Abstract
The control of flowering time is essential for reproductive success and has a major effect on seed and fruit yield and other important agricultural traits in crops. Nuclear factors Y (NF-Ys) are transcription factors that form heterotrimeric protein complexes to regulate gene expression required for diverse biological processes, including flowering time control in plants. However, to our knowledge, there has been no report on mutants of individual NF-YA subunits that promote early flowering phenotype in plants. In this study, we identified SlNF-YA3b, encoding a member of the NF-Y transcription factor family, as a key gene regulating flowering time in tomato. Knockout of NF-YA3b resulted in an early flowering phenotype in tomato, whereas overexpression of NF-YA3b delayed flowering in transgenic tomato plants. NF-YA3b was demonstrated to form heterotrimeric protein complexes with multiple NF-YB/NF-YC heterodimers in yeast three-hybrid assays. Biochemical evidence indicated that NF-YA3b directly binds to the CCAAT cis-elements of the SINGLE FLOWER TRUSS (SFT) promoter to suppress its gene expression. These findings uncovered a critical role of NF-YA3b in regulating flowering time in tomato and could be applied to the management of flowering time in crops.
Introduction
Flowering is an important transition in flowering plants from vegetative to reproductive growth. In agricultural production, flowering is not only an important stage for the transfer of genetic material from parental plants to their offspring but also a prerequisite to producing fruits and seeds [[4]]. The timing of floral transition plays an essential role in the control of plant fertility, plant yield quality, and other important agricultural traits in crops.
In Arabidopsis, the FLOWERING LOCUS T (FT) protein is widely believed to be the key component of the elusive flowering hormone florigen, which plays a pivotal role in flowering time control. FT serves as the long-distance signal that is expressed in the phloem cells of leaf veins and transported from leaves to the shoot apex to induce the initiation of floral primordia in Arabidopsis [10, 19]. Heading date 3a (Hd3a), the rice ortholog of Arabidopsis FT, plays a similar role in the induction of flowering in rice [48]. In the shoot apex, FT generates a protein complex with a 14-3-3 protein and FLOWERING LOCUS D (FD) to promote flowering [49, 55]. Tomato SINGLE FLOWER TRUSS (SFT) is a homolog of Arabidopsis FT and its overexpression in transgenic tomato plants leads to early flowering, as is the case for FT overexpression in Arabidopsis [50]. In addition, SFT has an effect on the development of flower and inflorescence morphology in tomato. The sft mutant plants not only have a delayed flowering phenotype but also develop a single inflorescence with flowers having large sepals [31].
NF-Y transcription factors are sometimes referred to as CCAAT-binding factors (CBFs) or hemo-activator proteins (HAPs). They are a highly prevalent and evolutionary conserved class of transcription factors that are found in yeast, animals, and plants [11]. According to the presence of different structural features, NF-Y subunits can be classified into three major groups, NF-YA (HAP2/CBF-B), NF-YB (HAP3/CBF-A), and NF-YC (HAP5/CBF-C) [24]. The tomato genome has the most NF-Y genes, containing 10 NF-YAs, 29 NF-YBs, and 20 NF-YCs [28]. In plants, NF-YB and NF-YC subunits form dimers through the interaction between their histone folding domains (HFDs) [15, 42], subsequently recruiting NF-YA to form an NF-Y heterotrimeric protein complex [20]. NF-YA can recognize and bind to specific CCAAT-box cis-elements of promoters and enhancers from target genes [6]. Several studies have demonstrated that NF-Ys are essential for the development of symbiotic root nodules, flavonoid biosynthesis, photomorphogenesis, photosynthesis, abscisic acid (ABA)-regulated seed germination, response to stress, and reproductive development [7, 22, 23, 25, 40, 41, 43].
The NF-Y transcription factor family plays a crucial role in flowering regulation [3, 16, 24, 41, 54]. It has been demonstrated that overexpression of a series of individual NF-Y subunits, including NF-YA1/4/8, NF-YB1/2/3, and NF-YC1/2/3/4/9, can change flowering times in transgenic plants [7, 16–18, 22, 23, 47, 62]. The DNA binding domain of CONSTANS (CO) is homologous to that of NF-YA, and various NF-YB and NF-YC subunits can interact with CO to form NF-Y/CO heterotrimeric complexes that mediate CO-regulated flowering processes [1, 14, 24, 58]. Further studies suggest that the NF-YB/NF-YC dimer interacts with CO, Heading Date1 (HD1), and other proteins that contain the structural domain of TIMING of CAB EXPRESSION1 (CCT), and this distinctive structure of CCT allows its specific targeting to the conserved CCACA motif from the promoters of its target genes to regulate flowering [8, 44]. Furthermore, the C-terminal CCT structural domain of CO forms a complex with NF-YB/NF-YC that recognizes multiple cis-elements in the FT promoter, and the N-terminal tandem B-box structural domain exhibits a head-to-tail oligomeric conformation to form a homopolymer and mediates FT activation, and these multivalent bindings give the CO-NF-Y complex high affinity and specificity [35, 60]. Overexpression of AtNF-YA1 and AtNF-YA4 may compete with CO for binding to the NF-YB/NF-YC dimer, resulting in reduced transcript levels of FT and causing delayed flowering [26, 33, 37]. Moreover, overexpression of AtNF-YC1 and AtNF-YC2 results in elevated transcript levels of FT and accelerated flowering process, and, vice versa, mutations in the AtHAP3b (an NF-YB gene) gene lead to down-regulated transcript levels of FT and delayed flowering time [16]. In rice, OsNF-YB11 (DTH8/Ghd8/LHD1) suppresses the expression of flowering-associated genes and delays photoperiod-induced flowering [12, 53]. Overexpression of HvNF-YB1 in barley is found to promote early flowering. In wheat, NF-Y interacts with Vernalization Gene 2 (VRN2) and Constans 2 (CO2) in vivo and in vitro, playing an important role in integrating vernalization and photoperiodic signals to regulate the flowering time [29]. In addition, NF-Y transcription factors participate in the gibberellic acid (GA) signaling pathway-mediated flowering time control and the microRNA-mediated age-dependent flowering time regulation [17, 52, 61, 62].
Previous studies on the effect of NF-Y on flowering time have mainly been focused on model plants such as Arabidopsis and rice. To our knowledge, there have been few studies on the effect of NF-YA transcription factors on flowering time control in tomato. Here, we report that NF-YA3b negatively regulated flowering time by binding to the CCAAT cis-element of the SFT promoter. As compared to wild-type (WT) tomato plants, NF-YA3b knockout lines had significantly earlier flowering time, and the SFT transcript level was also significantly up-regulated. We demonstrated that NF-YA3b was recruited by multiple NF-YB/NF-YC heterodimers to generate heterotrimeric complexes in yeast three-hybrid (Y3H) assays. The direct binding of NF-YA3b to the CCAAT cis-element of the SFT promoter was verified in the yeast one-hybrid (Y1H) system, the dual luciferase reporter system, and the electrophoretic mobility shift assay (EMSA). These findings demonstrate the critical role of NF-YA3b in regulating flowering time in tomato and provide an opportunity for using the NF-YA3b gene as a target of genetic manipulation for flowering time management in crops using the CRISPR/Cas9 system.
Results
Characterization of SlNF-YA3b
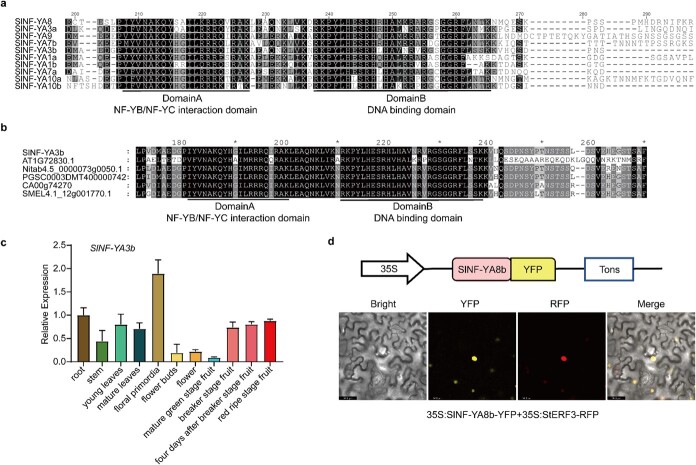
In our previous work, we have shown that NF-Y plays a crucial role in regulating flavonoid biosynthesis in tomato [51]. To understand whether the NF-YA gene family might participate in the regulation of other important physiological and developmental processes, we performed a systematic bioinformatic analysis of the NF-YA family and investigated their possible effects on the regulation of tomato flowering. There are 10 NF-YA genes in the tomato genome. Protein sequence alignment of the 10 NF-YA members (SlNF-YA1a/1b/3a/3b/7a/7b/8/9/10a/10b) in tomato using MEGA10 and GeneDoc software revealed the presence of a highly conserved amino acid region among the NF-YA members (Fig. 1a). This conserved NF-YA region comprises the NF-YB/NF-YC interaction domain (Domain A) and the DNA binding domain (Domain B), separated by a short spacer of 9–10 amino acids that are also highly conserved (Fig. 1a). The presence of the conserved NF-YA region and its Domain A and Domain B structures are also found in the NF-YA counterparts from other plant species, including Arabidopsis, pepper, eggplant, tobacco, and potato (Fig. 1b).
Figure 1.
Characterization of SlNF-YA3b. a, b Protein sequence alignments of 10 tomato NF-YA members (a) and representatives of NF-YA counterparts from other plant species (b). A highly conserved NF-YA region comprises Domain A and Domain B, which are underlined. Protein sequences of the 10 tomato NF-YA members (a) and their counterparts from other plants were obtained from GenBank and other databases, including protein sequences for tomato (SlNF-YA3b, Solyc12g009050.1), Arabidopsis (AT1G72830.1), pepper (CA00g74270), eggplant (SMEL4.1_12g001770.1), tobacco (Nitab4.5_0000073g0050.1), and potato (PGSC0003DMT400000742). c Relative expression levels of NF-YA3b in different tissues of WT plants. Each statistic is displayed as a mean value ± standard error (n = 3). d Subcellular localization of an NF-YA3b-YFP fusion protein. Diagram of the construct used for subcellular localization (upper panel). TNOS, transcription termination sequence of the Nopaline Synthase (NOS) gene. Potato Ethylene Responsive Factor 3 (StERF3) tagged with an RFP served as a nuclear localization marker (StERF3-RFP) and was co-expressed transiently with NF-YA3b-YFP in tobacco leaves. Confocal microscopy was used to capture the fluorescence images. Scale bars, 36.8 μm.
In this study, we focused on SlNF-YA3b (Solyc12g009050), because it was found to be expressed highly in the floral primordia (Fig. 1c) and this expression pattern would imply a possible role of SlNF-YA3b in tomato flowering time control. The open reading frame of SlNF-YA3b is 762 bp long and encodes a polypeptide of 253 amino acids. The SlNF-YA3b gene expression profile was investigated using different tissues of WT tomato ‘Ailsa Craig’ (AC) plants. SlNF-YA3b was found to be expressed in all tissues tested, with the highest level of expression in the floral primordia (Fig. 1c). The expression levels of SlNF-YA3b were found to decrease drastically in flower buds and flowers, suggesting a role of SlNF-YA3b in the induction and early development of floral primordia, but not in the formation of flowers and young fruits. To investigate the subcellular localization of SlNF-YA3b, we expressed a yellow fluorescent protein (YFP)-tagged fusion protein of NF-YA3b under the CaMV35S promoter (35S:SlNF-YA3b-YFP) transiently in tobacco leaves, along with the co-expression of the nuclear localization marker of potato Ethylene Responsive Factor 3 (StERF3), which was tagged with the red fluorescent protein (RFP) under the CaMV35S promoter (35S:StERF3-RFP). As observed by confocal microscopy, the yellow fluorescent signal of SlNF-YA3b-YFP was detected in the nuclei and was found to be overlapped completely with the red fluorescent signal of the nucleus localization maker (StERF3-RFP) (Fig. 1d). The full-length coding sequence (CDS) of NF-YA3b was cloned into pGBKT7 vector, which was used for transformation of yeast AH109 strain to test the transcriptional activity of NF-YA3b. Transformed yeast cells with NF-YA3b-BD were grown on SD/−Trp−His medium with X-α-gal, whereas yeast cells containing the empty pGBKT7 vector did not grow well on the same medium. This result suggests that NF-YA3b has transcriptional activity to drive the expression of the HIS3 selection marker (Supplementary Data Fig. S1). This nucleus localization pattern of SlNF-YA3b protein and the transcriptional activity of NF-YA3b are consistent with its function as a transcription factor.
SlNF-YA3b negatively regulates flowering time in tomato
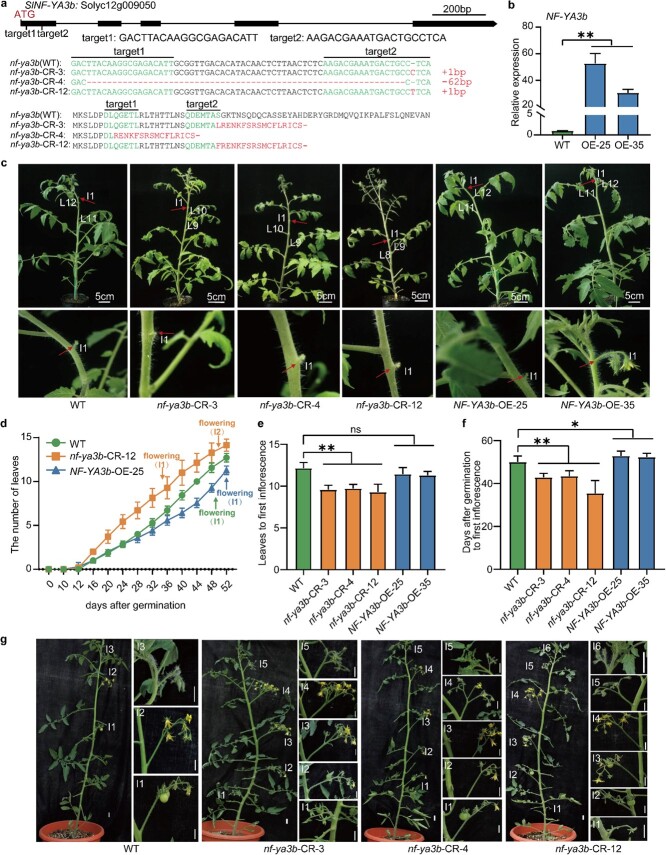
To explore the function of SlNF-YA3b in tomato, we created both SlNF-YA3b knockout mutant transgenic lines and overexpression lines in the tomato AC genetic background. The SlNF-YA3b knockout lines were created using CRISPR/Cas9 technology. Two targets were designed on the first exon of SlNF-YA3b. Analysis of genomic DNA sequences in the SlNF-YA3b knockout lines (CR-3, CR-4, and CR-12) showed that 1 bp was inserted in nf-ya3b-CR-3, 62 bp were deleted in nf-ya3b-CR-4, and 1 bp was inserted in nf-ya3b-CR-12, compared with the gDNA sequence of the WT plants (Fig. 2a). The insertion or deletion of nucleotide bases in the genomic DNA of SlNF-YA3b in the SlNF-YA3b knockout lines (CR-3, -4, and -12) would result in a change in the open reading frame of SlNF-YA3b, leading to the loss of the function of NF-YA3b (Fig. 2a). For the overexpression lines (OE-25 and OE-35), the expression levels of NF-YA3b were ~52- and ~30-fold higher in OE-25 and OE-35, respectively, than that in AC plants (Fig. 2b).
Figure 2.
Flowering time phenotypes of NF-YA3b transgenic tomato lines. a Generation of NF-YA3b mutant lines (CR-3, CR-4, and CR-12) using the CRISPR/Cas9 system. Two sgRNAs (target1 and 2) used for CRISPR were designed to target exon 1 of the NF-YA3b gene, which contains five exons (upper panel). The nucleotide sequences of the two sgRNAs (target1 and 2) in the recipient plant (WT) of the nf-ya3b mutant and the knockout lines (nf-ya3b CR-3, 4, 12) are shown in green letters, while the CRISPR-edited sequences of the NF-YA3b gene are indicated by red dash symbols for base deletions and red letters for insertions (middle panel). The deduced amino acid sequences of the NF-YA3b gene in the nf-ya3b mutant and the knockout lines are shown in the lower panel. b Relative expression levels of NF-YA3b in NF-YA3b-OE lines relative to the WT. The expression level of NF-YA3b in WT was set at 1.0. c Eight-week-old WT, nf-ya3b-CR, and NF-YA3b-OE plants. WT, NF-YA3b knockout, and NF-YA3b-OE lines are shown. The positions of the first inflorescences (I1) and leaf (L) numbers are denoted. High-magnification images of the first inflorescence (I1) from WT, nf-ya3b-CR lines, and NF-YA3b-OE lines (lower panel). d Flowering time of nf-ya3b-CR lines (CR-12), NF-YA3b-OE lines (OE-25), and WT plants. The arrows indicating the first and second inflorescences’ development, respectively, represent flowering (I1) and flowering (I2). e, f Flowering time was measured relative to the number of real leaves (e) and relative to the number of days after seed germination (f) below the first inflorescences in WT, nf-ya3b-CR lines, and NF-YA3b-OE lines. Each statistic is displayed as a mean value ± standard error (n = 7). ns, not statistically significant; *P < 0.05; **P < 0.01 (t-test). g Ten-week-old WT and nf-ya3b-CR plants. The positions of the inflorescences are indicated with I-numbers.
Under normal growth conditions, the NF-YA3b knockout and overexpression lines had significant differences in flowering time from the WT plants (Fig. 2c–f and Supplementary Data Figs S2 and S3). We counted the number of real leaves under the first inflorescence and assessed the period of flowering (days) between seed germination and the first inflorescence’s development. The average number of real leaves under the first inflorescence was 9–10 in the NF-YA3b knockout mutant lines, compared with 12–13 in the WT plants (Fig. 2c and e). The days after seed germination for the inflorescence’s development were notably fewer in NF-YA3b knockout mutant lines than in the WT plants. In WT plants, it took around 48 days after seed germination for the inflorescence’s development, whereas in the NF-YA3b knockout lines it took just 38–44 days (Fig. 2d and f and Supplementary Data Fig. S2). Additionally, there were more inflorescences (five or six inflorescences) in the NF-YA3b knockout lines compared with the three inflorescences in the WT plants with the same growth time (Fig. 2g). Taken together, these results showed that SlNF-YA3b knockout results in early flowering, whereas SlNF-YA3b overexpression in tomato AC plants leads to late flowering (Fig. 2c–f and Supplementary Data Fig. S3). The number of real leaves under the first inflorescence showed no difference between the WT and NF-YA3b-OE lines, but the number of days after seed germination for the first inflorescence’s development were increased in the NF-YA3b-OE lines (Fig. 2c–f and Supplementary Data Fig. S3). The phenotypic observation of SlNF-YA3b transgenic lines suggested that SlNF-YA3b functions as a flowering time repressor in tomato.
Figure 3.
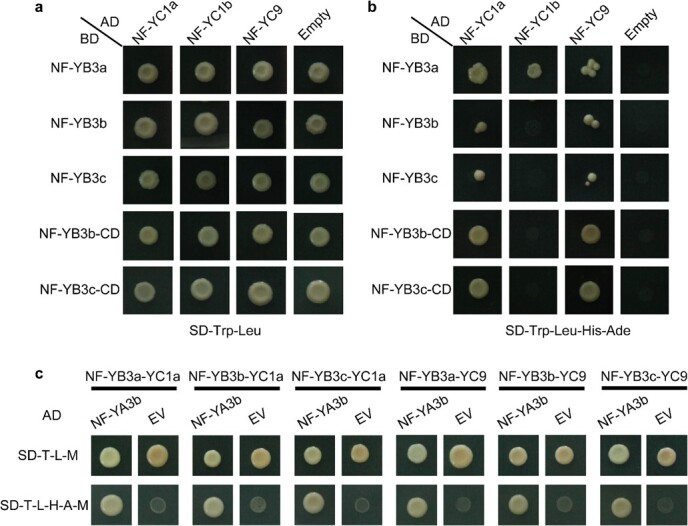
Interaction of NF-YA3b with NF-YB/NF-YC heterodimers and assembly of NF-Y complexes in yeast cells. a, b Y2H experiments for interactions among NF-YB3a/3b/3c and NF-YC1a/1b/9. NF-YB3b-CD and NF-YB3c-CD represent the truncated versions of NF-YB3b and NF-YB3c, respectively. The negative control was an empty pGADT7 vector. c Y3H experiments for interactions between NF-YA3b and the NF-YB3a-YC1a, NF-YB3b-YC1a, NF-YB3c-YC1a, NF-YB3a-YC9, NF-YB3b-YC9, and NF-YB3c-YC9 heterodimers. The negative control was an empty pGADT7 vector (EV).
SlNF-YA3b was recruited by NF-YB/NF-YC heterodimers, assembling NF-Y complexes
Previous studies revealed that the motifs of NF-YB and NF-YC associate with each other in the cytoplasm, becoming a heterodimer, which subsequently translocates to the nucleus and recruits an NF-YA subunit to form the mature NF-Y heterotrimeric complex [20]. We selected three NF-YB family members (NF-YB3a/3b/3c) and three NF-YC family members (NF-YC1a/1b/9) that have reportedly been related to the regulation of flowering time in Arabidopsis to verify whether NF-YA3b is recruited by the interactions between the above NF-YBs and NF-YC using yeast hybrid experiments, respectively.
Analysis of toxicity tests found that the full-length NF-YB3b and NF-YB3c proteins are toxic to yeast cells (Supplementary Data Table S3). Therefore, the full-length and truncated versions of NF-YB3b and NF-YB3c were used in Y2H experiments (Fig. 3a). NF-YB3a interacted with NF-YC1a/9 and NF-YC1b. In addition, truncated versions of NF-YB3b/3c interacted with NF-YC1a/9 but not with NF-YC1b (Fig. 3a). Subsequently, we used Y3H assays to test whether the six heterodimers formed by NF-YB3a, NF-YB3b, and NF-YB3c interacting with NF-YC1a and NF-YC9 could recruit SlNF-YA3b and form the complete NF-Y complex. Yeast cells expressing SlNF-YA3b alone and the six heterodimers were cultured on yeast transformation and interaction-selection media (Fig. 3c). However, yeast cells expressing SlNF-YA3b alone and NF-YB subunits did not grow on SD/−Trp/−Leu/−His/−Ade medium (Supplementary Data Fig. S4). The above results suggested that NF-YA3b does not interact with NF-YB subunits without NF-YC subunits. However, NF-YA3b could be recruited by the NF-YB/NF-YC heterodimers to assemble into the higher-order NF-Y complexes.
NF-Y complexes do not change SlSFT promoter activity
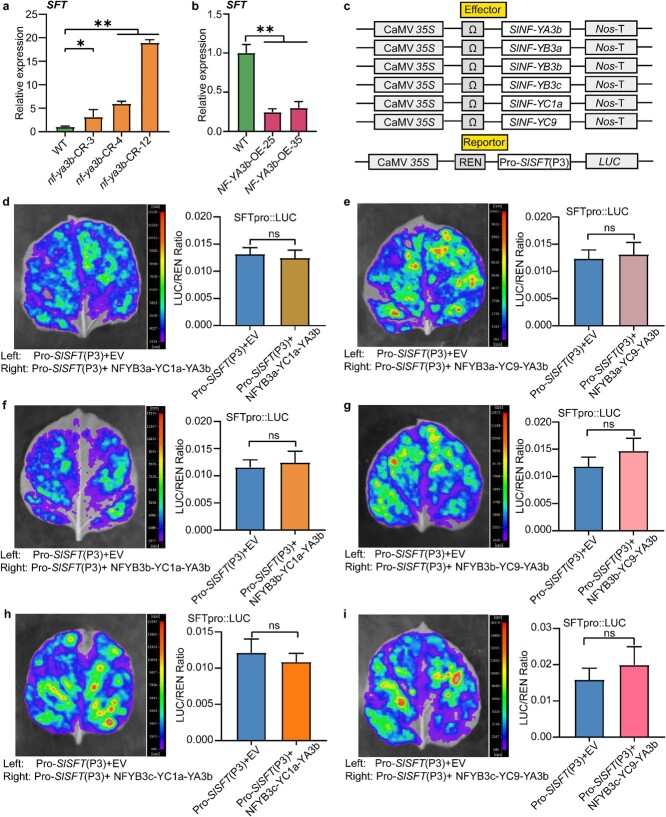
SFT is the ortholog of Arabidopsis FT and is considered to be the putative florigen in tomato [21, 32]. There are ~20 true leaves below the first inflorescence in the sft mutant compared with only 10–12 leaves in the control plants [36]. Thus, sft is considered a late-flowering mutant of tomato [36]. Overexpression of SFT led to early flowering, developing the first inflorescence after three to five real leaves [31]. In this work, nf-ya3b mutant lines and SFT overexpression lines were found to have similar phenotypes in flowering time (Fig. 2). Therefore, we speculated that NF-YA3b may negatively regulate the expression of SFT. To test this idea, we detected the expression levels of the SFT gene in the nf-ya3b mutant lines. SFT expression was significantly up-regulated in the nf-ya3b mutant lines (Fig. 4a) and down-regulated in the NF-YA3b-OE lines (Fig. 4b), suggesting that SlNF-YA3b controls tomato flowering time by inhibiting SFT expression.
Figure 4.
Effect of NF-Y complexes on SlSFT promoter activity. a, b Relative expression levels of SFT in nf-ya3b-CR lines (a) and NF-YA3b-OE lines (b). SFT gene expression in WT plants was set at 1. n = 3; *P < 0.05; **P < 0.01 (t-test). c Dual-luciferase reporter assays. NF-YA3b, NF-YB3a/3b/3c, and NF-YC1a/9 were expressed from pGreenII 62-SK with the CaMV 35S promoter and used as the effector. Pro-SlSFT (P3):LUC served as the reporter and was expressed from pGreenII 0800-LUC. Pro-SlSFT (P3) was a fragment of the SFT promoter containing boxes 1–7, which is 4150 bp in length upstream of the translation start site (TSS). d–i Representative images of luciferase activity (left) and ratios of firefly luciferase (LUC)/Renilla luciferase (REN) activities (right). In the control, the empty vector (EV) was used as the effector. Co-expression of the reporter Pro-SlSFT (P3):LUC with different effector vectors (NFYB3a-YC1a-YA3b, NFYB3a-YC9-YA3b, NFYB3b-YC1a-YA3b, NFYB3b-YC9-YA3b, NFYB3c-YC1a-YA3b, and NFYB3c-YC9-YA3b) was examined in N. benthamiana leaves. Values are expressed as mean ± standard error (n = 8); ns, not statistically significant (t-test).
Previous studies have shown that the NF-Y complex functions by recognizing and binding the key cis-regulatory elements on the target gene promoter by the NF-YA subunit [24]. Our results of Y3H experiments showed that SlNF-YA3b could be recruited by the NF-YB/NF-YC heterodimer to form the NF-Y complex (Fig. 3). To verify whether the NF-Y complex could regulate SFT expression, we performed dual-luciferase reporter experiments. Co-expression of SFTpro:LUC with any NF-Y complex had no significant effect on LUC activity compared with LUC activity in transgenic plants expressing the empty vector (Fig. 4c–i), suggesting that NF-Y complexes do not change SFT promoter activity.
SlNF-YA3b binds to the CCAAT element of the SFT promoter and represses its expression
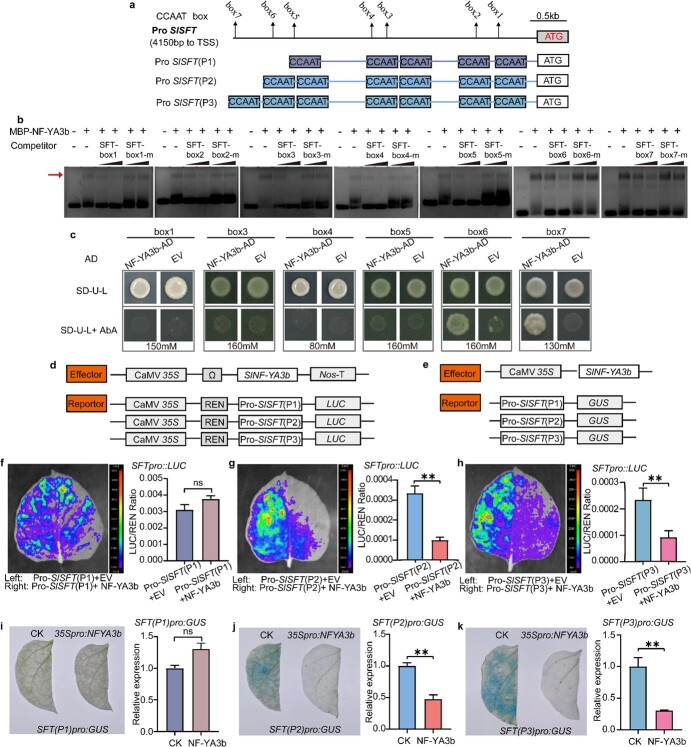
Previous findings have shown that the NF-YA subunit of NF-Y complexes is the protein component that specifically binds to the CCAAT box of the promoters of target genes [38]. Seven CCAAT cis-elements (boxes 1–7) were identified in the 4150-bp genomic DNA fragment upstream of the SFT promoter (Fig. 5a). To find out whether NF-YA3b could bind to the CCAAT cis-element of the SFT promoter in vitro, we performed EMSA assays. In these assays, NF-YA3b protein was expressed in and purified from Escherichia coli. Seven oligonucleotides of 39 bp each, containing the core CCAAT box 1, 2, 3, 4, 5, 6, and 7 sequences from the SFT promoter, respectively, were labeled with the fluorescent dye 6-carboxyfluorescein (FAM), which absorbs 495-nm light and emits a 517-nm signal. Unlabeled oligonucleotides were used as the competitive binding targets. To assess the specificity of binding, mutant oligonucleotides where the CCAAT cis-element was changed to CCCCC or AAAAA were used as unlabeled mutant oligonucleotide competitors. When NF-YA3b protein was incubated with FAM-labeled probes corresponding to sequences of boxes 1, 2, 3, 4, 5, 6, and 7, DNA–protein complexes were detected with retarded mobility on EMSA, suggesting that NF-YA3b binds to the seven CCAAT cis-elements (boxes 1–7) of the SFT promoter (Fig. 5b). When an excessive and increasing amount of unlabeled competitive probes was added to the assays, the signals of the DNA–protein complexes decreased or even disappeared, whereas when the same amount of mutant oligonucleotides (SFT boxes 1/2/3/4/5/6/7-m) was added, the signals of the DNA–protein complexes did not change (Fig. 5b), suggesting that NF-YA3b recognizes and binds to the specific DNA sequence of CCAAT in the 4150-bp genomic DNA fragment upstream of the SFT promoter. In conclusion, these results indicate that NF-YA3b directly binds to the seven CCAAT cis-elements in the 4150-bp genomic DNA fragment upstream of the SFT promoter in vitro.
Figure 5.
Specific binding of SlNF-YA3b to the CCAAT cis-elements of the SFT promoter. a Schematic diagram of the 4150-bp SFT promoter region. Seven CCAAT (boxes 1–7) cis-elements were identified in the 4150-bp fragment of the SFT promoter. The translation start codon (ATG) is indicated. TSS, translation start site. Pro SlSFT (P1), Pro SlSFT (P2), and Pro SlSFT (P3) constitute a genomic fragment of the SFT promoter containing boxes 1–5, boxes 1–6, and boxes 1–7, respectively. They are 3589, 3627, and 4150 bp in length upstream of the TSS. b EMSAs for binding of NF-YA3b to the CCAAT cis-elements in boxes 1/2/3/4/5/6/7 of the SFT promoter. NF-YA3b protein was incubated with FAM-labeled box 1/2/3/4/5/6/7 probes, and the mobilities of the protein–oligonucleotide probe complexes on non-denaturing gels are indicated with red arrows. The specific unlabeled competitors (SFT-box1/2/3/4/5/6/7) or unlabeled mutant oligonucleotide competitors (SFT-box1/2/3/4/5/6/7-m) were added to the incubation mixtures of lanes 3, 4, 5, and 6, by a 10- and 30-fold molar excess. −, absence; +, presence. c Y1H experiments for the binding of NF-YA3b to the CCAAT cis-element at different positions of the SFT promoter. Seven constructs containing individual CCAAT cis-elements were used in the assays. The negative control was an empty pGADT7 vector (EV). d Dual-luciferase reporter assays. NF-YA3b was expressed from pGreenII 62-SK under the CaMV 35S promoter and served as an effector. Pro SlSFT (P1), Pro SlSFT (P2), and Pro SlSFT (P3) were cloned into the pGreenII 0800-LUC vector and used to drive the expression of the LUC reporter. e GUS gene expression assays. The full-length CDS of NF-YA3b was cloned into pHellsgate8 for expression of NF-YA3b protein as the effector. Three SFT promoter fragments of 3589 bp (SFT-P1), 3627 bp (SFT-P2), and 4150 bp (SFT-P3) upstream of the translation start codon (ATG) were cloned into pMV2-GUS vector to drive GUS gene expression. f–h Representative images of luciferase activity (left) and ratios of LUC/REN activities (right) in N. benthamiana leaves. Note that SlNF-YA3b suppresses its transcriptional activity on SlSFT (P2) and SlSFT (P3) but does not change its transcriptional activity on SlSFT (P1). The values displayed are mean ± standard error (n = 8). ns, not statistically significant; **P < 0.01 (t-test). i–k Representative images of GUS activity staining (left) and relative GUS gene expression levels (right) in N. benthamiana leaves. In the control (CK), the empty pHellsgate8 vector was used to replace the effector for co-expression with SFT(P1)pro:GUS (i), SFT(P2)pro:GUS (j), and SFT(P3)pro:GUS (k) in N. benthamiana leaves. The GUS gene expression level in the control (CK) was set as 1.0. Data are mean ± standard error (n = 3). ns, not statistically significant, **P < 0.01 (t-test).
We also carried out Y1H assays to clarify whether NF-YA3b could bind to the CCAAT cis-element of the SFT promoter in yeast cells. When NF-YA3b and individual CCAAT cis-elements (boxes 1–7) were co-expressed in yeast cells, it was found that the SFT promoters containing box 6 and box 7 conferred antibiotic resistance in the presence of 160 and 130 mM aureobasidin A (AbA), respectively (Fig. 5c). In contrast, when the SFT promoters containing boxes 1, 3, 4, and 5, or no CCAAT cis-element (negative control) were used, the yeast cells were unable to grow in the presence of the antibiotic AbA (Fig. 5c), suggesting that NF-YA3b could not bind to the SFT promoter containing boxes 1, 3, 4, and 5, thus failing to activate the expression of the antibiotic resistance gene. The SFT promoter containing box 2 was ‘toxic’ to yeast cells for unknown reasons and yeast cells containing this construct were unable to grow on transformation-selection medium SD/−Ura−Leu without AbA (data not shown). Taken together, these Y1H assay results showed that NF-YA3b specifically binds to CCAAT boxes 6 and 7 of the SFT promoter.
To further determine whether NF-YA3b could bind to the CCAAT cis-elements of the SFT promoter to regulate its gene expression in planta, we conducted dual-luciferase reporter and GUS expression assays using NF-YA3b as the effector. NF-YA3b was cloned into the pGreenII 62-SK effector vector. Three SFT promoter fragments of 3589 bp (SFT-P1), 3627 bp (SFT-P2), and 4150 bp (SFT-P3) upstream of the translation start codon (ATG) were cloned into the pGreenII 0800 LUC reporter vector (Fig. 5a and d). The dual-luciferase reporter results showed that NF-YA3b did not change the expression levels of the Luc reporter gene under the SFT promoter SFT-P1, which contained boxes 1–5 (Fig. 5f). However, NF-YA3b was found to significantly suppress Luc reporter gene expression under either the SFT promoter SFT-P2 or SFT-P3, which contained boxes 1–6 and 1–7, respectively (Fig. 5g and h). Similarly, the results of the GUS activity assay were consistent with those of the dual-luciferase reporter assays (Fig. 5e). When SFT(P1)pro:GUS and 35Spro:NFYA3b were co-expressed, the GUS gene expression levels were not significantly different from the control, which co-expressed SFT(P1)pro:GUS with the empty pHellsgate8 (CK) vector (Fig. 5i). However, when SFT(P2)pro:GUS or SFT(P3)pro:GUS was co-expressed with 35Spro:NFYA3b (Fig. 5j and k), significantly decreased GUS gene expression levels were observed. Taken together, these findings indicate that NF-YA3b binds to CCAAT boxes 6 and 7 of the SFT promoter and acts as a transcriptional suppressor of SFT gene expression in planta.
Discussion
NF-Ys have been demonstrated to regulate a wide range of biological processes in plants [7, 22, 23, 25, 40, 41, 43]. One of these NF-Y-regulated processes is flowering time control in flowering plants [24, 41, 47]. NF-YA, a subunit of the NF-Y tri-protein complex, plays a pivotal role in the regulation of flowering time in plants [5, 6, 24, 41]. In Arabidopsis, overexpression of any of the NF-YA1/3/4/5/7/8/9/10 genes delays flowering [26, 37, 54, 62]. AtNF-YA2 and AtNF-YA6 are positive regulators of flowering by activating FT gene expression [47]. Extensive functional redundancy of the NF-YA family and the embryo lethality of multiple mutants dramatically limit the possibility of NF-YA mutants in studies on flowering time control [13, 37, 46]. Therefore, for single mutations of these NF-YA members, there have been no reports of a significantly early flowering phenotype. Here, we identified a tomato nf-ya3b mutant with an early flowering phenotype. Knockout of the NF-YA3b gene by CRISPR-Cas 9 technology was found to promote flowering in tomato. In contrast, overexpression of NF-YA3b led to delays in flowering in transgenic tomato plants. These results suggested that NF-YA3b is a negative flowering regulator in tomato, which agrees with the findings of previous studies in Arabidopsis [26, 37, 54]. Notably, our findings fill the gap of a single nf-ya3b mutant causing early flowering in tomato.
The NF-Y transcriptional activator is a heterotrimeric complex formed by three different subunits: NF-YA, NF-YB, and NF-YC [3, 5, 22]. NF-YB and NF-YC form heterodimers in the cytoplasm and subsequently translocate to the nucleus, recruiting NF-YA to form heterodimers [20]. The NF-YA/YB/YC complex regulates the expression of target genes through a mechanism by which the NF-YA subunit of the heterotrimeric complex recognizes and binds to the CCAAT cis-element of gene promoters [24]. In this study, the results from Y2H and Y3H assays showed that NF-YA3b was recruited by the NF-YB/NF-YC heterodimers, assembling into higher-order NF-Y heterotrimeric complexes (Fig. 3). However, in dual-luciferase experiments we found that these NF-Y complexes did not affect the expression of the luciferase (Luc) reporter gene under the SFT promoter (Fig. 4c–i). We speculate that these NF-Y complexes might regulate other target genes? involved in processes that are unrelated to flowering time control. Interestingly, the individual NF-YA3b could lead to more than 2-fold decreases in the expression of the SFT-Luc reporter gene (Fig. 5g–h). Consistently, numerous single NF-Y subunits have been shown to bind to the CCAAT cis-element in the absence of the other two NF-Y subunits [2, 7, 45, 56]. By binding to the CCAAT cis-element of AtXTH21, AtHAP5A (an NF-YC gene) affects freezing stress resistance in Arabidopsis [45]. The aleurone-specific NF-YB1, which is more abundant on the dorsal side, is critical in regulating rice grain fullness by stimulating the expression of the Sucrose Transporter (SUT) genes SUT1/3/4 [2]. In rice, NF-YC12 directly binds to the FLOURY ENDOSPERM6 (FLO6) and Glutamine Synthetase1 (OsGS1;3) promoters and regulates endosperm development [56]. In this study, NF-YA3b was demonstrated to bind to the SFT promoter without NF-YB and NF-YC subunits (Fig. 5).
NF-Y transcription factors are heterotrimeric complexes that can change their activities depending on the compositions of the three subunits. Therefore, there is wide variability in the biological activity of NF-Y in plants, and there are various possibilities for opposing regulations at a single DNA binding site [23, 26]. Numerous studies have demonstrated that positive regulators of FT expression are known as NF-Ys in Arabidopsis. AtNF-YA2, AtNF-YA6, AtNF-YB2, AtNF-YB3, AtNF-YC1, and AtNF-YC2 have been shown to promote flowering by inducing the expression of FT [16, 22, 47]. Additionally, there have been reports presenting strong evidence for the negative effect of NF-Ys on gene expression [39, 54, 57]. Overexpression of AtNF-YA1, AtNF-YA4, and AtNF-YB1 has been shown to down-regulate FT expression, resulting in delayed flowering [39, 54]. AtNF-YA4/5/7/9 have negative effects on the expression of several ABA-responsive genes by blocking the interaction between NF-YB/NF-YC dimers and bZIP family members [57]. AtNF-YA8 suppresses the expression of a subset of age-dependent genes, negatively regulating flowering time [62]. SlNF-YA10 negatively regulates ascorbate accumulation by binding to the SlGGP1 promoter and inhibiting its expression [9]. In this study, we provide genetic and biochemical evidence that NF-YA3b binds to the CCAAT cis-element of the SFT promoter and suppresses its expression (Fig. 5). SFT is the florigen gene in tomato, which positively regulates flowering time. The sft mutant is known to produce flowers later than WT tomato plants [32, 36]. Consistent with this notion of SFT in flowering control, there has been a report showing that overexpression of SFT results in earlier flowering after three to five true leaves in tomato [30]. Consistent with the conclusion of these reports, our data showed that the SlNF-YA3b knockout lines exhibited early flowering (Fig. 2) and the expression level of the SFT gene was significantly up-regulated (Fig. 4a). Taken together, these results imply that NF-YA3b acts as a transcriptional repressor of SFT, negatively controlling flowering time in tomato. Remarkably, to our knowledge, this is the first direct demonstration that the individual NF-Y subunit directly binds to the SFT promoter and functions as a repressor of SFT transcription.
A model is proposed based on our findings that NF-YA3b controls flowering time by repressing SFT expression in tomato (Fig. 6). In this model, NF-YB and NF-YC form heterodimers in the cytoplasm and subsequently move to the nucleus, recruiting NF-YA3b to form heterodimers. NF-YA3b directly binds to the CCAAT cis-element of the SFT promoter and suppresses its gene expression, leading to late flowering in tomato. We speculate that the NF-YB/YC/YA3b heterotrimeric protein complex may bind to the promoters of other target genes, participating in the regulation of gene expression in other physiological and developmental pathways. There also exists a possibility that loss of function of NF-YA3b may promote flowering through a different mechanism by which the up-regulation of SFT expression is caused via regulation by transcription activation mediated by CO or other transcription factors. In summary, the results from this work have advanced our understanding of the regulatory mechanism involved in tomato flowering, which could be applied to crop improvement and germplasm innovation.
Figure 6.
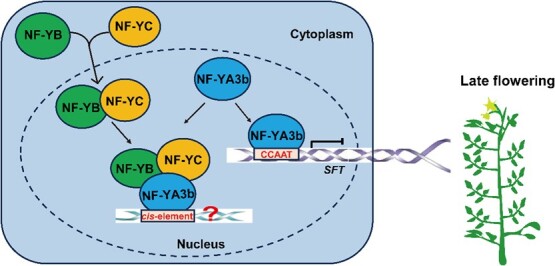
A model for the regulation of tomato flowering by NF-YA3b. In this model, NF-YB and NF-YC form heterodimers in the cytoplasm and move to the nucleus, where they recruit NF-YA3b to form heterotrimer protein complexes. The NF-YB/YC/YA3b complex may bind to the promoters of other target genes and be involved in the regulation of other pathways. On the other hand, NF-YA3b may bind directly to the CCAAT cis-element of the SFT promoter and suppress its gene expression, leading to late flowering in tomato.
Materials and methods
Plant materials and growth conditions
In this study, the tomato variety ‘Ailsa Craig’ (AC) served as the wild-type (WT) and was used for background plants in stable transformation of tomato. For Agrobacterium-mediated transient transformation experiments, Nicotiana benthamiana was utilized. All plants were grown in the greenhouse under conditions that included 23°C ambient temperature, 60–75% relative humidity, and a photoperiod of 16 h of natural daylight followed by 8 h of darkness.
Subcellular localization
The total cDNA of AC plants was used as a template for amplification of the CDS of SlNF-YA3b excluding the stop codon. The amplified SlNF-YA3b CDS was fused with the coding sequence for YFP to generate 35S:SlNFYA3b-YFP for expression of the NF-YA3b-YFP fusion protein for subcellular localization experiments. Plasmid 35S:StERF3-RFP, which expressed the fusion protein of potato Ethylene Responsive Factor 3 (StERF3)–RFP, served as a nuclear localization marker. 35S:SlNFYA3b-YFP and 35S:StERF3-RFP were expressed together in tobacco leaves. Two days after infiltration with Agrobacterium strains carrying the appropriate plasmids, leaves expressing fluorescent fusion proteins were obtained using a confocal laser scanning microscope (Leica TCS-SPE).
RNA extraction and gene expression profiling
Total RNAs were extracted from various tomato tissues using the TRIzol® 117 reagent (Invitrogen). A reverse transcription kit (Vazyme, Nanjing, China) was utilized to generate single-stranded cDNA using 2 μg of total RNA. Gene expression assays by qPCR analysis were performed using the ChamQ SYBR Color qPCR Master Kit (Vazyme, Nanjing, China). All data had three biological replicates and were analyzed. The internal control was the expression of the Actin gene (Solyc11g005330). Supplementary Data Table S1 lists the primer sequences used in real-time PCR.
Vector construction and tomato genetic transformation
For the generation of the SlNF-YA3b overexpression construct, the CDS of SlNF-YA3b was amplified from AC and connected to the pHellagate 8 vector driven by the CaMV35S promoter. For the generation of the SlNF-YA3b CRISPR/Cas9 construct, two sgRNAs in the first exon of SlNF-YA3b were designed by CRISPR-direct web (http://crispr.dbcls.jp) and linked to pTX041 vector following a method described previously [59]. SlNF-YA3b CRISPR/Cas9 and overexpression constructs were introduced into the WT tomato AC plants using an Agrobacterium-mediated stable transformation system.
Yeast two-hybrid assays
The Matchmaker GAL4-based Yeast Two-Hybrid System (Clontech, CA, USA) was used for verifying protein–protein interactions. The full-length CDSs of NF-YB3a/3b/3c and truncated CDS of NF-YB3b/3c were amplified and linked to pGBKT7 vectors. The full-length CDSs of NF-YC1a/1b/9 and NF-YA3b were amplified and then linked into pGADT7 vectors. Saccharomyces cerevisiae strain AH109 was co-transformed with the pairs of plasmids. Yeast cells were cultured on transformation-selection (SD/−Leu−Trp) and interaction-selection (SD/−Leu-Trp−Ade−His) medium to screen for protein–protein interactions. Specific primers (Supplementary Data Table S1) were used to construct the plasmids.
For transcriptional activation assays in yeast, the full-length CDS of NF-YA3b was cloned into pGBKT7 vector, and the resulting construct was used for transformation of yeast AH109 strain. The empty vector pGBKT7 and the combination of pGBKT7-53 + pGADT7-RecT vectors were used as the negative and positive controls, respectively. Transformed yeast cells were grown on SD/−Trp, SD/−Trp−His, and SD/−Trp−His with X-α-gal media.
Yeast three-hybrid assays
Y3H assays were conducted using the pBridge system (Clontech) to test the interactions between NF-YA3b and NF-YBs-YCs (including NF-YB3a-YC1a, NF-YB3b-YC1a, NF-YB3c-YC1a, NF-YB3a-YC9, NF-YB3b-YC9, and NF-YB3c-YC9). There are two multiple cloning sites in the pBridge system. The full-length CDSs of NF-YCs and NF-YBs were separately linked to the pBridge vectors, while the NF-YA3b CDS was linked to pGADT7 vector. The pairs of plasmids were co-transformed into yeast strain AH109, and the transformed yeast cells were cultured on SD/−Leu−Trp−Met before selecting them for protein interactions on SD/−Leu−Trp−Ade−His−Met media.
Yeast one-hybrid assays
For Y1H assays, the Matchmaker Gold One-Hybrid Library Construction & Screening Kit (Clontech) was utilized. There are seven CCAAT cis-elements in the 4.3-kb SFT promoter. The promoter fragments containing each CCAAT cis-element with surrounding sequences were amplified and inserted into pAbAi. The NF-YA3b CDS was linked to pGADT7. Yeast strain Y1H Gold was co-transformed with the pairs of plasmids. Yeast cells were cultured on SD/−Ura−Leu and selected for promoter activities on selection medium (SD/−Ura−Leu) containing different concentrations of AbA.
Protein expression and electrophoretic mobility shift assay
The CDS of NF-YA3b was amplified and then ligated into pET15d-MBP, expressing the recombinant protein with a maltose-binding protein (MBP) tag and 6-His tags. The plasmid was subsequently transformed into E. coli DE3 cells. The proteins were purified following the method described previously [27].
Two SFT promoter fragments each containing a CCAAT cis-element (box 6 and box 7) were used as oligonucleotide probes. The probes were labeled with FAM and synthesized by Tianyi (Wuhan, China). Mutant probes contained CCCCC to replace CCAAT in box 6 or GGGGG to replace ATTGG in box 7 and were used as competitor probes in EMSA, following a previously described method [34].
Dual-luciferase reporter assays for transcription activities
The CDS of NF-YA3b was cloned into the pGreenII 62-SK effector vector under the CaMV35S promoter. Three genomic fragments of 3589 bp (containing boxes 1–5 cis-elements), 3627 bp (containing boxes 1–6 cis-elements), and 4150 bp (containing boxes 1–7 cis-elements) upstream of the start codon from the SFT promoter were inserted into pGreenII 0800-LUC reporter vector. The pairs of constructs were transformed into Agrobacterium tumefaciens strain GV2260 cells with the helper plasmid pSoup19. Co-expression of the effector and reporter vectors in tobacco leaves was performed via Agrobacterium-mediated transformation following a method described previously [51]. Tecan’s Infinite 200 Pro microplate reader and the Dual-Luciferase Reporter Assay System (Promega, USA) were used to measure the activities of firefly luciferase (LUC) and Renilla luciferase (REN). Luciferin (1 mM, Gold Biotech, Olivette, MO, USA) was sprayed onto leaves of N. benthamiana to detect firefly LUC activity using the NightSHADE LB 985 system (Berthold, Bad Wildbad, Germany).
GUS activity assay
The full-length CDS of NF-YA3b was cloned into pHellsgate8 for expression of the effector. Three SFT promoter fragments of 3589 bp (SFT-P1), 3627 bp (SFT-P2), and 4150 bp (SFT-P3) upstream of the start codon were cloned into pMV2-GUS to drive the expression of the GUS reporter. The reporter and effector constructs were co-expressed in N. benthamiana leaves in the Agrobacterium-mediated transient expression system. Three days after Agrobacterium infiltration, the leaves were submerged in GUS staining buffer with 2 mM X-glucuronide at 37°C for 24 h, followed by washing with 95% ethanol. Relative expression levels of the GUS gene were measured by qRT–PCR.
Statistical analysis
GraphPad Prism 8.0 and Excel were both utilized for statistical analysis. Student’s t-test was employed to compare groups in pairs. Two categories have been defined for statistically significant differences: P < 0.05 and P < 0.01.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32272743), the Science and Technology Planning Project of Guangxi (GuikeAA22068088-1), the Science and Technology Major Project of Zhumadian (ZMDSZDZX2022005), and the earmarked fund for China Agriculture Research System (CARS-23-A13).
Contributor Information
Dedi Zhang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Kangna Ji, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Jiafa Wang, College of Horticulture, Northwest A&F University, Yangling 712100, Shaanxi, China.
Xinyu Liu, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Zheng Zhou, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Rong Huang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Guo Ai, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Yan Li, Zhumadian Academy of Agricultural Sciences, Zhumadian 463000, China.
Xin Wang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Taotao Wang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Yongen Lu, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Zonglie Hong, Department of Plant Sciences, University of Idaho, Moscow, ID 83844, USA.
Zhibiao Ye, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Junhong Zhang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China.
Author contributions
D.Z., K.J., J.W., X.L., Z.Z., R.H., G.A., X.W., T.W., Z.H., Z.Y., Y.L., and J.Z designed the experiments. D.Z., K.J., J.W., X.L., Z.Z., and R.H. conducted the experiments and analyzed the data. D.Z. and J.Z wrote the manuscript. D.Z., J.W., Y.L., X.W., T.W., Y.L., Z.H., Z.Y., and J.Z revised the manuscript.
Data availability
The data on which this article is based can be found in this article and its online supplement. The sequences of the genes presented in this study can be accessed in the Sol Genomics Network (http://solgenomics.net/) under the following accession numbers: SlNF-YA3b, Solyc12g009050; SlNF-YA1a, Solyc01g008490; SlNF-YA1b, Solyc11g065700; SlNF-YA3a, Solyc03g121940; SlNF-YA7a, Solyc02g069860; SlNF-YA7b, Solyc10g079150; SlNF-YA8, Solyc08g062210; SlNF-YA9, Solyc01g087240; SlNF-YA10a, Solyc01g006930; SlNF-YA10b, Solyc10g081840; SlNF-YB3a, solyc04g054150; SlNF-YB3b, solyc07g065500; SlNF-YB3c, solyc12g006120; SlNF-YC1a, solyc03g110860; SlNF-YC1b, solyc03g111450; SlNF-YC1d, solyc06g072040; SlNF-YC9, solyc01g079870; SFT, Solyc03g063100; Actin, Solyc11g005330. The protein sequences used for multiple sequence alignment analysis are listed in Supplementary Data Table S2.
Conflict of interest
The authors state that there is no conflict of interest.
Supplementary data
Supplementary data are available at Horticulture Research online.
References
- 1. Adrian J, Farrona S, Reimer JJ. et al. Cis-regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bai AN, Lu XD, Li DQ. et al. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016;26:384–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ben-Naim O, Eshed R, Parnis A. et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–76 [DOI] [PubMed] [Google Scholar]
- 4. Blumel M, Dally N, Jung C. Flowering time regulation in crops – what did we learn from Arabidopsis? Curr Opin Biotechnol. 2015;32:121–9 [DOI] [PubMed] [Google Scholar]
- 5. Cai XN, Ballif J, Endo S. et al. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007;145:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calvenzani V, Testoni B, Gusmaroli G. et al. Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One. 2012;7:e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao S, Kumimoto RW, Gnesutta N. et al. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26:1009–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaves-Sanjuan A, Gnesutta N, Gobbini A. et al. Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants. Plant J. 2021;105:49–61 [DOI] [PubMed] [Google Scholar]
- 9. Chen WF, Hu TX, Ye J. et al. A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the d-mannose/l-galactose pathway in tomato. Hortic Res. 2020;7:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corbesier L, Vincent C, Jang SH. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–3 [DOI] [PubMed] [Google Scholar]
- 11. Coustry F, Maity SN, Sinha S. et al. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J Biol Chem. 1996;271:14485–91 [DOI] [PubMed] [Google Scholar]
- 12. Dai XD, Ding YN, Tan LB. et al. LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J Integr Plant Biol. 2012;54:790–9 [DOI] [PubMed] [Google Scholar]
- 13. Fornari M, Calvenzani V, Masiero S. et al. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLoS One. 2013;8:e82043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gnesutta N, Kumimoto RW, Swain S. et al. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell. 2017a;29:1516–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gnesutta N, Saad D, Chaves-Sanjuan A. et al. Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol Plant. 2017b;10:645–8 [DOI] [PubMed] [Google Scholar]
- 16. Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci. 2012;13:3458–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hou XL, Zhou JN, Liu C. et al. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun. 2014;5:4601. [DOI] [PubMed] [Google Scholar]
- 18. Hwang YH, Kim SK, Lee KC. et al. Functional conservation of rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC proteins in the regulation of flowering time. Plant Cell Rep. 2016;35:857–65 [DOI] [PubMed] [Google Scholar]
- 19. Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–4 [DOI] [PubMed] [Google Scholar]
- 20. Kahle J, Baake M, Doenecke D. et al. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin β and importin 13. Mol Cell Biol. 2005;25:5339–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42:459–63 [DOI] [PubMed] [Google Scholar]
- 22. Kumimoto RW, Adam L, Hymus GJ. et al. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–23 [DOI] [PubMed] [Google Scholar]
- 23. Kumimoto RW, Zhang Y, Siefers N. et al. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63:379–91 [DOI] [PubMed] [Google Scholar]
- 24. Laloum T, De Mita S, Gamas P. et al. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:594–5 [DOI] [PubMed] [Google Scholar]
- 25. Lee HS, Fischer RL, Goldberg RB. et al. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA. 2003;100:2152–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A. et al. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One. 2012;7:e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li G, Wang J, Zhang C. et al. L2, a chloroplast metalloproteinase, regulates fruit ripening by participating in ethylene autocatalysis under the control of ethylene response factors. J Exp Bot. 2021;72:7035–48 [DOI] [PubMed] [Google Scholar]
- 28. Li S, Li K, Ju Z. et al. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics. 2016;17:1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang MX, Hole D, Wu JX. et al. Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta. 2012;235:779–91 [DOI] [PubMed] [Google Scholar]
- 30. Lifschitz E, Ayre BG, Eshed Y. Florigen and anti-florigen – a systemic mechanism for coordinating growth and termination in flowering plants. Front Plant Sci. 2014;5:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lifschitz E, Eshed Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J Exp Bot. 2006;57:3405–14 [DOI] [PubMed] [Google Scholar]
- 32. Lifschitz E, Eviatar T, Rozman A. et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu TF, Zhou TT, Lian MT. et al. Genome-wide identification and characterization of the AREB/ABF/ABI5 subfamily members from Solanum tuberosum. Int J Mol Sci. 2019;20:311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lv XC, Zeng XL, Hu HM. et al. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell. 2021;33:1182–95 [DOI] [PubMed] [Google Scholar]
- 36. Molinero-Rosales N, Latorre A, Jamilena M. et al. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta. 2004;218:427–34 [DOI] [PubMed] [Google Scholar]
- 37. Mu JY, Tan HL, Hong SL. et al. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6:188–201 [DOI] [PubMed] [Google Scholar]
- 38. Nardini M, Gnesutta N, Donati G. et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–43 [DOI] [PubMed] [Google Scholar]
- 39. Nelson DE, Repetti PP, Adams TR. et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA. 2007;104:16450–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pereira SLS, Martins CPS, Sousa AO. et al. Genome-wide characterization and expression analysis of citrus NUCLEAR FACTOR-Y (NF-Y) transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance. PLoS One. 2018;13:e0199187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petroni K, Kumimoto RW, Gnesutta N. et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24:4777–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Romier C, Cocchiarella F, Mantovani R. et al. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–45 [DOI] [PubMed] [Google Scholar]
- 43. Sato H, Suzuki T, Takahashi F. et al. NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 2019;180:1677–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen CC, Liu HY, Guan ZY. et al. Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell. 2020;32:3469–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi HT, Ye TT, Zhong B. et al. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014;203:554–67 [DOI] [PubMed] [Google Scholar]
- 46. Siefers N, Dang KK, Kumimoto RW. et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siriwardana CL, Gnesutta N, Kumimoto RW. et al. NUCLEAR FACTOR Y, subunit a (NF-YA) proteins positively regulate flowering and act through FLOWERING LOCUS T. PLoS Genet. 2016;12:e1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamaki S, Matsuo S, Wong HL. et al. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–6 [DOI] [PubMed] [Google Scholar]
- 49. Taoka K, Ohki I, Tsuji H. et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–5 [DOI] [PubMed] [Google Scholar]
- 50. Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17:2661–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang JF, Li GB, Li CX. et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 2021;229:3237–52 [DOI] [PubMed] [Google Scholar]
- 52. Wei Q, Ma C, Xu YJ. et al. Control of chrysanthemum flowering through integration with an aging pathway. Nat Commun. 2017;8:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei XJ, Xu JF, Guo HN. et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wenkel S, Turck F, Singer K. et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wigge PA, Kim MC, Jaeger KE. et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–9 [DOI] [PubMed] [Google Scholar]
- 56. Xiong YF, Ren Y, Li W. et al. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J Exp Bot. 2019;70:3765–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamamoto A, Kagaya Y, Toyoshima R. et al. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009;58:843–56 [DOI] [PubMed] [Google Scholar]
- 58. Yang J, Xie ZY, Glover BJ. Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol. 2005;165:623–32 [DOI] [PubMed] [Google Scholar]
- 59. Yang TX, Deng L, Zhao W. et al. Rapid breeding of pink-fruited tomato hybrids using the CRISPR/Cas9 system. J Genet Genomics. 2019;46:505–8 [DOI] [PubMed] [Google Scholar]
- 60. Zeng XL, Lv XC, Liu R. et al. Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation. J Integr Plant Biol. 2022;64:731–40 [DOI] [PubMed] [Google Scholar]
- 61. Zhang C, Jian M, Li W. et al. Gibberellin signaling modulates flowering via the DELLA-BRAHMA-NF-YC module in Arabidopsis. Plant Cell. 2023;35:3470–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao H, Lin K, Ma L. et al. Arabidopsis NUCLEAR FACTOR Y A8 inhibits the juvenile-to-adult transition by activating transcription of MIR156s. J Exp Bot. 2020;71:4890–902 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data on which this article is based can be found in this article and its online supplement. The sequences of the genes presented in this study can be accessed in the Sol Genomics Network (http://solgenomics.net/) under the following accession numbers: SlNF-YA3b, Solyc12g009050; SlNF-YA1a, Solyc01g008490; SlNF-YA1b, Solyc11g065700; SlNF-YA3a, Solyc03g121940; SlNF-YA7a, Solyc02g069860; SlNF-YA7b, Solyc10g079150; SlNF-YA8, Solyc08g062210; SlNF-YA9, Solyc01g087240; SlNF-YA10a, Solyc01g006930; SlNF-YA10b, Solyc10g081840; SlNF-YB3a, solyc04g054150; SlNF-YB3b, solyc07g065500; SlNF-YB3c, solyc12g006120; SlNF-YC1a, solyc03g110860; SlNF-YC1b, solyc03g111450; SlNF-YC1d, solyc06g072040; SlNF-YC9, solyc01g079870; SFT, Solyc03g063100; Actin, Solyc11g005330. The protein sequences used for multiple sequence alignment analysis are listed in Supplementary Data Table S2.