Abstract
We isolated cDNA clones for two nuclear-encoded, organellar members of the Arabidopsis hsp70 gene family, mtHsc70-2 (AF217458) and cpHsc70-2 (AF217459). Together with the completion of the genome sequence, the hsp70 family in Arabidopsis consists of 14 members unequally distributed among the five chromosomes. To establish detailed expression data of this gene family, a comprehensive reverse transcriptase-polymerase chain reaction analysis for 11 hsp70s was conducted including analysis of organ-specific and developmental expression and expression in response to temperature extremes. All hsp70s showed 2- to 20-fold induction by heat shock treatment except cpHsc70-1 and mtHsc70-1, which were unchanged or repressed. The expression profiles in response to low temperature treatment were more diverse than those evoked by heat shock treatment. Both mitochondrial and all cytosolic members of the family except Hsp70b were strongly induced by low temperature, whereas endoplasmic reticulum and chloroplast members were not induced or were slightly repressed. Developmentally regulated expression of the heat-inducible Hsp70 in mature dry seed and roots in the absence of temperature stress suggests prominent roles in seed maturation and root growth for this member of the hsp70 family. This reverse transcriptase-polymerase chain reaction analysis establishes the complex differential expression pattern for the hsp70s in Arabidopsis that portends specialized functions even among members localized to the same subcellular compartment.
Hsp70s comprise one subset of heat shock proteins that are induced by a rapid increase of temperature. In eukaryotes, hsp70s are encoded by a highly conserved multi-gene family whose proteins function in all major subcellular compartments of the cell. Numerous studies have elucidated hsp70 chaperone functions under stress conditions and in protein metabolism. Hsp70 binds and releases unfolded/non-native proteins, thereby helping polypeptides undergo productive folding. Hsp70 can prevent aggregation of denatured proteins (Sheffield et al., 1990) and refold stress-denatured proteins (Gaitanaris et al., 1990; Lee et al., 1995; Glover and Lindquist, 1998; Goloubinoff et al., 1999). It is also involved in translation (Nelson et al., 1992), translocation processes (Gao et al., 1991; Brodsky, 1996; Bush and Meyer, 1996), and steroid receptor function (Morishima et al., 2000). In addition, cytosolic hsp70s may act as negative repressors of heat shock factor (HSF)-mediated transcription either by themselves or in a hsp90-associated multi-chaperone complex (Shi et al., 1998; Zou et al., 1998).
Partial genomic sequences for three cytosolic members of the Arabidopsis hsp70 gene family were first described more than 10 years ago (Wu et al., 1988). Since then, several additional hsp70 sequences have been added to the gene database (http://www.ncbi.nlm.nih.gov/GenBank/). With the completion of genome sequencing, 12 full-length Arabidopsis hsp70 sequences are available in the database, five genes encoding cytosolic proteins, three encoding endoplasmic reticulum (ER) luminal members, and two each for plastid or mitochondrion-localized proteins. Although each member of the hsp70 gene family shares a highly conserved structure and action mechanism, there is accumulating evidence that various members of the hsp70 family play distinct roles in growth and development of plants. First, they are targeted to various subcellular compartments where vastly different metabolic processes occur. Second, sequence analysis classifies hsp70s into subfamilies that may be linked to different functions. Third, expression profiles of individual members of the hsp70 family differ under various conditions and stimuli. Important questions yet to be resolved include how different functions are allocated to each member and to what extent members of the family within a single subcellular compartment are functionally distinct and/or redundant.
Functional analysis including determination of expression patterns of hsp70s in other organisms has been instrumental in providing information on the diverse roles of hsp70s (Flaherty et al., 1990; Nelson et al., 1992; Freeman and Morimoto, 1996; Zhu et al., 1996; James et al., 1997; Glover and Lindquist, 1998; Goloubinoff et al., 1999; Mogk et al., 1999). In plants, comprehensive expression analysis of hsp70s has been limited (Duck et al., 1989; Denecke et al., 1991; Wang and Lin, 1993; DeRocher and Vierling, 1995; Dudley et al., 1997; Li et al., 1999) because the entire complement of hsp70 genes has not been available or known for any plant. The expression of 10 spinach hsp70 genes was studied in different temperature regimes (Li et al., 1999). In response to heat shock treatment, all 10 members of the spinach hsp70 gene family were induced by 1 h at 37°C and declined to preheat shock levels by 2 to 4 h at 37°C. In contrast, the response of spinach hsp70 genes to cold treatment was not similar to the heat shock response. There was no synchronized induction of spinach hsp70 genes in response to cold treatment, albeit, several members of the family were induced by 48 to 168 h at 5°C (Li et al., 1999). Three cytosolic hsp70s (PsHSP71.2, PsHSP71.0, and PsHSP70b) in pea similarly were shown to be differentially regulated (DeRocher and Vierling, 1995). PsHSP71.0 was found to be expressed constitutively, whereas PsHSP70b was weakly expressed under normal conditions but strongly induced by heat shock. In vegetative tissues, PsHSP71.2 was expressed only upon heat shock. PsHSP71.2 was also expressed in zygotic organs of developing pea seeds, and the PsHSP71.2 protein was abundant during seed development but disappeared within 72 h after the onset of imbibition. In contrast, PsHSC71.0 and PsHSP70b were expressed in both maternal and zygotic organs throughout seed development.
Similar to pea, expression of only three Arabidopsis cytosolic hsp70s has been examined, whereas available sequence information indicates that Arabidopsis has five cytosolic hsp70s, (Wu et al., 1988, 1994). At-Hsc70-1/Hsp70-1 was shown to be expressed in leaves at normal temperature and further induced by heat shock. The mRNA for Hsp70-2, located 1.5 kb downstream from At-Hsc70-1/Hsp70-1, was not detected at normal temperature or during heat shock. At-Hsc70-3/Hsp70-3 mRNA was found to be present at very low levels and showed no induction by heat shock. In addition, At-Hsc70-1/Hsp70-1 was also highly expressed at normal temperature in root, stem, and flower but not detected in green or yellow siliques (Wu et al., 1994).
Reverse transcriptase (RT)-PCR is a powerful method for expression analysis of gene families because amplification from mRNAs can be highly specific and quantification of expression signals can be rapidly performed (McDowell et al., 1996; Wang et al., 1999). To better define the physiological roles of hsp70s in plant growth and development, mRNA levels for 11 Arabidopsis hsp70 genes were quantified using RT-PCR. Here, we report a comprehensive analysis of the expression for most of the Arabidopsis hsp70s including organ-specific expression, developmental regulation, and expression in response to temperature extremes. The data reveal that several members of the Arabidopsis hsp70 family show distinct expression patterns, allowing predictions of when and where function of each hsp70 is expected to become physiologically important. These data are necessary to devise experimental strategies to assess phenotypes of loss-of-function mutants and transgenic plants that over-/under-express individual hsp70s.
RESULTS
Arabidopsis Hsp70s Are Encoded by a Gene Family
Arabidopsis contains genes encoding five cytosolic hsp70s, three binding proteins (BiPs) (hsp70 homologs in the ER), two plastid hsp70s, and two mitochondrial hsp70s. Including the sequences of two organellar hsp70s cloned in this study (“Materials and Methods”), full-length sequences for 12 Arabidopsis hsp70 genes are now available in the database either from cDNA or genomic sequence, in addition to two truncated hsp70 sequences (Table I). One of the two truncated sequences, Hsp70t-1 (AC058785), has no corresponding expressed sequence tag (EST) clone in the database, whereas the other sequence, Hsp70t-2, has a corresponding EST clone (AI996202). This suggests that Hsp70t-1 may not be expressed or if expressed, is so under conditions not included in the construction of EST libraries. Although Neighbor-Joining analysis suggests that Hsp70t-1 belongs to the cytosolic group, there is not enough information to predict subcellular localization for either Hsp70t-1 or Hsp70t-2. From the genome sequencing database, we identified a mitochondrial hsp70 and refer to it as mtHsc70-1 (AL035538) and a chloroplast hsp70 as cpHsc70-1 (AL078637). We cloned cDNAs for a second mitochondrial hsp70 and a second chloroplast hsp70 and named them mtHsc70-2 (AF217459) and cpHsc70-2 (AF217458).
Table I.
Arabidopsis Hsp70 gene family
| Proposed Name | Original Name | Subcellular Location | No. of Amino Acids | Introns | GenBank Accession No. | Clone Type | Reference |
|---|---|---|---|---|---|---|---|
| Hsc70-1 | At-Hsc70-1 | Cytosol | 651 | 1 | AL162971 | BAC | Bevan et al. (2000b)a |
| Hsp70-1 | X74604 | cDNA | Wu et al. (1994) | ||||
| Hsc70-2 | Hsp70-2 | Cytosol | 653 | 1 | AL162971 | BAC | Bevan et al. (2000b)a |
| Hsc70-3 | At-Hsc70-3 | Cytosol | 649 | 1 | AC011436 | BAC | Lin et al. (1999)a |
| Hsp70-3 | Y17053 | cDNA | Hsieh et al. (1998) | ||||
| Hsp70 | Hsp70 | Cytosol | 650 | 1 | AP002055 | BAC | Nakamura (2000)a |
| AJ002551 | cDNA | Hinderhofer et al. (1998)a | |||||
| Hsp70b | – | Cytosol | 646 | 0 | AC010924 | BAC | Liu et al. (1999)a |
| BiP-1 | – | ER lumen | 669 | 5 | AF262043 | BAC | Wilson (2000)a |
| D89341 | Genomic | Koizumi and Sano (1997) | |||||
| BiP-2 | – | ER lumen | 668 | 5 | AB017067 | BAC | Nakamura (1999)a |
| D89342 | Genomic | Koizumi and Sano (1997) | |||||
| BiP-3 | – | ER lumen | 659 | 4 | AC000106 | BAC | Osborne et al. (1997)a |
| mtHsc70-1 | – | Mitochondrion matrix | 666 | 5 | AL035538 | BAC | Bevan et al. (1999b)a |
| mtHsc70-2 | Hsc70-5 | Mitochondrion matrix | 682 | 5 | AL353994 | BAC | Bevan et al. (2000a)a |
| AF217458 | cDNA | Vierling et al. (2000)a | |||||
| cpHsc70-1 | – | Plastid stroma | 718 | 7 | AL078637 | BAC | Bevan et al. (1999a)a |
| cpHsc70-2 | Hsc70-7 | Plastid stroma | 718 | 7 | AB024032 | TAC | Nakamura (1999)a |
| AF217459 | cDNA | Sung et al. (2000)a | |||||
| Hsp70t-1 | – | Unknown | 617 | 1 | AC058785 | BAC | Lin et al. (2000)a |
| Hsp70t-2 | – | Unknown | 563 | 0 | AC006223 | BAC | Lin et al. (2000)a |
The sequences for BiP-3, Hsp70t-1, and Hsp70t-2 were identified from database searches after RT-PCR analyses were completed. The nucleotide sequence for BiP-3 is quite divergent from BiP-1 and BiP-2. However, their amino acid sequences showed remarkably high homology. Hsp70t-1 (617 amino acids) and Hsp70t-2 (563 amino acids) are truncated at their C-terminal ends and their subcellular localizations have not been determined. BAC, Bacterial artificial chromosome; TAC, transformation-competent artificial chromosome.
Indicates direct submissions to GenBank. These entries are not included in “Literature Cited.”
We propose a new nomenclature for hsp70 genes in Arabidopsis to clarify and establish consistency for this gene family. All gene names used in this study are listed in Table I along with accession numbers. For the remainder of this article, we will use the gene names proposed in Table I.
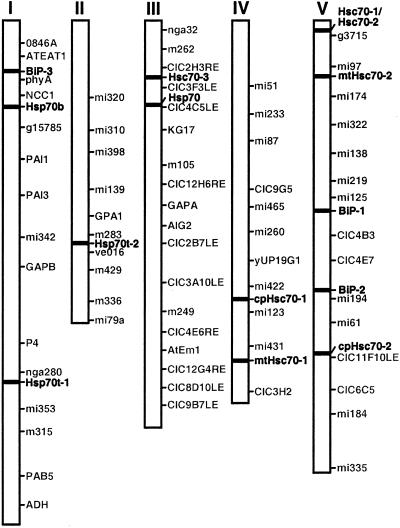
Hsp70 genes are found on all five chromosomes (Fig. 1). Six hsp70s representing members localized to the major subcellular compartments; two cytosolic (Hsc70-1 and Hsc70-2), two ER (BiP-1, BiP-2), one chloroplast (cpHsc70-2), and one mitochondrial member (mtHsc70-2) are found on chromosome 5. Of these six hsp70 genes on chromosome 5, only two cytosolic members (Hsc70-1 and Hsc70-2) are present in tandem. Chromosome 2 harbors only one hsp70 gene, Hsp70t-2.
Figure 1.
Chromosomal location of hsp70 genes in Arabidopsis. Hsp70 genes were positioned on a sequence map created by the Arabidopsis Genome Initiative (AGI, http://www.Arabidopsis.org/agi.html). This map contains molecular markers as well as genetic markers.
The intron-exon structure of the hsp70 genes in Arabidopsis is distinctive and different for genes encoding proteins targeted to different subcellular locations (Table I). Arabidopsis hsp70 genes encoding protein targeted to the same subcellular compartment are highly conserved in the number of introns and the length of exons (Table I; data not shown) indicating they are likely products of gene duplication events. For example, four cytosolic hsp70 genes (Hsc70-1, Hsc70-2, Hsc70-3, Hsp70) have one intron each, and their corresponding exons are the same size. The fifth cytosolic member, Hsp70b, has no intron like many of the strongly heat inducible hsp70 genes in other organisms. A new hsp70 member for the ER, BiP-3, has four introns, whereas the other two BiP genes (BiP-1 and BiP-2) have three introns each. There is no conservation in the length of exons between the first two BiP genes and BiP-3, indicating that BiP-3 probably arose from a different evolutionary lineage. All organellar members have more introns than cytosolic members.
Plastid and Mitochondrial Hsp70s Are Highly Conserved
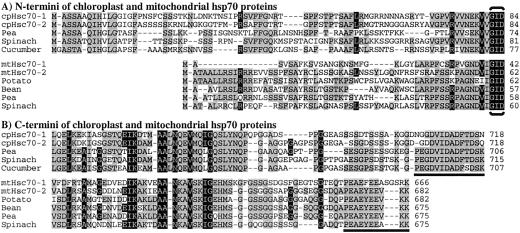
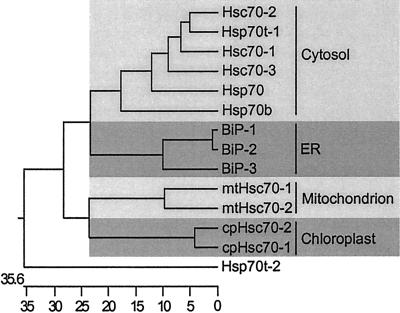
The two mitochondrial hsp70s, mtHsc70-1 and mtHsc70-2, encode proteins of 666 and 682 amino acids with predicted pI of 5.17 and 5.60, respectively. The two plastid hsp70s, cpHsc70-1 and cpHsc70-2 encode proteins of 718 amino acids each with predicted pI of 5.03 and 4.96, respectively. These organellar hsp70s were aligned with full-length organellar hsp70s from other plant species, revealing strong amino acid sequence conservation. The Arabidopsis plastid hsp70 proteins are 88% identical to each other and 81% to 85% identical with chloroplast hsp70 proteins from other plants. The two mitochondrial hsp70s are 78% identical to each other and 76% to 86% identical to those of other species. Sequence alignment also revealed that mtHsc70-1 differs from mtHsc70-2 and other plant mitochondria hsp70 proteins in the N and C termini (Fig. 2). Arabidopsis mtHsc70-1 has deletions of several amino acids in the N-terminal signal peptide region and an insertion of three amino acids in the C-terminal end compared with other plant mitochondrial hsp70 proteins. This unique C terminus of mtHsc70-1 may indicate alternative suborganellar localization or specialized cochaperone interaction that is different from that of mtHsc70-2 and other known mitochondrial hsp70s. The C terminus of organellar hsp70s is highly conserved and can be used as a predictive localization motif for organellar hsp70 proteins (Guy and Li, 1998). The C termini of the Arabidopsis plastid hsp70s and the mitochondrial hsp70s also contain these conserved motifs (underlined residues in Fig. 2). As noted in Table I, the proposed subcellular localization for the 12 full-length Arabidopsis hsp70s was consistent with Neighbor-Joining analysis (Fig. 3) and C-terminal sequence motifs. The general branching pattern of their dendrogram is also in agreement with previous phylogenic analyses of hsp70s in yeast, plants, and other organisms (Boorstein et al., 1994; Guy and Li, 1998).
Figure 2.
Sequence alignment of chloroplast and mitochondrial hsp70 proteins. Arabidopsis plastid hsp70s (cpHsc70-1, cpHsc70-2) were aligned with chloroplast hsp70s from pea, spinach, and cucumber. Arabidopsis mitochondrial hsp70s (mtHsc70-1, mtHsc70-2) were aligned with mitochondrial hsp70s from potato, bean, pea, and spinach. Black shading indicates consensus residues common to both chloroplast and mitochondrial hsp70s. Gray shading indicates consensus residues specific in either chloroplast or mitochondrial hsp70s. Brackets indicate the beginning of the N-terminal, highly conserved ATP-binding motif. Underlined residues are C-terminal signature motifs for organelle localization.
Figure 3.
Rooted Neighbor-Joining analysis using the CLUSTAL program in DNASTAR for protein sequences of the Arabidopsis hsp70 gene family. Full-length protein sequences of 12 Arabidopsis hsp70 proteins and two truncated sequences (Hsp70t-1, Hsp70t-2) were used in this analysis. The scale at the bottom represents the branch distance as the number of changes in character states between neighbors. Each shaded area represents subcellular localization; from the top, cytosol, ER, mitochondrion, and chloroplast.
Optimization of RT-PCR
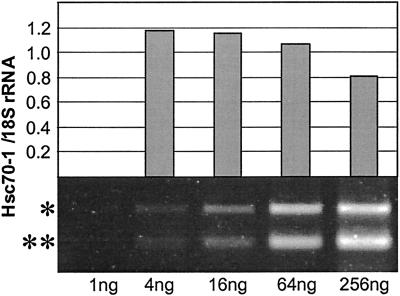
Robust analysis of hsp70 expression required gene-specific primers for the 11 hsp70s examined in this study (Table II). Primers were designed to produce PCR products with different lengths to conduct multiplex RT-PCR. However, multiplex RT-PCR could not be used because of unequal and biased amplification of different sequences. Therefore RT-PCR reactions for each gene were analyzed individually. The conditions for RT-PCR were optimized to produce unsaturated PCR product accumulation that retained a linear relationship with the original transcript levels in all samples. A range of 1 to 256 ng of total RNA was tested and 16 and 64 ng of total RNA were found to generate unsaturated RT-PCR product accumulation for each gene through 25 cycles of PCR. As an example, RT-PCR signals of Hsc70-1 over a range of 1 to 256 ng of total RNA are shown (Fig. 4). Two RT-PCR signals were generated for each sample; one for the individual hsp70 gene and one for 18S rRNA as an internal loading control. When the two signals were not saturated in the sample, the ratios of the two signals over a range of total RNA concentrations were reasonably constant. Total RNA concentrations of 16 and 64 ng consistently yielded the same ratio and also gave stoichiometric increases of RT-PCR signals (Fig. 4). For the present analysis, 16 ng of total RNA was used for all reactions. RNA samples were treated with DNase I to eliminate DNA contamination. However, even without DNase I treatment, no amplification products from DNA contamination were detected for any of the genes (data not shown). The gene specificity of RT-PCR was confirmed by sequencing all RT-PCR products.
Table II.
Oligonucleotide primers used in RT-PCR
| Gene | Primer | Sequence |
|---|---|---|
| Hsc70-1 | CG256F | 5′-TGCCTACGGTCTTGACAA-3′ |
| CG257R | 5′-ACCTGGATCAACACACCG-3′ | |
| Hsc70-2 | CG301F | 5′-TGGCCTTTCACTATCATC-3′ |
| CG302R | 5′-TAGAAGTCAGCTCCACCA-3′ | |
| Hsc70-3 | CG268F | 5′-CCCTTCACGCTCAAATCT-3′ |
| CG269R | 5′-TCCTCCAGCGGTTTCAAG-3′ | |
| Hsp70 | CG258F | 5′-TCAAGCGGATAAGAGTCACT-3′ |
| CG259R | 5′-CTCGTCCGGGTTAATGCT-3′ | |
| Hsp70b | CG284F | 5′-TGTCGGAGTTTGGATGAAT-3′ |
| CG285R | 5′-CTGTCTCAAGTCCAAGGCTA-3′ | |
| BiP-1 | CG260F | 5′-ACTAAGATGAAGGAGACAGCT-3′ |
| CG261R | 5′-ACTTGGTGCTGACTACTTAGA-3′ | |
| BiP-2 | CG262F | 5′-ACTAAGATGAAGGAGACGACC-3′ |
| CG263R | 5′-TTGGTGCTGACTGCTTAAG-3′ | |
| mtHsc70-1 | CG234F | 5′-GCTGCTGCACTATCATATGG-3′ |
| CG235R | 5′-CACGGAGGATACCACCTT-3′ | |
| mtHsc70-2 | CG266F | 5′-CGTTTCCTCTCCTTTCTCA-3′ |
| CG267R | 5′-TTTGGCTAGGTCTATTCCC-3′ | |
| cpHsc70-1 | CG247F | 5′-GGTGATCCTTGTTGGTGG-3′ |
| CG213R | 5′-ATCTCAACGCTTGTCTGTC-3′ | |
| cpHsc70-1 | CG264F | 5′-AGTGCCTTCTTCGGTACA-3′ |
| CG265R | 5′-GGACACTCAAGCTTAACATTATT-3′ |
F, Forward primer; R, reverse primer.
Figure 4.
RT-PCR optimization. Equivalent increase of duplex Hsc70-1 and 18S rRNA signal in the range of 1 to 256 ng of total RNA was tested. The 16-ng amount of total RNA was selected for subsequent experiments. *, RT-PCR band for Hsc70-1. **, RT-PCR band for 18S rRNA.
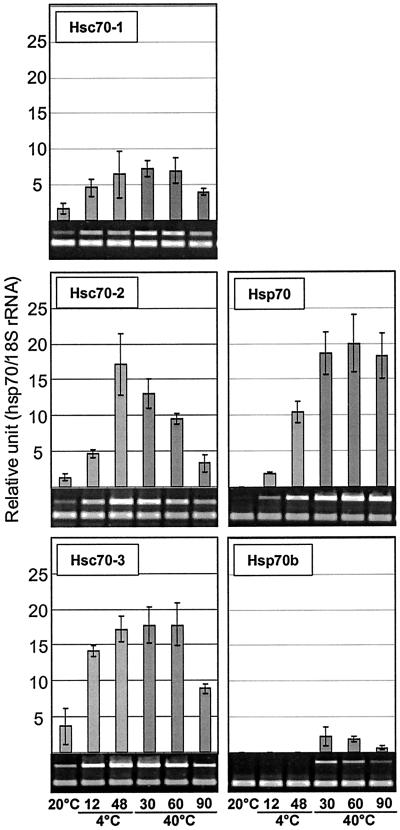
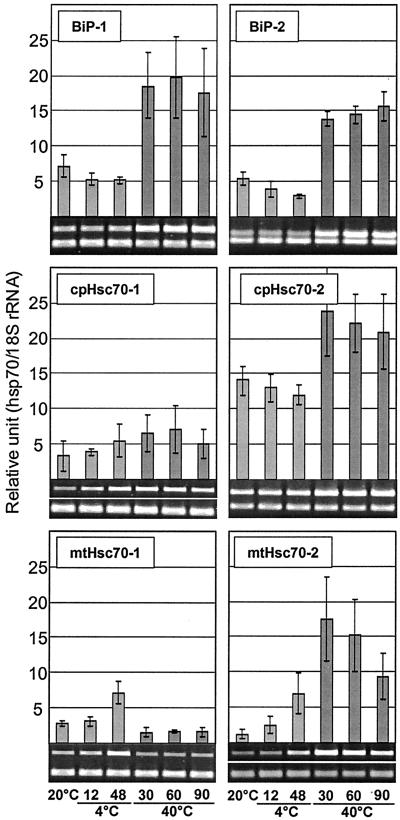
Diverse Responses of Arabidopsis Hsp70s to Temperature Extremes
Expression profiles for hsp70 genes during heat shock or cold acclimation were determined on plants that were exposed to 40°C for 30, 60, and 90 min of heat shock treatment and to 4°C for 12 and 48 h of low temperature treatment. Control plants were kept at 20°C. The most specific response to temperature extremes was that of Hsp70b (Fig. 5). The Hsp70b transcript was detectable only during heat treatment and was not detected during any other treatment or developmental stage or in any organ. Except for mtHsc70-1 and cpHsc70-1, all members of the family showed induction of 2- to 20-fold by 30 min at 40°C (Figs. 5 and 6). The induction of Hsc70-1 and BiP expression in response to heat shock was in good agreement with previous findings by other laboratories using hybridization-based techniques (Wu et al., 1988, 1994; Koizumi, 1996).
Figure 5.
Response of cytosolic hsp70s to temperature extremes. Two-week-old Arabidopsis plants were subjected to 4°C for 12 and 48 h for low temperature treatment or 40°C for 30, 60, and 90 min for heat shock treatment. Control plants (C) were incubated at 20°C, simultaneously. Signal values obtained from each gene were normalized with the 18S rRNA signal value, and the resulting mean values were presented as relative units. Error bar represents sd.
Figure 6.
Response of organellar hsp70s to temperature extremes. Empty space indicated by a white line in the gel pictures of cpHsc70-1 and mtHsc70-2 was cut out to achieve uniform spatial arrangement of the images.
Despite the strong and nearly universal induction by heat shock, repression kinetics of hsp70s were quite diverse. Three classes of repression kinetics could be discerned: rapid, within 30 to 60 min at 40°C (Hsc70-2); moderate, 60 to 90 min at 40°C (Hsc70-1, Hsc70-3, Hsp70b); and slow, 90 min or more at 40°C (Hsp70, cpHsc70-2, BiP-1, BiP-2, mtHsc70-2) (Figs. 5 and 6).
Several hsp70s were also induced during low temperature treatment, but responsiveness to cold was limited to cytosolic and mitochondrial hsp70s. Hsc70-1 and Hsc70-3 were induced 3- to 5-fold within 12 h at 4°C, whereas Hsc70-2 and Hsp70 were induced 10-fold or more by 48 h at 4°C. mtHsc70-1 and mtHsc70-2 showed approximately a 2-fold increase after 48 h at 4°C, whereas the transcript levels of both ER and chloroplast members (BiP-1, BiP-2, cpHsc70-1, and cpHsc70-2) showed little or no change (Figs. 5 and 6). The lack of obvious change in gene expression of BiP-1 and BiP-2 at low temperature is similar to the response of a spinach BiP gene (Anderson et al., 1994).
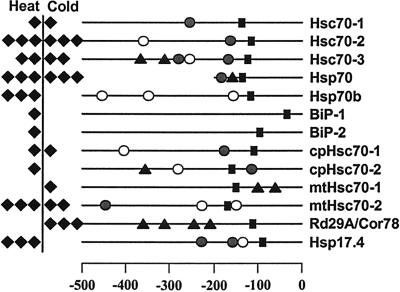
Two interesting aspects of the temperature response of Arabidopsis hsp70s are the absence of heat induction of mtHsc70-1 and the induction of Hsp70b exclusively by heat shock. The absence of heat inducible expression in mtHsc70-1 appears to be due to the absence of heat shock elements in the promoter, as the promoters for 11 Arabidopsis hsp70 genes were examined for the presence of two major temperature responsive cis-elements, heat shock element (HSE), and C-repeat or dehydration responsive element (CRT/DRE) (Fig. 7). HSE has been linked with the heat inducible expression of many heat shock genes (Czarnecka et al., 1989). CRT/DRE is known to be associated with drought- and cold-inducible expression of many genes (Yamaguchi-Shinozaki and Shinozaki, 1994). Overall, the expression profiles of hsp70 genes and the presence of cis-elements in the promoters are in good agreement. Hsp70s that showed strong induction by heat shock contain multiple HSE elements (Fig. 7), but no functional HSE was found in the promoter of mtHsc70-1 (Fig. 7). One or more CRT/DRE were found in the promoters for strongly cold-inducible members such as Hsc70-3, Hsp70, and mtHsc70-2. In contrast, there are exceptions for the presence of cis-elements and induction of hsp70s by temperature stress. HSE was not found in BiP-1 and BiP-2 where heat induction was clearly observed, and CRT/DREs were not found in the promoter of the strongly cold-inducible member, Hsc70-2. A CRT/DRE, conversely, was found in cpHsc70-2, yet cold induction was not detected. HSE and CRT/DRE are the best characterized cis-elements for heat and cold induction of hsp70 genes, but heat and cold induction of hsp70 genes clearly results from the function of a complex array of cis-elements. For example, heat induction of BiPs without an HSE can be explained by the presence of multiple C1 elements that are critical for the unfolded protein response (Wooden et al., 1991). Cold induction of Hsc70-2 without CRT/DRE similarly could possibly be explained by the presence of abscisic acid responsive elements.
Figure 7.
Predicted cis elements in the promoters of Arabidopsis hsp70 genes. Promoter sequences for 11 Arabidopsis hsp70 genes, a cold-inducible gene (Rd29A/Cor78), and a heat-inducible gene (Hsp17.4) were analyzed. The numbers at the bottom indicate the number of nucleotides upstream to the translation initiation codon, ATG. Induction fold of each gene in response to heat and cold are indicated as solid diamonds; one diamond, less than 5-fold; two diamonds, 5- to 10-fold; and three diamonds, more than 10-fold. ●, Perfect HSE (nTTCnnGAAnnTTCn or nGAAnnTTCnnGAAn). ○, Imperfect HSE. ▴, The core sequence of CRT/DRE (CCGAC). ▪, TATA box. Only a small portion (approximately 200 bp) of the promoter region for Hsp70 was available at the time of analysis.
Specific Members of Arabidopsis Hsp70s Are Induced during Seed Maturation and Germination
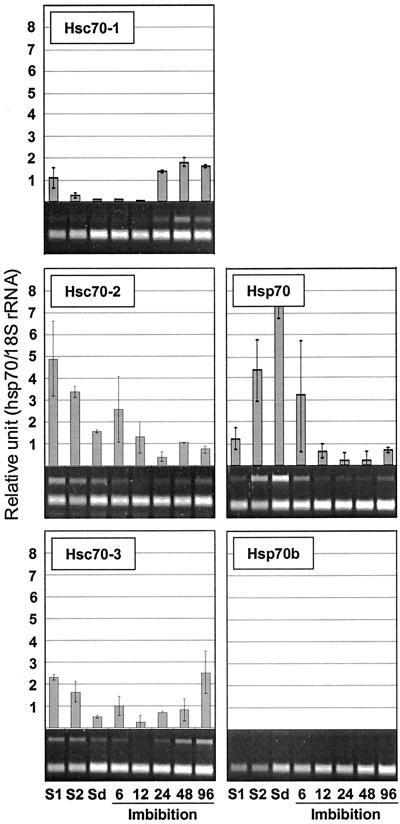
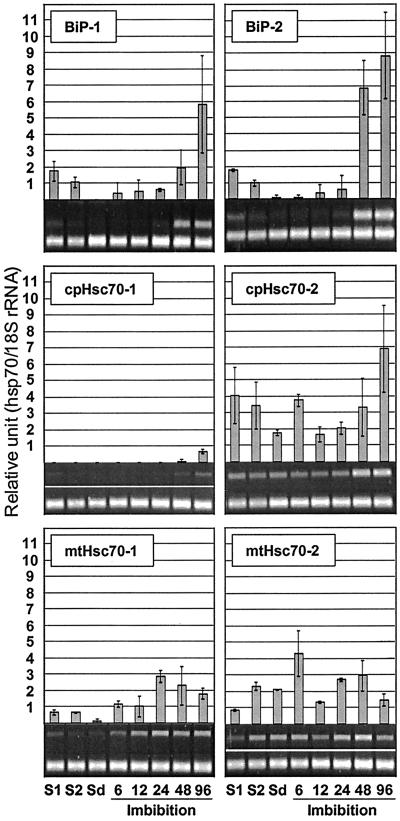
Transcript levels of hsp70 genes in green silique (7 days after pollination [DAP]), yellow silique (14 DAP), and dry seed were analyzed. Hsp70 showed the greatest induction (8-fold) of the family during seed maturation and desiccation (Fig. 8). Transcript levels of mtHsc70-2 also rose during this period but to a lesser extent (Fig. 9), whereas the transcript levels for Hsc70-1, Hsc70-2, Hsc70-3, BiP-1, BiP-2, cpHsc70-2, and mtHsc70-1 were diminished. The transcripts of Hsp70b and cpHsc70-2 were not detectable during this stage of development (Figs. 8 and 9).
Figure 8.
Expression of cytosolic hsp70s during seed maturation and germination. Silique samples (S1, S2) were harvested from 5- and 6-week-old plants. S1, Silique at 7 DAP; S2, silique at 14 DAP; Sd, mature dry seed. Samples were also harvested at 6, 12, 24, 48, and 96 h after imbibition.
Figure 9.
Expression of organellar hsp70s during seed maturation and germination.
Previous analyses from our laboratory showed induction of hsp70 genes around 2 d of imbibition (data not shown). When samples were taken at 6, 12, 24, 48, and 96 h of imbibition and analyzed, transcripts of Hsp70 were found to disappear within 24 h after the onset of imbibition. Hsp70b was not detected at any time point during imbibition and germination (Fig. 8). Depending on the timing of peak expression during germination, members of the family could be divided into three classes; early, intermediate, and late. The early class showed peak expression at 6 h after imbibition, and Hsc70-2 is indicative of this class. The intermediate class showed peak expression between 6 and 24 h after imbibition, and the two mitochondrial members (mtHsc70-1, mtHsc70-2) belong to this class. The late class showed peak expression at 24 and 96 h of imbibition and Hsc70-1, Hsc70-3, BiP-1, BiP-2, cpHsc70-1, and cpHsc70-2 belong to this class. Noteworthy of this class was the very low expression level of BiP-1 and BiP-2 in mature seed, which was followed by very strong induction at 48 and 96 h of imbibition.
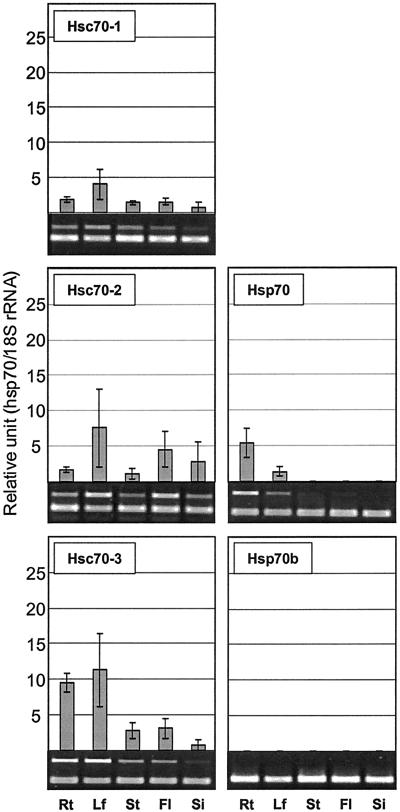
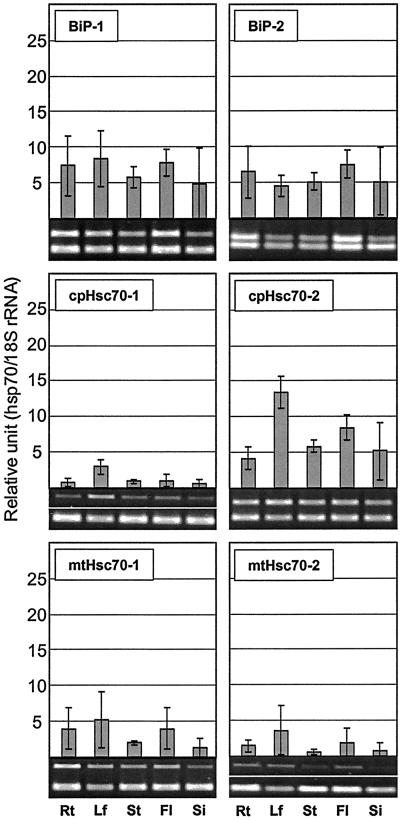
Hsp70s Are Differentially Expressed in the Organs of Arabidopsis
The expression of Arabidopsis hsp70 genes was analyzed to determine whether individual members of the family were expressed in particular organ(s). Hsp70 transcripts were abundant in root but barely detectable in other organs (Fig. 10). Hsc70-3 and mtHsc70-1 were also detected at higher levels in root. In contrast, Hsp70b was not detected in any organ in the absence of heat shock. Transcripts for Hsc70-1, Hsc70-2, Hsc70-3, cpHsc70-1, and cpHsc70-2 were detected at higher levels in leaf than other organs (Figs. 10 and 11). Two ER members, BiP-1 and BiP-2, were abundantly present in all organs tested. However, BiP transcript levels were slightly increased in floral tissues (Fig. 11). Unfortunately transcript levels of mtHsc70-2 were too low to resolve organ specific expression. Transcript levels for all members appeared to diminish in young silique at 3 DAP (Figs. 10 and 11).
Figure 10.
Expression of cytosolic hsp70s in different organs. Rt, Root; Lf, leaf; St, stem; Fl, flower; Si, silique at 3 DAP.
Figure 11.
Expression of organellar hsp70s in different organs. Rt, Root; Lf, leaf; St, stem; Fl, flower; Si, silique at 3 DAP.
DISCUSSION
Temperature Response
Previous expression studies for plant hsp70s demonstrated induction in 2 h of heat shock either at 37°C or 40°C (Wu et al., 1988, 1994; DeRocher and Vierling, 1995; Koizumi, Wu et al., 1996; Li et al., 1999). Most Arabidopsis hsp70s reached peak induction within 30 min of heat shock exposure, and the rapid response of hsp70 genes to heat shock is not limited to Arabidopsis, as the induction of spinach hsp70 genes was detected as early as 5 min after heat shock (Q. B. Li and C. L. Guy, unpublished data).
After initial heat induction Arabidopsis hsp70s showed rapid and diverse repression profiles. The repression of some hsp70 genes (Hsc70-2, mtHsc70-2) starts as early as 60 min after the onset of heat shock, whereas others (BiP-1, BiP-2, cpHsc70-2) remained at an induced level 90 min after heat shock. Repression patterns for Arabidopsis BiP previously analyzed (Koizumi, 1996) were consistent with the present results and demonstrate that BiP genes have a relatively slower repression system than for other plant hsp70s.
In contrast, induction by low temperature treatment was limited to cytosolic and mitochondrial members of hsp70s in Arabidopsis. Except for Hsp70b, all cytosolic hsp70s showed strong induction by low temperature treatment. The reason why the cytosolic members are strongly induced at low temperature is not clear, but it may be related to increased demand for molecular chaperone function at low temperature. The temperature responses suggest that cytosolic and ER hsp70 genes are responsible for molecular chaperone activity under heat stress, and mainly cytosolic hsp70s are required under low temperature stress in Arabidopsis.
Roles of Hsp70s in Seed Maturation
Induction of BiP expression in Arabidopsis seems to occur earlier during seed development than in pumpkin, rice, and wheat (Hatano et al., 1997; Muench et al., 1997). BiPs in all three plants were induced in the middle stages (1–2 weeks after pollination) of seed development where rapid cell expansion and accumulation of seed storage proteins occurs but decreased rapidly toward the end of seed maturation (Hatano et al., 1997; Muench et al., 1997; DuPont et al., 1998). Two Arabidopsis BiP genes were induced at or before the time of pollination. The major seed storage protein in Arabidopsis (12S protein) begins to accumulate around 1 week after pollination (Wehmeyer et al., 1996). Whether the high transcript levels of the two BiPs in flowers were a prerequisite for the rising flux of seed storage proteins through the ER of floral organ cells requires further investigation.
We have shown the expression of all hsp70 genes except for Hsp70 and mtHsc70-2 was repressed in siliques during the later stages of seed development (7 and 14 DAP). This implies diminishing roles of hsp70 genes during seed development. Based on the overall low levels of hsp70 expression, siliques (including developing seeds) may be one of the most sensitive organs to heat stress. The decline in the expression of hsp70 genes in seeds alternatively may be compensated for by the increased expression of other chaperones and stress proteins during late seed maturation, including the small Hsps, Hsp101, and LEA proteins.
Hsp70 showed a striking expression pattern during seed maturation and germination where it was absent in flowers and young siliques but present at high levels in dry seed. Hsp70 transcripts accumulated during the later stages of seed development and/or during desiccation. Hsp70 transcripts subsequently and rapidly disappeared during germination. Similar expression patterns were observed for cytosolic hsp70s of pea and a mungbean hsp70 (Wang and Lin, 1993; DeRocher and Vierling, 1995). This pattern of Hsp70 expression also closely follows the expression patterns of small heat shock proteins (Wehmeyer et al., 1996) and LEA genes (Raynal et al., 1999) in Arabidopsis, suggesting that Hsp70 expression may be regulated by a common mechanism that regulates these classes of genes during development (Galau and Hughes, 1987; Almoguera et al., 1998).
The presence of Hsp70 transcripts in mature dormant seed makes it a preserved mRNA (Harris and Dure, 1978). The reason for the presence of Hsp70 transcript as a preserved mRNA remains unclear, but there are two possibilities. During imbibition and germination, the resumption of protein synthesis may require immediate production of the cytosolic chaperone for efficient protein biogenesis. The Hsp70 mRNA alternatively could serve as an immediate source of Hsp70 for translation if imbibition occurred during a period of high temperature exposure.
Roles of Hsp70s in Seed Germination
In the early stages of germination, disaggregation of protein bodies and use of storage proteins has to be efficiently maintained to cope with increased demand for amino acids and energy for organogenesis. In the latter stages, a substantial transformation takes place in seedlings during the formation of the photosynthetic apparatus and conversion of plastids to chloroplasts, which is manifested by the greening of seedlings approximately 48 to 96 h after imbibition. The chaperone activities of hsp70s may be needed in two important aspects of protein metabolism during germination, which explains the induction of many hsp70s. First, proteins that are unfolded or misfolded during seed desiccation could be susceptible to aggregation during seed imbibition (i.e. rehydration of proteins), and hsp70 chaperones may need to be present in every compartment of the cell as soon as the cells become rehydrated to minimize the toxic effects of protein aggregation. Second, the initiation of active synthesis and translocation of proteins must be protected to ensure optimal function of metabolic processes during germination. The copious amounts of Hsp70 transcript in dry seed may serve as a reservoir for rapid access to molecular chaperone activity during the initial stages of storage protein use. In the same context, induction of Hsc70-2 within the first 6 h of imbibition appears just as important as Hsp70 for the initial stages of germination since Hsp70 transcripts rapidly disappeared in the first 12 h of imbibition.
Five hsp70s, Hsc70-1, Hsc70-3, BiP-1, BiP-2, and cpHsc70-2, were strongly induced by 96 h after imbibition in this study. The induction of these genes coincides with the greening of cotyledons and could be involved in the use of storage proteins in cotyledons, formation of photosynthetic apparatus, and the developmental conversion of plastids to chloroplasts. In the case of BiP induction, specific roles for BiP proteins during germination have been elucidated in pumpkin where induction accompanied the degradation of seed storage proteins (Hatano et al., 1997). It was concluded that BiP functioned in the degradation of seed storage proteins by assisting in folding and assembly of newly synthesized hydrolytic enzymes responsible for the degradation of seed storage protein (Hatano et al., 1997), and the same may be true for Arabidopsis.
Organ-Specific Expression of Hsp70 Genes in Arabidopsis
High expression in root and very low or no expression of Hsp70 in other organs suggest a specific role of Hsp70 in root growth or function. Other hsp70s also expressed in root are Hsc70-3, BiP-1, BiP-2, and mtHsc70-1. When BiP gene expression was reduced in tobacco with an antisense approach, root formation of transgenic shoot cuttings was compromised, suggesting a role of BiP in root formation (Leborgne-Castel et al., 1999). In fact, the expression of the two BiP genes analyzed in this study was ubiquitous, indicating vital roles of BiP genes in whole-plant cellular metabolism. The signal of cpHsc70-1 was higher in leaf and very low elsewhere, suggesting a specific role of cpHsc70-1 in chloroplast. Transcripts of cpHsc70-2 were higher in all organs compared with cpHsc70-1, suggesting a general role(s) of cpHsc70-2 in all forms of plastids.
In summary, we have shown that the expression patterns of hsp70 genes, one of the most highly conserved gene families, are distinct, and in many cases differential expression pattern(s) can be linked to major physiological or developmental processes occurring in plants. Based on the expression patterns, a role for Hsp70 can be ascribed to seed maturation and germination, Hsp70b exclusively to heat stress, and the cytosolic/mitochondrial hsp70s to cold stress. It will be challenging to investigate how the chaperone activities of hsp70 proteins translate into specific physiological roles in the context of plant cell function and in the larger context of plant growth and development. We will use the information obtained from this study to devise experimental strategies to assess the major phenotypes of transgenic plants that over-/under-express individual hsp70s.
MATERIALS AND METHODS
Plant Growth and Harvest
Arabidopsis ecotype Columbia seeds were sown on water-soaked filter paper (no. 1; Whatman, Clifton, NJ) for germination experiments. For other experiments, plants were grown in a commercial soil mix (Fafard mix no. 2) containing Canadian sphagnum peat, perlite, and vermiculite, watered every third day, and fertilized once a week with a commercial fertilizer (Peter's 20-20-20). Plants were grown at 20°C with a photoperiod of 15-h light/9-h dark in growth cabinets. The irradiance was approximately 150 μmol m−2 s−1 at canopy height and was provided by incandescent bulbs and cool-white fluorescent tubes. Samples were harvested and flash-frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Isolation of cDNA Clones for Arabidopsis Hsp70s
A full-length cDNA clone for a plastid hsp70 (cpHsc70-2) was isolated from Arabidopsis cDNA libraries (obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus) by PCR amplification. In these libraries, Arabidopsis cDNAs are size fractionated (1–2 kb, 2–3 kb, 3–6 kb) and inserted at the EcoRI site in Lambda ZAP II (Stratagene, La Jolla, CA) phagemid. A forward primer (CG202; 5′-CCCAGTCACGACGTTGTAAAA-3′) was generated for the Lambda Zap II phagemid vector. A reverse primer (CG205; 5′-GCTGCCAACAAATCA CATTA-3′) was generated from a partial EST clone (T43623) of a plastid hsp70 that was identified by a conserved C-terminal motif. Amplified cDNAs were cloned into pCR2.1 vector (Invitrogen, Carlsbad, CA). The cDNA was sequenced in both directions and confirmed in its entirety. In addition, we used a PCR strategy to obtain genomic fragments of all related genes to identify an Arabidopsis homolog of plant mitochondria hsp70. Primers were designed to encode sequences with exact identity between Escherichia coli DnaK (K01298) and the Sacchromyces cerevisiae mitochondria hsp70, SSC1 (M27229). The forward primer was derived from NGDAWV (amino acids 98–103) of DnaK and the reverse primer from EAAEKA (amino acid 264–269). Primers were fully redundant and had additional 5′-restriction sites. PCR was performed using Arabidopsis (ecotype Columbia) genomic DNA. Two distinct 586-bp genomic fragments having ends with an exact match to the primers were amplified. Sequencing revealed both fragments encoded 166 amino acids, interrupted at amino acid 114 by an 88-bp intron having both consensus donor, and acceptor splice sites. With these genomic fragments, we screened a cDNA library prepared from Arabidopsis heat shock RNA (Helm and Vierling, 1989) and obtained a full-length cDNA clone of a mitochondria hsp70 (mtHsc70-2). A detailed screening method was described previously (Schirmer et al., 1994).
RNA Isolation and RT-PCR
Organ-specific expression of hsp70s was analyzed for roots, stems, leaves, flowers (0 DAP), siliques at 3 DAP from 4-week-old plants, siliques at 7 DAP from 5-week-old plants, and siliques at 14 DAP from 6-week-old plants. To examine changes in hsp70 expression during germination, intact seedlings at 0 to 96 h after imbibition were collected in liquid nitrogen and stored at −80°C. Hsp70 expression in response to temperature extremes was also examined in plants that were exposed to 4°C for 12 and 48 h and to 40°C for 30, 60, and 90 min. Control plants were kept at 20°C. Temperature treatment was initiated 2 h after the onset of the light period so that all samples would be harvested within the light period.
Samples were ground in liquid nitrogen, and total RNA isolated according to manufacturer's protocol using Trizol (Life Technologies/Gibco-BRL, Cleveland). The amount of total RNA was determined by UV spectrophotometry. Total RNA (1 μg) was treated with one unit of DNase I (Sigma, St. Louis) for 15 min at room temperature prior to RT-PCR to remove residual DNA contamination. Using commercial RT-PCR beads (Amersham-Pharmacia Biotech, Uppsala), aliquots of total RNA were reverse transcribed into cDNA with random primer, d(N)6, then amplified with gene specific primers (Table I) in the same tube. When resuspended in 25 μL, a RT-PCR bead generated a reaction solution containing 2.0 units of Taq DNA polymerase, 10 mm Tris-HCl (pH 9.0), 60 mm KCl, 1.5 mm MgCl2, 200 μm of each dNTP, and 1 unit of Moloney murine leukemia virus reverse transcriptase. The cDNAs produced by reverse transcription were amplified with a pair of gene specific primers (10 pmol for each primer) for each gene. For each RT-PCR reaction, a plant 18S internal standard (Ambion, Austin, TX) was included as a loading control. With this standard, a pair of 18S rRNA specific primers and a pair of competitive primers were mixed at the ratio of 2:8 (18S rRNA primers: competitive primers) to generate unsaturated RT-PCR signals over the concentration range of total RNA used in this experiment. PCR reactions for all genes were subjected to 25 cycles at 95°C (30 s), 52°C (45 s), and 72°C (90 s) with GeneAmp PCR System 2400 (Perkin-Elmer Applied Biosystems, Foster City, CA). For the analysis of temperature response and organ specific expression, three rounds of RT-PCR were conducted with three independently isolated total RNA samples. For the analysis of differential expression during seed maturation and germination, two rounds of RT-PCR were conducted with two independently isolated total RNA samples. Twenty microliters from each PCR reaction was fractionated by 1.5% (w/v) agarose gel in Tris-acetate EDTA buffer and stained with 0.5% (w/v) ethidium bromide. The ethidium bromide stained gels were digitally photographed with an IS-1000 Digital Imaging System (Alpha Innotech Corporation, San Leandro, CA). Scion Image for Windows (Scion, http://www.scioncorp.com) program was used to quantify the intensity of the ethidium bromide stained DNA bands from the negative images of the gels.
ACKNOWLEDGMENTS
We thank Dale Haskell and Fatma Kaplan for critical review of this manuscript. We appreciate L.C. Hannah for providing access to PCR/documentation facilities in his laboratory, N. Koizumi for providing cDNA clone for Arabidopsis BiP, and the Arabidopsis Biological Resource Center for providing Arabidopsis cDNA libraries and EST clones for a full length Hsc70-1, a partial chloroplast hsp70 (cpHsc70-2), and cDNA libraries.
Footnotes
This work was supported by the Florida Agricultural Experiment Station and University of Florida Plant Molecular and Cellular Biology Program, by the U.S. Department of Agriculture National Research Initiative (grant nos. 9800877 and 200000687 to C.L.G.), and by National Research Initiative funds and State of Arizona Hatch funds (to E.V.). This research was approved for publication as Florida Agricultural Experiment Station Journal Series no. R–08014.
LITERATURE CITED
- Almoguera C, Prieto-Dapena P, Jordano J. Dual regulation of a heat shock promoter during embryogenesis: stage-dependent role of heat shock elements. Plant J. 1998;13:437–446. doi: 10.1046/j.1365-313x.1998.00044.x. [DOI] [PubMed] [Google Scholar]
- Anderson JV, Li QB, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994;104:1359–1370. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the hsp70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- Bush GL, Meyer DI. The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their roles in protein translocation. J Cell Biol. 1996;135:1229–1237. doi: 10.1083/jcb.135.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka E, Key JL, Gurley WB. Regulatory domains of the Gmhsp17.5 heat shock promoter soybean. Mol Cell Biol. 1989;9:3457–3463. doi: 10.1128/mcb.9.8.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Goldman MHS, Demolder J, Seurinck J, Botterman J. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–1035. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocher A, Vierling E. Cytoplasmic HSP70 homologues of pea: differential expression in vegetative and embryonic organs. Plant Mol Biol. 1995;27:441–456. doi: 10.1007/BF00019312. [DOI] [PubMed] [Google Scholar]
- Duck N, McCormick S, Winter J. Heat shock protein hsp70 cognate gene expression in vegetative and reproductive organs of Lycopersicon esculentum. Proc Natl Acad Sci USA. 1989;86:3674–3678. doi: 10.1073/pnas.86.10.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley P, Wood CK, Pratt JR, Moore AL. Developmental regulation of the plant mitochondrial matrix located HSP70 chaperone and its role in protein import. FEBS Lett. 1997;417:321–324. doi: 10.1016/s0014-5793(97)01311-2. [DOI] [PubMed] [Google Scholar]
- Dupont FM, Hurkman WJ, Tanaka CK, Chan R. BiP, HSP70, NDK, and PDI in wheat endosperm: I. Accumulation of mRNA and protein during grain development. Physiol Plant. 1998;103:70–79. [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90 hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Gaitanaris GA, Papavassiliou AG, Rubock P, Silverstein SJ, Gottesman ME. Renaturation of denatured λ repressor requires heat shock proteins. Cell. 1990;61:1013–1020. doi: 10.1016/0092-8674(90)90066-n. [DOI] [PubMed] [Google Scholar]
- Galau GA, Hughes DW. Coordinate accumulation of homologous transcripts of seven cotton Lea gene families during embryogenesis and germination. Dev Biol. 1987;123:213–221. doi: 10.1016/0012-1606(87)90443-x. [DOI] [PubMed] [Google Scholar]
- Gao BC, Biosca J, Craig EA, Greene LE, Eisenberg E. Uncoating of coated vesicles by yeast hsp70 proteins. J Biol Chem. 1991;266:19565–19571. [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, hsp70 and hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Li QB. The organization and evolution of the spinach stress 70 molecular chaperone gene family. Plant Cell. 1998;10:539–556. doi: 10.1105/tpc.10.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B, Dure L. Developmental regulation in cotton seed germination: polyadenylation of stored messenger RNA. Biochemistry. 1978;8:3250–3256. doi: 10.1021/bi00609a012. [DOI] [PubMed] [Google Scholar]
- Hatano K, Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I. A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 1997;38:344–351. doi: 10.1093/oxfordjournals.pcp.a029172. [DOI] [PubMed] [Google Scholar]
- Helm KW, Vierling E. An Arabidopsis cDNA clone encoding a low molecular weight heat shock protein. Nucleic Acids Res. 1989;17:7995. doi: 10.1093/nar/17.19.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Wang YC, Lin BL. At-hsc70-3 encodes a cytosolic hsp70 in Arabidopsis thaliana. Plant Physiol. 1998;117:1525. [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Koizumi N. Isolation and responses to stress of a gene that encodes a luminal binding protein in Arabidopsis thaliana. Plant Cell Physiol. 1996;37:862–865. doi: 10.1093/oxfordjournals.pcp.a029023. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Sano H. Isolation of two genes (accession nos. D89341 and D89342) encoding luminal binding protein from Arabidopsis thaliana. Plant Physiol. 1997;113:664. [Google Scholar]
- Leborgne-Castel N, Jelitto-Van Dooren EP, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplamic reticulum stress. Plant Cell. 1999;11:459–470. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Li QB, Haskell DW, Guy CL. Coordinate and non-coordinate expression of the stress 70 family and other molecular chaperones at high and low temperature in spinach and tomato. Plant Mol Biol. 1999;39:21–34. doi: 10.1023/a:1006100532501. [DOI] [PubMed] [Google Scholar]
- McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 1996;111:699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275:18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kDa heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Raynal M, Guilleminot J, Gueguen C, Cooke R, Delseny M, Gruber V. Structure, organization and expression of two closely related novel Lea (late-embryogenesis-abundant) genes in Arabidopsis thaliana. Plant Mol Biol. 1999;40:153–165. doi: 10.1023/a:1026403215270. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield WP, Shore GC, Randall SK. Mitochondrial precursor protein: effects of 70-kilodalton heat shock protein on polypeptide folding, aggregation, and import competence. J Biol Chem. 1990;265:11069–11076. [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lin BL. The disappearance of an hsp70 species in mung bean seed during germination: purification and characterization of the protein. Plant Mol Biol. 1993;21:317–329. doi: 10.1007/BF00019947. [DOI] [PubMed] [Google Scholar]
- Wang SM, Khandekar JD, Kaul KL, Winchester DJ, Morimoto RI. A method for the quantitative analysis of human heat shock gene expression using multiplex RT-PCR assay. Cell Stress Chaperones. 1999;4:153–161. doi: 10.1379/1466-1268(1999)004<0153:amftqa>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E. Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 1996;112:747–757. doi: 10.1104/pp.112.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooden SK, Li LJ, Navarro D, Qadri I, Pereira L, Lee AS. Transactivation of the grp78 promoter by malfolded proteins, glycosylation block, and calcium ionophore is mediated through a proximal region containing a CCAAT motif which interacts with CTF/NF-I. Mol Cell Biol. 1991;11:5612–5623. doi: 10.1128/mcb.11.11.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Caspar T, Browse J, Lindquist S, Somerville C. Characterization of an hsp70 cognate gene family in Arabidopsis. Plant Physiol. 1988;88:731–740. doi: 10.1104/pp.88.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, Wang C, Chen J, Lin BL. Isolation of a cDNA encoding a 70 kDa heat-shock cognate protein expressed in vegetative tissues of Arabidopsis thaliana. Plant Mol Biol. 1994;25:577–583. doi: 10.1007/BF00043887. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by Hsp90 (Hsp90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]