whether mitochondria are tethered to the PM, whether Num1 is clustered on the PM, whether the Num1 FFAT motif is present, and the degree of enrichment of PI(4)P on the daughter cell PM compared to the mother in the indicated strains. Y = yes, N = no. “Y*” means that the indicated trait is present but significantly reduced compared to wild type. For the bud enriched PI(4)P category, “+++” = an average daughter:mother fluorescence ratio of GFP-PHOsh2 ≥ 5, “+” = between 5 and 3.25, and “–” = <3.25. For categories measured using P4M, “+” = an average daughter:mother fluorescence ratio of P4M > 1.5, and “–” = between 1 and 1.5.
Our previous work has established that mitochondria are required for the formation of Num1 clusters (Kraft and Lackner, 2017). Therefore, one explanation for the previous results is that mitochondria are solely required to cluster Num1 and the presence of mitochondria near the PM is not essential to maintain standard PI(4)P distribution. To test this hypothesis, we employed the PAN system to artificially cluster different Num1 alleles at the PM. While cells expressing PAN still showed an enrichment of the PI(4)P biosensor P4M on the daughter cell PM, the enrichment was less pronounced than in wild type cells (∼1.5-fold enrichment in PAN compared to ∼2-fold in wild type; Fig. 6, C and D; and Fig. S4 C). This likely reflects that the PAN system does not fully recapitulate all of the wild type functions of Num1. To test whether clustering of Num1, mitochondrial tethering, or ER tethering are required to maintain daughter cell PM PI(4)P enrichment in PAN cells, we expressed variants of PAN lacking various functional regions of Num1. PAN-CC contains only the CC domain of Num1 and is capable of tethering mitochondria but not the ER because it lacks the C-terminal FFAT motif (Schmit et al., 2018). PAN∆CC contains all of Num1 except the CC and PH domains and does not tether mitochondria. PAN∆FFAT contains all of Num1 except the FFAT motif and PH domain and tethers mitochondria but not ER. While all PAN variants formed clusters on the PM as expected, none of them were capable of maintaining the enrichment of PI(4)P on the daughter cell PM (Fig. 6, C–F). Therefore, we conclude that the formation of a tripartite contact site between mitochondria, the ER, and PM is required to maintain the wild type distribution of PI(4)P. Importantly, these results do not rule out the possibility that the presence of mitochondria alters the conformation and/or function of Num1 clusters rather than the mitochondrial compartment itself being directly required for maintaining PI(4)P distribution.
Finally, to test whether recruitment of mitochondria to the PM in a Num1-independent matter altered PI(4)P distribution, we used artificial mitochondria–PM tethers that stably anchor mitochondria to the PM in the absence of Num1 (White et al., 2022). These systems link the GFP-tagged mitochondrial binding domain of Mdv1, a protein involved in mitochondrial division, or the transmembrane domain of Tom70, a subunit of the mitochondrial import machinery, to the PM by interaction with a α-GFP nanobody fused to an eisosome component (Neupert and Herrmann, 2007; Tieu et al., 2002; White et al., 2022; Fig. S4 E). Despite forming cortically tethered mitochondrial networks, both artificial tethers failed to show enrichment of PI(4)P on the daughter cell PM (Fig. S4 F). Taken together, these experiments demonstrate that Num1’s role as a mitochondria–ER–PM tether plays a unique role in regulating PI(4)P distribution.
Enhancing Num1 clustering affects Scs2 localization and PI(4)P distribution
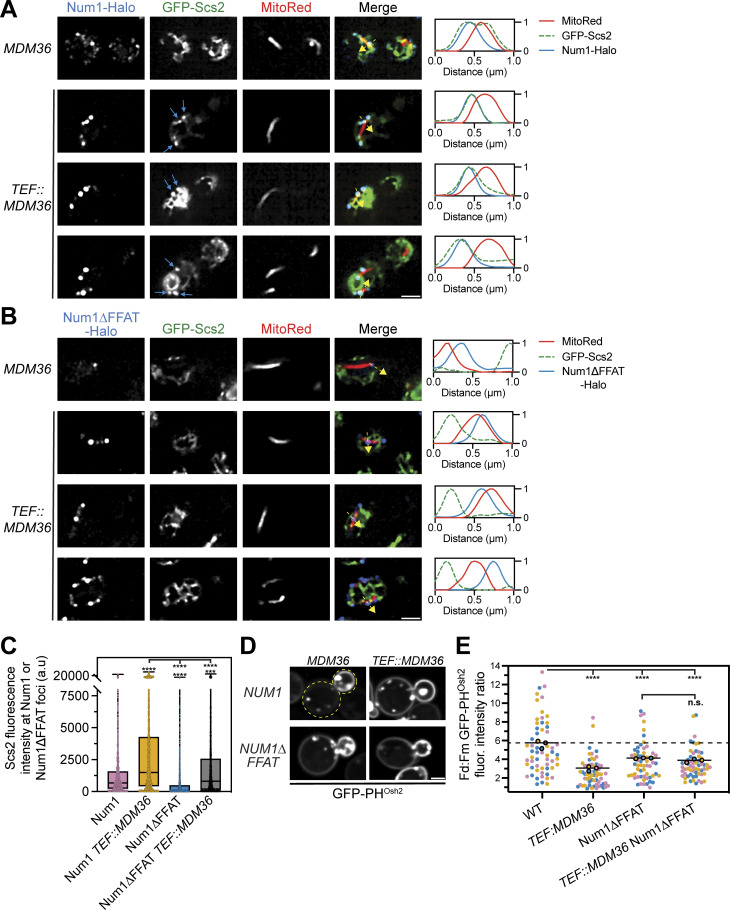
In yeast, at least 6 FFAT motif-containing proteins, including Num1, have been shown to interact with Scs2 in vivo, and there are >50 proteins with predicted FFAT motifs that have not been directly tested (Omnus et al., 2020; Slee and Levine, 2019). All FFAT motif-containing proteins are predicted to interact with the MSP domain of Scs2; therefore, competition for binding to Scs2 is a likely mechanism that regulates FFAT motif-containing protein localization and function. While we have already demonstrated that disrupting the Num1–Scs2 interaction altered PI(4)P distribution, we were curious if increasing the interaction would also produce an effect (Fig. 6 B). We hypothesized that the enhanced Num1 clusters generated by Mdm36 overexpression would bind more Scs2 (Omer et al., 2020; Fig. S2, A and B). To test this, we first examined the localization of Scs2 relative to mitochondria and Num1 or Num1∆FFAT in cells overexpressing Mdm36 via the strong TEF promoter using super-resolution microscopy. Remarkably, overexpression of Mdm36 resulted in large punctate accumulations of Scs2 that colocalized with Num1 (Fig. 7 A, blue arrows). The accumulations of Scs2 were not seen in cells expressing Num1∆FFAT, indicating that this phenotype is due to the Num1–Scs2 interaction (Fig. 7 B). Quantification of this data revealed a substantial increase in the Scs2 signal that colocalized with Num1 upon Mdm36 overexpression (Fig. 7 C). While we also observed an increase in Scs2 fluorescence at Num1∆FFAT foci upon Mdm36 overexpression using this quantification method, this could potentially be explained by the increased size and abundance of Num1∆FFAT clusters, which would increase the chance of coincidental overlapping signals. Regardless, the amount of Scs2 at Num1 clusters was significantly higher than the amount of Scs2 at Num1∆FFAT clusters in both wild type and Mdm36 overexpression conditions (Fig. 7 C). The enhanced recruitment of Scs2 to Num1 clusters upon Mdm36 overexpression is reminiscent of the behavior of other contact sites, including those between mitochondria and vacuoles, whose size has been shown to be regulated by the expression level of the molecular tethers (Elbaz-Alon et al., 2014; Hönscher et al., 2014).
Figure 7.
Overexpression of Mdm36 enhances recruitment of Scs2 to Num1 foci and alters PI(4)P distribution. (A) Fluorescence micrographs of cells expressing genomically tagged Num1-Halo with GFP-Scs2 and MitoRed in a wild type or Mdm36 overexpression background. Mdm36 overexpression was achieved by replacing the endogenous promoter with the strong TEF promoter. Individual channels are shown in grayscale. Images are single slices from the top of cells. Three representative examples of the Mdm36 overexpression strain are shown. The blue arrows indicate punctate accumulations of Scs2 that colocalize with Num1. The dashed yellow arrows mark the location analyzed in the accompanying linescans to the right of the micrographs. Scale bar, 2 µm. (B) Identical to A except cells expressed Num1∆FFAT-Halo. (C) Quantification of A and B. Briefly, the Num1-Halo or Num1∆FFAT-Halo signal from a single slice from a Z-stack at the top of a cell was used to generate a mask and measure the GFP-Scs2 signal at individual Num1 foci. Each dot in the box and whisker plot represents the GFP-Scs2 fluorescence measured at an individual Num1 foci and the black line indicates the mean. See Materials and methods for a detailed description of the quantification methodology. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (**** = P < 0.0001, *** = P < 0.001). Statistical analyses without brackets are in comparison to Num1 with endogenous Mdm36 expression levels and those indicated with brackets are in comparison to Num1 with overexpressed Mdm36. (D) Fluorescence micrographs of cells in the indicated genetic backgrounds expressing GFP-PHOsh2. Yellow dashed lines indicate cell outlines. Scale bar, 2 µm. (E) Quantification of D performed and presented identically to Fig. 5 J. The WT and Num1∆FFAT data are replicated from Fig. 5 J and Fig. 6 B to aid visual comparison.
We next tested whether enhanced recruitment of Scs2 to Num1 foci plays a role in regulating PI(4)P distribution by examining the distribution of GFP-PHOsh2 in cells overexpressing Mdm36. Remarkably, overexpression of Mdm36 in cells expressing full-length Num1 resulted in a substantial decrease in the bud:mother PI(4)P enrichment (Fig. 7, D and E). Interestingly, in cells expressing Num1∆FFAT, there was no significant difference in the bud:mother PI(4)P enrichment between cells overexpressing Mdm36 and cells expressing Mdm36 at endogenous levels (Fig. 7, D and E). Thus, the change in PI(4)P distribution upon Mdm36 overexpression is linked to the Num1–Scs2 interaction and not solely the enhanced clustering of Num1. Changing the amount of Scs2 present at mitochondria–ER–PM contact sites should change the available pool of Scs2 that can interact with other FFAT-motif-containing proteins. Thus, one potential explanation for these results is that enhancing the Num1–Scs2 interaction alters the regulation of other Scs2-dependent processes, such as PI(4)P metabolism.
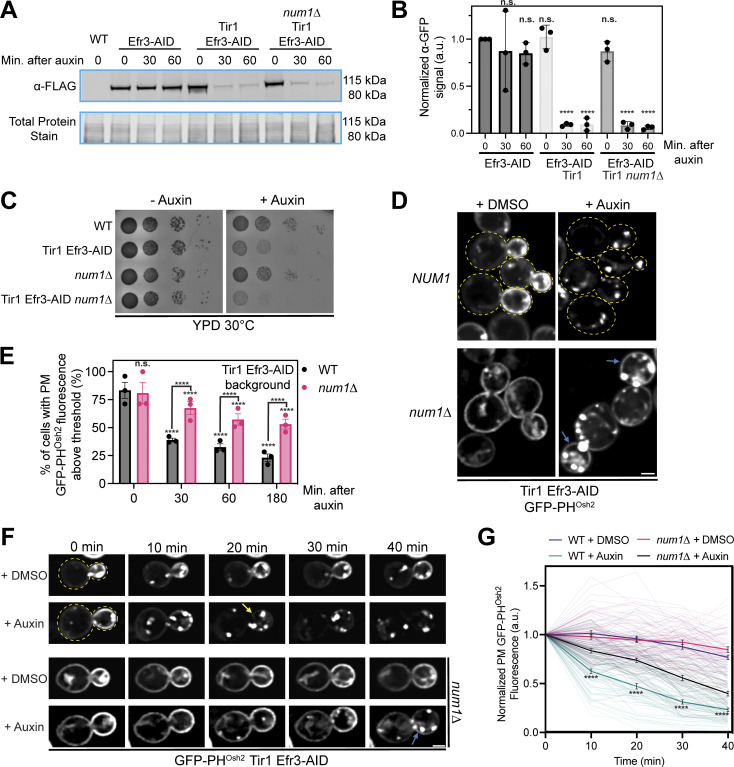
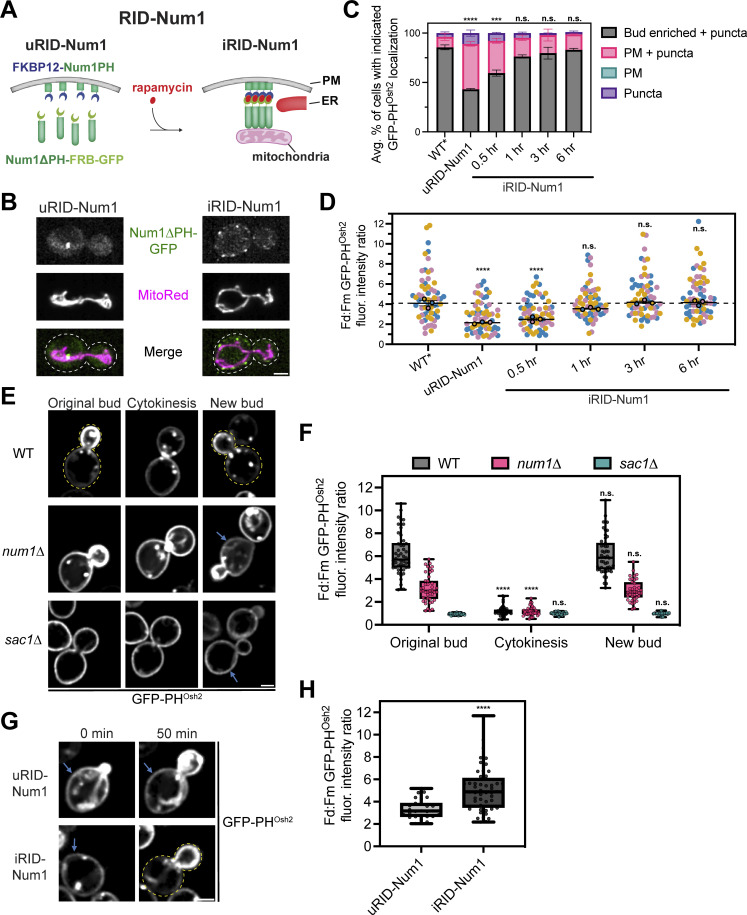
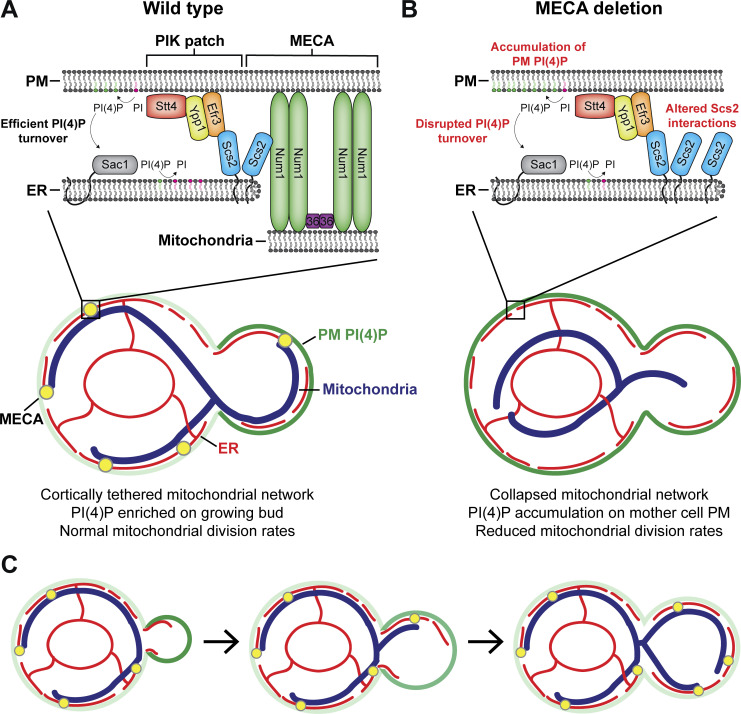
Loss of PM PI(4)P after PIK patch inactivation is slower in Num1 mutants
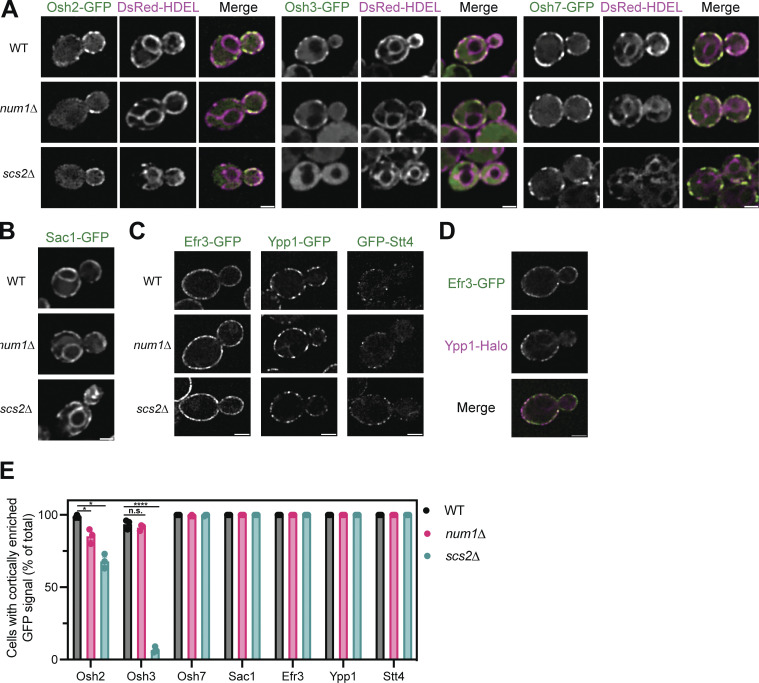
We next wanted to determine the mechanism by which Num1-mediated mitochondria–ER–PM contacts influence PI(4)P distribution. The distribution of any given lipid species is determined by the rates of synthesis, degradation, and transport to or from other membranes. Therefore, we first examined the localization of several proteins known to be involved in PI(4)P metabolism in Num1 and Scs2 mutants. Members of the Osh/ORP family of proteins have been localized to ER–PM contact sites and have been reported to influence PI(4)P distribution, likely by directly binding and transporting PI(4)P (Schulz et al., 2009; Stefan et al., 2011; Moser von Filseck et al., 2015). Loss of Num1 had no obvious effect on the cortical localization of Osh3 or Osh7 (Fig. S5, A and E). Interestingly, num1∆ cells displayed a small, but quantifiable, decrease in cells displaying cortical enrichments of Osh2 (Fig. S5, A and E). Loss of Scs2 caused a significant decrease in the cortical localization of Osh2 and Osh3, with the latter being almost completely lost from the cell cortex, and no obvious change to Osh7 localization (Fig. S5, A and E). These results are consistent with the presence of a FFAT motif in Osh2 and Osh3 but not Osh7 (Loewen and Levine, 2005; Slee and Levine, 2019). Next, we examined whether loss of Num1 or Scs2 influenced the localization of the PI(4)P phosphatase Sac1 or the assembly of PIK patches on the cell cortex. Loss of Num1 or Scs2 caused no obvious change in the localization of Sac1 or the PIK patch components Efr3, Ypp1, and Stt4 (Fig. S5, B–E). Interestingly, we observed that each PIK patch component displayed a slightly different localization pattern, with Efr3 being more diffuse along the PM than Ypp1 or Stt4 (Fig. S5, C and D). Additional work is still needed to understand the mechanism by which PIK patches form and the relative stoichiometry of the subunits. In summary, Num1 plays a minor role, if any, in controlling the localization of Osh2 and has no effect on Osh3, Osh7, Sac1, or PIK patch components.
Figure S5.
Determining the localization of Osh2, Osh3, Osh7, Sac1, Efr3, Ypp1, and Stt4 in num1∆ and scs2∆ mutants. (A) Fluorescence micrographs of yeast cells expressing Osh2-GFP, Osh3-GFP, or Osh7-GFP with the ER marker DsRed-HDEL in wild type, num1∆, or scs2∆ backgrounds. Osh2 and Osh7 are tagged at the genomic locus while Osh3 is expressed as a second copy from a CEN/ARS plasmid under control of the PHO5 promoter. Individual channels are shown in grayscale. Scale bar, 2 µm. (B) Fluorescence micrographs of cells expressing genomically tagged Sac1-GFP in wild type, num1∆, or scs2∆ backgrounds. Scale bar, 2 µm. (C) Super resolution fluorescence micrographs of cells expressing Efr3-GFP, Ypp1-GFP, or GFP-Stt4 in wild type, num1∆, or scs2∆ backgrounds. Efr3 and Ypp1 are tagged at the genomic locus while Stt4 is expressed as a second copy from a CEN/ARS plasmid under control of the CPY promoter. Scale bar, 2 µm. (D) Super-resolution fluorescence micrographs of cells expressing Efr3-GFP and Ypp1-Halo. Individual channels are shown in grayscale. Scale bar, 2 µm. (E) Quantification of the imaging data from A–D. For each protein in each genetic background, a central slice from a Z-stack was thresholded and manually scored for the presence of cortical enrichments of the indicated protein. Cortical enrichments were defined as punctate or elongated accumulations of fluorescent signal near the cell periphery. Each dot represents the average of one imaging replicate containing 100 cells. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (* = P < 0.05, **** = P < 0.0001, n.s. = not significant).
Given that PIK patch localization is normal and PI(4)P is readily detectable on the PM in Num1 mutants (Fig. 5, G and H; and Fig. S5, C–E), we reasoned that PI(4)P synthesis was unlikely to be perturbed. Therefore, we wanted to examine whether PI(4)P turnover is altered in Num1 mutants. The only PI(4)P phosphatase in Saccharomyces cerevisiae, Sac1, is confined to the ER membrane and is thought to hydrolyze PI(4)P that has been transported to the ER from the PM or Golgi membranes (Zewe et al., 2018). To test whether Sac1-mediated PI(4)P hydrolysis was disrupted in Num1 mutants, we developed a system in which we could conditionally inactivate the synthesis of PI(4)P on the PM and use the loss of PI(4)P from the PM as a proxy to measure Sac1 activity. Analogous experiments have been performed using chemical inhibitors of the mammalian homolog of Stt4 (Doyle et al., 2024). Specifically, we used the auxin-inducible degradation (AID) system to conditionally inactivate PIK patches by degrading Efr3 (Nishimura et al., 2009). PIK patches require Efr3 for formation, and the loss of Efr3 results in a loss of the PM pool of PI(4)P (Baird et al., 2008). First, we tested the efficacy of the Efr3-AID system by immunoblotting. In cells expressing both Efr3-AID and Tir1, the plant-specific F-box protein that targets AID-tagged proteins for degradation, the addition of auxin caused a significant decrease in Efr3 levels within 30 min in both wild type and num1∆ cells (Fig. 8, A and B). Additionally, as expected for an essential gene, cells expressing Tir1 and Efr3-AID showed a growth defect when grown on media containing auxin (Baird et al., 2008; Fig. 8 C). Finally, upon auxin treatment, Efr3-AID–expressing cells progressively lost the PM pool of PI(4)P, as measured by a change in the localization of GFP-PHOsh2 (Fig. 8, D and E). The Golgi pool of PI(4)P, which is synthesized by Pik1, remained visible throughout auxin treatment, matching previous reports that utilized temperature-sensitive alleles of Stt4 (Baird et al., 2008). Taken together, these results demonstrate that the Efr3-AID system efficiently inactivates PIK patches upon auxin treatment. With this system in hand, we then examined whether PI(4)P turnover is altered in Num1 mutants. Interestingly, in comparison to wild type cells, the fraction of num1∆ cells that retained PI(4)P on the PM after Efr3-AID depletion was significantly higher (Fig. 8 E). To further assess the kinetics of PI(4)P loss from the PM upon PIK patch activation, we used 4D confocal microscopy to monitor individual cells during the course of auxin treatment. As expected, auxin treatment resulted in a rapid (within 10–20 min) decrease in the levels of PM PI(4)P that was not seen in control cells (Fig. 8, F and G). Remarkably, loss of PI(4)P from the PM was significantly slower in num1∆ cells (Fig. 8, F and G). These results are consistent with a model in which Num1-mediated mitochondria–ER–PM contacts positively influence Sac1-mediated PI(4)P turnover.
Figure 8.
Loss of PM PI(4)P upon inactivation of PIK patches is slowed in num1∆ cells. (A) Efficient degradation of Efr3-AID via the auxin-inducible degradation system. Western blot analysis of Efr3-AID protein levels upon addition of auxin for the indicated time lengths. (B) Quantification of the Western blot in A and two other replicates. For each replicate, the intensity of the Efr3-AID band was quantified and normalized to the signal from the respective total protein stain. Data points were then normalized to the Efr3-AID signal at time 0 of cells that did not express Tir1. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistical analyses are in comparison to the Efr3-AID signal at time 0 of cells that did not express Tir1 condition. (C) 10-fold serial dilutions of the indicated strains were spotted on YPD + DMSO or YPD + 1 mM auxin medium and grown at 30°C for 2 days. The image is a representative example of three biological replicates. (D) Fluorescence micrographs of cells expressing Tir1, Efr3-AID, and GFP-PHOsh2 in a wild type or num1∆ background after DMSO or auxin treatment for 3 h. Yellow dashed lines indicate cell outlines. Blue arrows indicate PM-localized GFP-PHOsh2 signal that is visible after auxin treatment. Scale bar, 2 µm. (E) Quantification of the data from D. Cells were grown to mid log phase, treated with auxin, and imaged at the indicated timepoints. Each dot represents the average percent of cells that contained PM-localized GFP-PHOsh2 fluorescence above a threshold value from one imaging replicate. The threshold value was calculated by averaging multiple measurements of the PM of cells that contained no GFP-PHOsh2 prior to auxin treatment. Imaging replicates contained between 34 and 74 cells per replicate for a total of 128–214 cells per timepoint. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). Asterisks immediately above the bars are in comparison to the 0 timepoint of the respective genetic background. (F) Loss of PM PI(4)P upon inactivation of PIK patches is slower in num1∆ cells. Cells were grown to mid-log phase, adhered to a ConA treated confocal dish, and imaged before and at 10-min intervals after the introduction of DMSO or 1 mM auxin. Images are fluorescent micrographs displaying the progressive loss of GFP-PHOsh2 from the PM only in auxin treated cells. Yellow dashed lines indicate the cell outlines. The yellow arrow indicates the frame in which GFP-PHOsh2 signal is no longer detected at the PM. The blue arrow indicates clear GFP-PHOsh2 signal on the PM after 40 min of auxin treatment. Scale bar, 2 μm. (G) Quantification of the data in F. Each transparent line represents the normalized PM fluorescence intensity of a single cell through the course of the movie. PM fluorescence measurements were taken from the region that had the highest signal at each timepoint. Dark lines represent the averages for each condition and error bars represent SEM. Each condition contains data from 42 to 67 cells over three movies. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). The statistical comparisons are between WT + auxin and num1∆ + auxin at each indicated time point. Source data are available for this figure: SourceData F8.
The formation of mitochondria–ER–PM contact sites influences PM PI(4)P distribution during bud growth
Due to a lack of sufficiently sensitive assays, little is known about the kinetics of how MCS formation translates to cellular changes. To begin to answer these questions, we developed a tool that uses a rapamycin-inducible dimerization system to rapidly induce the formation of Num1-mediated mitochondria–ER–PM contact sites, allowing us to monitor downstream effects of contact site formation (Harper et al., 2023). In a rapamycin-resistant fpr1∆ tor1-1 strain, we expressed Num1∆PH-FRB from the endogenous Num1 locus in addition to an exogenously expressed copy of the Num1 PH domain fused to an FKBP12 tag. Upon addition of rapamycin, Num1∆PH-FRB and FKBP12-Num1PH dimerize to reconstitute a full Num1 molecule, which will begin to associate with other reconstituted Num1 molecules and form mitochondria–ER–PM contact sites (Fig. 9, A and B). We refer to this system as RID-Num1 for rapamycin-inducible dimerization of Num1. In a recent publication, we provide a full description of the functionality of RID-Num1 (Harper et al., 2023). To briefly summarize, in cells expressing RID-Num1, cortical, ER-associated, Num1-mediated mitochondrial tethering points form within 5 min of rapamycin addition and stably anchor the mitochondrial network to the cell cortex over time. Thus, the addition of rapamycin to cells expressing RID-Num1 rapidly induces the formation of Num1-mediated mitochondria–ER–PM contact sites.
Figure 9.
Inducing mitochondria–ER–PM tethering restores polarized PI(4)P distribution. (A) A cartoon representation of the rapamycin-inducible Num1 clustering system (RID-Num1). The Num1PH domain is tagged with FKBP12 and expressed from the TEF promoter while Num1∆PH-FRB-GFP is expressed from the endogenous NUM1 locus. The addition of rapamycin causes the components to dimerize and self-associate to form mitochondria–ER–PM tethering points. uRID-Num1 = uninduced RID-Num1, iRID-Num1 = induced RID-Num1. (B) Micrographs depicting Num1-RID cells before (uRID) and after (iRID) addition of rapamycin. Individual channels are shown in grayscale. Dashed white lines indicate cell outlines. Scale bar, 2 µm. (C and D) For these experiments, a variant of the RID system was used that expressed Num1∆PH-FRB rather than Num1∆PH-FRB-GFP. In addition, WT refers to the rapamycin resistant background harboring the tor1-1 fpr1∆ mutations and is denoted as WT*. In panel C, quantification of the percentage of cells in the indicated genetic backgrounds showing the GFP-PHOsh2 localization patterns depicted in Fig. 5 G before and after addition of rapamycin to RID-Num1 cells is shown. Quantification was performed and is presented identically to Fig. 5 H. In panel D, quantification of the GFP-PHOsh2 enrichment of daughter cells compared to mother cells before and after the introduction of rapamycin to Num1-RID cells is shown. Quantification was performed and is presented identically to Fig. 5 J. The dashed black line depicts the average GFP-PHOsh2 enrichment in WT* cells. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, *** = P < 0.001, **** = P < 0.0001). All statistical analyses are in comparison to the WT* condition and, for C, is comparing the Bud enriched + puncta category. (E) PI(4)P distribution changes throughout the cell cycle. Images are representative frames at the indicated cell cycle stages (original bud, cytokinesis, new bud) from Video 4. Cells expressing GFP-PHOsh2 in the indicated genetic backgrounds were grown to mid-log phase, adhered to a ConA treated confocal dish, and imaged every 10 min over the course of 3 h. The time of cytokinesis was approximated by the “snapping off” motion of the daughter cell and repositioning adjacent to the mother cell (Yeh et al., 1995). Scale bar, 2 µm. (F) Quantification of the data from E. The PI(4)P enrichment ratio was measured as described in Fig. 5 I for individual cells at the three different cell cycle stages indicated. The error bars on the box and whisker plots extend to the minimum and maximum values while the box extends from the 25th to 75th percentiles. To determine statistical significance, an ordinary one-way ANOVA with multiple comparisons was used (n.s. = not significant, **** = P < 0.0001). All statistics are in comparison to the Original bud category for their respective genetic background. Data are from at least three movies per condition. (G) RID-Num1 cells expressing GFP-PHOsh2 were grown to mid-log phase, adhered to ConA treated confocal dish, and imaged before and every 10 min after treatment with DMSO or rapamycin. Scale bar, 2 µm. (H) Quantification of the data in G. The PI(4)P enrichment ratio was measured as described in Fig. 5 I 50 min after rapamycin treatment. Only cells that began with very small or no buds at the beginning of the movie were selected for analysis. The error bars on the box and whisker plots extend to the minimum and maximum values while the box extends from the 25th to 75th percentiles. To determine statistical significance, an unpaired t test was used (**** = P < 0.0001).
To test how the formation of mitochondria–ER–PM contact sites regulates the distribution of PI(4)P, we examined the localization of GFP-PHOsh2 before and after the addition of rapamycin. Within 30 min of treatment, cells began to display an increase in the polarized distribution of PI(4)P between the mother and daughter cells, which gradually increased to wild type levels between 1 and 3 h after the contact sites were formed (Fig. 9, C and D). Therefore, despite the contact site being rapidly reformed (within 5 min), the resulting effect on PI(4)P distribution is not realized until significantly later. We were initially puzzled by the slow rate of rescue, as lipid trafficking at MCSs is thought to be a rapid process on the scale of seconds to minutes (Reinisch and Prinz, 2021).
To better understand our results, we wanted to characterize how PM PI(4)P distribution is regulated throughout the yeast cell cycle. To do so, we took 3-h long 4D confocal movies of wild type, num1∆, and sac1∆ cells expressing GFP-PHOsh2 and analyzed the PI(4)P distribution of cells that started with a small bud, progressed through cytokinesis, and began growing a new bud. Consistently, in wild type and num1∆ cells, small budded cells displayed a polarized distribution of PI(4)P that was lost by the time of cytokinesis (Fig. 9, E and F; and Video 4). Upon growth of a new bud, PI(4)P polarity was reestablished to the same level as the original bud (Fig. 9, E and F; and Video 4). The primary distinction between wild type and num1∆ cells was that num1∆ cells retained a larger pool of mother cell PM PI(4)P that persisted throughout the cell cycle (Fig. 9 E and Video 4). In contrast to both wild type and num1∆ cells, sac1∆ cells displayed uniform PI(4)P signal between the mother and bud throughout the cell cycle. Thus, PI(4)P polarity is established as a new bud forms in a manner that is dependent on Sac1 and is lost prior to cytokinesis.
Video 4.
PI(4)P localization changes throughout the cell cycle. 4D confocal microscopy of cells in the indicated genetic backgrounds expressing GFP-PHOsh2. Each cell comes from a unique movie and is tiled to aid visual comparison. Movies are a single slice from the center of a Z-stack. Time is indicated in hour:min format. Still images from this video are depicted in Fig. 9 E. Scale bar, 2 µm. The frame rate is 10 fps. In the labels, the lowercase “d” is used to represent the typical ∆ symbol to indicate gene deletions. The num1∆ movie stops at 2 h and 30 min.
Given these results, the heterogeneity we observe in our quantifications of PI(4)P distribution may be a consequence of changes in the degree of PI(4)P enrichment as the cell nears cytokinesis (Fig. 5 J). With this in mind, we revisited our RID-Num1 experiments. The initial experiments analyzed an asynchronous population of cells at static time points after rapamycin addition (Fig. 9, C and D). To more accurately identify when PI(4)P polarity is reestablished upon formation of Num1-mediated mitochondria–ER–PM contacts, we took 4D confocal movies and analyzed the distribution of PI(4)P in individual RID-Num1 cells upon the addition of rapamycin. We limited the analysis to cells that began with either no bud or very small buds and measured the PI(4)P enrichment ratio on the growing bud as the movie progressed. Our results indicate that, within 50 min, most rapamycin-treated RID-Num1 cells established a PI(4)P enrichment ratio that resembled wild type (Fig. 9, G and H). Given that rapamycin is added to an asynchronous cell population, it will take 1–3 h for all cells in the population to undergo a full cell cycle, which may explain the gradual rescue observed in our initial experiments (Fig. 9, C and D). Thus, the rescue likely occurs rapidly but can be difficult to measure from bulk populations as PI(4)P polarity is regulated throughout the cell cycle.
Discussion
In this work, we set out to clarify the molecular mechanism by which the core component of MECA, Num1, contacts the ER and identify functions that require this contact site. We provide extensive evidence that Num1 contains an Scs2-interacting FFAT motif, composed of AAs 2128–2140, that mediates the interaction between Num1 and the ER. By identifying the molecular link between Num1 and the ER, we have filled the last gap in our knowledge of the mechanism by which Num1 forms a tripartite MCS.
What are the unique functions mediated by Num1’s multiple tethering activities? By characterizing the Num1∆FFAT allele, we identified a means to separate the role of Num1 as a mitochondria–ER tether from its role as a mitochondria-PM tether. We found that Num1-mediated mitochondria–ER tethering is not required for mitochondria–PM tethering or dynein anchoring. Unexpectedly, however, we found that Num1-mediated mitochondria–ER contacts are required to maintain the wild type rate of mitochondrial division. In cells lacking Num1–ER contacts, the rate of mitochondrial division is severely reduced despite cells maintaining reticular, cortically tethered mitochondrial networks (Fig. 3, A and B).
What is the mechanism by which Num1–ER contacts regulate mitochondrial division? Previous studies have established a role for mitochondria–ER MCSs in regulating mitochondrial division, potentially via localizing factors that promote the constriction of mitochondrial tubules and/or facilitate the recruitment of the mitochondrial division machinery (Friedman et al., 2011). A simple explanation, therefore, is that the Num1–Scs2 interaction increases the amount of mitochondria–ER contact, which increases the likelihood of forming ER-mediated mitochondrial constrictions that lead to division events. An attractive and testable hypothesis is that mitochondria–ER contact sites have a general function in promoting mitochondrial dynamics. In future work, we will test whether the extent of mitochondria–ER contact regulates mitochondrial division by examining division rates in a host of mitochondria–ER contact site mutants. Alternatively, there may be a unique feature of Scs2-mediated mitochondria–ER contacts that promotes mitochondrial division. Of note, loss of the VAP proteins, the mammalian Scs2 homologs, also reduces mitochondrial division rates (Boutry and Kim, 2021). Thus, VAP-mediated mitochondria–ER contacts play a conserved role in regulating mitochondrial division.
We also identified a novel function of the tripartite mitochondria–ER–PM MCS in the distribution of PI(4)P (Fig. 10, A and B). Through an analysis of Num1 alleles that lack specific tethering functions, we ascertained that Num1’s function as a tripartite mitochondria–ER–PM contact site is required to maintain the strong polarization of PI(4)P on the daughter cell PM (Fig. 6, A–D). These results support recent studies implicating ER–PM contact sites as key regulators of PI(4)P metabolism and add mitochondria as an unexpected player in PI(4)P homeostasis (Manford et al., 2012; Omnus et al., 2020; Stefan et al., 2011).
Figure 10.
The mitochondria–ER–PM contact site mediated by Num1 regulates the distribution of PI(4)P and mitochondrial dynamics. (A) MECA forms a tripartite contact site that tethers mitochondria to the PM and ER. Under wild type circumstances, PIK patches localize adjacent to MECA contact sites and synthesize PI(4)P (depicted as green lipids) from the precursor PI (depicted as magenta lipids). PI(4)P is then presented to the ER-localized Sac1 phosphatase where it is hydrolyzed into PI. Wild type levels of PI(4)P synthesis and turnover result in a robust enrichment of PI(4)P on the PM of the growing bud. (B) Loss of MECA results in a collapse of the mitochondrial network and PI(4)P accumulation on the mother cell PM. Loss of the Num1–Scs2 interaction may alter PI(4)P distribution by changing the amount of Scs2 available to interact with FFAT motif containing proteins. Additionally, loss of MECA may disrupt PI(4)P turnover by preventing PI(4)P from being properly presented to Sac1. Loss of MECA also severely perturbs mitochondrial division rates. (C) Changes in PM PI(4)P levels coincide with the formation of new mitochondria–ER–PM contact sites. Newly formed buds contain little cortical ER, have yet to inherit mitochondria, and maintain high levels of PM PI(4)P. As the cell cycle progresses, more cortical ER as well as mitochondria are inherited, MECA contact sites form, and bud PM PI(4)P levels begin to decrease. When the cell is near cytokinesis, the cortical ER and mitochondrial network are stably anchored via MECA and other MCSs, and the amount of PI(4)P on the bud PM decreases to a level that is comparable to the mother cell PM. The black arrows are used to indicate progression through the cell cycle. Darker shades of green represent higher concentrations of PM PI(4)P.
By what mechanism does MECA influence PI(4)P distribution? PI(4)P polarity on the PM is dependent on both new PI(4)P synthesis, as loss of PIK patch activity results in a complete loss of PI(4)P from the PM, and Sac1-mediated PI(4)P turnover, as loss of Sac1 results in a uniform distribution of PI(4)P between the daughter and mother cell PM (Fig. 8 D; and Fig. 9, E and F; Baird et al., 2008). One mechanism to generate a polarized distribution of PI(4)P that is consistent with these observations is to spatially regulate PI(4)P turnover between the mother and daughter cells. In line with this, a recently proposed model suggests that S. cerevisiae maintains PI(4)P polarity due to differences in the extent of ER–PM MCSs in the mother versus daughter cell (Omnus et al., 2020). In comparison to the mother cell, new buds have very little cortical ER, and the amount of ER in contact with the PM increases as the bud grows (West et al., 2011). As new ER–PM MCSs form in the growing bud, PI(4)P transport from the PM to ER may increase, resulting in a gradual decrease of PI(4)P levels on the PM until a steady state level similar to that of the mother cell is reached. In support of this, we documented the progressive loss of PI(4)P enrichment on the PM of the growing bud as the cell approaches cytokinesis (Fig. 9, E and F). Interestingly, Num1 foci are concentrated in the mother cell, and new Num1 foci form in the daughter cell at a later stage of the cell cycle when mitochondria are inherited (Kraft and Lackner, 2017). Thus, the progressive formation of new, stable mitochondria–ER–PM contact sites also coincides with the loss of PM PI(4)P polarity as the cell approaches cytokinesis (Fig. 10 C). While ER–PM contact sites have been implicated in PI(4)P transport and turnover (Manford et al., 2012; Omnus et al., 2020; Stefan et al., 2011; Thomas et al., 2022), our work suggests that mitochondria–ER–PM contact sites also play a role. In future experiments, we will examine the temporal relationship between the formation of new ER–PM and mitochondria–ER–PM contact sites in the bud and the loss of PI(4)P polarity. Importantly, our work does not rule out alternative models in which PI(4)P polarity is influenced via secretion of PI(4)P-containing vesicles, diffusion barriers between the mother and daughter cell PM, or changes in PI(4,5)P2 homeostasis.
We propose that the function of Num1 in regulating PI(4)P distribution is to enhance the efficiency of Sac1-mediated PI(4)P turnover by promoting the spatial organization of the membranes and protein machineries involved. In support of this, the clearance of PI(4)P from the PM upon PIK patch inactivation is slower but not abolished in num1∆ mutants (Fig. 8, F and G). Additionally, loss of both Num1 and Scs2 phenocopies the PI(4)P distribution defect seen upon loss of Scs2 alone, suggesting that Num1 operates in the previously characterized Scs2-dependent PI(4)P turnover pathways (Fig. 5 J). Finally, either loss or enhancement of Num1 clustering results in disrupted PI(4)P homeostasis (Fig. 7 E and Fig. 6 B). Based on these observations, we suggest that the size and localization of Num1 clusters may be specifically tuned to maintain wild type PI(4)P turnover levels. Num1 assembles into remarkably stable clusters that contain upwards of ∼40 molecules and persist throughout the cell cycle (Kraft and Lackner, 2017; Lackner et al., 2013; Omer et al., 2020). Num1, Efr3, and several Osh proteins that are implicated in PI(4)P transport all contain FFAT motifs and interact with Scs2 (Baird et al., 2008; Omnus et al., 2020; Slee and Levine, 2019). The optimal arrangement of Num1 clusters may locally regulate the interactions made by Scs2 and the organization of PIK patch components in a manner that favors efficient PI(4)P turnover (Fig. 10, A and B).
Why do cells utilize a mitochondrial MCS to regulate PI(4)P metabolism? Several recent lines of evidence support that PI(4)P may play a larger role in regulating mitochondrial biology than previously thought. Imaging studies in both mammalian tissue culture and Drosophila suggest that PI(4)P-containing membranes play a role in facilitating mitochondrial division, though the mechanism is unknown (Boutry and Kim, 2021; Nagashima et al., 2020; Terriente-Felix et al., 2020). Further, while stable pools of PI(4)P on mitochondrial membranes have yet to be reported, the precursor PI is highly enriched, and the artificial recruitment of a PI4KIIIα to mitochondria is sufficient to drive the synthesis of PI(4)P (Pemberton et al., 2020; Zewe et al., 2020). While likely not present in high abundance, PI4-kinase isoforms, including Stt4, have been detected in proteomic studies of isolated mitochondria (Hung et al., 2017; Morgenstern et al., 2017; Reinders et al., 2006). Therefore, it is possible that transient pools of PI(4)P are synthesized and rapidly turned over, making detection difficult. Lastly, mitochondria are involved in the metabolism of lipids, such as phosphatidylserine, that are known to be involved in PI(4)P-dependent counter-transport mechanisms at other organelles (Moser von Filseck et al., 2015). It is highly likely that cells have mechanisms to transport lipids to or from mitochondria that have yet to be fully described.
The culmination of our work suggests that the primary function of MECA is the formation of a cortical hub that orchestrates communication between three separate organelles (Fig. 10 A). By modulating the degree of tethering or the spatial distribution of the contact site, cells can rapidly communicate changes in physiological and environmental states or the presence of stressors throughout the organelle network. Therefore, Num1 is a prime example of a molecular tether that integrates signals from multiple organelles to control their spatial distribution and function. While a direct mammalian homolog of Num1 has not been identified, it is notable that several cell types, including neurons and pancreatic acinar cells, show extensive mitochondria–PM contact (Johnson et al., 2003; Perkins et al., 2010). Future studies will be required to determine which features of the crosstalk between mitochondria–ER–PM contact sites, mitochondrial dynamics, and PI(4)P metabolism are conserved.
Materials and methods
Strains and plasmids
Tables S1, S2, and S3 list all yeast strains, all plasmids, and select primers used in this study, respectively. The wild type background strain was W303 (ade2–1; leu2–3; his3–11, 15; trp1–1; ura3–1; can1–100) (Ralser et al., 2012). All new yeast strains were generated via transformation or mating, followed by sporulation and tetrad analysis. All new plasmids were generated via standard molecular biology techniques and relevant portions were confirmed via sequencing. Primer sets for gene deletions are named F1 and R1 and primer sets for C-terminal gene tagging are named F5 and R3. The plasmids used for gene deletions and tagging were previously described (Janke et al., 2004; Longtine et al., 1998; Sheff and Thorn, 2004). All strains and constructs used in this study are available upon request.
The Num1∆FFAT and Num1(F2135A) alleles were generated via homologous recombination by transformation with two PCR products, one amplified from either pRS306-GPD-GFP-Num1∆FFAT(2095-2562) or pRS306-GPD-GFP-Num1(2095-2562)(F2135A) and the other from genomic DNA of strains expressing Num1-3XHA (LLY271), Num1-GFP (LLY153), or Num1-Halo (LLY4131). Successful mutations were confirmed via sequencing.
N-terminal GFP-fusions of Scs2 and Mdm36 at the genomic locus were generated by transforming a wild type (LLY92) or mdm36∆ (LLY401) strain, respectively, with three PCR products containing a yEGFP cassette, KanMX resistance cassette, and the coding sequence of the gene generated with primers 1729, 1730, 1732, 1733, 1735, and 1736 for Scs2 and 300 and 1831–1835 for Mdm36. Successful integrations were confirmed via colony PCR.
pRS305-Pho88-mCherry::LEU was a gift from J. Brickner (Northwestern University, Evanston, IL, USA) and was digested with ClaI to direct integration at the LEU2 locus. pFA6a-yoHalo::CaUra3 (#743; LLEC) was a gift from J. Nunnari (University of California, Davis, Davis, CA, USA). pRS424-GFP-2xPH(Osh2) (#36095; Addgene) and pRS416-GFP-Stt4 were gifts from Scott Emr (Cornell University, Ithica, NY, USA). GFP-P4M-SidM and mCherry-P4M-SidM (#51469 and #51471, respectively; Addgene) were gifts from T. Balla (National Institutes of Health, Bethesda, MD, USA). pRS416-pPHO5-GFP-Osh3 was a gift from Tim Levine and Sean Munro (plasmid # 58839; Addgene; MRC Laboratory of Molecular Biology, Cambridge, UK) (Levine and Munro, 2001).
Imaging
For most imaging in this study, cultures were grown to mid-log phase (OD600 of 0.5–1.0) in synthetic complete + 2% wt/vol dextrose media with 2× adenine (SCD) at pH 6.4. Cells were concentrated by centrifugation and imaged on a 4% wt/vol agarose pad on a depression slide. For the 4D confocal data present in Figs. 3, 4, 8, and 9, cells were adhered to concanavalin A-treated confocal dishes. Dishes were prepared as previously described (Casler and Glick, 2019). Briefly, 250 µl of 2 mg/ml concanavalin A in water was added directly to the coverslip of a confocal dish for 10 min. Dishes were washed with water and allowed to dry completely. Cells were adhered by pipetting 250 µl of a mid-log phase cell culture onto the coverslip and incubating for 15 min. Cells were gently washed with media to remove non-adhered cells and the dish was filled with 2–3 ml of media.
Imaging was conducted on three separate confocal microscopes. Microscope 1 was a Leica Spinning Disk Confocal System fitted with a CSU-X1 spinning disk head (Yokogawa). Images were captured using A PLAN APO 63× (1.32 NA objective; Leica) and an Evolve 512 Delta EMCCD camera (Photometrics). A step size of 0.3 µm was used for all figures. Images were captured with Metamorph (Molecular Devices) and deconvolved using AutoQuant X3’s (Media Cybernetics) iterative, constrained three-dimensional deconvolution method. Microscope 1 was used to capture images shown in Fig. 2, B, C, E, and H; Fig. S1, A and C; and Fig. S2 A. Microscope 2 was a Leica SP8 confocal equipped with HyD detectors and a 63× 1.4NA oil immersion objective. No images from this system are displayed in figures but they were used for some of the quantification of PI(4)P biosensors in Figs. 5, 6, and 7. Microscope 3 was a Nikon SoRa Spinning Disk Confocal System equipped with a 60× 1.42 NA oil immersion objective and a Hamamatsu ORCA Fusion Digital CMOS camera. Images were captured with a 0.25–0.3 µm step size and deconvolved using NIS Elements software. Microscope 3 was used to capture images shown in Fig. 1; Fig. 2, D and F; Figs. 3, 4, 5, 6, 7, 8, and 9; Fig. S1, B and D; and Figs. S3, S4, and S5. All images displayed are deconvolved and whether they are projections or single slices is indicated in the figure legends. For super-resolution images shown in Fig. 5 D; Fig. 7, A and B; and Fig. S5, C and D, the same Nikon SoRa system was used with the SoRa disk in place, which enables optical photon reassignment. Images from the Nikon SoRa were deconvolved using the NIS-Elements (Nikon) software.
HaloTag labeling
Labeling and imaging of HaloTag-tagged fusion proteins were performed as previously described with slight modifications (Casler et al., 2019). JFX650 HaloTag ligand (Grimm et al., 2021) was added from a 1 mM stock in DMSO to a log-phase yeast culture in SCD pH 6.4 media to a final concentration of 1 μM, and labeling was performed for 30 min at 24°C with shaking. Excess dye was removed by filter washing the cells by pushing 6 ml of fresh medium through a 0.22 μm syringe filter. Washed cells were resuspended by pipetting on the filter, and cells were resuspended in SCD, pH 6.4, and imaged. All HaloTag fusion proteins in this work were conjugated to JFX650 for visualization.
Quantification of fluorescence microscopy data
Summarized below are the methods used to quantify fluorescence microscopy data in this study. All images were quantified using Fiji (Schindelin et al., 2012).
ER localization
The amount of fluorescent protein signal that was masked by an ER marker (quantifications displayed in Fig. 1, G and I; and Fig. 2, G and I) was quantified in FIJI as follows: first, the central four slices of a Z-stack were used to make an average projection. Next, a background value, measured from regions of the frame that did not contain cells, was subtracted from the channel of interest. The signal from the ER marker was smoothed using a Gaussian blur filter and used to generate a mask, and the signal from the channel of interest was measured before and after subtraction of the mask. Each dot on the graphs in the figures indicated above represents the percentage of fluorescent signal that was masked by the ER for a single cell. The three imaging replicates are displayed in different colors and the average of each replicate is indicated by an outlined dot of the appropriate color. The horizontal line represents the average of the three imaging replicate averages. At least 50 cells total were measured per condition.
Mitochondrial morphology
Mitochondrial morphology (quantifications displayed in Fig. 3 B and Fig. 4 E) was quantified as follows: Z-stacks were max projected, and the mitochondrial morphology of individual cells was classified as either reticular or collapsed. Reticular cells had at least 1/3 of the cell cortex containing mitochondria, while collapsed cells had <1/3. For Fig. 4 E, an additional netted category was scored. Netted cells contained hyperfused mitochondrial networks with at least three clearly resolvable fenestrations. Data are from at least 100 cells per replicate from three biological replicates. Each dot in the graph represents the percentage of cells displaying the indicated mitochondrial morphology from a single imaging replicate and the bars represent the average of the three replicates.
Num1-mediated mitochondrial tethering points
The number of Num1-mediated mitochondrial tethering points (quantifications displayed in Fig. 3 D and Fig. S2 B) was quantified as follows: first, Z-stacks from 2–3-min videos were max projected and thresholded so that only the punctate population of Num1 was visible. The thresholding step removes the diffuse pool of Num1 on the cell cortex that does not form functional mitochondrial tethering points, making it easier to identify true tethering points (Kraft and Lackner, 2017). Next, individual cells were manually scored for the total number of Num1-mediated mitochondrial tethering points. To be defined as a tethering point, the Num1 foci needed to persist above the threshold value and be adjacent to a mitochondrion for at least 90 s. If multiple Num1 foci were too close to easily resolve in the max projections, full Z-stacks were examined to determine the correct number of foci. Three imaging replicates were performed. At least 50 cells were measured per condition.
Scs2 fluorescence intensity at Num1 foci
The intensity of Scs2 fluorescence at Num1 foci (quantification displayed in Fig. 7 C) was measured as follows: first, an individual slice from the top of a cell was chosen. Then, the Num1 signal was smoothed with a Gaussian blur filter, thresholded so that only punctate signal remained, and used to generate a mask. The “Analyze Particles” tool in Fiji was used to automatically draw ROIs around the Num1 foci. The Scs2 fluorescence intensity at each region of the Num1 mask was quantified. Only foci that were more than four pixels were analyzed. Each dot in the graph represents the Scs2 fluorescence intensity at a single Num1 focus. Between 805 and 1,754 foci were measured per condition over three imaging replicates.
Localization of PI(4)P biosensors
The cellular localization of the PI(4)P probes GFP-PHOsh2 and P4M (quantifications displayed in Fig. 5 H, Fig. 9 C, and Fig. S4 B) was quantified as follows: Z-stacks of cells expressing either GFP-PHOsh2, GFP-P4M, or mCherry-P4M were thresholded and individual cells were manually scored as one of four mutually exclusive categories. Representative images of the four categories, “Bud enriched + puncta,” “PM + puncta,” “PM,” or “Puncta,” are displayed in Fig. 5 E and Fig. S4 A. The Bud enriched + puncta category included cells that displayed intracellular punctate signal above the threshold value as well as enrichment of the PI(4)P probe on the daughter cell PM. Enrichment of the PI(4)P probe on the daughter cell PM versus the mother cell PM was defined as cells displaying a ratio of the fluorescence intensity of the PI(4)P probe on the PM of the daughter cells to that of the mother cell >2.5 for GFP-PHOsh2 and 1.5 for P4M. The PM + puncta category was defined as cells that displayed PM localized PI(4)P biosensor fluorescence that did not show significant enrichment on the daughter cell versus the mother cell (ratio <2.5 for GFP-PHOsh2 and <1.5 for P4M) as well as intracellular punctate signal. The PM category was defined as cells that showed PM localized fluorescence of the PI(4)P biosensor without intracellular punctate signal above the threshold value. As previously noted, many cells falling into this category did display very weak intracellular punctate structures that were below the threshold used for this quantification (Stefan et al., 2011). Finally, the “Puncta” category was defined as cells that showed intracellular punctate accumulations of the PI(4)P biosensors but not appreciable PM signal. The bars represent the average of the percentage of cells displaying each category measured from three imaging replicates. Each imaging replicate consisted of at least 50 cells and the error bars represent the SEM.
To avoid complications from analyzing cells at very different stages of the cell cycle, only cells with medium bud sizes were analyzed. In addition, to avoid problems from oversaturation, only cells with moderate expression, as determined by fluorescence intensity, of the PI(4)P probes were analyzed (Wills et al., 2018).
Mother–daughter PI(4)P enrichment
Quantification of the ratio of PI(4)P biosensor fluorescence intensity on the PM of the daughter cell compared with the mother cell (quantifications displayed in Fig. 5 J; Fig. 6, B and D; Fig. 7 E; Fig. 9, D, F, and H; and Fig. S4, C and F) was performed as follows: first, regions of the cell cortex devoid of intracellular punctate signal were identified in both the mother and daughter cell. Next, a square region of interest was drawn over the chosen regions of the cell cortex and the fluorescence intensity was quantified (Fig. 5 G). Quantifications were performed on individual slices from the center of a Z-stack. Ratios above one (daughter cell intensity:mother cell intensity) indicate enrichment of the localization of the PI(4)P biosensors on the daughter cell PM. Each dot in the graphs indicated above represents the daughter cell PI(4)P biosensor enrichment ratio measured for an individual cell. The three imaging replicates are displayed in different colors and the average of each replicate is indicated by an outlined dot of the appropriate color. Each imaging replicate contains measurements from 20 cells. Black bars represent the average and error bars represent the SEM of the averages from the three replicates.
Mitochondrial dynamics
Quantification of mitochondrial division and fusion rates (quantifications displayed in Fig. 4, B and C; and Fig. S3 A) was performed as follows: 10-min timelapse videos of cells in various genetic backgrounds expressing the mitochondrial matrix marker MitoRed were captured at 30 s/frame. Individual movies were max-projected, and mitochondrial division and fusion events from individual cells were manually identified. Division events were only counted when two new mitochondrial tips formed from a previously connected tubule. Fusion events were only counted when two clearly distinct mitochondrial tubules fused and remained fused for at least one additional frame. Once putative division or fusion events were identified, the full volume of the Z-stack was examined to ensure that only true events were counted. Each dot in the graph represents the number of division or fusion events measured for an individual cell. The three imaging replicates are displayed in different colors and the average of each replicate is indicated by an outlined dot of the appropriate color. Black bars represent the average of the three replicates.
Coimmunoprecipitation
Coimmunoprecipitation experiments were performed by growing cells in 50 ml of YPD to an OD600 of ∼0.8, washing once with water to remove the media, and freezing the cell pellets in liquid nitrogen. Frozen pellets were thawed by resuspension in 300 µl IPLB (20 mM HEPES, 150 mM KOAc, 2 mM Mg(Ac)2, 1 mM EGTA, and 0.6 M Sorbitol, pH 7.4) + 1× protease inhibitors (539131; Millipore) and 0.1% Triton X-100. Resuspended pellets were lysed by the addition of glass beads and vortexed for 1.5 min seven times with 1 min on ice between vortexes. The lysates were transferred to a fresh tube and cell debris was removed by spinning in a microcentrifuge at 17,000 × g at 4°C for 30 min. GFP-tagged proteins were then immunoprecipitated by adding 25 µl of α-GFP beads (130-091-125; Miltenyi Biotec) to 250 µl of cell lysate followed by incubation on ice for 30 min. Samples were mixed three times by inversion during the incubation. α-GFP beads were isolated on µ columns (Miltenyi Biotec) using a magnetic µMACS separator (Miltenyi Biotec) which had previously been equilibrated with 250 µl IPLB + 1× protease inhibitors and 0.1% Triton X-100. Columns were washed three times with 800 µl IPLB + 1× protease inhibitors and 0.1% Triton X-100 followed by two additional washes with 500 µl IPLB. Samples were eluted with 50 µl MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea). Final samples were boiled for 5 min and spun at 17,000 × g in a microcentrifuge prior to loading for SDS-PAGE and Western blot analysis.
Western blots
For Fig. 8 A, cells were grown to mid-log phase in YPD with 1 mM auxin added for the indicated length of time. Cells were harvested by centrifugation and whole-cell extracts were prepared by NaOH lysis and trichloroacetic acid precipitation. Pellets were resuspended in 50 µl MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea) and analyzed by SDS-PAGE using a 10% Bis-tris gel (NP0302BOX; Invitrogen). After transfer, blots were analyzed with Revert Total Protein Stain (LI-COR Biosciences) according to the manufacturer’s protocol. Blots were then washed and probed with an α-FLAG (F3165; Sigma-Aldrich) antibody at a 1:2,000 dilution. A secondary goat α-mouse antibody (AC2135; Azure Biosystems) was used at a 1:15,000 dilution for detection. Blots were imaged on the Odyssey Infrared Imaging System (LI-COR Biosciences) and quantified using ImageStudio (LI-COR Biosciences). Molecular weight standards are PageRuler Plus Prestained Protein Ladder (26619; Thermo Fisher Scientific).
For the co-IP experiments shown in Fig. 1 D and Fig. 5 F, Western blots were performed the same way except samples were run on a 3–8% tris-acetate SDS-PAGE gel (EA03752BOX; Invitrogen), no total protein stain was used, and blots were probed with α-GFP (AE011; Abclonal) and α-HA (12CA5; Sigma-Aldrich) antibodies at 1:2,000 dilutions. Secondary goat α-rabbit (AC2128; Azure Biosystems) and goat α-mouse antibodies (AC2135 or AC2129; Azure Biosystems) were used at a 1:15,000 dilution for detection.
Immunoprecipitation–mass spectrometry (IP-MS)
For the IP-MS pulldowns of Num1, wild type yeast cells and cells expressing Num1-GFP were grown at 30°C in YPD to an OD600 of ∼1.0. For each immunoprecipitation, 500 OD units of cells were used. Cells were pelleted by centrifugation, washed once with water, and resuspended in an equivalent volume of IPLB (20 mM HEPES, 150 mM KOAc, 2 mM Mg(Ac)2, 1 mM EGTA, and 0.6 M Sorbitol, pH 7.4). The cell suspension was added dropwise to liquid nitrogen to create cell pellets and stored at −80°C. Cell lysis was performed using a freezer mill with a frequency of 30 frequency/s with three 2-min runs followed by a 1-min run. Protease inhibitors were added to a concentration of 1× to the cell lysates. Cell debris was removed by pelleting at 500 × g for 10 min at 4°C in a tabletop centrifuge. Crosslinking was performed by adding DSP to a final concentration of 1 mM from a 100 mM stock in DMSO. The reaction was incubated on ice for 30 min and quenched by the addition of Tris to a final concentration of 100 mM. To solubilize membranes, Triton-X 100 was added to a concentration of 1% and the lysates were cleared by spinning at 12,000 × g for 10 min at 4°C. Immunoprecipitation was performed as detailed above. Following elution of the samples with 50 µl MURB, samples were frozen in liquid nitrogen and stored at −80°C. The samples were boiled for 10 min prior to being loaded on a 4–20% Novex Tris-Glycine WedgeWell Gel (Invitrogen), and the gel was run for 7 min at 200 V. The gel was rinsed with water, stained for 1 h with Bio-Safe Coomassie G-250 Stain (Bio-Rad), and washed with distilled water for 2 × 1 h. The protein band from each sample was then cut from the gel and placed in a microfuge tube. 500 μl of distilled water was added to each tube and the tubes were stored at 4°C. Samples were submitted to the Northwestern Proteomics Core Facility for in-gel trypsin digestion and LC-MS/MS–based protein identification.
Growth assays
Serial dilution growth assays (shown in Fig. 3 G and Fig. 8 C) were performed by growing cells in a YPD medium overnight at 30°C. From the saturated cultures, 0.2 OD600 units were pelleted in a microcentrifuge (3,000 × g for 1 min) and resuspended in 400 µl water. 10-fold serial dilutions were spotted onto plates made of the indicated media and grown at the indicated temperatures for 2 days prior to imaging. YPD medium consists of 1% yeast extract, 2% peptone, and 2% dextrose. YPEG medium consists of 1% yeast extract, 2% peptone, 3% ethanol, and 3% glycerol. Auxin was added to a final concentration of 1 mM from a 500 mM stock in DMSO.
Online supplemental material
In Fig. S1, additional images of ER and PAN or PAN∆FFAT clusters are provided (related to Fig. 2). Fig. S2 characterizes the number of Num1-mediated mitochondrial tethering points in strains lacking combinations of Scs2 and Mdm36 (related to Fig. 3). Fig. S3 demonstrates that the change in mitochondrial division rates upon loss of the Num1–Scs2 interaction is not influenced by tagging Num1. Fig. S3 also demonstrates that loss of the Num1–Scs2 interaction does not alter the localization of Fzo1 or Dnm1 (related to Fig. 4). Fig. S4 provides evidence that an additional PI(4)P biosensor, GFP-P4M, also shows altered localization upon loss of Num1 (related to Fig. 5). In addition, Fig. S4 shows that artificial tethering of mitochondria to the PM is not sufficient to rescue the PI(4)P distribution defect seen upon loss of Num1. Fig. S5 demonstrates that loss of Num1 does not grossly alter the localization of Osh2, Osh3, Osh7, Sac1, or PIK patch components Efr3, Ypp1, and Stt4. Video 1 shows that Num1 tethers mitochondrial and ER membranes over time (still images are shown in Fig. 3 E). Video 2 shows that Num1∆FFAT tethers mitochondrial but not ER membranes over time (still images are shown in Fig. 3 F). Video 3 shows mitochondrial dynamics are altered upon loss of the Num1–Scs2 interaction (still images are shown in Fig. 4 A). Video 4 shows the localization of the PI(4)P biosensor, GFP-PHOsh2, throughout the cell cycle in wild type, num1∆, and sac1∆ cells (still images are shown in Fig. 9 E). Table S1 lists all of the yeast strains used in this study. Table S2 lists all of the plasmids used in this study. Table S3 lists critical oligonucleotides used in this study. Data S1 includes the full dataset from the Num1 IP-MS pulldown experiments depicted in Fig. 5 B.
Supplementary Material
lists all of the yeast strains used in this study.
lists all of the plasmids used in this study.
lists critical oligonucleotides used in this study.
includes the full dataset from the Num1 IP-MS pulldown experiments depicted in Fig. 5 B.
is the source file for Fig. 1.
is the source file for Fig. 5.
is the source file for Fig. 8.
Acknowledgments
We thank members of the Lackner lab for feedback on the manuscript and critical scientific discussions. We also thank Northwestern’s Cell Biology Supergroup and the Wignall-Lackner Cell Biology Group for constructive feedback on the project. Additional thanks to Jessica Hornick and Tong Zhang, who provided assistance with the fluorescence microscopy. The JFX650 dye (Grimm et al., 2021) used in the HaloTag imaging experiments was a kind gift from Luke Lavis at Janelia.
All microscopy was performed at the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), supported by the Chemistry for Life Processes Institute, the Northwestern University Office for Research, the Department of Molecular Biosciences, and the Rice Foundation. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by National Cancer Institute CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from National Institutes of Health Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. J.C. Casler is supported by the National Institutes of Health and the National Institute of General Medical Sciences grant 1F32GM145160-01. C.S. Harper was supported by the National Institutes of Health Biotechnology Training Program 5T32GM008449-25 and the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1842165. A.J. White was supported by the National Institutes of Health and the National Institute of General Medical Sciences grant T32 GM008382. H.L. Anderson was supported by the American Heart Association Predoctoral Fellowship 19PRE3481053. L.L. Lackner is supported by the National Institutes of Health and the National Institute of General Medical Sciences grant R01GM120303.
Author contributions: J.C. Casler: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing - original draft, Writing - review & editing; C.S. Harper: Conceptualization, Investigation; A.J. White: Investigation; H.L. Anderson: Conceptualization; L.L. Lackner: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Data availability
The data are available from the corresponding author upon reasonable request.
References
- Abrisch, R.G., Gumbin S.C., Wisniewski B.T., Lackner L.L., and Voeltz G.K.. 2020. Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J. Cell Biol. 219:201911122. 10.1083/jcb.201911122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, H.L., Casler J.C., and Lackner L.L.. 2022. Hierarchical integration of mitochondrial and nuclear positioning pathways by the Num1 EF hand. Mol. Biol. Cell. 33:ar20. 10.1091/mbc.E21-12-0610-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Baird, D., Stefan C., Audhya A., Weys S., and Emr S.D.. 2008. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 183:1061–1074. 10.1083/jcb.200804003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla, T. 2013. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., and Shaw J.M.. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298–304. 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry, M., and Kim P.K.. 2021. ORP1L mediated PI(4)P signaling at ER-lysosome-mitochondrion three-way contact contributes to mitochondrial division. Nat. Commun. 12:5354. 10.1038/s41467-021-25621-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler, J.C., and Glick B.S.. 2019. Visualizing secretory cargo transport in budding yeast. Curr. Protoc. Cell Biol. 83:e80. 10.1002/cpcb.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler, J.C., Papanikou E., Barrero J.J., and Glick B.S.. 2019. Maturation-driven transport and AP-1-dependent recycling of a secretory cargo in the Golgi. J. Cell Biol. 218:1582–1601. 10.1083/jcb.201807195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, K.L., Studer S.L., Jensen R.E., and Sesaki H.. 2007. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 12:363–375. 10.1016/j.devcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Chao, J.T., Wong A.K.O., Tavassoli S., Young B.P., Chruscicki A., Fang N.N., Howe L.J., Mayor T., Foster L.J., and Loewen C.J.R.. 2014. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 158:620–632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Chen, S., Novick P., and Ferro-Novick S.. 2012. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 14:707–716. 10.1038/ncb2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bel, L.M., and Brill J.A.. 2018. Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic. 19:301–318. 10.1111/tra.12554 [DOI] [PubMed] [Google Scholar]
- Doyle, C.P., Rectenwald A., Timple L., and Hammond G.R.V.. 2024. Orthogonal targeting of SAC1 to mitochondria implicates ORP2 as a major player in PM PI4P turnover. Contact (Thousand Oaks). 7:25152564241229272. 10.1177/25152564241229272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso, A., and Brickner J.H.. 2017. Epigenetic transcriptional memory. Curr. Genet. 63:435–439. 10.1007/s00294-016-0661-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz-Alon, Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Farkasovsky, M., and Küntzel H.. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003–1014. 10.1083/jcb.131.4.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky, M., and Küntzel H.. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152:251–262. 10.1083/jcb.152.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, J.B., Xie L., Casler J.C., Patel R., Tkachuk A.N., Falco N., Choi H., Lippincott-Schwartz J., Brown T.A., Glick B.S., et al. 2021. A general method to improve fluorophores using deuterated auxochromes. JACS Au. 1:690–696. 10.1021/jacsau.1c00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammermeister, M., Schödel K., and Westermann B.. 2010. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2443–2452. 10.1091/mbc.e10-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, G.R.V., Fischer M.J., Anderson K.E., Holdich J., Koteci A., Balla T., and Irvine R.F.. 2012. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 337:727–730. 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, G.R.V., Machner M.P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205:113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, C.S., Casler J.C., and Lackner L.L.. 2023. Temporal control of contact site formation reveals a relationship between mitochondrial division and Num1-mediated mitochondrial tethering. Mol. Biol. Cell. 34:ar108. 10.1091/mbc.E23-05-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., and Shaw J.M.. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373. 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, P.C., Bharat T.A.M., Wozny M.R., Boulanger J., Miller E.A., and Kukulski W.. 2019. Tricalbins contribute to cellular lipid flux and form curved ER-PM contacts that are bridged by rod-shaped structures. Dev. Cell. 51:488–502.e8. 10.1016/j.devcel.2019.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher, C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Hung, V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., and Ting A.Y.. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife. 6:e24463. 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Johnson, P.R., Dolman N.J., Pope M., Vaillant C., Petersen O.H., Tepikin A.V., and Erdemli G.. 2003. Non-uniform distribution of mitochondria in pancreatic acinar cells. Cell Tissue Res. 313:37–45. 10.1007/s00441-003-0741-1 [DOI] [PubMed] [Google Scholar]
- Kakimoto, Y., Tashiro S., Kojima R., Morozumi Y., Endo T., and Tamura Y.. 2018. Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci. Rep. 8:6175. 10.1038/s41598-018-24466-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, S., Tamura Y., Kojima R., Bala S., Asai E., Michel A.H., Kornmann B., Riezman I., Riezman H., Sakae Y., et al. 2018. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J. Cell Biol. 217:959–974. 10.1083/jcb.201704119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker, T., Scholz D., Förtsch J., and Westermann B.. 2013. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J. Cell Sci. 126:2924–2930. 10.1242/jcs.126045 [DOI] [PubMed] [Google Scholar]
- Kormanec, J., Schaaff-Gerstenschläger I., Zimmermann F.K., Perečko D., and Küntzel H.. 1991. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol. Gen. Genet. 230:277–287. 10.1007/BF00290678 [DOI] [PubMed] [Google Scholar]
- Kornmann, B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, L.M., and Lackner L.L.. 2017. Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J. Cell Biol. 216:3061–3071. 10.1083/jcb.201702022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, L.L. 2019. The expanding and unexpected functions of mitochondria contact sites. Trends Cell Biol. 29:580–590. 10.1016/j.tcb.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M.A., Ferguson K.M., O’Brien R., Sigler P.B., and Schlessinger J.. 1995. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 92:10472–10476. 10.1073/pnas.92.23.10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir, M., Grzybek M., Majkowski M., Rajesh S., Kaur J., Whittaker S.B.M., Coskun Ü., and Overduin M.. 2015. Structural basis of dynamic membrane recognition by trans-Golgi network specific FAPP proteins. J. Mol. Biol. 427:966–981. 10.1016/j.jmb.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Levine, T.P., and Munro S.. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 12:1633–1644. 10.1091/mbc.12.6.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T.P., and Munro S.. 2002. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12:695–704. 10.1016/S0960-9822(02)00779-0 [DOI] [PubMed] [Google Scholar]
- Loewen, C.J.R., and Levine T.P.. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280:14097–14104. 10.1074/jbc.M500147200 [DOI] [PubMed] [Google Scholar]
- Loewen, C.J.R., Young B.P., Tavassoli S., and Levine T.P.. 2007. Inheritance of cortical ER in yeast is required for normal septin organization. J. Cell Biol. 179:467–483. 10.1083/jcb.200708205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Manford, A.G., Stefan C.J., Yuan H.L., Macgurn J.A., and Emr S.D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Miller, R.K., and Rose M.D.. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377–390. 10.1083/jcb.140.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, M., Stiller S.B., Lübbert P., Peikert C.D., Dannenmaier S., Drepper F., Weill U., Höß P., Feuerstein R., Gebert M., et al. 2017. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19:2836–2852. 10.1016/j.celrep.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck, J., Čopič A., Delfosse V., Vanni S., Jackson C.L., Bourguet W., and Drin G.. 2015. INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 349:432–436. 10.1126/science.aab1346 [DOI] [PubMed] [Google Scholar]
- Murley, A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2:e00422. 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S.E., and Levine T.P.. 2016. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 1861:952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Nagashima, S., Tábara L.C., Tilokani L., Paupe V., Anand H., Pogson J.H., Zunino R., McBride H.M., and Prudent J.. 2020. Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science. 367:1366–1371. 10.1126/science.aax6089 [DOI] [PubMed] [Google Scholar]
- Neupert, W., and Herrmann J.M.. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749. 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Nishimura, K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nunnari, J., Marshall W.F., Straight A., Murray A., Sedat J.W., and Walter P.. 1997. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 8:1233–1242. 10.1091/mbc.8.7.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, S., Greenberg S.R., and Lee W.L.. 2018. Cortical dynein pulling mechanism is regulated by differentially targeted attachment molecule Num1. Elife. 7:e36745. 10.7554/eLife.36745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, S., Brock K., Beckford J., and Lee W.L.. 2020. Overexpression of Mdm36 reveals Num1 foci that mediate dynein-dependent microtubule sliding in budding yeast. J. Cell Sci. 133:jcs246363. 10.1242/jcs.246363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omnus, D.J., Cadou A., Thomas F.B., Bader J.M., Soh N., Chung G.H.C., Vaughan A.N., and Stefan C.J.. 2020. A heat-sensitive Osh protein controls PI4P polarity. BMC Biol. 18:28. 10.1186/s12915-020-0758-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton, J.G., Kim Y.J., Humpolickova J., Eisenreichova A., Sengupta N., Toth D.J., Boura E., and Balla T.. 2020. Defining the subcellular distribution and metabolic channeling of phosphatidylinositol. J. Cell Biol. 219:e201906130. 10.1083/jcb.201906130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, G.A., Tjong J., Brown J.M., Poquiz P.H., Scott R.T., Kolson D.R., Ellisman M.H., and Spirou G.A.. 2010. The micro-architecture of mitochondria at active zones: Electron tomography reveals novel anchoring scaffolds and cristae structured for high-rate metabolism. J. Neurosci. 30:1015–1026. 10.1523/JNEUROSCI.1517-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping, H.A., Kraft L.M., Chen W., Nilles A.E., and Lackner L.L.. 2016. Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J. Cell Biol. 213:513–524. 10.1083/jcb.201511021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon, E., Sere Y.Y., Chauhan N., Johansen J., Sullivan D.P., Dittman J.S., Rice W.J., Chan R.B., Di Paolo G., Beh C.T., and Menon A.K.. 2018. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol. 16:e2003864. 10.1371/journal.pbio.2003864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser, M., Kuhl H., Ralser M., Werber M., Lehrach H., Breitenbach M., and Timmermann B.. 2012. The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: Insights into ancestry and physiology of a laboratory mutt. Open Biol. 2:120093. 10.1098/rsob.120093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, J., Zahedi R.P., Pfanner N., Meisinger C., and Sickmann A.. 2006. Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5:1543–1554. 10.1021/pr050477f [DOI] [PubMed] [Google Scholar]
- Reinisch, K.M., and Prinz W.A.. 2021. Mechanisms of nonvesicular lipid transport. J. Cell Biol. 220:e202012058. 10.1083/jcb.202012058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A., and Levine T.P.. 2004. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279:44683–44689. 10.1074/jbc.M401583200 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit, H.L., Kraft L.M., Lee-Smith C.F., and Lackner L.L.. 2018. The role of mitochondria in anchoring dynein to the cell cortex extends beyond clustering the anchor protein. Cell Cycle. 17:1345–1357. 10.1080/15384101.2018.1480226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, T.A., Choi M.G., Raychaudhuri S., Mears J.A., Ghirlando R., Hinshaw J.E., and Prinz W.A.. 2009. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 187:889–903. 10.1083/jcb.200905007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano, L., De Matteis M.A., Emr S., Giordano F., Hajnóczky G., Kornmann B., Lackner L.L., Levine T.P., Pellegrini L., Reinisch K., et al. 2019. Coming together to define membrane contact sites. Nat. Commun. 10:1287. 10.1038/s41467-019-09253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen R.E.. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699–706. 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai, N., Yifrach E., van Roermund C.W.T., Cohen N., Bibi C., IJlst L., Cavellini L., Meurisse J., Schuster R., Zada L., et al. 2018. Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9:1761. 10.1038/s41467-018-03957-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Slee, J.A., and Levine T.P.. 2019. Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum. Contact (Thousand Oaks). 2:1–21. 10.1177/2515256419883136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan, C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., and Emr S.D.. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401. 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Stradalova, V., Blazikova M., Grossmann G., Opekarová M., Tanner W., and Malinsky J.. 2012. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PLoS One. 7:e35132. 10.1371/journal.pone.0035132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, K.A., Jochem A., Le Vasseur M., Lewis S., Paulson B.R., Reddy T.R., Russell J.D., Coon J.J., Pagliarini D.J., and Nunnari J.. 2019. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol. 218:1352–1368. 10.1083/jcb.201808044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Punch J.J., and Lee W.L.. 2009. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 8:3182–3190. 10.4161/cc.8.19.9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Germain B.S., and Lee W.L.. 2012. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J. Cell Biol. 196:743–756. 10.1083/jcb.201112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente-Felix, A., Wilson E.L., and Whitworth A.J.. 2020. Drosophila phosphatidylinositol-4 kinase fwd promotes mitochondrial fission and can suppress Pink1/parkin phenotypes. PLoS Genet. 16:e1008844. 10.1371/journal.pgen.1008844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F.B., Omnus D.J., Bader J.M., Chung G.H.C., Kono N., and Stefan C.J.. 2022. Tricalbin proteins regulate plasma membrane phospholipid homeostasis. Life Sci. Alliance. 5:1–23. 10.26508/lsa.202201430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak V., Naylor K., and Nunnari J.. 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158:445–452. 10.1083/jcb.200205031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., and Walter P.. 2006. Eisosomes mark static sites of endocytosis. Nature. 439:998–1003. 10.1038/nature04472 [DOI] [PubMed] [Google Scholar]
- West, M., Zurek N., Hoenger A., and Voeltz G.K.. 2011. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193:333–346. 10.1083/jcb.201011039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A.J., Harper C.S., Rosario E.M., Dietz J.V., Addis H.G., Fox J.L., Khalimonchuk O., and Lackner L.L.. 2022. Loss of Num1-mediated cortical dynein anchoring negatively impacts respiratory growth. J. Cell Sci. 135:jcs259980. 10.1242/jcs.259980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills, R.C., Goulden B.D., and Hammond G.R.V.. 2018. Genetically encoded lipid biosensors. Mol. Biol. Cell. 29:1526–1532. 10.1091/mbc.E17-12-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., Skibbens R.V., Cheng J.W., Salmon E.D., and Bloom K.. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700. 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.W., Mendrola J.M., Audhya A., Singh S., Keleti D., DeWald D.B., Murray D., Emr S.D., and Lemmon M.A.. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 13:677–688. 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Zaman, M.F., Nenadic A., Radojičić A., Rosado A., and Beh C.T.. 2020. Sticking with it: ER-PM membrane contact sites as a coordinating nexus for regulating lipids and proteins at the cell cortex. Front. Cell Dev. Biol. 8:675. 10.3389/fcell.2020.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe, J.P., Wills R.C., Sangappa S., Goulden B.D., and Hammond G.R.V.. 2018. SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. Elife. 7:e35588. 10.7554/eLife.35588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zewe, J.P., Miller A.M., Sangappa S., Wills R.C., Goulden B.D., and Hammond G.R.V.. 2020. Probing the subcellular distribution of phosphatidylinositol reveals a surprising lack at the plasma membrane. J. Cell Biol. 219:e201906127. 10.1083/jcb.201906127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists all of the yeast strains used in this study.
lists all of the plasmids used in this study.
lists critical oligonucleotides used in this study.
includes the full dataset from the Num1 IP-MS pulldown experiments depicted in Fig. 5 B.
is the source file for Fig. 1.
is the source file for Fig. 5.
is the source file for Fig. 8.
Data Availability Statement
The data are available from the corresponding author upon reasonable request.